Abstract
The molecular chaperone Hsp90 is an essential eukaryotic protein that makes up 1–2% of all cytosolic proteins. Hsp90 is vital for the maturation and maintenance of a wide variety of substrate proteins largely involved in signaling and regulatory processes. Many of these substrates have also been implicated in cancer and other diseases making Hsp90 an attractive target for therapeutics. Hsp90 is a highly dynamic and flexible molecule that can adapt its conformation to the wide variety of substrate proteins with which it acts. Large conformational rearrangements are also required for the activation of these client proteins. One driving force for these rearrangements is the intrinsic ATPase activity of Hsp90, as seen with other chaperones. However, unlike other chaperones, studies have shown that the ATPase cycle of Hsp90 is not conformationally deterministic. That is, rather than dictating the conformational state, ATP binding and hydrolysis shifts the equilibrium between a pre-existing set of conformational states in an organism-dependent manner. In vivo Hsp90 functions as part of larger heterocomplexes. The binding partners of Hsp90, co-chaperones, assist in the recruitment and activation of substrates, and many co-chaperones further regulate the conformational dynamics of Hsp90 by shifting the conformational equilibrium towards a particular state. Studies have also suggested alternative mechanisms for the regulation of Hsp90’s conformation. In this review, we discuss the structural and biochemical studies leading to our current understanding of the conformational dynamics of Hsp90 and the role that nucleotide, co-chaperones, post-translational modification and clients play in regulating Hsp90’s conformation. We also discuss the effects of current Hsp90 inhibitors on conformation and the potential for developing small molecules that inhibit Hsp90 by disrupting the conformational dynamics.
Introduction
Hsp90 is an essential eukaryotic protein making up 1–2% of all cytosolic proteins, and it is a member of the class of proteins known as molecular chaperones. Molecular chaperones are required for maintaining the correctly folded state of proteins within the cell. Hsp60 (GroEL) and Hsp70 (DnaK) represent two well studied members of this class of proteins. Both Hsp60 and Hsp70 interact with nascent polypeptide chains and promote their folding by interacting with hydrophobic surfaces on substrate proteins (Gomez-Puertas et al., 2004; Kurt et al., 2006; Rudiger et al., 1997; Weissman et al., 1995). Once substrates are bound, the chaperone undergoes subsequent rounds of ATP hydrolysis causing conformational changes within the chaperone (Young et al., 2004). These conformational changes lead to the release of the substrate either into the cytosol by Hsp70 or into the lumen of the Hsp60 folding machine where proper folding can occur (Walter & Buchner, 2002).
Similarly, Hsp90 binds and hydrolyzes ATP and is believed to interact with hydrophobic surfaces on substrate (client) proteins. Unlike other chaperones, Hsp90 appears to act largely at later stages of the folding pathway (McLaughlin et al., 2002). Unlike the substrates of other chaperones, Hsp90 clients have already achieved a partially folded or almost fully folded conformation before interacting with Hsp90 (Jakob et al., 1995; McLaughlin et al., 2002) suggesting that Hsp90 must adapt its conformation to match each client or that different conformations recognize different clienys. Conformational changes within Hsp90 then induce subtle conformational rearrangements within the client protein facilitating the binding of ligands or partner proteins (Richter & Buchner, 2001; Young et al., 2001; Zhao et al., 2005). Hsp90 interacts with clients that are found primarily in signaling and regulatory processes including steroid hormone receptors, kinases, and transcription factors (Caplan et al., 2007; Pearl & Prodromou, 2006; Picard, 2006; Richter & Buchner, 2001). For a complete list of the currently identified Hsp90 client proteins see http://www.picard.ch/downloads/Hsp90interactors.pdf. Many of Hsp90’s client proteins, including p53, Cdk4, c-src and v-src, are oncogenic or otherwise required for cell proliferation, making Hsp90 an attractive target for anti-cancer therapeutics (Neckers, 2007). Inhibition of Hsp90 with small molecules such as geldanamycin and its derivatives has been shown to be antitumorigenic, and several of these compounds are currently in clinical trials (Chiosis et al., 2003; Neckers & Ivy, 2003; Solit et al., 2008; Workman, 2004). Hsp90 has also been implicated in other diseases including Alzheimers (Dickey et al., 2008), vascular disease (Shah et al., 1999), and viral diseses (Geller et al., 2007). In the case of RNA viruses, capsid proteins are clients of Hsp90, and Hsp90 inhibitors reduce viral replication because capsid proteins become misfolded and are targeted for degradation. The viruses also appear unable to bypass the requirement for Hsp90 through mutation making Hsp90 an attractive anti-viral target (Geller et al., 2007). Hsp90 also plays a role outside of protein folding by buffering phenotypic variation and allowing these variations to present themselves only under conditions of stress. When mutations that reduce the cellular levels of Hsp90 were studied in Drosophila, unusual developmental morphologies were observed. These altered morphologies were the result of an increased expression of cryptic genetic variations. With even lower levels of Hsp90 expression, more and more genetic variations became expressed (Rutherford & Lindquist, 1998). In nature, transient changes in the expression level of Hsp90 would allow for the selection of new phenotypic traits assisting in the evolution of the organism. The ability of Hsp90 to buffer genetic variation has also been observed in Arabidopsis thaliana indicating that Hsp90’s role in evolution may be conserved across species (Sangster et al., 2008).
Hsp90 has been shown to be a highly flexible and dynamic molecule, and large conformational changes have been observed in the presence of nucleotide. While it is clear that the conformational dynamics of Hsp90 play an important role in recognizing and activating client protiens, the conformational changes and determinants of substrate recognition associated with Hsp90 function remain unclear. It is also of tremendous importance to understand the various means for regulating the dynamics and therefore function of Hsp90. The exact nature of the conformations of Hsp90, their function, and regulation has become an area of intense study in recent years, and, as discussed below, much progress has been made in understanding the conformational dynamics of Hsp90 and the role that these dynamics play in chaperone function.
Architecture of Hsp90
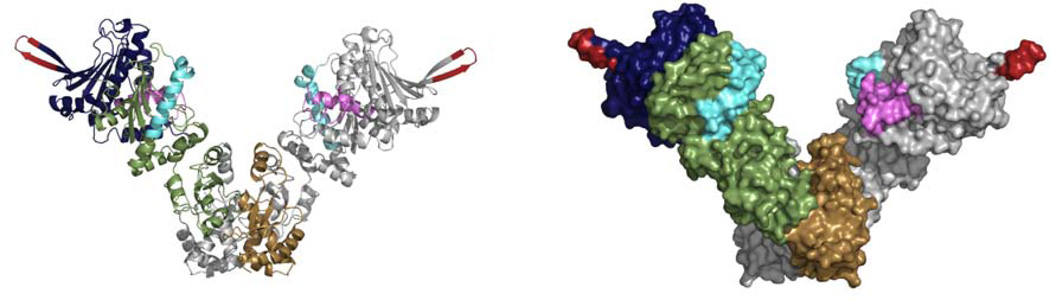
Hsp90 is highly conserved from bacteria to eukaryotes with ~50% sequence similarity between E. coli and humans. In humans, Hsp90 exists mainly as four isoforms with two cytosolic forms (Hsp90α, Hsp90β), a mitochondrial version (Trap1) and a form in the endoplasmic reticulum (Grp94). All versions of Hsp90 exist as obligate dimers consisting of three domains per monomer (Figure 1). The C-terminal domain (CTD, brown) is the site of dimerization, the middle domain (MD, green) has been implicated in client binding, and ATP binds to the N-terminal domain (NTD, blue) (Pearl & Prodromou, 2006). The nucleotide binding pocket in the NTD is the most conserved aspect of Hsp90 across species, and has been identified as related to the GHKL superfamily of ATPases, which include MutL and DNA gyrase (Bergerat et al., 1997; Dunbrack et al., 1997). As with other GHKL superfamily proteins, binding and hydrolysis of nucleotide leads to local and global conformational changes in the protein. For example, the binding of nucleotide leads to dimerization of the N-terminal domains and these changes represent a key step in the chaperone cycle of Hsp90. Other important conserved structural features include the lid (Figure 1, purple), a helix-loop-helix motif adjacent to the nucleotide binding pocket (Prodromou et al., 1997; Stebbins et al., 1997), and the catalytic loop (Figure 1, cyan), a region between the NTD and MD that is essential for catalysis (Meyer et al., 2003). These regions are also involved in the conformational dynamics of Hsp90, and the details of these structural changes and their importance in function are discussed in the following sections.
Figure 1.
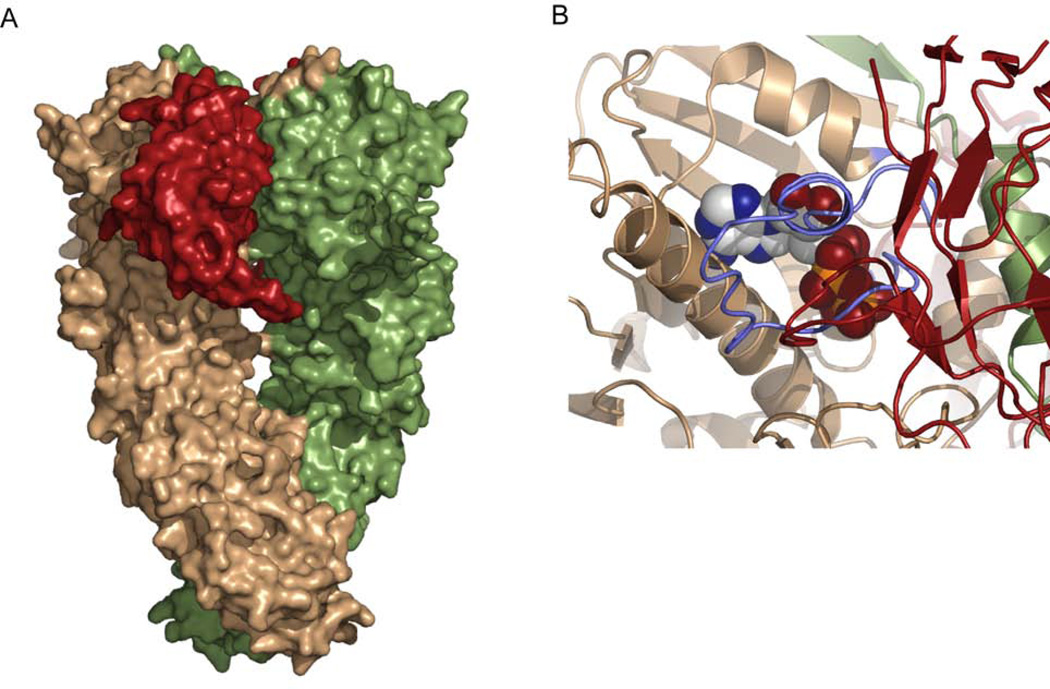
Hsp90 exists as a homodimer with three domains per monomer. The crystal structure of the bacterial homolog, HtpG (PDB code 2IOQ), is shown as both a cartoon representation and a surface representation. The domains of one monomer are colored blue (NTD), green (MD), and brown (CTD). The other monomer is colored grey. The NTD binds nucleotide and contains a variable charged loop (red) and the conserved lid region (purple) which is adjacent to the nucleotide binding pocket. The middle domain contains the conserved catalytic loop (cyan) which is essential for ATP hydrolysis. The CTD provides the constitutive dimer interface.
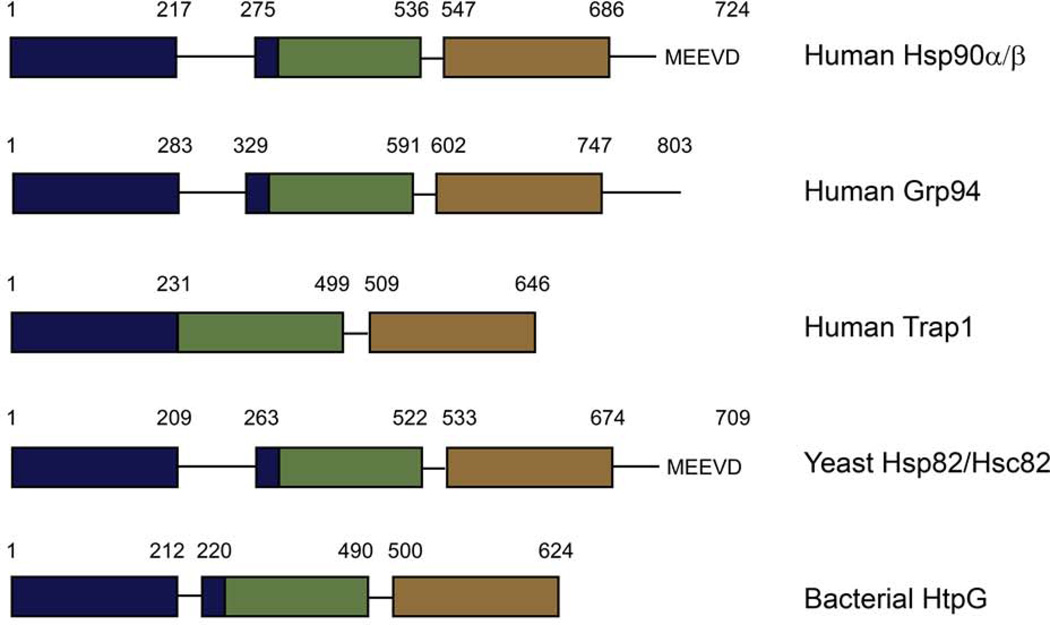
Despite the high similarity, there are important differences in the overall domain organization of the different Hsp90s (Figure 2). A repetitive charged region (Figure 1, red) that varies in length is present at the C-terminal end of the NTD in yeast and cytosolic higher eukaryotic Hsp90s as well as in Grp94. This charged region is also present in the bacterial protein HtpG, but it is significantly shorter in length. This region was originally believed to be a flexible tether between the two domains, but crystal structures have shown structured elements on either side of this charged region that are part of the NTD leaving only a few amino acids as the linker between the NTD and MD (Ali et al., 2006; Huai et al., 2005; Shiau et al., 2006; Soldano et al., 2003). Though the charged region is dispensible in yeast, it has been implicated in the binding of substrates. Truncation mutants containing the N-terminal domain and the charged region show higher affinity for peptide and non-native substrates than mutants lacking the charged region (Scheibel et al., 1999). The yeast and cytosolic higher eukaryotic proteins also have a C-terminal MEEVD peptide that is important in the binding of co-chaperones. Co-chaperones are helper proteins that form complexes with Hsp90 and are thought to function in client protein loading and in regulating the ATPase cycle and therefore the conformation of Hsp90. Many co-chaperones also function independently of Hsp90 as chaperones or have other functions in the cell. The co-chaperones Hsp70, Hop and FKBP52, amount others, contain TPR-domains that bind Hsp90 via the C-terminal MEEVD. Currently no co-chaperones have been identified for Trap1, Grp94 or HtpG, suggesting that they have evolved alternate methods for controlling the chaperone cycle.
Figure 2.
Domain architecture for different Hsp90 homologs. The domains are colored with the N-terminal domain (NTD) in blue, the middle domain (MD) in green, and the C-terminal domain (CTD) in brown.
