Abstract
The tetracycline-controlled Tet-Off and Tet-On gene expression systems are used to regulate the activity of genes in eukaryotic cells in diverse settings, varying from basic biological research to biotechnology and gene therapy applications. These systems are based on regulatory elements that control the activity of the tetracycline-resistance operon in bacteria. The Tet-Off system allows silencing of gene expression by administration of tetracycline (Tc) or tetracycline-derivatives like doxycycline (dox), whereas the Tet-On system allows activation of gene expression by dox. Since the initial design and construction of the original Tet-system, these bacterium-derived systems have been significantly improved for their function in eukaryotic cells. We here review how a dox-controlled HIV-1 variant was designed and used to greatly improve the activity and dox-sensitivity of the rtTA transcriptional activator component of the Tet-On system. These optimized rtTA variants require less dox for activation, which will reduce side effects and allow gene control in tissues where a relatively low dox level can be reached, such as the brain.
Keywords: Tet-On system, Tet-Off system, Transcription regulation, Doxycycline, Gene expression, rtTA, tTA, TetR.
1. Introduction
Gene therapy applications can be complicated by adverse side or off-target effects of the transgene product. The ability to turn transgene expression on and off or to modulate the expression level may therefore significantly improve the safety of gene therapy approaches. Several regulatory mechanisms have been developed that allow both quantitative and temporal control of gene expression in eukaryotic cells by an exogenous effector molecule [1]. We here focus on the Tet-Off and Tet-On systems that allow modulation of a gene-of-interest (G.O.I.) by administration or withdrawal of tetracyclines [2-4]. Tetracyclines constitute a diverse family of chemical compounds with 4 hydrocarbon rings and different functional groups. The prototype tetracycline (Tc) and derivatives like doxycycline (dox) have been widely used in humans as antibiotic. The Tet-Off and Tet-On systems are based on the Tet repressor protein (TetR) and tet operator (tetO) DNA elements that control the Tn10-encoded tetracycline resistance operon of Escherichia coli.
1.1. The Bacterial Tet Operon
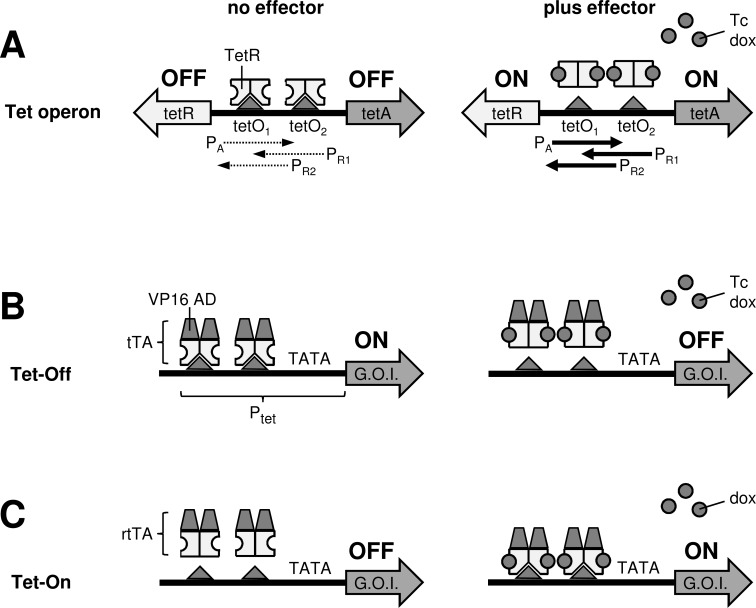
Tetracyclines are antibiotics that bind to the bacterial 30S ribosomal subunit and inhibit bacterial protein synthesis and growth. Bacteria have developed different mechanisms of tetracycline resistance: tetracycline efflux, ribosome protection and tetracycline modification [5]. In Gram-negative bacteria, tetracycline resistance is mostly mediated by efflux of the drug by the membrane protein TetA [6, 7] and the TetA level is controlled by the tetracycline-responsive repressor protein TetR. In the Tn10-encoded Tet operon, the tetA and tetR genes are oriented in opposite direction and their expression is regulated at the transcription level by a shared regulatory region (Fig. 1A). This regulatory region consists of overlapping promoters PA and PR1/PR2 (driving tetA and tetR transcription, respectively) and two superimposed tetO operator sequences [7, 8]. The TetR proteins form dimers that recognize and bind the tetO elements, thus suppressing the activity of the PA and PR promoters and shutting off TetA and TetR production [9]. The high affinity of TetR for the tetO sequence allows efficient repression of TetA, which is important because even low TetA levels are disadvantageous for bacterial cells in the absence of tetracycline [10]. Binding of tetracycline triggers a conformational change in the TetR dimer that prevents tetO binding. As a consequence, TetR will no longer suppress the PA and PR promoters and TetA and TetR will be produced. The high affinity of TetR for the tetracycline allows activation of TetA production at very low tetracycline levels that do not yet block protein synthesis [4]. The low affinity of TetR for non-operator sequences results in a high specificity, which implies that TetR can be used to selectively regulate transgene expression in organisms with much larger genomes than bacteria [7].
Fig. (1).
Tc-controlled regulation of gene expression. (A) Tn10 tet operon. In E. coli, TetR binds as a dimer to the tetO1 and tetO2 sites in the Tn10 tet operon. This interaction blocks the activity of the underlying promoters (PA, PR1 and PR2) and inhibits transcription of the tetA and tetR genes. Binding of Tc or dox triggers a conformational switch in TetR that prevents tetO binding and results in the activation of TetA and TetR production. (B) The Tet-Off system. Fusion of TetR to the activation domain of the herpes simplex virus VP16 protein (VP16 AD) resulted in the Tc-controlled transcriptional activator (tTA). Binding of tTA to the Ptet promoter that consists of 7 tetO sequences fused to a minimal TATA-box containing eukaryotic promoter, activates expression of the downstream positioned gene-of-interest (G.O.I.) Binding of Tc or dox induces a conformational change in the TetR domain of tTA, which prevents tetO binding and switches gene expression off. (C) The Tet-On system. The reverse-tTA (rtTA) variant exhibits a reverse phenotype and does not bind tetO in the absence of an effector. Binding of dox triggers a conformational switch in rtTA, which allows tetO binding. Subsequent activation of the Ptet promoter drives expression of the downstream positioned gene. The initial version of rtTA had a low affinity for Tc and was not activated by this compound.
1.2. The Eukaryotic Tet-Off System
Gossen et al. described in 1992 how the TetR and tetO components of the Tn10-encoded Tet operon can be used to develop a regulatory mechanism for controlled gene expression in mammalian cells. In this Tet-Off system (Fig. 1B), the 127-amino acid (127-aa) transcription activation domain (AD) of the herpes simplex virus VP16 protein was fused to the 207-aa TetR, which resulted in the tetracycline-controlled transcriptional activator (tTA) [11]. Furthermore, a tetracycline-responsive promoter (Ptet) was constructed by fusing 7 tetO sequences to a minimal TATA-box containing eukaryotic promoter. This promoter sequence was derived from the CMV immediate early gene and lacked enhancer sequences. In the absence of tetracycline, tTA dimers will bind the tetO sites in Ptet and activate expression of the downstream positioned transgene [11]. Tc or dox binding induces a conformational change in the TetR domain of tTA that prevents tetO binding, thus switching gene expression off.
