DELLA proteins physically and genetically interact with CO to modulate flowering under long-days in Arabidopsis.
Abstract
Gibberellin (GA) and photoperiod pathways have recently been demonstrated to collaboratively modulate flowering under long days (LDs). However, the molecular mechanisms underlying this collaboration remain largely unclear. In this study, we found that GA-induced expression of FLOWERING LOCUS T (FT) under LDs was dependent on CONSTANS (CO), a critical transcription factor positively involved in photoperiod signaling. Mechanistic investigation revealed that DELLA proteins, a group of crucial repressors in GA signaling, physically interacted with CO. The DELLA-CO interactions repressed the transcriptional function of CO protein. Genetic analysis demonstrated that CO acts downstream of DELLA proteins to regulate flowering. Disruption of CO rescued the earlier flowering phenotype of the gai-t6 rga-t2 rgl1-1 rgl2-1 mutant (dellap), while a gain-of-function mutation in GA INSENSITIVE (GAI, a member of the DELLA gene) repressed the earlier flowering phenotype of CO-overexpressing plants. In addition, the accumulation of DELLA proteins and mRNAs was rhythmic, and REPRESSOR OF GA1-3 protein was noticeably decreased in the long-day afternoon, a time when CO protein is abundant. Collectively, these results demonstrate that the DELLA-CO cascade inhibits CO/FT-mediated flowering under LDs, which thus provide evidence to directly integrate GA and photoperiod signaling to synergistically modulate flowering under LDs.
To maximize reproductive success and seed production, plants determine the most appropriate time to flower by monitoring internal and external environment changes. In Arabidopsis (Arabidopsis thaliana), approximately 180 genes are involved in flowering-time control. These occur in a network of six flowering pathways: the vernalization pathway, the photoperiod pathway, the gibberellin (GA) pathway, the age pathway, the autonomous pathway, and the ambient temperature pathway (Fornara et al., 2010). The photoperiod pathway monitors seasonal changes in day length to regulate flowering time (Fornara et al., 2010; Song et al., 2013). The vernalization and ambient temperature pathways control flowering in response to changes in temperature (Fornara et al., 2010; Kim et al., 2009). The GA, autonomous, and age pathways affect flowering time in response to the internal developmental status (Fornara et al., 2010; Kim et al., 2009).
As a long-day (LD) plant, flowering of Arabidopsis is accelerated by LDs and delayed by short-day (SD) conditions, which controls the photoperiod pathway. Numerous studies have defined the core of the photoperiod pathway comprises GIGANTEA (GI), CYCLING DOF FACTORs (CDFs), CONSTANS (CO), and FLOWERING LOCUS T (FT; Fornara et al., 2010; Kobayashi and Weigel, 2007; Fornara et al., 2009). At the end of the day, light promotes GI interaction with the F-box ubiquitin ligase FLAVIN-BINDING, KELCH REPEAT, F-BOX1 (FKF1), increasing its stability under LDs (Song et al., 2014; Sawa et al., 2007). SCFFKF1 degrades CDFs, which are a family of transcription factors that repress flowering by down-regulating CO expression in the leaves, through the 26S proteasome (Sawa et al., 2007; Imaizumi et al., 2005). CO directly activates transcription of the FT gene, which encodes a floral-inductive long-distance signal and thus determines flowering time (Samach et al., 2000; Corbesier et al., 2007). Besides being regulated by the photoperiodic pathway, expression of CO is also modulated by the circadian clock, with its mRNA peaking late in the day (Suárez-lópez et al., 2001). At the posttranscriptional level, CO protein stability is regulated by various signaling pathways. For example, the blue-light photoreceptor Crypotochrome and red/far-red light photoreceptor Phytochrome A stabilize CO in the evening, whereas the red/far-red light photoreceptor Phytochrome B facilitates CO degradation in the morning or in darkness (Valverde et al., 2004). In addition, the RING finger E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC1 promotes ubiquitin-mediated proteolysis of CO in darkness (Liu et al., 2008; Jang et al., 2008). Moreover, FKF1, ZEITLUPE, GI, and HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 are also involved in CO stability (Song et al., 2012, 2014; Lazaro et al., 2012).
GA are a class of critical plant hormones that function as essential growth regulators to mediate diverse aspects of developmental processes (Sun and Gubler, 2004; Fleet and Sun, 2005; Sun, 2008). Genetic screening has identified several molecular components involved in GA perception and signaling: the GA receptors GA INSENSITIVE DWARF1 (GID1a, b, and c), a group of repressor proteins DELLA (GA INSENSITIVE [GAI], RGA, RGA-LIKE1 [RGL1], RGL2, and RGL3), and the F-box ubiquitin ligase SLEEPY1 (SLY1; Peng et al., 1997; Silverstone et al., 1998; Dill and Sun, 2001; McGinnis et al., 2003; Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006). GID1 and SLY1 (as an SCFSLY1 complex) recruit DELLA proteins for ubiquitination and degradation, leading to activation of various transcriptional factors in the presence of GA (Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006; Dill et al., 2004; Harberd, et al., 2009; Claeys et al., 2014). Recently, several studies demonstrated that GA are essential for floral induction under both SDs and LDs. Mutations affecting GA synthesis fail to flower in SDs but show relatively weak late-flowering phenotypes under LDs (Wilson et al., 1992), suggesting that GA play their most important function in flowering under SDs. However, recent studies strongly indicated that GA are also involved in flowering time control under LDs. For example, GA have critical roles in promoting the transcription of FT, TWIN SISTER OF FT (TSF), and SQUAMOSA PROMOTER BINDING-LIKE (SPL) genes in response to LDs (Galvão et al., 2012; Porri et al., 2012; Posé et al., 2012). The gid1a, b, and c triple mutant exhibited a remarkably late flowering phenotype under LDs (Griffiths et al., 2006; Willige et al., 2007). Additionally, DELLA proteins directly bind to SPLs and inhibit transcriptional activation of MADS box genes and miR172 under LDs (Yu et al., 2012).
Accumulating evidence has indicated that the photoperiod and GA pathways coordinate to modulate flowering under LDs (Galvão et al., 2012; Porri et al., 2012; Reeves and Coupland, 2001; Hou et al., 2014; Nguyen et al., 2015). Very recently, Xu et al. (2016) reported that DELLA proteins physically interact with CO, indicating a direct association between the photoperiod and GA pathways. However, the biological significance of the DELLA-CO physical interactions remains largely unclear. Further molecular and genetic investigations are needed to elucidate the exact molecular mechanisms underlying the photoperiod and GA signaling to synergistically modulate flowering. In this study, we found that GA-induced expression of FT was compromised in the co-2 mutant under LDs. DELLA proteins were found to directly interact with the CO transcription factor and repress its transcriptional activity. Additional genetic analysis revealed that CO acts downstream of DELLA proteins and that DELLA represses flowering partially through CO. Thus, we propose that DELLA proteins act as upstream components of CO to modulate flowering under LDs.
