Abstract
Background
Diet has a recognized effect in shaping gut microbiota. Many studies link an increase in Prevotella to high-fibre diet, while Bacteroides abundance is usually associated with the consumption of animal fat and protein-rich diets. Nevertheless, closely related species and strains may harbour different genetic pools; therefore, further studies should aim to understand whether species of the same genus are consistently linked to dietary patterns or equally responsive to diet variations. Here, we used oligotyping of 16S rRNA gene sequencing data to exploit the diversity within Prevotella and Bacteroides genera in faecal samples of omnivore and non-omnivore subjects from a previously studied cohort.
Results
A great heterogeneity was found in oligotype composition. Nevertheless, different oligotypes within the same genus showed distinctive correlation patterns with dietary components and metabolome. We found that some Prevotella oligotypes are significantly associated with the plant-based diet but some are associated with animal-based nutrients, and the same applies to Bacteroides. Therefore, an indiscriminate association of Bacteroidetes genera with specific dietary patterns may lead to an oversimplified vision that does not take into account sub-genus diversity and the different possible responses to dietary components.
Conclusions
We demonstrated that Prevotella and Bacteroides oligotypes show distinctive correlation patterns with dietary components and metabolome. These results substantiate a current oversimplification of diet-dependent microbe-host associations and highlighted that sub-genus differences must be taken into account when planning gut microbiota modulation for health benefits
Electronic supplementary material
The online version of this article (doi:10.1186/s40168-016-0202-1) contains supplementary material, which is available to authorized users.
Keywords: Gut microbiota, Oligotyping, Plant-based diet, Omnivore diet
Background
The important contribution of the gut microbiota to human health and disease is widely recognized [1]. Habitual diet plays a key role in shaping the gut microbial community composition [2–6], and sudden dietary changes are reflected in a prompt response of the gut symbionts [7]. Bacteroidetes are among the most abundant bacterial groups found in the gut, and many genera within this phylum are recognized to be diet-responsive; in particular, high-fibre diet is commonly associated with an increase in Prevotella, while Bacteroides abundance is linked to the consumption of diet rich in animal fat and protein [3–5, 7, 8]. Nevertheless, closely related species and strains may harbour different genetic pools [9–11].
No studies have so far posed the question of whether species of the same genus are consistently linked to dietary patterns or equally responsive to diet variations. The high genetic diversity found between and within Prevotella spp. might explain the highly different behaviour observed when considering the genus-level identification [11]. In fact, only some of the subjects reported an increase of this genus after administration of a fibre-rich diet [8], while P. copri was supposed to drive chronic inflammation in HIV patients [12] and to induce insulin resistance [13]. Therefore, species and even strain variability have to be considered in order to understand how to modulate the gut microbiota for therapeutic purposes [11]. The current knowledge is limited by the common use of a de novo clustering approach, relying on the clustering of sequences into operational taxonomic units (OTUs) at 97 % of similarity, often resulting in phylogenetically mixed units [14] and leading to taxonomic assignment at the genus level [15]. This approach may fail to resolve ecologically important differences between closely related organisms [16]. On the contrary, even small differences in ribosomal RNA (rRNA) sequence can predict substantial genomic and ecological variation [14, 17]. Oligotyping is an alternative approach, which decomposes a given taxon, or 97 % OTU, into high-resolution units (“oligotypes”) by considering the nucleotide positions identified as the most information-rich and allows resolution at the species level and below [16].
In a recent cohort study, we investigated the associations of microbiota and metabolome with habitual diets, and the OTU-based analyses of the 16S rRNA gene sequences yielded the usual associations of Prevotella with plant-based and Bacteroides with animal-based diets [2]. Gut Prevotella oligotypes were recently supposed to be host-specific and suggested as possible markers useful to track the origin of faecal contamination [18, 19]. Here, we explored the diversity of Bacteroidetes genera in faecal samples of omnivore and non-omnivore subjects of the previously studied cohort [2] using oligotyping of 16S rRNA gene sequencing data and highlight for the first time that heterogeneity within Prevotella and Bacteroides prevent a comprehensive association of these genera with specific dietary patterns.
