Abstract
Eukaryotic Argonaute proteins play primary roles in miRNA and siRNA pathways that are essential for numerous developmental and biological processes. However, the functional roles of the four ZmAGO1 genes have not yet been characterized in maize (Zea mays L.). In the present study, ZmAGO1a was identified from four putative ZmAGO1 genes for further characterization. Complementation of the Arabidopsis ago1‐27 mutant with ZmAGO1a indicated that constitutive overexpression of ZmAGO1a could restore the smaller rosette, serrated leaves, later flowering and maturation, lower seed set, and darker green leaves at late stages of the mutant to the wild‐type phenotype. The expression profiles of ZmAGO1a under five different abiotic stresses indicated that ZmAGO1a shares expression patterns similar to those of Argonaute genes in rice, Arabidopsis, and wheat. Further, variation in ZmAGO1a alleles among diverse maize germplasm that resulted in several amino acid changes revealed genetic diversity at this locus. The present data suggest that ZmAGO1a might be an important AGO1 ortholog in maize. The results presented provide further insight into the function of ZmAGO1a.
Keywords: AGO1, functional complementation, maize, ZmAGO1a
Edited by: Hailing Jin, University of California, Riverside, USA
Abbreviations
- 35S
CaMV35S promoter
- AGO
Argonaute protein
- Col 0
Arabidopsis thaliana (L.) ecotype Colombia
- DUF
domain of unknown function
- HMM
Hidden Markov Model
- Low N
low nitrate
- Low P
low phosphate
- Mid
middle domain
- miRNA
microRNA
- MS
Murashige and Skoog nutrient
- Mu
Mu transposon
- NCBI
the National Center for Biotechnology Information
- NJ
neighbor‐joining
- PAZ
Piwi‐Argonaute‐Zwille
- PEG
polyethylene glycol
- piRNAs
Piwi‐interacting RNA
- PPFD
photosynthetic photon flux density
- qRT‐PCR
quantitative real time PCR
- QTL
quantitative trait loci
- RISC
RNA‐induced silencing complex
- RNAi
RNA interference
- siRNA
small interference RNAs
- RNAs
short or small RNAs
- wt
wild‐type
INTRODUCTION
Argonaute (AGO) proteins are key mediators of all small‐RNA‐guided gene‐silencing processes involving ≈ 21–26 nt short RNAs (sRNAs) that are cleaved from double‐stranded or partially double‐stranded RNAs by the RNase III enzyme Dicer (Baulcombe 2004; Meister 2013). As key components of the RNA‐induced silencing complex (RISC), Argonaute proteins guide sRNAs to specific targets through sequence complementarity, leading to cleavage of target mRNA sequences, translational repression, or chromatin modification (Baumberger and Baulcombe 2005). Through their crucial roles in the regulation of eukaryotic gene expression, AGO proteins broadly participate in numerous biological processes, including developmental timing, cell differentiation, cell proliferation, cell death, metabolic control, immunity, transposon silencing, alternative splicing, and DNA repair, among other processes (Wei et al. 2012).
Argonautes are large proteins, approximately 90–100 kDa, consisting of one variable N‐terminal domain and conserved C‐terminal PAZ (Piwi‐Argonaute‐Zwille), Mid, and PIWI domains (Tolia and Joshua‐Tor 2007; Hutvagner and Simard 2008). AGO protein families generally can be classified into three groups in view of their phylogenetic relationships and capacity to bind to sRNAs (Vaucheret 2008). These include the AGO proteins that bind to miRNAs and siRNAs, the PIWI proteins that bind to piRNAs, and the proteins that bind to secondary siRNAs that have only been described in C. elegans (Yigit et al. 2006). As for crop plants, a total of 19 Argonaute genes in rice (Oryza sativa) and 15 in tomato (Solanum lycopersicum) have been identified (Kapoor et al. 2008; Bai et al. 2012). Among Argonaute proteins characterized so far, AGO1 plays essential roles in miRNA and siRNA pathways (Vaucheret 2008). AtAGO1 mediates cleavage of target mRNAs, which is the main proposed mechanism of plant miRNA action. The AtAGO1‐RISC complex has also been found to repress translation initiation (Iwakawa and Tomari 2013). Indeed, complementation analysis of ago1 mutant plants revealed that the catalytic residues of AtAGO1 are required to restore the normal wild‐type phenotype to the ago1 mutant, suggesting that the Slicer activity of AtAGO1 is essential for plant development (Carbonell et al. 2012). Complementation of Arabidopsis ago1 mutants with genes from other higher plant species that also encode AGO1 and with the other alleles of ZmAGO1 should be an effective way to examine the function of the ZmAGO1 genes we identified.
The organization of genes encoding AGO1 in the genome differs among species, even though AGO1 plays important roles in similar important developmental, immunity, and other biological processes. For example, the Arabidopsis genome contains a single AGO1 gene, but the protein it encodes shares some functional redundancies with other AGO proteins. For example, AtAGO10 shows functional redundancies with AGO1 during at least some aspects of development (Lynn et al. 1999). In contrast, four AGO1 genes have been found in rice that exhibit some functional redundancies and specialization (Wu et al. 2009). In previous work on maize AGO proteins, 18 AGO genes were identified in the maize genome by Hidden Markov Model (HMM) analysis (Qian et al. 2011). The temporal‐spatial expression patterns of 17 of these AGO genes were analyzed to identify meiosis‐specific AGO proteins in maize. A meiosis‐expressed AGO protein (ZmAGO18b) was specifically expressed during meiosis in pollen mother cells in maize and was enriched in the tapetum and germ cells of meiotic anthers (Zhai et al. 2014). Maize AGO104 was found to be required for the generation of male and female spores through meiosis, and to act in repression of somatic fate in germ cells (Singh et al. 2011). Four maize AGO1 genes have been predicted in other studies (Qian et al. 2011; Zhai et al. 2014), but it has been unclear whether these genes could be annotated as encoding functional AGO1 proteins. Here, we focused on ZmAGO1a to analyze its expression pattern in response to abiotic stress, its functional complementation of an Arabidopsis ago1 mutant, and the DNA sequence variation in this gene among diverse maize germplasm. The data and results presented here provide further insights into the expression and function of the AGO1 protein and the sequence diversity within this gene in maize.