RESULTS
GA-Induced Expression of FT Is Dependent on CO
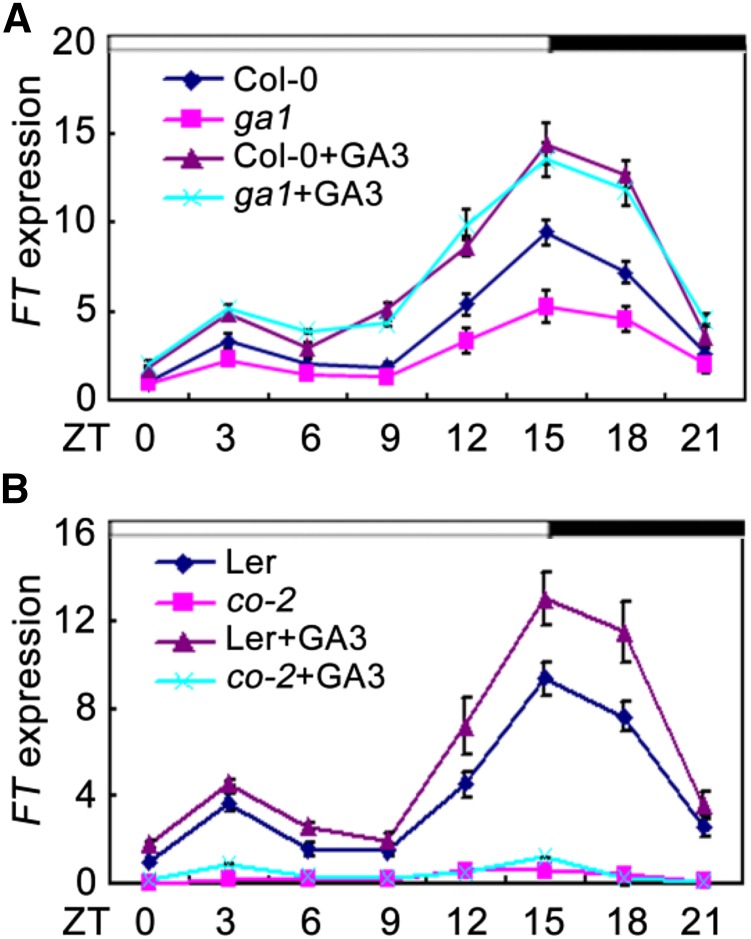
Previous studies have indicated that DELLA proteins act as repressors of flowering and that their GA-dependent degradation contributes to induction of flowering under LDs (Galvão et al., 2012). Molecular and genetic analysis has shown that GA and DELLA regulate flowering partially through modulating the expression of FT (Galvão et al., 2012; Porri et al., 2012). Consistent with previous studies, we also noticed that FT transcripts were strongly reduced in a GA-deficient mutant ga1 (Columbia-0 [Col-0]), and those reductions can be fully rescued by exogenous GA3 (Fig. 1A; Hou et al., 2014). These findings collectively suggest that GA positively regulates the expression of FT; however, little is known of how GA affect FT expression. Samach et al. (2000) demonstrated that the CO protein is a crucial positive regulator for FT expression under LDs. We thus queried whether GA-induced expression of FT under LDs required CO. To test this possibility, we analyzed the expression of FT in co-2 mutant and wild type (Landsberg erecta [Ler]) in response to GA3. As expected, the expression levels of FT were induced by GA3 in wild type; however, exogenous application of GA3 was almost unable to induce FT expression in the co-2 mutant plants (Fig. 1B). These observations indicated that the GA-induced expression of FT is dependent on CO.
Figure 1.
GA-induced expression of FT. qRT-PCR analysis of FT expression in response to GA3 in ga1 mutant (A) and in co-2 mutant (B) under LDs. The IPP2 gene was used as an internal control. Total RNA was extracted from 10-d-old seedlings. Time is expressed as hours from dawn. Error bars indicate sd from three independent RNA extracts.
DELLA Physically Interact with CO
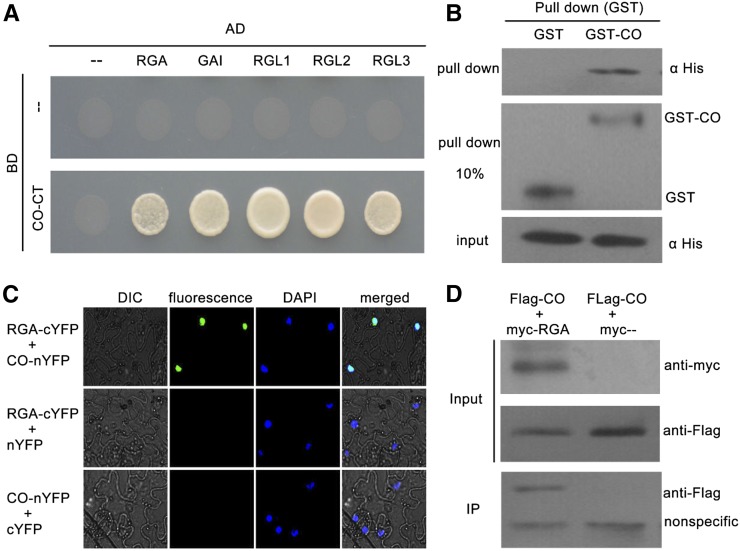
Having confirmed that CO may be involved in GA signaling-regulated flowering, we sought to determine how CO participates in GA signaling. Consistent with previous studies, we also found that CO transcription and CO protein abundance were not regulated by the GA pathway (Supplemental Fig. S1; Galvão et al., 2012; Xu et al., 2016). Recently, several transcription factors have been found to physically interact with DELLA repressors to regulate various GA-mediated physiological processes (Yu et al., 2012; Davière et al., 2008; de Lucas et al., 2008; Feng et al., 2008; Hong et al., 2012; Wild et al., 2012). Therefore, we hypothesized that CO may physically interact with DELLA proteins to mediate GA-signaled flowering. To test this possibility, we fused the CO protein with deleted activation domains (deleted amino acids 1-175) to the Gal4 DNA-binding domain of the bait vector (BD-CO-CT), and introduced the full-length coding sequences of DELLA proteins into the prey vector (AD-DELLA). Then, the interactions between CO and DELLA proteins were assayed using the yeast two-hybrid system. As shown in Figure 2A, CO protein interacted with all five DELLA proteins in yeast cells. We also confirmed the interaction of CO with RGA by performing in vitro pull-down assay. The pull-down results showed that the GST-fused CO could retain His-RGA, whereas GST alone could not (Fig. 2B). In agreement with our results, Xu et al. (2016) showed the DELLA protein GAI physically interacts with CO in yeast cells and binds to CO in vitro.
