Summary
Adaptive evolution plays a large role in generating the phenotypic diversity observed in nature, yet current methods are impractical for characterizing the molecular basis and fitness effects of large numbers of individual adaptive mutations. Here we used a DNA barcoding approach to generate the genotype-to-fitness map for adaptation-driving mutations from a Saccharomyces cerevisiae population experimentally evolved by serial transfer under limiting glucose. We isolated and measured the fitness of thousands of independent adaptive clones, and sequenced the genomes of hundreds of clones. We found only two major classes of adaptive mutations: self-diploidization, and mutations in the nutrient-responsive Ras/PKA and TOR/Sch9 pathways. Our large sample size and precision of measurement allowed us to determine that there are significant differences in fitness between mutations in different genes, between different paralogs, and even between different classes of mutations within the same gene.
Introduction
Adaptive evolution is a major driving force behind the observed phenotypic diversity in nature (Darwin, 1872; reviewed in Givnish, 2015 and Soulebeau et al., 2015), and is of key importance to many problems of biomedical interest, including cancer (Greaves and Maley, 2012; Korolev et al., 2014; Landau et al., 2013; Nowell, 1976) and the emergence of drug resistance (Davies and Davies, 2010; Palmer and Kishony, 2013; Pennings, 2012; Toprak et al., 2012). To further understand the process of adaptation, it is essential to obtain a large, statistically representative number of individual adaptive events and determine their fitness effects and molecular nature.
While there are many methods for identifying instances of adaptive evolution in natural populations, they are not suitable for a comprehensive analysis of the spectrum of mutations that drive adaptation. Indeed, methods that infer selection in natural populations (reviewed in Lachance and Tishkoff, 2013; Oleksyk et al., 2010; Stinchcombe and Hoekstra, 2008; Vitti et al., 2013) are typically unable to identify adaptive mutations with single base-pair resolution, much less quantify the fitness effects of single adaptive mutations. Mechanistic studies can be conducted in genetically tractable systems where one can measure the fitness effects of a set of engineered mutations (Bank et al., 2014; Bozek et al., 2014; Fowler and Fields, 2014; Giaever et al., 2002; Hietpas et al., 2013; De Meester et al., 2002; Rich et al., 2016; Sliwa and Korona, 2005; Warringer et al., 2011; Weinreich et al., 2006). However, mutations studied in such systems are typically limited to a small, artificial, and predominantly deleterious subset of possible mutations, e.g. whole-gene knock-out mutations or deep mutational scanning of one or a few genomic regions.
In principle, microbial experimental evolution provides an excellent framework for the comprehensive study of adaptive mutations due to the ease of both identifying adaptive mutations, and assaying their fitness by pairwise competition. Two experimental evolution approaches for identifying large numbers of independent beneficial mutations are to either sequence multiple isolates from populations evolved under identical conditions (e.g., Barrick et al., 2009; Gresham et al., 2008; Kryazhimskiy et al., 2014; Kvitek and Sherlock, 2011; Tenaillon et al., 2012; reviewed in Dettman et al., 2012 and Long et al., 2015), or to conduct whole-population, whole-genome sequencing at multiple time-points during the evolution (Herron and Doebeli, 2013; Kvitek and Sherlock, 2013; Lang et al., 2013). However, these approaches are limited to identifying only a subset of high frequency and easy to sequence mutations. Moreover, separating the adaptive mutations from those that are merely hitchhiking remains a challenge (Voordeckers and Verstrepen, 2015). For example, in many studies the sequenced clones were isolated after hundreds or thousands of generations to ensure the presence of adaptive mutations, resulting in multiple mutations per clone (Barrick et al., 2009; Kryazhimskiy et al., 2014; Tenaillon et al., 2012). This makes it difficult to distinguish adaptive mutations from hitchhikers and also precludes the measurement of the fitness effects of individual beneficial mutations in isolation. By contrast, whole-population genome sequencing provides us only with the trajectories of easy to sequence mutations that rise to high frequencies (>1%), at which time they tend to be present in clones with multiple mutations, and their behavior is driven by complex clonal interference dynamics (Desai and Fisher, 2007; Herron and Doebeli, 2013; Kvitek and Sherlock, 2013); this prevents both the identification of very low frequency yet beneficial mutations and the precise estimation of their individual or marginal selective effects. Finally, fitness measurements are typically done in a low throughput, pairwise fashion, precluding generation of a comprehensive genotype-to-fitness map.
Here, we use our lineage tracking method (Levy et al., 2015) to solve these technological limitations and characterize both the genetic basis and fitness effects of hundreds of independent adaptive mutations in a laboratory evolution experiment using S. cerevisiae. Using DNA barcodes as neutral markers to track the frequencies of ~500,000 independent lineages during an evolution experiment, Levy et al. (2015) identified ~25,000 lineages that gained an adaptive mutation within the first 168 generations of evolution. We have now isolated thousands of clones from a single early time point in those experiments—a point at which we expect most adaptive lineages to carry single adaptive mutations—and identified their DNA barcodes. We then pooled these clones and monitored their barcode frequencies during short-term pooled growth. This allowed us to assign a fitness value to each of the clones, within the context of a single experiment. We then selected and sequenced the genomes of hundreds of known adaptive clones with varying fitness effects, as well as many neutral clones. Combining the sequencing and fitness measurements, we linked the molecular targets of adaptation to their fitness effects and thus built a comprehensive genotype-to-fitness map of the mutations that drove initial adaptive evolution in this system. Our results show that initial adaptation under these conditions is overwhelmingly driven by two distinct classes of mutations, which together explain the bimodal distribution of fitness effects observed in Levy et al. (2015).
Results
Isolation of thousands of evolved clones, and parallel measurement of their fitness
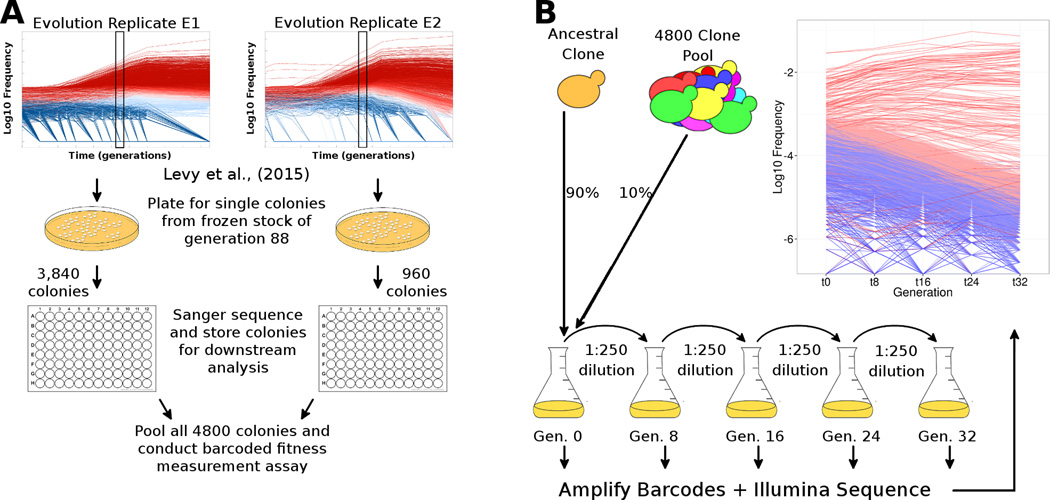
We isolated 4,800 random, single-colony-derived clones from frozen population samples taken at generation 88 from the Levy et al. (2015) experimental evolutions (Figure 1a, Table S1): 3,840 clones were from evolution replicate E1 and 960 clones from replicate E2. Those evolutions were performed by serial transfer in limiting glucose conditions, such that the populations grew for 8 generations each 48-hour growth/dilution cycle. The sampling generation and number of clones were chosen specifically to both maximize the fraction of clones with only a single adaptive mutation and allow fitness measurement assays to be cost-effective (see Methods and Resources, “M&R”). We unambiguously determined the barcode sequence for 4,149 of the clones via Sanger sequencing (M&R) and identified 4,009 unique barcodes with 140 duplicates, consistent with random sampling from the Levy et al. (2015) data.
