ABSTRACT
Proteins of the LCP (LytR, CpsA, Psr) family have been shown to inherit important roles in bacterial cell wall biosynthesis. However, their exact function in the formation of the complex cell wall structures of the Corynebacteriales, including the prominent pathogens Mycobacterium tuberculosis and Corynebacterium diphtheriae, remains unclear. Here, we analyzed the role of the LCP proteins LcpA and LcpB of Corynebacterium glutamicum, both of which localize at regions of nascent cell wall biosynthesis. A strain lacking lcpB did not show any growth-related or morphological phenotype under the tested conditions. In contrast, conditional silencing of the essential lcpA gene resulted in severe growth defects and drastic morphological changes. Compared to the wild-type cell wall, the cell wall of this mutant contained significantly less mycolic acids and a reduced amount of arabinogalactan. In particular, rhamnose, a specific sugar component of the linker that connects arabinogalactan and peptidoglycan, was decreased. Complementation studies of the lcpA-silencing strain with several mutated and truncated LcpA variants suggested that both periplasmic domains are essential for function whereas the cytoplasmic N-terminal part is dispensable. Successful complementation experiments with proteins of M. tuberculosis and C. diphtheriae revealed a conserved function of LCP proteins in these species. Finally, pyrophosphatase activity of LcpA was shown in an in vitro assay. Taken together, our results suggest that LCP proteins are responsible for the transfer of arabinogalactan onto peptidoglycan in actinobacterial species and support a crucial function of a so-far-uncharacterized C-terminal domain (LytR_C domain) which is frequently found at the C terminus of the LCP domain in this prokaryotic phylum.
IMPORTANCE About one-third of the world's population is infected with Mycobacterium tuberculosis, and multiple-antibiotic resistance provokes the demand for novel antibiotics. The special cell wall architecture of Corynebacteriales is critical for treatments because it is either a direct target or a barrier that the drug has to cross. Here, we present the analysis of LcpA and LcpB of the closely related Corynebacterium glutamicum, the first of which is an essential protein involved in cell wall biogenesis. Our work provides a comprehensive characterization of the impact of LCP proteins on cell wall biogenesis in this medically and biotechnologically important class of bacteria. Special focus is set on the two periplasmic LcpA domains and their contributions to physiological function.
INTRODUCTION
The LCP protein family was named after its first three described representatives, LytR (1), CpsA (2), and Psr (3), and comprises a class of transmembrane proteins that share a characteristic extracellular domain, the LCP domain (InterPro no. IPR004474 [4]). A phylogenetic analysis revealed that LCP proteins are present in all Gram-positive bacteria whereas they are not found in most Gram-negative bacteria (5). Often, bacterial genomes encode several (up to 11) LCP proteins, which all follow a common structural organization consisting of a short N-terminal cytoplasmic domain, a transmembrane fragment with 1 to 3 transmembrane helices, and a putative extracellular part carrying the LCP domain (5). In LCP proteins of actinobacteria, the LCP domain is often accompanied by a C-terminal domain (LytR_C domain; InterPro no. IPR027381) of unknown function.
The LytR protein of Bacillus subtilis was originally thought to act as a cell wall-related attenuator, because disruption of this gene led to increased transcription of adjacent genes (1). However, recent results suggested that these observations were secondary effects due to severe alterations of cell wall biogenesis (6). Kawai et al. provided several pieces of evidence that LCP proteins are enzymes required for the transfer of anionic cell wall polymers from their membrane-linked precursors onto cell wall peptidoglycan (6). With B. subtilis, LCP proteins were isolated in a search for proteins that interacted with MreB, a prokaryotic actin homolog (6). Another study showed that in Staphylococcus aureus LCP proteins are required for the transfer of capsular polysaccharides onto the glycan strands of peptidoglycan (7). A strain of this bacterium lacking all three LCP proteins was found to release type 5 capsular polysaccharides and wall teichoic acids into the medium (7, 8). A recent study of Actinomyces oris suggested that an LCP protein is involved in protein glycosylation, namely, the transfer of glycan from a polyprenol lipid-linked glycan to GspA (glycosylated surface-linked protein A) prior to cell wall anchoring (9). Overall, these data point to an involvement of LCP proteins in the transfer of saccharide compounds from a lipid-polyprenol carrier to peptidoglycan or proteins.
Most investigations of LCP proteins published to date were performed in Firmicutes. Furthermore, no study so far has addressed the function of the LytR_C domain that is commonly found in actinobacterial LCP proteins. One highly interesting order in this phylum is represented by the Corynebacteriales, including prominent human pathogens such as Mycobacterium tuberculosis and Corynebacterium diphtheriae. The diseases caused by these bacteria can be difficult to treat, especially in the case of tuberculosis, because the bacteria harbor a cell wall with a very special architecture that makes them resistant against standard antibiotic treatments (10). In the cell walls of these species, the peptidoglycan is linked to arabinogalactan, which is itself esterified with mycolic acids, forming a large mycolyl-arabinogalactan-peptidoglycan complex (11). Furthermore, lipomannan and lipoarabinomannan (LAM) are present (11). The mycolic acids form a second lipid bilayer which constitutes a diffusion barrier similar to the outer membrane of Gram-negative bacteria (11) (for a recent review on the structure and assembly of the mycobacterial cell wall, see reference 12). As corynebacteria and mycobacteria have no teichoic acids within their cell walls and only mycobacteria harbor capsule-like structures (13), the function of LCP proteins in these species and their relation to cell wall biosynthesis remains unclear. A recent study revealed that the LCP protein CpsA of Mycobacterium marinum plays an important role in cell wall assembly and that it is essential for proliferation in macrophages and virulence in zebrafish (14). Corynebacterium glutamicum is a well-established model species for cell wall-related studies in the Corynebacteriales because it shares the complex cell envelope organization with its pathogenic relatives (15–17). Furthermore, C. glutamicum is one of the main species used in the biotechnological industry, especially for the production of amino acids (18). The transport of products across the complex cell wall can become an issue in the development of biotechnological production strains. The genome of C. glutamicum carries two LCP protein genes, one of which, the cg3210 gene, was recently found to be a target of IpsA, a regulator of cell wall biosynthesis (15). In this study, we address the general importance of the two LCP proteins in C. glutamicum for cell wall biogenesis, with a special focus on the impact of the LytR_C domain.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. The C. glutamicum type strain ATCC 13032 was used as the wild type. For growth experiments with the promoter exchange strains, as first preculture, 5 ml brain heart infusion broth (BHI; Difco Laboratories, Detroit, MI) supplemented with 100 mM sodium gluconate (for strain SilG-lcpA) was inoculated with a single colony from a fresh agar plate and incubated overnight, with shaking, at 30°C. The cells were washed once in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4; pH 7.3) and used to inoculate a second preculture, consisting of 750 μl CGXII minimal medium (19) supplemented with 3,4-dihydroxybenzoate (30 mg/liter) as an iron chelator and glucose, sodium gluconate, myo-inositol, or a mixture thereof as carbon source, to an optical density at 600 nm (OD600) of 0.5. After 24 h of cultivation at 30°C and 1,200 rpm in a BioLector system (m2p-labs, Baesweiler, Germany) in 48-well FlowerPlates, 15-μl aliquots of the cultures were used to inoculate the main cultures in another FlowerPlate containing the same medium as the second preculture. All cloning was performed in Escherichia coli DH5α cells cultivated at 37°C in lysogeny broth (LB) (20) with 50 μg/ml kanamycin. E. coli BL21(DE3) was used for protein production.
Recombinant DNA work and construction of deletion mutants.
Routine methods, such as PCR, DNA restriction, and ligation, were performed using standard protocols (20–22). The oligonucleotides used in this study were obtained from Eurofins Genomics (Ebersberg, Germany) and are listed in Table S2 in the supplemental material. Gibson assembly was performed according to methods described elsewhere (23). All plasmids were sequenced to exclude unwanted mutations (Eurofins Genomics, Ebersberg, Germany). The ΔlcpB mutant of C. glutamicum and the strains with the exchanged promoter in front of lcpA were constructed via a two-step homologous recombination protocol described previously (24). For further details regarding plasmid and mutant construction, see Text S1 in the supplemental material.
Conditional gene silencing system for C. glutamicum.
The elucidation of the function of essential genes is often challenging, as no deletion mutant can be obtained which allows the analysis of a phenotype and might give hints toward the function of the respective genes. For these cases, gene silencing systems can be used to study the effects of reduced levels of the respective genes. For other organisms, there are well-established systems, such as the TetR-based approach (25). For C. glutamicum, this system did not appear to be efficient enough in earlier studies. Therefore, we used two well-characterized, native promoters of C. glutamicum for the silencing approach. The first was the promoter of the gluconate kinase GntK, which is activated in the presence of gluconate (26). The second was the promoter of the inositol phosphate synthase Ino1, which is repressed in the presence of myo-inositol (15). Both promoters turned out to be very efficient in downregulating our gene of interest. From transcriptome sequencing (RNA-seq) data, we estimate that about 2/3 of all C. glutamicum genes have a higher expression level than the normal lcpA expression level and about 90% of all C. glutamicum genes have an apparently higher expression level than the silenced lcpA expression level. This suggests that the majority of C. glutamicum genes can potentially be silenced with the system described here. Besides studying essential genes in C. glutamicum, the promoters might also prove to be useful tools for metabolic engineering approaches.
Fluorescence microscopy and colocalization studies.
For recording fluorescence microscopy with staining, the cells were centrifuged and resuspended in PBS containing 100 ng/ml Hoechst 33342 and 300 ng/ml Nile red stains (Sigma-Aldrich, Munich, Germany). After 5 to 10 min of incubation at room temperature, the cells were analyzed on agarose pads using a Zeiss Axio Imager M2 microscope that was equipped with an AxioCam MRm camera and a Plan-Apochromat 100×, 1.40-numerical aperture (NA) phase-contrast oil immersion objective. Cells with fluorescent proteins were imaged directly without any staining. For this purpose, a colony from a fresh agar plate was used to inoculate a preculture of 5 ml BHI with kanamycin (25 µg/ml), which was then cultivated with shaking at 30°C overnight. On the following day, 100 μl of this culture was used to inoculate a main culture consisting of 5 ml CGXII medium with 2% (wt/vol) glucose, kanamycin, and 10 μM isopropyl β-d-1-thiogalactopyranoside (IPTG). Digital images were acquired and analyzed with AxioVision 4.8 software (Zeiss, Göttingen, Germany) at 4 or 24 h after inoculation of the main culture. Functionality of the Venus-lcpA fusion protein was proven by complementation studies with the SilG-lcpA strain.
For colocalization experiments, C. glutamicum ATCC 13032 cells carrying plasmids pAN6-venus-lcpA or pAN6-eyfp-lcpB were grown as described above. After 4 h (OD600 of 1), aliquots of 200 μl were transferred for bioorthogonal labeling (27) into the wells of a microtiter plate and incubated for 5 min at 30°C and 850 rpm. One microliter of azido–d-alanine or d-alanine (1 M each) was added, and the cells were shaken for 3 min. The cells were harvested (5,000 × g, 25°C, 2 min), washed with PBS, and resuspended in 200 μl PBS. After addition of 1 μl of dibenzylcylcooctyne-PEG4-ATTO425 (1 mM in dimethyl sulfoxide [DMSO]; DBCO-PEG4-Atto425; synthesized by Jena Bioscience, Jena, Germany), cells were incubated at 30°C and 80 rpm for 20 min in the dark. Cells were harvested, washed three times with PBS containing 0.1% Tween 20, and resuspended in PBS. Cells were analyzed on agarose pads using a Zeiss Axio Imager M1 microscope equipped with an AxioCam HRm camera and a Plan-NeoFluar 100×, 1.30-NA phase-contrast oil immersion objective. Digital images were analyzed with AxioVision 4.6 software (Zeiss, Göttingen, Germany).