Figure 2.
Physical interactions between DELLA proteins and CO. A, Yeast two-hybrid (Y2H) analysis of DELLA-CO interactions. Interaction was indicated by the ability of cells to grow on selective media lacking Leu, Trp, His, and adenine. The Gal4 DNA binding domain (BD) and activation domain (AD) were used as negative controls. The pictures were taken 3 d after incubated at 28°C. B, In vitro GST pull-down assay for CO and RGA interaction. Soluble GST and GST-CO fusion proteins were extracted and immobilized to glutathione affinity resin. Purified GST, GST-CO were incubated with the His-RGA fusion protein from Escherichia coli cell lysate for 2 h at 4°C. The interaction was determined by western blot using anti-His antibody. The purified GST and GST-CO were diluted 10 times (pull down 10%) and detected with anti-GST antibody (middle). C, BiFC assay showing the fluorescence complementations of the cYFP fused with RGA and thenYFP fused with CO. 4’,6-Diamidino-2-phenylindole (DAPI) staining marks the nucleus. D, Co-IP assay for CO and RGA interaction. Flag-fused CO and Myc-fused RGA were transiently coexpressed in tobacco leaves. All infected leaves treated with 10 μm MG132 and 20 μm paclobutrazol for 8 h were used for Co-IP. MYC-RGA and MYC tag were immunoprecipitated with anti-MYC M2 agarose beads and detected with anti-FLAG antibodies. Protein input for Flag-CO proteins in immunoprecipitated complexes was also detected and shown.
To further determine whether CO interacts with DELLA proteins in plant cells, we used the bimolecular fluorescence complementation (BiFC) assay for analysis. Full-length DELLA proteins were fused to the C-terminal region of yellow fluorescent protein (DELLA-cYFP), and full-length CO protein was fused to the N-terminal region of YFP (CO-nYFP). When CO-nYFP was coinfiltrated with DELLA-cYFP in tobacco (Nicotiana tabacum) leaves, strong YFP fluorescence was detected in the nuclei (Fig. 2C; Supplemental Fig. S2). No fluorescence was detected in the negative controls (Fig. 2C; Supplemental Fig. S2). In addition, interaction between RGA and CO was also confirmed by a coimmunoprecipitation (Co-IP) assay in tobacco (Fig. 2D). Consistent with this, Xu et al. (2016) showed three DELLA proteins (RGA, GAI, and RGL1) colocalized with CO in the same nuclear bodies, further supporting the idea that CO interact with DELLA proteins in plant cells. Taken together, these results demonstrated that CO interacts with DELLA proteins in vitro and in vivo, indicating that CO functions as a target of DELLA proteins.
To characterize which domain of DELLA proteins is responsible for interacting with CO, the DELLA protein RGA was divided into N-terminal parts (amino acids 1-199) containing the DELLA motif and C-terminal parts (amino acids 200-587) containing two Leu zipper domains. Moreover, we also deleted the DELLA motif of RGA (deleted amino acids 44-60). The directed yeast two-hybrid analysis revealed that deleting the N-terminal residues or DELLA motif of RGA did not affect the physical interaction with CO (Supplemental Fig. S3). However, deletion of the C-terminal parts of RGA eliminated this interaction (Supplemental Fig. S3). Thus, C-terminal parts, but not the DELLA motif or N-terminal parts of RGA, contribute to the interaction between RGA and CO.
DELLA Inhibit the Transcriptional Function of CO
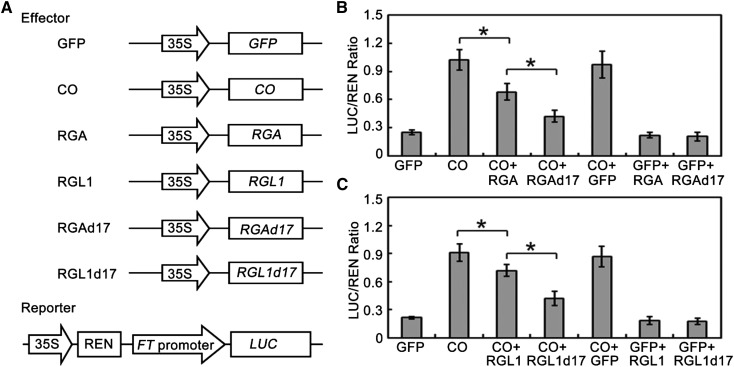
As transcriptional regulators, DELLA proteins lack a DNA binding domain and have been shown to exert their function mainly by inhibiting transcription factor activity through protein-protein interactions (Yu et al., 2012; Davière et al., 2008; de Lucas et al., 2008; Feng et al., 2008; Hong et al., 2012; Wild et al., 2012). Having ascertained that DELLA proteins directly interact with CO, we hypothesized that DELLA might affect the transcriptional function of CO. To test this possibility, we further performed transient expression assays in Col-0 wild-type Arabidopsis mesophyll protoplasts (Yoo et al., 2007). As FT is a direct target of CO (Samach et al., 2000), the FT promoter was fused to the Luciferase (LUC) gene as a reporter (Fig. 3A). The effector constructs had a CO or DELLA gene driven by the cauliflower mosaic virus (CaMV) 35S promoter (Fig. 3A). Consistent with the previous study (Samach et al., 2000), expression of CO dramatically activated expression of LUC driven by the FT promoter (Fig. 3, B and C). However, coexpression of RGA or RGL1 with CO repressed LUC expression in comparison with expression of CO alone (Fig. 3, B and C). This supported the hypothesis that DELLA proteins affect the transcriptional function of CO. Previous studies have indicated that DELLA proteins are subjected to GA-induced proteolysis in normal conditions, and the DELLA motif is essential for this process (Dill et al., 2001). To further test the effect of DELLA protein abundance on CO transcriptional activity, coexpression of RGAd17 (GA-insensitive form of RGA) or RGL1d17 (GA-insensitive form of RGL1) with CO was performed. Our results indicated that RGAd17 and RGL1d17 displayed a stronger repression of transcriptional activity of CO compared with RGA and RGL1 (Fig. 3, B and C). As negative controls, coexpression of RGA, RGL1, RGAd17, or RGL1d17 with GFP did not significantly affect the LUC/REN ratio in comparison with expression of GFP alone (Fig. 3, B and C). Together, these results suggest that DELLA proteins interact with CO and inhibit its transcriptional function to activate FT expression.
Figure 3.
DELLA proteins repress the transcriptional activity of CO. A, Schematic of the reporter and effectors used in the transient transactivation assays. B and C, Transient dual-luciferase reporter assays show that the activation of FT expression by CO is repressed by DELLA protein RGA (B) and RGL1 (C). Error bars indicate sd from three biological replicates; statistics by Student’s t test; *P < 0.05.
