Abstract
The extended-spectrum β-lactamase gene blaCTX-M-2 is mainly associated with ISCR1 embedded in complex sul1-type integrons, but information on the genetic context of plasmids harboring the ISCR1-blaCTX-M-2 module remains limited. In this study, a blaCTX-M-2-harboring plasmid (pYD786-1) belonging to the sequence type 2 (ST2)-IncHI2 plasmid type and isolated from an Escherichia coli ST410 clinical strain was sequenced and analyzed. pYD786-1 belongs to the APEC-O1-R-type IncHI2 plasmids, which are widely distributed in human, poultry, and livestock strains. It contains a multidrug resistance mosaic region (MRR) consisting of a Tn21::In2 transposon backbone augmented by acquisition of duplicate ISCR1-blaCTX-M-2 modules. Tn2411, a Tn21::In2 precursor, likely played a role in the generation of the MRR in pN13-01290_23, the putative progenitor plasmid of pYD786-1, found in a foodborne Salmonella strain. Tn21/Tn2411::In::ISCR1-blaCTX-M-2 derivatives, including pYD786-1, have been identified in strains from Europe, South America, and the United States, suggesting potential global dissemination of the blaCTX-M-2 modules mediated by this vehicle.
INTRODUCTION
CTX-M-type β-lactamases are the most common extended-spectrum β-lactamases (ESBLs) worldwide and include five major subgroups (CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25) (1). In the United States, CTX-M-15 of the CTX-M-1 group is the predominant ESBL in Escherichia coli, especially among the epidemic sequence type 131 (ST131) strains, followed by CTX-M-14 of the CTX-M-9 group (2–5). CTX-M-2 was identified in Argentina in 1989, first in different Salmonella serovars and then in diverse species of Enterobacteriaceae (1). Although it is not observed as frequently as CTX-M-1- and CTX-M-9-group enzymes, CTX-M-2 is endemic in South America (1, 6), caused a regional outbreak in Japan (7), circulates in animal and human strains in some European countries (8), and has been reported in North America (4, 5). In South America and Europe, blaCTX-M-2 is associated mainly with complex sul1-type integrons containing ISCR1 (9, 10), while in Japan most cases are associated with ISEcp1 (7, 11). Only a few studies have examined the incompatibility (Inc) types of the plasmids carrying blaCTX-M-2 (7, 8, 11, 12). In a survey of human and poultry isolates from Europe, blaCTX-M genes were found to be associated with IncHI2-type plasmids, and all blaCTX-M-2-carrying plasmids belonged to the IncHI2 APEC-O1-R type (8, 13, 14). Besides blaCTX-M, IncHI2 plasmids can recruit multiple resistance genes, including blaCMY-2 (accession no. KT347600.1) (15), blaVIM (accession no. LN555650.1) (16), blaIMP, armA, qnrA, oqxAB, fosA3, and mcr-1 (12, 17–21). So far, no IncHI2 plasmid carrying blaCTX-M-2 has been characterized fully. Here we report a novel IncHI2 plasmid carrying two copies of blaCTX-M-2 in tandem.
(Part of this study was presented at ASM Microbe 2016, June 2016, Boston, MA.)
MATERIALS AND METHODS
A total of 760 ESBL-producing E. coli strains collected between 2009 and 2014 were screened for fosfomycin resistance by plating on Mueller-Hinton agar plates containing 128 μg/ml of fosfomycin and 25 μg/ml of glucose-6-phosphate. A total of 10 strains stably resistant to fosfomycin were identified as a result. E. coli transformants with fosfomycin resistance could be obtained for 2 of these 10 strains by electroporation. One of the 2 strains harbored fosA3 (22), and the other harbored fosA6 (23). E. coli YD786 is an extraintestinal pathogenic E. coli (ExPEC) ST410 strain isolated from the urine of a female patient admitted to a hospital in Pennsylvania in 2012. It was sequenced and assembled as previously described (23), and it carries four plasmids, including the blaCTX-M-2-bearing plasmid pYD786-1 (accession no. KU254578.1) and the fosA6-carrying plasmid pYD786-2 (accession no. KU254579.1). Replicon sequence typing (RST) and plasmid double-locus sequence typing (pDLST) were conducted in silico based on a previously described scheme (12, 24; https://cge.cbs.dtu.dk/services/PlasmidFinder/; https://cge.cbs.dtu.dk/services/pMLST/). Nucleotide BLAST searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi) were used to find homologues of IncHI2 plasmid replication regions. Easyfig 2.0 was used to compare pYD786-1 to related plasmids. A phylogenetic tree was constructed by ClustalW alignment (http://www.genome.jp/tools/clustalw/).
