Abstract
The stringent response is a central adaptation mechanism that allows bacteria to adjust their growth and metabolism according to environmental conditions. The functionality of the stringent response is crucial for bacterial virulence, survival during host invasion as well as antibiotic resistance and tolerance. Therefore, specific inhibitors of the stringent response hold great promise as molecular tools for disarming and pacifying bacterial pathogens. By taking advantage of the valine amino acid auxotrophy of the Bacillus subtilis stringent response-deficient strain, we have set up a High Throughput Screening assay for the identification of stringent response inhibitors. By screening 17,500 compounds, we have identified a novel class of antibacterials based on the 4-(6-(phenoxy)alkyl)-3,5-dimethyl-1H-pyrazole core. Detailed characterization of the hit compounds as well as two previously identified promising stringent response inhibitors – a ppGpp-mimic nucleotide Relacin and cationic peptide 1018 – showed that neither of the compounds is sufficiently specific, thus motivating future application of our screening assay to larger and more diverse molecular libraries.
The stringent response is a central adaptation mechanism that adjusts bacterial growth and metabolism to environmental conditions. In response to various stress stimuli, RelA/SpoT Homologue (RSH) proteins modulate the intracellular concentration of the nucleotide alarmone guanosine (penta)tetraphosphate or (p)ppGpp1. An increased level of (p)ppGpp effectuates the adaptation to stress conditions via a global rewiring of the cellular metabolism and transcriptional program, e.g. by upregulating the production of amino acid biosynthesis enzymes upon amino acid starvation2.
In the most commonly used bacterial model organism – γ-proteobacterium Escherichia coli – the stringent response is orchestrated by two multi-domain ‘long RSH’ enzymes: RelA3 and SpoT4. Their activity is regulated by different sets of stress signals. RelA has strong ribosome-dependent (p)ppGpp synthetic activity that is triggered upon amino acid starvation via RelA directly sensing the deacylated tRNA in the ribosomal A-site5,6,7,8,9. As we have shown using an in vitro biochemical system another activator of RelA is its product ppGpp10, though the physiological significance of this effect is not yet clear. The other E. coli RSH, SpoT, possesses both (p)ppGpp synthetic and hydrolytic activities11,12. The weak synthetic activity of SpoT is induced by a variety of signals including fatty acid13, iron14 and carbon-source11 starvation. Constitutive (p)ppGpp hydrolysis by SpoT is crucial for counteracting the toxic effects of (p)ppGpp overproduction, and therefore disruption of the spoT gene in the presence of an intact copy of the relA gene renders E. coli non-viable11.
Phylogenetic analysis of the RSH protein family has shown that RelA and SpoT have a very limited evolutionary distribution1. In the majority of bacterial species, including the well-studied model organism Bacillus subtilis, ‘long’ RSHs are represented by the protein Rel, the progenitor of RelA and SpoT1. Like SpoT, Rel has both synthetic and hydrolytic activities, and like RelA, its synthetic activity is stimulated by starved ribosomes harbouring a deacylated tRNA in the A-site15,16. In addition to the above-mentioned ‘long’ multi-domain RSHs – RelA, Rel and SpoT – there are more than twenty subfamilies of ‘short’ single-domain RSHs that possess only the synthetic (Small Alarmone Synthetase, SAS) or hydrolytic (Small Alarmone Hydrolase, SAH) domain1. B. subtilis possesses two SAS proteins: SAS1 (synonyms: YjbM and RelQ) and SAS2 (synonyms: YwaC and RelP)17,18,19. While under normal growth conditions SAS enzymes contribute to basal (p)ppGpp levels20, cell wall stress stimuli such as treatment with cell wall-active antibiotics or alkaline shock induce expression of SAS via transcriptional up-regulation, and the resultant increase in (p)ppGpp levels orchestrates the response to stress17,21.
The functionality of the (p)ppGpp-mediated regulatory system is crucial for bacterial virulence22, survival during host invasion21 and antibiotic tolerance23. The alarmone (p)ppGpp was recently proposed to be the primary driver behind the formation of antibiotic-tolerant phenotypic variants in clonal bacterial populations, known as persister cells24. All this, in combination with the absence of a cytoplasmic RSH-mediated stringent response system in eukaryotes1,25, makes the enzymes involved in (p)ppGpp metabolism promising new targets for drug discovery, as inhibitors of the stringent response would act as anti-virulence agents. Disarming the pathogens, and targeting bacterial virulence – rather than killing bacteria – is believed to be a promising strategy due to lower selection pressure leading to slower emergence of resistance26.
The first steps towards the development of a specific and potent inhibitor of the stringent response have already been taken with the development of a nucleotide-based RSH inhibitor, Relacin27, and the anti-biofilm peptide 1018 that was suggested to inhibit the stringent response by binding (p)ppGpp and promoting its degradation28. However, Relacin is rather inefficient – it requires sub-mM concentrations27,29 – and 1018 has a strong bacteriotoxic effect; the concentration range in which it transitions from merely dispersing biofilms to killing bacteria is approximately 10-fold28.
