Abstract
The import of proteins containing the peroxisomal targeting signal 1 (PTS1) into the Leishmania glycosome is dependent on the docking of the PTS1-loaded LdPEX5 cytosolic receptor with LdPEX14 on the glycosome surface. Here we show that, in the absence of PTS1, LdPEX5 is a tetramer that is stabilized by two distinct interaction domains; the first is a coiled-coil motif encompassing residues 277 to 310, whereas the second domain is localized to residues 1 to 202. By using microcalorimetry, surface plasmon resonance, and size exclusion chromatography techniques, we show that PTS1 peptide binding to LdPEX5 tetramers promotes their dissociation into dimeric structures, which are stabilized by a coiled-coil interaction. Moreover, we demonstrated that the resulting LdPEX5-PTS1 complex is remarkably stable and exhibits extremely slow dissociation kinetics. However, binding of LdPEX14 to LdPEX5 modulates the LdPEX5-PTS1 affinity as it decreases the thermodynamic dissociation constant for this latter complex by 10-fold. These changes in the oligomeric state of LdPEX5 and in its affinity for PTS1 ligand upon LdPEX14 binding may explain how, under physiological conditions, LdPEX5 can function to deliver and unload its cargo to the protein translocation machinery on the glycosomal membrane.
The human protozoan pathogen Leishmania represents evolutionary ancient organisms that diverged early from the eukaryotic cell lineage. In addition to a number of unique biochemical and metabolic features, these parasites contain glycosomes, a microbody organelle that is evolutionarily related to the peroxisomes (41, 42), which compartmentalize a multitude of indispensable pathways. These include glycolysis, purine salvage, pyrimidine and ether-lipid biosynthesis, and β-oxidation of fatty acids (41, 20). A number of studies have demonstrated that the correct targeting of proteins to the glycosome is crucial for parasite viability, especially in the case of glycolytic enzymes (2, 3, 27). Consequently, the glycosome biogenesis machinery has attracted considerable attention as a potential drug target since chemotherapeutic agents that disrupt or block the import of proteins into the glycosome impair multiple biosynthetic and energy-producing pathways in these parasites (52, 23).
Glycosomal matrix proteins, such as the peroxisome, are encoded by nuclear genes and are synthesized by cytosolic polyribosomes. Proteins destined for these microbody organelles are sorted on the basis of two major classes of topogenic sequences known as peroxisomal targeting signal 1 (PTS1) and PTS2. PTS1 is the most commonly occurring targeting signal. It consists of a C-terminal tripeptide with the sequence Ser-Lys-Leu (SKL) or Ala-Lys-Leu (AKL) or variants thereof (4, 36). PTS2 is a more degenerate signal comprised of an N-terminal nonapeptide with the motif R/K-L/I/V-X5-H/E-A/L located within the first 20 to 30 residues of a protein (54). Analogous signal sequences are used for the targeting of proteins to the glycosomes of Leishmania spp. (51) and Trypanosoma brucei (4). In peroxisomes, nascent polypeptides containing the PTS1 or PTS2 signals are selectively bound by the cytosolic receptor proteins peroxin 5 (PEX5) or PEX7, respectively (46, 54). These cargo-laden receptor complexes move to the peroxisomal membrane surface, where they interact with a docking complex containing the membrane proteins PEX13 and PEX14, hence facilitating translocation across the peroxisomal membrane (47). Studies in yeast and mammalian cells suggest that translocation of PTS1 proteins into the peroxisome involves cycling of PEX5 between the cytosol and the peroxisome matrix (12, 16). PEX5 import into the peroxisome may be required to unload the PTS1 ligand (56). Whether glycosome biogenesis in kinetoplastids involves a similar cycling of the Leishmania donovani PEX5 (LdPEX5) and T. brucei PEX5 (TbPEX5) is unclear (15, 29). Subcellular fractionation experiments have shown that the bulk of LdPEX5 and TbPEX5 is cytosolic. However, a small portion of them (5 to 10%) sediments with the glycosomes. Whether this fraction of protein is surface associated or present in the glycosomal compartment has not been established.
LdPEX5, like other PEX5 homologues, is a bidomain protein. The C-terminal half of PEX5 is composed of six to seven tetratricopeptide repeats (TPRs) that adopt helical hairpin structures that cluster together to form the PTS1-binding pocket (21, 24, 29, 50). The N-terminal region of LdPEX5 appears to mediate diverse intramolecular and/or intermolecular interactions being important for PTS1 protein import (47). Mapping studies have demonstrated that the N-terminal region of LdPEX5 contains critical sequence elements required for interaction with PEX13 and PEX14 (1, 6-8, 18, 19, 22, 30, 44, 55). Mutagenesis and biochemical experiments have further established that interaction of PEX5 with the peroxisomal/glycosomal translocational machinery in human, Saccharomyces cerevisiae, plant, and T. brucei systems is, at least, mediated by the motif WXXXY/F conserved in all PEX5 proteins (6, 8, 40, 45, 49). In LdPEX5, however, this diaromatic motif does not appear to be required for interaction with LdPEX14 since LdPEX5 mutants in which these motifs have been sequentially mutated or deleted did not affect the LdPEX5-LdPEX14 interaction (K. P. Madrid and A. Jardim, submitted for publication).
In the absence of PTS1, the Leishmania (29), human (50), and Hansenula (5) PEX5 is known to form a tetrameric structure. Deletion mutagenesis experiments have shown that fragments of the human PEX5 encompassing residues 1 to 251 form tetramers. In contrast, the N-terminal fragment of the Leishmania PEX5, corresponding to residues 1 to 391, forms large soluble complexes with a mass >2 MDa (29). Whether the oligomeric state of PEX5 is important for its function in the sorting and import of newly synthesized protein from the cytosol to the peroxisome and/or glycosome has not been previously demonstrated. Recent studies by Boteva et al. (5) have shown that the Hansenula PEX5 undergoes a tetramer-to-monomer dissociation in acidic environment. These authors suggested that this process may be important for the release of PTS1 proteins in the peroxisome matrix. However, whether a similar mechanism is at play in mammalian peroxisome or Leishmania glycosome biogenesis is unclear.
Numerous studies have established that PEX5 not only interacts with PEX13 and PEX14 but also regulates the PEX13-PEX14 association (22, 45, 55). Several reports have suggested that the PTS1 ligand may participate in controlling these oligomeric interactions. Indeed, in vitro binding studies revealed that PTS1 actively enhances and decreases the affinity of PEX5 for PEX14 and PEX13, respectively (44, 55). These observations have led to a model suggesting that these opposite effects may facilitate the release of PTS1 proteins at the inner surface of peroxisome and may enhance PEX5 recycling to the cytosol in association with PEX13 (44). However, the effect of PEX14 or PEX13 on the PEX5-PTS1 affinity has not been extensively investigated.
It is known that a pivotal first step in the translocation of protein across the peroxisomal membrane is dependent on PTS1-bound PEX5 interactions with PEX13 and PEX14. Furthermore, mutations that alter the cellular levels of PEX14 are detrimental to peroxisome biogenesis and lead to the accumulation of PTS1 and PTS2 proteins in the cytosol (47, 54). PEX14 is also a vital factor for glycosome biogenesis. Creation of PEX14 functional mutants in T. brucei by using an RNA interference strategy resulted in a conditional lethal phenotype (23). Analysis of protein targeting in these mutants revealed that both PTS1 and PTS2 proteins accumulated in the cytosol, suggesting that TbPEX14 likely functions as a convergence point for TbPEX5 and TbPEX7 in protein translocation into the glycosome (39).
In the present study, we show that the LdPEX5 tetramer is composed of a dimer of dimers that dissociates upon binding to PTS1 ligand. Moreover, we demonstrate that these dimers are stabilized by a coiled-coil motif and that assembly of these dimers into a higher-order tetramer is stabilized by a second interaction domain that is localized to the first 202 residues. Here we also report that LdPEX5 and PTS1 form a high-affinity and stable complex that exhibits a slow dissociation rate. However, when LdPEX14 binds to LdPEX5, the association affinity between LdPEX5 and PTS1 is decreased 10-fold. These oligomeric changes may explain how, under physiological conditions, LdPEX5 can function to deliver and unload its cargo to the protein translocation machinery on the glycosomal membrane.
MATERIALS AND METHODS
Expression and purification of recombinant XPRT, LdPEX14, and LdPEX5 proteins.
The L. donovani xanthine phosphoribosyltransferase (XPRT) was overexpressed in low-phosphate buffer by using Escherichia coli Sφ609 cells transformed with pBAce-XPRT vector, and the enzyme was purified by ion-exchange chromatography as previously described (28). LdPEX14 containing an N-terminal hexahistidine and S tags was overexpressed by using E. coli ER2566 strain transformed with the pET30b(+)-LdPEX14 construct, and the protein purified on Ni2+-nitrilotriacetic acid resin as described previously (30). Expression and purification of the His6-CT-ldpex5 was performed as described previously (29).
LdPEX5 was expressed as a C-terminal fusion protein by cloning the LdPEX5 open reading frame into the NdeI/NotI sites of the pTYB12 vector (New England Biolabs) in frame with a chitin-binding domain, generating the pTYB12-LdPEX5 expression construct. The expression construct for the internal deletion mutant ldpex5 Δ269-291, which lacks residues 269 to 291, was generated by digesting pTYB12-LdPEX5 with the endonuclease AatII (New England Biolabs) to drop out a 60-bp AatII fragment, treating the linearized plasmid with T4 DNA polymerase (Invitrogen) in the presence of 0.1 mM dTTP (Invitrogen) for 30 min at 20°C, and religating the construct with T4 DNA ligase. The pTYB12-ldpex5 1-202, for expression of an ldpex5 fragment encompassing residues 1 to 202, was constructed by cloning the LdPEX5 NdeI/EcoRI fragment into the corresponding sites of the pTYB12 vector. The pTYB12-ldpex5 268-303 vector for expression of the peptide encompassing residues 268 to 303 of LdPEX5 was constructed by cloning a PCR fragment encoding these residues into the NdeI/EcoRI sites of the pTYB12 plasmid. All expression constructs were verified by automated DNA sequence analysis.
Cultures of E. coli ER2566 transformed with pTYB12-LdPEX5, pTYB12-ldpex5 Δ269-291, or pTYB12-ldpex5 1-202 were grown to an optical density at 600 nm of ∼1.2 in Luria broth supplemented with 50 μg of ampicillin/ml and induced for 5 h at 20°C with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cell pellets were suspended in 30 ml of buffer (20 mM Tris-HCl [pH 8.0]) containing a protease inhibitor cocktail and lysed with two passes through a French press. NaCl was added to the lysates to a final concentration of 500 mM and clarified by centrifugation at 14,000 rpm for 30 min at 4°C. Supernatants were loaded onto a chitin resin column (2.0 by 5.0 cm) (New England Biolabs) equilibrated with 40 mM Tris-500 mM NaCl (pH 8.0) (TS buffer) at 4°C at a flow rate of 0.5 ml/min. The column was washed with 100 ml of TS buffer and then rinsed with two column volumes of TS buffer containing 50 mM dithiothreitol (DTT). The resin was resuspended in 1.5 column volumes of TS buffer with 50 mM DTT, followed by incubation for 48 h at 4°C to promote protein splicing of the chitin-binding domain-LdPEX5 fusion protein. LdPEX5 was eluted with two column volumes of 40 mM Tris-HCl (pH 8.0)-500 mM NaCl, concentrated by using a 5 NMWL centrifugal filter unit (Millipore), quantified by the method of Gill and von Hippel (26), and frozen in aliquots at −80°C.
