Abstract
West syndrome (WS), defined by the triad of infantile spasms, pathognomonic hypsarrhythmia and developmental regression, is a rare epileptic disease affecting about 1:3500 live births. To get better insights on the genetic of this pathology, we exome-sequenced the members of a consanguineous family affected with isolated WS. We identified a homozygous variant (c.1825G>T/p.(Ala609Ser)) in the GUF1 gene in the three affected siblings. GUF1 encodes a protein essential in conditions that counteract faithful protein synthesis: it is able to remobilize stuck ribosomes and transiently inhibit the elongation process to optimize protein synthesis. The variant identified in the WS family changes an alanine residue conserved in all eukaryotic organisms and positioned within the tRNA-binding moiety of this nuclear genome-encoded mitochondrial translational elongation factor. Yeast complementation assays show that the activity of GUF1A609S is modified in suboptimal environments. We suggest a new link between improper assembly of respiratory chain complexes and WS.
Introduction
Infantile spasms (IS) are an age-dependant epileptic syndrome characterized by clinical spasms occurring in clusters and abnormal pattern on electroencephalography (EEG).1, 2 Their incidence ranges from 2 to 5/10 000 per live births.3 West syndrome (WS) is one of the well-defined epileptic syndromes. It associates IS with a typical EEG pattern called hypsarrhythmia consisting of random, high-voltage, nonsynchronous spikes and slow waves and developmental delay. Despite improvements of early diagnosis and appropriate treatment, prognosis of IS/WS remains poor; 75–90% of patients show cognitive impairment, while 50–60% of affected children have recurrent seizures at 5 years of age.4, 5 The etiology of IS/WS is highly heterogeneous.6 They occur as isolated feature or as part of more complex disease syndrome. They typically are the consequence of acquired events, such as hypoxemia or intracranial hemorrhage, or determined by genetic factors. The most frequent genetic causes of IS are mutations in TSC1 (about 9% of all IS/WS patients, OMIM#191100), CDKL5, ARX and STXBP1 genes, as well as multiple genomic imbalances, the commonest being Pallister–Killian syndrome (tetrasomy 12p; OMIM#601803) and 1p36 deletion (OMIM#607872).7, 8, 9, 10, 11, 12, 13 These and other genetic causes such as genes encoding proteins involved in inborn errors of metabolism were recently reviewed in Paciorkowski et al.14 IS/WS also occur with a lower prevalence as a feature of other genetic diseases, such as Down syndrome.14 This heterogeneity often impedes the identification of the etiology of IS/WS within the clinical practice.15 Despite early clinical diagnosis and appropriate treatment, prognosis remains a major concern with frequent intellectual deficiency (75–90%) and recurrence of seizures later in life (50–60%).16 Definite etiological diagnosis can give information about the prognosis and is mandatory for accurate genetic counseling. Recent studies based on whole-exome sequencing identified de novo genetic alterations in GABRB3, CACNA1A, CHD2, FLNA, GABRA1, GRIN1, GRIN2B, HNRNPU, IQSEC2, MTOR, NEDD4L, STXBP1, CASK and ALG13 and recessive variants in PNPO and ADSL.17, 18 These studies suggested that many other genes underlying IS/WS remain to be found.
We report on a consanguineous family with three siblings (a female and two males) affected by WS and severe neurological impairment. They had no malformations or other clinical features, and both biochemical screening and array comparative genomic hybridization results were normal. The family structure prompted us to use whole-exome sequencing to find the causative gene.
Materials and methods
Detailed patients' description
Familial history
There was a family history of death, in the first week of life of a paternal aunt and uncle from unknown causes, and of epilepsy without developmental delay in a different paternal aunt and a paternal cousin.
Patient IV-1
This boy was born at the thirty-sixth week of gestation after an uneventful pregnancy, with weight 2670 g (−1.5 SD), length 42 cm (−3 SD) and occipital frontal circumference (OFC) 36 cm (+2 SD). He had hyaline membrane disease with favorable outcome but had feeding difficulties and periods of lethargy. EEG and transcranial sonography were normal at birth. At 5 months of age, systematic pediatric examination disclosed severe hypotonia with poor eye contact that had not been noticed by the parents. EEG showed hypsarrhythmia and electroclinical spasms were video-recorded (Supplementary Figure S1 and S2). He had no dysmorphic features. Multiple simultaneous or successive antiepileptic drug attempts (Vigabatrin, Prednisolone, Tetracosactide, Nitrazepam, Phenytoine, Topiramate, Levetiracetam, Lamotrigine, Oxcarbamazepine, Rufinamide), as well as Pyridoxal and Pyridoxal phosphate, were unsuccessful, and evolution was marked by persistence of spasms and myoclonia. Neurological outcome was also poor. At last follow-up, at 6 years of age, he had profound psychomotor delay with no babbling, severe axial hypotonia with spastic tetraparesis, no ability to handle or grasp objects and he displayed some dystonic fits and choreoathetotic movements. Eye contact was partially preserved.
Brain MRI performed at epilepsy onset was normal, but diffuse cortical atrophy was present at 2 years of age (Supplementary Figure S3). Electromyography and fundoscopy did not reveal any anomaly. Blood karyotype did not show any chromosomal imbalance. Metabolic dosages in blood included: ammonemia, lactate, pyruvate, amino-acid chromatography, organic acid chromatography, pipecolic acid, SAICAR (succinyl-5-aminoimidazole-4-carboxamide-1-ribose-5′-phosphate), testing for congenital disorders of glycosylation, dosages of beta-glycosydase (GM1) and beta-hexosaminidase (GM2) in leucocytes, dosages of palmitoyl thio-esterase-1 and tripeptidyl peptidase-1 in leukocytes were within normal range. In the CSF, glucose, lactate (1050 μmol/l) and pyruvate (67 μmol/l) dosages were normal, whereas dosages of neurotransmitter metabolite analysis showed a slight decreased level of homo vanillic acid, which was considered non-significant (HVA: 258 nmol/l (normal range: 429–789 nmol/l), MHPG: 54 nmol/l (normal range: 33–71 nmol/l), HVA/MHPG ratio: 4.8 (normal range: 5–15)). No variants were found in the CDKL5 and ARX genes.
