Abstract
The amphibian model Xenopus, has been used extensively over the past century to study multiple aspects of cell and developmental biology. Xenopus offers advantages of a non-mammalian system, including high fecundity, external development, and simple housing requirements, with additional advantages of large embryos, highly conserved developmental processes, and close evolutionary relationship to higher vertebrates. There are two main species of Xenopus used in biomedical research, Xenopus laevis and Xenopus tropicalis; the common perception is that both species are excellent models for embryological and cell biological studies, but only Xenopus tropicalis is useful as a genetic model. The recent completion of the Xenopus laevis genome sequence combined with implementation of genome editing tools, such as TALENs (transcription activator-like effector nucleases) and CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR associated nucleases), greatly facilitates the use of both Xenopus laevis and Xenopus tropicalis for understanding gene function in development and disease. In this paper, we review recent advances made in Xenopus laevis and Xenopus tropicalis with TALENs and CRISPR-Cas and discuss the various approaches that have been used to generate knockout and knock-in animals in both species. These advances show that both Xenopus species are useful for genetic approaches and in particular counters the notion that Xenopus laevis is not amenable to genetic manipulations.
Keywords: CRISPR-Cas, TALENs, J strain, Xenopus laevis, Xenopus tropicalis, knock-in, human disease model
Introduction
With the advent of customizable genome editing technologies, such as TALENs (transcription activator-like effector nucleases) and CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR associated), it is now possible to model human genetic disorders in any animal or cellular system, including previously non-genetic models such as Xenopus laevis (Harrison et al., 2014; Peng et al., 2014). These advances provide researchers with a broader range of animal models, allowing them to choose the most experimentally tractable and biologically relevant system in which to test the function of specific disease-associated genes. In this review, we discuss the application of new genome editing technologies in both Xenopus laevis and Xenopus tropicalis, and how these advances will enhance our understanding of the molecular mechanisms underlying human disease.
Xenopus has many experimental advantages that make it a well-suited model for the study and functional characterization of candidate genes involved in human disease. The Xenopus tropicalis genome contains orthologues of 79% of the identified human disease genes (Hellsten et al., 2010; Khokha, 2012). Both Xenopus species have been used to study all aspects of vertebrate embryology, such as gastrulation, axis development and organ formation, dating back to the nineteenth century. These studies have provided insight into highly conserved members of major signal transduction pathways, for instance, BMPs and Wnts (Bier and De Robertis, 2015; Hikasa and Sokol, 2013). Notably, Sir John Gurdon (2012) and Professor Tim Hunt (2001) were awarded Nobel prizes for their ground-breaking research performed in Xenopus laevis (Gurdon, 2013; Hunt, 2002). The ease with which X. laevis and X. tropicalis embryos can be cultured in simple buffers and raised to adulthood enables the use of this model in most laboratory settings. Due to the large number of progeny that can be obtained from a single mating, Xenopus provides ample embryonic tissue for a wide range of experimental assays, including phenotypic analyses, RNAseq, ChIPseq and proteomics (Chung et al., 2014; Onjiko et al., 2015; Peshkin et al., 2015; Wühr et al., 2014; Yanai et al., 2011). Because of their relatively large size and ability to develop in culture, Xenopus embryos are particularly amenable to studies focused on manipulating gene function via microinjection of morpholinos, DNA constructs, translated capped RNA, and protein. Overexpression or misexpression of wildtype or dominant negative proteins were facilitated by simple and efficient transgenesis approaches, by a variety of means, including restriction enzyme mediated integration (REMI), I-SceI meganuclease or transposon mediated insertion of constructs into the Xenopus genome, and are reviewed elsewhere (Buchholz, 2012; Ishibashi et al., 2012b; 2012c; Kelley et al., 2012). In addition, the ability to fate map cell lineages from various embryonic blastomeres and tissue regions in Xenopus has greatly enhanced our understanding of the developmental, genetic, and evolutionary origin of adult structures and organs, which is essential for determining disease etiology (Chalmers and Slack, 2000; M. C. Lane and Sheets, 2006). These experimental advantages, together with their rapid external development, detailed temporal staging atlas, and relative transparency facilitate gene function assessment, making both Xenopus species versatile model systems for disease research and phenotypic drug screening (Harland and Grainger, 2011; Schmitt et al., 2014).
Among aquatic vertebrate model animals, Xenopus excels by having comparable organ development and morphology to mammalian systems, but with the added benefit of being able to regenerate adult tissues, such as optic nerve, lens, spinal cord and limb tissue (Blitz et al., 2006; Muñoz et al., 2015; Slack et al., 2008). Xenopus animals and oocytes are used extensively to understand normal organ function and disease in humans (Labonne and Zorn, 2015), including cardiac congenital heart disorders and heterotaxy (Boskovski et al., 2013; Duncan and Khokha, 2016; Fakhro et al., 2011; Kaltenbrun et al., 2011; Langdon et al., 2012; 2007), gastrointestinal and pancreatic diseases (Kofent and Spagnoli, 2016; Pearl et al., 2009; 2011; Salanga and Horb, 2015; Womble et al., 2016), endocrine functions and disorders (Buchholz, 2015), kidney disease (Lienkamp, 2016), lung development (Rankin et al., 2011; 2015; Wallmeier et al., 2014), cancer (Chernet and Levin, 2013; Cross and Powers, 2009; Hardwick and Philpott, 2015; Haynes-Gilmore et al., 2014; Van Nieuwenhuysen et al., 2015; Wylie et al., 2015), ciliopathies (Kim et al., 2010; Klos Dehring et al., 2013; Ma et al., 2014), orofacial defects (Dickinson, 2016), and neurodevelopmental disorders (Erdogan et al., 2016; Pratt and Khakhalin, 2013). Looking forward, Xenopus is poised to take advantage of the new developments in genomics and genome engineering to better understand the molecular mechanisms underlying human disease (Harland and Grainger, 2011; Labonne and Zorn, 2015).
While genetics is available in fish and mice as surrogate systems for understanding human biology and disease, the development of Xenopus genetics offers a number of advantages not found in other organisms. Unlike the mouse, Xenopus embryos can be produced in large numbers and are accessible throughout their development, simplifying phenotypic screening at embryonic stages. Xenopus shares surprising similarities with humans both at the level of its genome and its anatomy. The frog genome has long regions in which genes exhibit remarkably similar syntenic relationships to those found in the human genome (Amodeo et al., 2015; Blitz, 2012; Blitz et al., 2013; Bodart and Duesbery, 2006; Davidson, 1973; Grant et al., 2015; Hellsten et al., 2010; Paranjpe et al., 2013; Roe et al., 1985; Showell et al., 2011; Uno et al., 2013). In many cases, orthologous genes are found in equivalent regions of the human and frog genomes, in which the order of genes along the chromosomes is largely conserved. In fact, the vast majority of the breaks in synteny are from single exon genes identified by automated ORF prediction algorithms but not supported by EST evidence (Blitz, 2012; Geach et al., 2012; Krylov and Tlapakova, 2015; Macha et al., 2012; Pollet and Mazabraud, 2006; Showell and Conlon, 2007). Therefore, shared synteny may be even more prevalent than is currently thought. Moreover, while it is well established that mice are genetically tractable, mice are difficult for live imaging or biochemistry. Thus by combining genome editing with the advantages of both Xenopus species, researchers have the unique opportunity to integrate systems level genomic and proteomic analyses with quantitative live imaging of cell behaviors in genetically approachable vertebrate model systems.
Xenopus laevis versus Xenopus tropicalis
Currently, there are two main Xenopus species used in biomedical research, Xenopus laevis and Xenopus tropicalis. Historically, X. laevis has been the predominant Xenopus species studied since the 1950s due to its large size, ability to ovulate year round, and experimental robustness. These animals can be grown between 18–23°C in basic salt solutions, in relatively simple aquatic housing. They are the animal of choice for many biomedical researchers, including those using the Xenopus oocyte model to study ion channel electrophysiology, cell protein biochemistry, cell cycle biology, and cytoskeletal dynamics, as well as those employing the Xenopus embryo model to study development (Dubaissi and Papalopulu, 2011; Kay and Peng, 1991; Maksaev and Haswell, 2015; Mitchison et al., 2015). One of the drawbacks often cited with X. laevis is its poor genetic tractability, which was thought due to its allotetraploid genome and relatively long generation time of 10–12 months; thus, most researchers have had little interest in the genetic background of X. laevis and have purchased outbred genetically heterogeneous frogs from commercial vendors. However, in the past 5 years, an inbred strain of X. laevis, known as the J strain, has become available through the National Xenopus Resource and the European Xenopus Resource Centre (Gantress et al., 2003; Pearl et al., 2012; Tochinai and Katagiri, 1975), and the J strain draft genome sequence is now available on Xenbase (http://www.xenbase.org; X. laevis Genome Project Consortium). This newly sequenced strain will allow for more genetic analyses in X. laevis and when combined with recent advances in genome editing technologies, has made it possible for essentially any lab to make mutants through targeted reverse genetics. In more recent years, X. tropicalis has become increasingly used as a genetic model because it offers the same embryological benefits as X. laevis, but has a shorter generation time of 5–7 months, a smaller size, and a diploid genome. However, water conditions are slightly different for X. laevis and X. tropicalis, and hence they require separate housing systems, making it more expensive for individual labs to maintain colonies of both species. In light of these considerations, most researchers focus on one model species, but both species offer unique advantages that are being exploited by the wider Xenopus community.