To purify the ldpex5 268-303 fragment, the peptide was cleaved from the fusion partner with 50 mM DTT in 40 mM Tris-HCl (pH 8.0) at 4°C for 48 h. The eluate was applied to a Q Sepharose column (1 by 5 cm) equilibrated with 40 mM Tris-HCl (pH 8.0) to remove the DTT, and the peptide was eluted with 10 column volumes of 0.1% trifluoroacetic acid. Fractions containing peptide were concentrated by lyophilization. Peptide purity was assessed by reversed-phase high-pressure liquid chromatography on a C8 column developed with a 0.1% trifluoroacetic acid-acetonitrile chromatography system. The integrity of the peptide was determined by mass spectroscopy on a Ciphergen IIc instrument with dihydroxybenzoic acid as the matrix. The observed mass of the peptide was 4,514 Da.
LdPEX5-XPRT interactions.
High-binding flat-bottom 96-well microtiter plates (Perkin-Elmer, Meriden, Conn.) were coated with 1 μg of purified XPRT/well in 100 μl of PBS (pH 8.0) for 16 h at 4°C. Unbound protein was removed by two washes with PBS, and plates were blocked with 200 μl of 2% milk powder in PBS for 45 min at 20°C. Plates were washed with PBS-0.05% Tween 20 and then incubated for 2 h at 20°C with various concentrations of LdPEX5 (0.4 to 850 nM) diluted in 100 μl of PBS-0.05% Tween 20-2% adult bovine serum (ABS) in the absence or presence of 4 μg of His6-S-LdPEX14. Unbound protein was removed by three washes with 200 μl of PBS-0.05% Tween 20 and bound LdPEX5 was quantitated by indirect enzyme-linked immunosorbent assay (ELISA) with LdPEX5-specific rabbit antisera (1:5,000) and goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:5,000) (Sigma) diluted in PBS-0.05% Tween 20-2% ABS.
For control experiments designed to determine whether the binding of LdPEX14 to LdPEX5 alters the ability of anti-LdPEX5 antibodies to bind LdPEX5, wells were coated with 1 μg of LdPEX5 in 100 μl of PBS for 16 h at 4°C. Unbound LdPEX5 was removed, and wells were blocked with 200 μl of 1% milk powder in PBS. Sets of triplicate wells were incubated with increasing concentration of LdPEX14 (0.8 to 800 nM) diluted in PBS-0.05% Tween 20-1% ABS or dilution buffer alone for 2 h at 20°C. Wells were washed five times with 300 μl of PBS-0.05% Tween 20 to remove unbound LdPEX14. A 100-μl portion of a 1:5,000 dilution of anti-LdPEX5 antisera was added to each well, followed by incubation at 20°C for 1 h. The amount of primary antibody bound to LdPEX5 was measured by using a goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:5,000) diluted in PBS-0.05% Tween 20-2% ABS.
For LdPEX5-LdPEX14 and ldpex5 Δ269-291-LdPEX14 interaction assays, microtiter plates were coated with 1 μg of His6-S-LdPEX14/well in 100 μl of PBS for 16 h at 4°C. Unbound protein was removed, and plates blocked with 200 μl of 2% milk powder in PBS for 45 min at 25°C. Microtiter plates were rinsed and incubated with increasing concentrations of LdPEX5 or ldpex5 Δ269-291 proteins (0.4 to 860 nM) diluted in 100 μl of 2% ABS-0.05% Tween 20-PBS for 2 h at 25°C. Bound LdPEX5 or ldpex5 Δ269-291 was quantitated as described above.
ELISAs were developed by using ABTS [2,2′azinobis(3-ethylbenzthiazolinesulfonic acid)] as the chromogenic substrate. Color development was measured on a Benchmark microplate reader (Bio-Rad) at 405 nm, and the data were analyzed by using ORIGIN 7.0 software (Microcal Software, Northampton, Mass.).
Gel permeation chromatography analysis of LdPEX5.
Gel permeation chromatography was performed on Beckman-Coulter System Gold equipped with a Bio-Sil SEC 450-5/Bio-Sil SEC 250-5 column (7.8 by 150 mm or 7.8 by 300 mm) or a Bio-Sil SEC 250-5 column (7.8 by 300 mm or 7.8 by 600 mm) (Bio-Rad) or with a Superdex 200 column equilibrated with 25 mM Tris-HCl-100 mM NaCl-2 mM DTT (pH 8.0) at a flow rate of 0.4 or 0.25 ml/min. Then, 25 μg of LdPEX5, ldpex5 Δ269-291, ldpex5 203-391, ldpex5 290-391, or ldpex5 1-202 was injected, and the column eluant was monitored at 280 nm. The column was calibrated by using a protein standard mixture containing thryoglobulin (660 kDa), ferritin (440 kDa), bovine catalase (250 kDa), bovine immunoglobulin G (IgG) (160 kDa), ovalbumin (45 kDa), equine myoglobin (17 kDa), and vitamin B12 (1.4 kDa).
Peptide synthesis.
The synthetic peptide CNDRYRDLRHILILRDGEATRYPAKL (AKL peptide) corresponding to the C-terminal 26 residues of the L. donovani XPRT was synthesized with an N-terminal cysteine to facilitate covalent coupling. This peptide was synthesized at the Protein Microchemistry Centre at the University of Victoria by using a Leucine Pam resin and Fmoc (9-fluorenylmethoxy carbonyl)-HOBT chemistry. Peptide was cleaved from the resin by using trifluoromethanesulfonic acid and purified by C18 reversed-phase chromatography using a 0.1% trifluoroacetic acid-acetonitrile system. Sequence of the purified peptide was confirmed by matrix-assisted laser desorption ionization-time of flight mass spectroscopy.
Circular dichroism analysis.
Lyophilized ldpex5 268-303 peptide was dissolved in 20 mM sodium phosphate (pH 7.2)-100 mM NaCl buffer or 20 mM sodium phosphate (pH 7.2)-100 mM NaCl-50% trifluoroethanol at a concentration of 1 mg/ml. The secondary structure of ldpex5 268-303 was determined with a JASCO 710 spectropolarimeter. Spectra were recorded from 260 to 200 nm in a 0.1-cm quartz cuvette at room temperature. The helical content of the peptide was calculated from the molar ellipticity at 222 nm by the method of Wu et al. (57).
ITC analysis of LdPEX5-PTS1 interaction.
Isothermal titration calorimetry (ITC) experiments were performed at 30°C with a MicroCal VP-ITC isothermal titration calorimeter (MicroCal, Northampton, Mass.). The reference cell was filled with degassed 20 mM sodium phosphate (pH 7.5)-120 mM NaCl-3 mM β-mercaptoethanol buffer (SSM buffer), and the 1.43-ml sample cell was filled with a degassed 60 μM solution of LdPEX5, His6-CT-ldpex5 or ldpex5 Δ269-291 diluted in SSM buffer. Typically, 30 to 40 5-μl injections of 1.0 mM AKL peptide in SSM buffer were performed at 6-min intervals, and the enthalpy (ΔH0), stoichiometry of binding (n), and dissociation constant (Kd) was determined by using ORIGIN 7.0 software.
SPR analysis.
All surface plasmon resonance (SPR) experiments were performed on a Biacore 3000 Instrument (Biacore, Inc., Piscataway, N.J.). The amine coupling kit containing N-hydroxysuccinimide (NHS) and N-ethyl-N′-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC), 2-(2-pyridinyldithio)ethaneamine hydrochloride (PDEA), and PIONEER sensor chips CM4 were purchased from Biacore. SPR experiments were carried out at 25°C. The data collection rate was set to 10 Hz. The running buffer was HEPES-buffered saline (HBS; 10 mM HEPES, 150 mM NaCl, 3 mM ETDA, 0.005% Tween 20 [pH 7.5]). LdPEX5, ldpex5 Δ269-291, and His6-CT-ldpex5 proteins were dialyzed against HBS (which was also used for all dilutions) in order to minimize changes in bulk refractive index upon sample injection.
All steps in the immobilization process were carried out at a flow rate of 5 μl/min. The AKL peptide was immobilized on two surfaces of CM4 sensor chips by using the standard ligand thiol coupling procedure and AKL peptide (10 mM, pH 2.0) freshly dissolved in 10 mM acetic acid (pH 4.0) to a final concentration of 1 mM. AKL injections were manually controlled to couple ∼10 and ∼100 resonance units (RUs). A control surface was prepared similarly by replacing AKL peptide by a running buffer injection (10-μl injection).
Steady-state analysis of LdPEX5-PTS1 interaction by SPR.
Experiments were conducted at a flow rate of 20 μl/min, except for the His6-CT-ldpex5 injections for which the flow rate was reduced to 10 μl/min in order to limit material consumption. LdPEX5 and ldpex5 Δ269-291 were injected (60 μl) in duplicate at concentrations varying from 3.7 to 2700 nM on both AKL peptide and control surfaces. In the case of His6-CT-ldpex5, concentrations varying from 11.1 to 8180 nM were injected on the AKL peptide and control surfaces (120 μl). For each series of PEX5 injections, five buffer injections were performed before any PEX5 injection, and five additional buffer injections were evenly interspaced between the PEX5 injections. Regeneration was achieved between each injection by two 25-μl pulses (100 μl/min) of 6 M guanidine hydrochloride (GdnHCl), followed by an EXTRACLEAN procedure and a buffer injection (100 μl, 100 μl/min) to avoid any GdnHCl carryover.
Biacore data preparation and analysis.
Data were prepared by using the double referencing method (45). For kinetic analysis, the data were globally analyzed by using the SPR evolution software package (54). For steady-state analysis, the apparent thermodynamic dissociation constants were determined by plotting the control corrected plateau value (response units at equilibrium [plateau] [Req]) versus the injected concentration of the various LdPEX5 mutant chains. The experimental data generated with both AKL surfaces were globally fit with a simple interaction model: Req = Rmax i × [C/(C + Kd)], where C is the injected LdPEX5 chain concentration, Rmax i is the maximum amount of LdPEX5 that can be bound to the ith surface (i = 1, 2), and Kd is the thermodynamic dissociation constant of the interaction. The fitting procedure was performed in Microsoft Excel by nonlinear regression with Rmax i and Kd as floating parameters. Based on the Rmax i values, data from high- and low-loaded surfaces were normalized to be displayed as shown in Fig. 6.