Accession number(s).
The nucleotide sequence of pYD786-1 has been deposited in the GenBank database under accession number KU254578.1.
RESULTS AND DISCUSSION
Comparative analysis of the IncHI2 plasmid backbone.
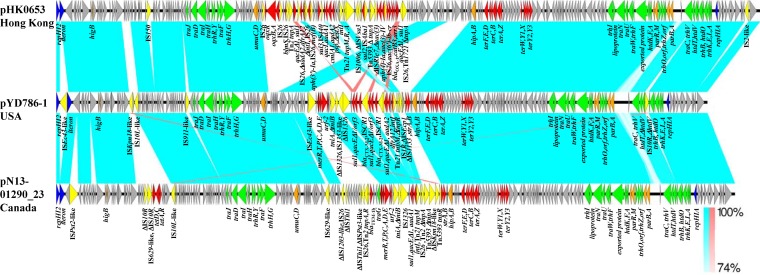
pYD786-1 is a 226.6-kb circular plasmid that possesses an IncHI2 plasmid backbone composed of replication (repHIA, repHI2, and iterons), partitioning (parAB and parMR), conjugation (tra, trh, and htd genes), and tellurium resistance (terABCDEFWXYZ) gene regions, complete or partial toxin-antitoxin systems (hipA/hipB and higB), and a umuCD operon involved in SOS mutagenesis (Fig. 1). Twenty-four fully sequenced IncHI2 plasmids were available in GenBank as of 5 April 2016, and they could be divided into three subtypes based on phylogenetic analysis of the replication regions, including repHIA (876 bp), repHI2 (1,089 bp), and its iterons (1,425 to 1,930 bp long) (see Fig. S1 in the supplemental material). Of these 24 plasmids, 15 plasmids clustered with the R478 type and 8 plasmids, including pYD786-1, to the APEC-O1-R type, whereas 1 plasmid, from Salmonella enterica strain CFSAN00205, was distinct. All of the R478-type plasmids were assigned to ST1 by pDLST, including two ST1-like plasmids (pRH-R178 and p34977-263.138kb; smr0199 is absent) (see Fig. S1). In contrast, in the APEC-O1-R group, 5 plasmids, including pYD786-1, belonged to ST2 or were closest to ST2 (one nucleotide difference in smr0018), and 3 plasmids from China belonged to ST3. The R478 plasmid was the first IncHI2 plasmid identified, from a clinical strain of Serratia marcescens in 1969 (13), while pAPEC-O1-R was recovered from an avian ExPEC strain in 2006 (14); both were recovered in the United States. Four of the APEC-O1-R-type (ST2-IncHI2) plasmids were from North America, and the other four were from China, including plasmids from Salmonella spp., E. coli, and Shigella spp. from human, poultry, and swine (see Fig. S1). ST3-IncHI2 plasmids are reportedly widely disseminated in Chinese poultry and livestock farms, and they carry fosA3, blaCTX-M-9-group genes (blaCTX-M-14/65/27), and/or oqxAB (17, 19). In previous studies, ST1-IncHI2 plasmids were associated with blaCTX-M-9 in Europe, while ST2- or APEC-O1-R-type IncHI2 plasmids linked to blaCTX-M-2 were found in Salmonella spp. from Belgium (poultry) and French Guiana (human feces), suggesting their distribution in Europe and South America (8, 12). pYD786-1 is the only fully sequenced blaCTX-M-2-bearing ST2-IncHI2 plasmid whose backbone is closely related to those of the other 7 APEC-O1-R-type plasmids (see Fig. S1). It has a replication region identical to those of pN13-01290_23, pHK0653, 1205p1, pEC5207, and pAPEC-O1-R, except that pHK0653, 1205p1, and pEC5207 have an 81-bp fragment deletion in the iterons and pAPEC-O1-R has a nucleotide substitution in repHIA. Comparison of three sequences (pYD786-1, pN13-01290_23, and pHK0653) revealed conservation of the backbone, including the tellurium resistance gene cluster, and substantial differences in the multidrug resistance mosaic region (MRR), reflecting the common origin of an APEC-O1-R-type IncHI2 plasmid and the contributions of various subsequent lateral gene transfer events (Fig. 1).