DELLA-Repressed Flowering under LDs Requires Functional CO
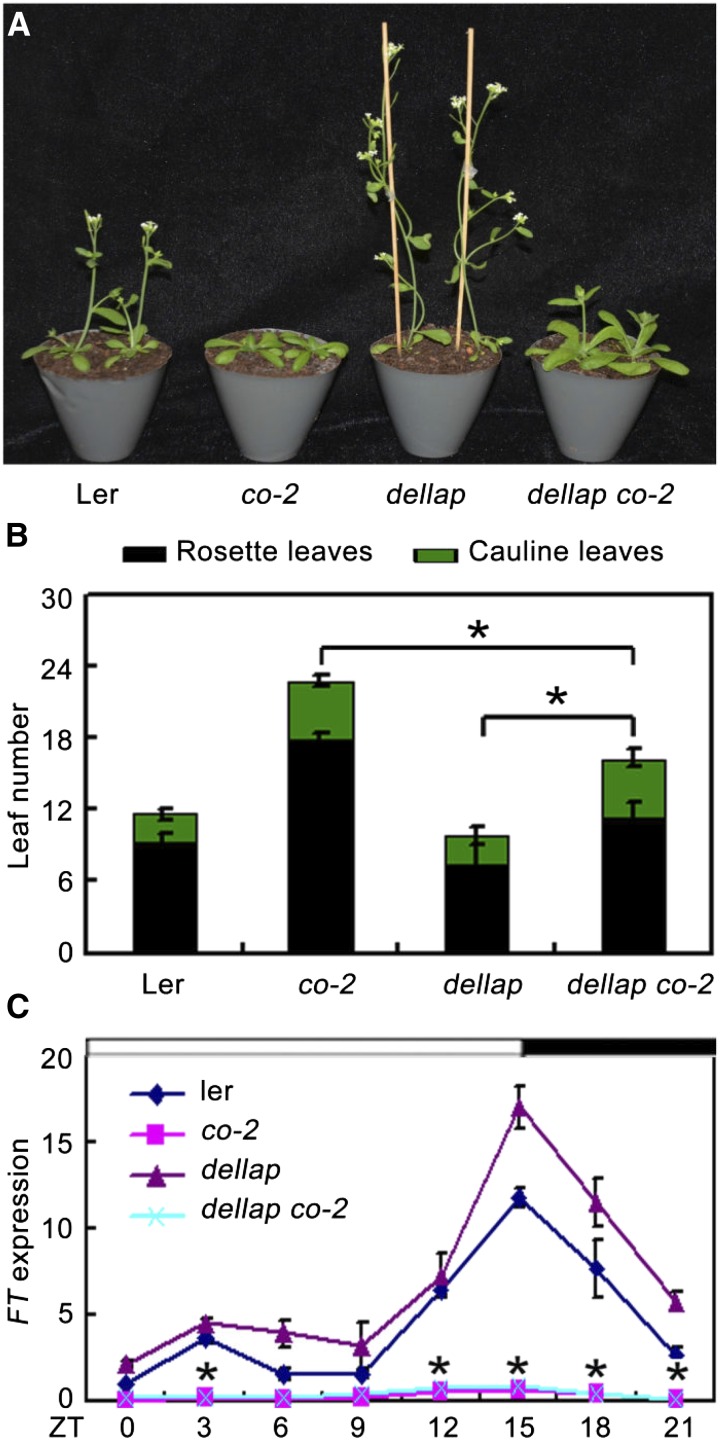
As DELLA form a complex with CO and affect its transcriptional functions, we further asked whether DELLA genetically interact with CO to mediate GA-regulated flowering. To test this, we analyzed the flowering phenotype of the gai-t6 rga-t2 rgl1-1 rgl2-1 co-2 mutant plants (dellap co-2), which was generated by crossing gai-t6 rga-t2 rgl1-1 rgl2-1 (dellap; Ler) with co-2. Consistent with previous studies, the dellap mutant exhibited early flowering, while the co-2 (Ler) mutant exhibited late flowering compared with the wild type (Fig. 4, A and B; Supplemental Fig. S4; Yu et al., 2012; Putterill et al., 1995). Interestingly, dellap co-2 plants flowered with 11.3 ± 1.3 rosette leaves and 4.7 ± 0.7 cauline leaves under LDs, later than both the dellap mutant (7.4 ± 0.6 rosette leaves, 2.4 ± 0.5 cauline leaves) and wild type (9.3 ± 0.7 rosette leaves, 2.5 ± 0.5 cauline leaves) but earlier than the co-2 single mutant plants (17.9 ± 3.1 rosette leaves, 4.8 ± 0.8 cauline leaves; Fig. 4B). Consistent with the flowering phenotype, expression levels of FT in dellap plants were increased compared with those in the wild type (Fig. 4C). However, FT transcripts in the dellap co-2 plants displayed lower expression levels, similar with that in co-2 single mutant plants (Fig. 4C). Collectively, these results demonstrated that the flowering time of dellap co-2 is closer to that of co-2 and much later than dellap and wild type, indicating that the early-flowering phenotype of dellap requires functional CO and that DELLA represses flowering partially through CO.
Figure 4.
co-2 partially rescued the earlier flowering phenotype of the dellap mutant. A, Plants of Ler, co-2, dellap, and dellap co-2 observed 25 d after germination under LDs. All plants are in Ler background. B, Flowering time of Ler, co-2, dellap, and dellap co-2 plants under LD conditions. Data are mean of at least 15 plants. Error bars indicate the sd of total leaf number. *Difference between dellap and dellap co-2 is highly significant (Student’s t test; P < 0.01). C, qRT-PCR analysis of FT expression in 10-d-old Ler and various mutant seedlings under LDs. The IPP2 gene was used as an internal control. Error bars indicate sd from three independent experiments. *Difference between dellap and dellap co-2 is highly significant (Student’s t test; P < 0.01).
