Abstract
Klebsiella pneumoniae is a gram-negative bacterium that causes numerous diseases, including pneumonia and urinary tract infections. An increase in multidrug resistance has complicated the treatment of these bacterial infections, and although tigecycline shows activity against a broad spectrum of bacteria, resistant strains have emerged. In this study, the whole genomes of two clinical and six laboratory-evolved strains were sequenced to identify putative mutations related to tigecycline resistance. Of seven tigecycline-resistant strains, seven (100%) had ramR mutations, five (71.4%) had lon mutations, one (14.2%) had a ramA mutation, and one (14.2%) had an rpsJ mutation. A higher fitness cost was observed in the laboratory-evolved strains but not in the clinical strains. A transcriptome analysis demonstrated high expression of the ramR operon and acrA in all tigecycline-resistant strains. Genes involved in nitrogen metabolism were induced in the laboratory-evolved strains compared with the wild-type and clinical strains, and this difference in nitrogen metabolism reflected the variation between the laboratory-evolved and the clinical strains. Complementation experiments showed that both the wild-type ramR and the lon genes could partially restore the tigecycline sensitivity of K. pneumoniae. We believe that this manuscript describes the first construct of a lon mutant in K. pneumoniae, which allowed confirmation of its association with tigecycline resistance. Our findings illustrate the importance of the ramR operon and the lon and rpsJ genes in K. pneumoniae resistance to tigecycline.
Introduction
Klebsiella pneumoniae is a gram-negative bacterium of the Enterobacteriaceae family [1] that can cause numerous diseases, including pneumonia, urinary tract infections, septicemia, and pyogenic live abscesses [2]. Resistance of K. pneumoniae to carbapenems is increasing worldwide [3], and this rise in multidrug resistance has limited the available treatment options for this bacterium, which currently include only colistin, tigecycline, aminoglycosides, and fosfomycin [4]. Moreover, strains resistant to tigecycline have been reported [5–8].
Tigecycline belongs to the glycylcycline family of antibiotics, which consists of drugs modified from minocycline, and has bacteriostatic activity against a broad spectrum of gram-positive and gram-negative bacteria [9]. Compared with tetracycline, tigecycline exhibits increased affinity for the ribosome due to its interaction with 16S rRNA, and this increased affinity proves helpful for overcoming TetM-mediated resistance [10]. Resistance to tigecycline is mainly attributed to overproduction of the AcrAB-TolC efflux pump, which is regulated by RamA in K. pneumoniae [7]. ramA transcription is de-repressed by the ramR mutation in K. pneumoniae [5], and both rarA and marA provide alternate pathways for RamA-independent tigecycline resistance [11]. Moreover, a mechanism for tigecycline resistance independent of the AcrAB-TolC pump has also been identified; mutations in rpsJ encoding ribosomal protein S10 and kpgABC encoding a putative transporter are associated with AcrAB-TolC-independent tigecycline resistance [6, 8].
In this study, we combined whole-genome sequencing and RNA-Seq to identify putative mutations related to tigecycline resistance in both clinical and laboratory-evolved strains of K. pneumoniae. Mutations in the ramR, lon, ramA and rpsJ genes were observed in the tigecycline-resistant strains. In addition, the fitness costs associated with the mutants were detected to predict the risk of the bacteria spreading in the environment. A transcriptome analysis demonstrated that the ramR locus was highly expressed in all tigecycline-resistant strains. To confirm the role of ramR and lon in tigecycline resistance in K. pneumoniae, we performed a complementation experiment and constructed a knockout strain.
Materials and Methods
Bacterial isolates and antimicrobial susceptibility testing
The bacteria evaluated in this study included the clinical isolates XH209 and XH210 [12], the laboratory-evolved mutants XH211-XH216 and two gene-knockout mutants (ramR XH872 and lon XH889) of K. pneumoniae (Table 1). Strain XH209 was isolated from the blood of a patient in Hangzhou China who was at the beginning of tigecycline treatment, and a strain isolated after the patient received tigecycline treatment was denoted XH210. The MICs were determined by broth microdilution with cation-adjust Mueller-Hinton (MH) broth or by Etest (bioMérieux, Marcy l'Etoile, France) on MH agar, and the results were interpreted according to the CLSI or EUCAST breakpoints. The bacteria were cultured in Luria-Bertani (LB) or MH (Oxoid, Basingstoke, UK) medium at 37°C. Hygromycin and apramycin were added to the media to final concentrations of 100 mg/L and 50 mg/L, respectively, as necessary.
Table 1. Strains and plasmids used in this study.
| Strain/plasmid | Isolate day | Parental strain | genotype | Other genetic changes | Reference |
|---|---|---|---|---|---|
| XH209 | 0 | NA | NA | NA | this study |
| XH210 | 1 | XH209 | ramR Q122* | NA | this study |
| XH211 | 13 | XH209 | ramA Q72L, lon Q317* ramR Δ190 bp (322–511) | cspE N57K | this study |
| XH212 | 13 | XH209 | PramR +G, lon D445V, rpsJ V57L | tetA I235F, 300 kb dup | this study |
| XH213 | 13 | XH209 | ramR A40T, lon R33W, rpoC Δ18 bp (634–651) | eutL E95Q | this study |
| XH214 | 13 | XH209 | ramR L58P, rpoC G336A | this study | |
| XH215 | 13 | XH209 | ramR Q135*, lon Δ 9 bp (791–799), rpoC S263Y | yfiR C89Y hypo K302T | this study |
| XH216 | 13 | XH209 | ramR S29*, lon N417K | Mobile element protein G12E | this study |
| XH490 | 1 | XH209 | ramR Q122* | ND | this study |
| XH491 | 1 | XH209 | ramR T42Ins (8 bp) | ND | this study |
| XH492 | 1 | XH209 | ramR S137* | ND | this study |
| XH493 | 1 | XH209 | ramR A49Ins (8 bp) | ND | this study |
| XH494 | 1 | XH209 | ramR M1V | ND | this study |
| XH495 | 1 | XH209 | ramR W185* | ND | this study |
| XH496 | 1 | XH209 | ramR F45Del (8 bp) | ND | this study |
| XH497 | 1 | XH209 | ramR A2FS | ND | this study |
| XH498 | 1 | XH209 | ramR F45Ins (8 bp) | ND | this study |
| XH499 | 1 | XH209 | ramR R107H | ND | this study |
| XH500 | 1 | XH209 | ramR W89L | ND | this study |
| XH501 | 1 | XH209 | ramR A37V | ND | this study |
| XH502 | 1 | XH209 | ramR W89* | ND | this study |
| XH503 | 1 | XH209 | ramR T119P | ND | this study |
| XH504 | 1 | XH209 | ramR K5FS | ND | this study |
| XH505 | 1 | XH209 | ramR A105G | ND | this study |
