The UDP-glucosyltransferase Bradi5g03300 conjugates the Fusarium mycotoxin deoxynivalenol into deoxynivalenol-3-O-glucose and confers resistance to primary infection by Fusarium graminearum.
Abstract
Fusarium head blight (FHB) is a cereal disease caused by Fusarium graminearum, a fungus able to produce type B trichothecenes on cereals, including deoxynivalenol (DON), which is harmful for humans and animals. Resistance to FHB is quantitative, and the mechanisms underlying resistance are poorly understood. Resistance has been related to the ability to conjugate DON into a glucosylated form, deoxynivalenol-3-O-glucose (D3G), by secondary metabolism UDP-glucosyltransferases (UGTs). However, functional analyses have never been performed within a single host species. Here, using the model cereal species Brachypodium distachyon, we show that the Bradi5g03300 UGT converts DON into D3G in planta. We present evidence that a mutation in Bradi5g03300 increases root sensitivity to DON and the susceptibility of spikes to F. graminearum, while overexpression confers increased root tolerance to the mycotoxin and spike resistance to the fungus. The dynamics of expression and conjugation suggest that the speed of DON conjugation rather than the increase of D3G per se is a critical factor explaining the higher resistance of the overexpressing lines. A detached glumes assay showed that overexpression but not mutation of the Bradi5g03300 gene alters primary infection by F. graminearum, highlighting the involvement of DON in early steps of infection. Together, these results indicate that early and efficient UGT-mediated conjugation of DON is necessary and sufficient to establish resistance to primary infection by F. graminearum and highlight a novel strategy to promote FHB resistance in cereals.
Fusarium head blight (FHB) is one of the more devastating diseases of small-grain cereals (Kazan et al., 2012). Besides direct losses due to alteration of grain filling, FHB constitutes a health threat due to the production of mycotoxins by the causal pathogens, which are harmful for humans and animals (Rocha et al., 2005; Yazar and Omurtag, 2008). One of the main FHB causal agents is the ascomycete fungus Fusarium graminearum (teleomorph Gibberella zeae). This fungus produces mycotoxins mostly belonging to type B trichothecenes (TCTBs) but also the estrogenic compound zearalenone (Yazar and Omurtag, 2008). TCTBs include deoxynivalenol (DON), nivalenol, and their acetylated derivatives 3-acetyldeoxynivalenol, 15-acetyldeoxynivalenol (15-ADON), 4-acetylnivalenol (or fusarenon X), and 4,15-acetylnivalenol (Yazar and Omurtag, 2008). An F. graminearum strain is generally characterized by its chemotype and produces one main TCTB. Genes encoding enzymes involved in TCTB biosynthesis have been identified, and most of them belong to the Tri5 cluster (Kimura et al., 2003), named after the Tri5 gene encoding the trichodiene synthase enzyme, which catalyzes the first committed step of the biosynthetic pathway. TCTBs are sesquiterpene secondary metabolites that inhibit protein synthesis in eukaryotic cells (Rocha et al., 2005). Animal toxicity is well described, and TCTBs have been shown to induce altered appetite but also immunotoxic effects (Pestka, 2010). In plants, the application of high concentrations of toxins induced the production of reactive oxygen species, apoptosis-like processes such as nuclear DNA laddering, chlorotic and necrotic lesions, and root growth inhibition (Masuda et al., 2007; Desmond et al., 2008).
The relationship between the ability to produce TCTBs and pathogenicity has been investigated using mutant strains impaired in the Tri5 gene and thus unable to produce TCTBs. In wheat (Triticum aestivum), a tri5 mutant strain was unable to efficiently colonize the spike from a single inoculation site, whereas in barley (Hordeum vulgare), both a wild-type strain and the near-isogenic tri5 mutant strain were limited in their progression (Maier et al., 2006). These contrasting results were interpreted as likely reflecting the specificity of resistance mechanisms of the host plants and led to the conclusion that TCTBs generally can be considered as aggressiveness factors.
The resistance of small-grain cereals to FHB has been investigated extensively. No monogenic resistance has been identified, but more than 100 quantitative trait loci have been described so far (Buerstmayr et al., 2009). A number of studies using DON-producing strains of F. graminearum have tried to decipher the biological functions leading to quantitative resistance to FHB in wheat or barley. Some secondary metabolism pathways were frequently found to be associated with resistance, such as the induction of the phenylpropanoid pathway and metabolites such as Phe, p-coumarate, and sinapate (Bollina et al., 2011; Kumaraswamy et al., 2011a, 2011b) as well as the production of conjugates of flavonoids/isoflavonoids like naringenin and kaempferol (Kumaraswamy et al., 2011a, 2011b). The induction of jasmonate-regulated allene oxide synthase and 12-oxo-phytodienoic acid reductase (Gottwald et al., 2012; Kugler et al., 2013; Xiao et al., 2013), and the related metabolites linoleic and linolenic acids,jasmonates, and traumatic acid (Bollina et al., 2011; Kumaraswamy et al., 2011a), have been described as markers of quantitative resistance to FHB. Other common features were associated with defense responses such as the induction of β-1,3-glucanases, chitinases, and thaumatin-like proteins, the scavenging of reactive oxygen species, and xenobiotic detoxification by family 1 UDP-glycosyltransferases, glutathione S-transferases, cytochrome P450-monooxygenases, and transporter detoxification (Boddu et al., 2006; Golkari et al., 2007; Jia et al., 2009; Gardiner et al., 2010; Foroud et al., 2012; Kugler et al., 2013; Xiao et al., 2013). Other phenomena that have been linked to resistance to FHB include increased photosynthesis and carbohydrate metabolism involving chloroplast oxygen-evolving enhancer proteins, NAD(P)+-binding proteins, and the pentose phosphate pathway (Kugler et al., 2013; Zhang et al., 2013). The biosynthesis and metabolism of riboflavin also have been associated with FHB resistance (Kugler et al., 2013).
Although a full understanding of the genetic basis of such resistance reactions is challenging, in particular due to the genetic diversity of the cultivars, a common feature is the relationship between partial resistance and the production on infected cereal spikes of deoxynivalenol-3-O-glucose (D3G), now clearly considered a resistance-related metabolite (Lemmens et al., 2005; Kumaraswamy et al., 2011a, 2011b; Kugler et al., 2013). Glucosylation is a well-known step in detoxification processes in plants, leading to more hydrophilic and generally less toxic compounds. This reaction is catalyzed by UDP-glucosyltransferases (UGT) that are encoded by the multigene family 1 of glycosyltransferases in plants (Ross et al., 2001). These enzymes mediate the transfer of glycosyl residues from nucleotide sugars to acceptor aglycones, thus regulating properties of the acceptors such as their bioactivity, water solubility, and transport within the cell and throughout the organism (Gachon et al., 2005). Candidate genes encoding UGTs able to conjugate DON into D3G have been identified in Arabidopsis (Arabidopsis thaliana [UGT73C5]; Poppenberger et al., 2003), barley (Hv13248; Schweiger et al., 2010; Shin et al., 2012), wheat (TaUGT3 [Lulin et al., 2010] and TaUGT12887 [Schweiger et al., 2013b]), and Brachypodium distachyon (Bradi5g03300; Schweiger et al., 2013a). Conjugation of DON into D3G by the corresponding UGTs was only shown in heterologous species, such as Saccharomyces cerevisiae (Schweiger et al., 2010, 2013a, 2013b), Arabidopsis (Poppenberger et al., 2003; Shin et al., 2012), or wheat (Li et al., 2015). In that most recent study, heterologous overexpression of the barley UGT gene Hv13248 in wheat varieties susceptible to FHB was shown to result in higher resistance in both greenhouses and field conditions (Li et al., 2015). However, that study only investigated DON conjugation after direct application of the toxin and not during infection by F. graminearum, thereby preventing the establishment of a direct correlation between the in planta conjugation of DON into its glucoside during infection and resistance to the fungal pathogen (Li et al., 2015).
In the last decade, B. distachyon has emerged as a model plant species for small-grain cereals because of its interesting characteristics: a small size, a short life cycle avoiding vernalization under long-day conditions, a small genome, a routine genetic transformation, and the availability of a number of genetic and genomic resources (Mur et al., 2011). B. distachyon has been shown to be a host species for many cereal pathogens (Peraldi et al., 2011, 2014; Mandadi and Scholthof, 2012; Falter and Voigt, 2014; Sandoya and de Oliveira Buanafina, 2014; Fitzgerald et al., 2015). In particular, it has been shown to exhibit typical FHB symptoms following infection by F. graminearum and to accumulate DON in infected spikes (Peraldi et al., 2011; Pasquet et al., 2014), demonstrating the potential of this model plant species to conduct functional genomics of FHB resistance. In the recent study of Schweiger et al. (2013a), a phylogenetic analysis of B. distachyon UGT protein sequences was performed to identify potential orthologous sequences to UGT73C5 from Arabidopsis (Poppenberger et al., 2003) and HvUGT13248 from barley (Schweiger et al., 2010), which are known to be able to glucosylate DON. This work identified the B. distachyon UGT Bradi5g03300 as a potential functional homolog of barley HvUGT13248 and demonstrated that the enzyme was able to glucosylate DON using S. cerevisiae as a heterologous system (Schweiger et al., 2013a). The corresponding gene also was shown to be differentially induced following infection by F. graminearum strains producing DON or not (Schweiger et al., 2013a).
Here, we report the functional analysis of the UGT-encoding Bradi5g03300 gene in B. distachyon as a model host infected with F. graminearum. By using Bradi5g03300 mutant or overexpressing lines, we demonstrate that early in planta conjugation of DON into D3G is functionally linked to root tolerance to the mycotoxin as well as to spike resistance toward FHB. A detailed analysis of how overexpression impacts the infection provides in planta evidence that DON is crucial in the early establishment of the fungal pathogen in the model cereal species B. distachyon.
RESULTS
Identification of B. distachyon Lines Mutated in the Bradi5g03300 Gene
UGTs exhibit two important domains in their primary sequence. The first domain, localized at the N terminus of the protein, is involved in the interaction between the protein and its substrate but is not conserved at the amino acid level (Osmani et al., 2009). The second domain, located at the C terminus, is involved in binding of the sugar donor and is named the plant secondary product glycosyltransferase (PSPG) box. This 44-amino acid motif is highly conserved in all family 1 plant UGTs (Gachon et al., 2005). The PSPG box of the Bradi5g03300 UGT is encoded by exons 2 and 3 and is located between amino acids 345 and 388 of the protein (Supplemental Fig. S1). Screening of the TILLING mutant collection of B. distachyon (BRACHYTIL; Dalmais et al., 2013) allowed us to search for mutants in the Bradi5g03300 gene. This was performed using specific primers (3300_N2Ft1 and 3300_N2Rt1) allowing the amplification of a 964-bp DNA fragment surrounding the PSPG box-encoding region (see “Materials and Methods”; Supplemental Fig. S1). Fifteen potential mutant families were identified (Supplemental Table S1). Following sequencing of the corresponding mutant alleles, six were shown to carry mutations located in the region encoding the PSPG box, of which three (8637-12, 6829-7, and 6491-12) could potentially impact the protein activity (Supplemental Table S1). In mutant line 8637-12, a nonsynonymous substitution of Ser-368 by Phe (S368F) was identified. In lines 6829-7 and 6491-12, Trp residues at positions 345 and 366, respectively, were replaced by a stop codon (W345* and W366*), resulting in both cases in the production of a truncated protein. As a control line for mutant family 6829, we selected a line, named 6829-3, carrying a wild-type Bradi5g03300 allele. No control lines could be obtained for the two other mutant families.
Developmental criteria were observed and quantified: seed germination time and rate, emergence of the first three leaves, heading date, overall size of the mature plant, and yield. For the three TILLING mutant lines, no differences were observed as compared with the wild-type Bd21-3 line or with the control line 6829-3 (Supplemental Fig. S2).
