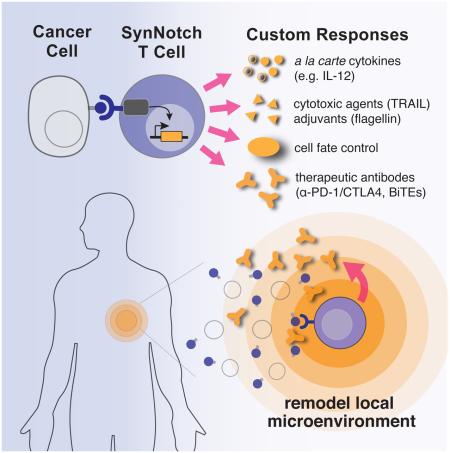
Abstract
Redirecting T cells to attack cancer using engineered chimeric receptors provides powerful new therapeutic capabilities. But the effectiveness of therapeutic T cells is constrained by the endogenous T cell response: certain facets of natural response programs can be toxic, whereas other responses, such as the ability to overcome tumor immunosuppression, are absent. Thus, the efficacy and safety of therapeutic cells could be improved if we could custom sculpt immune cell responses. Synthetic Notch receptors induce transcriptional activation in response to recognition of user-specified antigens. We show that synNotch receptors can be used to sculpt custom response programs in primary T cells: they can drive a la carte cytokine secretion profiles, biased T cell differentiation, and local delivery of non-native therapeutic payloads, such as antibodies, in response to antigen. SynNotch T cells can thus be used as a general platform to recognize and remodel local microenvironments associated with diverse diseases.
Graphical abstract
INTRODUCTION
Immune cells have evolved the remarkable ability to monitor and sense abnormalities in the body – including pathogenic insult or divergence from homeostasis, and in response they initiate protective and restorative programs (Chovatiya and Medzhitov, 2014). For example, T cells can traffic through the body, sense disease, and initiate a potent response to eliminate infections or cancer (Lämmermann and Sixt, 2008). T cells also have the capacity to generate long-lived memory to disease, in principle, preventing recurrence over the course of years or a lifetime (Kalos et al., 2011; Maus et al., 2014; Mueller et al., 2013). The ability to survey and respond to disease makes T cells an attractive platform for cell therapies.
Currently, major advances in engineering T cell therapeutics for treatment of cancer have focused on redirecting the native T cell response against disease cells. T cells can be engineered to recognize novel disease antigens using tumor-specific T cell receptors (TCRs) or chimeric antigen receptors (CARs) (Figure 1A) (Barrett et al., 2014a; Gill and June, 2015; June et al., 2009; Maus et al., 2014; Miller and Sadelain, 2015). While these redirected T cell therapies have been extremely successful for a limited set of cancers, their effectiveness remains constrained by the nature of endogenous T cell response programs. First, T cell response programs are multi-faceted, and not all facets are beneficial in a given disease context. Particular subresponses can contribute to toxic side effects (Barrett et al., 2014b; Brentjens et al., 2013; Grupp et al., 2013; Porter et al., 2011), which in some cases can be as dangerous as the disease itself (Davila et al., 2014; Lamers et al., 2006; Magee and Snook, 2014; Morgan et al., 2010; Zhao et al., 2009). Moreover, native T cell response programs lack certain ideal properties (Barrett et al., 2014b). For example, even when redirected to recognize tumors, T cells have limited capabilities to overcome the immunosuppressive microenvironment of the tumor (Gajewski et al., 2013).
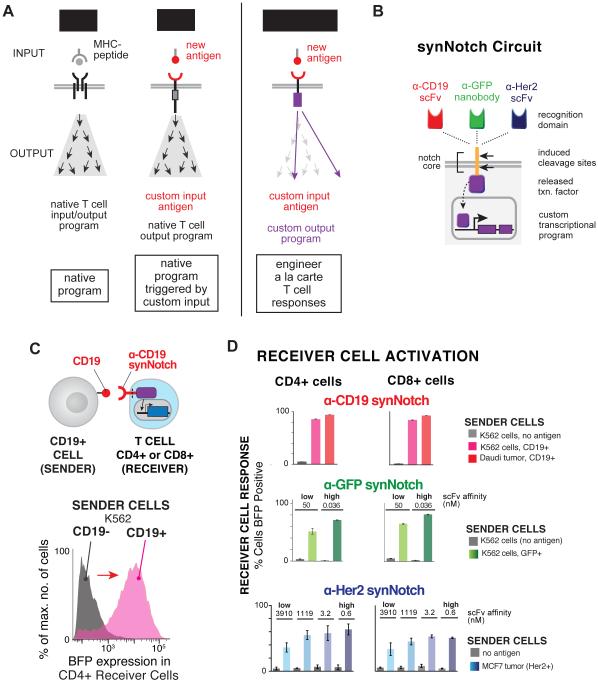
Figure 1. Engineering Antigen Triggered T cell Responses with Diverse SynNotch Receptors.
(A) TCRs and CARs activate kinase-based signaling cascades that drive the native T cell activation program providing little control over reshaping the T cell response. synNotch receptors recognize cell-surface antigens (e.g. disease related antigens) and directly regulate custom transcriptional programs with more precise control over the T cell response. Thus, in principle, synNotch receptors could be used to engineer a la carte responses.
(B) synNotch receptors have a custom ligand binding domain that detects a cell- surface antigen of interest (e.g. scFvs targeted to CD19 or Her2 or nanobodies to GFP), the core regulatory region of Notch that controls proteolysis, and a cytoplasmic orthogonal transcription factor (e.g. Gal4 VP64). The corresponding response elements for the orthogonal transcription factor controlling custom transcriptional programs are also engineered into the T cell.
(C) CD4+ AND CD8+ primary human T cells were engineered with the α-CD19 synNotch Gal4VP64 receptor and 5× Gal4 response elements controlling the expression of a BFP reporter. Histogram showing selective induction of the BFP reporter in α-CD19 synNotch receptor receiver CD4+ T cells in response to stimulation with sender cells with CD19- or CD19+ K562s.
(D) CD4+ AND CD8+ primary human T cells were engineered with either the α- CD19, α-GFP (LaG17 or LaG16_2) nanobody, or α-Her2 (scFv affinity variants) synNotch Gal4VP64 receptors and 5× Gal4 response elements controlling the expression of a BFP reporter. The percentages of synNotch T cells that upregulate the BFP reporter after 24 hours of stimulation with the indicated sender cells is given (n ≥ 3 for all conditions, error bars = SEM).
Thus, for future T cell therapies, it would be ideal to be able to sculpt new classes of behaviors that are induced by user-specified environmental cues (e.g. tumor antigens) but drive customized cellular programs – programs that are both edited to minimize toxicity, and expanded to incorporate new, non-native responses that increase therapeutic efficacy (Figure 1A). If cell-based therapies are to be used more broadly to treat diseases beyond terminal cancers such as autoimmunity, new precision cellular control mechanisms such as these must be developed to improve safety (Fischbach et al., 2013; Lim, 2010).
Recently developed synthetic Notch (synNotch) receptors provide a potential way to flexibly sculpt customized and inducible immune cell responses. SynNotch receptors contain the core regulatory domain from the cell-cell signaling receptor Notch, but have synthetic extracellular recognition domains (e.g. single chain antibodies) and synthetic intracellular transcriptional domains (Gordon et al., 2015; Morsut et al., 2016). When it engages the cognate antigen, the synNotch receptor undergoes induced transmembrane cleavage, akin to native Notch activation (Bray, 2006; Selkoe and Kopan, 2003), thereby releasing the intracellular transcriptional domain to enter the nucleus and activate expression of target genes regulated by the cognate upstream cis-activating promoter. Thus, synNotch circuits can be used to generate synthetic cell response programs in which a customized antigen recognition event can drive a customized gene expression program (Figure 1B).
Here we have built synNotch receptors capable of sensing disease-related and orthogonal inputs, and linked their activity to customized gene regulation in primary human T cells (Figure 1A). T cells engineered with synNotch receptors have robust and highly controlled custom behaviors. We show that synNotch receptors can drive T cells to produce custom a la carte cytokine profiles, undergo defined differentiation programs, and locally deliver natural and synthetic therapeutic payloads (e.g. cytotoxic proteins, antibodies, bispecific engagers, immune stimulators, immune supressors), all bypassing the requirement of canonical T cell activation. We also show that T cells encoding synNotch circuits are able to specifically target solid tumors in vivo to locally deliver their custom payloads.
The wide-range of synNotch applications shown here demonstrates their versatility as a platform to customize and refactor T cell function. The unprecedented programmability of synNotch cells to recognize and remodel microenvironments associated with diverse diseases could have applications in the design of therapeutic cells to treat a broad range of diseases including and extending beyond cancer and autoimmunity.
RESULTS
synNotch Receptors Can Drive Antigen-Induced Transcription in CD4+ and CD8+ Human Primary T Lymphocytes
The native Notch receptor has three critical components: 1) the ligand-binding epidermal growth factor (EGF) repeats, 2) the core regulatory region that controls cleavage of the receptor during activation, and 3) the Notch intracellular domain (NICD) that is released and regulates transcription (Gordon et al., 2007; Mumm et al., 2000; Selkoe and Kopan, 2003). To build a synNotch receptor platform that allows for fully customizable receptor sensing and transcriptional regulation, we utilized the Notch core regulatory region that controls ligand-dependent cleavage and activation as a minimal scaffold, but then append a customized input recognition (extracellular) and output transcriptional (intracellular) module (Figure 1B) (Morsut et al., 2016). The Notch core regulatory region includes the Lin12-Notch repeats (LNRs) that control the accessibility of the S2 cleavage site to metalloproteases, the heterodimerization domains (HD), and transmembrane domain (TMD) that contains the γ-secretase cleavage site required for release of the Notch intracellular domain (NICD) (Selkoe and Kopan, 2003). The extracellular EGF repeats, normally involved in recognition of the natural ligand Delta, were removed and replaced with a single-chain variable fragment (scFv) directed towards the cancer antigen CD19 or Her2 or nanobodies to orthogonal antigens, such as surface displayed GFP (Figure 1B and 1C). The NICD that is normally required for transcriptional regulation was replaced with the Gal4 DNA binding domain fused to the tetrameric viral transcriptional activator domain, VP64 (Lecourtois and Schweisguth, 1998; Struhl and Adachi, 1998). This general approach can be used to engineer synNotch receptors to any surface antigen of interest and link receptor activity to a customized cellular output controlled by orthogonal transcription factors and their associated response elements. A range of other extracellular and intracellular domains were also shown to function with synNotch (Morsut et al., 2016).
To show that synNotch receptors can function in relevant cell types for cell-based therapy, we engineered primary human CD4+ and CD8+ T cells with synNotch receptors directed towards the cancer-related antigens CD19 and Her2 or to an orthogonal antigen – surface displayed GFP. CD4+ and CD8+ T cells were engineered to express each synNotch receptor and the associated promoter (5× Gal4 response elements) controlling expression of a BFP reporter gene (Figure 1C and 1D). CD4+ and CD8+ T cells engineered with the α-CD19 synNotch receptor drove BFP reporter expression in 80 to 90% of the T cells within 24 hours of co-culture with cells expressing the cognate ligand CD19 -- either Daudi B cell lymphoblast tumors, which naturally express CD19, or K562 myelogenous leukemia cells with ectopically expressed CD19 (Figure 1A-D and Figure S1A-C). These T cells did not show BFP expression when unstimulated or treated with cells that did not express the cognate CD19 antigen. These data show that synNotch receptors can function in a controlled and antigen-dependent manner in primary T cells, and can detect natural levels of antigen on the surface of cancer cells. The synNotch receptors also have equivalent function in CD4+ and CD8+ T cells, which are often used in together for T cell immunotherapies (Figure 1D and 1C and Figure S1A-S2C).
For the cancer antigen Her2, an affinity panel of scFvs exists with which we made a series of receptors with different affinities for Her2 (Liu et al., 2015). This allowed us to determine if there are affinity requirements for synNotch receptors to function and whether affinity for ligand is parameter that could be tuned to shape the magnitude of the customized T cell response. The scFvs span nearly five orders of magnitude of affinity from 0.6 nM to 3.91 µM (Liu et al., 2015). CD4+ and CD8+ T cells were engineered with the four affinity variants of the α-Her2 synNotch receptor and the same BFP reporter as described above. All of the α-Her2 synNotch receptors were expressed in T cells and could drive expression of the BFP reporter in response to MCF7 breast cancer cells, a low Her2 expressing breast cancer cell line (Figure 1D and Figure S1D-S2E) (Liu et al., 2015). This suggests that even cancer antigens expressed at minimal levels can be recognized and efficiently activate synNotch receptors. The α-Her2 synNotch receptors in the nanomolar affinity range all drove reporter expression in 50-60% of T cells, but the lowest affinity receptor only activated 35% of the T cells Figure 1D and Figure S1E). These data show that changing the affinity of the synNotch receptors towards the target antigen is a viable approach to titrate and control synNotch-mediated cellular response programs.