Figure 1. Experimental procedures to select and measure fitness of evolved clones.
(A) Schematic of isolation and identification of individual evolved yeast clones. We isolated 4,800 single colonies from generation 88 across both replicate evolution experiments from Levy et al. (2015), determined their lineage barcodes, and stored them individually. (B) Schematic of barcoded fitness measurement assay. We grew all 4,800 colonies individually (not shown) and pooled them. The pool was mixed with an ancestral clone at a 1:9 ratio and the mixture was propagated for 32 generations in four independent batches (2–3 replicates per batch). At each transfer (every 8 generations), we isolated DNA, amplified the barcodes and conducted high-throughput sequencing to estimate the frequency trajectory of each barcode. The inset graph shows the frequency trajectory of all lineages with fitness > −1%, where adaptive lineages are colored in red (darker red lineages are more fit) and neutral lineages are colored in blue. Fitness was estimated using 24 generations of data from these frequency trajectories (M&R). Raw data for the sampled clones and their fitness measurements are in Tables S1–S3.
To measure the fitness, s, of each of these clones, we conducted fitness measurements in a single pooled assay (Figure 1b, Tables S2, S3). We grew each of the 4,800 clones independently in liquid media and then pooled equal volumes of their saturated cultures; this pool was then frozen as a stock culture to use for all subsequent fitness measurements described in this work, unless specified otherwise. For each assay, we re-grew the pool from a frozen stock of ~108 cells, then mixed it 1:9 with a population of the ancestral clone. We then propagated this mixed population by serial transfer through four 8-generation cycles for a total of 32 generations under conditions identical to the original evolution experiment (Figure 1b); the starting population size for each cycle was ~5×107 cells, large enough to minimize drift. This design allowed us to measure the fitness relative to the ancestor of each of the 4,800 clones in the pool without allowing substantial further adaptive evolution during the propagation. These measurements were conducted with 2–3 biological replicates across each of 4 different experimental batches (experiments conducted on different days).
The frequency of each barcode was measured after each transfer cycle by Illumina sequencing (M&R). We detected 3,883 of the 4,009 unique lineage barcodes; clones carrying the 126 missing barcodes may not have recovered from the frozen stock in high enough numbers to establish and thus were not present in the pool used for the fitness measurements. We used the frequency measurements from three of the four 8-generation cycles (for a total of 24 generations of data) to estimate the fitness of the 3,883 clones. Details of the fitness estimation, and extensive analysis of the fitness measurement errors and the batch effects are in the M&R and Figure S1–Figure S5. The distribution of fitness effects for all sampled lineages is shown in Figure S6.
The fitness values (s) reported throughout this work are the inverse variance weighted mean and sample standard error of the mean across the four batches of fitness measurements, and are quoted, following convention, as percent per generation. The fitness measurements are consistent across replicates within batches (Figure 2a, Figure S4) and between batches, although not to the same extent (Figure 2b–c, Figure S5). Sources of error between the replicates and batches include counting noise, caused by the growth/bottleneck dynamics of the assay itself, and from sampling and sequencing the DNA from the population, as well as intrinsic experimental noise. In addition, there appear to be systematic deviations among the batches. Batch 2 showed the largest systematic deviations (Figure S5), on the order of 6.5% for high fitness lineages (s > 5%) rather than the 1–2% deviations for all other batches (Figure 2b–c), which may be due to the slightly different measurement protocol used for this batch when compared to the other batches (M&R).
Figure 2. Fitness measurements are consistent across replicates and techniques.
(A) Comparison of fitness values for individual barcoded clones obtained from independent replicate assays conducted in the same experimental batch. (B–C) Comparison of fitness values for individual barcoded clones obtained from independent experimental batches (averaged over all replicates within a batch) of the fitness measurement assay. For A-C, a small number of lineages with extreme fitness estimates in at least one replicate (s < −5% or s > 20%) are not shown for increased resolution. A comparison of our fitness measurements using the 4,800 clone pool and (D) fitness measurements from a 500 clone pool, (E) to their barcode lineage fitness measurements from the Levy et al. (2015) lineage tracking estimates, and (F) the pairwise fluorescence competition assay measurements, from Levy et al. (2015). The solid lines on all panels are Y=X while X and Y error bars show the fitness measurement errors (M&R). For each panel, we report the mean and standard deviation of the difference in fitness for each comparison, grouped by low and high fitness clones. Systematic differences between measurements appear to be lower in low-fitness clones compared to high fitness clones, but the measurements are generally consistent throughout. We conducted extensive validation of our fitness estimation methodology, highlighted in Figures S1–S5.
Some deviations across the batches might be caused by slight differences in the growth conditions between batches or may be induced by different population compositions during the latter growth cycles of the fitness assay. We considered the possibility that a few lineages present at a substantial frequency in the pool (13 lineages at 1%-8% frequency) could drive non-linear effects at the latter growth cycles of the assay. To investigate this, we created a pool of 500 of the barcoded clones, providing us with a biological replicate of pooling, and specifically avoiding the introduction of the anomalously large lineages. We performed the fitness assay as for the larger pool and found that the fitness estimates remained largely unchanged (Figure 2d) with similar systematic batch deviations, on the order of ~3.2% for high fitness lineages (s > 5%) (see M&R). This indicates that most of the among-batch variation is likely to be driven by biological variability and not variation in the pool composition or a few anomalously large lineages. Overall, the systematic effects appear to be small compared to the measured fitness values, and our analysis below controls for the batch effect in all pairwise comparisons.
Our fitness measurements are consistent with those of Levy et al. (2015) as reported for both the lineage tracking fitness estimates (Figure 2e) and pairwise competition assays of single clones against a YFP-marked ancestor (Figure 2f). We suspect that the deviations in fitness in these assays when compared to our 4,800 pool estimates are largely due to batch effects, though we cannot rule out fitness differences due to frequency dependent effects as the adaptive clones begin each of these assays at a different starting frequency (see Levy et al. 2015 and M&R for details).
Note, a number of lineages were classified as adaptive by Levy et al. (2015), while our isolated clones from those lineages proved to be neutral, and vice versa (highlighted in Figure 2e). This is expected: adaptive mutants in lineages called adaptive by Levy et al. should generally comprise the majority but not all of the cells in their lineages. Thus, there will be instances where the sampled isolate from a lineage does not have the adaptive mutation. Conversely, some sampled isolates from lineages called neutral by Levy et al. will have acquired an adaptive mutation late enough in the evolution that the lineage was not classified as adaptive. The pooled-clone fitness measurements conducted in this study were thus critical for assigning fitness effects to our isolated clones (see below).
We determined that 59% of our 3,883 sampled lineages were adaptive (defined as s > 0% with 99% confidence); we refer to these clones as “adaptive”, and the clones falling outside the 99% confidence level as “neutral”. This 59% adaptive fraction is similar to the Levy et al. (2015) estimate of 50% adaptive lineages at generation 88.