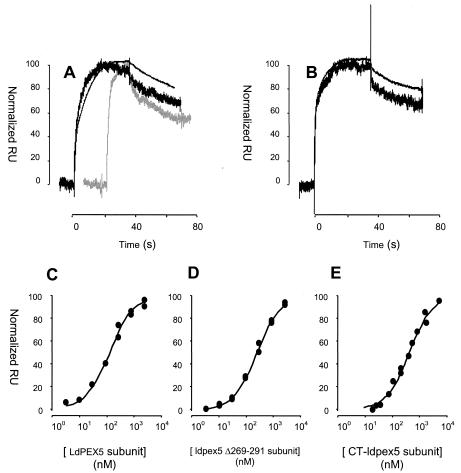
FIG. 6.
Analysis of LdPEX5-AKL PTS1 peptide interaction by SPR. (A) Control-corrected and normalized sensorgrams corresponding to the injection of 300 nM LdPEX5 (black) or His6-CT-ldpex5 (gray) over the 10-RU AKL surface (thick curves) and the 100-RU AKL surface (thin curve). (B) Control-corrected and normalized sensorgrams corresponding to the injection of 300 nM ldpex5 Δ269-291 over the 10-RU AKL surface (thick curve) and the 100-RU AKL surface (thin curve). (C to E) Control-corrected and normalized plateau RUs (RU at the equilibrium plateau) obtained when various concentrations of LdPEX5 (C), ldpex5 Δ269-291 (D), or His6-CT-ldpex5 (E) solutions were injected over the two different density AKL peptide surfaces. The solid lines correspond to the fit when the data generated on both surfaces are globally analyzed with a simple model.
RESULTS
LdPEX5 oligomerization.
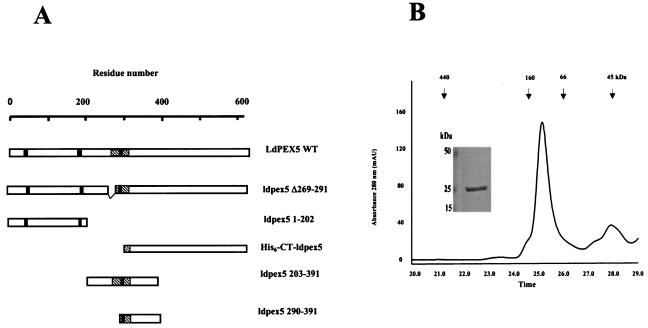
Previous studies of PEX5 from Leishmania sp., human, and Hansenula polymorpha (5, 29, 50) have demonstrated that these receptor proteins form homotetrameric structures. In the human PEX5, the portion of the protein responsible for tetramer formation has been localized to residues 1 to 251 (50). Using deletion mutants, we have previously demonstrated that oligomerization of LdPEX5 is dependent on domains located in the N-terminal region spanning residues 1 to 391 (NT-ldpex5-His6) (29). However, in contrast to the human PEX5 N-terminal fragment, NT-ldpex5-His6 formed large oligomeric structures with a mass higher than 2 MDa. To further characterize the sequence elements that control LdPEX5 oligomerization, we dissected this NT-ldpex5-His6 by examining the quaternary structure of two new recombinant proteins corresponding to residues 1 to 202 (ldpex5 1-202) and residues 203 to 391 (ldpex5 203-391) (Fig. 1A).
FIG. 1.
Oligomerization domains of LdPEX5. (A) Schematic diagram of the LdPEX5 protein variants used in the present study. The hatched regions in each construct represents the coiled-coil motif, while the black bars denote the three WXXXY/F pentapeptide repeats that are conserved in all PEX5 proteins. (B) The oligomeric state of ldpex5 1-202 was determined on a Bio-Sil SEC 250-5 (7.8 by 600 mm) column as described in Materials and Methods. One-minute fractions were collected, and the proteins in each fraction were analyzed by Coomassie blue-stained SDS-PAGE (numbers at the bottom of the gel correspond to the elution times on the trace). The inset represents a gel analysis of the ∼132-kDa peak eluting at 25 min. SDS-PAGE gel electrophoresis of this peak reveals that it is composed of a protein with a subunit molecular mass of ∼25 kDa. (C) The quaternary structure of the fragments ldpex5 203-391 (thick trace) and ldpex5 290-391 (thin trace) was determined by analysis of 25 μg of each protein on a Superdex 200 HR10/30 column equilibrated with 50 mM Tris (pH 8.0)-100 mM NaCl-2 mM DTT (pH 8.0) at a flow rate of 0.25 ml/min. The inset shows an SDS-PAGE analysis of ldpex5 290-391 (lane 1) and ldepx5 203-391 (lane 2) protein preparations used to perform the gel permeation chromatography experiment. Gel permeation columns were calibrated with protein standard containing a mixture of thyroglobulin (660 kDa), ferritin (440 kDa), bovine IgG (160 kDa), bovine serum albumin (66 kDa), ovalbumin (45 kDa), β-lactoglobulin (35 kDa), or equine myoglobin (17 kDa).
Analysis of the quaternary structure of these fragments by gel permeation chromatography on a Superdex 200 column revealed that ldpex5 1-202 eluted with an apparent mass of ∼132 kDa (Fig. 1B), a finding consistent with this N-terminal fragment forming hexamers since the theoretical mass of this fragment is calculated to be 22.3 kDa. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) confirmed that the 132-kDa peak was composed of subunits that migrated with an apparent molecular mass of ∼25 kDa (Fig. 1B, inset). The ldpex5 203-391 fragment, which has a calculated molecular mass of 21.7 kDa, eluted with an apparent mass of ∼52 kDa, which is diagnostic of this fragment forming a dimer (Fig. 1C). It should be noted that on SDS-PAGE ldpex5 203-391 migrates anomalously, with an apparent mass of ∼37 kDa (Fig. 1C). This unusual behavior may be attributed to the acidic nature of this protein which has a calculated pI of 4.3 (29). A smaller protein fragment corresponding to residues 290 to 391 (ldpex5 290-391), with a calculated mass of 11.9 kDa that migrated on SDS-PAGE with an apparent molecular mass of ∼12 kDa also formed a dimer that eluted with a mass of ∼24 kDa (Fig. 1C). It should be stressed that, although the fragments ldpex5 203-391 and ldpex5 290-391 both contain a single cysteine residue, it is unlikely that dimerization was due to disulfide bond formation since (i) all LdPEX5 proteins were stored in 3 mM DTT after intein cleavage from the chitin-binding domain fusion partner and (ii) the gel permeation chromatography analysis was performed in a mobile phase containing 2 mM DTT. The findings from these mapping studies with various portions of the LdPEX5 protein suggest that the region encompassing residues 1 to 391 contains several oligomerization domains that are important not only for stabilizing the tetrameric structure of the full-length LdPEX5 but also for modulating the LdPEX5 quaternary structure upon binding the PTS-1 ligand.
Further in silico analysis of LdPEX5 using the COILS algorithm (37) revealed two putative coiled-coil regions spanning residues 59 to 91 and 277 to 310, respectively. Secondary structure predictions using the GORIV algorithm of Combet et al. (10) also indicated that the two regions have a high propensity for adopting an α-helical structure. The first putative coiled-coil region is rich in glutamine and alanine residues, a sequence that is not consistent with the classical coiled-coil motif. When the weighting option of the COILS program, which gives a more favorable weighting to positions a and d is selected, the probability of this region to adopt a coil structure drops from 85% to <10%, an indication that this region is probably not a coiled coil (38). In contrast, the second coiled-coil motif corresponding to the protein sequence DAYVKEMDMAANDVEDWAQEYAEMQERLQKVTNS was predicted with 100% probability with or without the COILS weighting option.
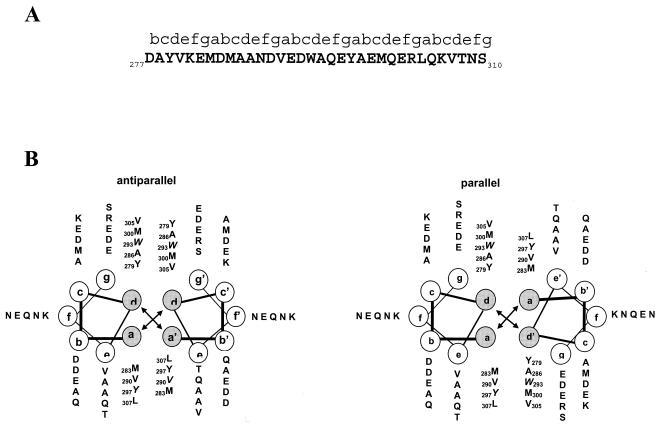
The classical coiled-coil structure consists of a repeating seven-residue heptad unit (denoted by the letter sequence abcdefg) that is stabilized by hydrophobic interactions between nonpolar residues at positions a and d in the heptad as the helices wrap around each other forming a left-handed supercoil (58) (Fig. 2A). Positions a and d in the LdPEX5 coiled coil are occupied primarily by the bulky hydrophobic residues tryptophan, tyrosine, leucine, valine, and methionine. This creates an amphipathic helix with a hydrophobic face that facilitates a tight helical packing in a knobs-in-holes manner (Fig. 2B) (11). This coiled-coil is predicted to contain five heptads; however, the first heptad starts in the b register since proline 276, a known helix-disrupting residue, would be placed in the a position. Analysis of this coiled-coil motif on a helical wheel diagram suggested that packing of these coils in either an antiparallel or a parallel conformation was equally possible (Fig. 2B).
FIG. 2.
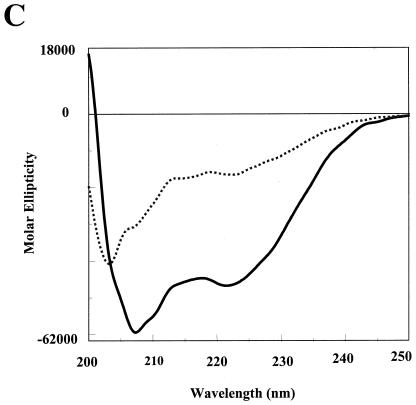
In silico analysis of the LdPEX5 coiled coil. (A) Primary sequence for the predicted LdPEX5 coiled coil (residues 277 to 310). Above the amino acid sequence is the designation of the heptad repeats using the abcdefg nomenclature. (B) The hydrophobic core contacts, a↔a′ and d↔d′ for the parallel orientation or the a↔d′ and a′↔d for the antiparallel arrangement, that stabilize the coiled-coil homodimer packing are illustrated by plotting the primary sequence on helical wheel diagrams. (C) The secondary structure of ldpex5 268-303 was determined by circular dichroism on a JASCO 710 spectropolarimeter with a 0.1-cm cylindrical quartz cuvette. Peptide was dissolved at a concentration of 1 mg/ml in 40 mM sodium phosphate 100 mM NaCl (pH 7.2; dashed line) or 50% trifluoroethanol-40 mM sodium phosphate-100 mM NaCl (pH 7.2; solid line). Spectra represent the average of 10 scans from 200 to 250 nm, and the buffer baseline has been subtracted from each spectra. The uncorrected percent helicity was calculated with the [θ]222 by the method of Wu et al. (55) using the following equation: % helix = {([θ]222 + 2,000)/(−37,400 + 2,000)} × 100.