Inbred J strain X. laevis and Nigerian X. tropicalis frogs can be purchased from the National Xenopus Resource (NXR) in the US or the European Xenopus Resource Center (EXRC) in the UK. These resource centers were established in the past 10 years to serve as the main stock centers for the Xenopus community for inbred, transgenic and mutant lines (Pearl et al., 2012). Their recent advances in husbandry have optimized X. laevis maintenance, and it is now possible to perform in vitro fertilization from males as young as 4 months old, which has decreased the time required to raise lines of X. laevis (Horb, M.E. unpublished). In addition, these stock centers have worked to improve the cryopreservation of X. laevis and X. tropicalis sperm, allowing subsequent matings and the sharing of colony lines among the community. These improvements bring Xenopus more in line with other model systems and enable the proper maintenance and distribution of different strains and lines from both species.
Complete sequencing of both X. laevis and X. tropicalis genomes has revealed larger regions of synteny, and presumably the sharing of a longer common evolutionary history, with humans than those of other popular model systems (Hellsten et al., 2010). X. tropicalis is the only diploid species in the Xenopus genus, and its genome is comprised of twenty chromosomes, whereas X. laevis is an allotetraploid species that arose from the interspecific hybridization of two diploid species, and its genome consists of 36 chromosomes (Matsuda et al., 2015; Uno et al., 2013). There are nine pairs of homeologous chromosomes in X. laevis, which are named Long (L) and Short (S) that functionally segregate as two separate diploid genomes (Matsuda et al., 2015; Tymowska, 1991). A new nomenclature for the X. laevis chromosomes was recently established based on their phylogenetic relationship and length, such that the homeologous chromosomes are named Xla1L and Xla1S through Xla9L and Xla9S (Matsuda et al., 2015). The first eight pairs of X. laevis chromosomes correspond to those of X. tropicalis, whereas the ninth chromosome pair contains fusions of chromosomes 9 and 10 from X. tropicalis and thus, they are also named Xla9_10L and Xla9_10S to emphasize their phylogenetic relationship. As the name implies, S chromosomes are shorter than L chromosomes, and this is due to loss of genes on the S chromosomes; it has been postulated that at least 17%, but perhaps as much as 50%, of the genes in X. laevis remain in a diploid/singleton state due to loss of genes on the S chromosomes (Hellsten et al., 2007; Uno et al., 2013), making the generation of mutants relatively simple for such target genes. These genome annotation advances, together with the recently completed Xenopus ORFeome project (Grant et al., 2015) that identified orthologues of 2,724 human genes associated with an Online Mendelian Inheritance in Man (OMIM)-recognized disease, makes X. laevis an ideal model to study vertebrate development and human disorders.
All of the benefits discussed above demonstrate that both Xenopus species are excellent model systems that will be greatly enhanced with emerging genome-editing methodology. These revolutionizing technologies will allow for the rapid creation of mutant frogs to model human diseases, providing an abundant source of material for functional studies. Utilizing genome editing methods in Xenopus laevis and Xenopus tropicalis will provide a cost effective platform to rapidly identify, validate, and characterize genes involved in human diseases, which will ultimately provide more detailed mechanistic insight to guide new therapeutic strategies. Below, we describe the use of TALENs and CRISPR-Cas based methods in Xenopus, and highlight some of the emerging applications of these methods to understand human diseases.
Genome Editing Tools in Xenopus
In the past few years numerous methods have been generated to efficiently edit the genomes of almost any cell type or organism, including the amphibian models Xenopus laevis and tropicalis, through the use of Zinc Finger Nucleases (ZFNs), Transcription Activator-Like Effector Nucleases (TALENs) and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR-Cas) nuclease systems (Harrison et al., 2014; Peng et al., 2014). The nucleases are targeted to a specific region of interest in the genome through the microinjection of sequence-specific RNA constructs, and induce double/single-stranded breaks in the DNA sequence. Upon recognizing this damage, the cell uses non-homologous end joining (NHEJ) or homology-directed repair (HDR) to repair the break. NHEJ produces random insertions or deletions (indels) into the target site, thereby causing potential DNA coding frame shifts and altered protein translation. HDR, on the other hand, uses a template sequence to repair the broken DNA strands; this enables the insertion of exogenous DNA sequences, such as fluorescent reporter transgenes to track gene expression and cell lineage, or point mutations mimicking known human single nucleotide polymorphisms (SNP) associated with a disease trait. As both TALENs and CRISPR-Cas systems have proven to be more versatile than ZFNs, we will focus on these more recently described gene-editing methods that have revolutionized basic biology as well as biomedical and other translational research.
TALENs in Xenopus
Originally identified in the plant pathogen Xanthomonas, TALE proteins are transcriptional activators that specifically bind and regulate plant gene expression upon infection (Joung and Sander, 2013; Kim and Kim, 2014). The TALE structure comprises a central domain harboring specific repeating units of 33–35 amino acids that target individual DNA bases (Bogdanove and Voytas, 2011). These repeat variable di-residue (RVD) domains enable TALE proteins to target almost any genomic region of interest. Site-specific TALE proteins can be tethered to endonucleases to modify genome sequence, or to transcriptional effector proteins, such as VP16 and KRAB, to regulate gene activity in most eukaryotic organisms tested to date. To induce DNA breaks and modify genome sequences, two TALEN arms are required to recognize and bind the DNA sequence with a small 15–25 base spacer region in between. This spacer allows the dimerization of the tethered Fok1 endonuclease catalytic domains, thereby enabling its enzymatic function. Cloning individual TALENs is a multi-day process that involves piecing together individual RVDs to create custom repeat arrays targeted to a specific DNA sequence (Cermak et al., 2011).
The first use of TALEN-induced mutations in Xenopus were performed in X. tropicalis, demonstrating efficient somatic and germline mutagenesis (Ishibashi et al., 2012a; Lei et al., 2012). The initial study targeted eight Xenopus genes involved in human disease, and showed that TALENs induced mutations in F0 embryos with a high efficiency at all eight loci (Lei et al., 2012). In particular, they demonstrated that injection of TALENs targeting the pancreatic transcription factor ptf1a resulted in pancreatic agenesis in F0 tadpoles, mimicking the phenotype seen in humans and confirming previous morpholino knockdown phenotypes observed in Xenopus (Afelik et al., 2006; Jarikji et al., 2007; Sellick et al., 2004), with efficient germline transmission of mutations. In addition, they showed that TALEN mutagenesis is more efficient and less toxic when compared directly to ZFNs, with no detectable off-target effects (Lei et al., 2012). Another study by the Chen lab showed that TALEN-mediated disruption of the n-myc downstream regulated 1 (ndgr1) gene in X. tropicalis displayed a similar phenotype to that observed using morpholino knockdown of the gene in X. laevis (Zhang et al., 2013). More recently, a comprehensive study by the Grainger lab demonstrated that X. tropicalis is a useful model for understanding the developmental basis of human eye disorders. They performed an extensive characterization of different TALEN-induced mutations in the pax6 gene and identified several different phenotypes in both F0 and F1 frogs (Nakayama et al., 2015). This group found that partial loss of function of pax6 in F1 animals resulted in froglets with an underdeveloped iris, a phenotype similar to that observed in human aniridia. Collectively, these initial studies demonstrated that TALEN-mediated genome editing works efficiently in F0 X. tropicalis, and that these mutations are transmitted through the germline.
Xenopus not only provides a platform for the study of genes involved in congenital malformations, but also for the study of genes involved in developmental processes and diseases at later stages. Recent studies by the Shi and Buchholz labs demonstrated that TALEN-mediated genome editing is an effective method to induce mutations in genes involved in hormonal control of metamorphosis in F0 X. tropicalis tadpoles, specifically in thyroid hormone receptor, alpha (thra) gene (Choi et al., 2015; Wen et al., 2015; Wen and Shi, 2015). Choi et al. showed that F0 phenotype analysis and germline transmission of the TALEN-induced mutations are facilitated by injection into one cell of two-cell embryos. The Vleminckx lab used TALENs to create mutations in the X. tropicalis tumor suppressor gene adenomatous polyposis coli (apc), which is implicated as the initiating mutation in many colorectal cancers, including familial adenomatous polyposis (FAP) syndrome (Van Nieuwenhuysen et al., 2015). These studies demonstrated F0 tadpoles derived from embryos injected with apc-specific TALENs develop intestinal hyperplasia and other neoplasms commonly observed in FAP patients within 6 weeks providing a useful model for the rapid testing of chemical compounds to treat FAP. These results therefore demonstrate that it is possible to utilize the mosaic nature of TALEN-induced mutagenesis for studying advanced developmental events as well as tumorigenesis in Xenopus tadpoles.
Circumventing embryonic lethality in F0 animals
One of the potential problems in generating mutant Xenopus lines using TALENs is that, in some instances, the mutations result in embryonic lethality, thus limiting analysis to the F0 generation. Ideally, the best way to overcome this issue would be to induce mutations only in germ cells and not in somatic tissue. In Xenopus, there are two methods used to create such germ cell-specific mutants. The first is to limit the translation of injected mRNA to germ cells. Several maternal mRNAs have specific germ cell restricted translation due to the 3’UTR region, such as the gene ddx25 (Kataoka et al., 2006). Recent work from the Yaoita lab showed that the ddx25 3’UTR is sufficient for largely restricting translation of TALEN-injected mRNAs to the germ cells (Nakajima and Yaoita, 2015a). They further demonstrated that bi-allelic TALEN-induced mutations are present in the F0 adult germ cells and transmitted to the F1 generation, with limited mutations found in other organs, thus defining a method for creating germ cell-specific mutants using TALENs. Another method to produce germ cell-specific mutations is to transplant primordial germ cells from a mutant embryo into a wild-type host embryo. In Xenopus, this can be done by transplanting a portion of the vegetal hemisphere, which contains the germ plasm of the early developing embryo, at the blastula stage (Yang et al., 2015). Recent work from the Cho lab showed that this method works well in X. tropicalis to produce F0 adults that are wild-type in the soma, but contain bi-allelic mutations in the germ cells (Blitz and Cho personal communication); adults produced using this method can be mated to create F1 null progeny. These two methods will help speed up the process of analyzing null mutations and allow for more detailed studies of mutations that are embryonic lethal. In addition, because Xenopus have long life spans (over 10 years), the mutant adults can be used for many years.
