Abstract
In vivo labeling of DNA with thymidine and thymidine analogs has long been a cornerstone of replication studies. Unfortunately, yeast lack a thymidine salvage pathway and thus do not incorporate exogenous thymidine. Specifically, yeast neither efficiently take up exogenous thymidine from their growth media, nor phosphorylate it to thymidylate, the precursor of dTTP. We have overcome these problem in fission yeast by expressing the human equilibrative nucleoside transporter 1 (hENT1) along with herpes simplex virus thymidine kinase (tk). hENT1 tk cells are healthy and efficiently incorporate exogenous thymidine and thymidine analogs. We present protocols for labeling DNA with tritiated thymidine, for in situ detection of incorporated BrdU by immunofluorescence, for double labeling with CldU and IdU, for CsCl gradient separation of IdU labeled DNA and for using hENT1 and tk as both positive and negative selection markers.
Keywords: fission yeast, Schizosaccharomyces pombe, DNA labeling, thymidine incorporation, BrdU, CldU, IdU, human equilibrative nucleoside transporter 1 (hENT1), thymidine kinase (tk)
Introduction
Incorporation of exogenous thymidine into the DNA of living cells is a widely used tool in the study of DNA replication. Thymidine incorporation experiments take advantage of the almost ubiquitous nature of the thymidine salvage pathway, which directly converts thymidine (TdR, thymine deoxyriboside) to thymidylate (dTMP, deoxythymidine monophosphate), whence it enters the cellular nucleotide pool and is incorporated into DNA. Fungi are unusual among the eukaryotes in that they lack a thymidine salvage pathway. There are two enzymes in particular lacking in fungi that prevent them from incorporating exogenous thymidine and thymidine analogs. The first is thymidine kinase (tk), which is required to phosphorylate thymidine to dTMP; the second is a plasma membrane thymidine transporter required to allow efficient entry of thymidine into cells.
Expression of herpes simplex virus thymidine kinase (tk) is sufficient to allow incorporation of thymidine into yeast DNA. This strategy has been used to label DNA in both budding and fission yeast [1-4](J. Leatherwood, personal communication). The ability of tk to phosphorylate toxic thymidine analogs has also been used in counter selection strategies [2, 3, 5]. However, the rate limiting step for thymidine incorporation in tk expressing yeast is entry of thymidine into the cell, not the rate of DNA synthesis [6](our unpublished results). Thus, these strains are not useful for kinetic studies of replication, or for techniques requiring high levels of incorporation. Budding yeast mutants are available that take up exogenous thymidine at higher rate, but they are quite sick [3].
To allow for efficient incorporation of exogenous thymidine in the fission yeast Schizosaccharomyces pombe, we have introduced the human equilibrative nucleoside transporter 1 (hENT1) into an established tk expressing background [5]. hENT1 is a passive nucleoside transporter that allows nucleosides, but not nucleotides or bases, to diffuse across the plasma membrane [7]. hENT1 has been shown to catalyze the diffusion of exogenous thymidine into budding yeast cells [8]. We cloned a full length hENT1 cDNA into an expression vector containing the strong constitutive adh1 promoter. This vector was integrated in the existing adh1:tk background to create the adh1:tk adh1:hENT1 strain yFS240 [5]. yFS240 is healthy and grows with the same doubling time as the wild-type yFS105 (data not shown). Susan Forsburg and colleagues have developed similar strains [9]. As described below, these strains provide useful tools for the in vivo labeling of DNA, and the accompanying tk plasmids provide positive and negative selection markers for knockout/knockin and plasmid shuffle strategies.
Methods
General Methods
Growth media and general techniques have been described [10, 11]. The following strains were used: yFS105 h- leu1-32 ura4-D18; yFS233 h- leu1-32 ura4-D18 ade6-210 his7-366 pJL218 (his7 adh1:tk); yFS240 h- leu1-32 ura4-D18 ade6-210 his7-366 pJL218 (his7 adh1:tk) pFS181 (leu1 adh1:hENT1); yFS284 h- leu1-32 ura4-D18 ade6-210 his7-366 cdc25-22 pJL218 (his7 adh1:tk) pFS181 (leu1 adh1:hENT1). The pJL218 integrant used in these strains is not integrated at his7; presumably it is integrated at a random, non-homologous locus. Furthermore, crosses with this integrant occasionally produce His-, FUdR sensitive progeny. The adh1:tk construct does not appear to be affected by the loss of the his7 gene.
