Abstract
In a fibroblast colony model of corneal stromal development, we asked how physiological tension influences the patterning dynamics of fibroblasts and the orientation of deposited extracellular matrix (ECM). Using long-term live-cell microscopy, enabled by an optically accessible mechanobioreactor, a primary human corneal fibroblast colony was cultured on three types of substrates: a mechanically biased, loaded, dense, disorganized collagen substrate (LDDCS), a glass coverslip, and an unloaded, dense, disorganized collagen substrate (UDDCS). On LDDCS, fibroblast orientation and migration along a preferred angle developed early, cell orientation was correlated over long distances, and the colony pattern was stable. On glass, fibroblast orientation was poorly correlated, developed more slowly, and colony patterns were metastable. On UDDCS, cell orientation was correlated over shorter distances compared with LDDCS specimens. On all substrates, the ECM pattern reflected the cell pattern. In summary, mechanically biasing the collagen substrate altered the early migration behavior of individual cells, leading to stable emergent cell patterning, which set the template for newly synthesized ECM.
Introduction
In vertebrate animals, there is substantial, unequivocal, and long-established evidence that applied mechanical forces are necessary for appropriate patterning, growth, maintenance, and remodeling of musculoskeletal tissues.1–8 In culture, it is known that mechanical stimulation can alter vertebrate fibroblast orientation,9,10 extracellular matrix (ECM) molecule synthesis,11–14 migration,15 growth,16,17 and differentiation.18 Mechanical force is also critical to the development and refinement of tendon constructs grown under mechanical stress.19 In the ocular system, reduction of pressure during development markedly slows the expansion of the corneoscleral envelope, resulting in a small eye.20 It is becoming clear that mechanical forces are potent guidance cues for many developing collagenous tissues.
The cornea is an integral part of the tough ocular tunic and its stromal ECM is a structurally complex mesenchymal tissue that is highly ordered on the nanoscale. The organizational control is necessary to permit the cornea to function mechanically and to act as the principal refracting, transparent optical component in the eye.21 In humans, the stromal ECM has evolved a highly regular arrangement of collagen fibrils with nearly monodisperse diameters (30.8 ± 0.8 nm) and regular spacing (∼31.0 nm, surface-to-surface) that persists in aligned layers over length scales much larger than the cells themselves.22 Evidently, exquisite local and global control mechanisms must be applied and integrated over time and space to synthesize and then organize the continuous collagen network. Unfortunately, very little is known about how the cornea (or any other connective tissue) is built.23 Corneal development is an excellent model to investigate the process of organized matrix production.24
In culture, primary human corneal fibroblasts (PHCFs) form highly proliferative synthetic colonies in the presence of fetal bovine serum and stabilized vitamin C.25,26 In addition, it has been shown that corneal stromal keratocytes retain neural crest progenitor plasticity and can recapitulate multiple tissues even after overt differentiation.27 PHCF colonies cultured in a scaffold-free, unloaded tissue engineering model have been shown to synthesize a thick, locally organized, corneal stromal analog.25
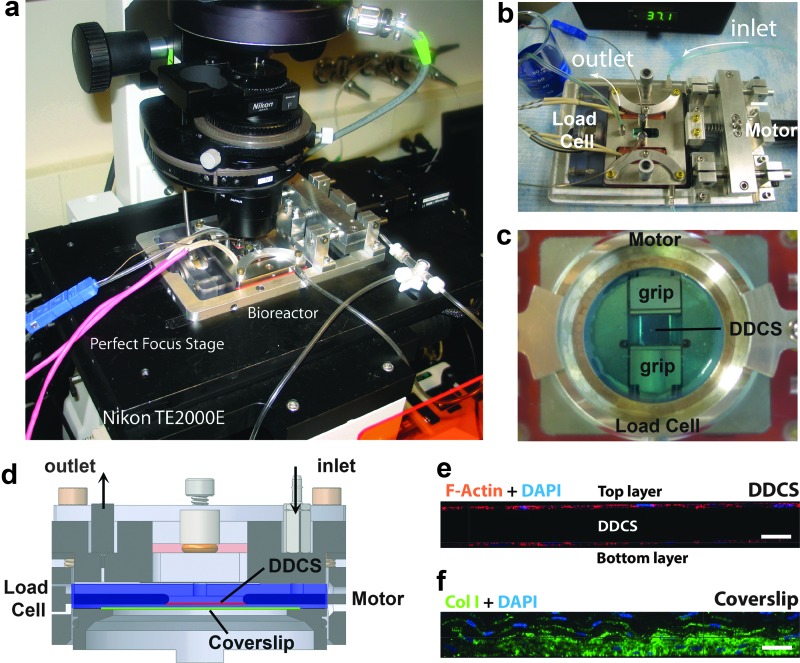
In this investigation, we have combined a PHCF culture system with a custom mechanobioreactor28 to open a remarkable series of windows into the minute-by-minute behavior of human fibroblasts while they initially populate common culture substrates, self-organize, and then synthesize ECM. The device (Fig. 1a, b) permits sustained long-term optical and mechanical probing of dual-cell colonies grown on a dense, disorganized collagen substrate (DDCS) that is suspended between two tissue grips or grown directly on the coverslip below (Fig. 1c, d). The mechanobioreactor also permits direct uniaxial mechanical stimulation of the DDCS at any time during the experimental series. Viable cell culture conditions can be maintained for sufficient time (weeks) to permit confluent culture formation on both sides of the suspended substrate and on the glass coverslip, which then synthesize collagenous ECM (Fig. 1e, f).
FIG. 1.
Experimental apparatus. (a) The custom mechanobioreactor was mounted on a Nikon TE2000E inverted microscope outfitted with a perfect focus (PFS®) automated stage for the duration of the experiment (up to 2 weeks). (b) The mechanobioreactor allowed the perfusion of cell culture media through the inlet and outlet port and temperature was maintained at 37°C. (c) Inside the culture chamber, the DDCS was clamped between two grips positioned near the glass. (d) The bioreactor cross section shows the position of cells cultured on the DDCS (red line) between the grips as well as the glass coverslip (green line) below the DDCS inside the culture chamber (blue shading). The motor on the inlet side of the chamber allowed the application of mechanical strain to the DDCS. Transverse section of a confocal immunofluorescence image of the tissue at the end of the experiment showed (e) multilayers of cells and collagen extracellular matrix on glass coverslip and (f) single layer of cells on the top and bottom of the DDCS. DDCS, dense, disorganized collagen substrate. Color images available online at www.liebertpub.com/tea
The dynamic PHCF migration data we present here have been derived from live cell imaging experiments, which generated a vast number of images (58,576 images), constituting 448 days of continuous imaging if laid end to end. Additional static images for the correlation analysis comprise large-scale spatial mosaics taken across seven experimental series.
Materials and Methods
Experimental design
In all experiments, a dense, disorganized collagen substrate (DDCS; 6 mm width and 40 ± 5 μm thickness) was mounted to the mechanobioreactor grips to obtain a grip-to-grip length of 8.0 ± 0.5 mm. The bioreactor was filled with cell culture medium enriched with 10% fetal bovine serum, 1% antibiotic/antimycotic (mixture of 10 mg/mL streptomycin and 25 μL/mL amphotericin B), and dissolved CO2 and stabilized to 37°C. One million (1000 cells/μL) PHCFs, extracted from a 12-year-old donor cornea, suspended in cell culture medium were injected into the bioreactor through one of the two inlet ports. The cell culture medium was continuously supplied to the bioreactor at a rate of 8 μL/min, as described in Paten et al.28 On day 3, 0.05 mg/mL ascorbic acid was added to the medium.
In the loaded DDCS (LDDCS) experimental series (n = 3 with no live stain; n = 1 with live stain), a 0.01 N uniaxial load was applied to mechanically bias the DDCS before the addition of the PHCFs. To monitor the culture behavior, the principal imaging method was label-free differential interference contrast (DIC). DIC was chosen for its high sensitivity to index of refraction gradients (edges) and for its short exposure time. Live-cell fluorescence imaging (LCFI) was used on one experiment. At the end of day 6, an additional 6% strain was applied to the 0.01 N loaded samples to determine if increased strain altered the patterning. After day 12, the samples were removed from the mechanobioreactor for postprocessing. The experimental series comprised a cell coculture grown on both the LDDCS and a glass coverslip beneath the DDCS. Both cultures were monitored during the full experimental time course by continuous imaging at 6-min increments and at multiple locations.
In the unloaded DDCS (UDDCS) control series (n = 3), no load was applied to the DDCS. The DDCS was left to sag in the mechanobioreactor. Once every day, the UDDCS was briefly raised into the focal plane by applying the minimal tension that is sufficient to flatten the surface to image one location at the center of the specimen. The unloaded control series was run for 1 week to permit time for the PHCFs to reach confluence and establish a pattern. At the end of the experiment, a large panoramic image of the entire substrate was taken.