Therefore, there is a need for more potent and selective stringent response inhibitors, motivating the current High Throughput Screening (HTS) project. Our HTS strategy is based on the following considerations. First, we opted for a whole-cell assay instead of an enzyme-based one, since inefficient cellular uptake is one of the main challenges in the discovery of novel antibacterials30,31. Second, we chose a phenotype-based screening approach – a strategy designed for the identification of compounds that target a specific pathway rather than antibacterials in general32.
Results
Screening strategy for the identification of B. subtilis Rel inhibitors relying on amino acid auxotrophy
We chose the Gram-positive bacterium B. subtilis to be used in the screening process because the chances of identifying biologically active compounds in Gram-positive bacteria are considerably higher than in Gram-negative bacteria30. To improve the selectivity of the HTS for the inhibition of long ribosomal RSH Rel – the primary driver of acute stringent response – we used a B. subtilis strain lacking functional SAS RelQ and RelP (ΔSAS strain)17. Moreover, SAS enzymes can be refractory to inhibitors of Rel, e. g. Enterococcus faecalis RelQ is insensitive to ppGpp analogue Relacin29, thus potentially masking the effect of Rel inhibition.
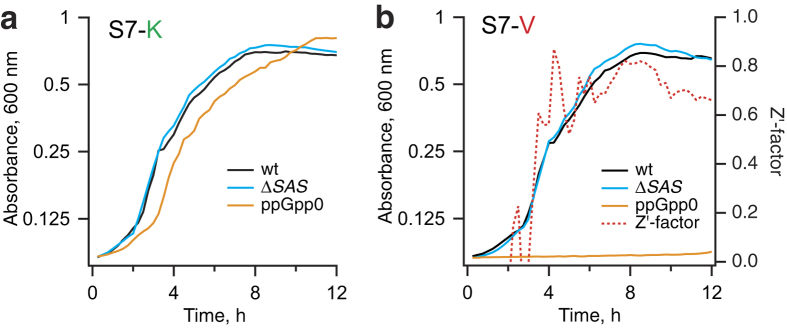
Our screening assay relies on the amino acid auxotrophy phenotype characteristic of (p)ppGpp-deficient (ppGpp0) strains11,33. Specifically, we exploit the auxotrophy of ppGpp0 B. subtilis for branched-chain amino acids – valine, leucine and isoleucine33 – by using a combination of two drop-out variants of S7 growth medium34: S7 lacking valine (S7-V) and S7 lacking lysine (S7-K). Wild type, ΔSAS and ppGpp0 B. subtilis (ΔSASΔrel) strains grew efficiently on S7-K (Fig. 1a). However, omission of valine completely inhibits the growth of the ppGpp0 strain without significantly affecting the growth of the isogenic wild type and the ΔSAS strains (Fig. 1b). The growth assay differentiates between ΔSAS and ppGpp0 strains with a Z’-factor35 exceeding 0.5 when the OD600 of the ΔSAS strain reaches 0.25 and above, and is readily adaptable to the 384-well screening format (Supplementary Fig. S1).
Figure 1. Valine autotrophy of ppGpp0 B. subtilis can be exploited for the detection of Rel inhibition.
B. subtilis strain 168 (wt), its isogenic strain lacking Small Alarmone Synthetases (SAS) YjbM and YwaC (ΔSAS; RIK100217) and a ppGpp-deficient strain lacking both SAS and long RSH Rel (ppGpp0; RIK100317) were grown in S7 minimal medium supplemented with 1% glucose and a set of 19 amino acids lacking either L-lysine (a; S7-K) or L-valine (b; S7-V). (a) In S7 medium lacking lysine (S7-K) the ppGpp0 strain grows in a similar way to the isogenic wild type and ΔSAS. (b) Due to valine autotrophy, ppGpp0 B. subtilis is unable to grow in S7-V while the ΔSAS strain grows as well as the wild type. Traces show arithmetic mean OD600 values of four technical replicates; means and standard deviations of OD600 readings were used to calculate the Z’-factor35.
Two-stage HTS for Rel inhibitors
To identify potential Rel inhibitors, we screened for a conditional growth inhibition of the ΔSAS strain in S7-V medium, but not in S7-K medium. The conditionality of the growth inhibition is used to sieve out generally cytotoxic compounds. S7-V-specific growth inhibition can, potentially, occur for several reasons. It may be due to the inhibition of the stringent response – the desired outcome – or be due for an unrelated reason, e.g. the compound is directly targeting an enzyme in valine biosynthesis leading to the absence of valine or is affecting either GTP biosynthesis or GTP sensing thus phenocopying ppGpp0 33,36,37,38,39. Even the desired outcome – inhibition of (p)ppGpp accumulation – could, potentially, be brought about not via inhibition of Rel’s (p)ppGpp synthesis activity (the desired hit) but via activation of its hydrolytic activity; cross-talk between the two active sites of the enzyme40 can further complicate matters. Therefore, it is impossible to estimate a priori the proportion of false positive hits generated by the screen and it is crucial to directly test the efficiency of identified compounds against the desired molecular target, i. e. the (p)ppGpp synthesis activity of B. subtilis Rel.