RT-qPCR.
For reverse transcriptase quantitative PCR (RT-qPCR), C. glutamicum wild type and the promoter mutants were in principle cultivated as described above, but the cultivation volumes were increased to 20 ml (precultures) or 50 ml (main cultures). The cells were cultivated in baffled shake flasks (100-ml and 500-ml volumes) at 30°C and 130 rpm. Aliquots of 25 ml of cells of the main cultures were harvested by centrifugation (4,120 × g, 10 min, 4°C) at an OD600 of 5 using precooled 50 ml Falcon tubes filled with 25 g crushed ice. The cell pellets were subsequently frozen in liquid nitrogen and stored at −70°C. The preparation of total RNA was performed using the RNeasy kit from Qiagen (Hilden, Germany) with DNA on column digest. To remove any residual DNA, an additional DNase treatment was performed as follows: 5 to 10 μg RNA from the kit purification was mixed with 5 μl RDD buffer and 8 μl (20 U) DNase (Qiagen, Hilden, Germany) in a total volume of 50 μl and incubated at 30°C for 30 min followed by 70°C for 10 min to inactivate the DNase. Afterwards, the RNA was purified by phenol-chloroform extraction and resuspended in 15 μl RNase-free H2O. RNA integrity was assessed using gel electrophoresis. For reverse transcription, 1 μg RNA in a total volume of 10 μl was mixed with 1 μl random primers (0.5 μg/μl; Life Technologies, Darmstadt, Germany) and incubated at 70°C for 10 min. The samples were immediately cooled on ice and then mixed with the following solutions: 4 μl first-strand buffer (5×), 2 μl 1,4-dithiothreitol (DTT; 0.1 M), 1 μl deoxynucleoside triphosphates (10 mM each), 2 μl SuperScript III reverse transcriptase (200 U/μl; Life Technologies, Darmstadt, Germany). For each RNA sample, a no-amplification control (NAC) was prepared that contained all the above-mentioned substances except for the reverse transcriptase. The samples were incubated at 42°C for 50 min, followed by incubation at 70°C for 15 min to inactivate the reverse transcriptase. RNA was removed from the samples by addition of 1 μl RNase H (5 U/μl; NEB, Frankfurt, Germany) and incubation at 37°C for 20 min. The samples were mixed with 179 μl H2O to reach a concentration of about 5 ng/μl cDNA for the qPCR. qPCR was performed in a qTOWER real-time PCR cycler (Analytic Jena, Jena, Germany) using innuMix qPCR master mix SyGreen (Analytic Jena, Jena, Germany). The 20-μl reaction mixtures contained 10 μl master mix, 0.5 μM each primer, and 2 μl cDNA. Measurements were performed with three biological and two technical replicates each. For further details regarding quality controls, referencing, and the PCR protocol, please refer to Text S1 in the supplemental material.
Transmission electron microscopy.
In preparation for transmission electron microscopy (TEM) analysis, bacteria were fixed with 3% (vol/vol) glutaraldehyde (Agar Scientific, Wetzlar, Germany) in PBS for at least 4 h, washed in 0.1 M Soerensen's phosphate buffer (Merck, Darmstadt, Germany), and postfixed in 1% OsO4 in 17% sucrose buffer. After fixation, bacteria were embedded in 2.5% agarose (Sigma, Steinheim, Germany), then rinsed in 17% sucrose buffer and deionized water, and dehydrated via an ethanol series (30%, 50%, 70%, 90%, 100%) for 10 min each and the last step thrice. The dehydrated specimens were incubated in propylene oxide (Serva, Heidelberg, Germany) for 30 min in a mixture of Epon resin (Sigma) and propylene oxide (1:1) for 1 h and finally in pure Epon for 1 h. Samples were embedded in pure Epon. Epon polymerization was performed at 37°C for 12 h and then at 80°C for 48 h. Ultrathin sections (70 to 100 nm) were cut with a diamond knife and picked up on Cu/Rh grids. Negative staining by uranyl acetate and lead citrate (all from EMS, Munich, Germany) was performed to enhance TEM contrast. The specimens were viewed using a Philips EM400T electron microscope, operated at 60 kV.
Scanning electron microscopy.
For scanning electron microscopy (SEM), bacteria were fixed with 3% (vol/vol) glutaraldehyde (Agar Scientific, Wetzlar, Germany) in PBS for at least 4 h, washed in 0.1 M Soerensen's phosphate buffer (Merck, Darmstadt, Germany) for 15 min, and dehydrated by incubating consecutively in an ascending acetone series (30%, 50%, 70%, 90%, and 100%) for 10 min each and the last step thrice. The samples were critical point dried in liquid CO2 and then sputter coated (Sputter Coater EM SCD500; Leica, Wetzlar, Germany) with a 10-nm gold/palladium layer. Samples were analyzed using an environmental scanning electron microscope (ESEM XL 30 FEG, FEI, Philips, Eindhoven, Netherlands) with a 10-kV acceleration voltage in a high-vacuum environment.
Cell wall preparations.
Cell walls were essentially prepared as described previously (28). In brief, cells were cultivated as described for the isolation of material from culture supernatants (see Text S1 in the supplemental material), harvested at an OD600 of 15 to 20, frozen, and lyophilized. Crude cell walls were prepared as follows. Cells were mixed with glass beads in distilled water and disrupted using a Fast Prep 24 apparatus (MP Biomedicals, Eschwege, Germany). Cell disruption was controlled by phase-contrast microscopy. Crude cell walls were obtained by centrifugation at 48,000 × g for 20 min at 4°C.
SDS-purified cell walls were prepared as described previously (28, 29). Crude cell walls were resuspended in double-distilled water and added dropwise to the same volume of a stirring, boiling 8% (wt/vol) SDS solution. After boiling for a further 30 min, the solution was allowed to cool to room temperature and centrifuged at 48,000 × g for 20 min at 20 to 25°C. To get rid of any residual SDS, the sediment was washed five times with double-distilled water. SDS-purified cell walls were further purified by protease treatment.
To remove covalently bound protein, 1 g of SDS-purified cell walls was resuspended in 10 mM Tris-HCl (pH 7.5); 10 ml proteinase K (ProtK; final concentration, 200 μg/ml) and NaN3 (final concentration, 0.2 mM) were added, and the mixture was incubated with shaking (100 to 130 rpm) overnight at 37°C. SDS-purified, ProtK-treated cell walls were obtained by centrifugation (48,000 × g for 20 min), washed three times with double-distilled water, and lyophilized.
Analysis of cell wall components.
The extraction of arabinogalactan was performed essentially as described previously (30): 50 mg of SDS-purified, ProtK-treated cell walls was mixed in a centrifuge tube (16 by 100 mm) with a Teflon cap, using a small magnetic stir bar and 5 ml of 50 mM H2SO4, and left stirring for 4 days at 37°C and 850 rpm. Afterwards, the suspension was transferred to 15-ml Falcon tubes and centrifuged for 15 min at 3,000 × g and 25°C. The supernatant was transferred to a new tube, mixed with a few granules of Ba(OH)2, and incubated for 1 h at room temperature. After 15 min of centrifugation at 3,000 × g and 25°C, the supernatant was transferred to a new tube and dried in a Speed-Vac (Savant, Farmingdale, NY), representing arabinogalactan. The pellet from the first centrifugation was washed two times with double-distilled water and subsequently lyophilized (representing arabinogalactan-extracted cell walls).
For sugar analysis, arabinogalactan was hydrolyzed with 2 M HCl for 3 h at 100°C (31) and analyzed by thin-layer chromatography (TLC). A standard containing free sugars was treated in the same way. Hydrolysates of 0.2 mg SDS-purified, ProtK-treated cell walls and 20 nmol of sugar (standard) were applied per spot. TLC was carried out on silica 60 plates (Merck, Darmstadt, Germany) according to methods described elsewhere (32). In brief, plates were run in acetonitrile:water (85:15, vol/vol) twice and developed with diphenylamine/aniline. Spots were quantified with a ChemiDoc analyzer (Bio-Rad, Munich, Germany). To elucidate the efficiency of the arabinogalactan extraction, arabinogalactan-extracted cell walls were hydrolyzed (2 M HCl, 16 h, 100°C), and the hydrolysate was analyzed by TLC.
Preparation and detection of mycolic acid methyl esters.
Preparation and detection of mycolic acid methyl esters was essentially performed as described previously (33). In brief, SDS-purified, ProtK-treated cell walls were subjected to acidic methanolysis; 10 mg of SDS-purified, ProtK-treated cell walls was incubated with 600 μl methanol-toluene-sulfuric acid (30:15:1, vol/vol/vol) for 16 h at 50°C. After cooling to room temperature, 400 μl of petroleum ether (60 to 80°C) was added and the mixture was shaken vigorously. After phase separation, mycolic acid methyl esters were released into the petroleum ether phase, dried under a stream of nitrogen, and analyzed by TLC. This was performed on silica 60 plates (Merck, Darmstadt, Germany) in toluene-acetone (97:3, vol/vol) developed with phosphomolybdic acid (3% in isopropanol). Ammonium vapor was used to destain the yellow background.
Pyrophosphatase assay.
For the pyrophosphatase activity assay, E. coli BL21(DE3) was transformed with pET-TEV-lcpAΔTM, pET-TEV-lcpAΔTM-D88A, pET-TEV-lcpAΔTM-R138A, and pET-TEV-lcpAΔTMR257A, and also pET-TEV-cgtR10 (negative control). A colony from a fresh agar plate was used to inoculate a preculture of 50 ml LB medium with kanamycin and cultivated overnight at 37°C and 120 rpm. A 500-ml volume of LB medium with kanamycin was inoculated with 10 ml from the preculture and cultivated at 37°C and 100 rpm until an OD600 of ∼0.5 was reached. Protein production was induced with 100 μM IPTG, and the cells were further cultivated at 16°C until the next morning. The pellet from a 250-ml culture was resuspended in TNI20 buffer (100 mM Tris-HCl [pH 8.0], 500 mM NaCl, 20 mM imidazole) with Complete, Mini, EDTA-free protease inhibitor (Roche, Basel, Switzerland), and cells were broken by passing three times through a French press. The cell extract was centrifuged, and the supernatant was used for Ni-nitrilotriacetic acid affinity chromatography. The protein was eluted with TNI400 (TNI20 with 400 mM imidazole). The buffer was exchanged to GF buffer (20 mM Tris-HCl [pH 8.0], 200 mM NaCl) using PD10 columns (GE Life Sciences, Munich, Germany), and the proteins were concentrated to at least 5 mg/ml with Amicon Ultra centrifugal filters (Merck Millipore, Cork, Ireland). Gel filtration was performed at a flow rate of 0.5 ml/min in GF buffer using a Superdex 200 Increase 10/300 column connected to an Äkta Pure25 system. For the gel filtration under reducing conditions, 5 mM DTT was added to the protein sample, which was then incubated for 1 h on ice, prior to the gel filtration using GF buffer supplemented with 1 mM DTT. The molecular mass of the protein was estimated by comparison with standard proteins of known molecular masses.