Plasmid Constructions
To create the adh1:tk shuttle vectors, the minimal ars1 sequence [12] was amplified with the oligos NR74 (5'-CTCCCTAGGTCTATAATTATAGCTAAAAATTG-3') and NR75 (5'-CTCCCTAGGTAGGCATTTTGTTTAGTTAAAG-3'), cut with AvrII and cloned into AvrII cut pLIT28 (New England Biolabs) to create pFS253. The adh1:tk HindIII-KpnI fragment from pJL218, with the KpnI site blunted by T4 DNA polymerase, was cloned into HindIII, EcoRV cut pFS253 to create pFS254. The ura4 adh1:tk shuttle vector pFS118 (previously known as pNR228) was created by cloning the ura4 HindIII genomic fragment into HindIII cut pFS254. The ade6 adh1:tk shuttle vector pFS119 (previously known as pNR210) was created by cloning the ade6 StuI-SpeI genomic fragment into BglII-SpeI cut pFS254, with the BglII site blunted by T4 DNA polymerase.
To make the episomal adh1:hENT1 expression vector pFS177, hENT1 was amplified from the IMAGE consortium cDNA clone # 610324 with the oligos NR140 (5'-CAGCATGCGGCCGCGAGCTCTCACACAATTGCCCGGAA-3') and NR141 (5'-GCGAGATCTCATATGACAACCAGTCACCAG-3'), cut with BglII and SacI, and cloned into the BamHI and SacI cut adh1 expression vector pART1 [13]. To make the integrating adh1:hENT1 expression vector pFS181, the SspI-SacI adh1:hENT1 fragment from pFS177 was cloned into the SnaBI and SacI cut leu1 integration vector pJK148 [14].
The adh1:tk/kanMX6 cassette, contained in plasmid pFS255, was created by taking the HindIII/BglII adh1:tk fragment from pFS118, blunting it with T4 DNA polymerase, and cloning it into BglII cut pFA6a-kanMX6, also blunted with T4 DNA polymerase.
Plasmid maps and sequences are available from the Forsburg Lab vector database at pingu.salk.edu/~forsburg/vectors.html.
Labeling with 3H-Thymidine
Quantitation of the rate of thymidine incorporation using 3H-thymidine is a sensitive to measure the rate of bulk DNA synthesis. Isolation of total cellular nucleic acid provides a quick way to isolate the incorporated label; methyl-3H-thymidine is a specific for DNA because it cannot be incorporated in to RNA without loss of the methyl-3H label. Strategies designed to remove unincorporated counts by permeablizing and washing fixed cells were found to result in unacceptably high background counts.
adh1:tk adh1:hENT1 cells (yFS240) growing exponentially in YES at 30°C were labeled with 5 μCi/ml 3H TdR for the indicated amount of time. 2 OD units of cells were taken at each timepoint. (OD units are a measure of cell number calculated as the optical density of the culture at 600 nm times the volume of the culture in milliliters. Thus a 20 ml culture at an OD600 of 0.5 contains 10 OD units of cells. 1 OD unit is about 2×107 cells.) Samples were pelleted, resuspended in 200 μl lysis buffer (1% SDS 1xTE), and lysed by vortexing with an equal volume of 0.5 μm glass beads for 5 minutes in a 1.5 ml screw cap microfuge tube. The lysate was recovered by puncturing the bottom of the tube with a needle and spinning the lysate into a new tube. The soluble lysate was extracted with 1xTE saturated 1:1 phenol:chloroform until the interface was clear (approximately 3 times). Total nucleic acid was precipitated by the addition of 2 volumes of ethanol, resuspended in water, and the recovered radioactivity was quantitated by scintillation counting. For DNase treatment, samples were resuspended in restriction digestion buffer, incubated with 0.5 mg/ml DNase I (or no DNase I) for 30 minutes at 37°C, ethanol precipitated, and then resuspended and counted (Figure 1A). For the temperature-shift experiments, cdc25-22ts adh1:tk adh1:hENT1 cells (yFS284) were grown at 25°C. Samples were taken every 30 minutes. At 60 minutes, half of the culture was transferred to 35°C for the remainder of the experiment (Figure 1B). Each data point is the average of 3 experiments +/− standard deviation. A strain lacking hENT1, or the hENT1 strain treated with 10μM NBTI (Sigma N2255) , a hENT1 inhibitor, incorporated 3H-thymidine at approximately 2% the rate (data not shown). The unexpected incorporation of 3H-thymidine by G2 arrested cells is discussed below.