1.3. The Eukaryotic Tet-On System
If only transient expression of the gene-of-interest is aimed for, a disadvantage of the Tet-Off system is that Tc or dox has to be administered continuously outside this small time window, while long-term exposure to these effectors is often undesirable. Another disadvantage is that the tetracycline has to be removed to activate gene expression. Although this can be achieved relatively easily in small cell culture experiments by thorough washing of the cells and replacement of the culture medium, it is more problematic when large cell cultures are used and in animal experiments. In vivo, the biological half-life of the effector will determine the kinetics of induction. In 1995, Gossen et al. developed the Tet-On system (Fig. 1C), a regulatory system that allows activation of gene expression by the addition - instead of removal - of dox [12]. For this, random mutation and phenotype screening in E. coli were first used to select a TetR variant that functions in a reverse fashion, i.e. binds tetO in the presence and not in the absence of the effector. This reverse-TetR (rTetR) differed at four amino acid positions from the original protein (Fig. 2). The high resolution TetR-Tc crystal structure that is available for the TetR class D protein, which shares 63% sequence identity with the TetR class B protein that was used to construct the Tet systems, indicated that these amino acids do not contact Tc directly and are not part of the tetO DNA binding domain (DBD). Subsequent fusion of the VP16 activation domain resulted in a reverse-tTA (rtTA) that binds Ptet and activates transcription exclusively in the presence of dox [12]. Unfortunately, the 4 amino acid
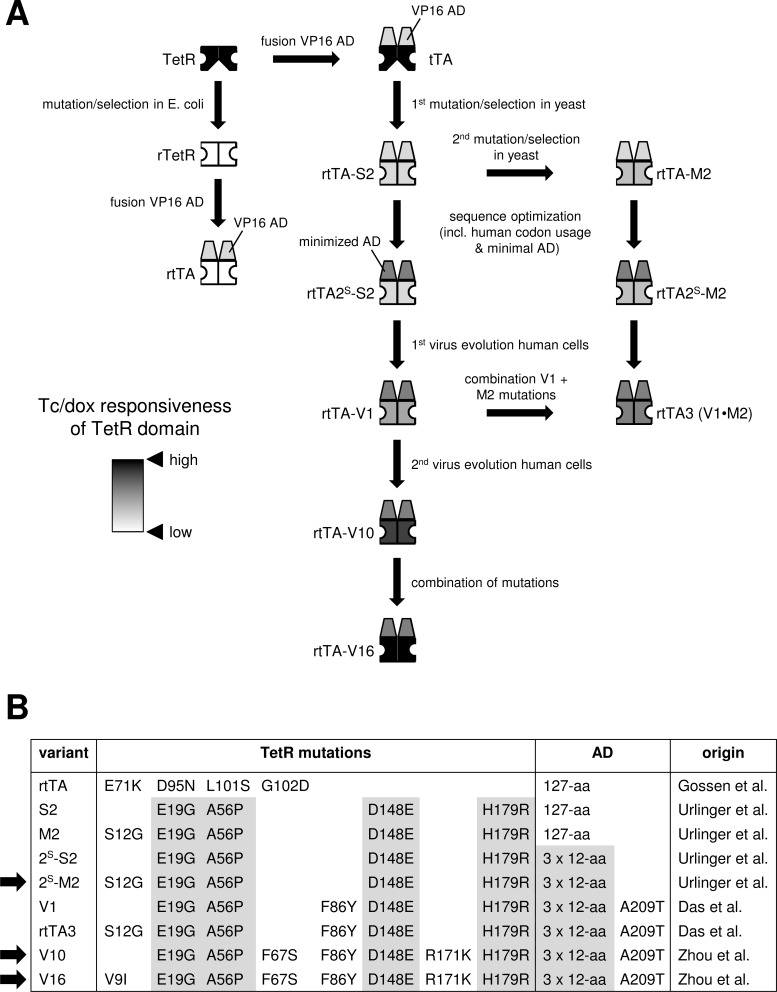
Fig. (2).
Construction and optimization of the Tet-On system. (A) Schematic overview of the development of rtTA variants by rational design, random mutation and screening in E. coli and yeast (S. cerevisiae) and virus evolution in human cells. See text for details. The color of the TetR domain reflects the effector responsiveness (white, low Tc/dox response; black, high Tc/dox response). (B) Mutations in the TetR moiety and activation domain (AD) in different rtTA variants. The activity of the variants indicated with an arrow was directly compared in transiently transfected and stably transduced cells (Figs. 5, 6).
substitutions in the TetR domain of rtTA that resulted in the reverse phenotype significantly reduced the sensitivity toward the effector. As a consequence, rtTA was only poorly activated by Tc and required 100-fold more dox for maximal induction than what is needed for complete tTA inhibition [3].
2. Improvements of the TET-On system
Optimal control of gene expression requires a Tet-On system with low background activity in the absence of dox and high activity in the presence of the effector. Important characteristics of the Tet-On system that influence background activity are the intrinsic, rtTA-independent activity of the pTet promoter, which is due to the presence of binding sites for cellular transcription factors, and the residual binding of rtTA to pTet in the absence of dox. The pTet promoter has been optimized through systematic modification, which resulted in novel promoter configurations that minimized intrinsic background activity while maintaining high induced expression [13, 14]. We will here describe the different strategies that have been used to improve the rtTA component of the Tet-On system.
Random mutagenesis of tTA followed by a functional screen for the reverse, dox-inducible phenotype in the yeast S. cerevisiae resulted in a novel rtTA variant, rtTA-S2, with 4 amino acid substitutions in the TetR domain [15] (Fig. 2). Two of the selected mutations (E19G in the DBD and A56P in the helix connecting the DBD with the core) were shown to reverse the phenotype, whereas the other mutations (D148E and H179R; positioned in the core, close to amino acids involved in effector binding and dimerization) enhanced the activity. Notably, all 4 amino acid changes differed from the rTetR mutations identified in the TetR mutation/screening in E. coli, which were used for the development of the first generation rtTA. A second round of mutagenesis and screening in S. cerevisiae yielded an additional DBD mutation (S12G) that increased the activity of rtTA-S2, in particular at low dox concentrations. The rtTA-S2 variant and the S12G-mutated derivative rtTA-M2 were optimized by substitution of the 127-aa VP16 AD with 3 repeats of the 12-aa minimal AD. This AD minimization removed potential targets for interactions with cellular transcription factors, which reduced toxic squelching effects [16]. This truncation of the VP16 moiety also reduced the number of potential epitopes that may elicit a cellular immune response in animal applications. Furthermore, the rtTA coding sequence was optimized for human codon usage and putative toxic mRNA elements were removed: splice donor and acceptor sites, motifs for endonuclease cleavage sites and sequences inducing hairpin structures [15]. These studies resulted in the rtTA2S-S2 and rtTA2S-M2 variants that demonstrated increased stability and reduced background activity in the absence of dox when compared with the original rtTA. The rtTA2S-M2 variant (a.k.a. Tet-On Advanced, Clontech) demonstrated the highest sensitivity toward dox and functioned at a 10-fold lower dox concentration than the original rtTA and rtTA-S2. However, further improvement of the Tet-On system was desirable. For example, rtTAs with an increased dox-sensitivity will require less dox, which will avoid cellular side effects due to the inhibitory effect of high tetracycline levels on mitochondrial translation [17]. Furthermore, such rtTAs will be beneficial for applications in tissues where a relatively low dox level can be reached, such as the brain.
3. Gene therapy applications in the CNS require a dox-sensitive TET-On system
Whereas immune reactions directed against the viral vector or transgene may limit systemic gene therapy applications [18], gene therapy in immunologically-privileged sites, like the central nervous system (CNS), are not restricted by such deleterious immune responses [19]. Unfortunately, the main neurodegenerative diseases, i.e. Alzheimer and Parkinson’s disease, are usually not inherited diseases of monogenetic origin and cannot currently be treated by a gene correction approach. Instead, gene therapy approaches based on neurotrophic factor delivery, aiming to halt or reduce neuronal cell death, are envisaged. Specifically, nerve growth factor (NGF) has been delivered in the brain of patients with Alzheimer disease either via direct intracerebral injection of an AAV vector or via transplantation of genetically-modified cells [20] and neurturin, a member of the glial cell line-derived neurotrophic factor (GDNF) family ligands, was administered to advanced Parkinson’s disease patients using an AAV vector [21]. Because adverse effects of uncontrolled delivery of neurotrophic factors have been described [22-24], the ability to modulate transgene expression is crucial for these applications [25, 26].
Several inducible or repressible genetic systems to regulate intracerebral delivery of GDNF, which is envisaged for the treatment of Parkinson’s disease [27] (https://clinical trials.gov/ct2/show/NCT01621581), have been described. In a preclinical study, the mifepristone-regulated Gene Switch system provided neuroprotective effects of GDNF at a clinically-acceptable dose of mifepristone [28]. However, this system was tightly regulated only in a 2-vector configuration [29], which limits applicability and clinical feasibility. The rapamycin-regulated system also allowed tight regulation of GDNF transgene expression, but the level of expression was low when compared with expression driven by a constitutively active promoter [30].
Efficient on/off kinetics with a single-cassette AAV-Tet-Off vector in the rat brain has been demonstrated [31]. Complete extinction of GDNF transgene expression required a dox concentration of ~0.3 µg/ml in the blood plasma, which corresponds to a clinically-acceptable dose. However, a very high dose of viral vector, well above that approved in current clinical trials, was used in this study. Using an auto-regulated AAV-Tet-On vector containing the rtTA2S-M2 transactivator (AAV-tetbidi-On) [32], a transgene expression efficiency could be reached that was only slightly lower than the level obtained with a constitutively active CMV enhancer/promoter [33]. GDNF delivered by this vector in the brain of rats injected with 6-hydroxydopamine to model Parkinson’s disease, enhanced the function of dopaminergic neurons and resulted in behavioral improvements. In this study, the vector dose injected into the rat brain was 5 x 108 viral genomes, which is approximately equivalent to 2 x 107 viral genomes/mm3, the dose approved for the AAV-neurturin clinical trial [34]. However, this and other studies [35] demonstrated that daily administration of a very high dox dose was required to induce rtTA2S-M2 driven transgene expression in the brain, which is mainly due to the blood-brain barrier that causes a relatively low dox concentration in the cerebrospinal fluid (CSF) [36-38]. For example, in the rat/AAV-tetbidi-On study, administration of 600 mg/l dox in drinking water, which resulted in a dox concentration of ~5.3 µg/ml in blood and ~0.4 µg/ml in CSF, was required to activate gene expression in the brain (unpublished data). Considering the putative toxic side effects of dox, long-term administration of high dox doses should however be avoided. Therefore, new rtTA variants that are more active at lower dox levels are desirable. Because of the antibiotic activity of dox, such new rtTAs should preferably be active at sub-antimicrobial dox doses that do not elicit adverse effects (such as affecting the gut flora) or lead to antibiotic resistance. Brain applications of the Tet-On system could also benefit from rtTAs that are responsive to the tetracycline-derivative minocycline (Mc), because Mc has an increased bioavailability and a greater lipophilicity, which results in a better tissue penetration [39]. In addition, Mc and dox exhibit anti-inflammatory properties which could be of interest when treating neurodegenerative diseases [40, 41].
