Abstract
Purpose
To explore the genetic landscape of tumors from patients enrolled on the BOLERO-2 trial to identify potential correlations between genetic alterations and efficacy of everolimus treatment. The BOLERO-2 trial has previously demonstrated that the addition of everolimus to exemestane prolonged progression-free survival by more than twofold in patients with hormone receptor–positive, human epidermal growth factor receptor 2–negative, advanced breast cancer previously treated with nonsteroidal aromatase inhibitors.
Patients and Methods
Next-generation sequencing was used to analyze genetic status of cancer-related genes in 302 archival tumor specimens from patients representative of the BOLERO-2 study population. Correlations between the most common somatic alterations and degree of chromosomal instability, and treatment effect of everolimus were investigated.
Results
Progression-free survival benefit with everolimus was maintained regardless of alteration status of PIK3CA, FGFR1, and CCND1 or the pathways of which they are components. However, quantitative differences in everolimus benefit were observed between patient subgroups defined by the exon-specific mutations in PIK3CA (exon 20 v 9) or by different degrees of chromosomal instability in the tumor tissues.
Conclusion
The data from this exploratory analysis suggest that the efficacy of everolimus was largely independent of the most commonly altered genes or pathways in hormone receptor–positive, human epidermal growth factor receptor 2–negative breast cancer. The potential impact of chromosomal instabilities and low-frequency genetic alterations on everolimus efficacy warrants further investigation.
INTRODUCTION
Everolimus, an oral mammalian target of rapamycin (mTOR) inhibitor, has antitumor activity in multiple cancer types. The double-blinded, randomized, placebo-controlled, phase III trial BOLERO-2 (NCT00863655) demonstrated that everolimus plus exemestane substantially improved progression-free survival (PFS) compared with placebo plus exemestane in hormone receptor–positive (HR+), human epidermal growth factor receptor 2–negative (HER2−) advanced breast cancer that recurred and/or progressed during or after nonsteroidal aromatase inhibitor therapy (median PFS, 7.8 v 3.2 months, respectively; hazard ratio [HR], 0.45; 95% CI, 0.38 to 0.54; P < .001).1 Everolimus was associated with similar risk reduction in progression across patient subgroups predefined by age, race, previous treatment, and disease characteristics.
Development of NextGen Sequencing (NGS) techniques allows for extensive exploration of tumor genetic landscapes, including HR+, HER2− breast cancer.2-9 However, correlations between such genetic alteration data and clinical outcome in well-defined large clinical trial cohorts of any cancer have yet to be explored. We sequenced exons of cancer-related genes using NGS technology in archival tumor specimens from a subset of patients in BOLERO-2 and explored potential associations between PFS benefit from everolimus and genetic alterations in phosphoinositide 3-kinase (PI3K) and fibroblast growth factor receptor (FGFR) pathway genes. Furthermore, we developed an estimation metric for chromosomal instability using NGS data from a target gene panel and explored its correlation with everolimus PFS benefit. To our knowledge, this is the first report of the use of broad NGS data to explore predictive biomarkers in a pivotal oncology trial.
PATIENTS AND METHODS
BOLERO-2 Trial and NGS Patient Subgroup
BOLERO-21 included 724 patients with HR+, HER2− advanced breast cancer who were randomly assigned in a ratio of 2:1 to everolimus plus exemestane or placebo plus exemestane. Tumor samples from 302 patients had NGS data available for evaluation. All patients included in the present analysis consented to evaluation of their archival tumor samples.
Tumor Sample and Biomarker Assays
Tumor DNA was extracted from formalin-fixed, paraffin-embedded archival tumor tissue sections. Tumor DNA preparation, library construction, hybrid capture, and sequencing were previously described.10 Samples with a minimum unique coverage of 250× were evaluated in the subsequent data analysis. Somatic genetic alterations were reported after we filtered for known germline variants present in public databases (Data Supplement). Only somatic mutations predicted with our variant annotation pipeline were included in these analyses.
Scoring of Chromosomal Instability From NGS Data
A scoring metric for measuring chromosomal instability (CIN) in tumor samples based on genomic structural aberrations reported in the NGS data was calculated for each sample (Data Supplement). The CIN score comprises the number of rearrangements and the number and magnitude of copy-number changes between neighboring genes on the chromosome (Data Supplement).
Statistical Analyses
Somatic alterations were summarized by using descriptive statistics. PFS in patients whose tumors harbored altered versus wild-type status for the indicated biomarker in each treatment arm was estimated by using Kaplan-Meier method. A Cox proportional hazards model was used to test the interaction between treatment and biomarkers and to compute HRs and the associated 95% CI for the biomarker-defined subpopulations in each treatment arm, adjusted for clinical covariates if any were significantly imbalanced between treatment arms and varied by comparison (Data Supplement). All P values presented were nominal without adjustment for multiple testing. A hyperactive PI3K pathway was defined as at least one mutation in PIK3CA, PTEN, AKT1, or PIK3R1 or as low PTEN expression by means of immunohistochemistry with an H-score < 10. The CIN score was dichotomized into a binary variable by a cutoff point, and the one that maximized the difference in treatment effect between the two subgroups was identified by a grid search of a range of values.
RESULTS
BOLERO-2 NGS Cohort and Genetic Alterations
Evaluable data were obtained from 302 samples. In the NGS subgroup, 244 (81%) were from primary tumors, 57 (19%) were from metastatic lesions, and one was of unknown status. This represented 42% of the BOLERO-2 trial population.
This NGS subgroup included 209 (43.1%) of 485 patients from the everolimus arm and 93 (38.9%) of 239 patients from the placebo arm. Demographic and baseline clinical characteristics were similar between the NGS and trial populations and between the treatment arms of the NGS subgroup (Table 1). The PFS benefit with everolimus in the NGS subgroup was comparable to that in the overall trial population (Table 1). This finding suggested that the NGS subgroup was representative of the BOLERO-2 trial population and, more importantly, that the integrity of random assignment was preserved in the NGS subgroup.
