The 5′UTR intron of GGT1 determines maximum transcript abundance and affects RNA polymerase II binding.
Abstract
Photorespiration is essential for the detoxification of glycolate and recycling of carbon to the Calvin Benson Bassham cycle. Enzymes participating in the pathway have been identified, and investigations now focus on the regulation of photorespiration by transporters and metabolites. However, regulation of photorespiration on the gene level has not been intensively studied. Here, we show that maximum transcript abundance of Glu:glyoxylate aminotransferase 1 (GGT1) is regulated by intron-mediated enhancement (IME) of the 5′ leader intron rather than by regulatory elements in the 5′ upstream region. The intron is rich in CT-stretches and contains the motif TGTGATTTG that is highly similar to the IME-related motif TTNGATYTG. The GGT1 intron also confers leaf-specific expression of foreign promoters. Quantitative PCR analysis and GUS activity measurements revealed that IME of the GGT1 5′UTR intron is controlled on the transcriptional level. IME by the GGT1 5′UTR intron was at least 2-fold. Chromatin immunoprecipitation experiments showed that the abundance of RNA polymerase II binding to the intron-less construct is reduced.
A central step in the evolution of photosynthesis was the evolution of the CO2-fixing enzyme Rubisco 3.5 billion years ago (Sage, 2004). Rubisco accepts O2 as an alternative substrate. O2 and CO2 compete for binding to the same catalytic center. Even though the affinity of Rubisco for CO2 is 100 times higher than for O2 (Jordan and Ogren, 1984), every fourth reaction that is catalyzed by Rubisco is an oxygenation reaction (Sharkey, 2001). The fixation of O2 leads to the formation of 3-phosphoglycerate and 2-phosphoglycolate. The subsequent dephosphorylation of the latter produces the toxic intermediate glycolate, which needs to be detoxified in the pathway of photorespiration (Stabenau and Winkler, 2005; Peterhänsel et al., 2010). Since the pathway of photorespiration was resolved by an EMS mutant screen in Arabidopsis (Arabidopsis thaliana; Somerville and Ogren, 1979; Somerville, 2001), intensive research has been done on the characterization of the different enzymes involved. To date, the focus of research turns toward the regulation of photorespiration on different levels, such as transport (Eisenhut et al., 2013) and metabolites (Timm et al., 2013). However, little is known about the coordinated regulation of photorespiration on gene level.
A central compartment of higher plant photorespiration is the peroxisome in which glycolate is metabolized by the key enzyme glycolate oxidase (Igamberdiev and Lea, 2002). The product of this reaction, glyoxylate, is the substrate of Glu:glyoxylate aminotransferase (GGT; Liepman and Olsen, 2003). In Arabidopsis, two peroxisome located GGT isoforms exist (Liepman and Olsen, 2003; Igarashi et al., 2003). Even though GGT1 (At1g23310) and GGT2 (At1g70580) show 97.9% similarity on protein level, the major function in photorespiration can be attributed to GGT1. This is indicated by the observation that the severe growth phenotype of ggt1 knockout mutants (Igarashi et al., 2003; Verslues et al., 2007; Dellero et al., 2015) cannot be rescued by GGT2. However, the phenotype can be alleviated by the addition of Suc and salvaged by high CO2 concentrations (Igarashi et al., 2003), implying a direct connection to photorespiration. Analyses of ggt1 knockout mutants in the last decade have underlined the importance of photorespiration for detoxifying glycolate and recycling carbon to the Calvin Benson Bassham cycle. Despite the high energy costs of this process, photorespiration has been found to play a central role in the control of amino acid biosynthesis and metabolism. It has been estimated that ammonia fixed by photorespiration is 50 times higher than the primary assimilation of nitrogen that is fixed by nitrate reduction (Keys, 2006). Igarashi et al. (2006) confirmed a direct interaction of amino acid accumulation and photorespiration. Recently, it was shown that ggt1 mutants have a 50 times lower CO2-fixation rate compared to the wild type (Dellero et al., 2015). As a consequence, nitrogen assimilation became limited by low net CO2 assimilation rates, which subsequently led to a decrease of leaf Rubisco content. In addition, GGT1 was found to have an indirect effect on stress and abscisic acid (ABA) response due to the accumulation of hydrogen peroxide in ggt1 knockout mutants (Verslues et al., 2007). The authors assumed that the reduced sensitivity of ggt1 mutants toward stress and ABA was caused by the high level of hydrogen peroxide and, therefore, led to perturbation of the basal stress response of the plants.
However, nothing is known about the regulation of GGT1 on gene level. Eukaryotic gene regulation is not only driven by cis-elements in the 5′ upstream region of genes but is also mediated by elements and features in the untranslated regions and other noncoding sequences (Barrett et al., 2012). According to the TAIR (Huala et al., 2001) and Athena databases (O’Connor et al., 2005), GGT1 contains a 5′ upstream region of approximately 1,500 bp relative to the transcription initiation start. This region is terminated by the next upstream lying gene. A characteristic of the GGT1 gene structure is an intron in its 5′UTR. 5′ proximal introns of genes have been found to be associated with intron-mediated enhancement (Callis et al., 1987; Mascarenhas et al., 1990). Introns have been shown to be required for high expression of many genes in several organisms including plants (Callis et al., 1987), mammals (Furger et al., 2002), Saccharomyces cerevisiae (Moabbi et al., 2012), nematodes (Okkema et al., 1993), and insects (Jiang et al., 2015). Moreover, 79% of all Arabidopsis genes contain introns (Arabidopsis Genome Initiative, 2000). Thus, introns are thought to be of great importance for gene regulation through mechanism like alternative splicing (Nilsen and Graveley, 2010).
Based on this, organ specificity and diurnal of the GGT1 gene was investigated by analyzing the two characteristics in both 5′ upstream deletion::gusA mutants and a mutant lacking the intron in the GGT1 5′UTR (GGT1 Δ5I::gusA). Data revealed that GGT1 transcription was elevated by intron-mediated enhancement by its 5′UTR intron (5I). GGT1 5I was able to substitute the endogenous 5′UTR intron of the second GGT isoform GGT2 and mediated leaf expression of the chimeric construct. Intron-mediated enhancement (IME) of GGT1 5I was associated with higher abundance of RNA polymerase II on the gusA coding sequence.
RESULTS
Transgenic GGT1::gusA and Endogenous GGT1 Transcripts Display the Same Regulation
To investigate regulatory features of the GGT1 promoter, the GGT1 (At1g23310) 5′ upstream sequence (1,454 bp) as well as the 5′UTR (604 bp) was cloned upstream of the gusA gene into the pSAG vector (Adwy et al., 2015), resulting in GGT1::gusA. GusA, formally known as uidA, encodes for the bacterial reporter protein β-glucuronidase. Transgenic lines were produced by floral dip and independent transformation events were used for further analyses. Both endogenous GGT1 (Fig. 1A) and transgenic gusA mRNA (Fig. 1B) levels were analyzed for two parameters: (1) tissue specific expression in leaves and roots and (2) diurnal regulation. GAPDH mRNA was measured as a control (Fig. 1C). Leaf and root material used to determine tissue-specific expression was harvested from hydroponically grown plants because both tissue types could be harvested separately in sufficient amounts. Diurnal transcript abundance was studied in 16-d-old plants.
Figure 1.
GusA expression levels driven by the GGT1 5′ upstream sequence in transgenic plants resemble regulatory features of the endogenous GGT1. Relative transcript levels of (A) endogenous GGT1, (B) transgenic GGT1::gusA, and (C) GAPDH levels were measured in leaf and root tissue throughout the day. Arabidopsis transgenic plants were grown under short-day conditions for either 6 weeks in hydroponical cultures (tissue specificity) or 16 d on MS plates (diurnal). Plant material was harvested either 1 h after illumination (tissue specificity) or at the time points given. Data for diurnal regulation were generated from plant material presented in Figure 3D. Data represent 8–10 individual transformation events ± sd. Student’s t test significance levels are as followed: **P ≤ 0.05, ***P ≤ 0.01. 1hL, 1 h light; 7hL, 7 h light.
Figure 1 shows that endogenous GGT1 mRNA was much more abundant in leaves over roots (Fig. 1A, tissue specificity). This pattern was also observed for gusA mRNA levels, whereas GAPDH levels were identical in both tissues (Fig. 1, A and B, tissue specificity). GGT1 mRNA transcript abundance was higher in the morning (1hL) compared to the end of the day (7hL) (Fig. 1A, diurnal). GusA mRNA levels showed an identical diurnal regulation (Fig. 1B, diurnal). Again, control mRNA levels of GAPDH were identical at both time points (Fig. 1B, diurnal).
In summary, the GGT1::gusA construct reproduced the expression patterns of the endogenous GGT1 and, thus, was a suitable tool to investigate regulatory elements of the GGT1 gene.
5′ Upstream Deletion Mutants Did Not Identify a Region of Positive GGT1 Gene Regulation
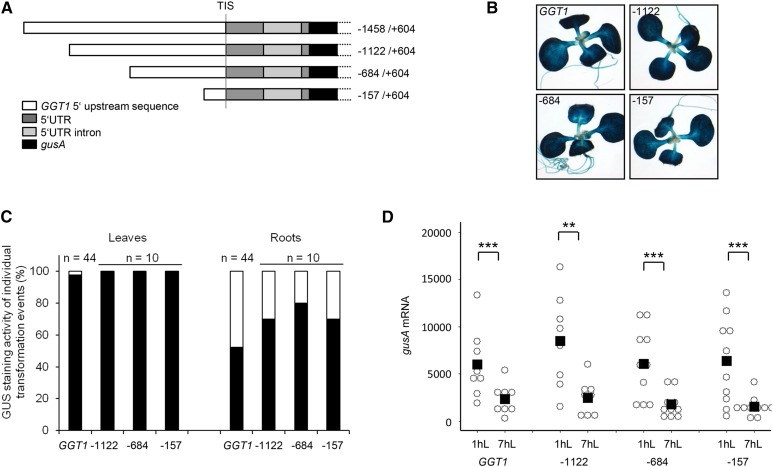
In a second step, the influence of 5′ upstream sequences on the regulation of the GGT1 gene was tested. Therefore, three sequential 5′ upstream deletions mutants (5′UDMs) at positions −1122, −684, and −157 relative to the transcription initiation site were generated (Fig. 2A).