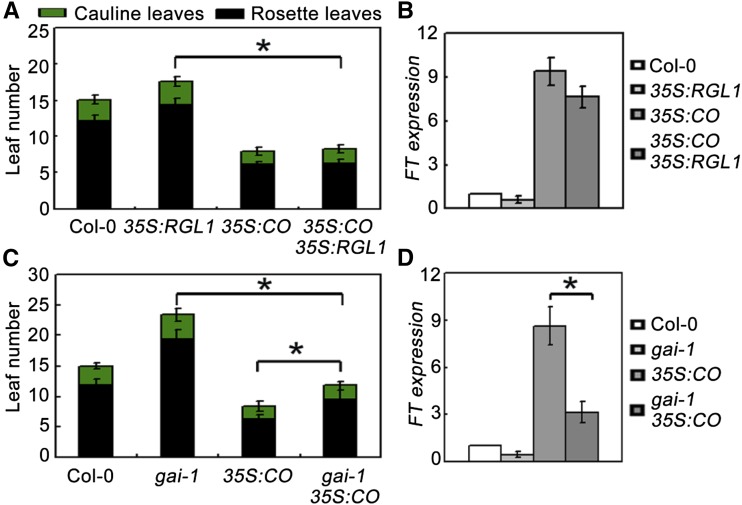
To further elucidate the genetic interaction between DELLA and CO, we generated transgenic plants overexpressing RGL1 and CO (Col-0 background) driven by the CaMV 35S promoter. One line of 35S:RGL1 (line 4) and one line of 35S:CO (line 3) was selected for further study (Supplemental Fig. S5, A and B). Under LDs, overexpression of RGL1 resulted in slightly delayed flowering, while plants overexpressing CO exhibited a much earlier flowering phenotype compared with the wild type (Fig. 5A; Samach et al., 2000). However, plants constitutively expressing RGL1 and CO simultaneously (genetic crossing of 35S:RGL1 to 35S:CO) exhibited early flowering similar to 35S:CO plants (Fig. 5, A and B; Supplemental Figs. S5 and 6), which appears to be in conflict with the notion that DELLA repress CO. If DELLA inhibits the function of CO, one should expect that the earlier flowering phenotype of 35S:CO should be compromised by 35S:RGL1. However, our finding that DELLA protein abundance affects CO transcriptional activity (Fig. 3) may help to reconcile these discrepancies. To further determine this, we generated a gain-of-function gai-1 (Col-0) mutant from the gai-1 (Ler) allele (a GA-insensitive mutant) through backcrossing gai-1 (Ler) with Col-0 three times. Indeed, overexpression of CO in this gai-1 (Col-0) background (genetic crossing of 35S:CO to gai-1 [Col-0]) conferred plants flowering significantly later than 35S:CO plants under LDs (Fig. 5C; Supplemental Figs S5 and 7). This finding was corroborated by quantitative real-time PCR (qRT-PCR), which showed that FT transcripts were enhanced in 35S:CO but reduced in 35S:CO gai-1 plants (Fig. 5D).
Figure 5.
A gain-of-function mutation (gai-1) of GAI represses the earlier flowering phenotype of CO-overexpressing plants. A and C, Flowering phenotypes of various gene types under LDs. All plants are in Col-0 background. Error bars indicate the SD of total leaf number; statistics by Student’s t test; *P < 0.01. B and D, qRT-PCR analysis of FT expression levels in various plants. The ACTIN2 gene was used as an internal control. Total RNA was extracted from 10-d-old plants at ZT 16 grown under LD. Error bars indicate sd from three independent RNA extracts; statistics by Student’s t test; *P < 0.05.
Taken together, our results indicate that DELLA represses flowering partially through the CO/FT-mediated pathway.
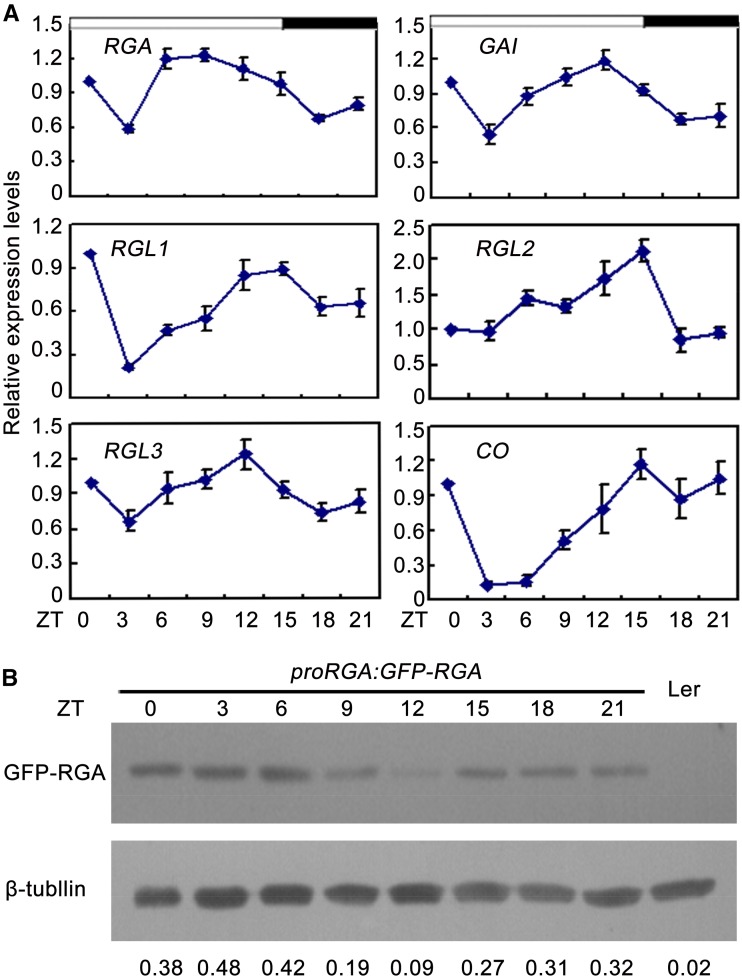
The Expression of DELLA mRNA and Accumulation of RGA Protein Are Rhythmic
Previous studies demonstrated CO acts between the circadian clock and the control of flowering (Suárez-lópez et al., 2001; Song et al., 2015). Recently, Arana et al. (2011) revealed DELLA proteins participate in controlling rhythmic growth of wild-type hypocotyls under SDs. To examine whether DELLA are rhythmic, their expression profiles were investigated under LDs. qRT-PCR analysis showed that DELLA were rhythmically expressed, peaking at the LD afternoon (zeitgeber time [ZT] 12 to 15; Fig. 6A). Consistent with the previous study (Suárez-lópez et al., 2001), the transcripts of CO varied during a 24-h period, showing peak levels between ZT12 and ZT21 (Fig. 6A). Having ascertained that the expression of DELLA mRNA is rhythmic, we further queried whether the accumulation of DELLA protein is also rhythmically regulated. Initially, we analyzed expression profiles of GFP-RGA in proRGA:GFP-RGA transgenic plants. qRT-PCR analysis showed that GFP-RGA was rhythmically expressed, which is similar to the expression profile of endogenous RGA (Supplemental Fig. S8). Then, we examined the accumulation pattern of RGA protein in proRGA:GFP-RGA transgenic plants. As shown in Figure 6B, the RGA protein was strongly decreased at ZT9, ZT12, and ZT15 under LDs when CO protein was abundant (Fig. 6B; Valverde et al., 2004). Taken together, these results indicate that DELLA proteins were rhythmically regulated at both transcriptional and posttranslational levels.
Figure 6.