We also tested whether T cells could be engineered to recognize orthogonal surface proteins by constructing synNotch receptors that recognized surface expressed GFP. We used two α-GFP nanobodies that have low (Kd = 50 nM) or high affinity (Kd = 0.036 nM) (Fridy et al., 2014). These receptors stimulated reporter expression upon exposure to K562 cells expressing surface GFP (but not with cells lacking the antigen). The resulting transcriptional response was similar to that observed for the α-CD19 synNotch, highlighting the modularity of the synNotch platform (Figure 1D and Figure S1F-S1H). The higher affinity α-GFP nanobody synNotch receptor drove a greater fraction of T cells to upregulate reporter expression upon exposure to K562 sender cells, when compared to the lower affinity receptor, yet both receptors activated gene expression in over half of the T cells (Figure 1D and Figure S1G). These results, much like the α-Her2 synNotch receptor affinity panel, suggest that varying the affinity of the synNotch ligand-binding domain can fine-tune the magnitude of the cellular response.
synNotch Receptors Can Drive Customized T Cell Cytokine Profiles
Immune cells and tissues throughout the body secrete soluble proteins known as cytokines to communicate and regulate cell behavior and shape the overall immune response. The specific cytokine profile is critical for eradicating pathogens and tumors (Dranoff, 2004; Iwasaki and Medzhitov, 2015). In many cases different sets of cytokines have the opposite effect of suppressing the immune response (Sakaguchi et al., 2008). Moreover, in certain scenarios where cytokines could boost immunity towards cancers or suppress damaging inflammation in an autoimmune setting, these cytokines are absent (Dranoff, 2004; O'Shea et al., 2002). Ideally we would like to precisely shape what cytokines therapeutic T cells secrete. However, when T cells are activated through CARs or the natural T cell receptor, there is little control over the cytokines that are produced and often cytokine profile depends on the disease and activation context as well as the receptor characteristics (Figure 2A and 2B) (Dotti et al., 2014; Gill and June, 2015). For many T cell therapies, it may be beneficial to bias towards the production of specific cytokines to tailor the immune response for the specific disease or therapeutic need.
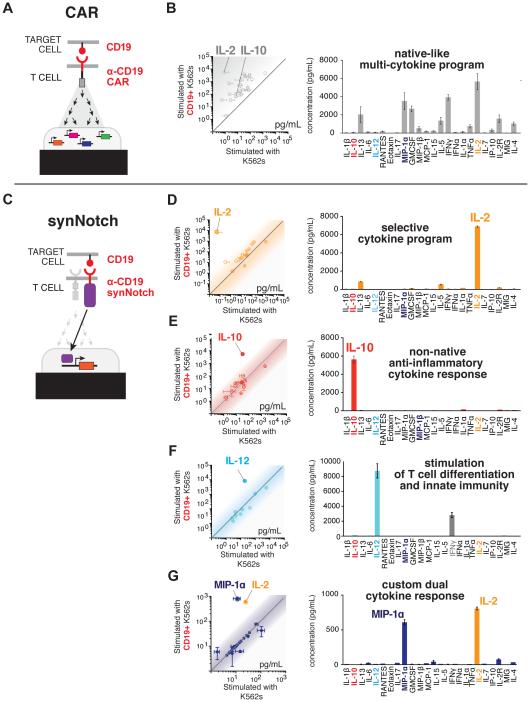
Figure 2. synNotch Receptors can Drive Antigen-Induced Custom Cytokine Programs.
(A) CAR activation drives CD4+ T cells to produce a diverse set of cytokines.
(B) A scatter plot showing the level (pg/mL) of 24 cytokines (see panel C for list of cytokines) produced by primary human α-CD19 CAR CD4+ T cells activated with target CD19+ K562 cells (y-axis) or negative control CD19- K562s (x-axis) after 24 hours of stimulation (n=3, error bars = SEM). The level of 24 cytokines produced by CD4+ α-CD19 CAR T cells stimulated by target CD19+ K562s (n=3, error bars = SEM).
(C) CD4+ T cells were engineered with the α-CD19 synNotch Gal4VP64 receptor and the corresponding response elements controlling the expression of either IL- 2, IL-10, IL-12, or combined IL-2/MIP-1α. The cells were co-cultured with target CD19+ K562s or CD19- non-target K562s.
(D) Scatterplots showing the production of synNotch regulated cytokines in response to CD19+ vs. CD19- K562 stimulation (n=3, error bars = SEM). The level of cytokines produced by α-CD19 synNotch T cells driving IL-2, IL-10, Flexi IL-12, or IL-2/MIP-1α production in response to CD19+ K562 cells is given. Only the synNotch regulated cytokines were produced above background levels. For synNotch CD4+ T cells driving IL-12 production, IFNγ was also produced as IL-12 can cause CD4+ T cells to differentiate into Th1 IFNγ-producing T cells (n=3, error bars = SEM).
With this in mind, we engineered CD4+ T cells with the α-CD19 synNotch receptor and the corresponding transcriptional response element controlling the expression of a single cytokine. Under these conditions, T cells selectively produced only a defined “a la carte” cytokine profile in response to the CD19 antigen (Figure 2C-G). synNotch receptors drove high-level production of the T cell stimulatory cytokine IL-2 with no basal secretion prior to antigen sensing (Figure S2A-S2D). The amount of IL-2 produced by synNotch activation is similar to what is produced in response to CAR or TCR stimulation (α-CD3/α-CD28 beads) of T cells (Figure S2D). Unlike normal stimulation of T cells through the TCR pathway, synNotch-driven cytokine production is independent of T cell activation, shown by the lack of upregulation of the canonical activation marker CD69 (Figure S2E).
We also engineered synNotch T cells to produce the immunosuppressive cytokine IL-10 (Figure 2E and Figure S2F-S2H), the immunostimulatory cytokine IL-12 (Figure 2F), and a combined cytokine and chemokine program (Figure 2G). These results highlight the ability of synNotch receptors to control a spectrum of therapeutically relevant cytokines that could be useful for diverse disease types. synNotch controlled production of IL-10 or other suppressive cytokines could be used to locally suppress inflammation in autoimmune or inflammatory disease (O'Garra et al., 2008). Locally concentrated IL-12 production, or that of other immune stimulatory factors, in tumors could drive potent innate immune responses to cancer while reducing the chances of highly toxic effects that have been observed with systemic administration of IL-12 (Lacy et al., 2009). Combined cytokine and chemokine programs such as the local production of IL-2 and MIP-1α could both enhance T cell survival and proliferation and recruit more T cells and innate immune cells such as macrophages to the tumor, respectively. The customizability and precision of synNotch circuits in T cells should allow for local control of immune system functions that rely on autocrine and paracrine cellular communication to effectively coordinate a response to disease.
synNotch Receptors Can Drive Antigen-Dependent Skewing of T cell Differentiation to the Anti-Tumor Th1 Fate
Another way to precisely shape the output of a therapeutic T cell is to control its differentiation. Beyond producing protein effectors like cytokines, T cells undergo specific differentiation programs important for mounting an effective subtype immune response. These subtype differentiation programs are normally determined by the mechanism of T cell activation, the cytokine milleu, and ultimately the regulation of master regulator transcription factors that initiate the specific T cell fate (O'Shea and Paul, 2010). T helper cell 1 (Th1) or T helper cell 2 (Th2) T cells are two canonical CD4+ T cell fate choices that are controlled by the master regulator transcription factors, Tbet and GATA3, respectively (Figure 3A) (O'Shea and Paul, 2010). Th1 cells are important for cellular immunity towards pathogens and cancer whereas Th2 cells are involved in stimulation of antibody production (Kara et al., 2014). In many diseases, the local environment skews the differentiation of T cells along the wrong path such that they are rendered ineffective. This is especially true in cancer where T cells can be pushed into a suppressive phenotype, hampering the immune response and leading to tumor expansion (Amarnath et al., 2011; Zou, 2006).
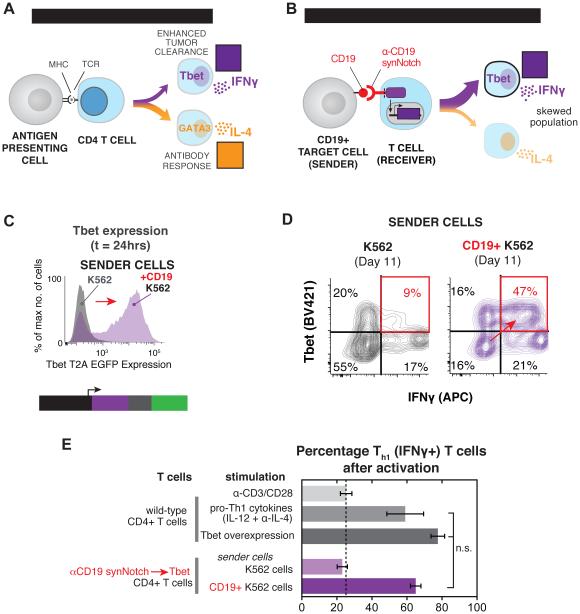
Figure 3. synNotch Receptors Can Drive Antigen-Dependent Skewing of T Cell Differentiation to the Anti-Tumor Th1 Fate.
(A) When CD4+ T cells are activated through engagement of pathogen-derived peptides presented by MHC molecules on antigen-presenting cells they differentiate into particular T cell subtypes depending on the infection. Th1 and Th2 are canonical CD4+ T cell fates that drive different immune responses. Th1 cells express the transcription factor Tbet, produce IFNγ, and aid in cellular immunity and tumor clearance. Th2 cells produce IL-4, an important cytokine for stimulation of antibody production by B cells.
(B) CD4+ α-CD19 synNotch T cells were engineered to regulate the expression Tbet and thus Th1 fate choice by T cells. The synNotch T cells were co-cultured with target CD19+ or non-target CD19- K562 cells for 11 days to determine if synNotch driven Tbet expression could skew CD4+ T cells to Th1 fate in a CD19- dependent manner.
(C) Histograms showing the selective expression of Tbet T2A EGFP after 24 hours of CD4+ α-CD19 synNotch T cells with CD19+ K562s (representative of at least 3 experiments).
(D) Two dimensional dot plots of intracellular stained CD4+ α-CD19 synNotch Gal4VP64 T cells for Tbet and IFNγ after 11 days of culture with either CD19+ or CD19- K562s. T cells were stimulated with PMA/Ionomycin for 4 hrs prior to staining to drive cytokine production (representative of at least 3 experiments).
(E) The percentage of IFNγ+ (Th1) T cells after 11 days of the indicated treatment (n ≥ 3 for all treatments, error bars = SEM, significance determined by Student’s t-test, n.s. p≥0.05).
Given the importance of T cell fate choice for cancer clearance, we wanted to determine if synNotch receptors could skew T cells to differentiate into IFNγ producing Th1 cells, important for anti-cancer immunity. IFNγ is critical for activation of innate immune cells that aid in tumor clearance, such as macrophages and dendritic cells, and direct exposure of cancer cells to IFNγ can enhance their susceptibility to the cytotoxic T cells. To skew T cell differentiation, we engineered CD4+ T cells with the α-CD19 synNotch receptor that controlled the expression of the Th1 transcription factor, Tbet (Figure 3B). Since ectopic expression of Tbet is known to be sufficient to drive the Th1 fate choice in CD4+ T cells, we reasoned that synNotch could provide local, antigen-dependent control over Th1 fate regulation by regulating the levels of Tbet in response to the tumor antigen CD19 (Sundrud et al., 2003).
To test this, the engineered primary CD4+ T cells were co-cultured with either CD19+ target K562 cells or CD19-control K562 cells for 11 days to induce differentiation. As comparative controls, a matched population of CD4+ T cells was either treated with a cocktail of Th1 differentiation agents (IL-12 and α-IL-4) or subject to Tbet constitutive overexpression. These two conditions allowed for the comparison of synNotch driven Th1 differentiation to previous gold standards in the field. The engineered synNotch CD4+ T cells induce the GFP reporter of Tbet expression specifically in response to CD19 within 24 hours of stimulation (Figure 3C and Figure S3A). After 11 days of long-term co-culture with CD19+ K562s, the T cells were stimulated with Phorbal myristate acetate (PMA) and ionomycin for intracellular cytokine staining to reveal whether the T cells had become Th1 cells. For the T cells that had been stimulated with CD19+ K562 cell for 11 days, >60% were found to be IFNγ+ Th1 cells (Figure 3D). This magnitude of skewed differentiation was equivalent to what was observed with treatment with the Th1 differentiation cocktail and constitutive Tbet overexpression (Figure 4E and Figure S3A-S3E). Thus, synNotch receptors can be used to skew T cells to the anti-tumor Th1 fate, and could in principle be used to skew T cells to many of the known T cell fates (e.g. Th2, Treg, Th17) as long as the expression of the defining master regulator transcription factor is sufficient for fate determination (O'Shea and Paul, 2010).
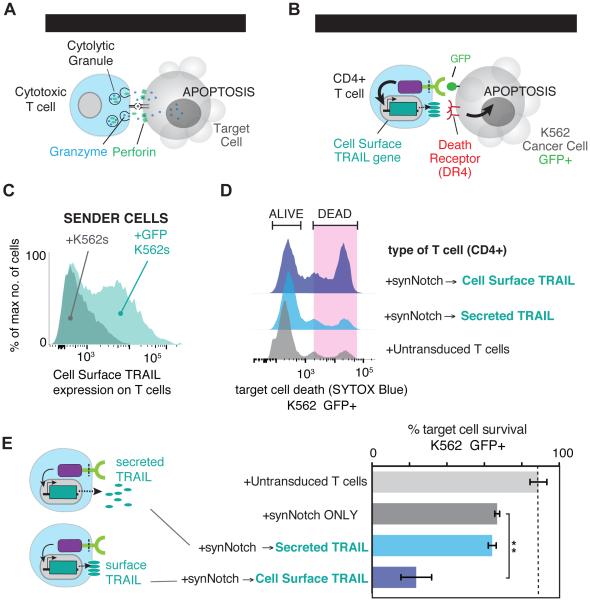
Figure 4. Customized Killer T Cells – synNotch Driven TRAIL Production.