Global conformational changes in the full-length protein
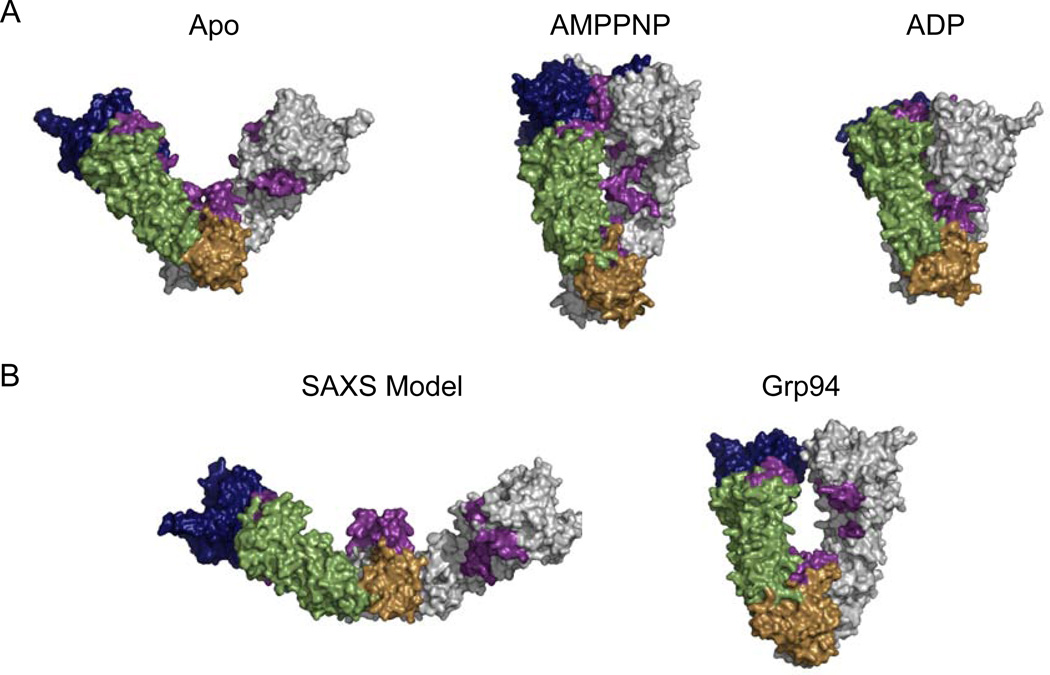
Structural studies of Hsp90 have shown the molecular chaperone to be a highly dynamic and flexible molecule, and the conformational variability of Hsp90 is essential in its interaction with and activation of client proteins. As mentioned above, Hsp90 interacts with a wide-variety of substrate proteins and the large differences in both size and shape amongst these client proteins indicates that Hsp90 must adapt its conformation according to the conformation of the client protein. The ability to adapt structurally may be one mechanism by which Hsp90 is able to recognize such a wide variety of client proteins. A crystal structure of the bacterial Hsp90 shows the apo (nucleotide-free) protein in an open V-like conformation with only the CTDs dimerized (Figure 3) (Shiau et al., 2006). In this structure hydrophobic surfaces are presented along the inner cleft of the dimer suggesting a means of substrate recognition (Figure 3, purple, residues 8–23, 326–342, 368–384, and 586–612; Hsp82 numbering). A SAXS study of apo HtpG revealed a much more extended conformation in solution (Figure 3) (Krukenberg et al., 2008). It is possible that both conformations are functionally relevant as a means of differentiating between client proteins. For example, the extended structure may bind to larger client proteins such as telomerase whereas the crystal structure can more tightly interact with smaller substrates such as steroid hormone receptors. An additional apo conformation that is remarkably similar to the crystal structure of the endoplasmic reticulum homolog, Grp94, has also been described for HtpG (Figure 3). Interestingly, in a citrate synthase aggregation assay the Grp94-like state prevents the aggregation of citrate synthase whereas the extended state has no effect, further supporting the idea that different Hsp90 conformations recognize different client proteins.
Figure 3.
Full-length structures of Hsp90. The NTD is shown in blue, the MD in green and the CTD in brown. The second monomer of the biological dimer is shown in grey. In all structures exposed hydrophobic surfaces are highlighted in purple. A) Surface representations of the apo and ADP HtpG structure (Shiau et al., 2006); 2IOQ and 2IOP) and the yeast AMPPNP-bound structure (Ali et al., 2006). B) Surface representations of the apo HtpG solution state (Krukenberg et al., 2008) and the Grp94 crystal structure (Dollins et al., 2007).
The apo conformation of Hsp90 is also variable across species. SAXS and EM studies of human Hsp90 (hHsp90), yeast Hsp90 (yHsp90), and pig Hsp90 show a conformation that is even more extended than the conformation seen in HtpG (Bron et al., 2008). The mitochondrial Grp94 also exists in a similar extended conformation in solution (Krukenberg et al, in press). An additional conformation similar to the HtpG crystal structure was also observed for yHsp90, hHsp90, pig Hsp90 (Bron et al., 2008). The structural differences between prokaryotic and eukaryotic Hsp90 may be indicative of the interaction with different client proteins or of differing methods of regulating the conformational cycle.
While structural variability exists for Hsp90 in the absence of nucleotide, it is known that nucleotide also plays an important role in regulating the conformational dynamics of Hsp90. The ATP binding pocket of Hsp90 is unique to the GHKL superfamily of proteins and is characterized as a Bergerat fold (Dutta & Inouye, 2000), and the ATPase activity of Hsp90 is essential for function‥ Due to the similarity of the ATPase domain to the GHKL superfamily, a conformational cycle, similar to MutL and DNA Gyrase, with Hsp90 acting as a molecular clamp after NTD dimerization was originally proposed (Dutta & Inouye, 2000). Structural studies of full-length Hsp90 demonstrate that NTD dimerization does indeed occur across species. In the presence of the non-hydrolyzable ATP analog, AMPPPNP, and the co-chaperone p23, which binds the NTD, yHsp90 was crystallized in a closed conformation where the NTDs are dimerized (Figure 3) (Ali et al., 2006). In this state, the hydrophobic surfaces are less well exposed than in the apo state but still accessible on the outer edges of the dimer cleft (Ali et al., 2006) suggesting that the molecular clamp analogy is not the most appropriate description of Hsp90’s interaction with client proteins. The closed state does not allow sufficient room between the monomers for a client protein and the placement of the hydrophobic surfaces indicates that clients bind to the outer edges of the dimer cleft. This hypothesis was confirmed by an EM reconstruction of Hsp90 bound to a client protein that shows the client bound to the side of the dimer (Vaughan et al., 2006). Because the AMPPNP-bound state was crystallized in the presence of a co-chaperone, it was unknown if this state was accessible through nucleotide binding alone. Negative-stain EM and SAXS studies reveal that the AMPPNP-bound state is accessible in the absence of co-chaperone and HtpG and hHsp90 also sample the closed AMPPNP-bound state (Krukenberg et al., 2008; Southworth & Agard, 2008).
In the presence of ADP, an even more compact structure was observed for HtpG (Figure 3) (Shiau et al., 2006). In this compact state the hydrophobic surfaces are buried in the core of the protein suggesting a means for client protein release. The compact ADP-bound crystal structure was initially quite controversial, especially because it has not been observed by SAXS. Recent negative-stain EM and cross-linking studies, however, demonstrate the presence of the compact ADP state in HtpG, yHsp90, and hHsp90 suggesting that this state is functionally relevant (Southworth & Agard, 2008). In eukaryotes the ADP state is only observed in the presence of small levels of cross-linker highlighting the transient nature of the conformation. The relevance of the ADP-state is further supported by cross-linking studies of Grp94 (Chu et al., 2006). Of the known structures, the observed cross-links are only compatible with the compact ADP crystal state. The state was most likely not seen by SAXS due to the higher concentrations of the SAXS experiments. The higher concentration appears sufficient to disrupt the compact conformation. Hsp90 may self-chaperone in order to open the ADP state back to the apo state resetting the conformational cycle.
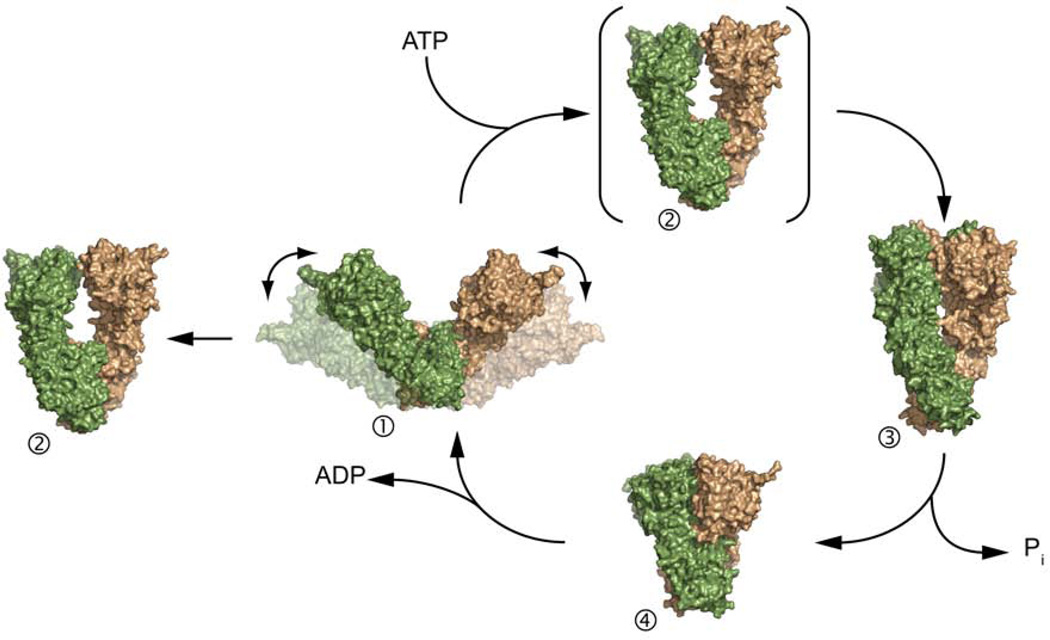
These structures illustrate the amazing flexibility of Hsp90 with the NTD moving by ~50Å between each state and point to a conserved conformational cycle. An additional conformation, intermediate to the open apo and closed AMPPNP-bound states observed in other homologs, is observed in the full-length crystal structure of nucleotide bound Grp94 (Figure 3) (Dollins et al., 2007). The structures of Hsp90 taken together suggest a nucleotide-dependent conformational cycle (Figure 5). In this cycle, Hsp90 begins in an open state with a variable open angle. The addition of ATP then causes a conformational shift to a more closed state. The Grp94 crystal structure may represent an intermediate between the extended and closed states, or the Grp94 state may be a catalytically inactive state. The cycle continues with the hydrolysis of ATP and the conversion to a more compact state where all hydrophobic surfaces are now buried, thus releasing client proteins/co-chaperones. The compact state would then open and return to the extended conformation to begin the cycle again. While the evidence implys a conserved conformational cycle across homologs especially HtpG, yHsp90, and hHsp90, the degree of conservation with other homologs including Grp94 and Trap1 remains an open question.
Figure 5.
The structures of Hsp90 suggest a nucleotide-dependent conformational cycle. In this cycle, the binding of ATP shifts the conformation from an open structure (1) to a NTD-dimerized closed structure (3). The Grp94 crystal structure (2) may represent and intermediate along this path or non-catalytic conformation. The hydrolysis of ADP causes a further compaction of the protein (4) and the release of nucleotide restarts the conformational cycle.
Local structural changes in the NTD due to nucleotide binding
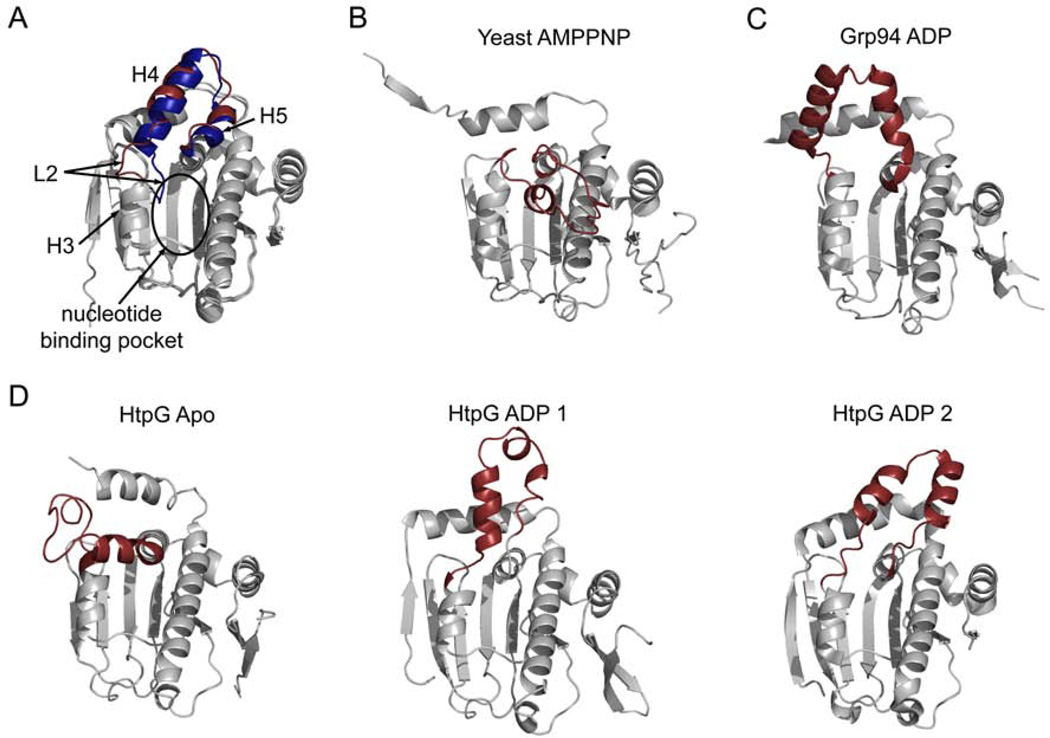
While the crystal structures correspond to remarkably large changes in Hsp90’s conformation, the individual domain structure remains largely conserved. The flexibility of Hsp90 comes mainly from rigid body motions centered at the NM (between the NTD and MD) and MC (between the MD and CTD) interfaces. There are, however, important local changes that occur in the NTD and these local changes facilitate the global rearrangements discussed above. The largest local changes occur in the lid (residues 94–124, Hsp82 numbering, Figure 1, purple) which consists of a helix-loop-helix positioned near the binding pocket and these motions communicate the nucleotide state of Hsp90 to the rest of the protein especially the domain interfaces. Crystal structures have revealed that the lid is highly variable in its orientation, and molecular dynamics simulations with the human NTD confirm that the most significant local changes upon nucleotide binding involve the lid (Colombo et al., 2008). In one orientation seen in cytosolic human Hsp90 (hHsp90) loop 2 of the lid (L2, see Figure 4A, blue) is displaced by more than 9Å into the nucleotide binding pocket potentially blocking the binding of ligand (Stebbins et al., 1997). In an alternate crystal form of the hHsp90 NTD and in crystal structures of the isolated yeast (yHsp90) NTD, L2 is moved away from the binding pocket allowing nucleotide to bind (Figure 4A, red) (Prodromou et al., 1997; Stebbins et al., 1997). A dramatically different arrangement for the lid is seen in the yHsp90 full-length structure with the non-hydrolyzable AMPPNP bound (Ali et al., 2006). In this conformation the lid now covers the nucleotide binding pocket trapping the bound AMPPNP (Figure 4B). This lid position also exposes hydrophobic surfaces that are buried in other lid conformations, and this hydrophobic surface forms part of the NTD dimerization interface stabilizing the closed AMPPNP-bound state. The endoplasmic reticulum homolog, Grp94, has a unique 5 amino acid insertion in the lid (residues 182–186) leading to a structural rearrangement of the lid in response to nucleotide that is different from other Hsp90 homologs (Figure 4C). Although the lid position is unique in nucleotide-bound Grp94, this conformation also leads to the exposure of a large predominantly non-polar surface (Immormino et al., 2004) as seen in AMPPNP-bound yHsp90. The hydrophobic surface may also serve as an interaction site for NTD dimerization in Grp94. In the absence of nucleotide the isolated NTD of Grp94 overlays well with the yeast and human conformations shown in Figure 4A (blue) (Dollins et al., 2005). In the full-length Grp94 structure bound to nucleotide the lid becomes disordered (Dollins et al., 2007).
Figure 4.
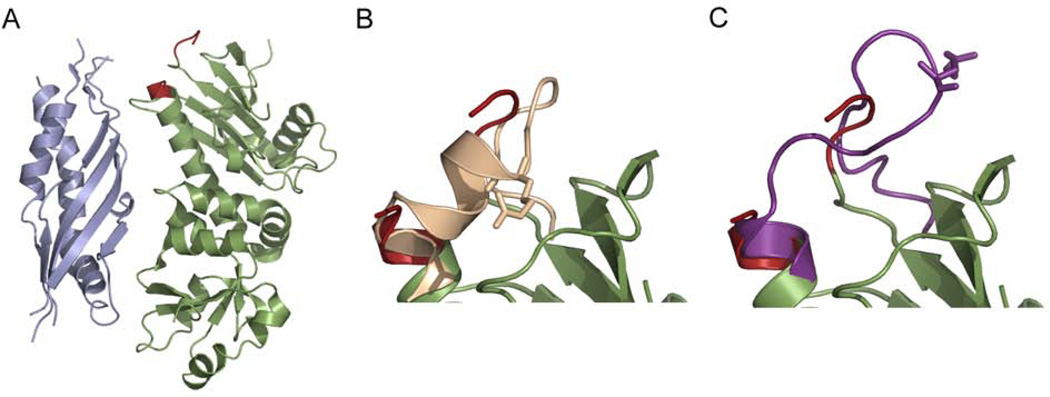
Variation in the N-terminal domain of Hsp90 occurs mostly in the lid. All structures are of the N-terminal domain of Hsp90 with the lid highlighted in red or blue. A) Two conformations of the isolated hHsp90 NTD (1YER and 1YES). In one conformation (blue) loop 2 (L2) extends into the nucleotide binding pocket. Large motions in the lid can also be seen in B) the NTD from the full length yeast structure (2CG9), and C) the isolated NTD of Grp94 bound to ADP (1TBW). D) Three additional lid conformations are observed in crystals of HtpG (2IOR, 2IOQ, and 2IOP).