For the pyrophosphatase assay, 0.18 mg protein was mixed with 1 mM geranyl-pyrophosphate (GPP) and either 10 mM MgCl2 or 5 mM EDTA in 20 mM Tris-HCl (pH 8.0; 100 μl, final volume). The reaction mixtures were incubated overnight at 30°C. Controls lacking either protein or substrate or both were also included. For analysis, the samples were diluted 1:5 with water and filtered through Amicon Ultra centrifugal filters to remove the protein.
Liquid chromatography-mass spectrometry (LC-MS) analysis was performed using an ultrahigh-performance LC (uHPLC) 1290 Infinity system coupled to a 6130 quadrupole LC-MS system (Agilent, Santa Clara, CA). LC separation was carried out with a Kinetex 1.7 μm C18 100-Å-pore-size column (50 mm by 2.1 mm [internal diameter]; Phenomenex, Torrance, CA) at 50°C. For elution, 0.1% acetic acid (solvent A) and acetonitrile supplemented with 0.1% acetic acid (solvent B) were applied as the mobile phases at a flow rate of 0.5 ml/min. A gradient was used, where solvent B was increased stepwise: minute 0 to 6, 10% to 30%; minute 6 to 7, 30% to 50%; minute 7 to 8, 50% to 100%; minute 8 to 8.5, 100% to 10%. The mass spectrometer was operated in negative electrospray ionization (ESI) mode, and data acquisition was performed in selected ion-monitoring mode with m/z 233 corresponding to [M − H]− of geranyl monophosphate. An authentic geranyl monophosphate standard was obtained from Sigma-Aldrich (Munich, Germany).
RESULTS
Domain structure and conservation of C. glutamicum LCP proteins.
The genome of C. glutamicum strain ATCC 13032 contains two genes encoding LCP proteins with different domain organizations. Here, these proteins are designated LcpA (Cg0847/NCgl0708) and LcpB (Cg3210/NCgl2802).
The gene lcpA is located in close proximity to genes involved in cell wall biosynthesis, such as wbbL1 (cg0848; encoding rhamnosyltransferase) and rmlA2 (cg0849; encoding GDP-mannose pyrophosphorylase) (Fig. 1A). This organization is conserved among corynebacteria and mycobacteria (Fig. 1A). The coding region of lcpB is flanked upstream by the gene cg3209, which encodes a metal-dependent membrane protease, the amidotransferase GatA; pheA, which encodes a prephenate dehydratase involved in the biosynthesis of aromatic amino acids; and phoE, which encodes a phosphoglycerate mutase (see Fig. S1A in the supplemental material). The downstream region of lcpB contains several genes for uncharacterized hypothetical proteins and is not conserved.
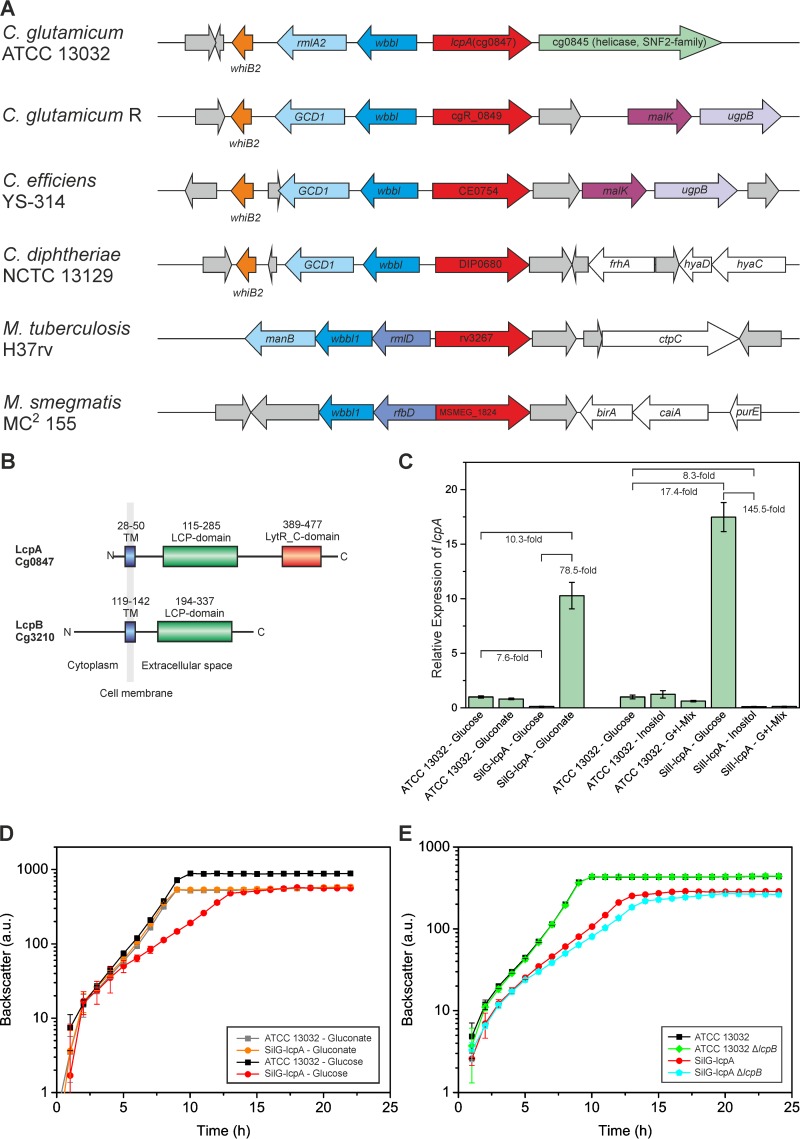
FIG 1.
Overview of LCP proteins in C. glutamicum and their impact on cellular growth. (A) Phylogenetic conservation of lcpA among related species (red arrows). Neighboring conserved genes are colored accordingly. White indicates other, nonconserved proteins of known function, and gray represents hypothetical or uncharacterized proteins. Data were obtained from reference 45. The corresponding figure for lcpB is presented in Fig. S1A in the supplemental material. (B) Domain organization of LcpA and LcpB. (C) Silencing of lcpA expression. Given are relative expression levels of lcpA in different strains and on different media, determined by RT-qPCR. G+I-Mix is a carbon source mixture of 1% (wt/vol) glucose and 1% (wt/vol) myo-inositol (final concentrations). The averages and standard deviations of results for three biological replicates with two technical replicates each are presented. (D) Growth of the lcpA-silencing strain SilG-lcpA on glucose or gluconate as carbon source in comparison to the wild type. Cells were cultivated in complex media with gluconate followed by two consecutive cultivations in CGXII minimal media with the indicated carbon source. The averages and the standard deviations of results for three biological replicates of the second CGXII culture are presented. (E) Impact of lcpA silencing and/or lcpB deletion on growth of C. glutamicum. The same cultivation procedure was used as that described for panel D, with glucose as carbon source.
LcpA consists of a short N-terminal cytoplasmic region followed by one transmembrane helix (Fig. 1B). Besides the characteristic LCP domain (InterPro no. IPR004474), it contains an additional uncharacterized C-terminal domain of unknown function (InterPro no. IPR027381; LytR_C domain) which is often found in combination with the LCP domain. LcpB does not contain a LytR_C domain, but it has an unusually long cytoplasmic region of 118 amino acids that is very rich in proline, arginine, and glutamine, resulting in a positive net charge (Fig. 1B). This cytoplasmic region seems to be unique for just a few corynebacteria. The obvious differences in domain structure suggest that these two proteins have distinct functions in C. glutamicum. In M. tuberculosis the situation is even more complex, as the genome encodes a total of 4 proteins with an LCP domain, three of which also have a LytR_C domain (Rv3484, Rv3267, and Rv0822) and one of which has just the LCP domain (Rv3840) (see Fig. S1B in the supplemental material).
lcpA is an essential gene in C. glutamicum.
Previous studies in different organisms revealed that deletion of LCP genes—if possible—often leads to altered growth and/or morphology, indicating a potential role for these proteins in cell wall biogenesis. To elucidate whether a deletion of LCP proteins has an impact on C. glutamicum, deletion plasmids were constructed and gene deletion by double crossover was performed. Deletion mutants were readily obtained for lcpB, whereas deletion as well as inactivation of lcpA failed in several different methodological attempts. Therefore, the lcpA gene likely encodes an essential protein in C. glutamicum.
Development of a conditional gene-silencing system for C. glutamicum.
To enable the conditional downregulation of lcpA, a gene-silencing system for application in C. glutamicum was developed. The native lcp promoter was exchanged with (i) the promoter of the gluconate kinase gene (gntK; cg2732) (26), which is repressed in the absence of gluconate in the medium, and (ii) the promoter of the inositol phosphate synthase gene (ino1; cg3323) (15), which can be downregulated by the addition of myo-inositol. The relative expression levels of lcpA were measured in the resulting strains to determine the regulatory strength and the dynamic range of the two systems. The expression level of lcpA was very similar in the wild type when it was cultivated on either glucose or gluconate (Fig. 1C). Cultivation of the SilG-lcpA strain (Silencing strain with Gluconate switch for lcpA) on glucose led to a nearly 8-fold decrease in lcpA expression below that of the wild type on glucose. When the same strain was cultivated on gluconate, the expression of lcpA was increased about 10-fold above that in the wild type on glucose. This resulted in a dynamic range of this system of about 80-fold. For the SilI-lcpA strain (Silencing strain with Inositol switch for lcpA), similar results were obtained (Fig. 1C). Here, even a mixture of glucose and myo-inositol (1% [wt/vol] each) was sufficient for 8-fold downregulation of lcpA.
Characterization of strains with decreased LCP protein levels.
After proving the functionality of the gene-silencing system, the SilG-lcpA strain was used for investigating the effects of decreased LcpA levels. First of all, the growth of the silencing strain was compared with that of the wild type under different conditions. In CGXII medium with gluconate, SilG-lcpA grew comparable to the wild type (growth rates of 0.61 ± 0.01 h−1 and 0.61 ± 0.01 h−1 [means ± standard deviations]) (Fig. 1D). When gluconate was exchanged with glucose as carbon source, significantly reduced lcpA expression was achieved, resulting in a strongly decreased growth rate and a lower final backscatter (growth rates of 0.28 ± 0.02 h−1 for SilG-lcpA and 0.58 ± 0.02 h−1 for the wild type) (Fig. 1D). Colony counting revealed a decreased viability of SilG-lcpA compared to the wild type (see Fig. S2 in the supplemental material). No growth defect of the ΔlcpB mutant was observed on glucose minimal medium (Fig. 1E). However, when lcpB was deleted in the SilG-lcpA strain, an even more pronounced growth defect was observed (growth rate, 0.30 ± 0.01 h−1 for SilG-lcpA versus 0.26 ± 0.01 h−1 for SilG-lcpA ΔlcpB) (Fig. 1E). These data suggest nonidentical but possibly partially overlapping functions of LcpA and LcpB.