Although inducing mutations in both homeologs in the allotetraploid X. laevis is, in principal, more difficult to achieve, several recent studies have revealed that it is possible to generate highly efficient gene knockouts in both homeologs in X. laevis. The complication with X. laevis is that, if the alloalleles are thought to be functionally redundant, mutations must be induced in both homeologs on the S and L chromosomes. The first reports of the effectiveness of TALENs in X. laevis showed that a single TALEN pair can be designed to target both homeologous genes, and they revealed that mutations can be detected as early as the morula stage of development (Sakane et al., 2013; Suzuki et al., 2013). Because sequence differences do exist between the alloalleles, it is also possible to design TALENs to target only one of the homeologous genes. In one study, injection of two different pairs of TALENs, targeting each alloallele, into X. laevis embryos was found to induce mutations in each homeologous gene (Nakade et al., 2015). This study also demonstrated mutation of the individual homeolog was not possible when the specific TALEN binding site contained three mismatches to the endogenous sequence, highlighting the specificity of TALENs. Although these results showed that it is possible to induce mutations in both alloalleles in X. laevis, either separately or together, they also revealed that generating mutations in all four copies is not efficient enough to produce null F0 frogs due to the delay in initiating NHEJ in all cells.
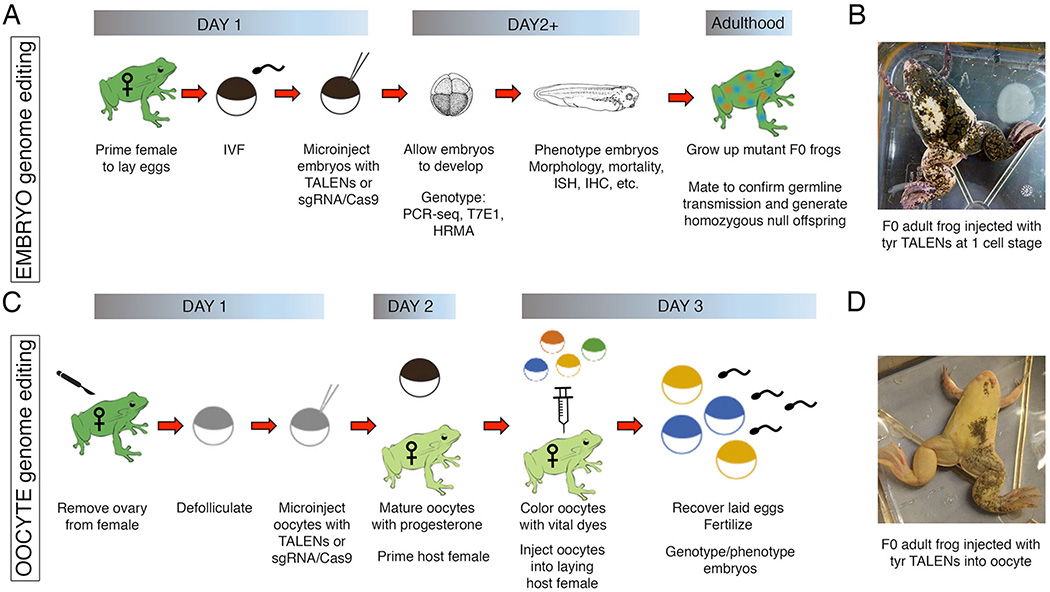
Using a well-established Xenopus method called the oocyte-host transfer technique, which was developed to knockdown maternal transcripts, it is possible to induce mutations more rapidly and efficiently than traditional embryo injections. Therefore, using the oocyte-host transfer (OHT) technique combined with genome editing will result in less mosaic F0 animals and more efficient germ line transmission of mutations (Fig. 1). In this method, heterologous mRNAs are injected into Xenopus oocytes, which are then cultured in vitro for 24–48 hours, transferred into an ovulating host female, and the laid eggs are subsequently fertilized in vitro (Olson et al., 2012). Using this technique, the Yaoita lab demonstrated that TALEN-mediated gene disruption is more efficient when injected into oocytes rather than embryos (Nakajima and Yaoita, 2015b). Similarly, we found that a single TALEN pair targeting both tyrosinase homeologs was able to generate almost complete albinism in F0 animals when injected into X. laevis oocytes (Ratzan et al., 2016); conversely, injection of these same TALENs into embryos resulted in mosaic F0 frogs, with only small patches of albinism (Fig. 1). Furthermore, the F0 oocyte-injected adults laid albino eggs, and when mated with sibling males, produced 50–75% albinism in the F1 generation; in contrast, all offspring from sibling-mated F0 embryo-injected adults failed to produce albino offspring, and very few mutations were recovered in the F1 generation (Ratzan et al., 2016).
Figure 1. Genome editing using Xenopus embryos or oocytes.
A comparison of the steps required to generate Xenopus mutants using either embryo injection or oocyte-host transfer methods. (A) For the embryo injection method, TALEN or CRISPR-Cas9 capped mRNA and/or protein is microinjected into fertilized embryos at the one-cell stage. The embryos are genotyped to confirm editing efficiency using PCR-sequencing, T7 endonuclease assays, or high-resolution melt analysis. The F0 mosaic embryos are allowed to develop, and gene function is analyzed using a host of established assays, including in situ hybridization (ISH) and immunohistochemistry (IHC). These embryos, once grown to adulthood, can be tested for germline transmission to generate subsequent mutant lines. (B) An image of a mutant frog generated from embryos injected at the one-cell stage with TALEN mRNAs targeting the tyrosinase gene shows mosaic pigmentation throughout the skin. (C) For the oocyte-host transfer method, stage VI oocytes are surgically removed from an adult female frog, manually defollicated, and microinjected with TALENs or CRISPR-Cas9 capped mRNA and/or protein. The oocytes are then matured using progesterone and colored with vital dyes for visualization; the coloring of oocytes is not necessary if implanted into an albino female. The oocytes are then transferred into pre-primed host females and subsequently laid to incorporate the jelly coat that is essential for in vitro fertilization with sperm. The resulting embryos are genotyped and phenotyped as previously described. (D) An image of a mutant frog generated from oocytes injected with the same TALENs as in panel B, targeting the tyrosinase gene, shows more dramatic levels of albinism than the embryo-injected frog, thereby confirming more efficient mutagenesis.
An alternative method to induce genetic mutations prior to fertilization has recently been published by the Gurdon lab (Miyamoto et al., 2015a; 2015b; 2013). To forego oocyte transfer back into a host female and therefore abolish further surgery, they used intracytoplasmic sperm injection (Miyamoto et al., 2015a). In this procedure, X. laevis oocytes are extracted, enzymatically defolliculated and subsequently injected with TALEN mRNA (Fig. 1); these late stage oocytes are then matured by addition of progesterone and subsequently injected with a sperm mixture (to negate the need of a jelly coat for normal in vitro fertilization methods). This method has the benefits of being less technically challenging by eliminating manual oocyte defolliculation and reintroduction of injected oocytes in the host female. In their surviving embryos Miyamoto observed a similar striking mutation efficiency for the tyrosinase and pax6 genes (between 80–90%) with all four alloalleles being targeted. Both oocyte methods are highly efficient at inducing mutations early in development, and thus allows researchers to study gene function in an F0 generation. This thereby enables the rapid assessment of disease-causing gene mutations without the need to establish mutant lines whilst also allowing the study of those genes that are essential for embryo or sexual maturation. In conclusion, these results show that TALENs work efficiently in X. tropicalis and X. laevis, and that whilst homeologous X. laevis genes can be independently targeted with two different pairs of TALENs, OHT is the most efficient method to generate mutations in all alloalleles in F0 animals.
CRISPR-Cas in Xenopus
Another genome editing technique that works well in Xenopus is CRISPR-Cas, and it has become more widely used due to its simpler design. In prokaryotes there are chromosomal loci that harbor repetitive DNA sequences, termed CRISPR elements, and adjacent to these elements there are endonuclease gene coding regions, termed CRISPR-associated genes (Cas) (Bolotin et al., 2005; Jansen et al., 2002; Jinek et al., 2012; Makarova et al., 2015; Wright et al., 2016). This combination provides the bacteria with an acquired adaptive immune system capable of specifically targeting nucleic acid sequences of invading viruses or plasmids (Bhaya et al., 2011; Horvath and Barrangou, 2010; Karginov and Hannon, 2010). Researchers have exploited the programmable nature of this DNA targeting nuclease to mutate specific genetic loci of interest in many model systems including Xenopus (Blitz et al., 2013; Cho et al., 2013; Cong et al., 2013; Doudna and Charpentier, 2014; Guo et al., 2014; Harrison et al., 2014; Jinek et al., 2012; Mali et al., 2013a; Nakayama et al., 2013; Peng et al., 2014). To induce double strand breaks in Xenopus using CRISPR-Cas, one or more single guide RNAs (sgRNA) targeting a gene of interest is injected into Xenopus embryos together with the Cas endonuclease, either as mRNA or protein. This method is especially time saving when targeting multiple loci in the same embryo, because the sgRNAs can be generated by PCR in a single day, in contrast to the multi-day process for cloning TALENs. However, unlike TALENs, sgRNAs can only be designed to target regions containing a protospacer adjacent motif (PAM) site, limiting the regions that can be targeted.