To experimentally assess the in silico prediction, we examined the secondary structure of the peptide ldpex5 268-303, which corresponds to residues 268 to 303 of LdPEX5 (ahg268AMTSPENGDPDAYVKEMDMAANDVEDWAQEYAEMQER303; the first three residues [in lowercase letters] were derived from the chitin-binding protein fusion partner), a peptide previously expressed for mapping of the LdPEX5-LdPEX14 interaction domain (Madrid and Jardim, submitted). Spectra of ldpex5 268-303 recorded at 20°C in aqueous solvent at pH 7.2 showed two minima at 208 and 222 nm, bands that are diagnostic of helical structure (Fig. 2C). The percent helicity calculated by the method of Wu et al. (57) using the ellipticity at 222 nm indicated that in aqueous solutions ldpex5 268-303 was estimated to contain 32% α-helix structure. However, this value is likely an underestimation of the helical content since aromatic amino acid side chain absorbance can significantly decrease the ellipticity at 222 nm (33). It has been estimated that the presence of three aromatic residues, as in the case of ldpex5 268-303, can account for an ∼20% error in the estimation of the helical content (33). Moreover, the presence of two proline residues, which are not predicted to be part of the LdPEX5 coiled-coil motif, likely leads to N-terminal fraying of the peptide, giving rise to some random coil structure that may account for this reduction.
The propensity of ldpex5 268-303 to adopt a helical conformation was further examined by measuring the circular dichroism spectra in 50% trifluoroethanol, a solvent that is known to stabilize the helical structure. Under these conditions, the helical content of this peptide is calculated to be 125%. Again, this estimation of helical structure has not been corrected for the aromatic residue side chain absorbance which, in the presence of organic solvents, increases the negative ellipticity at 222 nm, resulting in an overestimation (125%) of helical content of this peptide (34).
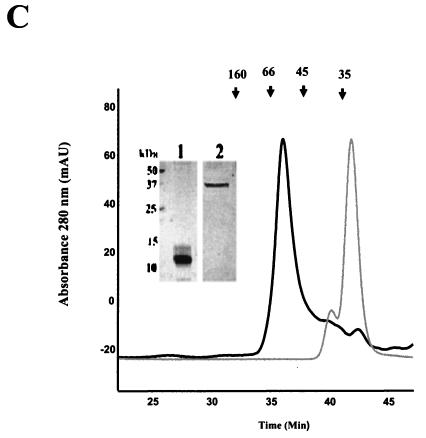
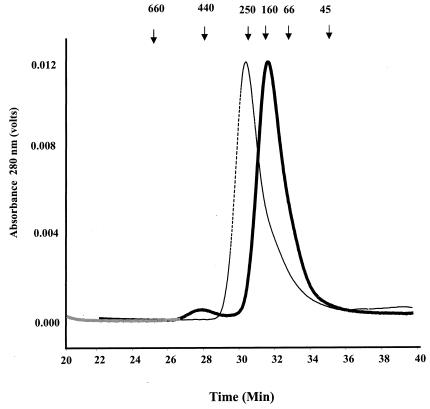
To determine whether the predicted coiled-coil region was important for LdPEX5 tetramer formation, we overexpressed and purified a variant of the LdPEX5 protein, ldpex5 Δ269-291, which lacks the residues 269 to 291 (Fig. 1A). This internal deletion removed only two of the five heptads of the coiled-coil motif. This deletion was designed to disrupt the latter coiled coil, while attempting to minimize global structural changes in the ldpex5 structure. Gel permeation analyses of wild-type LdPEX5 and ldpex5 Δ269-291 revealed that these proteins eluted with apparent molecular masses of ∼270 and ∼140 kDa, respectively. These results are consistent with LdPEX5 forming a tetramer and ldpex5 Δ269-291 being dimeric (Fig. 3), since the calculated molecular masses for the LdPEX5 and ldpex5 Δ269-291 are 69.7 and 67.5 kDa, respectively.
FIG. 3.
Quaternary structure analysis of LdPEX5 proteins. The oligomeric structure of recombinant LdPEX5 proteins and ldpex5 fragments overexpressed in E. coli were assessed by high-pressure liquid gel permeation chromatography. A total of 25 μg of LdPEX5 (thin trace) or ldpex5 Δ269-291 (thick trace) was injected onto a Bio-Sil column equilibrated with the mobile phase 50 mM Tris (pH 8.0)-100 mM NaCl-2 mM DTT at a flow rate of 0.4 ml/min, and protein elution was monitored spectrophotometrically at 280 nm. Gel permeation columns were calibrated with a protein standard containing a mixture of thyroglobulin (660 kDa), ferritin (440 kDa), bovine catalase (250 kDa), bovine IgG (160 kDa), bovine serum albumin (66 kDa), ovalbumin (45 kDa), and equine myoglobin (17 kDa).
Analysis of the LPEX5-PTS1 interactions by ITC.
Earlier studies with the L. donovani XPRT, a glycosomal enzyme, have demonstrated that LdPEX5 and His6-CT-ldpex5, the C-terminal fragment corresponding to the TPR domain, selectively recognize and bind the AKL PTS1 signal of XPRT with nanomolar affinity as determined by ELISA-based (29) or far-Western assays (unpublished data). To further examine the LdPEX5-PTS1 interaction, we decided to use a synthetic peptide corresponding to the C-terminal 26 amino acids of XPRT (AKL peptide). Intact XPRT was not used in ITC or SPR studies since the recombinant protein is known to be tetrameric in solution (unpublished data). It should be noted that the interaction of LdPEX5 with LdXPRT has been previously demonstrated to be dependent on the topogenic signal Ala-Lys-Leu, and removal of these residues results in a complete loss of LdPEX5-LdXPRT association (29). This quaternary structure may further complicate the analysis and interpretation of the PTS1 signal sequence interaction with LdPEX5 by these biophysical techniques.
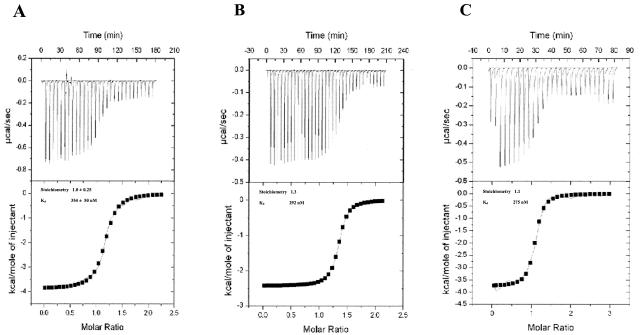
To further characterize the LdPEX5-PTS1 interaction, we used ITC to determine the stoichiometry, the number of AKL PTS1 peptide molecules bound to each LdPEX5 subunit, and the Kd for the LdPEX5-AKL PTS1, ldpex5 Δ269-291-AKL PTS1, and His6-CT-ldpex5-AKL PTS1 complexes (9, 31). The Kd values determined by ITC for the AKL peptide interacting with LdPEX5, ldpex5 Δ269-291, and His6-CT-ldpex5 were 354, 292, and 275 nM, respectively (Fig. 4). The Kd value for LdPEX5 is in good agreement with the value previously determined by fluorescence polarization (30). Moreover, fluorescence polarization experiments revealed that in solution His6-CT-ldpex5 bound the fluorescein-labeled AKL PTS1 peptide (30) with a Kd value of 204 ± 31 nM (data not shown), further validating our ITC results.
FIG. 4.
ITC analysis of LdPEX5-PTS1 interaction. (A) 60 μM LdPEX5, (B) 60 μM ldpex5 Δ269-291, and (C) 26 μM His6-CT-ldpex5 in degassed 20 mM sodium phosphate (pH 7.5)-120 mM NaCl-3 mM β-mercaptoethanol were titrated at a constant temperature of 30°C against a 1 mM solution of AKL peptide delivered at 6-min intervals from a syringe. The top panel for each LdPEX5 protein represents the thermogram showing the amount of heat released (in microcalories per second) after each injection. The bottom panel is the corresponding plot for the heat of reaction per injection (in kilocalories per mole of peptide) as a function of the [peptide]/[LdPEX5] ratio. The binding association constant Ka (Kd = 1/Ka), the stoichiometry (n), and the enthalpy (ΔH0) were determined after fitting the data by using the model for a one type of binding site.
For all LdPEX5 variants, the molar ratio of peptide-LdPEX5 subunit was measured to be ∼1:1. It should be noted that all ITC experiments with the AKL peptide were performed under reducing conditions, and after the titrations the reaction mixtures were analyzed by reversed-phase chromatography to confirm that no disulfide AKL peptide dimers were present (data not shown). The ITC profiles for the binding of AKL PTS1 to all three LdPEX5 variants were comparable regardless of the oligomeric state, which suggests that binding occurs without any major allosteric effect. The absence of cooperativity was further confirmed by Hill plot analysis of the ITC and fluorescence polarization data, which both yielded a linear relationship with a Hill factor of 1.
Effect of PTS1 ligand on LdPEX5 quaternary structure.
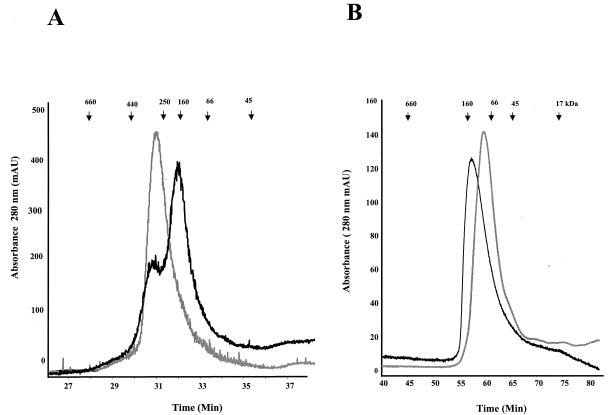
ITC experiments indicated that LdPEX5 and ldpex5 Δ269-291 were capable of binding the AKL PTS1 peptide with comparable kinetics and that each LdPEX5 subunit bound a single AKL peptide. This series of experiments, however, did not provide any information on the oligomeric state of the resulting AKL peptide containing complexes. To further characterize the quaternary structures of LdPEX5-AKL PTS1 and ldpex5 Δ269-291-AKL PTS1, these complexes were analyzed by gel permeation chromatography. A comparison of the elution profile of LdPEX5 with the profile of LdPEX5-AKL mixture revealed a shift in the elution of LdPEX5 from a complex with a mass of ∼270 kDa (tetramer) to a complex with a mass of ∼140 kDa, a size consistent with the AKL-bound complex being composed of two LdPEX5 subunits (Fig. 5A). Similar experiments conducted with ldpex5 Δ269-291 revealed that incubation of this mutant receptor protein with the AKL peptide induced a ldpex5 Δ269-291 dimer-to-monomer transition (Fig. 5B).
FIG. 5.
