Abstract
Transcription factors are key players in the control of the activation or repression of gene expression programs in response to environmental stimuli. The study of regulatory networks taking place in fungal pathogens is a promising research topic that can help in the fight against these pathogens by targeting specific fungal pathways as a whole, instead of targeting more specific effectors of virulence or drug resistance. This review is focused on the analysis of regulatory networks playing a central role in the referred mechanisms in the human fungal pathogens Aspergillus fumigatus, Cryptococcus neoformans, Candida albicans, Candida glabrata, Candida parapsilosis, and Candida tropicalis. Current knowledge on the activity of the transcription factors characterized in each of these pathogenic fungal species will be addressed. Particular focus is given to their mechanisms of activation, regulatory targets and phenotypic outcome. The review further provides an evaluation on the conservation of transcriptional circuits among different fungal pathogens, highlighting the pathways that translate common or divergent traits among these species in what concerns their drug resistance, virulence and host immune evasion features. It becomes evident that the regulation of transcriptional networks is complex and presents significant variations among different fungal pathogens. Only the oxidative stress regulators Yap1 and Skn7 are conserved among all studied species; while some transcription factors, involved in nutrient homeostasis, pH adaptation, drug resistance and morphological switching are present in several, though not all species. Interestingly, in some cases not very homologous transcription factors display orthologous functions, whereas some homologous proteins have diverged in terms of their function in different species. A few cases of species specific transcription factors are also observed.
Keywords: transcription factor, antifungal drug resistance, pathogenesis, immune system evasion, Candida species, Cryptococcus neoformans, Aspergillus fumigatus
Introduction
Infections caused by fungal pathogens have become a relevant threat to human health as their prevalence has continuously increased over the past decades (Perlroth et al., 2007; Miceli et al., 2011; Miceli and Lee, 2011). Three genera are particularly significant as human pathogens: fungi belonging to the Aspergillus spp. and yeasts from Cryptococcus spp. and Candida spp. Infections caused by these pathogens are especially severe in immunocompromised patients, particularly HIV-infected, cancer and transplant patients (Sims et al., 2005; Chauhan et al., 2006; Pongpom et al., 2015; Schmalzle et al., 2016).
Infection niches and mechanisms diverge according to specific traits of each organism. Infections by Aspergillus fumigatus start in the pulmonary epithelia and evolve into allergic bronchopulmonary aspergillosis, aspergilloma, invasive pulmonary aspergillosis and hematogenously disseminated aspergillosis (Brown and Goldman, 2016). On the other hand, infections by the pathogenic yeast Cryptococcus neoformans are primarily pulmonary, persisting for long periods of time, and then spreading to the central nervous system (CNS) (Hole and Wormley, 2016). On the contrary, infections by Candida spp. are established in mucosal and cutaneous surfaces, translating into systemic infections with high tissue burden if able to invade and reach the bloodstream (Pfaller and Diekema, 2007; Azie et al., 2012; Montagna et al., 2013; Papon et al., 2013).
The severity of infections caused by fungal pathogens is associated with a concerted interplay between antifungal drug resistance, virulence and immune system evasion features. Therefore, it is pertinent not only to study the referred mechanisms, but also the transcriptional networks controlling such traits. This knowledge is crucial to identify new therapeutic targets, while ultimately helping to overcome fungal infections.
This review will focus on the transcriptional regulation of antifungal drug resistance, virulence and immune system evasion mechanisms employed by Aspergillus fumigatus, Cryptococcus neoformans and the four most prevalent Candida species: Candida albicans, Candida glabrata, Candida parapsilosis, and Candida tropicalis. This comprehensive analysis aims to identify and compare conserved transcriptional regulators among the indicated organisms, while also contributing to find additional uncharacterized homologs, whose functional analysis may bring further light to these multifactorial processes. This inter-species comparison will provide a better understanding of the regulatory networks applied by fungal pathogens to regulate crucial features for their pathogenic nature and of their variability and evolution among the considered species.
Transcription factors described as relevant regulators of drug resistance, virulence traits and host immune evasion among A. fumigatus, C. neoformans, and Candida spp. were selected and used to study the variability of regulatory networks governing these processes. Resorting to the Phylome Database (http://phylomedb.org/), the phylomes of each transcription factor were then used to search for protein phylogenies with the objective of identifying predicted homologs in the remaining species. BLASTp analysis was used to complement the Phylome DB data by searching for homologous sequences in the studied species (considering as threshold E < 10−50). The amino acid sequences of the studied transcriptional regulators in A. fumigatus, C. neoformans and Candida spp. were retrieved from the Aspergillus Genome Database (http://www.aspergillusgenome.org/), Cryptococcus neoformans TF Phenome Database (http://tf.cryptococcus.org/), Candida Genome Database (http://www.candidagenome.org/), and EnsemblFungi (http://fungi.ensembl.org/) (for C. tropicalis), respectively. For tree representation, the MEGA 7 software (http://www.megasoftware.net/) was used to perform multiple sequence alignments and tree visualization. Combining this bioinformatics approach with already described information for characterized transcription factors and their regulatory targets, an inter-species comparison of the transcriptional networks governing important traits such as drug resistance, virulence and immune evasion in yeast and fungal pathogens is presented in this review.
Drug resistance transcription regulators
In order to overcome drug resistance it is essential to understand the structure of the transcription networks regulating this phenomenon, as it implies a complex regulatory circuit in order to activate the most appropriate response according to distinct stimuli.
One of the major regulators of drug resistance in C. glabrata is the transcription factor Pdr1. C. glabrata Pdr1 is a Zn(2)-Cys(6) DNA binding protein responsible for the activation of drug resistance genes, such as the multidrug resistance transporters Cdr1 and Pdh1 (also known as Cdr2), via pleiotropic drug response elements (PDRE) (Vermitsky et al., 2006; Caudle et al., 2011; Paul et al., 2011). Gain-of-Function (GOF) mutations in the CgPdr1 transcription factor have been found in clinical isolates to be responsible for increased CgPdr1 activity and consequent constitutive high expression of the ABC drug efflux pumps, as well as its positive autoregulation (Tsai et al., 2006; Ferrari et al., 2009; Paul et al., 2011). Although typical regulatory targets of Pdr1 include the ATP-Binding Cassette (ATP) efflux pumps Cdr1 and Pdh1, it was also found to activate the expression of efflux pumps from the Major Facilitator Superfamily (MFS), including Qdr2 and Tpo3 (Costa et al., 2013, 2014), thus reaffirming its role as a major regulator of drug resistance in C. glabrata. No proteins displaying significant homology in the remaining studied species were identified by phylome search.
Drug resistance regulation is a complex process that must be controlled according to the specific stress exerted over fungal pathogens, activating or repressing key pathways to better express the desired response. As such, a negative Zn(2)-Cys(6) regulator of azole resistance, the transcription factor Stb5, was also addressed as a relevant regulator of drug resistance in C. glabrata. As a result of its negative regulation, Stb5 overexpression leads to a higher susceptibility toward azole drugs, while its deletion causes a small increase in azole resistance (Noble et al., 2013). Also, expression analysis showed that Stb5 shares many transcriptional targets with Pdr1, such as Cdr1, Pdh1, and Yor1, but working as a pleitropic drug resistance repressor (Noble et al., 2013). Homologous proteins were identified in C. parapsilosis (uncharacterized, encoded by ORF CPAR2_109760) and in C. albicans (Stb5) in the phylome analysis. Additionally, the C. tropicalis protein encoded by ORF CTRG_04421 was identified as a C. glabrata Stb5 homolog by BLASTp search. C. albicans Stb5 shares the Zn(2)-Cys(6) DNA binding domain found in C. glabrata Stb5, and despite the fact that its role and regulation mechanisms are still poorly characterized, it has been shown to be repressed by Hap43 (Singh et al., 2011).
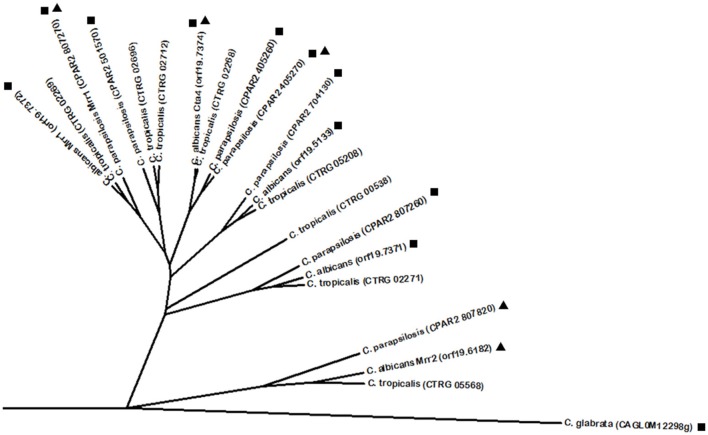
C. albicans has its own master regulator of drug resistance Tac1, a Zn(2)-Cys(6) DNA binding activator of drug-responsive genes such as the ABC multidrug resistance transporters Cdr1 and Cdr2 (Coste et al., 2004) by binding the drug response element (DRE) (Coste et al., 2009). Onward with the transcriptional control of Tac1 over Cdr1 and Cdr2 expression, changes in this transcription factor gene were described to modulate its function and consequently add an extra layer of regulation in its network. Several substitutions and small deletions were found to increase Tac1 function (Coste et al., 2007), while chromosomal rearrangements in chromosome 5 lead to loss of heterozigosity resulting in Tac1 dosage adjustments by overexpression of its encoding gene (Coste et al., 2007; Selmecki et al., 2008). There is evidence supporting positive autoregulation of Tac1 (Liu et al., 2007; Znaidi et al., 2007). Altogether, evidence shows that TAC1 controls its target genes by increasing its own expression or by GOF mutations (Coste et al., 2006, 2007, 2009). Despite having similar functions and regulating similar gene targets, Tac1 was not found to share sequence homology with C. glabrata Pdr1, according to the Phylome database. Instead, Tac1 presents high similarity with several uncharacterized proteins encoded by other CTG clade Candida spp. These include C. parapsilosis proteins encoded by ORFs CPAR2_303510, CPAR2_303520, and CPAR2_303500 and C. tropicalis proteins encoded by ORFs CTRG_05307, CTRG_05306, and CTRG_05308. Interestingly, phylome analysis highlights two other C. albicans regulators close to Tac1: the Zn(2)-Cys(6) transcription factors Znc1 and Hal9. These findings are intriguing, given that Znc1 is required for yeast cell adherence to silicone substrate (Finkel et al., 2012) and Hal9 is induced by Mnl1 under weak acid stress (Ramsdale et al., 2008), and therefore do not display a conserved function with Tac1, despite their predicted homology. C. albicans carries yet another major regulator of multidrug resistance transporters in the transcription factor Mrr1, an activator of the MFS multidrug transporter Mdr1, leading to acquisition of multidrug resistance in azole resistant clinical isolates (Morschhäuser et al., 2007). As observed for C. glabrata Pdr1 and C. albicans Tac1 transcriptional regulators, Mrr1 is a Zn(2)-Cys(6) transcription factor and its gene sequence is subjected to GOF mutations responsible for increased protein activity (Dunkel et al., 2008). As described above for Tac1, Mrr1 also appears to be auto-regulated (Schubert et al., 2011). Additionally, it was found to be induced by Hap43 (Singh et al., 2011). Showing some level of functional conservation with the previous regulators is also the transcription factor Mrr2, seen to control the expression of Cdr1 in C. albicans (Schillig and Morschhäuser, 2013). Mrr1 and Mrr2 do not present significant homology to each other, but interestingly, in the search for Mrr1 and Mrr2 homologs using PhylomeDB, several common hits were identified in Candida spp. (Figure 1). Among them is a closely related C. parapsilosis Zn(2)-Cys(6) homolog (named Mrr1) described to be upregulated in azole resistant strains, probably leading to the upregulation of C. parapsilosis Mdr1 (Silva et al., 2011). Similarly to what is described in C. albicans, the upregulation of C. parapsilosis Mrr1 and Mdr1 is correlated with point mutations in the MRR1 gene (Silva et al., 2011). However, beyond these homologs, BLASTp analysis revealed an array of additional proteins that show some similarity with Mrr1 and Mrr2. Interestingly, C. parapsilosis Mrr1 was also found to share sequence similarity with C. albicans Mrr2 (Figure 1). Additionally, other regulators not primarily related to drug resistance display sequence similarity with Mrr1 and Mrr2, namely C. albicans Cta4 (Coste et al., 2008), a transcription factor involved in nitrosative stress resistance (Chiranand et al., 2008). Nevertheless, it is relevant to point out that Cta4 expression in S. cerevisiae was seen to contribute for azole drug resistance (Coste et al., 2008). It is interesting to see that Mrr1 and Mrr2 display some level of similarity not only with other regulators (e.g., Cta4), but also with several uncharacterized proteins, both in C. albicans and other Candida spp.
Figure 1.
Phylogenetic analysis of the C. albicans Mrr1 and Mrr2 homologs. Phylome predicted homologs of Mrr1 are marked with (■). Phylome predicted homologs of Mrr2 are marked with (▲). Unmarked branches represent additional proteins showing some degree of similarity identified by BLASTp (E < 10−50). The tree was constructed using the Molecular Evolutionary Genetics Analysis (MEGA 7) software (Kumar et al., 2016). Multiple alignments of the amino acid sequences were calculated by ClustalW algorithm (Sneath and Sokal, 1973). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method (Jones et al., 1992) and are in the units of the number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1).
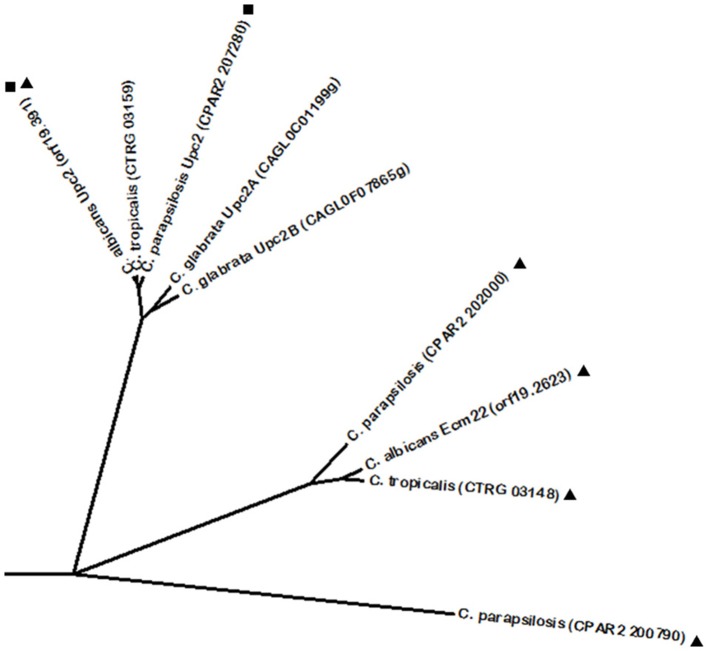
Resistance to azole drugs has often been attributed to the upregulation of ergosterol biosynthetic genes, given that these drugs act by inhibiting the activity of Erg11, thus leading to ergosterol depletion in the fungal plasma membrane (Kelly et al., 1995; Ghannoum and Rice, 1999; Kanafani and Perfect, 2008). In this context, the transcription factor Upc2 is an important player in azole drug resistance, being a transcriptional activator of ergosterol biosynthetic genes in C. albicans, but also of the MFS multidrug transporter encoding gene MDR1 (Silver et al., 2004; MacPherson et al., 2005; Dunkel et al., 2008; Heilmann et al., 2010; Synnott et al., 2010). C. albicans Upc2 phylome analysis revealed a C. parapsilosis Upc2 homolog. In fact, C. parapsilosis Upc2 displays a conserved function, as it was described to confer resistance against azole drugs and to regulate the ergosterol pathway in hypoxia (Guida et al., 2011). Despite no other homologs were identified at the Phylome DB, C. glabrata harbors two Upc2 regulators known to participate in the same process. C. glabrata Upc2A, but not Upc2B, displays the prominent role in the resistance against azoles and sterol biosynthesis inhibitors (Nagi et al., 2011). However, Upc2B was shown to regulate the expression of Erg2 and Erg3 from the ergosterol biosynthetic pathway, whereas both Upc2A and Upc2B are required for expression of the sterol transporter Aus1 (Nagi et al., 2011). Given the conserved role of these proteins with the Upc2 proteins from C. albicans and C. parapsilosis, their phylogenetic proximity was evaluated using BLASTp analysis, through which a predicted C. tropicalis Upc2 was also found to share significant sequence similarity with the rest of the Upc2 proteins (Figure 2). Interestingly, Upc2 phylome analysis did not reveal Ecm22 as a possible homolog; however, reciprocal phylome analysis unveiled Upc2 as an Ecm22-related protein. Several uncharacterized proteins in other CTG clade species were further predicted to share homology with C. albicans Ecm22 (Figure 2). Additionally, there is also an identified Ecm22 protein in C. neoformans, however, it does not share significant homology with C. albicans Ecm22. In most fungi, regulation of sterol biosynthesis is based on well conserved Sterol Regulatory-Element Binding Proteins (SREBP), usually harboring a basic helix-loop-helix (bHLH) leucine zipper domain. However, it is interesting to note that this system has been disrupted in yeasts such as S. cerevisiae and Candida species (Maguire et al., 2014). In these species, the role of SREBPs in sterol biosynthesis has been replaced by the Zn(2)-Cys(6) Upc2 proteins, which are structurally unrelated to SREBPs. Maguire and colleagues proposed that Upc2 arose in the common ancestor of the Saccharomycotina and was created by duplication of another zinc finger protein gene, although it diverged too much from its orthologs in other species, such as A. fumigatus (Maguire et al., 2014).
Figure 2.
Phylogenetic analysis of the C. albicans Upc2 and Ecm22 homologs. Phylome predicted homologs of Upc2 are marked with (■). Phylome predicted homologs of Ecm22 are marked with (▲). Unmarked branches represent additional proteins showing some degree of similarity identified by BLASTp (E < 10−50). The tree was constructed using the Molecular Evolutionary Genetics Analysis (MEGA 7) software (Kumar et al., 2016). Multiple alignments of the amino acid sequences were calculated by ClustalW algorithm (Sneath and Sokal, 1973). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method (Jones et al., 1992) and are in the units of the number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1).
Upregulation of ergosterol synthesis upon azole drug exposure is controlled in A. fumigatus by the transcription factor SrbA, encoding a bHLH protein belonging to the SREBP family. As stated previously, this family comprises the main regulators of sterol biosynthesis in yeasts outside of the Saccharomycotina lineage. SrbA is responsible for mediating azole drug resistance by activating the expression of cyp51, the ERG11 ortholog in this pathogen (Mellado et al., 2005; Willger et al., 2008; Moye-Rowley, 2015). Additionally, it has a secondary role in the maintenance of cell polarity, therefore directing hyphal growth (Willger et al., 2008). SrbA controls the expression of a target gene with which it shares sequence similarity: srbB. SrbB is another transcriptional regulator that together with SrbA co-regulates heme biosynthesis and sterol demethylation genes. However, it acquired new functions as it also regulates hypoxia response and virulence genes (Chung et al., 2014). Phylome analysis did not provide any possible SrbA or SrbB homologous proteins in any of the remaining species addressed in this work. Nonetheless, C. neoformans harbors the transcription factor Sre1, another bHLH protein that despite not being found to share a phylogenetic relationship with SrbA or SrbB was found to be required for azole drug resistance (Chun et al., 2007; Bien et al., 2009; Jung et al., 2015). Like A. fumigatus SrbA, this is due to Sre1 involvement in the expression of genes required for ergosterol biosynthesis (Chang et al., 2007; Willger et al., 2008). Additionally, Sre1 is also involved in virulence, as it was found to be important for the establishment and growth of yeast cells in the brain; as well as being involved in the regulation of genes involved in iron uptake (Chang et al., 2007).