(A) CD8+ cytotoxic T cells recognize infected cells via their TCR and directly kill the infected cell by creating pores in the cell with perforin allowing for the delivery of granzymes that initiate programmed cell death.
(B) CD4+ T cells were engineered with the α-GFP synNotch controlling the expression of the apoptotic regulator TRAIL in response to surface GFP.
(C) Histograms showing the selective expression of surface TRAIL after 24 hours of CD4+ α-GFP synNotch T cell incubation with surface GFP+ K562s.
(D) Histograms showing surface GFP+ K562 cell death via uptake of the dead stain SYTOX blue after 24hr. co-culture with the indicated T cell type (T cell:Target Cell Ratio = 1:1).
(E) Percentage target cell survival calculated from replicate data shown in panel D (n=4, error bars = SEM, significance determined by Student’s t-test ** p≤0.01)
SynNotch Driven T cell Delivery of Custom Therapeutics - TRAIL Production
Another important component of future T cell therapeutics is to engineer T cells with new capabilities that allow them to deliver customized therapeutic payloads, even ones that are non-native. Natural CD8+ T cells or CAR T cells directly recognize infected cells or cancer cells and kill them through the delivery of lytic granules (Figure 4A) (Stinchcombe and Griffiths, 2007). As a proof of principle experiment, we asked whether we could engineer CD4+ T cells – a T cell subset that is minimally cytotoxic – into a customized synthetic “killer T cell” by designing it to produce an apoptosis-inducing payload. We used the α-GFP nanobody synNotch receptor and response elements controlling production of TNF-related apoptosis-inducing ligand (TRAIL), an inducer of apoptosis and a cancer therapeutic (Figure 4B) (Johnstone et al., 2008; Lemke et al., 2014). T cells normally do not produce TRAIL upon TCR stimulation, therefore, if synthetically expressed in a controlled manner, this could aid in their cytotoxic activity (Figure S4A). Soluble forms of TRAIL are effective at killing the highly susceptible colon cancer cell line HCT116, but for other cancer lines like K562 cells, soluble TRAIL does not induce apoptosis even at high doses (Figure S5A-D) (Kim et al., 2000; Park et al., 2009). However, a recent study showed that if TRAIL is delivered in a membrane anchored form (e.g., a supported lipid bilayer or liposome), it is more effective at inducing apoptosis, even for resistant cancer cells such as K562 cells (Nair et al., 2015). Therefore, we engineered the CD4+ T cells to produce one of two TRAIL variants: 1) a secreted form of TRAIL fused to the GCN4 trimeric leucine zipper (LZ-TRAIL) known to be more potent than soluble monomeric TRAIL, or 2) a natural surface displayed TRAIL (Figure S4E-S4H) (Walczak et al., 1999).
synNotch T cells driving TRAIL production were co-cultured with TRAIL-resistant K562 cells to determine if T cells were an effective delivery platform that enhanced the apoptotic effects of TRAIL. synNotch T cells drove cell surface TRAIL expression (Figure 4C) and LZ-TRAIL secretion within 24 hours of co-culture (Figure S4F-H), but only cell surface TRAIL initiated K562 cell death, indicated by their uptake of the live/dead stain SYTOX blue. In contrast, synNotch T cells that secreted LZ-TRAIL were not effective at killing the resistant K562 cells, consistent with recent studies (Figure 4D-E). Overall, these findings suggest that synNotch T cells can be efficient and effective delivery agents for therapeutics such as TRAIL and potentially other biologics that are ineffective or toxic when systemically delivered.
SynNotch T cells can Drive Antigen-dependent Production of Antibodies and BiTEs
To go beyond sculpting natural T cell behavior with synNotch circuits, we also wanted to show the ability of synNotch T cells to drive non-natural therapeutic responses that could have a wide-range of applications for treatment of disease. Antibody therapies for cancer that help to elicit an anti-tumor immune response by releasing the breaks on T cells are having unprecedented curative effects in patients with cancers that were otherwise considered terminal (Sharma and Allison, 2015). These antibodies known as ‘checkpoint blockade’ inhibitors target inhibitory receptors on T cells such as programmed cell death-1 (PD-1) (Pembrolizumab) and cytotoxic-T-lymphocyte-associated protein 4 (CTLA-4) (Tremelimumab) or their ligands. While these antibodies are often effective, many patients do not show a response or have significant adverse effects from treatment (Postow et al., 2015). A potential way to increase the effectiveness of the therapeutic antibodies and reduce adverse events is to have T cells locally produce the antibodies in tumors. These antibodies could enhance the activity of therapeutic T cells and at the same time elicit a response from endogenous tumor-infiltrating T cells. Thus, we engineered primary human CD4+ T cells with the anti-GFP synNotch receptor controlling production of both Pembrolizumab and an scFv directed towards CTLA-4. The T cells reliably produced high levels of the antibodies and the secreted antibodies bound and coated antigen-expressing targets (Figure 5A). We also have engineered T cells to singly produce Pembrolizumab and Tremelimumab at high levels (Figure S5A-5C). Therefore, synNotch T cells are a viable platform for the controlled delivery of important clinical administered antibodies, both individually and in complementary combinations.
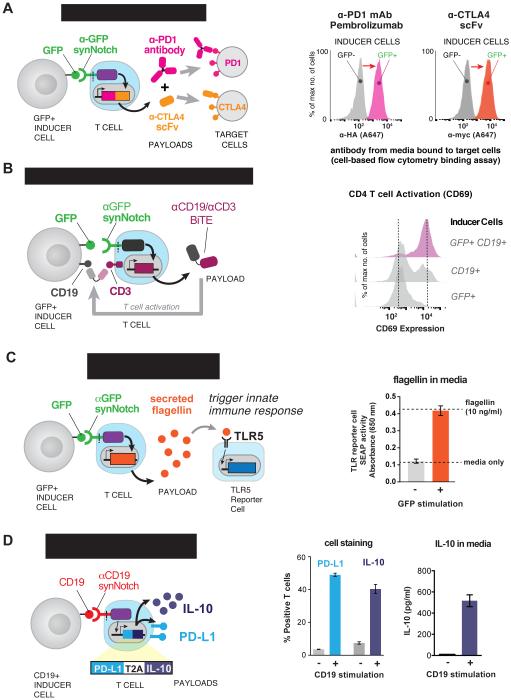
Figure 5. synNotch T Cell Delivery of Immunotherapeutics – Antibodies, Adjuvants, and Immunosuppressive Agents.
(A) CD4+ T cells were engineered with the α-GFP synNotch controlling the expression of Pembrolizumab (α-PD-1 HA) and a myc-tagged α-CTLA4 scFv (both antibodies were expressed as a single transcript with a T2A sequence between). After 24 hours of stimulation of the T cells with surface GFP+ or GFP- K562s, the supernatant was collected and used to stain PD-1+ or CTLA4+ K562s. Secreted antibody binding to target cells was monitored via flow cytometry after secondary staining with α-HA A647 (α-PD-1) or α-myc A647 (CTLA-4 scFV) antibodies (representative of 3 replicates).
(B) CD4+ T cells were engineered with the α-GFP synNotch receptor controlling the expression of Blinatumomab, an α-CD19/CD3 bi-specific antibody that retargets T cells to CD19+ tumors. Histograms showing CD69 (activation marker) expression on the synNotch T cells after co-culture with surface GFP+ only, CD19+ only, or surface GFP+CD19+ K562s. The T cells strongly activate in the presence of the surface GFP+CD19 K562s and a small percentage of the T cells activate when incubated with CD19+ only K562s due to low levels of basal leakage of Blinatumomab expression (representative of 3 independent experiments).
(C) CD4+ T cells were engineered with the α-GFP synNotch receptor controlling the expression of Flagellin. Supernatant was harvested from the T cells after co- culture with surface GFP+ or GFP- K562s and added to hTLR5 HEK-blue secreted alkaline phosphatase (SEAP) reporter cells. After 24 hours SEAP activity was monitored and the level of Flagellin in the supernatant was measured a purified Flagellin standard (n=3, error bars = SD).
(D) CD4+ T cells were engineered with the α-CD19 synNotch receptor controlling the expression of PD-L1 and IL-10. Quantification of the percentage of synNotch T cells that express PD-L1 and intracellular IL-10 after co-culture with CD19+ or CD19- K562s for 24 hrs is given. The amount of IL-10 in the supernatant was also determined by ELISA (n=3, error bars = SEM).
This approach is also effective for the production of bi-specific antibodies (e.g. bispecific T cell engagers -- BiTEs) that simultaneously bind tumor antigens and the CD3 chain of the TCR, effectively redirecting T cell activity toward tumor cells (Figure 5B) (Nagorsen et al., 2012). synNotch T cells were engineered to produce the anti-CD19/CD3 BiTE, Blinatumomab, in response to anti-GFP synNotch stimulation. When stimulated with GFP+CD19+ K562 cells, the resulting Blinatumomab production led to canonical TCR activation of the T cells, assayed by upregulation of the activation marker CD69 (Figure 5B). These synNotch → BiTE T cells required the presence of combinatorial antigens to activate -- both the synNotch antigen (GFP) and the tumor antigen (CD19) targeted by the BiTE (Figure 5B). Therefore, this is yet another way to engineer T cells with AND logic where T cells must sense multiple antigens to activate (Kloss et al., 2013; Roybal et al., 2016). By confining BiTE production to the tumor, synNotch controlled T cells could potentially reduce the toxicity of bi-specific antibodies that are targeted to tumor antigens that are also expressed on bystander tissues.
SynNotch T cells can Drive Antigen-dependent Production of Adjuvants
Adjuvants are another class of therapeutics that being actively studied for cancer therapy. Microbial products have long been used as adjuvants and have a history of stimulating immune response to tumors (Temizoz et al., 2016). There are many microbial products that are genetically encodable, including the bacterial flagellum component, flagellin, a product of genes such as FliC from S. Typhi (Figure 5C). Flagellin can enhance an immune response to cancer by stimulating innate immune cells that express Toll-like receptors (TLR) such as TLR5 that recognize flagellin (Akira and Takeda, 2004; Leigh et al., 2014). These innate cells include professional antigen presenting cells (APCs) such as dendritic cells that though TLR5 simulation can drive immune responses that are otherwise suppressed in the tumor microenvironment. The anti-GFP synNotch receptor reliably controlled the production of Flagellin, and the amount of secreted Flagellin (~10 ng/mL) strongly stimulated a TLR5 expressing reporter cell line (Figure 5C). This again shows the ability of synNotch T cells to control the production of a therapeutic agent that could have substantial toxicity if given systemically, but which when produced locally could significantly enhance the immune response to solid tumors.
SynNotch T cells can Drive Antigen-dependent Production of Immunosuppressive Agents
synNotch T cells may also have important applications outside of cancer, where they can play an immunosuppressive role in an autoimmune setting. Therefore, we engineered synNotch T cells that drive the simultaneously production of paracrine inhibitory signal such as the cytokine, IL-10, and the T cell inhibitory ligand, PD-L1, which could drive cell-to-contact inhibition (Chen, 2004; Ouyang et al., 2011). The anti-CD19 synNotch receptor was able to drive both suppressive agents in response to stimulation with CD19+ K562s (Figure 5D and Figure S5D and S5E). All of these examples of synNotch receptor T cells controlling non-native synthetic T cell responses highlight the vast possibilities to use synNotch engineered cells to produce a spectrum of therapeutic agents that could enhance the effectiveness of the therapeutic cells or help to reeducate and modulate a tissue or disease environment to restore homeostasis and natural function.
In vivo Expression of Cytokine in a Solid Tumor via synNotch Receptor Engineered T cells
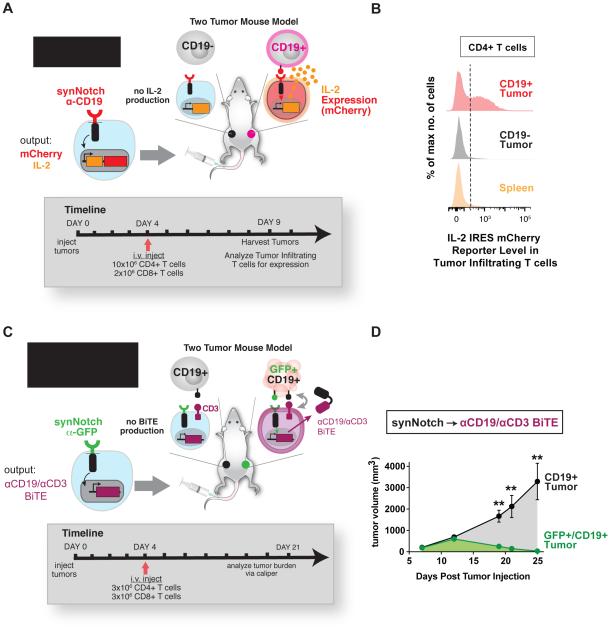
Since synNotch receptors could precisely regulate a spectrum of T cell responses in vitro, we wanted to determine whether the receptors could enable T cells to selectively produce a custom payload such as the cytokine IL-2 in a synNotch ligand-expressing solid tumor. For these experiments, we established a bilateral K562 xenograft solid tumor model in immunocompromised NOD scid IL-2Rγ−/− (NSG) mice where a non-target CD19- tumor and a target CD19+ tumor were implanted subcutaneously in the left flank and right flank, respectively (Figure 6A). The tumors were allowed to establish for four days and then we intravenously (i.v.) injected CD4+ and CD8+ T cells engineered with the α-CD19 synNotch receptor and response elements in control of IL-2 expression and an IRES mCherry reporter (Figure 6A).