Methods
We used the oligotyping technique to explore differences within Bacteroides and Prevotella populations in omnivore (n = 44) and non-omnivore (self-declared ovo-lacto vegetarians and vegans, n = 93) subjects of a previously studied cohort [2]. Subject recruitment, DNA extraction, sequencing of the V1–V3 regions of rRNA gene, and metabolomics were carried out as recently described [2]. Raw reads were quality-filtered as follows: reads were trimmed at the first ambiguous base or when the average quality score dropped below 25 within a 50-bp-long window, and reads shorter than 500 bp and with >1 primer mismatch were discarded. High-quality reads were aligned to the Greengenes alignment template by using PyNAST 1.2.1 [20], and alignment was further trimmed to equal length by using the o-smart-trim script included in the oligotyping package v.1.0 [15]. In order to avoid biases due to different sequencing depths, all samples were rarefied at 4500 reads after raw read quality filtering. Taxonomy assignment of all the reads was carried out by using the RDP classifier [21]. Reads assigned to Prevotella or Bacteroides genera were extracted, and entropy analysis and oligotyping were carried out separately as described by the developers. High-entropy positions were chosen to compute the oligotypes (-C option): 106, 121, 137, 272, 349, 462, 502, 544, 549, 739, 746, 778, 851 and 98 and 251, 319, 432, 440, 462, 463, 471, 477, 502, 526, 528, 549, 578, 746, 778, 847 and 851 for Prevotella and Bacteroides dataset, respectively. To minimize the impact of sequencing errors, we required an oligotype to be represented by at least 100 reads (-M option). Moreover, rare oligotypes present in less than 10 samples were discarded (-s option). These parameters led to 61 (omnivores n = 17; non-omnivores n = 44) and 136 (omnivores n = 44; non-omnivores n = 92) samples left in Prevotella and Bacteroides dataset, respectively. BLASTn (match/mismatch scores: 1, −2; gap cost: linear) was used to query the representative sequences against the NCBI nr database, and the top hit was considered for taxonomic assignment. Statistical analyses and plotting were carried out in R environment (https://www.r-project.org). Pairwise Wilcoxon tests were used in order to determine significant differences in specific oligotype abundance according to diets. Pairwise Spearman’s correlations between oligotype relative abundance and diet or metabolome were computed and plotted by using the psych and made4 packages. A cladogram of representative sequences was generated using MrBayes (http://mrbayes.sourceforge.net) and depicted using the Interactive Tree of Life (http://itol.embl.de).
Results
The relative abundance of Prevotella and Bacteroides ranged from 86.7 to 0.08 % and from 58 to 0.13 %, respectively, regardless of the habitual diet. Since no significant differences in Prevotella and Bacteroides oligotype profiles between vegans and vegetarians were detected, the two vegetable-based diets were grouped together (as non-omnivore). A total of 24 and 51 oligotypes were identified for Prevotella and Bacteroides, respectively, but the number of different oligotypes detected was not related to the relative abundance of the genus in the faecal samples (Additional file 1: Figure S1). Oligotype representative sequences and BLASTn results are reported in Additional file 2: Table S1. High inter- and intra-individual diversity was found, with some diet-specific signatures (Additional file 3: Figure S2). Prevotella oligotypes P5, P11, P12 and P14 were clearly more abundant in omnivores, while P1, P10 and P19 prevailed in non-omnivores (Fig. 1a). Consistently, P12 showed positive correlations with animal-origin nutrients and foodstuffs (Fig. 2a) and with urinary trimethylamine oxide (TMAO) concentration (Additional file 4: Figure S3A). On the contrary, P1, P10 and P19 were strongly correlated with vegetable-based diets (Fig. 2a) and to faecal short-chain fatty acid (SCFA) levels (Additional file 4: Figure S3A). Besides showing a similar correlation pattern, P10 and P19 had higher sequence similarity, differing from all the others (Additional file 5: Figure S4A); they were identified as P. loescheii by best BLASTn match. All the other Prevotella oligotypes matched P. copri sequences, although none with 100 % identity (Additional file 5: Figure S4A and Additional file 2: Table S1A). A higher number of oligotypes was found within Bacteroides (Additional file 3: Figure S2B and Additional file 2: Table S1B), and 10 different species were identified, some showing exact matches with sequences in the nr database (Additional file 5: Figure S4B and Additional file 2: Table S1B). Bacteroides oligotype B1, B2 and B6 had higher abundance in non-omnivores, and B44 was found only in non-omnivores. On the contrary, B23, B27, B28 and B29 prevailed in omnivores (Fig. 1b); showed positive correlations with animal fats, proteins and animal-origin foodstuffs (Fig. 2b); and consistently with urinary TMAO (Additional file 4: Figure S3B). In particular, B29 was associated with fish, meat and preserved meat consumption, while B27 was specifically associated with fish. Finally, the co-occurrence/exclusion pattern highlights that some oligotypes frequently co-occur, e.g. Prevotella oligotypes P10 and P19 (P < 0.05), and are mutually exclusive with P5, P11 and P12, associated with an omnivore diet (Additional file 6: Figure S5).
