Abstract
Leber congenital amaurosis (LCA) represents the most severe form of inherited retinal dystrophies with an onset during the first year of life. Currently, 21 genes are known to be associated with LCA and recurrent mutations have been observed in AIPL1, CEP290, CRB1 and GUCY2D. In addition, sequence analysis of LRAT and RPE65 may be important in view of treatments that are emerging for patients carrying variants in these genes. Screening of the aforementioned variants and genes was performed in 64 Danish LCA probands. Upon the identification of heterozygous variants, Sanger sequencing was performed of the relevant genes to identify the second allele. In combination with prior arrayed primer extension analysis, this led to the identification of two variants in 42 of 86 cases (49%). Remarkably, biallelic RPE65 variants were identified in 16% of the cases, and one novel variant, p.(D110G), was found in seven RPE65 alleles. We also collected all previously published RPE65 variants, identified in 914 alleles of 539 patients with LCA or early-onset retinitis pigmentosa, and deposited them in the RPE65 Leiden Open Variation Database (LOVD). The in silico pathogenicity assessment of the missense and noncanonical splice site variants, as well as an analysis of their frequency in ~60 000 control individuals, rendered 864 of the alleles to affect function or probably affect function. This comprehensive database can now be used to select patients eligible for gene augmentation or retinoid supplementation therapies.
Introduction
Leber congenital amaurosis (LCA; OMIM 204000) is a group of rare inherited retinal dystrophies that affects ~1 in 50 000 individuals worldwide.1, 2 LCA is characterized by a severe visual impairment in the first year of life, sensory nystagmus, amaurotic pupils and markedly diminished or extinguished rod and cone electroretinograms (ERG).3, 4 LCA is predominantly inherited in an autosomal recessive manner, although autosomal dominant inheritance has also been reported a few times.4 To date, mutations in 21 genes underlie at least 70% of LCA cases.5, 6 These genes encode proteins that exert their functions in diverse processes and pathways in the retina, such as photoreceptor morphogenesis (CRB1, CRX, GDF6), retinal differentiation (OTX2), phototransduction (AIPL1, GUCY2D, RD3), retinoid cycle (LRAT, RDH12, RPE65), guanine synthesis (IMPDH1), signal transduction (CABP4, KCNJ13), outer segment phagocytosis (MERTK), coenzyme NAD biosynthesis (NMNAT1) or intra-photoreceptor ciliary transport processes (CEP290, IQCB1, LCA5, RPGRIP1, SPATA7, TULP1).4, 6 Mutations in many of these genes can also give rise to early-onset retinal dystrophies.
The RPE65 and CHM gene augmentation trials in patients with LCA7, 8, 9 and choroideremia,10 respectively, have been milestones in the retinal therapeutics field. An oral retinoid-based treatment was also reported to be beneficial for individuals with RPE65 or LRAT mutations.11 Preclinical gene augmentation studies in animal models due to mutations in several other genes have also demonstrated safety and efficacy.12 However, the identification of molecular genetic causes is a prerequisite to assess the patients' eligibility in receiving suitable treatments. Various techniques, including classical genetic linkage analysis, identity-by-descent (IBD) mapping, Sanger sequencing of candidate genes or next-generation sequencing, have been used for gene discovery and diagnostic methods in LCA. Previous studies in LCA revealed the intronic CEP290 variant c.2991+1655A>G as the most frequent in the Caucasian population, explaining ~12% of cases in Caucasian populations originating from North-West Europe.13, 14, 15 In addition, the AIPL1 variant p.(W278*) in exon 6, a conspicuous cluster of CRB1 variants in exons 7 and 9, and the GUCY2D exon 12 variant p.(R768W) together may explain ~14% of the cases.16, 17 Therefore, prescreening of these 5 exons potentially identifies 26% of the mutations, serving as a cost- and time-effective genotyping procedure for LCA. Finally, screening of RPE65 and LRAT should uncover the cause of disease in 7 and 1% of LCA cases, respectively.4 The identification of individuals with mutations in these genes is very relevant in view of the above-mentioned treatment options.
By using a combination of different genotyping techniques, we identified the underlying mutations in approximately half of the Danish LCA probands, and most significantly found RPE65 variants to be the most prevalent causes of LCA and early-onset RP. In addition, we collected all published RPE65 variants and completed the RPE65 Leiden Open Variation Database. These results are essential for genetic counseling and for the selection of individuals who are eligible for therapeutic interventions.
Materials and methods
Subjects and clinical evaluation
A total of 86 unrelated Danish families comprising 80 isolated and 6 multiplex families diagnosed with LCA were ascertained in this study. The diagnosis of LCA was based on clinical history, a full eye exam including slit-lamp and retinal examination, full-field electroretinography (ffERG), visual acuity, refraction, visual field and optical coherence tomography (OCT) in some cases. The diagnostic criteria were based on a congenital or early infantile onset of a generalized retinal dystrophy with substantially reduced cone and rod ffERG responses. Many patients showed nystagmus and/or hypermetropia, but these clinical signs were not compulsory for the diagnosis.18, 19 In this study, we consider LCA and early-onset retinitis pigmentosa as a continuum of phenotypes, as they are clinically and genetically overlapping and the true age at disease onset can be difficult to establish. Research procedures were in accordance with institutional guidelines and adhered to the tenets of the Declaration of Helsinki. An ethics committee approval was obtained and all samples were acquired after written informed consent was obtained from the patient or, in the case of children, their legal guardians.
Genetic analysis
Genomic DNA of participating individuals was isolated from peripheral leukocytes using a standard salting out procedure.20 A candidate gene approach and an arrayed primer-extension (APEX) analysis were performed in several probands before recurrent mutation screening. Complete genotyping methods for the affected individuals are listed in Supplementary Table S1.
Sanger sequencing in selected LCA-associated genes
Sanger sequencing based on a candidate gene approach was performed in the LCA-associated genes AIPL1, CRB1, CRX, GUCY2D, LRAT, RDH12, RPE65, RPGRIP1 and TULP1 by a commercial supplier (Seqlab, Göttingen, Germany) using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) on a ABI 3730XL platform (Applied Biosystems).