Fabrication of DDCS
Pepsin-extracted type I atelo-collagen solution (PureCol; Advanced Biomatrix) was mixed with 10 × phosphate-buffered saline (PBS) and neutralized to obtain collagen concentration of 2.4 mg/mL. Then, 10 mL of the solution was transferred into a 10,000 MWCO dialysis cassette (8–12 mL capacity) and left in a 37°C humidified incubator to polymerize for 3 h. After polymerization, the cassette was immersed in a 40% polyethylene glycol solution (MW = 35,000) at least overnight to dehydrate the substrate. The DDCS was cut into 6 × 15-mm strips. Immediately before starting the cell culture experiment in the mechanobioreactor, the DDCS strips were sterilized by immersing in 70% ethanol for 15 min. Subsequently, the strips were rinsed in sterile de-ionized (DI) water, 1× phosphate buffered saline (PBS), and 1× Dulbecco's modified Eagle medium (DMEM) to remove traces of ethanol. The DDCS surface comprised randomly oriented collagen fibrils with D-periodic banding (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/tea). The mechanical properties of the DDCS have been previously characterized.28,29
Extraction of PHCFs
PHCFs were extracted from a 12-year-old donor cornea using our standard cell extraction protocol.25 Briefly, the cornea was separated from the sclera using a sterilized razor blade and washed three times by 1× PBS in a sterilized dish. Then, the epithelium and endothelium layers were gently removed from the cornea using sterilized razor blades. The corneal stroma was cut into small pieces (2 × 2 mm) and washed three times in 1 × PBS before placing the pieces into six-well plates. Sterilized DMEM enriched with 10% fetal bovine serum (FBS) and 1% antibiotic (antibiotic/antimycotic; mixture of 10 mg/mL streptomycin and 25 μL/mL amphotericin B; Cellgro, Fisher Scientific) was added to the wells, and then the plates were stored in an incubator for 2 weeks. Every day, half of the DMEM volume was gently extracted from each well and replaced with fresh enriched DMEM. When the PHCFs reached confluence in the six-well plates, they were trypsinized, cultured in sterilized dishes, and expanded for 2 weeks. After three passages, the PHCFs were moved into sterilized culture flasks until the day of the experiment. For the experiment, the cells were detached from the flask by trypsinization and diluted to a final cell concentration of 1000 cells/μL. Only passages 3–6 were used in all experiments.
Detailed imaging protocol and LCFI
Detailed imaging protocol: The mechanobioreactor was mounted on an inverted microscope (Nikon TE 2000E; Nikon Instruments) outfitted with a focal plane stabilized (Nikon PFS®), automatic stage, and shutter control through NIS Elements Advanced Research software (version 3.2; Nikon), DIC/Nomarski imaging optical train, and epifluorescence package (comprising a Lumen Dynamics X-Cite 120 illumination system and UV-2A filter cube with EX 330–380, DM 400, BA 420, ET-DAPI filter cube). DIC imaging was conducted with an exposure time of 400 ms. For the LDDCS experiments, images were taken at multiple locations on the DDCS and on the glass coverslip. The image locations were chosen arbitrarily. However, there was always one local window near the center of the construct. The images were taken every 6 min for the experimental period. In at least one experimental series, panoramic images of the LDDCS and coverslip constructs were taken using DIC every day. For the UDDCS experiments, a similar setting was used. However, the images were only taken once every day at the center of the DDCS after it was briefly raised into the focal plane. Occasionally, a bubble would evolve or enter the chamber, obstructing an imaging window. Typically, this was immediately rectified by elevating one side of the bioreactor and gently flowing additional media through the system and letting the bubble out through the outlet.
Live-cell fluorescence imaging: The PHCF nuclei were labeled with Hoechst 33342 (trihydrochloride, trihydrate; 10 mg/mL solution in water) as follows: 1 μg/mL Hoechst 33342 solution in cell culture media was injected into the bioreactor chamber and left for 30 min without imaging. After the staining procedure, the chamber was flushed gently with preheated DMEM supplemented with 10% FBS, 1% antibiotic, and 0.05 mg/mL ascorbic acid and dissolved CO2. The PHCF cell staining procedure was repeated every 3 days to augment the fluorescent signal from the stained nuclei. While the dye flushing procedure has some potential to disrupt the system due to the increased shear rates, the total time of exposure to the higher shears was small in comparison with the length of the experiment (1.5 h of a total of 288 h or 0.5%).
Furthermore, there were three experiments that did not undergo chamber flushing, for which the orientation results were consistent with the results of the fluorescence experiments. Thus, it is unlikely that the dye procedure affected the cell organization or patterning behavior. When imaging, in addition to DIC, fluorescence images (excitation: 342 nm; emission: 483 nm) were also taken every hour at the same locations as the DIC on both the DDCS and on the glass coverslip. During the experiment, a long-pass filter (45 mm diameter long-pass UV Filter #54-052; Edmund Optics) was positioned above the condenser to minimize fluorescence excitation during DIC illumination.
Assessments
Dynamic cell culture metrics were extracted from the images acquired by both DIC and LCFI. The metrics extracted were cell speed, migration direction, and trajectories extracted from LCFI (Cell Speed, Migration Direction, and Trajectories from LCFI section), as well as cell orientation and directional correlation length from DIC (Analysis of Cell Orientation and Directional Autocorrelation from DIC section).
At the end of the experimental series, the cell-generated constructs deposited on both the LDDCS and coverslip were assessed for the production of important fibroblast and matrix components, including collagen type I, collagen type III, fibronectin, smooth muscle actin (SMA), and F-actin, using indirect immunofluorescence (IF) (Indirect Immunofluorescence section).
Cell speed, migration direction, and trajectories from LCFI
From the LCFI experiment, average cell speed and migration direction were calculated using particle imaging velocimetry (PIV) and cell trajectories were manually extracted.
Particle imaging velocimetry: For the PIV-based measurements, images (1392 × 1040; 8 bit; tiff format) where cell nuclei were fluorescently labeled using Hoechst 33342 taken from six locations, three on the coverslip and three on the DDCS, were preprocessed through a custom Matlab (version R2014b) algorithm using imadjust(), rgb2gray(), and imresize() to generate smaller cleaner images for processing (669 × 500; 8 bit; gray scale). To produce the vector fields, single two-image sequences (spaced 1 h apart) were imported into Fiji (imageJ-64bit) and processed with the Trackmate-2.5.4 plug-in. Contrast was increased through pixel saturation of 0.1–0.2%. The centers of cell nuclei were detected using a Laplacian of Gaussian filter with an estimated diameter size of 15 μm and a threshold between 0.2 and 0.5 based on the quality of the image. Spots were edited manually to ensure all cells were tracked and no particles had been selected.
The Simple LAP Tracker with linking and gap-closing max distances set to 45 μm was used to track the cell movement. Again, manual editing of obvious erroneous tracks ensured accurate results. Tracks were exported to an XML file for analysis. The cell displacements were imported into a custom postprocessing program written in Matlab (version R2014b). The displacements were used to extract a number of parameters from the cell migration fields, including average cell speed, average cell velocity, and maximum cell velocity. In addition, the individual cell displacement data were interpolated to generate local velocity fields for display. Supplementary Figure S2 demonstrates how sequential images were converted into display formats: (1) Windrose plots of cell direction, frequency, and velocity; (2) velocity vector field plots, overlaid on the original image; and (3) velocity direction heat maps overlaid with the velocity vector field.
Cell trajectories: Dynamic cell trajectories were measured in local windows either manually or using a field analysis algorithm in conjunction with PIV. Trajectories could only be obtained on the LCFI culture experiment. The individual cell trajectory data were extracted by manually locating the center of 10 cells over three 10-h windows during the culture period. The field data were extracted twice each day for an hour period each time.
Analysis of cell orientation and directional autocorrelation from DIC
Cell orientation: A set of linear transforms and filters were used to find the orientation of the cell population in a DIC image.30 Before the algorithm may be employed, the DIC images were cropped and made square. The spatial frequency spectrum was then computed using a two-dimensional discrete Fourier transform (MATLAB, 2015). A two-dimensional bandpass filter was used to remove noise, uneven lighting, and other irrelevant frequencies. A Radon transform was applied to the filtered frequency spectrum to create a sinogram. The angle corresponding to the peak of the sinogram was selected to be the most significant orientation of the cell population in the image.
Spatiotemporal analysis of cell orientation: The spatiotemporal analysis result presented in Figure 2 was generated from smaller DIC images (459 × 343 μm2) at three to five randomly chosen locations on each LDDCS (nspecimen = 4, nlocation = 15) and coverslip (nspecimen = 3, nlocation = 10) specimen every hour throughout the 12-day experiment. For each location, cell orientation was obtained using the method described above where a 198 × 198 μm2 central region of each image was used and a Gaussian filter centered at 27.4 μm−1 with a full-width half-maximum of 31.8 μm−1. Cell orientation was plotted in a polar plot where the radial distance is time. The angular variance at each time point was calculated using the definition of variance in circular statistics for a bimodal distribution,31,32
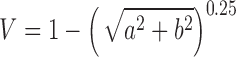 , where
, where 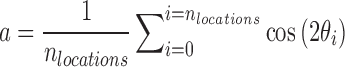 and
and 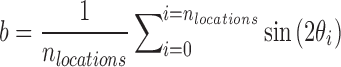 . In circular statistics, angular variance varies from 0 to 1, where
. In circular statistics, angular variance varies from 0 to 1, where 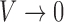 signifies an anisotropic orientation distribution, while
signifies an anisotropic orientation distribution, while 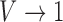 suggests a random distribution.
suggests a random distribution.