RESULTS
Identification of Argonaute 1 genes in maize
Four putative Zea mays ZmAGO1 genes (Table 1), encoding both PAZ and PIWI domains, were predicted using an online DELTA‐BLAST tool against the maize genome (Zea mays, (TaxId:4577)) at http://blast.ncbi.nlm.nih.gov/Blast.cgi. The predicted results were consistent with previous studies (Qian et al. 2011; Zhai et al. 2014) that identified GRMZM2G361518, which had been designated ZmAGO1d by Qian et al. (2011), but was named ZmAGO1f by Zhai et al. (2014). The genomic location, intron/exon structure, and cDNA length of this gene have been reported previously (Qian et al. 2011; Zhai et al. 2014), but its genomic location has changed due to updates to the maize genome sequence. Based on the existing functional annotations, four OsAGO1 genes from rice, one AtAGO1 gene from Arabidopsis, and one TaAGO1 gene from wheat were chosen for analysis of their phylogenetic relationships with the four predicted putative ZmAGO1 genes. Phylogenetic analyses revealed a close evolutionary relationship among the predicted Zea mays AGO1 genes with those from rice, wheat, and Arabidopsis (Figure 1A). Among these AGO‐encoding genes, those from monocot species, ZmAGO1a, OsAGO1a, and TaAGO1b, were clustered together, indicating their relatively closer evolutionary relationship. Motif Scan (See Materials and Methods.) was used to search for conserved motifs across these known and putative AGO1 genes. Motif scanning revealed that these genes, except for OsAGO1c and TaAGO1b, all contain PIWI, PAZ, DUF1785, and Gly‐rich domains (Table 1). The four AGO1 genes in rice appear to be functionally redundant in the miRNA pathway (Wu et al. 2009). The genomic organization, phylogeny, and conserved domains of the ZmAGO1 genes suggested that the maize genes might share a function similar to that of the rice AGO1 genes. The expression profiles of four ZmAGO1 genes (Figure 1B) among different organs or tissues including seedling leaves and roots, young ear, pollinating tassels, and flowering silks revealed that ZmAGO1a had the highest expression levels across those observed organs or tissues, indicating it is the primary member of the families. In addition, the phylogenetic and expression profiling analyses suggest that functional characterization should focus initially on ZmAGO1a.
Table 1.
Conserved motifs among AGO1 gene homologs across maize, rice, wheat, and Arabidopsis
| Gene name | Accession number | Amino acid sequence regions of conserved motifs | |||||
|---|---|---|---|---|---|---|---|
| PIWI | PAZ | DUF1785 | Gly‐rich | Gln‐rich | Pro‐rich | ||
| AtAGO1 | AT1G48410 | 678–999 | 389–526 | 336–388 | 13–108 | 33–168 | – |
| ZmAGO1a | GRMZM2G441583 | 728–1050 | 438–575 | 385–437 | 8–148 | – | 48–181 |
| ZmAGO1b | AC209206.3_FG011 | 723–1042 | 424–570 | 371–423 | 8–132 | 180–201 | – |
| ZmAGO1c | GRMZM2G039455 | 708–1029 | 418–555 | 365–417 | 13–126 | – | – |
| ZmAGO1d | GRMZM2G361518 | 653–974 | 360–498 | 307–359 | 20–86 | – | – |
| OsAGO1a | LOC_Os02g45070 | 709–1030 | 419–556 | 366–418 | 29–130 | – | – |
| OsAGO1b | LOC_Os04g47870 | 746–1067 | 456–593 | 403–455 | 25–164 | 212–233 | – |
| OsAGO1c | LOC_Os02g58490 | 638–959 | 351–485 | 298–350 | – | 32–66 | – |
| OsAGO1d | LOC_Os06g51310 | 669–990 | 379–516 | 326–378 | 10–55 | – | – |
| TaAGO1b | AGB34310.1 | 497–818 | 207–344 | 154–206 | – | – | – |
Figure 1.
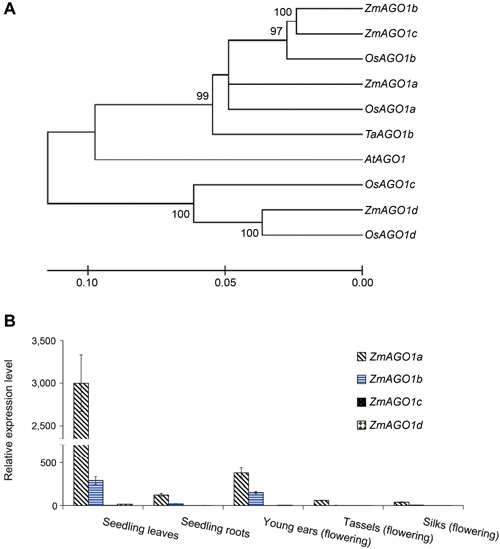
Phylogenetic analysis (A) of four predicted Zea mays AGO1 genes (ZmAGO1a‐1d) and their expression pattern (B) in selected organs in different developmental stages (A) AtAGO1, Arabidopsis AtAGO1; ZmAGO1a,1b, 1c and 1d were AGO1 genes in maize; OsAGO1a, 1b,1c, 1d were AGO1 genes in rice; TaAGO1b, wheat AGO1b. (B) Seedling leaves, 3‐week maize seeding leaves; Seedling roots, 3‐week maize seeding roots; Young ear, maize immature ear in flowering; Tassels, maize flowering male inflorescence; Silks, maize flowering (silking) silks in young ear. Note: The AGO1 gene accessions are listed in Table 1.