Figure 2.
The 5′ upstream deletions could not identify regulatory cis-elements for tissue specificity, diurnal expression, and maximum promoter activity in the GGT1 promoter. A, Schematic overview over the different GGT1 5′ upstream deletion constructs. B, GUS staining of 16-d-old transgenic plants. The images show a representative seedling of the GUS staining pattern of plants originating from 10 individual transformation events of each construct (Supplemental Fig. S2). C, Tissue specificity of GGT1::gusA and 5′ deletion constructs as observed in the individual transformation events. Black bar, expression in leaves/roots; white bar, no expression. Relative gusA mRNA transcript levels of 16-d-old plants were quantified by qPCR (D, diurnal). Individual transformation events of each construct are visualized by open circles while the mean of the data are indicated by a black square in the dot plot. The mean represents the average of 8-10 individual transformation events. Student’s t test significance levels are as followed: **P ≤ 0.05, ***P ≤ 0.01. 1hL, 1 h light; 7hL, 7 h light; TIS, transcription initiation start.
After cloning the constructs into the GUS expression vector pSAG, stable Arabidopsis plants were produced by Agrobacterium-mediated transformation. Again independent transformation events were used for analyzing tissue specificity by GUS staining and diurnal regulation of gusA mRNA levels, respectively. Supplemental Figures S1 and S2 summarize all individual transformation events that have been generated and used in the data set of Figure 2.
GGT1::gusA plants showed a deep blue GUS staining in leaves and cotyledons in 98% of all tested transformation events, while root expression was only visible in 50% of the plants and was generally much weaker (Fig. 2, B and C). Similar to the pattern found in GGT1::gusA plants, the GUS staining pattern in leaves was identical in the 5′UDM constructs. Root expression was detected in 70 to 80% of the individual transformation events of the 5′ UDMs (Fig. 2B and C). Thus, expression in leaves and roots was not disturbed in 5′UDMs.
Figure 2D illustrates diurnal mRNA levels of GGT1::gusA and the three 5′UDMs. Importantly, none of the 5′UDMs showed significant changes in diurnal regulation compared to the full length construct (Fig. 2D). For clarity, individual data on the diurnal rhythm of the individual transformation events are summarized in Supplemental Table S1.
Taken together, deletion of the 5′ upstream sequence did not identify sequences required for GGT1 gene regulation.
The 5′UTR Intron of GGT1 Is Involved in the Regulation of Maximum Transcript Abundance
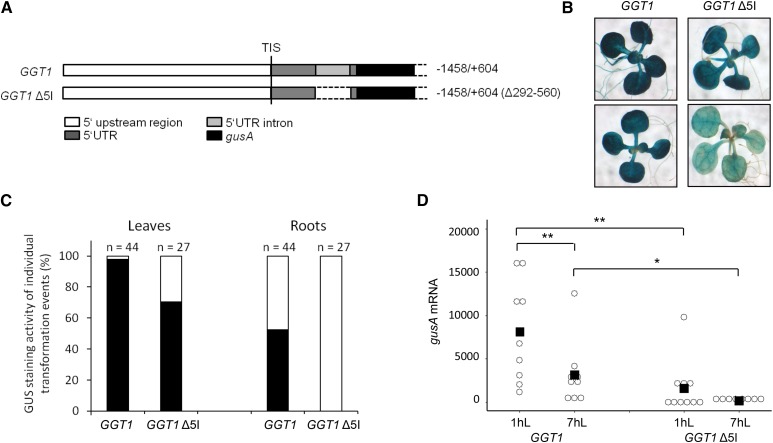
Bioinformatic analysis of the GGT1 genomic sequence revealed that GGT1 contains an intron in its 5′UTR. It is known that introns that are located close to the transcription initiation site enhance transcription via a mechanism called IME (Callis et al., 1987; Mascarenhas et al., 1990; Parra et al., 2011). Thus, we hypothesized that the 5′UTR intron (5I) of GGT1 has a positive influence on GGT1 transcript levels. To test this hypothesis, the intron was deleted from the 5′UTR sequence of GGT1::gusA, resulting in GGT1 Δ5I::gusA (Fig. 3A). After stable transformation into Arabidopsis and selection of independent transformation events, GGT1::gusA and GGT1 Δ5I::gusA lines were analyzed for tissue specificity and diurnal expression. Supplemental Figures S1 and S3 summarize all individual transformation events that have been generated and used in the data set of Figure 3.
Figure 3.
GGT1 expression is enhanced by its 5′UTR intron (5I). A, Schematic overview over the GGT1 5I deletion construct in comparison to the nonmutated 5′UTR region. B, GUS staining of 16-d-old plants. The images show a representative seedling of the GUS staining pattern of plants originating from 10 individual transformation events of each construct (Supplemental Fig. S3). C, Plot of GUS levels in leaves and roots of individual transformation events as observed after GUS staining. Black bar, Expression in leaves/roots; white bar, no expression. Relative gusA mRNA transcript levels of 16-d-old transgenic plants were quantified by qPCR (D, diurnal). Arabidopsis plants were grown under short-day conditions for 16 d on MS plates. Individual transformation events of each construct are visualized by open circles while the mean of the data are indicated by a black square in the dot plot. The mean represents the average of 7–10 individual transformation events. Student’s t test significance levels are as followed: *P ≤ 0.1, **P ≤ 0.05. 1hL, 1 h light; 7hL, 7 h light; TIS, transcription initiation start.
Figure 3B illustrates the tissue specificity of both constructs. It was obvious that transgenic plants carrying the GGT1 Δ5I::gusA construct showed less intense GUS staining in leaves compared to GGT1::gusA plants. Secondly, expression in leaves and roots was qualitatively analyzed (Fig. 3C). It was observed that only 70% of the 27 individual transformation events of Ggt1 Δ5I::gusA but 98% of the 44 Ggt1::gusA transformation events showed GUS activity in leaves. Furthermore, GUS staining was not observed in roots of GGT1 Δ5I::gusA plants, while 50% of the roots from GGT1::gusA transformed plants stained for GUS. However, staining of GGT1 Δ5I::gusA plants was very inhomogeneous in general. To validate the influence of GGT1 5I on gene expression further, all experiments were performed using only those plants that showed a deeper blue staining (Supplemental Fig. S1, Ggt1 Δ5I::gusA plants 18–27).
In a next step, changes in diurnal expression were analyzed in the transgenic plants. Figure 3D shows gusA mRNA levels of GGT1::gusA and GGT1 Δ5I::gusA plants 1 h and 7 h after illumination. Diurnal regulation was observed for both constructs. One hour after illumination, gusA mRNA levels were approximately 5.1-fold higher in GGT1::gusA plants compared to the Ggt1 Δ5I::Gus plants. For clarity, individual data on the diurnal rhythm of the individual transformation events are summarized in Supplemental Table S1
Taken together, the data suggested a role of GGT1 5I in the enhancement of promoter activity as well as the regulation of maximum transcript abundance.
GGT1 5I Drives Leaf Expression of GGT2 When Substituted for the GGT2 5′UTR Intron
In Arabidopsis, a second GGT isoform exists, GGT2 (At1g70580; Igarashi et al., 2003, Liepman and Olsen, 2003). GGT1 and GGT2 show 97.9% similarity on protein level and share the same gene structure. According to in silico predictions, both isoforms include an intron in their 5′UTR. A prediction by the TAIR database (Huala et al., 2001) revealed four different splice forms of GGT2 that (1) differ in the length of the 5′UTR and (2) in the position and length of the 5′UTR intron (Supplemental Fig. S4). To identify the most abundant splice form of GGT2, splice form specific primers were designed that allowed to distinguish between the four splice forms on cDNA level (Supplemental Fig. S4). cDNA of both 16-d-old and 5-week-old plants was used to exclude any developmental effects on the abundance of the splice forms. In this experiment, splice form 1 was determined as the most abundant splice form at the developmental stages tested (Supplemental Fig. S5).
Publications by Igarashi et al. (2003), Verslues et al. (2007), and Dellero et al. (2015) have shown that a ggt1 knockout led to a severe growth phenotype that could be rescued by high CO2 concentrations as expected for a gene or enzyme involved in photorespiration. Thus, GGT2 could not fully compensate for the loss of GGT1 in photorespiration. Expression of the gusA gene under the control of the GGT2 5′upstream region and the endogenous 5′UTR showed that the GGT2 promoter was mainly active in the vascular system and in cotyledons (Fig. 4C). However, GGT1 and GGT2 both contain an intron in their 5′UTR. Thus, we wanted to test the hypothesis whether GGT1 5I would be able to enhance expression of GGT2 in leaves and roots when substituted for the endogenous GGT2 5I (Fig. 4A). On the other hand, we also aimed testing the functionality of the GGT2 5I using a GGT2 Δ5I mutant and an equivalent intron swap mutant to the one described above in which the endogenous GGT1 5I was substituted by GGT2 5I (Fig. 4A).
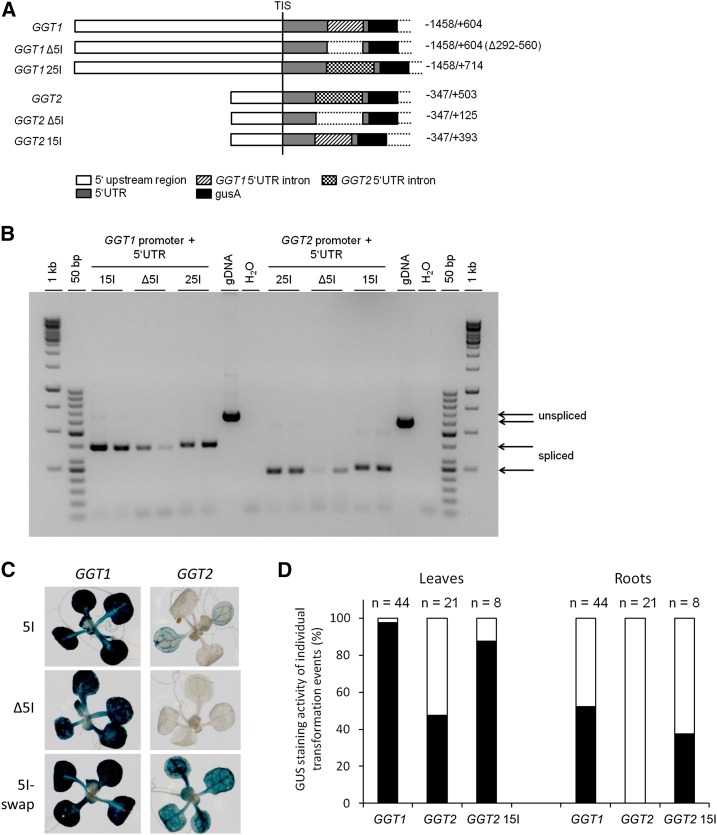
Figure 4.