Table 1.
Demographic and Baseline Characteristics in the BOLERO-2 NGS Subgroup and Overall Study Population
| Characteristic | NGS Subgroup |
Overall Study Population |
||||||
|---|---|---|---|---|---|---|---|---|
| Everolimus (n = 209) |
Placebo (n = 93) |
Everolimus (n = 485) |
Placebo (n = 239) |
|||||
| No. | % | No. | % | No. | % | No. | % | |
| Age, years | ||||||||
| Median (range) | 62 (56-70) | 60 (55-66) | 62 (56-69) | 61 (55-66) | ||||
| Race | ||||||||
| White | 152 | 73 | 68 | 73 | 361 | 74 | 186 | 78 |
| Asian | 43 | 21 | 23 | 25 | 98 | 20 | 45 | 19 |
| Other | 14 | 6 | 2 | 2 | 27 | 6 | 8 | 3 |
| Eastern Cooperative Oncology Group performance status | ||||||||
| 0 | 139 | 67 | 64 | 69 | 292 | 60 | 142 | 59 |
| 1 | 64 | 31 | 27 | 29 | 175 | 36 | 84 | 35 |
| 2 | 3 | 1 | 2 | 2 | 9 | 2 | 7 | 3 |
| Visceral disease | 113 | 54 | 47 | 51 | 271 | 56 | 135 | 56 |
| No. of metastatic sites | ||||||||
| 1 | 71 | 34 | 29 | 31 | 154 | 32 | 64 | 27 |
| 2 | 61 | 29 | 30 | 32 | 149 | 31 | 84 | 35 |
| ≥ 3 | 76 | 36 | 34 | 37 | 180 | 37 | 91 | 38 |
| Previous sensitivity to hormone therapy | 180 | 86 | 77 | 83 | 409 | 84 | 201 | 84 |
| Previous endocrine treatment | ||||||||
| Letrozole or anastrozole | 209 | 100 | 93 | 100 | 485 | 100 | 239 | 100 |
| Tamoxifen | 96 | 46 | 45 | 48 | 230 | 47 | 119 | 50 |
| Fulvestrant | 34 | 16 | 14 | 15 | 80 | 17 | 39 | 16 |
| PFS events | 127 | 78 | 293 | 197 | ||||
| Median PFS, months (95% CI) | 7.0 (6.7 to 8.5) | 4.0 (2.6 to 4.2) | 7.8 (6.9 to 8.5) | 3.2 (2.8 to 4.1) | ||||
| Hazard ratio (95% CI) | 0.44 (0.33 to 0.59) | 0.44 (0.37 to 0.53) | ||||||
Abbreviations: NGS, next-generation sequencing; PFS, progression-free survival.
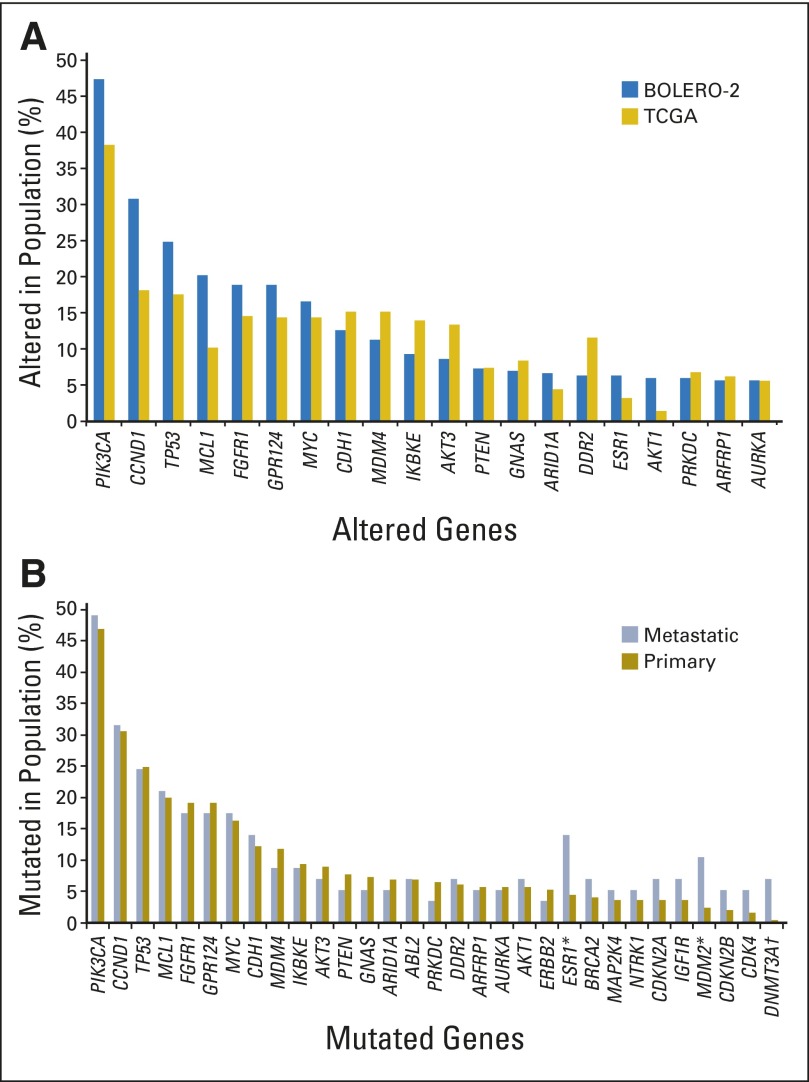
The genetic profile of the patients in the NGS subgroup was largely consistent with data from > 400 HR+, HER2− breast cancer tumors in The Cancer Genome Atlas network (TCGA) report (Fig 1A).3 This included most of the frequently altered genes and the relationships between mutations in genes in the same signaling pathways. The genes most frequently altered were PIK3CA (47.6%), CCND1 (31.3%), TP53 (23.3%), and FGFR1 (18.1%; Fig 1A; Data Supplement).