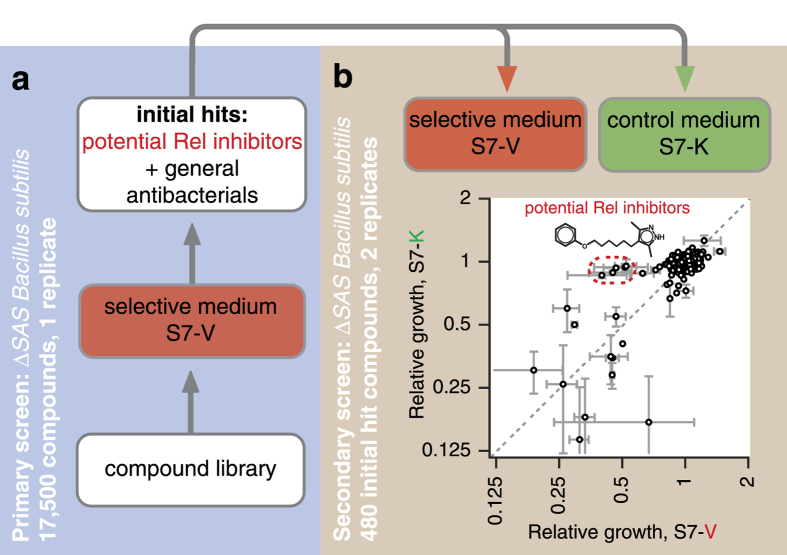
We based our HTS assay on the protocol by Zlitni and colleagues41. Frozen stock of the B. subtilis ΔSAS strain was used as a starting material by diluting the cell suspension ten times in the respective medium. For the primary screen we used the LCBU Screening Set of 17,500 synthetic drug-like organic compounds in S7-V medium at a final concentration of 10 μM in the presence of 0.9% DMSO (Fig. 2a). The growth of the ΔSAS strain was scored after 9 hours at 37 °C using the absorbance at 600 nm as a readout. Compounds inhibiting bacterial growth were identified using two complementary approaches: MScreen42 and our custom pipeline (see Methods for details). MScreen calculates mean value for all sample wells plate-by-plate and rates compounds according to their relative Z-score35. Our pipeline identifies and ranks local outliers using median absolute deviation (MAD) of absorbance values within a sliding window as a measure of local variability (see Supplementary Fig. S2 for the graphical output of the in-house program). The 300 top scoring candidates identified by the two approaches were pooled together yielding a final list of 480 hits candidate compounds after the removal of duplicates (see Supplementary Fig. S3 for a comparison of the two approaches).
Figure 2. Two-stage HTS for Rel inhibitors.
B. subtilis ΔSAS was grown on 384-well plates in the presence of 10 μM of each test compound. OD600 values were scored after 9 hours of growth at 37 °C. (a) The primary screen was performed in S7-V medium using 17,500 compounds selected from a set of small molecule libraries (ChemBridge). Compounds that inhibited B. subtilis growth in S7-V were scored using MScreen42 and a custom R script (see Methods), the lists of initial hit compounds identified by the two approaches were pooled and analysed by the secondary screen. (b) In the secondary screen 480 initial hits from the primary screen were screened in both S7-V (selective medium) and S7-K (control medium for identification of general antibacterials) in two replicates. Results fall into the following three categories: selective inhibitors causing valine auxotrophy, e.g. via inhibition of Rel; general antibacterials; and random fluctuations causing growth retardation. Five compounds, all sharing the common 4-(6-(phenoxy)alkyl)-3,5-dimethyl-1H-pyrazole moiety (see Table 1), selectively inhibited growth in S7-V, but not in S7-K (encircled with a red dashed line).
In the secondary screening we tested the 480 initial hits for growth retardation of ΔSAS B. subtilis in both S7-V (the selective medium) and S7-K (the control medium for identification of general antibacterials) (Fig. 2b). Since the readout – growth retardation – is prone to the generation of spurious hits, we performed two replicates of the secondary screening. The two replicates showed satisfactory reproducibility with Pearson’s product-moment correlation coefficient above 0.8 (Supplementary Fig. S4).