Patient IV-2
Patient IV-1's sister had intrauterine growth retardation. She was born at the thirty-fifth week of gestation by cesarean section after an uneventful pregnancy, with weight 1400 g (<−3 SD), length 44 cm (−2 SD), and OFC 29.5 cm (−2.5 SD). The first days of life were characterized by hyaline membrane disease with favorable outcome. Neonatal EEG was performed because of the early onset of epilepsy in the previous child; cranial ultrasonography and fundoscopy were normal. Careful pediatric follow-up was performed and slight hypotonia with good eye contact was noticed before the first epileptic manifestations. Spasms onset started at 5 months of life. EEG showed hypsarrhythmia, and electroclinical spasms were recorded (Supplementary Figure S1). Epilepsy onset was associated with psychomotor arrest. At examination, she displayed axial hypotonia, alteration of eye contact and no dysmorphic features. Brain MRI was normal. Lymphocyte karyotype, mutation screening of the ARX gene, CSF lactate and pyruvate dosages were normal (lactate: 1110 μmol/l, pyruvate: 85 μmol/l, lactate/pyruvate ratio: 13), and CSF neurotransmitter metabolite analysis showed slight decreased level of vanillic acid, which was considered non-significant (HVA: 283 nmol/l (normal range: 295–932 nmol/l), MHPG: 51 nmol/l (normal range: 4–50 nmol/l), HVA/MHPG ratio: 1.2 (normal range: 1.11–3.48)). No antiepileptic treatment (Vigabatrin, Clonazepam, Prednisolone and Levetiracetam) was effective. Neurological outcome was very poor. She died at 1 year of age from SUDEP (sudden unexpected death in epilepsy) during a H1N1 virus and respiratory syncytial virus co-infection.
Patient IV-3
Patient IV-1's younger brother was born at the thirty-seventh week of gestation after an uneventful pregnancy, with weight 3350 g (mean), length 50 cm (+1 SD) and OFC 35.5 cm (+1 SD). Hypotonia and psychomotor delay had been noticed since the first days of life with preserved eyes contact. Neonatal transcranial sonography and EEG were normal. First spasms occurred when he was 4 months with psychomotor delay. EEG showed hypsarrythmia and electroclinical spasms were recorded (Supplementary Figure S1). He had no dysmorphic feature. Epilepsy could not be controlled despite treatment with Vigabatrin, and neurological outcome was poor. At last follow-up, he was 15 months of age, spasms and myoclona occurred several times per day. He displayed severe hypotonia with slight spasticity, fluctuating eye contact, no babbling and no ability to handle or grasp objects. No dystonic fit was noticed until now. Lymphocyte karyotype, array CGH, blood amino-acid chromatography and CSF lactate and pyruvate dosages (lactate: 1090 μmol/l, pyruvate: 101 μmol/l, lactate/pyruvate ratio: 11). CSF neurotransmitter metabolite analysis showed normal values: HVA 528 nmol/l (normal range: 310–1328 nmol/l), MHPG 66 nmol/l (normal range: 30–168 nmol/l), HVA/MHPG ratio: 8 (normal range: 5–15). No brain MRI was performed.
Exome sequencing and analysis
All samples used in this study were collected with appropriate informed consent and approval of the local ethics committees. We performed exome sequencing on both parents and the three affected siblings. Exomes were captured using the Agilent Sureselect Human All Exon V4 Enrichment Kit (Santa Clara, CA, USA) and multiplex sequenced on two Illumina Hiseq lanes (San Diego, CA, USA). Sequencing reads were aligned to the reference human genome using BWA,19 and variants were called with GATK20 using a Minimum Confidence Calling and Emitting score of 30, Base Quality Score of 17 and Mapping Quality score of 20. Prior to variant calling, each sample was screened for duplicates using PICARD tools (http://picard.sourceforge.net), base quality score recalibration and InDel realignment with GATK.21 We reached a mean bait coverage of 87-fold, with 76% of the nucleotides with at least a 20-fold coverage on average. We filtered out synonymous variant, as well as the variants previously recorded in dbSNP, the 1000 genome project, the HapMap projects and our in-house exome variant server (>1000 exomes performed) with MAF>0.1%. The remaining variants were sorted according to their inheritance pattern with VCFtools22 and Snpeff (http://snpeff.sourceforge.net/). The c.1825G>T, p.(Ala609Ser) variant was submitted to the GUF1 locus-specific database of the Leiden Open Variation Database (www.lovd.nl/GUF1). Of note, the Exome Aggregation Consortium (ExAC), Cambridge, MA, USA (http://exac.broadinstitute.org) (accessed March 2015) uncovered 288 missense or loss-of-function variants in GUF1 of which only chr4:44691873 G>C p.(Gly406Arg) was (i) deleterious according to SIFT or Polyphen; (ii) not found in homozygosity; and (iii) evolutionary constrained. We acknowledge that we could, however, not exclude cases of compound heterozygosity.