Fig 1.
(A) Alteration frequency in BOLERO-2 versus The Cancer Genome Atlas (TCGA) estrogen receptor–positive (ER+), human epidermal growth factor receptor 2–negative (HER2−) breast cancer cohort. Genes most frequently altered in the BOLERO-2 next-generation sequencing (NGS) subgroup and corresponding frequency in patients with ER+, HER2− breast cancer samples from TCGA were compared.3 Genes with alteration frequency > 5% (ie, altered in at least 15 patients) in the BOLERO-2 NGS cohort are shown. (B) Alteration frequency in metastatic versus primary tumors in the BOLERO-2 NGS cohort. Genetic alteration rates of ESR1, MDM2, and DNMT3A were different between metastatic versus primary tumors at statistically significant levels. *P ≤ .05; †P ≤ .005.
The genetic profiles of metastatic and primary tumor samples from BOLERO-2 were generally similar, with statistically significantly increased mutation rates observed only for ESR1, MDM2, and DNMT3A (Fig 1B). The increased mutation rate of ESR1 in metastatic samples is likely associated with their development of resistance to hormonal therapies.11
Correlations Between Everolimus Benefit and Genetic Alterations in the PI3K/mTOR Pathway
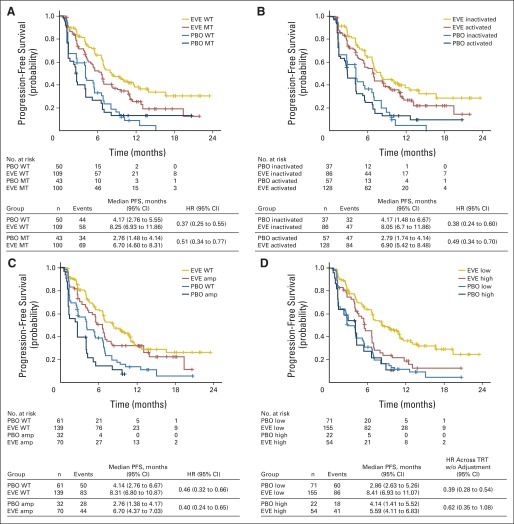
We tested the hypothesis that tumors dependent on a hyperactive PI3K/mTOR pathway might yield greater treatment benefit from everolimus by examining the effect of PIK3CA mutations on PFS benefit. All PIK3CA mutations were previously reported, with almost 90% occurring in exon 9 (32.9%) or 20 (53.9%). Median PFS was longer in patients with wild-type PIK3CA in both treatment arms than in those with mutations, with a slightly greater reduction in risk of progression with everolimus versus placebo. For wild-type PIK3CA, the HR was 0.37; (95% CI, 0.25 to 0.55), and for mutated PIK3CA, the HR was 0.51 (95% CI, 0.34 to 0.77; interaction P = .35). These findings suggested that PIK3CA mutations only minimally affected the efficacy of everolimus (Fig 2A, Table 2). Similar results were observed in patients with PI3K pathway activity that was either normal (HR, 0.38; 95% CI, 0.24 to 0.50) or hyperactive (HR, 0.49; 95% CI, 0.34 to 0.70; interaction P = .5; Fig 2B). When patients were assigned to subgroups on the basis of mutations in PIK3CA exon 20 (kinase domain) or exon 9 (helical domain), PFS benefit from everolimus appeared to be greater in those with exon-9 mutations (HR, 0.26; 95% CI, 0.12 to 0.54) than in those with exon-20 mutations (HR, 0.56; 95% CI, 0.31 to 1.00, Table 2).
Fig 2.
Plots of Kaplan-Meier estimates of progression-free survival (PFS) by treatment arm for patient subgroups in the BOLERO-2 next-generation sequencing population. Subgroups were defined by gene mutation (MT) versus wild-type (WT), amplification (amp), chromosomal instability (CIN) score low or high, or pathway activity. (A) PIK3CA pathway status. (B) PI3K pathway status. (C) Cell-cycle control genes. (D) CIN score in which the 75th percentile was used as the cutoff. EVE, everolimus; HR, hazard ratio; PBO, placebo; TRT, treatment; w/o, without.
Table 2.
PFS Benefit in Patient Subgroups Defined by Overall and Exon-Specific Mutation Status of PIK3CA
| PIK3CA and Study Arm | No. | PFS Events | Median PFS (months) | 95% CI | Hazard Ratio | 95% CI |
|---|---|---|---|---|---|---|
| Wild type | ||||||
| Placebo | 50 | 44 | 4.2 | 2.8 to 5.6 | 0.37 | 0.25 to 0.55 |
| Everolimus | 109 | 58 | 8.3 | 6.9 to 11.9 | ||
| All mutations | ||||||
| Placebo | 43 | 34 | 2.8 | 1.5 to 4.1 | 0.51 | 0.34 to 0.77 |
| Everolimus | 100 | 69 | 6.7 | 4.6 to 8.3 | ||
| Mutation in exon 20 | ||||||
| Placebo | 21 | 17 | 4.1 | 1.7 to 6.8 | 0.56 | 0.31 to 1.00 |
| Everolimus | 56 | 38 | 6.8 | 4.4 to 9.6 | ||
| Mutation in exon 9 | ||||||
| Placebo | 15 | 13 | 1.5 | 1.3 to 2.6 | 0.26 | 0.12 to 0.54 |
| Everolimus | 32 | 22 | 5.2 | 2.7 to 6.9 |
Abbreviation: PFS, progression-free survival.
Correlations between mTOR mutations and enhanced clinical benefit from mTOR inhibitors were suggested in a few case studies involving NGS analyses of tumor samples obtained from patients in whom extraordinary clinical efficacy was observed.12,13 We were able to confirm or predict 10 somatic mutations in mTOR; this allowed only case studies in our analysis. The results appear to agree with reported observations that patients with mTOR mutations may derive greater benefit from everolimus (Data Supplement). However, a definitive conclusion awaits more systematic investigation of functionally well-characterized somatic mutations in larger patient cohorts. Overall, although mutations in PIK3CA or the PI3K/AKT/mTOR pathway as a whole were unlikely to be associated with everolimus efficacy, these results suggest that mutations in certain PIK3CA or mTOR functional domains might render a tumor more sensitive to mTOR blockade in relatively small subsets of patients.