FIG 1.
Comparison of plasmid pYD786-1 and the related IncHI2 plasmids pHK0653 (accession no. KT334335.1) and pN13-01290_23 (accession no. CP012931.1) (rearranged and inversely displayed for comparison of structures). Genes with different functions are color coded as follows: blue, plasmid replication genes; orange, plasmid stability, partitioning, and maintenance genes; green, conjugal transfer genes; red, resistance genes; yellow, mobile genetic elements; and light gray, genes with other functions. Cyan shading indicates shared backbone regions and antimicrobial resistance regions with high degrees of homology (74% to 100% identity of nucleotide sequences).
Genetic analysis of the MRR of pYD786-1.
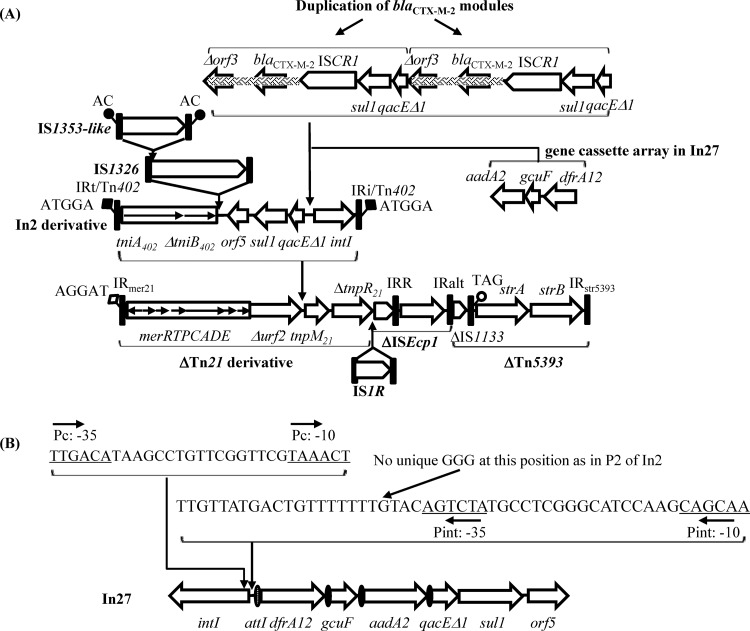
The MRR of pYD786-1 is composed of a Tn21-derived transposon carrying a mercuric resistance gene operon, followed by an ISEcp1 transposition unit and a Tn5393::IS1133 element characterized by the presence of the strA-strB streptomycin resistance genes (Fig. 1 and 2). Insertion of IS1R truncates tnpR of Tn21 and ISEcp1 at the 5′ end, while ISEcp1 disrupts IS1133 at the 5′ end. The Tn21 derivative apparently underwent the following insertion events, as reported previously (25): (i) a Tn402-derived class 1 integron inserted into urf-2 of Tn21 and generated two direct repeats (DRs), and (ii) the insertion of IS1326 disrupted tniB, followed by the transposition of IS1353, which interrupted IS1326, thus creating a nested complex structure almost identical to that of In2-bearing Tn21 (Fig. 2A). However, In2 contained only one gene cassette, aadA1, while in pYD786-1, dfrA12-gcuF-aadA2 gene cassettes were incorporated into the integron, presumably by replacing aadA1 with IntI1-mediated recombination or by homologous recombination with similar integrons, thus generating In27 (Fig. 2A) (26–28). In27 in pYD786-1 contains a strong Pc promoter (PcS) (−35 sequence, TTGACA; −10 sequence, TAAACT) (Fig. 2B) identical to that of In4 in Tn1696 (accession no. U12338.1), in contrast to the weak Pc promoter (PcW) (−35 sequence, TGGACA; −10 sequence, TAAGCT) in In2, as demonstrated experimentally by Jové et al. (29). Uniquely, In2 has extra GGG residues in the P2 region which are absent in the other integrons mentioned above, thus creating a functional P2 promoter for gene cassettes (Fig. 2B) (29), while In4 has a 19-bp duplication located in the attI1 region. At least the following 3 Pc promoters are associated with In27, indicating possible recombination events: PcH1 (−35 sequence, TGGACA; −10 sequence, TAAACT) (accession no. EU780013.1), from Uruguay (6); PcS (accession no. EF592571.1), from French Guiana (8); and PcW (accession no. EF219134.3) in pJIE137, from Australia. However, the evolutionary process of the Pc/P2 promoters still remains obscure. In27 in pYD786-1 is inserted in an unusual location, as is In2 in Tn21, and the targeted positions of IS1326 and IS1353 are also identical to those for In2 in Tn21, indicating that the Tn21-like::In27 region in pYD786-1 is a descendant of In2-bearing Tn21 and shares a common evolutionary lineage with In0, In2, and In5 (25, 30). In addition, pYD786-1 captured an ISCR1-blaCTX-M-2 module characterized by partially duplicated 3′ conserved segments (3′-CS), as described previously (6, 31). Uniquely, however, the ISCR1-blaCTX-M-2-3′-CS module was duplicated, generating two copies of blaCTX-M-2 in tandem and thus forming an unusual complex class 1 integron (Fig. 2A).