Figure 1.
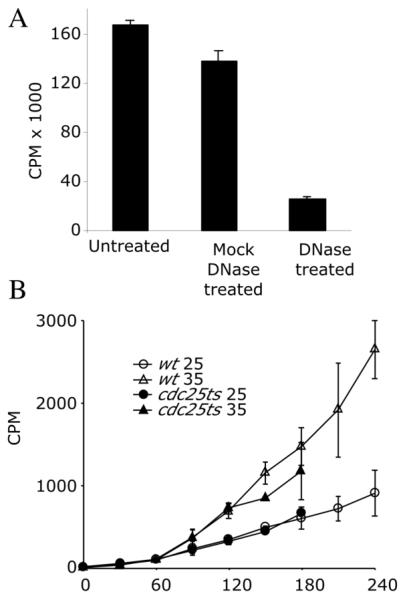
Labeling with tritiated thymidine
A) adh1:tk adh1:hENT1 cells (yFS240) were labeled for two hours with 5 μCi/ml 3H TdR. Total nucleic acid was prepared, treated as indicated and incorporation of 3H TdR was measured by scintillation counting.
B) adh1:tk adh1:hENT1 cells (yFS240) or adh1:tk adh1:hENT1 cdc25-22ts cells (yFS284) were labeled for 60 minutes at 25°C, and then half the culture was shifted to 35°C to arrest the cells in G2. Samples were taken at the indicated times.
All points represent the average of 3 experiments +/− standard deviation.
Labeling with Halogenated Thymidine Analogs
In addition to utilizing exogenous thymidine, hENT1 tk cells readily incorporate 5-halogenated analogs of thymidine, such as CldU, BrdU and IdU, in which the 5-methyl group is replaced by the halogen chlorine, bromine, or iodine, respectively. These halogenated thymidine analogs are useful because there appears to be no significant enzymatic discrimination between them and thymidine. In BrdU, in particular, the bromine has steric and electrostatic properties that are very similar to a methyl group. These analogs are recognized by several commercially available monoclonal antibodies raised against BrdU. Of these antibodies, some recognize CldU, but not IdU, and others vice versa, allowing CldU and IdU to be used for double labeling experiments. The differences in molecular weight between thymidine and its halogenated analogs also allow halogenated DNA to be separated from unlabeled DNA by CsCl density gradient centrifugation.
adh1:tk adh1:hENT1 cells (yFS240) growing exponentially in YES at 30°C were labeled with 5-chloro, 2'-deoxyuridine (CldU), 5-bromo, 2'-deoxyuridine (BrdU) or 5-iodo, 2'-deoxyuridine (IdU) at a final concentration of 500 nM. The optimal concentration was determined by titration during both single S phase and multiple S phase exposure. For single S phase exposure (4 hours for a synchronous cultures), 500 nM BrdU gave as strong a signal as higher concentrations, without effecting the subsequent mitosis. 2 mM BrdU caused a significant, heterogeneous delay of mitosis. For multiple S phase exposure (12 hours for asynchronous cultures), 70 nM BrdU showed no detectable slowing of cell growth; 100 nM BrdU caused occasional elongation, suggestive of DNA damage and checkpoint activation; 500 nM BrdU caused significant cell death. Interestingly at the higher doses, most cells appeared to die without elongating, suggesting that the lethality is due not to accumulating DNA damage, but rather a general toxicity, possibly a disruption of major grove DNA-protein interactions.