Yeast complementation
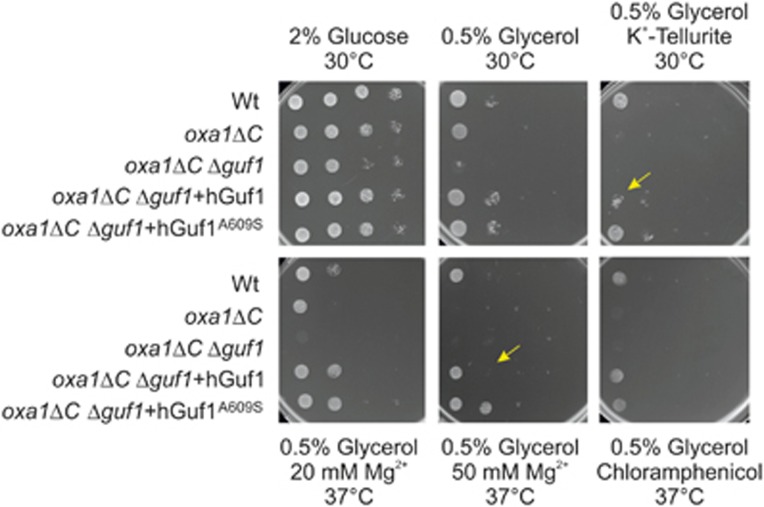
We used cDNA from a WS patient and from a control as template to amplify the coding sequences of the normal and variant human GUF1, hGUF1 and hGUF1A609S, respectively. These sequences were cloned into the pYX232 yeast expression plasmid (Novagen, EMD Millipore, Billerica, MA, USA) under control of a constitutive triosephosphate isomerase promoter. These plasmids were transformed into Δguf1 yeast strains. The resulting strains were grown to logarithmic growth phase, and serial dilutions were spotted onto agar plates containing 0.5% glycerol. Upon incubation at elevated growth temperatures (37 °C), the Δguf1 strains showed a mild growth defect confirming earlier studies.23 The growth defect was further aggravated by overexpression of either hGUF1 or hGUF1A609S, indicating that both variants interfere with mitochondrial translation machinery with similar degrees. Next we evaluated whether the expression of the human Guf1 proteins influences mitochondrial protein expression. We isolated mitochondria by subcellular fractionation of control, Δguf1 and Δguf1-overexpressing hGUF1 or hGUF1A609S cells and analyzed the levels of the mitochondrially encoded protein Cox2. As Cox2 is a central part of cytochrome c oxidase (Complex IV), a decrease of its level would indicate a severe defect in respiration. We found, however, no changes in the levels of the Cox2 proteins. To assess mitochondrial protein synthesis more directly, we incubated mitochondria in the presence of ATP, amino acids and radiolabeled [35S]-methionine for 10 min at either 15 or 30 °C. Mitochondria were then reisolated, and newly synthesized translation products were visualized by autoradiography as described in Bauerschmitt et al.23 Again, no differences were observed between Δguf1 and Δguf1-overexpressing hGUF1 or hGUF1A609S cells, all strains showing a similar translation pattern. Whereas deletion of GUF1 leads to almost no detectable defects in yeast, Guf1 activity is critical in the oxa1ΔC mutant.23 This strain lacks part of the C-terminal ribosome-binding domain of the mitochondrial Oxa1 insertase (residues 332–402). It shows no obvious defect in the biogenesis of mitochondrial translation products24, 25 but exhibits a strong synthetic growth defect with Δguf1 at high temperatures and on non-fermentable carbon sources. We used this synthetic defect to analyze the activity of the human Guf1 variant protein in yeast mitochondria. To this end, we transformed the oxa1ΔC Δguf1 double mutant with plasmids expressing either hGuf1 or hGuf1A609S and tested the growth of the resulting strains in a drop dilution assay. The strong synthetic effect observed with the oxa1ΔC Δguf1 double mutant at higher temperatures was completely suppressed upon expression of both hGuf1 and hGuf1A609S. This shows that both variant proteins, when expressed in yeast, are targeted to mitochondria where they are able to take over the function of yeast Guf1. To eventually detect subtle differences between hGuf1 or hGuf1A609S in the above complementation assays, we compared the fitness of oxa1ΔC Δguf1+hGuf1 and oxa1ΔC Δguf1+hGuf1A609S in a competition assay. Both strains were grown in the same culture over a 12-day period while the proportion of each mutant was assessed. To discriminate both strains, we transformed the oxa1ΔC Δguf1+hGuf1 cells with a plasmid harboring a uracil marker and the oxa1ΔC Δguf1+hGuf1A609S mutant with a plasmid containing a leucine marker. Both mutants were pregrown to log phase in glycerol-containing medium and inoculated at equal proportions into the common culture. The culture was repeatedly diluted to promote continuous cell growth. Aliquots were taken after 0, 3, 6, 9 and 12 days, and the proportion of each mutants was determined by plating different volumes on uracil- or leucine-deficient plates. Even after prolonged growth (equivalent to about 70 generations) and using an increased granularity of the measurement, the proportion of both strains remained equal, indicating that both mutants grew with identical rates. We then tested complementation of oxa1ΔC Δguf1 by the two variants in suboptimal environments, that is, 0.5% glycerol plates supplemented with potassium tellurite, magnesium or chloramphenicol (see main text for details).
Results
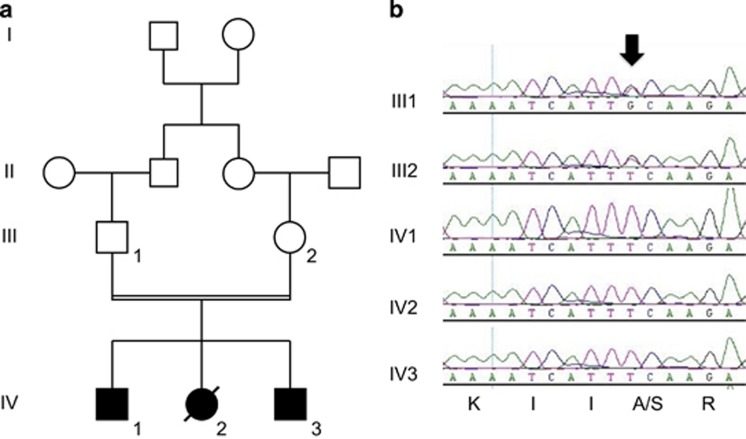
We describe the characterization of a consanguineous pedigree with three siblings affected with WS suggestive of a recessive monogenic disorder (Figure 1a). The parents were healthy first cousins from Algerian origin. There was no previous family history of severe epileptic or neurocognitive disorder. The phenotypes of the three siblings (and relatives) are summarized in Table 1. In summary, the three siblings had normal EEG and cranial ultrasonography at birth. Hypotonia preceded the first spasms, which set in at 5, 5 and 4 months of age in patient IV-1, IV-2 and IV-3, respectively. EEG at 5 months showed hypsarrhythmia, as well as electroclinical spasms in the three siblings (Supplementary Figures S1 and S2). Epilepsy onset was associated with psychomotor arrest and poor neurological outcome. Antiepileptic drug treatments, sometimes simultaneous or successive, were inefficient (Table 1). Levels of blood metabolites, as well as CSF lactate and pyruvate concentrations, were normal. We recorded, however, a slight decrease in homovanillic acid levels in patients IV-1 and IV-2. Brain MRI of patient IV-1 revealed a diffuse cortical atrophy (Supplementary Figure S3 and Table 1). Of note, patient IV-2 died at 1 year of age from SUDEP during a H1N1 virus and respiratory syncytial virus co-infection.
Figure 1.
(a) Four generation pedigree of the consanguineous family with three siblings affected with WS. (b) Sequencing fluorograms showing the alanine to serine variant of GUF1 codon 609 (black arrow) in the heterozygote and homozygote state in the two parents and their affected siblings, respectively. Codon translation is indicated at the bottom.