GGT1 5I drives leaf expression of GGT2 when substituted for GGT2 5I. A, Schematic overview over the different constructs. B, PCR confirming the correct splicing of the introns located in the 5′UTRs. C, GUS staining of 16-d-old plants. The images show a representative seedling of the GUS staining pattern of plants originating from 10 individual transformation events of each construct (Supplemental Fig. S6). D, Plot of GUS levels in leaves and roots of individual transformation events as observed by GUS staining. Black bar, Expression in leaves/roots; white bar, no expression. 15I, GGT1 5′UTR intron; 25I, GGT2 5′UTR intron; Δ5I, deletion of the 5′UTR intron; Gdna, genomic DNA; TIS, transcription initiation start.
The chimeric constructs GGT1 25I::gusA (endogenous 5I substituted by GGT2 5I) and GGT2 15I::gusA (endogenous 5I substituted by GGT1 5I) were amplified in a series of overlapping extension PCRs (Supplemental Fig. S10). Stable Arabidopsis plants were generated, and individual transformation events were selected. Supplemental Figures S1 and S6 summarize all individual transformation events that have been generated and used in the data set of Figure 4 and Figure 5. All transgenic lines were analyzed for both tissue specificity and diurnal regulation.
Figure 5.
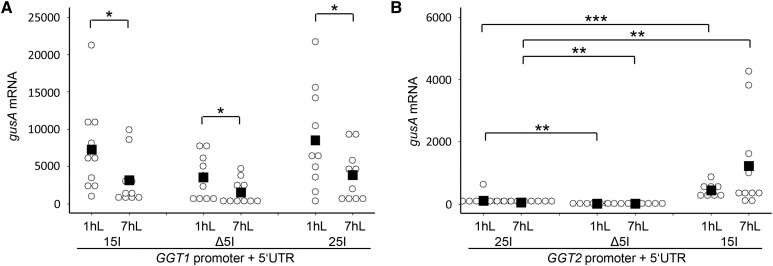
Quantification of the IME effect in the chimeric GGT constructs. Relative gusA mRNA transcript levels of 16-d-old transgenic plants were quantified by qPCR in the diurnal rhythm. Individual transformation events of each construct are visualized by open circles, while the mean of the data are indicated by a black square in the dot plot. The mean represents the average of 7–10 individual transformation events. Student’s t test significance levels are as followS: *P ≤ 0.1, **P ≤ 0.05, ***P ≤ 0.01. 15I, GGT1 5′UTR intron; 25I, GGT2 5′UTR intron; Δ5I, deletion of the 5′UTR intron; 1hL, 1 h light; 7hL, 7 h light.
Before analyzing the plants, it was tested whether the transgenic introns were correctly spliced from the 5′UTRs in the chimeric constructs. Figure 4B shows the amplification of the splice products from cDNA 1 h after illumination. The natural 5′UTR introns of GGT1 and GGT2 were fully spliced as were the introns in the chimeric constructs. However, splicing was partially incomplete in GGT2 15I::gusA plants. This was indicated by a faint PCR product at approximately 500 bp that reflected the unspliced GGT2 15I version. Because the GGT1 5I is 110 bp shorter than the GGT2 5I, the unspliced PCR product was smaller than that of our genomic DNA control used in this experiment (Fig. 4B, unspliced GGT2 sequence).
First, the GUS staining pattern was analyzed in the different constructs (Fig. 4C). As observed before, a strong GUS expression was visible in the GGT1::gusA construct, while the intensity was slightly reduced in the intron-less version GGT1 Δ5I::gusA (Figs. 3B and 4C). GUS staining of the plant carrying the substitution of the endogenous GGT1 5I by GGT2 5I was indistinguishable from the intensity observed for GGT1::gusA plants (Fig. 4C). The deletion of GGT2 5I completely abolished GUS staining in all tissues (Fig. 4C). However, the substitution of GGT2 5I by GGT1 5I led to (1) expression of the GGT2 promoter in leaves and in roots and (2) further enhancement of GGT2 expression in the vasculature (Fig. 4C). Because the observation that GGT2 is only weakly expressed in leaves contradicts an observation by Igarashi et al. (2006), we additionally proved successful transformation of the individual transformation events used in this experiment. For this, the bacterial neomycin phosphotransferase (nptII) gene that confers kanamycin resistance of the transgenic lines was amplified by qPCR. All lines showed a good expression of nptII (Supplemental Fig. S7). Thus, the weak GUS staining of GGT2::gusA and GGT2 Δ5I::gusA lines was not due to an inefficient transformation.
Qualitative analysis of GUS staining in leaves and roots underlined the activation of the GGT2 promoter by GGT1 5I in leaves and roots of GGT2 15I::gusA transgenic lines (Fig. 4D). While 52% of the 21 individual GGT2 transgenic lines showed weak GUS staining in leaves, 87% of the transgenic lines carrying GGT2 15I::gusA showed a significant increase in staining in this tissue type (Fig. 4D). In roots, GGT2 expression was activated in 38% of the transgenic lines carrying GGT2 15I::gusA, while no staining was observed in roots of GGT2::gusA lines (Fig. 4D).
In a next step, the diurnal regulation of the two GGT::gusA constructs was analyzed in comparison to their respective intron deletion mutants and chimeric intron swap constructs. The enhancing effect of GGT1 5I in the chimeric GGT2 15I::gusA lines was also confirmed on the transcript level for diurnal regulation (Fig. 5B). As indicated by the GUS staining, gusA mRNA transcript was hardly detectable in GGT2::gusA and GGT2 Δ5I::gusA lines (Figs. 4C and 5B). In GGT2 15I::gusA plants, mRNA levels were elevated 10- and 21.1-fold compared to GGT2::gusA 1 h and 7 h after illumination, respectively. This was approximately 0.1-fold of the gusA mRNA transcript levels in GGT1::gusA plants 1 h after illumination. Quantitative qPCRs data revealed that GGT2 5I can substitute for the endogenous GGT1 5I to a certain extent (Fig. 5). However, this effect could not be seen on the level of GUS staining (Fig. 4; Supplemental Fig. S6). For clarity, individual data on the diurnal rhythm of the individual transformation events are summarized in Supplemental Table S1
In summary, GGT1 5I drove expression of GGT2 in leaves when substituted for the endogenous GGT2 5I. However, GGT2 5I was only shown to significantly mediated enhancement of its endogenous promoter.
CT-Stretches Are Highly Abundant in the 5′UTRs and the Imbedded Intron Sequences of GGT1 and GGT2
The mechanism of IME is still largely unknown, and many different models have been proposed since its discovery in plants 1987 by Callis and colleagues (Callis et al., 1987; Gallegos and Rose, 2015). Along with this, the aim was to define DNA sequence motifs that are linked to IME. Here, T-rich sequences have been proposed to be important for controlling gene expression (Huang et al., 1997). Furthermore, CT-richness in both the 5′UTR and the enhancing intron has been reported in spinach and petunia (Bolle et al., 1994; Mun et al., 2002). Bioinformatic analysis of enriched motifs in enhancing introns with the IMEter software (Rose et al., 2008) revealed two similar motifs in Arabidopsis with the consensus sequences TTNGATYTG (Rose et al., 2008) and CGATT (Parra et al., 2011).
Thus, the GGT1 and GGT2 5′UTRs and imbedded intron sequences were screened for the existence of CT-rich regions and motifs predicted by the IMEter software. Figure 7 shows that both GGT sequences are highly enriched in CT-rich regions both in the 5′UTR and the intron. A motif similar to TTNGATYTG (TGTGATTTG) was found at the promoter proximal site of GGT1 5I. However, GGT1 5I did not contain the IME-related sequence motif CGATT. In contrast to GGT1, two CGATT motifs are located in the GGT2 5′UTR intron sequence, one motif at the beginning of the intron and one at the end of the intron (Fig. 6).
Figure 7.