4. A dox-controlled HIV-1 variant as a tool to improve the TET-On system
We used the Tet-On system to generate an HIV-1 variant that replicates exclusively in the presence of dox [42]. The construction of this unique virus and optimization of the Tet-On system by means of spontaneous virus evolution will be described in detail below. This conditionally replicating virus was also proposed as a novel approach toward a safe live-attenuated HIV vaccine, a topic that will be introduced first.
4.1. Live-attenuated HIV Vaccine
Live-attenuated virus (LAV) variants have proven to be an effective vaccine strategy against several viral diseases (e.g. small pox, polio, measles). The principle of a LAV vaccine is that the attenuated, non-pathogenic virus can replicate to a limited extent, which does not cause disease, but elicits a potent immune response that protects against a subsequent challenge with the wild-type, pathogenic virus. Live-attenuated human immunodeficiency virus (HIV) variants have also been considered as vaccine for the protection of humans against HIV infection. Because such studies are not possible in humans, the animal model of simian immunodeficiency virus (SIV) infection in macaques was used. Attenuated virus strains were constructed by deletion of accessory viral functions that stimulate in vivo replication, but are not absolutely required for virus replication, like the nef gene and particular long-terminal repeat (LTR) promoter domains. Vaccination of macaques with attenuated, non-pathogenic SIV strains did indeed induce effective protection against a challenge with wild-type, pathogenic SIV [43-45]. However, SIV, like HIV and other retroviruses, stably integrates in the genome of target cells and is never cleared upon infection. Because of this persistence, in combination with continuous low-level virus replication and the error-prone viral replication machinery, the attenuated vaccine virus can revert to virulence by the accumulation of mutations that improve replication. Evolution of the vaccine virus to a pathogenic variant did indeed cause disease in a minority of the vaccinated animals [46-49]. Similar observations with HIV confirmed the genetic instability of attenuated virus strains. For instance, an HIV∆3 variant with deletions in nef, vpr and LTR regained replication capacity in long-term cell culture infections by acquisition of compensatory mutations elsewhere in the viral genome [50]. There is also some in vivo evidence as some of the long-term survivors of the Sydney Blood Bank Cohort, all infected with a naturally Nef-deleted HIV-1 variant, did eventually progress to AIDS [51]. The enormous evolutionary capacity of HIV poses a serious safety risk for the application of live-attenuated HIV-based vaccines in humans. Such undesirable evolution of the vaccine virus can be prevented by blocking continuous replication of the attenuated virus after vaccination using the Tet-On system.
4.2. A Conditionally Replicating HIV Variant
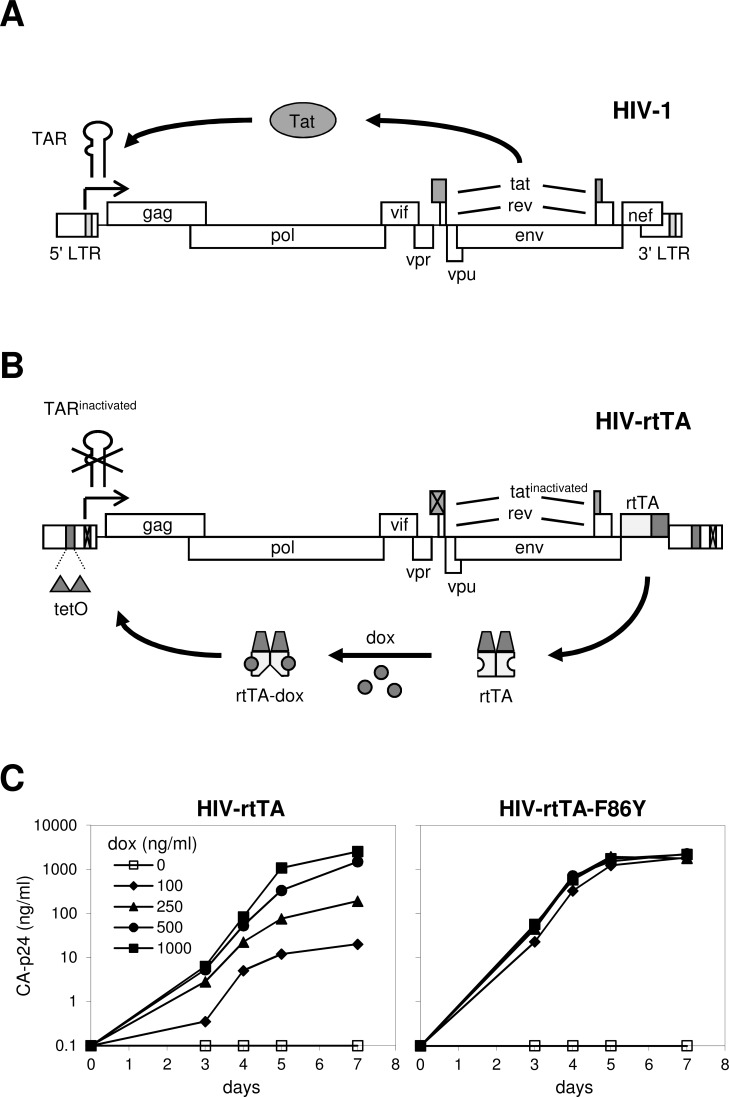
To improve the safety of live-attenuated HIV vaccine candidates, we and others previously presented a conditionally replicating HIV variant, with replication that can be switched on and off at will by the administration and removal of an exogenous effector, respectively [42, 52-55]. In this approach, the effector-dependence feature (on/off switch) will make it possible to allow temporal replication of the vaccine virus to the extent needed for induction of protective immunity. Subsequent effector-withdrawal will stop viral replication, which should prevent evolution of the vaccine virus toward a pathogenic variant. We used the Tet-On system to construct a conditionally replicating HIV-1 variant that was named HIV-rtTA. HIV-1 transcription and gene-expression are normally driven by the 5’ LTR promoter and stimulated by binding of the viral Tat protein to the transacting responsive (TAR) RNA element that is present at the 5’ end of the nascent viral transcripts (Fig. 3A). This auto-regulatory loop results in the production of high levels of RNA and protein to support virus replication. In HIV-rtTA, the Tat-TAR transcription activation mechanism was functionally replaced by the rtTA-tetO components of the Tet-On system [42, 55]. The Tat-TAR mechanism was inactivated through mutations in the TAR sequence that prevent Tat binding and an amino acid substitution in Tat that prevents trans-activation of transcription (Fig. 3B). The Tet-On system was integrated into the viral genome by replacing the accessory nef gene by the gene encoding the optimized rtTA2S-S2 variant and inserting tet operator (tetO) sites in the viral LTR promoter. In the presence of dox, rtTA will bind to the tetO-LTR promoter and activate viral transcription. HIV-rtTA replication can thus be controlled by the administration of dox (Fig. 3C). Dox-dependent HIV-rtTA replication was shown in T cell line infections in vitro, ex vivo in human lymphoid tissue [56] and in vivo in a humanized mouse model [57, 58]. Application in humans may require additional safety measures. For instance, we inserted a second drug-dependent control mechanism into HIV-rtTA [59]. Alternatively, one may consider deletion of the integrase (IN) function, which will likely prevent persistence of the vaccine virus. A dox-controlled SIV-rtTA variant [60, 61] was employed in vaccination studies in macaques to study whether continuous low-level replication of the vaccine virus is required for protection and to determine the protective correlates that are induced by an effective vaccine [62].
Fig. (3).
The dox-dependent HIV-rtTA virus. (A) Transcription of wild-type HIV-1 is activated by the binding of the viral Tat protein to the TAR hairpin structure that is present at the 5’ end of nascent RNA transcripts. This Tat-TAR axis thus controls viral gene expression and replication. (B) In HIV-rtTA, Tat and TAR are inactivated by mutation and functionally replaced by the rtTA-tetO components of the Tet-On system, by insertion of the rtTA gene at the site of the nef gene and insertion of tetO sequences in the LTR-promoter region. Transcription of this HIV-1 variant is activated by the binding of rtTA to the tetO-LTR promoter. Because rtTA binds the tetO sites exclusively in the presence of dox, HIV-rtTA does not replicate in the absence of this effector. (C) Dox-controlled replication of HIV-rtTA strains with the original rtTA-2S-S2 gene (left panel) and the F86Y-mutated variant (right panel) in SupT1 T-cells at different dox-concentrations. Virus replication is monitored by measuring the viral capsid protein (CA-p24) in the culture supernatant. Reproduced from [67] (© 2004, the American Society for Biochemistry and Molecular Biology).
4.3. Evolution of HIV-rtTA
Genetic diversity is continuously generated during HIV replication, which is primarily due to the error-prone reverse transcription process. Faster replicating variants will subsequently be selected by outcompeting the original virus. Thus, the virus can rapidly evolve and enhance its replication capacity. We anticipated that during in vitro culturing of HIV-rtTA in human T cell lines this designer virus may adapt the integrated components of the Tet-On system, which are largely derived from E. coli, for their new function to support virus replication in human cells.