Correlations Between Everolimus Benefit and Genetic Alterations in FGFR and Cell-Cycle Control Genes
We next examined potential correlations between everolimus-derived PFS benefit and aberrations in genes-encoding proteins involved in FGFR signaling and cell-cycle regulation. These are two oncogenic pathways that are frequently mutated and that are under intensive therapeutic investigations in estrogen receptor–positive (ER+), HER2− breast cancer.14 Similar to the observations with the PI3K pathway, amplification of CCND1 or genetic alterations in cell-cycle control genes CCND1, CDK4, CDK6, and CDKN2A had minimal effect on PFS gain with everolimus (Fig 2C).
The genetic alterations in FGFR1 and FGFR2 were mutually exclusive (Data Supplement). The PFS benefit with everolimus appeared similar between FGFR1-amplified and wild-type cohorts, with a median PFS gain of approximately 4 months in each cohort (Table 3). However, the sample size and number of events in the FGFR1-amplified subgroup were too small to meet statistical significance.
Table 3.
PFS by Treatment Arm for Patients With FGFR1 Wild Type Versus Amplification
| Group | No. | PFS Events | Median PFS (months) | 95% CI | Hazard Ratio Across Treatments Without Adjustment | 95% CI |
|---|---|---|---|---|---|---|
| Placebo, wild type | 70 | 58 | 4.0 | 2.6 to 5.3 | 0.43 | 0.31 to 0.6 |
| Everolimus, wild type | 166 | 96 | 8.3 | 6.8 to 10.7 | ||
| Placebo, amplification | 22 | 19 | 2.8 | 1.5 to 4.4 | 0.39 | 0.21 to 0.72 |
| Everolimus, amplification | 35 | 24 | 6.8 | 4.7 to 8.1 |
Abbreviation: PFS, progression-free survival.
Only seven patients, all in the everolimus arm, had confirmed, activating somatic mutations in FGFR2. Median PFS for these patients was 2.7 months (range, 1.3 to 3.9 months), which was significantly shorter than the 7.0-month median PFS for everolimus-treated patients in the NGS subgroup. (Additional details are given in the Data Supplement).
Correlations Between Everolimus Benefit and Chromosomal Instability
Increased chromosomal instability and tumor heterogeneity are thought to be associated with treatment resistance in several solid tumors.15-17 To assess the effect of chromosomal instability of the tumors on the benefit derived from everolimus, and, more importantly, to complement the single-gene or pathway-focused analyses and to include low-frequency genetic alterations, we derived and evaluated a CIN scoring system using the NGS data from the targeted gene panel (see Discussion and Data Supplement).
When the NGS subgroup was dichotomously assigned to subgroups by high versus low CIN scores, those with lower scores appeared to have a larger PFS benefit. A search for an optimal cut point showed that the 75th percentile yielded the maximal difference in HR between high– versus low–CIN score subgroups (Fig 2D; Data Supplement). Patients with CIN score below the 75th percentile derived a median PFS gain of 5.5 months from adding everolimus to exemestane (median PFS 8.4 v 2.9 months for everolimus v placebo; HR, 0.39; 95% CI, 0.28 to 0.54; interaction P = .17). In comparison, the median PFS gain was 1.5 months in patients with a CIN score above the 75th percentile (median PFS 5.6 v 4.1 months for everolimus v placebo; HR, 0.62; 95% CI, 0.35 to 1.08). These data suggested that tumors with lower chromosomal instability might derive greater benefit from the addition of everolimus. The effect of chromosomal instability on PFS was only evident in patients in the everolimus arm and not those in the placebo arm, suggesting that the potential association between CIN and PFS was everolimus-specific (Fig 2D).
DISCUSSION
To our knowledge, this analysis represents the first attempt to explore correlations between clinical efficacy and broad genetic alterations—both sequence variations and copy-number variations—in tumor tissues characterized by means of massive parallel sequencing of many cancer-related genes (Data Supplement) in a phase III trial of a common solid tumor.
Several observations with implications for improving our understanding of the PI3K/mTOR and related signaling pathways, potential clinical utilities, and development of new therapeutics emerged from this analysis. Although similar to findings from other reported analyses,18 the lack of appreciable effect of activating PIK3CA mutations or PI3K-pathway activity (defined by genetic alterations and PTEN level) on everolimus benefit highlighted the oversimplification of both the hypothesis and the analytic method of pooling all genetic alterations in this frequently mutated gene or hyperactive pathway. Our data suggested that PIK3CA exon-9 mutations were associated with more benefit from everolimus than exon-20 mutations were. This exon-specific, or protein functional domain–specific, effect is consistent with the results from a phase II neoadjuvant study in patients with ER+, HER2− breast cancer who were randomly assigned to receive letrozole or letrozole plus everolimus.19 A difference of approximately two-fold was observed in the antiproliferative effect between treatments in patients with PIK3CA exon-9 mutations compared with a nearly identical effect in patients with PIK3CA exon-20 mutations.19 Data from > 400 Cancer Cell Line Encyclopedia cell lines20 showed significantly increased phospho-AKT levels in cell lines with PIK3CA exon-20 mutations versus exon-9 mutations, whereas mTOR activities, indicated by phospho-S6 level, were similar (Data Supplement). This finding suggested that activating mutations in the kinase domain may limit the efficacy of everolimus by means of AKT-mediated tumor survival. It was also shown that only exon-20 mutations directly enhanced PI3K activity, whereas exon-9 mutations might gain the ability to interact with insulin receptor substrate-1 independent of the p85 PIK3R1 (regulatory subunit), thereby rewiring the oncogenic signaling pathway.21 More importantly, this observation suggests the necessity of a clinical evaluation of PIK3CA mutations according to their specific functional effect. The same principle should apply to analysis of mutations in genes encoding proteins with multiple functional domains (eg, mTOR) and different components of an oncogenic pathway.