Virulence transcriptional regulators
Biofilm formation and tissue invasion regulators
The ability of fungal pathogens to cause disease relies upon an array of strategies to colonize surfaces and invade host tissues. The establishment of biofilms is one of the main virulence traits displayed by human pathogens. The development of biofilms in medical devices represents a relevant risk factor for patients, as such devices serve as reservoirs and entry points for potential infections (Douglas, 2003; Kojic and Darouiche, 2004; Martinez and Casadevall, 2015). Once inside the host, the development of biofilms allows pathogens to overcome environmental stresses, such as drug exposure and immune system attack, while also resulting in the establishment of persistent infections (Jabra-Rizk et al., 2004; Fanning and Mitchell, 2012).
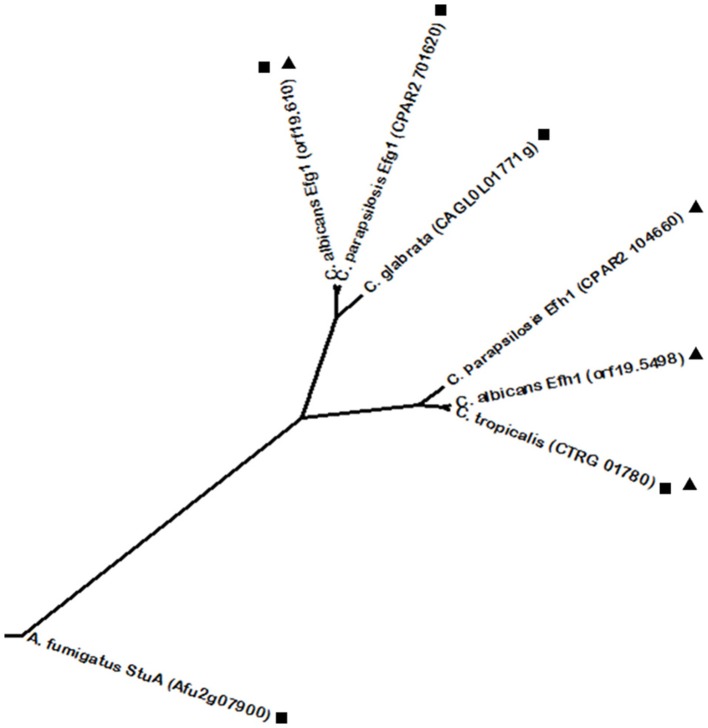
Generally, biofilm formation is among the most studied subjects in virulence and is based on adherence and morphogenic phenomena, including hyphae formation. Besides biofilm formation, another key virulence factor is invasion of non-phagocytic host cells, as it represents the ability of the pathogen to disseminate and infect the host. Epithelial cell invasion by Candida spp. has been correlated with the production of lytic enzymes, such as secreted aspartyl proteinases (SAPs) that digest the surface of epithelial tissue thus enabling tissue invasion (Schaller et al., 1999, 2003). In the case of C. albicans, hyphae development is also associated with non-phagocytic host cell invasion, as Sap4-6 enzymes are regarded as hyphal-associated proteins (Schweizer et al., 2000; Korting et al., 2003) and hyphae are found within epithelial cells, whereas yeast forms are found on their surface or between them (Scherwitz, 1982; Ray and Payne, 1988). For this reason, hyphal form is considered to be the invasive form of C. albicans. Additionally, hyphae were also associated with a more efficient induction of epithelial cell endocytosis, a process in which epithelial cells are stimulated to produce pseudopods that internalize the pathogen (Park et al., 2005). Interestingly, the role of hyphae in the invasion of endothelial tissue appears to be more complex, as distinct tissues endocytose preferentially hyphae, while other endothelial cell linings are as easily crossed by yeast cells (Klotz et al., 1983; Jong et al., 2001; Lossinsky et al., 2006). Yeast-to-hyphae transition is a well-studied feature in C. albicans. It can be activated by a variety of conditions, mostly by stress factors, such as changes in pH and temperature (e.g., 37°C) and nitrogen starvation that promote hyphal growth (Kadosh and Johnson, 2005). One of the most powerful factors to induce hyphae formation is the presence of serum, as its nutrients are usually unavailable to C. albicans cells, thus constituting a stress condition and therefore inducing hyphal growth. The same principle was verified when using N-acetylglucosamine (Glc-NAc), a poor source of carbon and nitrogen capable of inducing hyphae formation (Mattia et al., 1982; Kadosh and Johnson, 2005). The C. albicans positive regulators Efg1 and Cph1 regulate a defined core set of genes required for hyphal growth, including ECE1, HYR1, HWP1, and ALS3. These genes encode mainly cell wall-associated proteins, involved in processes including hyphal-cell elongation and adhesion to host tissues (Birse et al., 1993; Bailey et al., 1996; Staab et al., 1996, 1999; Hoyer et al., 1998). It is understandable that the transcriptional regulators controlling hyphae formation are responsible for the expression of cell wall related genes, given that yeast-hyphae transition implicates morphological alterations that require cell wall remodeling (Chaffin et al., 1998; Sohn et al., 2003). Analyzing the phylogenetic relationship between C. albicans transcription factor Efg1 and its closest homologs, the C. parapsilosis Efg1 and A. fumigatus StuA transcription factors are highlighted (Figure 3). All three homologs contain an APSES DNA binding domain, which contributes to their close relationship. In turn, C. parapsilosis Efg1 was found to be a morphological switch regulator, similarly to its C. albicans ortholog (Connolly et al., 2013). In turn, A. fumigatus StuA controls adhesion and virulence by regulation of the uge3 gene, encoding for uridine diphosphate (UDP)-glucose-epimerase which is essential for adherence through mediating the synthesis of galactosaminogalactan (Lin et al., 2015). Other than regulating conidiophore morphology, whole genome transcriptional analysis identified StuA as regulating secondary metabolite biosynthesis genes, the catalase gene cat1 and morphogenesis genes (Sheppard et al., 2005). Interestingly, a C. glabrata homolog (encoded by ORF CAGL0L01771g) was identified, although C. glabrata is unable to develop true hyphae. Furthermore, C. albicans Efg1 has a paralog, Efh1. Efh1 is also an APSES protein but with a minor role compared to Efg1, which is also the case of the C. parapsilosis Efg1 ortholog (Doedt et al., 2004; Connolly et al., 2013). However, Phylome DB shows that Efh1 homologs are restricted to Candida spp., with homologous proteins in both C. tropicalis and C. parapsilosis (Figure 3). As expected given their paralogous status, C. albicans Efg1 and Efh1 were found to be phylogenetically related with each other by phylome analysis. Interestingly, only one single C. tropicalis protein (encoded by ORF CTRG_01780) was found to be homologous to both C. albicans Efg1 and Efh1, while the single C. glabrata protein was found to share homology only with Efg1 (Figure 3). Relative to C. albicans Cph1, phylome analysis found it to be closely related to the C. parapsilosis Cph1 protein. C. tropicalis also features a Cph1 protein, despite it was not considered to have a homology relationship at the Phylome DB. Nevertheless, BLASTp shows a high degree of sequence homology between the Cph1 proteins from C. albicans and C. tropicalis. All Cph1 proteins belong to the STE-like transcription factor family. As observed for the yeast model S. cerevisiae, in which the Cph1 ortholog is the Ste12 transcription factor, other species also have Ste12 homologs from Cph1. Accordingly, C. glabrata Ste12 was described as being required for nitrogen starvation induced filamentation and to have a role in virulence (Calcagno et al., 2003). Despite its functional conservation, C. glabrata Ste12 was not predicted to be a Cph1 homolog by phylome analysis, indicating that their sequences have somewhat diverged. However, BLASTp comparison shows a significant degree of sequence homology between the two proteins. According to the Phylome DB, no homologs were predicted for C. glabrata Ste12, nonetheless, BLASTp analysis revealed an additional C. glabrata protein, encoded by ORF CAGL0H02145g, presenting significant homology. Moreover, A. fumigatus contains a SteA protein, also belonging to the STE-like family, that showed significant homology to C. glabrata Ste12 by BLASTp analysis, but this transcription factor is still uncharacterized. C. neoformans also harbors a Ste12 transcription factor, though it was not found to share an evolutionary link to the other STE-like family proteins in the previously considered species. This fact, together with the knowledge that C. neoformans Ste12 is associated with this pathogen's particular trait of capsule formation (Chang et al., 2001) and melanin production through the expression of the LAC1 gene (Jung et al., 2015), seems to indicate that the presence of a STE-like domain can be the only trait shared with the remaining proteins. Capsule and melanin production in C. neoformans are regulated by a cyclic AMP-dependent protein kinase A (Pka) signaling pathway. Pka is composed of a catalytic (Pka1) and a regulatory subunit (Pkr1) (D'Souza et al., 2001). Mutant strains lacking Pka1 do not produce a capsule under normal conditions, whereas disruption of Pkr1 results in capsule overproduction and hypervirulence, providing evidence of the importance of this pathway in C. neoformans virulence (D'Souza et al., 2001; D'Souza and Heitman, 2001). This may be explained by the regulation by Pka1 of glucan synthesis-related genes, important for the production of the capsule, such as FKS1, AGS1, AGN1, KRE6, KRE61, and SKN1 (O'Meara and Alspaugh, 2012). This pathway is responsible for the activation of the Ste12α transcription factor, involved in mating, since pka1 mutants restored a mating phenotype by overexpression of this transcription factor (D'Souza et al., 2001). The Ras signal transduction pathway was also previously shown to be involved in C. neoformans virulence (Alspaugh et al., 2000). The virulence of a C. neoformans ras1 mutant was attenuated and the induction of the RAS pathway and capsule formation have been associated with growth in minimal media (Alspaugh et al., 1997, 2000). Ras1 is a major C. neoformans Ras protein found to contribute to high-temperature growth and invasive growth, which are essential features for proliferation inside the host (Alspaugh et al., 2000). The Ras1/Cdc24 and Ras1/Cdc42 pathways are required for thermotolerance and actin cytoskeleton regulation, whereas Ras1/cAMP governs mating and invasive growth (Alspaugh et al., 2000; Waugh et al., 2003; Nichols et al., 2007). Initially, Ras1 absence does not result in defects in capsule or melanin production and the lack of virulence is attributed to lack of growth at 37°C (Alspaugh et al., 2000). Nevertheless, Ras1 was later shown to have a role in capsule formation induced by serum, however this was not considered as a major mechanism through which Ras1 signaling affects virulence (Zaragoza et al., 2003; Haynes et al., 2011). In the case of C. neoformans, much less is known about its adhesion and invasion strategies. In vitro, it was shown to adhere and be internalized by pulmonary epithelial cells, thus resulting in host cell damage (Barbosa et al., 2006). Upon reaching the brain, C. neoformans cells were found to cross endothelial cell lining by transcytosis, however, such process appears to cause minimal cell damage (Chrétien et al., 2002; Chen et al., 2003; Chang et al., 2004). Although not strictly required, the presence of a capsule appears to enhance initial adherence to endothelial cells and the rate of transcytosis (Chen et al., 2003; Chang et al., 2006). However, this effect seems to be dependent on the endothelial tissue in question (Ibrahim et al., 1995). Nevertheless, evidence suggests that C. neoformans capsule is bound by a receptor mainly present in brain endothelial cells, thus potentiating brain tissue invasion (Filler and Sheppard, 2006). Another well characterized pathway of biofilm formation is based on the C. albicans regulators Tec1 and Bcr1 (Schweizer et al., 2000; Nobile and Mitchell, 2005). Tec1 is a positive regulator of morphogenesis belonging to the TEA/ATSS family that is predominantly expressed during hyphal growth and is required for hyphae formation during serum induction, during macrophage evasion after phagocytosis and for expression of the aspartyl proteinase genes SAP4-6 (Schweizer et al., 2000). Tec1 is in turn regulated by Efg1 (Lane et al., 2001a). Moreover, Tec1 expression is directly regulated by Cph2 (Lane et al., 2001a,b). Bcr1 is a C2H2 zinc finger transcriptional activator of cell-surface protein and adhesion genes such as the previously referred ECE1, ALS3, HWP1, and HYR1 (Nobile and Mitchell, 2005; Nobile et al., 2006). Bcr1 was found to relay a signal within the hyphal developmental network, being positively regulated by Tec1 (Nobile and Mitchell, 2005; Nobile et al., 2006). Starting with the analysis of Tec1 phylogenetic relationships, one close homolog was identified in C. parapsilosis (encoded by ORF CPAR2_805930). Despite not showing a homology relationship according to phylome analysis, the A. fumigatus AbaA transcription factor shares the TEA/ATTS domain and also presents a related function. AbaA regulates the specific A. fumigatus feature of conidiation by activating the expression of the velvet regulators veA and velB (Park et al., 2012), but similarly to the role of Tec1, AbaA also controls adherence, a trait that correlates with conidiatian in A. fumigatus (Lin et al., 2015). Additionally, AbaA activates the expression of wetA, a regulator with a predicted role in hyphal growth (Tao and Yu, 2011). Regarding the phylogenetic relationships of Bcr1, close homologs were identified in the CTG clade species, including C. parapsilosis Bcr1, also involved in biofilm formation (Ding and Butler, 2007; Ding et al., 2011) and an uncharacterized C. tropicalis homolog (ORF CTRG_00608).
Figure 3.
Phylogenetic analysis of the C. albicans Efg1 and Efh1 homologs. Phylome predicted homologs of Efg1 are marked with (■). Phylome predicted homologs of Efh1 are marked with (▲). The tree was constructed using the Molecular Evolutionary Genetics Analysis (MEGA 7) software (Kumar et al., 2016). Multiple alignments of the amino acid sequences were calculated by ClustalW algorithm (Sneath and Sokal, 1973). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method (Jones et al., 1992) and are in the units of the number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1).
Also related with biofilm formation regulation is the C. glabrata transcription factor Cst6. It is a bZIP transcription factor involved in the negative regulation of Epa6, the major adhesin found in C. glabrata biofilms (Riera et al., 2012). Additionally, Cst6 also accumulates other roles as demonstrated by the control exerted over the carbonic anhydrase Nce103 in response to carbon dioxide (Cottier et al., 2013). Although no close homologs were predicted in the phylome analysis, the Rca1/Cst6 C. albicans transcription factor plays a role related to that of C. glabrata Cst6, as it was characterized as a regulator of hyphal formation through the transcription factor Efg1 and positive control of hyphal genes including GWP1, ECE1, HGC1, and ALS3 (Vandeputte et al., 2012). Additionally, it was also found to control CO2 sensing by regulating the expression of the carbonic anhydrase Nce103 (Cottier et al., 2012) and antifungal drug resistance (negative regulation of azole and echinocandin drug response, whereas positive regulation of 5-flucytosine response), associated to the regulatory control of cell wall genes (Vandeputte et al., 2012). Additionally, reciprocal phylome analysis of Rca1 identified one predicted homolog in C. parapsilosis (ORF CPAR2_109540). Furthermore, BLASTp analysis revealed an additional C. tropicalis protein (encoded by ORF CTRG_04281) showing high degree of homology with C. albicans Rca1.
Similarly to what is observed in Candida, A. fumigatus conidia (yeast form cells) and hyphae are known to induce their own endocytosis by alveolar epithelial cells through pseudopod engulfment (DeHart et al., 1997; Paris et al., 1997; Zhang et al., 2005). Hyphae development of A. fumigatus in alveolar cells occurring after internalization results in no detectable damage to the host cell (Wasylnka and Moore, 2003). Interestingly, conidia endocytosis was also found to result in pneumocyte apoptosis inhibition, therefore showing the importance of host cell invasion in A. fumigatus infections (Berkova et al., 2006). The pulmonary epithelium is also penetrated by hyphae, contributing to the subsequent invasion of endothelial tissue by passing from the abluminal to the luminal surface of endothelial cells or by hyphae fragments that enter the bloodstream and disseminate to other organs by invading endothelial cells (Filler and Sheppard, 2006). More recently, a key regulator of biofilm formation in A. fumigatus, SomA, was identified (Lin et al., 2015). SomA controls conidiation primarily by acting in the expression of flbB, a bZIP transcription factor which controls the expression of other regulatory genes such as brlA, medA, and stuA, thus having a central role in the network regulating biofilm formation and adherence in A. fumigatus (Lin et al., 2015). SomA also takes part on the regulation of uge3 expression (previously referred) and the spore hydrophobin RodA, which provides adherence (Thau et al., 1994; Lin et al., 2015). Phylome analysis did not reveal any protein in the remaining species addressed in this study that shares significant homology with A. fumigatus SomA. As stated previously, one of the regulatory genes controlled by SomA is the transcription factor brlA, encoding a C2H2 zinc finger protein that represents a central regulator for the asexual development and controls the formation of vesicles required for conidiation processes (Lin et al., 2015). BrlA induces the expression of the previously referred abaA and wetA regulatory genes, which induce differentiation of spore forming cells and the subsequent maturation of conidia (Yu, 2010). A. fumigatus MedA is another transcription factor regulated by SomA that together with it regulates BrlA expression. As a result, MedA is a positive regulator of conidiation (Adams et al., 1988). Interestingly, BLASTp analysis revealed a C. neoformans protein, encoded by ORF CNAG_03859, with high homology to MedA. Together with StuA (also regulated by SomA), MedA regulates adhesion and virulence in A. fumigatus by regulation of the uge3 gene (Lin et al., 2015).
Involved in the regulation of biofilm formation is also the C. neoformans Znf2 transcription factor. This transcription factor contains a C2H2 zinc finger domain and is responsible for control of filamentation, but also of the expression of an important adhesin in C. neoformans, Cfl1 (Wang et al., 2012). This adhesin is involved in cell adhesion and biofilm formation. Searching for possible Znf2 homologs using the Phylome DB, the A. fumigatus ZafA transcription factor was identified. Interestingly, ZafA has acquired a distinct function in A. fumigatus: it is a zinc-responsive regulator, found to be required for A. fumigatus virulence by regulating zinc homeostasis (Moreno et al., 2007). The C. albicans transcription factor Csr1 was also found to share similarity with Znf2. It shares the C2H2 zinc finger domain and is also involved in filamentous growth regulation by regulating the expression of, for instance, HWP1 (Kim et al., 2008; Nobile et al., 2009; Finkel et al., 2012), therefore showing not only sequence similarity but also functional conservation. In turn, Csr1 phylome analysis unveiled homologous proteins in C. parapsilosis, C. tropicalis, and C. glabrata encoded by ORFs CPAR2_403080, CTRG_03883, and CAGL0J05060g, respectively.