FIG 2.
Schematic map of multidrug resistance mosaic region (MRR) in pYD786-1. (A) Evolution of the Tn21-like module in pYD786-1. The integration locations of the integron, IS1326, the IS1353-like element, and the gene cassette array are indicated by vertical arrows. Duplication of the blaCTX-M-2 module is also indicated. DNA fragments shaded with basketwork showed 99% identity to those in the chromosome of Kluyvera ascorbata (accession no. AJ272538.2). The predicted open reading frames (ORFs) and insertion sequences are indicated by bold arrows and annotated above or below the maps, with arrowheads showing the direction of transcription. Putative direct repeat (DR) sequences are given to indicate the boundaries. Paired filled/unfilled squares or circles represent DRs of transposition units. The features shown are not necessarily drawn to scale. (B) Features of In27 and its promoter region in pYD786-1. attI and 59-be of the gene cassette are indicated by a filled oval. The promoters Pc (for the dfrA12 gene) and Pint (for the intI1 gene) are underlined. The position of unique GGG nucleotides in P2 of In2 is indicated. The position of and direction of transcription from the promoters are indicated by vertical and horizontal arrows, respectively.
Genetic context of ISCR1-blaCTX-M-2-bearing integrons.
The ISCR1-blaCTX-M-2 module has been reported worldwide for various complex class 1 integrons, such as In35, In0, In2, In4, In27, In37, In131, In132, and many other types (1, 32). Most of these were identified in strains from Argentina or its adjacent countries in South America, whereas some were from Europe (6, 32). In Argentina, which is the major cradle of blaCTX-M-2, the most common vehicle of blaCTX-M-2 in Enterobacteriaceae is an In35::ISCR1::blaCTX-M-2 element carrying the gene cassette array aac(6′)-Ib-blaOXA-2-orfD. For instance, InV117 (PcS) in pAS1 was recovered from an endemic Vibrio cholerae strain, InS21 (PcS) in pS21 from an outbreak Salmonella enterica strain, In116 (PcIn116) (−35 sequence, TTGACA; −10 sequence, TGAACT) in pM16 from Morganella morganii, and In35 in pMAR12 from Proteus mirabilis (33).
Only a few reports have described the genetic contexts of ISCR1-blaCTX-M-2-bearing integrons. One such combination is Tn1696-like::In35::ISCR1::blaCTX-M-2. Tn1696 is a close relative of Tn21 that is always associated with In4. InV117 is located downstream of a Tn1696-like element. InV117 has >99.8% identity with InS21, In116, and In35, indicating their common origin and possible association with a Tn1696-like element (33). A fully sequenced multidrug resistance IncA/C2 plasmid (accession no. CP007636.1), harbored by V. cholerae outbreak strain 2012EL-2176 from Haiti (18), contains a structure closely related to that of Tn1696-like::In35::ISCR1::blaCTX-M-2, although arr-3-dfrA27-aadA16 gene cassettes in In152 (PcH1) replaced those in In35, indicating their coevolution in this lineage.
