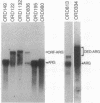
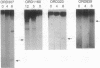
Abstract
In the yeast Saccharomyces cerevisiae, as in other eukaryotes, some regions of the genome have a much higher level of meiotic gene conversion than others. Previous deletion analysis indicated that the sequence necessary for the high level of gene conversion within the ARG4 region defined an initiation site located between positions -316 and -37 [relative to the first base pair (+1) of the ARG4 coding sequence] of the ARG4 promoter. To test whether this sequence is sufficient to promote gene conversion in a novel chromosomal context, we inverted on the chromosome various DNA fragments including the implicated region and the ARG4 coding sequence. Surprisingly, these inversions resulted in the loss of the normal recombination properties and double-strand-break formation associated with this process. By Northern analysis, we found that a transcript traverses the ARG4 initiation site in these inversion mutants but not in the wild type. When transcription through this region was prevented by a transcription terminator, the activity of the initiation site and the formation of double-strand breaks were restored. From these results and from complementary deletion analysis in the normal ARG4 orientation, we conclude that the activity of the ARG4 initiation site requires protection from transcriptional interference.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell. 1982 Jul;29(3):939–944. doi: 10.1016/0092-8674(82)90456-1. [DOI] [PubMed] [Google Scholar]
- Alani E., Padmore R., Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990 May 4;61(3):419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Beacham I. R., Schweitzer B. W., Warrick H. M., Carbon J. The nucleotide sequence of the yeast ARG4 gene. Gene. 1984 Sep;29(3):271–279. doi: 10.1016/0378-1119(84)90056-8. [DOI] [PubMed] [Google Scholar]
- Cao L., Alani E., Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990 Jun 15;61(6):1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciaramella M., Rocco V., Pulitzer J. F. Foreign transcriptional enhancers in yeast. II. Interplay of the polyomavirus transcriptional enhancer and Saccharomyces cerevisiae promoter elements. Nucleic Acids Res. 1988 Sep 26;16(18):8869–8886. doi: 10.1093/nar/16.18.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A., Bloom K. Genetic manipulation of centromere function. Mol Cell Biol. 1987 Jul;7(7):2397–2405. doi: 10.1128/mcb.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschman J. E., Durbin K. J., Winston F. Genetic evidence for promoter competition in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Nov;8(11):4608–4615. doi: 10.1128/mcb.8.11.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M., Borts R. H., Haber J. E. Meiotic gene conversion and crossing over between dispersed homologous sequences occurs frequently in Saccharomyces cerevisiae. Genetics. 1987 Feb;115(2):233–246. doi: 10.1093/genetics/115.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J., Dobson M. J., Roberts N. A., Kingsman A. J., Kingsman S. M. Factors affecting heterologous gene expression in Saccharomyces cerevisiae. Gene. 1985;33(2):215–226. doi: 10.1016/0378-1119(85)90096-4. [DOI] [PubMed] [Google Scholar]
- Nicolas A., Treco D., Schultes N. P., Szostak J. W. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature. 1989 Mar 2;338(6210):35–39. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J. Transcriptional interference and termination between duplicated alpha-globin gene constructs suggests a novel mechanism for gene regulation. Nature. 1986 Aug 7;322(6079):562–565. doi: 10.1038/322562a0. [DOI] [PubMed] [Google Scholar]
- Resnick M. A., Stasiewicz S., Game J. C. Meiotic DNA metabolism in wild-type and excision-deficient yeast following UV exposure. Genetics. 1983 Aug;104(4):583–601. doi: 10.1093/genetics/104.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross L. O., Treco D., Nicolas A., Szostak J. W., Dawson D. Meiotic recombination on artificial chromosomes in yeast. Genetics. 1992 Jul;131(3):541–550. doi: 10.1093/genetics/131.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Rudolph H., Koenig-Rauseo I., Hinnen A. One-step gene replacement in yeast by cotransformation. Gene. 1985;36(1-2):87–95. doi: 10.1016/0378-1119(85)90072-1. [DOI] [PubMed] [Google Scholar]
- Russo P., Li W. Z., Hampsey D. M., Zaret K. S., Sherman F. Distinct cis-acting signals enhance 3' endpoint formation of CYC1 mRNA in the yeast Saccharomyces cerevisiae. EMBO J. 1991 Mar;10(3):563–571. doi: 10.1002/j.1460-2075.1991.tb07983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultes N. P., Szostak J. W. A poly(dA.dT) tract is a component of the recombination initiation site at the ARG4 locus in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jan;11(1):322–328. doi: 10.1128/mcb.11.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultes N. P., Szostak J. W. Decreasing gradients of gene conversion on both sides of the initiation site for meiotic recombination at the ARG4 locus in yeast. Genetics. 1990 Dec;126(4):813–822. doi: 10.1093/genetics/126.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Sapolsky R. J., Davis R. W. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1988 May;8(5):2184–2194. doi: 10.1128/mcb.8.5.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Struhl K., Davis R. W. Transcription of the his3 gene region in Saccharomyces cerevisiae. J Mol Biol. 1981 Nov 5;152(3):535–552. doi: 10.1016/0022-2836(81)90267-9. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco D., Schultes N. P., Szostak J. W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989 Mar 2;338(6210):87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco D., Szostak J. W. Extensive 3'-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991 Mar 22;64(6):1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989 Feb 24;56(4):619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Keil R. L., Roeder G. S. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987 Mar 27;48(6):1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- White M. A., Detloff P., Strand M., Petes T. D. A promoter deletion reduces the rate of mitotic, but not meiotic, recombination at the HIS4 locus in yeast. Curr Genet. 1992 Feb;21(2):109–116. doi: 10.1007/BF00318468. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]