The AKL PTS1 Peptide induces quaternary structural changes in LdPEX5. (A) Purified LdPEX5 (30 μg in 50 mM Tris [pH 8.0], 100 mM NaCl) was incubated in the absence (thin trace) or in the presence of a 40-fold molar (thick traces) excess of AKL peptide at 20°C for 1 h and then injected onto a sizing column composed of a tandemly plumbed Bio-Sil SEC 450-5/Bio-Sil SEC 250-5 (7.8 by 150 mm or 7.8 by 300 mm) equilibrated with 25 mM Tris-100 mM NaCl (pH 8.0)-2 mM DTT at a flow rate of 0.4 ml/min. Protein elution was monitored at 280 nm. (B) ldpex5 Δ269-291 in the absence (thin trace) or presence (thick trace) of an excess of AKL peptide incubated at 30°C for 3 h was analyzed on a Bio-Sil SEC 250-5 (7.8 by 600 mm) column by using a mobile phase of 25 mM Tris-100 mM NaCl (pH 8.0)-2 mM DTT at a flow rate of 0.25 ml/min. Quaternary structure of ldpex5 Δ269-291 in the presence of PTS1 was performed on the reaction mixture after the ITC analysis.
Analysis of LPEX5-PTS1 interactions by SPR.
To examine the LdPEX5-AKL PTS1 interaction in real-time and to obtain information on the kinetics interaction and dissociation of the LdPEX5 variants binding to the AKL PTS1 ligand, we used an SPR-based biosensor (the Biacore) approach. The AKL synthetic peptide was covalently coupled to the biosensor chip in an oriented manner via the thiol group of the engineered N-terminal cysteine. Binding of LdPEX5 to the AKL peptide was detected as a change in RUs, whereas LdPEX5 was injected at fixed concentrations over the PTS1 and control surfaces in a continuous fashion (the wash-on phase in Biacore terminology). The LdPEX5 solution was then replaced with buffer, and dissociation of LdPEX5:AKL complex was monitored (the wash-off phase). Regeneration of the surfaces, i.e., dissociation of the remaining complexes, was achieved via 6 M GdnHCl injections.
Biacore sensor surfaces from CM4 chips are composed of long carboxy-methylated dextran chains harboring functional carboxyl group to allow covalent immobilization of ligand to the surface. The flexibility of these dextran chains permits the reconstitution of multimeric complexes in which more than one immobilized molecule can participate in the interaction with its injected partner, giving rise to an avidity phenomena. An example of that situation was given in De Crescenzo et al. (14). In that study, the authors demonstrated that a single molecule of transforming growth factor β (TGF-β; a covalent homodimer) can bind to two TGF-β type II receptor (TβRIIED) molecules immobilized on the biosensor surface. The increased binding avidity is due to the multiplicity of contacts between one TGF-β molecule and two TβRIIED receptor domains, which stabilize the complex. This effect is most evident when the dissociation phases of sensorgrams in which TβRIIED is immobilized on the biosensor and TGF-β injected are compared to those in which the TGF-β is immobilized and the TβRIIED is injected. The effect was observed even when a low amount of TβRIIED was captured, thus demonstrating the high flexibility of the dextran chains.
Figure 6 shows sensorgrams control corrected and normalized to arbitrary units corresponding to injections of LdPEX5, ldpex5 Δ269-291, and His6-CT-ldpex5 subunits at 300 nM over the 10 RU- and 100 RU-loaded AKL surfaces. The resulting sensorgrams indicate that LdPEX5, ldpex5 Δ269-291, and His6-CT-ldpex5 subunits bound to AKL peptide with apparently similar association and dissociation rates (Fig. 6A and B). This observation strongly suggests that LdPEX5 tetramers and ldpex5 Δ269-291 dimers dissociate upon AKL binding. Indeed, if both tetrameric and dimeric structures had been preserved after AKL binding, an avidity situation, i.e., LdPEX5 oligomers involved in simultaneous binding to at least two AKL peptides, would have occurred at the biosensor surface. This would have resulted in drastically different dissociation profiles, as reported for TGF-β-TβRIIED interaction (14).
This avidity effect would have also resulted in noticeable changes in the dissociation profiles of LdPEX5 and ldpex5 Δ269-291 as the AKL surface density is increased since the probability for the multimeric PEX5 complexes to be engaged in more than one interaction with the surface AKL would have increased. However, this was not observed (Fig. 6B), strongly suggesting that LdPEX5 tetramers and ldpex5 Δ269-291 dimers dissociate upon AKL binding.
Global analysis trials using a simple model were conducted for each LdPEX5 mutant set of sensorgrams. This approach resulted in poor fits, as monitored by the nonrandom distribution of the residuals (data not shown). This deviation from a simple model is not likely due to “crowding effects” (43), mass transport limitation (13), or avidity effect (see above) since similar deviations were observed for experimental data sets generated at high flow rate (100 μl/min) on both 10 RU- and 100 RU-loaded AKL surfaces (data not shown). This complex mode of binding is, however, consistent with our gel permeation data, which revealed a change in the quaternary structure of LdPEX5 and ldpex5 Δ269-291 complexes upon AKL binding.
A steady-state analysis approach was used to determine the apparent thermodynamic dissociation constants (Kd) for each LdPEX5-AKL PTS1 interaction. Kd values of 140 ± 20, 257 ± 34, and 620 ± 100 nM for LdPEX5, ldpex5 Δ269-291, and His6-CT-ldpex5 were estimated by plotting the control-corrected plateau values for the 10 and 100 RU surfaces as a function of the LdPEX5, ldpex5 Δ269-291, and His6-CT-ldpex5 subunit concentrations (Fig. 6C to E). These Kd values are in good agreement with those obtained by ITC (Fig. 4) and fluorescence polarization techniques (30); the slight differences in Kd may be attributed to the fact that ITC experiments were performed at 30°C, whereas Biacore experiments were performed at 25°C. The minor twofold difference in the Kd values measured for LdPEX5 by using ITC and SPR may be attributed to the fact that the tetrameric structure of LdPEX5 would facilitate accumulation of LdPEX5 at the biosensor surface prior to the dissociation of the tetramer. This view is supported by the complex kinetics observed during the wash-on phase of the sensorgram. Furthermore, the good correlation between results generated on the 10 RU- and 100 RU-loaded AKL surfaces and a single site binding model, in addition to the similarity of the Kd values determined for the various LdPEX5 mutants (fourfold difference only between His6-CT-ldpex5 and LdPEX5) reinforces our conclusion that an avidity situation did not occur at the biosensor surface.
LdPEX14 alters the LdPEX5-PTS1 interaction affinity.
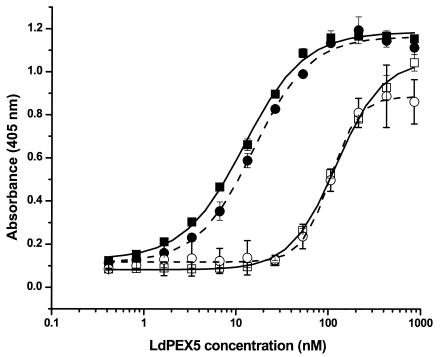
SPR experiments demonstrated that the LdPEX5-AKL PTS1 peptide complex was stable (slow dissociation rate, see Fig. 6A and B). This observation suggested that release of PTS1 ligands is dependent on the recruitment of additional glycosome biogenesis proteins. A likely candidate that may facilitate this event is LdPEX14, since the initial contact of the LdPEX5-PTS1 complex with the glycosome involves this protein. We used an ELISA-based assay to test whether LdPEX14 recruitment of LdPEX5-PTS1 complex can modulate the LdPEX5-PTS1 interaction affinity. In these experiments, recombinant LdXPRT, immobilized on microtiter plates, was incubated with various concentrations of LdPEX5 or ldpex5 Δ269-291 in the absence or presence of LdPEX14. The amount of LdPEX5 that bound to the LdXPRT was quantitated by using anti-LdPEX5 specific antisera. The binding of LdPEX5 is known to be dependent on the C-terminal AKL tripeptide of LdXPRT since a mutant form of XPRT lacking the PTS1 tripeptide does not bind LdPEX5 (29). Kd values measured for LdPEX5 and ldpex5 Δ269-291 binding to LdXPRT were 12.2 ± 0.7 and 14.1 ± 1.4 nM, respectively (Fig. 7). However, in the presence of LdPEX14 the affinity of LdPEX5 and ldpex5 Δ269-291 for LdXPRT decreased by ∼10-fold (Kd values of 122.3 ± 5.5 and 104 ± 4.3 nM for LdPEX5 and ldpex5 Δ269-291, respectively). The addition of LdPEX14 alone to LdXPRT-coated microtiter plates confirmed that these two proteins did not directly interact, since no detectable LdPEX14 binding was observed by using anti-LdPEX14-specific antisera (data not shown). Control experiments also confirmed that there was no cross-reactivity between anti-LdPEX5 antisera with LdXPRT or LdPEX14 proteins.
FIG. 7.
LdPEX14 modulates the LdPEX5-XPRT interaction affinity. Microtiter plates were coated with 1 μg of wild-type LdXPRT/well for 16 h at 4°C in 100 μl of PBS and blocked with 2% milk powder in PBS. Twofold serial dilutions (0.4 to 860 nM) of LdPEX5 (▪), LdPEX5 plus 4 μg of His6-S-LdPEX14 (□), ldpex5 Δ269-291 (•), or ldpex5 Δ269-291 plus 4 μg of His6-S-LdPEX14/well (○)/well in 100 μl of 2% ABS in PBS-0.1% Tween 20 was added to the LdXPRT-coated wells, followed by incubation for 2 h at 20°C. LdPEX5 or ldpex5 Δ269-291 bound to the LdXPRT on the microtiter plates was quantitated by using anti-LdPEX5 primary antibody, goat anti-rabbit horseradish peroxidase-conjugated secondary antibody, and ABTS as the chromogenic substrate. Kd for LdPEX5 with XPRT was determined by using ORIGIN software version 7.0 (Microcal Software).
That the shift in the LdPEX5-LdXPRT binding affinity observed in the presence of LdPEX14 was not due to a decrease in anti-LdPEX5 antibody binding is supported by control ELISAs performed with LdPEX5 immobilized on microtiter plates. These experiments showed that preincubation of LdPEX5 with LdPEX14 concentrations as high as 4 μg/100 μl resulted in only ∼10 to 15% decrease in color development (optical densities at 405 nm of 0.5 or 0.59 in the presence or absence of LdPEX14, respectively), suggesting that association of LdPEX14 with LdPEX5 has a minimal steric effect on the binding of anit-LdPEX5 antibodies. To further validate that LdPEX14 binding to LdPEX5 was responsible for the modulation of the LdPEX5-LdXPRT binding affinity, we performed an additional control experiment with an N-terminal mutant of LdPEX14 that does not bind LdPEX5 (Madrid and Jardim, submitted). These experiments showed that in the presence of high concentrations of this ldpex14 mutant the Kd for the LdPEX5-LdXPRT association was ∼12 nM, which is essentially identical to the affinities measured in the absence of LdPEX14, further confirming that association of LdPEX14 to LdPEX5 dramatically decreases its affinity for PTS-1 ligands. That LdPEX14 induced a comparable modulatory effect on the LdPEX5-LdXPRT and ldpex5 Δ269-291-LdXPRT interactions suggested that LdPEX14 was capable of binding LdPEX5 and ldpex5 Δ269-291 with similar affinities. ELISAs confirmed that LdPEX14 bound LdPEX5 and ldpex5 Δ269-291 with Kd values of 14 ± 8 and 23 ± 7 nM, respectively.