4.3.1. Optimization of the tetO-LTR Promoter Region
We did frequently observe specific deletions in the viral tetO-LTR promoter [63, 64]. In several independent cultures, we observed deletion of 6 of the 8 introduced tetO elements, which was followed by a deletion of 14 or 15 nt in the spacer between the remaining tetO sites. Strikingly, the new spacing between the remaining tetO elements resembles the spacing between these sites in the original E. coli Tn10 element. The new 2ΔtetO-LTR configuration significantly improved HIV-rtTA replication. Analysis of the promoter activity revealed that 2ΔtetO-LTR was less active than the original 8tetO-LTR and similarly active as the wild-type HIV-1 LTR, which indicates that HIV-1 requires a fine-tuned level of transcription
for efficient replication. This modulation was perhaps triggered by acquired mutations in rtTA that increased the transcriptional activity (see section below). Alternatively, the tetO region may have been truncated because 8 copies of the tetO sequence may have a negative effect on virus replication. Sequence repeats can indeed trigger recombination events [65, 66] or cause aberrant folding of the viral RNA, which may reduce viral replication fitness.
4.3.2. Optimization of rtTA
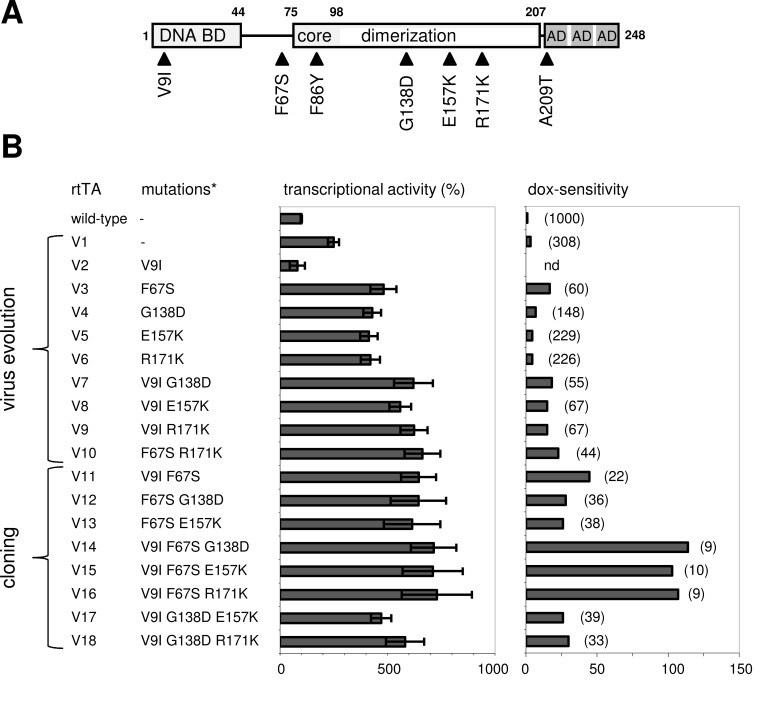
We also observed mutations in the rtTA gene upon long term replication of HIV-rtTA. Initially, a Phe to Tyr substitution was observed at amino acid position 86 (F86Y) in the TetR core domain (Fig. 4A), at a position that directly interacts with Tc in the TetR-Tc crystal structure [67]. This mutation coincided with an Ala to Thr substitution at position 209 (A209T) in one of the minimal AD sequences. Subsequent analyses demonstrated that the F86Y mutation significantly improved HIV-rtTA replication, in particular al lower dox levels (Fig. 3C), whereas the A209T mutation had no effect. To test the impact in the context of the standard Tet-On system, the capacity to activate transcription of Ptet-reporter constructs was measured in human cell lines transfected with rtTA-expressing and Ptet-luciferase plasmids.
Fig. (4).
Optimization of the Tet-On system through virus evolution. (A) Amino acid substitutions observed in rtTA-2S-S2 (wild-type) upon long-term culturing of HIV-rtTA in human SupT1 T-cells. (B) The transcriptional activity (at 1000 ng/ml dox) and dox-sensitivity of the wild-type, naturally evolved (V1-V10) and constructed (V11-V18) rtTA variants was measured in HeLa X1/6 cells that contain chromosomally integrated copies of the Ptet-luciferase reporter construct. The wild-type rtTA activity at 1000 ng/ml dox was set at 100%. The activity measured at different dox concentrations was used to calculate the dox concentration that each rtTA variant needs to reach an activity comparable to that of the wild-type rtTA at 1000 ng/ml dox. These concentrations are indicated between brackets in the right panel (nd, not determined), and were used to calculate the dox-sensitivity for each rtTA variant (dox-sensitivity of wild-type rtTA set at 1). *, V1-V18 carry the F86Y and A209T mutations in addition to the shown mutations. Reproduced from [69] (© 2006, Zhou et al.).
The F86Y mutation was found to increase the transcriptional activity and dox-sensitivity of rtTA, whereas the A209T mutation had again no effect. Importantly, the mutations did not affect the background transcriptional activity in the absence of dox. The rtTA activity and dox-sensitivity of rtTAF86Y+A209T (named rtTA-V1; Fig. 2) was further increased by introducing the S12G mutation that is typical for rtTA2S-M2. When compared with rtTA2S-S2, the new rtTAS12G+F86Y+A209T variant (a.k.a. rtTA3 [68]) was 5-fold more active at high dox levels and required 25-fold less dox to be similarly active (25-fold more sensitive to dox).
To further improve the Tet-On system by virus evolution, we started multiple cultures of HIV-rtTA-V1, which contained both the improved rtTA-V1 gene and the optimized 2ΔtetO-LTR promoter configuration. Upon long term culturing, we identified several additional mutations in the TetR domain of the rtTA protein that were selected in independent cultures: a V9I substitution in the DBD; F67S, G138D and R171K substitutions at positions that do not directly bind Tc in the TetR-Tc crystal structure, but are in close proximity to the binding pocket and may indirectly influence binding of the effector; and an E157K substitution in a flexible loop region that was not solved in the crystal structure (V2-V6 variants; Fig. 4). The V9I mutation appeared both as an individual mutation (V2) and in combination with G138D, E157K or R171K (V7-V9). A combination of F67S and R171K was also observed (V10). Except for the V2 variant, the new variants demonstrated higher activity and dox sensitivity than V1 when the capacity to activate pTet-driven reporter expression was analyzed (Fig. 4B). The V10 double mutant showed the highest activity and dox-sensitivity [69]. Several additional rtTA variants were constructed in which the observed mutations were combined (V11-V18; Fig. 4B). The triple mutant V16, in which the V10 mutations were combined with V9I, demonstrated the highest activity and dox-sensitivity. V16 was seven-fold more active at high dox levels and 100-fold more sensitive to dox than rtTA2S-S2, and showed no activity without dox when the activity was measured upon transfection of rtTA plasmids into cells with an integrated pTet-reporter construct (Fig. 4B). The mutations also improved the activity with other non-dox effectors and restored the responsiveness to Tc [69]. The mutations did not affect the intracellular rtTA protein level, indicating an improved intrinsic rtTA activity and effector-sensitivity [67, 69]. The improved rtTA characteristics likely result from an improved interaction with the effector molecule and/or the tetO DNA elements.
5. Comparison of novel TET-On systems
To identify which Tet-On system is optimal for specific cell culture applications, we recently compared the optimized rtTA variants. For this, we tested the rtTA2s-M2 (abbreviated as M2; a.k.a. Tet-On advanced, Clontech), V10 (a.k.a. Tet-On 3G) and V16 variants in several experimental settings in some frequently used cell types.