Fig. 1.
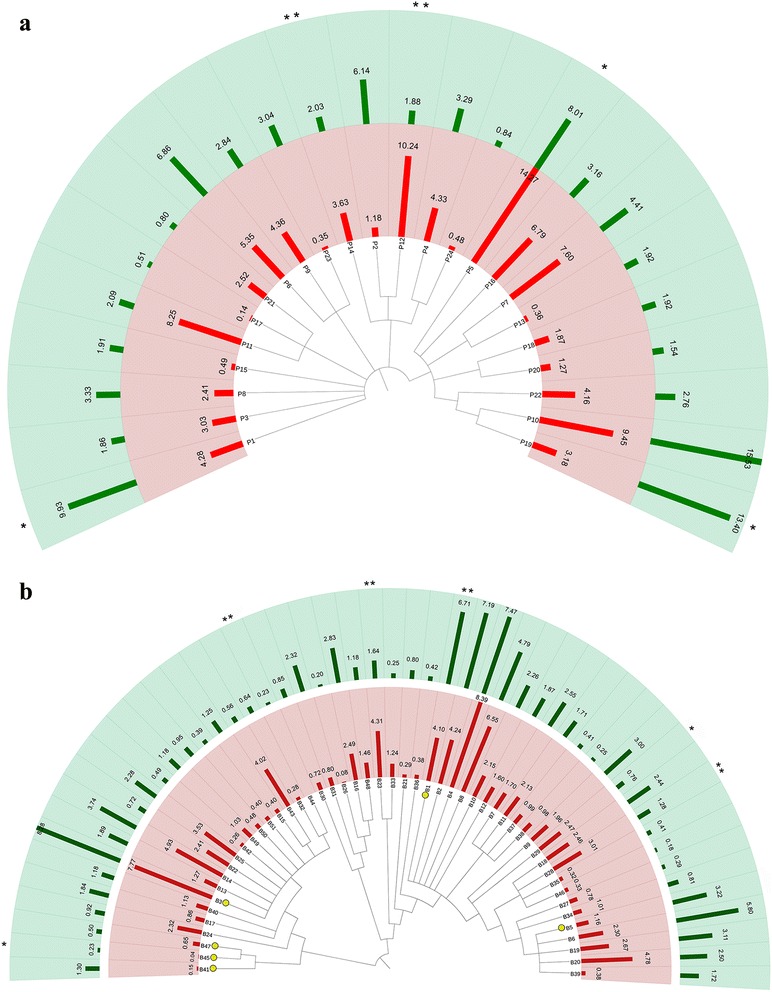
Differences in oligotype composition between omnivores and non-omnivores. Dendrogram of Prevotella (a) and Bacteroides (b) oligotype representative sequences and their average relative abundance in omnivores (inner circle) and non-omnivores (outer circle). Asterisks denote oligotypes significantly different in abundance between the two groups (*P < 0.05; **P < 0.01). Yellow circles denote oligotypes showing 100 % match with sequences in the NCBI nr database
Fig. 2.
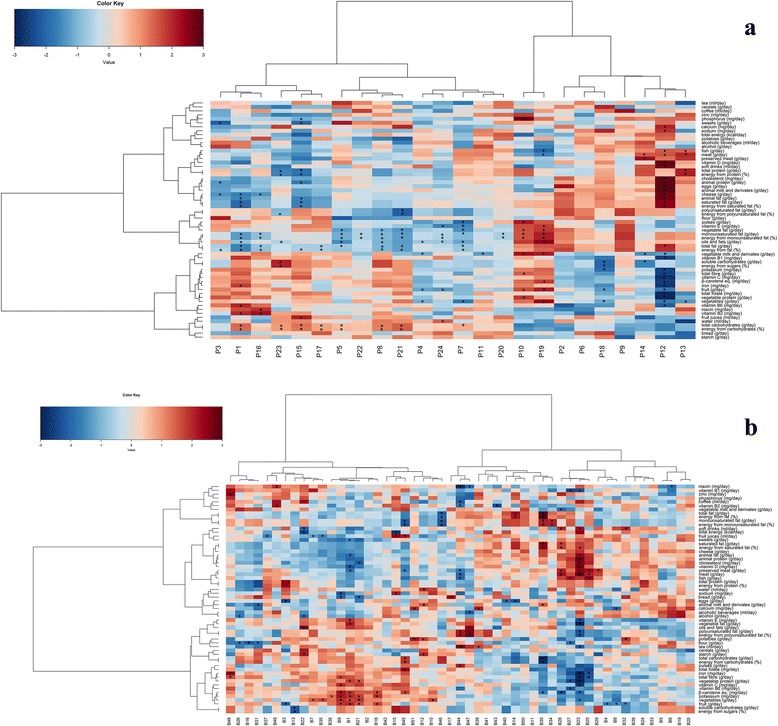
Correlation patterns between oligotypes and diet. Correlation between Prevotella (a) and Bacteroides (b) oligotypes and dietary data. Heatplot showing Spearman’s correlations between oligotypes and dietary data. Rows and columns are clustered by Euclidean distance and Ward linkage hierarchical clustering. The intensity of the colours represents the degree of association between oligotypes and foods/nutrients as measured by Spearman’s correlations. Asterisks denote significant correlations after P value corrections (P < 0.05)
Discussion
Gut microbiota is a complex consortium of microbial species and strains [9]. However, analyses based on OTU clustering methodology often result in genus-level classification and may underestimate this diversity [16]. This oversimplified vision does not take into account the possible different responses that species or strains belonging to the same genus may have to dietary components [8, 10]. The oligotyping approach was proposed as a powerful tool allowing resolution at the species level and below. It was successfully used to explain the microbial complexity of the oral microbiota [15] and to associate diversity within Blautia and Prevotella genera to different animal hosts [13, 18]. Moreover, differentiations in faecal Bacteroides oligotype composition were previously observed in Western and agrarian populations [14]. Here, we oligotyped Bacteroides and Prevotella 16S rRNA gene sequences in human faecal samples and demonstrated that specific oligotypes within these genera may be associated with the habitual diet. Consistently, oligotypes associated with an omnivore diet were positively correlated to TMAO levels, a metabolite derived from carnitine and choline catabolism and related to the development of cardiovascular diseases [22]. On the contrary, those linked to a vegetable-based diet showed positive correlations with