Whole-genome sequencing
To determine the genetic basis of adaptation we conducted whole-genome sequencing for 418 of the 3,883 unique barcoded clones with assigned fitness estimates (M&R). These included 333 adaptive clones, consisting of nearly every sampled clone with s > 5% and many lower fitness clones (0% < s < 5%). To understand the spectrum of neutral mutations we also sequenced 85 neutral clones. Our sequenced clones thus covered the entire range of observed fitness values (Figure S6, blue bars). We obtained ~20x average and 5x minimum coverage for each clone. We called SNPs and short indels using a GATK based pipeline and manual curation, and larger structural variants were identified with CLC Genomics Workbench (M&R). Sanger sequencing of 57 randomly chosen mutations that passed manual curation revealed no false positives (M&R). Across all clones (adaptive and neutral), we identified a total of 445 mutations (Table 1, Table S4 and Data File S1), including 352 point mutations, 44 insertion/deletion events, 4 chromosomal aneuploidy events, and 45 transposable element (TE) insertion events. A total of 211 clones (188 adaptive clones) have more than one mutation.
Table 1. Summary of mutations observed in this study.
Mutations are tabulated by gene in different subsets of clones. “In/Del” stands for short “insertion/deletion” events in coding regions, while “TE” stands for “transposable element”. The column labeled “# Clones with no mutations outside locus” should be interpreted as e.g.: three adaptive haploid clones with CYR1 mutations, two of which have no mutations outside CYR1 and 237 adaptive diploid clones, 102 of which have no mutations aside from diploidy. See Table S4 and Data File S1 for details for all the mutations used in this study. See Figure S7 for validation that the diploidy mutations arose early in the evolution experiment.
| Class | Locus | # Clones |
# Clones with no mutations outside locus |
Synonymous | Missense | Nonsense | Coding In/Del |
Coding TE insertion |
Noncoding SNV |
Noncoding In/Del |
Noncoding TE insertion |
Chromosome Amplification |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neutral Diploid | ||||||||||||
| All | 3 | 1 | 1 | 1 | 1 | |||||||
| Neutral Haploid | ||||||||||||
| All | 82 | 34 | 6 | 25 | 1 | 8 | 20 | 3 | 11 | |||
| Adaptive Diploid | ||||||||||||
| All | 237 | 102 | 31 | 77 | 8 | 4 | 3 | 43 | 7 | 5 | 4 | |
| Adaptive Haploid with nutrient response pathway mutations | ||||||||||||
| IRA1 | 32 | 14 | 9 | 11 | 11 | 1 | ||||||
| IRA2 | 12 | 4 | 9 | 2 | 1 | |||||||
| GPB1 | 4 | 1 | 1 | 2 | 1 | |||||||
| GPB2 | 15 | 8 | 2 | 8 | 5 | |||||||
| PDE2 | 11 | 5 | 2 | 3 | 5 | 1 | ||||||
| RAS2 | 1 | 1 | ||||||||||
| TFS1 | 1 | 1 | ||||||||||
| CYR1 | 3 | 2 | 3 | |||||||||
| TOR1 | 1 | 1 | 1 | |||||||||
| KOG1 | 1 | 1 | 1 | |||||||||
| SCH9 | 1 | 1 | ||||||||||
| All other loci |
14 | 33 | 3 | 2 | 2 | 15 | 1 | 11 | ||||
| All | 82 | 14 | 64 | 29 | 25 | 4 | 15 | 1 | 11 | |||
| Adaptive Haploid without nutrient response pathway mutations | ||||||||||||
| All | 14 | 3 | 2 | 8 | 2 | 2 | 1 | 6 | 2 | |||
| All Adaptive Haploid (sum of above two classes) | ||||||||||||
| All | 96 | 3 | 16 | 72 | 31 | 27 | 5 | 21 | 1 | 13 | ||
Self-diploidization is an adaptive mechanism
In 83 adaptive clones, we observed the surprising presence of unambiguous heterozygous mutations, suggesting that many of the clones were diploid. To validate this, and to measure the frequency of diploidy, we developed a high throughput method to determine the ploidy of all 4,800 sampled clones, based on Upshall et al. (1977) (see M&R). This method takes advantage of the stronger growth inhibition at 25°C of diploid cells compared to haploid cells in media containing benomyl; our assay was 99% concordant with flow cytometry ploidy analysis of a sample of ~800 clones. Of the 4,800 clones, 43% from evolution E1 and 60% from evolution E2 were diploid (Table S1). We also performed mating assays (M&R) for ~1,200 randomly chosen clones, including haploids and diploids from both E1 and E2, and found that every clone behaved as a MATα strain (the mating type of the founding ancestor). Thus all of the diploids apparently arose via self-diploidization to generate MATα /MATα diploids, rather than by mating type switching and subsequent mating between haploids of opposite mating types. Such self-diploidization has been observed to be beneficial in a prior glucose-limited evolution experiment (Gerstein et al., 2006).
Of our whole-genome sequenced clones, 240 were diploid, of which the vast majority—237 (99%)—were measured as adaptive, with an average fitness benefit of 3.6% ± 0.6%. This included 12 clones used for the pairwise competition assays in Levy et al. (2015), which had an average fitness benefit of 3.5% in that assay, validating diploidy as an adaptive mutation. Aside from three diploid clones carrying an extra copy of chromosome 11 (discussed below), there was no significant difference in the fitness of adaptive diploid clones that contained no additional mutations (n=102), as compared to either diploids with additional mutations that do not alter protein sequence (n=53), or diploids containing additional mutations (i.e., missense, nonsense and insertion/deletion) that do alter protein sequence (n=79) (3.4% vs. 3.2% vs. 4.2%; P > 0.1; ANOVA). This strongly suggests that diploidy is the only driving adaptive mutation in most or all of these clones. Three of the sequenced adaptive diploid clones contained an extra copy of chromosome 11, which conferred a significant fitness advantage beyond diploidy alone (s = 7.6% ± 0.6%; P ≤ 0.0001; ANOVA test for each of the 4 batches of fitness measurements). One additional diploid clone contained an extra copy of chromosome 12, but was not significantly more fit than the average diploid (s = 4.6%, P > 0.1).
Of the 1,649 lineages that we determined to be diploids, 451 (27%) had been previously determined by the lineage tracking analysis of Levy et al. (2015)—without any knowledge of ploidy—to be lineages that were adaptive, with roughly the same fitness values, across both replicate evolution experiments. This suggests that many of these lineages were already self-diploidized by the time they were present in the barcoded population used to found the replicate evolutions; potentially the self-diploidization occurred during the transformation process itself when the barcodes were introduced into the cells. To investigate this, we measured the frequency of diploids throughout the Levy et al. replicate evolutions, and determined that at time zero the frequency of diploidy was low (~1%; Figure S7). We also conducted additional 200-generation evolution experiments using the experimental conditions of Levy et al. (2015) but using an isogenic non-barcoded haploid ancestral population (i.e., that had not undergone transformation) and found that < 0.1% of sampled clones were diploid at generation 88, indicating that spontaneous self-diploidization under our adaptive growth conditions is a rare event. The possibility of transformation-induced diploidy prevents us from accurately estimating a mutation rate for self-diploidization, but it is clear that whole-genome duplication alone is beneficial under our growth conditions with a fitness effect of ~3.4%.