SSCP and APEX analysis
Single-strand conformation polymorphism (SSCP) analysis was applied to 52 index patients as described previously21 followed by subsequent Sanger sequencing of exons with aberrant bands in AIPL1, CEP290, CRX, CRB1, GUCY2D, LRAT, RDH12, RPE65, RPGRIP and TULP1. Thereafter, APEX LCA microarray analysis, versions 2007–2012 (Asper Ophthalmics, Tartu, Estonia) was performed in the 15 remaining unsolved cases.22 This microarray-based genotyping method allowed the simultaneous analysis of previously identified disease-associated sequence variants in the LCA-associated genes AIPL1, CEP290, CRB1, CRX, GUCY2D, IQCB1, LCA5, LRAT, MERTK, RDH12, RD3, RPE65, RPGRIP1, SPATA7 and TULP1. The methods employed in this analysis have been described elsewhere (www.asperbio.com; Asper Biotech, Ltd).23
LCA recurrent mutation screening
The screening was performed in cases carrying one or no variant identified using Sanger sequencing of selected LCA-associated genes or APEX analysis, and 19 additional cases that had not been previously analyzed. Recurrent mutation screening was performed in 13 amplicons of four LCA-associated genes. The CEP290 c.2991+1655A>G variant was assessed by utilizing the amplification-refractory mutation system (ARMS) as described by den Hollander et al.13 In addition, AIPL1 exon 6, GUCY2D exon 12 and CRB1 exons 7 and 9 were amplified and sequenced by the Sanger method. Primers were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/) and are listed in Supplementary Table S2. Purified PCR products were analyzed in either sense or antisense direction using dye termination chemistry (BigDye Terminator, version 1.1, on a 3730 DNA analyzer; Applied Biosystems, Inc.). All identified sequence variants were validated with independent PCR reactions.
Sequence analysis of RPE65 and LRAT
All coding exons and flanking intronic sequences of RPE65 and LRAT were amplified by PCR, purified and analyzed using Sanger sequencing as described above. PCR primers and conditions are available upon request.
Ethnically matched control frequencies
The allele frequency of each identified variant in the Danish population was assessed using whole-exome data of 2000 ethnically matched individuals from the Danish exome study.24
Pathogenicity interpretation of sequence variants
To determine the pathogenicity of missense variants, we used the following in silico prediction methods: Sorting Tolerant from Intolerant (SIFT, http://sift.jcvi.org/), Polymorphism Phenotyping v2 (PolyPhen-2; http://genetics.bwh.harvard.edu/pph2/) and PhyloP (http://genome.ucsc.edu/cgi-bin/hgTrackUi?g=cons46way) the latter of which assesses the evolutionary conservation at the nucleotide level. We also assessed the allele frequency data of identified variants derived from an available ‘control' exome data set provided by NHLBI Exome Variant Server (EVS, http://evs.gs.washington.edu/EVS/) and the Danish exome study.24 For the RPE65 variants deposited in the RPE65 LOVD, we also assessed their prevalence using the Exome Aggregate Consortium (ExAC) database (http://exac.broadinstitute.org). Five computational prediction programs, SpliceSite finder-like,25 MaxEntScan,26 NNSPLICE,27 Gene Splicer28 and Human Splicing Finder29 were used to determine the pathogenic effect of noncanonical splice variants.
Sequence variants nomenclature
All identified variants are described as recommended by the Human Genome Variation Society (HGVS) nomenclature guidelines, considering nucleotide +1 as the A of start codon (ATG).30 When the nucleotide position is indicated, the GenBank cDNA reference sequences were used as follows: AIPL1 (NG_008474.1); CEP290 (NG_008417.1); CRB1 (NG_008483.1); GUCY2D (NG_009092.1); LRAT (NG_009110.1); RDH12 (NG_008321.1); RPE65 (NG_008472.1); RPGRIP1 (NG_008933.1); and TULP1 (NG_009077.1).31
RPE65 Leiden open variation database variant depositions
RPE65 variants were collected from all articles published until 2014 (Supplementary Tables S5–S7). To assess the pathogenicity of missense variants, we combined three prediction programs, that is, PhyloP, SIFT and PolyPhen-2, as well as the allele count in the ExAC database that contains exome data of ~60 000 individuals (http://exac.broadinstitute.org). A PhyloP score of ≥2.5 was employed as one of the criteria to consider a variant to be potentially disease causative. Subsequently, we classified missense variants as ‘probably no functional effect', ‘variants of unknown significance' or ‘affects function'. They were considered to have ‘probably no functional effect' when none of the prediction programs described the variant as damaging and/or if the ExAC allele count was ≥18. The latter cutoff number was chosen as the three most frequent pathologic RPE65 variants, p.(R91W), p.(Y368H) and c.11+5G>A, representing 11.6, 8.0 and 7.2% of the published 864 autosomal recessive RPE65 function-affecting variants, respectively, were found in 10, 8 and 11 alleles in the ExAC database. Statistical analysis of the most frequent RPE65 variant c.11+5G>A, with a total allele count of 120 956 in the ExAC database for this position, results in a 95% confidence interval between 0.3 × 10−4 and 1.4 × 10−4. From this we can infer that any variant with an allele frequency >1.4 × 10−4 can be considered to be benign with 95% certainty. Assuming the maximum number of alleles present in the ExAC database (122 972 alleles), benign variants are expected to be present in ≥18 alleles. The pathogenicity of a variant was considered as ‘variants of unknown significance' if one of three prediction programs described the variant as damaging and the allele count in ExAC was <18. Finally, a variant was considered to ‘affect function' when at least two prediction programs described the variant as damaging and the ExAC allele count was <18.
Results
Molecular genetic analysis
Genotyping analysis before recurrent mutation screening
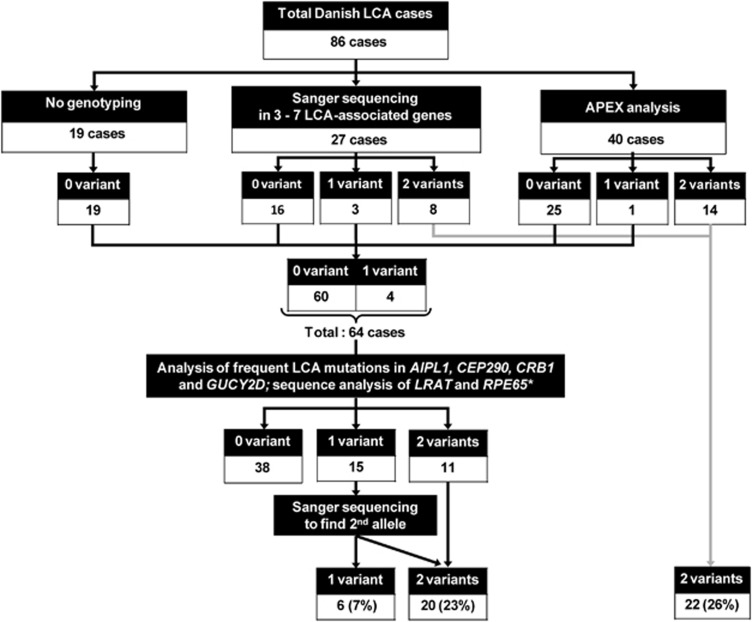
For diagnostic purposes, genotyping analysis using Sanger sequencing of selected LCA-associated genes and/or APEX analyses were performed in 27 and 40 LCA cases, respectively (Figure 1). Using the selected Sanger sequencing approach, two function-affecting variants were identified in eight cases and one variant in three cases. In addition, APEX analysis detected two variants in 14 cases and one variant in one case. Subsequently, 41 cases without any identified variants were included in our recurrent mutation screening.