The transcriptional regulation of biofilm formation is complex, being dependent on a diversity of environmental conditions. As a result, biofilm regulatory networks also feature negative regulators that ensure a tight control of this process. Two of the most well characterized negative regulators of biofilm formation are the C. albicans regulators Nrg1 and Rfg1 (Braun et al., 2001; Khalaf and Zitomer, 2001; Murad et al., 2001a). Nrg1 is a C2H2 zinc finger transcription factor that acts together with the general corepressor Tup1 to suppress hyphal growth and expression of hypha-specific genes, which are derepressed as a result of Nrg1 downregulation in typical filamentation conditions (Braun et al., 2001; Murad et al., 2001a,b; Kadosh and Johnson, 2005). Nrg1 also represses the expression of chlamydospore formation genes, by repressing CSP1 and CSP2, two specific chlamydospore related genes (Palige et al., 2013). As for Rfg1, it is a HMG domain negative regulator of several genes that were previously induced by filamentation inducing conditions, indicating that this transcription factor is required for hyphae derepression even under such stimuli (Kadosh and Johnson, 2005). Both RGF1 and NRG1 negatively regulate the expression of the hyphae-specific genes ALS3, ECE1, and HWP1, however, their regulons do not completely overlap (Kadosh and Johnson, 2001, 2005), indicating a distinct function of each regulator in control of hyphae formation in C. albicans. It is noteworthy to point out that Nrg1 negatively regulates another transcription factor, Ume6, required for hyphal extension, which is also associated with virulence (Banerjee et al., 2008, 2013). Analyzing the phylogenetic relationships of Nrg1 with its homologous proteins, one identified homolog was C. tropicalis Nrg1, sharing the C2H2 zinc finger domain and with a conserved role in filamentation repression (Zhang et al., 2016). Additional uncharacterized homologs were found in C. parapsilosis, C. tropicalis and C. glabrata, encoded by ORFs CPAR2_300790, CTRG_00608, and CAGL0G08107g, respectively. C. neoformans also harbors a Nrg1 protein, conserving the C2H2 domain but with a more specialized role, as Nrg1 was found to be an activator of capsule formation in C. neoformans (O'Meara and Alspaugh, 2012). This specialized function can be the result of a divergent phylogenetic relationship, which could justify why it was not identified as a homolog of C. albicans Nrg1 in phylome analysis. C. neoformans Nrg1 was found to be responsible for capsule formation since mutants in its encoding gene showed a defect in capsule induction. This transcription factor is activated downstream of the cAMP-PKA cascade (O'Meara and Alspaugh, 2012). For the case of Rfg1, phylome analysis only revealed one homolog, an uncharacterized C. parapsilosis protein, encoded by ORF CPAR2_801100.
The C. glabrata transcription factor Ace2 was found to be a negative regulator of virulence in this pathogenic yeast since its inactivation leads to an increase in the ability of C. glabrata to cause disease by almost 200-fold (Kamran et al., 2004), thus being regarded as a major virulence regulator in this yeast. Ace2 was also found to regulate the expression of CTS1, EGT2, TAL1, and TDH3 genes, involved in cell separation and biofilm formation processes, which may be related with the hypervirulence phenotype (Stead et al., 2010). Searching for possible homologs, an additional C. glabrata protein, Swi5, was identified by phylome analysis. Swi5 is mostly uncharacterized, but it appears to have a conserved function, given that Swi5 mutants display increased fungal burdens in mouse lungs and brain (MacCallum et al., 2006). Despite not being identified by phylome analysis, C. albicans also contains an Ace2 transcription factor; however, its sequence appears to have diverged too much for a phylogenetic relationship to be fully established. Nevertheless, it conserves the C2H2 zinc finger domain as well as a related role in regulation of a wide variety of pathways in C. albicans, including regulation of morphogenesis, cell separation, adherence and virulence (Kelly et al., 2004). Furthermore, Ace2 appears to play distinct functions in the regulation of such traits: its absence results in hyperfilamentation and hypervirulence (Kelly et al., 2004; MacCallum et al., 2006), however, it was found to be required for filamentous growth under hypoxic conditions (Mulhern et al., 2006) and to act as positive regulator of biofilm formation during normoxia (Stichternoth and Ernst, 2009). Related with these roles, it was found to be a positive regulator of the cell wall genes DSE1 and SCW11 (Kelly et al., 2004). Additionally, it also plays a role as regulator of antifungal drug resistance against antimycin A (Stichternoth and Ernst, 2009). Concordantly, a C. parapsilosis Ace2 homolog conserves the C2H2 domain and was found to be a biofilm regulator (Holland et al., 2014). BLASTp analysis unveiled yet another Ace2 homolog in C. tropicalis encoded by ORF CTRG_03073. Despite not sharing significant homology, A. fumigatus also harbors an Ace2 protein, sharing the C2H2 zinc finger domain. As the remaining regulators, A. fumigatus Ace2 is involved in the regulation of several mechanisms, ranging from conidiophore development, pigment production and virulence (Ejzykowicz et al., 2009). Additionally, the lack of Ace2 results in increased invasion capacity and virulence, translated into increased pulmonary fungal burden. The higher virulence phenotype is related with Ace2 control over ppoC, ecm33, and ags3 expression (Ejzykowicz et al., 2009).
Host adaptation regulators
Despite the ability to adhere and form biofilms, there is a much larger set of features that determines the degree of damage caused by a pathogen upon infecting the host. Such traits can be conserved among fungal pathogens, or they can be specific according to the specific characteristics of each pathogen. One general virulence factor is the ability to metabolize available sources of nitrogen. Nitrogen source utilization affects morphological transitions and virulence factor production that confer a competitive advantage for survival, proliferation and colonization (Lee et al., 2013; Ene et al., 2014). One conserved family of nitrogen utilization transcriptional regulators is the Gat1 family. The best characterized is Gat1 from C. albicans. It is a transcription factor involved in nitrogen catabolite repression and utilization of isoleucine, tyrosine and tryptophan as sole nitrogen sources (Limjindaporn et al., 2003). Accordingly, C. albicans Gat1 regulates the expression of nitrogen associated genes, including GAP1, UGA4, DAL5, and MEP2 (Limjindaporn et al., 2003; Dabas and Morschhäuser, 2007). Phylome analysis reveals a Gat1 homolog in C. parapsilosis. Interestingly, A. fumigatus transcription factor AreA, which does not display sequence homology to C. albicans Gat1, is similarly involved in nitrogen catabolite repression and nitrate utilization, also showed to contribute to virulence (Hensel et al., 1998; Lamarre et al., 2008). Curiously, C. neoformans also expresses a Gat1 protein, but it was not found to have a role in virulence, according to a phenotypic screening (Jung et al., 2015), and was not found to be phylogenetically related to Gat1. Subsequent BLASTp search showed the existence of a C. tropicalis homolog encoded by ORF CTRG_03831. Nevertheless, the presence of Gat1 proteins is markedly conserved among fungal pathogens, which is specially reinforced by the conservation of the GATA DNA binding domain present in these proteins.
Another general feature that correlates with several virulence traits is the ability to activate different cellular pathways in response to pH changes. Rim101 from C. albicans is known to regulate the induction of alkaline expressed genes and repress acid expressed genes at alkaline pH (Ramon et al., 1999). Additionally, it is part of the regulatory circuit that control hyphae stimulation in response to alkaline pH (Davis et al., 2000). Phylome analysis predicts one closely related homolog in C. parapsilosis, encoded by ORF CPAR2_700450. Furthermore, BLASTp indicates another homolog in C. tropicalis, Rim101. C. neoformans also has an identified Rim101 regulator, but interestingly, it was found to regulate capsule maintenance (O'Meara and Alspaugh, 2012), thus showing some level of specialization in this pathogen and reinforcing the hypothesis of a divergent evolution when compared to the remaining Rim101 proteins. Additionally, C. neoformans Rim101 is activated after phosphorylation by Pka1 (O'Meara et al., 2010). Low nitrogen and glucose concentration are also inducers of capsule formation by activation of the cAMP pathway. Additionally, the A. fumigatus transcription factor PacC displays a similar role in this fungus, once it also regulates alkaline responsive genes, including the dehydrin-like dprB (Wong Sak Hoi et al., 2011; Brown and Goldman, 2016). Furthermore, it was found to play a role in host invasion capacity (Bertuzzi et al., 2014) and is also involved in cell wall biogenesis (Brown and Goldman, 2016). Likely, all proteins contain a C2H2 zinc finger domain that is conserved among all species. Interestingly, the S. cerevisiae Rim101 was found to play an additional role in weak acid stress tolerance (Mira et al., 2009).
Following the same principle, adaptation to weak acid stress is another relevant factor in the establishment of infection, especially in niches where such conditions are felt, as in the vaginal tract. One characterized transcriptional regulator of weak acid resistance resistance is C. albicans War1, a Zn(2)-Cys(6) transcription factor required for resistance to weak organic acids such as sorbate (Lebel et al., 2006). It acts similarly to the S. cerevisiae War1 protein that governs weak acid stress response (Schüller et al., 2004). Looking for War1 homologs in other fungal pathogens, phylome analysis shows homologs in each of the CTG clade Candida spp. (encoded by ORFs CPAR2_110360 and CTRG_04350 in C. parapsilosis and C. tropicalis, respectively), closely related to that of C. albicans. Additionally, one War1 homolog was also identified in C. glabrata, uncharacterized until now. Additionally, no War1 homologs were found in C. neoformans, while two A. fumigatus uncharacterized homologs were identified (encoded by ORFs Afu7g01640 and Afu8g00950).
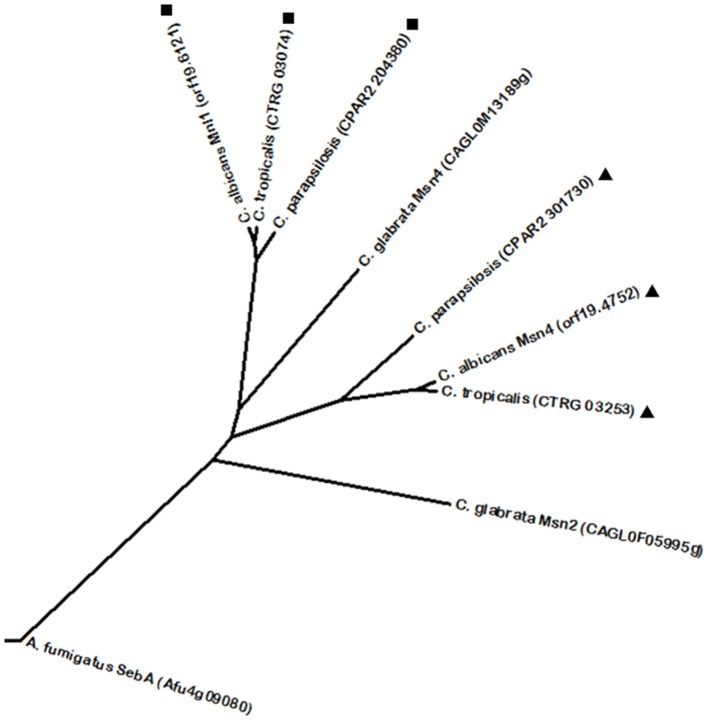
One of the most well characterized regulators of weak acid stress response in fungal pathogens is C. albicans Mnl1. It is a Zn(2)-Cys(6) transcription factor required for adaptation to weak acid stress, activating a subset of genes that are repressed by the previously mentioned Nrg1, through SLE (STRE-like) elements (Hope et al., 2004; Ramsdale et al., 2008). Mnl1 is considered to be related with the yeast conserved Msn2/4 proteins. In fact, C. albicans Mnl1 is also known as the Msn2 correspondent in this yeast. Similar to S. cerevisiae Msn2/4 that are involved in the general stress response, C. albicans Mnl1 is required for the induction of stress response genes via SLE elements (Martínez-Pastor et al., 1996; Ramsdale et al., 2008). Mnl1 phylome analysis revealed homologous proteins in the other CTG clade species (Figure 4). Furthermore, C. albicans also features a Msn4 protein. Similarly to Mnl1, phylome analysis shows Msn4 homologs in the remaining CTG clade species (Figure 4), however, Msn4 does not appear to be a significant stress response regulator, unlike its S. cerevisiae homolog (Nicholls et al., 2004), and was found to be induced during biofilm formation (Nobile and Mitchell, 2005). Interestingly, despite the fact that C. glabrata harbors Msn2/4 proteins involved in oxidative stress resistance by activating the expression of the catalase gene CTA1 (Cuéllar-Cruz et al., 2008), these regulators were not found to share a phylogenetic relationship with its C. albicans counterparts. Likewise, A. fumigatus SebA is a transcription factor described to be involved in response to oxidative stress and heat shock (Dinamarco et al., 2012), thus displaying some level of functional conservation as well, despite not showing a significant homology.
Figure 4.
Phylogenetic analysis of the C. albicans Mnl1 and Msn4 homologs. Phylome predicted homologs of Mnl1 are marked with (■). Phylome predicted homologs of Msn4 are marked with (▲). Unmarked branches represent additional proteins showing some degree of similarity identified by BLASTp (E < 10−50). The tree was constructed using the Molecular Evolutionary Genetics Analysis (MEGA 7) software (Kumar et al., 2016). Multiple alignments of the amino acid sequences were calculated by ClustalW algorithm (Sneath and Sokal, 1973). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method (Jones et al., 1992) and are in the units of the number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1).
Phenotypic switching regulators
Another virulence factor displayed by the yeast C. albicans is the stochastic phenotypic switch known as white-opaque transition (Slutsky et al., 1987; Soll, 1992; Lin et al., 2013). The two cell types differ in shape, gene expression profile, virulence features and colony appearance (Zordan et al., 2007). Opaque cells are the sexually competent form of C. albicans, as they present a much higher mating efficiency than white cells (Miller and Johnson, 2002). Despite white-opaque switching occurring spontaneously every 104 generations (Rikkerink et al., 1988), it can be induced by specific environmental conditions, such as high CO2 concentration, use of GlcNAc as carbon source, genotoxic stresses and oxidative stress (Kolotila and Diamond, 1990; Ramírez-Zavala et al., 2008; Alby and Bennett, 2009; Huang et al., 2010). However, temperature changes from 25°C to 37°C promote the reverse transition, from opaque to white cells (Slutsky et al., 1987; Srikantha and Soll, 1993). The ability to switch between different phenotypes also constitutes and advantageous trait to enhance its adaptation to host environments (Guan and Liu, 2015), thus affecting a variety of virulence traits (Slutsky et al., 1987; Soll, 1992). Opaque cells are able to colonize skin and escape macrophage detection; whereas white cells are more prone to cause bloodstream infections (Kvaal et al., 1999; Lachke et al., 2003; Lohse and Johnson, 2008). Similar phenomena have been described for C. parapsilosis, C. tropicalis, and C. glabrata (Lachke et al., 2000; Laffey and Butler, 2005; Moralez et al., 2014). Investigating possible homologous proteins in other fungal species (including C. neoformans and A. fumigatus) can help to unveil putative specialization events in related proteins from organisms not known to stochastically change their phenotype in this manner. Phenotypic switching is mainly controlled in C. albicans by Wor1, regarded as master regulator of this pathway (Zordan et al., 2006; Huang et al., 2010). Consistent with the related switching process occurring in C. parapsilosis and C. tropicalis, one Wor1 homolog was found in each species, encoded by ORFs CPAR2_805000 and CTRG_03345, respectively. Interestingly, phylome analysis also revealed one A. fumigatus Wor1 homolog, encoded by ORF Afu6g04490. Additionally, no homologs were found in C. neoformans. Despite Wor1 being the master regulator of white-opaque switching in C. albicans, other transcriptional regulators are known to be part of the network controlling this phenomenon. As part of the Wor1 regulon, there are two additional transcription factors involved in white-opaque switching: Wor2 and Czf1. The expression of both transcriptional regulators is directly induced by Wor1, while in turn Wor2 and Czf1 both activate Wor1, creating yet another series of positive feedback loops in the opaque state (Zordan et al., 2007). More recently, two new transcription factors designated Wor3 and Wor4 were added to the existing network governing white-opaque switching. Similarly to what is verified for the previously mentioned regulators, the ectopic expression of Wor3 induced white-opaque switching and is correlated with Wor1, Wor2, and Czf1 (Lohse et al., 2013). As a unique feature, Wor3 was proposed as a member of a different family of DNA-binding proteins. As for Wor4, it was found to be located upstream of Wor1, as its ectopic expression is sufficient to induce white-opaque switching (Lohse and Johnson, 2016). The predicted regulon of this newly discovered transcriptional regulator highly correlates with the ones from Wor1 and Wor2, indicating that Wor4 is integrated in the already described network. Taken together, these transcription factors form an integrated regulatory network with each regulator controlling the expression of the others in the phenotypic switching pathway (Lohse and Johnson, 2016). Analyzing the presence of homologous proteins in other fungal species, similar results are obtained for each regulator. Wor2 phylome analysis identified one closely related protein in C. parapsilosis, encoded by ORF CPAR2_405400. In the case of Czf1 there is one closely related protein in C. tropicalis, encoded by ORF CTRG_03771. A similar situation is found for Wor3, with predicted homologs in C. parapsilosis and C. tropicalis, encoded by ORFs CPAR2_202450 and CTRG_00711, respectively. As for Wor4, one homolog was also identified in each of C. parapsilosis and C. tropicalis, encoded by ORFs CPAR2_808100 and CTRG_05581, respectively. Additionally, in C. albicans, a relationship between phenotypic switching and filamentous growth regulators was uncovered, as in white cells Efg1 represses Wor1 in a Wor2-dependent manner. At the same time, in opaque cells Wor1, Wor2, and Czf1 were found to repress Efg1 (Zordan et al., 2007; Lin et al., 2013).