Twelve compounds displayed equally pronounced inhibitory effects in both S7-V and S7-K, suggesting that these are general antibacterials and are not of interest as potential stringent response inhibitors (Supplementary Table 1). However, five compounds have a moderate inhibitory effect on B. subtilis growth in S7-V (growth retardation of approximately 50%) while they have virtually no effect on growth in S7-K (Fig. 2b, encircled in red dashed line). The identified compounds are: 4-[6-(4-chlorophenoxy)hexyl]-3,5-dimethyl-1H-pyrazole (1, C302), 4-[6-(2,5-dimethylphenoxy)hexyl]-3,5-dimethyl-1H-pyrazole (2, C318), 3,5-dimethyl-4-[6-(2-methylphenoxy)hexyl]-1H-pyrazole (3, C303), 4-[6-(3,5-dimethylphenoxy)hexyl]-3,5-dimethyl-1H-pyrazole (4, C285), and 4-[4-(2-tert-butylphenoxy)butyl]-3,5-dimethyl-1H-pyrazole (5, C385) (Table 1). Importantly, all compounds share the same core, 4-(6-alkyl)-3,5-dimethyl-1H-pyrazole, and do not possess any of the characteristics of Pan Assay Interference compounds (PAINS) – a group of structurally diverse compounds displaying nonspecific activity against a wide range of unrelated target proteins43,44.
Table 1. Structure and growth inhibition efficiency of the hit compounds.

Our two-stage screen resulted in five hits with a common structural core, 4-(6-(phenoxy)alkyl)-3,5-dimethyl-1H-pyrazole. Based on dose-response analysis, none of the compounds inhibit growth specifically in S7 medium lacking valine (S7-V) when compared to the medium lacking lysine (S7-K). Dose-response analyses were performed in three technical replicates. IC50: half-maximal inhibitory concentration; CI95%: 95% confidence interval.
Derivatives 4-(6-(phenoxy)alkyl)-3,5-dimethyl-1H-pyrazole are general antibacterials
To investigate the specificity of the five identified hit compounds against the stringent response, we characterized their efficiency in dose-response assays (Supplementary Fig. S5). Disappointingly, as the concentrations increase all of the compounds efficiently inhibit B. subtilis growth not just in the selective S7-V medium, but also in the control S7-K medium. The half maximal inhibitory concentrations (IC50) range from 4.5 to 14.5 μM, and the concentration used for the HTS (10 μM) is located on the slope of the dose-response curve where the effect is extremely sensitive to assay conditions such as composition of the growth medium.
To confirm that Rel is not the primary target of the hit compounds, we have tested a representative hit compound – C302 – against wild type, ΔSAS and ppGpp0 B. subtilis strains in S7 medium supplemented with the full set of 20 amino acids in order to mitigate the potential metabolic differences between the strains (Fig. 3). Growth of all strains is inhibited by C302 with an IC50 around 20–25 μM, though ppGpp0 B. subtilis is marginally more sensitive than the other two strains. Finally, we directly tested the effect of C302 on ppGpp synthesis by using purified B. subtilis Rel activated by ‘starved’ ribosomal complexes carrying deacylated tRNA in the A-site; we observed no inhibition (Supplementary Fig. S6). Taken together, our data suggest that rather than acting as specific Rel inhibitors, the derivatives of 4-(6-(phenoxy)alkyl)-3,5-dimethyl-1H-pyrazole act as general antibacterials.
Figure 3. Growth inhibition efficiency of compound C302 against B. subtilis ppGpp-synthetase mutants.
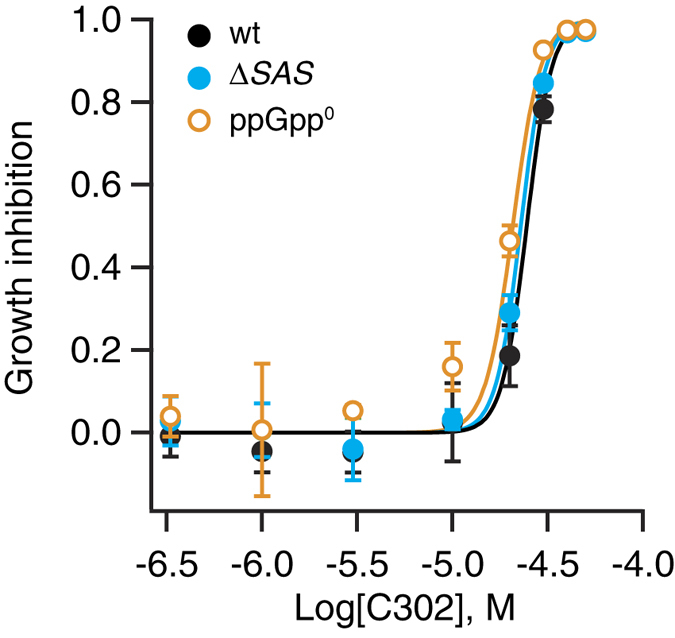
A representative of the HTS hit compounds, compound C302 (4-[6-(4-chlorophenoxy)hexyl]-3,5-dimethyl-1H-pyrazole), was serially diluted in DMSO and titrated into liquid cultures of B. subtilis wild-type (wt) strain, mutant strain lacking YjbM and YwaC (ΔSAS), and ppGpp-deficient (ppGpp0) strains. The growth conditions are identical to those used for HTS with the exception of the S7 medium being supplemented with the full set of 20 amino acids and 1.3% DMSO. Half-maximal inhibitory concentrations are 24.5 μM (CI95% 23.3–25.9) for wt, 22.8 μM (CI95% 21.8–23.9) for ΔSAS and 20.7 μM (CI95% 19.3–22.3) for ppGpp0. The experiments were performed in three replicates and error bars (too small to be seen for some of the points) indicate standard error of the mean.