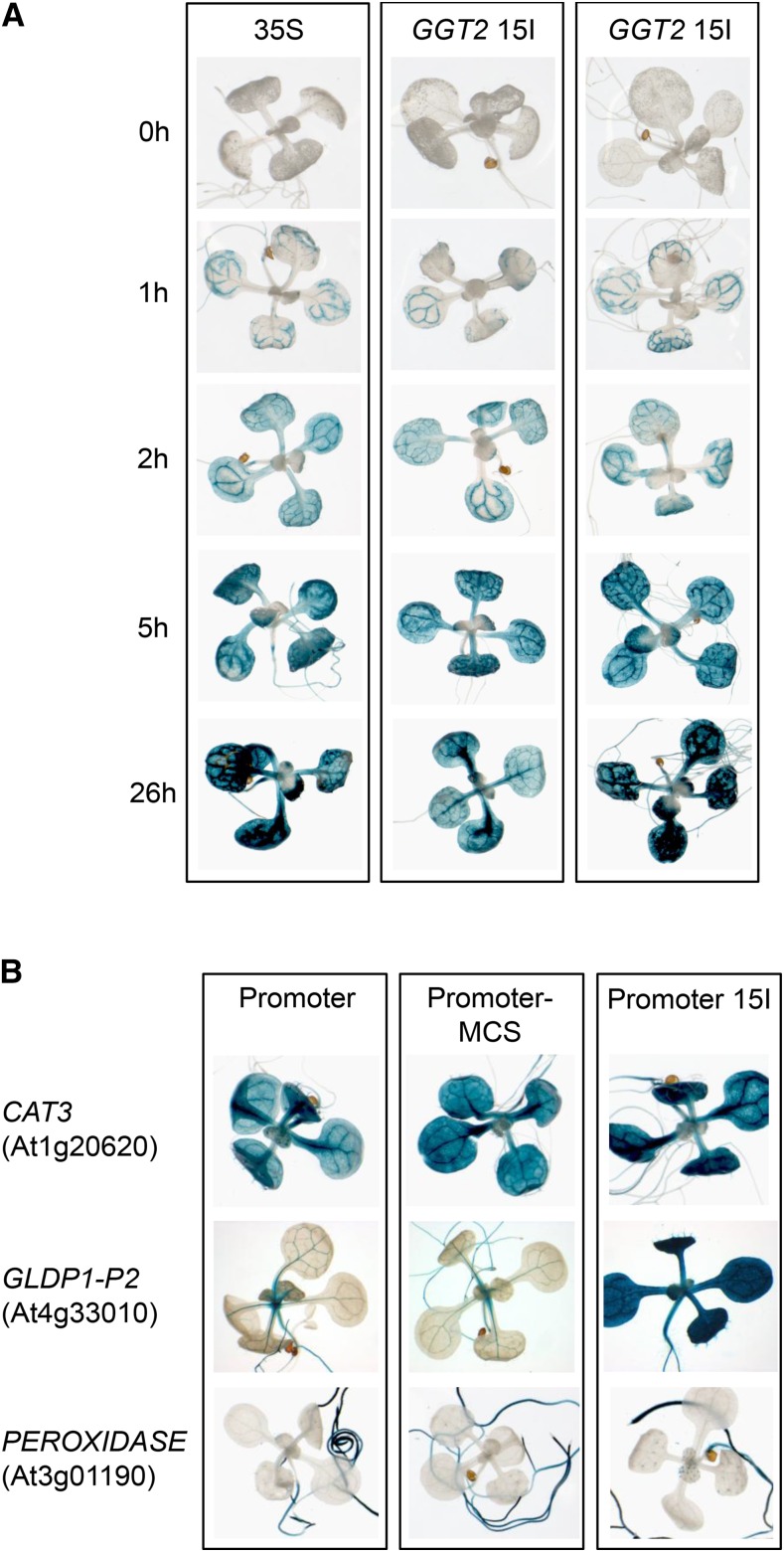
The 5′UTR intron of GGT1 can drive leaf expression of individual promoters. A, Time dependence of GUS staining of the GGT2 15I swap construct indicating leaf-specific expression of the GGT2 when the endogenous 5I is substituted by GGT1 5I. GUS staining was stopped by exchanging the GUS staining solution by 70% ethanol at the time points indicated. The 35S promoter was used as a control to observe the process of GUS staining driven by promoter that drives leaf expression. B, GUS staining of 16-d-old plants. The images show a representative seedling of the GUS staining pattern of plants originating from 8–16 individual transformation events of each construct (Supplemental Fig. S8). 15I, GGT1 5′UTR intron; MCS, multiple cloning site.
Figure 6.
IME related motives in the GGT1 and GGT2 5′UTRs and their imbedded introns. Gray, 5′UTR sequence; white, 5′UTR intron sequence; black box, C/T-stretches; dashed box, TTNGATYTG motif (Rose et al., 2008); underlined, CGATT motif (Parra et al., 2011). As an indication of C/T nucleotide accumulation, only C/T stretches of a nucleotide length of n ≥ 4 have been marked.
Introns that functions in intron-mediated enhancement are located in close vicinity to the transcription initiation site (TIS; Rose, 2004). Rose et al. (2008) developed the IMEter software that is based on structural differences between introns that are located close to TIS compared to introns that are located further downstream in the gene sequence. The bioinformatic analysis revealed that the 5′UTR introns of GGT1 and GGT2 have IMEter scores of 12.62 and 17.30, respectively, according to the version v2.1 of the IMEter software (Parra et al., 2011). Hence, according to a correlation by Rose et al. (2008), an increase in mRNA abundance of approximately 4- to 5-fold can be predicted for the GGT 5Is.
In summary, the existence of both important IME features in the complete 5′UTR sequences, and the high IMEter scores of the GGT 5Is additionally point toward a function of these introns in intron-mediated enhancement.
The GGT1 5′UTR Intron Changes Tissue-Specific Expression of Selected Promoters
The intron swap experiment showed that GGT1 5I drives leaf expression of the GGT2 promoter (Fig. 4C). We wanted to analyze this phenomenon in more detail. First, GUS staining was analyzed side by side with plants carrying 35S::gusA. The 35S promoter is known to be strongly expressed in most tissue types of Arabidopsis (Odell et al., 1985). Thus, it is a suitable tool to study the GUS staining pattern in a time-dependent manner. Furthermore, a ferricyanide concentration of 2.5 mm instead of 1 mm was used in the GUS staining buffer. The increase in the concentration of ferri- and ferrocyanide further accelerates the formation of the blue, insoluble GUS reaction product (Vitha et al., 2007) and, hence, minimizes the diffusion of the primary reaction product 5-bromo-4-chloro-3-indolyl from the side of its production. Thus, if a GUS staining in GGT2 15I plants is observed in leaves in the presence of a 2.5 mm ferri- and ferrocyanide, it can be excluded that 5-bromo-4-chloro-3-indolyl has diffused from the vascular system to the surrounding leaf tissue.
Figure 7A shows the GUS staining pattern of plants carrying 35S::gusA and GGT2 15I::gusA. GUS staining in 35S::gusA plants was visible after 1 h at 37°C. However, the staining was first restricted to the vascular system of the plants. After 2 h at 37°C, a weak GUS staining was also visible in leaves and roots, respectively. From here, GUS staining intensity successively increased with time. An identical time-dependent staining pattern was observed for two individual GGT2 15I::gusA lines (Fig. 7A).
Thus, the leaf expression observed in GGT2 15I::gusA plants is most likely due to an activation of the promoter by GGT1 5I rather than a matter of 5-bromo-4-chloro-3-indolyl diffusion from the vascular system to the surrounding leaf tissue.
Secondly, the influence of GGT1 5I on the tissue specificity on three other promoters was tested. Introns involved in IME are known to increase expression of foreign promoters (Callis et al., 1987). To test whether GGT1 5I is able to drive leaf expression of other promoters than GGT2, the promoter and 5′UTRs of three additional genes were cloned into the GUS expression vector. CATALASE3 (At1g20620, Zimmermann et al., 2006) and GLDP1-P2 (At4g33010, Adwy et al., 2015) are known to be mainly expressed in the vascular system, while the expression of a PEROXIDASE (At3g01190, Winter et al., 2007) is restricted to roots.
First, the tissue-specific expression of the three genes was tested in 16-d-old seedlings (Fig. 7B). GLDP1-P2 showed the reported expression pattern in the vascular system (Adwy et al., 2015), while CAT3 was expressed in the vascular system but also weakly in the entire leaf tissue as described by Zimmermann and colleagues (2006). As expected, the PEROXIDASE was proofed to be root-specific (Winter et al., 2007). The cloning of GGT1 5I downstream of the three genes necessitated the insertion of a multiple cloning site (MCS) into the linker sequence between the 5′UTRs and gusA. Plants carrying the promoter-MCS::gusA constructs showed a similar tissue-specific expression and GUS staining intensity when compared to the respective promoter::gusA lines (Fig. 7B).
The effect of GGT1 5I on the foreign promoters was very different. CAT3 15I::gusA and CAT3::gusA lines showed no difference in the intensity of GUS staining (Fig. 7B; Supplemental Fig. S8). As observed in GGT2 15I::gusA lines, GGT1 5I activated leaf expression of GLDP1-P2, which is exclusively expressed in the vascular system (Adwy et al., 2015). However, GGT1 5I was not able to drive expression of the PEROXIDASE in leaves but enhanced its expression in roots. Interestingly, 50% of the transformation events of PEROXIDASE 5I::gusA showed a staining in trichomes (Fig. 7B; Supplemental Fig. S8).
The data indicate once more that IME and its effect in gene expression and tissue specificity is not only depending on the intron itself but is more likely determined by sequences both in the promoter and the intron.
The 5′UTR Intron of GGT1 Enhances Expression by IME on the Transcriptional Level
IME can take place on various levels of gene expression, including both transcription and posttranscriptional processes (Samadder et al., 2008; Callis et al., 1987). To test which levels were affected by GGT1 and GGT2 5I mediated enhancement, gusA unspliced RNA levels as well as GUS protein activity were measured in samples originating from plant material analyzed in Figures 4 and 5.
Table I summarizes the percentages of unspliced RNA, mRNA, and GUS activity detected in GGT1::gusA and GGT2::gusA plants compared to both their intron-less versions and the 5′UTR intron swap constructs. For this, values of GGT1::gusA and GGT2::gusA were set to 100%. Relative unspliced RNA levels and GUS activity data are shown in Supplemental Figure S9.
Table I. Intron-mediated enhancement of GGT1 5I affects the transcriptional level.