Figure 6. In vivo Local Expression of Cytokines and Bi-specifc Tumor- targeted Antibodies in Solid Tumors Via synNotch T cells.
(A) NSG mice were subcutaneously injected with CD19- non-target K562s and target CD19+ in the left and right flank, respectively. α-CD19 synNotch T cells in control of IL-2 iRES mCherry expression were injected into the mice after tumors were established and tumors were harvested at the indicated time point to determine whether the synNotch T cells had infiltrated the tumor and expression of IL-2 and mCherry reporter was induced.
(B) Histograms of IL-2 IRES mCherry reporter levels in tumor and spleen infiltrated CD4+ synNotch T cells injected i.v. showing selective expression of the mCherry reporter in target CD19+ tumors (data representative of 3 replicate mice).
(C) NSG mice were subcutaneously injected with CD19- non-target K562s and target CD19+ in the left and right flank, respectively. α-GFP synNotch T cells in control of Blinatumomab (α-CD19/CD3 BiTE) expression were injected i.v. into the mice 4 days after tumor implantation. The tumors were measured by caliper over for 25 days.
(D) Bilateral CD19+ and GFP/CD19+ K562 tumor growth curves in mice treated with CD4+ and CD8+ T cells engineered with the α-GFP synNotch receptor controlling Blinatumomab (α-CD19/CD3 BiTE) expression. The dual antigen GFP/CD19+ tumor is selectively cleared (n=5 mice, error = SEM, significance determined by Student’s t-test ** p≤0.01).
After six days the tumors were harvested and the tumor infiltrating CD4+ and CD8+ T cells were analyzed for expression of the mCherry reporter (Figure 6A – 6B and Figure S6A-6C). Only the T cells localized in the target CD19 tumor expressed IL-2 IRES mCherry, and the reporter level was similar to what was observed for the same T cells when stimulated in vitro (Figure S6C and S6D). While the frequency of T cells was not high in the tumor (Figure S6E), the activity of the T cells was highly specific to the target tumor (Figure 6B and Figure S6B and S6C). In addition to i.v. injection of synNotch → IL-2 T cells, we also directly injected the T cells into non-target and target tumors. The tumors were then harvested and analyzed via flow cytometry two days after injection and also showed selective expression of the IL-2 reporter in the target tumor at similar levels to matched in vitro stimulated T cells (Figure S6D and S6F). While the ability of synNotch T cells to infiltrate these tumors could still be improved, these data clearly show that synNotch T cells can selectively induce production of a therapeutic agent in a target solid tumor. Thus, synNotch engineered T cells may prove effective for the delivery of a wide-range of genetically encodable therapeutics that could benefit from local delivery both to enhance effectiveness and reduce toxicity of systemic administration.
SynNotch T cell Production of Bi-specific Antibodies to Drive Solid Tumor Clearance
We then asked if synNotch T cells could drive clearance of a solid tumor in vivo, using synNotch induced local production of the anti-CD19/CD3 BiTE, Blinatumomab. To test these cells, a bilateral subcutaneous tumor model was established in NSG mice, similar to the model used for localized IL-2 secretion. However, in this model we wanted to determine if T cells could distinguish a single antigen bystander tumor expressing the BiTE antigen CD19 from a tumor expressing the synNotch antigen (GFP – priming antigen) and BiTE antigen (CD19 – killing antigen). To perform these experiments, we established a CD19+ single antigen tumor on the left flank and a GFP+CD19+ dual antigen tumor on the right flank. After the tumors had established, we i.v. injected primary human CD4+ and CD8+ T cells engineered with the anti-GFP synNotch circuit controlling the expression of Blinatumomab (Figure 6C and 6D). The synNotch T cells should only activate their cytotoxic program in the GFP+CD19+ tumor due to GFP induced synNotch activity and Blinatumomab production. Tumor growth was monitored by caliper, and we observed the selective clearance of the dual antigen tumor over the course of 25 days (Figure 6D). synNotch T cells are thus capable of producing biologically relevant levels of a therapeutic that can effectively drive tumor clearance. We observed minimal slowed growth of the single antigen tumor, indicating that BiTE expression was reliably and spatially gated by synNotch activity (Figure 6D and Figure S6G). Overall, synNotch receptors are viable controllers of therapeutic delivery and are able to provide a spatially defined responses to disease in vivo.
DISCUSSION
Hacking Immune Cell Function with synNotch Circuits: A Programmable Platform for the Generation of Smarter Cellular Therapeutics
Most immune therapy strategies today focus on modulating the immune system to enhance its ability to attack cancer. Even canonical CAR T cell therapy, one of the most radical forms of immune therapy, primarily involves redirecting the native T cell response towards cancer. Here we describe a fundamentally different approach that involves hacking the function of immune cells. We show how synNotch circuits allow one to construct radically new antigen-driven response programs in immune cells that can supplement and extend beyond the endogenous response capabilities of these cells.
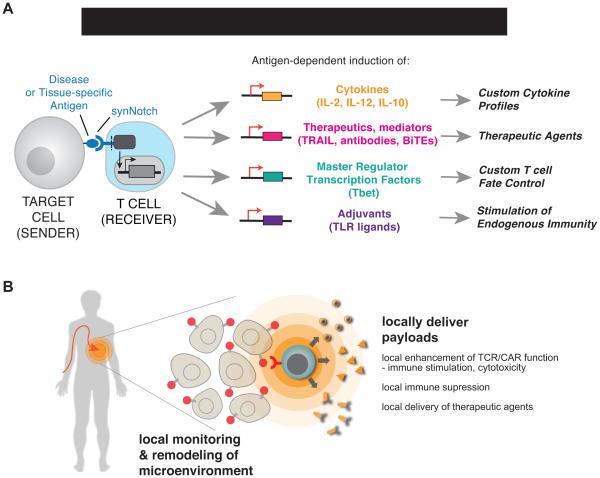
Building these novel response programs using synNotch represents a strategy of refactoring T cell responses. Refactoring is a term used in computer programming and synthetic biology, and describes rebuilding a complex program or system from the bottom up in a more modular and logical way that allows for a higher degree of control, predictability and extensibility (the ability to extend the system to perform more complex functions) (Smanski et al., 2014). In particular, it is important that synNotch circuits allow one to control T cell sensing and response in a manner that can be completely independent from canonical TCR or co-stimulatory receptors of the immune system. Thus, this approach removes constraints of the endogenous system, allowing programming of more diverse responses that, in principle, have both a higher degree of controllability as well as new capabilities. SynNotch circuits can be used to control diverse classes of responses, both natural and non-natural, as shown in Figure 7A.
Figure 7. SynNotch Circuits Allow Versatile Reprogramming of T cells to Monitor and Selectively Modulate Their Microenvironment.
(A) synNotch receptors can drive diverse behaviors in primary human T cells. We show that synNotch receptors can drive custom cytokine production profiles, effectively deliver non-native therapeutics, and control T cell differentiation, all in an antigen-dependent and T cell activation independent manner.
(B) synNotch are sufficient to target T cells in vivo to locally produce a therapeutic payload. Future T cell therapies could utilize synNotch receptors to target T cells to disease-related or tissue-specific antigens for local delivery of therapeutics that are ineffective or toxic as systemically administered agents in humans.
This approach of more deeply rewiring cell response programs is particularly powerful when combined with T cells and other immune cells, because T cells have the intrinsic ability to autonomously traffic through the body, to infiltrate tissues, and to survey their microenvironment for their cognate molecular signals. Since synNotch circuits are used to create synthetic programs to detect and respond to tissue based microenvironmental signals, this is an ideal pairing of a cell type and molecular sensor system, allowing the engineering of what amounts to living, programmable nanodevices that can carry out highly localized actions in the complex environment of the body (Figure 7B)
synNotch Receptors could be used to improve current T cell therapies
An immediate application of synNotch circuits could be to improve existing T cell therapies. synNotch receptors can act as an additional environmental sensing system for TCR and CAR T cell therapies and help to modulate the activity of the cells to improve their effectiveness and safety profile. We have demonstrated that synNotch receptors can be used to control expression of directly cytotoxic factors (CARs, BiTEs). But, in addition, an important use of synNotch receptors could be the control the local production of immune stimulatory factors, such as IL-12 or other innate immune adjuvants, in TIL or engineered T cell therapies. This type of local delivery and enhancement may be particularly important, given, that factors like IL-12 are potent at driving anti-tumor immunity, but are too toxic for systemic administration (Lacy et al., 2009). The inability of many of the current T cell therapies to infiltrate and eliminate solid tumors could be greatly improved by utilizing synNotch receptors to help the T cells to prime the local disease environment to make it more susceptible to both the cellular therapeutic and the endogenous immune response (Tang et al., 2016). We have also shown that synNotch circuits can be used to locally produce combinations of checkpoint blockade antibodies, showing that a host of different factors could be produced to both enhance immune response and to overcome the immunosupresive blockade raised by many tumors.
Beyond Cancer: synNotch T cells as remodelers of Disease Microenvironments
Because T cells can traffic throught he body and have access to many disease tissues, synNotch T cells could in principle by used to address diverse disease needs beyond cancer, especially localized diseases that require spatial sensing and localized microenvironment remodeling. The examples of customized T cell behavior described here could be used to help T cells control and disrupt diverse diseases. For example, synNotch engineered cells could also be deployed for the control of autoimmune or inflammatory diseases. In this case, however, the engineered cells would now be engineered to have a suppressive role that could promote immune tolerance. While T regulatory cells can in principle be used towards this goal, there may be distinct control and predictability advantages in using a refactored synNotch T cell to sense auto-antigens and deliver a customized and controlled set of immunosuppressive factors. synNotch engineered cells may have even broader applications as general delivery agents that can locally produce any genetically encoded bio-therapeutic that has systemic toxicity (Figure 7B).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Source of Primary Human T cells
Blood was obtained from Blood Centers of the Pacific (San Francisco, CA) as approved by the University Institutional Review Board. Primary CD4+ and CD8+T cells were isolated from anonymous donor blood after apheresis (described in METHOD DETAILS).
Animal Model Details
Animal studies were conducted with the UCSF Preclinical Therapeutics Core under a protocol approved by the UCSF Institutional Animal Care and Use Committee. NOD scid gamma (NSG) (female, 8~12 weeks old, Jackson Laboratory #005557) mice were used for all in vivo mouse experiments.
METHOD DETAILS
synNotch Receptor and Response Element Construct Design
synNotch receptors were built by fusing the CD19 scFv (Grupp et al., 2013), Her2 set of scFvs (Liu et al., 2015), LaG17 (lower affinity), or LaG16_2 (high affinity) GFP nanobody (Fridy et al., 2014) to the mouse Notch1 (NM_008714) minimal regulatory region (Ile1427 to Arg1752) and Gal4VP64. All synNotch receptors contain an n-terminal CD8α signal peptide (MALPVTALLLPLALLLHAARP) for membrane targeting and a myc-tag (EQKLISEEDL) for easy determination of surface expression with α-myc A647 (cell-signaling #2233). The receptors were cloned into a modified pHR’SIN:CSW vector containing a PGK promoter for all primary T cell experiments. The pHR’SIN:CSW vector was also modified to make the response element plasmids. Five copies of the Gal4 DNA binding domain target sequence (GGAGCACTGTCCTCCGAACG) were cloned 5’ to a minimal CMV promoter.
The human IL-2, IL-10, flexi IL-12 (Anderson et al., 1997), MIP-1α, Tbet, or TRAIL, and PD-L1_T2A_IL-10 codon optimized mRNA sequence was cloned into a MCS downstream of the Gal4 inducible promoter and 5’ of an IRES mCherry reporter. All constructs were cloned via In fusion cloning (Clontech #ST0345)).
Primary Human T cell Isolation and Culture
Primary CD4+ and CD8+ T cells were isolated from anonymous donor blood after apheresis by negative selection (STEMCELL Technologies #15062 & 15023). T cells were cryopreserved in RPMI-1640 (UCSF cell culture core) with 20% human AB serum (Valley Biomedical Inc., #HP1022) and 10% DMSO. After thawing, T cells were cultured in human T cell medium consisting of X-VIVO 15 (Lonza #04-418Q), 5% Human AB serum and 10 mM neutralized N-acetyl L- Cysteine (Sigma-Aldrich #A9165) supplemented with 30 units/mL IL-2 (NCI BRB Preclinical Repository) for all experiments.
Lentiviral Transduction of Human T cells
Pantropic VSV-G pseudotyped lentivirus was produced via transfection of Lenti-X 293T cells (Clontech #11131D) with a pHR’SIN:CSW transgene expression vector and the viral packaging plasmids pCMVdR8.91 and pMD2.G using Fugene HD (Promega #E2312). Primary T cells were thawed the same day, and after 24 hours in culture, were stimulated with Dynabeads Human T-Activator CD3/CD28 (Life Technologies #11131D) at a 1:3 cell:bead ratio. At 48 hours, viral supernatant was harvested and the primary T cells were exposed to the virus for 24 hours. At day 4 post T cell stimulation, Dynabeads were removed and the T cells expanded until day 9 when they were rested and could be used in assays. T cells were sorted for assays with a FACs ARIA II.