Construction of Lines Overexpressing the Bradi5g03300 Gene
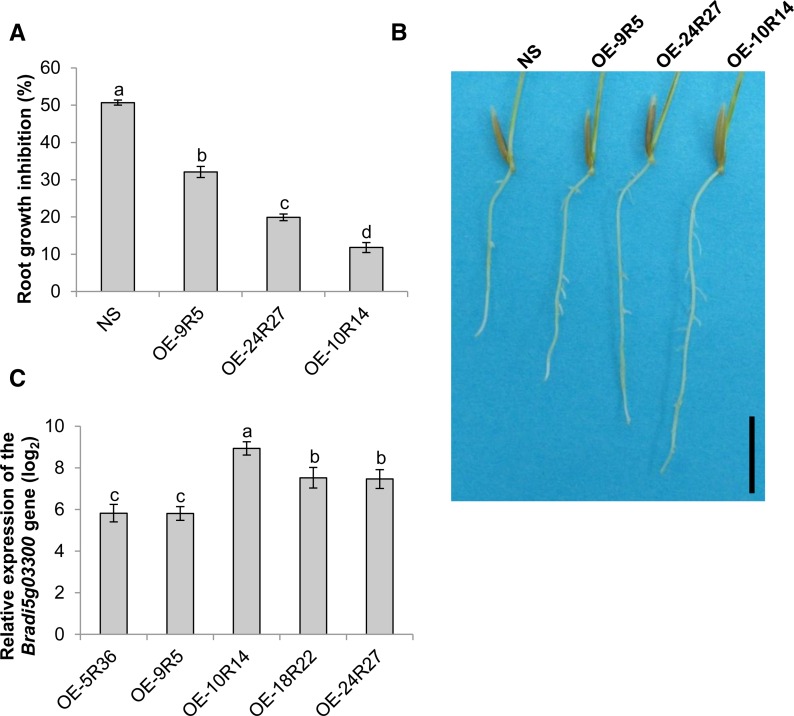
In order to obtain lines overexpressing the Bradi5g03300 gene, a pIPKb002-derived binary vector was constructed (Himmelbach et al., 2007; see “Materials and Methods”). The resulting construct, carrying both the Bradi5g03300 complementary DNA (cDNA) under the control of the Zea mays ubiquitin promoter and a hygromycin resistance cassette for the selection of transformants, was named pIPKb002-Bradi5g03300 and used for Agrobacterium tumefaciens-mediated transformation of B. distachyon Bd21-3 embryogenic calli. Five independent homozygous lines, each carrying the corresponding transfer DNA as a single insertion locus, were obtained (Supplemental Fig. S3). Quantitative reverse transcription (qRT)-PCR experiments were conducted to identify lines exhibiting a high level of Bradi5g03300 overexpression in spikelets (Table I; Supplemental Fig. S4A). The overexpression strength was shown to be similar in leaves (Supplemental Fig. S4B) and roots (Fig. 2C; Supplemental Fig. S4B). For further functional analyses, three overexpressing lines (OE-9R5, OE-24R27, and OE-10R14) differing in overexpression rate were used. In addition, a null segregant obtained during the same transformation experiment was kept as a control.
Table I. B. distachyon lines used in this study.
| Line Name | Line Type | Impact on the Bradi5g03300 Gene |
|---|---|---|
| Bd21-3 | Wild type | Wild-type allele |
| 6829-3 | TILLING mutant | Wild-type allele |
| 6829-7 | TILLING mutant | Truncated protein (stop codon), W345* |
| 6491-37 | TILLING mutant | Truncated protein (stop codon), W366* |
| 8637-12 | TILLING mutant | Amino acid substitution (PSPG box), S368F |
| OE-5R36 | Transformant (pIKB002-Bradi5g03300) | Overexpression |
| OE-9R5 | Transformant (pIKB002-Bradi5g03300) | Overexpression |
| OE-18R22 | Transformant (pIKB002-Bradi5g03300) | Overexpression |
| OE-24R27 | Transformant (pIKB002-Bradi5g03300) | Overexpression |
| OE-10R14 | Transformant (pIKB002-Bradi5g03300) | Overexpression |
Figure 2.
Overexpression of Bradi5g03300 increases root tolerance to DON. A, Percentage of root growth inhibition by 50 μm DON measured on 7-d-old seedlings of control null segregant (NS) and overexpressing lines (n > 30; error bars represent se, and different letters indicate significant differences between conditions; Newman and Keuls test, P ≤ 0.001). B, Photograph showing typical root development of plants from control and overexpressing lines grown for 7 d on agar medium containing 50 μm DON. Bar = 1 cm. C, Relative expression of the Bradi5g03300 gene in roots of overexpressing lines OE-9R5, OE-24R27, and OE-10R14. The relative quantity of Bradi5g03300 gene transcripts compared with the Bd21-3 wild-type line was calculated using the comparative cycle threshold (Ct) method (2−∆∆Ct). The B. distachyon UBC18 and ACT7 genes (Bradi4g00660 and Bradi4g41850) were used as endogenous controls to normalize the data for differences in input RNA between the different samples. Data represent mean values of three independent biological experiments, and error bars represent sd (different letters indicate significant differences between conditions; Student’s t test, P ≤ 0.05).
As for TILLING mutant lines, no effects on plant growth or phenotype were observed for the three selected overexpressing lines at different stages of shoot development (Supplemental Fig. S2). These lines, therefore, represent appropriate genetic tools with which to specifically investigate the importance of this UGT in response to DON and F. graminearum.
Mutation and Overexpression in the Bradi5g03300 Gene Alters Root Tolerance to DON
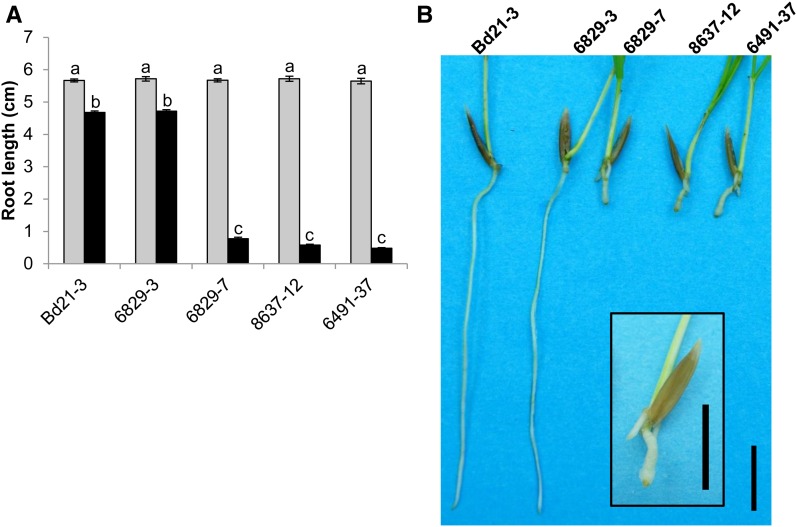
DON has already been reported to preferentially affect root growth of wheat grown on agar medium (Masuda et al., 2007). To determine the involvement of the Bradi5g03300 UGT in the DON effect on root growth, seeds from the three mutant lines, the 6829-3 control line, and the Bd21-3 wild-type line were germinated on agar medium with or without DON. While no difference of root development was observed between the lines on agar medium without DON, strong phenotypes were observed on 10 μm DON. Whereas root growth inhibition was around 18% for the Bd21-3 wild-type and 6829-3 control lines, it reached 90% for the three mutant lines (6829-7, 8637-12, and 6491-37; Fig. 1; Newman and Keuls test, P ≤ 0.001). Similar tests were performed on overexpressing lines, but DON concentration in the medium was increased to 50 µm in order to obtain more pronounced effects. The overexpressing lines exhibited root tolerance to DON (Fig. 2, A and B) with an apparent positive correlation with the level of Bradi5g03300 overexpression (Fig. 2C). The OE-10R14 line revealed nearly full resistance to DON at this concentration, the reduction of root growth reaching only 10%, whereas the null segregant showed 50% root growth inhibition (Fig. 2, A and B).
Figure 1.
Mutations in Bradi5g03300 dramatically increase root sensitivity to DON. A, Root length of Bd21-3 wild-type, 6829-3 control, and three mutant lines on medium with (black bars) or without (gray bars) 10 μm DON, measured on 7-d-old seedlings (n > 30; error bars represent se, and different letters indicate significant differences between conditions; Newman and Keuls test, P ≤ 0.001). B, Photograph showing typical root development of plants from wild-type, control, and mutant lines grown for 7 d on agar medium containing 10 μm DON. The inset shows a magnification of the 6829-7 mutant plant. Bars = 1 cm.
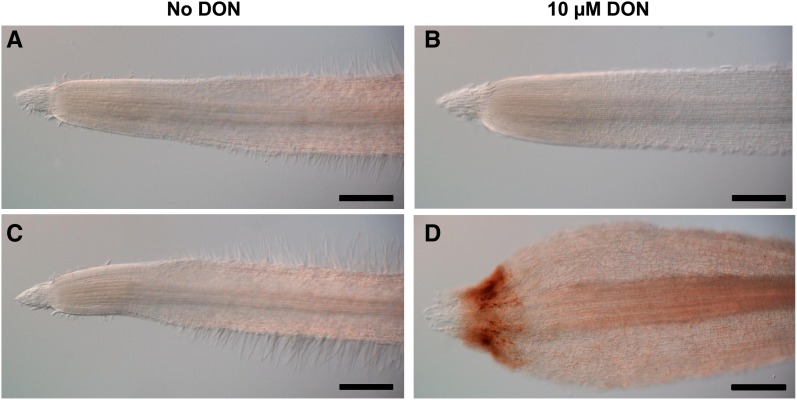
The strong root growth inhibition observed in all the mutant lines was accompanied by an important swelling of the root apex grown with 10 μm DON (Fig. 3A). DON treatment of roots from the wild-type line did not induce major changes, apart from an apparent decrease of root hair development (Fig. 3B). In contrast, for the 6829-7 mutant line, although root apex morphology was similar to that of the wild-type line in the absence of the mycotoxin (Fig. 3C), DON induced important changes (Fig. 3D). These included a marked reduction of the root apical meristem, an apparent disorganization of dividing cells, and a global enlargement of cells at the root tip (Fig. 3D). No such changes were observed in the root apex of the OE-10R14 line, even on a high DON concentration (Supplemental Fig. S5).
Figure 3.
DON induces root apex disorganization in Bradi5g03300 mutant lines but not in the Bd21-3 wild-type line. Root apices are shown for the wild-type line Bd21-3 (A and B) and the mutant line 6829-7 (C and D) on agar medium without (A and C) or with 10 μm DON (B and D). Bars = 300 µm.
The Bradi5g03300 UGT Gene Is Involved in Susceptibility/Resistance to F. graminearum Infection
The above observations show that TILLING mutants and overexpressing lines affect root responses to DON in an opposite manner. To test the impact of DON on spike susceptibility/resistance to FHB, spray inoculations with a DON-producing F. graminearum strain (FgDON+) were performed, and both the primary infection and the fungal spread were assessed in spikes. In parallel, the fungal spread also was evaluated in individual florets using point inoculations with spores of the same F. graminearum strain (Miedaner et al., 2003).