| XH466 | XH210 | XH210 /pCR2.1-T vector | this study | ||
| XH468 | XH210 | XH210 /pCR2.1-ramR | this study | ||
| XH539 | XH211 | XH211 /pCR2.1-T vector | this study | ||
| XH540 | XH211 | XH211 /pCR2.1-lon | this study | ||
| XH583 | XH211 | XH211 /pCR2.1-ramR | this study | ||
| XH585 | XH211 | XH211 /pCR2.1-lon-ramR | this study | ||
| XH541 | XH212 | XH212 /pCR2.1-T vector | this study | ||
| XH593 | XH212 | XH212 /pCR2.1-ramR | this study | ||
| XH542 | XH212 | XH212 /pCR2.1-lon | this study | ||
| XH587 | XH212 | XH212 /pCR2.1-lon-ramR | this study | ||
| XH396 | XH213 | XH213 /pCR2.1-T vector | this study | ||
| XH398 | XH213 | XH213 /pCR2.1-ramR | this study | ||
| XH579 | XH213 | XH213 /pCR2.1-lon | this study | ||
| XH589 | XH213 | XH213 /pCR2.1-lon-ramR | this study | ||
| XH448 | XH214 | XH214 /pCR2.1-T vector | this study | ||
| XH452 | XH215 | XH215 /pCR2.1-T vector | this study | ||
| XH450 | XH214 | XH214 /pCR2.1-ramR | this study | ||
| XH581 | XH215 | XH215 /pCR2.1-lon | this study | ||
| XH591 | XH215 | XH215 /pCR2.1-lon-ramR | this study | ||
| XH456 | XH216 | XH216 /pCR2.1-T vector | this study | ||
| XH454 | XH215 | XH215 /pCR2.1-ramR | this study | ||
| XH544 | XH216 | XH216 /pCR2.1-lon | this study | ||
| XH568 | XH216 | XH216 /pCR2.1-lon-ramR | this study | ||
| XH872 | XH209 | ΔramR::apr | this study | ||
| XH889 | XH209 | Δlon::apr | this study | ||
| XH478 | XH209 | XH209 /pACBSR-Hyg | this study | ||
| plasmid | |||||
| pCR2.1-T vector | Thermo Fisher Scientific | ||||
| pCR2.1-ramR | pCR2.1-T vector carrying wild-type ramR | this study | |||
| pCR2.1-lon | pCR2.1-T vector carrying wild-type lon | this study | |||
| pCR2.1-lon-ramR | pCR2.1-T vector carrying wild-type ramR and lon | this study | |||
| pIJ773 | Template for amplification of the apramycin resistance gene | Pep Charusanti | |||
| pACBSR-Hyg | A p15A replicon plasmid containing an arabinose-inducible λ-Red recombinase and hygromycin resistance selection marker | Pep Charusanti |
Note:
*: stop codon;
Δ: Deletion; Ins: Insertion; Del: Deletion; FS: frame shift;
Laboratory evolution of tigecycline-resistant mutants
Six independent single colonies of K. pneumoniae XH209 were grown overnight at 37°C, and the cultures were diluted in LB broth with a serially increasing concentration of tigecycline. The concentration of tigecycline was started at a value equal to 1/2 MIC and doubled every 24 h. The overnight cultures were stored at -80°C for further experiments and analysis [13].
The overnight cultures of K. pneumoniae XH209 were plated on LB plates containing 4 mg/L tigecycline. Mutants were randomly selected from the plates after incubation at 37°C for 24 h and then streaked onto LB plates. The colonies were stored in LB medium with 15% glycerol. The ramR gene of the mutants was amplified by PCR and Sanger sequencing [14].
Homology modeling
RamR structure homology modeling was performed with the SWISS-MODEL workspace using the structure of RamR from Salmonella Typhimurium (PDB ID: 3VVX) as a template [15]. The 3D structure of the RamR protein was visualized using the PyMOL molecular graphics system, and the positions of the mutations were labeled with the corresponding amino acids.
Whole-genome DNA sequencing and analysis
Bacteria from a single colony were cultured overnight at 37°C in MH broth. Genomic DNA was extracted using a QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA) following the protocol recommended by the manufacturer. Agarose gel electrophoresis and a NanoDrop spectrophotometer were used to determine the quality and quantity of the extracted genomic DNA, respectively. The 300-bp library used for Illumina paired-end sequencing was constructed using 5 μg of genomic DNA from the two clinical strains and six laboratory-evolved mutants. In addition, an 8-kb mate-pair library was prepared for XH209 to complete its genome [16]. The raw Illumina data were de novo assembled using IDBA-Hybrid [17]. The pre-assembled contigs were arranged into scaffolds using SSPACE [18], and gaps within the scaffolds were closed with GapFiller [19]. Mapping and SNP detection were performed using the CLC Genomics Workbench (CLC bio, Aarhus, Denmark). The regions containing the detected SNPs were amplified by PCR using the primers listed in S1 Table. The PCR products were sent to Biosune (Hangzhou, China) for Sanger sequencing.
Growth rate measurement
Four independent cultures of each strain were grown overnight and diluted to 1:1000 in LB, and four replicates of each culture were aliquoted into a flat-bottom 96-well plate. The plate was incubated at 37°C with agitation, and the OD600 of each culture was determined every 5 min for 16 h using a BioTEK Synergy plate reader (BioTEK, Winooski, VT, USA). The growth rate was estimated based on the OD600 curves using R script [20].
RNA-Seq and transcript analysis
The wild-type and mutant strains were grown overnight in 2 mL of LB broth at 37°C. The overnight cultures were diluted 1:100 in 50 mL of LB broth and incubated at 37°C with shaking for 2 h. The bacteria were pelleted at 4°C, and after grinding in liquid nitrogen, total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). Then, 10 U of RNase-free DNase I (Promega, Mannheim, Germany) was added to the samples, and the RNA was purified through phenol-chloroform extraction. The RNA quality and quantity were determined by 1.0% formaldehyde denaturing agarose gel electrophoresis and a NanoDrop ND-1000 spectrophotometer, respectively. rRNA removal and RNA sequencing were performed as previously described [21] by staff at Zhejiang Tianke (Hangzhou, China). The raw data from the samples were analyzed using Subread [22, 23], and the raw counts of each sample were normalized and processed using the EdgeR Bioconductor package [24]. Genes with adjusted p-values (BH method) less than 0.05 and presenting at least two-fold differences in expression were considered to be differentially expressed.
Complementation experiment
Plasmids carrying wild-type ramR or lon were constructed and then introduced into laboratory-evolved resistant strains of K. pneumoniae by electroporation. Briefly, a region including the open reading frame of ramR or lon, derived from the XH209 genome sequence, was cloned into the pCR 2.1 vector (Invitrogen, USA). The plasmid containing the ramR gene and/or lon gene was then transferred into the resistant strains. The empty vector (pCR 2.1 vector) was also introduced into the resistant strains as a control. The MIC for tigecycline of the transformants were determined by broth microdilution with MH broth.