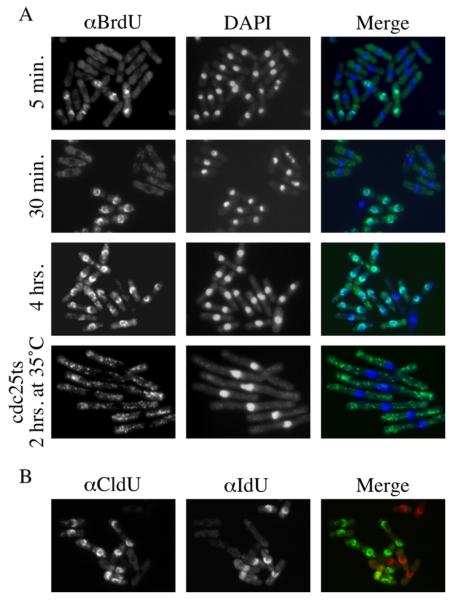
Labeled cells were prepared for indirect immunofluorescence [11]. 10 OD units of cells were fixed by adding 1/10th volume of 30% formaldehyde to culture medium with agitation for 1 hr. After fixation, the cells were pelleted and washed twice with PEM (100 mM PIPE pH 6.9, 1 mM EGTA, 1mM MgSO4) and once with PEMS (1.2 M Sorbitol in PEM). The pellets were resuspended in 1 ml of 300 μg/ml Zymolyase-20T in PEMS and incubated for 30 minutes at 37°C. The cells were again washed three times with PEMS and resuspended in 1 ml of 0.1% Triton X-100 in PEMS for 30 seconds at room temperature. They were centrifuged and resuspended in PEMS with 0.1 N HCl, 0.5% Triton X-100 on ice. After 10 minutes of incubation, the cells were washed once with PEMS, resuspended in 1 ml PEMS, heat-treated at 97°C for 10 minutes and chilled on ice. Next, the cells were centrifuged, resuspended with PEMBAL (1% BSA, 0.1M L-Lysine HCl in PEM) and incubated in room temperature for 30 minutes. The cells were again pelleted and resuspended in 10 μl PEMBAL, to which 10 μl of mouse (Becton-Dickinson, B44) or rat (Abcam, BU1/75(ICR1)) anti-BrdU antibody was added. The mouse antibody cross-reacts well with IdU, but only weakly with CldU; the rat antibody cross-reacts well with CldU, but only weakly with IdU. After 4 hrs of incubation with the primary antibody at room temperature, the cells were washed three times with PEMBAL and resuspended with 50 μl PEMBAL containing 1 μl Alexa-488 conjugated goat anti-mouse IgG or Alexa-594 conjugated anti-rat IgG secondary antibody (Molecular Probes) for 4 hrs at room temperature. For double labeling, the primary antibodies were combined, as were the secondary antibodies. For positive control of nuclear staining, the cells were washed once with PBS containing 10 μg/ml DAPI. The cells were washed three times in PBS and visualized by epifluorescence microscopy (Figure 2).
Figure 2.
Labeling with halogenated analogs of thymidine.
A) adh1:tk adh1:hENT1 cells (yFS240) were labeled with BrdU for the indicated amount of time. The brief pulses labeled only binucleate cells, confirming that the nuclear labeling is confined to S phase. Both nuclear and mitochondrial DNA is evident in the 4 hour labeling. The mitochondria are heavily labeled in yFS284 cells arrested in G2 at a cdc25-22 block.
B) adh1:tk adh1:hENT1 cells (yFS240) were labeled separately for 4 hours with CldU or IdU, combined and stained simultaneously with anti-CldU (Abcam BU1/75(ICR1) anti-BrdU) and anti IdU (Becton-Dickinson B44 anti-BrdU) antibodies. The one cell that stained with both antibodies was not permeablized and stained non-specifically.
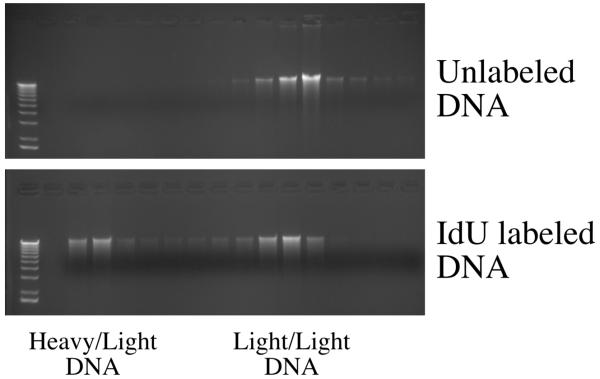
CsCl density gradient fractionation of IdU labeled DNA
Density gradient separation of pulse-labeled DNA is a powerful technique for measuring the timing of specific sequences during S phase [15]. While 13C and 15N are the most commonly used density labels, they are quite expensive, and BrdU has been used successfully as an alternative [3]. Density shift experiments using 13C and 15N labeled cells achieves a maximum molecular weight difference of 27 dalton/bp between heavy and light DNA. For CldU, BrdU and IdU, the difference is 10, 33 and 56 , respectively, assuming 50% G+C content. We used high concentrations (70 μM) of IdU to maximize the density difference. The toxicity of this level of IdU may account for the fact that only about half of the DNA in the IdU labeled sample has IdU incorporated (Figure 3). However, the DNA that does have IdU incorporated moves all the way to the bottom of the gradient, suggesting that it would be possible to use less IdU, or to use BrdU, either of which would expected to be less toxic. It may also be possible to separate IdU-labeled DNA from CldU-labeled DNA in double label experiments.