Adaptive haploid clones typically carry a single adaptive mutation
Of the 418 clones we sequenced, 178 were haploid, of which 96 were adaptive and 82 neutral. We found a significant excess in the total number of mutations in adaptive haploid clones compared to neutral haploid clones (1.95 vs. 0.94 mutations per clone; P = 0.00004; ANOVA; Table 1); note, the observed number of mutations in neutral clones (0.94 per clone) is higher than the expected 0.5 events per clone after 88 generations, based on the mutation rate estimates of Levy et al. (2015). The source of this excess is unknown, though it is possible that mutations may have been induced by transformation of the DNA barcodes. It has been speculated that transformation is mutagenic (Giaever et al., 2002; Shortle et al., 1984), and would be consistent with the transformation-induced diploidy hypothesized above.
The adaptive clones have, on average, almost exactly one additional mutation compared to neutral clones, suggesting that they indeed carry only a single adaptive mutation. The adaptive haploid clones also have a significantly larger proportion of protein sequence altering mutations (i.e. missense, nonsense or insertion/deletion mutations) (73%) when compared to the neutral clones (46%) (Table 1; P = 0.0001, Fisher’s exact test), strongly suggesting that the additional mutations in the adaptive clones impact protein function.
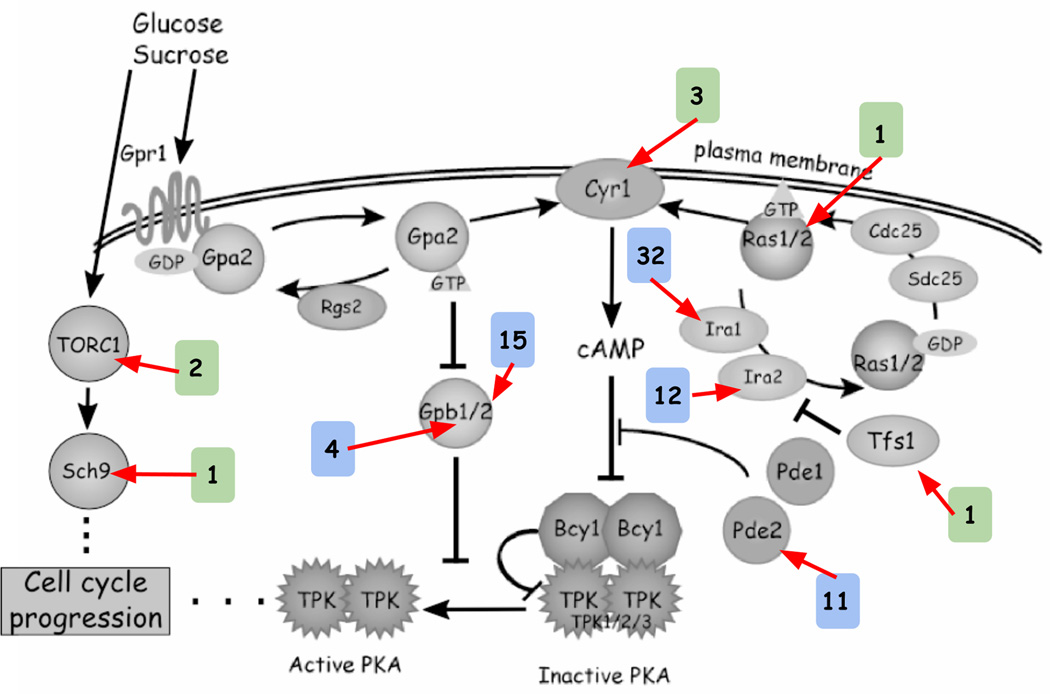
Adaptive haploids are enriched for mutations in the nutrient response pathways
A hallmark of adaptive mutations in laboratory evolution experiments is the finding of recurrent mutations within genes or pathways, which is unlikely under neutral evolution. We define candidate adaptive targets as those loci with at least two independent adaptive mutations among our sequenced clones. None of the protein-altering mutations found in the neutral clones occurred in the same gene; by contrast, 77 of the 135 (57%) protein-altering mutations in the adaptive clones were found in recurrently mutated genes (P = 10−11, Fisher’s exact test). All of these 77 mutations were found in clones with different barcodes and are thus independent. The recurrent mutations in the adaptive clones occurred in 6 genes (IRA1, IRA2, GPB1, GPB2, PDE2, CYR1), all of which are in the Ras/PKA pathway and are known to regulate yeast cell growth in response to glucose availability (reviewed in Conrad et al., 2014). A number of identical mutations occurred independently more than once: single mutations in CYR1, GPB1, and GPB2 and two different mutations in IRA1 each occurred twice independently, while a single mutation in PDE2 occurred independently four times. Mutations in this pathway have been identified as adaptive in previous glucose-limited yeast evolution experiments (e.g. Kao and Sherlock, 2008; Wenger et al., 2011, reviewed in Long et al., 2015), with selective effects of ~10% - 25% per generation in chemostats. We also observed one mutation in each of three different genes belonging to the TOR/Sch9 pathway (TOR1, KOG1, SCH9), which also integrates nutrient availability information with growth. We did not observe recurrent mutations in any other genes or pathways.
A total of 82 of our 96 (85%) sequenced adaptive haploid clones contained a mutation in either the Ras/PKA or TOR/Sch9 pathways (Figure 3, Table 1); 36 of these 82 clones had no other identified mutations, strongly indicating for these clones (and implying for the other clones) that the mutation in the Ras/PKA or TOR/Sch9 pathway gene is the causal adaptive mutation. We also note that four diploid clones (not included in the 82 described above) also carried mutations in the nutrient response pathway genes. Of the remaining adaptive haploid clones that did not have mutations in the Ras/PKA or TOR/Sch9 pathways, 3 were clones for which we were unable to identify any mutations, and 11 had mutations that did not appear to affect other nutrient response pathways (Table 2). We do not find any evidence for adaptive copy number changes in any of our haploid clones.
Figure 3. Schematic of the Ras/PKA and TOR/Sch9 pathways in yeast and the number of adaptive mutations found per gene.
The colored boxes denote the number of independent haploid lineages observed in our dataset with mutations in a particular gene. Blue boxes indicate mutations in negative regulators of cell cycle progression, while green boxes indicate mutations in positive regulators. Modified from Kao and Sherlock (2008) Figure S1.
Table 2. Mutations in adaptive haploid clones without a nutrient response pathway mutation.
| Lineage ID |
Fitness | Gene 1 | Gene 2 | Gene 3 | Gene 4 | Gene 5 |
|---|---|---|---|---|---|---|
| 7538 | 8.2% |
ERG1; missense; H220Y |
THI3; nonsense; S18* |
|||
| 7953 | 10.0% | |||||
| 13183 | 13.9% | |||||
| 14688 | 2.3% |
STE3; missense; I141T |
RXT2; missense; N63Y |
|||
| 18152 | 13.9% |
KTI12; missense; K208Q |
||||
| 21863 | 3.2% | |||||
| 26598 | 4.0% | YLR157W–E; TE insertion |
||||
| 53054 | 1.6% |
ATG17; Upstream SNV; 2655 bp A/G |
||||
| 60700 | 8.5% |
SSK2; nonsense; E702* |
||||
| 88494 | 6.4% |
NCL1; Upstream SNV; 374 bp T/A |
||||
| 225103 | 2.1% | YKL068W–A upstream TE insertion |
IES3; missense; N241T |
LAA1; Synonymous; T623T |
SEC4; Synonymous; N65N |
|
| 254044 | 2.2% |
TCB2; Upstream SNV; 99 bp C/A |
BAT1; Upstream SNV; 498bp A/T |
LEU4; missense; I264V |
DMA1; frameshift; 989bp |
SUP51 CYR1 Upstream TE insertion |
| 262917 | 2.6% |
FPK1; frameshift; 2113bp |
POP4; Upstream SNV; 246 bp C/G |
|||
| 304483 | 3.1% | YOL014W; missense; L65M |
BEM2; missense; D2054Y |
PAU16; Upstream SNV; 835 bp AT/TG |
In genes known to be positive regulators of the Ras/PKA and TOR/Sch9 pathways (RAS2, CYR1, TOR1, KOG1, SCH9 and TFS1) we identified only missense mutations, and for each of these genes there were only 1 to 3 clones with such mutations (Table 1). By contrast, in genes encoding negative regulators of the Ras/PKA pathway (IRA1, IRA2, GPB1, GPB2 and PDE2) many of the mutations were likely inactivating (insertion/deletion and nonsense) and mutations in these genes were observed much more frequently, with 4 to 32 mutant clones per gene (Table 1). These results suggest that most adaptive mutations in the positive regulator genes increase or modify activity (hypermorphic) and thus have a small mutational target size, while those in negative regulator genes of the nutrient response pathway decrease or abolish activity (hypomorphic). As expected of clones with hypermorphic mutations in the Tor/Sch9 pathway, those clones had increased rapamycin resistance (data not shown).