Gene knockout
Mutant ramR and lon genes were constructed as previously described [25]. In brief, the pIJ773 plasmid was used as the template for amplification of an apramycin resistance cassette, and the pACBSR-Hyg plasmid was used for arabinose-inducible λ–Red recombination. The knockout cassette was amplified from the FRT-flanked ApraR cassette of pIJ773 using response primers (Table 1). The PCR-amplified knockout cassette was then transformed into K. pneumoniae XH209+PACBSR-Hyg, and the transformants were screened overnight in LBApra at 37°C. The loss of pACBSR-Hyg was screened by streaking onto LBApra and low-salt LB + hygromycin plates overnight at 37°C. PCR and Sanger sequencing were performed to confirm the correct insertion of the knockout cassette.
Results
Clinical strains and in vitro selection of mutants with tigecycline resistance
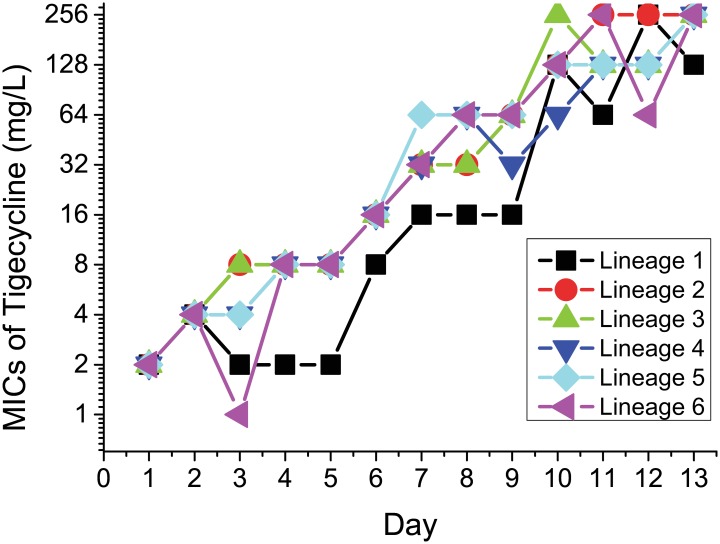
We obtained two K. pneumoniae strains that were isolated from the blood of a patient during tigecycline treatment. The K. pneumoniae tigecycline MICs increased from 2 mg/L (XH209) to 8 mg/L (XH210). Six independent colonies of XH209 were also selected at increased concentrations of tigecycline. After 13 days of serial passage (every 24 h), we obtained six tigecycline-resistant mutants, and the observed resistance to tigecycline increased in a step-wise manner (fold-increases compared with the MIC of K. pneumoniae XH209) following tigecycline passaging (Fig 1). The MICs of the six selected mutants ranged from 64 to 256 mg/L.
Fig 1. Resistance to tigecycline increased in a stepwise manner (as a fold increase over the MIC of K. pneumoniae XH209) following serial passage in tigecycline.
Putative resistance mutations and their fitness costs
The whole genomes of the clinical isolates and in vitro selection mutants were sequenced to identify mutations that are potentially responsible for resistance to tigecycline. The most commonly observed SNPs were nucleotide substitutions resulting in amino acid changes or stop codons (Table 2). A mutation in ramR was found in seven strains. In K. pneumoniae, the ramR gene encodes a repressor of ramA, which is known to be associated with resistance to tigecycline and ciprofloxacin [5]. RamA is a positive global regulator of the AcrAB efflux system [14]. Five strains harbored a lon mutation, and a mutation in rpoC was detected in three strains. In addition, a 287-kb duplication was observed in the genome of K. pneumoniae XH212. The biological fitness costs of the clinical isolates and in vitro selection mutants were also measured based on their relative growth rates compared with that of XH209. Fitness costs ranging from 2% to 58% were observed in most of the strains, and the costs showed a good correlation with the lag time. Notably, the clinical isolates showed the lowest fitness costs.
Table 2. Characterization of laboratory-evolved tigecycline-resistant K. pneumoniae and single-step selection of K. pneumoniae mutants.
| Strain | Parental strain | Putative mutation(s) causing reduced susceptibility to TGC | Other genetic changes | TGC MIC (mg/L) | TGC MIC (mg/L) +PaβN (50 mg/L) | Relative growth rate | Lag time (min) | TET | CHL | AK | CTX | CIP | IPM | NI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| XH209 | NA | NA | NA | 2 | 2 | 1 | 120 | >256 | >256 | 2 | >256 | 0.75 | 4 | 64 |
| XH210 | XH209 | ramR Q122* | NA | 16 | 8 | 0.98 | 113.44 | >256 | >256 | 2 | >256 | 4 | 4 | 192 |
| XH211 | XH209 | ramA Q72L, lon Q317* ramR Δ190 bp (322–511) | cspE N57K | 128 | 8 | 0.42 | 232 | >256 | >256 | 1 | 1.5 | 3 | 0.125 | 3 |
| XH212 | XH209 | PramR +G, lon D445V, rpsJ V57L | tetA I235F, 300 kb dup | >256 | 256 | 0.59 | 183.75 | >256 | >256 | 1.5 | >256 | 6 | 4 | 16 |
| XH213 | XH209 | ramR A40T, lon R33W, rpoC Δ18 bp (634–651) | eutL E95Q | >256 | 64 | 0.46 | 223.44 | >256 | >256 | 0.5 | 128 | 3 | 4 | 8 |