FGFR1 amplification has been associated with invasiveness of breast cancer and resistance to endocrine therapy.22,23 In our analysis, tumors with FGFR1 and FGFR2 amplification or activating mutations appeared to derive less PFS benefit from everolimus possibly because of reduced dependence on mTOR signaling and/or increased expression of mTOR substrate 4E-BP1. 4E-BP1 is encoded by EIF4EBP1 located between FGFR1 and GPR124 on chromosomal region 8p12. FGFR1 and GPR124 are coamplified in almost all NGS-assessed BOLERO-2 tumor samples and TCGA cohorts, which should result in coamplification and, presumably, increased expression of EIF4EBP1 in FGFR1-amplified tumor samples. CCND1 and RPS6KB2 (encoding S6 kinase, another mTOR substrate) and genes encoding FGF3, FGF4, and FGF19 are colocalized on chromosomal region 11q13. Coamplification of 11q13 and 8p12 has been observed in some breast cancer cell lines and primary breast tumors.24-26 Chromosomal colocalization and, often, coamplification of genes that encode mTOR substrates, FGFR family members, and cyclin D1 may provide an intriguing explanation for the observed lessened PFS benefit with everolimus in FGFR1-amplified tumors. Consistent with our observation, a recent report suggests that TKI258, an FGFR inhibitor, produced greater disease stabilization in patients with HR+, HER2−, FGFR1-amplified breast cancer than in those with wild-type FGFR1.27 Similarly, in a phase I study of E-3810, an FGFR1 and VEGFR inhibitor, four of eight patients with breast cancer (mostly ER+, HER2−) with FGFR1 or FGF4 amplification had a sustained partial response.28 These observations may support an exploration of combination therapies in the subset of HR+, HER2− breast cancer with altered FGFRs to further improve clinical effectiveness.
Chromosomal instability has been associated with poor clinical outcome and treatment response in a number of solid and hematologic malignancies.16,29 Limited studies demonstrated that lower chromosomal instability might be associated with a better outcome with taxanes in the adjuvant setting in ER+ breast cancer.30 Our analysis demonstrated, for the first time, a potential correlation between the efficacy of a targeted therapy and chromosomal instability. Although experimental evidence is awaited to demonstrate a direct mechanistic link between more stable chromosomes and higher sensitivity to everolimus, a plausible explanation may be that higher chromosomal instability leads to more tumor heterogeneity, giving rise to polyclonal populations in tumors and thereby making them prone to treatment resistance. In addition, the placebo-controlled trial design enabled us to conclude that the CIN score did not equally affect exemestane monotherapy, and, therefore, it was likely to be associated with everolimus efficacy.
Various metrics for estimating chromosomal instability have been previously developed based on gene-expression signatures and genome-wide mutation profiles of tumors; examples include CINGEC (Chromosomes Instability Genome Event Count), GII (Genome Instability Index), CIN70 (70-Gene Chromosomal Instability), and CINSARC (Complexity Index in Sarcoma).15,17,31-35 Although the whole-genome and transcriptome assays are the presently preferred methods for measurement of chromosomal instability, development of an equally valid scoring metric by using data acquired from a medium-sized, pan-cancer gene panel such as ours provides a beneficial expansion of the utility of the NGS data increasingly collected in oncology practice. By comparing CIN scores derived from whole-exome and gene subset data in the ER+, HER2− breast cancer cohort from TCGA, we demonstrated that chromosomal instability can be estimated by using CIN scores calculated with data from a targeted gene panel of random gene compositions (Data Supplement). Furthermore, substantial enrichment of mutations in p53 pathway genes in the high–CIN score subgroup supports the observations in the literature associating loss-of-function of TP53 with higher chromosomal instability in tumors36-38 (Data Supplement). With these additional assessments, we believe that the estimation of chromosomal instability using CIN score based on data from a targeted gene panel can be potentially useful for routine application in exploring predictive signals for drug efficacy. It is also worth pointing out that, although CIN scores are correlated with mutations in genes in the p53 pathway or the number of genetic alterations in four common oncogenic drivers, FGFR1/2, PIK3CA, and CCND1 (Data Supplement), only a moderately reduced PFS benefit from everolimus was observed in patient subgroups defined by mutations in p53 pathway genes or by having at least two genetic alterations in the four aforementioned genes (Data Supplement). We believe that the observation suggests that, as a generic scoring scheme, the CIN score accounts for genetic alterations of all frequencies in an unbiased fashion. However, the CIN score needs further validation on additional patient cohorts before it is applied in clinical settings.
The strengths of this exploratory biomarker analysis include the ability to generate NGS data from a large subset of representative patients from the randomized, placebo-controlled trial and to assess their effect on clinical benefit by testing single and multiple biomarker hypotheses and by using a CIN score derived from broad genetic alterations. Nonetheless, at least two methodologic limitations and challenges, likely of general implication in this type of analysis, are worth note. First, subgroup analysis based on low-frequency genetic alterations suffers from statistical challenges and the risks of overfitting the biomarkers to the trial population, as exemplified by the lack of statistically significant signals in the correlative analyses of PFS and mTOR, FGFR1, FGFR2, and PIK3CA exon 9– and exon 20–specific mutations. To circumvent this inherent limitation of NGS-based biomarker analysis, successful correlative analyses require development of methods for effectively clustering genetic variations in a hypothesis-free fashion, such as assessing overall genomic instability of a tumor sample. These will enable inclusion of genes that may not fit any obvious biologic hypothesis or have low population frequency, revealing new biologic mechanisms or ascertainment of multiple genetic lesions. Second, definitive identification of somatic alterations and, more importantly, evaluation of the functional impact of many somatic mutations, especially missense mutations, are critical for effective usage of genetic data in correlative analysis. Because the exact functional-effect evaluations often depend on time- and resource-demanding laboratory experiments, especially for genes that have a broad mutation spectrum or mutations affecting different protein function domains (eg, TSC1/2), we expect this to be one of the major limiting factors for correlative analyses based on broad NGS data in the coming years.