Analysis of DELLA transcript and protein accumulation under LD photoperiod. A, qRT-PCR of DELLA and CO relative to IPP2 under LDs. Total RNA was extracted from 10-d-old Ler seedings. Error bars indicate sd from three independent RNA extracts. B, Western-blot analysis of GFP-RGA protein accumulation. The blots from seedlings of 10-d-old wild type (Ler) or homozygous proRGA:GFP-RGA transgenic plants. An anti-GFP mouse antibody (Sigma-Aldrich) was used to detect GFP-RGA. β-Tubulin was used as an internal control, and GFP-RGA/β-tubulin ratios were calculated and shown. Western-blot experiments were repeated at least three times, and similar results were obtained. Time is expressed as hours from dawn.
DELLA and CO Antagonistically Regulate Expression of Multiple Flowering-Related Genes
Acting as two important pathways in flowering time control, the photoperiod and GA pathways have been shown to regulate multiple flowering-related genes, including SPLs, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), TSF, and FRUITFULL (FUL; Porri et al., 2012; Yu et al., 2012; Jung et al., 2012). To further investigate whether DELLA-CO interactions affect expression of these genes, their expression was monitored in co-2, dellap, and dellap co-2 mutants by qRT-PCR analysis. As shown in Supplemental Figure S9, their expression was decreased in co-2 and increased in dellap compared with that in the wild type (Supplemental Fig. S9). Furthermore, their transcripts in dellap co-2, which were more than those in co-2, were significantly less than those in dellap (Supplemental Fig. S9). Taken together, these results indicate that DELLA and CO proteins also antagonistically affect the expression of several other flowering-related genes.
DISCUSSION
Recently, numerous studies have demonstrated that the photoperiod and GA pathways act synergistically to promote flowering in response to inductive LDs. For example, Porri et al. (2012) found that GA are required for FT and TSF expression in the vascular tissue under LDs. Similarly, Galvão et al. (2012) showed that GA regulate flowering through controlling the expression of FT, TSF, and SPLs in a day length-specific manner. Additionally, BOTRYTIS SUSCEPTIBLE1 INTERACTORs (a group of interacting partners of DELLA proteins) interact with CO, which inhibit CO to target FT (Nguyen et al., 2015). However, the exact molecular mechanisms underlying interactions of the photoperiod and GA pathways remain limited. Investigating specific crosstalk between these two critical flowering-promoted pathways will provide new insights into our understanding of floral transition.
As crucial repressors of the GA pathway, DELLA proteins were previously reported to regulate GA-mediated responses through physically interacting with several transcription factors. For example, GLABRA1, PHYTOCHROME INTERACTING FACTORs, MYC2, GLABRA3, ENHANCER OF GLABRA3, Class I TCP, BRASSINAZOLE-RESISTANT1, ETHYLENE INSENSITIVE3, AUXIN RESPONSE FACTOR6, and SPLs, were reported as targets of DELLA proteins to regulate diverse aspects of GA-mediated processes (Yu et al., 2012; Davière et al., 2008, 2014, 2016; de Lucas et al., 2008; Feng et al., 2008; Hong et al., 2012; Wild et al., 2012; Qi et al., 2014; Bai et al., 2012; An et al., 2012; Oh et al., 2014). Furthermore, our study and Xu et al. (2016) found that the B-box protein CO also interacts with DELLA (Fig. 2), indicating that CO may also function as a target of DELLA. Consistently, DELLA inhibits the transcriptional activity of CO to regulate its target gene FT (Fig. 3). As CO is a critical regulator of the photoperiod pathway, the DELLA-CO physical interactions may integrate GA and photoperiod signaling to regulate flowering under LDs.
Further genetic analysis demonstrated that CO may act downstream of DELLA proteins to regulate flowering. As shown in Figure 4, the flowering time of dellap co-2 quintuple mutants, which was earlier than the co-2 single mutant, was significantly later than that of dellap plants, indicating that disruption of CO partially rescued the earlier flowering phenotype of dellap. Consistently, the expression of FT was remarkably reduced in dellap co-2 quintuple mutant compared with that in dellap (Fig. 4). These results demonstrate that DELLA proteins repress flowering required functional CO. Moreover, a gain-of-function mutation in GAI (gai-1) repressed the earlier flowering phenotype of CO-overexpressing plants (Fig. 5). All those genetic results together with the finding that DELLA proteins repress the transcriptional activity of CO support the notion that CO acts downstream of DELLA proteins to regulate flowering.
The expression analysis showed that the FT expression was abolished in the dellap co-2 quintuple mutants (Fig. 4), which was similar with that in the co-2 single mutants, indicating that the GA-promoted FT-mediated flowering pathway is blocked in both dellap co-2 and co-2 mutant background. However, phenotypic analysis showed that the flowering time of dellap co-2 was still earlier than that of the co-2 single mutant plants (Fig. 4), suggesting that other DELLA-repressed flowering-related targets may contribute to the induction of flowering in dellap co-2. Consistently, our expression analysis showed that the expression of multiple flowering-related genes was increased in the dellap co-2 quintuple mutants compared with that in the co-2 single mutants (Supplemental Fig. S9). Moreover, Yu et al. (2012) found that DELLA proteins directly interact with SPLs and repress flowering partially through inactivating miR172 and MADS box genes under LDs. Recently, Li et al. (2016) showed that DELLA proteins act as corepressors to regulate flowering by interacting with FLOWERING LOCUS C. All those previous findings and our results in this study demonstrate a complex regulation for GA- and DELLA-modulated flowering. Future studies are needed to illustrate the relationships among these GA- and DELLA-signaled flowering pathways.
As repressors of GA signaling, DELLA proteins are subjected to GA-induced proteolysis in normal conditions (Dill et al., 2001). In this study, we observed that RGAd17 and RGL1d17 showed a stronger repression of the transcriptional activity of CO than RGA and RGL1 (Fig. 3). Consistent with this, the early flowering phenotype of 35S:CO was compromised in the gai-1 (Col-0) background but not in the 35S:RGL1 background (Fig. 5). It is possible that DELLA may repress CO-mediated flowering in a dose-dependent manner. Interestingly, Xu et al. (2016) recently reported that DELLA protein RGA represses the interaction of CO with NF-YB2 in a dose-dependent manner. The results suggest that the tight regulation of DELLA dose may also be critical for flowering. Interestingly, as shown in Figure 6B, we found that the accumulation of RGA protein was rhythmically regulated. RGA was strongly decreased in ZT12 and ZT15, when CO protein was abundant (Fig. 6B; Valverde et al., 2004). It is thus possible that the rhythmically regulated degradation of DELLA in the LD afternoon releases CO and subsequently activates FT expression. Consistent with these, the expression of FT was strongly induced at the end of the day (Figs. 1 and 4; Suárez-López et al., 2001). Further studies elucidating the mechanisms underlying how DELLA proteins are rhythmically regulated may shed new light on the molecular basis of DELLA- and GA-modulated flowering.