Relacin and 1018 do not pass the valine auxotrophy test for specific inhibitors of the stringent response in B. subtilis
We have benchmarked our assay against the two previously reported inhibitors of the stringent response: the anti-biofilm peptide 101828 and the ppGpp-based nucleotide Relacin27. When tested against the ΔSAS strain, the addition of up to 2 mM of Relacin does not inhibit the bacterial growth in either S7-V or S7-K medium (Supplementary Fig. S7). Similarly, the compound has little effect on the growth of the ppGpp0 strain (Supplementary Fig. S7). Peptide 1018 efficiently inhibits B. subtilis growth in the S7-V medium with half of the maximal inhibitory concentration IC50 = 0.4 μM (CI 95%: 0.36–0.44), whereas in S7-K medium the bacterium is slightly less sensitive to the inhibitor (IC50 = 0.6 μM; CI 95%: 0.56–0.63) – a similar effect has lead to the identification of the compounds in the current HTS (Fig. 4). Moreover, 1018 has essentially the same growth inhibition efficiency on the ppGpp0 strain in S7-K (IC50 = 0.53 μM, CI95%: 0.48–0.59), underscoring the ppGpp-independent nature of the antibacterial effect.
Figure 4. Peptide 1018 does not pass the selection criteria for a selective Rel inhibitor of the current auxotrophy-based HTS.
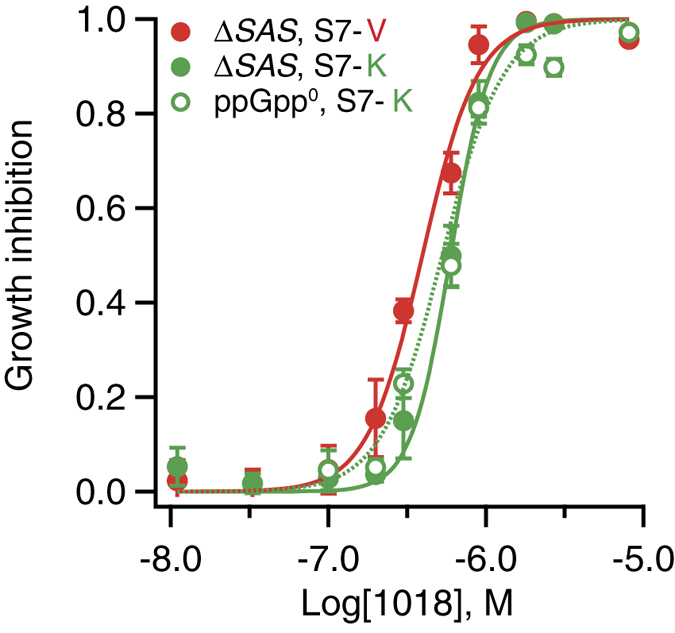
Dose-response curves of the B. subtilis ΔSAS strain in S7-K and the B. subtilis ppGpp0 strain in S7-K in the presence of increasing concentrations of peptide 1018. Peptide 1018 has the same growth inhibitory efficiency against ΔSAS and ppGpp0 strains in S7-K with an IC50 of 0.6 μM (CI95% 0.56–0.63) and 0.53 μM (CI95% 0.48–0.59), respectively. In S7-V medium the ΔSAS strain is slightly more sensitive to the peptide treatment with an IC50 of 0.4 μM (CI95% 0.36–0.44)). The experiments were performed in three replicates and error bars (too small to be seen for some of the points) indicate standard error of the mean.
Discussion
Despite relying on a robust, virtually “all-or-nothing” auxotrophic phenotype of the ppGpp0 B. subtilis strain, our HTS project did not yield a selective stringent response inhibitor. A class of compounds – derivatives of 4-(6-(phenoxy)alkyl)-3,5-dimethyl-1H-pyrazole – did display preferential activity against B. subtilis growing in the medium lacking valine, as compared to the medium lacking lysine in a single-concentration HTS assay. However, subsequent dose-response assays revealed a dominant general antibacterial effect. Further derivatization and structure activity relationship analysis is necessary to assess the potential of 4-(6-(phenoxy)alkyl)-3,5-dimethyl-1H-pyrazoles as antibacterials. We have used our screening assay to test two promising stringent response inhibitors – a ppGpp-mimic nucleotide, Relacin,27 and an antibiofilm peptide, 101828 – and neither of the two pass the stringent selection criteria used in the current study. The efficiency of both compounds against B. subtilis is virtually the same for ppGpp-deficient and wild type strains (Fig. 4, Supplementary Fig. S7). The quest for the discovery of specific and potent stringent response inhibitors continues.