In summary, we have demonstrated that, in BOLERO-2, the efficacy of everolimus was well maintained in patient subgroups defined by their tumors' status of the most commonly altered genes and/or pathways. Our results, however, also revealed some potential quantitative differences in the efficacy of everolimus among patients with tumors of different genetic status in specific PIK3CA exons, FGFR2, or mTOR, as well as with different degrees of chromosomal instabilities. The biologic and therapeutic implications of these observations and the methodologic issues raised in this analysis should be investigated further.
Supplementary Material
Acknowledgment
We thank all the patients enrolled onto BOLERO-2 who consented for this exploratory analysis and who provided tumor tissues. We also acknowledge our colleagues Adnan Derti, Creton Kalfoglou, Michael Morrissey, and Francois Ringeisen for their contribution to this work or their critical input on this article. Foundation Medicine performed next-generation sequencing. We thank Jerome F. Sah and Shalini Murthy (ProEd Communications) for editorial assistance.
GLOSSARY TERMS
- everolimus:
see RAD001.
- fibroblast growth factor receptor (FGRF):
a family of tyrosine kinase receptors that has four members: FGFR1 to 4. Like other tyrosine kinase receptors, intracellular signaling follows receptor dimerization in response to ligand binding. This leads to autophosphorylation of the tyrosine molecules in the intracellular tyrosine kinase region of the receptor.
- gene amplification:
the presence of multiple copies of a gene or genes that leads to overexpression of that gene or gene.
- genomic:
the scientific discipline in which multiple genes, gene products, or regions of the genome are analyzed by using large-scale, high-throughput molecular approaches directed at DNA and RNA. This definition is a deviation from that of the original term, which meant an analysis of the whole genome.
- mammalian target of rapamycin (mTOR):
a member of a protein complex, along with raptor and GβL, that is used by cells to sense nutrients in the environment. mTOR is a serine or threonine kinase that is activated by AKT and that regulates protein synthesis on the basis of nutrient availability. It was discovered when rapamycin, a drug used in transplantation, was shown to block cell growth presumably by blocking the action of mTOR.
- NextGen Sequencing:
a non-Sanger rapid DNA sequencing method that can be done with greater speed, developed after the first methodologic articles describing relatively rapid DNA sequencing produced by Sanger et al (1977).
- PI3K/PTEN/AKT pathway:
a signal transduction pathways involving the signaling molecules PI3K, PTEN, and AKT. PI3K generates phosphorylated inositides at the cell membrane, which are required for the recruitment and activation of the serine kinase AKT. PTEN is a lipid phosphatase that counteracts the effect of PI3K. Accordingly, mutated PI3K and AKT act as dominant oncogenes, whereas PTEN is a tumor suppressor gene.
- RAD001:
an orally active derivative of rapamycin, RAD001 (also known as everolimus) is an inhibitor of mammalian target of rapamycin. See mammalian target of rapamycin (mTOR).
- somatic mutation:
a change in the genotype of a cancer cell. This is distinguished from a germline mutation, which is a change in the genotype of all the normal cells in a patient's body. Germline mutations may be passed to offspring, but somatic mutations may not.
Footnotes
See accompanying editorial on page 393
Supported by Novartis Pharmaceuticals (for medical editorial assistance).
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
Presented in part at the 49th Annual Meeting of the American Society of Clinical Oncology, Chicago, IL, May 31-June 4, 2013.
Authors' disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
Clinical trial information: NCT00863655.
AUTHOR CONTRIBUTIONS
Conception and design: Gabriel N. Hortobagyi, David Chen, Martine Piccart, Hope S. Rugo, Shaker Dakhil, Alan Huang, Tetiana Taran, David Lebwohl, José Baselga
Provision of study materials or patients: Gabriel N. Hortobagyi, Martine Piccart, Hope S. Rugo, Howard A. Burris III, Mario Campone, Fabienne Lebrun, José Baselga
Collection and assembly of data: David Chen, Hope S. Rugo, Howard A. Burris III, Alejandra T. Perez, Ines Deleu, Mikhail Shtivelband, Norikazu Masuda, Shaker Dakhil, Ian Anderson, Douglas M. Robinson, Tetiana Taran, Thomas Bachelot, Fabienne Lebrun
Data analysis and interpretation: Gabriel N. Hortobagyi, David Chen, Martine Piccart, Hope S. Rugo, Howard A. Burris III, Kathleen I. Pritchard, Mario Campone, Shinzaburo Noguchi, Alejandra T. Perez, Norikazu Masuda, Shaker Dakhil, Douglas M. Robinson, Wei He, Abhishek Garg, E. Robert McDonald III, Hans Bitter, Alan Huang, Tetiana Taran, David Lebwohl
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Correlative Analysis of Genetic Alterations and Everolimus Benefit in Hormone Receptor–Positive, Human Epidermal Growth Factor Receptor 2–Negative Advanced Breast Cancer: Results From BOLERO-2
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Gabriel N. Hortobagyi
Consulting or Advisory Role: Pfizer, Antigen Express, Novartis, Galena, Genentech, Amgen
Research Funding: Novartis (Inst)
Travel, Accommodations, Expenses: Novartis
David Chen
Employment: Novartis Pharmaceuticals
Stock or Other Ownership: Novartis Pharmaceuticals