Complementation of Arabidopsis ago1‐27 with ZmAGO1a
ZmAGO1a has a gene structure typical of AGO1 genes (Figure 2A). The full‐length ZmAGO1a cDNA from Ye478, an important temperate inbred maize line from China, was isolated to construct a constitutive overexpression vector driven by the CaMV35S (35S) promoter (Figure 2B) in an Arabidopsis ago1‐27 mutant in the Columbia (Col) background (Figure 2C). Plants derived from three independent transformation events, lines 16, 19 and 21, the Arabidopsisago1‐27 mutant, and Arabidopsis Col wild‐type (wt) plants were grown in the same culture medium and under the same conditions (Liu et al. 2014) to compare their phenotypes (Figure 2C, D, E). As in a previous study (Morel et al. 2002), the ago1‐27 mutant developed smaller rosettes with darker green, serrated, narrower leaves; initiated flowering in 7–12 d; and produced around ≈ 50% of the seeds with typical wild type (Figure 2C, D, E; Table 2). At the seedling stage (Figure 2C), ZmAGO1a overexpression lines had larger leaves with shallower serrated leaf edges compared to the ago1‐27 mutant, but its leaf color was darker green than that of wild type (Figure 2C; Table 2). All 35S::ZmAGO1a‐transformed lines had significant differences in leaf length and width from the ago1‐27 mutant, indicating larger leaves and rosettes than in the mutant (Table 2). As shown by the leaf length/width ratio, the mutant had narrower leaves than the wild type and the 35S::ZmAGO1a‐transformed lines. Most of the phenotypic traits of the 35S::ZmAGO1a‐transformed lines, except for leaf color at the seedling stage, were comparable to those of wild type. However, the darker green leaf color disappeared at the flowering stage (Figure 2D; Table 2). ZmAGO1a overexpression lines 16, 19, and 21 appeared more similar to wild type at flowering time in terms of plant height, leaf number, and leaf color. However, these lines matured about 12 d earlier, and had higher silique numbers per plant and fertile seeds compared with the ago1‐27 mutant (Figure 2D, E; Table 2). Thus, constitutive overexpression of ZmAGO1a in the ago1‐27 mutant could nearly rescue the hypomorphicago1‐27 mutant phenotype.
Figure 2.
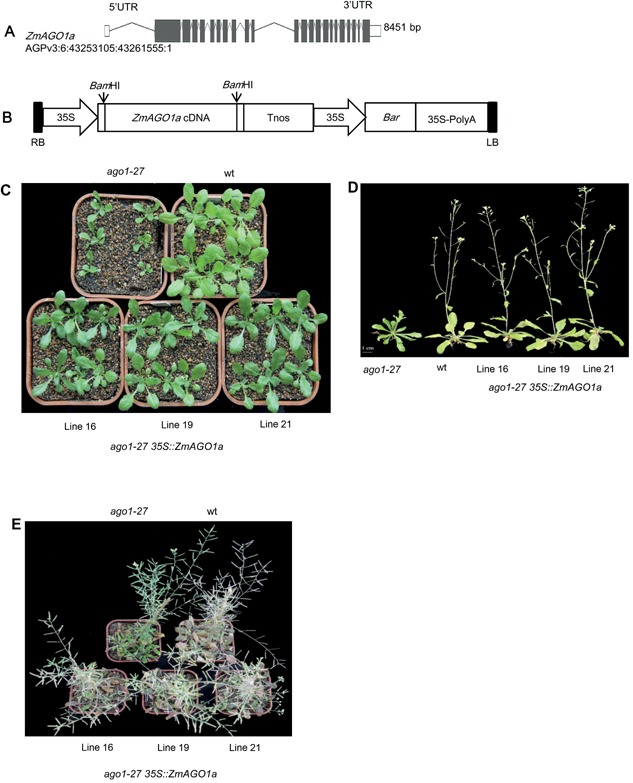
The effect of constitutive expression of ZmAGO1a in ago1‐27 of Arabidopsis (A) ZmAGO1a gene structure. Empty rectangles, untranslated region (5′ or 3′ UTR); Black rectangles, exons; Fold lines, introns; 8451bp, total genomic DNA length; AGPv3:6:43253105:43261555:1, maize genome ver3 physical region annotation. (B) The p35S::ZmAGO1a construct in which the ZmAGO1a cDNA was expressed under the control of CaMV35S promoter (35S) and NOS terminator (Tnos). Hollow arrows with the text “35S” inside indicated CaMV35S promoter. Bar, glufosinate ammonium resistance selection marker; 35S‐PloyA, CaMV35S polyA signal sequence; LB and RB, T‐DNA left and right borders. Vertical arrows indicated the restriction sites. (C) Phenotypic comparison of seedlings among the wild type, ago1‐27 and ago1‐27 carrying p35S::ZmAGO1a construct. (D) Phenotypic comparison at the flowering stage among the wild type, ago1‐27 and ago1‐27 carrying p35S::ZmAGO1a construct (note: the ago mutant flowered later). (E) Phenotypic comparison at the mature state among the wild type, ago1‐27 and ago1‐27 carrying p35S::ZmAGO1a construct (note: the ago mutant matured later.) C–E: wt, wild type Arabidopsis thaliana (L.) Colombia (Col‐0); ago1‐27, Arabidopsis thaliana (L.) ago1‐27 mutant in the Col‐0 background; line 16, 19, and 21: three independent homozygous ago1‐27 T3 lines transformed with 35S::ZmAGO1a.
Table 2.