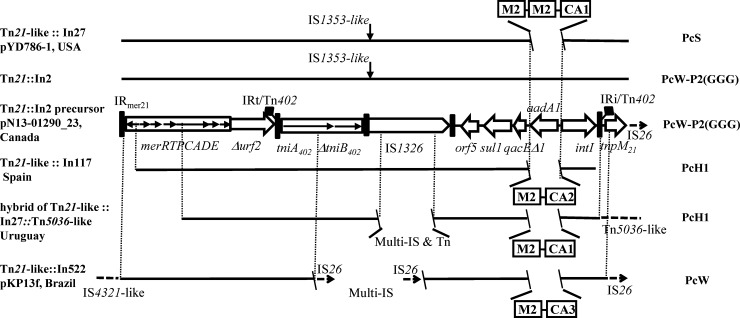
It is worth noting that Tn21 derivatives, which are involved in the formation of MRRs in plasmids, including pYD786-1, pN13-01290_23, and pHK0653 (Fig. 1), are also associated with the ISCR1-blaCTX-M-2 module. pN13-01290_23 from Canada harbors a precursor of Tn21::In2, Tn2411 (26), but lacks IS1353 (Fig. 3). Similar derivatives of Tn21::In2-like transposons bearing the ISCR1-blaCTX-M-2 module have been identified in at least 4 strains from North America, South America, and Europe (Fig. 3).
FIG 3.
Schematic diagram of the Tn21::In2 transposon and its derivatives bearing ISCR1-blaCTX-M-2. Tn2411 in pN13-01290_23, a precursor of Tn21::In2, was used as the template. Solid lines indicate sequences identical to the template. Vertical arrows indicate the insertion site of the IS1353-like element. Sequences between the double slash were exchanged. The Pc promoters of the integrons are indicated on the right. M2, blaCTX-M-2 module as shown in Fig. 2A. “CA” designations indicate the gene cassettes, as follows: CA1, aadA2-gcuF-dfrA12; CA2, aadA1-estX; and CA3, aadA2-cmlA1-dfrA15. The sequences used in the diagram are available under the following GenBank accession numbers: pYD786-1, KU254578.1; Tn21::In2, AF071413.3; pN13-01290_23, CP012931.1; In117, DQ125241.2; Tn21-like::In27::Tn5036-like, EU780013.1; and pKP13f, CP004000.1.
In117, which is embedded in Tn2411, was identified in an E. coli strain from the feces of a healthy volunteer in Spain in 2003 (34). Compared to In2, In117 acquired an estX cassette, thus generating the estX-aadA1a cassette array and an intermediate Pc promoter (PcH1). A hybrid transposon in Klebsiella pneumoniae strain 12836, isolated in Uruguay in 2005, contains an integron identical to In27 in pYD786-1, except for a nucleotide difference in Pc (PcH1 versus PcS) (6). The hybrid likely underwent multiple recombination events, causing the loss of a large part of IS1326 in the middle but still leaving an intact left inverted repeat (IRL) as evidence of the lack of transposition of IS1353. Moreover, the Tn21 tnp region was replaced by that of a Tn5036-like transposon, presumably via homologous recombination, and formed a hybrid of Tn21-like and Tn5036-like transposons (6). In the IncH plasmid pKP13f, harbored by a KPC-producing K. pneumoniae strain in Brazil (35), the Tn21-like transposon is interrupted by multiple copies of IS26, which is suggestive of extensive genetic exchange resulting in a complicated MRR and a partial sequence of IS1326. The dfrA15-cmlA1-aadA2 gene cassettes comprise the variable region of In522 in pKP13f.
Overall, we observed several instances where the ISCR1-blaCTX-M-2 module was linked to an ancestral tnp-mer transposon backbone of Tn21::In2 or its precursor, suggesting an early combination of three types of mobile genetic elements (MGEs) in association with blaCTX-M-2. Frequent exchanges in the variable regions of class 1 integrons with various Pc promoters suggest the contributions of functional gene cassettes to the dissemination of resistance genes. blaCTX-M-2-bearing APEC-O1-R-type IncHI2 plasmids have been identified from human and poultry Salmonella strains in Europe, suggesting transmission via the food chain (8). pYD786-1 is likely a descendant of the APEC-O1-R-type plasmid pN13-01290_23, which is also carried by a Salmonella strain, providing another example of foodborne pathogens serving as a potential source of resistance genes and MGEs.