Patents, Royalties, Other Intellectual Property: Novartis Pharmaceuticals
Martine Piccart
Consulting or Advisory Role: Amgen, Astellas Pharma, AstraZeneca, Bayer, Eli Lilly, Invivis, MSD, Novartis, Pfizer, Genentech, sanofi-aventis, Symphogen, Synthon, Verastem
Research Funding: Amgen (Inst), Astellas Pharma (Inst), AstraZeneca (Inst), Bayer (Inst), Eli Lilly (Inst), Invivis (Inst), MSD (Inst), Novartis (Inst), Pfizer (Inst), Genentech (Inst), sanofi-aventis (Inst), Symphogen (Inst), Synthon (Inst), Verastem (Inst)
Hope S. Rugo
Research Funding: Novartis Pharmaceuticals (Inst), Pfizer (Inst), Eli Lilly (Inst), Genentech (Inst)
Travel, Accommodations, Expenses: Novartis, Pfizer, Genentech
Howard A. Burris III
No relationship to disclose
Kathleen I. Pritchard
Honoraria: sanofi-aventis, AstraZeneca, Pfizer, Roche, Amgen, Novartis, GlaxoSmithKline, Boehringer Ingelheim, Genomic Health, Eisai
Consulting or Advisory Role: sanofi-aventis, AstraZeneca, Pfizer, Roche, Amgen, Novartis, GlaxoSmithKline, Boehringer Ingelheim, Genomic Health, Eisai
Travel, Accommodations, Expenses: sanofi-aventis, AstraZeneca, Pfizer, Roche, Amgen, Novartis, GlaxoSmithKline, Boehringer Ingelheim, Genomic Health, Eisai
Mario Campone
Honoraria: Novartis, Servier, Menarini, AstraZeneca
Consulting or Advisory Role: Novartis, SERVIER, Menarini
Speakers' Bureau: Novartis, Amgen
Research Funding: Novartis (Inst)
Travel, Accommodations, Expenses: Novartis
Shinzaburo Noguchi
Honoraria: Novartis, AstraZeneca, Taiho Pharmaceutical, Takeda, Daiichi Sankyo, sanofi-aventis, Pfizer, Chugai, Eisai, Sysmex
Consulting or Advisory Role: Novartis, AstraZeneca, Taiho Pharmaceutical
Research Funding: Novartis, AstraZeneca, Taiho Pharmaceutical, Takeda, Daiichi Sankyo, sanofi-aventis, Bristol-Myers Squibb, Pfizer, Eisai, Chugai, Sysmex
Patents, Royalties, Other Intellectual Property: Sysmex
Alejandra T. Perez
Research Funding: Novartis (Inst), Celgene (Inst), Genentech (Inst), Duke University Medical Center (Inst)
Ines Deleu
Research Funding: Novartis (Inst), Amgen (Inst), Roche (Inst), Vifor Pharma (Inst)
Travel, Accommodations, Expenses: Novartis
Mikhail Shtivelband
No relationship to disclose
Norikazu Masuda
Honoraria: Chugai, Eisai, AstraZeneca
Shaker Dakhil
No relationship to disclose
Ian Anderson
No relationship to disclose
Douglas M. Robinson
Employment: Novartis Pharmaceuticals
Stock or Other Ownership: Novartis Pharmaceuticals
Patents, Royalties, Other Intellectual Property: Novartis Pharmaceuticals
Wei He
Employment: Novartis Pharmaceuticals
Abhishek Garg
Employment: Novartis Institute of Biomedical Research
E. Robert McDonald III
Employment: Novartis Institute for BioMedical Research
Stock or Other Ownership: Novartis
Research Funding: Novartis
Patents, Royalties, Other Intellectual Property: Novartis
Hans Bitter
Employment: Novartis
Stock or Other Ownership: Novartis
Alan Huang
Employment: Novartis
Stock or Other Ownership: Novartis
Tetiana Taran
Employment: Novartis
Stock or Other Ownership: Novartis
Thomas Bachelot
Consulting or Advisory Role: Roche, Novartis
Research Funding: Roche (Inst), Novartis (Inst)
Travel, Accommodations, Expenses: Roche, Novartis
Fabienne Lebrun
No relationship to disclose
David Lebwohl
Employment: Novartis Pharmaceuticals
Stock or Other Ownership: Novartis
José Baselga
Honoraria: Novartis, Infinity
Consulting or Advisory Role: Novartis, Verastem, Infinity
REFERENCES
- 1.Baselga J, Campone M, Piccart M, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbareschi M, Buttitta F, Felicioni L, et al. Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin Cancer Res. 2007;13:6064–6069. doi: 10.1158/1078-0432.CCR-07-0266. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dumont AG, Dumont SN, Trent JC. The favorable impact of PIK3CA mutations on survival: An analysis of 2587 patients with breast cancer. Chin J Cancer. 2012;31:327–334. doi: 10.5732/cjc.012.10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellis MJ, Ding L, Shen D, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heikkinen T, Greco D, Pelttari LM, et al. Variants on the promoter region of PTEN affect breast cancer progression and patient survival. Breast Cancer Res. 2011;13:R130. doi: 10.1186/bcr3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones N, Bonnet F, Sfar S, et al. Comprehensive analysis of PTEN status in breast carcinomas. Int J Cancer. 2013;133:323–334. doi: 10.1002/ijc.28021. [DOI] [PubMed] [Google Scholar]
- 8.Loi S, Haibe-Kains B, Majjaj S, et al. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor-positive breast cancer. Proc Natl Acad Sci U S A. 2010;107:10208–10213. doi: 10.1073/pnas.0907011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stephens PJ, Tarpey PS, Davies H, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toy W, Shen Y, Won H, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45:1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyer G, Hanrahan AJ, Milowsky MI, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voss MH, Hakimi AA, Pham CG, et al. Tumor genetic analyses of patients with metastatic renal cell carcinoma and extended benefit from mTOR inhibitor therapy. Clin Cancer Res. 2014;20:1955–1964. doi: 10.1158/1078-0432.CCR-13-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ades F, Zardavas D, Bozovic-Spasojevic I, et al. Luminal B breast cancer: Molecular characterization, clinical management, and future perspectives. J Clin Oncol. 2014;32:2794–2803. doi: 10.1200/JCO.2013.54.1870. [DOI] [PubMed] [Google Scholar]