Complementation by constitutive overexpression of ZmAGO1a in Arabidopsis ago1‐27 mutant
| Line | Genotype | Phenotypes on leaves | Growing days (d) | Silique and seed nos. per plant | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Leaf No. | Margin serration | Stages with dark green leaves | Length (cm) | Width (cm) | Length/width | Days to flower | Days to maturity | Silique count | Seed count | ||
| ago1‐27 | Columbia AtAGO1 mutant | 7 | +++ | vegetative and reproductive stages | 1.14 ± 0.03 C* | 0.53 ± 0.02 C* | 2.2 ± 0.09 A* | wt + 12 | wt + 12 | 32.4 ± 1.83 B* | 1458 ± 82.5 B* |
| 16 | ago1‐27 with 35S::ZmAGO1a | 10 | + | only at vegetative stage | 1.34 ± 0.03 B* | 0.80 ± 0.02 B* | 1.7 ± 0.05 B* | wt + 2 | Wt + 1 | 46.4 ± 1.50 A* | 2088 ± 67.6 A* |
| 19 | ago1‐27 with 35S::ZmAGO1a | 10 | + | only at vegetative stage | 1.49 ± 0.03 A* | 0.92 ± 0.03 A* | 1.6 ± 0.03 B* | wt + 1 | Wt + 1 | 46.6 ± 1.40 A* | 2097 ± 63.0 A* |
| 21 | ago1‐27 with 35S::ZmAGO1a | 10 | + | only at vegetative stage | 1.50 ± 0.04 A* | 0.90 ± 0.02 A* | 1.7 ± 0.02 B* | wt + 1 | Wt + 1 | 47.4 ± 0.87 A* | 2133 ± 39.2 A* |
| wt | Columbia C0 (wild type) | 11 | − | Control | 1.58 ± 0.03 A* | 1.00 ± 0.02 A* | 1.6 ± 0.04 B* | wt | wt | 48.0 ± 3.89 A* | 2160 ± 174.9 A* |
“*” Values followed by different letters are significantly different according to Tukey's HSD (Honestly Significant Difference) test at a significance level of 0.05.
To provide a full view of the functional complementation of Arabidopsis ago1‐27 by ZmAGO1a, profiles of the restored phenotypes of transformants from the T1 and T2 generations are shown in Figure 3. Of ∼ 2,300 transformed T1 seeds screened, 21 bialaphos‐resistant plants were recovered and confirmed for the presence of the integrated transgene using polymerase chain reaction (PBR). The phenotypes of eight of 21 T1 lines were fully rescued (Figure 3A), and 13 of them exhibited partially restored phenotypes for leaf color, leaf morphology, or both (Figure 3A). The phenotype of four T2 lines (Figure 3B) including three selected T3 homozygous lines (Figure 2C, D, E) was well restored. The result shows that the functional complementation was quite stable both within individual lines and across generations.
Figure 3.
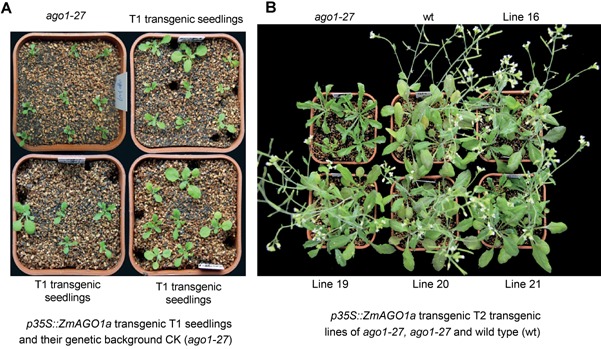
Overexpression of ZmAGO1a in Arabidopsisago1‐27 (T1 and T2 generations) and its rescue effect on phenotype (A) p35S::ZmAGO1a transgenic T1 seedlings and their genetic background CK (ago1‐27). (B) Phenotyping comparison of T2 transgenic lines (line 16, 19, 20, and 21) at genetic background of ago1‐27 with ago1‐27 and wild type (wt. Col‐0). ago1‐27 was also Col‐0 background but with ago1 mutation.
Expression of ZmAGO1a among ZmAGO1a transformants and expression of endogenous AtAGO1 in the ago1‐27 mutant and wild type
To analyze the expression of ZmAGO1 and native AtAGO1, quantitative analyses of relative mRNA expression levels of the AGO1 gene were performed on each independent ZmAGO1a transformant Arabidopsis line, and on the Arabidopsis ago1‐27 mutant and wild type. The expression of ZmAGO1 relative to alpha‐Tubulin 4, the internal reference gene, was over 2000‐fold higher in the Arabidopsis ZmAGO1a transformant lines 16, 19, and 21, compared to the same internal reference gene in the ago1‐27 mutant and wild‐type Arabidopsis (Figure 4A). These results indicate that the ZmAGO1a transgene was expressed at high levels across different tissues when transformed into the Arabidopsis ago1‐27 background. However, the expression of endogenous AtAGO1 was 3.7‐fold higher in ago1‐27 than in Col wild type (Figure S1). This result was consistent with a previous study suggesting that the single‐nucleotide mutation in AtAGO1 results in compensatory overexpression of the AtAGO1a mRNA and AtAGO1 protein in the ago1‐27 background (Mallory et al. 2009). Such mRNA overexpression was also observed among the Arabidopsis ZmAGO1a overexpression lines.
Figure 4.
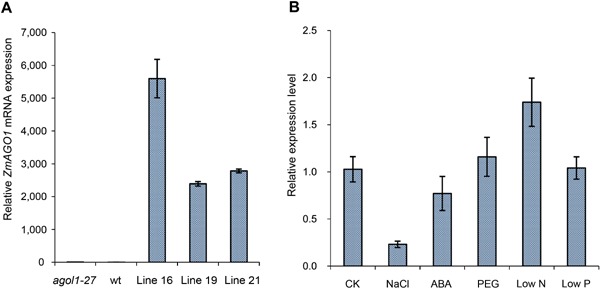
Relative expression levels of ZmAGO1a (A) in different Arabidopsis genotypes and relative expression level of ZmAGO1a in response to different abiotic stresses in 3‐weeks‐old maize seedling leaves (A) wt, wild type Arabidopsis thaliana (L.) Colombia (Col‐0); ago1‐27, Arabidopsis thaliana (L.) ago1‐27 mutant in the Col‐0 background; lines 16, 19, and 21, three independent ago1‐27 lines transformed with 35S::ZmAGO1a. (B) CK, control; NaCl, salt stress (NaCl, 200 mM); ABA, 100 mΜ ABA; PEG, dehydration (PEG‐6000; 20% w/v); LN, low nitrate (0.04 mM NO3 −); LP, low phosphorus (2.5 µM KH2PO4).