More recently, sequences for another 9 APEC-O1-R-type plasmids were released (as of 19 July 2016), including plasmid 180-PT54, carried by a foodborne E. coli O157 strain from the United Kingdom (ST4-IncHI2) (accession no. CP015833.1), and 3 MCR-1-encoding plasmids, identified from foodborne E. coli strains in China (pHNSHP45-2; ST3-IncHI2) (20) and Italy (pS38; ST4-IncHI2) (21) and from a human E. coli strain in Saudi Arabia (pSA26-MCR-1; ST4-IncHI2) (accession no. KU743384.1). pHNSHP45-2 is almost identical to pHK0653 (coverage, 95%; identity, >99%), except for 3 mosaic modules, ISApI1-mcr-1, ΔISEcp1-blaCTX-M-14-IS903B-fosA3, and floR-orf-ISCR2 in the MRR, probably generated by 3 genetic events (20). pS38 is genetically closest to 180-PT54 (coverage, 95%; identity, >99%), including an almost identical Tn21-like transposon. Like pN13-01290_23, the 180-PT54 plasmid also harbors a precursor of Tn21 with an intact IS1326 element which is lost in the Tn21-like transposon in pS38. Besides pYD786-1, pS38 and the 180-PT54 plasmid are genetically closest to pN13-01290_23 (coverage, 83 to 85%; identity, >99%), providing more evidence of global dissemination of this lineage of plasmids by the food chain.
Conclusions.
We identified a duplication of the blaCTX-M-2 module in an ST2-IncHI2 plasmid, captured by an ancient Tn21::In2 transposon which likely underwent subsequent exchanges of gene cassettes and acquisition of various insertion sequences and transposons. Alignment of the blaCTX-M-2 modules carried by different Tn21::In2 progenitor derivatives identified in Europe and South America suggests an early dissemination of ISCR1-blaCTX-M-2 by this vehicle, together with Tn1696-like::In35::ISCR1::blaCTX-M-2 elements or their homologues.
Supplementary Material
ACKNOWLEDGMENTS
Q.G. was supported by grants 81673479, 81402976, and 81120108024 from the National Natural Science Foundation of China and by grant 16PJD010 from the Shanghai Pujiang Program. N.S. was funded through a Public Health England/University of Oxford Clinical Lectureship. The effort of Y.D. was supported by research grants from the National Institutes of Health (grants R01AI104895 and R21AI123747).
Y.D. has served on advisor boards for Shionogi, Meiji, Tetraphase, and Achaogen, received speaking fees from Merck, and received research funding from Merck and The Medicines Company for studies unrelated to this work. All other authors declare no potential conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01173-16.
REFERENCES
- 1.Bonnet R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother 48:1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson JR, Urban C, Weissman SJ, Jorgensen JH, Lewis JS II, Hansen G, Edelstein PH, Robicsek A, Cleary T, Adachi J, Paterson D, Quinn J, Hanson ND, Johnston BD, Clabots C, Kuskowski MA, AMERECUS Investigators. 2012. Molecular epidemiological analysis of Escherichia coli sequence type ST131 (O25:H4) and blaCTX-M-15 among extended-spectrum-β-lactamase-producing E. coli from the United States, 2000 to 2009. Antimicrob Agents Chemother 56:2364–2370. doi: 10.1128/AAC.05824-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen LF, Freeman JT, Nicholson B, Keiger A, Lancaster S, Joyce M, Woods CW, Cook E, Adcock L, Louis S, Cromer AL, Sexton DJ, Anderson DJ. 2014. Widespread dissemination of CTX-M-15 genotype extended-spectrum-β-lactamase-producing Enterobacteriaceae among patients presenting to community hospitals in the southeastern United States. Antimicrob Agents Chemother 58:1200–1202. doi: 10.1128/AAC.01099-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qureshi ZA, Paterson DL, Peleg AY, Adams-Haduch JM, Shutt KA, Pakstis DL, Sordillo E, Polsky B, Sandkovsky G, Bhussar MK, Doi Y. 2012. Clinical characteristics of bacteraemia caused by extended-spectrum β-lactamase-producing Enterobacteriaceae in the era of CTX-M-type and KPC-type β-lactamases. Clin Microbiol Infect 18:887–893. doi: 10.1111/j.1469-0691.2011.03658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park YS, Adams-Haduch JM, Shutt KA, Yarabinec DM III, Johnson LE, Hingwe A, Lewis JS II, Jorgensen JH, Doi Y. 2012. Clinical and microbiologic characteristics of