Figure 1.
Genotyping workflow in this study. Eighty-six individuals with LCA were included in this study. Twenty-seven cases were sequenced for variants in several LCA-associated genes, 40 cases were analyzed using APEX analysis and 19 cases had not been analyzed. Thereafter, 60 patients carrying no variants and 4 patients with one variant were included in the screening of common LCA mutations in AIPL1 exon 6, CEP290 c.2991+1655A>G variant, CRB1 exons 7 and 9, GUCY2D exon 12 and all coding exons of RPE65 and LRAT. Together with the previous genotyping analysis, this screening yielded one variant in 6 cases and two variants in 42 cases.
LCA recurrent mutation screening and second allele sequencing
Sixty-four individuals carrying no or one variant were included in the screening for the presence of frequent LCA mutations. Two function-affecting variants were identified in 11 cases and 15 carried only one variant. Additional Sanger sequencing to find a second allele identified additional variants in nine of the latter individuals. Taken together, the LCA recurrent mutation screening followed by a search for a second allele by Sanger sequencing identified two variants in 20 cases (23%) and one variant in six cases (7%), as shown in Figure 1.
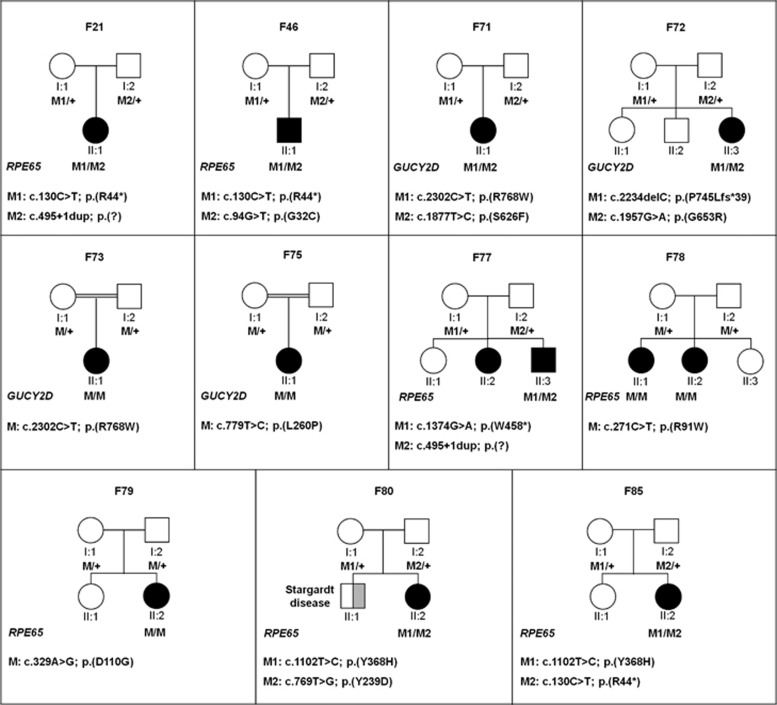
Interestingly, based on the number of cases with two causal variants, this study revealed the RPE65 gene as the most frequently mutated gene in this Danish LCA cohort (16%). Out of 46 identified variants, 17 are novel variants (Table 1 and Supplementary Table S4) with the 3 most frequent alleles being c.2991+1655A>G in CEP290 (8 alleles), p.(W278*) in AIPL1 (7 alleles) and, surprisingly, a novel p.(D110G) variant in RPE65 (7 alleles). Segregation analysis in 11 families in whom parents and/or siblings were available showed that the unaffected parents and/or siblings each carried one of the two variants (Figure 2).
Table 1. Overview of function-affecting variants identified in Danish LCA patients.
| Allele 1 | Allele 2 | Allele 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Fam. ID | Gene | Nucleotide change | Amino acid change | Nucleotide change | Amino acid change | Gene | Nucleotide change | Amino acid change | Segregation analysis |
| 1 | GUCY2D | c.2513G>C | p.(R838P) | + | + | NA | |||
| 2 | CRB1 | c.2549G>T | p.(G850V) | c.4006G>A | p.(V1340I) | ND | |||
| 4 | CEP290 | c.2991+1655A>G | p.(C998*)/p.=a | c.2991+1655A>G | p.(C998*)/p.=a | ND | |||
| 5 | CEP290 | c.2991+1655A>G | p.(C998*)/p.=a | + | + | NA | |||
| 14 | RPE65 | c.329A>G | p.(D110G) | + | + | GUCY2D | c.1093C>T | p.(R365W) | NA |
| 17 | CEP290 | c.2991+1655A>G | p.(C998*)/p.=a | c.5364+2T>G | p.(?) | ND | |||
| 20 | CRB1 | c.2290C>T | p.(R764C) | c.2843G>A | p.(C948Y) | ND | |||
| 21 | RPE65 | c.495+1dup | p.(?) | c.130C>T | p.(R44*) | Yes | |||
| 23 | CEP290 | c.2991+1655A>G | p.(C998*)/p.=a | c.2119_2123dupCAGCT | p.(Q709Sfs*11) | ND | |||
| 26 | GUCY2D | c.2302C>T | p.(R768W) | c.1877T>C | p.(S626F) | ND | |||
| 27 | CEP290 | c.2991+1655A>G | p.(C998*)/p.=a | c.2119_2123dupCAGCT | p.(Q709Sfs*11) | ND | |||
| 28 | AIPL1 | c.834G>A | p.(W278*) | + | + | NA | |||
| 30 | CEP290 | c.2991+1655A>G | p.(C998*)/p.=a | c.5364+2T>G | p.(?) | AIPL1 | c.244C>T | p.(H82Y) | ND |
| 33 | RPE65 | c.1543C>T | p.(R515W) | c.130C>T | p.(R44*) | ND | |||
| 38 | RPE65 | c.329A>G | p.(D110G) | c.11+5G>A | p.(?) | ND | |||
| 42 | RPE65 | c.329A>G | p.(D110G) | c.11+5G>A | p.(?) | ND | |||
| 43 | AIPL1 | c.834G>A | p.(W278*) | c.834G>A | p.(W278*) | ND | |||
| 46 | RPE65 | c.94G>T | p.(G32C) | c.130C>T | p.(R44*) | Yes | |||
| 49 | GUCY2D | c.2302C>T | p.(R768W) | c.1694T>C | p.(F565S) | ND | |||
| 51 | RPE65 | c.1328T>C | p.(V443A) | c.1102T>C | p.(Y368H) | ND | |||
| 52 | RPE65 | c.11+5G>A | p.(?) | c.1102T>C | p.(Y368H) | ND | |||
| 55 | RPE65 | c.11+5G>A | p.(?) | c.1088C>G | p.(P363R) | ND | |||
| 58 | GUCY2D | c.2135_2136delinsTC | p.(E712V) | c.3043+5G>A | p.(?) | ND | |||
| 59 | AIPL1 | c.815G>C | p.(R272P) | + | + | NA | |||
| 62 | AIPL1 | c.834G>A | p.(W278*) | c.834G>A | p.(W278*) | ND | |||
| 63 | AIPL1 | c.834G>A | p.(W278*) | c.834G>A | p.(W278*) | ND | |||