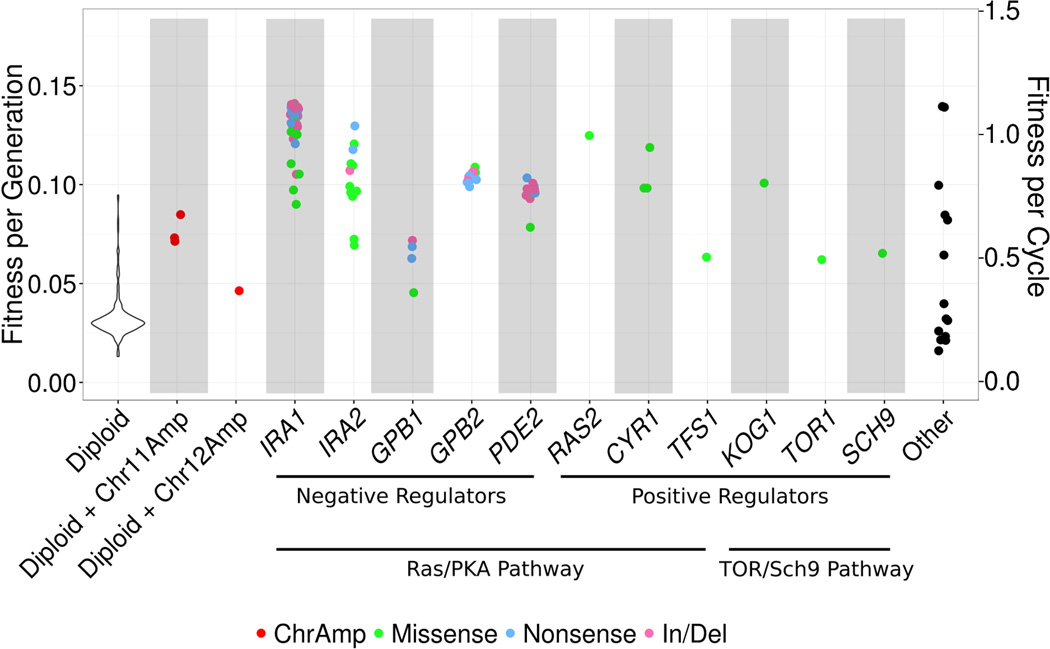
The fitness effect of a mutation is dependent on the gene and the mutation type
We integrated our genotype data with our fitness estimates to study the distribution of fitness effects for all of our major mutation classes, generating a genotype-to-fitness map for the initial driver mutations in our evolution experiment (Figure 4). As the fitness benefits may not necessarily be gained during exponential growth, we also provide an additional y-axis on the plot, showing the fitness per growth cycle (a factor of 8 larger). We found that most diploid clones have a fitness advantage close to the mean for diploids without other mutations (~3.4%) with variations consistent with counting noise (Figure S3), again suggesting that these clones have functionally identical adaptive mutations – that is, solely diploidy. By contrast, lineages with mutations in the Ras/PKA and TOR/Sch9 nutrient response pathways have fitness benefits ranging from 5% to 15%, depending on the gene and type of mutation, suggesting a lack of functional equivalency between different adaptive mutations within these nutrient response pathway genes. Together, the diploidy (s ~3.4%) and nutrient response pathway mutations (s ~5–15%) explain the two major fitness classes observed in Levy et al. (2015) (Figure 3b of that work) and in our fitness measurement assays (see Figure S6).
Figure 4. The fitness spectrum (genotype-to-fitness map) of evolved clones with different adaptive mutations.
The inverse variance weighted fitness averaged across all batches and replicates is plotted. Mutations are colored by their molecular basis (i.e. chromosomal amplification, insertion/deletion, nonsense or missense). The “other” class includes the 14 adaptive haploid clones for which we did not identify a nutrient response pathway mutation. Within-batch standard deviations (not shown for clarity) are ≤ 1% for > 90% of clones with nutrient response pathway mutations, while between-batch standard deviations are ~2% for all clones. To highlight the effect of single mutations on fitness, the six diploid clones with nutrient response pathway mutations are not shown. We show per-cycle fitness (8 generations per cycle) as a secondary y-axis (right side), as the fitness benefit of these mutations may not exclusively be due to changes in per-generation fermentative growth rate, but due to changes in other parts of the growth cycle such as growth lag, diauxic shift, aerobic growth, or increased viability after stationary phase. Figure S6 shows the distribution of fitness effects of our 4,800 sampled and 418 sequenced clones.
We conducted a number of ANOVA tests for the effects of gene identity, mutation type, and the presence of additional coding mutations on the fitness of our clones containing nutrient response pathway mutations. We found significant effects of both gene identity (P < 10−7; ANOVA), and mutation type (P < 10−3; ANOVA after controlling for gene effects for three of four batches) on the fitness of these lineages. These differences can even be found between paralogs: the 32 mutations in IRA1 confer a significantly greater fitness advantage, on average, than the 12 mutations in its paralog IRA2 (12.9% vs 10.2%) (P < 0.05; ANOVA), and mutations in GPB2 confer a significantly greater fitness advantage than mutations in GPB1 (10.4% vs 6.2%) (P < 10−4; ANOVA). In addition, missense mutations in IRA1 confer a significantly lower fitness benefit than nonsense or insertion/deletion mutations within the same gene (P ≤ 0.05, ANOVA for three of four batches).
The fitness distribution for lineages carrying mutations in GPB2 is remarkably narrow within replicates (standard deviation < 1% per generation across all replicates), particularly when compared to other nutrient response pathway genes such as IRA1 (standard deviation of 1–3% per generation). Note, this variation in GPB2 is substantially less than the average variation observed between replicates and batches for high fitness lineages (Figure 2). One possible explanation is that every mutation in GPB2 completely abolishes gene function; alternatively, partial loss of GPB2 function may still lead to the same level of Ras/PKA pathway activation as a complete loss of function, resulting in these highly consistent fitness estimates. In either case, the lack of fitness variation among the lineages with mutations in GPB2 demonstrates the precision of our fitness estimates and further suggests that the fitness differences observed between replicates and batches (Figure 2) may be due to biological variation in fitness due to slight differences in conditions rather than estimation error.
We also tested for the presence of additional adaptive mutations in the adaptive haploid clones containing nutrient response pathway mutations. We found that the 32 clones with both a nutrient response pathway mutation and an additional protein sequence altering mutation do not have a significantly different fitness than the 50 clones with a nutrient response pathway mutation alone (P < 0.05 for only one of the 4 batches; ANOVA controlling for gene and mutation type).