SCFA, derived from carbohydrate fermentation and associated with beneficial outcomes for the host, such as anti-inflammatory and anti-carcinogenic effects [23, 24]. Our results highlighted how an indiscriminate association of a whole genus with a specific dietary pattern may lead to an oversimplified vision of the correlations between gut microbiota and diet, which does not take into account the diversity existing within the same genus or even within the same species [9, 11] and the possible different responses of these variants to dietary components [8, 10].
Conclusions
Sub-genus-level diversity can play a key role in establishing the interconnection between gut microbiome, diet, microbial metabolome and host response, and this should be taken into account in diet-based intervention studies aiming at changing gut microbiota structure and functions in order to gather specific health benefits. The consistency of the correlative patterns between oligotypes, diet and metabolome suggests the intriguing presence of a diet-driven adaptive selection, associated with different metabolic and enzymatic profiles. Different Prevotella and Bacteroides oligotypes, although identified as the same species, showed differential relative abundance in omnivores and non-omnivores and a consistent correlation pattern with foodstuffs/nutrients and metabolome, highlighting a possible distinct behaviour below the genus level never demonstrated before and the presence of different putative strains with diverse metabolic potential, possibly selected by the habitual diet.
Acknowledgements
Not applicable.
Funding
This study was supported by the Italian Ministry of University and Research (MIUR) programme PRIN 2010–2011 (grant number 2010WZ2NJN). Francesca De Filippis was supported by a grant from Regione Campania within the programme POR Campania FSE 2007/2013–2014/2020.
Availability of data and materials
The 16S rRNA gene sequences are available at the Sequence Read Archive of NCBI (accession number SRP042234).
Authors’ contributions
FDF, MG and DE conceived and designed the experiments. FDF, NP and LL performed the experiments. FDF and DE analysed the data. FDF wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of (a) Azienda Sanitaria Locale (Bari) (protocol N.1050), (b) Azienda Ospedaliera Universitaria of Bologna (protocol N.0018396), (c) the Province of Parma (protocol N.22884) and (d) the University of Torino (protocol N.1/2013/C).