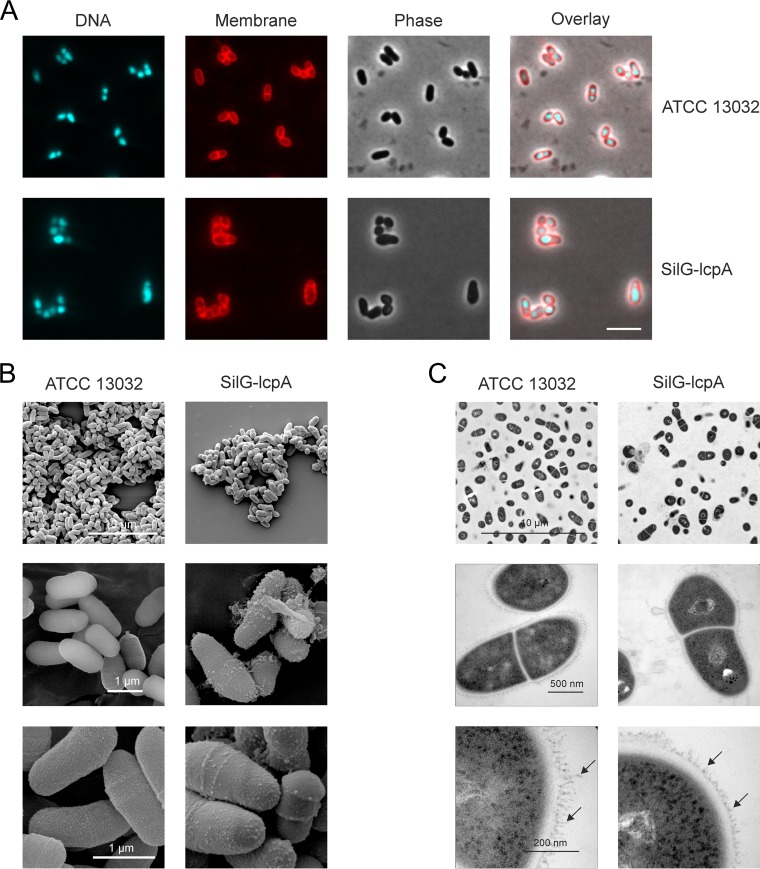
The cell morphology was analyzed by fluorescence microscopy. Whereas SilG-lcpA displayed wild-type morphology on gluconate-containing medium, it showed severely altered cell morphology after cultivation on glucose (Fig. 2A). The cells seemed to be swelling, losing their coryneform shape, and formed larger cell clusters. The morphology was further studied by SEM and TEM (Fig. 2B and C). In the SEM pictures, several bubbles were visible on the surface of the SilG-lcpA strain which were absent in the wild-type sample. Furthermore, it was apparent on several pictures that numerous cells were losing large pieces of their cell envelope. The TEM pictures also showed the more irregular cell shape of the mutant and the dissociation of cell wall material from the bacterial surface. Furthermore, fringe-like structures were visible on the surface of wild-type cells, possibly representing mycolic acids or arabinogalactan, which appeared to be much shorter in the mutant (Fig. 2C, lower panel, arrows).
FIG 2.
Microscopic pictures of C. glutamicum wild type (ATCC 13032) and the lcpA-silencing strain SilG-lcpA. (A) Fluorescence microscopy after the second cultivation (2 times 24 h) in CGXII minimal medium with glucose as carbon source with staining of DNA (Hoechst 33342) and membranes (Nile red). Scale bar, 5 μm. (B and C) SEM (B) and TEM (C) pictures at different magnifications (×2,500, ×20,000, and ×35,000 for SEM as well as ×2,800, ×46,000, and ×130,000 for TEM). Arrows point toward fringe-like structures that are much shortened in the SilG-lcpA strain.
LCP proteins of C. glutamicum are localized at regions of nascent cell wall biosynthesis.
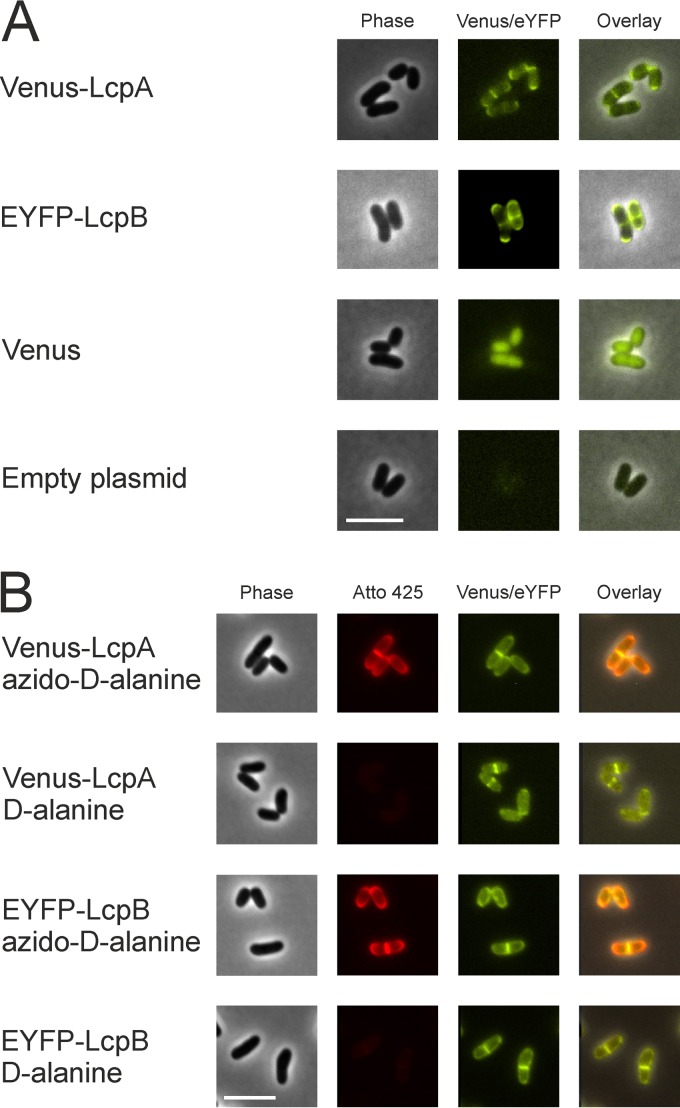
LCP proteins are known to contain at least one transmembrane helix, suggesting their location in the cytoplasmic membrane. In order to assess their localization within the cytoplasmic membrane, N-terminal translational fusions of LcpA and LcpB to fluorescent proteins (Venus or enhanced yellow fluorescent protein [EYFP]) were constructed and analyzed by fluorescence microscopy. Interestingly, both proteins were located in the cell membrane at regions of strong cell wall biosynthesis, such as the poles and the division plane (Fig. 3A). This was confirmed by labeling newly synthesized peptidoglycan using azido–d-alanine and DBCO-PEG4-Atto425 (Fig. 3B). As a control, this finding was compared to that with a homogenously distributed membrane protein, which only showed a slightly stronger fluorescence signal at the division plane due to the doubled amount of membrane in this region (see Fig. S3 in the supplemental material). In contrast, LcpA and LcpB were clearly enriched at cell poles and septa. Furthermore, the functionality of the Venus-LcpA fusion protein was tested by complementation studies in strain SilG-lcpA. The fusion protein complemented equally as well as the nonlabeled variant (see Fig. S4 in the supplemental material).
FIG 3.
Colocalization of Venus-LcpA and EYFP-LcpB with newly synthesized peptidoglycan in C. glutamicum ATCC 13032. The cells carrying a plasmid encoding Venus N-terminally fused to LcpA (pAN6-venus-lcpA) or EYFP N-terminally fused to LcpB (pAN6-eyfp-lcpB) were cultivated in CGXII minimal medium with 2% (wt/vol) glucose as carbon source and 10 μM IPTG. (A) Localization of Venus-LcpA and EYFP-LcpB. Strains with an empty plasmid and a plasmid harboring only Venus served as controls. (B) Colocalization of Venus-LcpA and EYFP-LcpB with newly synthesized peptidoglycan. After 4 h of cultivation, newly synthesized peptidoglycan was visualized by bioorthogonal labeling (click chemistry with azido–d-alanine and DBCO-PEG4-Atto425). Cells were either incubated for 3 min with azido–d-alanine or with d-alanine as control. For better contrast, fluorescence of Atto 425 (excitation at 436 nm, emission at 484 nm) is shown in red. Scale bars, 5 μm.
Impact of reduced lcpA expression on C. glutamicum cell wall composition.
As LcpA seemed to be involved in cell wall formation, we next analyzed putative changes of the different cell wall components, in order to gain further information regarding the function of this protein. Cell walls were isolated and analyzed as described in Material and Methods.
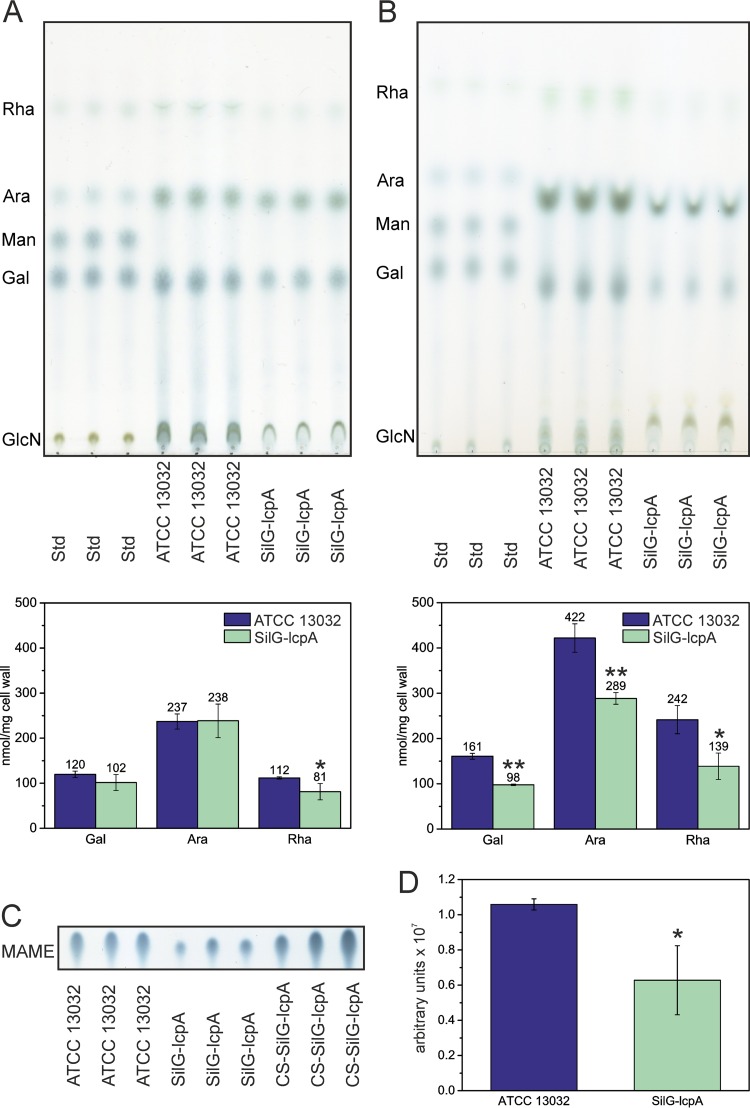
The two strains did not reveal any significant differences with respect to the peptidoglycan cross-linking and amidation (data not shown). We next isolated specifically the arabinogalactan component of the cell wall (Fig. 4A). Quantification of the isolated sugar components revealed that the arabinogalactan isolated from the SilG-lcpA mutant was significantly reduced in rhamnose but not in arabinose and galactose, thereby suggesting that LcpA likely acts in the enzymatic step linking the rhamnose linker of the nascent arabinogalactan to peptidoglycan. We also analyzed how much arabinogalactan remained in the extracted cell walls. Indeed, a large proportion of the arabinogalactan was not extracted, and a clear difference between the wild type and the SilG-lcpA mutant was observed. The mutant cell walls contained significantly less arabinogalactan than the wild type (Fig. 4B), indicating that the mutant cell walls had an overall decreased amount of arabinogalactan within their cell envelopes.