Since our HTS assay is calibrated against a complete genetic disruption of the ppGpp synthesis in the cell, the potential hit compounds are expected to be both efficient and specific. Unfortunately, such molecules were absent in the screening library employed in the current study. This is a common problem: conventional chemical libraries used for HTS projects are ill-suited for antibiotic discovery since, as judged by molecular weight and polarity, natural antibiotics poorly fit the molecular profile of the drug-like molecules used to populate the chemical libraries30,45. Development of targeted HTS libraries with validated bacterial permeability could considerably improve the hit rate. However, we are not aware of the existence of such libraries. Therefore, application of the HTS assay described in the current report to alternative sources of compounds such as natural products46 is a possible next step.
Methods
Bacillus subtilis strains and growth conditions
Construction of the B. subtilis 168-based strain RIK1002 lacking Small Alarmone Synthetases (SAS) YjbM and YwaC (ΔSAS; trpC2 ΔyjbM ywaC::spc) and the ppGpp-deficient strain RIK1003 additionally lacking Rel (ppGpp0; trpC2 ΔyjbM ywaC::spc relA::erm) is described in Nanamiya et al.17. Liquid cultures were grown at 37 °C in S7 minimal medium supplemented with 1% glucose and amino acids as per Nicholson and Seltow47. The medium was prepared as described in Nanamiya et al.17 except L-glutamate was substituted with a full set of 20 naturally-occurring amino acids as per Cutting and Horn34; L-valine (S7-V) or L-lysine (S7-K) were omitted to generate selective media for screening. Cultures were grown with aeration for the preparation of cell stocks and without aeration during HTS. To prepare the bacterial stock for inoculation for HTS B. subtilis strains RIK1002 and RIK1003 were grown from a single fresh colony to OD600 1.7 in S7 medium supplemented with the full set of 20 amino acids (S7), diluted to OD600 0.25 in S7-V or S7-K, supplemented with 8% DMSO, aliquoted, snap frozen in liquid nitrogen and stored at −70 °C prior to the screen for not longer than 6 weeks.
Primary and secondary HTS
The HTS procedure was based on the protocol by Zlitni and colleagues41. The primary screen used a LCBU Screening Set of 17,500 compounds selected from a set of small molecule libraries (ChemBridge) dispensed using Echo®-(Labcyte) directly into 384-well plates yielding 56 plates with 320 compounds per plate with the first two and last two columns reserved for controls, 80 nl of 10 mM compound in 100% DMSO was added per well, and 80 nl of 100% DMSO was used in control wells. Frozen stocks of RIK1002 were thawed on ice, diluted 10 times with S7-V medium and 80 μl was dispensed per well on library plates (final concentration of compounds: 10 μM and DMSO of 0.9%). The ppGpp0 strain RIK1003 served as a positive control (10 wells, 80 μl per well) and ΔSAS strain RIK1002 as a negative control (16 wells, 80 μl per well). The first and the last column contained pure medium as a contamination control. Plate lids were sealed with parafilm, stacked by four and incubated at 37 °C for 9 hours. Evaporation from the plates during the incubation at 37 °C was countered in two ways. First, plates were stacked by four and each stack was topped with an extra plate filled with water. Second, a water bath was placed in the incubator. Growth was measured at OD600. For the secondary screen using a combination of S7–V and S7–K media, 480 compounds identified as possible hits (see below, HTS data analysis) during the primary screen were tested in two replicates. As an extra precaution against the edge effects the two outer-most columns of each plate were left empty.
HTS data analysis
Statistical analysis of HTS results followed the guidelines of Malo and colleagues48. Primary screen data were analysed with a custom script in the R programming language49 and with MScreen42. The R script compares the read out of the individual well with the median of a window of wells, calculated in median absolute deviations (MAD) (the code is provided in the Supplementary Information). MScreen compares the read out of the individual well with the average of sample wells on the whole plate, calculated in standard deviations (SD)42. The 300 top candidates identified by the two programs were pooled together yielding, after removal of duplicates, a final list of 480 hits used for the secondary screen. The performance of the two approaches is compared in Supplementary Fig. S3. Identity of the hit compounds was confirmed by liquid chromatography-mass spectrometry (LC-MS).