The structural variability of the lid is even more pronounced in the bacterial homolog HtpG. In structures of the isolated NM domain the lid of HtpG (residues 96–126) is largely disordered (Huai et al., 2005). From the portion of the lid that is ordered in the crystal structure it can be seen that the addition of ADP causes H3 and H4 to partially unwind. Molecular dynamics simulations also show a loss of lid structure for hHsp90 when ATP is bound. The lid loses α-helical content and gains flexibility as a consequence of interactions with the nucleotide (Colombo et al., 2008). This is in contrast to other GHKL family members such as MutL where the lid becomes more ordered in the presence of nucleotide (Ban et al., 1999). Two additional lid positions are seen in different crystals of ADP-bound HtpG (Figure 4D) (Shiau et al., 2006).
As observed with the yHsp90 AMPPNP-bound structure and the Grp94 structure, rearrangements in the lid region lead to larger conformational rearrangements in the protein such as NTD dimerization. Other global conformational rearrangements have also been proposed as resulting from rearrangements in the lid. The full length structure of apo HtpG supports this idea. In this structure the lid adopts a unique position orthogonal to the NTD β-sheet as opposed to parallel to the β-sheet as seen in other structures (Figure 4) (Shiau et al., 2006). This lid conformation precludes ATP binding by positioning residue F123 where the base and sugar of the nucleotide would be positioned, blocking access to the binding pocket. If the lid is positioned as seen in the ADP-bound form of HtpG, steric clashes occur between the NTD and MD in the apo structure suggesting that the binding of nucleotide must cause the lid to become disordered or the rearrangement of the NTD in relation to the MD. Simulations also show that local conformational changes in the lid region are coupled to larger conformational changes. While the lid becomes more flexible upon the binding of ATP, Essential Dynamics and Covariance analysis of the simulations shows that the rest of the NTD becomes more constrained with a stronger interaction network across the entire NTD (Colombo et al., 2008).
The role of conformation in regulating the ATPase cycle of Hsp90
The ATPase cycles of cytosolic yeast and human Hsp90
While the structural studies provide a convincing picture of the Hsp90 conformational cycle and its dependence on nucleotide, many questions remain about the molecular mechanism of Hsp90 and the connections between conformation, ATPase activity, and client activation. All Hsp90 homologs studied so far have ATPase activity and this activity is essential for the functioning of Hsp90 in the cell. The ATPase activity varies between homologs, but overall this activity is low especially in comparison to other known enzymes (Table 1). The precise regulation of the ATPase rate is also essential. Mutant Hsp90s with either a faster (T22I) or slower (T101I) ATPase rate result in temperature sensitive growth phenotypes and are unable to fully activate glucorticoid receptor (GR) in vivo (Nathan & Lindquist, 1995; Prodromou et al., 2000). Using biochemical and structural studies, the pathways involved in the hydrolysis of ATP as well as key structural factors involved in the regulation of the cycle are beginning to be understood.
Table 1.
Kinetic parameters of ATP hydrolysis for homologs of Hsp90 at 25°C
| Homolog | Km (µM) | ATPase rate (s−1) |
|---|---|---|
| hHsp90a | 190 | 0.0009 |
| yHsp90b | 100 | 0.0015 |
| HtpGc,f | 250 | 0.011 |
| Trap1d | 15.4 | 0.0027 |
| Grp94e | 92 | 0.006 |
Parameters were taken from the following
measured at 30°C
From the crystal structures of the isolated NTD and the full-length protein, it is clear that local conformational changes in the lid region as well as global changes leading to NTD dimerization occur in the presence of nucleotide. The structural studies alone do not reveal the role of conformation in regulating the ATPase cycle. Thorough biochemical characterization of the kinetic cycle of yHsp90 shows that NTD dimerization is required for attaining the correct catalytic conformation and this conformational change determines the rate of hydrolysis. Mutant yHsp90 lacking the C-terminal dimerization domain has a significantly impaired ATPase rate (Prodromou et al., 2000; Richter et al., 2001) supporting a role for NTD dimerization in the hydrolysis of ATP. This was confirmed with an Hsp90 heterodimer containing only one NTD. The heterodimer has one third the ATPase rate of the wild-type (WT) protein (Richter et al., 2001). This data together with results showing the rate-limiting step to be a conformational change upon ATP binding (Hessling et al., 2009; Weikl et al., 2000) suggest that NTD dimerization is the rate-limiting step. A recent FRET study reveals that upon ATP binding a slow transition occurs to an intermediate open state. This intermediate open state may be characterized by the lid folding over the nucleotide binding pocket and exposing hydrophibic surfaces important for dimerization as discussed above. NTD dimerization then occurs in a second slow transition to another intermediate state. A final conformational change to the catalytically active state occurs and ATP is then rapidly hydrolyzed (Hessling et al., 2009). The structures of the intermediate states have yet to be determined and it will be interesting to see if the Grp94 structure captures the open intermediate as proposed in Figure 5.
Further biochemical studies of yHsp90 have dissected the regions of the NTD that are important for controlling the ATPase rate. The N-terminus and the lid regulate the rate of NTD dimerzation and therefore the rate of ATP hydrolysis. Removal of the lid abolishes all ATPase activity, but, surprisingly, a heterodimer lacking the lid on only one NTD (lidless) has a substantially increased ATPase rate (Richter et al., 2006). Structural studies reveal that the lid serves to inhibit NTD dimerization and the increase in activity in the lidless heterodimer is due to an increase in NTD dimerization (Hessling et al., 2009; Richter et al., 2006). The increased dimerization promoted by the lidless heterodimer is dependent upon the first 24 amino acids of the N-terminus. In the closed yeast AMPPNP structure these residues form the dimerization interface. When these 24 amino acids are deleted from the lidless NTD, the ATPase activity of the heterodimer is inhibited (Richter et al., 2006) presumably because the site of NTD dimerization has been disrupted.
Initial biochemical studies of hHsp90 suggested that the human protein had a substantially different hydrolysis mechanism without dimerization of the NTDs. Recent structural and biochemical studies have shown that the molecular mechanism of hydrolysis is largely conserved between yHsp90 and hHsp90 although important differences in the regulation of the cycle exist. A mechanism that depends upon NTD dimerization predicts a degree of cooperativity between the two NTDs, but initial kinetic studies of hHsp90 indicated that the two ATP binding sites do not interact cooperatively (McLaughlin et al., 2004). The dimerization of the NTDs and the structure of the lid in the AMPPNP-bound state also suggest that ATP is trapped in the binding pocket during hydrolysis. Nucleotide exchange studies have confirmed this hypothesis for yHsp90 (Weikl et al., 2000), but in hHsp90 ATP can freely exchange throughout the hydrolysis cycle (Richter et al., 2008) bringing into question the conserved nature of the ATPase cycle.
As discussed above, a negative-stain EM comparison of yHsp90 and hHsp90 in the presence of nucleotide demonstrate that both homologs populate the closed conformation as seen in the AMPPNP-bound yeast structure (Southworth & Agard, 2008). Recent biochemical studies provide additional evidence that the ATPase mechanisms are indeed conserved between yHsp90 and hHsp90. Like yHsp90, C-terminal dimerization and the presence of both NTDs is required for the ATPase activiy of hHsp90 (Richter et al., 2008; Vaughan et al., 2009). Removal of the lid in hHsp90 also leads to a complete lose of activity in the homodimer, while the heterodimer between the lidless mutant and WT shows a strong increase in the ATPase activity (Richter et al., 2008). As expected for a mechanism requiring NTD dimerization, point mutations that increase NTD dimerization also show an increase in ATPase activity for both yeast and human Hsp90 (Vaughan et al., 2009).
hHsp90 also populates at least one intermediate conformation between the open and closed states observed by EM and the rate-limiting step for hydrolysis is a conformational change upon the binding of nucleotide further supporting the idea of a conserved mechanism (McLaughlin et al., 2004).
The observed differences for yHsp90 and hHsp90 lie in the regulation of the cycle instead of in the underlying mechanism. hHsp90 has a higher activation energy leading to a more transient closed state than the yeast protein (Richter et al., 2008). Negative-stain EM analysis of hHsp90 supports this idea. Unlike yHsp90 the closed AMPPNP-bound state of hHsp90 can only be seen by EM in the presence of very low levels of cross-linker (Southworth & Agard, 2008). The transient nature of the closed state also explains hHsp90’s inability to trap ATP during the hydrolysis cycle and the lack of cooperativity between the two ATP binding sites. yHsp90 and hHsp90 may have evolved different levels of regulation because of the different client proteins that they encounter. The slower rate of ATP hydrolysis by hHsp90 may indicate that in humans Hsp90 clients require longer interaction times with Hsp90 in order to be fully activated. Also, the transient nature of the closed state for hHsp90 suggests that co-chaperones or other means of regulation may play a larger role in humans than in yeast.
The ATPase cycles of HtpG, Grp94, and Trap1
The ATPase cycle of the bacterial HtpG has been less well characterized, but it appears that it shares the same hydrolysis mechanism as both yHsp90 and hHsp90. HtpG progresses from a similar open to closed state in the presence of nucleotide as is seen for both yHsp90 and hHsp90 (Krukenberg et al., 2008; Southworth & Agard, 2008). Also, hydrogen-deuterium exchange mass spectrometry (HX-MS) experiments have demonstrated that an intermediate conformation exists before the closed state and the conversion from the intermediate to closed state is the rate-limiting step in the hydrolysis reaction. Like yHsp90, HtpG traps the nucleotide during the cycle (Graf et al., 2009) suggesting that the closed state is less transient than observed for hHsp90. This is supported by negative-stain EM and SAXS experiments where the closed state is readily observable in the presence of AMPPNP (Krukenberg et al., 2008; Southworth & Agard, 2008).
It is less clear whether or not the ER homolog, Grp94, or the mitochondrial homolog, Trap1 follow the same pathway as other Hsp90s. For Grp94, ATP binding proceeds via a one-step mechanism unlike other homologs, and NTD dimerization has not been directly measured. Like other homologs, the rate-limiting step is either hydrolysis or a conformational change prior to hydrolysis, and ATP is not trapped by Grp94 during hydrolysis (Frey et al., 2007). Like hHsp90 this may be due to a transient closed state. . For Trap1, hydrolysis or a conformational change is also rate-limiting, and like Grp94 nucleotide is not trapped during hydrolysis. Based on tryptophan fluorescence binding of ATP to Trap1 is a two-step process (Leskovar et al., 2008). Given the structural and sequence similarities of Grp94 and Trap1 to other Hsp90s it appears most likely that the overall hydrolysis cycle is conserved across homologs. The differences are therefore most likely due to differences in the apo structures or the regulation of the cycle.
The importance of dimerization
NTD and CTD dimerization are required aspects of Hsp90’s function, and it is essential to understand the implications of dimerization on the molecular mechanism of Hsp90. CTD dimerization is required for hydrolysis mainly as a way of facilitating the additional dimerization of the NTDs, and dimerization at the NTD is required to stabilize the catalytically active conformation of the NTD and MD. Hsp90 missing only the CTD retains WT ATPase activity as long as the two monomer are connected by a cross-linker (Wegele et al., 2003) demonstrating that the NTD and MD are sufficient for catalysis. Cross-linked NTDs lacking the MD exhibit 100 fold lower ATPase activity than WT (Wegele et al., 2003), supporting a catalytic role for the MD. Residues in the MD near the interface with the NTD (the catalytic loop, Hsp82 residues 375–388) are essential for the hydrolysis of ATP (Meyer et al., 2003), and NTD dimerization serves to orient the catalytic loop so that hydrolysis can occur. Catalysis at both NTDs is not required for full activity as a heterodimer containing one WT NTD and one NTD that is unable to bind ATP is fully active (Richter et al., 2001) supporting the hypothesis that NTD dimerization establishes the catalytically active NM conformation.
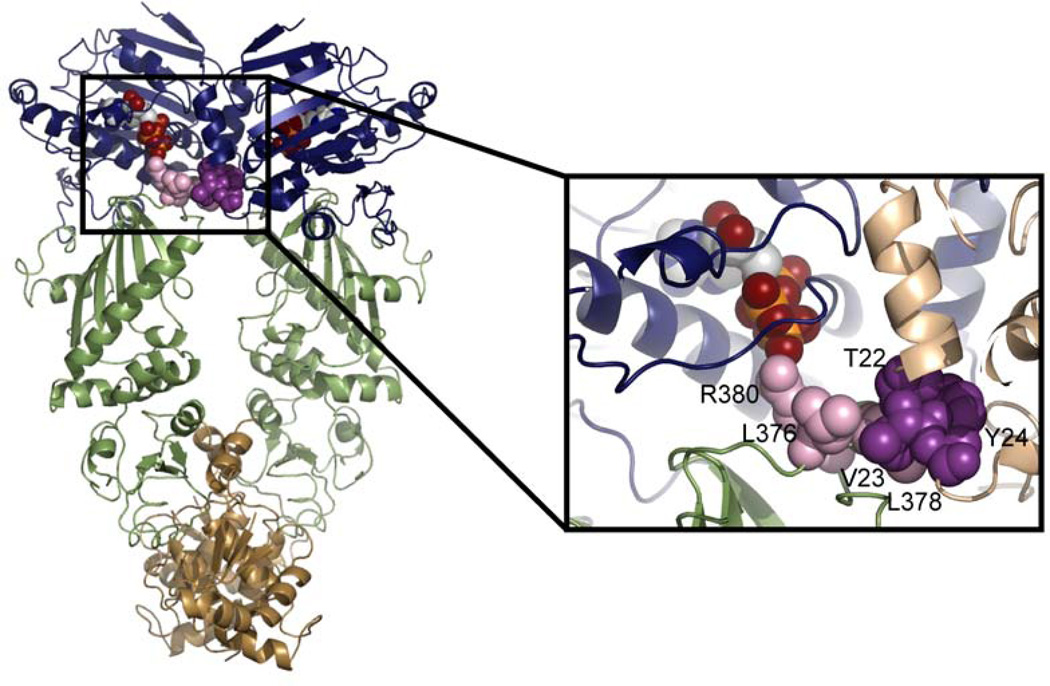
Recent evidence confirms that interactions between the two monomers are required for attaining the catalytic conformation and this conformation may be distinct from the conformation seen in the yeast AMPPNP-bound crystal structure. An interaction network between the NTD of one monomer and the MD catalytic loop of the other monomer has been described (Figure 6) (Cunningham et al., 2008). By examining the ATPase activity of a series of yHsp90 heterodimers, it was shown that the loop containing T22, V23, and Y24 (Hsp82 numbering) forms a hydrophobic network with residues L376, L378, and R380 (part of the MD catalytic loop) of the opposing monomer. Disruption of this network through mutagenesis leads to a loss in ATPase activity. R380, has previously been shown to be essential for the ATPase activity of yHsp90 (Meyer et al., 2003). In the AMPPNP-bound structure of yHsp90, R380 is the only residue contacting the γ-phosphate, and the hydrophobic network may serve to stabilize this interaction. MD simulations confirmed the importance of these hydrophobic contacts by showing the stabilization of the interaction network in the presence of ATP(Morra et al., 2009). The involvement of the middle domain in the binding of nucleotide is further supported by HX-MS data showing increased protection of the MD catalytic loop upon the addition of ATP (Graf et al., 2009). This protection could be accounted for by residues 22–24 packing against the arginine containing loop in the opposite monomer (Figure 6).
Figure 6.
A hydrophobic interaction network is formed between the MD catalytic loop and the NTD of opposing monomers. The NTD is shown in blue, the MD in green and the CTD in brown. ATP and the residues involved in the interaction network are shown as spheres with residues from the NTD shown in dark purple and residues from the MD shown in lavender. In the expanded view, the second monomer is shown in beige.
Both the biochemical evidence and the simulations suggest that the inter-protamer hydrophobic network serves to stabilize the hydrolysis competent state and that this conformation is different then the one seen in the yeast AMPPNP-bound crystal structure. A mutation on the examined NTD loop, T22F, causes an increase in the ATPase activity of yHsp90 but a phenylalanine at this position would cause steric clashes within the yeast AMPPNP-bound crystal state (Cunningham et al., 2008). An alternate conformation observed in the simulations optimally accommodates the T22F mutation (Morra et al., 2009). The increased HX-MS protection of the MD catalytic loop in the presence of ATP also supports the idea that the catalytic conformation is not the one observed in the yHsp90 AMPPNP-bound crystal structure.