Table 1. Summary of phenotypic features presented by the three affected siblings.
| Patient | IV-1 | IV-2 | IV-3 |
|---|---|---|---|
| Sex | Male | Female | Male |
| Born after weeks of gestation (weeks) | 36 | 35a | 37 |
| Weight (g) | 2670 | 1400 | 3350 |
| Length (cm) | 42 | 44 | 50 |
| OFC (cm) | 36 | 29.5 | 35.5 |
| Hyaline membrane disease with favorable outcome | Yes | Yes | No |
| Neonatal EEG | Yes | Yes | No |
| Neonatal cranial ultrasonography | Normal | Normal | Normal |
| Spasms onset (months) | 5 | 5 | 4 |
| EEG at 5 months | Hypsarrhythmia and electroclinical spasms | Hypsarrhythmia and electroclinical spasms | Hypsarrhythmia and electroclinical spasms |
| Hypotonia | No hypotonia during the first months of life Severe hypotonia with poor eye contact at 5 months and at last follow-up | Slight hypotonia with good eye contact before first epileptic manifestations Severe hypotonia at last follow-up | Hypotonia with preserved eye contact since the first days of life Severe hypotonia at last follow-up |
| Unsuccesful epileptic treatment with | Multipleb | Multiplec | Vigabratin |
| Brain MRI | Normal at 5 months Major diffuse cortical atrophy at 2 years | Normal at 5 months | Not performed |
| Dysmorphic features | No | No | No |
| Karyotype | Normal | Normal | Normal |
| Array-CGH | Not performed | Not performed | Normal |
| Variants in ARX (Sanger seq) | None | None | Not performed |
| Variants in CDKL5 (Sanger seq) | None | Not performed | Not performed |
| Metabolic dosage in blood | Normal | Normal | Normal |
| Lactate and pyruvate in CSF | Normal | Normal | Normal |
| Homovanillic acid in CSF (nmol/l) | 258 | 283 | 528 |
By cesarean section.
Vigabatrin, Prednisolone, Tetracosactide, Nitrazepam, Phenytoine, Topiramate, Levetiracetam, Lamotrigine, Oxcarbamazepine, Rufinamide, as well as Pyridoxal and Pyridoxal phosphate.
Vigabatrin, Clonazepam, Prednisolone and Levetiracetam.
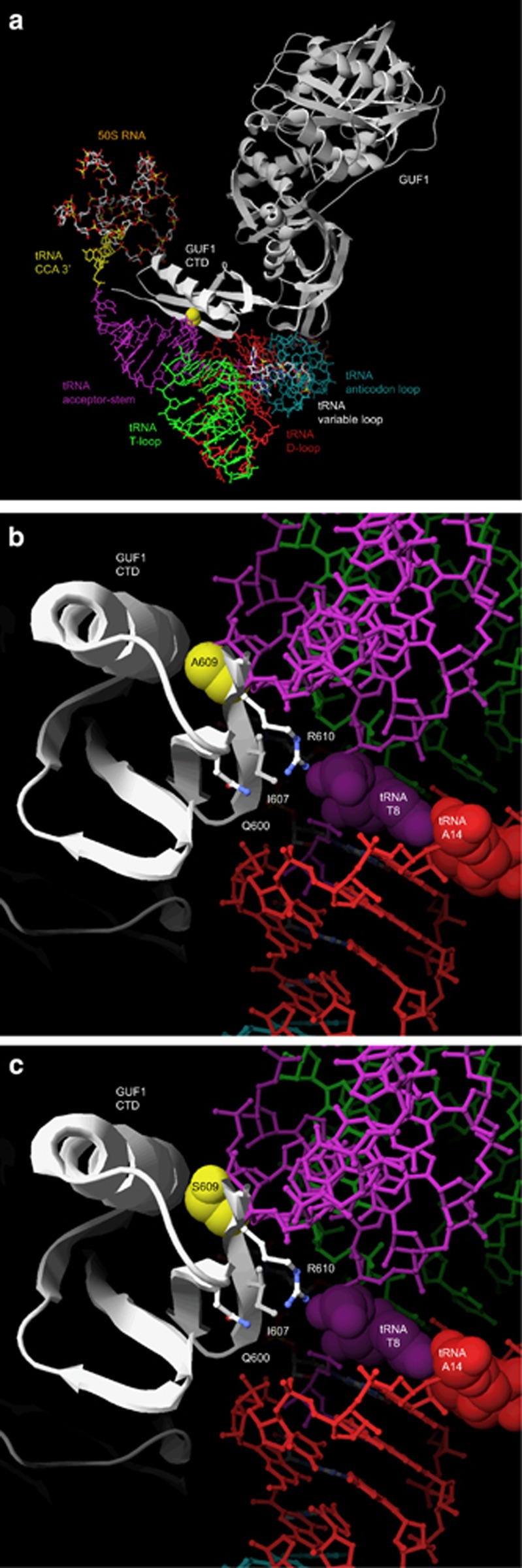
We performed exome sequencing on both parents and the three affected siblings. After filtering known and synonymous variants and sorting according to inheritance pattern, we found a single unknown variant that segregated with the disease and putatively affected the protein it encodes (SIFT score=0.05;26 Mutation Taster disease-causing prediction: P=1.0). This variant was homozygote in the three siblings and heterozygote in the parents and thus was compatible with an autosomal-recessive inheritance of the disease. We found no variant compatible with a de novo variant in the three affected assuming a possible germline mosaicism of one of the parents. The identified variant is a G to T transversion (chromosome 4 at nucleotide 44,697,741 [hg19]) in the 15th exon of the GUF1 gene (GTPase of unknown function 1; r.(2032g>u) (c.1825G>T) in NM_021927.2), which substitutes an alanine into a serine codon at position 609 (p.(Ala609Ser)). We confirmed by Sanger sequencing the heterozygosity and homozygosity of this variant in the parents and siblings, respectively (Figure 1b). GUF1 encodes a universally conserved GTPase in mitochondria and chloroplasts, as well as in prokaryotes where it is named EF4 or LepA. We used an X-ray structure of EF4, a cryo-EM model of EF4 bound to an elongating ribosome with a back-translocated tRNA and Swiss-PdbViewer27, 28, 29 to model a three-dimensional representation of the human GUF1 bound to a tRNA (Figure 2a). The variant we identified in the WS family substitutes an alanine residue of the C-terminal Domain (CTD) that is conserved in all eukaryotes from budding yeast to human and the majority of prokaryotes (A609; an A residue is found at this position in 497 out of 539 EF4/GUF1 alignments; Supplementary Figure S4). The CTD, which is not found in other translational GTPases,30 participates in the binding of the tRNA molecule via a beta-sheet28 (Figure 2b and c).
Figure 2.