Figure 3.
CsCl density gradient fractionation of IdU labeled DNA
DNA from adh1:tk adh1:hENT1 cells (yFS240) labeled or unlabeled with IdU was fractionated on CsCl density gradients. DNA from the fractions was recovered by ethanol precipitation, run on a 1% TAE agarose gel and visualized by EtBr staining. The gradient fractions, from denser to lighter, were matched by refractive index, and loaded from left to right on the gel.
adh1:tk adh1:hENT1 cells (yFS240) growing exponentially in YES at 30°C were labeled for three hours in 70 μM IdU. 50 OD units of cells were harvested. DNA was isolated as described above and resuspended in 200 μl of TE with 20μg/ml RNase A. A CsCl solution of refractive index 1.4021 (approximately 1.28 g/ml CsCl) was prepared in TE of refractive index of 1.3329 (adjusted by adding 10 mM Tris). Refractive indices were measured using a Reichert-Jung Leica Mark II Abbe Refractometer. The DNA sample was placed in a 5 ml Beckman quickseal centrifuge tube which was then filled with CsCl solution and sealed. Gradients were formed by spinning for in an NVT90 rotor in a Beckman L8-55M Ultracentrifuge at 50 krpm for 20 hours. Gradients were manually fractionated into approximately 250μl fractions by dripping out of the bottom of the punctured tube. The refractive index was taken using 100μl of each fraction. 125μl of each fraction was diluted to 500 μl with water, and the DNA was precipitated by addition of 1μl of glycogen and 1 ml of ethanol. The pellets were washed with 70% ethanol, dried and resuspended in 21μl of TE. Seven μl of each fraction was run on a 1% agarose gel (Figure 3).
Positive and Negative Selection for tk and hENT1
Negative selection makers are useful for plasmid shuffle experiments and for the knockin of site-directed mutations into a genomic locus. ura4 can be selected against with 5-fluoroorotic acid. However, in fission yeast, this selection is not very tight. Better results are obtained by selecting against adh1:tk using 5-fluoro-deoxyuridine (FdU, more commonly called FUdR, for 5-fluorouracil deoxyriboside, Sigma F0503)[5]. (N.B. FUdR is 5-fluoro-2'-deoxyuridine; 5-fluoro-5'-deoxyuridine, an anti-cancer drug, is not specifically toxic to tk+ cells.) FUdR is phosphorylated by tk to 5-fluorodeoxyuridylate (F-dUMP). F-dUMP is a non-competitive inhibitor of thymidylate synthase, the enzyme that methylates dUMP to make dTMP. Thus, F-dUMP starves cells for dTTP. adh1:tk cells growing in YES are killed by 100 nM FUdR, but are unaffected by 1 nM FUdR; tk- cells are unaffected by 100 μM FUdR (data not shown). At 10 nM FUdR, adh1:tk cells grow slowly and are elongated by a rad3 dependent checkpoint (data not shown). The presence of adh1:hENT1 does not affect the sensitivity of cells to FUdR. The selection is strong enough that if adh1:tk is integrated between repeated sequences, excision mutants are readily obtained (data not shown). One advantage of the pJL218 integration used to construct yFS240 is that it does not readily excise, presumably because it is integrated in a non-homologous locus.
The toxicity of FUdR to adh1:tk adh1:hENT1 cells can be completely suppressed by supplying exogenous 500 nM thymidine. This rescue demonstrates that hENT1 cells can grow normally on entirely exogenous thymidine. However, higher levels of exogenous thymidine causes a toxicity of its own, presumably by upsetting cellular nucleotide pools. 1 μM thymidine slows growth somewhat, roughly doubling the growth rate of adh1:tk adh1:hENT1 cells; 100 μM prevents cell growth. 1 mM exogenous thymidine has no effect on cells that lack tk or hENT1.