Expression profiles of ZmAGO1a in response to diverse abiotic stresses in maize
To generate an expression profile of ZmAGO1a to analyze the role(s) of this gene during abiotic stress responses, five diverse abiotic stresses, including dehydration (polyethylene glycol (PEG) treatment), salt (NaCl), ABA, low nitrate (low N) and low phosphate (low P), were applied at the seedling stage as described in Materials and Methods. Young leaves, the organs with the highest level of ZmAGO1aexpression at this stage, were chosen for analysis of ZmAGO1a expression in response to these abiotic stresses.
ZmAGO1a was differentially expressed in leaves under different stress conditions. ZmAGO1a expression was slightly upregulated under low N, low P, and PEG treatment, increasing by 1.74‐, 1.04‐, and 1.16‐fold, respectively, (Figure 4B). A previous analysis in rice indicated that expression of the OsAGO1a and OsAGO1b genes was also slightly upregulated in seedlings under various abiotic stresses (Kapoor et al. 2008). Our results indicate that ZmAGO1a expression was obviously downregulated under salt stress (NaCl), while the previous study indicated that expression of both OsAGO1a and OsAGO1d were slightly downregulated in rice under salt and drought stresses (Kapoor et al. 2008). However, the effects of NaCl and PEG stress treatments on the expression of ZmAGO1a revealed in this study were consistent with those of previous studies in maize (Qian et al. 2011).
DNA sequence variation in ZmAGO1a genes
To gain insight into the diversity of the ZmAGO1a gene among lines within Zea mays, sequence variation in the ZmAGO1a gene among 87 lines representing both temperate and tropical maize germplasm was analyzed. In total, 105 SNPs were detected (Table S1). Most of the identified SNPs, 79 out of 105, were located in introns; however, 26 SNPs were located in exons (Table 3). Seven of 26 SNPs located in coding regions could result in amino acid changes. The allele frequencies of these variants, including those that resulted in protein sequence changes and those that did not, ranged from 5.75% to 36.78%, indicating that these variants were not rare alleles.
Table 3.
DNA sequence variation in the coding region of ZmAGO1a among resequenced 87 lines representing broad tropical and temperate maize germplasm
| Mutation type | DNA site | SNP | Amino acid site | Domain | Amino acid change | Nos. of variants (Tropical/Temperate) | Variant frequencies |
|---|---|---|---|---|---|---|---|
| Missense | 1567 | G‐A | 8 | Gly‐rich | G 8 D | 5 (5/0) | 5.75% |
| 1828 | G‐A | 95 | Gly‐/Pro‐rich | R 95 H | 32 (11/21) | 36.78% | |
| 1873 | G‐T | 110 | Gly/Pro‐rich | G 110 V | 7 (3/4) | 8.05% | |
| 2118 | G‐C | 192 | – | A 192 P | 8 (3/5) | 9.20% | |
| 6348 | G‐T | 770 | PIWI | Q 770 H | 32 (11/21) | 36.78% | |
| 6821 | A‐G | 870 | PIWI | T 870 A | 32 (11/21) | 36.78% | |
| 7219 | G‐C | 925 | PIWI | E 925 Q | 7 (3/4) | 8.05% | |
| Subtotal of missense mutation | 123 (47/76) | ||||||
| Synonymous | 1829 | T‐C | 95 | Gly‐/Pro‐rich | – | 21 (16/5) | 24.14% |
| 2015 | A‐G | 157 | Pro‐rich | – | 18 (14/4) | 20.69% | |
| 2288 | T‐C | 248 | – | – | 21 (16/5) | 24.14% | |
| 2294 | G‐T | 250 | – | – | 29 (20/9) | 33.33% | |
| 2410 | G‐A | 263 | – | – | 32 (11/21) | 36.78% | |
| 2545 | A‐G | 308 | – | – | 21 (16/5) | 24.14% | |
| 2584 | A‐G | 321 | – | – | 29 (20/9) | 33.33% | |
| 2605 | A‐C | 328 | – | – | 29 (20/9) | 33.33% | |
| 3001 | T‐G | 408 | DUF1785 | – | 29 (20/9) | 33.33% | |
| 6046 | C‐T | 695 | – | – | 29 (20/9) | 33.33% | |
| 6158 | C‐T | 733 | PIWI | – | 28 (19/9) | 32.18% | |
| 6270 | C‐T | 744 | PIWI | – | 32 (11/21) | 36.78% | |
| 6273 | A‐G | 745 | PIWI | – | 28 (20/8) | 32.18% | |
| 7245 | T‐C | 933 | PIWI | – | 32 (11/21) | 36.78% | |
| 7447 | A‐G | 970 | PIWI | – | 32 (11/21) | 36.78% | |
| 8018 | A‐G | 1071 | – | – | 29 (20/9) | 33.33% | |
| 8024 | G‐A | 1073 | – | – | 29 (20/9) | 33.33% | |
| 8048 | G‐A | 1081 | – | – | 21 (16/5) | 24.14% | |
| 8075 | C‐T | 1090 | – | – | 32 (11/21) | 36.78% | |
| Subtotal of synonymous mutation | 521 (312/209) | ||||||
| Total | 644 (359/285) | ||||||
Note: the reference allele for both DNA and protein sequence numbering was B73 ZmAGO1a.
DISCUSSION
Possible functional redundancy of ZmAGO1 genes
In contrast with Arabidopsis, which has a single AGO1 gene, maize possesses four AGO1 homologs, as does rice (Wu et al. 2009). Five maize AGO1 genes were identified based on HMM analysis of sequences coding for conserved PAZ and PIWI domains in a previous report (Qian et al. 2011). However, our analysis and another parallel analysis (Zhai et al. 2014) with a more stringent model indicated that only four of the maize AGO1 genes likely have biological activity. Consistent with previous reports (Qian et al. 2011; Zhai et al. 2014), our phylogenetic analysis showed that the four ZmAGO1 homologs were highly conserved and were closely related to other rice and wheat AGO1 homologs (Figure 1). Both some redundancy and some possible functional specialization among AGO1 genes in rice has been suggested by sequencing miRNAs identified by gel retardation assays of AGO1 immunoprecipitates from the in vitro interaction of AGO and miRNAs (Wu et al. 2009). However, the specialized functions and biological roles of each particular AGO1 are still unknown in maize. Our expression profiles data showed that the ZmAGO1a was the highest expression in diverse organs, suggesting its' primary role among family members (Figure 1B). The phylogeny and similar organization of AGO1‐encoding homologs in rice and maize suggest that functional redundancy of ZmAGO1 genes might be conserved. Among the four maize AGO1 genes, ZmAGO1a, which is very closely related to ZmAGO1b and ZmAGO1c and more conserved with AtAGO1 and OsAGO1, was selected for further analysis of gene expression and complementation analysis.