| XH214 | XH209 | ramR L58P, rpoC G336A | 64 | 32 | 0.74 | 189.06 | >256 | >256 | 0.75 | >256 | 2 | 1 | 128 | |
| XH215 | XH209 | ramR Q135*, lon Δ9 bp (791–799), rpoC S263Y | yfiR C89Y hypo K302T | 256 | 64 | 0.56 | 187.19 | >256 | >256 | 0.75 | 64 | 3 | 2 | 4 |
| XH216 | XH209 | ramR S29*, lon N417K | Mobile element protein G12E | 64 | 16 | 0.55 | 241.25 | >256 | >256 | 0.75 | 128 | 2 | 6 | 4 |
| XH490 | XH209 | ramR Q122* | ND | 16 | ND | 0.98 | 123.75 | >256 | >256 | 1.5 | >256 | 3 | 8 | 128 |
| XH491 | XH209 | ramR T42Ins (8 bp) | ND | 8 | ND | 0.98 | 120 | >256 | >256 | 1 | >256 | 3 | 6 | 128 |
| XH492 | XH209 | ramR S137* | ND | 16 | ND | 0.98 | 113.44 | >256 | >256 | 1 | >256 | 3 | 3 | 192 |
| XH493 | XH209 | ramR A49Ins (8 bp) | ND | 16 | ND | 0.97 | 232 | >256 | 64 | 1 | >256 | 3 | 3 | >512 |
| XH494 | XH209 | ramR M1V | ND | 16 | ND | 0.99 | 183.75 | >256 | >256 | 1 | >256 | 2 | 4 | 96 |
| XH495 | XH209 | ramR W185* | ND | 16 | ND | 0.97 | 223.44 | >256 | 32 | 1.5 | >256 | 4 | 6 | 128 |
| XH496 | XH209 | ramR F45Del (8 bp) | ND | 16 | ND | 0.88 | 189.06 | >256 | >256 | 1.5 | >256 | 2 | 1 | 32 |
| XH497 | XH209 | ramR A2FS | ND | 8 | ND | 0.98 | 187.19 | >256 | >256 | 1.5 | >256 | 2 | 6 | 64 |
| XH498 | XH209 | ramR F45Ins (8 bp) | ND | 16 | ND | 0.99 | 241.25 | >256 | >256 | 1.5 | >256 | 2 | 3 | 128 |
| XH499 | XH209 | ramR R107H | ND | 16 | ND | 0.99 | 123.75 | >256 | >256 | 1.5 | >256 | 2 | 3 | 64 |
| XH500 | XH209 | ramR W89L | ND | 16 | ND | 0.98 | 123.75 | >256 | >256 | 2 | >256 | 3 | 2 | 96 |
| XH501 | XH209 | ramR A37V | ND | 8 | ND | 0.99 | 122.5 | >256 | >256 | 1.5 | >256 | 3 | 4 | 128 |
| XH502 | XH209 | ramR W89* | ND | 8 | ND | 0.97 | 115.31 | >256 | >256 | 1.5 | >256 | 3 | 6 | 128 |
| XH503 | XH209 | ramR T119P | ND | 8 | ND | 0.97 | 114.38 | >256 | >256 | 1.5 | >256 | 3 | 4 | 192 |
| XH504 | XH209 | ramR K5FS | ND | 8 | ND | 0.98 | 123.44 | >256 | >256 | 1.5 | >256 | 3 | 4 | 128 |
| XH505 | XH209 | ramR A105G | ND | 8 | ND | 0.97 | 124.06 | >256 | >256 | 1.5 | >256 | 3 | 4 | 128 |
TGC: tigecycline; TET: tetracycline; CHL: chloramphenicol; AK: amikacin; CTX: cefotaxime; CIP: ciprofloxacin; IPM: imipenem;NI: nitrofurantoin. Note: NA: not found;ND: not detect;
*: Stop codon;
Δ: Deletion; Ins: Insertion; Del: Deletion; FS: Frameshift;
Up-regulation of the ram locus in tigecycline-resistant mutants
In this study, we selected the genes exhibiting at least a two-fold change in expression level in the mutants compared with the wild-type XH209 strain. In total, seven (0.14%), 118 (2.42%), 82 (1.68%), 69 (1.41%), 55 (1.13%), 73 (1.50%) and 30 (0.61%) genes had increased expression in XH210, XH211, XH212, XH213, XH214, XH215 and XH216, respectively. Two (0.04%), 44 (0.90%), 152 (3.11%), 47 (0.96%), 87 (1.78%), 41 (0.84%) and 78 (1.60%) genes showed reduced expression in these strains, respectively. The up-regulation of seven genes was observed in all seven strains. The annotations and reads per kilobase per million mapped reads (RPKM) values are listed in Table 3. These genes can be divided into two groups: one group includes the ram locus (the ramR-romA-ramA genes) and the efflux pump acrA, and the other group includes gsiA and entE. The gsiA gene encodes an ATP-binding protein of a glutathione importer [26], and EntE is an enzyme involved in the enterobactin biosynthesis pathway [27].
Table 3. Differentially expressed genes in laboratory-evolved strains compared with wild-type and clinical strains.
| Gene | Gene | Product | XH210 | XH211 | XH212 | XH213 | XH214 | XH215 | XH216 |
|---|---|---|---|---|---|---|---|---|---|
| up-regulated | |||||||||
| LQ47_01505 | phospholipid ABC transporter substrate-binding protein | 2.2* | 4.0 | 4.3 | 3.5 | 3.4 | 3.5 | 3.8 | |
| LQ47_01510 | ABC transporter substrate-binding protein | 2.1* | 4.0 | 4.2 | 3.9 | 3.1 | 3.6 | 4.1 | |
| LQ47_01515 | phospholipid ABC transporter substrate-binding protein | 2.8* | 4.5 | 4.7 | 4.5 | 4.1 | 4.5 | 4.9 | |