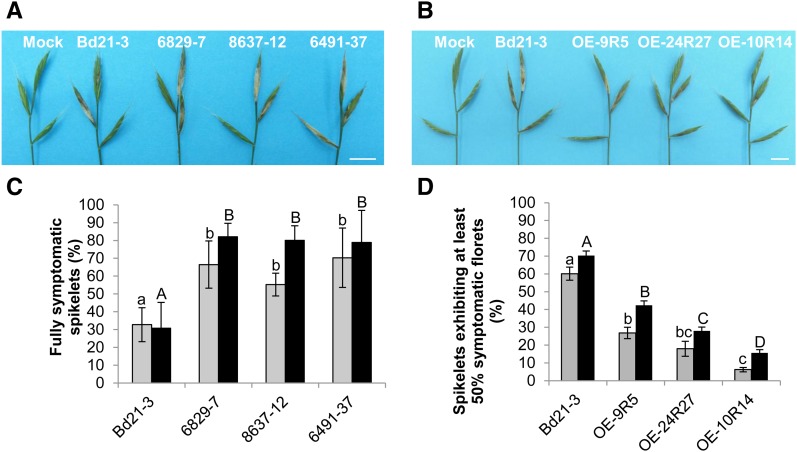
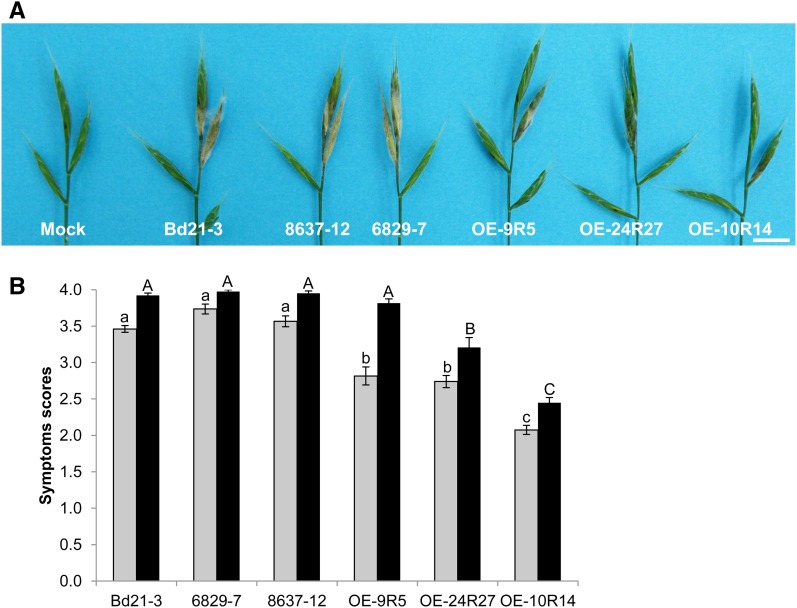
The 6829-7, 8637-12, and 6491-37 mutant lines as well as the Bd21-3 wild-type line and the 6829-3 control line were first spray inoculated by FgDON+. In this experiment, an inoculated spikelet was considered symptomatic if it exhibited full bleaching. Symptoms were scored 7 and 10 d after spraying. In the mutant lines, a significant increase of spikelets exhibiting bleaching symptoms was observed compared with the wild-type line (Fig. 4A). Seven days after spray inoculation (dai), the Bd21-3 wild-type line exhibited bleaching symptoms on 30% of the inoculated spikelets on average (Fig. 4C, gray bars). In contrast, this percentage reached 50% to 60% of overall inoculated spikelets in mutant lines. This differential between the wild-type line and the three mutant lines was significant (Newman and Keuls test, P ≤ 0.01), but no significant difference was observed between the three mutant lines (Fig. 4C). Increased susceptibility of the mutant lines was still observed at 10 dai (Fig. 4C, black bars). At this stage, almost all inoculated spikelets of the mutant lines were fully symptomatic, whereas only up to half of the spikelets of the wild-type and control lines showed extensive bleaching.
Figure 4.
Mutation or overexpression of the Bradi5g03300 gene increases spike susceptibility or resistance, respectively, to F. graminearum following spray inoculation. A and B, Typical FHB symptoms observed on mutant lines (A) or overexpressing lines (B) at 7 dai by the FgDON+ strain. Bars = 1 cm. C, Percentage of spikelets exhibiting FHB symptoms on the entire inoculated spikelet at 7 dai (gray bars) and 10 dai (black bars) by fungal strain FgDON+ (n > 30; error bars represent sd, and different letters indicate significant differences between conditions; Newman and Keuls test, P ≤ 0.01). D, Percentage of spikelets exhibiting FHB symptoms on half or more of the florets of the inoculated spikelet at 7 dai (gray bars) and 10 dai (black bars) by fungal strain FgDON+ (n > 30; error bars represent sd, and different letters indicate significant differences between conditions; Newman and Keuls test, P ≤ 0.01).
Similar experiments were conducted on the three overexpressing lines, OE-9R5, OE-24R27, and OE-10R14. Considering the potential resistance level of these lines, the scoring method was adapted: an inoculated spikelet was considered symptomatic if it exhibited bleaching symptoms on half or more of its florets (approximately 10 florets per spikelet). At 7 dai, strong differences in the extent of symptoms could be observed on overexpressing lines as compared with the Bd21-3 wild-type line (Fig. 4B). For the wild-type line, 60% of the inoculated spikelets were found to be symptomatic 7 dai, reaching 70% at 10 dai (Fig. 4D). In contrast, the overexpressing lines exhibited a strong reduction of symptomatic spikelets at 7 dai (between 5% and 25%) and 10 dai (between 15% and 42%; Fig. 4D). As shown for root tolerance to DON, the OE-10R14 line was the most resistant line, exhibiting only very minor bleaching symptoms 7 and 10 dai (Fig. 4, B and D).
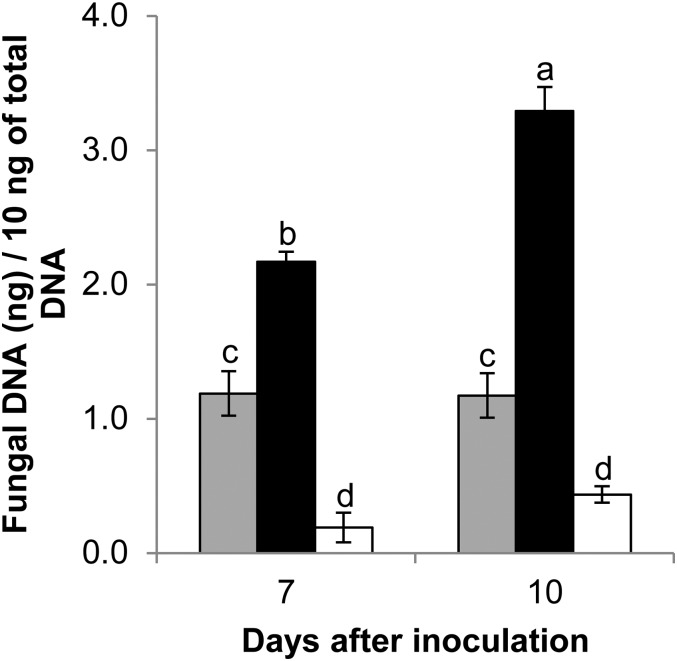
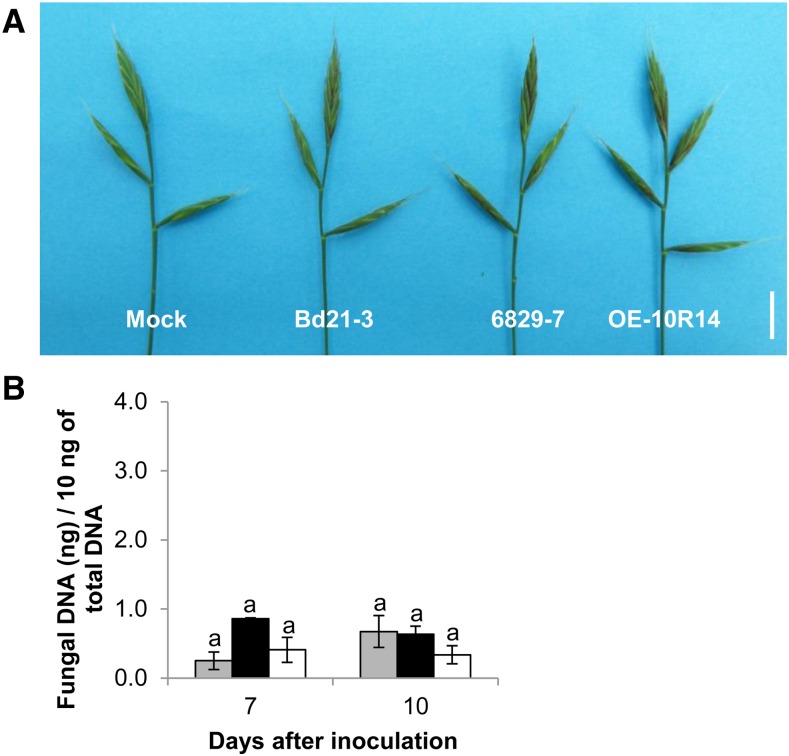
To assess the impact of Bradi5g03300 function on fungal development more directly, fungal DNA was quantified by qRT-PCR 7 and 10 dai of the Bd21-3 wild-type line, the 6829-7 mutant line, and the OE-10R14 line. Tissues of the 6829-7 mutant line exhibited a significantly higher accumulation of fungal DNA after inoculation with FgDON+ (Fig. 5, black bars). At 7 and 10 dai, fungal DNA had accumulated, respectively, to 2.1 and 2.8 times higher in the mutant line than in the Bd21-3 wild-type line (Fig. 5, gray bars; Newman and Keuls test, P ≤ 0.01). By contrast, after infection of the OE-10R14 line by FgDON+, 5.3 and 2.7 times less fungal DNA was detected in comparison with Bd21-3 at 7 and 10 dai, respectively (Fig. 5, white bars). Thus, the results on fungal DNA abundance in the different lines were in accordance with the macroscopic observations (Fig. 4).
Figure 5.
Bradi5g03300 contributes to resistance to FgDON+ colonization. Quantification of fungal DNA by qRT-PCR at 7 and 10 dai of spikes by the FgDON+ strain is shown. Data represent mean values of three independent biological experiments, and error bars show sd (different letters indicate significant differences between conditions; Newman and Keuls test, P ≤ 0.001). Gray bars, Bd21-3; black bars, 6829-7; and white bars, OE-10R14.
To date, DON production has been correlated mainly with the ability of fungal colonization through the rachis in bread wheat (Maier et al., 2006) and, more recently, in B. distachyon (Pasquet et al., 2014). In order to precisely determine the role of the Bradi5g03300 gene on fungal spread in infected spikes, point inoculations by the FgDON+ strain were performed on two mutant lines (8637-12 and 6829-7) as well as on the three overexpressing lines (OE-9R5, OE-24R27, and OE-10R14), and symptoms were compared with those obtained on the Bd21-3 wild-type line at 7 and 14 dai. Both the Bd21-3 wild-type line and the 8637-12 and 6829-7 mutant lines exhibited bleaching symptoms on the entire inoculated spikelets that progressed to the most adjacent spikelet (Fig. 6A). In contrast, 7 d after point inoculation, the three overexpressing lines exhibited a high level of resistance to F. graminearum. The bleaching symptoms decreased (OE-9R5 line) or even were not observed (OE-24R27 and OE-10R14 lines) on these lines in comparison with the symptoms observed on the Bd21-3 line (Fig. 6A). These observations were supported by scoring of the symptoms at 7 and 14 dai, using a scoring scale to evaluate the extent of spikelet and spike colonization (see “Materials and Methods”). No significant difference was observed between the mutant lines and the Bd21-3 wild-type line, but the differential between the overexpressing lines and the wild-type line was highly significant at 7 dai (Fig. 6B; Newman and Keuls test, P ≤ 0.001). At 14 dai, lines OE-24R27 and OE-10R14 continued to exhibit less symptoms than Bd21-3, while symptoms on OE-9R5 were similar to those of the parental line (Fig. 6B). As observed for root tolerance to DON, the higher resistance rate was observed for the OE-10R14 line, exhibiting the strongest Bradi5g03300 overexpression.
Figure 6.
The Bradi5g03300 gene is involved in the control of spike colonization by the FgDON+ strain. A, Typical FHB symptoms on the different lines 7 d after point inoculation by the FgDON+ strain. Bar = 1 cm. B, Quantification of FHB symptoms using a scoring scale (see “Materials and Methods”) at 7 (gray bars) and 14 (back bars) d after point inoculation by fungal strain FgDON+ (n > 30; error bars represent se, and different letters indicate significant differences between conditions; Newman and Keuls test, P ≤ 0.001).
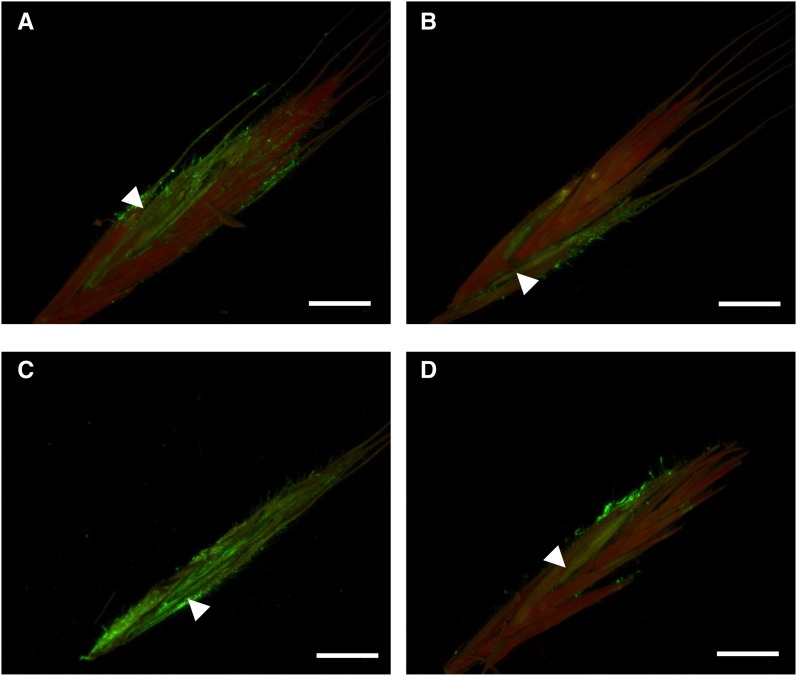
In order to better characterize the observed phenotypes during fungal infection, the wild-type line and the overexpressing line OE-10R14 showing the greatest resistance phenotype were point inoculated with a transformed FgDON+ strain constitutively expressing GFP. While a normal spikelet colonization was observed in the wild-type Bd21-3 line between 3 and 7 dai (Fig. 7, A and C), the fungal strain appeared to be restricted close to the inoculation site in the overexpressing line (Fig. 7, B and D).