Cancer Cell Lines
The cancer cell lines used were K562 myelogenous leukemia cells (ATCC #CCL-243), Daudi B cell lymphoblasts (ATCC #CCL-213), MCF7 adenocarcinoma breast cancer cells (ATCC #HTB22), and HCT116 colon cancer cells (ATCC #CCL-247). K562s were lentivirally transduced to stably express human CD19 at equivalent levels as Daudi tumors. CD19 levels were determined by staining the cells with α-CD19 APC (Biolegend #302212). K562s were also transduced to stably express surface GFP (GFP fused to the PDGF transmembrane domain). All cell lines were sorted for expression of the transgenes.
In vitro Stimulation of synNotch T cells
For all in vitro synNotch T cell stimulations, 2×105 T cells were co-cultured with sender cells at a 1:1 ratio. After mixing the T cells and sender cells in round bottom 96-well tissue culture plates, the cells were centrifuged for 1 min at 400×g to force interaction of the cells and the cultures were analyzed at 24 hours for reporter expression or expression of custom gene induction via flow cytometry with a BD LSR II. All flow cytometry analysis was performed in FlowJo software (TreeStar).
Luminex MAGPIX Cytokine Quantification
Primary CD4+ T cells expressing the α-CD19 synNotch Gal4VP64 receptor and 5× Gal4 response elements controlling either human IL-2, IL-10, IL- 12, or IL-2/MIP-1α expression were stimulated as described above with K562 myelogenous leukemia cells (CD19- or CD19+). As references, CD4+ T cells expressing the α-CD19 4-1BBζ CAR were stimulated along with untransduced T cells stimulated with α-CD3/CD28 Dynabeads at a 1:3 ratio. The supernatant was collected at 24 hours and analyzed with a Luminex MAGPIX (Luminex Corp.) Human Cytokine Magentic 25-plex Panel (Invitrogen ref#LHC0009M) according to the manufacturer’s protocol. All cytokine levels were calculated based on standard curves with Her2 software (Luminex Corp.).
IL-2 Intracellular Cytokine Staining and CD69 Staining
synNotch T cells controlling IL-2 production were assayed to determine if they basally produced IL-2 by intracellular cytokine stain (ICS). The synNotch T cells and untransduced T cell controls were cultured for 6 hours in the presence of GolgiPlug (BD Biosciences #555029). The T cells were then stained with α-IL-2 FITC (BD #340448) with a BD Biosciences ICS kit (#555028). The levels of IL- 2 were analyzed via flow cytometry with a BD LSRII.
To assess whether synNotch receptors activated the T cells, the T cells were stained after stimulation for the activation marker CD69. CD69 expression was determined by staining the cells with α-CD69 APC (Biolegend #310910).
synNotch Driven T cell Differentiation
Primary human CD4+ T cells were stimulated with Dynabeads Human T- Activator CD3/CD28 as described above. To differentiate T cells into the Th1 subset during the activation, the cells were cultured as described above but with the addition of 2.5 ng/mL recombinant IL-12 (R&D Systems) and 12.5 µg/mL α- IL-4 clone MP4-25D2 (BD Pharmigen #554481). IL-12 and α-IL-4 were added at least twice weekly. In parallel, primary CD4 T cells were lentivirally transduced to express human Tbet T2A mCherry (TBX21, NCBI #EAW94804.1) and cultured normally in T cell medium supplemented with IL-2. CD4+ T cells expressing the α-CD19 synNotch Gal4VP64 receptor and 5× Gal4 response elements controlling Tbet T2A GFP expression were cultured in the presence of CD19- or CD19+ K562 sender cells 24 hours after viral transduction. The synNotch T cells were cultured in the presence of K562s in T cell medium supplemented with IL-2. All T cells were cultured for 11 to 14 days and then subject to intracellular cytokine staining (ICS) to determine the percentage of Th1 T cells.
For ICS, the T cells were first treated with 50 ng/mL Phorbal myristate acetate and 1 µg/mL ionomycin (both from Sigma) for 6 hours in the presence of GolgiPlug. The T cells were then stained with α-Tbet BV421 (Biolegend #644816) and α-IFNγ APC (Biolegend #502512). The levels of Tbet and IFNγ were analyzed via flow cytometry with a BD LSRII.
Sensitivity of Cancer Cell lines to Recombinant TRAIL and synNotch Driven TRAIL Production in Primary T cells
HCT116 colon cancer cells and K562s were treated with recombinant TRAIL (from 1 to 200 ng/mL, 1:2 dilution series) for 24 hours (RND Systems #375-TEC-010). The cells were then harvested and stained with the live/dead stain, SYTOX Blue (Thermo Scientific #S34857) and the fraction of dead cells was determined by flow cytometry on a BD LSR II. The level of the death receptor 4 (DR4) expressed by K562s was assessed by staining with α-TRAIL R1 (DR4) APC (Biolegend #307208).
For synNotch driven TRAIL cytotoxicity assays, primary human CD4+ T cells were transduced to express the α-GFP nanobody (LaG17) synNotch Gal4VP64 receptor and 5× Gal4 response elements controlling the expression of LZ-TRAIL or cell surface wild-type TRAIL. The synNotch TRAIL killer cells were co-cultured with surface GFP+ or GFP- K562s for 24 hrs and death was determined by staining with SYTOX Blue. Surface levels of TRAIL was determined by staining T cells with α-TRAIL (CD253) APC (Biolegend #308210). Production and secretion of LZ-TRAIL was determined by TRAIL ELISA (R&D systems #DTRL00).
Antibody Response Element Construct Design
The pembrolizumab (http://www.drugbank.ca/drugs/DB09037), tremelimumab (http://www.google.com/patents/US20090074787), and blinatumomab (http://www.drugbank.ca/drugs/DB09052) codon optimized mRNA sequences were cloned into a MCS downstream of the Gal4 inducible promoter via Infusion cloning. Both antibody constructs consist of (from n-terminus to c- terminus): an n-terminal human IgG heavy chain signal peptide (MDWTWRVFCLLAVTPGAHP) for secretion, a myc-tag for easy detection of binding to target cells with α-myc AF647, the antibody heavy chain sequence, a furin cleavage site and T2A peptide, a human IgG light chain signal peptide (MAWSPLFLTLITHCAGSWA), a HA-tag as an alternative detection means, and the antibody light chain sequence. The Blinatumomab construct contained an n- terminal human IgK signal peptide (MDMRVLAQLLGLLLLCFPGARC). The dual α-PD-1/α-CTLA-4 construct consists of (from n-terminus to c-terminus): an n- terminal human IgG heavy chain signal peptide (MDWTWRVFCLLAVTPGAHP) for secretion, the pembrolizumab heavy chain sequence, a furin cleavage site and T2A peptide, a HA-tag for detection of binding with α-HA A647 (CST #3444S), the Pembrolizumab light chain sequence, a Furin cleavage site and P2A peptide, a human IgK signal peptide, a myc-tag, and the α-CTLA-4 scFv sequence.
synNotch Driven Antibody Production
Primary human CD4+ T cells were transduced to express the α-GFP nanobody (LaG17) synNotch Gal4VP64 receptor and a second vector expressing a user-defined antibody under the control of a 5× Gal4 response element. 2 × 105 transduced T cells were co-cultured 1:1 with GFP+ or GFP− K562s for 24, 48, or 72 hours to induce antibody production and secretion. Cell culture supernatant was harvested and stored for analysis at −80°C.
To detect secreted antibody in cell culture supernatant, a cell-based binding assay was employed. K562 cells were lentivirally transduced to stably express human PD-1 or human CTLA-4, so that these cells could serve as ‘target cells’ for cell-based binding assays to detect antibody secreted from synNotch T cells. 5 × 104 target cells were blocked with human BD Fc Block (BD #564220) prior to incubation with cell culture supernatant from co-cultures of synNotch antibody-producing T cells and inducer K562 cells. Following incubation with cell culture supernatant, target cells were stained with α-myc AF647 to detect bound antibody (which was myc-tagged). Flow cytometry was performed to quantify the fluorescence intensity (which correlated with amount of antibody produced by synNotch T cells) of antibody-labeled target cells.
To quantify the amount of secrete antibody in cell culture supernatant, a standard curve was generated for the cell-based binding assay. E6-1 Jurkat T cells (ATCC #TIB-152) were lentivirally transduced to create a stable cell line that constitutively secretes α-PD-1 pembrolizumab. Supernatant was harvested from a culture of pembrolizumab-secreting Jurkat T cells. The concentration of α-PD- 1 in the supernatant was determined via human IgG4 ELISA (Life Technologies #991000). The supernatant with a known concentration of α-PD-1 was then serially diluted and incubated with PD-1+ K562 target cells to generate a standard curve. Target cell binding of supernatants from co-cultures of synNotch antibody-producing T cells and inducer K562 cells was then compared to the standard curve to determine the concentration of α-PD-1 produced by synNotch T cells co-cultured with different inducer cells.
synNotch Driven Blinatumomab Production
Primary human CD4+ T cells were transduced with the α-GFP nanobody (LaG17) synNotch Gal4VP64 receptor and 5× Gal4 response elements controlling α-CD19/CD3 BiTE, Blinatumomab expression. synNotch BiTE T cells were stimulated with either surface GFP+ or GFP- K562s for 24 hours and supernatant was harvested. The T cells were also collected and stained with α- CD69 APC (Biolegend #310910) to determine if they were activated.
synNotch Driven Flagellin Production
Primary human CD4+ T cells were transduced with the α-GFP nanobody (LaG17) nanobody synNotch Gal4VP64 receptor and 5× Gal4 response elements controlling S. Typhi FliC expression. 2 × 105 transduced T cells were co-cultured 1:1 with GFP+ or GFP− K562s for 24 hours to induce flagellin production. Cell culture supernatant was harvested 24 hours after stimulation and added to wells containing HEK-Blue hTLR5 reporter cells (Invivogen) that express secreted alkaline phosphatase (SEAP) under control of a TLR5 inducible NF-κB promoter. Recombinant FliC (Invivogen) was utilized as a positive control. Twenty-four hours later supernatant was harvested from HEK-TLR5 cells and alkaline phosphatase activity quantified in a colorimetric assay using QUANTIBLUE detection reagent (Invivogen) with a Flexstation III (Molecular Devices).
synNotch Driven PD-L1 and IL-10 Production
Primary human CD4+ T cells were transduced with the α-CD19 synNotch Gal4VP64 receptor and 5× Gal4 response elements controlling human PD-L1 T2A IL-10 expression. The synNotch T cells were stimulated with either surface CD19+ or CD19- K562s for 24 hr and supernatant was harvested for IL-10 ELISA analysis (eBiosciences #BMS215/2). The T cells were also collected and stained separately for intracellular IL-10 with α-IL-10 APC (Biolegend #501410) and for surface PD-L1 with α-PD-L1 BV421 (Biolegend #329714).
Xenograft Tumor Model, Cell Isolation, and Flow Cytometry
Animal studies were conducted with the UCSF Preclinical Therapeutics Core under a protocol approved by the UCSF Institutional Animal Care and Use Committee. NOD scid gamma (NSG) (female, 8~12 weeks old, Jackson Laboratory #005557) mice were used for all in vivo mouse experiments. Primary CD4+ and CD8+ T cells expressing the α-CD19 synNotch Gal4VP64 receptor and 5× Gal4 response elements controlling human IL-2 IRES mCherry were sorted and used in the experiments.
For in vivo synNotch driven localized IL-2 production experiments, NSG mice were injected on day 0 with 5×106 CD19- and CD19+ K562s subcutaneously on the left flank and right flank of the mice, respectively. The tumors were allowed to establish for 4 days and T cells were injected via the tail vein (i.v.) on day 4 or intratumoral on day 8. The T cells were suspended in PBS for all injections. CD4+ and CD8+ synNotch T cells were injected at a 1:1 ratio. For i.v. injections, 6×106 total T cells were injected, and for intratumoral injections, 5×105 total T cells were injected.
Tumors were harvested at day 10 into RPMI supplemented with 1% FBS (UCSF Cell Culture Core). The tumors were then minced by razor blade and digested for an hour in RPMI with 0.1 mg/mL DNase (Roche #10104159001) and 0.2 mg/mL collagenase P (Roche # 11249002001) at 37°C. After incubation, the digested tumors were passed over a 75 µm cell strainer and the tumor cells were collected by centrifugation. The cells were then treated with red blood cell lysis buffer (Biolegend #420301) and washed with PBS. The tumors were then stained with a LIVE/DEAD Green (Thermo Scientific #34969) and α-CD4 A647 (BD 557707) and α-CD8 BV786 (BD #563823) to analyze the tumor infiltrating T cells. Expression of IL-2 IRES mCherry was assessed in the CD4+ and CD8+ T cell populations with a BD LSR II.
For in vivo synNotch driven localized Blinatumomab experiments, NSG mice were injected on day 0 with 5×106 CD19+ only and surface GFP/CD19+ K562s subcutaneously on the left flank and right flank of the mice, respectively. The tumors were allowed to establish for 4 days. Primary human CD4+ and CD8+ T cells (6 × 106 total T cells 1:1 CD4+ and CD8+ T cells) transduced with the α-GFP nanobody (LaG17) synNotch Gal4VP64 receptor and 5× Gal4 response elements controlling the α-CD19/CD3 BiTE, Blinatumomab, expression or untransduced T cells were injected via the tail vein (i.v.) on day 4. Tumor growth was monitored by caliper for 25 days after tumor cell implantation.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical significance was determined by Student’s t-test (two-tailed) unless otherwise noted. All statistical analysis was performed with Prism 7a (Graphpad) and p values are reported (n.s. = p > 0.05, * = p ≤ 0.05, ** = p ≤ 0.01, *** = p ≤ 0.001, **** = p ≤ 0.0001). All error bars represent either S.E.M. or S.D.