(a) Three-dimensional representation of the human GUF1 (light gray) and its CTD (white) in contact with a tRNA in the A-site of the ribosome. The different domains of the tRNA are color-coded: CCA 3′ (yellow), acceptor stem (purple), T-loop (green), D-loop (red), and anticodon loop (turquoise). The A609 residue is highlighted in yellow. (b) Three-dimensional model of the GUF1 tRNA-binding moiety (white) bound to the tRNA acceptor stem (purple) and D-loop (red). The T8 and A14 nucleotidic positions that are unconstrained in human mitochondrial tRNAs are emphasized (see text for details). The alanine 609 residue (highlighted in yellow) is located on the external beta-strand within the KIIARETV stretch (defined as ‘contact 2' in Connell et al28), close to the evolutionary conserved glutamine (Q600), branched hydrophobic (I607) and Arginine (R610) residues. Their sidechains form a tightly packed motif that might be important for the proper recognition of the tRNA in the region located between the acceptor stem and the D-loop. The T-loop and part of the anticodon loop are indicated in green and turquoise, respectively. (c) The addition of a hydroxyl group at position 609 (p.(A609S) variant; compare with panel (b)) results in a bulkier residue, which might interfere with the tRNA acceptor stem by modifying the ‘contact 2' moiety. For color codes, see panel (b).
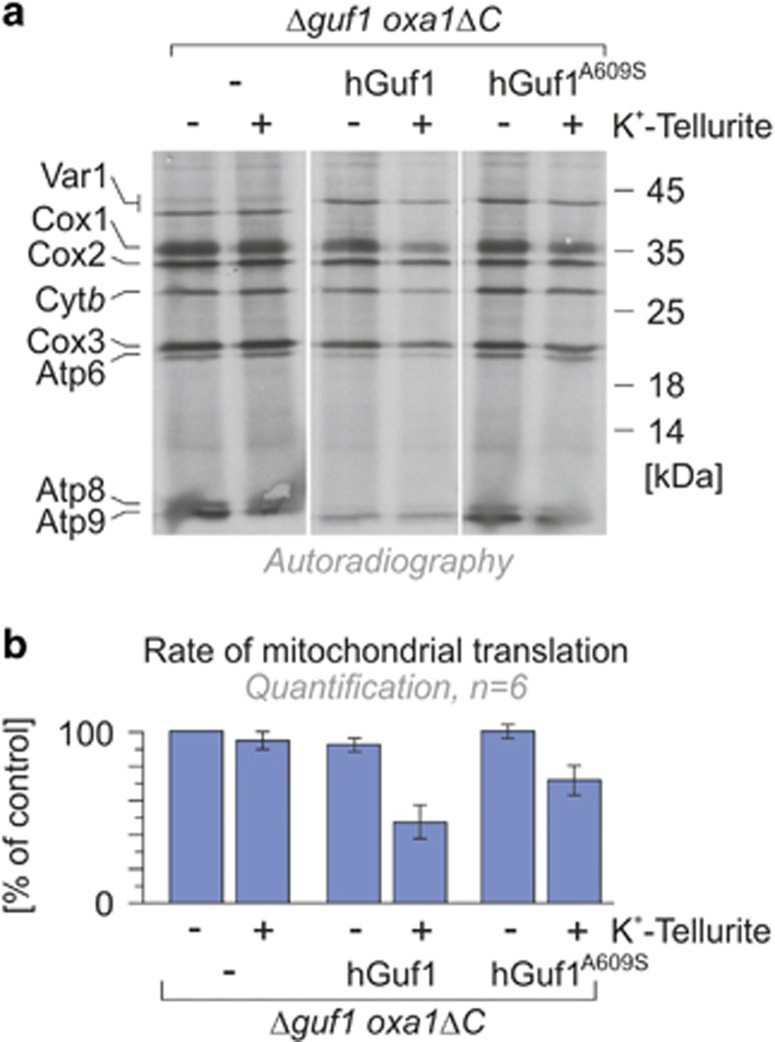
GUF1 is able to back-translocate the tRNA on ribosomes and as increasing amount of the protein resulted in reduced translational error rates, it was suggested that EF4 controls the fidelity of translation.30 The extreme conservation across phyla is ‘paradoxically associated with an apparent lack of phenotype in deletion mutants'.31 For example, while E. coli cells lacking the EF4 gene have no ostensible defects in cell growth or protein export under normal medium and temperature, they show defective growth under stress conditions32 (see below). Similarly, EF4 is one of the 10 genes essential to the survival of Helicobacter pylori in low pH environment.33 In metazoan, the lack of EF4/GUF1 was associated with mitochondrial dysfunction, in particular disruption of mitochondrial respiratory chain complex IV.34, 35 These and other results induced Zhang and Qin31 to suggest that ‘under special ionic stress, for example, extreme pH environments for bacteria or proton gradient around a mitochondrion, the translation process is much more dependent on EF4'. To assess the functional effect of the c.1825G>T variant, we used a complementation approach (see Materials and Methods). In budding yeasts, GUF1 is important for mitochondrial protein synthesis under suboptimal conditions.23 Yeasts that lack the GUF1 (Δguf1) elongation factor show a mild growth defect under extreme growth-limiting concentrations of non-fermentable carbon sources and low and high temperature. On the contrary, overexpression of Guf1 exhibits a pronounced negative effect,23 consistent with the observation that overproduction of LepA stalls translation in bacteria.30 We found that overexpression of hGUF1 and hGUF1A609S in Δguf1 had comparable effect on growth, mitochondrial protein expression and translation (Supplementary Figure S5A and D and Supplementary Materials). Likewise, hGUF1 and hGUF1A609S were both able to suppress the strong synthetic growth defect of the oxa1ΔC Δguf1 double mutant on plates and in direct competition assays (Figure 3, Supplementary Figure S6A and B and Supplementary Materials). Recent results suggest that EF4 is essential in conditions that counteract faithful protein synthesis such as upon exposure to magnesium salts36 and potassium tellurite.32 In these suboptimal environments, Guf1/LepA activity remobilizes stuck ribosomes and transiently inhibits the elongation process to optimize protein synthesis.36, 37 We therefore assessed complementation on plates supplemented with potassium tellurite, magnesium or low levels of chloramphenicol. Under these conditions, respiratory growth of the oxa1ΔC Δguf1 double mutant was strictly dependent on the expression of the human Guf1 protein (Figure 3). Interestingly, when compared with hGUF1, expression of hGUF1A609S showed an increased resistance to some stress conditions such as exposure to tellurite or high magnesium concentrations combined with growth-limiting concentrations of non-fermentable carbon sources (Figure 3). These results encouraged us to assess mitochondrial protein synthesis in suboptimal environments. The tellurite-sensitive oxa1ΔC Δguf1 double mutant effectively synthesizes mitochondrial proteins both in the presence and absence of tellurite. Whereas overexpression of hGUF1 in the double mutant strain was sensitive to this oxidizing compound, complementation with the hGUF1A609S variant showed an intermediate phenotype (Figure 4). Thus tellurite exposure blocks protein synthesis in a GUF1-dependent manner in agreement with the notion that LepA counteracts protein synthesis to prevent the misincorporation of non-cognate amino acids in stressful conditions.36, 37 The improved growth observed with the p.(A609S) variant suggests that this variant is less proficient than wild-type GUF1 in this stalling and hence possibly in limiting misincorporation.