Figure 7.
Overexpression of Bradi5g03300 reduces spikelet colonization by the FgDON+ strain. Micrographs show longitudinally sectioned spikelets at 3 (A and B) and 7 (C and D) d post inoculation with the GFP-tagged F. graminearum FgDON+ strain, visualized with epifluorescence illumination. A and C, Wild-type Bd21-3 ecotype. B and D, OE-10R14 line overexpressing the Bradi5g03300 gene. White arrowheads indicate the inoculated florets. Bars = 500 μm.
To verify that the observed differences are not due to increased basal defenses in the OE-10R14 line, the expression of a subset of four defense genes was investigated during an infection time course following point inoculation of the FgDON+ strain. Three of these genes encode pathogenesis-related proteins (PR2 [Blümke et al., 2015], chitinase, and PR9 [Pasquet et al., 2014]), and the fourth one codes for Phe ammonia lyase, the first committed step of the phenylpropanoid pathway (Pasquet et al., 2014). The results showed that the overexpressing line OE-10R14 did not exhibit higher expression of defense genes than the wild-type Bd21-3 line (Supplemental Fig. S6). In contrast, a lowered induction of defense genes could be observed at some but not all later time points of infection (Supplemental Fig. S6), in correlation with a reduced fungal development in the overexpressing line (Fig. 6).
The Bradi5g03300 UGT Conjugates DON into D3G in B. distachyon
The results above demonstrate that mutant lines exhibit increased root sensitivity to DON and spikelet susceptibility to FgDON+ and that overexpressing lines show increased root tolerance to DON and spikelet resistance to FgDON+. A glucosylated form of DON, D3G, has been detected in wheat and barley (Lemmens et al., 2005; Kumaraswamy et al., 2011a). In order to evaluate the role of the Bradi5g03300 UGT in DON conjugation in planta, a quantification of DON, D3G, as well as 15-ADON, a minor TCTB mycotoxin produced by the FgDON+ strain, was conducted following inoculation. The 6829-7 and 8637-12 mutant lines and the three overexpressing lines were used and compared with the Bd21-3 wild-type line, and one mutant and one overexpressing line were selected for a more detailed time-course analysis of mycotoxin production during the infection.
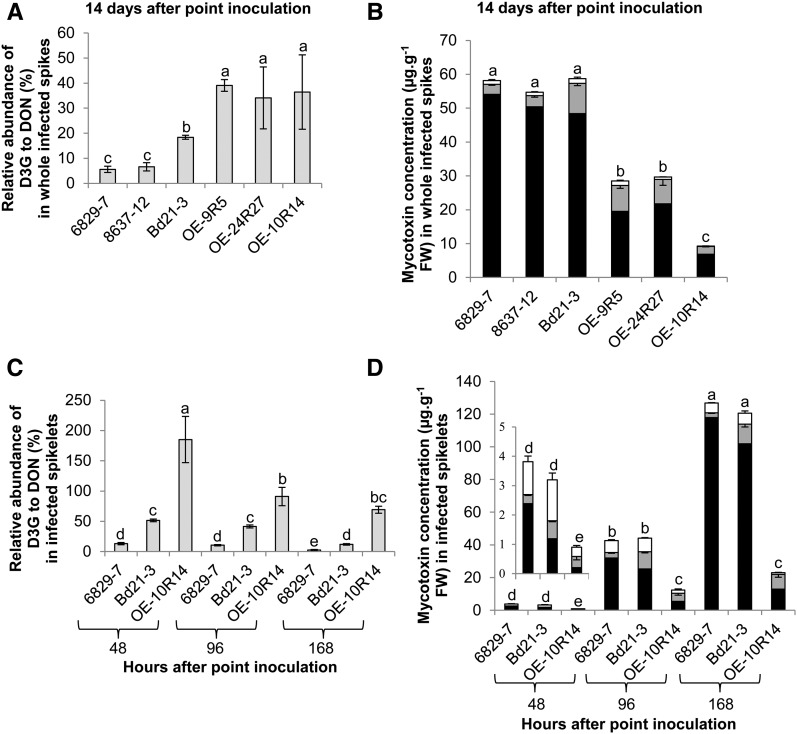
First, the relative abundance of D3G to DON was measured at 14 dai in whole infected spikes of the different lines. The results revealed important differences between the lines. This value was 5.6% and 6.6% for the 6829-7 and 8637-12 mutant lines, respectively, and 18.4% for Bd21-3 (Fig. 8A; Student’s t test, P ≤ 0.05). In contrast, it was significantly enhanced in all the overexpressing lines, with an average increase of 40% over the wild-type value (Fig. 8A). Absolute quantification of DON, D3G, and 15-ADON in all lines was then performed at the same infection time point. An average of 56 μg g−1 fresh weight of total DON (DON + D3G) and much lower quantities of 15-ADON (around 1 μg g−1 fresh weight) were quantified in infected spikes of the Bd21-3 line and the two mutant lines (Fig. 8B). No significant difference in total DON amount was observed between these three lines. In contrast, the quantities of total DON in infected spikes of overexpressing lines were drastically reduced, with only 27.2, 29, and 9.2 μg g−1 fresh weight in the OE-9R5, OE-24R27, and OE-10R14 lines, respectively (Fig. 8B; Student’s t test, P ≤ 0.05).
Figure 8.
The Bradi5g03300 UGT glucosylates DON in planta and controls total DON produced. A, Relative abundance of D3G to DON in whole spikes 14 d after infection. B, DON (black bars), 15-ADON (white bars), and D3G (gray bars) absolute quantification in whole spikes 14 d after infection. C, Relative abundance of D3G to DON in infected spikelets during an infection time course. D, DON (black bars), 15-ADON (white bars), and D3G (gray bars) absolute quantification in infected spikelets during an infection time course. Data represent mean values of three independent biological experiments, and error bars represent sd (different letters indicate significant differences between conditions for the total quantities of DON [DON and D3G]; Student’s t test, P ≤ 0.05). FW, Fresh weight.
Next, to better understand the dynamics of D3G formation during fungal infection, quantification was performed over a shorter time scale during infection of spikelets by FgDON+. Three time points were considered: 48, 96, and 168 h after inoculation (hai). The calculation of the relative abundance of D3G to DON showed important differences between the 6829-7, Bd21-3, and OE-10R14 lines as early as 48 hai. Indeed, this value was only 13.2% in the 6829-7 mutant line, 51.4% in the Bd21-3 wild-type line, but as high as 185.1% in the OE-10R14 line (Fig. 8C). The value remained at a lower level during the time-course experiment in the 6829-7 mutant line: 10.5% and 2.5% at 96 and 168 hai, respectively (Fig. 8C). In the Bd21-3 wild-type line, we observed the same tendency, with a decrease of the relative abundance of D3G between 96 and 168 hai from 41.5% to 11.8% (Fig. 8C). In the OE-10R14 line, although the value decreased from the very high value observed at 48 hai, it remained as high as 69% even at 168 hai (Fig. 8D). The values in the three different lines were always significantly different from each other at the same infection time point (Student’s t test, P ≤ 0.05). Absolute quantification of DON, D3G, and 15-ADON in infected spikelets of the three lines was then performed during the same infection time course. For the three time points, no significant difference in total DON produced could be detected between the 6829-7 mutant line and the Bd21-3 wild-type line (Fig. 8D). In contrast, we observed a drastic reduction of total DON in the OE-10R14 line, with only 0.6 µg g−1 fresh weight at 48 hai, 10.3 µg g−1 fresh weight at 96 hai, and 22 µg g−1 fresh weight at 168 hai, as compared with infected spikelets of the Bd21-3 wild-type line, containing 1.8, 35.8, and 113.9 µg g−1 fresh weight, respectively, at the same time points.
Schweiger et al. (2013a) recently demonstrated that expression of the Bradi5g03300 cDNA in the heterologous S. cerevisiae system conferred resistance to DON and allowed the conversion of DON into D3G. To more precisely determine that the Bradi5g03300 UGT was acting on DON in vitro, the enzyme was produced by heterologous expression in Escherichia coli as a His fusion protein (Supplemental Fig. S7, A and B). The estimated apparent Km value toward DON was determined to be 32.5 ± 3.6 µm (Supplemental Fig. S7C). No glucosylation of scopoletin, a 6-methoxy-7-hydroxycoumarin often used as a reference substrate in UGT assays (Lim et al., 2003), was detected (data not shown).
Mutation or Overexpression of the Bradi5g03300 Gene Does Not Modify the Interaction with an F. graminearum Strain Unable to Produce DON
The interaction between B. distachyon and an F. graminearum strain unable to produce DON (FgDON−) impaired in the Tri5 gene encoding the first committed enzyme of the trichothecene biosynthetic pathway (Cuzick et al., 2008) has been described in two independent studies (Pasquet et al., 2014; Blümke et al., 2015). Both showed that the mutant strain is largely delayed in symptom development following point inoculation of B. distachyon spikes (Pasquet et al., 2014; Blümke et al., 2015). In order to determine whether the differences observed between the mutant and overexpressing lines following inoculation with the FgDON+ strain are strictly correlated with DON conjugation, spray inoculations of the same three lines were performed with the FgDON− strain. The resulting symptoms were observed 7 and 10 dai, and fungal DNA was quantified. No striking differences in symptoms could be observed 7 and 10 dai of spikes of the 6829-7 mutant line and the OE-10R14 expressing line compared with the Bd21-3 wild-type line (Fig. 9A). Quantification of fungal DNA on the same biological material confirmed the absence of significant differences between the three lines during infection by FgDON− (Fig. 9B).
Figure 9.
Bradi5g03300 is not involved in the control of FgDON− colonization following spray inoculation. A, Brown symptoms observed 10 dai by the FgDON− strain. Bar = 1 cm. B, Quantification of fungal DNA by quantitative PCR (qPCR) at 7 and 10 dai of spikes of the wild-type line Bd21-3 (gray bars), the 6829-7 mutant line (black bars), and the overexpressing line OE-10R14 (white bars). Data represent mean values of three independent biological experiments, and error bars represent sd (different letters indicate significant differences between conditions; Newman and Keuls test, P ≤ 0.001).