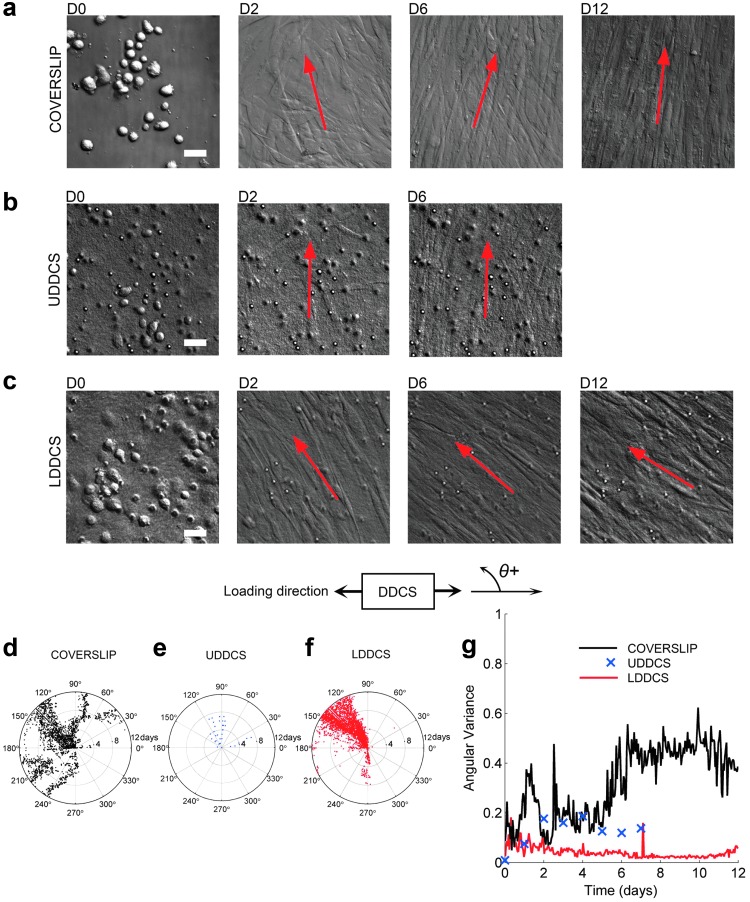
FIG. 2.
Local culture dynamics: orientation. Representative DIC image sequence of PHCF culture on (a) coverslip, (b) UDDCS, and (c) LDDCS shows that locally the cells align parallel to each other in a preferred direction. However, the LDDCS sample achieved the local alignment earlier in the experiment. The red arrows in (a–c) represent the calculated principal cell orientation of the corresponding image. The cell orientation angle on (d) glass coverslip (three experiments, 10 locations) shifts over the course of the experiment (12 days; time increases in radial direction), while cell orientation on (f) LDDCS (four experiments, 15 locations) was more stable. Cell orientation on (e) UDDCS (three experiments, five locations) over the course of 7 days was less stable than LDDCS. (g) The variance of cell orientation angles with respect to time in culture also confirms the enhanced stability of cell orientation on LDDCS compared with the coverslip specimens. Scale bars: 100 μm. DIC, differential interference contrast; LDDCS, loaded, dense, disorganized collagen substrate; UDDCS, unloaded, dense, disorganized collagen substrate. PHCF, primary human corneal fibroblast. Color images available online at www.liebertpub.com/tea
Directional autocorrelation: To study cell patterning on each type of surface, large panoramic DIC images of at least 1300 × 1300 μm2 at the center of the specimen were obtained at day 7. A cell orientation map from the panoramic DIC image was obtained by dividing the image into smaller 198 × 198 μm2 regions (Supplementary Fig. S3). For each region, cell orientation was obtained using the method described above. From the orientation map, directional autocorrelation was calculated.33,34 The general expression for a two-dimensional normalized cross-correlation is
 |
where 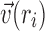 is the unit vector representing the general direction of cells at ri. Since the magnitude of all vectors is 1,
is the unit vector representing the general direction of cells at ri. Since the magnitude of all vectors is 1, 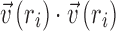 = 1. Therefore, Equation 1 can be simplified as follows:
= 1. Therefore, Equation 1 can be simplified as follows:
 |
Equation 2 was then modified to account for the diametrically bimodal orientation distribution of cells (i.e., cell oriented at 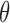 and
and 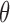 + 180° are equivalent).
+ 180° are equivalent).
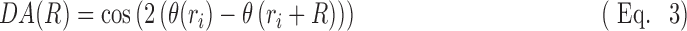 |
For each panoramic DIC image, 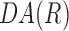 was calculated for all ri for
was calculated for all ri for 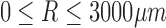 . Using Equation 3, when the cell orientation at two points is the same,
. Using Equation 3, when the cell orientation at two points is the same, 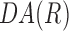 = 1, and when the difference between cell orientation is 90°,
= 1, and when the difference between cell orientation is 90°, 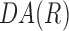 = −1. If cell orientation is random at R, the mean of
= −1. If cell orientation is random at R, the mean of 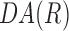 is close to 0. The following exponential equation,
is close to 0. The following exponential equation,
 |
was curve fitted to the 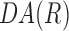 data points by minimizing the mean squared error (Supplementary Fig. S4). The variable, Ro, represents the characteristic correlation length of the cell orientation. A large value of Ro implies that cell orientation persists at a longer distance. Statistical differences for characteristic correlation length were determined using the Tukey–Kramer post hoc analysis of variance with a significance level of 5% in MATLAB.
data points by minimizing the mean squared error (Supplementary Fig. S4). The variable, Ro, represents the characteristic correlation length of the cell orientation. A large value of Ro implies that cell orientation persists at a longer distance. Statistical differences for characteristic correlation length were determined using the Tukey–Kramer post hoc analysis of variance with a significance level of 5% in MATLAB.
Indirect immunofluorescence
At the end of the experiment, the samples were fixed and IF was performed as previously described.35 Briefly, the constructs were fixed in 4% paraformaldehyde and incubated at 4°C overnight with the primary antibody—anti-type III collagen (1:40; Southern Biotech), anti-SMA (1:50; Dako North America), type I collagen (1:50; Abcam), and fibronectin (1:800; Sigma)—diluted in 1% bovine serum albumin (BSA) +0.1% Triton-X. The constructs were then washed and incubated overnight at 4°C with the corresponding secondary antibody—donkey anti-goat IgG (1:200, type III collagen), donkey anti-mouse IgG (1:200, SMA), donkey anti-rabbit IgG (1:200, type I collagen), and donkey anti-mouse IgM (1:200, fibronectin)—diluted in 1% BSA +0.1% Triton-X. Phalloidin–rhodamine (Invitrogen), which binds to the cell F-actin, was also used. Constructs were counterstained with TOPRO-3 iodide (1:1000; Invitrogen), a marker of cell nuclei. Negative controls, where the primary antibody was omitted, were run with all experiments. Constructs were washed, mounted with Vectashield Mounting Media (Vector Laboratories), observed, and photographed using a confocal TCS-SP2 Leica microscope (Leica Microsystems). When imaging type I collagen, the boundary between the cell-deposited collagen and the DDCS was easily distinguishable, even though the antibody stained both bovine and human collagen. This is because the density and general structure of the DDCS collagen were very distinctive.
Results
Substrate mechanics and biasing
The DDCS is a hydrated, viscoelastic, nonlinear collagenous biomaterial, which has an equilibrium elastic modulus similar to the native corneal stroma (1–5 MPa36), but possesses a surface comprising random fibril orientations (Supplementary Fig. S1). The glass coverslip provides an infinite modulus control. The small, static (0.01 N), uniaxial tensile force applied before cell seeding produced a strain of ∼1% and an initial stress magnitude of 40 kPa (the stress in the adult cornea is equibiaxial, static, and has a magnitude of ∼25 kPa). Due to the viscoelastic nature of the DDCS, the stress level is expected to decrease when the substrate was kept at a constant strain (stress relaxation). Therefore, we expect that the stress level was brought closer to or below the physiological stress level in the cornea. After day 6, an additional strain of 6% was applied to substantially strengthen the mechanical guidance cue.