Iron response regulators
Iron availability is also known to play a crucial role in the establishment of infection. Hosts resist microbial infection by maintaining a low level of free iron to restrict pathogen growth (Hsu et al., 2011). This is achieved by producing transferrin or lactoferrin (Schrettl and Haas, 2011). Therefore, the capacity of a certain pathogen to invade the host is also dependent on its ability to overcome iron deprivation and express iron uptake systems. Among iron uptake and homeostasis regulators, transcription factors belonging to the Hap family can be found in more than one fungal or yeast species. One of the best characterized cases is A. fumigatus HapX, a bZIP negative regulator of iron-consuming pathways (e.g., heme biosynthesis, respiration, TCA cycle and amino acid metabolism) that is required for adaptation to iron depletion, while acting as an activator of the siderophore iron uptake pathway, a known virulence factor (Schrettl et al., 2010). Consequently, the HapX mediated iron limitation stress response was interlinked to primary metabolism, oxidative stress and virulence (Brown and Goldman, 2016). Unveiling a more complex role in the regulation of iron homeostasis, HapX was also described to be involved in response to iron excess (Gsaller et al., 2014). Additionally, it was found to be activated by the previously referred regulator SrbA during hypoxia (Blatzer et al., 2011). These observations are in accordance with one of its possible orthologs in C. glabrata, Yap5, also described to play a role in both iron excess and iron deprivation conditions (Merhej et al., 2015, 2016). This knowledge indicates a wide-spread role of these regulators in iron sensing, acting as both activators and repressors of gene expression according to differential iron availability. Moreover, in C. glabrata, the activation of iron uptake in iron limiting conditions seems to involve the Aft1 transcription factor (ORF CAGL0H03487g), as in S. cerevisiae (Srivastava et al., 2015). Searching for possible homologs by phylome analysis, no HapX homolog was predicted in the remaining species. However, C. albicans harbors the Hap43 transcription factor, a bZIP negative regulator required for low iron response. Such as A. fumigatus HapX, Hap43 is responsible for the repression of genes involved in iron-dependent pathways involved in mitochondrial respiration and iron-sulfur cluster assembly (Hsu et al., 2011). Additionally, its role in the regulation of iron acquisition under low iron conditions seems to be more complex, given its action as a positive regulator in iron-limiting conditions (Hsu et al., 2011; Singh et al., 2011). Due to functional conservation, phylome analysis was also performed for C. albicans Hap43. As a result, one uncharacterized C. parapsilosis protein, encoded by ORF CPAR2_209090, was predicted to be a Hap43 homolog (Merhej et al., 2016). Likewise, BLASTp predicts a highly homologous protein in C. tropicalis encoded by ORF CTRG_04121. In a complementary approach, one of the best studied cases of iron homeostasis in iron replete conditions is the A. fumigatus negative regulator SreA (Schrettl et al., 2008). SreA is a GATA transcription factor that negatively regulates siderophore biosynthesis and other iron acquisition genes in the presence of high iron concentrations, including the iron permease FtrA, the ferroxidase FetC and the siderophore-biosynthetic protein SidA (Schrettl et al., 2008). Interestingly, SreA also negatively regulates the previously referred HapX transcription factor (Blatzer et al., 2011). Searching for possible related proteins in other species, no homologous proteins were predicted by phylome analysis. Nevertheless, C. albicans Sfu1 is a nice candidate function-wise, playing a function similar to that of SreA. The regulatory relationship observed between A. fumigatus SreA and HapX is maintained in C. albicans by the negative regulation of Sfu1 over Hap43 (Hsu et al., 2011). In fact, reciprocal Sfu1 phylome analysis revealed A. fumigatus SreA as a predicted homolog. Beyond SreA, uncharacterized homologs were also identified in C. parapsilosis and C. tropicalis, encoded by ORFs CPAR2_700810 and CTRG_03356, respectively.
Host immune evasion transcription regulators
Upon infection, human pathogens encounter several barriers that need to be overcome, such as tissue barriers and immune responses. In order to establish infection, fungal pathogens take advantage of the virulence traits analyzed so far, but such traits also include evading the host's cellular immune response. Fungal pathogens display diverse immune evasion strategies, including antigen masking to avoid recognition and persistence/active escaping from phagocytic cells (Netea et al., 2006; Erwig and Gow, 2016). Depending on the strategy applied by each pathogen, distinct sets of genes need to be expressed, uncovering complex regulatory networks according to different environmental conditions.
It should be noted that an additional immune evasion mechanism is known to occur in the yeast C. albicans. The capacity of this yeast to undergo yeast-to-hyphae transition, whose regulation is discussed and analyzed in the “Biofilm formation and tissue invasion” section, is described to be an active mechanism for macrophage rupture and evasion after phagocytosis (McKenzie et al., 2010; Lewis et al., 2012a; Rudkin et al., 2013; Bain et al., 2014), and to inhibit macrophage cell division during mitosis (Lewis et al., 2012b).
Oxidative stress regulators
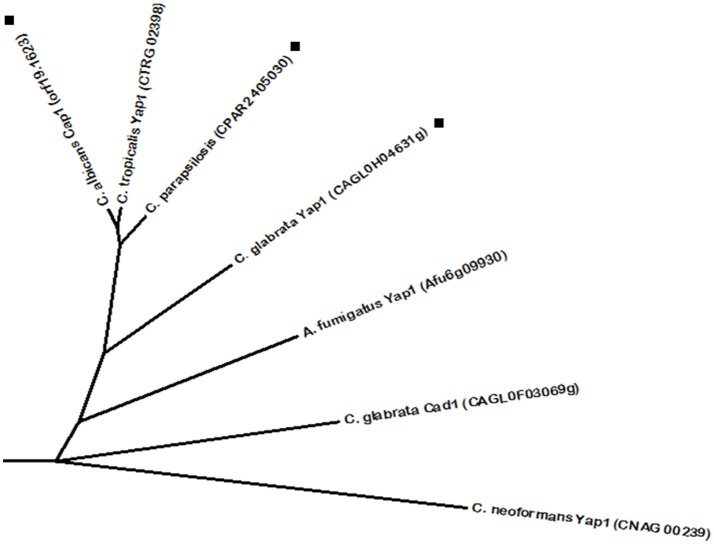
When the immune system response is activated, macrophages, neutrophils, and other phagocytic cells act against fungal pathogens by producing high levels of reactive oxygen species (ROS) and nitric oxide (NO), which results in oxidative and nitrosative stress, respectively (Brown et al., 2009). For this reason, the activation of anti-oxidant responses is a prime strategy upon internalization by phagocytes. The C. albicans bZIP regulator Cap1 is one of the most well characterized AP1-like transcription factors. It is responsible for the activation of antioxidant systems, carbohydrate metabolism and energy generation (Limjindaporn et al., 2003). Within its action in defense against ROS, Cap1 directly activates several genes from distinct pathways of antioxidant scavenging, including glutathione S-transferase reactions, superoxide dismutases and the Cat1 catalase (Enjalbert et al., 2006; Wang et al., 2006). As for carbohydrate and energy metabolism, Cap1 upregulates the expression of enzymes involved in NADPH production, which is involved in several redox cycles against ROS (Müller, 2004; Pócsi et al., 2004). Furthermore, a cluster of mitochondrial respiratory genes are expressed in a Cap1-dependent manner, which is consistent with the knowledge that mitochondrial function is required for oxidative stress tolerance in yeast (Grant et al., 1997). Additionally, Cap1 was found to be involved in drug resistance, as it regulates the expression of the multidrug transporter Mdr1 (Mogavero et al., 2011). As depicted in Figure 5, phylome analysis has identified one C. parapsilosis protein and C. glabrata Yap1 as phylogenetically related to C. albicans Cap1. Accordingly, C. glabrata Yap1 also has a conserved role in response to oxidative stress, being involved in the induction of conserved genes encoding antioxidant effectors, such as the Cta1 catalase (Cuéllar-Cruz et al., 2008; Roetzer et al., 2011). Just as its C. albicans ortholog, C. glabrata Yap1 also regulates the expression of the multidrug transporter Flr1, ortholog of the C. albicans multidrug transporter Mdr1, particularly in response to 4-nitroquinoline-N-oxide (4-NQO), benomyl and cadmium chloride (Chen et al., 2007). However, there is no evidence to suggest regulation of Flr1 by Yap1 in azole drug resistance (Chen et al., 2007). C. glabrata Yap1 has a paralog, Cad1, which was recently found to bind the promoter region of the cadmium response Tna1 protein and the vacuolar transporter Ycf1; however, no changes were detected in Ycf1 expression upon deletion of Cad1; nor any transcriptome changes in response to cadmium (Merhej et al., 2016). Interestingly, C. tropicalis also encodes a Yap1 protein, found to be closely related to its CTG clade counterparts. Likewise, A. fumigatus also possesses a Yap1 transcription factor that despite being encoded by a filamentous fungus is more similar to the proteins from Candida spp., than the Yap1 protein from C. neoformans. As for the A. fumigatus ROS sensing transcription factor Yap1, it was also described to regulate catalase gene expression (cta1 and cta2) and the thioredoxin antioxidant pathway, thus protecting against neutrophil killing (Lessing et al., 2007; Leal et al., 2012). Likewise, C. neoformans Yap1 is also involved in the regulation of antioxidant genes, such as thioredoxins and glutathione peroxidases (Paul et al., 2015). Accordingly, the thioredoxin system is known to be required for the response against oxidative stress. It is composed by two thioredoxin proteins and one thioredoxin reductase. The absence of these two thioredoxin proteins, Trx1 and Trx2, leads to growth defect and sensitivity to multiple stresses, while Trx2 is especially important for nitric oxide stress. These findings highlight a high degree of function conservation among Yap1 proteins in several fungal species.
Figure 5.
Phylogenetic analysis of the C. albicans Cap1 homologs. Phylome predicted homologs of Cap1 are marked with (■). Unmarked branches represent additional proteins showing some degree of similarity identified by BLASTp (E < 10−50). The tree was constructed using the Molecular Evolutionary Genetics Analysis (MEGA 7) software (Kumar et al., 2016). Multiple alignments of the amino acid sequences were calculated by ClustalW algorithm (Sneath and Sokal, 1973). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method (Jones et al., 1992) and are in the units of the number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1).
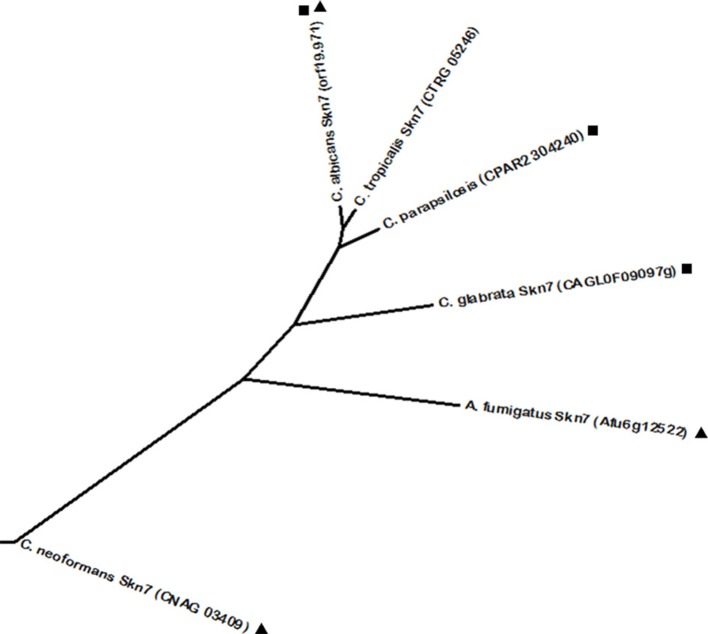
Another conserved regulator of oxidative stress resistance in fungi is Skn7. C. albicans Skn7 was described as required for hydrogen peroxide resistance in a phenotypic screening (Homann et al., 2009). Phylome analysis revealed as Skn7 predicted homologs one uncharacterized protein in C. parapsilosis (encoded by ORF CPAR2_304240) and the C. glabrata Skn7 (Figure 6). C. glabrata Skn7 is involved in hydrogen peroxide response by inducing the expression of the thioredoxins Trx2, Trr1, Tsa1, and the catalase Cta1 (Cuéllar-Cruz et al., 2008; Saijo et al., 2010). Interestingly, there is interdependence of both C. glabrata Yap1 and Skn7 over the regulation of a set of genes—such as TRR1, GPX2, PKH2, TSA1, and CTA1. Additionally, besides Yap1 and Skn7, the transcription factors Msn2 and Msn4 are also involved in the regulation of oxidative stress through regulation of the Cta1, in a concerted action between these four transcription factors (Cuéllar-Cruz et al., 2008). The described interplay of regulatory networks suggests that C. glabrata expression of oxidative stress protective genes is well adapted for when it faces a host-pathogen interaction (Roetzer et al., 2011). The C. tropicalis Skn7 transcription factor was found to be closely related to its CTG clade homologs, which is reinforced by a high sequence homology determined by BLASTp. Furthermore, A. fumigatus and C. neoformans also express Skn7 transcription factors. Interestingly, reciprocal phylome analysis revealed a phylogenetic relationship between these regulators and the Skn7 protein from C. albicans. Curiously, A. fumigatus Skn7 was found to be closer to the remaining yeast proteins than C. neoformans Skn7, located significantly further away from the remaining proteins. This is probably due to the fact that while A. fumigatus Skn7 has a conserved role in mediating resistance to peroxides (Lamarre et al., 2007), C. neoformans Skn7 was found to diverge from the remaining proteins as it specialized in the C. neoformans specific trait of melanin production through the activation of the LAC1 gene (Jung et al., 2015). Melanin accumulates in the cell wall of C. neoformans having a protective role against oxidative and temperature stresses. In fact, melanin is an effective antioxidant, protecting C. neoformans cells against oxygen and nitrogen oxidants (Wang and Casadevall, 1994). Nevertheless, all Skn7 proteins have a conserved heat shock factor (HSF) DNA binding domain.
Figure 6.
Phylogenetic analysis of the C. albicans Skn7 and C. neoformans Skn7 homologs. Phylome predicted homologs of C. albicans Skn7 are marked with (■). Phylome predicted homologs of C. neoformans Skn7 are marked with (▲). Unmarked branches represent additional proteins showing some degree of similarity identified by BLASTp (E < 10−50). The tree was constructed using the Molecular Evolutionary Genetics Analysis (MEGA 7) software (Kumar et al., 2016). Multiple alignments of the amino acid sequences were calculated by ClustalW algorithm (Sneath and Sokal, 1973). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method (Jones et al., 1992) and are in the units of the number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1).
Other than the Yap1 and Skn7 families, there are additional less studied regulators of oxidative stress response in A. fumigatus and C. neoformans, AtfA and Atf1, respectively. AtfA is a transcription factor that targets antioxidant-related genes such as catalase encoding catA and dehydrin-like encoding dprA, which mediate cellular defense against oxidative stress (Hagiwara et al., 2014). This transcription factor has been also identified in Saccharomyces species, but its role in A. fumigatus needs further assessment (Hong et al., 2013). As for Atf1, it is described to be required for oxidative stress induction of the thioredoxin genes TRX1 and TRX2 in C. neoformans (Missall and Lodge, 2005). Interestingly, Atf1 was also found to repress melanin and capsule formation, as null Atf1 mutants show increased capsule and melanin production. Since Atf1 is regulated by Can2, Pka1, and Rim101, it is possible that once again the cAMP pathway might be involved in this network (Kim et al., 2010). Additionally, Atf1 was also found to play a role in thermotolerance and drug resistance, given that its absence was seen to increase resistance against amphotericin B and fluconazole (Kim et al., 2010). In this case, phylome analysis predicted AtfA and Atf1 as homologs, thus showing a high degree of sequence similarity, translated in their functional conservation. Curiously, the Sko1 transcription factor from C. albicans was found to share sequence similarity with A. fumigatus AtfA, by phylome analysis. Given this finding, other potential Sko1 proteins were searched in other Candida spp. Sko1 phylome predicted one uncharacterized C. tropicalis homolog (encoded by ORF CTRG_04352) and the C. glabrata Sko1 protein. All proteins contain a bZIP domain, but given the knowledge concerning C. albicans Sko1 it is possible that these proteins have a participation in additional roles (e.g., cell wall stress and virulence) while still maintaining activity in oxidative stress response, as it was described for C. albicans Sko1 control over the dehydrogenase Ifd4 (Alonso-Monge et al., 2010; Singh et al., 2011).
Nitrosative stress regulators
In what concerns resistance to nitrosative stress, one of the best studied cases is the C. albicans regulator Cta4, a Zn(2)-Cys(6) zinc finger positive regulator of nitrosative stress response. It is upregulated upon nitric oxide exposure and it was found to be required for the expression of the nitric oxide dioxygenase Yhb1, required for NO consumption and detoxification, by directly binding the regulatory region of its gene (Ullmann et al., 2004; Chiranand et al., 2008). In C. glabrata, the transcription factor Yap7 was shown to exert control over Yhb1 by strongly inhibiting its expression in a direct manner; a regulatory association also verified in S. cerevisiae (Merhej et al., 2015). Another described player in nitrosative stress response in pathogenic yeasts is C. neoformans Yap4, an activator of the thioredoxin genes in C. neoformans in response to nitrosative stress, especially Trx2 (Missall and Lodge, 2005). Just as the Yap1 case, C. neoformans Yap4 contains a bZIP domain, however, Yap4 phylome did not reveal any homologs in the studied species. Nevertheless, there are known Yap4 proteins in some Candida spp., namely C. albicans Cap4 and C. glabrata Yap4/6. These proteins remain uncharacterized, therefore it would be interesting to assess if their role has diverged, given that no relationship was identified with the nitrosative stress regulator C. neoformans Yap4. Using phylome analysis, homology relationships among C. albicans Cap4, C. glabrata Yap4/6 and the C. parapsilosis protein encoded by ORF CPAR2_11470 were identified.
Amino acid starvation regulators
One additional factor also thought to play a role in phagocyte persistence and evasion by pathogenic yeasts is the reprogramming of carbohydrate and amino acid metabolism. This has to do with the fact that the environment present inside phagocytic cells is often limiting in terms of nutrients, especially nitrogen sources (Pérez-delos Santos and Riego-Ruiz, 2016). In this context, the bZIP transcription factor Gcn4 was identified in C. albicans as playing a key role in amino acid control response and was found to be expressed upon neutrophil phagocytosis (Fradin et al., 2005). It activates the transcription of amino acid biosynthetic genes (HIS4, HIS7, LYS1, LYS2, and ARO4) via Gcn4-response elements (GCRE) (Tripathi et al., 2002). Among others, Gcn4 plays a role in the biosynthetic pathway of arginine, which in turn is involved in the production of CO2 and urea; products that induce filamentation inside macrophages as an escape mechanism and neutralization of the acidic pH of the phagolysosome (Ghosh et al., 2009; Vylkova et al., 2011). This is further supported by the observation that Gcn4 is required for Efg1-dependent filament induction by amino acid starvation, but not by serum (Tripathi et al., 2002). C. albicans Gcn4 is closely related to proteins found in other CTG clade species, including a C. tropicalis protein encoded by ORF CTRG_02060 and a C. parapsilosis Gcn4 protein. Additionally, phylome analysis also revealed the C. glabrata Gcn4 regulator as being closely related. Moreover, A. fumigatus harbors a regulator with a related function: CpcA. Despite not showing enough similarity with Gcn4, CpcA was also found to be a transcriptional activator of amino acid biosynthesis and to play a role in virulence in this filamentous fungus (Krappmann et al., 2004).
Gliotoxin and melanin production regulators
An additional host immune evasion mechanism is the production of toxins by some pathogens. This is particularly true for A. fumigatus, in which the C2H2 zinc finger containing transcription factor MtfA is involved in the expression of gliotoxin genes, gliZ and gliP, and its biosynthesis, as well as protease activity in the secretome and conidiation (Smith and Calvo, 2014). Gliotoxins have been described as possessing anti-inflammatory and immunosuppressive activities toward the host immune effector cells, including neutrophils and macrophages (Stanzani et al., 2005; Orciuolo et al., 2007; Scharf et al., 2012). No homologs in the additional fungal pathogens considered in this review are predicted by phylome analysis, which can be correlated with the specialized function of MtfA in gliotoxin production regulation.