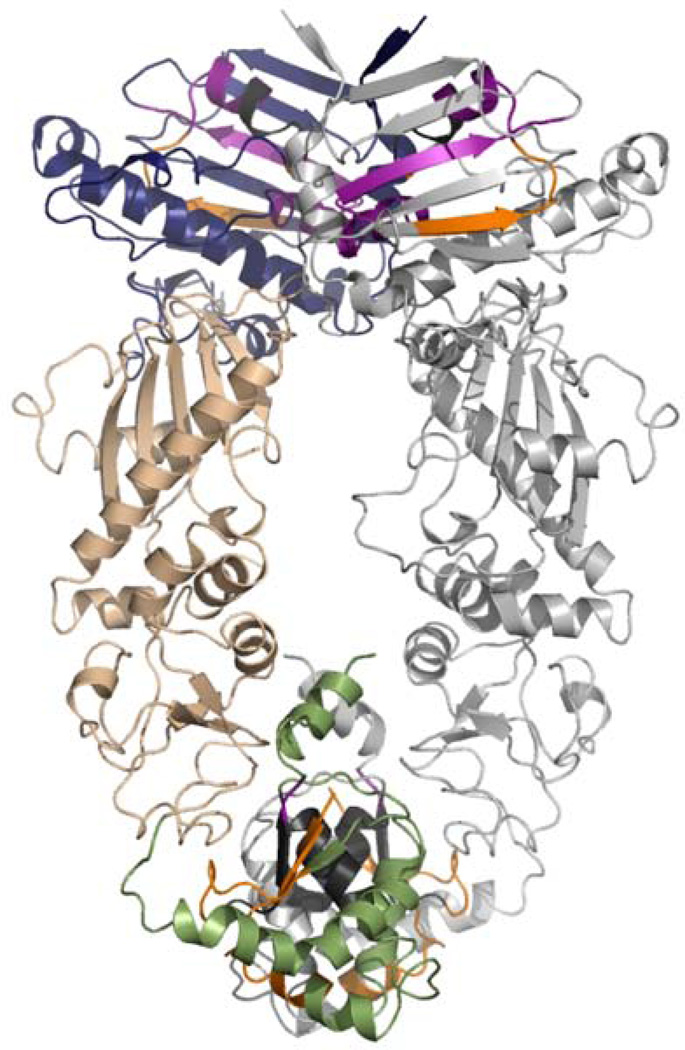
Interaction networks in Hsp90 must also extend beyond the NTD and MD. CTD dimerization facilitates NTD dimerization through large conformational changes at the MC interface that convert Hsp90 from the open state to the closed catalytic state. The MD rotates by 80° between the apo HtpG crystal state and the yeast AMPPNP-bound crystal state. For NTD dimerization to occur nucleotide binding must propagate a signal over the entire length of the protomer. In support of this hypothesis, MD simulations of full-length Hsp90 found an increase in communication efficiency between residues in the NTD and CTD upon the addition of nucleotide (Morra et al., 2009). Interestingly, while both ATP- and ADP-bound Hsp90 show residues communicating over a distance of 80Å, each nucleotide activates a different set of communicating residues between the NTD and CTD as shown in Figure 7. These different pathways may determine the different conformations of Hsp90 in the presence of ATP or ADP, and further mutational studies could be used to elucidate each networks role in establishing Hsp90’s conformation.
Figure 7.
Molecular dynamics simulations have shown different long range communication networks between the NTD and CTD in the presence of ATP or ADP. Residues that participate in long-range communication are shown on the yeast AMPPNP-bound crystal structure. The NTD is shown in blue, the MD in beige, and the CTD in green with the second monomer shown in grey. Long-range interactions observed only in the presence of ATP are shown in purple. Interactions seen only in the presence of ADP are shown in orange. Interactions observed with both ATP and ADP are shown in dark grey.
Hsp90 exists in a dynamic equilibrium between conformational states
As discussed, the structures of Hsp90 suggest a deterministic mechanism for the role of ATP in the conformational cycle (Figure 5) much like has been demonstrated for other chaperones. The crystal structures, however, present only static representations of the conformation of Hsp90 under different conditions. Additional experiments that examine the structure of Hsp90 in solution or that capture the dynamics of the protein show that unlike many other ATPases, Hsp90 is stochastic in nature. Instead of nucleotide binding/hydrolysis determining the conformation, Hsp90 exists in a dynamic equilibrium between different conformational states, and nucleotide binding/hydrolysis shifts the equilibrium between the already existing conformations.
MD simulations of the NTD showed that the lid spontaneously converts from the nucleotide free conformation to the nucleotide bound conformation (Colombo et al., 2008) suggesting that a continuum of conformations may exist and nucleotide serves to stabilize one conformation over another. Kinetic studies provided the first biochemical evidence that Hsp90 exists as a mixture of states in the presence of nucleotide and the relative amounts of different conformations is homolog specific. Analysis of the yHsp90 ATPase cycle shows that upon ATP binding approximately 80% of the molecules shift to a presumably closed state (Weikl et al., 2000). Negative-stain EM and SAXS provide structural evidence that bound to AMPPNP, yHsp90 is largely in the closed conformation seen in the yeast AMPPNP-bound crystal structure but a small population of the open conformation remains even with saturating nucleotide (Southworth & Agard, 2008). The same studies demonstrate that AMPPNP-bound hHsp90 remains largely in the open state and the closed state can only be observed in the presence of low levels of cross-linker. Kinetic studies suggest that the mitochondrial Trap1 is also predominantly (70%) in a closed state in the presence of ATP (Leskovar et al., 2008), whereas Grp94 remains largely in an open state (97%) in the presence of nucleotide (Frey et al., 2007). SAXS data for Grp94 confirms that it is mostly in an open, extended state in the presence of nucleotide (Krukenberg et al, in press)‥ Interestingly, bacterial HtpG has tuned the nucleotide response so that maximal levels of both the open and closed conformations are present even with saturating amounts of nucleotide (Krukenberg et al., 2008; Southworth & Agard, 2008).
Homolog specific differences seen in the conformational equilibria may shed light upon the differences in ATPase activity that have been reported. At 37°C yHsp90 has the highest ATPase activity, HtpG has a moderate level of activity, and hHsp90 has the lowest activity (C. Cunningham, personal communication). The relative ATPase rates correlate well with the proportion of the closed state observed in solution. For yHsp90, the conformational equilibria in the presence of nucleotide can be shifted completely to the closed state by deleting the first eight amino acids (Δ8-Hsp90) (Mickler et al., 2009). The ATPase activity of this mutant is doubled in comparison to WT further supporting the hypothesis that differences in ATPase rates relate to the extent of the closed state present in solution.
Due to these results, the emerging picture of Hsp90’s conformational cycle must include a dynamic equilibrium between the open and closed states in the presence of nucleotide. Recent experiments indicate that the cycle is even more complex. As mentioned, FRET studies of yHsp90 describe two intermediate states between the previously described open and closed states, and based on calculations of the energy barriers between the states, all states are also accessible in the absence of nucleotide. The role of nucleotide is not to determine conformation but to lower the energy barriers between the states (Hessling et al., 2009; Mickler et al., 2009). Cryo-EM and SAXS data for the pig Hsp90 protein (Bron et al., 2008) reveal two open conformations for the apo protein. One conformation results in a structure with the same opening angle as the HtpG apo crystal structure but with the NTDs in a more extended conformation forming a V-like structure. In the second conformation the NTDs rotate away from one another giving a structure described as a ‘flying seagull’ because of the resulting bend in the NM domains of the protein. HtpG also has at least two apo conformations in solution. SAXS studies identified an extended conformation shown in Figure 3 (Krukenberg et al., 2008) and a conformation almost identical to the Grp94 crystal structure (Krukenberg et al, in press). Though the exact conformations for the apo states may not be conserved, the dynamic equilibria between apo conformations or nucleotide-bound conformations are conserved from bacteria to humans. In vivo, equilibria between nucleotide-free and nucleotide-bound states must also be taken into account because of the relatively weak affinity of ATP for Hsp90.
Interestingly, all of the observed states appear to be roughly iso-energetic further highlighting the stochastic nature of the chaperone cycle. This is important in terms of understanding where the energy derives from for activating client proteins. Currently, this remains an open question that requires further exploration. It will also be important to understand how co-chaperones and other means of regulation affect the conformational dynamics especially in the context of client activation. The role of co-chaperones in the chaperone cycle is discussed in the next section.
The role of co-chaperones in regulating the conformational cycle of Hsp90
While nucleotide provides one means for influencing the conformation of Hsp90, protein binding partners of Hsp90 also influence its conformational dynamics. yHsp90 and hHsp90 are found in large heterocomplexes with other proteins known as co-chaperones. Co-chaperones are essential for the correct function and regulation of Hsp90 in vivo, and an ever growing number have been discovered (Table 2). Co-chaperones also provide a potential method for the recognition of different client proteins by Hsp90. A minimal set of necessary co-chaperones has been established for the progesterone receptor (PR), the glucocorticoid receptor (GR), and the kinase Chk1 providing information about the role of different co-chaperones and about the specific requirements among client proteins. In vitro, GR requires the fewest number of co-chaperones (only Hsp70 and Hop) in addition to Hsp90 for the reconstitution of steroid binding activity (Arlander et al., 2006; Dittmar & Pratt, 1997; Kosano et al., 1998). For the in vitro reconstitution of PR, Hsp70, Hsp40, Hop and p23 are required in addition to Hsp90 (Kosano et al., 1998). The kinase Chk1 also requires Hsp90, Hsp70, Hsp40, and Hop for maximal activation, but unlike PR the kinase specific co-chaperone p50 is also required and p23 is not (Arlander et al., 2006). These differences in co-chaperone requirements may provide a mechanism for differentiating between client proteins.
Table 2.
Partial list of Hsp90 associated co-chaperones
| Protein | Function |
|---|---|
| Aha1 | stimulates ATPase activity* |
| p50 | blocks ATPase activity and is thought to be kinase specific* |
| Cpr6 | TPR-containing and peptidy-prolyl-isomerase (Johnson et al., 2007) |
| FKBP52 | TPR-containing and peptidy-prolyl-isomerase (Riggs et al., 2003) |
| GCUNC-45 | TPR-containing that blocks progression of cycle (Chadli et al., 2006) |
| Hop | TPR-containing and scaffolds Hsp70-Hsp90 interaction* |
| Hsp70 | recruits substrates to Hsp90* |
| NudC | CHORD-domain containing (Te et al., 2007) |
| p23 | blocks ATPase activity and traps client proteins in complex with Hsp90* |
| PP5 | TPR-containing and phosphatase* |
| Sse1 | part of Hsp90 complex in budding yeast (Liu et al., 1999) |
| Tah1 | TPR-containing (Zhao et al., 2005) |
For a list of additional co-chaperones see http://www.picard.ch/downloads/Hsp90interactors.pdf
References can be found in the text.
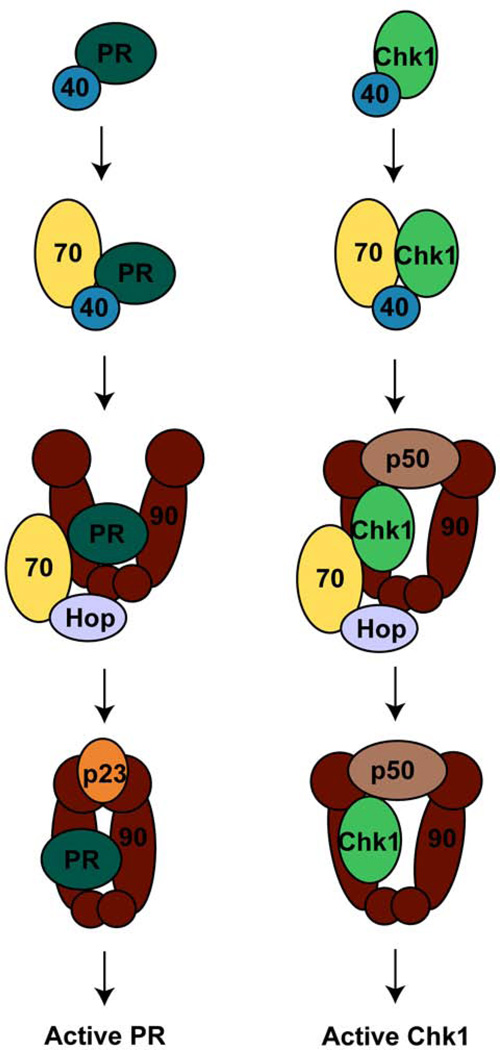
The reconstituted systems also reveal the formation of specific heterocomplexes at different stages throughout the chaperone cycle (Figure 8). Both Chk1 and PR bind Hsp40 to begin the cycle. Hsp40 brings the client into complex with Hsp70. The initial Hsp90 complex then forms with Hop mediating the interaction between Hsp70 and Hsp90 allowing the client protein to be transferred to Hsp90. In the case of Chk1, this initial complex also includes p50. Hsp70 dissociates from the intermediate complexes and the late-stage complexes form. For Chk1, this complex includes the kinase, Hsp90, and p50. The steroid hormone receptors are in complex with Hsp90 and p23 at this stage. Activated client proteins are then released from the Hsp90 complex (Felts et al., 2007; Hernandez et al., 2002; Kosano et al., 1998; Smith, 1993). The diagrams in Figure 8 detail the minimal systems required for activation, but numerous other co-chaperones have been identified and are believed to play important roles in the regulation of the chaperone cycle in vivo (Table 2). For example, the immunophilin FKBP52 is found in complex with Hsp90, GR/PR, and p23 in vivo; FKBP52 also increases the hormone binding ability of GR and is essential for the correct activation of PR in vivo (Chadli et al., 2008; Riggs et al., 2003).While some co-chaperones such as Hsp70 and Hsp40 are required for client recruitment, other co-chaperones regulate both the ATPase activity and the conformational dynamics of Hsp90.
Figure 8.
Proposed chaperone cycles for the activation of the progesterone receptor and the Chk1 kinase. The progesterone receptor (PR) or Chk1 are first bound by Hsp40 followed by recruitment to Hsp70. Next, a complex with Hsp90, Hop, and Hsp70 forms. This intermediate complex also includes the kinase-specific p50 in the case of Chk1. The mature complex is then formed when Hsp70 and Hop dissociate. The mature complex for PR requires the binding of the additional factor, p23.
p23 arrests the ATPase cycle of Hsp90
Since it was first identified in complex with Hsp90 and unactivated PR (Smith et al., 1990), p23 has been shown to bind Hsp90 directly and this binding is greatly enhanced by the presence of ATP (Grenert et al., 1999; Johnson & Toft, 1995; Sullivan et al., 1997). ADP inhibits complex formation (Johnson & Toft, 1995; Sullivan et al., 1997) suggesting that p23 stabilizes the ATP bound form of Hsp90 and hydrolysis of ATP subsequently releases p23 from the complex.
p23 binds directly to the NTD of Hsp90 although contacts are also made with Hsp90’s MD (Ali et al., 2006; Martinez-Yamout et al., 2006). The complex promotes or stabilizes the closed conformation and leads to the inhibition of the ATPase activity with inhibition observed for both yHsp90 and hHsp90 (McLaughlin et al., 2002; McLaughlin et al., 2006; Richter et al., 2004; Siligardi et al., 2004). Biochemical data demonstrate that p23 preferentially binds to the NTD dimerized state (McLaughlin et al., 2006; Prodromou et al., 2000; Richter et al., 2004), and a crystal structure of the full-length yeast protein bound to residues 1–134 of p23 (the complete protein is 160 residues). confirms that p23 stabilizes the closed AMPPNP-bound form of Hsp90 (Figure 9) (Ali et al., 2006)‥ The affinity of Hsp90 for p23 can be tuned with Hsp90 mutants that either increase or decrease NTD dimerization. The A107N and Δ8-Hsp90 mutants that show increased amounts of the closed state have increased affinity for p23 (Richter et al., 2004; Siligardi et al., 2004), wherase T101I and F349A that decrease the ability of Hsp90’s NTDs to dimerize interact more weakly with p23 (Siligardi et al., 2004). As expected, truncation of the NTD domain so that dimerization no longer occurs abolishes the binding of p23. These mutants could be used to better understand the exact role of p23 in Hsp90 mediated client protein activation by tuning the pathway’s dependence on p23.
Figure 9.