Dose-response analysis
For dose-response analysis, the HTS assay was performed in a 96 well microtiter plate format using 160 μl of ΔSAS B. subtilis cultures supplemented with increasing concentrations of test compound in the presence of 1.3% DMSO. Bacterial growth was measured at OD600 after 9 hours of incubation at 37 °C. Growth inhibition was calculated as 1 – (AS – AM)/(AUT-AM) where A stands for OD600 absorbance of the well and indexes S, M, UT indicate sample, medium and untreated control, respectively. Dose-response curves and IC50 values together with 95% confidence intervals were calculated in Prism 6 (GraphPad) using the variable slope Hill equation (Y = Bottom + (Top-Bottom)/(1 + 10^((LogIC50-X)*HillSlope)) where Y is the modelled response; Bottom is the lowest experimental growth inhibition value; Top is the highest experimental growth inhibition value; IC50 is the half-maximal inhibitory concentration; and X is the compound concentration. Relacin was synthesized as per Gaca et al. (2015)29. Peptide 1018 (VRLIVAVRIWRR-NH2) was ordered in lyophilized form from Storkbio Ltd (>95% pure).
B. subtilis Rel enzymatic assay
The ppGpp synthesis assay was performed as per Shyp et al. (2012)10 with B. subtilis Rel activated by B. subtilis ribosomes with dealylated tRNAPhe in the presence of 100 μM pppGpp and using 1 mM ATP and 300 μM 3H-GTP as substrates. For more details, see Supplementary Information.
Additional Information
How to cite this article: Andresen, L. et al. Auxotrophy-based High Throughput Screening assay for the identification of Bacillus subtilis stringent response inhibitors. Sci. Rep. 6, 35824; doi: 10.1038/srep35824 (2016).
Supplementary Material
Acknowledgments
We are grateful to Per-Anders Enquist from Laboratories for Chemical Biology Umeå for helpful discussions and Dominik Rejman for synthesizing Relacin. This work was supported by the by grant IUT2–22 from the Estonian Research Council (TT); European Regional Development Fund through the Centre of Excellence for Molecular Cell Engineering (VH and TT); Estonian Science Foundation (PUT37 to VH); Umeå University, the Swedish Research council Vetenskapsrådet (grant 2013–4680), Kempe and Ragnar Söderberg foundations (VH).
Footnotes
Author Contributions V.H. conceived the project and coordinated the study. L.A., V.V. and V.H. designed the experiments, wrote the code and analysed the data. L.A., V.V. and S.J. performed experiments with assistance of S.L. Y.T., S.L. and T.T. contributed tools and reagents. V.H. wrote the paper with contributions from L.A. and V.V.
References
- Atkinson G. C., Tenson T. & Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6, e23479 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauryliuk V., Atkinson G. C., Murakami K. S., Tenson T. & Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13, 298–309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashel M. & Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221, 838–841 (1969). [DOI] [PubMed] [Google Scholar]
- Laffler T. & Gallant J. spoT, a new genetic locus involved in the stringent response in E. coli. Cell 1, 27–30 (1974). [Google Scholar]
- Haseltine W. A. & Block R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci USA 70, 1564–1568 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine W. A., Block R., Gilbert W. & Weber K. MSI and MSII made on ribosome in idling step of protein synthesis. Nature 238, 381–384 (1972). [DOI] [PubMed] [Google Scholar]
- Arenz S. et al. The stringent factor RelA adopts an open conformation on the ribosome to stimulate ppGpp synthesis. Nucleic Acids Res 44, 6471–6481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A., Fernandez I. S., Gordiyenko Y. & Ramakrishnan V. Ribosome-dependent activation of stringent control. Nature 534, 277–280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland A. B. et al. Ribosome*RelA structures reveal the mechanism of stringent response activation. Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyp V. et al. Positive allosteric feedback regulation of the stringent response enzyme RelA by its product. EMBO Rep 13, 835–839 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H. et al. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem 266, 5980–5990 (1991). [PubMed] [Google Scholar]
- An G., Justesen J., Watson R. J. & Friesen J. D. Cloning the spoT gene of Escherichia coli: identification of the spoT gene product. J Bacteriol 137, 1100–1110 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfzadeh M., Keener J. & Nomura M. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc Natl Acad Sci USA 90, 11004–11008 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D., Albrecht C., Cashel M. & D’Ari R. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol Microbiol 56, 958–970 (2005). [DOI] [PubMed] [Google Scholar]
- Avarbock A. et al. Functional regulation of the opposing (p)ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochemistry 44, 9913–9923 (2005). [DOI] [PubMed] [Google Scholar]
- Avarbock D., Salem J., Li L. S., Wang Z. M. & Rubin H. Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene 233, 261–269 (1999). [DOI] [PubMed] [Google Scholar]
- Nanamiya H. et al. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol Microbiol 67, 291–304 (2008). [DOI] [PubMed] [Google Scholar]