Local dynamic behavior: proliferation, population, orientation, and migration
Proliferation and population
On the glass, the PHCFs proliferated extensively and populated the coverslip, eventually forming multiple layers in a few locations (e.g., Fig. 1f). The cells on the LDDCS grew to confluence, populated both surfaces of the substrate, but generally formed only one layer (Fig. 1e). In each experimental series, we were able to use label-free, long-term DIC imaging to open and continuously monitor multiple local windows on the dynamics of the cell culture system. In one representative experimental series, DIC imaging was overlaid with live-cell fluorescence imaging (LCFI-Hoechst nuclear stain) to capture the entire course of cell/colony movement in more quantitative detail (Supplementary Movies, S1 and S2). The dynamics of PHCFs indicated a highly active PHCF culture in which cell division, vesicle shedding, migration, and even cell death can be readily observed.
Local orientation dynamics
Local DIC images on the coverslip, UDDCS, and the LDDCS show markedly different initial morphology (day 2), but qualitatively similar final cell alignment uniformity and morphology (Fig. 2a–c). When the local orientation was tracked continuously across all experiments, the orientation angle did not generally converge to a consistent value on the glass or on the UDDCS (Fig. 2d, e), while on the surface of the LDDCS, the orientation converged toward 131.8° ± 5.5° (measured at day 6) (Fig. 2f). The application of the additional 6% strain after day 6 to the LDDCS only shifted the orientation angle to 133.4° ± 9.8° (measured at day 12) (Fig. 2f). The variance of the orientation angle was small and decreased consistently with time in culture only for the LDDCS specimens (Fig. 2g), indicating that the mechanical bias affected migration. Intriguingly, the PHCFs on the bottom surface of the LDDCS were nearly always oriented orthogonally to those on the top surface (Supplementary Movie S3). A similar effect has been noted in BHK fibroblasts and has been attributed to a form of rudimentary cellular vision.37 Cells cultured on UDDCS appeared to converge on inconsistent local orientations in the few windows we were able to obtain (shown in three windows in Supplementary Fig. S4 and Supplementary Movie S4).
Local migration dynamics
To more accurately investigate the influence of the substrate on local cell migration, an additional experiment was performed where the PHCF nuclei were fluorescently labeled (Supplementary Movies, S1 and S2). This experiment permitted the tracking of cell nuclei every hour over 12 days in three windows on the LDDCS and three windows on the glass coverslip. Velocity vector field maps of the colony (Fig. 3a, b) were generated for each window at each time point (Supplementary Movie S5). On the glass coverslip, a disorganized motion with variable velocity direction and magnitudes was observed early in the experiment, and by day 6, the velocity direction became bidirectional and the magnitudes decreased and became more uniform.
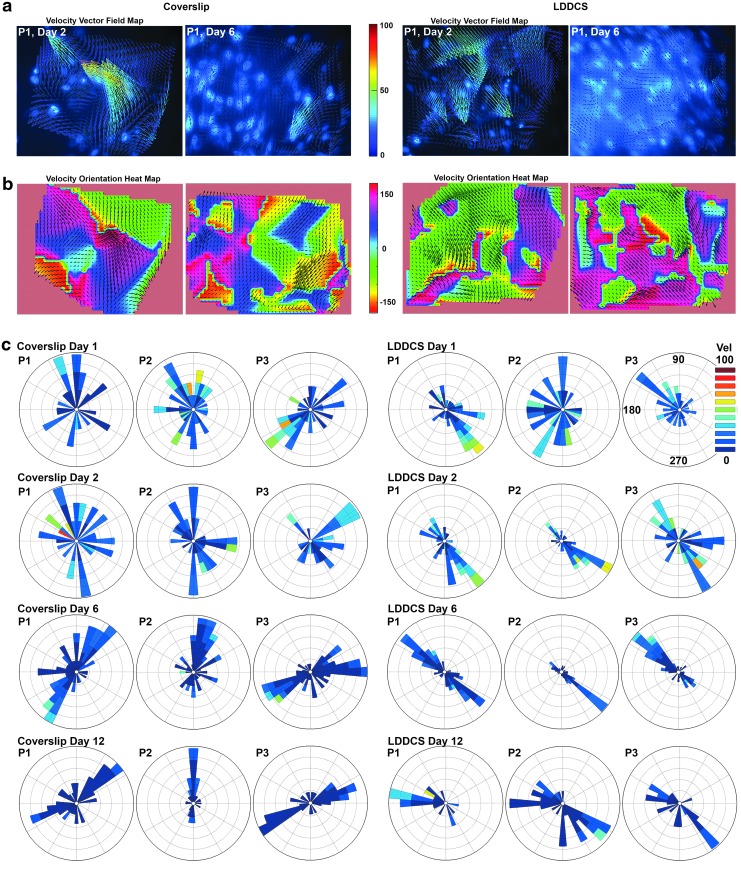
FIG. 3.
Local culture dynamics: velocity field. (a) Interpolated velocity vector field plots overlaid on the live nuclear stain images for a representative PHCF culture system on the glass coverslip and on LDDCS. The velocity vector field plots show that on the coverslip, at day 2, there was disorganized motion with variable velocity directions and magnitudes. On the LDDCS, however, it is already clear by day 2 that the cells had a preferred angle of migration. By day 6, the velocity fields on both the glass and the LDDCS have clearly slowed (shorter and bluer vector arrows) and have become bidirectional. The vector arrow's length reflects the velocity as does the color. Colorbar: velocity magnitude in μm/h. (b) Heat maps of the velocity direction overlaid with the velocity vector field for the culture system on the glass coverslip and on the LDDCS. The day 2 directional heat map of the coverslip shows many colors and multiple domains, indicating relatively random velocity directions and magnitudes. On the LDDCS, the heat map at day 2 shows large single-color domains that indicate that the culture migration angle is already fairly uniform and bidirectional. As the culture time progresses, the heat maps show that the LDDCS migration direction sharpens into a bidirectional flow of cells oriented on the same line at ∼135° and −45°. The coverslip culture still shows multiple velocity domains and colors. Colorbar: velocity direction in degrees. (c) Windrose plots of the velocity direction, speed, and frequency of the PHCF colony at three different positions on the coverslip and LDDCS and at four different time points. The windrose plots confirm the general trend from initial disorganized motion to a highly organized, bidirectional migration pattern on both the coverslip and on the DDCS. It is clear from the data, however, the DDCS induces a more rapid approach of the colony to a stable migration pattern and that the pattern of migration always converges to the preferred angle of ∼135° and −45°.
On the LDDCS, the magnitude of the velocity vectors also showed a decreasing trend (Fig. 3a), but the cells appeared to achieve the bidirectional motion earlier, as shown by the bimodal orientation distribution in day 2 (Fig. 3b, c). Consistent with the local orientation analysis from the DIC images, the direction of cell motion on LDDCS in the three windows analyzed converged to a similar orientation, along ∼135° and −45° (Fig. 3c, LDDCS day 6), while on the glass coverslip, the three windows showed cells migrating along different orientations (Fig. 3c). The average velocity magnitude (speed) on the coverslip and LDDCS declined from 8.7–19.7 μm/h to a stable value of 4.7–10.3 μm/h postconfluence and through matrix production (after day 6) (Fig. 4a). The decrease in cell speed of movement with time in culture has been attributed to the increase in the number of neighboring cells and therefore number of contacts with other cells.38 Similarly, in this experiment, the decrease in speed from seeding to day 6 was accompanied with the increase in the average number of cell nuclei, which can be viewed and tracked within the image (Fig. 4b). When cell speed stabilized at day 6, cell number also plateaued before it started to decrease on day 10. The decrease in the number of tracked nuclei near the end of this experiment may be caused by cell death from photobleaching or phototoxicity of fluorescence imaging.39 Individual cell tracking at days 2, 6, and 12 (Supplementary Movie S6) confirms the early onset of directional migration on the LDDCS. In one experiment, larger areas (3 × 3 panoramic window) were examined in an attempt to observe the initial migration dynamics of the PHCF colony. Supplementary Movie S7 clearly shows wave-type swarming invasion of fibroblasts, which are preferentially tracking at ∼135° before confluence. The movie also shows the early establishment of directional migration on the bottom side of the substrate at a nearly 90° angle to the top surface.
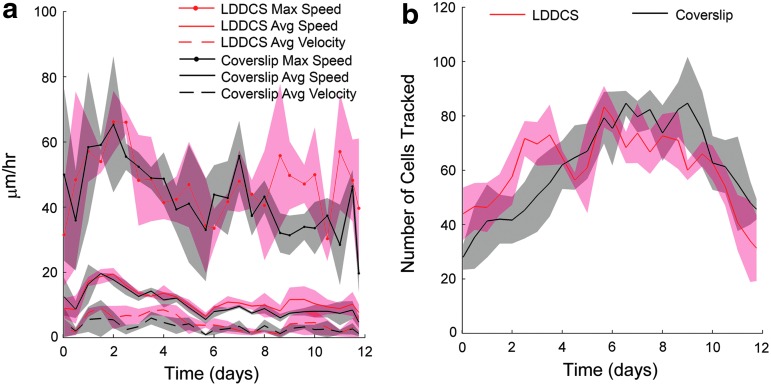
FIG. 4.