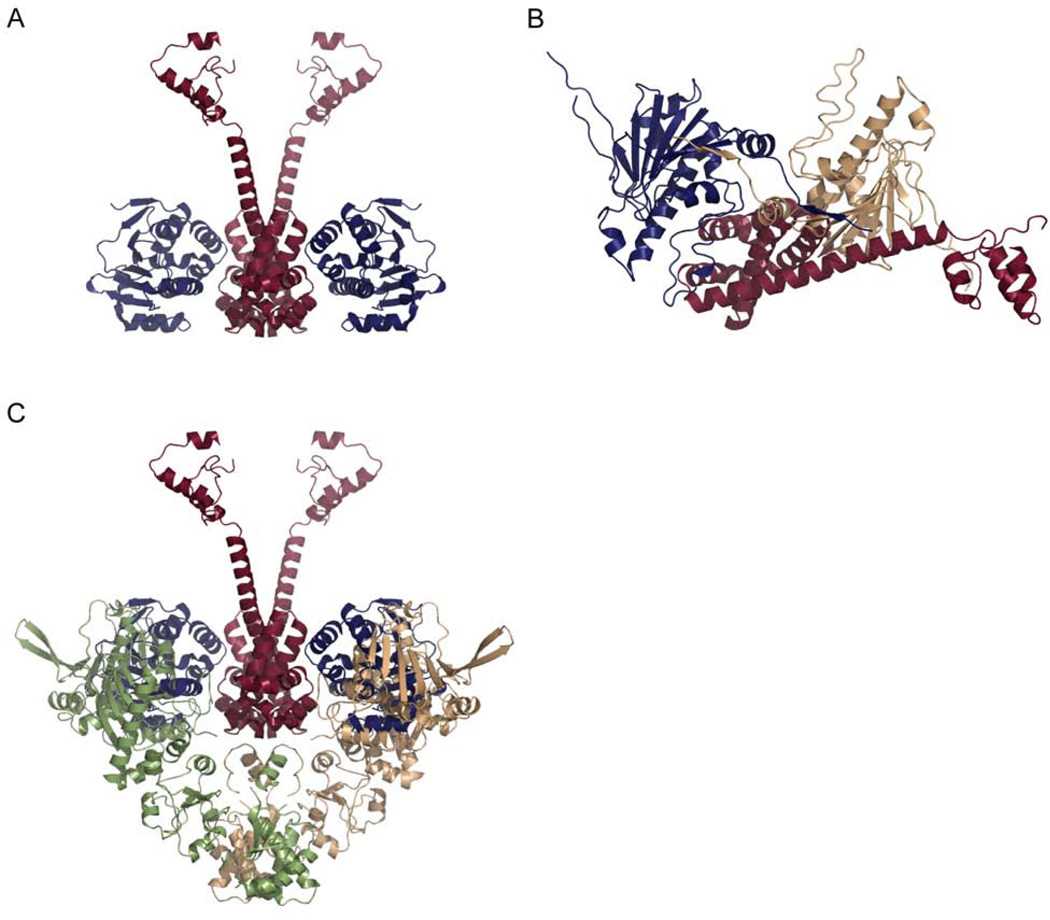
Crystal structure of the p23:Hsp90 complex. A) Residues 1–134 of yeast p23 were crystallized in complex with the full-length yeast Hsp90 (PDB code, 2CG9). The two monomers of Hsp90 are shown in brown and green while p23 is shown in red. B) An expanded view of the ATP binding pocket shows that the active site lid (shown in light blue) folds over the bound nucleotide when p23 is present. This conformation blocks access in and out of the ATP binding pocket.
The data evoke a model where p23 traps Hsp90 in an N-terminally dimerized state with a functional consequence of inhibiting the ATPase activity of Hsp90. As determined by kinetic studies, p23 uses a mixed inhibition mechanism, and the binding of p23 increases the Km for AMPPNP binding to Hsp90. If the p23:Hsp90 complex cannot bind AMPPNP once it is formed, the increase in Hsp90’s affinity for AMPPNP in the presence of p23 would result from the relatively higher affinity of p23 for the AMPPNP-bound Hsp90 rather than Hsp90 alone (McLaughlin et al., 2006). The motion of the lid over the nucleotide binding pocket when p23 binds (Figure 9) blocks access to the nucleotide binding pocket supporting this model. Though the exact mechanism of inhibition remains unclear, one hypothesis is that p23 traps Hsp90 in a pre-hydrolysis state. The crystal structure is unable to provide any insight into the mechanism of inhibition, but as suggested by the biochemical and computational evidence discussed above, the crystal structure may represent a catalytically inactive state and p23 may therefore prevent the necessary NM domain rearrangements required for hydrolysis.
Promoting the closed state of Hsp90 and inhibiting the ATPase activity has the functional consequence of stabilizing client proteins in a complex with Hsp90 and p23. This has been observed for both GR and PR (Dittmar & Pratt, 1997; Kosano et al., 1998). p23 may serve as a molecular timer for the activation of client proteins, and once clients are activated p23 is released, hydrolysis occurs, and client proteins dissociate. Either the slow hydrolysis of ATP in the presence of p23 or the dynamic dissociation of p23 allowing hydrolysis to occur could lead to dissociation of the p23:Hsp90 complex. There is evidence for both of these models suggesting that the actual mechanism may be a combination of both. In yeast, Δ8-Hsp90 (increased ATPase activity) hydrolyzes ATP even at saturating levels of bound p23 (Richter et al., 2004). Complete inhibition of hHsp90’s ATPase was observed in the presence of p23 but the binding of other co-chaperones may displace p23 and overcome inhibition (McLaughlin et al., 2006). In the absence of other co-chaperones stochastic fluctuations in the p23:Hsp90 complex, as observed in lysates (Johnson & Toft, 1995), could allow hydrolysis to occur and the chaperone cycle to continue‥ Also, the half-life (T1/2) of the human p23:Hsp90 complex is 40 seconds at 37°C whereas the T1/2 of hydrolysis is 7 minutes (McLaughlin et al., 2006) providing additional corroboration for the dynamic nature of the p23:Hsp90 complex.
Hop traps Hsp90 early in the conformational cycle
Unlike p23, the co-chaperone Hop functions early in the chaperone cycle. Hop is a TPR domain containing protein that binds as a dimer to the C-terminal MEEVD of Hsp90. Through this interaction, Hop facilitates the transfer of substrate from Hsp70 to Hsp90 (Wegele et al., 2006). Though the main interaction between Hop and Hsp90 is mediated by the C-terminus of Hsp90 other regions of Hsp90 are involved in the interaction. The affinity of Hop for Hsp90 decreases with increasing truncations of Hsp90’s N-terminus (Richter et al., 2003) suggesting that Hop’s interaction with Hsp90 spans the entire monomer. On the contrary, a recent biophysical study of Hop indicated that the MD and CTD of Hsp90 is sufficient to recapitulate WT Hop binding (Onuoha et al., 2008). Further structural studies will be required to reconcile this apparently conflicting evidence.
Hop binds Hsp90 in both the absence and presence of nucleotide although a small decrease in binding in the presence of ATP has been reported (Grenert et al., 1999). Like p23, Hop inhibits the ATPase activity of Hsp90, but unlike p23 this inhibition is strictly noncompetitive (Onuoha et al., 2008; Prodromou et al., 1999; Richter et al., 2003; Siligardi et al., 2004). Whereas p23 stabilizes Hsp90 in the closed conformation, Hop stabilizes Hsp90 in the open state and prevents conversion to the closed state (Johnson et al., 1998). Hop and p23 act at different steps in the chaperone cycle by recognizing/stabilizing different conformations of Hsp90; once Hsp90 has significantly populated the closed state the affinity for p23 is much higher than the affinity for Hop allowing the cycle to progress. As with p23, the binding affinity of Hop for Hsp90 can be modulated with Hsp90 mutants. As expected for a model where Hop binds the open state, mutants that promote the closed state show decreased affinity for Hop. Δ8-Hs90 and A107N, which have increased NTD dimerization, bind Hop less tightly (Onuoha et al., 2008; Richter et al., 2003) The inhibition of Hop can be reversed by the addition of other co-chaperones that presumable disrupt the binding of Hop to Hsp90. Both the co-chaperones Cpr6 and PP5, two TPR-containing co-chaperones that compete with Hop for binding to Hsp90, have been shown to reverse the inhibition of Hop providing a mechanism for the progression of the chaperone cycle (Prodromou et al., 1999).
All of this data points to a mechanism of inhibition where Hop binds Hsp90 in the open state and blocks the conversion to the closed state. A recent FRET study confirms thatin the presence of ATP and Hop, the conformational transition for Hsp90 from the open to the closed state no longer occurs (Hessling et al., 2009). While Hop undoubtedly stabilizes an open conformation of Hsp90, it is also likely that Hop binding in and of itself causes a conformational change in Hsp90 increasing the inhibitory potential of Hop. A SAXS study of the Hop:Hsp90 complex suggests a conformational change upon complex formation. The radius of gyrations (Rg) for Hsp90 and Hop alone are 62.2 and 55.2 Å respectively with corresponding maximum interatomic distances (Dmax) of 207 and 193 Å. The Hop:Hsp90 complex has an Rg of 61.5 Å and a Dmax of 204 Å (Onuoha et al., 2008). This decrease in the overall size of the complex compared to Hsp90 alone is indicative of a conformational change.
p50, a kinase specific co-chaperone, also traps Hsp90 in an open conformation
The co-chaperone p50 also inhibits the ATPase activity and affects the conformational dynamics of Hsp90, but its mechanism differs from both Hop and p23. p50 is a kinase specific co-chaperone and is thought to assist in the transfer of kinases to Hsp90 (Grammatikakis et al., 1999; Lee et al., 2002; Stepanova et al., 1996). Domain mapping has shown that p50’s N-terminal domain is critical for binding kinases whereas the middle and C-terminal domains interact with Hsp90 (Grammatikakis et al., 1999; Roe et al., 2004; Shao et al., 2003). Biochemical studies show that p50 binds to Hsp90 in a 1:1 ratio withthe interaction occuring largely at the NTD of Hsp90. The interaction also requires the charged loop and deletion of the CTD leads to a loss in affinity (Roe et al., 2004; Siligardi et al., 2002; Zhang et al., 2004). The crystal structure of the C-terminal region of p50 (residues 148–348) bound to the NTD of Hsp90 shows a dimer for p50 but in this configuration the dimer interface is small and may arise solely from crystal contacts (Roe et al., 2004). Biochemical studies suggest that p50 may initially bind Hsp90 as a monomer because the KD for dimerization (80 uM) is less than the KD for complex formation (4 uM) (Zhang et al., 2004). If this is the case, the increased local concentration of one p50 monomer bound to each Hsp90 NTD may lead to the dimerization of p50 even though the KD for p50 dimerization is relatively weak.
The crystal structure also indicates a mechanism for the inhibition of Hsp90’s ATPase activity. R167 of p50 points into the nucleotide binding pocket of Hsp90 and forms a hydrogen bond with the catalytic glutamate of Hsp90 (E33). Nucleotide still binds, but the catalytic water is no longer coordinated for hydrolysis (Roe et al., 2004). The crystal structure also reveals another potential means of inhibition where p50 blocks the conversion of Hsp90 from the open to the closed state. When the p50:Hsp90 dimer is docked onto one NTD of the yHsp90 AMPPNP-bound crystal structure, p50 sterically clashes with the second Hsp90 NTD (Figure 10B). Also, in the crystal, p50 forms a dimer between the two NTDs of Hsp90 potentially blocking Hsp90’s transition to the closed state (Figure 10A). When docked onto the full-length apo crystal form of HtpG, the p50 dimer fits perfectly into the cleft of the Hsp90 dimer potentially stabilizing Hsp90 in an open conformation (Figure 10C). p50 may therefore block the formation of the closed state, and be unable to bind once the closed state has formed. Biochemical studies using the Hsp90 mutants T22I and T101I confirm the preference of p50 for the open conformation of Hsp90 (Millson et al., 2004).
Figure 10.
Crystal structure of p50 bound to Hsp90. A) In the crystals the C-terminal region of p50 (red) and the NTD of Hsp90 (blue) form a heterotetramer with p50 forming a dimer between the two NTD domains of Hsp90. B) p50 binding is incompatible with the dimerized NTDs of the Hsp90 closed state. The two NTDs of Hsp90 are shown in blue and beige and they are shown in the configuration seen in the yeast AMPPNP-bound crystal state. Docking of the p50:Hsp90 crystal dimer onto one NTD of the closed state reveals steric clashes between p50 (red) and the second Hsp90 NTD (beige). C) The p50:Hsp90 heterotetramer from A is aligned with the two NTDs from the apo bacterial crystal structure suggesting a mechanism for blocking the closure of Hsp90. One monomer of the apo structure is shown in green and the other in beige. For Cdc37 to bind to this structure, the NTDs of the apo state would have to move as seen by the blue Hsp90 NTDs.
Interestingly, for p50 to bind to the full-length apo conformation a structural rearrangement must occur between the NTD and MD of Hsp90 (Figure 10C). Solution studies of the p50:Hsp90 complex also indicate that complex formation causes a conformational change in Hsp90. As measured by SAXS, both the Rg and Dmax of hHsp90 decrease when p50 binds. The Rg and Dmax shift from 64.6 and 219 Å respectively for Hsp90 alone to 62.7 and 200 Å in the presence of p50 (Zhang et al., 2004). HX-MS also shows changes in the protection of hHsp90 in the presence of p50 consistent with a conformational change. Two peptides (77–85 and 221–235, Hsp90β numbering) in the NTD of hHsp90 that were not perturbed in the p50:Hsp90 crystal structure show increased protection in the presence of p50. Increased protection in a middle domain peptide adjacent to the NTD also occurs when p50 binds. The stabilization pattern observed is consistent with an increased interaction between the NTD and MD in the presence of p50 but this interaction is incompatible with the NM domain orientation in the apo crystal structure (Phillips et al., 2007) implying that p50 binding causes a rearrangement of the Hsp90 NM domain.
Aha1 activates the ATPase of Hsp90
Unlike the other co-chaperones discussed, Aha1 stimulates the ATPase activity of Hsp90 (Lotz et al., 2003; Meyer et al., 2004; Panaretou et al., 2002). Aha1 binds mainly to the MD domain of Hsp90 and this interaction is mediated by the N-terminal portion of Aha1 (N-Aha1) (Lotz et al., 2003; Meyer et al., 2004; Siligardi et al., 2004). N-Aha1 has been crystallized in complex with the MD of Hsp90 (Meyer et al., 2004) providing the precise positioning of Aha1 on Hsp90. When aligned with either the closed crystal structure or the open crystal structure, Aha1 binds to Hsp90 at the edge of the dimer cleft approximately 90° from the dimer interface (Figure 11). Studies using gel filtration and NMR show that Aha1 competes with both Hop and p23 for binding to Hsp90 (Harst et al., 2005; Martinez-Yamout et al., 2006), indicating that Aha1 acts at multiple stages of the chaperone cycle to disrupt inhibition by other co-chaperones. Aha1 may displace Hop from early complexes and p23 from mature complexes stimulating the ATPase activity and allowing for the continuation of the chaperone cycle.
Figure 11.
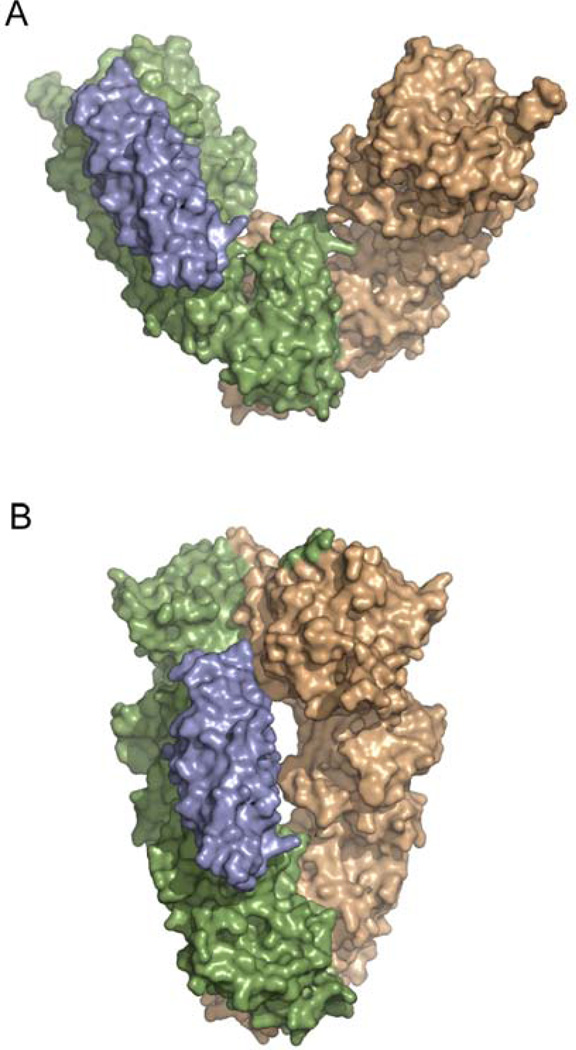
Aha1 binds Hsp90 on the edge of the cleft. By docking the N-Aha1:MD-Hsp90 complex on the full length apo (A) and ATP (B) crystal structures, the approximate orientation of Aha1 can be determined. Aha1 is colored in grey while one monomer of Hsp90 is green and the other monomer is beige.