The Kd values measured for the LdPEX5-LdXPRT and ldpex5 Δ269-291-LdXPRT interactions are in good agreement with previous studies (29) and confirm the ITC and SPR results, i.e., that disruption of the coiled-coil motif does not alter the affinity of AKL PTS1 for LdPEX5. The discrepancy in the binding constants measured for PTS1 peptide and full-length XPRT has been previously reported for other PEX5 proteins (25). It is possible that these differences are due to additional contacts between the PEX5 receptor and the PTS1-containing protein, which would result in tighter binding. Such a loss of affinity may also result from differences in the conformation of PTS1 peptide versus PTS1 motif within the context of the XPRT protein.
DISCUSSION
LdPEX5 is a critical cytosolic receptor required for the sorting and import of PTS1 proteins into the glycosomal matrix. To gain further insight into the role of LdPEX5 in glycosome biogenesis, we examined the quaternary structure of LdPEX5 and assessed how the binding of PTS1 to this receptor protein modulates its oligomeric state. Previous biochemical studies have demonstrated that the human, yeast, and Leishmania PEX5 proteins form tetramers in the absence of PTS1 (5, 29, 49). However, the issue of whether the quaternary structure of this receptor plays a significant role in protein targeting has not been previously reported.
In an effort to identify the LdPEX5 regions responsible for its quaternary structure, a computational analysis of the LdPEX5 primary sequence was performed. This in silico approach predicted, with strong probability, the presence of a single coiled-coil motif located in the region corresponding to residues 277 to 310. This putative coiled-coil motif is located immediately upstream of the first TPR repeat (Fig. 1) that participates in the formation of the PTS1 binding site (24, 29, 32). A similar examination of the human, S. cerevisiae, H. polymorpha, and T. brucei PEX5 sequences indicated that these proteins lack a comparable coiled-coil structure. This suggests that LdPEX5 may be structurally and functionally unique from other PEX5 receptors. The importance of this putative coiled-coil motif for LdPEX5 oligomerization was experimentally investigated by examining the quaternary structures of (i) various LdPEX5 deletion mutants and (ii) peptides corresponding to the putative coiled-coil motif (Fig. 2). Gel permeation chromatography results for ldpex5 Δ269-291, a deletion mutant, designed to eliminate two of the five predicted heptad units, indicated that the oligomeric state of LdPEX5 changed from tetramer to dimer upon removal of residues 269 to 291 (Fig. 3). These results are in agreement with the observations that the stability of a coiled-coil interaction is in part related to the length of each helix. Indeed, several studies have demonstrated that decreasing the number of heptad units from five to three can lead to a dramatic loss in affinity from picomolar to high micromolar (13, 53).
Further evidence that amino acids 277 to 310 modulate LdPEX5 oligomerization was provided by the fact that the peptides ldpex5 203-391 and ldpex5 290-391 form dimers (Fig. 1C). It is interesting that although ldpex5 290-391 only retained a small portion of the predicted coiled coil it still formed a dimeric structure. Examination of the ldpex5 290-391 sequence revealed that this protein still retained two TPRs (residues 323 to 391). The crystal structures of human (24) and T. brucei (32) PEX5 have shown that these TPR motifs form intra- and interhelical contacts that are stabilized by hydrophobic interaction. It is possible that dimerization the ldpex5 290-391 fragment may in part be stabilized by these intermolecular TPR interactions as previously suggested (24, 32). In the full-length PEX5 proteins these inter-TPR interactions are important for stabilizing the largely α-helical structure that forms the PTS-1 binding pocket (24). It is unlikely that the putative inter-TPR contacts found in ldpex5 290-391 would be involved in the oligomerization of larger ldpex5 fragments since previous studies have shown that the C-terminal portion of LdPEX5, His6-CT-ldpex5, a fragment encompassing amino acids 303 to 625, is known to be monomeric (29).
Analysis of the coiled-coil motif on a helical wheel diagram (Fig. 2B) revealed that the helical interface formed by residues at positions a and d, contained predominantly tryptophan, tyrosine, leucine, and methionine. Such clustering of larger nonpolar residues at the hydrophobic core of two-stranded α-helical coiled-coils is a thermodynamically favorable factor important for controlling protein folding and stability (35).
Coiled coils can pack into either (i) a parallel geometry that orients the N termini of the two coil subunits in the same direction, an architecture stabilized by a↔a′ and d↔d′ interhelical contacts, or (ii) an antiparallel geometry that orients the N termini of each coil in opposite directions, a configuration stabilized by a↔d′ and a′↔d contacts (59). Examination of the helical diagrams (Fig. 2B) for the LdPEX5 coiled-coil sequence suggests that the parallel and antiparallel packing geometries are both probable, although the antiparallel organization may be slightly favored since several electrostatic interactions and hydrogen bonds involving residues at positions g and g′ are possible (13).
Another feature of the LdPEX5 coiled coil is the presence of a centrally located WXXXY/F pentapeptide, a motif that in yeast (6), mammalian (48), T. brucei (8), and plant (40) PEX5 proteins has been demonstrated to be critical for interactions with PEX14 and PEX13. Nuclear magnetic resonance structures of one of the yeast PEX5 WXXXY/F motifs confirmed that this sequence adopted an α-helical conformation that placed both aromatic residues on the same helix face, creating an amphipathic structure that is cradled in a hydrophobic cleft upon binding to PEX14 or to the SH3 domain of PEX13 (17). Surprisingly, even though LdPEX5 contains three such repeats, site-directed mutagenesis, deletion mutagenesis, and biochemical measurements have established that none of these repeats are required for LdPEX14 interaction since manipulations of these sequences do not dramatically alter the LdPEX5-LdPEX14 affinity (Madrid and Jardim, submitted). The results reported here would further argue that the motif 293WAQEY297 is not required for LdPEX14 association since the critical aromatic residues are buried within the coiled-coil hydrophobic core. Interestingly, deletion of this coiled-coil motif resulted in an ldpex5 mutant protein that adopted a dimeric structure.
This observation, together with the ITC and gel permeation chromatography findings, demonstrated that the LdPEX5 tetramer is stabilized by two discreet oligomerization domains that have been mapped to the first 391 residues. The first of these domains corresponding to the coiled-coil motif spanning residues 277 to 310 is a region critical for dimer formation, and disruption of this motif, as in the lpdex5 Δ269-291 mutant, results in this protein adopting a dimeric rather than tetrameric structure in the absence of a PTS1 ligand. The second oligomerization domain is located within the first 202 residues, and expression of the N-terminal mutant ldpex5 1-202 revealed that this protein adopted a hexameric structure. It is plausible that ldpex5 1-202 forms a hexamer rather than a tetrameric state observed with the full-length LdPEX5 because removal of the C-terminal TPR domain eliminated steric hindrances that allow for the formation of a higher-order quaternary complex. These findings contrast with previous studies with the human PEX5 that reported that a single N-terminal region was responsible for tetramer formation (50). It is the interplay of these two domains that are responsible for homotetrameric structure formed by LdPEX5 and deletion of one or both of these domains results in the mutant LdPEX5 proteins forming dimers or monomers, as is the case for ldpex5 Δ269-291 and His6-CT-ldpex5, respectively.
A novel finding of the present study, one not previously observed with other PEX5 receptor proteins, was the observation that PTS1 ligands induced an oligomeric change in LdPEX5. The binding of AKL PTS1 peptide triggered an LdPEX5 tetramer-to-dimer and an ldpex5 Δ269-291 dimer-to-monomer dissociation. ITC experiments showed that His6-CT-ldpex5, which encompassed only the TPR domain, bound the AKL PTS1 peptide by one-to-one stoichiometry. Furthermore, ITC and SPR studies showed that the affinity for the AKL peptide was not influenced by the initial quaternary structure of LdPEX5 since similar Kd values were determined for the LdPEX5, ldpex5 Δ269-291, and His6-CT-ldpex5 mutants (Fig. 4). This is consistent with the X-ray crystallographic structure of the human PEX5 TPR domain determined by Gatto et al. (24), which showed a single PTS1 peptide bound within a groove formed by the TPR helical hairpin clusters. Although LdPEX5 is tetrameric, each subunit appears to function independently since no apparent cooperative effects for PTS1 binding were detected in ITC, fluorescence polarization (29), or SPR experiments (Fig. 4 and 6). Hill plot analysis of the ITC and fluorescence polarization data further confirmed the absence of cooperativity since these plots were linear with a Hill constant of 1. Recently, it has been reported that H. polymorpha PEX5 also undergoes quaternary structural cycling from a tetramer to monomer during the process of delivering PTS1 ligands to the peroxisomal matrix. In contrast to LdPEX5, HpPEX5 does not dissociate on binding PTS1, but rather the transition from a tetramer to a monomer is triggered by binding the intraperoxisomal protein HpPEX8 (56) to form a HpPEX5-HpPEX8 heterodimer that does not bind PTS1. Boteva et al. (5) have also suggested that dissociation may also occur in response to a shift in environmental pH from 7.2 to 6.0 (5).
The SPR studies also revealed that, in spite of the tetrameric and dimeric oligomerization state of LdPEX5 and ldpex5 Δ269-291, respectively, no avidity effects were observed regardless of the AKL peptide loading on the biosensor surface, suggesting that the tetrameric and dimeric structures of these proteins are rapidly disrupted upon AKL peptide binding (Fig. 6).
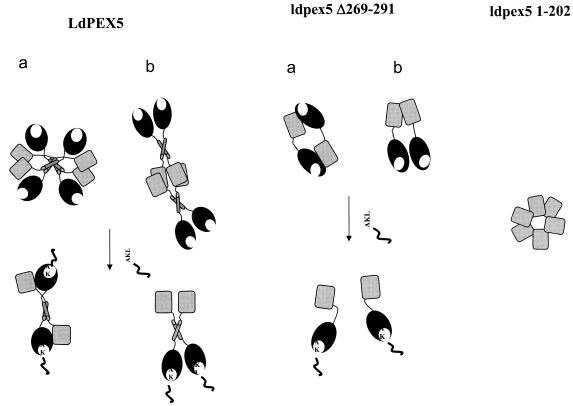
On the basis of the structural and biophysical data, two models for the LdPEX5 tetrameric structure are proposed (Fig. 8), in which the coiled-coil motif has an antiparallel or a parallel orientation. In the model with antiparallel orientation, the TPR domain, containing the PTS1-binding pocket, packs adjacent to and is proposed to form intermolecular contacts with the N-terminal domain of the neighboring LdPEX5 subunit and binding of PTS1 would disrupt these intermolecular interaction between the N- and C-terminal regions resulting in tetramer dissociation (Fig. 8, model a). In the model where the coiled-coil has parallel organization, the N termini of two LdPEX5 subunits are arranged in a head-to-head orientation that is stabilized by the interhelical hydrophobic interaction of the coiled-coil to form a dimeric structure. It is possible that this dimer may also be stabilized by addition of intersubunit protein-protein contacts formed between the N-terminal, as well as the C-terminal regions (Fig. 8, model b). In this model the stabilization of the LdPEX5 tetramer (dimer-of-dimers) involves primarily contacts between the oligomerization domains within residues 1 to 202.