Supplementary Material
Figure S1. synNotch Receptors Drive Antigen-dependent Custom Gene Regulation in Primary Human T cells (related to main text Figure 1).
(A) Two dimensional dot plots of CD4+ (left plot) and CD8+ (right plot) primary human T cells transduced with the α-CD19 synNotch Gal4VP64 receptor (myc tagged) and 5×Gal4 response elements controlling expression of BFP. The response element vector also contains a PGK promoter that drives constitutive expression of mCherry to identify the T cells with the inserted response elements. All BFP reporter expression analysis was performed on T cells that had both the synNotch receptor and the corresponding 5×Gal4 response elements controlling BFP expression (population outlined in red box).
(B) Histograms showing selective induction of the BFP reporter in synNotch receptor receiver CD8+ T cells in response to CD19+ K562s or Daudi Tumor cells compared to CD19- K562 control cells after 24 hours of co-culture (representative of ≥ 3 experiments).
(C) Histograms showing CD19 levels on K562s and Daudi tumor cells. Daudi tumors naturally express CD19 and K562s ectopically express CD19 at similar levels.
(D) Two dimensional dot plots similar to panel A of α-Her2 synNotch Gal4VP64 receptor variants (scFv ID and affinity is given) expressing CD4+ (top row) and CD8+ (bottom row) primary human T cells. Each column is for a particular α-Her2 scFv affinity variant. All BFP reporter expression analysis was performed on T cells in the red outlined gate as in panel A.
(E) Histograms showing selective induction of the BFP reporter in α-Her2 synNotch receptor receiver CD4+ and CD8+ T cells in response to surface Her2+ MCF7 breast cancer cells compared to no senders cells added after 24 hours of co-culture (representative of ≥ 3 experiments).
(F) Two dimensional dot plots similar to panel A for α-GFP nanobody synNotch Gal4VP64 receptor expressing CD4 (top row) and CD8 (bottom row) primary human T cells. Each column is for a particular α-GFP nanobody affinity variant (LaG17, LaG16-2). All BFP reporter expression analysis was performed on T cells in the red outlined gate as in panel A.
(G) Histograms showing selective induction of the BFP reporter in α-GFP (LaG17 or LaG16_2) synNotch receptor receiver CD8+ T cells in response to surface GFP+ compared to surface GFP- K562s after 24 hours of co-culture (representative of ≥ 3 experiments).
(H) Histograms showing total GFP level in K562 cancer cells expressing surface GFP compared to K562 GFP- controls.
Figure S2. synNotch Receptors Drive Customized Cytokine Profiles (related to main text Figure 2)
(A) CD4+ primary human T cells were engineered with the α-CD19 synNotch Gal4VP64 receptor and the associated 5×Gal4 response elements in control of IL-2 production.
(B) Two dimensional dot plot (left panel) of CD4+ primary human T cells transduced with the α-CD19 synNotch Gal4VP64 receptor and 5×Gal4 response elements controlling expression IL-2 expression IRES mCherry. The response element vector also contains a PGK promoter that drives constitutive expression of BFP to identify the T cells with the inserted response elements. T cells that had both the synNotch receptor and the corresponding 5×Gal4 response elements controlling IL-2 IRES mCherry expression were sorted and used for all corresponding assays in Figure 3 and S3. (orange shaded box). The left panel shows no basal induction of the IL-2 IRES mCherry reporter in dual positive T cells compared to untransduced T cells.
(C) Dot plots of intracellular cytokine stains for IL-2 are shown for unstimulated CD4+ and CD8+ T cells, unstimulated α-CD19 synNotch Gal4VP64 T cells controlling IL-2 production, and positive control T cells stimulated with PMA/ionomycin for 6 hours.
(D) The basal and stimulated IL-2 levels are given for supernatants harvested from untransduced CD4+ T cells, α-CD19 4-1BBζ CAR T cells, and α-CD19 synNotch Gal4VP64 T cells controlling IL-2 production (n = 4).
(E) CD69 levels (left column) and IL-2 IRES mCherry reporter levels in control CD4+ T cells stimulated with α-CD3/CD28 dynabeads and α-CD19 synNotch Gal4VP64 T cells controlling IL-2 production stimulated with CD19- or CD19+ K562s. CD69 is not upregulated on synNotch T cells upon stimulation with cognate antigen.
(F) CD4+ human primary T cells were engineered with the α-CD19 synNotch Gal4VP64 receptor and the associated 5×Gal4 response elements in control of IL-10 production.
(G) Equivalent data to panel B for CD4+ primary human T cells transduced with the α-CD19 synNotch Gal4VP64 receptor and 5×Gal4 response elements controlling expression of IL-10 IRES mCherry expression.
(H) Equivalent data to panel D for CD4+ primary human T cells transduced with the α-CD19 synNotch Gal4VP64 receptor and 5×Gal4 response elements controlling expression IL-10 IRES mCherry expression.
(I) Equivalent data to panel E for CD4 primary human T cells transduced with the α-CD19 synNotch Gal4VP64 receptor and 5×Gal4 response elements controlling expression IL-10 IRES mCherry expression.
Figure S3. synNotch Receptor Circuits can Skew T cell Differentiation (related to main text Figure 3).
(A) Quantification of replicate data from panel C in Figure 4. CD4+ T cells with α- CD19 synNotch Gal4VP64 receptor response elements controlling Tbet T2A GFP expression were co-cultured with CD19+ or CD19- K562s for 24 hrs. The percentage of T cells with Tbet T2A GFP expression is quantified showing Tbet was only upregulated when the T cells were exposed to CD19+ K562s (n=3).
(B) Representative dot plot of CD4 T cells stimulated with α-CD3/CD28 dynabeads and intracellularly stained for the Th1 cytokine IFNγ after 11 days in culture. The IFNγ+ positive gate is boxed in red.
(C) Representative dot plot similar to panel B for CD4+ T cells cultured for 11 days in Th1 differentiation conditions (IL-12 + α-IL-4) (n=4).
(D) Representative histograms of CD4 T cells with constitutive overexpression of Tbet T2A mCherry at 48 hours post transduction.
(E) Representative dot plot of intracellular stains of CD4+ T cells with constitutive overexpression of Tbet T2A mCherry for Tbet and IFNγ after 11 days of culture (n=6). Dual positive Tbet and IFNγ T cells are boxed in red.
Figure S4. synNotch Receptor Circuits Can be Used to Engineer Customized Killer T cells (related to main text Figure 4).
(A) Rested CD4+ and CD8+ T cells were restimulated for 24 hrs with α- CD3/CD28 dynabeads and stained for cell surface TRAIL. Neither T cell subset acutely upregulates TRAIL expression.
(B) Cancer cells vary in their sensitivity to recombinant TRAIL-mediated apoptosis. HCT116 colon cancers have the death receptors bound by TRAIL and are sensitive to low level TRAIL treatment. K562s express the death receptors for TRAIL but are insensitive to TRAIL treatment.
(C) Surface GFP- or GFP+ K562s or HCT116 cancer cells were treated with 0 to 200 ng/mL of recombinant TRAIL for 24 hrs and death was monitored via flow cytometry by staining with the dead stain SYTOX blue (n=4).
(D) Histogram of K562s stained for the death receptor 4 (DR4) bound by TRAIL. K562s express the receptor their resistance to TRAIL-mediated apoptosis.
(E) Representative dot plots of CD4+ T cells transduced with the LaG17 α-GFP nanobody synNotch Gal4VP64 and 5×Gal4 response elements controlling expression of leucine zipper TRAIL (LZ-TRAIL left panel) or full-length surface TRAIL (right panel). The level of the mCherry reporter of TRAIL expression is shown below for the dual positive T cells and control untranduced T cells.
(F) CD4+ T cells transduced with the LaG17 α-GFP nanobody synNotch Gal4VP64 and 5×Gal4 response elements controlling expression LZ-TRAIL were co-cultured with surface GFP- or GFP+ K562s for 24 hrs to determine if LZ- TRAIL is secreted only in response to GFP+ K562s.
(G) Histograms showing the level of mCherry reporter levels of TRAIL production in sorted synNotch CD4 LZ-TRAIL T cells shown in panel E were co-cultured with surface GFP= or GFP+ K562s. The reporter was exclusively activated in response to surface GFP+ K562s.
(H) TRAIL ELISA of supernatant from sorted synNotch CD4 LZ-TRAIL T cells co-cultured with either surface GFP- or GFP+ K562s for 24 hrs (n=2).
Figure S5. synNotch Receptor Circuits for Antigen-dependent Production of Antibodies, Adjuvants, and Immune Suppressive Agents (related to main text Figure 5).
(A) CD4+ T cells were engineered with the α-GFP synNotch controlling the expression of Pembrolizumab (α-PD-1 myc) or Tremelimumab (α-CTLA4 myc). After 24 hours of stimulation of the T cells with surface GFP+ or GFP- K562s, the supernatant was collected and used to stain PD-1+ or CTLA4+ K562s. Secreted antibody binding to target cells was monitored via flow cytometry after secondary staining with α-myc A647 (representative of 3 replicates).
(B) Histograms of PD-1+ K562s stained with Pembrolizumab antibody (myc tagged) at known concentrations (see material and methods). Pembrolizumab binding was monitored by secondary staining with α-myc A647. The MFI of stained PD-1+ K562 cells for each antibody dilution was plotted and fit via linear regression. The derived linear equation was used to quantify the amount of secreted antibody in panel C.
(C) CD4+ T cells were engineered with the α-GFP synNotch controlling the expression of Pembrolizumab (α-PD-1 myc). After 48 and 72 hours of stimulation of the T cells with surface GFP+ or GFP- K562s, the supernatant was collected and used to stain PD-1+ K562s. The calculated concentration of secreted Pembrolizumab is given at each time point based on the standard curve from panel B (representative of 3 replicates)
(D) CD4+ T cells were engineered with the α-GFP synNotch receptor controlling the expression of Flagellin. Supernatant was harvested from the synNotch T cells or untransduced T cells after co-culture with surface GFP+ or GFP- K562s and added to hTLR5 HEK-blue secreted alkaline phosphatase (SEAP) reporter cells. After 24 hours, SEAP activity was monitored and the level of Flagellin in the supernatant was measured a purified Flagellin standard (n=3, error bars = SD).
(E) Representative histograms of CD4+ T cells engineered with the α-CD19 synNotch receptor controlling the expression of PD-L1 and IL-10 after co-culture with CD19+ or CD19- K562s for 24 hrs. These data were used to calculate the percentage of cells expressing PD-L1 and intracellular IL-10 in Figure 5D (representative of 3 replicates).
Figure S6. synNotch Receptor T cells Locally Produce Cytokines and Therapeutic Agents in vivo (related to main text figure 6).
(A) Two dimensional dot plots of CD4+ (left plot) and CD8+ (right plot) primary human T cells transduced with the α-CD19 synNotch Gal4VP64 receptor (myc tagged) and 5×Gal4 response elements controlling expression of IL-2 IRES mCherry. The response element vector also contains a PGK promoter that drives constitutive expression of BFP to identify the T cells with the inserted response elements. T cells in the red shaded box were sorted and used for all in vivo and in vitro experiments.
(B) Histograms of IL-2 IRES mCherry reporter levels in tumor and spleen infiltrated CD8+ synNotch T cells injected i.v. showing selective expression of the mCherry reporter in target CD19+ tumors (data representative of the same 3 replicate mice in Figure 6B).
(C) Quantification of the percentage of tumor infiltrated CD4+ and CD8+ synNotch T cells that induced expression of the mCherry reporter of IL-2 expression from replicate data shown in Figure 6B and panel B (n=3, significance determined by Student’s t-test *** p<0.001 and **** p<0.0001).
(D) Quantification of the percentage CD4+ and CD8+ synNotch T cells that induced expression of the mCherry reporter of IL-2 expression after in vitro co- culture with CD19+ or CD19= K562s (n=3, significance determined by Student’s t-test *** p≤0.001 and **** p≤0.0001).
(E) Two dimensional dot plots showing the percentage of CD4+ and CD8+ T cells in representative single cell suspensions generated from the harvested spleen and CD19+ and CD19- tumors from a mouse i.v. injected with synNotch T cells controlling the expressing of IL-2 (same mouse analyzed in Figure 6A and 6B and panel B).
(F) CD4+ and CD8+ synNotch T cells were directly injected into both the CD19- and CD19+ tumor 8 days after tumor implantation. Tumors were harvested at day 10 and infiltrating T cells were analyzed for IL-2 mCherry reporter expression by flow cytometry. Histograms of IL-2 IRES mCherry reporter levels in tumor infiltrated CD4+ and CD8+ synNotch T cells injected intratumorally, showing selective expression of the mCherry reporter in target CD19+ tumors are given (data representative of 3 replicate mice).
(G) Bilateral CD19+ and GFP/CD19+ K562 tumor growth curves in mice treated with untransduced control CD4+ and CD8+ primary human T cells. The single antigen CD19+ tumor and dual antigen GFP/CD19+ tumor grew out at the same rate. The growth curve of the single antigen CD19+ from mice treated with CD4+ and CD8+ engineered with the α-GFP synNotch receptor controlling Blinatumomab (α-CD19/CD3 BiTE) expression is provided as a reference showing minimal slowed growth (n.s. at day 19) compared to control conditions (n=5 mice, error = SEM, significance determined by Student’s t-test n.s. = p≥0.05 ** p≤0.01).