| LQ47_08715 | nitrate/nitrite sensor protein NarX | -0.5 | 3.0 | 2.9 | 3.5 | 3.0 | 2.7 | 2.9 | |
| LQ47_08720 | nitrate/nitrite transporter NarK | -1.1 | 6.2 | 4.9 | 5.5 | 4.9 | 5.2 | 5.9 | |
| LQ47_08730 | nitrate reductase | -0.7 | 5.1 | 4.5 | 5.2 | 4.6 | 4.3 | 5.1 | |
| LQ47_08735 | nitrate reductase | -0.3 | 5.3 | 4.5 | 5.5 | 4.8 | 4.3 | 5.3 | |
| LQ47_08740 | nitrate reductase | -1.0 | 4.4 | 4.3 | 4.9 | 4.9 | 3.9 | 4.8 | |
| LQ47_08745 | nitrate reductase | 2.6* | 7.1 | 6.6 | 7.1 | 6.7 | 6.5 | 7.3 | |
| LQ47_16215 | sensor protein BasS/PmrB | 1.9* | 4.0 | 3.3 | 4.2 | 3.9 | 3.8 | 4.0 | |
| LQ47_22580 | transcriptional regulator | 3.6* | 4.9 | 6.4 | 5.0 | 4.9 | 5.1 | 5.6 | |
| down-regulated | |||||||||
| LQ47_02160 | hypothetical protein | -1.1* | -10.4 | -6.8 | -5.9 | -6.7 | -7.4 | -8.9 | |
| LQ47_04290 | hydrogenase 3 membrane subunit | -0.5 | -4.9 | -4.1 | -6.3 | -6.8 | -5.9 | -4.7 | |
| LQ47_04295 | hydrogenase 3 large subunit | -0.2 | -5.2 | -4.5 | -6.2 | -6.7 | -5.9 | -4.9 | |
| LQ47_04310 | formate hydrogenlyase maturation protein HycH | -0.6 | -5.5 | -3.7 | -6.2 | -5.9 | -6.1 | -4.6 | |
| LQ47_04315 | hydrogenase 3 maturation protease | -0.2 | -6.8 | -5.5 | -5.6 | -10.9 | -10.4 | -5.6 | |
| LQ47_04680 | fimbrial protein | 0.1 | -5.5 | -8.2 | -7.1 | -5.7 | -10.8 | -7.1 | |
| LQ47_09405 | formate dehydrogenase | -0.3 | -5.8 | -4.0 | -5.2 | -5.4 | -4.7 | -5.0 | |
| LQ47_09545 | acetoin reductase | -0.5 | -7.5 | -4.8 | -7.5 | -6.4 | -7.8 | -7.1 | |
| LQ47_09550 | acetolactate synthase | -0.4 | -6.3 | -4.5 | -6.3 | -7.9 | -6.7 | -5.3 | |
| LQ47_11630 | hypothetical protein | -3.9* | -5.1 | -10.3 | -7.7 | -10.3 | -6.2 | -7.8 | |
| LQ47_12010 | methionine synthase | -2.4 | -5.4 | -8.5 | -5.1 | -7.4 | -6.3 | -6.0 | |
| LQ47_23900 | 5,10-methylenetetrahydrofolate reductase | -1.5 | -4.8 | -5.7 | -6.0 | -4.6 | -6.1 | -5.2 | |
| Common | |||||||||
| LQ47_04775 | acrA | acriflavin resistance protein AcrA | 14.9 | 19.3 | 14.3 | 15.1 | 17.5 | 15.3 | 16.8 |
| LQ47_10655 | gsiA | glutathione ABC transporter ATP-binding protein | 16.4 | 6.3 | 6.2 | 7.0 | 6.9 | 13.4 | 14.6 |
| LQ47_17285 | entE | enterobactin synthase subunit E | 10.6 | 16.5 | 17.2 | 10.2 | 10.8 | 19.9 | 18.2 |
| LQ47_17585 | ramA | transcriptional regulator | 5.2 | 6.3 | 5.3 | 7.1 | 5.7 | 6.8 | 8.2 |
| LQ47_17590 | romA | beta-lactamase | 6.4 | 14.3 | 15.2 | 13.6 | 14.1 | 6.8 | 8.0 |
| LQ47_17595 | ramR | TetR family transcriptional regulator | 4.9 | 3.6 | 5.1 | 4.9 | 6.0 | 4.3 | 4.9 |
| LQ47_22005 | membrane protein | 4.2 | 3.8 | 5.1 | 5.3 | 3.6 | 5.3 | 4.9 |
*: differentially expressed genes in XH210, clinical isolate.
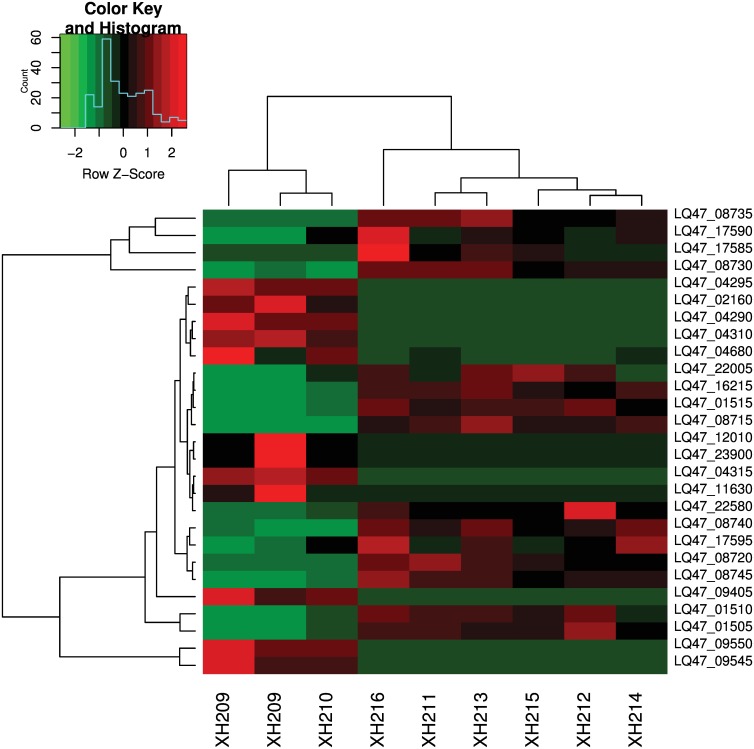
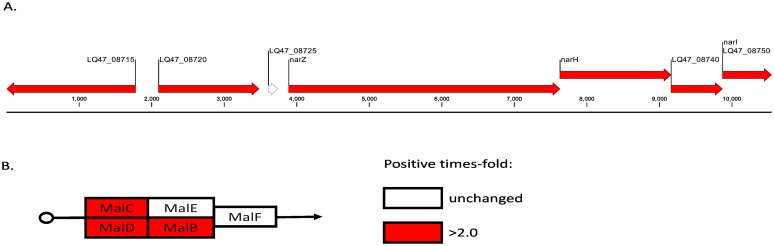
To investigate differences between the clinical and the laboratory-evolved strains, we selected genes that were differentially expressed in the laboratory-evolved strains but not in the clinical strains (Fig 2). A total of 23 genes were differentially expressed only in the laboratory strains, and these included 11 up-regulated and 12 down-regulated genes (Table 3). After mapping the genes to pathways, we found that several genes involved in nitrogen metabolism were up-regulated (Fig 3A). In addition, ABC transporters were also induced (Fig 3B).
Fig 2. Heatmap of differentially expressed genes in the laboratory-evolved strains but not in the clinical strains.
Fig 3. Comparison of the transcriptional profiles of genes in the laboratory-evolved strains with those of the wild-type and clinical strains.
A). Changes in the transcription of genes involved in nitrogen metabolism between the laboratory-evolved strains and the wild-type and clinical strains. B). ABC transporters were induced in the laboratory-evolved strains. All values show the fold-change differences. The genes depicted in white were not differentially regulated.