Several recent reports in the past couple of years have shown that CRISPR-Cas is highly efficient in producing mutations in both X. tropicalis and X. laevis (Blitz et al., 2013; Guo et al., 2014; Nakayama et al., 2013; Wang et al., 2015). Initial studies attempted to optimize the amounts of sgRNA and Cas9 mRNA required to produce efficient mutagenesis in the F0 generation with limited developmental defects. Generally, the amount of sgRNA required to produce efficient indels varies with each locus. For X. tropicalis, most loci require a range of 50–200 pg sgRNA, whereas other loci require up to 400 pg sgRNA; for X. laevis, 300–500 pg sgRNA is optimal, but greater for some loci. Interestingly, the amount of Cas9 mRNA required to induce indels varied among these studies. Two of the studies revealed that a relatively high amount (2.2–3 ng) of Cas9 mRNA is required to induce efficient mutagenesis (Blitz et al., 2013; Nakayama et al., 2013), whereas the third study showed that a much lower amount (300–500 pg) is sufficient (Guo et al., 2014).
These reported Cas9 dosage discrepancies may be due to the different Cas9 versions used in each study, which are identical at the amino acid level, but are only 80% identical at the nucleotide level (Cong et al., 2013; Mali et al., 2013b). As Xenopus codon usage slightly differs from that of humans, the use of rare codons may impact the translational efficiency of each Cas9 transcript, especially with the large size of the Cas9 protein. In addition, the two versions differ at their N- and C-termini: the Cong Cas9, used by the Chen lab, contains two nuclear localization sequences (NLS), one at each end, and a 3× FLAG tag at the N-terminus, whereas the Mali Cas9, used by Cho and Grainger labs, contains a single NLS at the C-terminus. Comparison of the codon usage showed that the Cong Cas9 used slightly different codons, some of which were more optimal for X. laevis. These differences may explain why a lower dose of the Cong Cas9 was required in Xenopus. As both versions were tested in the standard Xenopus vector, pCS2, they contain identical 5’ and 3’UTRs. These initial reports illustrated that CRISPR-Cas works efficiently in both X. laevis and X. tropicalis, but that care must be taken in selecting codon-optimized transcripts when performing mRNA injections of Cas9. An alternative to Cas9 mRNA is to use Cas9 protein, which eliminates the concerns of codon differences. The Khokha laboratory recently compared the efficacy of protein versus mRNA in Xenopus tropicalis and found that Cas9 protein was more effective than mRNA (Bhattacharya et al., 2015). They also reported Cas9 protein was able to induce mutations earlier in development than Cas9 mRNA and was less toxic. However, in light of the discussion of Cas9 codon usage, it is relevant to point out that they used Cas9 protein from PNA Bio, which contains only one NLS.
In addition to the technical importance, the Khokha group’s study also clearly demonstrated that X. tropicalis can be used to rapidly produce F0 phenotypes for human disease genes using CRISPR-Cas. As we have discussed, many candidate genes for human diseases have been identified, but little functional analysis of them has been possible, particularly outside a few model organisms. In this study, they designed sgRNAs to six human disease genes and showed that the F0 CRISPR mutants largely reproduced the expected disease phenotypes. Thus, with the increased efficiency afforded by Cas9 protein injection, F0 analysis was sufficient for functional screening of candidate genes involved in human disease, expanding the utility of the X. tropicalis model for this important application.
Beyond inducing genetic mutations, CRISPR-Cas can also be used to label specific chromosomal regions (Chen et al., 2013) and recent work by the Heald lab demonstrated how this can be applied to Xenopus (Lane et al., 2015). In this study they developed a technique that allows for the creation of a library of sgRNAs targeting a defined genomic region, which they call CRISPR EATING (everything available turned into new guides). The library is made by PCR amplification of specific genomic region followed by restriction digestion with an enzyme that cuts immediately 5’ to a PAM sequence resulting in various sized fragments; subsequent steps of adaptor ligations and restriction digests results in a mixture of 136 nt sgRNA fragments that contain a T7 RNA polymerase promoter for RNA transcription. The authors then complexed the sgRNAs with a recombinant catalytically inactive Cas9 fused to mNeon-Green (dCas9-Neon) to label specific chromosomal regions using Xenopus sperm nuclei in vitro. One of the main benefits of this approach is the ability to use Xenopus egg extracts and sperm nuclei to image genomic loci throughout the cell cycle. In addition to being useful for visualization of genomic loci, this approach can be adapted to generate mutations in larger regions of DNA that will be of benefit to making mutants in genes with many small exons.
Knock-in strategies in Xenopus
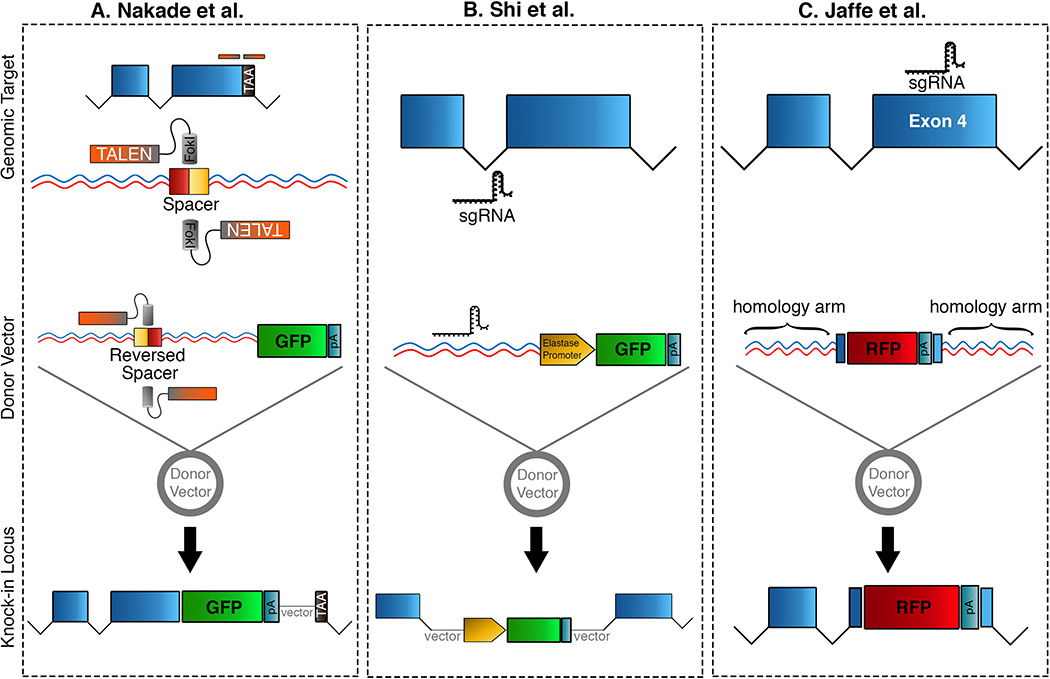
Several different methods have been used for many years to promote integration of exogenous DNA into the genome of Xenopus laevis or Xenopus tropicalis for the production of transgenic lines. These methods rely on random integration into the genome, and as such will not be discussed here, but we refer the reader to several excellent reviews that cover these methods in detail (Allen and Weeks, 2006; Chesneau et al., 2008; Love et al., 2011; Ogino et al., 2006; Takagi et al., 2013; Yergeau et al., 2009). In contrast, CRISPR-Cas and TALENs can be used for site-specific integration of exogenous DNA and three recent reports demonstrated that this can be accomplished in Xenopus. In the first study, they used TALENs to create F0 knock-in X. laevis tadpoles through a microhomology-mediated end joining (MMEJ) strategy, which they refer to as TALEN-mediated precise integration into target chromosome, or TAL-PITCh (Nakade et al., 2014). They used a single homology arm, containing the TALEN target site (with an inverted spacer sequence) to insert exogenous DNA into two different loci, no29 and fgk (Fig. 2A). In the first instance, they targeted the start codon of no29 to knock-in a no29-GFP fusion template, whereas for fgk they targeted the 3’ end to insert EGFP just before the stop codon to make an endogenous fusion. In both instances, they found integration at the 5’ junction was precise in most cases, but at the 3’ junction there were often deletions and insertions. In the second study, they used CRISPR-Cas to mediate integration of exogenous DNA in X. tropicalis and showed efficient germline transmission (Z. Shi et al., 2015). In contrast to traditional knock-in strategies that use homology-dependent integration, this study showed that targeted integration could be achieved independent of homology arms, as long as the sgRNA target site is included in the donor DNA (Fig. 2B). Although both methods showed that integration is imprecise and results in deletions and insertions around the target site, they illustrate that it is feasible to use TALENs and CRISPR-Cas for insertional mutagenesis.
Fig. 2. Integrating exogenous DNA into Xenopus using genetic editing tools.
Outline of the various knock-in strategies that have been employed to insert DNA into a targeted genomic locus in Xenopus. (A) Nakade et al. described the use of TALENs and microhomology-mediated end joining (MMEJ, TAL-PITCh) to integrate a fluorescent protein (eg. GFP) at the end of the coding region 5’ to the endogenous stop codon. (B) Shi et al. utilized CRISPR-Cas editing to insert plasmid DNA harboring a known pancreas tissue enhancer element (Elastase promoter) driving GFP, into the intron of their target gene. (C) Jaffe et al, used targeting constructs containing allele-specific homology arms to insert fluorescent proteins into an sgRNA-targeted exon, thereby visualizing cells in which specific gene function was abrogated. TAA; stop codon, FokI; Fok1 nuclease, GFP; green fluorescent protein, pA; poly-A tail, sgRNA; guide RNA for CRISPR.
While knock-in of exogenous DNA in F0 embryos is mosaic, this does not preclude all F0 studies as illustrated by a recent study by the Mitchell lab (Jaffe et al., 2016). In that study, they used an elegant knock-in approach to create mosaic mutant F0 X. laevis in c21orf59, a gene involved in cilia polarization in multi-ciliated cells (MCC) in Xenopus (Fig. 2C). As there are two c21orf59 alloalleles on chromosomes 2L and 2S, the Mitchell lab developed a strategy that allowed them to independently verify if one or both alloalleles were mutated in individual cells. The injection of a single sgRNA to target both gene homeologs was coupled with the incorporation of two different donor vectors harboring either BFP or RFP reporter genes. These vectors differed in their homology arm DNA sequence that would target them specifically to either the Xla2L or Xla2S alloallele. Using this approach, they identified those cells where one or both alloalleles was disrupted. Cells that expressed both BFP and RFP displayed a complete loss of cilia polarity, whereas cells that expressed only BFP or RFP displayed an intermediate phenotype. Such an approach takes good advantage of the different alloalleles in Xenopus laevis and illustrates a useful general method for knock-ins in Xenopus.