5.1. Transiently Transfected Cells
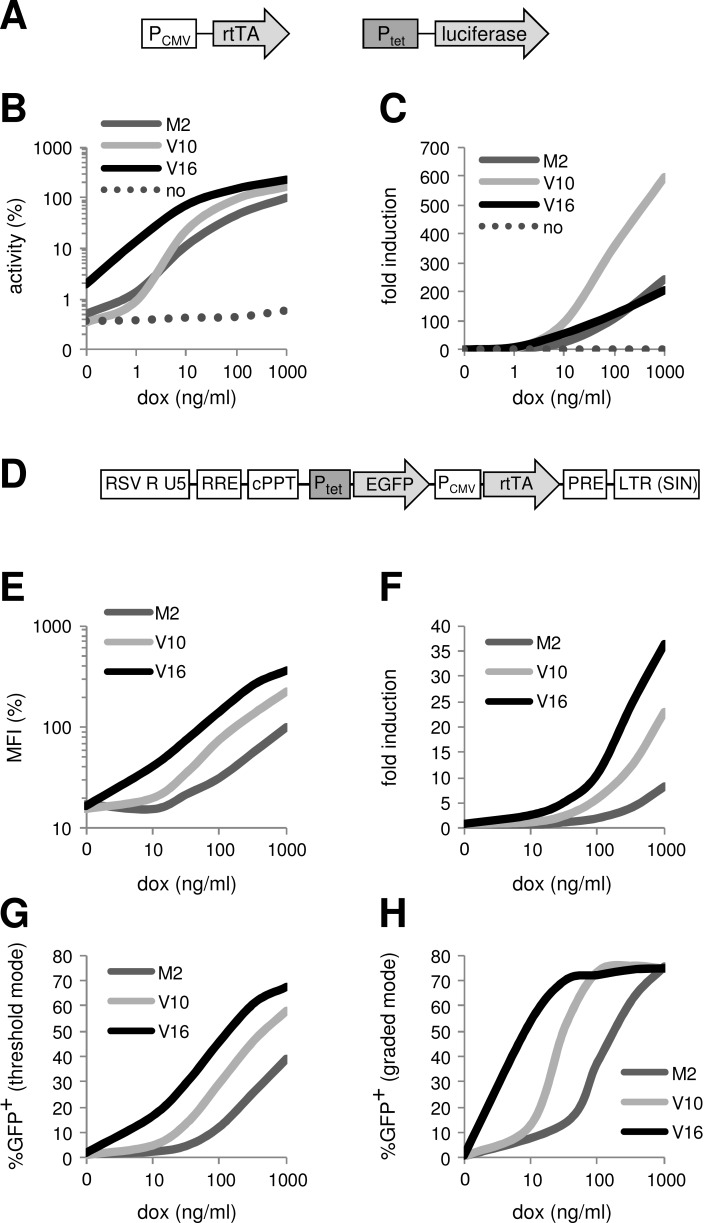
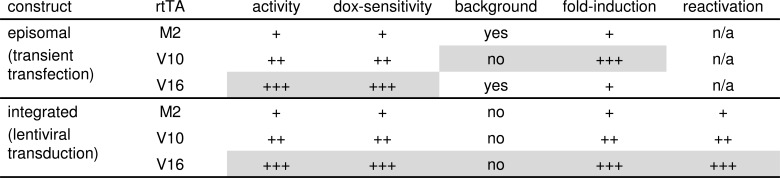
When rtTA activity was measured in cells upon transient transfection with the rtTA and Ptet-reporter plasmids (Fig. 5A), we consistently observed highest dox-induced activity for the V16 variant, intermediate activity for V10 and lowest activity for M2 (Fig. 5B) [70]. V16 activated reporter expression at very low dox levels, confirming its extreme dox sensitivity. However, V16 caused very low, but detectable background expression in the absence of dox, which becomes particularly noticeable when high amounts of DNA were transfected. M2 also demonstrated a low level of background activity, although to a lesser extent than V16. The V10 variant did not show any background activity and, as a consequence, showed superior fold-induction rates (Fig. 5C). Therefore, V10 will be the optimal system for most applications involving transiently transfected cells and other situations where the Tet-On components will be present episomally at relatively high concentrations (Fig. 6). When high transcriptional activity at low dox levels is required, for example when the cells do not tolerate a high dox level, the dox-sensitive V16 variant may be preferred.
Fig. (5).
Comparison of rtTA variants in transiently transfected and stably transduced cells. (A-C) Different human cell lines (HeLa cervix carcinoma and HEK293 embryonal kidney cells) were transfected with low and high amounts of CMV enhancer/promoter-driven PCMV-rtTA (either rtTA2S-M2 [M2], rtTA-V10 or rtTA-V16) and dox-rtTA activated Ptet-luciferase constructs (constructs shown in A), and cultured in the presence of 0 to 1000 ng/ml dox. (B) The luciferase level measured 2 days after transfection reflects the rtTA activity. The activity of M2 at 1000 ng/ml dox was set at 100%. The average of the values obtained for different experimental conditions (different cells and different amounts of DNA) [70] is shown to illustrate the differences between the rtTA variants (no, transfection of cells with the empty vector instead of the rtTA plasmid). The original data for each cell line and amount of DNA are presented in [70]. (C) Fold induction levels, calculated as the ratio between the rtTA activity at the indicated dox concentration and the activity in the absence of dox (0 ng/ml). (D-H) Different human cell lines (SupT1, HeLa, HepG2 hepatocellular carcinoma and SJNB-8 neuroblastoma cells) were transduced with a lentiviral vector containing Ptet-d2EGFP and PCMV-rtTA (M2, V10 or V16) cassettes (construct shown in D). Cells were cultured with dox for 3 days. GFP-positive (GFP+) cells were sorted and cultured without dox for 6 days to switch off GFP expression. The isolated transduced cells were subsequently cultured with different dox concentrations for 3 days, after which the intracellular GFP level was analyzed. (E) The mean fluorescent intensity (MFI) of the total cell population reflects the rtTA activity. The activity of M2 at 1000 ng/ml dox was set at 100%. The average of the values obtained with the different cells (original data published in [70]) is shown to illustrate the differences between the rtTA variants. (F) Fold induction levels, calculated as the ratio between the MFI at the indicated dox concentration and the MFI in the absence of dox (0 ng/ml). (G) The average percentage of GFP+ cells measured for the SupT1, HeLa and SJNB-8 cells that express GFP predominantly in a threshold mode (i.e. increasing the dox concentration activated GFP expression in more cells, rather than that it increased the fluorescence intensity of the GFP+ cells [89]; original data published in [70]). (H) The percentage of GFP+ cells measured for HepG2 cells that express GFP predominantly in a graded mode (i.e. increasing the dox concentration predominantly increased the fluorescence intensity of the GFP+ cells [89]).
Fig. (6).
Identification of the optimal Tet-On system for different applications. Direct comparison of the activity of the rtTA-2S-M2 (M2), rtTA-V10 and rtTA-V16 variants in transiently transfected and stably transduced cells (as shown in Fig. 5) revealed their quality with respect to different parameters. +, good; ++, better; +++, best; n/a, not applicable.
5.2. Stably Transduced Cells
Also when the rtTA and Ptet-reporter cassettes were stably integrated in the cellular genome through lentiviral vector transduction (Fig. 5D), V16 demonstrated highest, V10 intermediate and M2 lowest dox-induced activity (Fig. 5E) [70]. Since the background activity of all rtTAs was similarly low, V16 also showed the highest fold-induction, whereas V10 showed intermediate and M2 the lowest fold-induction (Fig. 5F). When gene expression in the transduced cells was reactivated by dox administration after a latency period without dox, V16 consistently showed more robust induction of gene expression than V10 and M2, resulting in the reactivation of gene expression in a larger fraction of the transduced cells (Fig. 5G) or in reactivation at a lower dox concentration (Fig. 5H), depending on the cell type. These results indicate that the integrated reporter gene construct is sensitive to transcriptional silencing and variegation effects upon dox withdrawal, which are likely caused by the cellular DNA sequences surrounding the integration site [71]. Apparently, the highly active V16 overcomes this gene repression more efficiently than the less active V10 and M2 variants. With this high reactivation capacity of V16, more laborious procedures to obtain high transgene expressing cells, such as the selection of clonal cell lines, may not be needed. Because of its high activity, dox-sensitivity and robust reactivation capacity, V16 will be the optimal variant for most applications in which the gene constructs are stably integrated in the cellular genome (Fig. 6). The high dox-sensitivity of V16 also allowed us to develop a sensitive bioassay for measuring the dox concentration in biological samples [72].
6. Optimal TET-On system for in vivo applications
The direct comparison of the new Tet-On systems was limited to the in vitro cell culture setting. In vivo application of the systems can be complicated by immune responses against rtTA, as reported in rodents and non-human primates [73-78]. These immune responses, both cellular and humoral, are dependent on the target tissue and vector delivery route (reviewed in [78]). The amino acid substitutions typical for M2, V10 and V16 do not affect any of the known HLA-A*0201 restricted CTL epitopes in rtTA [73]. In transgenic mice, toxic side-effects of tTA and rtTA have also been reported [79-83]. The severity of these effects depends on the genetic background of the animal and possibly also on the (r)tTA version used. For example, the complete VP16 AD in the original versions may be more toxic to cells due to squelching effects than the minimized AD in later versions. The S12G and V9I mutations in the DBD of M2 and V16, respectively, may affect the interaction of rtTA with cellular promoter regions and thus influence cellular gene expression, but whether an up or down effect is induced remains to be determined. Another obvious issue to take into consideration is that a more dox-sensitive rtTA variant, like V16, may demonstrate prolonged activity when the dox level drops after dox-withdrawal [84]. These limitations of the model system should be recognized and appropriate controls should be included in the analysis. Which rtTA variant will be optimal in complex in vivo situations is currently unknown and may depend on the cell and tissue type that is targeted for transgene expression. If only relatively low dox levels can be reached, for example in the brain due to the blood-brain barrier, the ultra dox-sensitive V16 variant will likely be the best candidate. Recent studies successfully applied V16 to control transgene expression in mice [85, 86] and non-human primates [62, 87]. In a direct comparison, V16 outperformed the M2 variant when used to control the expression of an AAV-delivered GDNF gene in the rat brain, resulting in therapeutically-relevant biological effects of the transgene at clinically-approved sub-antimicrobial dox doses [88].
Acknowledgements
The HIV-rtTA and Tet-On research in our lab was sponsored by research grants from the AIDS Fonds (grant 2005022 and 2007028), the Technology Foundation STW (applied science division of the Netherlands Organisation for Scientific Research NWO and the technology program of the Ministry of Economic Affairs, Utrecht, the Netherlands) and the EU FP7 Marie Curie IAPP (Industry Academia Partnerships and Pathways) BrainVectors (contract 286071).
CONFLICT OF INTEREST
Authors declare having potential competing financial interests. The rtTA V10 and V16 variants have been filed in patent WO2007058527.