ZmAGO1a rescues phenotypic defects and gene expression of Arabidopsis ago1 mutants
In this study, we observed that constitutive overexpression of ZmAGO1a could rescue most of the phenotypic defects of the ago1‐27 mutant, including rosette size, leaf size, leaf shape, flowering time, maturation time, and seed set (Figure 2). Some mutant phenotypes, such as leaves that were darker green and more serrated than wild type, could be partially restored to normal green color at later stages and shallower serrated leaf edges (Figure 2). The partially rescued phenotypes observed in these experiments could have been due to expression patterns specific to the promoter used. In our transformants, the ZmAGO1a gene was driven by the CaMV35S promoter so that it would be highly and constitutively expressed (Figure 4). In the same organ and tissue, the expression of endogenous AtAGO1 was also 3.7‐fold higher in the ago1‐27 mutant and 3.0‐fold higher in the ZmAGO1a overexpression transformants of ago1‐27 than in wild‐type (Col‐0) Arabidopsis. Our results indicated that constitutive overexpression of ZmAGO1a could rescue most of the defects in the mutant phenotype, but could not rescue the impaired expression of native AtAGO1. So, AtAGO1 and ZmAGO1a in the ZmAGO1a overexpression transformants in ago1‐27 might share some but not all functional roles. The unusual expression of these genes may or may not have any effect on their downstream functions, unless a direct effect on their own expression is suggested. The level of AtAGO1 mRNA is maintained and modulated by feedback regulation of miR168 (Vaucheret et al. 2006). Obviously, a trans‐modulation element could affect the expression of the endogenous AtAGO1 gene, but not that of exogenous ZmAGO1a, due to the absence of the effector within the expression cassette.
Variation in ZmAGO1a sequences in maize
The functional basis of the genetic control of traits by quantitative trait loci (QTL) is due mostly to variation within numerous genes in a species like maize, which is one of the most diverse crops at both the molecular and phenotypic levels (Buckler et al. 2006). Such variation can occur within genes of interest in crop plant populations that have been subjected to both natural and artificial selection for a long time. Sequencing of 87 tropical and temperate maize lines at 10× depth was performed to analyze possible variation in the ZmAGO1a DNA sequence among these lines. Some missense mutations were found among these lines (Table 3), suggesting that ZmAGO1a could possibly contribute to phenotypic diversity in maize, as the function of AGO1 also contributes to the phenotype of other grass species such as rice (Wu et al. 2009).
Reverse genetic analysis of ZmAGO1a
A reverse genetics approach, which seeks to draw conclusions from the phenotypes that arise after mutation of a gene of interest, as by knockout via gene tagging or knockdown through RNA interference (RNAi), could provide solid biological evidence for functional redundancy of ZmAGO1a. For example, rice AGO1 genes have been knocked down using RNAi (Wu et al. 2009). By screening a public Mu transposon‐tagged maize library (Settles et al. 2007), we recovered one line (seed stock: UFMu‐04885) in which the 3′‐UTR region of ZmAGO1a was tagged. No obvious phenotype was evident in this line after the line was selfed until the Mu‐tagged ZmAGO1a allele was homozygous (data not shown). After obtaining this Mu‐tagged ZmAGO1a allele, more independent knockouts of each of the other AGO1 genes will be required to further test the proposed functional redundancy of ZmAGO1a and the other AGO1 genes in maize.
CONCLUSIONS
The Argonaute1 (AGO1) protein plays key roles in miRNA and siRNA pathways that are essential for plant developmental and biological processes. We performed bioinformatics searches for maize genes encoding proteins containing conserved motifs consistent with AGO1 function and cloned the ZmAGO1a cDNA. The AGO1 function of ZmAGO1a was confirmed by complementation of the Arabidopsis ago1‐27 mutant phenotype with a ZmAGO1a transgene that restores the mutant phenotype to wild type. The expression pattern of this gene and that of the endogenous AtAGO1a gene in response to various abiotic stresses indicated that ZmAGO1a might play an important role in both development and responses to environmental change as a member of the AGO gene family in maize.
MATERIALS AND METHODS
Plant materials, culture, and stress treatment conditions
Arabidopsis thaliana (L.) ecotype Colombia (Col‐0) was used as the wild type. The mutant ago1‐27 in the Col ecotype, which developed rosettes with dark green and serrated leaves, had been described previously. The rosette size of this mutant is intermediate between that of stronger alleles and wild‐type plants, and this mutant initiates flowering 7–12 d after the wild type and is fertile (Morel et al. 2002).
After seed sterilization, plants were grown in sterile in 0.5× Murashige and Skoog (MS) nutrient agar medium in a short‐day (8 h light/16 h dark) regimen under a PPFD (photosynthetic photon flux density) of 70 µmol/ m2/s and constant temperature of 23 ± 1 °C. After 10 d, the plants were transferred to soil in a growth chamber with 16 h light: 8 h dark at 23 ± 1 °C.
Maize (Zea mays L. inbred lines B73 and Ye478) plants were grown in a greenhouse under 16 h light: 8 h dark at 28 °C (Xu et al. 2011). Three‐week‐old seedlings were subjected to one of the five following abiotic stress treatments: low phosphorus (2.5 µM KH2PO4), low nitrogen (0.04 mM NO3 –), salt stress (NaCl, 200 mM), dehydration (polyethylene glycol, PEG‐6000; 20% w/v), or ABA (100 mΜ). Seedling leaves were sampled at 24 h after the treatment. The controls were treated with normal nutrient solution with no additives as listed (Xu et al. 2011).