Local culture dynamics: speed and velocity distribution. The nucleus tracking analysis showed that (a) the distribution of cell speed as a function of length of time in culture follows a similar trend on LDDCS and coverslip. In both cases, the maximum and average cell speed decreased and stabilized around day 6. The average velocity, defined as the magnitude of the average velocity vector, was much lower than the average speed. This indicates the significant presence of cell bidirectional motion throughout the duration of the experiment. (b) The number of cell nuclei tracked in each window was also comparable on both substrates. Color images available online at www.liebertpub.com/tea
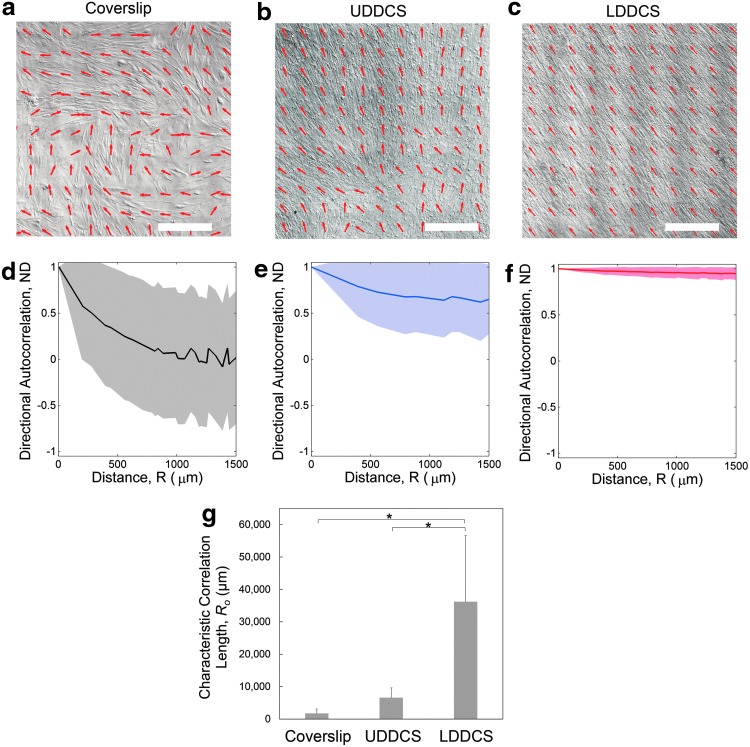
Global patterning behavior
Large-area panoramic differential interference contrast images and fast fourier transform-based cell orientation maps show differences in the colony patterning on LDDCS, UDDCS, and glass coverslip at day 6 (Fig. 5a–c). Autocorrelation analysis of cell orientation at the center of the specimen shows a more rapid decay of the directional correlation coefficient on glass and on the UDDCS compared with the LDDCS (Fig. 5d–f). The value of Ro for the LDDCS was statistically higher compared with the coverslip (p < 0.0001) as well as UDDCS (p = 0.0004) (Fig. 5g). This suggests that cell orientation on the LDDCS was significantly more uniform throughout the panoramic image analyzed. On the coverslip, discontinuities and abrupt changes in orientation of neighboring groups of cells were observed (Fig. 5a). This is consistent with the results of previous investigations by Elsdale and coworkers,40–42 which described the patterns of fibroblasts cultured to confluence as a patchwork of locally parallel fibroblasts. In contrast, the cell colony on the LDDCS specimens formed a consistent pattern with uniform orientation angle, without discontinuities.
FIG. 5.
Global organization and correlation length data. Representative mosaic DIC images of PHCFs on (a) glass coverslip, (b) UDDCS, and (c) LDDCS superimposed with the cell orientation field (red arrows) show the different patterning behavior of PHCFs on each substrate. Scalebar: 500 μm. The extent of variation in cell orientation was quantified by calculating the (d–f) directional autocorrelation (solid line and shading is average ± standard deviation). The correlation on glass coverslip dropped off rapidly with a large standard deviation, while the correlation on LDDCS stayed near one over long length scales. (g) The characteristic correlation length, found by fitting the directional autocorrelation data with an exponential decay equation, allowed statistical comparison of the directional autocorrelation using a numerical representation (n ≥ 3 for each group). *Statistically significant with p < 0.005. Color images available online at www.liebertpub.com/tea
ECM synthesis
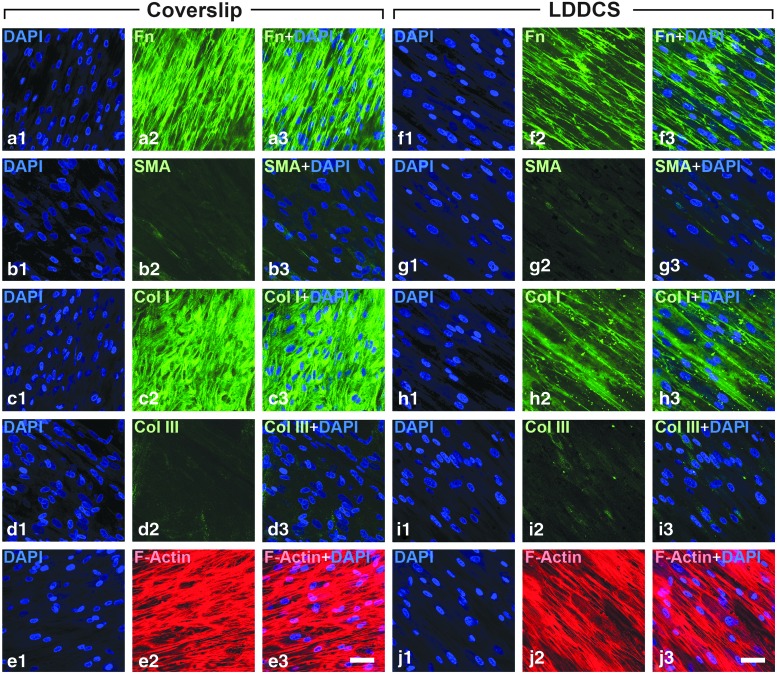
Because of the stability of the mechanobioreactor environment, it was possible to maintain the culture system long enough for the PHCFs to begin synthesis of an ECM. At the end of the experimental period, the constructs were examined for the expression of intracellular and extracellular components, which are indicators of a normal or fibrotic corneal fibroblast phenotype.25,43,44 We found that PHCFs on both substrates produced markers consistent with a normal, nonfibrotic culture system (Fig. 6). Fibronectin, collagen I, and F-actin were present, while collagen III and SMA were absent. Figure 6 also qualitatively shows the coalignment of the intra and ECM components with the direction of the cell long axis. We have observed this before in this PHCF culture system.26,45 The cell–matrix coalignment is striking on the LDDCS, where all intracellular matrix and ECM components are oriented in a similar direction. Alignment analysis on the immunostained images suggests that the ICM, ECM, and cell nuclei are coaligned (Supplementary Fig. S5). Figure 6 suggests that the PHCF population on the coverslip produced more ECM than the population on LDDCS. This is consistent with our previous data, which demonstrated that a collagen substrate may depress the assembly/retention of ECM in our PHCF culture system.45
FIG. 6.
Confocal immunofluorescence of cell internal and external matrix composition. The composition and organization of the cell internal matrix structure and secreted external matrix were imaged using confocal immunofluorescence at the end of the experiment. Three specimens from the coverslip and LDDCS group were stained for fibronectin [(a) coverslip; (f) LDDCS], smooth muscle actin (SMA) [(b) coverslip; (g) LDDCS], collagen I [(c) coverslip; (h) LDDCS], collagen III [(d) coverslip; (i) LDDCS], and F-actin [(e) coverslip; (j) mechanically biased DDCS]. Seguence of images: ×1 - Dapi Stain only; ×2 - Antibody stain for component; ×3 - Overlayed antibody and Dapi stain. Scale bars: 100 μm.
Discussion
During human development, a well-timed series of synthetic and cell migratory events gives rise to the cornea and the surrounding ocular tunic.46 Morphogenesis of the highly organized corneal stromal ECM is thought to occur following a radial migration of neural crest-derived mesenchymal cells into the 2-D planar space between the surface ectoderm (prospective corneal epithelium) and the lens surface.47 Very little is known about the myriad of guidance cues, which control the invasion, self-organization, and synthetic behavior of the invading cells, but our results suggest that the mechanical state of the ECM/cell complex is of paramount importance to these events.
Mechanically induced guidance cues in the DDCS
The PHCF patterning we observed is not consistent, in general, with the behavior of other cell systems when they are cultured on loaded substrates. Cells can align with the applied load direction10,48–50 or transverse to it.9,51–54 On a similar collagen-based substrate, stretched uniaxially to 30% static strain (much larger than our strain), fibroblasts reoriented to the load direction.49 The alignment fate of a collection of cells depends on a large number of parameters related to the material type, which could, in turn, be load sensitive. Unfortunately, it is nearly impossible to isolate them from one another.