MATERIALS AND METHODS
Materials and Arabidopsis Growth Conditions
The plant hormone GA3 was purchased from Sigma-Aldrich. Taq DNA polymerase was purchased from Takara Biotechnology. Other chemicals were obtained from Shanghai Sangon Biotechnology. Arabidopsis (Arabidopsis thaliana) dellap mutant seeds were obtained from Dr. Xingliang Hou (South China Botanical Garden, Chinese Academy of Sciences) and co-2 seeds were provided by Dr. Hongquan Yang (Shanghai Jiaotong University). Homozygous dellap was crossed with co-2 to generate dellap co-2 homozygous plants. To generate CO and RGL1 overexpression transgenic plants, full-length cDNAs of CO and RGL1 were cloned into a pOCA30 vector in the sense orientation behind a CaMV 35S promoter (Wang et al., 2015). Plants used in this study were derived from Arabidopsis Col-0 or Ler ecotypes. Arabidopsis plants were grown in growth chambers at 22°C under LDs (16-h light [100 μE m−2s−1]/8-h dark cycle) or SD conditions (8-h light [100 μE m−2s−1]/16-h dark cycle). Induction treatments with the plant hormone GA3 was performed as described in Galvão et al. (2012). Primers used for identification of mutants or clones are listed in Supplemental Table S1.
RNA Extraction and qRT-PCR
For real-time RT-PCR analysis, total RNA was extracted from Arabidopsis seedlings using the TriZol reagent (Invitrogen). A total of 1 μg of DNase-treated RNA was reverse transcribed in a 20-μL reaction mixture using Superscript II in accordance with manufacturer’s instructions (Invitrogen). Following the reaction, 1 μL of resultant cDNA was used as a template for qRT-PCR, using a SYBR Premix Ex Taq kit (Takara). At least three independent biological samples were conducted for each experiment. The IPP2 gene was used as an internal control in Figures 1, 4C, and 6A, while the ACTIN2 gene was used as an internal control in Figure 5, B and D, and Supplemental Figure S9. Primers used for qRT-PCR are listed in Supplemental Table S1.
Yeast Two-Hybrid Assay
The truncated CO CDSs were cloned into the bait vector pGBKT7 and full-length or truncated CDSs of DELLA proteins were cloned into the prey vector pGADT7. Yeast two-hybrid assay was performed as described previously (Hu and Yu, 2014). Primers used for generating various clones for yeast two-hybrid assays are listed in Supplemental Table S1.
BiFC Assay
Full-length CDS of CO was inserted into pFGC-nYFP vector to generate N-terminal in-frame fusions with N-YFP, while DELLA coding sequences were cloned into pFGC-cYFP vector to form C-terminal in-frame fusions with C-YFP (Hu et al., 2013a). All plasmids were introduced into Agrobacterium tumefaciens strain EHA105, and infiltration of tobacco (Nicotiana tabacum) leaves was performed as described previously (Hu et al., 2013a). Infected tissues were analyzed 48 h after infiltration under a confocal laser-scanning microscope (Olympus). Primers used for clones are listed in Supplemental Table S1.
Co-IP Analysis
For Co-IP assays, the full-length CDS of CO or RGA was amplified and cloned into tagging vectors behind the single FLAG or MYC tag in the sense orientation behind the CaMV 35S promoter (Hu et al., 2013a). Flag-fused CO and Myc-fused RGA were transiently coexpressed in tobacco leaves. All infected leaves were treated with 10 μm MG132 and 20 μm paclobutrazol (a GA biosynthesis inhibitor) 40 h after infiltration. After 8 h, those leaves were homogenized in an extraction buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.1% (w/v) Triton X-100, and 1× complete protease inhibitor cocktail (Roche). Then, MYC-fused RGA and MYC was immunoprecipitated using an anti-MYC rabbit antibody (Sigma-Aldrich), and coimmunoprecipitated proteins were detected using an anti-Flag mouse antibody (Sigma-Aldrich). Primers used for clones are listed in Supplemental Table S1.
Pull-Down Assay
Full-length CO and RGA cDNAs were cloned into pGEX-TX-1 (GE Healthcare) and pET-28a (Novagen), respectively. All plasmids were introduced into Escherichia coli BL21 cells (TransGen Biotech). GST, GST-CO, and His-RGA protein expression was induced by 0.1 mm isopropyl-β-thiogalactopyranoside. Soluble GST and GST-CO fusion proteins were extracted and immobilized to glutathione affinity resin (Thermo Fisher Scientific). For pull-down assays, His-RGA fusion protein from E. coli cell lysate was incubated with the immobilized GST and GST-CO fusion proteins in Pull-down buffer (50 mm Tris-HCl, pH 7.2, 150 mm NaCl, 10% glycerol, 0.1% Triton X-100, 1× protease inhibitor cocktail) for 2 h at 4°C. Proteins were eluted in the elution buffer, and the interaction was determined by western blot using anti-His antibody (Sigma-Aldrich).
Transient Transactivation Assay
To generate reporter constructs, a 2,675-bp region upstream of the start codon of FT was amplified and cloned into a pGreenII 0800-LUC vector (Hellens et al., 2005). To create the effector constructs, the corresponding cDNAs of RGA, RGL1, RGAd17, and RGL1d17 were amplified and cloned into pGreenII 62-SK vectors (Hellens et al., 2005). All primers used for generating these constructs are listed in Supplemental Table S1. Preparation of Arabidopsis mesophyll protoplasts from wild-type (Col-0) leaves and subsequent transfections were performed as described by Yoo et al. (2007). A dual-luciferase reporter assay system (Promega) was used to measure firefly LUC and renilla luciferase (REN) activities. The REN gene under the control of the CaMV 35S promoter and the LUC gene were in the pGreenII 0800-LUC vector (Hellens et al., 2005). Relative REN activity was used as an internal control, and LUC/REN ratios calculated.
Protein Analysis
To analyze the accumulation pattern of RGA protein, the proRGA:GFP-RGA transgenic plants were grown under LD for 10 d and harvested every 3 h. Total protein was extracted using an extraction buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.1% (w/v) Triton X-100, and 1× complete protease inhibitor cocktail (Roche). An anti-GFP mouse antibody (Sigma-Aldrich) was used to detect GFP-RGA. β-Tubulin was used as an internal control, and GFP-RGA/β-tubulin ratios were calculated and shown.
Accession Numbers
Arabidopsis Genome Initiative numbers for the genes discussed in this article are as follows: CO (At5g15840), RGA (At2g01570), GAI (At1g14920), RGL1 (At1g66350), RGL2 (At3g03450), RGL3 (At5g17490), FT (At1g65480), SOC1 (At2g45660), SPL3 (At2g33810), SPL4 (At1g53160), SPL5 (At3g15270), FUL (At5g60910), TSF (At4g20370), IPP2 (At3g02780), and ACT2 (At3g18780).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. CO transcription and protein abundance are not regulated by the GA pathway.