Aha1 stimulates the ATPase activity of Hsp90 by promoting or stabilizing a conformation of Hsp90’s NM domain that optimally positions the MD catalytic loop for catalysis (Figure 12). In the full-length apo crystal structure, the catalytic loop is positioned so that R380, part of the hydrophobic interaction network discussed above, makes contacts with the rest of the MD and is removed from contacts with the NTD (Figure 12B). In the closed AMPPNP-bound crystal state, the loop has less α-helical structure causing R380 to orient towards the NTD where it contacts the γ-phosphate of ATP (Figure 12C and Figure 6). Even though in the Aha1:Hsp90 complex parts of the catalytic loop are disordered, the orientation is much more consistent with the loop conformation found in the closed state (Figure 12). Interestingly, the R380A mutant, which has a decreased ATPase rate, is not activated by Aha1 (Meyer et al., 2003) implying that R380 is critical in the Aha1 mediated activation of Hsp90. The importance of the catalytic loop in Aha1 mediated activation is further highlighted by the insensitivity of the Hsp90 E381K (located on the catalytic loop) mutant to activation, even though the mutation alone has very little affect on the ATPase activity (Meyer et al., 2003). All of the evidence suggests that Aha1 promotes or stabilizes a conformation of Hsp90 that occurs after NTD dimerization and has increased interactions between the MD and NTD. Data showing that the Hsp90 F349A mutant, located at the NM interface and potentially involved in interdomain communication, is hypersensitive to Aha1 also supports this conclusion (Meyer et al., 2003; Siligardi et al., 2004).
Figure 12.
Aha1 binding to Hsp90 causes a rearrangement in the MD catalytic loop. A) N-Aha1 (grey) was crystallized in complex with the MD of Hsp90 (green). The catalytic loop, shown in red, is partially disordered in the crystal structure. B) The catalytic loop in the apo full length structure (beige) has more α-helical structure than in the Aha1:Hsp90 complex, and the catalytically required R380 (shown in sticks) forms contacts with the MD in the apo structure. C) In the closed AMPPNP-bound crystal structure the catalytic loop, shown in purple, orients R380 towards the NTD where it interacts with the γ-phosphate of ATP. The loop structure in the Aha1:Hsp90 complex is closely related to the loop structure in the closed state.
Though it is clear from these studies that NTD dimerization and rearrangement of the NM domain is important for Aha1’s effect on Hsp90, it is not clear if Aha1 simply stabilizes the catalytic conformation once it forms or if binding of Aha1 results in a conformational change in Hsp90 leading to catalysis. A recent FRET study suggests that Aha1 directly induces structural changes in yHsp90. Aha1 binding results in the rapid acceleration of conformational changes between the NTD and MD similar to the acceleration seen in the lidless Hsp90 mutant, which shows increased NTD dimerization. The FRET results also suggest that Aha1 binding allows Hsp90 to bypass the first conformational intermediate after nucleotide binding leading to an accumulation of the second NTD dimerized intermediate instead (Hessling et al., 2009). Given the dynamic nature of Hsp90, it most likely samples all of the states along the kinetic pathway and the binding of Aha1 serves to shift the equilibrium towards the catalytically active conformation thus increasing the ATPase activity of Hsp90.
Alternate methods for controlling the conformational cycle of Hsp90
As illustrated above, co-chaperones provide an elegant layer of regulation for the conformational cycle of Hsp90, and this regulation is essential for the proper function of cytosolic yeast and human Hsp90. Bacterial HtpG and the ER homolog Grp94, however, have no known co-chaperones. Given the importance of this additional level of regulation for the cytosolic yeast and human proteins, it is highly possible that HtpG and Grp94 have developed alternative means of regulation that extend beyond the effects of nucleotide binding. While this regulation has yet to be well defined for HtpG or Grp94, a few possible mechanisms have been suggested.
Recent work demonstrates a role for pH in controlling the conformation of HtpG (Krukenberg et al., in press). SAXS studies at varied pH reveal that apo HtpG exists in predominantly two conformational states, an extended state and a Grp94-like state, and the equilibrium between these two states is optimized so that maximal levels of both conformations are present at physiological pH. pH shifts in either direction drastically alter the relative populations of the two conformations. As discussed previously, these states appear to play different roles in the binding of client proteins. In the citrate synthase aggregation assay only the Grp94-like state is able to prevent the aggregation of citrate synthase (Krukenberg et al, in press). The use of pH to control the conformation of Hsp90 appears to be a uniquely bacterial trait. In vivo bacteria may use the pH equilibrium as a sensor to modulate HtpG’s conformation in times of metabolic stress. It is also possible that the finely tuned conformational equilibrium serves only to optimize the levels of both conformations in the absence of c-ochaperones, and each conformation binds a specific subset of client proteins.
An alternative mechanism for controlling conformation involving Ca2+ binding has been suggested for Grp94. Because the ER functions as a Ca2+ storage organelle, the role of Ca2+ in the function of Grp94 has been investigated, and studies defined Grp94 as a Ca2+ binding protein (Macer & Koch, 1988; Van et al., 1989). A surface plasmon resonance study suggests that Ca2+ binding alters the conformation of Grp94 (Ying & Flatmark, 2006), and a study investigating the effect of Ca2+ on peptide binding to Grp94 concluded that the divalent cation causes a conformational change in Grp94 promoting the binding of peptide (Biswas et al., 2007). As with pH and HtpG, Ca2+ binding may provide a means for Grp94 to access/stabilize alternative conformations that bind specific client proteins. Ca2+ could also be used to modulate Grp94 conformation in times of stress. More direct structural studies will be required to determine the exact nature and extent of these conformational changes.
The relationship between client protein binding and Hsp90 conformation
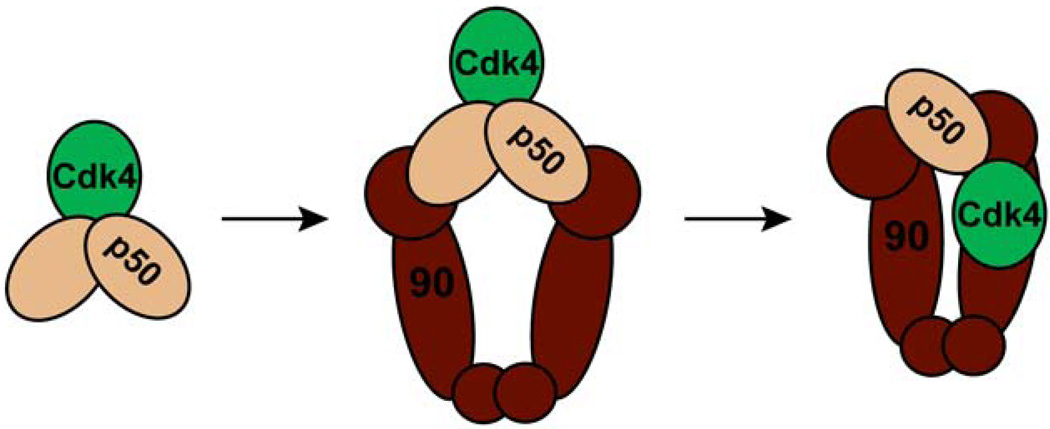
The conformational dynamics of Hsp90 are undoubtedly directly involved in the activation of client proteins. Changes in Hsp90 most likely confer structural changes upon the client protein itself. As another level of regulation, it is also possible that client proteins themselves affect the conformation of Hsp90. Client induced changes in Hsp90 may be one method that allows Hsp90 to differentiate between the wide-array of clients. Given the differences in how specific client proteins are activated, i.e. PR versus Chk1, the transfer of this information from the client to Hsp90 is essential. While the exact nature of the interaction between client proteins and Hsp90 remains largely a mystery there is emerging evidence to support this idea. Some of the first evidence for client proteins altering the conformation of Hsp90 came from studies investigating the stability of the p50:Hsp90 complex. Complexes formed by only Hsp90 and p50 are sensitive to high salt concentrations, but when a kinase is added, the complex becomes salt resistant (Hartson et al., 2000). Kinase binding may cause a structural rearrangement in Hsp90, p50 or both conferring salt resistance on the complex. The potential for client binding to change Hsp90’s conformation is not a trait unique to kinases. The ligand binding domain of GR stimulates the ATPase rate of hHsp90 (McLaughlin et al., 2002) suggesting that GR shifts the population of human Hsp90 towards the closed catalytic state. This correlates well with data suggesting that hHsp90 has such a low basal ATPase rate because it does not significantly populate the closed state. This conclusion is further supported by data showing that Hop inhibits the stimulated rate but has little effect on the basal rate (McLaughlin et al., 2002) and p23 much more dramatically inhibits hHsp90 in the presence of GR (McLaughlin et al., 2002) than in the absence. It will be of great interest to test other client proteins for their ability to stimulate the ATPase activity of Hsp90. If differences are seen between clients, this would support the hypothesis that ATPase stimulation is one method for Hsp90 recognition of specific client proteins. This most direct evidence for client binding influencing the conformation of Hsp90 comes from a 3D reconstruction of the p50:Hsp90:Cdk4 complex from negative-stain EM (Vaughan et al., 2006). The reconstruction shows an asymmetric complex with two Hsp90 monomers, one p50 monomer, and one Cdk4 kinase monomer. The Hsp90 dimer is in a conformation similar to the closed AMPPNP-bound state except the NTDs are not dimerized. One NTD hinges backwards away from the other and p50 binds between the two NTDs. Cdk4 interacts with the outer edge of the complex. The kinase contacts the Hsp90 MD with what appears to be the C-lobe, and the N-lobe contacts one Hsp90 NTD and p50. Previous studies, discussed above, indicate that p50 binds as a 1:1 complex with Hsp90 and this complex is locked in a conformation more closely related to the apo crystal structure. To reconcile these results, it has been suggested that the kinase initially binds a dimer of p50 and this complex is recruited to Hsp90 forming a symmetric complex. A structural rearrangement then occurs displacing one p50 monomer and leading to the asymmetric complex observed by EM (Figure13) (Vaughan et al., 2006). More studies will be required to test this hypothetical pathway and to determine the individual effects of p50 and Cdk4 on the conformation of Hsp90. While both p50 and Cdk4 may influence Hsp90’s conformation, most likely there is a synergistic effect on Hsp90’s dynamics when both are bound. It will also be important to understand how these conformational changes affect the structure of the kinase. The shift from a complex with two p50 monomers bound to one monomer may be the key step in the activation of the client. To test this hypothesis, further structural studies in combination with in vitro assays of kinase activation will be required.
Figure 13.
Proposed model for Hsp90:p50:Cdk4 complex formation. Initially the kinase Cdk4 binds to a dimer of p50. This complex is then recruited to a dimer of Hsp90 to form a symmetric complex. A conformational rearrangement the occurs and one monomer of p50 is displaced leading to the asymmetric complex observed by EM.
The effect of phosphorylation and acetylation on the conformational dynamics of Hsp90
Apart from co-chaperones and client proteins, post-translational modifications may provide additional levels of regulation for the dynamics of Hsp90. While it is clear that acetylation and phosphorylation are important aspects of Hsp90’s in vivo function, the precise role of these modifications in the chaperone cycle is not well understood. Hyperactylation of Hsp90 disrupts the binding of both client proteins and co-chaperones (Kekatpure et al., 2009; Kovacs et al., 2005; Scroggins et al., 2007; Yu et al., 2002). Regulation of Hsp90’s acetylation status occurs primarily through HDAC6 which binds directly to Hsp90 (Kekatpure et al., 2009), and inhibition of HDAC6 leads to the hyperacetylation of Hsp90 (Yu et al., 2002). Hsp90 contains a least two acetylation sites and one site as been mapped to K294 (Hsp82 numbering) at the junction of the NTD and MD (Scroggins et al., 2007). Given the location of this site, acetylation may lead to conformational changes in the NM domain disrupting the binding of clients and co-chaperones alike. Structural analysis of the acetylated protein will be required in order to investigate this possibility.
Phosphorylation also plays an important role in the function of Hsp90 and may be involved in conformational dynamics. As with acetylation, increased phosphorylation negatively regulates Hsp90’s interaction with client proteins (Adinolfi et al., 2003; Ogiso et al., 2004; Wandinger et al., 2006). The phosphatase PP5 directly associates with Hsp90 via a TPR-domain, and PP5 directly dephosphorylates Hsp90 (Wandinger et al., 2006). Evidence suggests that phosphorylation may be more complex with differing roles on Hsp90 function depending on the client protein involved. Phosphorylation at S225 and S254 in Hsp90 causes a decreased affinity for the aryl hydrocarbon receptor (AhR) (Ogiso et al., 2004). In another example, c-src directly phosphorylates Hsp90 on Y300 and unlike other clients this phosphorylation event is required for the binding of eNOS to Hsp90 (Duval et al., 2007). Further studies will be required to determine the effect of phosphorylation on the conformation of Hsp90.
Small molecules also shift the conformation of Hsp90
As discussed, the conformational dynamics of Hsp90 are essential for proper function. Also, multiple layers of regulation affect the conformation suggesting that small molecule inhibitors of Hsp90’s conformational dynamics may be potent inhibitors of chaperone function providing important therapeutics for Hsp90 related diseases. Currently, the majority of small molecule Hsp90 inhibitors compete for binding with ATP and were not designed to influence the conformation of Hsp90. There are, however, indications that some of these inhibitors function not only by blocking the binding of nucleotide but also by shifting the conformation of Hsp90 to a state that is incapable of client protein binding.
As would be expected for small molecules occupying the nucleotide binding pocket, inhibitor binding causes local changes in the lid of Hsp90 (Immormino et al., 2004; Stebbins et al., 1997). Binding of the inhibitor geldanamycin (GA) leads to a lid conformation of Hsp90 (Stebbins et al., 1997) similar to the conformation seen in the isolated apo hHsp90 NTD (Figure 3A, red). This conformation is unlike the lid state seen in the nucleotide bound complexes where hydrophobic surfaces are accessible for NTD dimerization. The local changes in the NTD induced by the inhibitor most likely correlate with changes in the global structure of the Hsp90 dimer. The GA-bound lid conformation suggests that GA and nucleotide have very different effects on the overall structure of Hsp90.
In vivo, the binding of GA to Hsp90 leads to the destabilization and degradation of client proteins (Mimnaugh et al., 1996; Schulte et al., 1995; Whitesell et al., 1994). In most cases the destabilization is due to the disruption of Hsp90:client complexes as observed for the kinases v-src and bcr-abl (An et al., 2000). GA also disrupts the binding of p23 (An et al., 2000; Sullivan et al., 1997) further suggesting that the inhibitor alters the normal conformation of Hsp90. The disruption of the p23:Hsp90 complex indicates that GA prefers to bind Hsp90 in an open state. This claim is further substantiated by evidence showing that preformed p23:Hsp90 complexes are less inhibitable than free Hsp90. GA also increases the presence of Hop and Hsp70 in pull-downs of Hsp90 (An et al., 2000; Sullivan et al., 1997). Though GA binds to an open conformation of Hsp90, this conformation is distinct from the conformation of apo Hsp90. As measured by SAXS, GA causes a decrease in the Rg of Hsp90 (Zhang et al., 2004). In HX-MS experiments, DMAG, a derivative of GA, and radicicol, an inhibitor similar to GA, cause changes in the protection of amide protons in all three domains of Hsp90 (Phillips et al., 2007). Radicicol also increases the protease sensitivity of Hsp90 as compared to the apo protein (Nishiya et al., 2009).
Other small molecule inhibitors that function via alternative mechanisms appear to also cause global conformational changes within Hsp90. Novobiocin is a small molecule inhibitor that binds to the CTD of Hsp90 (Marcu et al., 2000a; Marcu et al., 2000b; Soti et al., 2002). Like GA and radicicol, novobiocin inhibits the formation of Hsp90 complexes with client or p23, but unlike GA it also reduces the affinity of Hsp90 for TPR-containing co-chaperones that bind to the CTD (Allan et al., 2006; Yun et al., 2004). The disruption of these Hsp90 complexes suggests that novobiocin causes structural rearrangements within Hsp90 that are distinct from the changes seen with GA. Epigallocatechin gallate (ECGG) the active ingredient found in green tea also directly binds to Hsp90 and acts as an AhR antagonist. As opposed to disrupting the interaction of Hsp90 with AhR, ECGG functions by stabilizing the interaction between Hsp90 and AhR (Palermo et al., 2005). All of these compounds suggest that influencing the conformation of Hsp90 with small molecules is a viable strategy for the development of future therapeutics.