| 64 | CRB1 | c.4121_4130del10 | p.(A1374Nfs*20) | c.4121_4130del10 | p.(A1374Nfs*20) | ND | |||
| 65 | CRB1 | c.613_619delATAGGAA | p.(I205Dfs*13) | c.2401A>T | p.(K801*) | ND | |||
| 66 | CRB1 | c.584G>T | p.(C195F) | c.613_619delATAGGAA | p.(I205Dfs*13) | RPGRIP1 | c.2869G>A | p.(E957K) | ND |
| 67 | CEP290 | c.1219_1220delAT | p.(M407Efs*14) | c.2991+1655A>G | p.(C998*)/p.=a | ND | |||
| 68 | GUCY2D | c.1561C>T | p.(R521*) | c.1561C>T | p.(R521*) | ND | |||
| 69 | RDH12 | c.379G>T | p.(G127*) | c.379G>T | p.(G127*) | ND | |||
| 70 | RDH12 | c.2T>C | p.(M1T) | c.582C>G | p.(Y194*) | ND | |||
| 71 | GUCY2D | c.2302C>T | p.(R768W) | c.1877T>C | p.(S626F) | RPGRIP1 | c.2555G>A | p.(R852Q) | Yes |
| 72 | GUCY2D | c.1957G>A | p.(G653R) | c.2234delC | p.(P745Lfs*39) | Yes | |||
| 73 | GUCY2D | c.2302C>T | p.(R768W) | c.2302C>T | p.(R768W) | Yes | |||
| 74 | GUCY2D | c.2849C>T | p.(A950V) | + | + | NA | |||
| 75 | GUCY2D | c.779T>C | p.(L260P) | c.779T>C | p.(L260P) | Yes | |||
| 76 | GUCY2D | c.2849C>T | p.(A950V) | c.380C>T | p.(P127L) | NA | |||
| 77 | RPE65 | c.495+1dup | p.(?) | c.1374G>A | p.(W458*) | Yes | |||
| 78 | RPE65 | c.271C>T | p.(R91W) | c.271C>T | p.(R91W) | Yes | |||
| 79 | RPE65 | c.329A>G | p.(D110G) | c.329A>G | p.(D110G) | Yes | |||
| 80 | RPE65 | c.769T>G | p.(Y239D) | c.1102T>C | p.(Y368H) | Yes | |||
| 81 | RPGRIP1 | c.930+3A>G | p.(?) | c.3618-5del | p.(?) | ND | |||
| 82 | TULP1 | c.1495_1496dup | p.(D500Vfs*4) | c.1495_1496dup | p.(D500Vfs*4) | ND | |||
| 83 | CRB1 | c.613_619delATAGGAA | p.(I205Dfs*13) | c.2401A>T | p.(K801*) | ND | |||
| 85 | RPE65 | c.1102T>C | p.(Y368H) | c.130C>T | p.(R44*) | Yes | |||
| 86 | RPE65 | c.329A>G | p.(D110G) | c.329A>G | p.(D110G) | ND | |||
NA, not applicable; ND, not done.
Novel variants are marked in bold.
The deep-intronic CEP290 mutation c.2991+1655A>G in ~50% of the RNA results in the insertion of an intronic sequence in the mRNA that carries a stop mutation, and in ~50% of the RNA has no effect on splicing.
Figure 2.
Family pedigrees and segregation analysis. Pedigrees of patients for whom parent samples were available. All variants segregate as expected in the families.
Three variants, each identified in a heterozygous state in one LCA proband, AIPL1 c.286G>A; p.(V96I), AIPL1 c.286G>A; p.(Y134F) and GUCY2D c.2101C>T; p.(P701S), were identified in 2.8, 1.0 and 2.6% of 2000 ethnically matched Danish control individuals, respectively, ruling them out as highly penetrant LCA-causing alleles. In four probands with one or two causal variants in a given gene, we identified an additional variant in a different gene.
The benign GUCY2D p.(P701S) variant was identified in one proband, together with a likely pathogenic variant, that is, p.(A950V). However, as p.(P701S) is deemed benign and sequencing was not completely performed for all exons of GUCY2D, this case remains unsolved but a second mutation can be located in another exon.
Pathogenicity prediction of novel variants
Of the 46 identified variants in this study, 17 are novel, consisting of 8 missense, 5 splice site, 2 frameshift and 2 nonsense variants. None of the missense variants was present in 6503 individuals in the EVS database or in 2000 Danish control individuals. Based on in silico predictions, all missense variants except p.(V1340I) in CRB1 (found in F2) are deemed pathogenic (Table 2). The AIPL1 missense variant p.(R272P) affects a highly conserved amino acid in the tetratricopeptide (TPR) domain and may disrupt the interaction with the Hsp70 and Hsp90 molecular chaperones, crucial for facilitating retinal protein maturation.32 The CRB1 missense variant p.(G850V) affects a conserved glycine in the second laminin G domain, encoded by one of the most frequently mutated regions in CRB1. Three missense variants in RPE65 affect conserved amino acid residues located in the carotenoid oxygenase that is essential for its isomerohydrolase activity. By using several in silico tools, three out of five novel splice site variants in CEP290, GUCY2D, RPE65 and RPGRIP1 are predicted to affect normal splicing (Table 3). None of these splice site variants were found in 2000 Danish control individuals.