RNA levels and GUS activity of 16-d-old plants were determined 1 h after onset of light under short-day conditions. Values of relative RNA transcript levels originate from data generated from plant material investigated in Figure 5. GUS activity (nmol/min/mg protein) was measured by the production of the fluorescent 4-methylumbelliferone according to Jefferson et al. (1987). Data represent the average of 8–10 individual transformation events ± sd. 15I GGT1 5′UTR intron, 25I: GGT2 5′UTR intron, Δ5I: deletion of the 5′UTR intron. Values measured for GGT1 15I and GGT2 25I lines were set to 100%. Student’s t test significance levels are as follows: *P ≤ 0.1.
| Construct | gusA Unspliced RNA Accumulation (%) | gusA mRNA Accumulation (%) | GUS Activity (%) |
|---|---|---|---|
| GGT1 15I | 100 | 100 | 100 |
| GGT1 Δ5I | 56.3 ± 53.9 | 40.3 ± 39.2 | 60.2 ± 40.5 |
| GGT1 25I | 123.2 ± 81.2 | 96.9 ± 69.1 | 50.9 ± 43.3 |
| GGT2 25I | 100 | 100 | 100 |
| GGT2 Δ5I | 31.4 ± 35.0 | 15.2 ± 16.3 | 0 ± 0 * |
| GGT2 15I | 1265.7 ± 1436.5* | 1005.6 ± 505.7 | 323.0 ± 395.0 |
The analysis of gusA unspliced RNA accumulation in GGT1 Δ5I::gusA plants revealed that IME already takes place on transcriptional level (Table I). The effect of GGT1 5I IME is constant at the different levels investigated. We observed remaining gusA unspliced RNA, gusA mRNA and GUS activity levels of 56.3% ± 53.9, 40.3% ± 39.2, and 60.2% ± 40.5, respectively, in GGT1 Δ5I::gusA plants (Table I). Substitution of the endogenous GGT1 5I by GGT2 5I compensated for the loss of 5I both on gusA unspliced RNA and mRNA, but not on the level of GUS activity. We recorded data of 123.2% ± 81.2, 96.9% ± 69.1, and 50.9% ± 43.3 for GGT1 25I::gusA on gusA unspliced RNA, gusA mRNA, and GUS activity level, respectively (Table I). Thus, because the consequence of IME is always an increase in protein activity levels independently of the level IME takes place primarily, GGT2 5I was not able to substitute for the endogenous GGT1 5I.
In the case of GGT2, a reduction of gusA unspliced RNA accumulation was observed in GGT2 Δ5I::gusA lines (30.4% ± 35.0). Furthermore, the loss of GGT2 5I had a stronger effect on the levels of mRNA accumulation and GUS protein activity. While gusA mRNA levels of 15.2% ± 16.3 were still detectable in GGT2 Δ5I plants, no GUS activity was measurable (Table I). GGT1 5I strongly enhances expression of the GGT2 promoter. As observed for GGT2::gusA lines, expression was mainly enhanced on gusA unspliced RNA level. We measured an intron-mediated enhancement of 1265.7% ± 1436.5, 1005.6% ± 505.7, and 323.0% ± 395.0 on gusA unspliced RNA, gusA mRNA, and GUS activity levels, respectively (Table I). Thus, even though GGT1 5I mediates IME on all levels in its endogenous constellation, it fails to drive IME on all levels evenly in the GGT2 background.
In summary, the data indicated that IME by GGT1 5I mainly takes place on the level of transcription.
RNA Polymerase II Is a Possible Target of IME by Ggt1 5I
In the previous section, GGT1 5I IME was assign to transcription. It was assumed before that changes in chromatin modifications and RNA polymerase II (RNA Pol II) binding are associated with IME (Meinhart et al., 2005; Rose, 2008; Gallegos and Rose, 2015). To test the hypothesis that GGT1 5I had an influence on both the abundance of activating chromatin modifications RNA Pol II binding a chromatin immunoprecipitation (ChIP) experiment was performed using GGT1::gusA and GGT1 Δ5I::gusA lines. In this experiment, plants were grown for 5 weeks to produce sufficient leaf material for ChIP. To confirm that the loss of GGT1 5I also had an impact on gusA mRNA levels in 5-week-old plants, transcript levels were determined by qPCR from plants that were grown in parallel (Fig. 8B). Data confirmed the results shown for 16-d-old seedlings (Fig. 1).
Figure 8.
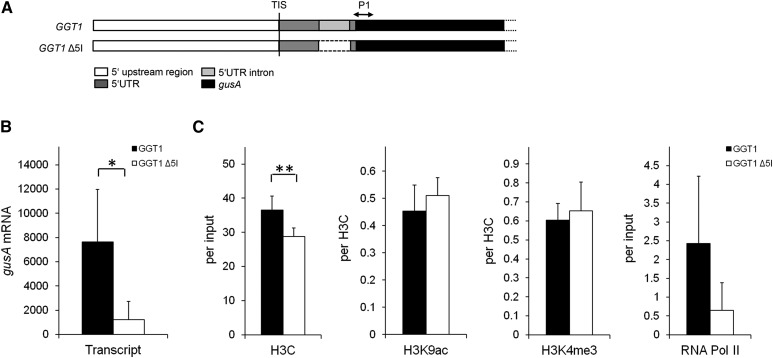
A lower abundance in RNA Pol II binding is a possible consequence of IME by GGT1 5I. A, Scheme of the position P1 (+146 bp downstream of ATG) measured on the gusA gene. B, Relative gusA mRNA levels in both GGT1::gusA and GGT1 Δ5I::gusA transgenic lines measured by qPCR. C, Chromatin analysis of GGT1::gusA and GGT1 Δ5I::gusA lines. Antibodies used were directed against the C terminus of histone 3 (H3C), histone 3 Lys 9 acetylation (H3K9ac), histone 3 Lys 4 trimethylation (H3K4me3), and RNA polymerase II (RNA Pol II). Plant material of 5-week-old plants was crosslinked 1 h after illumination. Data represent the average of four biological replicates ± se in which each sample is a pool of two individual transformation events which sums up to eight different lines used per construct. Detailed information on calculations is given in “Materials and Methods.” Student’s t test significance levels are as follows: *P ≤ 0.1, **P ≤ 0.05.
To discriminate between chromatin modifications and RNA Pol II abundance on the endogenous GGT1 locus and the reporter gene, the position P1 on the gusA gene was measured, which is centered +146 bp relative to the gusA ATG (Fig. 8A). ChIP analysis was performed for the activating modifications histone 3 Lys 9 acetylation (H3K9ac) and histone 3 Lys 4 trimethylation (H3K4me3; Santos-Rosa et al., 2002; Heintzman et al., 2007; Wang et al., 2009), as well as for the C terminus of histone 3 (H3C), which reflects nucleosome density at the position measured (Burlingame et al., 1985). In addition, an antibody directed against the C-terminal YSTPSPS repeat of the largest subunit of RNA Pol II was used (Nawrath et al., 1990).
Figure 8C summarizes the ChIP data recorded. Nucleosome density, expressed as H3C at P1, was lowered by 20% in the GGT1 Δ5I::gusA lines (37% of input) compared to GGT1::gusA (29% of input). Both activating modifications remained largely unchanged in GGT1 Δ5I::gusA plants compared to GGT1::gusA plants when standardized for H3C. Interestingly, an almost 4-fold reduction of the amount of RNA Pol II bound to P1 was observed between to GGT1::gusA and GGT1 Δ5I::gusA lines.
This suggests that RNA Pol II might be a possible target of IME by GGT1 5I on transcriptional level.
DISCUSSION
We studied regulatory features of the GGT1 promoter and found that the regulation of promoter strength could be attributed to the 5′UTR intron (5I) of GGT1 rather than being controlled by cis-elements in the 5′ upstream region of the gene. Furthermore, GGT1 5I was able to drive leaf expression of GGT2 and GLDP1-P2. Bioinformatics revealed high IMEter scores for GGT1 5I and GGT2 5I, respectively. Furthermore, the 5′UTR and the imbedded intron of both GGT1 and GGT2 contain IME-related motifs. We identified changes in RNA polymerase II binding as a characteristic of GGT1 5I-mediated enhancement on the transcriptional level.
GGT1 is an important component of plant photorespiration (Liepman and Olsen, 2003; Igarashi et al., 2003; Verslues et al., 2007). Loss-of-function mutants suffer from increased hydrogen peroxide concentrations and are sensitive to light (Verslues et al., 2007). Thus, tissue-specific expression in leaves and diurnal regulation are key regulatory features of the GGT1 promoter. Diurnal and tissue specific expression have been analyzed previously by Igarashi et al. (2006). In line with their results, we found that GGT1 transcripts were high in the morning and lower in the evening (Fig. 1A). In addition, diurnal expression was confirmed by the software tool “Diurnal” (Mockler et al., 2007). The analysis of transcript abundances in leaves and roots showed that GGT1 was much higher expressed in leaves over roots (Fig. 1, A and B). Quantitative data of endogenous and transgenic GGT1 expression were consistent with GUS staining intensities in leaves and roots in our study.
Expression of photorespiratory genes in roots has been investigated recently (Nunes-Nesi et al., 2014). Herein, the authors discuss a function of photorespiratory genes in heterotrophic tissues like roots. According to the Arabidopsis EFP Browser (Winter et al., 2007), GGT1 is expressed marginally in roots. Using the pep2pro proteome database (Baerenfaller et al., 2008), the normalized quantity spectra revealed values for about 50 and below 10 for leaves and roots of GGT1, respectively. This means that there is a low expression of GGT1 in roots. The corresponding normalized quantity spectra for GGT2 in both tissue types were below 4 and approximately 2, respectively. Thus, it can possibly be that GGT1 5I is important for root-specific expression in combination with its endogenous and the foreign GGT2 promoter. However, the function of GGT1 in roots remains to be elucidated.
Promoter Strength Is Mediated by the 5′UTR Intron of GGT1 Rather than by cis-Elements in the 5′ Upstream Region
Eukaryotic gene regulation is not only driven by cis-elements in the promoter of genes, but is also mediated by elements in untranslated regions like the 5′UTR and introns (Barrett et al., 2012). This holds true for GGT1 gene regulation.
Promoter strength of GGT1 is not determined by regulatory features in the 5′ upstream sequence of GGT1 (Fig. 2D). These features are attributed to GGT1 5I (Fig. 3D). However, diurnal regulation of GGT1 was impaired neither in the 5′UDMs nor in GGT1 Δ5I::gusA lines (Figs. 2D and 3D). Both the shortest 5′UDM (-157::gusA) and the GGT1 Δ5I::gusA construct still contain a putative CCA1-binding site motif −87 to −94 bp relative to the transcription initiation start of GGT1. The CCA1 motif has been associated with circadian clock function and, therefore, diurnal regulation (Michael and McClung, 2002). In contrast to the GGT1 5I deletion, the series of 5′ upstream deletions had no influence on maximum transcript abundance of GGT1 (Fig. 2D). This phenomenon has been described before. In Arabidopsis, the 5′ upstream region of FERREDOXIN A could be deleted up to −143 bp relative to the translation start without any changes in expression or tissue specificity (Vorst et al., 1993). In accordance with our finding, the Unc-54 gene in C.elegans functions with its promoter deleted but not without its introns (Okkema et al., 1993).