Xenopus resources for genome editing (Xenbase and NXR)
Xenbase, the Xenopus model organism bioinformatics database, is an invaluable resource for the design and application of reverse genetics in Xenopus (James-Zorn et al., 2015). Xenbase (http://www.xenbase.org) is a unique resource that serves as the central repository for all things related to Xenopus genomics. It provides a user-friendly interface to interrogate data related to a specific gene, and in particular, it provides essential genomic sequences and web-based tools for genome editing. Individual gene pages in Xenbase provide a wealth of information for a particular gene, including functional descriptions and expression profiles, associations with human disease, and links to other model organism databases. Individual gene pages also have information about the genes in both X. tropicalis and X. laevis, including both L and S homeologs in X. laevis. Each gene page provides a direct link to GBrowse, which allows visualization of the gene structure (exons and introns) and comparison of synteny with human and mouse genomes to confirm that the correct gene is being targeted. For an unannotated gene, a BLAST search can be performed directly in Xenbase to interrogate the respective genome and determine if the gene sequence is present. As annotation of the Xenopus genomes continues to be updated, often times a gene may not appear as annotated on the main gene page, but when viewed within GBrowse, the exon and intron information is properly annotated. Thus, it is critical to use the Xenbase BLAST function to identify the chromosomal location of each gene. This information is extremely useful when searching the X. laevis genome for both homeologs, because Xenbase will identify both genes on the L and S chromosomes. From GBrowse, one can then download the exon and intron sequences for an individual gene. Xenbase also provides useful links to several web-based genome editing tools for the identification of sgRNA or TALEN target sites. Thus, all of the information on Xenbase provides an essential platform to identify chromosomal locations of genes, and to design sgRNA and TALEN target sites.
Due to cost and space constraints, one of the difficulties within the Xenopus community is the raising and breeding of specific lines. In fact, most Xenopus laboratories are not experienced in breeding and maintaining mutant lines, particularly with the diploid X. tropicalis species that is ideal for genetic studies. For those unable to generate or breed their own frog lines, the National Xenopus Resource (NXR) offers a custom mutant service that will design, inject, and breed F0 or F1 X. tropicalis or X. laevis frogs. The NXR works closely with individual researchers to identify the specific region of a gene that should be mutated, and they offer multiple choices for genome editing, including TALEN and CRISPR sgRNA design and injection. In addition, for those researchers unable to maintain colonies of both X. laevis and X. tropicalis, or for those wishing to work with multiple mutant lines, the NXR provides a unique service called research facility service, which allows researchers to come to the NXR facility at the Marine Biological Laboratory (MBL) for short-term visits. Here, they can access the resources at the NXR, including the large number of different X. laevis and X. tropicalis strains and lines, and take advantage of the genome editing expertise at the NXR. The NXR also provides a service to raise and maintain animal lines at the MBL, thus allowing researchers to enhance the scope of their research. Lastly, a recent project funded by the NIH at the MBL, in collaboration with the NXR, is focused on creating 100–200 mutants for the Xenopus community. This new project works with individual researchers to generate CRISPR-Cas- and TALEN-mediated mutations in specific genes to create new Xenopus models of human disease.
Conclusions
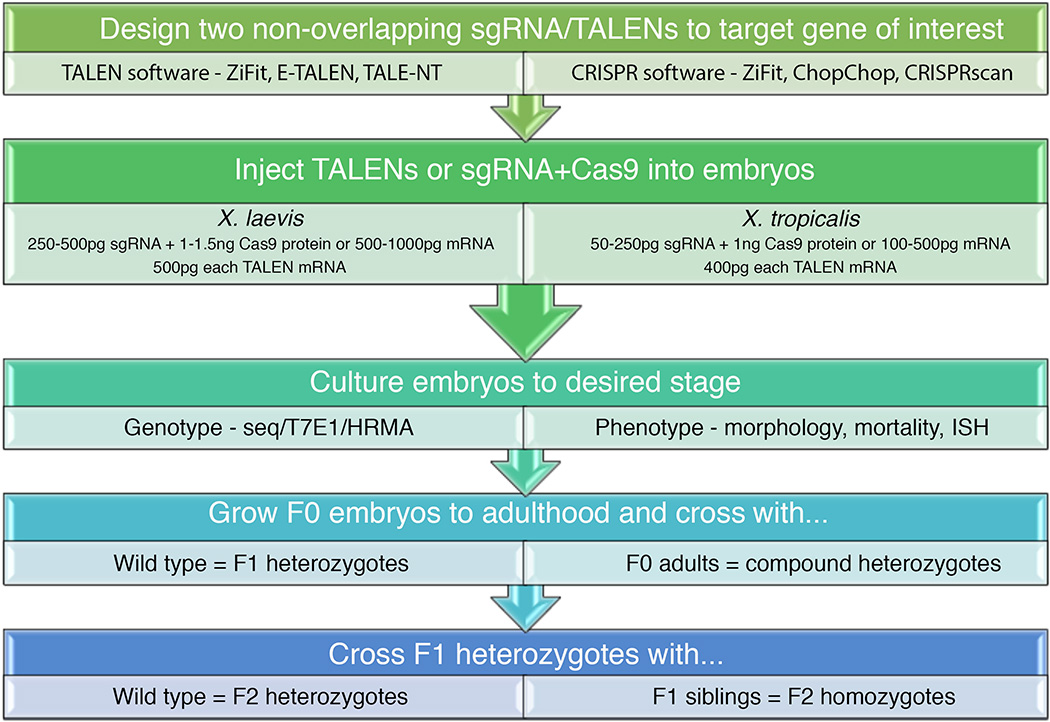
There is substantial and growing evidence that TALENs and CRISPR-Cas genome editing tools can now be used to manipulate endogenous genes in both X. tropicalis and X. laevis, providing researchers with the powerful ability to model a host of human disorders. In Figure 3, we outline the steps required to generate mutants in Xenopus using TALENs or CRISPR-Cas systems; however, a more detailed discussion of CRISPR-Cas protocols in Xenopus can be found in a recent review (Nakayama et al., 2014). The design and application of genome editing tools for Xenopus has been streamlined by incorporating online software to design target oligonucleotides and assess potential off-target effects in the Xenopus genome. These tools, combined with simple DNA extraction and PCR techniques that are already employed by most laboratories, will enable the generation and identification of mutant embryos in a fast and efficient manner, allowing the establishment of mutant lines (Fig. 3). TALENs and CRISPR-Cas can be used in a variety of ways in Xenopus to modify specific protein domains, rearrange chromosomal organization, or to introduce the equivalent human point mutations identified through genome-wide association studies (GWAS). In addition, unbiased studies of organ formation and function in Xenopus have been shown to reveal phenotypes similar to those observed in human diseases (Iwasaki and Thomsen, 2014; Pearl et al., 2011; Sojka et al., 2014); thus, the generation of new mutants via genome editing may lead to the identification of new disease candidates. Therefore, the genome-edited Xenopus model will be instrumental as an initial tool for understanding the components and pathways affected by genetic disorders in a highly conserved vertebrate in vivo environment, which is not yet achievable with primary cell cultures or mammalian models. Furthermore, when coupled with high-throughput assays that require tissue explants and/or large numbers of embryonic samples, the genome-edited Xenopus model should aid the discovery of chemical or gene therapeutics that may serve to treat human diseases.
Figure 3. Workflow for generating Xenopus mutants using TALENs and CRISPR-Cas9.
A schematic depicts the steps required to generate the gene editing tools to target a gene of interest, induce mutations in Xenopus embryos, perform subsequent assays to phenotype mosaic F0 embryos, and generate mutant lines. For more detailed information including web URLs we refer the reader to Xenbase (http://www.xenbase.org/other/static/CRISPr.jsp).
Acknowledgments
Funding
This work was supported by the National Institutes of Health (P40 OD010997 to M.E.H., R01 HD084409 to M.E.H., R01 HL112618 to P.T. and F.C., and R01 HL127640 to P.T. and F.C.; and the U.S. Environmental Protection Agency (G11E10367 to D.F.).