Table 2. The in silico predictions of the functional effects of the identified novel missense variants.
| Gene | cDNA | Amino acid | SIFT | PPH2 | PhyloP value | Final classification |
|---|---|---|---|---|---|---|
| AIPL1 | c.815G>C | p.(R272P) | Deleterious | Probably damaging | 1.09 | Affects function |
| CRB1 | c.2549G>T | p.(G850V) | Deleterious | Probably damaging | 5.37 | Affects function |
| CRB1 | c.4018G>A | p.(V1340I) | Tolerated | Benign | −0.04 | No functional effect |
| GUCY2D | c.1877C>T | p.(S626F) | Deleterious | Probably damaging | 3.03 | Affects function |
| GUCY2D | c.2135_2136delinsTC | p.(E712V) | Deleterious | Probably damaging | 4.56/0.45 | Affects function |
| GUCY2D | c.380C>T | p.(P127L) | Deleterious | Probably damaging | 2.87 | Affects function |
| RPE65 | c.94G>T | p.(G32C) | Deleterious | Probably damaging | 5.53 | Affects function |
| RPE65 | c.329A>G | p.(D110G) | Deleterious | Probably damaging | 4.73 | Affects function |
| RPE65 | c.1088C>G | p.(P363R) | Deleterious | Probably damaging | 5.77 | Affects function |
SIFT, http://sift.jcvi.org/.
Polymorphism Phenotyping v2 (PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/).
Table 3. The in silico predictions of the functional effects of the novel splice site variants.
| Splice site prediction tool | Splice Site Finder-like25 | MaxEntScan26 | NNSPLICE27 | GeneSplicer28 | HSF29 | |||
|---|---|---|---|---|---|---|---|---|
| Score range | (0–100) | (0–12) | (0–1) | (0–15) | (0–100) | |||
| Threshold | ≥70 | ≥0 | >0.4 | ≥0 | ≥60 | EVS | Final classification | |
| CEP290 | c.5364+2T>G | 77.40⇒— | 5.54⇒— | 0.68⇒— | 83.55⇒— | T=0/G=13006 | Affects function | |
| GUCY2D | c.3043+5G>A | 7.55⇒— | 0.53⇒— | 9.83⇒2.60 (−73.5%) | 80.99⇒68.82 (−15.0%) | T=0/A=13006 | Affects function | |
| RPE65 | c.495+1dup | 80.23⇒— | 6.71⇒— | 0.74⇒— | 92.09⇒— | A1=0/R=13006 | Affects function | |
| RPGRIP1 | c.3618−5del | 81.46⇒75.54 (−7.3%) | 5.98⇒6.71 (+12.2%) | 0.97⇒0.79 (−19.3%) | 82.94⇒78.83 (−5.0%) | A1=0/R=13006 | No functional effect | |
| RPGRIP1 | c.930+3A>G | 76.52⇒— | 8.40⇒0.05 (−99.4%) | 0.85⇒— | 96.23⇒80.67 (−16.2%) | G=33/A=12973 | No functional effect | |
Clinical findings
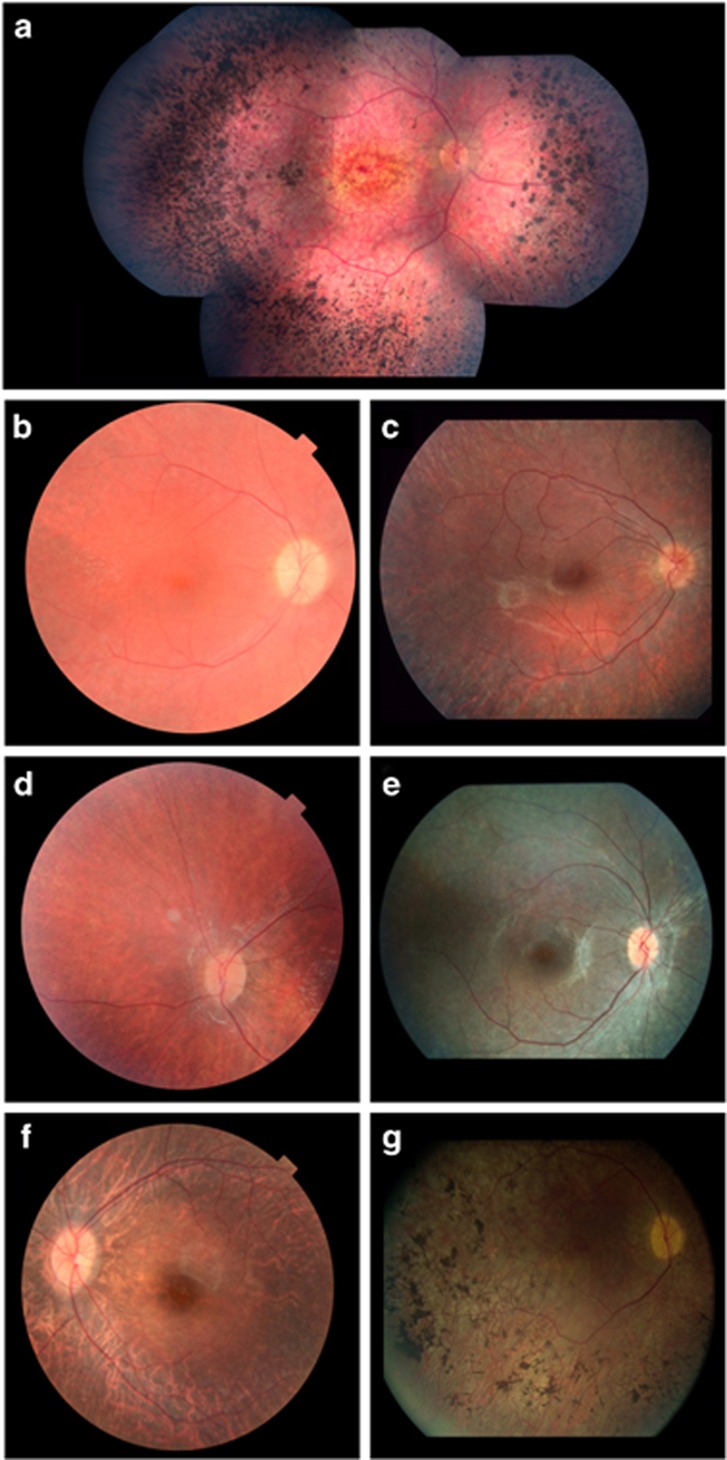
Clinical characteristics of the patients with novel variants are presented in Table 4 and Figure 3. Patients carrying CRB1 variants display an early-onset retinal degeneration with maculopathy and nummular pigmentations in the retina that are typical features for individuals with variants in CRB1.17 As reported for other cases with RPGRIP1 variants, patient 81 had poor vision in the first year of life with nystagmus and hypermetropic eyes (+5.5/+6.0).33 Early-onset visual impairment and nystagmus were also observed in patient 71 carrying GUCY2D variants who showed no ERG responses at the age of 1 year.34, 35
Table 4. Clinical phenotypes of affected individuals carrying two novel causal variants.