Durotaxis
In our system, cells were cultured on a prestrained substrate. In this study, mechanical forces were not externally applied to the cells. Instead, the cells must actively sense their mechanical environment. An active cell mechanosensing mechanism has been suggested where cells monitor the substrate resistance by applying pinching forces, mathematically modeled as compressive dipole forces at two anchor points, representing two focal adhesion points, on the substrate.55 The model predicts that the elastic energy exerted by these dipole compressive forces is minimized in the direction of applied load.55 Durotaxis predicts that cells cultured on a substrate with a stiffness gradient will move toward the stiffer region.56–58 If the guidance cue in durotaxis is consistent with active cell mechanosensing, the PHCFs should naturally reorient along the applied load direction (i.e., toward the direction of minimum strain energy and maximum stiffness).
Contact guidance
Uniaxial loading of a collagen gel will rotate collagen fibrils toward the load, possibly generating an appreciable contact guidance cue.59 We estimated the maximum directional change of any single fibril on the surface of the DDCS to be very small (∼2°). Human corneal stem cells respond to contact guidance cues by reorienting in the direction of aligned microstructures,60 and fibroblast orientation is generally proportional to the extent of anisotropy of collagen fibrils in a substrate.61 However, the contact guidance signal should be weak in our system.
Anisotropy in collagen density
In tension, the DDCS in our system possesses a high Poisson's ratio,29 which results in lateral contraction of the collagen network, which enhances the number of available cell adhesion sites and collagen density in the transverse direction by ∼13%. If the cells have a predilection for higher concentrations of collagen (i.e., higher binding site density), they may tend to orient in the transverse direction.
Electrotaxis
Collagenous tissues produce a shear piezoelectric signal when loaded,62,63 and for some corneal cells (epithelium), electrotaxis overrides contact guidance if the potential field is high enough.64 However, the magnitude of our mechanical bias was very small and piezoelectric signals dissipate very quickly under conditions of static load in ionic fluids. It is not clear how the piezoelectric behavior of collagen might influence the PHCFs, but their alignment near the maximum shear angle remains intriguing.
Boundary effects
The interface at the grips and the free edge of the DDCS present confining boundary conditions that affect the orientation of PHCFs.65–67 At the boundaries, the PHCF culture tends to coalign with the barrier or edge. However, boundary effects did not appear to propagate far into the tissue construct and the directional migration pattern was established well before boundary effect propagation could occur on the LDDCS.
PHCF migration dynamics
Collective migration of cells as cohesive colonies is a principal mechanism underlying modeling and remodeling events such as embryonic morphogenesis, wound repair, and cancer invasion (see reviews by Friedl and Gilmour68 and Rorth69). We have observed an organized wave-like migration before culture confluence, which includes bidirectional migration throughout the experimental period. The bidirectional migration observed on both the LDDCS and the coverslip does not appear to be fully consistent with published cell migration models. Escape and pursuit migration dynamics are difficult to fit to the patterning observed as it makes no allowance for antiparallel motion.70 Collective migration of a tightly connected cell mass as defined by Friedl and Gilmour68 is not likely responsible for the final patterning behavior of our PHCF colony. The migration dynamics are more consistent with the inclusive definition of Rorth69 and comprise some degree of streaming behavior where the cells are loosely associated, but move in an organized manner (see Supplementary Movie S7). This type of migration has been characterized in the context of contact inhibited locomotion counterbalanced by cell–cell attractive interactions.71 The bidirectional migration, which appears before confluence, prevents us from effectively categorizing the mode of colony migration that we observe in our in vitro system.
During human corneal stromal synthesis in vivo, NC lineage mesenchymal cells initially populate the prospective stromal space in waves and are eventually replaced by or differentiate into corneal keratocytes.47 Our in vitro culture system comprises primary human keratocytes, which have been transformed back into fibroblasts through exposure to fetal bovine serum. There are two potential factors that might explain the PHCF bidirectional migration: (1) when cultured in vitro, PHCFs are capable of forming multilayers and synthesizing copious ECM proteins. The presence of an ECM coating on the cell surface allows a cell to regard its neighboring cells as substrate and prevents contact inhibition of motility.40,72 It is possible that the fibroblasts' antiparallel, rather than collectively uniform, motion is due to the synthesis of an ECM coat, which can interfere with direct cell–cell contact and prevent migration as a tightly connected cell mass. If there is direct cell–cell communication, the contacts are necessarily dynamic and persist over large cell separation distances as has been observed in chick neural crest cells.73 Indeed, we have observed that PHCFs generate exceedingly long filopodial extensions, which could be used to signal neighboring cells (>100 μm; Supplementary Movies, S1 and S2). (2) Cadherin, a protein found in mechanically strong cell–cell adherens junctions between cells, has not been detected in corneal fibroblast cultures.74 Instead, they express connexin43, the protein associated with gap junctions,74 suggesting that PHCF cell–cell communication through mechanical signaling is not likely to be robust. Cell–cell contact facilitated by cadherin-based adherens junctions is an important element of collective cell migration, which enables the cells to migrate as a group.75 Thus, the role of the substrate and its mechanics is even more critical for PHCFs in this study since most likely the only mechanism to convey mechanical signals to neighboring cells, other than the gap junctions, is through the substrate/cell interaction.
PHCF patterning and orientation
On the mechanically biased DDCS, we observed a rapid rotation to a persistent, 131.8° ± 5.5°, orientation before confluence. This suggests that PHCFs are individually responsive directly to the slightly altered mechanical state of the DDCS. The individual cell response to the substrate leads to apparent emergent patterning before confluence, precluding any possible jamming effect. The stability of the patterning angle was robust on the DDCS. The orientation angle persisted globally, even after the strain was increased an additional 6%, as the cell orientation was still measured at 133.4° ± 9.8° by day 12 of the experiment. In addition, cells on the bottom surface of the LDDCS were always oriented orthogonal to the top surface. The failure of the PHCF system to reverse this symmetry is an open question. On the glass, the local orientation angle was established later than on the DDCS and the orientation angle could wander significantly over time (as seen in Fig. 2c). Thus, on glass, the orientation pattern is potentially the result of local jamming without pattern locking.
ECM synthesis and orientation
Given the experimental design, it was difficult to correlate cell motion with ECM production. However, we have observed that the cell's alignment matched that of the matrix produced, in general, and that the direction of motion of the cells was aligned with their orientation early in the experimental period. In Figure 6, one can readily appreciate the coalignment of the F-actin, fibronectin, and type I collagen with the orientation of the cells. Measurement of the cell nucleus major axis and ECM orientation showed that they are oriented within 5° (Supplementary Fig. S5). This coalignment is consistent with the results in other investigations.45,76
Regardless of when the ECM was synthesized, it appears that the cells are in constant motion along the direction of matrix orientation and that they are moving in an antiparallel manner. If we assume that the majority of the ECM is produced toward the end of the experimental period (hours 150–300), our estimates of average speed of the cells suggest that they have slowed to about 7.5 ± 2.5 μm/h (Supplementary Fig. S3). Continuous cell motion during matrix production supports the concept of a type of printing methodology where the cells may be generating matrix as they migrate, leaving behind ECM trails. However, the details of precisely how the matrix is assembled remain unknown. Our recent work suggests that shearing or extensional strain applied to condensed collagen solutions in the direction of cell motion is the most likely driver of collagen crystallization.77
Miscellaneous observations
The constant optical accessibility to the culture system permitted many observations worth noting, including the ability of the culture system to locally reorganize. In Supplementary Movie S8, the self-healing capacity of the PHCF culture system was captured when the PHCFs became bound up in a multilayer disorganized knot on a region of the coverslip. This kind of structure has been observed in TGF-β1-treated primary corneal keratocytes and is an indication of a stable local contraction caused by transformation to a myofibroblastic phenotype.78 However, in this case, the PHCF colony rapidly broke down the knot to the level of the bare glass surface, repopulated the area, reorganized, and continued on to ECM synthesis. Other observations include the prolific shedding of vesicle-like structures (seen at time 1–24 h in Supplementary Movie S8) and the extraordinary length of the extensions produced by the PHCFs (seen in Supplementary Movie S1 during the first 100 h). These extensions are potentially related to keratopodia recently identified by Young et al.24 in developing chick corneas.
Conclusions
It is clear that a small, static, physiological mechanical bias, applied to a collagen substrate, alters the perception of the substrate by the PHCFs, influencing their individual migration and orientation behavior. The effect of the bias operates on individual cells, leading rapidly to organized, antiparallel, collective migration behavior, which may appear to be emergent. It is unclear how the mechanical effect is communicated to the cells or why they appear to converge on an orientation angle of ∼135°. Unloading the DDCS led to significantly poorer correlation of cell orientations, suggesting that the applied mechanical bias is the guidance cue. On glass, cell orientation and migration are initially random, only becoming organized as the culture system becomes crowded, suggesting that local orientation is an uncontrolled jamming phenomenon and that it is metastable. This may be a result of the exceedingly high stiffness of glass, which prevented cell to cell interaction through substrate deformation. Further investigations are required to identify the significance of the mechanically controlled cellular migration behavior, which leads to organized matrix synthesis in vitro and in vivo. Combining the ability to directly observe cell migration, manipulate load conditions, and track matrix deposition in the mechanobioreactor may provide insight into the mechanism and mechanics of tissue patterning.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Eye Institute of the National Institutes of Health under NIH/NEI (R01) EY0015500. The authors would like to acknowledge and thank Ebraheim Ismail for providing the image of the DDCS surface in Supplementary Figure S1.