Bioinformatic analyses and identification of Argonaute1 genes
AtAGO1 protein sequences from the MaizeGDB database (http://www.maizegdb.org/) and the National Center for Biotechnology Information (NCBI) were used to perform blastp queries against the maize (Zea mays, (TaxId:4577)) NR (Non‐redundant protein sequence)) database at NCBI using the newly developed algorithm DELTA‐BLAST (Domain Enhanced Lookup Time Accelerated BLAST). Amino acid alignments were conducted using DNAman software (LynnonBioSoft, Vaudreuil, Quebec, Canada). A phylogenetic tree was constructed based on this amino acid alignment result using the neighbor‐joining (NJ) method in MEGA5.05 (http://www.megasoftware.net/) with a Poisson model, pairwise deletion, uniform rates among sites, and 1000 bootstrap replicates. Protein domains were analyzed using Motif Scan (http://hits.isb-sib.ch/cgi-bin/motif_scan).
Cloning the full‐length ZmAGO1a cDNA and construction of the overexpression vector
The primers for ZmAGO1a cDNA isolation were designed using Primer 5.0 (Premier Biosoft International, Palo Alto, CA, USA). The sense and antisense primers were 5′‐ggactctagaggatccCCCTTTCTCCGTCCGCAT‐3′ and 5′‐cggtacccggggatccCCCATCTTAATAGGAGTATATCGCA‐3′, respectively. PCR amplifications were performed using the HiFidelity PCR Kit (Tiangen Biotech Co., Ltd. Beijing, China). The temperature cycling program was 95 °C for 5 min, followed by 35 cycles of 95 °C for 15 s, 58 °C for 10 s, and 68 °C for 2 min, with a final extension at 68 °C for 10 min. PCR products were visualized on 1.0% agarose gels by ethidium bromide staining. The amplified fragment was subcloned into the CPB vector using the In‐Fusion HD Cloning Kit (Clontech Laboratories Inc., Mountain View, CA, USA). The vector was then isolated from Escherichia coli strain DH5α and transferred into A. tumefaciens strain GV3101 using the freeze‐thaw method (Clough and Bent 1998). The DNA sequence of the ZmAGO1a insertion in the plasmid construct in both E. coli and A. tumefaciens were verified by sequencing.
Plant transformation and identification of transformants
Arabidopsis was transformed using the Agrobacterium floral dip method following a reported method (Clough and Bent 1998). Plants of the mutant ago1‐27 in the Col‐0 background were used for transformation. Transgenic plants were screened on plates with 50 µg/mL glufosinate ammonium. The glufosinate‐resistant transformed plants were validated by PCR and with a strip test for detecting the bialaphos resistance protein encoded by the bar gene (QuickStix Kit for Liberty Link (bar) Cotton, EnviroLogix, Portland, ME, USA). T3 homozygous lines were used for all phenotyping and gene expression analysis.
Gene expression analysis
Quantitative (q)RT‐PCR primers were designed using Beacon Designer7 software (Premier Biosoft International, Palo Alto, CA, USA), and qRT‐PCR was performed using the Applied Biosystems 7500 Fast Real‐Time PCR System with the SYBR Premix Ex Taq II Master Mix (Takara Bio Inc., Dalian, China). The primers for analyzing ZmAGO1a expression profiles were 5′‐GCTTGACTTGCTGATGGTAATACT‐3′ and 5′‐CTGATGCTTGTTCGCCTTGA‐3′. The primer pairs for ZmAGO1b, ZmAGO1c and ZmAGO1d were 5′‐GCCCGAGGTCACGAAATAC‐3′ and 5′‐GGGTCCTGCCATACTTTGA‐3′, 5′‐TTCTATTGATGGTGCCTGTGC‐3′ and 5′‐CAGACAGAAGGCAAGCAAAACG‐3′, 5′‐TCCAGAAACGCCACCATA‐3′ and 5′‐ACGACAGTTCCAGGAAGTAT‐3′, respectively. The primers used to amplify GAPDH, the internal reference for expression analysis in maize, were 5′‐ACTGTTCATGCCATCACTGC‐3′ and 5′‐GAGGACAGGAAGCACTTTGC‐3′. Amplification was followed by a melting curve analysis and an appropriate blank was applied. The primers used to amplify AtAGO1 were 5′‐ATCAGACAGTGGCTCAAT‐3′ and 5′‐GACACGCTTCACATTCTC‐3′. The primers used to amplify Arabidopsis alpha‐Tubulin 4, the internal reference for expression analysis in Arabidopsis, were 5′‐GAGGGAGCCATTGACAACATCTT‐3′ and 5′‐GCGAACAGTTCACAGCTATGTTCA‐3′. The expression of both ZmAGO1a and AtAGO1 was analyzed in the mutant, wild type, and transformants using qRT‐PCR. The 2‐ΔΔCT method was used to estimate fold changes in the expression of each gene (Livak and Schmittgen 2001). Each data point represents four biological replicates with three technical replicates.
Analysis of DNA sequence variation in ZmAGO1a among maize populations
The resequencing data (mean Poisson‐distributed 10 × depth for each line) from 87 maize inbreds representing tropical (47 of 87) and temperate (40) lines (Table S1) were used to analyze DNA sequence variation in ZmAGO1a across lines. The data, SNP‐calling methods, and maize lines in our germplasm panel were described in our previous report (Xie et al. 2013). The resequencing data for ZmAGO1a have been deposited into GenBank at the NCBI under the accession numbers ranging from KJ958272 to KJ958358.
PCR confirmation of Mu insertions
The Mu insertion mutations (UFMu‐04885) in ZmAGO1a were submitted to the MaizeGDB database (http://www.maizegdb.org/documentation/uniformmu/). Confirmation of Mu insertions was performed according to McCarty et al. (2005), including primer sequences.