Mutation in ramR was dominant in the single-step tigecycline resistance evolution experiment
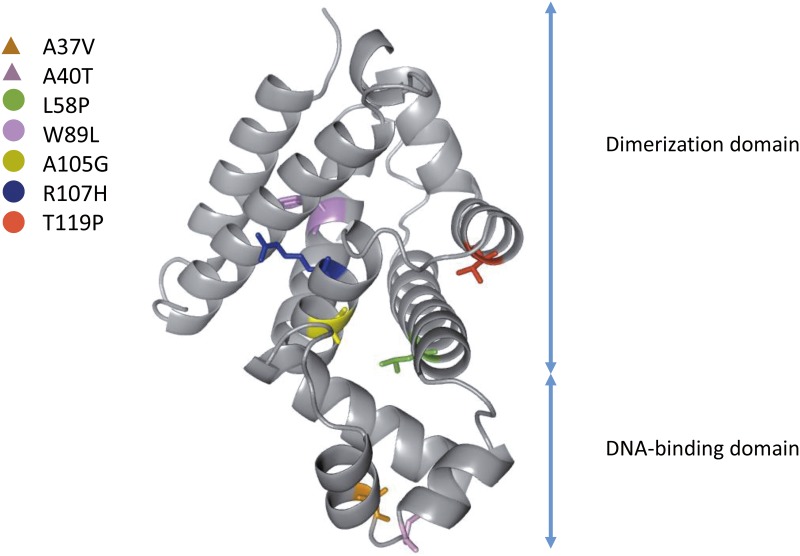
Twenty mutants were obtained through single-step evolution experiments. The Sanger sequencing results for the ramR gene in the mutants obtained from the single-step evolution experiments showed that 80% (16/20) of the strains harbored a mutation in ramR, including base substitutions, frameshifts, insertions and deletions (Table 2). The tigecycline MICs of the mutants ranged from 8 to 16 mg/L, and the fitness costs ranged from 1 to 12%. Notably, only one strain showed a fitness cost of 12%, whereas the fitness costs of the other strains were not greater than 3%. The structure of K. pneumoniae RamR was subjected to homology modeling using the SWISS-MODEL workspace, and the mutation sites in the structure were labeled (Fig 4): five mutations were found to be located in the dimerization domain, and two mutations were localized in the DNA-binding domain.
Fig 4. Homology modeling of K. pneumoniae RamR.
The mutation sites are mapped onto the structure of RamR, and the amino acids are labeled.
Tigecycline-resistant mutants showed cross-resistance to other antibiotics
To test the influence of the mutations on the response of these bacteria to other antibiotics, six different antibiotics belonging to several major classes (tetracycline, chloramphenicol, amikacin, cefotaxime, ciprofloxacin and imipenem) were tested (Table 2). The MICs of the mutants for ciprofloxacin increased from 0.75 mg/L to 2–6 mg/L, which might be caused by up-regulation of an efflux pump gene, acrA. Furthermore, XH211 became sensitive to beta-lactams, including cefotaxime and imipenem.
The role of ramR and lon in tigecycline resistance was confirmed by complementation and gene knockout
To confirm the roles of ramR and lon in tigecycline resistance, we cloned the wild-type ramR and lon genes into the pCR2.1-T vector and introduced the plasmids into the tigecycline-resistant mutants. Tigecycline sensitivity was restored in the XH210 strain carrying the ramR plasmid but not in bacteria carrying the empty vector. Analysis of the in vitro selection mutants revealed that the plasmid carrying ramR or lon only partially restored sensitivity to tigecycline (Table 4). It should be noted that the resistant mutant that presented only a partial restoration of sensitivity after transfection of the plasmid harboring both ramR and lon showed mutations in other genes, such as ramA, rpsJ and rpoC. This result might indicate the involvement of ramA, rpsJ and rpoC in tigecycline resistance.
Table 4. Complementation experiment.
| TGC MIC (mg/L) | Wild type | pCR2.1-T vector | pCR2.1-ramR | pCR2.1-lon | pCR2.1-lon-ramR | |
|---|---|---|---|---|---|---|
| Strain | Genotype | |||||
| XH210 | ramR Q122* | 8 | 8 | 4 | ||
| XH211 | ramA Q72L lon Q317* ramR Δ190 bp (322–511) | 128 | 64 | 16 | 4 | 16 |
| XH212 | rpsJ V57L lon D445V PramR +G | 256 | 256 | 256 | 128 | 64 |
| XH213 | ramR A40T lon R33W rpoC Δ18 bp (634–651) | 128 | 128 | 4 | 64 | 32 |
| XH214 | ramR L58P rpoC G336A | 64 | 64 | 8 | ||
| XH215 | ramR Q135* lon Δ9 bp (791–799) rpoC S263Y | 256 | 128 | 8 | 16 | 32 |
| XH216 | ramR S29* lon N417K | 64 | 64 | 32 | 64 | 32 |
Note:
*: stop codon;
Δ: deletion; bp: base pair; NA: no mutation.
The roles of ramR and lon in tigecycline resistance in K. pneumoniae were also verified by gene knockout. The ramR and lon genes were knocked out in the XH209 strain, and the resulting mutants displayed higher tigecycline resistance than the wild-type strain, although the tigecycline MIC of the ramR mutant was higher than that of the lon mutant (Table 5). Moreover, the relative growth rate of the ramR and lon mutants were measured, and both showed slower growth in MH medium compared with the wild-type strain.
Table 5. Tigecycline MICs and relative growth rates of K. pneumoniae XH209 and its isogenic mutants.
| Strain | Genotype | TGC MIC (mg/L) | Relative growth rate | |
|---|---|---|---|---|
| Broth | E-test | |||
| XH209 | wt | 2 | 1 | 100.0 |
| XH872 | ΔramR::apr | 16 | 12 | 93.6 |
| XH889 | Δlon::apr | 8 | 3 | 96.3 |
Discussion
In this study, we found that the MICs for tigecycline increased in a step-wise manner with the presence of mutations in the ramR operon and the lon and rpsJ genes. Our transcriptional analysis results showed that the ramR operon is highly expressed in all seven tigecycline-resistant K. pneumoniae strains, indicating that the ramR operon plays an important role in tigecycline resistance in K. pneumoniae. The ramR gene, located upstream of ramA, encodes a transcriptional repressor belonging to the TetR family, and a mutation in ramR leads to the overexpression of ramA [28, 29]. This regulation is achieved via the binding of RamR to the promoter of ramA [30]. Nonsynonymous mutations in ramR are reported with high frequency in tigecycline-non-susceptible K. pneumoniae clinical isolates [7]. We also identified base substitutions, insertions and deletions in the ramR gene (7/7) in K. pneumoniae, confirming these previous findings. These results indicate that ramR mutation is a common mechanism involved in tigecycline resistance.
The Lon protease is involved in the degradation of MarA in Escherichia coli [31]. A loss-of-function mutation in lon would lead to higher concentrations of MarA, which would increase expression of the AcrAB efflux pump. We detected three different types of point mutations in the lon gene, and complementation and gene knockout experiments demonstrated that lon mutants exhibited higher resistance to tigecycline than wild-type K. pneumoniae. Inactivation of lon is involved in the mechanism of tigecycline resistance in E. coli and S. Typhimurium [32, 33]. To the best of our knowledge, this study includes the first construction of a lon mutant in K. pneumoniae, which allowed confirmation of the association of mutations in this gene with tigecycline resistance. A transcript analysis showed that XH211, XH212, XH215 and XH216 presented higher expression levels of oqxAB compared with the wild-type strain. These results suggest that RarA and OqxAB play an important role in laboratory-evolved tigecycline-resistant strains [34], whereas the expression of oqxAB might be regulated by lon in all four strains that harbor lon mutations.