FIG 4.
LcpA is presumably involved in linking arabinogalactan and peptidoglycan. (A and B) Sugar content in arabinogalactan (A) and remaining sugar content in SDS-purified, ProtK-treated cell walls (B) after extraction of arabinogalactan. *, P < 0.05; **, P < 0.01 (two-sided t test compared to wild type). (C) Mycolic acids were liberated as mycolic acid methyl esters (MAME) from purified cell walls by acidic methanolysis. TLC was run in toluene:acetone (97:3, vol/vol) and developed with phosphomolybdic acid. CS-SilG-lcpA, material from the culture supernatant of SilG-lcpA. Results from three technical replicates of one sample each are presented. The full picture is provided in Fig. S5 in the supplemental material. (D) Relative amounts of MAMEs per milligram of cell wall. *, P < 0.05 (two-sided t test compared to wild type).
We next analyzed the mycolic acid contents of wild-type and mutant cells after liberating them as mycolic acid methyl esters (MAME) from purified cell walls. Consistently, we observed a reduced amount of mycolic acids in the SilG-lcpA mutant (Fig. 4C and D; see also Fig. S5 in the supplemental material). A reduced amount of mycolic acids is likely a direct consequence of the reduced arabinogalactan content in the SilG-lcpA mutant. In turn, the stronger decrease of mycolic acids compared to arabinogalactan could be a reason for the observed different extraction behavior of arabinogalactan in SilG-lcpA; in other words, arabinogalactan extraction was more complete in the mutant background. However, the remaining arabinogalactan in the wild-type and mutant backgrounds had similar ratios between galactose, arabinose, and rhamnose (Fig. 4B). By comparison of the residual sugar content of arabinogalactan-extracted cell walls about 60% of arabinogalactan remained in extracted cell walls of the wild type, whereas it was only 50% in SilG-lcpA (Fig. 4B).
Analysis of material from culture supernatants.
Other bacteria lacking LCP proteins have been described to release capsule material (34) or teichoic acids (8) into the supernatant. Furthermore, the electron microscopy pictures suggested impaired cell wall integrity of SilG-lcpA (Fig. 2). Therefore, solid material from the supernatant of the wild-type and SilG-lcpA cells was isolated by centrifugation. Given that the supernatant of the mutant culture contained much more material than the wild type (see Fig. S1C in the supplemental material), it was only possible to collect enough material from this culture for further analysis. Protein analysis by SDS-PAGE and matrix-assisted laser desorption–time of flight analysis revealed two major bands (see Fig. S1C) containing two trehalose corynomycolyl transferases (Cg0413 and Cg3182) (see Table S3 in the supplemental material), which are associated with the outer membrane. Lipid analysis of the material showed the presence of mycolic acid and fatty acid methyl esters (Fig. 4C; see also Fig. S5 in the supplemental material). In the arabinogalactan analysis, rhamnose, arabinose, and glucose (used as carbon sources and possibly derived from trehalose mycolates) as well as another so-far-unidentified sugar were found (see Fig. S6 in the supplemental material). However, in contrast to the whole-cell analysis, galactose was absent (see Fig. S6). Therefore, the material extracted from the culture supernatant clearly differed from material of whole cells, which would indicate a large amount of lysed cells.
Complementation studies with plasmid-carried lcpA and lcpB.
Complementation experiments with plasmid-carried lcpA under the control of an IPTG-inducible promoter were performed to confirm that the observed phenotype was in fact a result of silenced lcpA gene expression. Full complementation was already achieved by basal expression from the leaky Ptac (Fig. 5A, blue curve). Strong induction (with 100 μM IPTG) led to a growth rate lower than that of SilG-lcpA and a decreased final backscatter (data not shown). Interestingly, the effect of the silenced lcpA could be partially complemented by overexpressing lcpB, but only to a certain extent (see Fig. S1D in the supplemental material). Higher expression levels led to severe growth effects (see Fig. S1D). In contrast, the SilG-lcpA ΔlcpB strain could be fully complemented by plasmid-carried lcpA. Under the tested conditions, LcpB was not required for normal growth as long as sufficient amounts of LcpA were present (see Fig. S1E).
FIG 5.
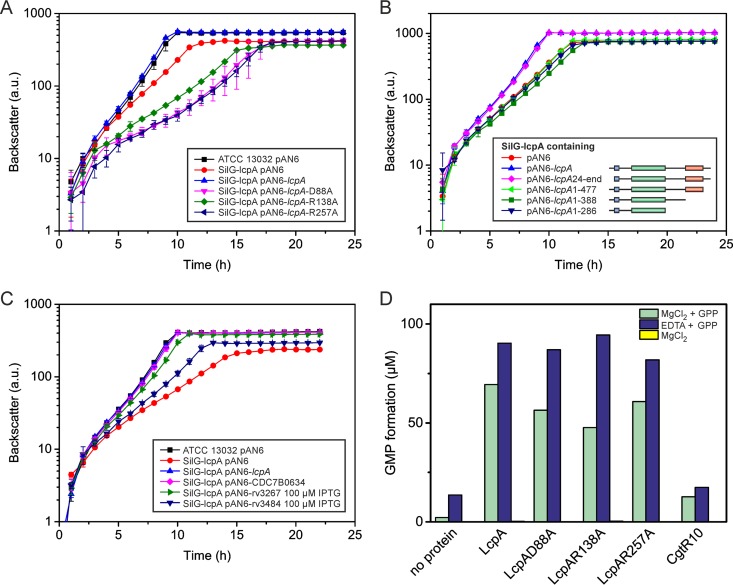
Complementation experiments and pyrophosphatase assay. (A) Complementation experiment of SilG-lcpA with LcpA variants carrying single mutations of conserved residues in the LCP domain (residues highlighted in Fig. S7 in the supplemental material). (B) Complementation experiment with N- and C-terminally trimmed LcpA variants. (C) Complementation experiment with homologous proteins of the related species C. diphtheriae and M. tuberculosis. Unless indicated otherwise, the growth experiments were performed without induction by IPTG. The mycobacterial proteins were induced to achieve maximal complementation. (D) Pyrophosphatase assay with different LcpAΔTM variants and CgtR10 as a negative control. The proteins were purified from E. coli and incubated in 20 mM Tris-HCl (pH 8.0) with the artificial substrate GPP (1 mM) and either MgCl2 or EDTA at 30°C overnight. The formation of GMP was quantified by LC-MS. No activity was measurable in the absence of the substrate (yellow bars, not visible). One representative result of two independent protein purifications is presented.
Mutational analysis of conserved amino acid residues in the LCP domain of LcpA.
The structural studies of Cps2A of Streptococcus pneumoniae with bound ligand and magnesium led to the identification of several amino acids that are likely crucial for enzymatic activity of the protein (6). Complementation studies in the SilG-lcpA strain with LcpA variants mutated at conserved residues were designed to show whether the catalytic center of LcpA is similar to Cps2A. Three positions of LcpA that are potentially involved in binding the pyrophosphate head group of the substrate (R138 and R257) or coordinating the magnesium ion (D88) (corresponding to D234, R267, and R362 in Cps2A or to D75 and R106 and R198 in B. subtilis TagU) were chosen for mutation based on sequence comparisons with Cps2A (6) (see Fig. S7 in the supplemental material). Remarkably, all three mutated variants failed to complement the growth defect of SilG-lcpA (Fig. 5A) and, interestingly, growth of the three strains was even worse than the control strain with the empty plasmid, suggesting critical roles of all three amino acids in LcpA function.
The C-terminal region of LcpA is important for its function in cell wall biogenesis.
To study the importance of different domains of LcpA for functionality, several truncated variants were cloned and tested for their ability to complement the phenotype of SilG-lcpA. The first variant (amino acids 24 to 518), lacking the N-terminal cytoplasmic part of the protein (Fig. 5B), fully complemented the SilG-lcpA phenotype, suggesting that the N-terminal part of the protein is not strictly required for function. In contrast, variants with truncations lacking 41, 130, or 232 amino acids of the C-terminal region were also tested and all of them lost the ability to complement the SilG-lcpA phenotype (Fig. 5B). This indicates the importance of the outer C-terminal part containing the LytR_C domain for LcpA function in C. glutamicum. Given the high conservation of a C-terminal motif (CXN), these amino acids seem to be crucial for LcpA function (see Fig. S7 in the supplemental material).
Complementation of SilG-lcpA with C. diphtheriae and M. tuberculosis LCP proteins.
Complementation studies with proteins of M. tuberculosis and C. diphtheriae were conducted to test for a similar function in cell wall biosynthesis, which would make them promising drug targets. Three proteins (CDC7B0634, Rv3267, and Rv3484) were chosen by homology searches for analysis, cloned into a C. glutamicum-compatible plasmid, and expressed in the SilG-lcpA strain (Fig. 5C). Different amounts of IPTG were tested to obtain optimal complementation. The C. diphtheriae protein already fully complemented the SilG-lcpA phenotype by leaky expression without any additional induction. In contrast, both mycobacterial proteins complemented only partially, even after induction with 100 μM IPTG, of which Rv3267 performed significantly better than Rv3484 (Fig. 5C).
Pyrophosphatase activity and oligomerization of LcpA.
To test whether LcpA also exhibits pyrophosphatase activity similar to TagT of Bacillus subtilis (6), the following proteins were analyzed: LcpAΔTM (LcpA lacking the transmembrane helix) and three variants mutated at the same residues that led to loss of complementation ability (D88A, R138A, and R257A). Gel filtration of LcpAΔTM suggested the formation of dimers under oxidizing conditions (theoretical molecular mass of monomer, 55.05 kDa; molecular mass according to gel filtration, ∼104 kDa) and the formation of presumably tetramers under reducing conditions (∼244 kDa) (see Fig. S8 in the supplemental material). Because the native substrate of LcpA is presently unknown, GPP was chosen as artificial substrate for the assay. To test whether the conversion to geranyl monophosphate (GMP) is magnesium dependent, the reaction was carried out either with addition of MgCl2 or in the presence of EDTA. All LcpA protein variants showed significant levels of pyrophosphatase activity, which was always higher in the presence of EDTA than upon addition of MgCl2 (Fig. 5D). This means that in contrast to other LCP proteins, the pyrophosphatase activity of LcpA does not seem to be magnesium dependent. The pyrophosphatase activity measured with the control protein (CgtR10) was within the range of the no-protein sample, excluding an unspecific activity resulting from the protein purification procedure.
Domain distribution of LCP and LytR_C in bacteria.