REFERENCES
- 1.Fussenegger M. The impact of mammalian gene regulation concepts on functional genomic research, metabolic engineering, and advanced gene therapies. Biotechnol. Prog. 2001;17(1):1–51. doi: 10.1021/bp000129c. [DOI] [PubMed] [Google Scholar]
- 2.Gossen M., Bujard H. Tetracyclines in the control of gene expression in eukaryotes. In: Nelson M., Hillen W., Greenwald R.A., editors. Tetracyclines in biology, chemistry and medicine. Basel: Birkhäuser Verlag; 2001. pp. 139–157. [DOI] [Google Scholar]
- 3.Baron U., Bujard H. Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol. 2000;327:401–421. doi: 10.1016/S0076-6879(00)27292-3. [DOI] [PubMed] [Google Scholar]
- 4.Berens C., Hillen W. Gene regulation by tetracyclines. Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes. Eur. J. Biochem. 2003;270(15):3109–3121. doi: 10.1046/j.1432-1033.2003.03694.x. [DOI] [PubMed] [Google Scholar]
- 5.Roberts M.C. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 2005;245(2):195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 6.Chopra I., Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001;65(2):232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertram R., Hillen W. The application of Tet repressor in prokaryotic gene regulation and expression. Microb. Biotechnol. 2008;1(1):2–16. doi: 10.1111/j.1751-7915.2007.00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meier I., Wray L.V., Hillen W. Differential regulation of the Tn10-encoded tetracycline resistance genes tetA and tetR by the tandem tet operators O1 and O2. EMBO J. 1988;7(2):567–572. doi: 10.1002/j.1460-2075.1988.tb02846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillen W., Berens C. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen T.N., Phan Q.G., Duong L.P., Bertrand K.P., Lenski R.E. Effects of carriage and expression of the Tn10 tetracycline-resistance operon on the fitness of Escherichia coli K12. Mol. Biol. Evol. 1989;6(3):213–225. doi: 10.1093/oxfordjournals.molbev.a040545. [DOI] [PubMed] [Google Scholar]
- 11.Gossen M., Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA. 1992;89(12):5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gossen M., Freundlieb S., Bender G., Müller G., Hillen W., Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268(5218):1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 13.Loew R., Heinz N., Hampf M., Bujard H., Gossen M. Improved Tet-responsive promoters with minimized background expression. BMC Biotechnol. 2010;10:81. doi: 10.1186/1472-6750-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agha-Mohammadi S., OMalley M., Etemad A., Wang Z., Xiao X., Lotze M.T. Second-generation tetracycline-regulatable promoter: repositioned tet operator elements optimize transactivator synergy while shorter minimal promoter offers tight basal leakiness. J. Gene Med. 2004;6(7):817–828. doi: 10.1002/jgm.566. [DOI] [PubMed] [Google Scholar]
- 15.Urlinger S., Baron U., Thellmann M., Hasan M.T., Bujard H., Hillen W. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl. Acad. Sci. USA. 2000;97(14):7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baron U., Gossen M., Bujard H. Tetracycline-controlled transcription in eukaryotes: novel transactivators with graded transactivation potential. Nucleic Acids Res. 1997;25(14):2723–2729. doi: 10.1093/nar/25.14.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moullan N., Mouchiroud L., Wang X., Ryu D., Williams E.G., Mottis A., Jovaisaite V., Frochaux M.V., Quiros P.M., Deplancke B., Houtkooper R.H., Auwerx J. Tetracyclines disturb mitochondrial function across eukaryotic models: A call for caution in biomedical research. Cell Reports. 2015;10(10):1681–1691. doi: 10.1016/j.celrep.2015.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.High K.A. The gene therapy journey for hemophilia: are we there yet? Blood. 2012;120(23):4482–4487. doi: 10.1182/blood-2012-05-423210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McPhee S.W., Janson C.G., Li C., Samulski R.J., Camp A.S., Francis J., Shera D., Lioutermann L., Feely M., Freese A., Leone P. Immune responses to AAV in a phase I study for Canavan disease. J. Gene Med. 2006;8(5):577–588. doi: 10.1002/jgm.885. [DOI] [PubMed] [Google Scholar]
- 20.Tuszynski M.H., Yang J.H., Barba D., U H.S., Bakay R.A., Pay M.M., Masliah E., Conner J.M., Kobalka P., Roy S., Nagahara A.H. Nerve growth factor gene therapy: Activation of neuronal responses in alzheimer disease. JAMA Neurol. 2015;72(10):1139–1147. doi: 10.1001/jamaneurol.2015.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marks W.J., Jr, Bartus R.T., Siffert J., Davis C.S., Lozano A., Boulis N., Vitek J., Stacy M., Turner D., Verhagen L., Bakay R., Watts R., Guthrie B., Jankovic J., Simpson R., Tagliati M., Alterman R., Stern M., Baltuch G., Starr P.A., Larson P.S., Ostrem J.L., Nutt J., Kieburtz K., Kordower J.H., Olanow C.W. Gene delivery of AAV2-neurturin for Parkinsons disease: a double-blind, randomised, controlled trial. Lancet Neurol. 2010;9(12):1164–1172. doi: 10.1016/S1474-4422(10)70254-4. [DOI] [PubMed] [Google Scholar]
- 22.Schlichtenbrede F.C., MacNeil A., Bainbridge J.W., Tschernutter M., Thrasher A.J., Smith A.J., Ali R.R. Intraocular gene delivery of ciliary neurotrophic factor results in significant loss of retinal function in normal mice and in the Prph2Rd2/Rd2 model of retinal degeneration. Gene Ther. 2003;10(6):523–527. doi: 10.1038/sj.gt.3301929. [DOI] [PubMed] [Google Scholar]
- 23.Nutt J.G., Burchiel K.J., Comella C.L., Jankovic J., Lang A.E., Laws E.R., Jr, Lozano A.M., Penn R.D., Simpson R.K., Jr, Stacy M., Wooten G.F., ICV GDNF Study Group. Implanted intracerebroventricular. Glial cell line-derived neurotrophic factor Randomized, double-blind trial of glial cell line-derived neurotrophic factor (GDNF) in PD. Neurology. 2003;60(1):69–73. doi: 10.1212/WNL.60.1.69. [DOI] [PubMed] [Google Scholar]
- 24.Manfredsson F.P., Tumer N., Erdos B., Landa T., Broxson C.S., Sullivan L.F., Rising A.C., Foust K.D., Zhang Y., Muzyczka N., Gorbatyuk O.S., Scarpace P.J., Mandel R.J. Nigrostriatal rAAV-mediated GDNF overexpression induces robust weight loss in a rat model of age-related obesity. Mol. Ther. 2009;17(6):980–991. doi: 10.1038/mt.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chtarto A., Bockstael O., Tshibangu T., Dewitte O., Levivier M., Tenenbaum L. A next step in adeno-associated virus-mediated gene therapy for neurological diseases: regulation and targeting. Br. J. Clin. Pharmacol. 2013;76(2):217–232. doi: 10.1111/bcp.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manfredsson F.P., Bloom D.C., Mandel R.J. Regulated protein expression for in vivo gene therapy for neurological disorders: progress, strategies, and issues. Neurobiol. Dis. 2012;48(2):212–221. doi: 10.1016/j.nbd.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Su X., Kells A.P., Huang E.J., Lee H.S., Hadaczek P., Beyer J., Bringas J., Pivirotto P., Penticuff J., Eberling J., Federoff H.J., Forsayeth J., Bankiewicz K.S. Safety evaluation of AAV2-GDNF gene transfer into the dopaminergic nigrostriatal pathway in aged and parkinsonian rhesus monkeys. Hum. Gene Ther. 2009;20(12):1627–1640. doi: 10.1089/hum.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tereshchenko J., Maddalena A., Bähr M., Kügler S. Pharmacologically controlled, discontinuous GDNF gene therapy restores motor function in a rat model of Parkinsons disease. Neurobiol. Dis. 2014;65:35–42. doi: 10.1016/j.nbd.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 29.Maddalena A., Tereshchenko J., Bähr M., Kügler S. Adeno-associated virus-mediated, mifepristone-regulated transgene expression in the brain. Mol. Ther. Nucleic Acids. 2013;2:e106. doi: 10.1038/mtna.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hadaczek P., Beyer J., Kells A., Narrow W., Bowers W., Federoff H.J., Forsayeth J., Bankiewicz K.S. Evaluation of an AAV2-based rapamycin-regulated glial cell line-derived neurotrophic factor (GDNF) expression vector system. PLoS One. 2011;6(11):e27728. doi: 10.1371/journal.pone.0027728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manfredsson F.P., Burger C., Rising A.C., Zuobi-Hasona K., Sullivan L.F., Lewin A.S., Huang J., Piercefield E., Muzyczka N., Mandel R.J. Tight Long-term dynamic doxycycline responsive nigrostriatal GDNF using a single rAAV vector. Mol. Ther. 2009;17(11):1857–1867. doi: 10.1038/mt.2009.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chtarto A., Yang X., Bockstael O., Melas C., Blum D., Lehtonen E., Abeloos L., Jaspar J.M., Levivier M., Brotchi J., Velu T., Tenenbaum L. Controlled delivery of glial cell line-derived neurotrophic factor by a single tetracycline-inducible AAV vector. Exp. Neurol. 2007;204(1):387–399. doi: 10.1016/j.expneurol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 33.Bockstael O., Chtarto A., Wakkinen J., Yang X., Melas C., Levivier M., Brotchi J., Tenenbaum L. Differential transgene expression profiles in rat brain, using rAAV2/1 vectors with tetracycline-inducible and cytomegalovirus promoters. Hum. Gene Ther. 2008;19(11):1293–1305. doi: 10.1089/hum.2008.099. [DOI] [PubMed] [Google Scholar]