Figure 3.
The hGUF1A609S variant exhibits increased activity in some suboptimal environments. The indicated strains were grown in full medium containing galactose to log phase. Serial 10-fold dilutions were spotted on YP plates containing 2% glucose, 0.5% glycerol or 0.5% glycerol supplemented with potassium tellurite, magnesium or chloramphenicol at the indicated concentrations and plates were incubated at 30 or 37 °C. The decreased ability of hGUF1 to complement the respiratory growth of the oxa1ΔC Δguf1 double mutant on a non-fermentable carbon source supplemented with a high concentration of magnesium or with potassium tellurite compared with hGUF1A609S is indicated by arrows.
Figure 4.
The oxa1ΔC Δguf1 double mutant effectively synthesizes mitochondrial proteins in the presence of tellurite, whereas double mutants overexpressing hGUF1 and hGUF1A609S are highly and moderately sensitive to this oxidizing compound, respectively. The strains described in Figure 3 were incubated with cycloheximide to block cytosolic translation, and mitochondria were isolated, incubated in the presence of ATP, amino acids and radiolabeled [35S]-methionine before visualization of newly synthesized translation products by autoradiography. Visualized bands are annotated on the left, while molecular weight markers are indicated on the right (a). The rate of mitochondrial translation is quantified in panel (b).
Collectively, these data show that, while the hGuf1A609S variant is still functional, its activity is modified under particular conditions (at least in yeasts).
Discussion
Epileptic seizures of various types have been reported in one- to two-thirds of patients with mitochondrial chain respiratory disease.38, 39 These so-called ‘mitochondrial epilepsies' are clinically and genetically heterogeneous diseases; disease-causing mutations have been reported in many of the 37 mitochondrially encoded genes and at least 31 nuclear genes (reviewed in Rahman40). Mutated genes encode different subunits of the respiratory chain or proteins that have a role in mitochondrial genome maintenance, translation and replication, as well as enzymes involved in the biosynthesis of coenzyme Q10 or the mitochondrial glutamate carrier SLC25A22 (reviewed in Rahman40). IS/WS are, however, rare within known mitochondrial epilepsies, and when they occur, they are usually not the main epileptic presentation (http://www.mitomap.org). For example, mutations in TSFM and RARS2, nuclear genes with a role in mitochondrial translation, were identified in patients with epilepsies but not with IS/WS.41, 42 According to the two larger studies in the field, WS accounts for 14–21% (8/56 and 10/48) of the patients with epilepsy and biochemically confirmed mitochondrial respiratory chain defects.43, 44 Conversely, in 13 out of the 17 children affected with IS the monitoring of body fluid metabolites suggested a defect in energy metabolism.45 Definite molecular confirmation could be completed only in a minority of patients: within the El Sabbagh's cohort, three patients had the m.8993T>G variant in MT-ATP6 (ATP synthase subunit 6) while a fourth one had a variant in the nuclear SDHA gene.46, 47 The m.8993T>G variant is commonly found in patients with Leigh (OMIM#256000) or NARP (neuropathy, ataxia and retinitis pigmentosa; OMIM#551500) syndromes. Leigh syndrome is an early-onset progressive neurodegenerative disorder associated with abnormal MRI T2 signals in the basal ganglia.48 Brain MRI revealed such abnormal hypersignals in the three WS patients who shared this variant. They also showed increased CSF lactate levels, a common feature in Leigh syndrome.47 Among the 13 patients with IS reported in Shah et al,45 a possibly causing variant was found only in two monozygotic twin sisters who had increased lactate plasma levels. They carried the m.3243A>G change in the tRNALeu1 gene, which is commonly associated with MELAS (mitochondrial myopathy, encephalopathy, lactate acidosis and stroke-like episodes; OMIM#540000) syndrome. In summary, nuclear defects affecting mitochondrial protein synthesis are common within combined respiratory chain deficiencies (about one-third of patients49) and could result in mitochondrial epilepsies.40, 50 They are, however, easily overlooked as an etiology of IS possibly because of the difficulty to correctly gauge the activity of the respiratory chain complexes, especially when no lactate and/or pyruvate increase is identified in blood and/or CSF.45, 51
We show that exome sequencing is a suitable strategy to identify causative genes in familial WS and potentially IS. Exploiting this approach, we identified a novel homozygous c.1825G>T variant in the universally conserved elongation factor GUF1 in three siblings affected with WS. The substituted residue, that is, A609, is located within the KIIARETV stretch (defined as ‘contact 2' in Connell et al28) of the external beta-strand of the CTD tRNA-binding moiety (Figure 2b). This beta-strand #5 contacts on one side the CTD alpha-helix #1 (numbering according to Connell et al28) and on the other side harbors the fully conserved Q600 (539/539 alignments), a branched hydrophobic residue (364/539 I and 175/539 V) and R610 (530/539; S in 9/539) whose three sidechains form a tightly packed motif likely to be important for the proper recognition of the tRNA acceptor stem and D-loop. The addition of a hydroxyl group at position 609 (p.(A609S) variant) will result in a bulkier residue, which might displace the beta-sheet relative to the alpha-helix if the serine is ‘looking' inside. Alternatively, as shown in Figure 2c, a S residue at this position turned toward the outside might interfere with the tRNA acceptor stem by modifying the ‘contact 2' moiety. Of note, a serine residue is the only other tolerated residue at this position (42 of the 539 alignments; Supplementary Figure S4). It is, however, only found in a subgroup of bacteria such as Bactericides, Chlorobium, Clostridium, Geobacillus, Listeria, Mycoplasma, Pseudomonas and Thermoanaerobacter with tRNAs that are restricted to exactly 13 nucleotides from the beginning of the acceptor stem to the end of the D-stem.52 This stretch is always followed by an A at position 14 that pairs with the strictly conserved T at position 8, a site located between the acceptor stem and the D-stem. This is in stark contrast to human mitochondrial tRNAs in which position 8 is non-constrained; any nucleotide can be positioned there and these do not necessarily form a perfect pair with the similarly non-conserved position 14 (Figure 2b). As the acceptor stem and D-stem regions have a role in binding to GUF1, we postulate that the greater variability within human mitochondrial tRNAs may result in modified interaction when the A609S mutation is present (Figure 2b and c).