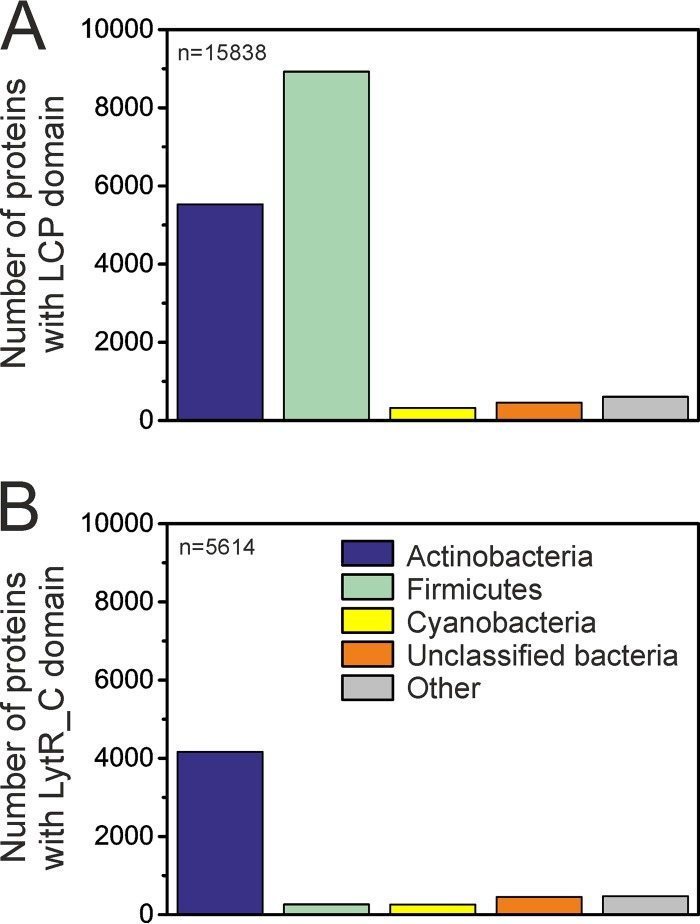
The complementation studies with mutated and truncated LcpA revealed that both domains are important for function. A more detailed analysis of the distribution of the two domains among bacteria showed that 56% of the LCP domains known to date are found in firmicutes and 35% in actinobacteria (Fig. 6A) (data from InterPro, 18 July 2016; 15,838 domain sequences in total). The distribution of the LytR_C domain differs significantly: 74% of the LytR_C domains belong to actinobacteria and only 5% to firmicutes (Fig. 6B) (5,614 proteins in total). Most (84%) of the latter are found in members of the genus Clostridium. This analysis of sequence data suggests a distinct function of the LytR_C domain in actinobacteria. The relative distribution of both domains among actinobacteria is comparable. Therefore, both domains are equally important for different members of this group.
FIG 6.
Distributions of the LCP (A) and the LytR_C (B) domains among bacteria. Whereas the LCP domain is found predominantly in firmicutes, the LytR_C domain is most widely spread among actinobacteria. Within firmicutes, 84% of the proteins with the LytR_C domain belong to clostridia. Color coding is the same for both diagrams. Data were obtained from InterPro, 18 July 2016. n represents the total number of the respective domains in the database on the given date.
The LCP and the LytR_C domains occur both separately and in combination. Among all LCP domain-containing proteins, the majority (∼76%) contain only an LCP domain, whereas 19% contain an additional LytR_C domain (5% in combination with other domains). In contrast, 53% of the LytR_C domains are found in association with an LCP domain, whereas 44% of LytR_C domain-containing proteins contain only the LytR_C domain. Solo LytR_C domains are also present in C. glutamicum (Cg3032) and M. tuberculosis (Rv0431 and Rv2700), but so far there has been only one study that reported Rv0431 was involved in vesiculogenesis (see Fig. S1B in the supplemental material) (35).
DISCUSSION
Relevance of LCP proteins in C. glutamicum.
LCP proteins are omnipresent in Gram-positive bacteria and appear to have an important function in cell wall biogenesis. However, recent studies suggest functional differences depending on the particular cell wall structure of the respective bacterial group. Here, we have presented physiological and biochemical analyses of two LCP proteins of C. glutamicum. Whereas a mutant lacking lcpB exhibited a wild-type-like phenotype, deletion of lcpA failed and silencing of lcpA expression resulted in a strong effect on cell growth and morphology. One LCP protein of M. marinum (MMAR_1274; Rv3267 in M. tuberculosis) has also been described as essential but has not been studied further to date (14). In contrast, an M. marinum cpsA mutant (MMAR_4966; Rv3484) displayed negative effects on cell wall biosynthesis and growth, leading to the hypothesis that CpsA might be responsible for the ligation of arabinogalactan to peptidoglycan (14). On a sequence level, LcpA is more similar to MMAR_1274 than to CpsA, favoring a function comparable to the uncharacterized essential protein. This was further supported by the better complementation of strain SilG-lcpA by Rv3267 expression compared to that with Rv3484 (Fig. 5C). B. subtilis survives a deletion of two of its three LCP proteins, but a triple deletion strain is not viable, proposing an essential but redundant function of these three proteins in transfer of anionic cell wall polymers onto peptidoglycan (6).
Both LCP proteins localize at sites of active cell wall biosynthesis.
Both LCP proteins of C. glutamicum are highly enriched in regions of nascent cell wall biosynthesis, which has also been demonstrated for S. pneumoniae harboring LCP proteins involved in capsule formation (34). In B. subtilis, LCP proteins were found to interact with MreB, a cytoskeleton protein which is probably responsible for their localization (6). The localization of the LCP proteins in C. glutamicum suggests that, analogous to MreB/TagTUV in B. subtilis, they might be part of the elongasome, which is required for polar growth, and be spatially organized by DivIVA (17, 36–38). However, given that C. glutamicum lacks MreB and that the cytoplasmic part of LcpA is dispensable for function, the localization likely appears within the membrane or in the periplasm, and additional proteins would be required to connect it to the elongasome. However, efforts at the identification of putative interaction partners of LcpA by copurification have failed so far (data not shown).
Both the LCP and the LytR_C domains of LcpA are essential for function.
With the exception of studies of M. marinum CpsA (14) and Anabaena sp. strain PCC 7120 All0187 (39), most studies of LCP proteins published to date have focused on those proteins containing only the LCP domain and not the LytR_C domain (6–9, 40). As described above, in actinobacteria the two domains frequently exist in combination within one protein, and the question arises whether both domains are required for LcpA function. Our complementation studies with truncated and mutated LcpA variants not only revealed the importance of the conserved aspartate and arginine residues located in the LCP domain but also suggested an important function for the LytR_C domain.
There are currently two possible explanations for the necessity of two domains in LcpA: (i) LcpA catalyzes one single reaction for which both domains are strictly required, or (ii) LcpA catalyzes at least two separate reactions which are catalyzed by either one of the domains. Two arguments supporting the latter case are that (i) both domains also quite frequently occur without the other one (6, 35) and (ii) although both domains appear to be important for LcpA function, mutations in the LCP domain are more detrimental to the growth of C. glutamicum than the absence of the LytR_C domain (Fig. 5A and B). Maybe a toxic intermediate produced by the LytR_C domain accumulates, which is further processed by the LCP domain under normal circumstances.
The oligomerization of LcpA observed here is a unique feature among LCP proteins characterized so far; ΔTM-Cps2A, for example, has been reported to be monomeric (6). Considering the fact that Cps2A lacks a LytR_C domain, it is possible that this domain is involved in dimerization or oligomerization. Given that LcpA forms dimers or tetramers, the effects of mutated LcpA expression (Fig. 5A) could be explained by a titration of functional LcpA by mutated, nonfunctional LcpA proteins through the formation of nonfunctional heterodimers. This would result in a further reduction of the small amounts of active LcpA (Fig. 5A) and consequently further impair growth. In contrast, the LytR_C truncated variants were possibly no longer able to form dimers and were just present as separate inactive proteins (Fig. 5B).
Interestingly, although the LcpA variants with mutations in the LCP domain failed to complement, the pyrophosphatase activity measured with the purified proteins was not affected. One reason for this could be that the artificial substrate GPP was used, and its catalysis might not be influenced by the mutations of the protein. Furthermore, the measured pyrophosphatase activity might differ from that of TagT (6) by being catalyzed by a different, magnesium-independent mechanism and possibly by being located within the LytR_C domain and not within the LCP domain. Of course, further studies are required to test these hypotheses.
Possible functions of LcpA in cell wall biogenesis.
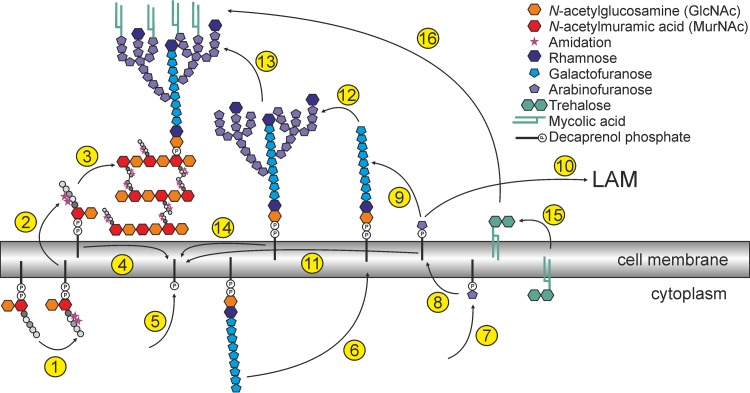
Known or proposed steps of cell wall biogenesis in C. glutamicum that are relevant to discuss the possible function of LCP proteins in this species are summarized in Fig. 7. Similar mechanisms for mycobacteria have been reviewed recently (12). It was proposed by Wang et al. (14) that the LCP protein CpsA of M. marinum is responsible for the transfer of arabinogalactan onto peptidoglycan. This hypothesis is based on data showing a reduced arabinogalactan content of the cell wall in the mutant (14).
FIG 7.
Schematic summary of known or proposed important steps of cell wall biogenesis in C. glutamicum. The peptidoglycan precursor Lipid II is composed of the carrier decaprenyl phosphate, GlcNAc-MurNAc and a pentapeptide (l-Ala, d-Glu, mDAP [meso diaminopimelic acid], d-Ala, d-Ala) (46). d-Glu and mDAP become amidated to a large extent in C. glutamicum (step 1). Recently, it was shown that the enzyme LtsA is responsible for the amidation of mDAP (46). The amidation of d-Glu is probably performed by Cg0299 and Cg0300, as shown for homologous proteins of Staphylococcus aureus (47, 48). Subsequently, Lipid II is translocated across the cell membrane (step 2), where the peptidoglycan polymerization takes place. The disaccharide-pentapetide is connected to the glycan chain by transglycosylation and transpeptidation (step 3), which is presumably performed by proteins of the penicillin-binding protein family and l,d-transpeptidases (49–52). Several candidates can be identified by homology searches (PBSs Cg3313, Cg2375, Cg2199, Cg0336, Cg0060, Cg2649, Cg2478, and Cg2987; l,d-transpeptidases Cg2720 [homologous to Rv0116c] and Cg0650). After transfer of the disaccharide-pentapeptide the carrier becomes recycled (step 4). The decaprenyl phosphate is synthesized de novo by UppS (Cg2508) (step 5) (53). Similar to the peptidoglycan precursors, also the arabinogalactan precursor is built on a decaprenol carrier. GlcNAc and rhamnose serve as linker, whereon the galactose chain is built and flipped to the periplasm by a so-far-unknown mechanism (step 6) (54, 55). Arabinose is provided as decaprenylmonophosphoryl-d-arabinose (DPA), another polyprenol-dependent precursor, involving the enzyme UbiA (step 7) (56). DPA is most likely reoriented to the periplasm by a transporter encoded by Cg0234 (Rv3789 in M. tuberculosis) (57) (step 8) and serves as arabinose donor both for arabinogalactan (step 9) and lipoarabinomannan (LAM) (step 10) before being recycled (step 11). Some chains of arabinogalactan are capped by rhamnose (step 12) (58). The arabinogalactan precursor is then presumably transferred onto peptidoglycan by the LcpA protein characterized in this study (59, 60) (step 13), and the decaprenol-phosphate becomes recycled (step 14). The mycolic acids are also synthesized within the cell, and most of them are transported across the membrane as trehalose monomycolates or trehalose dimycolates (61, 62) (step 15). Subsequently, the mycolic acids are transferred onto arabinogalactan by corynomycolic acid transferases (encoded by cop1 and cmt1 to cmt5) (step 16) (63).