Concluding remarks
Structural dynamics play an important role in the function of Hsp90, and understanding the complete conformational ensemble is imperative for defining Hsp90s molecular mechanism. Large conformational changes occur throughout the ATPase hydrolysis cycle, and the conformational state is not rigorously determined by the bound nucleotide. Instead nucleotide binding shifts a conformational equilibrium between states that are also sampled in the absence of nucleotide. In vivo Hsp90 will also sample nucleotide bound and unbound states simultaneously due to the weak interaction with ATP. For the cytosolic eukaryotic Hsp90s, co-chaperones regulate the conformational state of Hsp90 by shifting the equilibrium and the finely tuned regulation is essential for proper chaperone function. Other levels of regulation exist in the form of post-translational modifications such as phosphorylation and acetylation, but the exact nature of these effects is poorly understood. Other modifications will also require investigation to determine their importance in the regulation of Hsp90. Disrupting the conformational dynamics of Hsp90 provides an attractive target for new therapeutics for the treatment of cancer and potentially vascular disease and Alzheimer’s, and inhibitors currently in clinical trials show signs of altering the conformation of Hsp90 and thereby disrupting the binding of client proteins.
While a much clearer picture of the mechanism of Hsp90 has emerged over the past few years, many outstanding questions still exist and require investigation. To begin with, the exact nature of the interaction of client proteins with Hsp90 is largely uncharacterized, and it is essential to understand how client protein binding affects the conformation of Hsp90 and vice versa. This information is key to understanding how Hsp90’s conformational changes relate to the activation of client proteins. Another open question is the origin of the energy required for remodeling of the client protein. Because the ATPase activity is low and not strongly coupled to the conformational changes that occur, it is unclear where the force originates from to drive the conformational changes in Hsp90 and any consequent changes that occur in the client protein. Also, important differences exist between different homologs and each homolog has uniquely determined the relative amounts of each conformation present under different conditions. Understanding how the equilibria are established and the functional consequences for shifting the equilibria will provide critical information about the molecular mechanism of Hsp90. Ultimately, a more detailed understanding of Hsp90, its dynamics and interactions, will pave the way for the development of more targeted therapeutics.
Acknowledgments
We would like to thank Ulrike Böttcher, Timothy Street, and Daniel Southworth for their helpful discussions and critical reading of the manuscript.
References
- Adinolfi E, Kim M, Young MT, Di Virgilio F, Surprenant A. Tyrosine phosphorylation of HSP90 within the P2X7 receptor complex negatively regulates P2X7 receptors. J Biol Chem. 2003;278:37344–37351. doi: 10.1074/jbc.M301508200. [DOI] [PubMed] [Google Scholar]
- Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan RK, Mok D, Ward BK, Ratajczak T. Modulation of chaperone function and cochaperone interaction by novobiocin in the C-terminal domain of Hsp90: evidence that coumarin antibiotics disrupt Hsp90 dimerization. J Biol Chem. 2006;281:7161–7171. doi: 10.1074/jbc.M512406200. [DOI] [PubMed] [Google Scholar]
- An WG, Schulte TW, Neckers LM. The heat shock protein 90 antagonist geldanamycin alters chaperone association with p210bcr-abl and v-src proteins before their degradation by the proteasome. Cell Growth Differ. 2000;11:355–360. [PubMed] [Google Scholar]
- Arlander SJ, Felts SJ, Wagner JM, Stensgard B, Toft DO, Karnitz LM. Chaperoning checkpoint kinase 1 (Chk1), an Hsp90 client, with purified chaperones. J Biol Chem. 2006;281:2989–2998. doi: 10.1074/jbc.M508687200. [DOI] [PubMed] [Google Scholar]
- Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/s0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- Biswas C, Ostrovsky O, Makarewich CA, Wanderling S, Gidalevitz T, Argon Y. The peptide-binding activity of GRP94 is regulated by calcium. Biochem J. 2007;405:233–241. doi: 10.1042/BJ20061867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bron P, Giudice E, Rolland JP, Buey RM, Barbier P, Diaz JF, Peyrot V, Thomas D, Garnier C. Apo-Hsp90 coexists in two open conformational states in solution. Biol Cell. 2008;100:413–425. doi: 10.1042/BC20070149. [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Mandal AK, Theodoraki MA. Molecular chaperones and protein kinase quality control. Trends Cell Biol. 2007;17:87–92. doi: 10.1016/j.tcb.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Chadli A, Bruinsma ES, Stensgard B, Toft D. Analysis of Hsp90 cochaperone interactions reveals a novel mechanism for TPR protein recognition. Biochemistry. 2008;47:2850–2857. doi: 10.1021/bi7023332. [DOI] [PubMed] [Google Scholar]
- Chadli A, Graham JD, Abel MG, Jackson TA, Gordon DF, Wood WM, Felts SJ, Horwitz KB, Toft D. GCUNC-45 is a novel regulator for the progesterone receptor/hsp90 chaperoning pathway. Mol Cell Biol. 2006;26:1722–1730. doi: 10.1128/MCB.26.5.1722-1730.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiosis G, Lucas B, Huezo H, Solit D, Basso A, Rosen N. Development of purine-scaffold small molecule inhibitors of Hsp90. Curr Cancer Drug Targets. 2003;3:371–376. doi: 10.2174/1568009033481778. [DOI] [PubMed] [Google Scholar]
- Chu F, Maynard JC, Chiosis G, Nicchitta CV, Burlingame AL. Identification of novel quaternary domain interactions in the Hsp90 chaperone, GRP94. Protein Sci. 2006;15:1260–1269. doi: 10.1110/ps.052065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo G, Morra G, Meli M, Verkhivker G. Understanding ligand-based modulation of the Hsp90 molecular chaperone dynamics at atomic resolution. Proc Natl Acad Sci U S A. 2008;105:7976–7981. doi: 10.1073/pnas.0802879105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CN, Krukenberg KA, Agard DA. Intra- and intermonomer interactions are required to synergistically facilitate ATP hydrolysis in Hsp90. J Biol Chem. 2008;283:21170–21178. doi: 10.1074/jbc.M800046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey CA, Koren J, Zhang YJ, Xu YF, Jinwal UK, Birnbaum MJ, Monks B, Sun M, Cheng JQ, Patterson C, Bailey RM, Dunmore J, Soresh S, Leon C, Morgan D, Petrucelli L. Akt and CHIP coregulate tau degradation through coordinated interactions. Proc Natl Acad Sci U S A. 2008;105:3622–3627. doi: 10.1073/pnas.0709180105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KD, Pratt WB. Folding of the glucocorticoid receptor by the reconstituted Hsp90-based chaperone machinery. The initial hsp90.p60.hsp70-dependent step is sufficient for creating the steroid binding conformation. J Biol Chem. 1997;272:13047–13054. doi: 10.1074/jbc.272.20.13047. [DOI] [PubMed] [Google Scholar]
- Dollins DE, Immormino RM, Gewirth DT. Structure of unliganded GRP94, the endoplasmic reticulum Hsp90. Basis for nucleotide-induced conformational change. J Biol Chem. 2005;280:30438–30447. doi: 10.1074/jbc.M503761200. [DOI] [PubMed] [Google Scholar]
- Dollins DE, Warren JJ, Immormino RM, Gewirth DT. Structures of GRP94-nucleotide complexes reveal mechanistic differences between the hsp90 chaperones. Mol Cell. 2007;28:41–56. doi: 10.1016/j.molcel.2007.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbrack RL, Jr, Gerloff DL, Bower M, Chen X, Lichtarge O, Cohen FE. Meeting review: the Second meeting on the Critical Assessment of Techniques for Protein Structure Prediction (CASP2), Asilomar, California, December 13–16, 1996. Fold Des. 1997;2:R27–R42. doi: 10.1016/S1359-0278(97)00011-4. [DOI] [PubMed] [Google Scholar]
- Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- Duval M, Le Boeuf F, Huot J, Gratton JP. Src-mediated phosphorylation of Hsp90 in response to vascular endothelial growth factor (VEGF) is required for VEGF receptor-2 signaling to endothelial NO synthase. Mol Biol Cell. 2007;18:4659–4668. doi: 10.1091/mbc.E07-05-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felts SJ, Karnitz LM, Toft DO. Functioning of the Hsp90 machine in chaperoning checkpoint kinase I (Chk1) and the progesterone receptor (PR) Cell Stress Chaperones. 2007;12:353–363. doi: 10.1379/CSC-299.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Leskovar A, Reinstein J, Buchner J. The ATPase cycle of the endoplasmic chaperone Grp94. J Biol Chem. 2007;282:35612–35620. doi: 10.1074/jbc.M704647200. [DOI] [PubMed] [Google Scholar]
- Geller R, Vignuzzi M, Andino R, Frydman J. Evolutionary constraints on chaperone-mediated folding provide an antiviral approach refractory to development of drug resistance. Genes Dev. 2007;21:195–205. doi: 10.1101/gad.1505307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Puertas P, Martin-Benito J, Carrascosa JL, Willison KR, Valpuesta JM. The substrate recognition mechanisms in chaperonins. J Mol Recognit. 2004;17:85–94. doi: 10.1002/jmr.654. [DOI] [PubMed] [Google Scholar]
- Graf C, Stankiewicz M, Kramer G, Mayer MP. Spatially and kinetically resolved changes in the conformational dynamics of the Hsp90 chaperone machine. Embo J. 2009;28:602–613. doi: 10.1038/emboj.2008.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammatikakis N, Lin JH, Grammatikakis A, Tsichlis PN, Cochran BH. p50(cdc37) acting in concert with Hsp90 is required for Raf-1 function. Mol Cell Biol. 1999;19:1661–1672. doi: 10.1128/mcb.19.3.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenert JP, Johnson BD, Toft DO. The importance of ATP binding and hydrolysis by hsp90 in formation and function of protein heterocomplexes. J Biol Chem. 1999;274:17525–17533. doi: 10.1074/jbc.274.25.17525. [DOI] [PubMed] [Google Scholar]
- Harst A, Lin H, Obermann WM. Aha1 competes with Hop, p50 and p23 for binding to the molecular chaperone Hsp90 and contributes to kinase and hormone receptor activation. Biochem J. 2005;387:789–796. doi: 10.1042/BJ20041283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartson SD, Irwin AD, Shao J, Scroggins BT, Volk L, Huang W, Matts RL. p50(cdc37) is a nonexclusive Hsp90 cohort which participates intimately in Hsp90-mediated folding of immature kinase molecules. Biochemistry. 2000;39:7631–7644. doi: 10.1021/bi000315r. [DOI] [PubMed] [Google Scholar]
- Hernandez MP, Chadli A, Toft DO. HSP40 binding is the first step in the HSP90 chaperoning pathway for the progesterone receptor. J Biol Chem. 2002;277:11873–11881. doi: 10.1074/jbc.M111445200. [DOI] [PubMed] [Google Scholar]
- Hessling M, Richter K, Buchner J. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat Struct Mol Biol. 2009;16:287–293. doi: 10.1038/nsmb.1565. [DOI] [PubMed] [Google Scholar]
- Huai Q, Wang H, Liu Y, Kim H, Toft D, Ke H. Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding. Structure. 2005;13:579–590. doi: 10.1016/j.str.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Immormino RM, Dollins DE, Shaffer PL, Soldano KL, Walker MA, Gewirth DT. Ligand-induced conformational shift in the N-terminal domain of GRP94, an Hsp90 chaperone. J Biol Chem. 2004;279:46162–46171. doi: 10.1074/jbc.M405253200. [DOI] [PubMed] [Google Scholar]
- Jakob U, Lilie H, Meyer I, Buchner J. Transient interaction of Hsp90 with early unfolding intermediates of citrate synthase. Implications for heat shock in vivo. J Biol Chem. 1995;270:7288–7294. doi: 10.1074/jbc.270.13.7288. [DOI] [PubMed] [Google Scholar]
- Johnson BD, Schumacher RJ, Ross ED, Toft DO. Hop modulates Hsp70/Hsp90 interactions in protein folding. J Biol Chem. 1998;273:3679–3686. doi: 10.1074/jbc.273.6.3679. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Halas A, Flom G. Nucleotide-dependent interaction of Saccharomyces cerevisiae Hsp90 with the cochaperone proteins Sti1, Cpr6, and Sba1. Mol Cell Biol. 2007;27:768–776. doi: 10.1128/MCB.01034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, Toft DO. Binding of p23 and hsp90 during assembly with the progesterone receptor. Mol Endocrinol. 1995;9:670–678. doi: 10.1210/mend.9.6.8592513. [DOI] [PubMed] [Google Scholar]
- Kekatpure VD, Dannenberg AJ, Subbaramaiah K. HDAC6 Modulates Hsp90 Chaperone Activity and Regulates Activation of Aryl Hydrocarbon Receptor Signaling. J Biol Chem. 2009;284:7436–7445. doi: 10.1074/jbc.M808999200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kosano H, Stensgard B, Charlesworth MC, McMahon N, Toft D. The assembly of progesterone receptor-hsp90 complexes using purified proteins. J Biol Chem. 1998;273:32973–32979. doi: 10.1074/jbc.273.49.32973. [DOI] [PubMed] [Google Scholar]
- Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Krukenberg KA, Forster F, Rice LM, Sali A, Agard DA. Multiple conformations of E. coli Hsp90 in solution: insights into the conformational dynamics of Hsp90. Structure. 2008;16:755–765. doi: 10.1016/j.str.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt N, Rajagopalan S, Cavagnero S. Effect of hsp70 chaperone on the folding and misfolding of polypeptides modeling an elongating protein chain. J Mol Biol. 2006;355:809–820. doi: 10.1016/j.jmb.2005.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, Rao J, Fliss A, Yang E, Garrett S, Caplan AJ. The Cdc37 protein kinase-binding domain is sufficient for protein kinase activity and cell viability. J Cell Biol. 2002;159:1051–1059. doi: 10.1083/jcb.200210121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskovar A, Wegele H, Werbeck ND, Buchner J, Reinstein J. The ATPase cycle of the mitochondrial Hsp90 analog Trap1. J Biol Chem. 2008;283:11677–11688. doi: 10.1074/jbc.M709516200. [DOI] [PubMed] [Google Scholar]
- Liu XD, Morano KA, Thiele DJ. The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J Biol Chem. 1999;274:26654–26660. doi: 10.1074/jbc.274.38.26654. [DOI] [PubMed] [Google Scholar]
- Lotz GP, Lin H, Harst A, Obermann WM. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J Biol Chem. 2003;278:17228–17235. doi: 10.1074/jbc.M212761200. [DOI] [PubMed] [Google Scholar]
- Macer DR, Koch GL. Identification of a set of calcium-binding proteins in reticuloplasm, the luminal content of the endoplasmic reticulum. J Cell Sci. 1988;91(Pt 1):61–70. doi: 10.1242/jcs.91.1.61. [DOI] [PubMed] [Google Scholar]
- Marcu MG, Chadli A, Bouhouche I, Catelli M, Neckers LM. The heat shock protein 90 antagonist novobiocin interacts with a previously unrecognized ATP-binding domain in the carboxyl terminus of the chaperone. J Biol Chem. 2000a;275:37181–37186. doi: 10.1074/jbc.M003701200. [DOI] [PubMed] [Google Scholar]
- Marcu MG, Schulte TW, Neckers L. Novobiocin and related coumarins and depletion of heat shock protein 90-dependent signaling proteins. J Natl Cancer Inst. 2000b;92:242–248. doi: 10.1093/jnci/92.3.242. [DOI] [PubMed] [Google Scholar]
- Martinez-Yamout MA, Venkitakrishnan RP, Preece NE, Kroon G, Wright PE, Dyson HJ. Localization of sites of interaction between p23 and Hsp90 in solution. J Biol Chem. 2006;281:14457–14464. doi: 10.1074/jbc.M601759200. [DOI] [PubMed] [Google Scholar]
- Maruya M, Sameshima M, Nemoto T, Yahara I. Monomer arrangement in HSP90 dimer as determined by decoration with N and C-terminal region specific antibodies. J Mol Biol. 1999;285:903–907. doi: 10.1006/jmbi.1998.2349. [DOI] [PubMed] [Google Scholar]