Not every gene in the Ras/PKA pathway is a target of adaptation
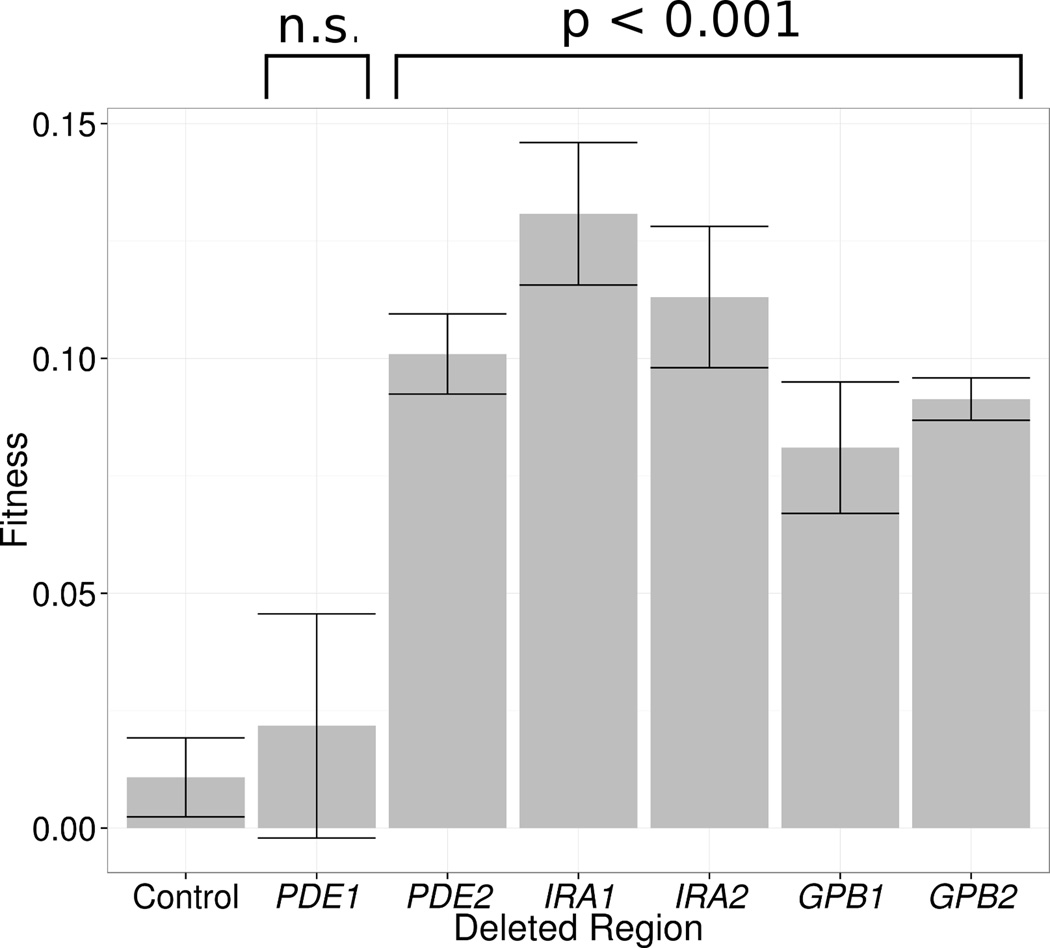
Among our sequenced adaptive clones, we found putative hypomorphic mutations in most of the negative regulators of the Ras/PKA pathway (IRA1, IRA2, GPB1, GPB2 and PDE2) but no mutations in PDE1. We hypothesized that PDE1 mutations did not confer a substantial fitness advantage, as Pde1 has a lower affinity for cAMP than Pde2 (Londesborough and Lukkari, 1980). To test this hypothesis, and to confirm that loss of any of the five negative regulators of the Ras/PKA pathway we observe as mutated is indeed adaptive, we constructed whole-gene deletions of IRA1, IRA2, GPB1, GPB2, PDE1, and PDE2, as well as the pseudogene YFR059C as a control, and assayed their fitness using fluorescence based pairwise competition assays (M&R). As predicted, we found that the fitness of the PDE1 deletion mutant was indistinguishable from neutrality, while deletion of the other genes was highly beneficial (Figure 5) with the fitness benefit roughly similar to that of the detected mutations in these genes.
Figure 5. Fitness of clones with synthetic whole-gene deletions in negative regulators of the Ras/PKA pathway.
Fitness was assayed using pairwise competitions against a fluorescently tagged ancestral clone. The plot shows the fitness for each of the constructed deletions and the error bars show standard error. P-values are for t-tests comparing the fitness of each gene deletion to the control, which is a deletion of the pseudogene YFR059C. n.s. stands for “non-significant”, meaning P > 0.05.
Discussion
One of the key goals of the study of adaptive evolution is to characterize the molecular basis and fitness effects of a comprehensive set of adaptation-driving mutations. We have overcome several challenges to achieve this goal: sampling a large number of independent clones without any bias for the type of adaptive event (e.g. point mutation vs. structural variant vs. epigenetic change), identifying adaptive events across the whole genome, and estimating the fitness effects of each of these mutations in a high-throughput manner, with high confidence and at a low cost per assay (~$0.07 per clone per replicate measurement). In addition, as exemplified by the small variation in the many independent fitness measurements for GPB2 mutants, our fitness measurements are both sensitive and precise.
By sampling adaptive mutations while they are still collectively a modest fraction of the population, we were able to identify the two major (and perhaps only) classes of adaptive mutations that drive early evolution in our experiment: (1) self-diploidization (s ~3.4%), and (2) presumably activating mutations in the Ras/PKA or TOR/Sch9 pathways (s ~5–15%). These two classes of mutations explain the fitness advantages of 319/333 (96%) of our sequenced adaptive clones, suggesting that in our system early-stage adaptation is driven by only a small number of mutational classes. We can also be certain that we didn’t miss a large class of difficult to identify adaptive events, such as mutations in repetitive regions, complex structural changes or epigenetic modifications.
We found a large number of recurring large-effect adaptive mutations in a small number of genes. In one case only a single member of a paralog pair, PDE2 and not PDE1, had any observed mutations. We confirmed that the reason we did not observe mutations in PDE1 was not due to insufficient sampling depth, but rather was due to PDE1 mutations not being adaptive under our experimental conditions. The results make us confident that we have generated a comprehensive map of the predominant adaptation-driving mutations in S. cerevisiae grown in one specific environment.
Note, we have not attempted to identify every potentially adaptive mutation in our experimental condition, rather we have identified most of the mutations that drive or are likely to drive the evolutionary dynamics of our system. In this system, with its well-mixed population, any adaptive mutation that is either too selectively weak or has a very low rate of occurrence cannot effectively drive the adaptive dynamics, because of clonal interference (Levy et al. 2015). For example, if the target sizes for adaptive mutations in two genes are k1 and k2 respectively, with selective advantages s1 and s2, then after a time T in a large population the ratio of the fractions of the population of the two classes of mutants are k1 exp(s1 T) and k2 exp(s2 T). If T=88 generations, as for our sampled clones, with s1 – s2 = 5%, and the same target sizes (k1 = k2), the mutant with 5% greater fitness benefit will be observed 100 times as often. However, the mutational target size is also important: if k2 were 100x larger than k1 (e.g., k2 includes many possible beneficial loss of function mutations while k1 includes only very few beneficial gain of function mutations), this compensates for the selective effect and mutations in the two genes will become comparable fractions of the population. Therefore, both selective advantage and the mutational target size are important in determining which mutations drive adaptive evolution.