- 15.Carter SL, Eklund AC, Kohane IS, et al. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 16.Ott K, Vogelsang H, Mueller J, et al. Chromosomal instability rather than p53 mutation is associated with response to neoadjuvant cisplatin-based chemotherapy in gastric carcinoma. Clin Cancer Res. 2003;9:2307–2315. [PubMed] [Google Scholar]
- 17.Szasz AM, Li Q, Eklund AC, et al. The CIN4 chromosomal instability qPCR classifier defines tumor aneuploidy and stratifies outcome in grade 2 breast cancer. PLoS One. 2013;8:e56707. doi: 10.1371/journal.pone.0056707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juric D, Baselga J. Tumor genetic testing for patient selection in phase I clinical trials: The case of PI3K inhibitors. J Clin Oncol. 2012;30:765–766. doi: 10.1200/JCO.2011.39.6390. [DOI] [PubMed] [Google Scholar]
- 19.Baselga J, Semiglazov V, van Dam P, et al. Phase II randomized study of neoadjuvant everolimus plus letrozole compared with placebo plus letrozole in patients with estrogen receptor-positive breast cancer. J Clin Oncol. 2009;27:2630–2637. doi: 10.1200/JCO.2008.18.8391. [DOI] [PubMed] [Google Scholar]
- 20.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burke JE, Perisic O, Masson GR, et al. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110alpha (PIK3CA) Proc Natl Acad Sci U S A. 2012;109:15259–15264. doi: 10.1073/pnas.1205508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jang MH, Kim EJ, Choi Y, et al. FGFR1 is amplified during the progression of in situ to invasive breast carcinoma. Breast Cancer Res. 2012;14:R115. doi: 10.1186/bcr3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner N, Pearson A, Sharpe R, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70:2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlsson E, Waltersson MA, Bostner J, et al. High-resolution genomic analysis of the 11q13 amplicon in breast cancers identifies synergy with 8p12 amplification, involving the mTOR targets S6K2 and 4EBP1. Genes Chromosomes Cancer. 2011;50:775–787. doi: 10.1002/gcc.20900. [DOI] [PubMed] [Google Scholar]
- 25.Kwek SS, Roy R, Zhou H, et al. Co-amplified genes at 8p12 and 11q13 in breast tumors cooperate with two major pathways in oncogenesis. Oncogene. 2009;28:1892–1903. doi: 10.1038/onc.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paterson AL, Pole JC, Blood KA, et al. Co-amplification of 8p12 and 11q13 in breast cancers is not the result of a single genomic event. Genes Chromosomes Cancer. 2007;46:427–439. doi: 10.1002/gcc.20424. [DOI] [PubMed] [Google Scholar]
- 27.Andre F, Bachelot T, Campone M, et al. Targeting FGFR with dovitinib (TKI258): Preclinical and clinical data in breast cancer. Clin Cancer Res. 2013;19:3693–3702. doi: 10.1158/1078-0432.CCR-13-0190. [DOI] [PubMed] [Google Scholar]
- 28.Dienstmann R, Andre F, Soria J, et al. Significant antitumor activity of E-3810, a novel FGFR and VEGFR inhibitor, in patients with FGFR1 amplified breast cancer. Ann Oncol. 2012;23:ix116–ix117. (abstr 319O) [Google Scholar]
- 29.Burrell RA, Juul N, Johnston SR, et al. Targeting chromosomal instability and tumour heterogeneity in HER2-positive breast cancer. J Cell Biochem. 2010;111:782–790. doi: 10.1002/jcb.22781. [DOI] [PubMed] [Google Scholar]
- 30.A'Hern RP, Jamal-Hanjani M, Szasz AM, et al. Taxane benefit in breast cancer–a role for grade and chromosomal stability. Nat Rev Clin Oncol. 2013;10:357–364. doi: 10.1038/nrclinonc.2013.67. [DOI] [PubMed] [Google Scholar]
- 31.Chin SF, Teschendorff AE, Marioni JC, et al. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 2007;8:R215. doi: 10.1186/gb-2007-8-10-r215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung TH, Mulligan G, Fonseca R, et al. A novel measure of chromosome instability can account for prognostic difference in multiple myeloma. PLoS One. 2013;8:e66361. doi: 10.1371/journal.pone.0066361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagarde P, Perot G, Kauffmann A, et al. Mitotic checkpoints and chromosome instability are strong predictors of clinical outcome in gastrointestinal stromal tumors. Clin Cancer Res. 2012;18:826–838. doi: 10.1158/1078-0432.CCR-11-1610. [DOI] [PubMed] [Google Scholar]
- 34.Chibon F, Lagarde P, Salas S, et al. Validated prediction of clinical outcome in sarcomas and multiple types of cancer on the basis of a gene expression signature related to genome complexity. Nat Med. 2010;16:781–787. doi: 10.1038/nm.2174. [DOI] [PubMed] [Google Scholar]
- 35.Lagarde P, Przybyl J, Brulard C, et al. Chromosome instability accounts for reverse metastatic outcomes of pediatric and adult synovial sarcomas. J Clin Oncol. 2013;31:608–615. doi: 10.1200/JCO.2012.46.0147. [DOI] [PubMed] [Google Scholar]
- 36.Campomenosi P, Assereto P, Bogliolo M, et al. p53 mutations and DNA ploidy in colorectal adenocarcinomas. Anal Cell Pathol. 1998;17:1–12. doi: 10.1155/1998/396371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dalton WB, Yu B, Yang VW. p53 suppresses structural chromosome instability after mitotic arrest in human cells. Oncogene. 2010;29:1929–1940. doi: 10.1038/onc.2009.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao C, Deng L, Henegariu O, et al. Chromosome instability contributes to loss of heterozygosity in mice lacking p53. Proc Natl Acad Sci U S A. 2000;97:7405–7410. doi: 10.1073/pnas.97.13.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.