Another important immune evasion mechanism displayed in C. neoformans is melanin synthesis, which is possible in the presence of exogenous dihydroxyphenols, catalyzed by a phenoloxidase (Almeida et al., 2015). Once yeast cells are phagocytosed, the production of melanin protects cells against the oxidative environment inside the phagolysosome (Panepinto and Williamson, 2006). According to a systematic functional profiling analysis, several transcription factors are assumed to be involved in the regulation of melanin production, including Mbs1 (Jung et al., 2015), however the particular pathways and role it regulates are still unknown. Despite melanin production is a specific trait of neurotropic fungi, such as C. neoformans, and is not known to occur in Candida spp. or A. fumigatus, investigating possible homologous proteins in other fungal species can help to shed light in this regard by unveilling putative related roles. Evaluating possible homology relationships of C. neoformans Mbs1 with other species, C. albicans Mbp1, Swi4 and Swi6 were identified as homologs by phylome analysis, despite not appearing to have a conserved function, given their involvement in G1/S cell cycle progression (Côte et al., 2009; Hussein et al., 2011). Additionally, one uncharacterized A. fumigatus protein, encoded by ORF Afu7g05620, was also identified as a homolog. Interestingly, one additional C. neoformans protein, encoded by ORF CND05520, was found to be phylogenetically related to Mbs1. Also, BLASTp unveiled the A. fumigatus protein encoded by ORF Afu3g13920 as a possible homolog. As referred, C. albicans Mbp1 was found to share significant similarity with C. neoformans Mbs1. Reciprocal phylome analysis did not reveal C. neoformans Mbs1 as a predicted homolog, instead, four proteins within Candida spp. were predicted as homologs, namely C. tropicalis Mbp1, a C. parapsilosis protein encoded by ORF CPAR2_102740 and two C. glabrata proteins encoded by ORFs CAGL0D01012g and CAGL0A04565g. Other than Candida spp. homologs, the same A. fumigatus protein found to share homology with C. neoformans Mbs1 was also identified to be related to C. albicans Mbp1 by reciprocal phylome analysis.
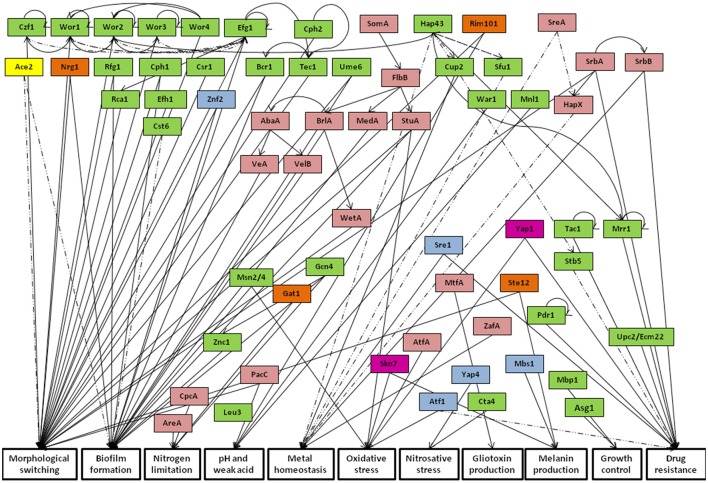
Conclusions and future perspectives
The study of drug resistance, virulence and immune evasion mechanisms in fungal pathogens has advanced considerably over the past years. However, the transcriptional control of such mechanisms has been studied to a different extent. The most studied transcriptional regulators in A. fumigatus, C. neoformans, and Candida spp. are those relevant to their virulence features. Nonetheless, this review highlights that major transcriptional regulators vary among species, leading to disperse information regarding regulatory networks in each pathogen.
The regulatory networks analyzed in this review are compiled in Figure 7. It is possible to observe that only the Yap1 and Skn7 transcription factors, involved in the response to oxidative stress, are conserved with similar functions in all the fungal pathogens considered in this review (Figure 7, purple). This fact points out to the primordial importance of being able to fight reactive oxygen species inside the host, and specifically, within macrophages, something that is common to all pathogens. Other transcription factors, such as Ace2, Ste12, Nrg1, Gat1, Upc2/Ecm22, and Rim101, were also found in most, but not all, of the considered species (Figure 7, yellow and orange), playing important roles in morphological switching, nutrient homeostasis, drug resistance and nitrosative and pH stress tolerance. It is interesting to point out, however, that most of the characterized transcription factors are specific for Candida species (Figure 7, green), C. neoformans (Figure 7, blue) or A. fumigatus (Figure 7, pink), an observation that highlights the divergence of regulatory control in these different fungal pathogens, suggesting that different molecular mechanisms have been adopted to tackle similar biological needs.
Figure 7.
Transcriptional networks of described transcription factors among A. fumigatus, C. neoformans and Candida spp. Transcriptional factors regulating distinct biological mechanisms are shown. Full lines represent positive regulation, dotted lines represent negative regulation. Colored boxes indicate the species that include characterized homologs of the indicated transcription factor, according to the following color code: Single species: Green, Candida spp.; Pink, A. fumigatus; Blue, C. neoformans; Multiple species: Orange, Candida spp. + C. neoformans; Yellow, Candida spp. + A. fumigatus; Purple, Candida spp. + A. fumigatus + C. neoformans.
Another interesting feature that comes from the observation of Figure 7 is that, in general, transcriptional control of a given mechanism is controlled by one or a few major regulators, that may also play complementary roles in other biological process. Examples of these occurences are transcription factors involved in morphology changes that are also often associated with biofilm formation, given that the two processes are associated. Common regulatory networks are also found in the establishment of biofilms and resistance to oxidative stress or drug resistance. The existence of regulatory circuits in which transcription factors do not regulate directly effector genes, but rather other transcription factors is also commonly verified. All these aspects translate complex intricated circuits with a series of interconnect relationships among regulators and the biological processes regulated by them. Regulatory network architecture is significantly complex, comprising multiple layers of regulation among expression control of transcription factors, effector genes and environmental conditions.
Differential knowledge concerning transcriptional control can be associated with the occurence of specific traits not known to occur in every fungal pathogen. These include gliotoxin production in filamentous fungi such as A. fumigatus and melanin production in neurotropic fungi such as C. neoformans.
The occurence of known or predicted homologous regulators among the different fungal pathogens considered in this review and the corresponding degree of functional conservation in infection-related processes is summarized in Table 1. To be highlighted is the presence of conserved transcriptional regulators that present divergent roles among species, participating in distinct mechanisms and regulating distinct targets. Examples of such ocurrences are the cases of Mbs1 and ZafA, which acquired new functions in C. neoformans and A. fumigatus, respectively, in comparison to their homologs. In some cases, this situation represents a specialization of a given protein to pathogen-specific pathway. The inverse situation is also observed, where distinct regulators in different species are found to participate in the control of similar features, such as the C. albicans Tac1 and the C. glabrata Pdr1 transcription factors, or C. albicans Efh1 and A. fumigatus StuA.
Table 1.
Conservation of function between described transcription factors in A. fumigatus, C. neoformans and Candida spp. and their correspondent homologs predicted by the Phylome DB.
| Function | Transcription factor | Similar function (homologs) | Distinct function (homologs) | Unknown function (homologs) | Similar function (non-homologs) |
|---|---|---|---|---|---|
| Drug resistance (efflux pumps expression) | Pdr1 (C. glabrata) | N.A. | N.A. | N.A. | Tac1 (C. albicans) |
| Mrr1 (C. albicans) | |||||
| Mrr2 (C. albicans) | |||||
| Stb5 (C. glabrata) | Stb5 (C. albicans) | N.A. | CPAR2_109760; CTRG_04421* | N.A. | |
| Tac1 (C. albicans) | N.A. | Znc1 (C. albicans) | CPAR2_303510; CPAR2_303520 | Pdr1 (C. glabrata) | |
| CPAR2_303500; CTRG_05307 | Mrr1 (C. albicans) | ||||
| Hal9 (C. albicans) | CTRG_05306; CTRG_05308 | Mrr2 (C. albicans) | |||
| Mrr1 (C. albicans) | Mrr1 (C. parapsilosis) | Cta4 (C. albicans) | orf19.5133; orf19.7371 | Pdr1 (C. glabrata) | |
| CPAR2_501570; CPAR2_405260 CPAR2_405270; CPAR2_704130 | Tac1 (C. albicans) Mrr2 (C. albicans) | ||||
| CPAR2_807260; CTRG_02269* | |||||
| CTRG_02696*; CTRG_02712* | |||||
| CTRG_02268*; CTRG_05208* | |||||
| CTRG_00538*; CTRG_02271* | |||||
| CPAR2_501570; CPAR2_405260 CPAR2_405270; CPAR2_704130 | |||||
| Mrr2 (C. albicans) | Mrr1 (C. parapsilosis) | Cta4 (C. albicans) | CPAR2_405270; CPAR2_807820 | Pdr1 (C. glabrata) | |
| CAGL0M12298g; CTRG_05568* | Tac1 (C. albicans) | ||||
| Mrr1 (C. albicans) | |||||
| Drug resistance (ergosterol biosynthesis) | Upc2 (C. albicans) | Upc2 (C. parapsilosis) | N.A. | Ecm22 (C. albicans) | N.A. |
| Upc2a (C. glabrata)* | Upc2 (C. tropicalis)* | ||||
| Upc2b (C. glabrata)* | |||||
| SrbA (A. fumigatus) | SrbB (A. fumigatus) | N.A. | N.A. | Sre1 (C. neoformans) | |
| Virulence (hyphal growth/biofilm formation) | Efg1 (C. albicans) | Efg1 (C. parapsilosis) | StuA (A. fumigatus) | CAGL0L01771g | N.A. |
| StuA (A. fumigatus) | |||||
| Efh1 (C. albicans) | CTRG_01780 | ||||
| Efh1 (C. albicans) | Efg1 (C. albicans) | N.A. | Efh1 (C. parapsilosis); CTRG_01780 | StuA (A. fumigatus) | |
| Cph1 (C. albicans) | Ste12 (C. glabrata) | N.A. | Cph1 (C. parapsilosis) | N.A. | |
| Cph1 (C. tropicalis)* | |||||
| Ste12 (C. glabrata) | Cph1 (C. albicans) | N.A. | CAGL0H02145g; SteA (A. fumigatus) | N.A. | |
| Tec1 (C. albicans) | N.A. | N.A. | CPAR2_805930 | AbaA (A. fumigatus) | |
| Bcr1 (C. albicans) | Bcr1 (C. parapsilosis) | N.A. | CTRG_00608 | N.A. | |
| Cst6 (C. glabrata) | N.A. | N.A. | N.A. | Rca1 (C. albicans) | |
| Rca1 (C. albicans) | N.A. | N.A. | CPAR2_109540; CTRG_04281* | N.A. | |
| SomA (A. fumigatus) | N.A. | N.A. | N.A. | N.A. | |
| BrlA (A. fumigatus) | N.A. | N.A. | N.A. | N.A. | |
| MedA (A. fumigatus) | CNAG_03859* | N.A. | |||
| Znf2 (C. neoformans) | Csr1 (C. albicans) | ZafA (A. fumigatus) | N.A. | N.A. | |
| Nrg1 (C. albicans) | Nrg1 (C. tropicalis) | N.A. | CPAR2_300790; CTRG_00608 | N.A. | |
| CAGL0G08107g | |||||
| Csr1 (C. albicans) | N.A. | N.A. | CPAR2_403080; CTRG_03883 | N.A. | |
| CAGL0J05060g | |||||
| Rfg1 (C. albicans) | N.A. | N.A. | CPAR2_801100 | N.A. | |
| Ace2 (C. glabrata) | Swi5 (C. glabrata) | N.A. | N.A. | Ace2 (C. albicans) | |
| Ace2 (A. fumigatus) | |||||
| Ace2 (C. albicans) | Ace2 (C. parapsilosis) | N.A. | CTRG_03073* | Ace2 (C. glabrata) | |
| Ace2 (A. fumigatus) | |||||
| Virulence (nitrogen use) | Gat1 (C. albicans) | N.A. | N.A. | Gat1; CTRG_03831* | AreA (A. fumigatus) |
| Virulence (pH and weak acid response) | Rim101 (C. albicans) | N.A. | N.A. | CPAR2_700450 | PacC (A. fumigatus) |
| Rim101 (C. tropicalis)* | |||||
| War1 (C. albicans) | N.A. | N.A. | CPAR2_110360; CTRG_04350 | N.A. | |
| War1 (C. glabrata) | |||||
| Afu7g01640; Afu8g00950 | |||||
| Mnl1 (C. albicans) | N.A. | N.A. | CPAR2_204380 | Msn4 (C. albicans) | |
| Msn2 (C. glabrata) | |||||
| CTRG_03074 | Msn4 (C. glabrata) | ||||
| SebA (A. fumigatus) | |||||
| Msn4 (C. albicans) | N.A. | N.A. | CPAR2_301730 | Mnl1 (C. albicans) | |
| Msn2 (C. glabrata) | |||||
| CTRG_03253 | Msn4 (C. glabrata) | ||||
| SebA (A. fumigatus) | |||||
| Phenotypic switching | Wor1 (C. albicans) | N.A. | N.A. | CPAR2_805000; CTRG_03345 | N.A. |
| Afu6g04490 | |||||
| Wor2 (C. albicans) | N.A. | N.A. | CPAR2_405400 | N.A. | |
| Czf1 (C. albicans) | N.A. | N.A. | CTRG_03771 | N.A. | |
| Wor3 (C. albicans) | N.A. | N.A. | CPAR2_202450; CTRG_00711 | N.A. | |
| Wor4 (C. albicans) | N.A. | N.A. | CPAR2_808100; CTRG_05581 | N.A. | |
| Iron homeostasis | HapX (A. fumigatus) | N.A. | N.A. | N.A. | Hap43 (C. albicans) |
| Hap43 (C. albicans) | N.A. | N.A. | CPAR2_209090; CTRG_04121* | N.A. | |
| SreA (A. fumigatus) | Sfu1 (C. albicans) | N.A. | N.A. | N.A. | |
| Sfu1 (C. albicans) | N.A. | N.A. | CPAR2_700810; CTRG_03356 | N.A. | |
| Oxidative stress resistance (antioxidant gene expression) | Cap1 (C. albicans) | Yap1 (C. glabrata) | N.A. | CPAR2_405030 | Yap1 (A. fumigatus) |
| Yap1 (C. tropicalis)* | Yap1 (C. neoformans) | ||||
| Skn7 (C. albicans) | Skn7 (C. glabrata) | N.A. | CPAR2_304240; Skn7 (C. tropicalis)* | Skn7 (A. fumigatus) | |
| Skn7 (A. fumigatus) | Skn7 (C. albicans) | Skn7 (C. neoformans) | N.A. | Skn7 (C. glabrata) | |
| AtfA (A. fumigatus) | Atf1 (C. neoformans) | Atf1 (C. neoformans) | N.A. | N.A. | |
| Sko1 (C. albicans) | AtfA (A. fumigatus) | N.A. | CTRG_04352; Sko1 (C. glabrata) | N.A. | |
| Nitrosative stress resistance | Cta4 (C. albicans) | N.A. | Mnl1 (C. albicans) | N.A. | N.A. |
| Msn4 (C. albicans) | |||||
| Yap4 (C. neoformans) | N.A. | N.A. | N.A. | N.A. | |
| Amino acid starvation | Gcn4 (C. albicans) | N.A. | N.A. | Gcn4 (C. parapsilosis) | CpcA (A. fumigatus) |
| CTRG_02060; Gcn4 (C. glabrata) | |||||
| Gliotoxin production | MtfA (A. fumigatus) | N.A. | N.A. | N.A. | N.A. |
| Melanin production | Mbs1 (C. neoformans) | N.A. | Mbp1 (C. albicans) | Swi6 (C. albicans); Afu7g05620 | N.A. |
| Swi4 (C. albicans) | Afu3g13920*; CND05520 | ||||
| Cell cycle | Mbp1 (C. albicans) | N.A. | N.A. | CPAR2_102740; CAGL0D01012g | N.A. |
| CAGL0A04565g; Afu7g05620 | |||||
| Mbp1 (C. tropicalis) |
N.A., Not Available;
Homologs identified by BLASTp.
The regulation of transcriptional networks is complex and presents significant variations among different fungal pathogens, either in terms of regulators themselves or their regulatory targets. The study of regulatory circuits should therefore be a prime strategy in the fight against fungal infections, allowing to develop better diagnostic and treatment approaches according to each pathogen's conserved or specific pathways.
Author contributions
PP, CC, MC, and DR reviewed the literature on the manuscript related subject, foccusing on Candida species, Cryptococcus neoformans and Aspergillus fumigatus, respectively. PP and MCT wrote most of the manuscript. MCT was the scientific coordinator of this work.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research conducted by the authors within the scope of this paper was supported by “Fundação para a Ciência e a Tecnologia” (FCT) [Contracts PTDC/BBB-BIO/4004/2014 and Ph.D. and post-doctoral grants to PP and MC (SFRD/BD/113631/2015 and SFRD/BD/116946/2016) and CC (SFRH/BD/100863/2014), respectively]. Funding received by iBB-Institute for Bioengineering and Biosciences from FCT-Portuguese Foundation for Science and Technology (UID/BIO/04565/2013) and from Programa Operacional Regional de Lisboa 2020 (Project No. 007317) is acknowledged.