The importance of both parameters may explain why we observed few candidate adaptive mutations in regulatory regions of Ras/PKA or TOR/Sch9 pathway genes. Indeed, we observed only one possible case, a transposon insertion upstream of the CYR1 gene, in a clone for which there were no other obvious adaptive mutations. Such mutations may therefore be rarer and/or confer a smaller selective advantage than changes to the actual protein sequences in our system and experimental condition.
The first key mutational event that we identified here was self-diploidization. The presence of a diploid fitness advantage in our growth condition is consistent with previous work showing that self-diploidization frequently fixes in yeast populations evolving under glucose limitation (Gerstein et al., 2006), but contrasts with the fitness disadvantage of diploids relative to haploids found under glucose limitation (Adams and Hansche, 1974; Zeyl et al., 2003) and no difference in fitness under nitrogen limitation (Hong and Gresham, 2014). Note however that these studies were performed in environmental conditions different from ours (chemostats vs. batch culture), which could significantly modify the relative fitness of haploids and diploids. This is consistent with a prior study which has found that the relative growth rates of haploid and diploid cells is highly dependent on both the specific strain genotype and the environment (Zörgö et al., 2013). A large body of work (reviewed in Otto, 2007) has sought an explanation for the evolution of diploidy in eukaryotes and the frequent polyploidization events in the evolutionary history of many organisms, including S. cerevisiae (Marcet-Houben and Gabaldón, 2015). Our work and that of Zörgö et al. (2013) suggest that diploidy may arise under some conditions due to a direct fitness advantage for diploids when compared to isogenic haploids. Further work is needed to determine the physiological basis for this fitness advantage and the generality of this advantage in other conditions.
The second major type of adaptive event targeted genes in the Ras/PKA and TOR/Sch9 nutrient response pathways. Previous work has shown that mutations in these pathways exhibit strong pleiotropic effects. For example, natural genetic variation present in many genes in the Ras/PKA pathway responds to selection for growth at 40°C (Parts et al., 2011). In addition, loss of function mutations in the TOR/Sch9 pathway result in an increased replicative lifespan (number of viable cell divisions per cell, Kaeberlein et al., 2005), as do mutants that decrease activity of the Ras/PKA pathway (Fabrizio et al., 2004; Lin et al., 2000). The study of the pleiotropic nature of fitness trade-offs (antagonistic pleiotropy), is critical to understanding adaptive evolution in the laboratory and in nature. Our DNA barcode based approach allows for the isolation and economic measurement of the individual fitness values of large pools of mutants, which will be of great use in investigating such evolutionary trade-offs.
In summary, we have conducted an in-depth survey of the molecular nature and associated fitness effects of the adaptive mutations in an evolving system, generating a genotype-to-fitness map for the mutations that drive the initial adaptive evolution. This approach opens the possibility of a far more in-depth understanding of adaptive evolution by de novo mutations and gives us a new way to assay the fitness landscapes in evolving systems comprehensively, economically, and precisely.
Supplementary Material
Acknowledgments
We wish to thank all members of the Petrov, Sherlock, and Fisher labs for useful discussions, and Michael Desai, Sergey Kryazhimskiy, Katja Schwartz, Dave Yuan, and Jake Cherry for technical help. We thank the Stanford Shared FACS facility for use of their flow cytometers, and the Stanford Center for Personalized Genomics and Medicine and NextSEQ for Illumina sequencing services.
Funding
SV is supported by NIH/NHGRI T32 HG000044 and the Stanford Center for Computational, Human and Evolutionary Genomics (CEHG); EE by NSF GFRP DGE-1247312; JC by NSF GRFP DGE-114747; AA by a Stanford Bio-X Bowes Fellowship; LH by NIH grant R01 GM110275 and a fellowship from CEHG; JB and SFL by the Louis and Beatrice Laufer Center; DSF by NSF PHY-1305433 and NIH R01 HG003328. The work was supported by NIH grants R01 HG003328 and GM110275 to GS and RO1 GM115919, GM10036601, and GM097415 to DAP. Data were collected on an instrument in the Shared FACS Facility obtained using NIH S10 Shared Instrument Grant RR027431.
Footnotes
Author Contributions
SV, BD, JB, SFL, DSF, GS and DAP conceived of the project and designed the experiments. SV, BD, YL, AA, JC, EE, KG-S, LH and JB conducted the experiments and analyzed the data. SV, BD, GS and DAP wrote the manuscript with substantial assistance from the other authors.
References
- Adams J, Hansche PE. Population studies in microorganisms. I. Evolution of diploidy in Saccharomyces cerevisiae . Genetics. 1974;76:327–338. doi: 10.1093/genetics/76.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank C, Hietpas RT, Jensen JD, Bolon DNA. A Systematic Survey of an Intragenic Epistatic Landscape. Mol. Biol. Evol. 2014;32:229–238. doi: 10.1093/molbev/msu301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. Genome evolution and adaptation in a long-term experiment with Escherichia coli . Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozek K, Wei Y, Yan Z, Liu X, Xiong J, Sugimoto M, Tomita M, Pääbo S, Pieszek R, Sherwood CC, et al. Exceptional evolutionary divergence of human muscle and brain metabolomes parallels human cognitive and physical uniqueness. PLoS Biol. 2014;12:e1001871. doi: 10.1371/journal.pbio.1001871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM, Zeebroeck G, Van Rubio-Texeira M, Thevelein JM. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae . FEMS Microbiol. Rev. 2014;38:254–299. doi: 10.1111/1574-6976.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin A, Heel K, Cowling WA, Nelson MN. An efficient high-throughput flow cytometric method for estimating DNA ploidy level in plants. Cytom. Part A. 2009;75:1015–1019. doi: 10.1002/cyto.a.20816. [DOI] [PubMed] [Google Scholar]
- Darwin C. The Origin of Species. London: John Murray; 1872. [Google Scholar]
- Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai MM, Fisher DS. Beneficial mutation selection balance and the effect of linkage on positive selection. Genetics. 2007;176:1759–1798. doi: 10.1534/genetics.106.067678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettman JR, Rodrigue N, Melnyk AH, Wong A, Bailey SF, Kassen R. Evolutionary insight from whole-genome sequencing of experimentally evolved microbes. Mol. Ecol. 2012;21:2058–2077. doi: 10.1111/j.1365-294X.2012.05484.x. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Pletcher SD, Minois N, Vaupel JW, Longo VD. Chronological aging-independent replicative life span regulation by Msn2/Msn4 and Sod2 in Saccharomyces cerevisiae. FEBS Lett. 2004;557:136–142. doi: 10.1016/s0014-5793(03)01462-5. [DOI] [PubMed] [Google Scholar]
- Fowler DM, Fields S. Deep mutational scanning: a new style of protein science. Nat. Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein AC, Chun H-JE, Grant A, Otto SP. Genomic convergence toward diploidy in Saccharomyces cerevisiae . PLoS Genet. 2006;2:e145. doi: 10.1371/journal.pgen.0020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gietz BRD, Woods RA, Peg DNA. Transformation of Yeast by Lithium Acetate / Single-Stranded Carrier DNA / Polyethylene Glycol Method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Givnish TJ. Adaptive radiation versus “radiation” and “explosive diversification”: why conceptual distinctions are fundamental to understanding evolution. New Phytol. 2015;207:297–303. doi: 10.1111/nph.13482. [DOI] [PubMed] [Google Scholar]
- Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D, Desai M, Tucker C, Jenq H, Pai D, Ward A, DeSevo C, Botstein D, Dunham M. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron MD, Doebeli M. Parallel Evolutionary Dynamics of Adaptive Diversification in Escherichia coli . PLoS Biol. 2013;11:e1001490. doi: 10.1371/journal.pbio.1001490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietpas RT, Bank C, Jensen JD, Bolon DNA. Shifting fitness landscapes in response to altered environments. Evolution. 2013;67:3512–3522. doi: 10.1111/evo.12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J, Gresham D. Molecular specificity, convergence and constraint shape adaptive evolution in nutrient-poor environments. PLoS Genet. 2014;10:e1004041. doi: 10.1371/journal.pgen.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, Steffen KK, Westman Ea, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kao KC, Sherlock G. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat. Genet. 2008;40:1499–1504. doi: 10.1038/ng.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev KS, Xavier JB, Gore J. Turning ecology and evolution against cancer. Nat. Rev. Cancer. 2014;14:371–380. doi: 10.1038/nrc3712. [DOI] [PubMed] [Google Scholar]
- Kryazhimskiy S, Rice DP, Jerison ER, Desai MM. Global Epistasis Makes Adaptation Predictable Despite Sequence-Level Stochasticity. Science. 2014;344:1519–1522. doi: 10.1126/science.1250939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitek DJ, Sherlock G. Reciprocal Sign Epistasis between Frequently Experimentally Evolved Adaptive Mutations Causes a Rugged Fitness Landscape. PLoS Genet. 2011;7:e1002056. doi: 10.1371/journal.pgen.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitek DJ, Sherlock G. Whole Genome, Whole Population Sequencing Reveals That Loss of Signaling Networks Is the Major Adaptive Strategy in a Constant Environment. PLoS Genet. 2013;9:e1003972. doi: 10.1371/journal.pgen.1003972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance J, Tishkoff Sa. Population Genomics of Human Adaptation. Annu. Rev. Ecol. Evol. Syst. 2013;44:123–143. doi: 10.1146/annurev-ecolsys-110512-135833. Vol 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau Da, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, Botstein D, Desai MM. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. 2013;500:571–574. doi: 10.1038/nature12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Blundell JR, Venkataram S, Petrov DA, Fisher DS, Sherlock G. Quantitative evolutionary dynamics using high-resolution lineage tracking. Nature. 2015;519:181–186. doi: 10.1038/nature14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae . Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Londesborough J, Lukkari TM. The pH and temperature dependence of the activity of the high Km cyclic nucleotide phosphodiesterase of bakers’ yeast. J. Biol. Chem. 1980;255:9262–9267. [PubMed] [Google Scholar]
- Long A, Liti G, Luptak A, Tenaillon O. Elucidating the molecular architecture of adaptation via evolve and resequence experiments. Nat. Rev. Genet. 2015 doi: 10.1038/nrg3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet-Houben M, Gabaldón T. Beyond the Whole-Genome Duplication: Phylogenetic Evidence for an Ancient Interspecies Hybridization in the Baker’s Yeast Lineage. PLOS Biol. 2015;13:e1002220. doi: 10.1371/journal.pbio.1002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meester L, Gomez A, Okamura B, Schwenk K. The monopolization hypothesis and the dispersal-gene flow paradox in aquatic organisms. Acta Oecologica. 2002;23:121–135. [Google Scholar]
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- Oleksyk TK, Smith MW, O’Brien SJ. Genome-wide scans for footprints of natural selection. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2010;365:185–205. doi: 10.1098/rstb.2009.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP. The Evolutionary Consequences of Polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Palmer AC, Kishony R. Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance. Nat. Rev. Genet. 2013;14:243–248. doi: 10.1038/nrg3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parts L, Cubillos FA, Warringer J, Jain K, Salinas F, Bumpstead SJ, Molin M, Zia A, Simpson JT, Quail MA, et al. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res. 2011;21:1131–1138. doi: 10.1101/gr.116731.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings PS. Standing genetic variation and the evolution of drug resistance in HIV. PLoS Comput. Biol. 2012;8:e1002527. doi: 10.1371/journal.pcbi.1002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich MS, Payen C, Rubin AF, Ong GT, Sanchez MR, Yachie N, Dunham MJ, Fields S. Comprehensive Analysis of the SUL1 Promoter of Saccharomyces cerevisiae . Genetics. 2016;203:191–202. doi: 10.1534/genetics.116.188037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D, Novick P, Botstein D. Construction and genetic characterization of temperature-sensitive mutant alleles of the yeast actin gene. Proc. Natl. Acad. Sci. U. S. A. 1984;81:4889–4893. doi: 10.1073/pnas.81.15.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliwa P, Korona R. Loss of dispensable genes is not adaptive in yeast. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17670–17674. doi: 10.1073/pnas.0505517102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulebeau A, Aubriot X, Gaudeul M, Rouhan G, Hennequin S, Haevermans T, Dubuisson J, Jabbour F. The hypothesis of adaptive radiation in evolutionary biology: hard facts about a hazy concept. Org. Divers. Evol. 2015;15:747–761. [Google Scholar]
- Stinchcombe JR, Hoekstra HE. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity (Edinb) 2008;100:158–170. doi: 10.1038/sj.hdy.6800937. [DOI] [PubMed] [Google Scholar]
- Tenaillon O, Rodriguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS. The Molecular Diversity of Adaptive Convergence. Science. 2012;335:457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- Toprak E, Veres A, Michel J-B, Chait R, Hartl DL, Kishony R. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat. Genet. 2012;44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upshall A, Giddings B, Mortimore ID. The Use of Benlate for Distinguishing Between Haploid and Diploid Strains of Aspergillus nidulans and Aspergillus terreus . J. Gen. Microbiol. 1977;100:413–418. [Google Scholar]
- Vitti JJ, Grossman SR, Sabeti PC. Detecting natural selection in genomic data. Annu. Rev. Genet. 2013;47:97–120. doi: 10.1146/annurev-genet-111212-133526. [DOI] [PubMed] [Google Scholar]
- Voordeckers K, Verstrepen KJ. Experimental evolution of the model eukaryote Saccharomyces cerevisiae yields insight into the molecular mechanisms underlying adaptation. Curr. Opin. Microbiol. 2015;28:1–9. doi: 10.1016/j.mib.2015.06.018. [DOI] [PubMed] [Google Scholar]
- Warringer J, Zörgö E, Cubillos FA, Zia A, Gjuvsland A, Simpson JT, Forsmark A, Durbin R, Omholt SW, Louis EJ, et al. Trait variation in yeast is defined by population history. PLoS Genet. 2011;7:e1002111. doi: 10.1371/journal.pgen.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- Wenger JW, Piotrowski J, Nagarajan S, Chiotti K, Sherlock G, Rosenzweig F. Hunger artists: yeast adapted to carbon limitation show trade-offs under carbon sufficiency. PLoS Genet. 2011;7:e1002202. doi: 10.1371/journal.pgen.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyl C, Vanderford T, Carter M. An evolutionary advantage of haploidy in large yeast populations. Science. 2003;299:555–558. doi: 10.1126/science.1078417. [DOI] [PubMed] [Google Scholar]
- Zörgö E, Chwialkowska K, Gjuvsland AB, Garré E, Sunnerhagen P, Liti G, Blomberg A, Omholt SW, Warringer J. Ancient Evolutionary Trade-Offs between Yeast Ploidy States. PLoS Genet. 2013;9:e1003388. doi: 10.1371/journal.pgen.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.