Using complementation assays, we further demonstrate that the activity of the GUF1A609S variant is modified in suboptimal conditions. GUF1 interacts genetically with MRPL36 (mitochondrial ribosomal protein L36), a gene with a crucial role in determining the rate of respiratory chain assembly.53 MRPL36 was similarly associated with LETM1 (PMID:19318571), a mitochondrial inner membrane protein, first identified in Wolf–Hirschhorn syndrome (OMIM#194190) and associated with abnormal electrical activity in the brain.54, 55 Definitive validation of the causative role of GUF1 variants in the phenotype could only be achieved through identification of more patients and engineering of animal models (a publication of a guf1 mouse knockout model was recently retracted34). We should also consider the possibility that only few genetic changes result in a hypomorphic form of this elongation factor, a hypothesis supported by the extreme conservation of the GUF1 gene and by our inability to find more WS families with GUF1 variants until now.
In conclusion, we suggest a link between improper assembly of respiratory chain complexes and IS/WS via the identification of a variant in a fidelity factor of mitochondrial translation.
Acknowledgments
We thank the family for its contribution. NB-K is a grantee of a young investigator grant from the University Hospital of Lyon (Hospices Civils de Lyon) and AAA is recipient of a scholarship from the Saudi Arabian National Guard Health Affairs. This work was supported by the Leenaards Foundation Prize (to AR), the Swiss National Science Foundation (to AR), the Deutsche Forschungsgemeinschaft (to JMH) and the Swiss Institute of Bioinformatics (to IX). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author contributions
AAA, NBK and AR conducted the sequencing and statistical analysis. VM and JMH performed the yeast assay. NG and IX modeled the protein. DV, JdB, CR, AL, VdP, PE, MT, DS and GL phenotyped the patients. AR wrote the manuscript with contributions from DS, JMH and GL. All authors commented on and approved the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Deprez L, Jansen A, De Jonghe P: Genetics of epilepsy syndromes starting in the first year of life. Neurology 2009; 72: 273–281. [DOI] [PubMed] [Google Scholar]
- Lux AL, Osborne JP: A proposal for case definitions and outcome measures in studies of infantile spasms and West syndrome: consensus statement of the West Delphi group. Epilepsia 2004; 45: 1416–1428. [DOI] [PubMed] [Google Scholar]
- Wong M, Trevathan E: Infantile spasms. Pediatr Neurol 2001; 24: 89–98. [DOI] [PubMed] [Google Scholar]
- Lagae L, Verhelst H, Ceulemans B et al: Treatment and long term outcome in West syndrome: the clinical reality. A multicentre follow up study. Seizure 2010; 19: 159–164. [DOI] [PubMed] [Google Scholar]
- Pellock JM, Hrachovy R, Shinnar S et al: Infantile spasms: a U.S. consensus report. Epilepsia 2010; 51: 2175–2189. [DOI] [PubMed] [Google Scholar]
- Pavone P, Striano P, Falsaperla R, Pavone L, Ruggieri M: Infantile spasms syndrome, West syndrome and related phenotypes: What we know in 2013. Brain Dev 2013; 36: 739–751. [DOI] [PubMed] [Google Scholar]
- Bahi-Buisson N, Nectoux J, Rosas-Vargas H et al: Key clinical features to identify girls with CDKL5 mutations. Brain 2008; 131: 2647–2661. [DOI] [PubMed] [Google Scholar]
- Chu-Shore CJ, Major P, Camposano S, Muzykewicz D, Thiele EA: The natural history of epilepsy in tuberous sclerosis complex. Epilepsia 2010; 51: 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrini R, Moro F, Kato M et al: Expansion of the first PolyA tract of ARX causes infantile spasms and status dystonicus. Neurology 2007; 69: 427–433. [DOI] [PubMed] [Google Scholar]
- Mignot C, Moutard ML, Trouillard O et al: STXBP1-related encephalopathy presenting as infantile spasms and generalized tremor in three patients. Epilepsia 2011; 52: 1820–1827. [DOI] [PubMed] [Google Scholar]
- Otsuka M, Oguni H, Liang JS et al: STXBP1 mutations cause not only Ohtahara syndrome but also West syndrome—result of Japanese cohort study. Epilepsia 2010; 51: 2449–2452. [DOI] [PubMed] [Google Scholar]
- Weaving LS, Christodoulou J, Williamson SL et al: Mutations of CDKL5 cause a severe neurodevelopmental disorder with infantile spasms and mental retardation. Am J Hum Genet 2004; 75: 1079–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutry-Kryza N, Ville D, Labalme A et al: Complex mosaic CDKL5 deletion with two distinct mutant alleles in a 4-year-old girl. Am J Med Genet A 2014; 164A: 2025–2028. [DOI] [PubMed] [Google Scholar]
- Paciorkowski AR, Thio LL, Dobyns WB: Genetic and biologic classification of infantile spasms. Pediatr Neurol 2011; 45: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne JP, Lux AL, Edwards SW et al: The underlying etiology of infantile spasms (West syndrome): information from the United Kingdom Infantile Spasms Study (UKISS) on contemporary causes and their classification. Epilepsia 2010; 51: 2168–2174. [DOI] [PubMed] [Google Scholar]
- Partikian A, Mitchell WG: Neurodevelopmental and epilepsy outcomes in a North American cohort of patients with infantile spasms. J Child Neurol 2010; 25: 423–428. [DOI] [PubMed] [Google Scholar]
- Michaud JL, Lachance M, Hamdan FF et al: The genetic landscape of infantile spasms. Hum Mol Genet 2014; 23: 4846–4858. [DOI] [PubMed] [Google Scholar]
- Epi KC, Epilepsy Phenome/Genome P Allen AS et al: De novo mutations in epileptic encephalopathies. Nature 2013; 501: 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R: Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R et al: A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011; 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E et al: The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G et al: The variant call format and VCFtools. Bioinformatics 2011; 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerschmitt H, Funes S, Herrmann JM: The membrane-bound GTPase Guf1 promotes mitochondrial protein synthesis under suboptimal conditions. J Biol Chem 2008; 283: 17139–17146. [DOI] [PubMed] [Google Scholar]