GGT1 and GGT2 Are Both Regulated by IME
GGT2 encodes the second isoform of the two GGTs that are located in peroxisomes in Arabidopsis (Liepman and Olsen, 2003). In our experiments, GGT2 expression was restricted to the cotyledons and the vasculature in 16-d-old GGT2::gusA plants (Fig. 4C). These data contradict the GUS expression pattern of GGT2 published by Igarashi and colleagues in which GUS expression was clearly visible in leaves and roots of 2-week-old plants (Igarashi et al., 2006). However, analysis of GGT2 expression in different databases, EIN3 seq browser (Chang et al., 2013), GeneAtlas (Kapushesky et al., 2010), and Genevestigator (Hruz et al., 2008) confirmed that GGT2 is much lower expressed in different tissues and developmental stages relative to GGT1, except during senescence. Furthermore, it was shown that a ggt1 knockout led to a severe growth phenotype, indicating that GGT2 could not fully compensate for the loss of GGT1 in photorespiration (Igarashi et al., 2003; Verslues et al., 2007; Dellero et al., 2015). GGT1 and GGT2 show 97.9% similarity on protein level. Thus, it is likely that expression of GGT2 in the leaf would replace GGT1 function in a ggt1 knockout. However, GGT2 transcripts were not induced in ggt1 mutant lines (Dellero et al., 2015). In addition, a ggt1 knockdown (SALK_064982) with a remaining GGT1 transcript level of 20% displays no photorespiratory phenotype (N. Lange, C. Peterhänsel, and M. Laxa, unpublished data) indicating that a fifth of the natural GGT1 activity is enough for normal growth.
We found that both GGT1 and GGT2 are regulated by their 5′UTR introns (Figs. 3–5). Both introns have high IMEter scores and contain CT-stretches as well as IME-related motifs (Fig. 6). Besides other features, the nucleotide composition of an intron rather than sequence motifs was suggested to be important for IME (Clancy and Hannah, 2002). CT-stretches have been identified as important elements for transcription in general (Bolle et al., 1994; Bolle et al., 1996; Huang et al., 1997). Importantly, CT-rich sequences were found in the 5′UTR and front sequence of the leader intron of petunia Adf1 whose expression in vegetative tissue is intron-dependent (Mun et al., 2002). In Arabidopsis, the effect of IME was enhanced by increasing the T-content within the intron (Rose, 2002). GGT1 5I contains the sequence TGTGATTTG that is highly similar to the IMEter consensus motif TTNGATYTG and not present in GGT2 5I (Rose et al., 2008). However, the second motif related to IME, CGATT (Parra et al., 2011), was found twice in GGT2 5I while it is missing in GGT1 5I (Fig. 6). Parra and colleagues showed that the addition of 11 copies of CGATT to the weakly enhancing Cor15α intron increased the enhancement by 4-fold relative to the control (Parra et al., 2011). Conversely, the deletion of this motif from the UBI10 intron only led to a 2-fold decrease of enhancement (Parra et al., 2011). Whether the IME-related motifs found in GGT1 5I and GGT2 5I play a significant role in the mechanism of IME, remains to be analyzed.
GGT1 5I Enhances the Expression of Foreign Promoters and Defines Tissue Specificity
GGT1 5I can drive leaf and root expression of GGT2 when substituted for GGT2 5I (Fig. 4). Furthermore, leaf expression was also observed for GLDP1-P2 15I::gusA lines (Fig. 7). In contrast, GGT1 5I was unable to drive leaf expression of the root-specific PEROXIDASE but induced the expression of the PEROXIDASE in trichomes in 50% of the transgenic lines (Fig. 7).
The ability of GGT1 5I to switch tissue-specific expression of foreign promoters showed that there is no specific link between introns and promoters for enhancement to take place. Introns can have a stronger influence on tissue specific expression than promoters have. In Arabidopsis, the first intron of UBIQUITIN10 disturbed the tissue-specific expression of CNGC2 and YAB3 and led to an expression of both genes in roots (Emami et al., 2013). Giani and colleagues found that the leader intron of rice beta-tubulin 4 regulates tissue-specific expression. They entitled the way of regulation with the term “intron dependent spatial expression,” or IDSE (Giani et al., 2009). Similar to our findings, the introns of Arabidopsis PRF2 and petunia Adh1 drive leaf expression of their respective promoters (Jeong et al., 2006; Mun et al., 2002).
The same intron can enhance expression of multiple genes (Emami et al., 2013; Callis et al., 1987). Independently from the ability of GGT1 5I to change tissue-specific expression, this holds true for GGT1 5I. With the exception of CAT3, GGT1 5I enhanced the promoter activity of all genes tested in this study (Figs. 4, 5, and 7).
The data underline the enormous spectrum of IME function in dependence of the intron sequence in combination with a specific promoter sequence.
GGT1 5I Mediated Enhancement Targets RNA Polymerase II on Transcriptional Level
GGT1 5I led to an enhancement on the transcriptional level in combination with its endogenous promoter in GGT1::gusA lines and the GGT2 promoter in GGT2 15I::gusA lines, respectively (Table I). IME targeting the level of transcription has been poorly described in the literature, but include mammals, yeast, and plants (Furger et al., 2002; Moabbi et al., 2012; Samadder et al., 2008). IME on the transcriptional level is rather weak, leading to fold changes that did not exceed 3-fold (Furger et al., 2002; Samadder et al., 2008). In line with this, we measured a 2- to 5.1-fold enhancement of gusA mRNA in stable GGT1::gusA lines compared to GGT1 Δ5I::gusA lines (Figs. 3D and 5A; Table I). However, GGT1 5I led to a strong transcriptional enhancement of 12.7-fold in GGT2 15I::gusA lines relative to GGT2::gusA lines (Fig. 5C, Table I). Callis and colleagues suggested that the weaker the promoter, the stronger the enhancement by introns (Callis et al., 1987). Recently, it was reported that the first intron of UBI10 led to different levels of enhancement when different promoters were used (Emami et al., 2013). In our case, it additionally needs to be taken into consideration that GGT1 5I changed the tissue specificity of GGT2 in the chimeric GGT2 15I::gusA plants (Fig. 4, C and D). Thus, the observed high level of induction on transcript level was also caused by GGT2 15I::gusA expression in leaves.
RNA Pol II and activating histone modifications have been associated with IME. The idea that histone modifications are linked to IME came from the observation that activating histone modifications are enriched at the 5′ boundaries of first introns in human genes (Bieberstein et al., 2012). Furthermore, Gallegos and Rose found a high similarity between the distribution of IMEter scores and activating histone modifications in Arabidopsis (Gallegos and Rose, 2015). The authors also suggested that introns might affect the chromatin state because introns have a tendency to be associated with fewer nucleosome compared to exons (Spies et al., 2009). However, no significant changes of activating histone modifications were found between GGT1::gusA and GGT1 Δ5I::gusA plants (Fig. 8B). But, the presence of GGT1 5I had an impact on the abundance of RNA Pol II binding to the gusA gene (Fig. 8B). Further experiments are necessary to unravel the molecular basis of how GGT1 5I affects RNA Pol II binding to a gene locus.
MATERIALS AND METHODS
Plant Material and Growth
Experiments were done with Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0). Col-0 was also used for stable transformations. Plants were grown on one-half-strength Murashige and Skoog (MS) medium including 1× vitamins and 0.7% agar (both Duchefa) in a Plate Percival (CU-41L5/D; CLF Plant Climatics) for 16 d under short-day conditions (115 µE, 8 h light/16 h darkness; 22/20°C). For the selection of positively transformed plants, kanamycin (25 µg/mL) was added to the medium. After 16 d, plants were either harvested for qPCR/ChIP analysis, transferred to soil (1 part soil and 1.5 parts sand), or placed in Araponics for hydroponical cultures. Plants were grown in Araponics in one-half-strength MS including 1× vitamins under oxygenous conditions. Plants were further grown under short-day conditions (115 µE, 8 h light/16 h darkness; 21/18°C; BB-XL3, CLF Plant Climatics) and harvested at the age of either 5 (ChIP experiments) or 6 (hydroponics) weeks. For prolonged darkness experiments, plates were transferred to darkness for 64 h before the onset of light at day 14.
RNA Isolation, cDNA Synthesis, and qPCR Analysis
Plant material was ground in liquid nitrogen. RNA was isolated with a RNA-DNA isolation method as follows: Ground plant material of either three to four 16-d-old plantlets or two spoons (∼40 mg) of 5-week-old plants was dissolved in DNA/RNA extraction buffer (0.05 m Tris-HCl, pH 7.6, 0.5% SDS). An equal volume of water-saturated phenol was added, and the suspension was mixed. After centrifugation, nucleic acids in the upper aqueous phase were precipitated with 3 m sodium acetate (pH 5.2) and 96% ethanol in an additional centrifugation step. The pellet was washed with 70% ethanol (5 min, top speed, 4°C) and resuspended in 100 µL distilled, deionized water. Two microliters were used for cDNA synthesis. After DNaseI treatment (EN0521; Thermo Scientific), a nonamer random primer was annealed and first-strand synthesis was performed using MMLV (M1705; Promega). Each experimental setup contained control samples lacking MMLV to ensure successful digestion of genomic DNA by the DNaseI treatment prior to cDNA synthesis. qPCR analysis was done on an ABI 7300 Real Time PCR System (Applied Biosystems). A typical 20 µL qPCR sample contained 2 µL of cDNA template, 0.4 µL of each primer (10 µm stock), and 7.2 µL Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen).
The PCR reaction was started with a carryover protection step at 50°C for 2 min followed by inactivation of uracil-DNA glycosylase at 95°C for 2 min. The main PCR program was 40 cycles of denaturation (95°C for 15 s) and annealing/elongation (60°C for 1 min). The run was finalized by recording the melting curves. Primers used for qPCR analysis are listed in Supplemental Table S2. Relative transcript levels were determined by quantifying the amount of mRNA and unspliced RNA according to an equation determined by a 1:4 dilution series of a cDNA standard.