Additional files
Bar plot of Prevotella (A) and Bacteroides (B) genera relative abundance ordered by size and line chart showing the number of different oligotypes identified in the same subject. (TIF 7240 kb )
Prevotella oligotypes representative sequences and BLASTn top hit. Table S1B. Bacteroides oligotypes representative sequences and BLASTn top hit. (DOCX 129 kb)
Pie charts showing the average relative abundance of Prevotella (A) and Bacteroides (B) oligotypes in omnivore (left side) and non-omnivore (right side) subjects. For clarity, only Bacteroides oligotypes showing >1 % abundance in at least 10 % of the subjects in each group are shown. (TIF 9270 kb)
Correlation between Prevotella (A) and Bacteroides (B) oligotypes and metabolome. Heatplot showing Spearman’s correlations between oligotypes, urinary TMAO and faecal SCFA levels. Rows and columns are clustered by Euclidean distance and Ward linkage hierarchical clustering. The intensity of the colours represents the degree of association between oligotypes and metabolites as measured by Spearman’s correlations. Asterisks denote significant correlations after P value corrections (P < 0.05). (TIF 8860 kb)
Heatplot showing the percent nucleotide identity between each pair of oligotypes within Prevotella (A) and Bacteroides (B) genera. Each oligotype is identified with the best match found in the NCBI nr database, with the percent of query coverage and the percent of identity. Row and columns are clustered by Horn distance and Ward linkage hierarchical clustering. (TIF 8620 kb)
Significant co-occurrence and co-exclusion relationships between Prevotella (A) and Bacteroides (B) oligotypes. Strong correlations are indicated by large circles, whereas weak correlations are indicated by small circles. The colours of the scale bar denote the nature of the correlation, with 1 indicating a perfectly positive correlation (dark blue) and −1 indicating a perfectly negative correlation (dark red) between two oligotypes. Only significant correlations (P < 0.05) are shown. (TIF 8840 kb)
References
- 1.Aron-Wisnewsky J, Clément K. The gut microbiome, diet, and links to cardiometabolic and chronic disorders. Nat Rev Nephrol. 2016;12(3):169–181. doi: 10.1038/nrneph.2015.191. [DOI] [PubMed] [Google Scholar]
- 2.De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65:1812–21. doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- 3.Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107(33):14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352(6285):565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JB, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 2015;22(6):971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Greenblum S, Carr R, Borenstein E. Extensive strain-level copy-number variation across human gut microbiome species. Cell. 2015;160(4):583–594. doi: 10.1016/j.cell.2014.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu M, McNulty NP, Rodionov DA, Khoroshkin MS, Griffin NW, Cheng J, et al. Genetic determinants of in vivo fitness and diet responsiveness in multiple human gut Bacteroides. Science. 2015;350(6256):aac5992. doi: 10.1126/science.aac5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ley RE. Gut microbiota in 2015: Prevotella in the gut: choose carefully. Nat Rev Gastroenterol Hepatol. 2016;13(2):69–70. doi: 10.1038/nrgastro.2016.4. [DOI] [PubMed] [Google Scholar]
- 12.Dillon SM, Lee EJ, Kotter CV, Austin GL, Gianella S, Siewe B, et al. Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T-cell activation in untreated HIV-1 infection. Mucosal Immunol. 2015;9:24–37. doi: 10.1038/mi.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535(7612):376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 14.Eren MA, Sogin ML, Morrison HG, Vineis JH, Fisher JC, Newton RJ, et al. A single genus in the gut microbiome reflects host preference and specificity. ISME J. 2015;9(1):90–100. doi: 10.1038/ismej.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eren MA, Borisy GG, Huse SM, Mark Welch JL. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci U S A. 2014;111(28):E2875–E2884. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eren AM, Maignien L, Sul WL, Murphy LG, Grim SL, Morrison HG, et al. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol. 2013;4(12):1111–1119. doi: 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaneveld JR, Lozupone C, Gordon JI, Knight R. Ribosomal RNA diversity predicts genome diversity in gut bacteria and their relatives. Nucleic Acids Res. 2010;38(12):3869–3879. doi: 10.1093/nar/gkq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koskey AM, Fisher JC, Eren AM, Ponce-Terashima R, Reis MG, Blanton RE, et al. Blautia and Prevotella sequences distinguish human and animal fecal pollution in Brazil surface waters. Environ Microbiol Rep. 2014;6(6):696–704. doi: 10.1111/1758-2229.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher JC, Eren AM, Green HC, Shanks OC, Morrison HG, Vineis JH, et al. Comparison of sewage and animal fecal microbiomes by using oligotyping reveals potential human fecal indicators in multiple taxonomic groups. Appl Environ Microbiol. 2015;81(20):7023–7033. doi: 10.1128/AEM.01524-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caporaso JG, Bittinger K, Bushman FD, De Santis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stock J. Gut microbiota: an environmental risk factor for cardiovascular disease. Atherosclerosis. 2013;229(2):440–442. doi: 10.1016/j.atherosclerosis.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 23.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12(10):661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 24.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The microbial metabolites, short-chain fatty acids, regulate the colonic Treg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The 16S rRNA gene sequences are available at the Sequence Read Archive of NCBI (accession number SRP042234).


