Abstract
Mutations in PINK1 (PTEN-induced putative kinase 1) cause early onset familial Parkinson's disease (PD). PINK1 accumulates on the outer membrane of damaged mitochondria followed by recruiting parkin to promote mitophagy. Here, we demonstrate that BCL2/adenovirus E1B 19-kDa interacting protein 3 (BNIP3), a mitochondrial BH3-only protein, interacts with PINK1 to promote the accumulation of full-length PINK1 on the outer membrane of mitochondria, which facilitates parkin recruitment and PINK1/parkin-mediated mitophagy. Inactivation of BNIP3 in mammalian cells promotes PINK1 proteolytic processing and suppresses PINK1/parkin-mediated mitophagy. Hypoxia-induced BNIP3 expression results in increased expression of full-length PINK1 and mitophagy. Consistently, expression of BNIP3 in Drosophila suppresses muscle degeneration and the mitochondrial abnormality caused by PINK1 inactivation. Together, the results suggest that BNIP3 plays a vital role in regulating PINK1 mitochondrial outer membrane localization, the proteolytic process of PINK1 and PINK1/parkin-mediated mitophagy under physiological conditions. Functional up-regulation of BNIP3 may represent a novel therapeutic strategy to suppress the progression of PD.
Keywords: hypoxia, mitochondria, parkin, Parkinson's disease, ubiquitin
Introduction
Mutations in PINK1 or parkin are linked to the early onset familial form of Parkinson's disease (PD),2 the most common neurodegenerative movement disorder (1–3). PINK1 encodes a putative serine/threonine kinase with an N-terminal mitochondrial localization sequence (3). Two forms of PINK1 are identified in the cell, including a 64-kDa full-length form and a 55-kDa proteolytic fragment lacking the N-terminal mitochondrial localization sequence (4–6). Parkin is a RING domain-containing E3 ligase (7–10). PINK1, parkin, and DJ-1 form an E3 ligase complex to promote the degradation of mis-/unfolded proteins (11). Recently, PINK1 and parkin were shown to play a critical role in the clearance of damaged mitochondria via a mitophagy-mediated mechanism (12–16). It is proposed that PINK1 is stabilized and accumulates on the mitochondrial outer membrane (MOM) upon mitochondrial depolarization. This subsequently recruits parkin to ubiquitinate MOM proteins followed by degradation of the damaged mitochondria via a mitophagy (15–18). However, the physiological regulatory mechanisms for PINK1-controlled mitophagy remain poorly understood.
BNIP3 is a mitochondrial BH3-only protein that induces cell death via activating BAX/BAK and opening of the mitochondrial permeability transition pore (19–22). BNIP3 is also a potent autophagy inducer through mechanisms independent of its pro-cell death activity (23–25). It has been shown to serve as an autophagy receptor for the binding of mitochondria to LC3-II on the autophagosome via its N-terminal LC3-interacting region (26, 27). BNIP3 may also regulate mitophagy through other mechanisms. A recent study shows that NIX, a BNIP3 homolog, interacts with parkin, resulting in its recruitment to depolarized mitochondria (28).
In the present study, we aimed to understand the regulatory mechanism of PINK1/parkin-mediated mitophagy. The results reveal that BNIP3 interacts with PINK1 to suppress its cleavage, resulting in the accumulation of 64-kDa full-length PINK1 on MOM, leading to increased parkin recruitment and enhanced mitochondrial clearance via mitophagy. Inactivation of BNIP3 in mammalian cells promotes PINK1 proteolytic processing and suppresses PINK1/parkin-regulated mitophagy. Hypoxia-induced endogenous BNIP3 expression results in increased levels of full-length PINK1 and mitophagy. Furthermore, expression of BNIP3 restores mitochondrial morphology and ATP production in PINK1 null Drosophila. This study identified a physiological regulatory mechanism of PINK1 and PINK1-mediated mitophagy.