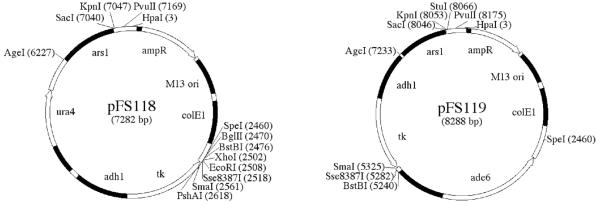
To facilitate plasmid shuffle experiments, we have constructed two episomal vectors that carry adh1:tk; pFS118 is marked with ura4, and pFS119 is marked with ade6 (Figure 4). The use of such vectors for plasmid shuffle screening of temperature-sensitive mutants has been described [5].
Figure 4.
adh1:tk based plasmid shuffle vectors
Unique restriction sites are marked that can be used for cloning without disrupting the function of the vectors. Sequence of the vectors is available at pingu.salk.edu/~forsburg/vectors.html
It is convenient to be able to both positively and negatively select for a marker. For instance, a gene can be knocked out with ura4 by selecting for Ura prototrophy, and then a site-directed mutant can be knocked back in by selecting for 5-FOA resistance. As described above adh1:tk can be negatively selected by using FUdR. The combination of adh1:tk and adh1:hENT1 can also be positively selected using sulfanilamide (SAA), methotrexate (MTX) and thymidine. Sulfanilamide inhibits dihydropteroate synthase, an enzyme required for dihydrofolate (DHF) synthesis; methotrexate inhibits dihydrofolate reductase (DHFR), the enzyme that reduces DHF to terahydrofolate (THF). THF is then methylated to N5, N10 methyeneTHF (mTHF), a 1-carbon donor in many biosynthetic reaction, including the synthesis of dTMP by thymidine synthase. However, thymidine synthase is the only enzyme that oxidizes mTHF back to DHF. Therefore, dTMP synthesis requires high levels of DHFR to regenerate THF, both for future dTMP synthesis and other mTHF dependent syntheses. Carefully selected levels of SAA and MTX can block enough THF synthesis to lethally inhibit dTMP synthesis while allowing sufficient THF accumulation for other mTHF dependent reactions. adh1:tk adh1:hENT1 cells can be rescued from SAA/MTX treatment by supplying exogenous thymidine for the synthesis of dTMP (Figure 3). The selection on SAA/MTX/TdR plates is not robust; adh1:tk adh1:hENT1 cells grow quite slowly and form only small colonies. Furthermore, a careful titration of the compounds is necessary. A two fold change in the concentration of SAA or MTX can cause the selection to fail. We found 5 mM SAA, 0.66 mM MTX, 0.5 mM TdR worked best. Colonies can be seen after 5-7 days (Figure 5).
Figure 5.
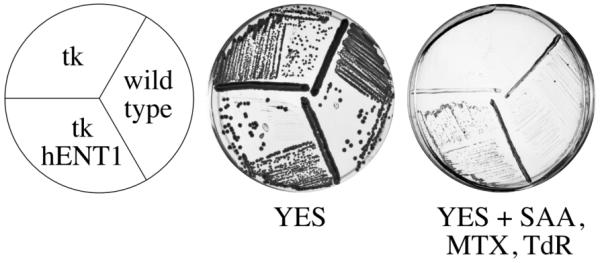
Positive selection for adh1:tk and adh1:hENT1
Wild-type cells (yFS105), adh1:tk cells (yFS233) or adh1:tk adh1:hENT1 cells (yFS240) were grown for 3 days at 30°C on YES or 5 days on YES + 5 mM SAA, 0.66 mM MTX, 0.5 mM TdR.
Because of the difficulty of selecting for adh1:tk and adh1:hENT1, we have created pFS255 containing a adh1:tk/kanMX6 cassette based on the commonly used kanMX6 cassette [16]. The primer binding sites of pFA6a-kanMX6 are preserved, so that the same oligos used to make kanMX6 knockouts can also be used to make adh1:tk/kanMX6 knockouts. adh1:tk/kanMX6 knockouts can then be used as a knockin target by selecting against adh1:tk as described above.