Overexpression of, But Not Mutation in, the Bradi5g03300 Gene Alters Primary Infection of F. graminearum
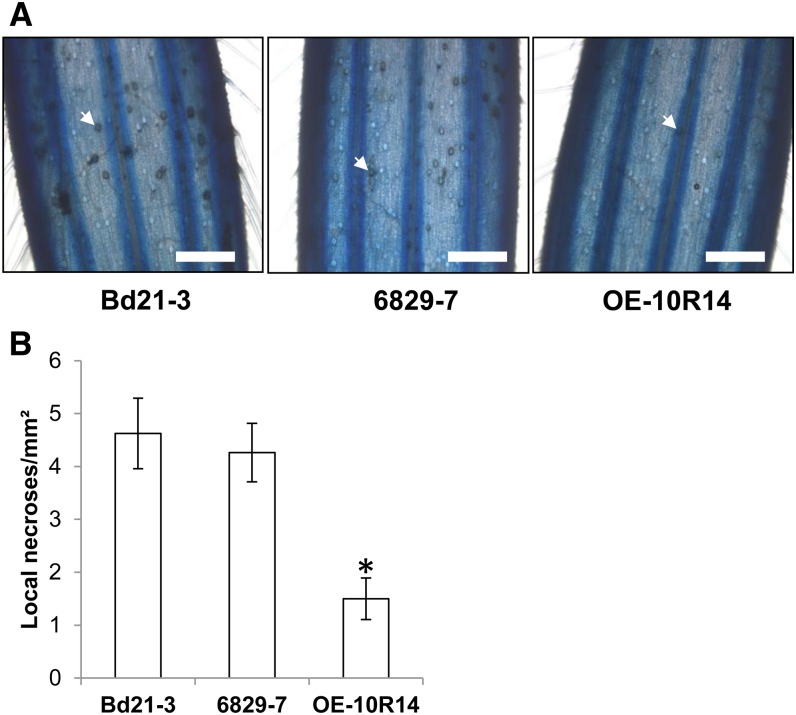
In the spray inoculation experiments, mutant and overexpressing lines were more susceptible and more resistant, respectively, to the fungal pathogen. In contrast, following point inoculations, which only allow estimation of the fungal colonization from the inoculation sites, the overexpressing lines remained highly resistant, but no significant differences were observed between the mutant lines and the wild-type line. To further explore the impact of the Bradi5g03300 UGT on the F. graminearum infection process, we set up a detached glumes assay (Rittenour and Harris, 2010) and observed the adaxial surface of infected lemma of mutant line 6829-7 and overexpressing line OE-10R14 as compared with those of the wild-type line Bd21-3 at 72 hai after Lactophenol Blue staining. Typical amber-brown discoloration of numerous circular cells, corresponding to the base of macrohairs, were observed on Bd21-3 infected lemma (Fig. 10A, left), described previously as favored target sites for fungal penetration (Peraldi et al., 2011). Although no significant difference was observed on the infected lemma in the mutant (Fig. 10A, middle), a strong decrease in necrosis was evident on OE-10R14 samples (Fig. 10A, right). Quantification of discoloration confirmed this effect in the overexpressing line (Fig. 10B), suggesting that early DON glucosylation in planta alters the potential of F. graminearum primary infection.
Figure 10.
Early DON glucosylation alters primary infection of F. graminearum. A, Detached infected lemmae of the Bd21-3, 6829-7, and OE-10R14 lines 72 hai showing amber-brown cells corresponding to F. graminearum infection sites at the base of macrohairs (white arrows). Bars = 1 mm. B, Quantification of amber-brown cells on detached infected lemmae (n ≥ 12) at 72 hai. The asterisk indicates a significantly different result (Student’s t test, P ≤ 0.01).
DISCUSSION
Mycotoxins are among the toxic compounds imposed exogenously on plants, and more specifically on small-grain cereals (Miller, 2008). TCTBs produced by plant pathogenic fungi belonging to the Fusarium genus are the most important mycotoxins worldwide (Foroud and Eudes, 2009). These sesquiterpene molecules, including DON, were shown to have phytotoxic effects on Arabidopsis (Masuda et al., 2007) and wheat (Desmond et al., 2008). Glycosylation of secondary metabolites by UGTs has been identified as one of the major detoxification steps of exogenous compounds in plants (Coleman et al., 1997; Messner et al., 2003; Lim and Bowles, 2004). Thus, the Arabidopsis UGT73C5 (Poppenberger et al., 2003) and HvUGT13248 in barley (Schweiger et al., 2010) were shown to be involved in DON glucosylation. Although very informative, these studies did not establish a causal role for the ability to conjugate DON into D3G in determining the resistance/susceptibility of plants to the DONproducing fungal pathogen. Here, we directly addressed the question of the role of the UGT encoded by Bradi5g03300 in the resistance of B. distachyon to F. graminearum.
The Bradi5g03300 UGT Is Involved in Root Tolerance to DON
The use of B. distachyon mutant and overexpressing lines in this study allowed us to establish a direct relationship between DON glucosylation and root sensitivity/tolerance to the mycotoxin. Mutant lines exhibited hypersensitivity to the mycotoxin (Fig. 1), whereas overexpressing lines showed a strong tolerance to DON, as compared with the wild-type line (Fig. 2). Masuda et al. (2007) previously reported the inhibitory effect of DON on root elongation in Arabidopsis and wheat plants. The authors mentioned an abnormal morphology of DON-treated Arabidopsis roots, including a reduction in root hair length and a relative disorganization of root cells (Masuda et al., 2007). In this work, we have shown that DON affected root cell organization in the apex, such that root apical meristematic area became difficult to distinguish in DON-treated mutants (Fig. 3). These effects were not observed in the wild-type line or in the overexpressing lines in the presence of the mycotoxin (Fig. 3; Supplemental Fig. S5). Such observations are reminiscent of phenotypes due to hormonal deregulation, in particular an auxin deficiency (Ubeda-Tomás et al., 2012). So far, F. graminearum has been shown to induce jasmonate and ethylene pathways (Ding et al., 2011; Gottwald et al., 2012), but there are no reports of a modulation of other plant hormone biosynthesis and/or signaling pathways.
The Bradi5g03300 UGT Is Involved in the Establishment of Early Resistance to F. graminearum
D3G has been established as a resistance-related metabolite in a number of studies on wheat (Gunnaiah et al., 2012) and barley (Kumaraswamy et al., 2011a, 2011b). Therefore, the Bradi5g03300 mutant and overexpressing lines were challenged with F. graminearum in order to determine whether DON glucosylation could be connected directly to FHB resistance. As described in previous studies on FHB, we used two infection assays, point inoculations and spray inoculations (Miedaner et al., 2003). Whereas the first approach allows the colonization of spikelets and spikes to be scored (type II resistance), the second also estimates primary infection (type I resistance). The lines overexpressing the Bradi5g03300 gene were shown to exhibit increased resistance to F. graminearum using both inoculation methods. Moreover, the level of resistance correlated well with the level of Bradi5g03300 overexpression (Fig. 4). Previous studies on wheat and barley have established that genes encoding UGTs are up-regulated following fungal infection (Lemmens et al., 2005; Boddu et al., 2007; Gottwald et al., 2012) or DON application (Gardiner et al., 2010; Lulin et al., 2010). However, in several transcriptomic studies, overall differences in gene expression were partly due to their different genetic backgrounds between FHB-resistant and -susceptible lines rather than differences in specific resistance loci (Jia et al., 2009). In a recent study using near-isogenic wheat lines, no differential induction of genes encoding UGTs could be detected after fungal infection (Xiao et al., 2013). While a lack of induction suggests that glucosylation of DON may not be associated with resistance to FHB in all genetic backgrounds, such evidence is only circumstantial. Here, we directly tested the importance of DON glucosylation by constructing and exploiting B. distachyon lines specifically altered in the function of the UGT encoded by Bradi5g03300. This allowed us to determine unambiguously the direct relationship between the in planta formation of D3G and resistance to FHB.
The mutant lines isolated through TILLING showed a significantly increased susceptibility in spray inoculation assays (Fig. 4). In contrast, no significant differences were observed between the mutants and the wild-type line after point inoculations (Fig. 6), a method specifically testing for symptoms spread along the spikes. So far, DON production has been reported as a virulence factor that contributes to the colonization of spikes in wheat but not in barley, a host plant exhibiting a high level of type II resistance (Bai et al., 2002; Maier et al., 2006). We have shown previously that an F. graminearum tri5 mutant strain unable to produce DON in B. distachyon was delayed in spike colonization (Pasquet et al., 2014). Together, these results suggest that DON is a virulence factor of F. graminearum for spike colonization of B. distachyon, but to a lesser extent than for wheat infection.
Two hypotheses can be proposed to explain the contrasting results obtained in the mutant lines with the two inoculation methods. The first one is that another B. distachyon UGT could, in the absence of a functional Bradi5g03300 UGT, glucosylate DON and, therefore, compensate any effect of the Bradi5g03300 gene mutation. In a recent study, another UGT-encoding gene localized in the same genomic region, Bradi5g02780, was shown to exhibit a slight activity on DON when expressed in a S. cerevisiae heterologous system (Schweiger et al., 2013a) and, therefore, could be a candidate. However, we have never observed any significant misregulation of this gene in the different mutant or overexpressing lines during infection (Supplemental Fig. S8); therefore, we have no evidence for this explanation. The second hypothesis is that DON may act as a virulence factor not only during the colonization phase, as we could show using a GFP-expressing strain (Fig. 7), but also for primary infection. Using a detached glumes assay to better synchronize the initial infection, a strong reduction of potentially successful fungal primary infection sites on the OE-10R14 line was observed (Fig. 10). Our results demonstrate, to our knowledge for the first time, that DON may be involved not only in spike colonization of wheat and barley, as already described (Jansen et al., 2005), but also in very early infection steps, which may be determinant for fungal establishment in plant tissues. These results are consistent with previous ones showing early induction of DON biosynthesis during host infection (Boenisch and Schäfer, 2011). Because of the complexity of the corresponding assays, type I resistance has been poorly investigated in the selection processes of FHB-resistant cereal crop varieties (Gosman et al., 2010). These aspects are worth reconsidering in efforts to improve resistance.
The Bradi5g03300 UGT Glucosylates DON in Planta during Infection by F. graminearum
Overexpression of the Arabidopsis UGT73C5 and barley Hv13248 genes in Arabidopsis (Poppenberger et al., 2003; Shin et al., 2012) and of the barley gene Hv13248 in wheat (Li et al., 2015) has been shown to increase DON conjugation to D3G following direct application of DON. However, in those studies, the conjugation of DON could not be or has not been demonstrated following F. graminearum infection (i.e. in the presence of fungus-derived mycotoxins). In this work, the selection of B. distachyon lines carrying mutations in the Bradi5g03300 gene, as well as the generation of lines overexpressing the gene, allowed us to establish the involvement of the UGT in DON conjugation in planta (Fig. 8). Indeed, we showed that mutant lines exhibited a 60% reduction of their D3G-DON ratios 14 d after inoculation as compared with the wild-type line. Residual amounts of D3G were detected in the mutant lines, suggesting the existence of another redundant glucosylation activity in these lines. In contrast, lines overexpressing the Bradi5g03300 gene showed a nearly 2-fold increase of their D3G-DON ratio. Similar results were obtained when analyzing samples from a time course covering 48 to 168 hai: significant differences were observed between the lines as early as 48 hai and were maintained throughout the infection time course. These results clearly demonstrate the involvement of the Bradi5g03300 UGT in DON conjugation in planta. Interestingly, a strong reduction of total DON produced in the overexpressing lines also was evident. Altogether, these results suggest either a very early resistance process leading to fungal growth inhibition/slowing down or fungal death in the overexpressing lines or a negative feedback control of DON production.
Is Bradi5g03300 Specific for DON?
The B. distachyon Bradi5g03300 UGT belongs to group L (subgroup L1) of secondary metabolism glycosyltransferases, which includes Arabidopsis UGT74 sequences and the IAGlu protein from Z. mays (Schweiger et al., 2013a). UGT74B1 was shown to be a thiohydroximate S-glycosyltransferase involved in glucosinolate biosynthesis (Grubb et al., 2004). Insertional mutants of the UGT74B1 gene exhibited strong developmental phenotypes linked to auxin accumulation (Grubb et al., 2004). Both Arabidopsis UGT74D1 (Jin et al., 2013; Tanaka et al., 2014) and UGT74E2 (Tognetti et al., 2010) and Z. mays IAGlu (Szerszen et al., 1994) were demonstrated to catalyze the glucosylation of one or several auxins such as indole-3-butyric acid, indole-3-acetic acid, and naphthaleneacetic acid. Arabidopsis lines overexpressing the UGT74D1 gene exhibited a mutant phenotype reminiscent of a deficiency in auxin content (Jin et al., 2013). Moreover, Dean and Delaney (2008) have shown that UGT74F1 and UGT74F2 both produced salicylic acid-2-O-β-d-Glc and that UGT74F2 formed a salicylic acid-Glc ester. Apart from metabolic differences, no phenotype was associated with the mutations in either of the two genes (Dean and Delaney, 2008). In our work, neither the mutant lines nor the overexpressing lines were shown to exhibit any developmental phenotype when the plants were grown under nonstress conditions in a growth chamber (Supplemental Fig. S2). These observations confirm that it is risky to assign the in vivo substrate specificity of a UGT solely based on phylogenetic analyses (Osmani et al., 2009). Moreover, they suggest that the Bradi5g03300 UGT is not essential for plant development, although this also could be due to partial functional redundancy with other UGTs. An alternative explanation may be that, following coevolution between the host plant and its pathogen, the Bradig03300 UGT specificity has adapted to specifically conjugate DON. Comparative metabolomic studies on wild-type, mutant, and overexpressing lines could help determine more precisely whether Bradi5g03300 is specific for DON or may fulfill another role in B. distachyon.