References
- Adams T. H., Boylan M. T., Timberlake W. E. (1988). brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54, 353–362. 10.1016/0092-8674(88)90198-5 [DOI] [PubMed] [Google Scholar]
- Alby K., Bennett R. J. (2009). Stress-induced phenotypic switching in Candida albicans. Mol. Biol. Cell 20, 3178–3191. 10.1091/mbc.E09-01-0040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida F., Wolf J. M., Casadevall A. (2015). Virulence-associated enzymes of Cryptococcus neoformans. Eukaryotic Cell 14, 1173–1185. 10.1128/EC.00103-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Monge R., Román E., Arana D. M., Prieto D., Urrialde V., Nombela C., et al. (2010). The Sko1 protein represses the yeast-to-hypha transition and regulates the oxidative stress response in Candida albicans. Fungal Genet. Biol. 47, 587–601. 10.1016/j.fgb.2010.03.009 [DOI] [PubMed] [Google Scholar]
- Alspaugh J. A., Cavallo L. M., Perfect J. R., Heitman J. (2000). RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol. Microbiol. 36, 352–365. 10.1046/j.1365-2958.2000.01852.x [DOI] [PubMed] [Google Scholar]
- Alspaugh J. A., Perfect J. R., Heitman J. (1997). Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 11, 3206–3217. 10.1101/gad.11.23.3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azie N., Neofytos D., Pfaller M., Meier-Kriesche H. U., Quan S. P., Horn D. (2012). The PATH (Prospective Antifungal Therapy) Alliance(R) registry and invasive fungal infections: update 2012. Diagn. Microbiol. Infect. Dis. 73, 293–300. 10.1016/j.diagmicrobio.2012.06.012 [DOI] [PubMed] [Google Scholar]
- Bailey D. A., Feldmann P. J., Bovey M., Gow N. A., Brown A. J. (1996). The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 178, 5353–5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J. M., Louw J., Lewis L. E., Okai B., Walls C. A., Ballou E. R., et al. (2014). Candida albicans hypha formation and mannan masking of beta-glucan inhibit macrophage phagosome maturation. MBio. 5:e01874. 10.1128/mBio.01874-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee M., Thompson D. S., Lazzell A., Carlisle P. L., Pierce C., Monteagudo C., et al. (2008). UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol. Biol. Cell 19, 1354–1365. 10.1091/mbc.E07-11-1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee M., Uppuluri P., Zhao X. R., Carlisle P. L., Vipulanandan G., Villar C. C., et al. (2013). Expression of UME6, a key regulator of Candida albicans hyphal development, enhances biofilm formation via Hgc1- and Sun41-dependent mechanisms. Eukaryotic Cell 12, 224–232. 10.1128/EC.00163-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa F. M., Fonseca F. L., Holandino C., Alviano C. S., Nimrichter L., Rodrigues M. L. (2006). Glucuronoxylomannan-mediated interaction of Cryptococcus neoformans with human alveolar cells results in fungal internalization and host cell damage. Microbes. Infect. 8, 493–502. 10.1016/j.micinf.2005.07.027 [DOI] [PubMed] [Google Scholar]
- Berkova N., Lair-Fulleringer S., Femenia F., Huet D., Wagner M. C., Gorna K., et al. (2006). Aspergillus fumigatus conidia inhibit tumour necrosis factor- or staurosporine-induced apoptosis in epithelial cells. Int. Immunol. 18, 139–150. 10.1093/intimm/dxh356 [DOI] [PubMed] [Google Scholar]
- Bertuzzi M., Schrettl M., Alcazar-Fuoli L., Cairns T. C., Muñoz A., Walker L. A., et al. (2014). The pH-responsive PacC transcription factor of Aspergillus fumigatus governs epithelial entry and tissue invasion during pulmonary aspergillosis. PLoS Pathog. 10:e1004413. 10.1371/journal.ppat.1004413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien C. M., Chang Y. C., Nes W. D., Kwon-Chung K. J., Espenshade P. J. (2009). Cryptococcus neoformans Site-2 protease is required for virulence and survival in the presence of azole drugs. Mol. Microbiol. 74, 672–690. 10.1111/j.1365-2958.2009.06895.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse C. E., Irwin M. Y., Fonzi W. A., Sypherd P. S. (1993). Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect. Immun. 61, 3648–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatzer M., Barker B. M., Willger S. D., Beckmann N., Blosser S. J., Cornish E. J., et al. (2011). SREBP coordinates iron and ergosterol homeostasis to mediate triazole drug and hypoxia responses in the human fungal pathogen Aspergillus fumigatus. PLoS Genet. 7:e1002374. 10.1371/journal.pgen.1002374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun B. R., Kadosh D., Johnson A. D. (2001). NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20, 4753–4761. 10.1093/emboj/20.17.4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. J., Haynes K., Quinn J. (2009). Nitrosative and oxidative stress responses in fungal pathogenicity. Curr. Opin. Microbiol. 12, 384–391. 10.1016/j.mib.2009.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. A., Goldman G. H. (2016). The contribution of Aspergillus fumigatus stress responses to virulence and antifungal resistance. J. Microbiol. 54, 243–253. 10.1007/s12275-016-5510-4 [DOI] [PubMed] [Google Scholar]
- Calcagno A. M., Bignell E., Warn P., Jones M. D., Denning D. W., Mühlschlegel F. A., et al. (2003). Candida glabrata STE12 is required for wild-type levels of virulence and nitrogen starvation induced filamentation. Mol. Microbiol. 50, 1309–1318. 10.1046/j.1365-2958.2003.03755.x [DOI] [PubMed] [Google Scholar]
- Caudle K. E., Barker K. S., Wiederhold N. P., Xu L., Homayouni R., Rogers P. D. (2011). Genomewide expression profile analysis of the Candida glabrata Pdr1 regulon. Eukaryotic Cell 10, 373–383. 10.1128/EC.00073-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffin W. L., López-Ribot J. L., Casanova M., Gozalbo D., Martínez J. P. (1998). Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol. Mol. Biol. Rev. 62, 130–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C., Bien C. M., Lee H., Espenshade P. J., Kwon-Chung K. J. (2007). Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol. Microbiol. 64, 614–629. 10.1111/j.1365-2958.2007.05676.x [DOI] [PubMed] [Google Scholar]
- Chang Y. C., Jong A., Huang S., Zerfas P., Kwon-Chung K. J. (2006). CPS1, a homolog of the Streptococcus pneumoniae type 3 polysaccharide synthase gene, is important for the pathobiology of Cryptococcus neoformans. Infect. Immun. 74, 3930–3938. 10.1128/IAI.00089-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C., Penoyer L. A., Kwon-Chung K. J. (2001). The second STE12 homologue of Cryptococcus neoformans is MATa-specific and plays an important role in virulence. Proc. Natl. Acad. Sci. U.S.A. 98, 3258–3263. 10.1073/pnas.061031998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C., Stins M. F., McCaffery M. J., Miller G. F., Pare D. R., Dam T., et al. (2004). Cryptococcal yeast cells invade the central nervous system via transcellular penetration of the blood-brain barrier. Infect. Immun. 72, 4985–4995. 10.1128/IAI.72.9.4985-4995.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan N., Latge J. P., Calderone R. (2006). Signalling and oxidant adaptation in Candida albicans and Aspergillus fumigatus. Nat. Rev. Microbiol. 4, 435–444. 10.1038/nrmicro1426 [DOI] [PubMed] [Google Scholar]
- Chen K. H., Miyazaki T., Tsai H. F., Bennett J. E. (2007). The bZip transcription factor Cgap1p is involved in multidrug resistance and required for activation of multidrug transporter gene CgFLR1 in Candida glabrata. Gene 386, 63–72. 10.1016/j.gene.2006.08.010 [DOI] [PubMed] [Google Scholar]
- Chen S. H., Stins M. F., Huang S. H., Chen Y. H., Kwon-Chung K. J., Chang Y., et al. (2003). Cryptococcus neoformans induces alterations in the cytoskeleton of human brain microvascular endothelial cells. J. Med. Microbiol. 52(Pt 11), 961–970. 10.1099/jmm.0.05230-0 [DOI] [PubMed] [Google Scholar]
- Chiranand W., McLeod I., Zhou H., Lynn J. J., Vega L. A., Myers H., et al. (2008). CTA4 transcription factor mediates induction of nitrosative stress response in Candida albicans. Eukaryotic Cell 7, 268–278. 10.1128/EC.00240-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrétien F., Lortholary O., Kansau I., Neuville S., Gray F., Dromer F. (2002). Pathogenesis of cerebral Cryptococcus neoformans infection after fungemia. J. Infect. Dis. 186, 522–530. 10.1086/341564 [DOI] [PubMed] [Google Scholar]
- Chun C. D., Liu O. W., Madhani H. D. (2007). A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog. 3:e22. 10.1371/journal.ppat.0030022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D., Barker B. M., Carey C. C., Merriman B., Werner E. R., Lechner B. E., et al. (2014). ChIP-seq and in vivo transcriptome analyses of the Aspergillus fumigatus SREBP SrbA reveals a new regulator of the fungal hypoxia response and virulence. PLoS Pathog. 10:e1004487. 10.1371/journal.ppat.1004487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly L. A., Riccombeni A., Grózer Z., Holland L. M., Lynch D. B., Andes D. R., et al. (2013). The APSES transcription factor Efg1 is a global regulator that controls morphogenesis and biofilm formation in Candida parapsilosis. Mol. Microbiol. 90, 36–53. 10.1111/mmi.12345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa C., Nunes J., Henriques A., Mira N. P., Nakayama H., Chibana H., et al. (2014). Candida glabrata drug:H+ antiporter CgTpo3 (ORF CAGL0I10384g): role in azole drug resistance and polyamine homeostasis. J. Antimicrob. Chemother. 69, 1767–1776. 10.1093/jac/dku044 [DOI] [PubMed] [Google Scholar]
- Costa C., Pires C., Cabrito T. R., Renaudin A., Ohno M., Chibana H., et al. (2013). Candida glabrata drug:H+ antiporter CgQdr2 confers imidazole drug resistance, being activated by transcription factor CgPdr1. Antimicrob. Agents Chemother. 57, 3159–3167. 10.1128/AAC.00811-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste A., Selmecki A., Forche A., Diogo D., Bougnoux M. E., d'Enfert C., et al. (2007). Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryotic Cell 6, 1889–1904. 10.1128/EC.00151-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste A. T., Crittin J., Bauser C., Rohde B., Sanglard D. (2009). Functional analysis of cis- and trans-acting elements of the Candida albicans CDR2 promoter with a novel promoter reporter system. Eukaryotic Cell 8, 1250–1267. 10.1128/EC.00069-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste A. T., Karababa M., Ischer F., Bille J., Sanglard D. (2004). TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryotic Cell 3, 1639–1652. 10.1128/EC.3.6.1639-1652.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste A. T., Ramsdale M., Ischer F., Sanglard D. (2008). Divergent functions of three Candida albicans zinc-cluster transcription factors (CTA4, ASG1 and CTF1) complementing pleiotropic drug resistance in Saccharomyces cerevisiae. Microbiology 154(Pt 5), 1491–1501. 10.1099/mic.0.2007/016063-0 [DOI] [PubMed] [Google Scholar]
- Coste A., Turner V., Ischer F., Morschhäuser J., Forche A., Selmecki A., et al. (2006). A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172, 2139–2156. 10.1534/genetics.105.054767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côte P., Hogues H., Whiteway M. (2009). Transcriptional analysis of the Candida albicans cell cycle. Mol. Biol. Cell 20, 3363–3373. 10.1091/mbc.E09-03-0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottier F., Leewattanapasuk W., Kemp L. R., Murphy M., Supuran C. T., Kurzai O., et al. (2013). Carbonic anhydrase regulation and CO(2) sensing in the fungal pathogen Candida glabrata involves a novel Rca1p ortholog. Bioorg. Med. Chem. 21, 1549–1554. 10.1016/j.bmc.2012.05.053 [DOI] [PubMed] [Google Scholar]
- Cottier F., Raymond M., Kurzai O., Bolstad M., Leewattanapasuk W., Jiménez-López C., et al. (2012). The bZIP transcription factor Rca1p is a central regulator of a novel sensing pathway in yeast. PLoS Pathog. 8:e1002485. 10.1371/journal.ppat.1002485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuéllar-Cruz M., Briones-Martin-del-Campo M., Cañas-Villamar I., Montalvo-Arredondo J., Riego-Ruiz L., Castaño I., et al. (2008). High resistance to oxidative stress in the fungal pathogen Candida glabrata is mediated by a single catalase, Cta1p, and is controlled by the transcription factors Yap1p, Skn7p, Msn2p, and Msn4p. Eukaryotic Cell 7, 814–825. 10.1128/EC.00011-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabas N., Morschhäuser J. (2007). Control of ammonium permease expression and filamentous growth by the GATA transcription factors GLN3 and GAT1 in Candida albicans. Eukaryotic Cell 6, 875–888. 10.1128/EC.00307-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D., Edwards J. E., Jr., Mitchell A. P., Ibrahim A. S. (2000). Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 68, 5953–5959. 10.1128/IAI.68.10.5953-5959.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeHart D. J., Agwu D. E., Julian N. C., Washburn R. G. (1997). Binding and germination of Aspergillus fumigatus conidia on cultured A549 pneumocytes. J. Infect. Dis. 175, 146–150. 10.1093/infdis/175.1.146 [DOI] [PubMed] [Google Scholar]
- Dinamarco T. M., Almeida R. S., de Castro P. A., Brown N. A., dos Reis T. F., Ramalho L. N., et al. (2012). Molecular characterization of the putative transcription factor SebA involved in virulence in Aspergillus fumigatus. Eukaryotic Cell 11, 518–531. 10.1128/EC.00016-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C., Butler G. (2007). Development of a gene knockout system in Candida parapsilosis reveals a conserved role for BCR1 in biofilm formation. Eukaryotic Cell 6, 1310–1319. 10.1128/EC.00136-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C., Vidanes G. M., Maguire S. L., Guida A., Synnott J. M., Andes D. R., et al. (2011). Conserved and divergent roles of Bcr1 and CFEM proteins in Candida parapsilosis and Candida albicans. PLoS ONE 6:e28151. 10.1371/journal.pone.0028151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doedt T., Krishnamurthy S., Bockmühl D. P., Tebarth B., Stempel C., Russell C. L., et al. (2004). APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 15, 3167–3180. 10.1091/mbc.E03-11-0782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas L. J. (2003). Candida biofilms and their role in infection. Trends Microbiol. 11, 30–36. 10.1016/S0966-842X(02)00002-1 [DOI] [PubMed] [Google Scholar]
- D'Souza C. A., Alspaugh J. A., Yue C., Harashima T., Cox G. M., Perfect J. R., et al. (2001). Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21, 3179–3191. 10.1128/MCB.21.9.3179-3191.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza C. A., Heitman J. (2001). Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25, 349–364. 10.1111/j.1574-6976.2001.tb00582.x [DOI] [PubMed] [Google Scholar]
- Dunkel N., Blass J., Rogers P. D., Morschhäuser J. (2008). Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol. Microbiol. 69, 827–840. 10.1111/j.1365-2958.2008.06309.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejzykowicz D. E., Cunha M. M., Rozental S., Solis N. V., Gravelat F. N., Sheppard D. C., et al. (2009). The Aspergillus fumigatus transcription factor Ace2 governs pigment production, conidiation and virulence. Mol. Microbiol. 72, 155–169. 10.1111/j.1365-2958.2009.06631.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene I. V., Brunke S., Brown A. J., Hube B. (2014). Metabolism in fungal pathogenesis. Cold Spring Harb. Perspect. Med. 4:a019695. 10.1101/cshperspect.a019695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert B., Smith D. A., Cornell M. J., Alam I., Nicholls S., Brown A. J., et al. (2006). Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell 17, 1018–1032. 10.1091/mbc.E05-06-0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwig L. P., Gow N. A. (2016). Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 1 14, 163–176. 10.1038/nrmicro.2015.21 [DOI] [PubMed] [Google Scholar]
- Fanning S., Mitchell A. P. (2012). Fungal biofilms. PLoS Pathog. 8:e1002585. 10.1371/journal.ppat.1002585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S., Ischer F., Calabrese D., Posteraro B., Sanguinetti M., Fadda G., et al. (2009). Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog. 5:e1000268. 10.1371/journal.ppat.1000268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filler S. G., Sheppard D. C. (2006). Fungal invasion of normally non-phagocytic host cells. PLoS Pathog. 2:e129. 10.1371/journal.ppat.0020129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel J. S., Xu W., Huang D., Hill E. M., Desai J. V., Woolford C. A., et al. (2012). Portrait of Candida albicans adherence regulators. PLoS Pathog. 8:e1002525. 10.1371/journal.ppat.1002525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin C., De Groot P., MacCallum D., Schaller M., Klis F., Odds F. C., et al. (2005). Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 56, 397–415. 10.1111/j.1365-2958.2005.04557.x [DOI] [PubMed] [Google Scholar]
- Ghannoum M. A., Rice L. B. (1999). Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 12, 501–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Navarathna D. H., Roberts D. D., Cooper J. T., Atkin A. L., Petro T. M., et al. (2009). Arginine-induced germ tube formation in Candida albicans is essential for escape from murine macrophage line RAW 264.7. Infect. Immun. 77, 1596–1605. 10.1128/IAI.01452-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C. M., MacIver F. H., Dawes I. W. (1997). Mitochondrial function is required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. FEBS Lett. 410, 219–222. 10.1016/S0014-5793(97)00592-9 [DOI] [PubMed] [Google Scholar]
- Gsaller F., Hortschansky P., Beattie S. R., Klammer V., Tuppatsch K., Lechner B. E., et al. (2014). The Janus transcription factor HapX controls fungal adaptation to both iron starvation and iron excess. EMBO J. 33, 2261–2276. 10.15252/embj.201489468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z., Liu H. (2015). The WOR1 5′ untranslated region regulates white-opaque switching in Candida albicans by reducing translational efficiency. Mol. Microbiol. 97, 125–138. 10.1111/mmi.13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guida A., Lindstädt C., Maguire S. L., Ding C., Higgins D. G., Corton N. J., et al. (2011). Using RNA-seq to determine the transcriptional landscape and the hypoxic response of the pathogenic yeast Candida parapsilosis. BMC Genomics 12:628. 10.1186/1471-2164-12-628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara D., Suzuki S., Kamei K., Gonoi T., Kawamoto S. (2014). The role of AtfA and HOG MAPK pathway in stress tolerance in conidia of Aspergillus fumigatus. Fungal Genet. Biol. 73, 138–149. 10.1016/j.fgb.2014.10.011 [DOI] [PubMed] [Google Scholar]
- Haynes B. C., Skowyra M. L., Spencer S. J., Gish S. R., Williams M., Held E. P., et al. (2011). Toward an integrated model of capsule regulation in Cryptococcus neoformans. PLoS Pathog. 7:e1002411. 10.1371/journal.ppat.1002411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann C. J., Schneider S., Barker K. S., Rogers P. D., Morschhäuser J. (2010). An A643T mutation in the transcription factor Upc2p causes constitutive ERG11 upregulation and increased fluconazole resistance in Candida albicans. Antimicrob. Agents Chemother. 54, 353–359. 10.1128/AAC.01102-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel M., Arst H. N., Jr., Aufauvre-Brown A., Holden D. W. (1998). The role of the Aspergillus fumigatus areA gene in invasive pulmonary aspergillosis. Mol. Gen. Genet. 258, 553–557. 10.1007/s004380050767 [DOI] [PubMed] [Google Scholar]
- Hole C., Wormley F. L., Jr. (2016). Innate host defenses against Cryptococcus neoformans. J. Microbiol. 54, 202–211. 10.1007/s12275-016-5625-7 [DOI] [PubMed] [Google Scholar]