Disclosure Statement
The custom mechanobioreactor used in this investigation has now been commercialized by Admet Corporation under the name BioTense®. However, no authors benefit financially from the commercialization agreement. Thus, no competing financial interests exist.
References
- 1.Wolff J. Das Gesetz der Transformation der Knochen. Berlin, Germany: Verlag von August Hirschwald, 1892 [Google Scholar]
- 2.Chalmers J., and Ray R.D. The growth of transplanted foetal bones in different immunological environments. J Bone Joint Surg Br Vol 44, 149, 1962 [Google Scholar]
- 3.Chamay A., and Tschantz P. Mechanical influences in bone remodeling. Experimental research on Wolff's law. J Biomech 5, 173, 1972 [DOI] [PubMed] [Google Scholar]
- 4.Cowin S.C. The mechanical and stress adaptive properties of bone. Ann Biomed Eng 11, 263, 1983 [DOI] [PubMed] [Google Scholar]
- 5.Cowin S.C. Bone stress adaptation models. J Biomech Eng 115, 528, 1993 [DOI] [PubMed] [Google Scholar]
- 6.Carter D.R., VanderMeulen M.C.H., and Beaupre G.S. Mechanical factors in bone growth and development. Bone 18, S5, 1996 [DOI] [PubMed] [Google Scholar]
- 7.Carter D., and Beaupre G. Skeletal Form and Function: Mechanobiology of Skeletal Development, Aging and Regeneration. Cambridge: Press Syndicate of the University of Cambridge, 2001 [Google Scholar]
- 8.Henderson J.H., and Carter D.R. Mechanical induction in limb morphogenesis: the role of growth-generated strains and pressures. Bone 31, 645, 2002 [DOI] [PubMed] [Google Scholar]
- 9.Buck R.C. Reorientation response of cells to repeated stretch and recoil of the substratum. Exp Cell Res 127, 470, 1980 [DOI] [PubMed] [Google Scholar]
- 10.Eastwood M., Mudera V.C., McGrouther D.A., and Brown R.A. Effect of precise mechanical loading on fibroblast populated collagen lattices: morphological changes. Cell Motil Cytoskeleton 40, 13, 1998 [DOI] [PubMed] [Google Scholar]
- 11.Parsons M., Kessler E., Laurent G.J., Brown R.A., and Bishop J.E. Mechanical load enhances procollagen processing in dermal fibroblasts by regulating levels of procollagen C-proteinase. Exp Cell Res 252, 319, 1999 [DOI] [PubMed] [Google Scholar]
- 12.Prajapati R.T., Chavally-Mis B., Herbage D., Eastwood M., and Brown R.A. Mechanical loading regulates protease production by fibroblasts in three-dimensional collagen substrates. Wound Repair Regen 8, 226, 2000 [DOI] [PubMed] [Google Scholar]
- 13.Carver W., Nagpal M.L., Nachtigal M., Borg T.K., and Terracio L. Collagen expression in mechanically stimulated cardiac fibroblasts. Circ Res 69, 116, 1991 [DOI] [PubMed] [Google Scholar]
- 14.Lambert C.A., Soudant E.P., Nusgens B.V., and Lapiere C.M. Pretranslational regulation of extracellular matrix macromolecules and collagenase expression in fibroblasts by mechanical forces. Lab Invest 66, 444, 1992 [PubMed] [Google Scholar]
- 15.Lin S.L., Yang J.C., Ho K.N., Wang C.H., Yeh C.W., and Huang H.M. Effects of compressive residual stress on the morphologic changes of fibroblasts. Med Biol Eng Comput 47, 1273, 2009 [DOI] [PubMed] [Google Scholar]
- 16.Butt R.P., Laurent G.J., and Bishop J.E. Collagen production and replication by cardiac fibroblasts is enhanced in response to diverse classes of growth factors. Eur J Cell Biol 68, 330, 1995 [PubMed] [Google Scholar]
- 17.Butt R.P., Laurent G.J., and Bishop J.E. Mechanical load and polypeptide growth factors stimulate cardiac fibroblast activity. Ann N Y Acad Sci 752, 387, 1995 [DOI] [PubMed] [Google Scholar]
- 18.Altman G.H., Horan R.L., Martin I., Farhadi J., Stark P.R., Volloch V., Richmond J.C., Vunjak-Novakovic G., and Kaplan D.L. Cell differentiation by mechanical stress. FASEB J 16, 270, 2002 [DOI] [PubMed] [Google Scholar]
- 19.Canty E.G., and Kadler K.E. Procollagen trafficking, processing and fibrillogenesis. J Cell Sci 118, 1341, 2005 [DOI] [PubMed] [Google Scholar]
- 20.Neath P., Roche S.M., and Bee J.A. Intraocular pressure-dependent and -independent phases of growth of the embryonic chick eye and cornea. Invest Ophthalmol Vis Sci 32, 2483, 1991 [PubMed] [Google Scholar]
- 21.Cox J.L., Farrell R.A., Hart R.W., and Langham M.E. The transparency of the mammalian cornea. J Physiol 210, 601, 1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meek K.M., and Leonard D.W. Ultrastructure of the corneal stroma: a comparative study. Biophys J 64, 273, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cowin S.C. How is a tissue built? J Biomech Eng 122, 553, 2000 [DOI] [PubMed] [Google Scholar]
- 24.Young R.D., Knupp C., Pinali C., Png K.M., Ralphs J.R., Bushby A.J., Starborg T., Kadler K.E., and Quantock A.J. Three-dimensional aspects of matrix assembly by cells in the developing cornea. Proc Natl Acad Sci U S A 111, 687, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo X., Hutcheon A.E., Melotti S.A., Zieske J.D., Trinkaus-Randall V., and Ruberti J.W. Morphologic characterization of organized extracellular matrix deposition by ascorbic acid-stimulated human corneal fibroblasts. Invest Ophthalmol Vis Sci 48, 4050, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren R., Hutcheon A.E., Guo X.Q., Saeidi N., Melotti S.A., Ruberti J.W., Zieske J.D., and Trinkaus-Randall V. Human primary corneal fibroblasts synthesize and deposit proteoglycans in long-term 3-D cultures. Dev Dyn 237, 2705, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lwigale P.Y., Cressy P.A., and Bronner-Fraser M. Corneal keratocytes retain neural crest progenitor cell properties. Dev Biol 288, 284, 2005 [DOI] [PubMed] [Google Scholar]
- 28.Paten J.A., Zareian R., Saeidi N., Melotti S.A., and Ruberti J.W. Design and performance of an optically accessible, low-volume, mechanobioreactor for long-term study of living constructs. Tissue Eng Part C Methods 17, 775, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Susilo M.E., Paten J.A., Sander E.A., Nguyen T.D., and Ruberti J.W. Collagen network strengthening following cyclic tensile loading. Interface Focus 6, 20150088, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mega Y., Robitaille M., Zareian R., McLean J., Ruberti J., and DiMarzio C. Quantification of lamellar orientation in corneal collagen using second harmonic generation images. Opt Lett 37, 3312, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mardia K.V., and Jupp P.E. Directional Statistics. New York: J. Wiley, 2000 [Google Scholar]
- 32.Sander E.A., and Barocas V.H. Comparison of 2D fiber network orientation measurement methods. J Biomed Mater Res Part A 88, 322, 2009 [DOI] [PubMed] [Google Scholar]
- 33.Gorelik R., and Gautreau A. Quantitative and unbiased analysis of directional persistence in cell migration. Nat Protoc 9, 1931, 2014 [DOI] [PubMed] [Google Scholar]
- 34.Fortin M.J., and Dale M.R.T. Spatial Analysis: A Guide for Ecologists. Cambridge, UK: Cambridge University Press, 1, 2005 [Google Scholar]
- 35.Zieske J.D., Hutcheon A.E., Guo X., Chung E.H., and Joyce N.C. TGF-beta receptor types I and II are differentially expressed during corneal epithelial wound repair. Invest Ophthalmol Vis Sci 42, 1465, 2001 [PubMed] [Google Scholar]
- 36.Ruberti J.W., Roy A.S., and Roberts C.J. Corneal biomechanics and biomaterials. Annu Rev Biomed Eng 13, 269, 2011 [DOI] [PubMed] [Google Scholar]
- 37.Albrecht-Buehler G. Rudimentary form of cellular “vision.” Proc Natl Acad Sci U S A 89, 8288, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abercrombie M., and Heaysman J.E. Observations on the social behaviour of cells in tissue culture. I. Speed of movement of chick heart fibroblasts in relation to their mutual contacts. Exp Cell Res 5, 111, 1953 [DOI] [PubMed] [Google Scholar]
- 39.Purschke M., Rubio N., Held K.D., and Redmond R.W. Phototoxicity of Hoechst 33342 in time-lapse fluorescence microscopy. Photochem Photobiol Sci 9, 1634, 2010 [DOI] [PubMed] [Google Scholar]