COMPETING INTERESTS
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
D.X. carried out the experiments. C.X. designed the study and analyzed data. H.Y. carried out transformation. Z.C. performed DNA sequence variation analysis. W.X.L. and Y.X. performed data analysis. D.X. drafted the paper, and C.X. and Y.X. wrote and revised the paper. All authors read and approved the final manuscript.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Table S1. Maize lines resequenced for analysis of DNA sequence variation
Figure S1. Relative expression levels of native AtAGO1 among wild type and ZmAGO1a transformed lines
ACKNOWLEDGEMENTS
This work was financially supported by the National Natural Science Foundation of China (31361140364 & 31171562), the National High Technology Research and Development Program of China (2012AA10A306) and The Agricultural Science and Technology Innovation Program (ASTIP) of CAAS to CX.
Xu D, Yang H, Zou C, Li WX, Xu Y, Xie C ( 2016) Identification and functional characterization of the AGO1 ortholog in maize. J Integr Plant Biol 58: 749–758
Available online on Feb. 5, 2016 at www.wileyonlinelibrary.com/journal/jipb
REFERENCES
- Bai M, Yang GS, Chen WT, Mao ZC, Kang HX, Chen GH, Yang YH, Xie BY ( 2012) Genome‐wide identification of Dicer‐like, Argonaute and RNA‐dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum . Gene 501: 52–62 [DOI] [PubMed] [Google Scholar]
- Baulcombe D ( 2004) RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC ( 2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler ES, Gaut BS, McMullen MD ( 2006) Molecular and functional diversity of maize. Curr Opin Plant Biol 9: 172–176 [DOI] [PubMed] [Google Scholar]
- Carbonell A, Fahlgren N, Garcia‐Ruiz H, Gilbert KB, Montgomery TA, Nguyen T, Cuperus JT, Carrington JC ( 2012) Functional analysis of three Arabidopsis ARGONAUTES using slicer‐defective mutants. Plant Cell 24: 3613–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF ( 1998) Floral dip: A simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ ( 2008) Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol 9: 22–32 [DOI] [PubMed] [Google Scholar]
- Iwakawa HO, Tomari Y ( 2013) Molecular insights into microRNA‐mediated translational repression in plants. Mol Cell 52: 591–601 [DOI] [PubMed] [Google Scholar]
- Kapoor M, Arora R, Lama T, Nijhawan A, Khurana JP, Tyagi AK, Kapoor S ( 2008) Genome‐wide identification, organization and phylogenetic analysis of Dicer‐like, Argonaute and RNA‐dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genomics 9: 451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WW, Tai HH, Li SS, Gao W, Zhao M, Xie CX, Li WX ( 2014) bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol 201: 1192–1204 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD ( 2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(T)(‐Delta Delta C) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK ( 1999) The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126: 469–481 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Hinze A, Tucker MR, Bouche N, Gasciolli V, Elmayan T, Lauressergues D, Jauvion V, Vaucheret H, Laux T ( 2009) Redundant and specific roles of the ARGONAUTE proteins AGO1 and ZLL in development and small RNA‐directed gene silencing. PLoS Genet 5: e1000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Settles AM, Suzuki M, Tan BC, Latshaw S, Porch T, Robin K, Baier J, Avigne W, Lai J, Messing J, Koch KE, Hannah LC ( 2005) Steady‐state transposon mutagenesis in inbred maize. Plant J 44: 52–61 [DOI] [PubMed] [Google Scholar]
- Meister G ( 2013) Argonaute proteins: Functional insights and emerging roles. Nat Rev Genet 14: 447–459 [DOI] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Béclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H ( 2002) Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post‐transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Cheng Y, Cheng X, Jiang H, Zhu S, Cheng B ( 2011) Identification and characterization of Dicer‐like, Argonaute and RNA‐dependent RNA polymerase gene families in maize. Plant Cell Rep 30: 1347–1363 [DOI] [PubMed] [Google Scholar]
- Settles AM, Holding D, Tan B, Latshaw S, Liu J, Suzuki M, Li L, O'Brien B, Fajardo D, Wroclawska E, Tseung CW, Lai J, Hunter C, Avigne W, Baier J, Messing J, Hannah LC, Koch K, Becraft P, Larkins B, McCarty D ( 2007) Sequence‐indexed mutations in maize using the UniformMu transposon‐tagging population. BMC Genomics 8: 116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Goel S, Meeley RB, Dantec C, Parrinello H, Michaud C, Leblanc O, Grimanelli D ( 2011) Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 23: 443–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolia NH, Joshua‐Tor L ( 2007) Slicer and the Argonautes. Nat Chem Biol 3: 36–43 [DOI] [PubMed] [Google Scholar]
- Vaucheret H ( 2008) Plant ARGONAUTES. Trends Plant Sci 13: 350–358 [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Mallory AC, Bartel DP ( 2006) AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell 22: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei KF, Wu LJ, Chen J, Chen YF, Xie DX ( 2012) Structural evolution and functional diversification analyses of argonaute protein. J Cell Biochem 113: 2576–2585 [DOI] [PubMed] [Google Scholar]
- Wu L, Zhang Q, Zhou H, Ni F, Wu X, Qi Y ( 2009) Rice microRNA effector complexes and targets. Plant Cell 21: 3421–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Weng J, Liu W, Zou C, Hao Z, Li W, Li M, Guo X, Zhang G, Xu Y, Li X, Zhang S ( 2013) Zea mays (L.) P1 locus for cob glume color identified as a post‐domestication selection target with an effect on temperate maize genomes. Crop J 1: 15–24 [Google Scholar]
- Xu Z, Zhong S, Li X, Li W, Rothstein SJ, Zhang S, Bi Y, Xie C ( 2011) Genome‐wide identification of microRNAs in response to low nitrate availability in maize leaves and roots. PLoS ONE 6: e28009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua‐Tor L, Mitani S, Simard MJ, Mello CC ( 2006) Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127: 747–757 [DOI] [PubMed] [Google Scholar]
- Zhai L, Sun W, Zhang K, Jia H, Liu L, Liu Z, Teng F, Zhang Z ( 2014) Identification and characterization of Argonaute gene family and meiosis‐enriched Argonaute during sporogenesis in maize. J Integr Plant Biol 56: 1042–1052 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Table S1. Maize lines resequenced for analysis of DNA sequence variation
Figure S1. Relative expression levels of native AtAGO1 among wild type and ZmAGO1a transformed lines