- Holland L. M., Schröder M. S., Turner S. A., Taff H., Andes D., Grózer Z., et al. (2014). Comparative phenotypic analysis of the major fungal pathogens Candida parapsilosis and Candida albicans. PLoS Pathog. 10:e1004365. 10.1371/journal.ppat.1004365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann O. R., Dea J., Noble S. M., Johnson A. D. (2009). A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 5:e1000783. 10.1371/journal.pgen.1000783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. Y., Roze L. V., Linz J. E. (2013). Oxidative stress-related transcription factors in the regulation of secondary metabolism. Toxins (Basel). 5, 683–702. 10.3390/toxins5040683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope W. W., Tabernero L., Denning D. W., Anderson M. J. (2004). Molecular mechanisms of primary resistance to flucytosine in Candida albicans. Antimicrob. Agents Chemother. 48, 4377–4386. 10.1128/AAC.48.11.4377-4386.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer L. L., Payne T. L., Bell M., Myers A. M., Scherer S. (1998). Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr. Genet. 33, 451–459. 10.1007/s002940050359 [DOI] [PubMed] [Google Scholar]
- Hsu P. C., Yang C. Y., Lan C. Y. (2011). Candida albicans Hap43 is a repressor induced under low-iron conditions and is essential for iron-responsive transcriptional regulation and virulence. Eukaryotic Cell 10, 207–225. 10.1128/EC.00158-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Yi S., Sahni N., Daniels K. J., Srikantha T., Soll D. R. (2010). N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 6:e1000806. 10.1371/journal.ppat.1000806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein B., Huang H., Glory A., Osmani A., Kaminskyj S., Nantel A., et al. (2011). G1/S transcription factor orthologues Swi4p and Swi6p are important but not essential for cell proliferation and influence hyphal development in the fungal pathogen Candida albicans. Eukaryotic Cell 10, 384–397. 10.1128/EC.00278-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim A. S., Filler S. G., Alcouloumre M. S., Kozel T. R., Edwards J. E., Jr., Ghannoum M. A. (1995). Adherence to and damage of endothelial cells by Cryptococcus neoformans in vitro: role of the capsule. Infect. Immun. 63, 4368–4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabra-Rizk M. A., Falkler W. A., Meiller T. F. (2004). Fungal biofilms and drug resistance. Emerging Infect. Dis. 10, 14–19. 10.3201/eid1001.030119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282. 10.1093/bioinformatics/8.3.275 [DOI] [PubMed] [Google Scholar]
- Jong A. Y., Stins M. F., Huang S. H., Chen S. H., Kim K. S. (2001). Traversal of Candida albicans across human blood-brain barrier in vitro. Infect. Immun. 69, 4536–4544. 10.1128/IAI.69.7.4536-4544.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K. W., Yang D. H., Maeng S., Lee K. T., So Y. S., Hong J., et al. (2015). Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nat. Commun. 6, 6757. 10.1038/ncomms7757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D., Johnson A. D. (2001). Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 21, 2496–2505. 10.1128/MCB.21.7.2496-2505.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D., Johnson A. D. (2005). Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16, 2903–2912. 10.1091/mbc.E05-01-0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamran M., Calcagno A. M., Findon H., Bignell E., Jones M. D., Warn P., et al. (2004). Inactivation of transcription factor gene ACE2 in the fungal pathogen Candida glabrata results in hypervirulence. Eukaryotic Cell 3, 546–552. 10.1128/EC.3.2.546-552.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanafani Z. A., Perfect J. R. (2008). Antimicrobial resistance: resistance to antifungal agents: mechanisms and clinical impact. Clin. Infect. Dis. 46, 120–128. 10.1086/524071 [DOI] [PubMed] [Google Scholar]
- Kelly M. T., MacCallum D. M., Clancy S. D., Odds F. C., Brown A. J., Butler G. (2004). The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol. Microbiol. 53, 969–983. 10.1111/j.1365-2958.2004.04185.x [DOI] [PubMed] [Google Scholar]
- Kelly S. L., Lamb D. C., Corran A. J., Baldwin B. C., Kelly D. E. (1995). Mode of action and resistance to azole antifungals associated with the formation of 14 α-methylergosta-8,24(28)-dien-3β,6α-diol. Biochem. Biophys. Res. Commun. 207, 910–915. 10.1006/bbrc.1995.1272 [DOI] [PubMed] [Google Scholar]
- Khalaf R. A., Zitomer R. S. (2001). The DNA binding protein Rfg1 is a repressor of filamentation in Candida albicans. Genetics 157, 1503–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. J., Kil M., Jung J. H., Kim J. (2008). Roles of Zinc-responsive transcription factor Csr1 in filamentous growth of the pathogenic Yeast Candida albicans. J. Microbiol. Biotechnol. 18, 242–247. [PubMed] [Google Scholar]
- Kim M. S., Ko Y. J., Maeng S., Floyd A., Heitman J., Bahn Y. S. (2010). Comparative transcriptome analysis of the CO2 sensing pathway via differential expression of carbonic anhydrase in Cryptococcus neoformans. Genetics 185, 1207–1219. 10.1534/genetics.110.118315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz S. A., Drutz D. J., Harrison J. L., Huppert M. (1983). Adherence and penetration of vascular endothelium by Candida yeasts. Infect. Immun. 42, 374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojic E. M., Darouiche R. O. (2004). Candida infections of medical devices. Clin. Microbiol. Rev. 17, 255–267. 10.1128/CMR.17.2.255-267.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolotila M. P., Diamond R. D. (1990). Effects of neutrophils and in vitro oxidants on survival and phenotypic switching of Candida albicans WO-1. Infect. Immun. 58, 1174–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korting H. C., Hube B., Oberbauer S., Januschke E., Hamm G., Albrecht A., et al. (2003). Reduced expression of the hyphal-independent Candida albicans proteinase genes SAP1 and SAP3 in the efg1 mutant is associated with attenuated virulence during infection of oral epithelium. J. Med. Microbiol. 52(Pt 8), 623–632. 10.1099/jmm.0.05125-0 [DOI] [PubMed] [Google Scholar]
- Krappmann S., Bignell E. M., Reichard U., Rogers T., Haynes K., Braus G. H. (2004). The Aspergillus fumigatus transcriptional activator CpcA contributes significantly to the virulence of this fungal pathogen. Mol. Microbiol. 52, 785–799. 10.1111/j.1365-2958.2004.04015.x [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvaal C., Lachke S. A., Srikantha T., Daniels K., McCoy J., Soll D. R. (1999). Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect. Immun. 67, 6652–6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachke S. A., Lockhart S. R., Daniels K. J., Soll D. R. (2003). Skin facilitates Candida albicans mating. Infect. Immun. 71, 4970–4976. 10.1128/IAI.71.9.4970-4976.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachke S. A., Srikantha T., Tsai L. K., Daniels K., Soll D. R. (2000). Phenotypic switching in Candida glabrata involves phase-specific regulation of the metallothionein gene MT-II and the newly discovered hemolysin gene HLP. Infect. Immun. 68, 884–895. 10.1128/IAI.68.2.884-895.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffey S. F., Butler G. (2005). Phenotype switching affects biofilm formation by Candida parapsilosis. Microbiology 151(Pt 4), 1073–1081. 10.1099/mic.0.27739-0 [DOI] [PubMed] [Google Scholar]
- Lamarre C., Ibrahim-Granet O., Du C., Calderone R., Latgé J. P. (2007). Characterization of the SKN7 ortholog of Aspergillus fumigatus. Fungal Genet. Biol. 44, 682–690. 10.1016/j.fgb.2007.01.009 [DOI] [PubMed] [Google Scholar]
- Lamarre C., Sokol S., Debeaupuis J. P., Henry C., Lacroix C., Glaser P., et al. (2008). Transcriptomic analysis of the exit from dormancy of Aspergillus fumigatus conidia. BMC Genomics 9:417. 10.1186/1471-2164-9-417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane S., Birse C., Zhou S., Matson R., Liu H. (2001a). DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 276, 48988–48996. 10.1074/jbc.M104484200 [DOI] [PubMed] [Google Scholar]
- Lane S., Zhou S., Pan T., Dai Q., Liu H. (2001b). The basic helix-loop-helix transcription factor Cph2 regulates hyphal development in Candida albicans partly via TEC1. Mol. Cell. Biol. 21, 6418–6428. 10.1128/MCB.21.19.6418-6428.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal S. M., Jr., Vareechon C., Cowden S., Cobb B. A., Latgé J. P., Momany M., et al. (2012). Fungal antioxidant pathways promote survival against neutrophils during infection. J. Clin. Invest. 122, 2482–2498. 10.1172/JCI63239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel K., MacPherson S., Turcotte B. (2006). New tools for phenotypic analysis in Candida albicans: the WAR1 gene confers resistance to sorbate. Yeast 23, 249–259. 10.1002/yea.1346 [DOI] [PubMed] [Google Scholar]
- Lee I. R., Morrow C. A., Fraser J. A. (2013). Nitrogen regulation of virulence in clinically prevalent fungal pathogens. FEMS Microbiol. Lett. 345, 77–84. 10.1111/1574-6968.12181 [DOI] [PubMed] [Google Scholar]
- Lessing F., Kniemeyer O., Wozniok I., Loeffler J., Kurzai O., Haertl A., et al. (2007). The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model. Eukaryotic Cell 6, 2290–2302. 10.1128/EC.00267-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L. E., Bain J. M., Lowes C., Gillespie C., Rudkin F. M., Gow N. A., et al. (2012a). Stage specific assessment of Candida albicans phagocytosis by macrophages identifies cell wall composition and morphogenesis as key determinants. PLoS Pathog. 8:e1002578. 10.1371/journal.ppat.1002578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L. E., Bain J. M., Lowes C., Gow N. A., Erwig L. P. (2012b). Candida albicans infection inhibits macrophage cell division and proliferation. Fungal Genet. Biol. 49, 679–680. 10.1016/j.fgb.2012.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limjindaporn T., Khalaf R. A., Fonzi W. A. (2003). Nitrogen metabolism and virulence of Candida albicans require the GATA-type transcriptional activator encoded by GAT1. Mol. Microbiol. 50, 993–1004. 10.1046/j.1365-2958.2003.03747.x [DOI] [PubMed] [Google Scholar]
- Lin C. H., Kabrawala S., Fox E. P., Nobile C. J., Johnson A. D., Bennett R. J. (2013). Genetic control of conventional and pheromone-stimulated biofilm formation in Candida albicans. PLoS Pathog. 9:e1003305. 10.1371/journal.ppat.1003305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. J., Sasse C., Gerke J., Valerius O., Irmer H., Frauendorf H., et al. (2015). Transcription factor soma is required for adhesion, development and virulence of the human pathogen Aspergillus fumigatus. PLoS Pathog. 11:e1005205. 10.1371/journal.ppat.1005205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. T., Znaidi S., Barker K. S., Xu L., Homayouni R., Saidane S., et al. (2007). Genome-wide expression and location analyses of the Candida albicans Tac1p regulon. Eukaryotic Cell 6, 2122–2138. 10.1128/EC.00327-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M. B., Hernday A. D., Fordyce P. M., Noiman L., Sorrells T. R., Hanson-Smith V., et al. (2013). Identification and characterization of a previously undescribed family of sequence-specific DNA-binding domains. Proc. Natl. Acad. Sci. U.S.A. 110, 7660–7665. 10.1073/pnas.1221734110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M. B., Johnson A. D. (2008). Differential phagocytosis of white versus opaque Candida albicans by Drosophila and mouse phagocytes. PLoS ONE 3:e1473. 10.1371/journal.pone.0001473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M. B., Johnson A. D. (2016). Identification and characterization of wor4, a new transcriptional regulator of white-opaque switching. G3 6, 721–729. 10.1534/g3.115.024885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossinsky A. S., Jong A., Fiala M., Mukhtar M., Buttle K. F., Ingram M. (2006). The histopathology of Candida albicans invasion in neonatal rat tissues and in the human blood-brain barrier in culture revealed by light, scanning, transmission and immunoelectron microscopy. Histol. Histopathol. 21, 1029–1041. [DOI] [PubMed] [Google Scholar]
- MacCallum D. M., Findon H., Kenny C. C., Butler G., Haynes K., Odds F. C. (2006). Different consequences of ACE2 and SWI5 gene disruptions for virulence of pathogenic and nonpathogenic yeasts. Infect. Immun. 74, 5244–5248. 10.1128/IAI.00817-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson S., Akache B., Weber S., De Deken X., Raymond M., Turcotte B. (2005). Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob. Agents Chemother. 49, 1745–1752. 10.1128/AAC.49.5.1745-1752.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire S. L., Wang C., Holland L. M., Brunel F., Neuvéglise C., Nicaud J. M., et al. (2014). Zinc finger transcription factors displaced SREBP proteins as the major Sterol regulators during Saccharomycotina evolution. PLoS Genet. 10:e1004076. 10.1371/journal.pgen.1004076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez L. R., Casadevall A. (2015). Biofilm formation by Cryptococcus neoformans. Microbiol. Spectr. 3:MB-0006-2014. 10.1128/microbiolspec.mb-0006-2014 [DOI] [PubMed] [Google Scholar]
- Martínez-Pastor M. T., Marchler G., Schüller C., Marchler-Bauer A., Ruis H., Estruch F. (1996). The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15, 2227–2235. [PMC free article] [PubMed] [Google Scholar]
- Mattia E., Carruba G., Angiolella L., Cassone A. (1982). Induction of germ tube formation by N-acetyl-D-glucosamine in Candida albicans: uptake of inducer and germinative response. J. Bacteriol. 152, 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie C. G., Koser U., Lewis L. E., Bain J. M., Mora-Montes H. M., Barker R. N., et al. (2010). Contribution of Candida albicans cell wall components to recognition by and escape from murine macrophages. Infect. Immun. 78, 1650–1658. 10.1128/IAI.00001-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado E., Garcia-Effron G., Buitrago M. J., Alcazar-Fuoli L., Cuenca-Estrella M., Rodriguez-Tudela J. L. (2005). Targeted gene disruption of the 14-α sterol demethylase (cyp51A) in Aspergillus fumigatus and its role in azole drug susceptibility. Antimicrob. Agents Chemother. 49, 2536–2538. 10.1128/AAC.49.6.2536-2538.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhej J., Delaveau T., Guitard J., Palancade B., Hennequin C., Garcia M., et al. (2015). Yap7 is a transcriptional repressor of nitric oxide oxidase in yeasts, which arose from neofunctionalization after whole genome duplication. Mol. Microbiol. 96, 951–972. 10.1111/mmi.12983 [DOI] [PubMed] [Google Scholar]
- Merhej J., Thiebaut A., Blugeon C., Pouch J., Ali Chaouche Mel A., Camadro J. M., et al. (2016). A network of paralogous stress response transcription factors in the human pathogen Candida glabrata. Front. Microbiol. 7:645. 10.3389/fmicb.2016.00645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miceli M. H., Díaz J. A., Lee S. A. (2011). Emerging opportunistic yeast infections. Lancet Infect. Dis. 11, 142–151. 10.1016/S1473-3099(10)70218-8 [DOI] [PubMed] [Google Scholar]
- Miceli M. H., Lee S. A. (2011). Emerging moulds epidemiological trends and antifungal resistance. Mycoses 54, e666–e678. 10.1111/j.1439-0507.2011.02032.x [DOI] [PubMed] [Google Scholar]
- Miller M. G., Johnson A. D. (2002). White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110, 293–302. 10.1016/S0092-8674(02)00837-1 [DOI] [PubMed] [Google Scholar]
- Mira N. P., Lourenço A. B., Fernandes A. R., Becker J. D., Sá-Correia I. (2009). The RIM101 pathway has a role in Saccharomyces cerevisiae adaptive response and resistance to propionic acid and other weak acids. FEMS Yeast Res. 9, 202–216. 10.1111/j.1567-1364.2008.00473.x [DOI] [PubMed] [Google Scholar]
- Missall T. A., Lodge J. K. (2005). Function of the thioredoxin proteins in Cryptococcus neoformans during stress or virulence and regulation by putative transcriptional modulators. Mol. Microbiol. 57, 847–858. 10.1111/j.1365-2958.2005.04735.x [DOI] [PubMed] [Google Scholar]
- Mogavero S., Tavanti A., Senesi S., Rogers P. D., Morschhauser J. (2011). Differential requirement of the transcription factor Mcm1 for activation of the Candida albicans multidrug efflux pump MDR1 by its regulators Mrr1 and Cap1. Antimicrob. Agents Chemother. 55, 2061–2066. 10.1128/AAC.01467-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagna M. T., Caggiano G., Lovero G., De Giglio O., Coretti C., Cuna T., et al. (2013). Epidemiology of invasive fungal infections in the intensive care unit results of a multicenter Italian survey (AURORA Project). Infection 41, 645–653. 10.1007/s15010-013-0432-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moralez A. T., França E. J., Furlaneto-Maia L., Quesada R. M., Furlaneto M. C. (2014). Phenotypic switching in Candida tropicalis: association with modification of putative virulence attributes and antifungal drug sensitivity. Med. Mycol. 52, 106–114. 10.3109/13693786.2013.825822 [DOI] [PubMed] [Google Scholar]
- Moreno M. A., Ibrahim-Granet O., Vicentefranqueira R., Amich J., Ave P., Leal F., et al. (2007). The regulation of zinc homeostasis by the ZafA transcriptional activator is essential for Aspergillus fumigatus virulence. Mol. Microbiol. 64, 1182–1197. 10.1111/j.1365-2958.2007.05726.x [DOI] [PubMed] [Google Scholar]
- Morschhäuser J., Barker K. S., Liu T. T., BlaB-Warmuth J., Homayouni R., Rogers P. D. (2007). The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 3:e164. 10.1371/journal.ppat.0030164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moye-Rowley W. S. (2015). Multiple mechanisms contribute to the development of clinically significant azole resistance in Aspergillus fumigatus. Front. Microbiol. 6:70. 10.3389/fmicb.2015.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulhern S. M., Logue M. E., Butler G. (2006). Candida albicans transcription factor Ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryotic Cell 5, 2001–2013. 10.1128/EC.00155-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S. (2004). Redox and antioxidant systems of the malaria parasite Plasmodium falciparum. Mol. Microbiol. 53, 1291–1305. 10.1111/j.1365-2958.2004.04257.x [DOI] [PubMed] [Google Scholar]
- Murad A. M., d'Enfert C., Gaillardin C., Tournu H., Tekaia F., Talibi D., et al. (2001a). Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42, 981–993. 10.1046/j.1365-2958.2001.02713.x [DOI] [PubMed] [Google Scholar]
- Murad A. M., Leng P., Straffon M., Wishart J., Macaskill S., MacCallum D., et al. (2001b). NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20, 4742–4752. 10.1093/emboj/20.17.4742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagi M., Nakayama H., Tanabe K., Bard M., Aoyama T., Okano M., et al. (2011). Transcription factors CgUPC2A and CgUPC2B regulate ergosterol biosynthetic genes in Candida glabrata. Genes Cells 16, 80–89. 10.1111/j.1365-2443.2010.01470.x [DOI] [PubMed] [Google Scholar]
- Netea M. G., Ferwerda G., van der Graaf C. A., Van der Meer J. W., Kullberg B. J. (2006). Recognition of fungal pathogens by toll-like receptors. Curr. Pharm. Des. 12, 4195–4201. 10.2174/138161206778743538 [DOI] [PubMed] [Google Scholar]
- Nicholls S., Straffon M., Enjalbert B., Nantel A., Macaskill S., Whiteway M., et al. (2004). Msn2- and Msn4-like transcription factors play no obvious roles in the stress responses of the fungal pathogen Candida albicans. Eukaryotic Cell 3, 1111–1123. 10.1128/EC.3.5.1111-1123.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols C. B., Perfect Z. H., Alspaugh J. A. (2007). A Ras1-Cdc24 signal transduction pathway mediates thermotolerance in the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 63, 1118–1130. 10.1111/j.1365-2958.2006.05566.x [DOI] [PubMed] [Google Scholar]