| Fam. ID | Gene | Origin | Gender | Age at disease presentation | Refraction LE/RE (Age) | Nystagmus | ERG (age) | Fundus | Other pathology |
|---|---|---|---|---|---|---|---|---|---|
| 2 | CRB1 | Denmark | F | 3 Years | +6.0/+5.5 (6) | No | Nearly extinct (6) | Early bull's eye maculopathy and later nummulate pigmentations and attenuated vessels | None |
| 21 | RPE65 | Denmark | F | Congenital | +7 (22) | Yes | Extinct (1) | Attenuated vessels, no hyperpigmentation | Epilepsia |
| 46 | RPE65 | Denmark | M | Congenital | −4.0/−4.0 (20) | Yes | Extinct (1) | Attenuated vessels, no hyperpigmentation | None |
| 55 | RPE65 | Denmark | F | Congenital | −1/+2 (1) | Yes | Extinct (1) | Attenuated vessels and muddy retinal pigmentation | None |
| 71 | GUCY2D | Denmark | F | Congenital | +6/+6 (1) | Yes | Extinct (1) | Attenuated vessels and muddy retinal pigmentation | None |
| 79 | RPE65 | Denmark | F | Congenital | −2/−3.25 (1) | Yes | Extinct cone response and reduced rod response (1) | Attenuated vessels and pale optic nerve | None |
| 81 | RPGRIP1 | Pakistan | F | 1 Year | +5.5/+6.0 (1) | Yes | Extinct (3) | Attenuated vessels and no hyperpigmentation | None |
| 86 | RPE65 | Jordan | M | 2 Years | −1.5/−2.0 (15) | Yes | Extinct (17) | Attenuated vessels, atrophic optic nerve and bone spicule pigmentations | None |
Figure 3.
Fundus pictures of LCA cases with novel variants. (a) The fundus of proband F2 with variants p.(G850V) and p.(V1340I) in CRB1, at the age of 28 years, showing central maculopathy, peripheral nummular pigmentations and attenuated vessels. F2 had a refraction of +6.0/+5.5 at the age of 6 years. (b) Fundus of patient F21-II:1 with variants c.495+1dup and p.(R44*) in RPE65, at the age of 17 years, showing attenuated vessels and no hyperpigmentation. At 22 years, the refraction was +7/+7. (c) Fundus of patient F46-II:1 with variants p.(G32C) and p.(R44*) in RPE65, at the age of 17 years, showing attenuated vessels and no hyperpigmentation. Refraction was −4.0/−4.0 at the age of 20 years. (d) Fundus picture of proband F55 with variants c.11+5G>A and p.(P363R) in RPE65, at the age of 6 years, showing attenuated vessels and slight muddy retinal pigmentation in the periphery. Refraction was −1/+2 at the age of 1 year. (e) Fundus picture of proband F81 with variants c.930+3A>G and c.3618-5del in RPGRIP1, at 9 years of age, showing attenuated vessels and no hyperpigmentation. Refraction was +5.5/+6.0 at the age of 1 year. (f) Fundus of patient F79-II:2 with a homozygous RPE65 p.(D110G) variant, at the age of 9 years, showing attenuated vessels and a pale optic nerve. Refraction was −2/−3.25 at the age of 1 year. (g) Fundus of proband F86 with a homozygous p.(D110G) variant in RPE65, at the age of 40 years. The fundus shows attenuated vessels, atrophic optic nerve and pigment aggregations. Refraction was −1.5/−2.0 at the age of 1 year.
Patients carrying two RPE65 variants presented with symptoms of nystagmus, lack of fixation, night blindness and/or orientation difficulties within the first year of age, except one patient who according to the anamnesis presented with visual impairment at 2 years of age. All patients showed extinct ERG responses and suffered from nystagmus and severe night blindness. The fundi were generally characterized by retinal degeneration and attenuated vessels with later signs of optic nerve atrophy and in some patients mottled or bone spicule pigmentations. Mean visual acuity at the age of 7 years for all RPE65-associated cases was 0.18 (SD 0.17). Most patients had severely constricted visual fields at the age of 10 years; however, a few patients had preserved visual fields at later stages. Similar phenotype data were described for individuals with RPE65 variants in several previous studies.36, 37
RPE65 LOVD variant depositions
Novel and previously described variants identified in this study have been listed in the respective LOVD for the mutated genes.38, 39, 40, 41, 42, 43 In addition, we collected all RPE65 variants that were published up to 2014. In total, we identified 914 variant alleles in 539 affected individuals that were described in 79 papers. In Supplementary Table S5, we list the 152 different variants according to their position in the RPE65 gene. Almost all variants were inherited in an autosomal recessive manner. Exceptions are p.(D477G), which was reported in a large autosomal dominant RP family,44 and one case showing uniparental isodisomy of chromosome 1 carrying c.11+5G>A.45 For the missense and noncanonical splice site variants, we show the in silico predictions of their pathogenic nature in Supplementary Tables S6 and S7, respectively. Based on the pathogenicity criteria described in the Materials and methods, 128 variants were designated ‘affects function', 10 with no functional effect and 12 as variants of unknown significance.
Missense variants that are present in ≥18 of ~60 000 individuals in ExAC (p.(A132T); p.(K294T); p.(N321K); p.(A434V); each of which are present in >300 alleles in ExAC) are considered as variants with no functional effect. Altogether, 231 and 144 individuals carried homozygous and compound heterozygous variants, respectively. The number of individuals with heterozygous variants is more difficult to assess as the RPE65 gene was not fully sequenced in many studies. For 6 individuals with one protein-truncating variant and 10 individuals with one probably function-affecting missense variant, the entire gene was analyzed by sequence analysis.
The three most frequent variants are c.271C>T; p.(R91W), c.1102T>C; (p.Y368H) and c.11+5G>A, identified in 100/864 (11.6%), 69/864 (8%) and 62/864 (7.2%) of the disease-causing alleles, respectively.
Discussion
In this study, disease-causing variants were identified in 49% (42/86) of the Danish LCA probands using different genotyping techniques. The recurrent mutation screening identified causal variants in 23% of the cases. The full potential of this approach was not used as APEX analysis and selective Sanger sequencing had previously been performed for diagnostic purposes in some cases. Assuming that these cases also had undergone the recurrent mutation analysis, the potential yield would have been 36% (Table 1), rendering it an inexpensive prescreening method for Danish LCA cases, and potentially also for other populations.
Sequence analysis of LRAT yielded no results but, remarkably, RPE65 was found to be the most frequently mutated gene (16%) in the Danish LCA cohort, followed by GUCY2D (10%), CEP290 (7%), CRB1 (7%), AIPL1 (4%), RDH12 (3%), TULP1 (1%), and RPGRIP1 (1%). We cannot rule out that as yet undiscovered LCA-associated genes are among frequently mutated genes in Danish LCA cases. Importantly, the most frequent RPE65 variant identified in this study was p.(D110G) (seven alleles), which hitherto was not described. It thereby is almost as frequent as the CEP290 variant c.2991+1655A>G, found in eight alleles, and equally frequent as the AIPL1 variant p.(W278*) in seven alleles. The p.(D110G) variant was found in individuals from Denmark and Jordan, and hence it remains to be investigated whether this is a founder mutation.