- Lemaire C, Guibet-Grandmougin F, Angles D, Dujardin G, Bonnefoy N: A yeast mitochondrial membrane methyltransferase-like protein can compensate for oxa1 mutations. J Biol Chem 2004; 279: 47464–47472. [DOI] [PubMed] [Google Scholar]
- Szyrach G, Ott M, Bonnefoy N, Neupert W, Herrmann JM: Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J 2003; 22: 6448–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S: Predicting deleterious amino acid substitutions. Genome Res 2001; 11: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RN, Blaha G, Bailey S, Steitz TA: The structure of LepA, the ribosomal back translocase. Proc Natl Acad Sci USA 2008; 105: 4673–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell SR, Topf M, Qin Y et al: A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat Struct Mol Biol 2008; 15: 910–915. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC: SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 1997; 18: 2714–2723. [DOI] [PubMed] [Google Scholar]
- Qin Y, Polacek N, Vesper O et al: The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell 2006; 127: 721–733. [DOI] [PubMed] [Google Scholar]
- Zhang D, Qin Y: The paradox of elongation factor 4: highly conserved, yet of no physiological significance? Biochem J 2013; 452: 173–181. [DOI] [PubMed] [Google Scholar]
- Shoji S, Janssen BD, Hayes CS, Fredrick K: Translation factor LepA contributes to tellurite resistance in Escherichia coli but plays no apparent role in the fidelity of protein synthesis. Biochimie 2010; 92: 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma JJ, Lie ALM, Nootenboom IC, Vandenbroucke-Grauls CM, Kusters JG: Identification of loci essential for the growth of Helicobacter pylori under acidic conditions. J Infect Dis 2000; 182: 1566–1569. [DOI] [PubMed] [Google Scholar]
- Piao YR, Jin ZH: Loss of Guf1 impairs sperm mitochondrial function and leads to male infertility. Biol Reprod 2015; 92: 117.25740543 [Google Scholar]
- Yang F, Gao Y, Li Z et al: Mitochondrial EF4 links respiratory dysfunction and cytoplasmic translation in Caenorhabditis elegans. Biochim Biophys Acta 2014; 1837: 1674–1683. [DOI] [PubMed] [Google Scholar]
- Pech M, Karim Z, Yamamoto H, Kitakawa M, Qin Y, Nierhaus KH: Elongation factor 4 (EF4/LepA) accelerates protein synthesis at increased Mg2+ concentrations. Proc Natl Acad Sci USA 2011; 108: 3199–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Chen C, Zhang H, Kaur J, Goldman YE, Cooperman BS: The conserved protein EF4 (LepA) modulates the elongation cycle of protein synthesis. Proc Natl Acad Sci USA 2011; 108: 16223–16228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debray FG, Lambert M, Chevalier I et al: Long-term outcome and clinical spectrum of 73 pediatric patients with mitochondrial diseases. Pediatrics 2007; 119: 722–733. [DOI] [PubMed] [Google Scholar]
- Khurana D, Salganicoff L, Melvin J et al: Epilepsy and respiratory chain defects in children with mitochondrial encephalopathies. Epilepsia 2008; 49: 1972. [DOI] [PubMed] [Google Scholar]
- Rahman S: Mitochondrial disease and epilepsy. Dev Med Child Neurol 2012; 54: 397–406. [DOI] [PubMed] [Google Scholar]
- Smeitink JA, Elpeleg O, Antonicka H et al: Distinct clinical phenotypes associated with a mutation in the mitochondrial translation elongation factor EFTs. Am J Hum Genet 2006; 79: 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edvardson S, Shaag A, Kolesnikova O et al: Deleterious mutation in the mitochondrial arginyl-transfer RNA synthetase gene is associated with pontocerebellar hypoplasia. Am J Hum Genet 2007; 81: 857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YM, Kang HC, Lee JS et al: Mitochondrial respiratory chain defects: underlying etiology in various epileptic conditions. Epilepsia 2008; 49: 685–690. [DOI] [PubMed] [Google Scholar]
- El Sabbagh S, Lebre AS, Bahi-Buisson N et al: Epileptic phenotypes in children with respiratory chain disorders. Epilepsia 2010; 51: 1225–1235. [DOI] [PubMed] [Google Scholar]
- Shah NS, Mitchell WG, Boles RG: Mitochondrial disorders: a potentially under-recognized etiology of infantile spasms. J Child Neurol 2002; 17: 369–372. [DOI] [PubMed] [Google Scholar]
- Horvath R, Abicht A, Holinski-Feder E et al: Leigh syndrome caused by mutations in the flavoprotein (Fp) subunit of succinate dehydrogenase (SDHA). J Neurol Neurosurg Psychiatry 2006; 77: 74–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desguerre I, Pinton F, Nabbout R et al: Infantile spasms with basal ganglia MRI hypersignal may reveal mitochondrial disorder due to T8993G MT DNA mutation. Neuropediatrics 2003; 34: 265–269. [DOI] [PubMed] [Google Scholar]
- Lee HF, Tsai CR, Chi CS, Lee HJ, Chen CC: Leigh syndrome: clinical and neuroimaging follow-up. Pediatr Neurol 2009; 40: 88–93. [DOI] [PubMed] [Google Scholar]
- Kemp JP, Smith PM, Pyle A et al: Nuclear factors involved in mitochondrial translation cause a subgroup of combined respiratory chain deficiency. Brain 2011; 134: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desguerre I, Hully M, Rio M, Nabbout R: Mitochondrial disorders and epilepsy. Rev Neurol 2014; 170: 375–380. [DOI] [PubMed] [Google Scholar]
- Desguerre I, Nabbout R, Dulac O: The management of infantile spasms. Arch Dis Child 2008; 93: 462–463. [DOI] [PubMed] [Google Scholar]
- Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J: tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 2009; 37: D159–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestele M, Vogel F, Reichert AS, Herrmann JM, Ott M: Mrpl36 is important for generation of assembly competent proteins during mitochondrial translation. Mol Biol Cell 2009; 20: 2615–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Zhao L, Clish CB, Clapham DE: Letm1, the mitochondrial Ca2+/H+ antiporter, is essential for normal glucose metabolism and alters brain function in Wolf-Hirschhorn syndrome. Proc Natl Acad Sci USA 2013; 110: E2249–E2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlickum S, Moghekar A, Simpson JC et al: LETM1, a gene deleted in Wolf-Hirschhorn syndrome, encodes an evolutionarily conserved mitochondrial protein. Genomics 2004; 83: 254–261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.