- Nobile C. J., Andes D. R., Nett J. E., Smith F. J., Yue F., Phan Q. T., et al. (2006). Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2:e63. 10.1371/journal.ppat.0020063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C. J., Mitchell A. P. (2005). Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 15, 1150–1155. 10.1016/j.cub.2005.05.047 [DOI] [PubMed] [Google Scholar]
- Nobile C. J., Nett J. E., Hernday A. D., Homann O. R., Deneault J. S., Nantel A., et al. (2009). Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 7:e1000133. 10.1371/journal.pbio.1000133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble J. A., Tsai H. F., Suffis S. D., Su Q., Myers T. G., Bennett J. E. (2013). STB5 is a negative regulator of azole resistance in Candida glabrata. Antimicrob. Agents Chemother. 57, 959–967. 10.1128/AAC.01278-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Meara T. R., Alspaugh J. A. (2012). The Cryptococcus neoformans capsule: a sword and a shield. Clin. Microbiol. Rev. 25, 387–408. 10.1128/CMR.00001-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Meara T. R., Norton D., Price M. S., Hay C., Clements M. F., Nichols C. B., et al. (2010). Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog. 6:e1000776. 10.1371/journal.ppat.1000776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orciuolo E., Stanzani M., Canestraro M., Galimberti S., Carulli G., Lewis R., et al. (2007). Effects of Aspergillus fumigatus gliotoxin and methylprednisolone on human neutrophils: implications for the pathogenesis of invasive aspergillosis. J. Leukoc. Biol. 82, 839–848. 10.1189/jlb.0207090 [DOI] [PubMed] [Google Scholar]
- Palige K., Linde J., Martin R., Böttcher B., Citiulo F., Sullivan D. J., et al. (2013). Global transcriptome sequencing identifies chlamydospore specific markers in Candida albicans and Candida dubliniensis. PLoS ONE 8:e61940. 10.1371/journal.pone.0061940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panepinto J. C., Williamson P. R. (2006). Intersection of fungal fitness and virulence in Cryptococcus neoformans. FEMS Yeast Res. 6, 489–498. 10.1111/j.1567-1364.2006.00078.x [DOI] [PubMed] [Google Scholar]
- Papon N., Courdavault V., Clastre M., Bennett R. J. (2013). Emerging and emerged pathogenic Candida species beyond the Candida albicans paradigm. PLoS Pathog. 9:e1003550. 10.1371/journal.ppat.1003550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris S., Boisvieux-Ulrich E., Crestani B., Houcine O., Taramelli D., Lombardi L., et al. (1997). Internalization of Aspergillus fumigatus conidia by epithelial and endothelial cells. Infect. Immun. 65, 1510–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Myers C. L., Sheppard D. C., Phan Q. T., Sanchez A. A., Edwards J. E., et al. (2005). Role of the fungal Ras-protein kinase A pathway in governing epithelial cell interactions during oropharyngeal candidiasis. Cell. Microbiol. 7, 499–510. 10.1111/j.1462-5822.2004.00476.x [DOI] [PubMed] [Google Scholar]
- Park H. S., Bayram O., Braus G. H., Kim S. C., Yu J. H. (2012). Characterization of the velvet regulators in Aspergillus fumigatus. Mol. Microbiol. 86, 937–953. 10.1111/mmi.12032 [DOI] [PubMed] [Google Scholar]
- Paul S., Doering T. L., Moye-Rowley W. S. (2015). Cryptococcus neoformans Yap1 is required for normal fluconazole and oxidative stress resistance. Fungal Genet. Biol. 74, 1–9. 10.1016/j.fgb.2014.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Schmidt J. A., Moye-Rowley W. S. (2011). Regulation of the CgPdr1 transcription factor from the pathogen Candida glabrata. Eukaryotic Cell 10, 187–197. 10.1128/EC.00277-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-delos Santos F. J., Riego-Ruiz L. (2016). Gln3 is a main regulator of nitrogen assimilation in Candida glabrata. Microbiology 162, 1490–1499. 10.1099/mic.0.000312 [DOI] [PubMed] [Google Scholar]
- Perlroth J., Choi B., Spellberg B. (2007). Nosocomial fungal infections epidemiology, diagnosis, and treatment. Med. Mycol. 45, 321–346. 10.1080/13693780701218689 [DOI] [PubMed] [Google Scholar]
- Pfaller M. A., Diekema D. J. (2007). Epidemiology of invasive candidiasis a persistent public health problem. Clin. Microbiol. Rev. 20, 133–163. 10.1128/CMR.00029-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pócsi I., Prade R. A., Penninckx M. J. (2004). Glutathione, altruistic metabolite in fungi. Adv. Microb. Physiol. 49, 1–76. 10.1016/S0065-2911(04)49001-8 [DOI] [PubMed] [Google Scholar]
- Pongpom M., Liu H., Xu W., Snarr B. D., Sheppard D. C., Mitchell A. P., et al. (2015). Divergent targets of Aspergillus fumigatus AcuK and AcuM transcription factors during growth in vitro versus invasive disease. Infect. Immun. 83, 923–933. 10.1128/IAI.02685-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Zavala B., Reuss O., Park Y. N., Ohlsen K., Morschhäuser J. (2008). Environmental induction of white-opaque switching in Candida albicans. PLoS Pathog. 4:e1000089. 10.1371/journal.ppat.1000089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon A. M., Porta A., Fonzi W. A. (1999). Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J. Bacteriol. 181, 7524–7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsdale M., Selway L., Stead D., Walker J., Yin Z., Nicholls S. M., et al. (2008). MNL1 regulates weak acid-induced stress responses of the fungal pathogen Candida albicans. Mol. Biol. Cell 19, 4393–4403. 10.1091/mbc.E07-09-0946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray T. L., Payne C. D. (1988). Scanning electron microscopy of epidermal adherence and cavitation in murine candidiasis: a role for Candida acid proteinase. Infect. Immun. 56, 1942–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera M., Mogensen E., d'Enfert C., Janbon G. (2012). New regulators of biofilm development in Candida glabrata. Res. Microbiol. 163, 297–307. 10.1016/j.resmic.2012.02.005 [DOI] [PubMed] [Google Scholar]
- Rikkerink E. H., Magee B. B., Magee P. T. (1988). Opaque-white phenotype transition: a programmed morphological transition in Candida albicans. J. Bacteriol. 170, 895–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roetzer A., Klopf E., Gratz N., Marcet-Houben M., Hiller E., Rupp S., et al. (2011). Regulation of Candida glabrata oxidative stress resistance is adapted to host environment. FEBS Lett. 585, 319–327. 10.1016/j.febslet.2010.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudkin F. M., Bain J. M., Walls C., Lewis L. E., Gow N. A., Erwig L. P. (2013). Altered dynamics of Candida albicans phagocytosis by macrophages and PMNs when both phagocyte subsets are present. MBio. 4:e00810–13. 10.1128/mBio.00810-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo T., Miyazaki T., Izumikawa K., Mihara T., Takazono T., Kosai K., et al. (2010). Skn7p is involved in oxidative stress response and virulence of Candida glabrata. Mycopathologia 169, 81–90. 10.1007/s11046-009-9233-5 [DOI] [PubMed] [Google Scholar]
- Schaller M., Bein M., Korting H. C., Baur S., Hamm G., Monod M., et al. (2003). The secreted aspartyl proteinases Sap1 and Sap2 cause tissue damage in an in vitro model of vaginal candidiasis based on reconstituted human vaginal epithelium. Infect. Immun. 71, 3227–3234. 10.1128/IAI.71.6.3227-3234.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller M., Korting H. C., Schäfer W., Bastert J., Chen W., Hube B. (1999). Secreted aspartic proteinase (Sap) activity contributes to tissue damage in a model of human oral candidosis. Mol. Microbiol. 34, 169–180. 10.1046/j.1365-2958.1999.01590.x [DOI] [PubMed] [Google Scholar]
- Scharf D. H., Heinekamp T., Remme N., Hortschansky P., Brakhage A. A., Hertweck C. (2012). Biosynthesis and function of gliotoxin in Aspergillus fumigatus. Appl. Microbiol. Biotechnol. 93, 467–472. 10.1007/s00253-011-3689-1 [DOI] [PubMed] [Google Scholar]
- Scherwitz C. (1982). Ultrastructure of human cutaneous candidosis. J. Invest. Dermatol. 78, 200–205. 10.1111/1523-1747.ep12506451 [DOI] [PubMed] [Google Scholar]
- Schillig R., Morschhäuser J. (2013). Analysis of a fungus-specific transcription factor family, the Candida albicans zinc cluster proteins, by artificial activation. Mol. Microbiol. 89, 1003–1017. 10.1111/mmi.12327 [DOI] [PubMed] [Google Scholar]
- Schmalzle S. A., Buchwald U. K., Gilliam B. L., Riedel D. J. (2016). Cryptococcus neoformans infection in malignancy. Mycoses. 59, 542–552. 10.1111/myc.12496 [DOI] [PubMed] [Google Scholar]
- Schrettl M., Beckmann N., Varga J., Heinekamp T., Jacobsen I. D., Jöchl C., et al. (2010). HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 6:e1001124. 10.1371/journal.ppat.1001124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M., Haas H. (2011). Iron homeostasis–Achilles' heel of Aspergillus fumigatus? Curr. Opin. Microbiol. 14, 400–405. 10.1016/j.mib.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M., Kim H. S., Eisendle M., Kragl C., Nierman W. C., Heinekamp T., et al. (2008). SreA-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 70, 27–43. 10.1111/j.1365-2958.2008.06376.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert S., Barker K. S., Znaidi S., Schneider S., Dierolf F., Dunkel N., et al. (2011). Regulation of efflux pump expression and drug resistance by the transcription factors Mrr1, Upc2, and Cap1 in Candida albicans. Antimicrob. Agents Chemother. 55, 2212–2223. 10.1128/AAC.01343-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller C., Mamnun Y. M., Mollapour M., Krapf G., Schuster M., Bauer B. E., et al. (2004). Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 706–720. 10.1091/mbc.E03-05-0322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer A., Rupp S., Taylor B. N., Röllinghoff M., Schröppel K. (2000). The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol. Microbiol. 38, 435–445. 10.1046/j.1365-2958.2000.02132.x [DOI] [PubMed] [Google Scholar]
- Selmecki A., Gerami-Nejad M., Paulson C., Forche A., Berman J. (2008). An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol. Microbiol. 68, 624–641. 10.1111/j.1365-2958.2008.06176.x [DOI] [PubMed] [Google Scholar]
- Sheppard D. C., Doedt T., Chiang L. Y., Kim H. S., Chen D., Nierman W. C., et al. (2005). The Aspergillus fumigatus StuA protein governs the up-regulation of a discrete transcriptional program during the acquisition of developmental competence. Mol. Biol. Cell 16, 5866–5879. 10.1091/mbc.E05-07-0617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. P., Miranda I. M., Guida A., Synnott J., Rocha R., Silva R., et al. (2011). Transcriptional profiling of azole-resistant Candida parapsilosis strains. Antimicrob. Agents Chemother. 55, 3546–3556. 10.1128/AAC.01127-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver P. M., Oliver B. G., White T. C. (2004). Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryotic Cell 3, 1391–1397. 10.1128/EC.3.6.1391-1397.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims C. R., Ostrosky-Zeichner L., Rex J. H. (2005). Invasive candidiasis in immunocompromised hospitalized patients. Arch. Med. Res. 36, 660–671. 10.1016/j.arcmed.2005.05.015 [DOI] [PubMed] [Google Scholar]
- Singh R. P., Prasad H. K., Sinha I., Agarwal N., Natarajan K. (2011). Cap2-HAP complex is a critical transcriptional regulator that has dual but contrasting roles in regulation of iron homeostasis in Candida albicans. J. Biol. Chem. 286, 25154–25170. 10.1074/jbc.M111.233569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutsky B., Staebell M., Anderson J., Risen L., Pfaller M., Soll D. R. (1987). “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169, 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. D., Calvo A. M. (2014). The mtfA transcription factor gene controls morphogenesis, gliotoxin production, and virulence in the opportunistic human pathogen Aspergillus fumigatus. Eukaryotic Cell 13, 766–775. 10.1128/EC.00075-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneath P. H. A., Sokal R. R. (1973). Numerical Taxonomy. San Francisco, CA: Freeman. [Google Scholar]
- Sohn K., Urban C., Brunner H., Rupp S. (2003). EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol. Microbiol. 47, 89–102. 10.1046/j.1365-2958.2003.03300.x [DOI] [PubMed] [Google Scholar]
- Soll D. R. (1992). High-frequency switching in Candida albicans. Clin. Microbiol. Rev. 5, 183–203. 10.1128/CMR.5.2.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantha T., Soll D. R. (1993). A white-specific gene in the white-opaque switching system of Candida albicans. Gene 131, 53–60. 10.1016/0378-1119(93)90668-S [DOI] [PubMed] [Google Scholar]
- Srivastava V. K., Suneetha K. J., Kaur R. (2015). The mitogen-activated protein kinase CgHog1 is required for iron homeostasis, adherence and virulence in Candida glabrata. FEBS J. 282, 2142–2166. 10.1111/febs.13264 [DOI] [PubMed] [Google Scholar]
- Staab J. F., Bradway S. D., Fidel P. L., Sundstrom P. (1999). Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283, 1535–1538. 10.1126/science.283.5407.1535 [DOI] [PubMed] [Google Scholar]
- Staab J. F., Ferrer C. A., Sundstrom P. (1996). Developmental expression of a tandemly repeated, proline-and glutamine-rich amino acid motif on hyphal surfaces on Candida albicans. J. Biol. Chem. 271, 6298–6305. 10.1074/jbc.271.11.6298 [DOI] [PubMed] [Google Scholar]
- Stanzani M., Orciuolo E., Lewis R., Kontoyiannis D. P., Martins S. L., St. John L. S., et al. (2005). Aspergillus fumigatus suppresses the human cellular immune response via gliotoxin-mediated apoptosis of monocytes. Blood 105, 2258–2265. 10.1182/blood-2004-09-3421 [DOI] [PubMed] [Google Scholar]
- Stead D. A., Walker J., Holcombe L., Gibbs S. R., Yin Z., Selway L., et al. (2010). Impact of the transcriptional regulator, Ace2, on the Candida glabrata secretome. Proteomics 10, 212–223. 10.1002/pmic.200800706 [DOI] [PubMed] [Google Scholar]
- Stichternoth C., Ernst J. F. (2009). Hypoxic adaptation by Efg1 regulates biofilm formation by Candida albicans. Appl. Environ. Microbiol. 75, 3663–3672. 10.1128/AEM.00098-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synnott J. M., Guida A., Mulhern-Haughey S., Higgins D. G., Butler G. (2010). Regulation of the hypoxic response in Candida albicans. Eukaryotic Cell 9, 1734–1746. 10.1128/EC.00159-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L., Yu J. H. (2011). AbaA and WetA govern distinct stages of Aspergillus fumigatus development. Microbiology 157(Pt 2), 313–326. 10.1099/mic.0.044271-0 [DOI] [PubMed] [Google Scholar]
- Thau N., Monod M., Crestani B., Rolland C., Tronchin G., Latgé J. P., et al. (1994). rodletless mutants of Aspergillus fumigatus. Infect. Immun. 62, 4380–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi G., Wiltshire C., Macaskill S., Tournu H., Budge S., Brown A. J. (2002). Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 21, 5448–5456. 10.1093/emboj/cdf507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H. F., Krol A. A., Sarti K. E., Bennett J. E. (2006). Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob. Agents Chemother. 50, 1384–1392. 10.1128/AAC.50.4.1384-1392.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann B. D., Myers H., Chiranand W., Lazzell A. L., Zhao Q., Vega L. A., et al. (2004). Inducible defense mechanism against nitric oxide in Candida albicans. Eukaryotic Cell 3, 715–723. 10.1128/EC.3.3.715-723.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeputte P., Pradervand S., Ischer F., Coste A. T., Ferrari S., Harshman K., et al. (2012). Identification and functional characterization of Rca1, a transcription factor involved in both antifungal susceptibility and host response in Candida albicans. Eukaryotic Cell 11, 916–931. 10.1128/EC.00134-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermitsky J. P., Earhart K. D., Smith W. L., Homayouni R., Edlind T. D., Rogers P. D. (2006). Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Mol. Microbiol. 61, 704–722. 10.1111/j.1365-2958.2006.05235.x [DOI] [PubMed] [Google Scholar]
- Vylkova S., Carman A. J., Danhof H. A., Collette J. R., Zhou H., Lorenz M. C. (2011). The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. MBio. 2:e00055–11. 10.1128/mBio.00055-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhai B., Lin X. (2012). The link between morphotype transition and virulence in Cryptococcus neoformans. PLoS Pathog. 8:e1002765. 10.1371/journal.ppat.1002765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cao Y. Y., Jia X. M., Cao Y. B., Gao P. H., Fu X. P., et al. (2006). Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free Radic. Biol. Med. 40, 1201–1209. 10.1016/j.freeradbiomed.2005.11.019 [DOI] [PubMed] [Google Scholar]
- Wang Y., Casadevall A. (1994). Susceptibility of melanized and nonmelanized Cryptococcus neoformans to nitrogen- and oxygen-derived oxidants. Infect. Immun. 62, 3004–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylnka J. A., Moore M. M. (2003). Aspergillus fumigatus conidia survive and germinate in acidic organelles of A549 epithelial cells. J. Cell Sci. 116(Pt 8), 1579–1587. 10.1242/jcs.00329 [DOI] [PubMed] [Google Scholar]
- Waugh M. S., Vallim M. A., Heitman J., Alspaugh J. A. (2003). Ras1 controls pheromone expression and response during mating in Cryptococcus neoformans. Fungal Genet. Biol. 38, 110–121. 10.1016/S1087-1845(02)00518-2 [DOI] [PubMed] [Google Scholar]
- Willger S. D., Puttikamonkul S., Kim K. H., Burritt J. B., Grahl N., Metzler L. J., et al. (2008). A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 4:e1000200. 10.1371/journal.ppat.1000200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Sak Hoi J., Lamarre C., Beau R., Meneau I., Berepiki A., Barre A., et al. (2011). A novel family of dehydrin-like proteins is involved in stress response in the human fungal pathogen Aspergillus fumigatus. Mol. Biol. Cell 22, 1896–1906. 10.1091/mbc.E10-11-0914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. H. (2010). Regulation of development in Aspergillus nidulans and Aspergillus fumigatus. Mycobiology 38, 229–237. 10.4489/MYCO.2010.38.4.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O., Fries B. C., Casadevall A. (2003). Induction of capsule growth in Cryptococcus neoformans by mammalian serum and CO2. Infect. Immun. 71, 6155–6164. 10.1128/IAI.71.11.6155-6164.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Tao L., Guan G., Yue H., Liang W., Cao C., et al. (2016). Regulation of filamentation in the human fungal pathogen Candida tropicalis. Mol. Microbiol. 99, 528–545. 10.1111/mmi.13247 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Liu R., Noordhoek J. A., Kauffman H. F. (2005). Interaction of airway epithelial cells (A549) with spores and mycelium of Aspergillus fumigatus. J. Infect. 51, 375–382. 10.1016/j.jinf.2004.12.012 [DOI] [PubMed] [Google Scholar]
- Znaidi S., De Deken X., Weber S., Rigby T., Nantel A., Raymond M. (2007). The zinc cluster transcription factor Tac1p regulates PDR16 expression in Candida albicans. Mol. Microbiol. 66, 440–452. 10.1111/j.1365-2958.2007.05931.x [DOI] [PubMed] [Google Scholar]
- Zordan R. E., Galgoczy D. J., Johnson A. D. (2006). Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc. Natl. Acad. Sci. U.S.A. 103, 12807–12812. 10.1073/pnas.0605138103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan R. E., Miller M. G., Galgoczy D. J., Tuch B. B., Johnson A. D. (2007). Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 5:e256. 10.1371/journal.pbio.0050256 [DOI] [PMC free article] [PubMed] [Google Scholar]