cephalosporin-resistant Escherichia coli at three centers in the United States. Antimicrob Agents Chemother 56:1870–1876. doi: 10.1128/AAC.05650-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marquez C, Labbate M, Raymondo C, Fernandez J, Gestal AM, Holley M, Borthagaray G, Stokes HW. 2008. Urinary tract infections in a South American population: dynamic spread of class 1 integrons and multidrug resistance by homologous and site-specific recombination. J Clin Microbiol 46:3417–3425. doi: 10.1128/JCM.00835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakano R, Nakano A, Abe M, Inoue M, Okamoto R. 2012. Regional outbreak of CTX-M-2 β-lactamase-producing Proteus mirabilis in Japan. J Med Microbiol 61:1727–1735. doi: 10.1099/jmm.0.049726-0. [DOI] [PubMed] [Google Scholar]
- 8.Garcia Fernandez A, Cloeckaert A, Bertini A, Praud K, Doublet B, Weill FX, Carattoli A. 2007. Comparative analysis of IncHI2 plasmids carrying blaCTX-M-2 or blaCTX-M-9 from Escherichia coli and Salmonella enterica strains isolated from poultry and humans. Antimicrob Agents Chemother 51:4177–4180. doi: 10.1128/AAC.00603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arduino SM, Roy PH, Jacoby GA, Orman BE, Pineiro SA, Centron D. 2002. blaCTX-M-2 is located in an unusual class 1 integron (In35) which includes Orf513. Antimicrob Agents Chemother 46:2303–2306. doi: 10.1128/AAC.46.7.2303-2306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Climaco EC, Minarini LA, da Costa Darini AL. 2010. CTX-M-producing Klebsiella spp. in a Brazilian hospital: what has changed in 6 years? Diagn Microbiol Infect Dis 68:186–189. doi: 10.1016/j.diagmicrobio.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 11.Kayama S, Shigemoto N, Kuwahara R, Oshima K, Hirakawa H, Hisatsune J, Jove T, Nishio H, Yamasaki K, Wada Y, Ueshimo T, Miura T, Sueda T, Onodera M, Yokozaki M, Hattori M, Ohge H, Sugai M. 2015. Complete nucleotide sequence of the IncN plasmid encoding IMP-6 and CTX-M-2 from emerging carbapenem-resistant Enterobacteriaceae in Japan. Antimicrob Agents Chemother 59:1356–1359. doi: 10.1128/AAC.04759-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Fernandez A, Carattoli A. 2010. Plasmid double locus sequence typing for IncHI2 plasmids, a subtyping scheme for the characterization of IncHI2 plasmids carrying extended-spectrum β-lactamase and quinolone resistance genes. J Antimicrob Chemother 65:1155–1161. doi: 10.1093/jac/dkq101. [DOI] [PubMed] [Google Scholar]
- 13.Gilmour MW, Thomson NR, Sanders M, Parkhill J, Taylor DE. 2004. The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid 52:182–202. doi: 10.1016/j.plasmid.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Johnson TJ, Wannemeuhler YM, Scaccianoce JA, Johnson SJ, Nolan LK. 2006. Complete DNA sequence, comparative genomics, and prevalence of an IncHI2 plasmid occurring among extraintestinal pathogenic Escherichia coli isolates. Antimicrob Agents Chemother 50:3929–3933. doi: 10.1128/AAC.00569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng H, Si HB, Zeng SY, Sun J, Fang LX, Yang RS, Liu YH, Liao XP. 2015. Prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in a farrowing farm: ST1121 clone harboring IncHI2 plasmid contributes to the dissemination of blaCMY-2. Front Microbiol 6:1210. doi: 10.3389/fmicb.2015.01210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falgenhauer L, Ghosh H, Guerra B, Yao Y, Fritzenwanker M, Fischer J, Helmuth R, Imirzalioglu C, Chakraborty T. 2015. Comparative genome analysis of IncHI2 VIM-1 carbapenemase-encoding plasmids of Escherichia coli and Salmonella enterica isolated from a livestock farm in Germany. Vet Microbiol 2015:S0378–1135(15)30020-1. doi: 10.1016/j.vetmic.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Fang L, Li X, Li L, Li S, Liao X, Sun J, Liu Y. 2016. Co-spread of metal and antibiotic resistance within ST3-IncHI2 plasmids from E. coli isolates of food-producing animals. Sci Rep 6:25312. doi: 10.1038/srep25312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folster JP, Katz L, McCullough A, Parsons MB, Knipe K, Sammons SA, Boncy J, Tarr CL, Whichard JM. 2014. Multidrug-resistant IncA/C plasmid in Vibrio cholerae from Haiti. Emerg Infect Dis 20:1951–1953. doi: 10.3201/eid2011.140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X, Liu W, Liu Y, Wang J, Lv L, Chen X, He D, Yang T, Hou J, Tan Y, Xing L, Zeng Z, Liu JH. 2014. F33: A−: B−, IncHI2/ST3, and IncI1/ST71 plasmids drive the dissemination of fosA3 and blaCTX-M-55/-14/-65 in Escherichia coli from chickens in China. Front Microbiol 5:688. doi: 10.3389/fmicb.2014.00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhi C, Lv L, Yu LF, Doi Y, Liu JH. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:292–293. doi: 10.1016/S1473-3099(16)00063-3. [DOI] [PubMed] [Google Scholar]