References
- Afelik S, Chen Y, Pieler T. Combined ectopic expression of Pdx1 and Ptf1a/p48 results in the stable conversion of posterior endoderm into endocrine and exocrine pancreatic tissue. Genes Dev. 2006;20:1441–1446. doi: 10.1101/gad.378706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen BG, Weeks DL. Using phiC31 integrase to make transgenic Xenopus laevis embryos. Nat Protoc. 2006;1:1248–1257. doi: 10.1038/nprot.2006.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodeo AA, Jukam D, Straight AF, Skotheim JM. Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proc Natl Acad Sci USA. 2015;112:E1086–E1095. doi: 10.1073/pnas.1413990112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Marfo CA, Li D, Lane M, Khokha MK. CRISPR/Cas9: An inexpensive, efficient loss of function tool to screen human disease genes in Xenopus. Dev Biol. 2015;408:196–204. doi: 10.1016/j.ydbio.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D, Davison M, Barrangou R. CRISPR-Cas Systems in Bacteria and Archaea: Versatile Small RNAs for Adaptive Defense and Regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- Bier E, De Robertis EM. EMBRYO DEVELOPMENT. BMP gradients: A paradigm for morphogen-mediated developmental patterning. Science. 2015;348:aaa5838–aaa5838. doi: 10.1126/science.aaa5838. [DOI] [PubMed] [Google Scholar]
- Blitz IL. Navigating the Xenopus tropicalis genome. Methods Mol Biol. 2012;917:43–65. doi: 10.1007/978-1-61779-992-1_4. [DOI] [PubMed] [Google Scholar]
- Blitz IL, Andelfinger G, Horb ME. Germ layers to organs: using Xenopus to study “later” development. Semin Cell Dev Biol. 2006;17:133–145. doi: 10.1016/j.semcdb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Blitz IL, Biesinger J, Xie X, Cho KWY. Biallelic genome modification in F(0) Xenopus tropicalis embryos using the CRISPR/Cas system. Genesis. 2013;51:827–834. doi: 10.1002/dvg.22719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodart J-FL, Duesbery NS. Xenopus tropicalis oocytes: more than just a beautiful genome. Methods Mol Biol. 2006;322:43–53. doi: 10.1007/978-1-59745-000-3_4. [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF. TAL Effectors: Customizable Proteins for DNA Targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology (Reading, Engl.) 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Boskovski MT, Yuan S, Pedersen NB, Goth CK, Makova S, Clausen H, Brueckner M, Khokha MK. The heterotaxy gene GALNT11 glycosylates Notch to orchestrate cilia type and laterality. Nature. 2013;504:456–459. doi: 10.1038/nature12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz DR. More similar than you think_ Frog metamorphosis as a model of human perinatal endocrinology. Dev Biol. 2015;408:188–195. doi: 10.1016/j.ydbio.2015.02.018. [DOI] [PubMed] [Google Scholar]
- Buchholz DR. Xenopus Protocols, Methods in Molecular Biology. Totowa, NJ: Humana Press; 2012. Tet-On Binary Systems for Tissue-Specific and Inducible Transgene Expression; pp. 265–275. [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers AD, Slack JM. The Xenopus tadpole gut: fate maps and morphogenetic movements. Development. 2000;127:381–392. doi: 10.1242/dev.127.2.381. [DOI] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li G-W, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B. Dynamic Imaging of Genomic Lociin Living Human Cells by an Optimized CRISPR/Cas System. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernet BT, Levin M. Transmembrane voltage potential is an essential cellular parameter for the detection and control of tumor development in a Xenopus model. Disease Models & Mechanisms. 2013;6:595–607. doi: 10.1242/dmm.010835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesneau A, Sachs LM, Chai N, Chen Y, Pasquier, Du L, Loeber J, Pollet N, Reilly M, Weeks DL, Bronchain OJ. Transgenesis procedures in Xenopus. Biol Cell. 2008;100:503–521. doi: 10.1042/BC20070148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim J-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Choi J, Suzuki K-IT, Sakuma T, Shewade L, Yamamoto T, Buchholz DR. Unliganded thyroid hormone receptor α regulates developmental timing via gene repression in Xenopus tropicalis. Endocrinology. 2015;156:735–744. doi: 10.1210/en.2014-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M-I, Kwon T, Tu F, Brooks ER, Gupta R, Meyer M, Baker JC, Marcotte EM, Wallingford JB. Coordinated genomic control of ciliogenesis and cell movement by RFX2. Elife. 2014;3:e01439. doi: 10.7554/eLife.01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross MK, Powers MA. Learning about cancer from frogs: analysis of mitotic spindles in Xenopus egg extracts. Disease Models & Mechanisms. 2009;2:541–547. doi: 10.1242/dmm.002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. Sequence organization in the genome of Xenopus laevis. Symp Soc Dev Biol. 1973;31:251–268. doi: 10.1016/b978-0-12-612975-5.50014-7. [DOI] [PubMed] [Google Scholar]
- Dickinson AJG. Using frogs faces to dissect the mechanisms underlying human orofacial defects. Semin Cell Dev Biol. 2016:1–10. doi: 10.1016/j.semcdb.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096–1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Dubaissi E, Papalopulu N. Embryonic frog epidermis: a model for the study of cell-cell interactions in the development of mucociliary disease. Disease Models & Mechanisms. 2011;4:179–192. doi: 10.1242/dmm.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AR, Khokha MK. Xenopus as a model organism for birth defects-Congenital heart disease and heterotaxy. Semin Cell Dev Biol. 2016;51:73–79. doi: 10.1016/j.semcdb.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdogan B, Ebbert PT, Lowery LA. Using Xenopus laevis retinal and spinal neurons to study mechanisms of axon guidance in vivo and in vitro. Semin Cell Dev Biol. 2016;51:64–72. doi: 10.1016/j.semcdb.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc Natl Acad Sci USA. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantress J, Maniero GD, Cohen N, Robert J. Development and characterization of a model system to study amphibian immune responses to iridoviruses. Virology. 2003;311:254–262. doi: 10.1016/s0042-6822(03)00151-x. [DOI] [PubMed] [Google Scholar]
- Geach TJ, Stemple DL, Zimmerman LB. Genetic analysis of Xenopus tropicalis. Methods Mol Biol. 2012;917:69–110. doi: 10.1007/978-1-61779-992-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant IM, Balcha D, Hao T, Shen Y, Trivedi P, Patrushev I, Fortriede JD, Karpinka JB, Liu L, Zorn AM, Stukenberg PT, Hill DE, Gilchrist MJ. The Xenopus ORFeome: A resource that enables functional genomics. Dev Biol. 2015;408:345–357. doi: 10.1016/j.ydbio.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Zhang T, Hu Z, Zhang Y, Shi Z, Wang Q, Cui Y, Wang F, Zhao H, Chen Y. Efficient RNA/Cas9-mediated genome editing in Xenopus tropicalis. Development. 2014;141:707–714. doi: 10.1242/dev.099853. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. The egg and the nucleus: a battle for supremacy. Development. 2013;140:2449–2456. doi: 10.1242/dev.097170. [DOI] [PubMed] [Google Scholar]
- Hardwick LJA, Philpott A. An oncologist׳s friend_ How Xenopus contributes to cancer research. Dev Biol. 2015;408:180–187. doi: 10.1016/j.ydbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland RM, Grainger RM. Xenopus research: metamorphosed by genetics and genomics. Trends Genet. 2011;27:507–515. doi: 10.1016/j.tig.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Jenkins BV, O'Connor-Giles KM, Wildonger J. A CRISPR view of development. Genes Dev. 2014;28:1859–1872. doi: 10.1101/gad.248252.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes-Gilmore N, Banach M, Edholm E-S, Lord E, Robert J. A critical role of non-classical MHC in tumor immune evasion in the amphibian Xenopus model. Carcinogenesis. 2014;35:1807–1813. doi: 10.1093/carcin/bgu100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, Blitz IL, Blumberg B, Dichmann DS, Dubchak I, Amaya E, Detter JC, Fletcher R, Gerhard DS, Goodstein D, Graves T, Grigoriev IV, Grimwood J, Kawashima T, Lindquist E, Lucas SM, Mead PE, Mitros T, Ogino H, Ohta Y, Poliakov AV, Pollet N, Robert J, Salamov A, Sater AK, Schmutz J, Terry A, Vize PD, Warren WC, Wells D, Wills A, Wilson RK, Zimmerman LB, Zorn AM, Grainger R, Grammer T, Khokha MK, Richardson PM, Rokhsar DS. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U, Khokha MK, Grammer TC, Harland RM, Richardson P, Rokhsar DS. Accelerated gene evolution and subfunctionalization in the pseudotetraploid frog Xenopus laevis. BMC Biol. 2007;5:31. doi: 10.1186/1741-7007-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikasa H, Sokol SY. Wnt signaling in vertebrate axis specification. Cold Spring Harb Perspect Biol. 2013;5:a007955. doi: 10.1101/cshperspect.a007955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Hunt T. Protein synthesis, proteolysis, and cell cycle transitions. Biosci. Rep. 2002;22:465–486. doi: 10.1023/a:1022077317801. [DOI] [PubMed] [Google Scholar]
- Ishibashi S, Cliffe R, Amaya E. Highly efficient bi-allelic mutation rates using TALENs in Xenopus tropicalis. Biol Open. 2012a;1:1273–1276. doi: 10.1242/bio.20123228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi S, Kroll KL, Amaya E. Xenopus Protocols, Methods in Molecular Biology. Totowa, NJ: Humana Press; 2012b. Generating Transgenic Frog Embryos by Restriction Enzyme Mediated Integration (REMI) pp. 185–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi S, Love NR, Amaya E. Xenopus Protocols, Methods in Molecular Biology. Totowa, NJ: Humana Press; 2012c. A Simple Method of Transgenesis Using I-Sce I Meganuclease in Xenopus; pp. 205–218. [DOI] [PubMed] [Google Scholar]