ACKNOWLEDGEMENTS
We would like to thank members of the Lim lab, Arthur Weiss and members of the Weiss lab for helpful discussions and comments on the manuscript. We thank the Preclinical Therapeutics Core at UCSF for help with in vivo studies and I. Sterin, C. Ghosh for technical assistance. This work was supported by a Jane Coffin Childs Memorial Fund Postdoctoral Fellowship A121505 (K.T.R.), a Human Frontiers of Science Program Postdoctoral Fellowship (L.M.), and NIH grants P50GM081879, R01 GM055040, R01 CA196277, and the Howard Hughes Medical Institute (W.A.L).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
K.R. and W.L. conceived of the study. K.R. performed and analyzed all experiments with the exception of the following. L.M. worked with K.R. to develop synNotch system. J.W. performed and analyzed antibody production experiments. L.R. aided in in vivo IL-2 experiments. L.R. and I.K. performed and analyzed Flagellin production experiments. J.C. performed IL-12 production experiments. W.W., and K.M. provided technical support. K.R. and W.L. wrote the paper.
REFERENCES
- Akira S, Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Amarnath S, Mangus CW, Wang JCM, Wei F, He A, Kapoor V, Foley JE, Massey PR, Felizardo TC, Riley JL, et al. The PDL1-PD1 axis converts human TH1 cells into regulatory T cells. Sci Transl Med. 2011;3:111ra120–111ra120. doi: 10.1126/scitranslmed.3003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R, Macdonald I, Corbett T, Hacking G, Lowdell MW, Prentice HG. Construction and biological characterization of an interleukin-12 fusion protein (Flexi-12): delivery to acute myeloid leukemic blasts using adeno-associated virus. Hum. Gene Ther. 1997;8:1125–1135. doi: 10.1089/hum.1997.8.9-1125. [DOI] [PubMed] [Google Scholar]
- Barrett DM, Singh N, Porter DL, Grupp SA, June CH. Chimeric antigen receptor therapy for cancer. Annu. Rev. Med. 2014a;65:333–347. doi: 10.1146/annurev-med-060512-150254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett DM, Teachey DT, Grupp SA. Toxicity management for patients receiving novel T-cell engaging therapies. Curr. Opin. Pediatr. 2014b;26:43–49. doi: 10.1097/MOP.0000000000000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Brentjens RJ, Davila ML, Rivière I, Park J, Wang X, Cowell LG, Bartido S, Stefanski J, Taylor C, Olszewska M, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med. 2013;5:177ra38–177ra38. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat. Rev. Immunol. 2004;4:336–347. doi: 10.1038/nri1349. [DOI] [PubMed] [Google Scholar]
- Chovatiya R, Medzhitov R. Stress, inflammation, and defense of homeostasis. Mol. Cell. 2014;54:281–288. doi: 10.1016/j.molcel.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila ML, Rivière I, Wang X, Bartido S, Park J, Curran K, Chung SS, Stefanski J, Borquez-Ojeda O, Olszewska M, et al. Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci Transl Med. 2014;6:224ra25–224ra25. doi: 10.1126/scitranslmed.3008226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti G, Gottschalk S, Savoldo B, Brenner MK. Design and development of therapies using chimeric antigen receptor-expressing T cells. Immunol. Rev. 2014;257:107–126. doi: 10.1111/imr.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. Nat. Rev. Cancer. 2004;4:11–22. doi: 10.1038/nrc1252. [DOI] [PubMed] [Google Scholar]
- Fischbach MA, Bluestone JA, Lim WA. Cell-based therapeutics: the next pillar of medicine. Sci Transl Med. 2013;5:179ps7–179ps7. doi: 10.1126/scitranslmed.3005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridy PC, Li Y, Keegan S, Thompson MK, Nudelman I, Scheid JF, Oeffinger M, Nussenzweig MC, Fenyö D, Chait BT, et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods. 2014;11:1253–1260. doi: 10.1038/nmeth.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski TF, Schreiber H, Fu Y-X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S, June CH. Going viral: chimeric antigen receptor T-cell therapy for hematological malignancies. Immunol. Rev. 2015;263:68–89. doi: 10.1111/imr.12243. [DOI] [PubMed] [Google Scholar]
- Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat. Struct. Mol. Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- Gordon WR, Zimmerman B, He L, Miles LJ, Huang J, Tiyanont K, McArthur DG, Aster JC, Perrimon N, Loparo JJ, et al. Mechanical Allostery: Evidence for a Force Requirement in the Proteolytic Activation of Notch. Dev. Cell. 2015;33:729–736. doi: 10.1016/j.devcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, Teachey DT, Chew A, Hauck B, Wright JF, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N. Engl. J. Med. 2013;368:1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015;16:343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone RW, Frew AJ, Smyth MJ. The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat. Rev. Cancer. 2008;8:782–798. doi: 10.1038/nrc2465. [DOI] [PubMed] [Google Scholar]
- June CH, Blazar BR, Riley JL. Engineering lymphocyte subsets: tools, trials and tribulations. Nat. Rev. Immunol. 2009;9:704–716. doi: 10.1038/nri2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalos M, Levine BL, Porter DL, Katz S, Grupp SA, Bagg A, June CH. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. 2011;3:95ra73–95ra73. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kara EE, Comerford I, Fenix KA, Bastow CR, Gregor CE, McKenzie DR, McColl SR. Tailored immune responses: novel effector helper T cell subsets in protective immunity. PLoS Pathog. 2014;10:e1003905. doi: 10.1371/journal.ppat.1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Fisher MJ, Xu SQ, el-Deiry WS. Molecular determinants of response to TRAIL in killing of normal and cancer cells. Clin. Cancer Res. 2000;6:335–346. [PubMed] [Google Scholar]
- Kloss CC, Condomines M, Cartellieri M, Bachmann M, Sadelain M. Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat. Biotechnol. 2013;31:71–75. doi: 10.1038/nbt.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy MQ, Jacobus S, Blood EA, Kay NE, Rajkumar SV, Greipp PR. Phase II study of interleukin-12 for treatment of plateau phase multiple myeloma (E1A96): a trial of the Eastern Cooperative Oncology Group. Leuk. Res. 2009;33:1485–1489. doi: 10.1016/j.leukres.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers CHJ, Sleijfer S, Vulto AG, Kruit WHJ, Kliffen M, Debets R, Gratama JW, Stoter G, Oosterwijk E. Treatment of metastatic renal cell carcinoma with autologous T-lymphocytes genetically retargeted against carbonic anhydrase IX: first clinical experience. J. Clin. Oncol. 2006;24:e20–e22. doi: 10.1200/JCO.2006.05.9964. [DOI] [PubMed] [Google Scholar]
- Lämmermann T, Sixt M. The microanatomy of T-cell responses. Immunol. Rev. 2008;221:26–43. doi: 10.1111/j.1600-065X.2008.00592.x. [DOI] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F. Indirect evidence for Delta- dependent intracellular processing of notch in Drosophila embryos. Curr. Biol. 1998;8:771–774. doi: 10.1016/s0960-9822(98)70300-8. [DOI] [PubMed] [Google Scholar]
- Leigh ND, Bian G, Ding X, Liu H, Aygun-Sunar S, Burdelya LG, Gudkov AV, Cao X. A flagellin-derived toll-like receptor 5 agonist stimulates cytotoxic lymphocyte-mediated tumor immunity. PLoS ONE. 2014;9:e85587. doi: 10.1371/journal.pone.0085587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke J, von Karstedt S, Zinngrebe J, Walczak H. Getting TRAIL back on track for cancer therapy. Cell Death Differ. 2014;21:1350–1364. doi: 10.1038/cdd.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WA. Designing customized cell signalling circuits. Nat. Rev. Mol. Cell Biol. 2010;11:393–403. doi: 10.1038/nrm2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jiang S, Fang C, Yang S, Olalere D, Pequignot EC, Cogdill AP, Li N, Ramones M, Granda B, et al. Affinity-Tuned ErbB2 or EGFR Chimeric Antigen Receptor T Cells Exhibit an Increased Therapeutic Index against Tumors in Mice. Cancer Res. 2015;75:3596–3607. doi: 10.1158/0008-5472.CAN-15-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee MS, Snook AE. Challenges to chimeric antigen receptor (CAR)-T cell therapy for cancer. Discov Med. 2014;18:265–271. [PubMed] [Google Scholar]
- Maus MV, Fraietta JA, Levine BL, Kalos M, Zhao Y, June CH. Adoptive immunotherapy for cancer or viruses. Annu. Rev. Immunol. 2014;32:189–225. doi: 10.1146/annurev-immunol-032713-120136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JFAP, Sadelain M. The Journey from Discoveries in Fundamental Immunology to Cancer Immunotherapy. Cancer Cell. 2015;27:439–449. doi: 10.1016/j.ccell.2015.03.007. [DOI] [PubMed] [Google Scholar]
- Morgan RA, Yang JC, Kitano M, Dudley ME, Laurencot CM, Rosenberg SA. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol. Ther. 2010;18:843–851. doi: 10.1038/mt.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsut L, Roybal KT, Xiong X, Gordley RM, Coyle SM, Thomson M, Lim WA. Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell. 2016;164:780–791. doi: 10.1016/j.cell.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SN, Gebhardt T, Carbone FR, Heath WR. Memory T cell subsets, migration patterns, and tissue residence. Annu. Rev. Immunol. 2013;31:137–161. doi: 10.1146/annurev-immunol-032712-095954. [DOI] [PubMed] [Google Scholar]
- Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- Nagorsen D, Kufer P, Baeuerle PA, Bargou R. Blinatumomab: a historical perspective. Pharmacol. Ther. 2012;136:334–342. doi: 10.1016/j.pharmthera.2012.07.013. [DOI] [PubMed] [Google Scholar]
- Nair PM, Flores H, Gogineni A, Marsters S, Lawrence DA, Kelley RF, Ngu H, Sagolla M, Komuves L, Bourgon R, et al. Enhancing the antitumor efficacy of a cell-surface death ligand by covalent membrane display. Proc. Natl. Acad. Sci. U.S.a. 2015;112:5679–5684. doi: 10.1073/pnas.1418962112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Garra A, Barrat FJ, Castro AG, Vicari A, Hawrylowicz C. Strategies for use of IL-10 or its antagonists in human disease. Immunol. Rev. 2008;223:114–131. doi: 10.1111/j.1600-065X.2008.00635.x. [DOI] [PubMed] [Google Scholar]
- O'Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea JJ, Ma A, Lipsky P. Cytokines and autoimmunity. Nat. Rev. Immunol. 2002;2:37–45. doi: 10.1038/nri702. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. Immunol. 2011;29:71–109. doi: 10.1146/annurev-immunol-031210-101312. [DOI] [PubMed] [Google Scholar]
- Park S-J, Kim M-J, Kim H-B, Sohn H-Y, Bae J-H, Kang C-D, Kim S-H. Cotreatment with apicidin overcomes TRAIL resistance via inhibition of Bcr-Abl signaling pathway in K562 leukemia cells. Exp. Cell Res. 2009;315:1809–1818. doi: 10.1016/j.yexcr.2009.02.024. [DOI] [PubMed] [Google Scholar]
- Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 2011;365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow MA, Callahan MK, Wolchok JD. Immune Checkpoint Blockade in Cancer Therapy. J. Clin. Oncol. 2015;33:1974–1982. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybal KT, Rupp LJ, Morsut L, Walker WJ, McNally KA, Park JS, Lim WA. Precision Tumor Recognition by T Cells With Combinatorial Antigen-Sensing Circuits. Cell. 2016;164:770–779. doi: 10.1016/j.cell.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Selkoe D, Kopan R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu. Rev. Neurosci. 2003;26:565–597. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161:205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smanski MJ, Bhatia S, Zhao D, Park Y, Woodruff L, Giannoukos G, Ciulla D, Busby M, Calderon J, Nicol R, et al. Functional optimization of gene clusters by combinatorial design and assembly. Nat. Biotechnol. 2014;32:1241–1249. doi: 10.1038/nbt.3063. B A. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JC, Griffiths GM. Secretory mechanisms in cell- mediated cytotoxicity. Annu. Rev. Cell Dev. Biol. 2007;23:495–517. doi: 10.1146/annurev.cellbio.23.090506.123521. [DOI] [PubMed] [Google Scholar]
- Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- Sundrud MS, Grill SM, Ni D, Nagata K, Alkan SS, Subramaniam A, Unutmaz D. Genetic reprogramming of primary human T cells reveals functional plasticity in Th cell differentiation. J. Immunol. 2003;171:3542–3549. doi: 10.4049/jimmunol.171.7.3542. [DOI] [PubMed] [Google Scholar]
- Tang H, Qiao J, Fu Y-X. Immunotherapy and tumor microenvironment. Cancer Lett. 2016;370:85–90. doi: 10.1016/j.canlet.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temizoz B, Kuroda E, Ishii KJ. Vaccine adjuvants as potential cancer immunotherapeutics. Int. Immunol. 2016;28:329–338. doi: 10.1093/intimm/dxw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat. Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Wang QJ, Yang S, Kochenderfer JN, Zheng Z, Zhong X, Sadelain M, Eshhar Z, Rosenberg SA, Morgan RA. A herceptin-based chimeric antigen receptor with modified signaling domains leads to enhanced survival of transduced T lymphocytes and antitumor activity. J. Immunol. 2009;183:5563–5574. doi: 10.4049/jimmunol.0900447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat. Rev. Immunol. 2006;6:295–307. doi: 10.1038/nri1806. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. synNotch Receptors Drive Antigen-dependent Custom Gene Regulation in Primary Human T cells (related to main text Figure 1).