Approval of Correct Splicing of the 5′UTR Introns
To verify correct splicing in the two intron swap constructs, DNA fragments were amplified from cDNA using the following primer combinations: GGT1 25I_fw: 5′-CTTCGTCAATTTCAGTGGCTC-3′; GGT2 15I_fw: 5′-GTCTTTCTTACTTGCCCTGC-3′; and GGT1 25I/GGT2 15I_rv: 5′-GGGGTTTCTACAGGACGTAAC-3′, with the latter binding to a sequence in the gusA gene. Splicing was verified by two different methods. First, DNA fragments were ligated into pCR4-Topo (Invitrogen), and positive clones were selected for plasmid isolation. Sequencing verified correct splicing of the different promoter::gusA constructs tested. Second, splicing was proofed by PCR.
Construction and Cloning of Promoter::gusA Constructs
The 5′ upstream and coding sequences were obtained from Phytozome v9.1. For construction of the 5′ deletion mutants, the GGT1 5′ upstream region was analyzed for putative cis-elements with the software program Athena (O’Connor et al., 2005). The GGT1 (At1g23310; TAIR Accession, Locus: 2028000) and GGT2 (At1g70580; TAIR Accession, Locus: 2026841) 5′UTR intron deletion (Δ5I) were generated by simple PCR using a primer that binds to the adjacent 5′ upstream region of the intron. In addition, the primer included the 3′ downstream region of the intron (44 bp upstream of the start codon). The 5I substitution of the corresponding GGT isoform was performed by overlapping extension PCR (Supplemental Table S3). To test the effect of GGT1 5I on tissue specificity, primers were designed to amplify the 5′ upstream sequences of CATALASE3 (At1g20620; TAIR Accession, Locus: 2034357), GLDP1-P2 (At4g33010; TAIR Accession, Locus: 2123777; Adwy et al., 2015), and a root-specific peroxidase (At3g01190; TAIR Accession, Locus: 2102087). Furthermore, cloning GGT1 5I downstream of the promoter sequences necessitated the introduction of an MCS into the linker region between the promoter and the gusA gene. This was realized by adding the MCS sequence to the respective reverse primers of each of the 5′ upstream sequences. CaMV 35S was cloned as a control to observe time-dependent GUS staining side by side with the GGT2 15I::Gus construct. DNA fragments were amplified with Phusion polymerase (F-530; Thermo Scientific), purified with the MSBSpin PCRapace Kit (Stratec Molecular) according to the manufacturer’s instructions, and cloned into pENTR/D-Topo (Invitrogen). Sequence accuracy was verified by sequencing. Cloned DNA sequences were recombined into the binary GUS expression vector pSAG (Adwy et al., 2015) with Gateway LR clonase II (Invitrogen). Plasmids were transformed in agrobacteria (GV3101::pMP90RK) by electroporation. Positive clones were selected and stored at −80°C for plant transformation. Primers used for cloning are listed in Supplemental Table S3.
Stable Arabidopsis Transformation
Stably transformed Col-0 plants were produced via floral dip according to Clough and Bent (1998). Agrobacteria (GV3101::pMP90RK) were grown to an OD600 of 1.0, collected by centrifugation, and resuspended in transformation medium (2.2 g/L MS medium, 5% Suc, 0.5 g/L MES, 0.03% Silwett L-77; pH 5.7). Arabidopsis flower buds were dipped for 6 min. Positive plants were selected by screening seeds on one-half-strength MS medium including 1× vitamins and kanamycin (25 µg/mL). Individual transformation events were further screened by a segregation analysis to identify single insertion lines according to the Mendelian Law as shown for the example of GGT1 Δ5I::gusA lines in Supplemental Table S1. However, plotting the x2 values of the segregation analysis to corresponding standardized gusA transcript levels of the individual transformation events revealed that there is no correlation between single insertion and transcript levels (Supplemental Fig. S11). This suggests that the positional effect does influence the individual lines at least to the same degree as multiple insertions might. Thus, we decided to test 7–10 transformation events per line to overcome positional and multiple insertion effects.
ChIP Analysis
ChIP analysis was performed according to Jaskiewicz et al. (2011) with minor modifications. Precleared chromatin was diluted 1:4 for ChIP analysis. Precipitated DNA was purified with the MSBSpin PCRapace Kit (Stratec Molecular) with minor modifications. An amount of 400 µL binding buffer was added to the de-crosslinked samples before they were loaded on the columns. After centrifugation, a second washing step with 500 µL binding buffer was included. DNA was eluted from the column with 80 µL elution buffer. Purified DNA was diluted 1:4 before qPCR analysis.
Antibodies used for ChIP analysis were as follows: histone 3 C terminus (H3C; 1 µL, ab1791; Abcam), histone 3 Lys 9 acetylation (H3K9ac; 5 µL, 07-352; Merck), histone 3 Lys 4 trimethylation (H3K4me3; 2.5 µL, 04-745; Merck), and RNA polymerase II (RNA Pol II; 7 µL, ab817; Abcam). An unrelated rabbit antiserum directed against starch branching protein from potato (Solanum tuberosum) was used as a control for background precipitation. Nucleosome density was expressed as percentage of input, with both acetylation and methylation as levels per H3C.
GUS Staining
Sixteen-day-old plantlets were stained for GUS activity by vacuum infiltration with GUS staining solution [GUS buffer containing 0.1 m Tris-HCl, pH 7.0, and 0.05 m NaCl, 100 mm hexacyanoferrate(III), 100 mm hexacyanoferrate(II), 50 mg/ml X-Gluc, 0.1% Triton X-100], followed by an incubation at 37°C for at least 12 h. Time-dependent GUS staining of 35S::gusA and GGT2 15I::gusA lines was done in the presence of 2.5 mm hexacyanoferrate. Chlorophyll was washed out with 70% ethanol, and plants were transferred to 10% glyercol. Images were taken using a binocular (Olympus SZ2-ILST) connected to a camera (Color View; Soft Imaging System) and visualized with the associated software program AnalySIS getIT Stereo.
GUS Activity Assay
GUS activity of 16-d-old plantlets was determined according to Jefferson et al. (1987) using the fluorogenic substrate 4-methylumbelliferyl β-d-glucuronide.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Diurnal expression of the individual transformation events used to generate the data in Figures 2D, 3D, and 5.
Supplemental Table S2. List of primers used for quantitative real-time PCR analysis.
Supplemental Table S3. Primer used for the amplification of the promoter::gusA constructs used in this study.
Supplemental Table S4. Segregation analysis of GGT1 Δ5I::gusA transgenic lines (F2-generation) used to generate the data in Figures 4 and 5.
Supplemental Figure S1. GUS staining of all available transformation events that have been generated for the constructs GGT1::gusA, GGT1 Δ5I::gusA, and GGT2::gusA.
Supplemental Figure S2. GUS staining of all transformation events that have been used to generate the quantitative qPCR data presented in Figure 2, C and D.
Supplemental Figure S3. GUS staining of all transformation events that have been used to generate the quantitative qPCR data presented in Figure 3, C and D.
Supplemental Figure S4. Alignment of the four predicted GGT2 splice forms (SF).
Supplemental Figure S5. Splice form 1 is the most abundant splice form of GGT2.
Supplemental Figure S6. GUS staining of all transformation events that have been used to generate the quantitative qPCR data presented in Figure 5.
Supplemental Figure S7. NptII expression indicates successful transformation of all transformation events chosen to generate the quantitative qPCR data presented in Figure 5.
Supplemental Figure S8. GUS staining of all transformation events that have been used to evaluate the effect of 5′UTR intron of GGT1 on the tissue specificity of CAT3, GLDP1-P2 (Adwy et al., 2015), and a root-specific PEROXIDASE.
Supplemental Figure S9. Relative gusA unspliced RNA transcript level and GUS activity in transgenic GGT1 and GGT2 lines.
Supplemental Figure S10. Scheme of the different steps of the overlapping extension PCRs performed to generate the fragments GGT1 25I and GGT2 15I, respectively.
Supplemental Figure S11. GusA mRNA levels standardized to GAPDH correlate to the corresponding x2-test values of the individual transformation events of GGT1 Δ5I::gusA transgenic plants.
Supplementary Material
Acknowledgments
We thank Julia Gunia and Valerie Schuchardt for technical assistance and Lisa Hagenau for the help with R Studio to generate the dot plots.