In conclusion, we have shown here that early and rapid conjugation of DON by a B. distachyon UGT is necessary and sufficient to establish resistance to F. graminearum, the FHB causal agent. Our results suggest that the mycotoxin is an important virulence factor for the fungus not only to colonize the plant tissues, as described previously (Jansen et al., 2005; Maier et al., 2006), but also for primary infection. These important results open new perspectives for innovative breeding strategies in cereal crops.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Brachypodium distachyon lines (Table I) were cultivated in a growth chamber under a 20-h light period at 23°C ± 2°C under fluorescent light (265 µE m−2 s−1 at the soil level and approximately 315 µE m−2 s−1 at the spike level). Prior to sowing, seeds were surface sterilized by incubation in a 0.6% sodium hypochlorite solution for 10 min with gentle shaking followed by three rinses in sterile distilled water. Sterilized seeds were subsequently incubated for 5 d at 4°C in the dark. Plants were grown routinely on a 2:1 mixture of compost (Tref terreau P1; Jiffy France SARL) and standard perlite (Sinclair) and soaked with an aqueous solution containing a carbamate fungicide (Previcur at 2 mL L−1; Bayer Crop Sciences) and a larvicide (Hortigard at 1 g L−1; Syngenta France). Plants were routinely watered in 2- to 3-d intervals using a standard nutritional solution and were never allowed to stand in water.
To study the overexpression rates of the Bradi5g03300 gene in roots, surface-sterilized seeds were pregerminated on filter paper soaked with sterile water for 3 d and then transferred under hydroponic conditions for 2 weeks in liquid one-quarter-strength Murashige and Skoog medium in trays to ensure darkness conditions for the root system. The medium was changed every 3 d.
Screening of the TILLING Mutant Collection
We screened the TILLING collection (http://urgv.evry.inra.fr/UTILLdb) available at the Institut Jean-Pierre Bourgin (Institut National de la Recherche Agronomique) for putative point mutations in a fragment of the Bradi5g03300 gene encompassing the PSPG box-encoding region. Screening was conducted at the Unité de Recherche en Génomique Végétale (Institut National de la Recherche Agronomique) as described by Dalmais et al. (2013). Specific primers used for the generation of the Bradi5g03300 PCR product were as follows: UGT33N1F2 (5′-GTCACTACCACCAAATATTTGGG-3′) and UGT33N1R1 (5′-CAAAGACCAGAAATGATGTAGGAGG-3′) for the first amplification and 3300_N2Ft1 (5′-GGTACTTGGTTGTTGATAGTTATGC-3′) and 3300_N2Rt1 (5′-GTGTGCTCCTTCGGCCC-3′) for the second amplification (Supplemental Fig. S1).
Binary Vector Construction
The Bradi5g03300 cDNA was amplified from spikelet cDNAs using the primers 5′-CGGGATCCCGATGGACAGCACAGGCAAATCGGTGATGGCGA-3′ and 5′-GGATATCCTTAACTTGACGAATACTTAGCAGCGAATTCAGCA-3′, adding a BamHI and an EcoRV restriction site, respectively (indicated in italics and underlines in the primer sequences). The PCR product was digested using the BamHI and EcoRV restriction enzymes, purified using a NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel EURL) using the manufacturer’s instructions, and then ligated into the pENTR1A plasmid linearized by the same restriction enzymes. The resulting pEntry-OE3300 plasmid was used to transfer a Bradi5g03300 cDNA fragment into the pIPKb002 binary vector (Himmelbach et al., 2007) by in vitro recombination using the Gateway LR Clonase II Enzyme mix according to the manufacturer’s recommendations (Invitrogen, Life Technologies).
B. distachyon Transformation
The pIPKb002-Bradi5g03300 binary vector was electroporated into Agrobacterium tumefaciens (AGL1 strain). The Bd21-3 wild-type line was genetically transformed using a method adapted from that described by Vogel and Hill (2008) and Alves et al. (2009). After selection of transformants in Murashige and Skoog medium (Murashige and Skoog, 1962) containing 40 mg L−1 hygromycin, segregation analysis was used to identify single-locus insertion lines in the T2 generation.
In Vitro Root Assays and Observations
For root tests in vitro, the palea of each seed was removed and seeds were surface sterilized by incubation in a 0.6% sodium hypochlorite solution for 5 min with gentle shaking followed by three rinses in sterile distilled water. Sterilized seeds were subsequently incubated for 5 d at 4°C in the dark. Seeds were sown on Murashige and Skoog medium with 3% saccharose and vitamins (100 mg L−1 myoinositol and 0.1 mg L−1 thiamine-HCl) in square petri dishes (12 × 12 cm) with or without DON (Sigma-Aldrich). During 2 d, petri dishes were placed in the dark and then transferred to light for 5 d (same photoperiod as described previously for plant growth). Roots were specifically protected from light by applying a piece of aluminum foil to cover the corresponding area of the petri dishes.
After 7 d of growth, roots were fixed during 24 h in an ethanol:acetic acid solution (3:1, v/v), washed for 20 min in 70% ethanol, and incubated overnight at room temperature in a chloral hydrate solution (8 g of chloral hydrate [Sigma], 2 mL of 50% glycerol, and 1 mL of water). Roots were observed by differential interference contrast using a macroscope (AZ100; Nikon). Images were captured with a Nikon RI1 video camera.
Fusarium graminearum Strains, Maintenance, and Spore Production
F. graminearum strains PH-1 (FgDON+) and Δtri5 (FgDON−; MU102 mutant strain; Cuzick et al., 2008) were maintained on potato dextrose agar plates. A PH-1 transformant expressing GFP under the control of the ToxA promoter region and resistant to geneticin was obtained through polyethylene glycol-mediated transformation of protoplasts of a plasmid derived from vector pCT74 (Lorang et al., 2001). The transformation procedure was as described before (Hua-Van et al., 2001), and geneticin used as a selection was incorporated directly onto the bottom plates at a final concentration of 100 mg L−1. To obtain fungal spores, 2- to 4-mm2 plugs from 15-d-old potato dextrose agar plates were inoculated in liquid mung bean medium (Bai and Shaner 1996; 10 plugs for 20 mL) and incubated at 150 rpm at room temperature for 5 to 6 d. The resulting spore suspension was then diluted 10 times in fresh liquid mung bean medium and further incubated for an additional 5 to 6 d under the same conditions. For pathogenicity assays, spores were further filtrated onto sterile Miracloth (Calbiochem) and resuspended in 0.01% Tween 20 at a final concentration of 105 spores mL−1.
Pathogenicity Assays
Point inoculation was performed by pipetting 300 spores (3 µL of a 105 spores mL−1 suspension) into a central floral cavity of the second spikelet starting from the top of the spike of different lines at midanthesis (approximately 30–35 d after sowing). Alternatively, whole plants were sprayed with the fungal spore suspension (1 × 105 conidia mL−1) until dripping.
Inoculated plants were covered with clear plastic bags whose interior had been sprayed with distilled water beforehand. The first 24-h inoculated heads were kept in the dark, then incubated with a photoperiod of 16 h of light and 8 h of darkness at 20°C with the same light intensities as those used for plant development (see “Plant Material and Growth Conditions”). Application of 0.01% Tween 20 was performed as the control condition for each inoculation experiment.
Evaluation of Symptoms
For spray inoculations, symptoms were observed at 7 and 10 d after spraying of the conidial suspension. In experiments with TILLING lines (6829-7, 6491-37, and 8637-12), a spikelet was considered as symptomatic if all florets of the spikelet were fully symptomatic. In experiments conducted with overexpressing lines (OE-9R5, OE-24R27, and OE-10R14), a spikelet was considered as symptomatic if at least half of its florets were symptomatic.
For point inoculations, symptoms were evaluated 7 and 14 d after inoculation with a scoring scale for each inoculated spike from 0 to 4 as follows: 0, no symptoms; 1, only the inoculated floret was symptomatic (most frequently, only browning); 2, extension of symptoms to additional florets of the inoculated spikelet (browning or bleaching); 3, symptoms appeared on the entire inoculated spikelet; and 4, extension of the symptoms to the whole inoculated spikelet and at least one adjacent spikelet.
Detached Glumes Assays
Detached glumes assays were conducted as described by Rittenour and Harris (2010) except that a drop of 50 µL of a 105 spores mL−1 suspension was used. Infected lemmae were stained as described by Pasquet et al. (2014).
RNA Extraction
Leaves from three 2-week-old plants or five spikelets from independent plants were ground in liquid nitrogen, and total RNA was extracted from 0.1 g of the resulting powder using TRIzol (Invitrogen, Life Technologies) followed by an RNase-free DNase I step (Ambion, Applied Biosystems) according to the manufacturers’ instructions. Total RNA was further purified using the NucleoSpin RNA Clean-up XS kit (Macherey-Nagel).
DNA Extraction, Southern-Blot Analysis, and Fungal DNA Quantification by qPCR
For Southern-blot analysis, genomic DNA was extracted from leaves collected from 3-week-old plantlets (Atoui et al., 2012). Ten micrograms of genomic DNA was digested with the either SacI or SalI restriction enzyme, separated by electrophoresis on 0.7% agarose gels, and transferred on nylon membranes using a vacuum blotter. A 712-bp hph-specific probe was obtained using hph1 (5′-CAGCGAGAGCCTGACCTATTGC-3′) and hph2 (5′-GCCATCGGTCCAGACGGCCGCGC-3′). DNA templates were 32P labeled using the rediprimeII kit (Amersham Biosciences). Hybridizations were conducted under standard conditions (Sambrook et al., 1989).
For quantification of F. graminearum DNA, 10 spikes spray-inoculated with either of the two strains used in this study were pooled per time point. Quantification of fungal DNA was realized by qPCR (see below) on 10 ng of total DNA using primers specific for the 18S ribosomal subunit-encoding genomic region (Mudge et al., 2006; Supplemental Table S2).
Real-Time PCR
cDNA synthesis was performed on 1 µg of total RNA using the ImProm-II reverse transcription system (Promega France) according to the manufacturer’s instructions. The resulting product was diluted 10 times in nuclease-free water. Primers were designed to amplify plant gene transcripts, including the reference genes Bradi4g00660 (UBC18) and Bradi4g41850 (ACT7; now referred to as ACT3-like under accession number XM_003578821 in the nucleotide National Center for Biotechnology Information database) as determined previously (Hong et al., 2008; Supplemental Table S2). qPCR was performed on 2 µL of the diluted cDNA product using 8 pmol of each specific primer and 10 µL of SYBRGreen Master Mix in a final volume of 20 µL. Reactions were performed in a Light Cycler LC480 real-time PCR system (Roche Diagnostics). All qRT-PCRs were carried out on biological triplicates, each in technical duplicate. The final Ct values were means of three values (biological triplicates), each corresponding to the mean of technical duplicates. The comparative Ct method was used to evaluate the relative quantities of each amplified product in the samples. The Ct was automatically determined for each reaction by the Light Cycler LC480 real-time PCR system set with default parameters. The specificity of the qRT-PCR was determined by melt-curve analysis of the amplified products using the standard method installed in the system.
Heterologous Expression of B. distachyon Bradi5g03300 UGT in Escherichia coli
A His-Bradi5g03300 expression plasmid was constructed using the pET-16b vector (Novagen, Merck Chimie). The Bradi5g03300 cDNA was amplified using specific primers 5′-GGAATTCCATATGGACAGCACAGGCAAATC-3′ and 5′-CGGGATCCCGTTATTAACTTGACGAATACTTAGCAG-3′. Both the PCR product and vector pET-16b (Novagen, Merck Millipore) were digested using BamHI and NdeI restriction enzymes (recognition sites underlined in the primer sequences) and ligated together to obtain the expression plasmid of the His-UGTBradi5g03300 fusion protein. Recombinant protein production was performed in E. coli BL21 (λDE3) strain transformed by thermoporation with the expression vector (pET-16b::His-Bradi5g03300). The transformed bacterial cells were grown at 37°C in Luria-Bertani medium containing 100 µg mL−1 ampicillin until they reached an optical density at 600 nm of 0.6. Isopropyl β-d-thiogalactoside (0.5 mm) was then added to induce the production of the His-Bradi5g03300 protein. The bacteria were grown at 18°C overnight, harvested by centrifugation, and resuspended in 6 mL of lysis buffer, pH 7 (50 mm Tris-HCl, pH 7, 300 mm NaCl, and 10 mm imidazole) containing protease inhibitor cocktail (cOmplete, Mini, EDTA-free [Roche Life Science]; one tablet for 50 mL of lysis buffer).