RpsJ is thought to act as a general target of tigecycline adaption and a marker for alterations in antibiotic resistance in bacteria [35]. The protein encoded by the rpsJ gene is a component of the 30S ribosomal subunit and participates in the formation of a BoxA-binding module [36]. Villa et al. reported an amino acid substitution of V57L in K. pneumoniae rpsJ [8], and our results confirmed the presence of this amino acid substitution in this gene. The V57L mutation might cause weaker binding of tigecycline to 16S rRNA, leading to tigecycline resistance [8]. The S10 mutation has also been reported in Enterococcus faecium, E. coli, Staphylococcus aureus, Streptococcus pneumoniae and Acinetobacter baumannii [35, 37, 38]. However, we did not achieve rpsJ knockout in K. pneumoniae. In addition, all attempts to achieve allelic replacement at this locus in E. coli, A. baumannii and E. faecium have failed [35, 39]. This failure could be due to the essential role of S10 in translation and transcription.
Overall, the dominant genetic mutations associated with tigecycline resistance in K. pneumoniae were found in the ramR, lon and rpsJ genes. Furthermore, the ramR locus was found to be highly expressed in all tigecycline-resistant strains. A higher fitness cost was observed in the laboratory-evolved strains but not in the clinical strains. We found differences in the transcriptional changes between the laboratory-evolved tigecycline-resistant mutants and the clinical tigecycline-resistant isogenic strains. Complementation experiments and knockout construction confirmed the roles of ramR and lon in tigecycline resistance in K. pneumoniae. We believe that we are the first to construct a lon mutant in K. pneumoniae, which allowed us to confirm its association with tigecycline resistance. These results suggest that the ramR operon and the lon and rpsJ genes play central roles in tigecycline resistance in K. pneumoniae.
Nucleotide Sequence Accession Numbers
The nucleotide sequences of XH209 have been deposited at DDBJ/EMBL/GenBank under the accession number CP009461. The whole-genome shotgun sequencing results for XH210, XH211, XH212, XH213, XH214, XH215 and XH216 have been deposited at DDBJ/EMBL/GenBank under the accession numbers JUGC00000000, JTEA00000000, JTEB00000000, JTGO00000000, JTJA00000000, JUBD00000000 and JUBE00000000, respectively.
Supporting Information
(DOCX)
Acknowledgments
We thank Dr. Pep Charusanti (University of California) for the plasmids used for gene knockout in K. pneumoniae. Part of this manuscript was presented as an abstract at the 7th International Congress of the Asia Pacific Society of Infection Control, Taipei, Taiwan, March 26–29, 2015.
Data Availability
The nucleotide sequences of XH209 have been deposited at DDBJ/EMBL/GenBank under the accession number CP009461. The whole-genome shotgun sequencing results for XH210, XH211, XH212, XH213, XH214, XH215 and XH216 have been deposited at DDBJ/EMBL/GenBank under accession numbers JUGC00000000, JTEA00000000, JTEB00000000, JTGO00000000, JTJA00000000, JUBD00000000 and JUBE00000000.
Funding Statement
This work was supported by the National Natural Science of China (81230039, 81401708 and 81101284), the Natural Science Foundation of Zhejiang Province, China (LY15H190004), and the Zhejiang Province Medical Platform Backbone Talent Plan (2016DTA003). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ramos PI, Picao RC, Almeida LG, Lima NC, Girardello R, Vivan AC, et al. Comparative analysis of the complete genome of KPC-2-producing Klebsiella pneumoniae Kp13 reveals remarkable genome plasticity and a wide repertoire of virulence and resistance mechanisms. BMC genomics. 2014;15:54 10.1186/1471-2164-15-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broberg CA, Palacios M, Miller VL. Klebsiella: a long way to go towards understanding this enigmatic jet-setter. F1000prime reports. 2014;6:64 10.12703/P6-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braykov NP, Eber MR, Klein EY, Morgan DJ, Laxminarayan R. Trends in resistance to carbapenems and third-generation cephalosporins among clinical isolates of Klebsiella pneumoniae in the United States, 1999–2010. Infection control and hospital epidemiology: the official journal of the Society of Hospital Epidemiologists of America. 2013;34(3):259–68. 10.1086/669523 . [DOI] [PubMed] [Google Scholar]
- 4.van Duin D, Kaye KS, Neuner EA, Bonomo RA. Carbapenem-resistant Enterobacteriaceae: a review of treatment and outcomes. Diagnostic microbiology and infectious disease. 2013;75(2):115–20. 10.1016/j.diagmicrobio.2012.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hentschke M, Wolters M, Sobottka I, Rohde H, Aepfelbacher M. ramR mutations in clinical isolates of Klebsiella pneumoniae with reduced susceptibility to tigecycline. Antimicrobial agents and chemotherapy. 2010;54(6):2720–3. 10.1128/AAC.00085-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen LE, Snesrud EC, Onmus-Leone F, Kwak YI, Aviles R, Steele ED, et al. IS5 element integration, a novel mechanism for rapid in vivo emergence of tigecycline nonsusceptibility in Klebsiella pneumoniae. Antimicrobial agents and chemotherapy. 2014;58(10):6151–6. 10.1128/AAC.03053-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheng ZK, Hu F, Wang W, Guo Q, Chen Z, Xu X, et al. Mechanisms of tigecycline resistance among Klebsiella pneumoniae clinical isolates. Antimicrobial agents and chemotherapy. 2014;58(11):6982–5. 10.1128/AAC.03808-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villa L, Feudi C, Fortini D, Garcia-Fernandez A, Carattoli A. Genomics of KPC-producing Klebsiella pneumoniae sequence type 512 clone highlights the role of RamR and ribosomal S10 protein mutations in conferring tigecycline resistance. Antimicrobial agents and chemotherapy. 2014;58(3):1707–12. 10.1128/AAC.01803-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noskin GA. Tigecycline: a new glycylcycline for treatment of serious infections. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2005;41 Suppl 5:S303–14. 10.1086/431672 . [DOI] [PubMed] [Google Scholar]