- 21.Zurfluh K, Klumpp J, Nuesch-Inderbinen M, Stephan R. 2016. Full-length nucleotide sequences of mcr-1-harboring plasmids isolated from extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolates of different origins. Antimicrob Agents Chemother 60:5589–5591. doi: 10.1128/AAC.00935-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alrowais H, McElheny CL, Spychala CN, Sastry S, Guo Q, Butt AA, Doi Y. 2015. Fosfomycin resistance in Escherichia coli, Pennsylvania, USA. Emerg Infect Dis 21:2045–2047. doi: 10.3201/eid2111.150750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Q, Tomich AD, McElheny CL, Cooper VS, Stoesser N, Wang M, Sluis-Cremer N, Doi Y. 2016. Glutathione-S-transferase FosA6 of Klebsiella pneumoniae origin conferring fosfomycin resistance in ESBL-producing Escherichia coli. J Antimicrob Chemother 71:2460–2465. doi: 10.1093/jac/dkw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Partridge SR, Brown HJ, Stokes HW, Hall RM. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob Agents Chemother 45:1263–1270. doi: 10.1128/AAC.45.4.1263-1270.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liebert CA, Hall RM, Summers AO. 1999. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev 63:507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall RM, Collis CM. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol 15:593–600. [DOI] [PubMed] [Google Scholar]
- 28.Partridge SR. 2011. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol Rev 35:820–855. doi: 10.1111/j.1574-6976.2011.00277.x. [DOI] [PubMed] [Google Scholar]
- 29.Jové T, Da Re S, Denis F, Mazel D, Ploy MC. 2010. Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS Genet 6:e1000793. doi: 10.1371/journal.pgen.1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown HJ, Stokes HW, Hall RM. 1996. The integrons In0, In2, and In5 are defective transposon derivatives. J Bacteriol 178:4429–4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toleman MA, Bennett PM, Walsh TR. 2006. Common regions e.g. orf513 and antibiotic resistance: IS 91-like elements evolving complex class 1 integrons. J Antimicrob Chemother 58:1–6. doi: 10.1093/jac/dkl204. [DOI] [PubMed] [Google Scholar]
- 32.Quiroga MP, Arduino SM, Merkier AK, Quiroga C, Petroni A, Argentinian Integron Study Group, Roy PH, Centron D. 2013. Distribution and functional identification of complex class 1 integrons. Infect Genet Evol 19:88–96. doi: 10.1016/j.meegid.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 33.Soler Bistue AJ, Martin FA, Petroni A, Faccone D, Galas M, Tolmasky ME, Zorreguieta A. 2006. Vibrio cholerae InV117, a class 1 integron harboring aac(6′)-Ib and blaCTX-M-2, is linked to transposition genes. Antimicrob Agents Chemother 50:1903–1907. doi: 10.1128/AAC.50.5.1903-1907.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valverde A, Canton R, Galan JC, Nordmann P, Baquero F, Coque TM. 2006. In117, an unusual In0-like class 1 integron containing CR1 and blaCTX-M-2 and associated with a Tn21-like element. Antimicrob Agents Chemother 50:799–802. doi: 10.1128/AAC.50.2.799-802.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramos PI, Picao RC, Almeida LG, Lima NC, Girardello R, Vivan AC, Xavier DE, Barcellos FG, Pelisson M, Vespero EC, Medigue C, Vasconcelos AT, Gales AC, Nicolas MF. 2014. Comparative analysis of the complete genome of KPC-2-producing Klebsiella pneumoniae Kp13 reveals remarkable genome plasticity and a wide repertoire of virulence and resistance mechanisms. BMC Genomics 15:54. doi: 10.1186/1471-2164-15-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.