As stated above, the mycobacterial proteins with the highest similarity to LcpA are not M. marinum CpsA (M. tuberculosis Rv3484), but Rv3267 (47% identity with LcpA) and MMAR_1274 (46% identity), which have been reported to be essential (14, 41). In contrast, CpsA (33% identity), Rv3484 (33% identity), and the third M. tuberculosis LCP protein with both an LCP domain and a LytR_C domain, Rv0822c (26% identity) (see Fig. S1B in the supplemental material), can be deleted (14, 41), which points toward distinct, nonredundant functions of CpsA and Rv3484 on one side and MMAR_1274 and Rv3267 on the other side. So far, nothing is known about MMAR_4858 (27% identity), the homolog of Rv0822c, but it is presumably also not essential.
In our studies, the silencing of lcpA led to a significant reduction of arabinogalactan content in the cell wall. These data, indeed, support the hypothesis that LcpA is responsible for the transfer of arabinogalactan onto peptidoglycan, which represents a central missing link in the mechanistic understanding of cell wall biosynthesis in corynebacteria and mycobacteria. This mechanism was furthermore corroborated by the observed pyrophosphatase activity and the significant reduction of rhamnose in the SilG-lcpA mutant. As a third piece of evidence, this strain also released a considerable amount of cell wall material into the supernatant during cultivation. Altogether, these data support a model where LcpA is involved in the covalent attachment of arabinogalactan to peptidoglycan in C. glutamicum.
Considering the domain structure of LcpA and the differences in functionality of LcpA and LcpB, further potential functions need to be considered in future studies. For A. oris, it has been shown that an LCP protein (containing only the LCP domain) is involved in a protein glycosylation pathway also, including a sortase which couples certain cell surface proteins to peptidoglycan (9). Here, the LCP protein is responsible for the transfer of a glycan polymer from its lipid-linked precursor onto the protein before it attaches to peptidoglycan. However, although C. glutamicum is predicted to contain a sortase (Cg3251), no sortase substrate has been identified so far (42). Therefore, given also that the A. oris protein lacks the LytR_C domain, a similar function is rather unlikely for LcpA.
While the manuscript was in revision, two further studies addressing the function of LCP proteins in the Corynebacteriales were published (43, 44), and these provided further evidence that LCP proteins are responsible for the ligation of arabinogalactan to peptidoglycan. However, these studies did not address a potential additional function of the C-terminal domain. Further studies are required to investigate whether arabinogalactan-peptidoglycan ligation is the only function of Lcp proteins in Corynebacteriales and to clarify the roles of LcpB and the C-terminal domain of LcpA in this context.
Conclusions.
The results presented in this study clearly show that LcpA has a very important role in cell wall biogenesis and integrity, as it is presumably responsible for the ligation of arabinogalactan to peptidoglycan, while both domains seem to be essential for function. Since the enzymes involved in the ligation of arabinogalactan to peptidoglycan were unknown to date, our data represent an important contribution to the mechanistic understanding of this missing link in corynebacterial cell wall biosynthesis. Our data emphasize this class of proteins as a potential drug target that is highly interesting for further investigation. Furthermore, the C. glutamicum SilG-lcpA strain offers an ideal platform strain for further functional and physiological studies of LCP proteins of Corynebacteriales, since we have shown that cross-species complementation with mycobacterial proteins is possible, and functionality of protein variants can easily be tested by growth studies. Altogether, our data underline the importance of LCP proteins in cell wall biogenesis in this medically and biotechnologically important class of bacteria and suggest an important role of the conserved C-terminal domain for functionality.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nicolai Kallscheuer for the help with LC-MS measurements, Mareike Hoß and Hiltrud Königs from the electron microscopy department of the UK Aachen for excellent SEM and TEM pictures, and also Kevin Titz for his contribution to this project during his master's thesis. We thank Jörn Kalinowski for the information about relative mRNA abundance of RNA-seq data. In addition, we thank Eva Hentschel for providing the picture of ChrS-EYFP. Furthermore, we thank Cornelia Gätgens and Christina Mack for excellent technical assistance and Andreas Küberl for critical reading of the manuscript.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00406-16.
REFERENCES
- 1.Lazarevic V, Margot P, Soldo B, Karamata D. 1992. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J Gen Microbiol 138:1949–1961. doi: 10.1099/00221287-138-9-1949. [DOI] [PubMed] [Google Scholar]
- 2.Cieslewicz MJ, Kasper DL, Wang Y, Wessels MR. 2001. Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J Biol Chem 276:139–146. doi: 10.1074/jbc.M005702200. [DOI] [PubMed] [Google Scholar]
- 3.Ligozzi M, Pittaluga F, Fontana R. 1993. Identification of a genetic element (psr) which negatively controls expression of Enterococcus hirae penicillin-binding protein 5. J Bacteriol 175:2046–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, Sangrador-Vegas A, Scheremetjew M, Rato C, Yong SY, Bateman A, Punta M, Attwood TK, Sigrist CJ, Redaschi N, Rivoire C, Xenarios I, Kahn D, Guyot D, Bork P, Letunic I, Gough J, Oates M, Haft D, Huang H, Natale DA, Wu CH, Orengo C, Sillitoe I, Mi H, Thomas PD, Finn RD. 2015. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res 43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hübscher J, Lüthy L, Berger-Bächi B, Stutzmann Meier P. 2008. Phylogenetic distribution and membrane topology of the LytR-CpsA-Psr protein family. BMC Genomics 9:617. doi: 10.1186/1471-2164-9-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawai Y, Marles-Wright J, Cleverley RM, Emmins R, Ishikawa S, Kuwano M, Heinz N, Bui NK, Hoyland CN, Ogasawara N, Lewis RJ, Vollmer W, Daniel RA, Errington J. 2011. A widespread family of bacterial cell wall assembly proteins. EMBO J 30:4931–4941. doi: 10.1038/emboj.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan YG, Kim HK, Schneewind O, Missiakas D. 2014. The capsular polysaccharide of Staphylococcus aureus is attached to peptidoglycan by the LytR-CpsA-Psr (LCP) family of enzymes. J Biol Chem 289:15680–15690. doi: 10.1074/jbc.M114.567669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan YG, Frankel MB, Dengler V, Schneewind O, Missiakas D. 2013. Staphylococcus aureus mutants lacking the LytR-CpsA-Psr family of enzymes release cell wall teichoic acids into the extracellular medium. J Bacteriol 195:4650–4659. doi: 10.1128/JB.00544-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu C, Huang IH, Chang C, Reardon-Robinson ME, Das A, Ton-That H. 2014. Lethality of sortase depletion in Actinomyces oris caused by excessive membrane accumulation of a surface glycoprotein. Mol Microbiol 94:1227–1241. doi: 10.1111/mmi.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mishra AK, Driessen NN, Appelmelk BJ, Besra GS. 2011. Lipoarabinomannan and related glycoconjugates: structure, biogenesis and role in Mycobacterium tuberculosis physiology and host-pathogen interaction. FEMS Microbiol Rev 35:1126–1157. doi: 10.1111/j.1574-6976.2011.00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eggeling L, Besra GS, Alderwick L. 2008. Structure and synthesis of the cell wall, p 267–294. In Burkovski A. (ed), Corynebacteria: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 12.Jankute M, Cox JA, Harrison J, Besra GS. 2015. Assembly of the mycobacterial cell wall. Annu Rev Microbiol 69:405–423. doi: 10.1146/annurev-micro-091014-104121. [DOI] [PubMed] [Google Scholar]
- 13.Daffe M, Crick DC, Jackson M. 2014. Genetics of capsular polysaccharides and cell envelope (glyco)lipids. Microbiol Spectr 2:MGM2-0021-2013. doi: 10.1128/microbiolspec.MGM2-0021-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q, Zhu L, Jones V, Wang C, Hua Y, Shi X, Feng X, Jackson M, Niu C, Gao Q. 2015. CpsA, a LytR-CpsA-Psr family protein in Mycobacterium marinum, is required for cell wall integrity and virulence. Infect Immun 83:2844–2854. doi: 10.1128/IAI.03081-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baumgart M, Luder K, Grover S, Gätgens C, Besra GS, Frunzke J. 2013. IpsA, a novel LacI-type regulator, is required for inositol-derived lipid formation in Corynebacteria and Mycobacteria. BMC Biol 11:122. doi: 10.1186/1741-7007-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seidel M, Alderwick LJ, Birch HL, Sahm H, Eggeling L, Besra GS. 2007. Identification of a novel arabinofuranosyltransferase AftB involved in a terminal step of cell wall arabinan biosynthesis in Corynebacterineae, such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J Biol Chem 282:14729–14740. doi: 10.1074/jbc.M700271200. [DOI] [PubMed] [Google Scholar]
- 17.Donovan C, Bramkamp M. 2014. Cell division in Corynebacterineae. Front Microbiol 5:132. doi: 10.3389/fmicb.2014.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eggeling L, Bott M. 2015. A giant market and a powerful metabolism: L-lysine provided by Corynebacterium glutamicum. Appl Microbiol Biotechnol 99:3387–3394. doi: 10.1007/s00253-015-6508-2. [DOI] [PubMed] [Google Scholar]
- 19.Keilhauer C, Eggeling L, Sahm H. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J Bacteriol 175:5595–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 21.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 22.van der Rest ME, Lange C, Molenaar D. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl Microbiol Biotechnol 52:541–545. doi: 10.1007/s002530051557. [DOI] [PubMed] [Google Scholar]
- 23.Gibson DG. 2011. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol 498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niebisch A, Bott M. 2001. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch Microbiol 175:282–294. doi: 10.1007/s002030100262. [DOI] [PubMed] [Google Scholar]