- Iwasaki Y, Thomsen GH. The splicing factor PQBP1 regulates mesodermal and neural development through FGF signaling. Development. 2014;141:3740–3751. doi: 10.1242/dev.106658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe KM, Grimes DT, Schottenfeld-Roames J, Werner ME, Ku T-SJ, Kim SK, Pelliccia JL, Morante NFC, Mitchell BJ, Burdine RD. c21orf59/kurly Controls Both Cilia Motility and Polarization. Cell Rep. 2016;14:1841–1849. doi: 10.1016/j.celrep.2016.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James-Zorn C, Ponferrada VG, Burns KA, Fortriede JD, Lotay VS, Liu Y, Brad Karpinka J, Karimi K, Zorn AM, Vize PD. Xenbase: Core features, data acquisition, and data processing. Genesis. 2015;53:486–497. doi: 10.1002/dvg.22873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, van Embden J, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Molecular Microbiology. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- Jarikji ZH, Vanamala S, Beck CW, Wright CVE, Leach SD, Horb ME. Differential ability of Ptf1a and Ptf1a-VP16 to convert stomach, duodenum and liver to pancreas. Dev Biol. 2007;304:786–799. doi: 10.1016/j.ydbio.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbrun E, Tandon P, Amin NM, Waldron L, Showell C, Conlon FL. Xenopus: An emerging model for studying congenital heart disease. Birth Defects Res. Part A Clin. Mol. Teratol. 2011;91:495–510. doi: 10.1002/bdra.20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K, Yamaguchi T, Orii H, Tazaki A, Watanabe K, Mochii M. Visualization of the Xenopus primordial germ cells using a green fluorescent protein controlled by cis elements of the 3' untranslated region of the DEADSouth gene. Mech Dev. 2006;123:746–760. doi: 10.1016/j.mod.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Kay BK, Peng HB. Xenopus laevis: Practical uses in cell and molecular biology. 1991 [PubMed] [Google Scholar]
- Kelley CM, Yergeau DA, Zhu H, Kuliyev E, Mead PE. Xenopus Protocols, Methods in Molecular Biology. Totowa, NJ: Humana Press; 2012. Xenopus Transgenics: Methods Using Transposons; pp. 231–243. [DOI] [PubMed] [Google Scholar]
- Khokha MK. Xenopus white papers and resources: Folding functional genomics and genetics into the frog. Genesis. 2012;50:133–142. doi: 10.1002/dvg.22015. [DOI] [PubMed] [Google Scholar]
- Kim H, Kim J-S. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- Kim SK, Shindo A, Park TJ, Oh EC, Ghosh S, Gray RS, Lewis RA, Johnson CA, Attie-Bittach T, Katsanis N, Wallingford JB. Planar cell polarity acts through septins to control collective cell movement and ciliogenesis. Science. 2010;329:1337–1340. doi: 10.1126/science.1191184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klos Dehring DA, Vladar EK, Werner ME, Mitchell JW, Hwang P, Mitchell BJ. Deuterosome-mediated centriole biogenesis. Dev Cell. 2013;27:103–112. doi: 10.1016/j.devcel.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofent J, Spagnoli FM. Xenopus as a model system for studying pancreatic development and diabetes. Semin Cell Dev Biol. 2016;51:106–116. doi: 10.1016/j.semcdb.2016.01.005. [DOI] [PubMed] [Google Scholar]
- Krylov V, Tlapakova T. Xenopus Cytogenetics and Chromosomal Evolution. Cytogenet Genome Res. 2015;145:192–200. doi: 10.1159/000406550. [DOI] [PubMed] [Google Scholar]
- Labonne C, Zorn AM. Modeling human development and disease in Xenopus. Dev Biol. 2015;408:179. doi: 10.1016/j.ydbio.2015.11.019. [DOI] [PubMed] [Google Scholar]
- Lane AB, Strzelecka M, Ettinger A, Grenfell AW, Wittmann T, Heald R. Enzymatically Generated CRISPR Libraries for Genome Labeling and Screening. Dev Cell. 2015;34:373–378. doi: 10.1016/j.devcel.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane MC, Sheets MD. Heading in a new direction: implications of the revised fate map for understanding Xenopus laevis development. Dev Biol. 2006;296:12–28. doi: 10.1016/j.ydbio.2006.04.447. [DOI] [PubMed] [Google Scholar]
- Langdon Y, Tandon P, Paden E, Duddy J, Taylor JM, Conlon FL. SHP-2 acts via ROCK to regulate the cardiac actin cytoskeleton. Development. 2012;139:948–957. doi: 10.1242/dev.067579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon YG, Goetz SC, Berg AE, Swanik JT, Conlon FL. SHP-2 is required for the maintenance of cardiac progenitors. Development. 2007;134:4119–4130. doi: 10.1242/dev.009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Guo X, Liu Y, Cao Y, Deng Y, Chen X, Cheng CHK, Dawid IB, Chen Y, Zhao H. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs) Proc Natl Acad Sci USA. 2012;109:17484–17489. doi: 10.1073/pnas.1215421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienkamp SS. Using Xenopus to study genetic kidney diseases. Semin Cell Dev Biol. 2016:1–8. doi: 10.1016/j.semcdb.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Love NR, Thuret R, Chen Y, Ishibashi S, Sabherwal N, Paredes R, Alves-Silva J, Dorey K, Noble AM, Guille MJ, Sasai Y, Papalopulu N, Amaya E. pTransgenesis: a cross-species, modular transgenesis resource. Development. 2011;138:5451–5458. doi: 10.1242/dev.066498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Quigley I, Omran H, Kintner C. Multicilin drives centriole biogenesis via E2f proteins. Genes Dev. 2014;28:1461–1471. doi: 10.1101/gad.243832.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macha J, Teichmanová R, Sater AK, Wells DE, Tlapakova T, Zimmerman LB, Krylov V. Deep ancestry of mammalian X chromosome revealed by comparison with the basal tetrapod Xenopus tropicalis. BMC Genomics. 2012;13:315. doi: 10.1186/1471-2164-13-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJM, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksaev G, Haswell ES. Expressing and Characterizing Mechanosensitive Channels in Xenopus Oocytes. Methods in cell biology. 2015;1309:151–169. doi: 10.1007/978-1-4939-2697-8_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013a;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Güell M, DiCarlo JE, Norville JE, Church GM. RNA-Guided Human Genome Engineering via Cas9. Science. 2013b;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Uno Y, Kondo M, Gilchrist MJ, Zorn AM, Rokhsar DS, Schmid M, Taira M. A New Nomenclature of Xenopus laevis Chromosomes Based on the Phylogenetic Relationship to Silurana/Xenopus tropicalis. Cytogenet Genome Res. 2015;145:187–191. doi: 10.1159/000381292. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Ishihara K, Nguyen P, Wühr M. Size Scaling of Microtubule Assemblies in Early Xenopus Embryos. Cold Spring Harb Perspect Biol. 2015;7:a019182–a019113. doi: 10.1101/cshperspect.a019182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Simpson D, Gurdon JB. Manipulation and in vitro maturation of Xenopus laevis oocytes, followed by intracytoplasmic sperm injection, to study embryonic development. J Vis Exp. 2015a:e52496–e52496. doi: 10.3791/52496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Suzuki K-IT, Suzuki M, Sakane Y, Sakuma T, Herberg S, Simeone A, Simpson D, Jullien J, Yamamoto T, Gurdon JB. The Expression of TALEN before Fertilization Provides a Rapid Knock-Out Phenotype in Xenopus laevis Founder Embryos. PLoS ONE. 2015b;10:e0142946. doi: 10.1371/journal.pone.0142946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Teperek M, Yusa K, Allen GE, Bradshaw CR, Gurdon JB. Nuclear Wave1 is required for reprogramming transcription in oocytes and for normal development. Science. 2013;341:1002–1005. doi: 10.1126/science.1240376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz R, Edwards-Faret G, Moreno M, Zuñiga N, Cline H, Larrain J. Regeneration of Xenopus laevis spinal cord requires Sox2/3 expressing cells. Dev Biol. 2015;408:229–243. doi: 10.1016/j.ydbio.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakade S, Sakuma T, Sakane Y, Hara Y, Kurabayashi A, Kashiwagi K, Kashiwagi A, Yamamoto T, Obara M. Homeolog-specific targeted mutagenesis in Xenopus laevis using TALENs. In Vitro Cell. Dev. Biol.-Animal. 2015;51:879–884. doi: 10.1007/s11626-015-9912-0. [DOI] [PubMed] [Google Scholar]
- Nakade S, Tsubota T, Sakane Y, Kume S, Sakamoto N, Obara M, Daimon T, Sezutsu H, Yamamoto T, Sakuma T, Suzuki K-IT. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat Commun. 2014;5:5560. doi: 10.1038/ncomms6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Yaoita Y. Development of a new approach for targeted gene editing in primordial germ cells using TALENs in Xenopus. Biol Open. 2015a;4:259–266. doi: 10.1242/bio.201410926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Yaoita Y. Highly efficient gene knockout by injection of TALEN mRNAs into oocytes and host transfer in Xenopus laevis. Biol Open. 2015b;4:180–185. doi: 10.1242/bio.201410009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Blitz IL, Fish MB, Odeleye AO, Manohar S, Cho KWY, Grainger RM. Cas9-based genome editing in Xenopus tropicalis. Meth Enzymol. 2014;546:355–375. doi: 10.1016/B978-0-12-801185-0.00017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Fish MB, Fisher M, Oomen-Hajagos J, Thomsen GH, Grainger RM. Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. Genesis. 2013;51:835–843. doi: 10.1002/dvg.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Fisher M, Nakajima K, Odeleye AO, Zimmerman KB, Fish MB, Yaoita Y, Chojnowski JL, Lauderdale JD, Netland PA, Grainger RM. Xenopus pax6 mutants affect eye development and other organ systems, and have phenotypic similarities to human aniridia patients. Dev Biol. 2015;408:328–344. doi: 10.1016/j.ydbio.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino H, McConnell WB, Grainger RM. High-throughput transgenesis in Xenopus using I-SceI meganuclease. Nat Protoc. 2006;1:1703–1710. doi: 10.1038/nprot.2006.208. [DOI] [PubMed] [Google Scholar]