Discussion
In an effort to establish a quantitative assay for replication in fission yeast, we created a strain that expresses both herpes thymidine kinase and human equilibrating nucleoside transporter 1. This strain readily incorporates exogenous H3-thymidine into DNA (Figure 1). To our surprise, arresting cells in G2 and thus preventing nuclear replication, did not inhibit the incorporation of H3-thymidine. From this result we conclude that the majority of the incorporation is into mitochondrial DNA. Consistent with this conclusion, cells lacking mtDNA incorporate at a much reduced rate and fail to incorporate during G2 (data not shown). Fission yeast cell can survive without mtDNA if they carry the mutation ptp1-1 [17]. However these cells are quite sick, and we have been unable to perform meaningful analyses with them. It is unclear why the rate of incorporation into mtDNA should be so high, given mtDNA comprises only about 6% of total cellular DNA [18]. However, we see a strong labeling of mtDNA by indirect immunofluorescence of incorporated BrdU (Figure 2).
The real strength of these labeling techniques seems to be the rapidity with which exogenous thymidine can be incorporated. We see incorporation of BrdU in a 5 minute pulse (Figure 2A) and can detect 3H TdR incorporation with in two minutes (data not shown). Such short pulse labelings should allow many experiments that have not previously been possible in fission yeast. Another particularly promising approach is the use of BrdU labeling for in situ localization of replication. The ability identify the sites of DNA replication and the proteins that localize to such sites has been a powerful techniques in metazoan cells that should now be possible in fission yeast.
Acknowledgements
We are grateful to Janet Leatherwood for providing the adh1:tk construct used in this study, to Susan Forsburg for discussing her hENT1 tk strains before publication and for making our plasmid information available on her web site, and to John Diffley, who’s idea to use hENT1 and tk together for DNA labeling in yeast motivated this work. This work was supported by the Chestnut Hill Charitable Foundation and a Worcester Foundation for Biomedical Research Annual Research Fund Innovation Grant. N.R. was supported by a Leukemia and Lymphoma Special Fellowship.
References
- 1.Lengronne A, et al. Nucleic Acids Res. 2001;29:1433–1442. doi: 10.1093/nar/29.7.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McNeil JB, Friesen JD. Mol Gen Genet. 1981;184:386–393. doi: 10.1007/BF00352510. [DOI] [PubMed] [Google Scholar]
- 3.Sclafani RA, Fangman WL. Genetics. 1986;114:753–767. doi: 10.1093/genetics/114.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vernis L, et al. Nucleic Acids Res. 2003;31:e120. doi: 10.1093/nar/gng121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiely J, et al. Genetics. 2000;154:599–607. doi: 10.1093/genetics/154.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vickers MF, et al. Mol Membr Biol. 2001;18:73–79. [PubMed] [Google Scholar]
- 7.Griffiths M, et al. Nat Med. 1997;3:89–93. doi: 10.1038/nm0197-89. [DOI] [PubMed] [Google Scholar]
- 8.Vickers MF, et al. Biochem J. 1999;339:21–32. [PMC free article] [PubMed] [Google Scholar]
- 9.Hodson JA, et al. Nucleic Acid Research. 31 in press. [Google Scholar]
- 10.Moreno S, et al. Methods Enzymol. Vol. 194. Academic Press; New York: 1991. pp. 795–823. 795795. [DOI] [PubMed] [Google Scholar]
- 11.Alfa C, et al. A Laboratory Course Manual. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1993. Experiments with Fission Yeast. [Google Scholar]
- 12.Clyne RK, Kelly TJ. EMBO J. 1995;14:6348–6357. doi: 10.1002/j.1460-2075.1995.tb00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLeod M, et al. EMBO J. 1987;6:729–736. doi: 10.1002/j.1460-2075.1987.tb04814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keeney JB, Boeke JD. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fangman WL, Brewer BJ. Annu Rev Cell Biol. 1991;7:375–402. doi: 10.1146/annurev.cb.07.110191.002111. [DOI] [PubMed] [Google Scholar]
- 16.Bähler J, et al. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 17.Haffter P, Fox TD. Genetics. 1992;131:255–260. doi: 10.1093/genetics/131.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bostock CJ. Biochim Biophys Acta. 1969;195:579–581. doi: 10.1016/0005-2787(69)90667-4. [DOI] [PubMed] [Google Scholar]