Supplemental Figure S2. BiFC assay showing the fluorescence complementations of the cYFP fused with DELLAs and the nYFP fused with CO.
Supplemental Figure S3. C-terminal parts of RGA contribute to the interaction between RGA and CO.
Supplemental Figure S4. Flowering time of Ler, co-2, dellap, and dellapco-2 under LDs.
Supplemental Figure S5. Overexpression lines for RGL1 and CO.
Supplemental Figure S6. Flowering time of Col-0, 35S:CO, 35S:RGL1, and 35S:CO 35S:RGL1 plants under LDs.
Supplemental Figure S7. Flowering time of Col-0, 35S:CO, gai-1, and gai-1 35S:CO plants under LDs.
Supplemental Figure S8. qRT-PCR of RGA and GFP-RGA expression.
Supplemental Figure S9. Expression of multiple flowering-related genes was coregulated by DELLA and CO.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Xingliang Hou (South China Botanical Garden, Chinese Academy of Sciences) and Dr. Hongquan Yang (Shanghai Jiaotong University) for sharing research materials. The authors also thank Hui Zhao (Yunnan University) for her assistance in the measure of firefly LUC and REN luciferase activities.
Glossary
- BiFC
bimolecular fluorescence complementation
- CaMV
cauliflower mosaic virus
- Co-IP
coimmunoprecipitation
- Col-0
Columbia-0
- GA
gibberellin
- LD
long day
- Ler
Landsberg erecta
- SD
short day
- ZT
zeitgeber time
References
- An F, Zhang X, Zhu Z, Ji Y, He W, Jiang Z, Li M, Guo H (2012) Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res 22: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana MV, Marín-de la Rosa N, Maloof JN, Blázquez MA, Alabadí D (2011) Circadian oscillation of gibberellin signaling in Arabidopsis. Proc Natl Acad Sci USA 108: 9292–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY (2012) Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 14: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H, De Bodt S, Inzé D (2014) Gibberellins and DELLAs: central nodes in growth regulatory networks. Trends Plant Sci 19: 231–239 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Davière JM, Achard P (2016) A pivotal role of DELLAs in regulating multiple hormone signals. Mol Plant 9: 10–20 [DOI] [PubMed] [Google Scholar]
- Davière JM, de Lucas M, Prat S (2008) Transcriptional factor interaction: a central step in DELLA function. Curr Opin Genet Dev 18: 295–303 [DOI] [PubMed] [Google Scholar]
- Davière JM, Wild M, Regnault T, Baumberger N, Eisler H, Genschik P, Achard P (2014) Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr Biol 24: 1923–1928 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dill A, Jung HS, Sun TP (2001) The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci USA 98: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Sun T (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet CM, Sun TP (2005) A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr Opin Plant Biol 8: 77–85 [DOI] [PubMed] [Google Scholar]
- Fornara F, de Montaigu A, Coupland G (2010) SnapShot: control of flowering in Arabidopsis. Cell 141: 550–550, 550.e1–550.e2 [DOI] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17: 75–86 [DOI] [PubMed] [Google Scholar]
- Galvão VC, Horrer D, Küttner F, Schmid M (2012) Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 139: 4072–4082 [DOI] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, Thomas SG (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd NP, Belfield E, Yasumura Y (2009) The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong GJ, Xue XY, Mao YB, Wang LJ, Chen XY (2012) Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24: 2635–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Zhou J, Liu C, Liu L, Shen L, Yu H (2014) Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat Commun 5: 4601. [DOI] [PubMed] [Google Scholar]
- Hu Y, Chen L, Wang H, Zhang L, Wang F, Yu D (2013a) Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J 74: 730–745 [DOI] [PubMed] [Google Scholar]
- Hu Y, Jiang L, Wang F, Yu D (2013b) Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25: 2907–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Yu D (2014) BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell 26: 4394–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297 [DOI] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Ju Y, Seo PJ, Lee JH, Park CM (2012) The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J 69: 577–588 [DOI] [PubMed] [Google Scholar]
- Kim DH, Doyle MR, Sung S, Amasino RM (2009) Vernalization: winter and the timing of flowering in plants. Annu Rev Cell Dev Biol 25: 277–299 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Weigel D (2007) Move on up, it’s time for change--mobile signals controlling photoperiod-dependent flowering. Genes Dev 21: 2371–2384 [DOI] [PubMed] [Google Scholar]
- Lazaro A, Valverde F, Piñeiro M, Jarillo JA (2012) The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24: 982–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ, An FY, Li WY, Ma MD, Feng Y, Zhang X, Guo HW (2016) DELLA proteins interact with FLC to repress flowering transition. J Integr Plant Biol 58: 642–655 [DOI] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M, et al. (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46: 880–889 [DOI] [PubMed] [Google Scholar]
- Nguyen KT, Park J, Park E, Lee I, Choi G (2015) The Arabidopsis RING domain protein BOI inhibits flowering via CO-dependent and CO-independent mechanisms. Mol Plant 8: 1725–1736 [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY (2014) Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3: e03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porri A, Torti S, Romera-Branchat M, Coupland G (2012) Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139: 2198–2209 [DOI] [PubMed] [Google Scholar]
- Posé D, Yant L, Schmid M (2012) The end of innocence: flowering networks explode in complexity. Curr Opin Plant Biol 15: 45–50 [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Qi T, Huang H, Wu D, Yan J, Qi Y, Song S, Xie D (2014) Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 26: 1118–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PH, Coupland G (2001) Analysis of flowering time control in Arabidopsis by comparison of double and triple mutants. Plant Physiol 126: 1085–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Estrada DA, Johnson RS, Kim SK, Lee SY, MacCoss MJ, Imaizumi T (2014) Distinct roles of FKF1, Gigantea, and Zeitlupe proteins in the regulation of Constans stability in Arabidopsis photoperiodic flowering. Proc Natl Acad Sci USA 111: 17672–17677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Ito S, Imaizumi T (2013) Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci 18: 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T (2015) Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol 66: 441–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Sun TP. (2008) Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arabidopsis Book 6: e0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Wang H, Hu Y, Pan J, Yu D (2015) Arabidopsis VQ motif-containing proteins VQ12 and VQ29 negatively modulate basal defense against Botrytis cinerea. Sci Rep 5: 14185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild M, Davière JM, Cheminant S, Regnault T, Baumberger N, Heintz D, Baltz R, Genschik P, Achard P (2012) The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 24: 3307–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EM, Maier A, Schwechheimer C (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR (1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Li T, Xu PB, Li L, Du SS, Lian HL, Yang HQ (2016) DELLA proteins physically interact with CONSTANS to regulate flowering under long days in Arabidopsis. FEBS Lett 590: 541–549 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu S, Galvão VC, Zhang YC, Horrer D, Zhang TQ, Hao YH, Feng YQ, Wang S, Schmid M, Wang JW (2012) Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24: 3320–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.