This study revealed the thus far highest prevalence for RPE65 variants in the Caucasian population. An equally high frequency of RPE65 variants (16.5%) was reported recently in the Indian population.46 Our findings facilitate the identification of Danish patients who are eligible for the AAV-RPE65 gene augmentation and/or 9-cis-retinoid supplementation therapies. By including the sequence analysis of RPE65 exons 1, 3, 4 and 10, carrying 19 variants, to the recurrent mutation screening (8 exons) for Danish LCA cases, the potential yield would increase to 45%. The clinical phenotype of patients with two RPE65 variants was relatively mild compared with several other more severe genetic types of LCA and in general similar to what was previously reported.36
In five individuals with LCA (F1, F5, F14, F28, F59), one heterozygous variant was identified upon sequence analysis of the entire coding regions of the corresponding genes. The GUCY2D variant p.(R838P) has been reported in previous LCA studies as an autosomal dominant mutation. In our study, this variant occurs in a sporadic case. In the absence of parental DNAs, we cannot test the possibility that it is a de novo variant. In the remaining cases, the identification of heterozygous variants could be attributed to one of the following possibilities. A deletion or duplication in the other allele may not be detected as copy number variant analysis was not performed. Our screening also does not cover deep-intronic regions, and thus variants in this part of the genome will not be identified.15, 47 Moreover, variants in regulatory elements located far away from the respective genes cannot be ruled out. Finally, the aggregate carrier frequency of variants in the most intensively studied genes in this study (AIPL1, CEP290, CRB1, GUCY2D, LRAT and RPE65) is estimated 1 in 25 individuals (Supplementary Table S3), meaning that three or four probands could carry a variant in one of these genes in this cohort of 86 families, without pathologic consequences. The latter may also explain the identification of more than two variants in different genes in four probands in this study. The identification of the remaining causal variants requires a more comprehensive analysis of all genes associated with LCA and early-onset RP, for example, using next-generation sequencing-based gene-panel sequencing, whole-exome or whole-genome sequencing.
By collecting all RPE65 variants reported in literature, we found RPE65 variants in 539 individuals, of whom 375 carried two variants. Among all reported variants, several variants tend to occur exclusively or in high frequency in certain ethnicities suggesting possible founder effects, for example, variant c.304G>T; p.(E102*) in the Greek population, c.1102T>C; p.(Y368H) in the Dutch population and c.271C>T; p.(R91W) in Saudi Arabia and Tunisia. Three variants (p.(R91W), p.(Y368H) and c.11+5G>A) together comprise 26.7% of all RPE65 variants identified to date. One splice site variant (c.94+10A>G) and 10 missense variants were designated benign. However, the latter missense variants are rare in LCA patients and in ExAC and therefore in the absence of functional assay data cannot completely be excluded to have functional effects.
Sixteen individuals carried only one pathogenic RPE65 variant, despite the fact that all exons and exon–intron boundaries were analyzed by Sanger sequencing. As described above and in Supplementary Table S3, we estimate the carrier frequency for RPE65 variants in the general population to be 1/158 individuals. Among the 516 individuals with one or two recessive RPE65 variants, three or four would be expected to carry such a variant in a heterozygous state by chance. The fact that 16 individuals were found to be heterozygous carriers of variants may point to the existence of ‘missed mutations'. These could be heterozygous deletions, deep-intronic variants or other variants affecting the expression of RPE65 mRNA. Individuals carrying two causal RPE65 variants are eligible for the above-mentioned novel therapies.
In summary, we identified the underlying mutations in a significant group of cases providing the most comprehensive genetic data of LCA in the Danish population. Most significantly, we identified RPE65 as the most frequently mutated LCA gene in Denmark, and this is of particular interest in view of the ongoing therapeutic trials using gene augmentation or 9-cis retinoid supplementation. Finally, we collected all published variants of the RPE65 gene and performed in silico assessment of the missense and noncanonical splice variants to predict their functional effects.
Acknowledgments
We thank Hanne Jensen and Susanne Timshel from the Kennedy Center who assisted with clinical and genetic counseling of the patients and Lonneke Haer-Wigman for statistical analysis in this paper. The research of GDNA was supported by the Directorate General for Higher Education (DIKTI) of the Ministry for National Education of Indonesia and the Radboud University Medical Center, Nijmegen, The Netherlands. The research of MA was supported by Pro-Retina, the Curing Retinal Blindness Foundation and by C Fasser. The research of MB and TR was supported by The Danish Association of the Blind.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Koenekoop RK: An overview of Leber congenital amaurosis: a model to understand human retinal development. Surv Ophthalmol 2004; 49: 379–398. [DOI] [PubMed] [Google Scholar]
- Stone EM: Leber congenital amaurosis - a model for efficient genetic testing of heterogeneous disorders: LXIV Edward Jackson Memorial Lecture. Am J Ophthalmol 2007; 144: 791–811. [DOI] [PubMed] [Google Scholar]
- Leber T: Über Retinitis pigmentosa und angeborene Amaurose. Albrecht von Graefe's Arch Klin Exp Ophthalmol 1869; 15: 25. [Google Scholar]
- den Hollander AI, Roepman R, Koenekoop RK, Cremers FP: Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog Retin Eye Res 2008; 27: 391–419. [DOI] [PubMed] [Google Scholar]
- Hamel CP: Gene discovery and prevalence in inherited retinal dystrophies. C R Biol 2014; 337: 160–166. [DOI] [PubMed] [Google Scholar]
- Retinal Information Network (RETNET). Available at https://sph.uth.edu/retnet.