- 34.Gasmi M., Brandon E.P., Herzog C.D., Wilson A., Bishop K.M., Hofer E.K., Cunningham J.J., Printz M.A., Kordower J.H., Bartus R.T. AAV2-mediated delivery of human neurturin to the rat nigrostriatal system: long-term efficacy and tolerability of CERE-120 for Parkinsons disease. Neurobiol. Dis. 2007;27(1):67–76. doi: 10.1016/j.nbd.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Vogel R., Amar L., Thi A.D., Saillour P., Mallet J. A single lentivirus vector mediates doxycycline-regulated expression of transgenes in the brain. Hum. Gene Ther. 2004;15(2):157–165. doi: 10.1089/104303404772679968. [DOI] [PubMed] [Google Scholar]
- 36.Karlsson M., Hammers S., Nilsson-Ehle I., Malmborg A.S., Wretlind B. Concentrations of doxycycline and penicillin G in sera and cerebrospinal fluid of patients treated for neuroborreliosis. Antimicrob. Agents Chemother. 1996;40(5):1104–1107. doi: 10.1128/aac.40.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yim C.W., Flynn N.M., Fitzgerald F.T. Penetration of oral doxycycline into the cerebrospinal fluid of patients with latent or neurosyphilis. Antimicrob. Agents Chemother. 1985;28(2):347–348. doi: 10.1128/AAC.28.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Centlivre M., Zhou X., Pouw S.M., Weijer K., Kleibeuker W., Das A.T., Blom B., Seppen J., Berkhout B., Legrand N. Autoregulatory lentiviral vectors allow multiple cycles of doxycycline-inducible gene expression in human hematopoietic cells in vivo. Gene Ther. 2010;17(1):14–25. doi: 10.1038/gt.2009.109. [DOI] [PubMed] [Google Scholar]
- 39.Saivin S., Houin G. Clinical pharmacokinetics of doxycycline and minocycline. Clin. Pharmacokinet. 1988;15(6):355–366. doi: 10.2165/00003088-198815060-00001. [DOI] [PubMed] [Google Scholar]
- 40.Blum D., Chtarto A., Tenenbaum L., Brotchi J., Levivier M. Clinical potential of minocycline for neurodegenerative disorders. Neurobiol. Dis. 2004;17(3):359–366. doi: 10.1016/j.nbd.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 41.McKeage K., Deeks E.D. Doxycycline 40 mg capsules (30 mg immediate-release/10 mg delayed-release beads): anti-inflammatory dose in rosacea. Am. J. Clin. Dermatol. 2010;11(3):217–222. doi: 10.2165/11204850-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 42.Verhoef K., Marzio G., Hillen W., Bujard H., Berkhout B. Strict control of human immunodeficiency virus type 1 replication by a genetic switch: Tet for Tat. J. Virol. 2001;75(2):979–987. doi: 10.1128/JVI.75.2.979-987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson R.P. Live attenuated AIDS vaccines: hazards and hopes. Nat. Med. 1999;5(2):154–155. doi: 10.1038/5515. [DOI] [PubMed] [Google Scholar]
- 44.Desrosiers R.C. Prospects for live attenuated HIV. Nat. Med. 1998;4(9):982. doi: 10.1038/1949. [DOI] [PubMed] [Google Scholar]
- 45.Mills J., Desrosiers R., Rud E., Almond N. Live attenuated HIV vaccines: a proposal for further research and development. AIDS Res. Hum. Retroviruses. 2000;16(15):1453–1461. doi: 10.1089/088922200750005976. [DOI] [PubMed] [Google Scholar]
- 46.Baba T.W., Jeong Y.S., Pennick D., Bronson R., Greene M.F., Ruprecht R.M. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science. 1995;267(5205):1820–1825. doi: 10.1126/science.7892606. [DOI] [PubMed] [Google Scholar]
- 47.Baba T.W., Liska V., Khimani A.H., Ray N.B., Dailey P.J., Penninck D., Bronson R., Greene M.F., McClure H.M., Martin L.N., Ruprecht R.M. Live attenuated, multiply deleted simian immunodeficiency virus causes AIDS in infant and adult macaques. Nat. Med. 1999;5(2):194–203. doi: 10.1038/5557. [DOI] [PubMed] [Google Scholar]
- 48.Chakrabarti L.A., Metzner K.J., Ivanovic T., Cheng H., Louis-Virelizier J., Connor R.I., Cheng-Mayer C. A truncated form of Nef selected during pathogenic reversion of simian immunodeficiency virus SIVmac239Deltanef increases viral replication. J. Virol. 2003;77(2):1245–1256. doi: 10.1128/JVI.77.2.1245-1256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whatmore A.M., Cook N., Hall G.A., Sharpe S., Rud E.W., Cranage M.P. Repair and evolution of nef in vivo modulates simian immunodeficiency virus virulence. J. Virol. 1995;69(8):5117–5123. doi: 10.1128/jvi.69.8.5117-5123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berkhout B., Verhoef K., van Wamel J.L., et al. Genetic instability of live-attenuated HIV-1 vaccine strains. J. Virol. 1999;73:1138–1145. doi: 10.1128/jvi.73.2.1138-1145.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zaunders J., Dyer W.B., Churchill M. The Sydney Blood Bank Cohort: implications for viral fitness as a cause of elite control. Curr. Opin. HIV AIDS. 2011;6(3):151–156. doi: 10.1097/COH.0b013e3283454d5b. [DOI] [PubMed] [Google Scholar]
- 52.Smith S.M., Khoroshev M., Marx P.A., Orenstein J., Jeang K.T. Constitutively dead, conditionally live HIV-1 genomes. Ex vivo implications for a live virus vaccine. J. Biol. Chem. 2001;276(34):32184–32190. doi: 10.1074/jbc.M101604200. [DOI] [PubMed] [Google Scholar]
- 53.Berkhout B., Marzio G., Verhoef K. Control over HIV-1 replication by an antibiotic; a novel vaccination strategy with a drug-dependent virus. Virus Res. 2002;82(1-2):103–108. doi: 10.1016/S0168-1702(01)00399-9. [DOI] [PubMed] [Google Scholar]
- 54.Das A.T., Zhou X., Vink M., Klaver B., Berkhout B. Conditional live virus as a novel approach towards a safe live attenuated HIV vaccine. Expert Rev. Vaccines. 2002;1(3):293–301. doi: 10.1586/14760584.1.3.293. [DOI] [PubMed] [Google Scholar]
- 55.Das A.T., Verhoef K., Berkhout B. A conditionally replicating virus as a novel approach toward an HIV vaccine. Methods Enzymol. 2004;388:359–379. doi: 10.1016/S0076-6879(04)88028-5. [DOI] [PubMed] [Google Scholar]
- 56.Kiselyeva Y., Ito Y., Lima R.G., Grivel J.C., Das A.T., Berkhout B., Margolis L.B. Depletion of CD4 T lymphocytes in human lymphoid tissue infected ex vivo with doxycycline-dependent HIV-1. Virology. 2004;328(1):1–6. doi: 10.1016/j.virol.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 57.Legrand N., van der Velden G.J., Ho Tsong Fang R., Douaisi M., Weijer K., Das A.T., Blom B., Uittenbogaart C.H., Berkhout B., Centlivre M. A doxycycline-dependent human immunodeficiency virus type 1 replicates in vivo without inducing CD4+ T-cell depletion. J. Gen. Virol. 2012;93(Pt 9):2017–2027. doi: 10.1099/vir.0.042796-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van der Velden Y.U., Kleibeuker W., Harwig A., Klaver B., Siteur-van Rijnstra E., Frankin E., Berkhout B., Das A.T. Construction of Nef-positive doxycycline-dependent HIV-1 variants using bicistronic expression elements. Virology. 2016;488:96–107. doi: 10.1016/j.virol.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 59.Das A.T., Baldwin C.E., Vink M., Berkhout B. Improving the safety of a conditional-live human immunodeficiency virus type 1 vaccine by controlling both gene expression and cell entry. J. Virol. 2005;79(6):3855–3858. doi: 10.1128/JVI.79.6.3855-3858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Das A.T., Klaver B., Harwig A., Vink M., Ooms M., Centlivre M., Berkhout B. Construction of a doxycycline-dependent simian immunodeficiency virus reveals a nontranscriptional function of tat in viral replication. J. Virol. 2007;81(20):11159–11169. doi: 10.1128/JVI.01354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Das A.T., Klaver B., Centlivre M., Harwig A., Ooms M., Page M., Almond N., Yuan F., Piatak M., Jr, Lifson J.D., Berkhout B. Optimization of the doxycycline-dependent simian immunodeficiency virus through in vitro evolution. Retrovirology. 