FIG. 8.
Models of the LdPEX5 quaternary structure. The diagram illustrates the possible quaternary architecture of the wild-type LdPEX5 and ldpex5 Δ269-291 recombinant proteins. In the absence of a PTS1 ligand, LdPEX5 is a tetramer composed of a dimer of dimers. Each dimer is stabilized by a coiled-coil interaction that has either an antiparallel (a) or a parallel (b) organization. The tetrameric structure is stabilized by contacts involving an oligomerization motif located in the N-terminal region between residues 1 and 202. The mutant ldpex5 Δ269-291, which has a disrupted coiled-coil motif, forms a dimer in the absence of PTS1. In the presence of PTS1 synthetic peptide, LdPEX5 undergoes a tetramer-to-dimer dissociation, and ldpex5 Δ269-291 dissociates from a dimer to a monomer. It is hypothesized that binding of PTS1 to the C-terminal TPR domain transduces a structural signal to the N-terminal domain that leads to oligomer dissociation.
Both models (Fig. 8) imply that binding of PTS1 to the C-terminal TPR domain transduces a structural change that alters the LdPEX5 protein-protein interaction, in turn leading to oligomer dissociation. This hypothesis is in agreement with our SPR results indicating that the AKL peptide-His6-CT-ldpex5 interaction deviates from a simple kinetic model. Such a deviation is not likely to be due to nonoptimized experimental conditions since we used an oriented approach to immobilize the AKL peptide and since experiments were performed at high flow rate and low ligand densities to eliminate any crowding and mass transport limitation effects. In the light of our other results, such a deviation may be due to a kinetically limiting conformational change. However, whether this putative conformational change alters intramolecular or intermolecular LdPEX5 contacts is unknown. Further experiments are needed to address this issue.
It should be noted that the partial deletion introduced into ldpex5 Δ269-291 did not appear to significantly alter the global architecture of the N- and C-terminal domains (29) since functional assays showed that this mutant protein was capable of binding PTS1 ligands and LdPEX14 with affinities comparable to that of the wild-type LdPEX5. These findings are consistent with the fact that ldpex5 Δ269-291 still adopts a dimeric structure and dissociates into monomers upon AKL peptide binding.
Implicit in this model is the suggestion that PTS1-laden LdPEX5 binds the glycosomal membrane-associated protein LdPEX14 as dimer and, once the PTS1 cargo is unloaded, LdPEX5 cycles back into the cytosol as tetramer in accordance with proposed PEX5 recycling models (12, 16). Dissociation of the LdPEX5 tetramer may expose additional contacts on LdPEX5 needed for enhanced LdPEX14 interaction or other uncharacterized components of the glycosome protein translocation machinery. Moreover, it is likely that the LdPEX5 tetramer-dimer interchange may provide a mechanism that regulates the interaction of LdPEX5 with membrane-associated LdPEX14. Such a regulatory mechanism would be important for maintaining the glycosomal PTS1 matrix protein import efficiency by preventing LdPEX5 devoid of a PTS1 cargo from forming a permanent association with the LdPEX14 docking complex on the glycosomal membrane. Alternatively, the dimeric form of LdPEX5 may expose hidden interaction domains that would permit other peroxins to bind and facilitate the release of LdPEX5 from LdPEX14, allowing the LdPEX5 tetramer form prior to recycling this receptor back into the cytosolic pool. Although PTS1 has not been previously demonstrated to alter the quaternary structure of other PEX5 receptors, it is known to modulate association of PEX5 with PEX13 and PEX14 (44, 50, 55). Whether the shuffling of the PEX5-PTS1 complex from PEX14 to PEX13 is important for the unloading of the protein cargo is unclear.
In Leishmania, experiments that mimic the glycosomal docking event showed that binding of the LdPEX5-PTS1 complex to LdPEX14 caused a 10-fold decrease in LdPEX5-PTS1 interaction affinity. This suggests that the LdPEX5-LdPEX14 contact may trigger PTS1 unloading, but whether this occurs at the cytosolic face of the glycosomal membrane or in the glycosomal matrix is unknown. SPR studies have demonstrated that the LdPEX5-PTS1 complex is extremely stable. This tight interaction is essential for effective sorting and trafficking of PTS1 matrix proteins from the cytosolic polyribosome to the glycosomal surface. However, once the LdPEX5-PTS1 complexes have reached their glycosomal destination, disruption of this tight interaction is necessary to facilitate unloading of the PTS1 cargo by binding to LdPEX14. It should be noted here that, unlike yeast and mammalian PEX14 proteins, which are integral peroxisome membrane proteins, LdPEX14 is a soluble protein that peripherally associates with glycosomal membrane (30). Wang et al. (56) have documented that release of PTS1 from H. polymorpha PEX5 is induced by formation of the HpPEX5-HpPEX8 heterodimeric complex in the peroxisome matrix.
In the present study we report that the mechanism for the delivery of the PTS1 protein may involve quaternary structural changes of the PEX5 receptor, which in turn influences the binding affinity of PEX5 for the PTS1 protein as the complex moves from the cytosol to the glycosomal membrane and interacts with PEX14.
Acknowledgments
This study was supported by grants from the Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council of Canada to A.J. and a Regroupements Stratégiques Centre grant from the Fonds de Recherche sur la Nature et les Technologies du Québec.
We thank J. Turnbull for assistance with the circular dichroism experiments.
REFERENCES
- 1.Albertini, M., R. Rehling, R. Erdmann, W. Girlasky, J. A. Kiel, M. Veenhuis, and W. H. Kunau. 1997. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell 89:83-92. [DOI] [PubMed] [Google Scholar]
- 2.Bakker, B. M., F. I. Mensonides, B. Teusink, P. van Hoek, P. A. Michels, and H. V. Westerhoff. 2000. Compartmentation protects trypanosomes from the dangerous design of glycolysis. Proc. Natl. Acad. Sci. USA 97:2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner, J., S. Helfert, P. Michels, and C. Clayton. 1998. Compartmentation of phosphoglycerate kinase in Trypanosoma brucei plays a critical role in parasite energy metabolism. Proc. Natl. Acad. Sci. USA 95:11596-11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, J., B. Swinkels, H. Dorsam, T. Prospero, S. Subramani, and C. Clayton. 1992. Glycosome assembly in trypanosomes: variations in the acceptable degeneracy of a COOH-terminal microbody targeting signal. J. Cell Biol. 119:1129-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boteva, R., A. Koek, N. V. Visser, A. J. Visser, E. Krieger, T. Zlateva, M. Veenhuis, and I. van der Klei. 2003. Fluorescence analysis of the Hansenula polymorpha peroxisomal targeting signal-1 receptor, Pex5p. Eur. J. Biochem. 270:4332-4338. [DOI] [PubMed] [Google Scholar]
- 6.Bottger, G., P. Barnett, A. T. J. Klein, A. Kragt, H. F. Tabak, and B. Distel. 2000. Saccharomyces cerevisiae PTS1 receptor Pex5p interacts with the SH3 domain of the peroxisomal membrane protein Pex13p in an unconventional, non-PXXP-related manner. Mol. Biol. Cell 11:3963-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brocard, C., G. Lametschwandtner, R. Koudelka, and A. Hartig. 1997. Pex14p is a member of the protein linkage map of Pex5p. EMBO J. 16:5491-5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choe, J., J. Moyersoen, C. Roach, T. L. Carter, E. Fan, P. A. M. Michels, and W. G. Hol. 2003. Analysis of the sequence motifs responsible for the interactions of peroxins 14 and 5, which are involved in glycosome biogenesis in Trypanosoma brucei. Biochemistry 42:10915-10922. [DOI] [PubMed] [Google Scholar]
- 9.Cliff, M. J., and J. E. Ladbury. 2003. A survey of the year 2002 literature on applications of isothermal titration calorimetry. J. Mol. Recognit. 16:383-391. [DOI] [PubMed] [Google Scholar]
- 10.Combet, C., C. Blanchet, C. Geourjon, and G. Deléage. 2000. NPS@: network protein sequence analysis. Trends Biochem. 25:147-150. [DOI] [PubMed] [Google Scholar]
- 11.Crick, F. H. C. 1953. The packing of alpha-helices: simple coiled coils. Acta Crystallogr. 6:685-697. [Google Scholar]
- 12.Dammai, V., and S. Subramani. 2001. The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell 105:187-196. [DOI] [PubMed] [Google Scholar]
- 13.De Crescenzo, G., J. R. Litowski, R. S. Hodges, and D. M. O'Connor-McCourt. 2003. Real-time monitoring of the interactions of two-stranded de novo designed coiled-coils: effect of chain length on the kinetic and thermodynamic constants of binding. Biochemistry 42:1754-1763. [DOI] [PubMed] [Google Scholar]
- 14.De Crescenzo, G., P. L. Pham, Y. Durocher, and D. M. O'Connor-McCourt. 2003. Transforming growth factor-β (TGF-β) binding to the extracellular domain of the type II TGF-β receptor: receptor capture on a biosensor surface using a new coiled-coil capture system demonstrates that avidity contributes significantly to high-affinity binding. J. Mol. Biol. 328:1173-1183. [DOI] [PubMed] [Google Scholar]
- 15.De Walque, S., J. A. Kiel, M. Veenhuis, F. R. Opperdoes, and P. A. Michels. 2000. Cloning and analysis of the PTS-1 receptor in Trypanosoma brucei. Mol. Biochem. Parasitol. 104:106-119. [DOI] [PubMed] [Google Scholar]