(A) Two dimensional dot plots of CD4+ (left plot) and CD8+ (right plot) primary human T cells transduced with the α-CD19 synNotch Gal4VP64 receptor (myc tagged) and 5×Gal4 response elements controlling expression of BFP. The response element vector also contains a PGK promoter that drives constitutive expression of mCherry to identify the T cells with the inserted response elements. All BFP reporter expression analysis was performed on T cells that had both the synNotch receptor and the corresponding 5×Gal4 response elements controlling BFP expression (population outlined in red box).
(B) Histograms showing selective induction of the BFP reporter in synNotch receptor receiver CD8+ T cells in response to CD19+ K562s or Daudi Tumor cells compared to CD19- K562 control cells after 24 hours of co-culture (representative of ≥ 3 experiments).
(C) Histograms showing CD19 levels on K562s and Daudi tumor cells. Daudi tumors naturally express CD19 and K562s ectopically express CD19 at similar levels.
(D) Two dimensional dot plots similar to panel A of α-Her2 synNotch Gal4VP64 receptor variants (scFv ID and affinity is given) expressing CD4+ (top row) and CD8+ (bottom row) primary human T cells. Each column is for a particular α-Her2 scFv affinity variant. All BFP reporter expression analysis was performed on T cells in the red outlined gate as in panel A.
(E) Histograms showing selective induction of the BFP reporter in α-Her2 synNotch receptor receiver CD4+ and CD8+ T cells in response to surface Her2+ MCF7 breast cancer cells compared to no senders cells added after 24 hours of co-culture (representative of ≥ 3 experiments).
(F) Two dimensional dot plots similar to panel A for α-GFP nanobody synNotch Gal4VP64 receptor expressing CD4 (top row) and CD8 (bottom row) primary human T cells. Each column is for a particular α-GFP nanobody affinity variant (LaG17, LaG16-2). All BFP reporter expression analysis was performed on T cells in the red outlined gate as in panel A.
(G) Histograms showing selective induction of the BFP reporter in α-GFP (LaG17 or LaG16_2) synNotch receptor receiver CD8+ T cells in response to surface GFP+ compared to surface GFP- K562s after 24 hours of co-culture (representative of ≥ 3 experiments).
(H) Histograms showing total GFP level in K562 cancer cells expressing surface GFP compared to K562 GFP- controls.
Figure S2. synNotch Receptors Drive Customized Cytokine Profiles (related to main text Figure 2)
(A) CD4+ primary human T cells were engineered with the α-CD19 synNotch Gal4VP64 receptor and the associated 5×Gal4 response elements in control of IL-2 production.
(B) Two dimensional dot plot (left panel) of CD4+ primary human T cells transduced with the α-CD19 synNotch Gal4VP64 receptor and 5×Gal4 response elements controlling expression IL-2 expression IRES mCherry. The response element vector also contains a PGK promoter that drives constitutive expression of BFP to identify the T cells with the inserted response elements. T cells that had both the synNotch receptor and the corresponding 5×Gal4 response elements controlling IL-2 IRES mCherry expression were sorted and used for all corresponding assays in Figure 3 and S3. (orange shaded box). The left panel shows no basal induction of the IL-2 IRES mCherry reporter in dual positive T cells compared to untransduced T cells.
(C) Dot plots of intracellular cytokine stains for IL-2 are shown for unstimulated CD4+ and CD8+ T cells, unstimulated α-CD19 synNotch Gal4VP64 T cells controlling IL-2 production, and positive control T cells stimulated with PMA/ionomycin for 6 hours.
(D) The basal and stimulated IL-2 levels are given for supernatants harvested from untransduced CD4+ T cells, α-CD19 4-1BBζ CAR T cells, and α-CD19 synNotch Gal4VP64 T cells controlling IL-2 production (n = 4).
(E) CD69 levels (left column) and IL-2 IRES mCherry reporter levels in control CD4+ T cells stimulated with α-CD3/CD28 dynabeads and α-CD19 synNotch Gal4VP64 T cells controlling IL-2 production stimulated with CD19- or CD19+ K562s. CD69 is not upregulated on synNotch T cells upon stimulation with cognate antigen.
(F) CD4+ human primary T cells were engineered with the α-CD19 synNotch Gal4VP64 receptor and the associated 5×Gal4 response elements in control of IL-10 production.
(G) Equivalent data to panel B for CD4+ primary human T cells transduced with the α-CD19 synNotch Gal4VP64 receptor and 5×Gal4 response elements controlling expression of IL-10 IRES mCherry expression.
(H) Equivalent data to panel D for CD4+ primary human T cells transduced with the α-CD19 synNotch Gal4VP64 receptor and 5×Gal4 response elements controlling expression IL-10 IRES mCherry expression.
(I) Equivalent data to panel E for CD4 primary human T cells transduced with the α-CD19 synNotch Gal4VP64 receptor and 5×Gal4 response elements controlling expression IL-10 IRES mCherry expression.
Figure S3. synNotch Receptor Circuits can Skew T cell Differentiation (related to main text Figure 3).
(A) Quantification of replicate data from panel C in Figure 4. CD4+ T cells with α- CD19 synNotch Gal4VP64 receptor response elements controlling Tbet T2A GFP expression were co-cultured with CD19+ or CD19- K562s for 24 hrs. The percentage of T cells with Tbet T2A GFP expression is quantified showing Tbet was only upregulated when the T cells were exposed to CD19+ K562s (n=3).
(B) Representative dot plot of CD4 T cells stimulated with α-CD3/CD28 dynabeads and intracellularly stained for the Th1 cytokine IFNγ after 11 days in culture. The IFNγ+ positive gate is boxed in red.
(C) Representative dot plot similar to panel B for CD4+ T cells cultured for 11 days in Th1 differentiation conditions (IL-12 + α-IL-4) (n=4).
(D) Representative histograms of CD4 T cells with constitutive overexpression of Tbet T2A mCherry at 48 hours post transduction.
(E) Representative dot plot of intracellular stains of CD4+ T cells with constitutive overexpression of Tbet T2A mCherry for Tbet and IFNγ after 11 days of culture (n=6). Dual positive Tbet and IFNγ T cells are boxed in red.
Figure S4. synNotch Receptor Circuits Can be Used to Engineer Customized Killer T cells (related to main text Figure 4).
(A) Rested CD4+ and CD8+ T cells were restimulated for 24 hrs with α- CD3/CD28 dynabeads and stained for cell surface TRAIL. Neither T cell subset acutely upregulates TRAIL expression.
(B) Cancer cells vary in their sensitivity to recombinant TRAIL-mediated apoptosis. HCT116 colon cancers have the death receptors bound by TRAIL and are sensitive to low level TRAIL treatment. K562s express the death receptors for TRAIL but are insensitive to TRAIL treatment.
(C) Surface GFP- or GFP+ K562s or HCT116 cancer cells were treated with 0 to 200 ng/mL of recombinant TRAIL for 24 hrs and death was monitored via flow cytometry by staining with the dead stain SYTOX blue (n=4).
(D) Histogram of K562s stained for the death receptor 4 (DR4) bound by TRAIL. K562s express the receptor their resistance to TRAIL-mediated apoptosis.
(E) Representative dot plots of CD4+ T cells transduced with the LaG17 α-GFP nanobody synNotch Gal4VP64 and 5×Gal4 response elements controlling expression of leucine zipper TRAIL (LZ-TRAIL left panel) or full-length surface TRAIL (right panel). The level of the mCherry reporter of TRAIL expression is shown below for the dual positive T cells and control untranduced T cells.
(F) CD4+ T cells transduced with the LaG17 α-GFP nanobody synNotch Gal4VP64 and 5×Gal4 response elements controlling expression LZ-TRAIL were co-cultured with surface GFP- or GFP+ K562s for 24 hrs to determine if LZ- TRAIL is secreted only in response to GFP+ K562s.
(G) Histograms showing the level of mCherry reporter levels of TRAIL production in sorted synNotch CD4 LZ-TRAIL T cells shown in panel E were co-cultured with surface GFP= or GFP+ K562s. The reporter was exclusively activated in response to surface GFP+ K562s.
(H) TRAIL ELISA of supernatant from sorted synNotch CD4 LZ-TRAIL T cells co-cultured with either surface GFP- or GFP+ K562s for 24 hrs (n=2).
Figure S5. synNotch Receptor Circuits for Antigen-dependent Production of Antibodies, Adjuvants, and Immune Suppressive Agents (related to main text Figure 5).
(A) CD4+ T cells were engineered with the α-GFP synNotch controlling the expression of Pembrolizumab (α-PD-1 myc) or Tremelimumab (α-CTLA4 myc). After 24 hours of stimulation of the T cells with surface GFP+ or GFP- K562s, the supernatant was collected and used to stain PD-1+ or CTLA4+ K562s. Secreted antibody binding to target cells was monitored via flow cytometry after secondary staining with α-myc A647 (representative of 3 replicates).
(B) Histograms of PD-1+ K562s stained with Pembrolizumab antibody (myc tagged) at known concentrations (see material and methods). Pembrolizumab binding was monitored by secondary staining with α-myc A647. The MFI of stained PD-1+ K562 cells for each antibody dilution was plotted and fit via linear regression. The derived linear equation was used to quantify the amount of secreted antibody in panel C.
(C) CD4+ T cells were engineered with the α-GFP synNotch controlling the expression of Pembrolizumab (α-PD-1 myc). After 48 and 72 hours of stimulation of the T cells with surface GFP+ or GFP- K562s, the supernatant was collected and used to stain PD-1+ K562s. The calculated concentration of secreted Pembrolizumab is given at each time point based on the standard curve from panel B (representative of 3 replicates)
(D) CD4+ T cells were engineered with the α-GFP synNotch receptor controlling the expression of Flagellin. Supernatant was harvested from the synNotch T cells or untransduced T cells after co-culture with surface GFP+ or GFP- K562s and added to hTLR5 HEK-blue secreted alkaline phosphatase (SEAP) reporter cells. After 24 hours, SEAP activity was monitored and the level of Flagellin in the supernatant was measured a purified Flagellin standard (n=3, error bars = SD).
(E) Representative histograms of CD4+ T cells engineered with the α-CD19 synNotch receptor controlling the expression of PD-L1 and IL-10 after co-culture with CD19+ or CD19- K562s for 24 hrs. These data were used to calculate the percentage of cells expressing PD-L1 and intracellular IL-10 in Figure 5D (representative of 3 replicates).
Figure S6. synNotch Receptor T cells Locally Produce Cytokines and Therapeutic Agents in vivo (related to main text figure 6).
(A) Two dimensional dot plots of CD4+ (left plot) and CD8+ (right plot) primary human T cells transduced with the α-CD19 synNotch Gal4VP64 receptor (myc tagged) and 5×Gal4 response elements controlling expression of IL-2 IRES mCherry. The response element vector also contains a PGK promoter that drives constitutive expression of BFP to identify the T cells with the inserted response elements. T cells in the red shaded box were sorted and used for all in vivo and in vitro experiments.
(B) Histograms of IL-2 IRES mCherry reporter levels in tumor and spleen infiltrated CD8+ synNotch T cells injected i.v. showing selective expression of the mCherry reporter in target CD19+ tumors (data representative of the same 3 replicate mice in Figure 6B).
(C) Quantification of the percentage of tumor infiltrated CD4+ and CD8+ synNotch T cells that induced expression of the mCherry reporter of IL-2 expression from replicate data shown in Figure 6B and panel B (n=3, significance determined by Student’s t-test *** p<0.001 and **** p<0.0001).
(D) Quantification of the percentage CD4+ and CD8+ synNotch T cells that induced expression of the mCherry reporter of IL-2 expression after in vitro co- culture with CD19+ or CD19= K562s (n=3, significance determined by Student’s t-test *** p≤0.001 and **** p≤0.0001).
(E) Two dimensional dot plots showing the percentage of CD4+ and CD8+ T cells in representative single cell suspensions generated from the harvested spleen and CD19+ and CD19- tumors from a mouse i.v. injected with synNotch T cells controlling the expressing of IL-2 (same mouse analyzed in Figure 6A and 6B and panel B).
(F) CD4+ and CD8+ synNotch T cells were directly injected into both the CD19- and CD19+ tumor 8 days after tumor implantation. Tumors were harvested at day 10 and infiltrating T cells were analyzed for IL-2 mCherry reporter expression by flow cytometry. Histograms of IL-2 IRES mCherry reporter levels in tumor infiltrated CD4+ and CD8+ synNotch T cells injected intratumorally, showing selective expression of the mCherry reporter in target CD19+ tumors are given (data representative of 3 replicate mice).
(G) Bilateral CD19+ and GFP/CD19+ K562 tumor growth curves in mice treated with untransduced control CD4+ and CD8+ primary human T cells. The single antigen CD19+ tumor and dual antigen GFP/CD19+ tumor grew out at the same rate. The growth curve of the single antigen CD19+ from mice treated with CD4+ and CD8+ engineered with the α-GFP synNotch receptor controlling Blinatumomab (α-CD19/CD3 BiTE) expression is provided as a reference showing minimal slowed growth (n.s. at day 19) compared to control conditions (n=5 mice, error = SEM, significance determined by Student’s t-test n.s. = p≥0.05 ** p≤0.01).