Glossary
- IME
intron-mediated enhancement
- TIS
transcription initiation site
- MCS
multiple cloning site
References
- Adwy W, Laxa M, Peterhänsel C (2015) A simple mechanism for the establishment of C₂-specific gene expression in Brassicaceae. Plant J 84: 1231–1238 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 [DOI] [PubMed] [Google Scholar]
- Barrett LW, Fletcher S, Wilton SD (2012) Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol Life Sci 69: 3613–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieberstein NI, Carrillo Oesterreich F, Straube K, Neugebauer KM (2012) First exon length controls active chromatin signatures and transcription. Cell Reports 2: 62–68 [DOI] [PubMed] [Google Scholar]
- Bolle C, Herrmann RG, Oelmüller R (1996) Different sequences for 5′-untranslated leaders of nuclear genes for plastid proteins affect the expression of the beta-glucuronidase gene. Plant Mol Biol 32: 861–868 [DOI] [PubMed] [Google Scholar]
- Bolle C, Sopory S, Lübberstedt T, Herrmann RG, Oelmüller R (1994) Segments encoding 5′-untranslated leaders of genes for thylakoid proteins contain cis-elements essential for transcription. Plant J 6: 513–523 [DOI] [PubMed] [Google Scholar]
- Burlingame RW, Love WE, Wang BC, Hamlin R, Nguyen HX, Moudrianakis EN (1985) Crystallographic structure of the octameric histone core of the nucleosome at a resolution of 3.3 A. Science 228: 546–553 [DOI] [PubMed] [Google Scholar]
- Callis J, Fromm M, Walbot V (1987) Introns increase gene expression in cultured maize cells. Genes Dev 1: 1183–1200 [DOI] [PubMed] [Google Scholar]
- Chang KN, Zhong S, Weirauch MT, Hon G, Pelizzola M, Li H, Huang SS, Schmitz RJ, Urich MA, Kuo D, et al. (2013) Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis eLife 2: e00675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy M, Hannah LC (2002) Splicing of the maize Sh1 first intron is essential for enhancement of gene expression, and a T-rich motif increases expression without affecting splicing. Plant Physiol 130: 918–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dellero Y, Lamothe-Sibold M, Jossier M, Hodges M (2015) Arabidopsis thaliana ggt1 photorespiratory mutants maintain leaf carbon/nitrogen balance by reducing RuBisCO content and plant growth. Plant J 83: 1005–1018 [DOI] [PubMed] [Google Scholar]
- Eisenhut M, Pick TR, Bordych C, Weber AP (2013) Towards closing the remaining gaps in photorespiration--the essential but unexplored role of transport proteins. Plant Biol (Stuttg) 15: 676–685 [DOI] [PubMed] [Google Scholar]
- Emami S, Arumainayagam D, Korf I, Rose AB (2013) The effects of a stimulating intron on the expression of heterologous genes in Arabidopsis thaliana. Plant Biotechnol J 11: 555–563 [DOI] [PubMed] [Google Scholar]
- Furger A, O’Sullivan JM, Binnie A, Lee BA, Proudfoot NJ (2002) Promoter proximal splice sites enhance transcription. Genes Dev 16: 2792–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos JE, Rose AB (2015) The enduring mystery of intron-mediated enhancement. Plant Sci 237: 8–15 [DOI] [PubMed] [Google Scholar]
- Gianì S, Altana A, Campanoni P, Morello L, Breviario D (2009) In trangenic rice, alpha- and beta-tubulin regulatory sequences control GUS amount and distribution through intron mediated enhancement and intron dependent spatial expression. Transgenic Res 18: 151–162 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, et al. (2001) The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res 29: 102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, An YQ, McDowell JM, McKinney EC, Meagher RB (1997) The Arabidopsis ACT11 actin gene is strongly expressed in tissues of the emerging inflorescence, pollen, and developing ovules. Plant Mol Biol 33: 125–139 [DOI] [PubMed] [Google Scholar]
- Igamberdiev AU, Lea PJ (2002) The role of peroxisomes in the integration of metabolism and evolutionary diversity of photosynthetic organisms. Phytochemistry 60: 651–674 [DOI] [PubMed] [Google Scholar]
- Igarashi D, Miwa T, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Ohsumi C (2003) Identification of photorespiratory glutamate:glyoxylate aminotransferase (GGAT) gene in Arabidopsis. Plant J 33: 975–987 [DOI] [PubMed] [Google Scholar]
- Igarashi D, Tsuchida H, Miyao M, Ohsumi C (2006) Glutamate:glyoxylate aminotransferase modulates amino acid content during photorespiration. Plant Physiol 142: 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskiewicz M, Peterhänsel C, Conrath U (2011) Detection of histone modifications in plant leaves. J Vis Exp 55: 3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong YM, Mun JH, Lee I, Woo JC, Hong CB, Kim SG (2006) Distinct roles of the first introns on the expression of Arabidopsis profilin gene family members. Plant Physiol 140: 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Huang C, Sun Q, Guo H, Cheng T, Peng Z, Dang Y, Liu W, Xu G, Xia Q (2015) The 5′-UTR intron of the midgut-specific BmAPN4 gene affects the level and location of expression in transgenic silkworms. Insect Biochem Mol Biol 63: 1–6 [DOI] [PubMed] [Google Scholar]
- Jordan DB, Ogren WL (1984) The CO2/O 2 specificity of ribulose 1,5-bisphosphate carboxylase/oxygenase: Dependence on ribulosebisphosphate concentration, pH and temperature. Planta 161: 308–313 [DOI] [PubMed] [Google Scholar]
- Kapushesky M, Emam I, Holloway E, Kurnosov P, Zorin A, Malone J, Rustici G, Williams E, Parkinson H, Brazma A (2010) Gene expression atlas at the European bioinformatics institute. Nucleic Acids Res 38: D690–D698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys AJ. (2006) The re-assimilation of ammonia produced by photorespiration and the nitrogen economy of C3 higher plants. Photosynth Res 87: 165–175 [DOI] [PubMed] [Google Scholar]
- Liepman AH, Olsen LJ (2003) Alanine aminotransferase homologs catalyze the glutamate:glyoxylate aminotransferase reaction in peroxisomes of Arabidopsis. Plant Physiol 131: 215–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas D, Mettler IJ, Pierce DA, Lowe HW (1990) Intron-mediated enhancement of heterologous gene expression in maize. Plant Mol Biol 15: 913–920 [DOI] [PubMed] [Google Scholar]
- Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P (2005) A structural perspective of CTD function. Genes Dev 19: 1401–1415 [DOI] [PubMed] [Google Scholar]
- Michael TP, McClung CR (2002) Phase-specific circadian clock regulatory elements in Arabidopsis. Plant Physiol 130: 627–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moabbi AM, Agarwal N, El Kaderi B, Ansari A (2012) Role for gene looping in intron-mediated enhancement of transcription. Proc Natl Acad Sci USA 109: 8505–8510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, McEntee C, Kay SA, Chory J (2007) The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol 72: 353–363 [DOI] [PubMed] [Google Scholar]
- Mun JH, Lee SY, Yu HJ, Jeong YM, Shin MY, Kim H, Lee I, Kim SG (2002) Petunia actin-depolymerizing factor is mainly accumulated in vascular tissue and its gene expression is enhanced by the first intron. Gene 292: 233–243 [DOI] [PubMed] [Google Scholar]
- Nawrath C, Schell J, Koncz C (1990) Homologous domains of the largest subunit of eucaryotic RNA polymerase II are conserved in plants. Mol Gen Genet 223: 65–75 [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR (2010) Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Florian A, Howden A, Jahnke K, Timm S, Bauwe H, Sweetlove L, Fernie AR (2014) Is there a metabolic requirement for photorespiratory enzyme activities in heterotrophic tissues? Mol Plant 7: 248–251 [DOI] [PubMed] [Google Scholar]
- O’Connor TR, Dyreson C, Wyrick JJ (2005) Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413 [DOI] [PubMed] [Google Scholar]
- Odell JT, Nagy F, Chua NH (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313: 810–812 [DOI] [PubMed] [Google Scholar]
- Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A (1993) Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135: 385–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Rose AB, Korf I (2011) Comparative and functional analysis of intron-mediated enhancement signals reveals conserved features among plants. Nucleic Acids Res 39: 5328–5337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterhänsel C, Horst I, Niessen M, Blume C, Kebeish R, Kürkcüoglu S, Kreuzaler F (2010) Photorespiration. The Arabidopsis Book 8: e0130, doi/10.1199/tab.0130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AB. (2002) Requirements for intron-mediated enhancement of gene expression in Arabidopsis. RNA 8: 1444–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AB. (2004) The effect of intron location on intron-mediated enhancement of gene expression in Arabidopsis. Plant J 40: 744–751 [DOI] [PubMed] [Google Scholar]
- Rose AB. (2008) Intron-mediated regulation of gene expression. Curr Top Microbiol Immunol 326: 277–290 [DOI] [PubMed] [Google Scholar]
- Rose AB, Elfersi T, Parra G, Korf I (2008) Promoter-proximal introns in Arabidopsis thaliana are enriched in dispersed signals that elevate gene expression. Plant Cell 20: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF. (2004) The evolution of C4 photosynthesis. New Phytol 161: 341–370 [DOI] [PubMed] [Google Scholar]
- Samadder P, Sivamani E, Lu J, Li X, Qu R (2008) Transcriptional and post-transcriptional enhancement of gene expression by the 5′ UTR intron of rice rubi3 gene in transgenic rice cells. Mol Genet Genomics 279: 429–439 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T (2002) Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411 [DOI] [PubMed] [Google Scholar]
- Sharkey TD. (2001) Photorespiration. eLS, http://dx.doi.org/10.1038/npg.els.0001292 [Google Scholar]
- Somerville CR. (2001) An early Arabidopsis demonstration. Resolving a few issues concerning photorespiration. Plant Physiol 125: 20–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL (1979) A phosphoglycolate phosphatase-deficient mutant of Arabidopsis. Nature 280: 833–836 [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB (2009) Biased chromatin signatures around polyadenylation sites and exons. Mol Cell 36: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabenau H, Winkler U (2005) Glycolate metabolism in green algae. Physiol Plant 123: 235–245 [Google Scholar]
- Timm S, Florian A, Wittmiß M, Jahnke K, Hagemann M, Fernie AR, Bauwe H (2013) Serine acts as a metabolic signal for the transcriptional control of photorespiration-related genes in Arabidopsis. Plant Physiol 162: 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Kim YS, Zhu JK (2007) Altered ABA, proline and hydrogen peroxide in an Arabidopsis glutamate:glyoxylate aminotransferase mutant. Plant Mol Biol 64: 205–217 [DOI] [PubMed] [Google Scholar]
- Vitha S, Bene K, Phillips J, Gartland K (2007) Histochemical Localization of β-Glucuronidase (GUS) Reporter Activity in Plant Tissues. Microscopy and Imaging Center, Texas A&M University, College Station, TX, http://microscopy.tamu.edu/lab-protocols/light-microscopy-protocols.html
- Vorst O, van Dam F, Weisbeek P, Smeekens S (1993) Light-regulated expression of the Arabidopsis thaliana ferredoxin A gene involves both transcriptional and post-transcriptional processes. Plant J 3: 793–803 [DOI] [PubMed] [Google Scholar]
- Wang X, Elling AA, Li X, Li N, Peng Z, He G, Sun H, Qi Y, Liu XS, Deng XW (2009) Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Heinlein C, Orendi G, Zentgraf U (2006) Senescence-specific regulation of catalases in Arabidopsis thaliana (L.) Heynh. Plant Cell Environ 29: 1049–1060 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.