- 16.Dodt, G., and S. J. Gould. 1996. Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J. Cell Biol. 135:1763-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douangamath, A., F. V. Filipp, A. T. Klein, P. Barnett, P. Zou, T. Voorn-Brouwer, M. C. Vega, O. M. Mayans, M. Sattler, B. Distel, and M. Wilmanns. 2002. Topography for independent binding of alpha-helical and PPII-helical ligands to a peroxisomal SH3 domain. Mol. Cell 10:1007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elgersma, Y., L. Kwast, Klein, T. A. Voorn-Brouwer, M. van den Berg, T. America, H. F. Tabak, and B. Distel. 1996. The SH3 domain of the Saccharomyces cerevisiae peroxisomal membrane protein Pex13p functions as a docking site for Pex5p, a mobile receptor for the import PTS1-containing proteins. J. Cell Biol. 135:97-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erdmann, R., and G. Blobel. 1996. Identification of Pex13p a peroxisomal membrane receptor for the PTS1 recognition factor. J. Cell Biol. 135:111-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fairlamb, A. H. 1989. Novel biochemical pathways in parasitic protozoa. Parasitology 99S:93-112. [DOI] [PubMed] [Google Scholar]
- 21.Fransen, M., C. Brees, E. Baumgart, J. C. Vanhooren, M. Baes, G. P. Mannaerts, and P. P. Van Veldhoven. 1995. Identification and characterization of the putative human peroxisomal C-terminal targeting signal import receptor. J. Biol. Chem. 270:7731-7736. [DOI] [PubMed] [Google Scholar]
- 22.Fransen, M., S. R. Terlecky, and S. Subramani. 1998. Identification of a human PTS1 receptor docking protein directly required for peroxisomal protein import. Proc. Natl. Acad. Sci. USA 95:8087-8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furuya, T., P. Kessler, A. Jardim, A. Schnaufer, C. Crudder, and M. Parsons. 2002. Glucose is toxic to glycosome-deficient trypanosomes. Proc. Natl. Acad. Sci. USA 99:14177-14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gatto, G. J., B. V. Geisbrecht, S. J. Gould, and J. M. Berg. 2000. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 7:1091-1094. [DOI] [PubMed] [Google Scholar]
- 25.Gatto, G. J., Jr., E. L. Maynard, A. L. Guerrerio, B. V. Geisbrecht, S. J. Gould, and J. M. Berg. 2003. Correlating structure and affinity for PEX5:PTS1 complexes. Biochemistry 42:1660-1666. [DOI] [PubMed] [Google Scholar]
- 26.Gill, S.C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 27.Guerra-Giraldez, C., L. Quijada, and C. E. Clayton. 2002. Compartmentation of enzymes in a microbody, the glycosome, is essential in Trypanosoma brucei. J. Cell Sci. 115:2651-2658. [DOI] [PubMed] [Google Scholar]
- 28.Jardim, A., S. E. Bergeson, S. Shih, N. Carter, R. W. Lucas, G. Merlin, P. J. Myler, K. Stuart, and B. Ullman. 1999. Xanthine phosphoribosyltransferase from Leishmania donovani: molecular cloning, biochemical characterization, and genetic analysis. J. Biol. Chem. 274:34403-34410. [DOI] [PubMed] [Google Scholar]
- 29.Jardim, A., W. Liu, E. Zheleznova, and B. Ullman. 2000. Peroxisomal targeting signal-1 receptor protein PEX5 from Leishmania donovani: molecular, biochemical, and immunocytochemical characterization. J. Biol. Chem. 275:13637-13644. [DOI] [PubMed] [Google Scholar]
- 30.Jardim, A., N. Rager, W. Liu, and B. Ullman. 2002. Peroxisomal targeting protein 14 (PEX14) from Leishmania donovani: molecular, biochemical, and immunocytochemical characterization. Mol. Biochem. Parasitol. 124:51-62. [DOI] [PubMed] [Google Scholar]
- 31.Jelesarov, I., and H. R. Bosshard. 1999. Isothermal titration calorimetry and differential scanning calorimetry as complementary tools to investigate the energetics of biomolecular recognition. J. Mol. Recognit. 12:3-18. [DOI] [PubMed] [Google Scholar]
- 32.Kumar, A., C. Roach, I. S. Hirsh, S. Turley, S. de Walque, P. A. Michels, and W. G. Hol. 2001. An unexpected extended conformation for the third TPR motif of the peroxin PEX5 from Trypanosoma brucei. J. Mol. Biol. 307:271-282. [DOI] [PubMed] [Google Scholar]
- 33.Krittani, C., and W. C. Johnson, Jr. 1997. Correcting the circular spectra of peptides for the contribution of absorbing side chains. Anal. Biochem. 253:57-64. [DOI] [PubMed] [Google Scholar]
- 34.Krittani, C., and W. C. Johnson, Jr. 2000. The relative order of helical propensity of amino acids changes with the solvent environment. Proteins 39:132-141. [DOI] [PubMed] [Google Scholar]
- 35.Kwok, S. C., and R. S. Hodges. 2003. Clustering of large hydrophobes in the hydrophobic core of two-stranded alpha-helical coiled-coils controls protein folding and stability. J. Biol. Chem. 278:35248-35254. [DOI] [PubMed] [Google Scholar]
- 36.Lametschwandtner, G., C. Brocard, M. Fransen, P. Van Veldhoven, J. Berger, and A. Hartig. 1998. The difference in recognition of terminal tripeptides as peroxisomal targeting signal 1 between yeast and human is due to different affinities of their receptor Pex5p to the cognate signal and to residues adjacent to it. J. Biol. Chem. 273:33635-33643. [DOI] [PubMed] [Google Scholar]
- 37.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 38.Lupas, A. 1996. Prediction and analysis of coiled-coil structures. Methods Enzymol. 266:513-525. [DOI] [PubMed] [Google Scholar]
- 39.Moyersoen, J. J. Choe, A. Kumar, F. G. Voncken, W. G. Hol, and P. A. Michels. 2003. Characterization of Trypanosoma brucei PEX14 and its role in the import of glycosomal matrix proteins. Eur. J. Biochem. 270:2059-2067. [DOI] [PubMed] [Google Scholar]
- 40.Nito, K., M. Hayashi, and M. Nishimura. 2002. Direct interaction and determination of binding domains among peroxisomal import factors in Arabidopsis thaliana. Plant Cell Physiol. 43:355-366. [DOI] [PubMed] [Google Scholar]
- 41.Opperdoes, F. R. 1987. Compartmentation of carbohydrate metabolism in trypanosomes. Annu. Rev. Microbiol. 41:128-151. [DOI] [PubMed] [Google Scholar]
- 42.Opperdoes, F. R., and P. Borst. 1977. Localization of nine glycolytic enzymes in a microbody-like organelle in Trypanosoma brucei: the glycosome. FEBS Lett. 80:360-364. [DOI] [PubMed] [Google Scholar]
- 43.O'Shannessy, D. J., and D. J. Winzor. 1996. Interpretation of deviations from pseudo-first order kinetic behavior in the characterization of ligand binding by biosensor technology. Anal. Biochem. 236:275-283. [DOI] [PubMed] [Google Scholar]
- 44.Otera, H., T. Harano, M. Honsho, K. Ghaedi, S. Mukai, A. Tanaka, A. Kawai, N. Shimizu, and Y. Fujiki. 2000. The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) type 1 transporter, translocates the Pex7p.PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. J. Biol. Chem. 275:21703-21714. [DOI] [PubMed] [Google Scholar]
- 45.Otera, H., K. Setoguchi, M. Hamasaki, T. Kumashiro, N. Shimizu, and Y. Fujiki. 2002. Peroxisomal targeting signal receptor Pex5p interacts with cargoes and import machinery components in a spatiotemporally differentiated manner: conserved Pex5p WXXXF/Y motifs are critical for matrix protein import. Mol. Cell. Biol. 22:1639-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parsons, M., T. Furuya, A. Pal, and P. Kessler. 2001. Biogenesis and function of peroxisomes and glycosomes. Mol. Biochem. Parasitol. 115:19-25. [DOI] [PubMed] [Google Scholar]
- 47.Purdue, P. E., and P. B. Lazarow. 2001. Peroxisome biogenesis. Annu. Rev. Cell Dev. Biol. 17:701-752. [DOI] [PubMed] [Google Scholar]
- 48.Rich, R. L., and D. G. Myszka. 2000. A survey of the year 2002 commercial optical biosensor literature. Curr. Opin. Biotechnol. 11:54-61. [DOI] [PubMed] [Google Scholar]
- 49.Saidowsky, J., G. Dodt, K. Kirchberg, A. Wegner, W. Nastainczyk, W. H. Kunau, and W. Schliebs. 2001. The di-aromatic pentapeptide repeats of the human peroxisome import receptor PEX5 are separate high-affinity binding sites for the peroxisomal membrane protein PEX14. J. Biol. Chem. 276:34524-34529. [DOI] [PubMed] [Google Scholar]
- 50.Schliebs, W., J. Saidowsky, B. Agianian, G. Dodt, F. W. Herberg, and W. H. Kunau. 1999. Recombinant human peroxisomal targeting signal receptor PEX5. Structural basis for interaction of PEX5 with PEX14. J. Biol. Chem. 274:5666-5673. [DOI] [PubMed] [Google Scholar]
- 51.Shih, S., H. Y. Hwang, D. Carter, P. Stenberg, and B. Ullman. 1998. Localization and targeting of the Leishmania donovani hypoxanthine-guanine phosphoribosyltransferase to the glycosome. J. Biol. Chem. 273:1534-1541. [DOI] [PubMed] [Google Scholar]
- 52.Sommer, J. M., and C. C. Wang. 1994. Targeting proteins to the glycosomes of African trypanosomes. Annu. Rev. Microbiol. 48:105-138. [DOI] [PubMed] [Google Scholar]
- 53.Su, J. Y., R. S. Hodges, and C. M. Kay. 1994. Effect of chain length on the formation and stability of synthetic alpha-helical coiled coils. Biochemistry 33:15501-15510. [DOI] [PubMed] [Google Scholar]
- 54.Subramani, S., A. Koller, and W. B. Snyder. 2000. Import of peroxisomal matrix and membrane proteins. Annu. Rev. Biochem. 69:399-418. [DOI] [PubMed] [Google Scholar]
- 55.Urquhart, A. J., D. Kennedy, S. J. Gould, and D. I. Crane. 2000. Interaction of Pex5p, the type 1 peroxisome targeting signal receptor, with the peroxisomal membrane proteins Pex14p and Pex13p. J. Biol. Chem. 275:4127-4136. [DOI] [PubMed] [Google Scholar]
- 56.Wang, D., N. V. Visser, M. Veenhuis, and I. J. van der Klei. 2003. Physical interactions of the peroxisomal targeting signal 1 receptor pex5p, studied by fluorescence correlation spectroscopy. J. Biol. Chem. 278:43340-43345. [DOI] [PubMed] [Google Scholar]
- 57.Wu, C. S., K. Ikeda, and J. T. Yang. 1981. Ordered conformation of polypeptide and proteins in acidc dodecyl sulfate solution. Biochemistry 20:566-570. [DOI] [PubMed] [Google Scholar]
- 58.Zhou, N. E., C. M. Kay, and R. S. Hodges. 1992. Synthetic model proteins. Positional effects of interchain hydrophobic interactions on stability of two-stranded alpha-helical coiled-coils. J. Biol. Chem. 267:2664-2670. [PubMed] [Google Scholar]
- 59.Zoetewey, D. L, B. P. Tripet, T. G. Kutateladze, M. J. Overduin, J. M. Wood, and R. S. Hodges. 2003. Solution structure of the C-terminal antiparallel coiled-coil domain from Escherichia coli osmosensor ProP. J. Mol. Biol. 334:1063-1076. [DOI] [PubMed] [Google Scholar]