- McLaughlin SH, Smith HW, Jackson SE. Stimulation of the weak ATPase activity of human hsp90 by a client protein. J Mol Biol. 2002;315:787–798. doi: 10.1006/jmbi.2001.5245. [DOI] [PubMed] [Google Scholar]
- McLaughlin SH, Sobott F, Yao ZP, Zhang W, Nielsen PR, Grossmann JG, Laue ED, Robinson CV, Jackson SE. The co-chaperone p23 arrests the Hsp90 ATPase cycle to trap client proteins. J Mol Biol. 2006;356:746–758. doi: 10.1016/j.jmb.2005.11.085. [DOI] [PubMed] [Google Scholar]
- McLaughlin SH, Ventouras LA, Lobbezoo B, Jackson SE. Independent ATPase activity of hsp90 subunits creates a flexible assembly platform. J Mol Biol. 2004;344:813–826. doi: 10.1016/j.jmb.2004.09.055. [DOI] [PubMed] [Google Scholar]
- Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, Piper PW, Pearl LH. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 2003;11:647–658. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- Meyer P, Prodromou C, Liao C, Hu B, Mark Roe S, Vaughan CK, Vlasic I, Panaretou B, Piper PW, Pearl LH. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. Embo J. 2004;23:511–519. doi: 10.1038/sj.emboj.7600060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickler M, Hessling M, Ratzke C, Buchner J, Hugel T. The large conformational changes of Hsp90 are only weakly coupled to ATP hydrolysis. Nat Struct Mol Biol. 2009;16:281–286. doi: 10.1038/nsmb.1557. [DOI] [PubMed] [Google Scholar]
- Millson SH, Truman AW, Wolfram F, King V, Panaretou B, Prodromou C, Pearl LH, Piper PW. Investigating the protein-protein interactions of the yeast Hsp90 chaperone system by two-hybrid analysis: potential uses and limitations of this approach. Cell Stress Chaperones. 2004;9:359–368. doi: 10.1379/CSC-29R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimnaugh EG, Chavany C, Neckers L. Polyubiquitination and proteasomal degradation of the p185c–erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. J Biol Chem. 1996;271:22796–22801. doi: 10.1074/jbc.271.37.22796. [DOI] [PubMed] [Google Scholar]
- Morra G, Verkhivker G, Colombo G. Modeling signal propagation mechanisms and ligand-based conformational dynamics of the Hsp90 molecular chaperone full-length dimer. PLoS Comput Biol. 2009;5:e1000323. doi: 10.1371/journal.pcbi.1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan DF, Lindquist S. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckers L. Heat shock protein 90: the cancer chaperone. J Biosci. 2007;32:517–530. doi: 10.1007/s12038-007-0051-y. [DOI] [PubMed] [Google Scholar]
- Neckers L, Ivy SP. Heat shock protein 90. Curr Opin Oncol. 2003;15:419–424. doi: 10.1097/00001622-200311000-00003. [DOI] [PubMed] [Google Scholar]
- Nishiya Y, Shibata K, Saito S, Yano K, Oneyama C, Nakano H, Sharma SV. Drug-target identification from total cellular lysate by drug-induced conformational changes. Anal Biochem. 2009;385:314–320. doi: 10.1016/j.ab.2008.11.034. [DOI] [PubMed] [Google Scholar]
- Ogiso H, Kagi N, Matsumoto E, Nishimoto M, Arai R, Shirouzu M, Mimura J, Fujii-Kuriyama Y, Yokoyama S. Phosphorylation analysis of 90 kDa heat shock protein within the cytosolic arylhydrocarbon receptor complex. Biochemistry. 2004;43:15510–15519. doi: 10.1021/bi048736m. [DOI] [PubMed] [Google Scholar]
- Onuoha SC, Coulstock ET, Grossmann JG, Jackson SE. Structural studies on the co-chaperone Hop and its complexes with Hsp90. J Mol Biol. 2008;379:732–744. doi: 10.1016/j.jmb.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Palermo CM, Westlake CA, Gasiewicz TA. Epigallocatechin Gallate Inhibits Aryl Hydrocarbon Receptor Gene Transcription through an Indirect Mechanism Involving Binding to a 90 kDa Heat Shock Protein. Biochemistry. 2005;44:5041–5052. doi: 10.1021/bi047433p. [DOI] [PubMed] [Google Scholar]
- Panaretou B, Siligardi G, Meyer P, Maloney A, Sullivan JK, Singh S, Millson SH, Clarke PA, Naaby-Hansen S, Stein R, Cramer R, Mollapour M, Workman P, Piper PW, Pearl LH, Prodromou C. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol Cell. 2002;10:1307–1318. doi: 10.1016/s1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- Phillips JJ, Yao ZP, Zhang W, McLaughlin S, Laue ED, Robinson CV, Jackson SE. Conformational dynamics of the molecular chaperone Hsp90 in complexes with a co-chaperone and anticancer drugs. J Mol Biol. 2007;372:1189–1203. doi: 10.1016/j.jmb.2007.04.059. [DOI] [PubMed] [Google Scholar]
- Picard D. Chaperoning steroid hormone action. Trends Endocrinol Metab. 2006;17:229–235. doi: 10.1016/j.tem.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Prodromou C, Panaretou B, Chohan S, Siligardi G, O’Brien R, Ladbury JE, Roe SM, Piper PW, Pearl LH. The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. Embo J. 2000;19:4383–4392. doi: 10.1093/emboj/19.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- Prodromou C, Siligardi G, O’Brien R, Woolfson DN, Regan L, Panaretou B, Ladbury JE, Piper PW, Pearl LH. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. Embo J. 1999;18:754–762. doi: 10.1093/emboj/18.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K, Buchner J. Hsp90: chaperoning signal transduction. J Cell Physiol. 2001;188:281–290. doi: 10.1002/jcp.1131. [DOI] [PubMed] [Google Scholar]
- Richter K, Moser S, Hagn F, Friedrich R, Hainzl O, Heller M, Schlee S, Kessler H, Reinstein J, Buchner J. Intrinsic inhibition of the Hsp90 ATPase activity. J Biol Chem. 2006;281:11301–11311. doi: 10.1074/jbc.M510142200. [DOI] [PubMed] [Google Scholar]
- Richter K, Muschler P, Hainzl O, Buchner J. Coordinated ATP hydrolysis by the Hsp90 dimer. J Biol Chem. 2001;276:33689–33696. doi: 10.1074/jbc.M103832200. [DOI] [PubMed] [Google Scholar]
- Richter K, Muschler P, Hainzl O, Reinstein J, Buchner J. Sti1 is a non-competitive inhibitor of the Hsp90 ATPase. Binding prevents the N-terminal dimerization reaction during the atpase cycle. J Biol Chem. 2003;278:10328–10333. doi: 10.1074/jbc.M213094200. [DOI] [PubMed] [Google Scholar]
- Richter K, Soroka J, Skalniak L, Leskovar A, Hessling M, Reinstein J, Buchner J. Conserved conformational changes in the ATPase cycle of human Hsp90. J Biol Chem. 2008;283:17757–17765. doi: 10.1074/jbc.M800540200. [DOI] [PubMed] [Google Scholar]
- Richter K, Walter S, Buchner J. The Co-chaperone Sba1 connects the ATPase reaction of Hsp90 to the progression of the chaperone cycle. J Mol Biol. 2004;342:1403–1413. doi: 10.1016/j.jmb.2004.07.064. [DOI] [PubMed] [Google Scholar]
- Riggs DL, Roberts PJ, Chirillo SC, Cheung-Flynn J, Prapapanich V, Ratajczak T, Gaber R, Picard D, Smith DF. The Hsp90-binding peptidylprolyl isomerase FKBP52 potentiates glucocorticoid signaling in vivo. Embo J. 2003;22:1158–1167. doi: 10.1093/emboj/cdg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe SM, Ali MM, Meyer P, Vaughan CK, Panaretou B, Piper PW, Prodromou C, Pearl LH. The Mechanism of Hsp90 regulation by the protein kinase-specific cochaperone p50(cdc37) Cell. 2004;116:87–98. doi: 10.1016/s0092-8674(03)01027-4. [DOI] [PubMed] [Google Scholar]
- Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. Embo J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Sangster TA, Salathia N, Lee HN, Watanabe E, Schellenberg K, Morneau K, Wang H, Undurraga S, Queitsch C, Lindquist S. HSP90-buffered genetic variation is common in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008;105:2969–2974. doi: 10.1073/pnas.0712210105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibel T, Siegmund HI, Jaenicke R, Ganz P, Lilie H, Buchner J. The charged region of Hsp90 modulates the function of the N-terminal domain. Proc Natl Acad Sci U S A. 1999;96:1297–1302. doi: 10.1073/pnas.96.4.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte TW, Blagosklonny MV, Ingui C, Neckers L. Disruption of the Raf-1-Hsp90 molecular complex results in destabilization of Raf-1 and loss of Raf-1-Ras association. J Biol Chem. 1995;270:24585–24588. doi: 10.1074/jbc.270.41.24585. [DOI] [PubMed] [Google Scholar]
- Scroggins BT, Robzyk K, Wang D, Marcu MG, Tsutsumi S, Beebe K, Cotter RJ, Felts S, Toft D, Karnitz L, Rosen N, Neckers L. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol Cell. 2007;25:151–159. doi: 10.1016/j.molcel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah V, Wiest R, Garcia-Cardena G, Cadelina G, Groszmann RJ, Sessa WC. Hsp90 regulation of endothelial nitric oxide synthase contributes to vascular control in portal hypertension. Am J Physiol. 1999;277:G463–G468. doi: 10.1152/ajpgi.1999.277.2.G463. [DOI] [PubMed] [Google Scholar]
- Shao J, Irwin A, Hartson SD, Matts RL. Functional dissection of cdc37: characterization of domain structure and amino acid residues critical for protein kinase binding. Biochemistry. 2003;42:12577–12588. doi: 10.1021/bi035138j. [DOI] [PubMed] [Google Scholar]
- Shiau AK, Harris SF, Southworth DR, Agard DA. Structural Analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- Siligardi G, Hu B, Panaretou B, Piper PW, Pearl LH, Prodromou C. Co-chaperone Regulation of Conformational Switching in the Hsp90 ATPase Cycle. J Biol Chem. 2004;279:51989–51998. doi: 10.1074/jbc.M410562200. [DOI] [PubMed] [Google Scholar]
- Siligardi G, Panaretou B, Meyer P, Singh S, Woolfson DN, Piper PW, Pearl LH, Prodromou C. Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. J Biol Chem. 2002;277:20151–20159. doi: 10.1074/jbc.M201287200. [DOI] [PubMed] [Google Scholar]
- Smith DF. Dynamics of heat shock protein 90-progesterone receptor binding and the disactivation loop model for steroid receptor complexes. Mol Endocrinol. 1993;7:1418–1429. doi: 10.1210/mend.7.11.7906860. [DOI] [PubMed] [Google Scholar]
- Smith DF, Faber LE, Toft DO. Purification of unactivated progesterone receptor and identification of novel receptor-associated proteins. J Biol Chem. 1990;265:3996–4003. [PubMed] [Google Scholar]
- Soldano KL, Jivan A, Nicchitta CV, Gewirth DT. Structure of the N-terminal domain of GRP94. Basis for ligand specificity and regulation. J Biol Chem. 2003;278:48330–48338. doi: 10.1074/jbc.M308661200. [DOI] [PubMed] [Google Scholar]
- Solit DB, Osman I, Polsky D, Panageas KS, Daud A, Goydos JS, Teitcher J, Wolchok JD, Germino FJ, Krown SE, Coit D, Rosen N, Chapman PB. Phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with metastatic melanoma. Clin Cancer Res. 2008;14:8302–8307. doi: 10.1158/1078-0432.CCR-08-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soti C, Racz A, Csermely P. A Nucleotide-dependent molecular switch controls ATP binding at the C-terminal domain of Hsp90. N-terminal nucleotide binding unmasks a C-terminal binding pocket. J Biol Chem. 2002;277:7066–7075. doi: 10.1074/jbc.M105568200. [DOI] [PubMed] [Google Scholar]
- Southworth DR, Agard DA. Species-dependent ensembles of conserved conformational states define the Hsp90 chaperone ATPase cycle. Mol Cell. 2008;32:631–640. doi: 10.1016/j.molcel.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- Stepanova L, Leng X, Parker SB, Harper JW. Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev. 1996;10:1491–1502. doi: 10.1101/gad.10.12.1491. [DOI] [PubMed] [Google Scholar]
- Sullivan W, Stensgard B, Caucutt G, Bartha B, McMahon N, Alnemri ES, Litwack G, Toft D. Nucleotides and two functional states of hsp90. J Biol Chem. 1997;272:8007–8012. doi: 10.1074/jbc.272.12.8007. [DOI] [PubMed] [Google Scholar]
- Te J, Jia L, Rogers J, Miller A, Hartson SD. Novel subunits of the mammalian Hsp90 signal transduction chaperone. J Proteome Res. 2007;6:1963–1973. doi: 10.1021/pr060595i. [DOI] [PubMed] [Google Scholar]
- Van PN, Peter F, Soling HD. Four intracisternal calcium-binding glycoproteins from rat liver microsomes with high affinity for calcium. No indication for calsequestrin-like proteins in inositol 1,4,5-trisphosphate-sensitive calcium sequestering rat liver vesicles. J Biol Chem. 1989;264:17494–17501. [PubMed] [Google Scholar]
- Vaughan CK, Gohlke U, Sobott F, Good VM, Ali MM, Prodromou C, Robinson CV, Saibil HR, Pearl LH. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol Cell. 2006;23:697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan CK, Piper PW, Pearl LH, Prodromou C. A common conformationally coupled ATPase mechanism for yeast and human cytoplasmic HSP90s. Febs J. 2009;276:199–209. doi: 10.1111/j.1742-4658.2008.06773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter S, Buchner J. Molecular chaperones--cellular machines for protein folding. Angew Chem Int Ed Engl. 2002;41:1098–1113. doi: 10.1002/1521-3773(20020402)41:7<1098::aid-anie1098>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Wandinger SK, Suhre MH, Wegele H, Buchner J. The phosphatase Ppt1 is a dedicated regulator of the molecular chaperone Hsp90. Embo J. 2006;25:367–376. doi: 10.1038/sj.emboj.7600930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegele H, Muschler P, Bunck M, Reinstein J, Buchner J. Dissection of the contribution of individual domains to the ATPase mechanism of Hsp90. J Biol Chem. 2003;278:39303–39310. doi: 10.1074/jbc.M305751200. [DOI] [PubMed] [Google Scholar]
- Wegele H, Wandinger SK, Schmid AB, Reinstein J, Buchner J. Substrate transfer from the chaperone Hsp70 to Hsp90. J Mol Biol. 2006;356:802–811. doi: 10.1016/j.jmb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Weikl T, Muschler P, Richter K, Veit T, Reinstein J, Buchner J. C-terminal regions of Hsp90 are important for trapping the nucleotide during the ATPase cycle. J Mol Biol. 2000;303:583–592. doi: 10.1006/jmbi.2000.4157. [DOI] [PubMed] [Google Scholar]
- Weissman JS, Hohl CM, Kovalenko O, Kashi Y, Chen S, Braig K, Saibil HR, Fenton WA, Horwich AL. Mechanism of GroEL action: productive release of polypeptide from a sequestered position under GroES. Cell. 1995;83:577–587. doi: 10.1016/0092-8674(95)90098-5. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v–src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci U S A. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman P. Combinatorial attack on multistep oncogenesis by inhibiting the Hsp90 molecular chaperone. Cancer Lett. 2004;206:149–157. doi: 10.1016/j.canlet.2003.08.032. [DOI] [PubMed] [Google Scholar]
- Ying M, Flatmark T. Binding of the viral immunogenic octapeptide VSV8 to native glucose-regulated protein Grp94 (gp96) and its inhibition by the physiological ligands ATP and Ca2+ Febs J. 2006;273:513–522. doi: 10.1111/j.1742-4658.2005.05084.x. [DOI] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- Young JC, Moarefi I, Hartl FU. Hsp90: a specialized but essential protein-folding tool. J Cell Biol. 2001;154:267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Guo ZS, Marcu MG, Neckers L, Nguyen DM, Chen GA, Schrump DS. Modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J Natl Cancer Inst. 2002;94:504–513. doi: 10.1093/jnci/94.7.504. [DOI] [PubMed] [Google Scholar]
- Yun BG, Huang W, Leach N, Hartson SD, Matts RL. Novobiocin induces a distinct conformation of hsp90 and alters hsp90-cochaperone-client interactions. Biochemistry. 2004;43:8217–8229. doi: 10.1021/bi0497998. [DOI] [PubMed] [Google Scholar]
- Zhang W, Hirshberg M, McLaughlin SH, Lazar GA, Grossmann JG, Nielsen PR, Sobott F, Robinson CV, Jackson SE, Laue ED. Biochemical and structural studies of the interaction of Cdc37 with Hsp90. J Mol Biol. 2004;340:891–907. doi: 10.1016/j.jmb.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, Boone C, Emili A, Houry WA. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]