- 25.Lutz R, Bujard H. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res 25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frunzke J, Engels V, Hasenbein S, Gätgens C, Bott M. 2008. Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol Microbiol 67:305–322. doi: 10.1111/j.1365-2598.2007.06020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegrist MS, Whiteside S, Jewett JC, Aditham A, Cava F, Bertozzi CR. 2013. d-Amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem Biol 8:500–505. doi: 10.1021/cb3004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braun V, Rehn K. 1969. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur J Biochem 10:426–438. [DOI] [PubMed] [Google Scholar]
- 29.Glauner B, Holtje JV, Schwarz U. 1988. The composition of the murein of Escherichia coli. J Biol Chem 263:10088–10095. [PubMed] [Google Scholar]
- 30.Draper P. 1971. The walls of Mycobacterium lepraemurium: chemistry and ultrastructure. J Gen Microbiol 69:313–324. doi: 10.1099/00221287-69-3-313. [DOI] [PubMed] [Google Scholar]
- 31.Schubert K, Reiml D, Accolas JP, Fiedler F. 1993. A novel type of meso-diaminopimelic acid-based peptidoglycan and novel poly(erythritol phosphate) teichoic acids in cell walls of two coryneform isolates from the surface flora of French cooked cheeses. Arch Microbiol 160:222–228. [DOI] [PubMed] [Google Scholar]
- 32.Gauch R, Leuenberger U, Baumgartner E. 1979. Quantitative determination of mono-, di- and trisaccharides by thin-layer chromatography. J Chromatogr 174:195–200. doi: 10.1016/S0021-9673(00)87050-8. [DOI] [Google Scholar]
- 33.Minnikin DE, Alshamaony L, Goodfellow M. 1975. Differentiation of Mycobacterium, Nocardia, and related taxa by thin-layer chromatographic analysis of whole organism methanolysates. J Gen Microbiol 88:200–204. doi: 10.1099/00221287-88-1-200. [DOI] [PubMed] [Google Scholar]
- 34.Eberhardt A, Hoyland CN, Vollmer D, Bisle S, Cleverley RM, Johnsborg O, Havarstein LS, Lewis RJ, Vollmer W. 2012. Attachment of capsular polysaccharide to the cell wall in Streptococcus pneumoniae. Microb Drug Resist 18:240–255. doi: 10.1089/mdr.2011.0232. [DOI] [PubMed] [Google Scholar]
- 35.Rath P, Huang C, Wang T, Wang T, Li H, Prados-Rosales R, Elemento O, Casadevall A, Nathan CF. 2013. Genetic regulation of vesiculogenesis and immunomodulation in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 110:E4790–E4797. doi: 10.1073/pnas.1320118110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letek M, Ordonez E, Vaquera J, Margolin W, Flardh K, Mateos LM, Gil JA. 2008. DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum. J Bacteriol 190:3283–3292. doi: 10.1128/JB.01934-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sieger B, Schubert K, Donovan C, Bramkamp M. 2013. The lipid II flippase RodA determines morphology and growth in Corynebacterium glutamicum. Mol Microbiol 90:966–982. doi: 10.1111/mmi.12411. [DOI] [PubMed] [Google Scholar]
- 38.Donovan C, Sieger B, Krämer R, Bramkamp M. 2012. A synthetic Escherichia coli system identifies a conserved origin tethering factor in Actinobacteria. Mol Microbiol 84:105–116. doi: 10.1111/j.1365-2958.2012.08011.x. [DOI] [PubMed] [Google Scholar]
- 39.Mella-Herrera RA, Neunuebel MR, Golden JW. 2011. Anabaena sp. strain PCC 7120 conR contains a LytR-CpsA-Psr domain, is developmentally regulated, and is essential for diazotrophic growth and heterocyst morphogenesis. Microbiology 157:617–626. doi: 10.1099/mic.0.046128-0. [DOI] [PubMed] [Google Scholar]
- 40.Rowe HM, Hanson BR, Runft DL, Lin Q, Firestine SM, Neely MN. 2015. Modification of the CpsA protein reveals a role in alteration of the Streptococcus agalactiae cell envelope. Infect Immun 83:1497–1506. doi: 10.1128/IAI.02656-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 42.Boekhorst J, de Been MW, Kleerebezem M, Siezen RJ. 2005. Genome-wide detection and analysis of cell wall-bound proteins with LPxTG-like sorting motifs. J Bacteriol 187:4928–4934. doi: 10.1128/JB.187.14.4928-4934.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grzegorzewicz AE, de Sousa-d'Auria C, McNeil MR, Huc-Claustre E, Jones V, Petit C, Angala SK, Zemanova J, Wang Q, Belardinelli JM, Gao Q, Ishizaki Y, Mikusova K, Brennan PJ, Ronning DR, Chami M, Houssin C, Jackson M. 2016. Assembling of the Mycobacterium tuberculosis cell wall core. J Biol Chem 291:18867–18879. doi: 10.1074/jbc.M116.739227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harrison J, Lloyd G, Joe M, Lowary TL, Reynolds E, Walters-Morgan H, Bhatt A, Lovering A, Besra GS, Alderwick LJ. 2016. Lcp1 is a phosphotransferase responsible for ligating arabinogalactan to peptidoglycan in Mycobacterium tuberculosis. mBio 7(4):e00972-16. doi: 10.1128/mBio.00972-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alm EJ, Huang KH, Price MN, Koche RP, Keller K, Dubchak IL, Arkin AP. 2005. The MicrobesOnline Web site for comparative genomics. Genome Res 15:1015–1022. doi: 10.1101/gr.3844805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levefaudes M, Patin D, de Sousa-d'Auria C, Chami M, Blanot D, Herve M, Arthur M, Houssin C, Mengin-Lecreulx D. 2015. Diaminopimelic acid amidation in Corynebacteriales: new insights into the role of LtsA in peptidoglycan modification. J Biol Chem 290:13079–13094. doi: 10.1074/jbc.M115.642843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Münch D, Roemer T, Lee SH, Engeser M, Sahl HG, Schneider T. 2012. Identification and in vitro analysis of the GatD/MurT enzyme-complex catalyzing lipid II amidation in Staphylococcus aureus. PLoS Pathog 8:e1002509. doi: 10.1371/journal.ppat.1002509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Figueiredo TA, Sobral RG, Ludovice AM, Almeida JM, Bui NK, Vollmer W, de Lencastre H, Tomasz A. 2012. Identification of genetic determinants and enzymes involved with the amidation of glutamic acid residues in the peptidoglycan of Staphylococcus aureus. PLoS Pathog 8:e1002508. doi: 10.1371/journal.ppat.1002508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Riegel P, Gutmann L, Mainardi JL. 2009. The beta-lactam-sensitive D,D-carboxypeptidase activity of Pbp4 controls the L,D and D,D transpeptidation pathways in Corynebacterium jeikeium. Mol Microbiol 74:650–661. doi: 10.1111/j.1365-2958.2009.06887.x. [DOI] [PubMed] [Google Scholar]
- 50.Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, Mainardi JL. 2008. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L,D-transpeptidation. J Bacteriol 190:4360–4366. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dubee V, Triboulet S, Mainardi JL, Etheve-Quelquejeu M, Gutmann L, Marie A, Dubost L, Hugonnet JE, Arthur M. 2012. Inactivation of Mycobacterium tuberculosis L,D-transpeptidase LdtMt1 by carbapenems and cephalosporins. Antimicrob Agents Chemother 56:4189–4195. doi: 10.1128/AAC.00665-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valbuena N, Letek M, Ordonez E, Ayala J, Daniel RA, Gil JA, Mateos LM. 2007. Characterization of HMW-PBPs from the rod-shaped actinomycete Corynebacterium glutamicum: peptidoglycan synthesis in cells lacking actin-like cytoskeletal structures. Mol Microbiol 66:643–657. doi: 10.1111/j.1365-2958.2007.05943.x. [DOI] [PubMed] [Google Scholar]
- 53.Grover S, Alderwick LJ, Mishra AK, Krumbach K, Marienhagen J, Eggeling L, Bhatt A, Besra GS. 2014. Benzothiazinones mediate killing of Corynebacterineae by blocking decaprenyl phosphate recycling involved in cell wall biosynthesis. J Biol Chem 289:6177–6187. doi: 10.1074/jbc.M113.522623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alderwick LJ, Radmacher E, Seidel M, Gande R, Hitchen PG, Morris HR, Dell A, Sahm H, Eggeling L, Besra GS. 2005. Deletion of Cg-emb in Corynebacterianeae leads to a novel truncated cell wall arabinogalactan, whereas inactivation of Cg-ubiA results in an arabinan-deficient mutant with a cell wall galactan core. J Biol Chem 280:32362–32371. doi: 10.1074/jbc.M506339200. [DOI] [PubMed] [Google Scholar]
- 55.Dianiskova P, Kordulakova J, Skovierova H, Kaur D, Jackson M, Brennan PJ, Mikusova K. 2011. Investigation of ABC transporter from mycobacterial arabinogalactan biosynthetic cluster. Gen Physiol Biophys 30:239–250. doi: 10.4149/gpb_2011_03_239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alderwick LJ, Dover LG, Seidel M, Gande R, Sahm H, Eggeling L, Besra GS. 2006. Arabinan-deficient mutants of Corynebacterium glutamicum and the consequent flux in decaprenylmonophosphoryl-d-arabinose metabolism. Glycobiology 16:1073–1081. doi: 10.1093/glycob/cwl030. [DOI] [PubMed] [Google Scholar]
- 57.Larrouy-Maumus G, Skovierova H, Dhouib R, Angala SK, Zuberogoitia S, Pham H, Villela AD, Mikusova K, Noguera A, Gilleron M, Valentinova L, Kordulakova J, Brennan PJ, Puzo G, Nigou J, Jackson M. 2012. A small multidrug resistance-like transporter involved in the arabinosylation of arabinogalactan and lipoarabinomannan in mycobacteria. J Biol Chem 287:39933–39941. doi: 10.1074/jbc.M112.400986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Birch HL, Alderwick LJ, Rittmann D, Krumbach K, Etterich H, Grzegorzewicz A, McNeil MR, Eggeling L, Besra GS. 2009. Identification of a terminal rhamnopyranosyltransferase (RptA) involved in Corynebacterium glutamicum cell wall biosynthesis. J Bacteriol 191:4879–4887. doi: 10.1128/JB.00296-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yagi T, Mahapatra S, Mikusova K, Crick DC, Brennan PJ. 2003. Polymerization of mycobacterial arabinogalactan and ligation to peptidoglycan. J Biol Chem 278:26497–26504. doi: 10.1074/jbc.M302216200. [DOI] [PubMed] [Google Scholar]
- 60.Hancock IC, Carman S, Besra GS, Brennan PJ, Waite E. 2002. Ligation of arabinogalactan to peptidoglycan in the cell wall of Mycobacterium smegmatis requires concomitant synthesis of the two wall polymers. Microbiology 148:3059–3067. doi: 10.1099/00221287-148-10-3059. [DOI] [PubMed] [Google Scholar]
- 61.Yamaryo-Botte Y, Rainczuk AK, Lea-Smith DJ, Brammananth R, van der Peet PL, Meikle P, Ralton JE, Rupasinghe TW, Williams SJ, Coppel RL, Crellin PK, McConville MJ. 2015. Acetylation of trehalose mycolates is required for efficient MmpL-mediated membrane transport in Corynebacterineae. ACS Chem Biol 10:734–746. doi: 10.1021/cb5007689. [DOI] [PubMed] [Google Scholar]
- 62.Grzegorzewicz AE, Pham H, Gundi VA, Scherman MS, North EJ, Hess T, Jones V, Gruppo V, Born SE, Kordulakova J, Chavadi SS, Morisseau C, Lenaerts AJ, Lee RE, McNeil MR, Jackson M. 2012. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat Chem Biol 8:334–341. doi: 10.1038/nchembio.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brand S, Niehaus K, Pühler A, Kalinowski J. 2003. Identification and functional analysis of six mycolyltransferase genes of Corynebacterium glutamicum ATCC 13032: the genes cop1, cmt1, and cmt2 can replace each other in the synthesis of trehalose dicorynomycolate, a component of the mycolic acid layer of the cell envelope. Arch Microbiol 180:33–44. doi: 10.1007/s00203-003-0556-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.