The recombinant protein was affinity purified using His-coupled Sepharose beads (HIS-Select Nickel Affinity Gel; Sigma-Aldrich), according to the manufacturer’s instructions, and desalted using PD-10 columns (GE Healthcare). The purified recombinant protein was quantified by the Bradford method (Bradford, 1976) with bovine serum albumin as the standard and separated by SDS-PAGE. After transfer to a polyvinylidene difluoride membrane with Tetra Blotting Module (Bio-Rad; http://www.bio-rad.com/en-uk/product/tetra-blotting-module), immunodetection of the His tag was performed using the monoclonal anti-His tag antibody produced in mouse (Sigma-Aldrich) at a 1:5,000 dilution. Antigen-antibody complexes were detected using horseradish peroxidase-conjugated anti-mouse secondary antibody (Pierce, Thermo Fisher Scientific) used at a 1:5,000 dilution.
Protein Production and Enzyme Assays
The glucosyltransferase activity assay mix was constituted of the following components: 1 µg of recombinant His fusion protein, 10 mm 2-mercaptoethanol, 50 mm Tris-HCl, pH 7, 0.1 mm radiolabeled UDP-[14C]Glc (4.625 MBq; Perkin Elmer), 1 mm UDP-Glc, and 0, 0.01, 0.1, 0.2, 0.4, and 1 mm concentrations of the acceptor substrate (dissolved in dimethyl sulfoxide for scopoletin and in methanol for DON). The reactions were carried out in 50 µL at 30°C for 1 h, stopped by adding 50 µL of methanol, and then stored at −20°C. Analysis of the reaction products was performed by thin-layer chromatography. An aliquot of each sample was spotted on a silica gel plate (DC-Fertigplatten SIL G-25 UV 254; Macherey-Nagel) and developed with methanol:chloroform (70:30, v/v) for scopoletin and with 1-butanol:1-propanol:ethanol:water (2:3:3:1, v/v/v/v) for DON. The location of each radioactive spot on the silica gel plate was determined using a PhosphorImager (Bio-Rad), and their respective intensities were determined by counting the specific 14C radioactivity integrated. D3G was used as a standard (Sigma-Aldrich).
Mycotoxin Extraction and Analysis
A total of 500 mg of fresh ground material (spikes or spikelets infected by PH1 strain) was extracted with 7 mL of acetonitrile:water (84:16, v/v) for 1 h at room temperature on a tube rotator (50 rpm). Before extraction, 0.5 µg of fusarenon X (4-acetyl-nivalenol; Romers Lab) was added in each sample as an internal standard. After centrifugation (5 min at 5,000g), the supernatant was purified on Trichothecene P Columns (P51 R-Biopharm), and 3 mL of filtrate was evaporated at 50°C dryness of nitrogen. The pellet was resuspended in 400 µL of methanol:water (50:50, v/v) and filtered through a 0.20-µm filter before analysis. DON, D3G, 15-ADON, and fusarenon X concentrations were determined using HPLC-tandem mass spectrometry analyses. These analyses were performed using the QTrap 2000 LC/MS/MS system (Applied Biosystems) equipped with the 1100 Series HPLC system (Agilent), a reverse-phase Kinetex 2.6-µm XB-C18 column (150 × 4.6 mm; Phenomenex) maintained at 45°C, and a TurboIonSpray electrospray ionization source. Solvent A consisted of methanol:water (10:90, v/v) and solvent B consisted of methanol:water (90:10, v/v). The flow rate was kept at 700 µL min−1 and was split so that 300 µL min−1 went to the electrospray source. Gradient elution was performed with the following conditions: 4 min with a linear gradient from 80% to 5% A, 4-min hold at 5% A, 1-min linear gradient from 5% to 80% A, and 80% A for 8-min postrun reconditioning. The injection volume was 10 µL. The electrospray interface was used in the negative ion mode at 400°C with the following settings: curtain gas, 20 p.s.i.; nebulizer gas, 30 p.s.i.; auxiliary gas, 70 p.s.i.; ion spray voltage, −4,200 V; declustering potential, −30 V; entrance potential, −10 V; collision energy, −30 eV; collision-activated dissociation gas, medium. Quantification was performed using external calibration with DON (Sigma-Aldrich), D3G (Sigma-Aldrich), 15-ADON (Sigma-Aldrich), and fusarenon X standard solutions, ranging from 10 to 1,000 ng mL−1.
Accession Numbers
Sequence data of the pIPKb002 vector used in this article can be found in the GenBank/EMBL data libraries under accession number EU161568.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Nucleotide and deduced amino acid sequences of the Bradi5g03300 gene.
Supplemental Figure S2. Phenotypes of the mutant line 6829-7 and overexpressing line OE-10R14 as compared with the wild-type line Bd21-3.
Supplemental Figure S3. Southern-blot analysis of B. distachyon transformed lines overexpressing the Bradi5g03300 gene.
Supplemental Figure S4. Relative expression of the Bradi5g03300 gene in spikelets and leaves of overexpressing lines OE-5R36, OE-9R5, OE-10R14, OE-18R22, and OE-24R27.
Supplemental Figure S5. DON does not induce root apex disorganization in Bradi5g03300-overexpressing lines.
Supplemental Figure S6. Relative expression of defense genes in wild-type and OE-10R14 lines after infection by F. graminearum.
Supplemental Figure S7. Control of the purified recombinant Bradi5g03300 UGT and Km determination toward DON.
Supplemental Table S1. Characteristics of the mutant families identified in the Bradi5g03300 gene following screening of the Bd21-3 TILLING collection.
Supplemental Table S2. List of primers used in qRT-PCR and qPCR experiments.
Supplementary Material
Acknowledgments
We thank G. Noctor for critical reading of the article.
Glossary
- FHB
Fusarium head blight
- TCTB
type B trichothecene
- DON
deoxynivalenol
- 15-ADON
15-acetyldeoxynivalenol
- D3G
deoxynivalenol-3-O-glucose
- qRT
quantitative reverse transcription
- cDNA
complementary DNA
- dai
days after spray inoculation
- hai
hours after inoculation
- qPCR
quantitative PCR
- Ct
cycle threshold
Footnotes
This work was supported by the Centre National de la Recherche Scientifique and an Attractivité grant from Université Paris-Sud (to M.D.) and by the Ministère de l’Enseignement Supérieur et de la Recherche (PhD fellowships to J.-C.P. and V.C.).
References
- Alves SC, Worland B, Thole V, Snape JW, Bevan MW, Vain P (2009) A protocol for Agrobacterium-mediated transformation of Brachypodium distachyon community standard line Bd21. Nat Protoc 4: 638–649 [DOI] [PubMed] [Google Scholar]
- Atoui A, El Khoury A, Kallassy M, Lebrihi A (2012) Quantification of Fusarium graminearum and Fusarium culmorum by real-time PCR system and zearalenone assessment in maize. Int J Food Microbiol 154: 59–65 [DOI] [PubMed] [Google Scholar]
- Bai GH, Desjardins AE, Plattner RD (2002) Deoxynivalenol-nonproducing Fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia 153: 91–98 [DOI] [PubMed] [Google Scholar]
- Bai GH, Shaner G (1996) Variation in Fusarium graminearum and cultivar resistance to wheat scab. Plant Dis 80: 975–979 [Google Scholar]
- Blümke A, Sode B, Ellinger D, Voigt CA (2015) Reduced susceptibility to Fusarium head blight in Brachypodium distachyon through priming with the Fusarium mycotoxin deoxynivalenol. Mol Plant Pathol 16: 472–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddu J, Cho S, Kruger WM, Muehlbauer GJ (2006) Transcriptome analysis of the barley-Fusarium graminearum interaction. Mol Plant Microbe Interact 19: 407–417 [DOI] [PubMed] [Google Scholar]
- Boddu J, Cho S, Muehlbauer GJ (2007) Transcriptome analysis of trichothecene-induced gene expression in barley. Mol Plant Microbe Interact 20: 1364–1375 [DOI] [PubMed] [Google Scholar]
- Boenisch MJ, Schäfer W (2011) Fusarium graminearum forms mycotoxin producing infection structures on wheat. BMC Plant Biol 11: 110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollina V, Kushalappa AC, Choo TM, Dion Y, Rioux S (2011) Identification of metabolites related to mechanisms of resistance in barley against Fusarium graminearum, based on mass spectrometry. Plant Mol Biol 77: 355–370 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Buerstmayr H, Ban T, Anderson JA (2009) QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: a review. Plant Breed 128: 1–26 [Google Scholar]
- Coleman J, Blake-Kalff M, Davies E (1997) Detoxification of xenobiotics by plants: chemical modification and vacuolar compartmentation. Trends Plant Sci 2: 144–151 [Google Scholar]
- Cuzick A, Urban M, Hammond-Kosack K (2008) Fusarium graminearum gene deletion mutants map1 and tri5 reveal similarities and differences in the pathogenicity requirements to cause disease on Arabidopsis and wheat floral tissue. New Phytol 177: 990–1000 [DOI] [PubMed] [Google Scholar]
- Dalmais M, Antelme S, Ho-Yue-Kuang S, Wang Y, Darracq O, d’Yvoire MB, Cézard L, Légée F, Blondet E, Oria N, et al. (2013) A TILLING platform for functional genomics in Brachypodium distachyon . PLoS ONE 8: e65503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean JV, Delaney SP (2008) Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana . Physiol Plant 132: 417–425 [DOI] [PubMed] [Google Scholar]
- Desmond OJ, Manners JM, Stephens AE, Maclean DJ, Schenk PM, Gardiner DM, Munn AL, Kazan K (2008) The Fusarium mycotoxin deoxynivalenol elicits hydrogen peroxide production, programmed cell death and defence responses in wheat. Mol Plant Pathol 9: 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Xu H, Yi H, Yang L, Kong Z, Zhang L, Xue S, Jia H, Ma Z (2011) Resistance to hemi-biotrophic F. graminearum infection is associated with coordinated and ordered expression of diverse defense signaling pathways. PLoS ONE 6: e19008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falter C, Voigt C (2014) Comparative cellular analysis of pathogenic fungi with a disease incidence in Brachypodium distachyon and Miscanthus × giganteus . BioEnergy Res 7: 958–973 [Google Scholar]
- Fitzgerald TL, Powell JJ, Schneebeli K, Hsia MM, Gardiner DM, Bragg JN, McIntyre CL, Manners JM, Ayliffe M, Watt M, et al. (2015) Brachypodium as an emerging model for cereal-pathogen interactions. Ann Bot (Lond) 115: 717–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foroud NA, Eudes F (2009) Trichothecenes in cereal grains. Int J Mol Sci 10: 147–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foroud NA, Ouellet T, Laroche A, Oosterveen B, Jordan MC, Ellis BE, Eudes F (2012) Differential transcriptome analyses of three wheat genotypes reveal different host response pathways associated with Fusarium head blight and trichothecene resistance. Plant Pathol 61: 296–314 [Google Scholar]
- Gachon CM, Langlois-Meurinne M, Saindrenan P (2005) Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci 10: 542–549 [DOI] [PubMed] [Google Scholar]
- Gardiner SA, Boddu J, Berthiller F, Hametner C, Stupar RM, Adam G, Muehlbauer GJ (2010) Transcriptome analysis of the barley-deoxynivalenol interaction: evidence for a role of glutathione in deoxynivalenol detoxification. Mol Plant Microbe Interact 23: 962–976 [DOI] [PubMed] [Google Scholar]
- Golkari S, Gilbert J, Prashar S, Procunier JD (2007) Microarray analysis of Fusarium graminearum-induced wheat genes: identification of organ-specific and differentially expressed genes. Plant Biotechnol J 5: 38–49 [DOI] [PubMed] [Google Scholar]
- Gosman N, Srinivasachary, Steed A, Chandler E, Thomsett M, Nicholson P (2010) Evaluation of type I Fusarium head blight resistance of wheat using non-deoxynivalenol-producing fungi. Plant Pathol 59: 147–157 [Google Scholar]
- Gottwald S, Samans B, Lück S, Friedt W (2012) Jasmonate and ethylene dependent defence gene expression and suppression of fungal virulence factors: two essential mechanisms of Fusarium head blight resistance in wheat? BMC Genomics 13: 369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb CD, Zipp BJ, Ludwig-Müller J, Masuno MN, Molinski TF, Abel S (2004) Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J 40: 893–908 [DOI] [PubMed] [Google Scholar]