- Steinchen W. et al. Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone. Proc Natl Acad Sci USA 112, 13348–13353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan A. et al. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet 4, e1000139 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaca A. O. et al. Basal levels of (p)ppGpp in Enterococcus faecalis: the magic beyond the stringent response. MBio 4, e00646–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T. et al. The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog 8, e1003016 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux Z. D., Svensson S. L., Gaynor E. C. & Swanson M. S. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev 74, 171–199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. Bacterial stress responses as determinants of antimicrobial resistance. J Antimicrob Chemother 67, 2069–2089 (2012). [DOI] [PubMed] [Google Scholar]
- Maisonneuve E. & Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell 157, 539–548 (2014). [DOI] [PubMed] [Google Scholar]
- Martini O., Irr J. & Richter D. Questioning of reported evidence for guanosine tetraphosphate synthesis in a ribosome system from mouse embryos. Cell 12, 1127–1131 (1977). [DOI] [PubMed] [Google Scholar]
- Rasko D. A. & Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov 9, 117–128 (2010). [DOI] [PubMed] [Google Scholar]
- Wexselblatt E. et al. Relacin, a novel antibacterial agent targeting the Stringent Response. PLoS Pathog 8, e1002925 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente-Nunez C., Reffuveille F., Haney E. F., Straus S. K. & Hancock R. E. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog 10, e1004152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaca A. O. et al. From (p)ppGpp to (pp)pGpp: Characterization of Regulatory Effects of pGpp Synthesized by the Small Alarmone Synthetase of Enterococcus faecalis. J Bacteriol 197, 2908–2919 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne D. J., Gwynn M. N., Holmes D. J. & Pompliano D. L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6, 29–40 (2007). [DOI] [PubMed] [Google Scholar]
- Tommasi R., Brown D. G., Walkup G. K., Manchester J. I. & Miller A. A. ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov 14, 529–542 (2015). [DOI] [PubMed] [Google Scholar]
- Farha M. A. & Brown E. D. Unconventional screening approaches for antibiotic discovery. Ann N Y Acad Sci 1354, 54–66 (2015). [DOI] [PubMed] [Google Scholar]
- Kriel A. et al. GTP dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes. J Bacteriol 196, 189–201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutting S. M. & Horn P. B. V. In Molecular biological methods for Bacillus (eds. Hardwood, & Cutting, ) (Cutting, New York, 1990). [Google Scholar]
- Zhang J. H., Chung T. D. & Oldenburg K. R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen 4, 67–73 (1999). [DOI] [PubMed] [Google Scholar]
- Ochi K., Kandala J. & Freese E. Evidence that Bacillus subtilis sporulation induced by the stringent response is caused by the decrease in GTP or GDP. J Bacteriol 151, 1062–1065 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J. M., Dromerick A. & Freese E. Response of guanosine 5′-triphosphate concentration to nutritional changes and its significance for Bacillus subtilis sporulation. J Bacteriol 146, 605–613 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinsmade S. R. & Sonenshein A. L. Dissecting complex metabolic integration provides direct genetic evidence for CodY activation by guanine nucleotides. J Bacteriol 193, 5637–5648 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriel A. et al. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol Cell 48, 231–241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg T., Mechold U., Malke H., Cashel M. & Hilgenfeld R. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected]. Cell 117, 57–68 (2004). [DOI] [PubMed] [Google Scholar]
- Zlitni S., Blanchard J. E. & Brown E. D. High-throughput screening of model bacteria. Methods Mol Biol 486, 13–27 (2009). [DOI] [PubMed] [Google Scholar]
- Jacob R. T. et al. MScreen: an integrated compound management and high-throughput screening data storage and analysis system. J Biomol Screen 17, 1080–1087 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baell J. & Walters M. A. Chemistry: Chemical con artists foil drug discovery. Nature 513, 481–483 (2014). [DOI] [PubMed] [Google Scholar]
- Dahlin J. L. & Walters M. A. How to Triage PAINS-Full Research. Assay Drug Dev Technol 14, 168–174 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea R. & Moser H. E. Physicochemical properties of antibacterial compounds: implications for drug discovery. J Med Chem 51, 2871–2878 (2008). [DOI] [PubMed] [Google Scholar]
- Harvey A. L., Edrada-Ebel R. & Quinn R. J. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov 14, 111–129 (2015). [DOI] [PubMed] [Google Scholar]
- Nicholson W. L. & Seltow P. In Molecular biological methods for Bacillus (eds. Hardwood, & Cutting, ) (Wiley: New York,, 1990). [Google Scholar]
- Malo N., Hanley J. A., Cerquozzi S., Pelletier J. & Nadon R. Statistical practice in high-throughput screening data analysis. Nat Biotechnol 24, 167–175 (2006). [DOI] [PubMed] [Google Scholar]
- A Language and Environment for Statistical Computing, R Core Team, R Foundation for Statistical Computing, Vienna, Austria https://www.R-project.org. (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.