- 40.Elsdale T., and Bard J. Cellular interactions in mass cultures of human diploid fibroblasts. Nature 236, 152, 1972 [DOI] [PubMed] [Google Scholar]
- 41.Elsdale T., and Foley R. Morphogenetic aspects of multilayering in Petri dish cultures of human fetal lung fibroblasts. J Cell Biol 41, 298, 1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elsdale T., and Wasoff F. Fibroblast cultures and dermatoglyphics: the topology of two planar patterns. Wilhelm Roux Arch Dev Biol 180, 121, 1976 [DOI] [PubMed] [Google Scholar]
- 43.Karamichos D., Guo X.Q., Hutcheon A.E., and Zieske J.D. Human corneal fibrosis: an in vitro model. Invest Ophthalmol Vis Sci 51, 1382, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karamichos D., Hutcheon A.E., and Zieske J.D. Transforming growth factor-beta3 regulates assembly of a non-fibrotic matrix in a 3D corneal model. J Tissue Eng Regen Med 5, e228, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saeidi N., Guo X., Hutcheon A.E., Sander E.A., Bale S.S., Melotti S.A., Zieske J.D., Trinkaus-Randall V., and Ruberti J.W. Disorganized collagen scaffold interferes with fibroblast mediated deposition of organized extracellular matrix in vitro. Biotechnol Bioeng 109, 2683, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Rahilly R. The prenatal development of the human eye. Exp Eye Res 21, 93, 1975 [DOI] [PubMed] [Google Scholar]
- 47.Sevel D., and Isaacs R. A re-evaluation of corneal development. Trans Am Ophthalmol Soc 86, 178, 1988 [PMC free article] [PubMed] [Google Scholar]
- 48.Henshaw D.R., Attia E., Bhargava M., and Hannafin J.A. Canine ACL fibroblast integrin expression and cell alignment in response to cyclic tensile strain in three-dimensional collagen gels. J Orthop Res 24, 481, 2006 [DOI] [PubMed] [Google Scholar]
- 49.Haston W.S., Shields J.M., and Wilkinson P.C. The orientation of fibroblasts and neutrophils on elastic substrata. Exp Cell Res 146, 117, 1983 [DOI] [PubMed] [Google Scholar]
- 50.Bell E., Ivarsson B., and Merrill C. Production of a tissue-like structure by contraction of collagen lattices by human fibroblasts of different proliferative potential in vitro. Proc Natl Acad Sci U S A 76, 1274, 1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neidlinger-Wilke C., Grood E., Claes L., and Brand R. Fibroblast orientation to stretch begins within three hours. J Orthop Res 20, 953, 2002 [DOI] [PubMed] [Google Scholar]
- 52.Hayakawa K., Sato N., and Obinata T. Dynamic reorientation of cultured cells and stress fibers under mechanical stress from periodic stretching. Exp Cell Res 268, 104, 2001 [DOI] [PubMed] [Google Scholar]
- 53.Wang H., Ip W., Boissy R., and Grood E.S. Cell orientation response to cyclically deformed substrates: experimental validation of a cell model. J Biomech 28, 1543, 1995 [DOI] [PubMed] [Google Scholar]
- 54.Dartsch P.C., Hammerle H., and Betz E. Orientation of cultured arterial smooth muscle cells growing on cyclically stretched substrates. Acta Anat 125, 108, 1986 [DOI] [PubMed] [Google Scholar]
- 55.Bischofs I.B., and Schwarz U.S. Cell organization in soft media due to active mechanosensing. Proc Natl Acad Sci U S A 100, 9274, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lo C.M., Wang H.B., Dembo M., and Wang Y.L. Cell movement is guided by the rigidity of the substrate. Biophys J 79, 144, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Isenberg B.C., Dimilla P.A., Walker M., Kim S., and Wong J.Y. Vascular smooth muscle cell durotaxis depends on substrate stiffness gradient strength. Biophys J 97, 1313, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hadjipanayi E., Mudera V., and Brown R.A. Guiding cell migration in 3D: a collagen matrix with graded directional stiffness. Cell Motil Cytoskelet 66, 121, 2009 [DOI] [PubMed] [Google Scholar]
- 59.Chandran P.L., and Barocas V.H. Affine versus non-affine fibril kinematics in collagen networks: theoretical studies of network behavior. J Biomech Eng 128, 259, 2006 [DOI] [PubMed] [Google Scholar]
- 60.Wu J., Du Y., Watkins S.C., Funderburgh J.L., and Wagner W.R. The engineering of organized human corneal tissue through the spatial guidance of corneal stromal stem cells. Biomaterials 33, 1343, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guido S., and Tranquillo R.T. A methodology for the systematic and quantitative study of cell contact guidance in oriented collagen gels. Correlation of fibroblast orientation and gel birefringence. J Cell Sci 105 (Pt 2), 317, 1993 [DOI] [PubMed] [Google Scholar]
- 62.Marino A.A., and Becker R.O. Piezoelectricity in hydrated frozen bone and tendon. Nature 253, 627, 1975 [DOI] [PubMed] [Google Scholar]
- 63.Minary-Jolandan M., and Yu M.F. Nanoscale characterization of isolated individual type I collagen fibrils: polarization and piezoelectricity. Nanotechnology 20, 085706, 2009 [DOI] [PubMed] [Google Scholar]
- 64.Rajnicek A.M., Foubister L.E., and McCaig C.D. Prioritising guidance cues: directional migration induced by substratum contours and electrical gradients is controlled by a rho/cdc42 switch. Dev Biol 312, 448, 2007 [DOI] [PubMed] [Google Scholar]
- 65.Kolodney M.S., and Elson E.L. Correlation of myosin light chain phosphorylation with isometric contraction of fibroblasts. J Biol Chem 268, 23850, 1993 [PubMed] [Google Scholar]
- 66.Baxter S.C., Morales M.O., and Goldsmith E.C. Adaptive changes in cardiac fibroblast morphology and collagen organization as a result of mechanical environment. Cell Biochem Biophys 51, 33, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Costa K.D., Lee E.J., and Holmes J.W. Creating alignment and anisotropy in engineered heart tissue: role of boundary conditions in a model three-dimensional culture system. Tissue Eng 9, 567, 2003 [DOI] [PubMed] [Google Scholar]
- 68.Friedl P., and Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol 10, 445, 2009 [DOI] [PubMed] [Google Scholar]
- 69.Rorth P. Fellow travellers: emergent properties of collective cell migration. EMBO Rep 13, 984, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Romanczuk P., Couzin I.D., and Schimansky-Geier L. Collective motion due to individual escape and pursuit response. Phys Rev Lett 102, 010602, 2009 [DOI] [PubMed] [Google Scholar]
- 71.Carmona-Fontaine C., Theveneau E., Tzekou A., Tada M., Woods M., Page K.M., Parsons M., Lambris J.D., and Mayor R. Complement fragment C3a controls mutual cell attraction during collective cell migration. Dev Cell 21, 1026, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bard J.B., and Hay E.D. The behavior of fibroblasts from the developing avian cornea. Morphology and movement in situ and in vitro. J Cell Biol 67, 400, 1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teddy J.M., and Kulesa P.M. In vivo evidence for short- and long-range cell communication in cranial neural crest cells. Development 131, 6141, 2004 [DOI] [PubMed] [Google Scholar]
- 74.Petridou S., and Masur S.K. Immunodetection of connexins and cadherins in corneal fibroblasts and myofibroblasts. Invest Ophthalmol Vis Sci 37, 1740, 1996 [PubMed] [Google Scholar]
- 75.Ilina O., and Friedl P. Mechanisms of collective cell migration at a glance. J Cell Sci 122, 3203, 2009 [DOI] [PubMed] [Google Scholar]
- 76.Wang J.H., Jia F., Gilbert T.W., and Woo S.L. Cell orientation determines the alignment of cell-produced collagenous matrix. J Biomech 36, 97, 2003 [DOI] [PubMed] [Google Scholar]
- 77.Paten J.A., Siadat S.M., Susilo M.E., Ismail E.N., Stoner J.L., Rothstein J.P., and Ruberti J.W. Flow induced crystallization of collagen: a potentially critical mechanism in early tissue formation. ACS Nano 10, 5027, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jester J.V., Barry-Lane P.A., Cavanagh H.D., and Petroll W.M. Induction of alpha-smooth muscle actin expression and myofibroblast transformation in cultured corneal keratocytes. Cornea 15, 505, 1996 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.