- Gunnaiah R, Kushalappa AC, Duggavathi R, Fox S, Somers DJ (2012) Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (Fhb1) contributes to resistance against Fusarium graminearum. PLoS ONE 7: e40695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Zierold U, Hensel G, Riechen J, Douchkov D, Schweizer P, Kumlehn J (2007) A set of modular binary vectors for transformation of cereals. Plant Physiol 145: 1192–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SY, Seo PJ, Yang MS, Xiang F, Park CM (2008) Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol 8: 112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua-Van A, Pamphile JA, Langin T, Daboussi MJ (2001) Transposition of autonomous and engineered impala transposons in Fusarium oxysporum and a related species. Mol Gen Genet 264: 724–731 [DOI] [PubMed] [Google Scholar]
- Jansen C, von Wettstein D, Schäfer W, Kogel KH, Felk A, Maier FJ (2005) Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc Natl Acad Sci USA 102: 16892–16897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Cho S, Muehlbauer GJ (2009) Transcriptome analysis of a wheat near-isogenic line pair carrying Fusarium head blight-resistant and -susceptible alleles. Mol Plant Microbe Interact 22: 1366–1378 [DOI] [PubMed] [Google Scholar]
- Jin SH, Ma XM, Han P, Wang B, Sun YG, Zhang GZ, Li YJ, Hou BK (2013) UGT74D1 is a novel auxin glycosyltransferase from Arabidopsis thaliana . PLoS ONE 8: e61705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Gardiner DM, Manners JM (2012) On the trail of a cereal killer: recent advances in Fusarium graminearum pathogenomics and host resistance. Mol Plant Pathol 13: 399–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Tokai T, O’Donnell K, Ward TJ, Fujimura M, Hamamoto H, Shibata T, Yamaguchi I (2003) The trichothecene biosynthesis gene cluster of Fusarium graminearum F15 contains a limited number of essential pathway genes and expressed non-essential genes. FEBS Lett 539: 105–110 [DOI] [PubMed] [Google Scholar]
- Kugler KG, Siegwart G, Nussbaumer T, Ametz C, Spannagl M, Steiner B, Lemmens M, Mayer KF, Buerstmayr H, Schweiger W (2013) Quantitative trait loci-dependent analysis of a gene co-expression network associated with Fusarium head blight resistance in bread wheat (Triticum aestivum L.). BMC Genomics 14: 728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaraswamy GK, Bollina V, Kushalappa A, Choo T, Dion Y, Rioux S, Mamer O, Faubert D (2011. b) Metabolomics technology to phenotype resistance in barley against Gibberella zeae. Eur J Plant Pathol 130: 29–43 [Google Scholar]
- Kumaraswamy KG, Kushalappa AC, Choo TM, Dion Y, Rioux S (2011. a) Mass spectrometry based metabolomics to identify potential biomarkers for resistance in barley against fusarium head blight (Fusarium graminearum). J Chem Ecol 37: 846–856 [DOI] [PubMed] [Google Scholar]
- Lemmens M, Scholz U, Berthiller F, Dall’Asta C, Koutnik A, Schuhmacher R, Adam G, Buerstmayr H, Mesterházy A, Krska R, et al. (2005) The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for Fusarium head blight resistance in wheat. Mol Plant Microbe Interact 18: 1318–1324 [DOI] [PubMed] [Google Scholar]
- Li X, Shin S, Heinen S, Dill-Macky R, Berthiller F, Nersesian N, Clemente T, McCormick S, Muehlbauer GJ (2015) Transgenic wheat expressing a barley UDP-glucosyltransferase detoxifies deoxynivalenol and provides high levels of resistance to Fusarium graminearum . Mol Plant Microbe Interact 28: 1237–1246 [DOI] [PubMed] [Google Scholar]
- Lim EK, Higgins GS, Li Y, Bowles DJ (2003) Regioselectivity of glucosylation of caffeic acid by a UDP-glucose:glucosyltransferase is maintained in planta . Biochem J 373: 987–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim EK, Bowles DJ (2004) A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J 23: 2915–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang JM, Tuori RP, Martinez JP, Sawyer TL, Redman RS, Rollins JA, Wolpert TJ, Johnson KB, Dickman MB, Ciufetti LM (2001) Green fluorescent protein is lighting up fungal biology. Appl Env Microb 67: 1987–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulin M, Yi S, Aizhong C, Zengjun Q, Liping X, Peidu C, Dajun L, Xiu-E W (2010) Molecular cloning and characterization of an up-regulated UDP-glucosyltransferase gene induced by DON from Triticum aestivum L. cv. Wangshuibai. Mol Biol Rep 37: 785–795 [DOI] [PubMed] [Google Scholar]
- Maier FJ, Miedaner T, Hadeler B, Felk A, Salomon S, Lemmens M, Kassner H, Schäfer W (2006) Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Mol Plant Pathol 7: 449–461 [DOI] [PubMed] [Google Scholar]
- Mandadi KK, Scholthof KB (2012) Characterization of a viral synergism in the monocot Brachypodium distachyon reveals distinctly altered host molecular processes associated with disease. Plant Physiol 160: 1432–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda D, Ishida M, Yamaguchi K, Yamaguchi I, Kimura M, Nishiuchi T (2007) Phytotoxic effects of trichothecenes on the growth and morphology of Arabidopsis thaliana. J Exp Bot 58: 1617–1626 [DOI] [PubMed] [Google Scholar]
- Messner B, Thulke O, Schäffner AR (2003) Arabidopsis glucosyltransferases with activities toward both endogenous and xenobiotic substrates. Planta 217: 138–146 [DOI] [PubMed] [Google Scholar]
- Miedaner T, Moldovan M, Ittu M (2003) Comparison of spray and point inoculation to assess resistance to fusarium head blight in a multienvironment wheat trial. Phytopathology 93: 1068–1072 [DOI] [PubMed] [Google Scholar]
- Miller JD. (2008) Mycotoxins in small grains and maize: old problems, new challenges. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 25: 219–230 [DOI] [PubMed] [Google Scholar]
- Mudge AM, Dill-Macky R, Dong Y, Gardiner DM, White RG, Manners JM (2006) A role for the mycotoxin deoxynivalenol in stem colonisation during crown rot disease of wheat caused by Fusarium graminearum and Fusarium pseudograminearum. Physiol Mol Plant Pathol 69: 73–85 [Google Scholar]
- Mur LA, Allainguillaume J, Catalán P, Hasterok R, Jenkins G, Lesniewska K, Thomas I, Vogel J (2011) Exploiting the Brachypodium Tool Box in cereal and grass research. New Phytol 191: 334–347 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Osmani SA, Bak S, Møller BL (2009) Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 70: 325–347 [DOI] [PubMed] [Google Scholar]
- Pasquet JC, Chaouch S, Macadré C, Balzergue S, Huguet S, Martin-Magniette ML, Bellvert F, Deguercy X, Thareau V, Heintz D, et al. (2014) Differential gene expression and metabolomic analyses of Brachypodium distachyon infected by deoxynivalenol producing and non-producing strains of Fusarium graminearum. BMC Genomics 15: 629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peraldi A, Beccari G, Steed A, Nicholson P (2011) Brachypodium distachyon: a new pathosystem to study Fusarium head blight and other Fusarium diseases of wheat. BMC Plant Biol 11: 100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peraldi A, Griffe LL, Burt C, McGrann GRD, Nicholson P (2014) Brachypodium distachyon exhibits compatible interactions with Oculimacula spp. and Ramularia collo-cygni, providing the first pathosystem model to study eyespot and ramularia leaf spot diseases. Plant Pathol 63: 554–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka JJ. (2010) Deoxynivalenol: mechanisms of action, human exposure, and toxicological relevance. Arch Toxicol 84: 663–679 [DOI] [PubMed] [Google Scholar]
- Poppenberger B, Berthiller F, Lucyshyn D, Sieberer T, Schuhmacher R, Krska R, Kuchler K, Glössl J, Luschnig C, Adam G (2003) Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J Biol Chem 278: 47905–47914 [DOI] [PubMed] [Google Scholar]
- Rittenour WR, Harris SD (2010) An in vitro method for the analysis of infection-related morphogenesis in Fusarium graminearum. Mol Plant Pathol 11: 361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha O, Ansari K, Doohan FM (2005) Effects of trichothecene mycotoxins on eukaryotic cells: a review. Food Addit Contam 22: 369–378 [DOI] [PubMed] [Google Scholar]
- Ross J, Li Y, Lim E, Bowles DJ (2001) Higher plant glycosyltransferases. Genome Biol 2: REVIEWS3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [Google Scholar]
- Sandoya GV, de Oliveira Buanafina MM (2014) Differential responses of Brachypodium distachyon genotypes to insect and fungal pathogens. Physiol Mol Plant Pathol 85: 53–64 [Google Scholar]
- Schweiger W, Boddu J, Shin S, Poppenberger B, Berthiller F, Lemmens M, Muehlbauer GJ, Adam G (2010) Validation of a candidate deoxynivalenol-inactivating UDP-glucosyltransferase from barley by heterologous expression in yeast. Mol Plant Microbe Interact 23: 977–986 [DOI] [PubMed] [Google Scholar]
- Schweiger W, Pasquet JC, Nussbaumer T, Paris MP, Wiesenberger G, Macadré C, Ametz C, Berthiller F, Lemmens M, Saindrenan P, et al. (2013. a) Functional characterization of two clusters of Brachypodium distachyon UDP-glycosyltransferases encoding putative deoxynivalenol detoxification genes. Mol Plant Microbe Interact 26: 781–792 [DOI] [PubMed] [Google Scholar]
- Schweiger W, Steiner B, Ametz C, Siegwart G, Wiesenberger G, Berthiller F, Lemmens M, Jia H, Adam G, Muehlbauer GJ, et al. (2013. b) Transcriptomic characterization of two major Fusarium resistance quantitative trait loci (QTLs), Fhb1 and Qfhs.ifa-5A, identifies novel candidate genes. Mol Plant Pathol 14: 772–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Torres-Acosta JA, Heinen SJ, McCormick S, Lemmens M, Paris MP, Berthiller F, Adam G, Muehlbauer GJ (2012) Transgenic Arabidopsis thaliana expressing a barley UDP-glucosyltransferase exhibit resistance to the mycotoxin deoxynivalenol. J Exp Bot 63: 4731–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerszen JB, Szczyglowski K, Bandurski RS (1994) iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science 265: 1699–1701 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Hayashi K, Natsume M, Kamiya Y, Sakakibara H, Kawaide H, Kasahara H (2014) UGT74D1 catalyzes the glucosylation of 2-oxindole-3-acetic acid in the auxin metabolic pathway in Arabidopsis. Plant Cell Physiol 55: 218–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti VB, Van Aken O, Morreel K, Vandenbroucke K, van de Cotte B, De Clercq I, Chiwocha S, Fenske R, Prinsen E, Boerjan W, et al. (2010) Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22: 2660–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás S, Beemster GT, Bennett MJ (2012) Hormonal regulation of root growth: integrating local activities into global behaviour. Trends Plant Sci 17: 326–331 [DOI] [PubMed] [Google Scholar]
- Vogel J, Hill T (2008) High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep 27: 471–478 [DOI] [PubMed] [Google Scholar]
- Xiao J, Jin X, Jia X, Wang H, Cao A, Zhao W, Pei H, Xue Z, He L, Chen Q, et al. (2013) Transcriptome-based discovery of pathways and genes related to resistance against Fusarium head blight in wheat landrace Wangshuibai. BMC Genomics 14: 197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazar S, Omurtag GZ (2008) Fumonisins, trichothecenes and zearalenone in cereals. Int J Mol Sci 9: 2062–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Fu J, Hiromasa Y, Pan H, Bai G (2013) Differentially expressed proteins associated with Fusarium head blight resistance in wheat. PLoS ONE 8: e82079 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.