2008;5:44. doi: 10.1186/1742-4690-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manoussaka M.S., Berry N., Ferguson D., Stebbings R., Robinson M., Ham C., Page M., Li B., Das A.T., Berkhout B., Almond N., Cranage M.P. Conditionally-live attenuated SIV upregulates global T effector memory cell frequency under replication permissive conditions. Retrovirology. 2013;10(1):59. doi: 10.1186/1742-4690-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marzio G., Verhoef K., Vink M., Berkhout B. In vitro evolution of a highly replicating, doxycycline-dependent HIV for applications in vaccine studies. Proc. Natl. Acad. Sci. USA. 2001;98(11):6342–6347. doi: 10.1073/pnas.111031498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marzio G., Vink M., Verhoef K., de Ronde A., Berkhout B. Efficient human immunodeficiency virus replication requires a fine-tuned level of transcription. J. Virol. 2002;76(6):3084–3088. doi: 10.1128/JVI.76.6.3084-3088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moumen A., Polomack L., Roques B., Buc H., Negroni M. The HIV-1 repeated sequence R as a robust hot-spot for copy-choice recombination. Nucleic Acids Res. 2001;29(18):3814–3821. doi: 10.1093/nar/29.18.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pathak V.K., Temin H.M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc. Natl. Acad. Sci. USA. 1990;87(16):6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Das A.T., Zhou X., Vink M., Klaver B., Verhoef K., Marzio G., Berkhout B. Viral evolution as a tool to improve the tetracycline-regulated gene expression system. J. Biol. Chem. 2004;279(18):18776–18782. doi: 10.1074/jbc.M313895200. [DOI] [PubMed] [Google Scholar]
- 68.Markusic D., Oude-Elferink R., Das A.T., Berkhout B., Seppen J. Comparison of single regulated lentiviral vectors with rtTA expression driven by an autoregulatory loop or a constitutive promoter. Nucleic Acids Res. 2005;33(6):e63. doi: 10.1093/nar/gni062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou X., Vink M., Klaver B., Berkhout B., Das A.T. Optimization of the Tet-On system for regulated gene expression through viral evolution. Gene Ther. 2006;13(19):1382–1390. doi: 10.1038/sj.gt.3302780. [DOI] [PubMed] [Google Scholar]
- 70.Das A.T., Zhou X., Metz S.W., Vink M.A., Berkhout B. Selecting the optimal Tet-On system for doxycycline-inducible gene expression in transiently transfected and stably transduced mammalian cells. Biotechnol. J. 2016;11(1):71–79. doi: 10.1002/biot.201500236. [DOI] [PubMed] [Google Scholar]
- 71.Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum. Gene Ther. 2005;16(11):1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- 72.Kleibeuker W., Zhou X., Centlivre M., Legrand N., Page M., Almond N., Berkhout B., Das A.T. A sensitive cell-based assay to measure the doxycycline concentration in biological samples. Hum. Gene Ther. 2009;20(5):524–530. doi: 10.1089/hum.2008.182. [DOI] [PubMed] [Google Scholar]
- 73.Ginhoux F., Turbant S., Gross D.A., Poupiot J., Marais T., Lone Y., Lemonnier F.A., Firat H., Perez N., Danos O., Davoust J. HLA-A*0201-restricted cytolytic responses to the rtTA transactivator dominant and cryptic epitopes compromise transgene expression induced by the tetracycline on system. Mol. Ther. 2004;10(2):279–289. doi: 10.1016/j.ymthe.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 74.Lena A.M., Giannetti P., Sporeno E., Ciliberto G., Savino R. Immune responses against tetracycline-dependent transactivators affect long-term expression of mouse erythropoietin delivered by a helper-dependent adenoviral vector. J. Gene Med. 2005;7(8):1086–1096. doi: 10.1002/jgm.758. [DOI] [PubMed] [Google Scholar]
- 75.Favre D., Blouin V., Provost N., Spisek R., Porrot F., Bohl D., Marmé F., Chérel Y., Salvetti A., Hurtrel B., Heard J.M., Rivière Y., Moullier P. Lack of an immune response against the tetracycline-dependent transactivator correlates with long-term doxycycline-regulated transgene expression in nonhuman primates after intramuscular injection of recombinant adeno-associated virus. J. Virol. 2002;76(22):11605–11611. doi: 10.1128/JVI.76.22.11605-11611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Latta-Mahieu M., Rolland M., Caillet C., Wang M., Kennel P., Mahfouz I., Loquet I., Dedieu J.F., Mahfoudi A., Trannoy E., Thuillier V. Gene transfer of a chimeric trans-activator is immunogenic and results in short-lived transgene expression. Hum. Gene Ther. 2002;13(13):1611–1620. doi: 10.1089/10430340260201707. [DOI] [PubMed] [Google Scholar]
- 77.Markusic D.M., de Waart D.R., Seppen J. Separating lentiviral vector injection and induction of gene expression in time, does not prevent an immune response to rtTA in rats. PLoS One. 2010;5(4):e9974. doi: 10.1371/journal.pone.0009974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Le Guiner C., Stieger K., Snyder R.O., Rolling F., Moullier P. Immune responses to gene product of inducible promoters. Curr. Gene Ther. 2007;7(5):334–346. doi: 10.2174/156652307782151461. [DOI] [PubMed] [Google Scholar]
- 79.McCloskey D.T., Turnbull L., Swigart P.M., Zambon A.C., Turcato S., Joho S., Grossman W., Conklin B.R., Simpson P.C., Baker A.J. Cardiac transgenesis with the tetracycline transactivator changes myocardial function and gene expression. Physiol. Genomics. 2005;22(1):118–126. doi: 10.1152/physiolgenomics.00016.2005. [DOI] [PubMed] [Google Scholar]
- 80.Barton M.D., Dunlop J.W., Psaltis G., Kulik J., DeGennaro L., Kwak S.P. Modified GFAP promoter auto-regulates tet-activator expression for increased transactivation and reduced tTA-associated toxicity. Brain Res. Mol. Brain Res. 2002;101(1-2):71–81. doi: 10.1016/S0169-328X(02)00170-5. [DOI] [PubMed] [Google Scholar]
- 81.Sisson T.H., Hansen J.M., Shah M., Hanson K.E., Du M., Ling T., Simon R.H., Christensen P.J. Expression of the reverse tetracycline-transactivator gene causes emphysema-like changes in mice. Am. J. Respir. Cell Mol. Biol. 2006;34(5):552–560. doi: 10.1165/rcmb.2005-0378OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Han H.J., Allen C.C., Buchovecky C.M., Yetman M.J., Born H.A., Marin M.A., Rodgers S.P., Song B.J., Lu H.C., Justice M.J., Probst F.J., Jankowsky J.L. Strain background influences neurotoxicity and behavioral abnormalities in mice expressing the tetracycline transactivator. J. Neurosci. 2012;32(31):10574–10586. doi: 10.1523/JNEUROSCI.0893-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morimoto M., Kopan R. rtTA toxicity limits the usefulness of the SP-C-rtTA transgenic mouse. Dev. Biol. 2009;325(1):171–178. doi: 10.1016/j.ydbio.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Anders K., Buschow C., Charo J., Blankenstein T. Depot formation of doxycycline impairs Tet-regulated gene expression in vivo. Transgenic Res. 2012;21(5):1099–1107. doi: 10.1007/s11248-011-9580-0. [DOI] [PubMed] [Google Scholar]
- 85.Sooksawate T., Isa K., Matsui R., Kato S., Kinoshita M., Kobayashi K., Watanabe D., Kobayashi K., Isa T. Viral vector-mediated selective and reversible blockade of the pathway for visual orienting in mice. Front. Neural Circuits. 2013;7:162. doi: 10.3389/fncir.2013.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tian X., Wang G., Xu Y., Wang P., Chen S., Yang H., Gao F., Xu A., Cao F., Jin X., Manyande A., Tian Y. An improved tet-on system for gene expression in neurons delivered by a single lentiviral vector. Hum. Gene Ther. 2009;20(2):113–123. doi: 10.1089/hum.2008.018. [DOI] [PubMed] [Google Scholar]
- 87.Kinoshita M., Matsui R., Kato S., Hasegawa T., Kasahara H., Isa K., Watakabe A., Yamamori T., Nishimura Y., Alstermark B., Watanabe D., Kobayashi K., Isa T. Genetic dissection of the circuit for hand dexterity in primates. Nature. 2012;487(7406):235–238. doi: 10.1038/nature11206. [DOI] [PubMed] [Google Scholar]
- 88.Chtarto A., Humbert-Claude M., Bockstael O., Das A.T., Boutry S., Breger L.S., Klaver B., Melas C., Barroso-Chinea P., Gonzalez-Hernandez T., Muller R.N., DeWitte O., Levivier M., Lundberg C., Berkhout B., Tenenbaum L. A regulatable AAV vector mediating GDNF biological effects at clinically-approved sub-antimicrobial doxycycline doses. Mol. Ther. Methods Clin. Dev. 2016;5:16027. doi: 10.1038/mtm.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Heinz N., Hennig K., Loew R. Graded or threshold response of the tet-controlled gene expression: all depends on the concentration of the transactivator. BMC Biotechnol. 2013;13:5. doi: 10.1186/1472-6750-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]