- Maguire AM, Simonelli F, Pierce EA et al: Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med 2008; 358: 2240–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge JW, Smith AJ, Barker SS et al: Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med 2008; 358: 2231–2239. [DOI] [PubMed] [Google Scholar]
- Hauswirth WW, Aleman TS, Kaushal S et al: Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial. Hum Gene Ther 2008; 19: 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLaren RE, Groppe M, Barnard AR et al: Retinal gene therapy in patients with choroideremia: initial findings from a phase 1/2 clinical trial. Lancet 2014; 383: 1129–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenekoop RK, Sui R, Sallum J et al: Oral 9-cis retinoid for childhood blindness due to Leber congenital amaurosis caused by RPE65 or LRAT mutations: an open-label phase 1b trial. Lancet 2014; 384: 1513–1520. [DOI] [PubMed] [Google Scholar]
- den Hollander AI, Black A, Bennett J, Cremers FP: Lighting a candle in the dark: advances in genetics and gene therapy of recessive retinal dystrophies. J Clin Invest 2010; 120: 3042–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander AI, Koenekoop RK, Yzer S et al: Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet 2006; 79: 556–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault I, Delphin N, Hanein S et al: Spectrum of NPHP6/CEP290 mutations in Leber congenital amaurosis and delineation of the associated phenotype. Hum Mutat 2007; 28: 416. [DOI] [PubMed] [Google Scholar]
- Coppieters F, Casteels I, Meire F et al: Genetic screening of LCA in Belgium: predominance of CEP290 and identification of potential modifier alleles in AHI1 of CEP290-related phenotypes. Hum Mutat 2010; 31: E1709–E1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yzer S, Fishman GA, Racine J et al: CRB1 heterozygotes with regional retinal dysfunction: implications for genetic testing of leber congenital amaurosis. Invest Ophthalmol Vis Sci 2006; 47: 3736–3744. [DOI] [PubMed] [Google Scholar]
- Bujakowska K, Audo I, Mohand-Said S et al: CRB1 mutations in inherited retinal dystrophies. Hum Mutat 2012; 33: 306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelsen M, Jensen H, Bregnhoj JF, Rosenberg T: Prevalence of generalized retinal dystrophy in Denmark. Ophthalmic Epidemiol 2014; 21: 217–223. [DOI] [PubMed] [Google Scholar]
- Bertelsen M, Jensen H, Larsen M, Lorenz B, Preising MN, Rosenberg T: Prevalence and diagnostic spectrum of generalized retinal dystrophy in Danish children. Ophthalmic Epidemiol 2013; 20: 164–169. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF: A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preising M, Op de Laak JP, Lorenz B: Deletion in the OA1 gene in a family with congenital X linked nystagmus. Br J Ophthalmol 2001; 85: 1098–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernant J, Kulm M, Dharmaraj S et al: Genotyping microarray (disease chip) for Leber congenital amaurosis: detection of modifier alleles. Invest Ophthalmol Vis Sci 2005; 46: 3052–3059. [DOI] [PubMed] [Google Scholar]
- Tonisson N, Kurg A, Kaasik K, Lohmussaar E, Metspalu A: Unravelling genetic data by arrayed primer extension. Clin Chem Lab Med 2000; 38: 165–170. [DOI] [PubMed] [Google Scholar]
- Lohmueller KE, Sparso T, Li Q et al: Whole-exome sequencing of 2,000 Danish individuals and the role of rare coding variants in type 2 diabetes. Am J Hum Genet 2013; 93: 1072–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MQ: Statistical features of human exons and their flanking regions. Hum Mol Genet 1998; 7: 919–932. [DOI] [PubMed] [Google Scholar]
- Yeo G, Holste D, Kreiman G, Burge CB: Variation in alternative splicing across human tissues. Genome Biol 2004; 5: R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, Haussler D: Improved splice site detection in Genie. J Comput Biol 1997; 4: 311–323. [DOI] [PubMed] [Google Scholar]
- Pertea M, Lin X, Salzberg SL: GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res 2001; 29: 1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C: Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 2009; 37: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE: Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 2000; 15: 7–12. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Brown GR, Hiatt SM et al: RefSeq: an update on mammalian reference sequences. Nucleic Acids Res 2014; 42: D756–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo-de-Quintana J, Evans RJ, Cheetham ME, van der Spuy J: The Leber congenital amaurosis protein AIPL1 functions as part of a chaperone heterocomplex. Invest Ophthalmol Vis Sci 2008; 49: 2878–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AO, Abu-Safieh L, Eisenberger T, Bolz HJ, Alkuraya FS: The RPGRIP1-related retinal phenotype in children. Br J Ophthalmol 2013; 97: 760–764. [DOI] [PubMed] [Google Scholar]
- Hanein S, Perrault I, Olsen P et al: Evidence of a founder effect for the RETGC1 (GUCY2D) 2943DelG mutation in Leber congenital amaurosis pedigrees of Finnish origin. Hum Mutat 2002; 20: 322–323. [DOI] [PubMed] [Google Scholar]
- Perrault I, Rozet JM, Gerber S et al: Spectrum of retGC1 mutations in Leber's congenital amaurosis. Eur J Hum Genet 2000; 8: 578–582. [DOI] [PubMed] [Google Scholar]
- Felius J, Thompson DA, Khan NW et al: Clinical course and visual function in a family with mutations in the RPE65 gene. Arch Ophthalmol 2002; 120: 55–61. [DOI] [PubMed] [Google Scholar]
- Jacobson SG, Aleman TS, Cideciyan AV et al: Defining the residual vision in leber congenital amaurosis caused by RPE65 mutations. Invest Ophthalmol Vis Sci 2009; 50: 2368–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiden Open Variation Database (LOVD) tubby like protein 1 (TULP1). Available at: http://databases.lovd.nl/shared/genes/TULP1.
- Leiden Open Variation Database (LOVD) retinal dehydrogenase 12 (RDH12). Available at: http://databases.lovd.nl/shared/genes/RDH12.
- Leiden Open Variation Database (LOVD) crumbs homolog 1 (Drosophila) (CRB1). Available at: http://databases.lovd.nl/shared/variants/CRB1.
- Leiden Open Variation Database (LOVD) guanylate cyclase 2D, membrane (retina-specific) (GUCY2D). Available at: http://databases.lovd.nl/shared/genes/GUCY2D.
- Leiden Open Variation Database (LOVD) centrosomal protein 290kDa (CEP290). Available at: http://databases.lovd.nl/shared/genes/CEP290.
- Leiden Open Variation Database (LOVD) aryl hydrocarbon receptor interacting protein-like 1 (AIPL1). Available at: http://databases.lovd.nl/shared/genes/AIPL1.
- Bowne SJ, Humphries MM, Sullivan LS et al: A dominant mutation in RPE65 identified by whole-exome sequencing causes retinitis pigmentosa with choroidal involvement. Eur J Hum Genet 2011; 19: 1074–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DA, McHenry CL, Li Y et al: Retinal dystrophy due to paternal isodisomy for chromosome 1 or chromosome 2, with homoallelism for mutations in RPE65 or MERTK, respectively. Am J Hum Genet 2002; 70: 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma A, Perumalsamy V, Shetty S, Kulm M, Sundaresan P: Mutational screening of LCA genes emphasizing RPE65 in South Indian cohort of patients. PLoS One 2013; 8: e73172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun TA, Mullins RF, Wagner AH et al: Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Hum Mol Genet 2013; 22: 5136–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.