- Olson DJ, Hulstrand AM, Houston DW. Maternal mRNA Knock-down Studies: Antisense Experiments Using the Host-Transfer Technique in Xenopus laevis and Xenopus tropicalis. Totowa, NJ: Humana Press; 2012. pp. 167–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onjiko RM, Moody SA, Nemes P. Single-cell mass spectrometry reveals small molecules that affect cell fates in the 16-cell embryo. Proc Natl Acad Sci USA. 2015;112:6545–6550. doi: 10.1073/pnas.1423682112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjpe SS, Jacobi UG, van Heeringen SJ, Veenstra GJC. A genome-wide survey of maternal and embryonic transcripts during Xenopus tropicalis development. BMC Genomics. 2013;14 doi: 10.1186/1471-2164-14-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl EJ, Bilogan CK, Mukhi S, Brown DD, Horb ME. Xenopus pancreas development. Dev Dyn. 2009;238:1271–1286. doi: 10.1002/dvdy.21935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl EJ, Grainger RM, Guille M, Horb ME. Development of Xenopus resource centers: the National Xenopus Resource and the European Xenopus Resource Center. Genesis. 2012;50:155–163. doi: 10.1002/dvg.22013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl EJ, Jarikji Z, Horb ME. Functional analysis of Rfx6 and mutant variants associated with neonatal diabetes. Dev Biol. 2011;351:135–145. doi: 10.1016/j.ydbio.2010.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Clark KJ, Campbell JM, Panetta MR, Guo Y, Ekker SC. Making designer mutants in model organisms. Development. 2014;141:4042–4054. doi: 10.1242/dev.102186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshkin L, Wühr M, Pearl E, Haas W, Freeman RM, Jr, Gerhart JC, Klein AM, Horb M, Gygi SP, Kirschner MW. On the Relationship of Protein and mRNA Dynamics in Vertebrate Embryonic Development. Dev Cell. 2015;35:383–394. doi: 10.1016/j.devcel.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollet N, Mazabraud A. Insights from Xenopus genomes. Genome Dyn. 2006;2:138–153. doi: 10.1159/000095101. [DOI] [PubMed] [Google Scholar]
- Pratt KG, Khakhalin AS. Modeling human neurodevelopmental disorders in the Xenopus tadpole: from mechanisms to therapeutic targets. Disease Models & Mechanisms. 2013;6:1057–1065. doi: 10.1242/dmm.012138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SA, Kormish J, Kofron M, Jegga A, Zorn AM. A gene regulatory network controlling hhex transcription in the anterior endoderm of the organizer. Dev Biol. 2011;351:297–310. doi: 10.1016/j.ydbio.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SA, Thi Tran H, Wlizla M, Mancini P, Shifley ET, Bloor SD, Han L, Vleminckx K, Wert SE, Zorn AM. A Molecular atlas of Xenopus respiratory system development. Dev Dyn. 2015;244:69–85. doi: 10.1002/dvdy.24180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzan W, Falco R, Salanga C, Salanga M, Horb ME. Generation of a Xenopus laevis F1 albino J strain by genome editing and oocyte host-transfer. Dev Biol. 2016:1–6. doi: 10.1016/j.ydbio.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe BA, Ma DP, Wilson RK, Wong JF. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J Biol Chem. 1985;260:9759–9774. [PubMed] [Google Scholar]
- Sakane Y, Sakuma T, Kashiwagi K, Kashiwagi A, Yamamoto T, Suzuki K-IT. Targeted mutagenesis of multiple and paralogous genes in Xenopus laevisusing two pairs of transcription activator-like effector nucleases. Dev Growth Differ. 2013;56:108–114. doi: 10.1111/dgd.12105. [DOI] [PubMed] [Google Scholar]
- Salanga MC, Horb ME. Xenopus as a Model for GI/Pancreas Disease. Curr Pathobiol Rep. 2015;3:137–145. doi: 10.1007/s40139-015-0076-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt SM, Gull M, Brändli AW. Advanced Drug Delivery Reviews. Advanced Drug Delivery Reviews. 2014;69–70:225–246. doi: 10.1016/j.addr.2014.02.004. [DOI] [PubMed] [Google Scholar]
- Sellick GS, Barker KT, Stolte-Dijkstra I, Fleischmann C, Coleman RJ, Garrett C, Gloyn AL, Edghill EL, Hattersley AT, Wellauer PK, Goodwin G, Houlston RS. Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat Genet. 2004;36:1301–1305. doi: 10.1038/ng1475. [DOI] [PubMed] [Google Scholar]
- Shi Z, Wang F, Cui Y, Liu Z, Guo X, Zhang Y, Deng Y, Zhao H, Chen Y. Heritable CRISPR/Cas9-mediated targeted integration in Xenopus tropicalis. The FASEB Journal. 2015;29:4914–4923. doi: 10.1096/fj.15-273425. [DOI] [PubMed] [Google Scholar]
- Showell C, Carruthers S, Hall A, Pardo-Manuel de Villena F, Stemple D, Conlon FL. A comparative survey of the frequency and distribution of polymorphism in the genome of Xenopus tropicalis. PLoS ONE. 2011;6:e22392. doi: 10.1371/journal.pone.0022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showell C, Conlon FL. Decoding development in Xenopus tropicalis. Genesis. 2007;45:418–426. doi: 10.1002/dvg.20286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack JMW, Lin G, Chen Y. The Xenopus tadpole: a new model for regeneration research. Cell Mol Life Sci. 2008;65:54–63. doi: 10.1007/s00018-007-7431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojka S, Amin NM, Gibbs D, Christine KS, Charpentier MS, Conlon FL. Congenital heart disease protein 5 associates with CASZ1 to maintain myocardial tissue integrity. Development. 2014;141:3040–3049. doi: 10.1242/dev.106518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K-IT, Isoyama Y, Kashiwagi K, Sakuma T, Ochiai H, Sakamoto N, Furuno N, Kashiwagi A, Yamamoto T. High efficiency TALENs enable F0 functional analysis by targeted gene disruption in Xenopus laevis embryos. Biol Open. 2013;2:448–452. doi: 10.1242/bio.20133855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi C, Sakamaki K, Morita H, Hara Y, Suzuki M, Kinoshita N, Ueno N. Transgenic Xenopus laevis for live imaging in cell and developmental biology. Dev Growth Differ. 2013;55:422–433. doi: 10.1111/dgd.12042. [DOI] [PubMed] [Google Scholar]
- Tochinai S, Katagiri C. Complete abrogation of immune response to skin allografts and rabbit erythrocytes in the early thymectomized Xenopus. Dev Growth Differ. 1975;17:383–394. doi: 10.1111/j.1440-169X.1975.00383.x. [DOI] [PubMed] [Google Scholar]
- Tymowska J. Polyploidy and cytogenetic variation in frogs of the genus Xenopus. In: Green DS, Session SK, editors. Amphibian Cytogenetics and Evolution. 1991. pp. 259–297. [Google Scholar]
- Uno Y, Nishida C, Takagi C, Ueno N, Matsuda Y. Homoeologous chromosomes of Xenopus laevis are highly conserved after whole-genome duplication. Heredity (Edinb) 2013;111:430–436. doi: 10.1038/hdy.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nieuwenhuysen T, Naert T, Tran HT, Van Imschoot G, Geurs S, Sanders E, Creytens D, Van Roy F, Vleminckx K. TALEN-mediated apc mutation in Xenopus tropicalis phenocopies familial adenomatous polyposis. Oncoscience. 2015;2:555–566. doi: 10.18632/oncoscience.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallmeier J, Al-Mutairi DA, Chen C-T, Loges NT, Pennekamp P, Menchen T, Ma L, Shamseldin HE, Olbrich H, Dougherty GW, Werner C, Alsabah BH, Köhler G, Jaspers M, Boon M, Griese M, Schmitt-Grohé S, Zimmermann T, Koerner-Rettberg C, Horak E, Kintner C, Alkuraya FS, Omran H. Mutations in CCNO result in congenital mucociliary clearance disorder with reduced generation of multiple motile cilia. Nature Publishing Group. 2014;46:646–651. doi: 10.1038/ng.2961. [DOI] [PubMed] [Google Scholar]
- Wang F, Shi Z, Cui Y, Guo X, Shi Y-B, Chen Y. Targeted gene disruption in Xenopus laevis using CRISPR/Cas9. Cell Biosci. 2015;5:15. doi: 10.1186/s13578-015-0006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Fu L, Guo X, Chen Y, Shi Y-B. Histone methyltransferase Dot1L plays a role in postembryonic development in Xenopus tropicalis. The FASEB Journal. 2015;29:385–393. doi: 10.1096/fj.14-252171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Shi Y-B. Unliganded thyroid hormone receptor α controls developmental timing in Xenopus tropicalis. Endocrinology. 2015;156:721–734. doi: 10.1210/en.2014-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womble M, Pickett M, Nascone-Yoder N. Frogs as integrative models for understanding digestive organ development and evolution. Semin Cell Dev Biol. 2016:1–14. doi: 10.1016/j.semcdb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Nuñez JK, Doudna JA. Biology and Applications of CRISPR Systems: Harnessing Nature's Toolbox for Genome Engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- Wühr M, Freeman RM, Jr, Presler M, Horb ME, Peshkin L, Gygi SP, Kirschner MW. Deep Proteomics of the Xenopus laevis Egg using an mRNA-Derived Reference Database. Curr Biol. 2014;24:1467–1475. doi: 10.1016/j.cub.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie LA, Hardwick LJA, Papkovskaia TD, Thiele CJ, Philpott A. Ascl1 phosphostatus regulates neuronal differentiation in a Xenopus developmental model of neuroblastoma. Disease Models & Mechanisms. 2015;8:429–441. doi: 10.1242/dmm.018630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev Cell. 2011;20:483–496. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Aguero T, King ML. The Xenopus Maternal-to-Zygotic Transition from the Perspective of the Germline. Curr. Top. Dev. Biol. 2015;113:271–303. doi: 10.1016/bs.ctdb.2015.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau DA, Johnson Hamlet MR, Kuliyev E, Zhu H, Doherty JR, Archer TD, Subhawong AP, Valentine MB, Kelley CM, Mead PE. Transgenesis in Xenopus using the Sleeping Beauty transposon system. Dev Dyn. 2009;238:1727–1743. doi: 10.1002/dvdy.21994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Guo X, Chen Y. Retinoic acid-activated Ndrg1a represses Wnt/β-catenin signaling to allow Xenopus pancreas, oesophagus, stomach, and duodenum specification. PLoS ONE. 2013;8:e65058. doi: 10.1371/journal.pone.0065058. [DOI] [PMC free article] [PubMed] [Google Scholar]