- 10.Jenner L, Starosta AL, Terry DS, Mikolajka A, Filonava L, Yusupov M, et al. Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(10):3812–6. 10.1073/pnas.1216691110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veleba M, Schneiders T. Tigecycline resistance can occur independently of the ramA gene in Klebsiella pneumoniae. Antimicrobial agents and chemotherapy. 2012;56(8):4466–7. 10.1128/AAC.06224-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hua X, Chen Q, Li X, Feng Y, Ruan Z, Yu Y. Complete genome sequence of Klebsiella pneumoniae sequence type 17, a multidrug-resistant strain isolated during tigecycline treatment. Genome announcements. 2014;2(6):e01337–14. 10.1128/genomeA.01337-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McAleese F, Petersen P, Ruzin A, Dunman PM, Murphy E, Projan SJ, et al. A novel MATE family efflux pump contributes to the reduced susceptibility of laboratory-derived Staphylococcus aureus mutants to tigecycline. Antimicrobial agents and chemotherapy. 2005;49(5):1865–71. 10.1128/AAC.49.5.1865-1871.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruzin A, Visalli MA, Keeney D, Bradford PA. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrobial agents and chemotherapy. 2005;49(3):1017–22. 10.1128/AAC.49.3.1017-1022.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamasaki S, Nikaido E, Nakashima R, Sakurai K, Fujiwara D, Fujii I, et al. The crystal structure of multidrug-resistance regulator RamR with multiple drugs. Nature communications. 2013;4:2078 10.1038/ncomms3078 . [DOI] [PubMed] [Google Scholar]
- 16.Gutman BA, Wang Y, Yanovsky I, Hua X, Toga AW, Jack CR Jr., et al. Empowering imaging biomarkers of Alzheimer's disease. Neurobiology of aging. 2014. 10.1016/j.neurobiolaging.2014.05.038 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng Y, Leung HC, Yiu SM, Chin FY. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28(11):1420–8. 10.1093/bioinformatics/bts174 . [DOI] [PubMed] [Google Scholar]
- 18.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27(4):578–9. 10.1093/bioinformatics/btq683 . [DOI] [PubMed] [Google Scholar]
- 19.Boetzer M, Pirovano W. Toward almost closed genomes with GapFiller. Genome biology. 2012;13(6):R56 10.1186/gb-2012-13-6-r56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linkevicius M, Sandegren L, Andersson DI. Mechanisms and fitness costs of tigecycline resistance in Escherichia coli. The Journal of antimicrobial chemotherapy. 2013;68(12):2809–19. 10.1093/jac/dkt263 . [DOI] [PubMed] [Google Scholar]
- 21.Hua X, Chen Q, Li X, Yu Y. Global transcriptional response of Acinetobacter baumannii to a subinhibitory concentration of tigecycline. International journal of antimicrobial agents. 2014;44(4):337–44. 10.1016/j.ijantimicag.2014.06.015 . [DOI] [PubMed] [Google Scholar]
- 22.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic acids research. 2013;41(10):e108 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30. 10.1093/bioinformatics/btt656 . [DOI] [PubMed] [Google Scholar]
- 24.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang TW, Lam I, Chang HY, Tsai SF, Palsson BO, Charusanti P. Capsule deletion via a lambda-Red knockout system perturbs biofilm formation and fimbriae expression in Klebsiella pneumoniae MGH 78578. BMC Res Notes. 2014;7:13 10.1186/1756-0500-7-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Xiang Q, Wang G, Wang H, Zhang Y. Optimizing expression and purification of an ATP-binding gene gsiA from Escherichia coli k-12 by using GFP fusion. Genetics and molecular biology. 2011;34(4):661–8. 10.1590/S1415-47572011005000043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gehring AM, Bradley KA, Walsh CT. Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry. 1997;36(28):8495–503. 10.1021/bi970453p . [DOI] [PubMed] [Google Scholar]
- 28.Abouzeed YM, Baucheron S, Cloeckaert A. ramR mutations involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrobial agents and chemotherapy. 2008;52(7):2428–34. 10.1128/AAC.00084-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenblum R, Khan E, Gonzalez G, Hasan R, Schneiders T. Genetic regulation of the ramA locus and its expression in clinical isolates of Klebsiella pneumoniae. International journal of antimicrobial agents. 2011;38(1):39–45. 10.1016/j.ijantimicag.2011.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baucheron S, Coste F, Canepa S, Maurel MC, Giraud E, Culard F, et al. Binding of the RamR repressor to wild-type and mutated promoters of the RamA gene involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrobial agents and chemotherapy. 2012;56(2):942–8. 10.1128/AAC.05444-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffith KL, Shah IM, Wolf RE Jr. Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Molecular microbiology. 2004;51(6):1801–16. . [DOI] [PubMed] [Google Scholar]
- 32.Linkevicius M, Anderssen JM, Sandegren L, Andersson DI. Fitness of Escherichia coli mutants with reduced susceptibility to tigecycline. The Journal of antimicrobial chemotherapy. 2016;71(5):1307–13. 10.1093/jac/dkv486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicoloff H, Andersson DI. Lon protease inactivation, or translocation of the lon gene, potentiate bacterial evolution to antibiotic resistance. Mol Microbiol. 2013;90(6):1233–48. 10.1111/mmi.12429 . [DOI] [PubMed] [Google Scholar]
- 34.De Majumdar S, Veleba M, Finn S, Fanning S, Schneiders T. Elucidating the regulon of multidrug resistance regulator RarA in Klebsiella pneumoniae. Antimicrobial agents and chemotherapy. 2013;57(4):1603–9. 10.1128/AAC.01998-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beabout K, Hammerstrom TG, Perez AM, Magalhaes BF, Prater AG, Clements TP, et al. The Ribosomal S10 Protein Is a General Target for Decreased Tigecycline Susceptibility. Antimicrobial agents and chemotherapy. 2015;59(9):5561–6. 10.1128/AAC.00547-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo X, Hsiao HH, Bubunenko M, Weber G, Court DL, Gottesman ME, et al. Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex. Molecular cell. 2008;32(6):791–802. 10.1016/j.molcel.2008.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cattoir V, Isnard C, Cosquer T, Odhiambo A, Bucquet F, Guerin F, et al. Genomic analysis of reduced susceptibility to tigecycline in Enterococcus faecium. Antimicrobial agents and chemotherapy. 2014. 10.1128/AAC.04174-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lupien A, Gingras H, Leprohon P, Ouellette M. Induced tigecycline resistance in Streptococcus pneumoniae mutants reveals mutations in ribosomal proteins and rRNA. The Journal of antimicrobial chemotherapy. 2015. 10.1093/jac/dkv211 . [DOI] [PubMed] [Google Scholar]
- 39.Cattoir V, Isnard C, Cosquer T, Odhiambo A, Bucquet F, Guerin F, et al. Genomic analysis of reduced susceptibility to tigecycline in Enterococcus faecium. Antimicrobial agents and chemotherapy. 2015;59(1):239–44. 10.1128/AAC.04174-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
The nucleotide sequences of XH209 have been deposited at DDBJ/EMBL/GenBank under the accession number CP009461. The whole-genome shotgun sequencing results for XH210, XH211, XH212, XH213, XH214, XH215 and XH216 have been deposited at DDBJ/EMBL/GenBank under accession numbers JUGC00000000, JTEA00000000, JTEB00000000, JTGO00000000, JTJA00000000, JUBD00000000 and JUBE00000000.