Results
BNIP3-PINK1 Interaction Inhibits Proteolytic Cleavage of PINK1
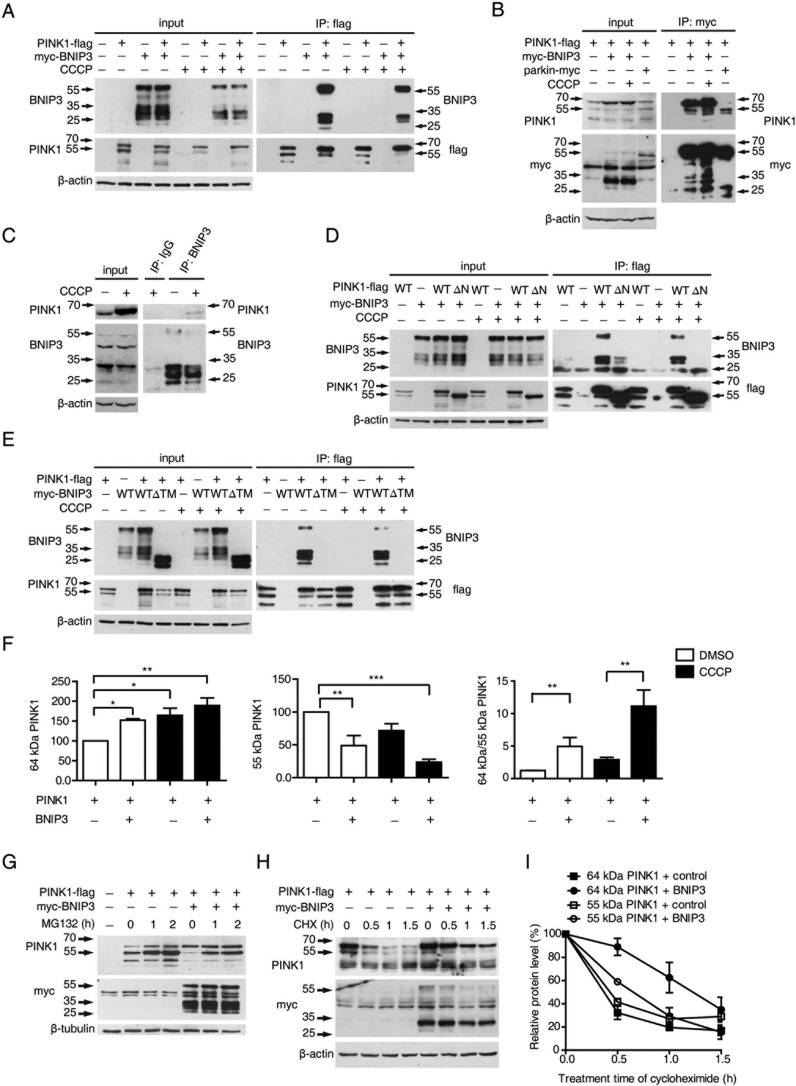
The endogenous interaction between BNIP3 and PINK1 was initially identified by a mass spectrometry analysis using mitochondrial proteins isolated from mouse brains (40). To verify the interaction between BNIP3 and PINK1, HEK293 cells expressing Myc-BNIP3 and PINK1-FLAG were lysed followed by co-immunoprecipitation assays. The results showed that immunoprecipitation of PINK1 co-precipitated BNIP3 with or without the mitochondrial uncoupling agent, carbonyl cyanide m-chlorophenylhydrazone (CCCP) (Fig. 1A). Conversely, immunoprecipitation of BNIP3 brought down PINK1 (Fig. 1B). As a positive control, PINK1 was co-precipitated with parkin (Fig. 1B). Consistently, endogenous PINK1 was also co-immunoprecipitated with endogenous BNIP3 when cells were treated with CCCP (Fig. 1C). Further analysis revealed that deletion of the N-terminal mitochondrial localization sequence of PINK1 (PINK1 ΔN) abolished its binding to BNIP3 (Fig. 1D). Moreover, deletion of the C-terminal transmembrane domain of BNIP3 (BNIP3 ΔTM) eliminated its binding to PINK1 (Fig. 1E). The results suggest that PINK1 and BNIP3 interact on mitochondrial membrane. Two PINK1 kinase-deficient mutants, G309D and D384N, continue to bind BNIP3, suggesting that the kinase activity of PINK1 is not required for its interaction with BNIP3 (supplemental Fig. 1).
FIGURE 1.
BNIP3-PINK1 interaction inhibits proteolytic cleavage of PINK1. A, co-immunoprecipitation of BNIP3 and PINK1. HEK293 cells expressing PINK1-FLAG or/and Myc-BNIP3 were immunoprecipitated (IP) with an anti-FLAG antibody followed by immunoblotting with anti-BNIP3 or an anti-FLAG tag. Input controls are shown on the left panel. B, full-length PINK1 interacts with BNIP3. HEK293 cells expressing PINK1-FLAG or/and Myc-BNIP3 or parkin-Myc were immunoprecipitated with an anti-Myc tag and immunoblotted with either anti-PINK1 or anti-Myc tag (to detect BINP3 or parkin). The parkin-PINK1 interaction is a control for co-immunoprecipitation. Input controls are shown on the left panel. C, co-immunoprecipitation of endogenous BNIP3 and PINK1. HEK293 cells with/without CCCP treatment were immunoprecipitated with either a control IgG or anti-BNIP3 followed by immunoblotting with anti-PINK1. Input controls are shown on the left panel. D, PINK1 MTS deletion impairs PINK1-BNIP3 interaction. HEK293 cells were co-transfected with Myc-BNIP3 with an empty plasmid (−), FLAG-tagged PINK1 (WT), or MTS-deleted PINK1 (ΔN) followed by CCCP treatment. Lysates were immunoprecipitated with an anti-FLAG tag followed by immunoblotting with anti-BNIP3 or anti-FLAG. Input controls are shown on the left panel. E, BNIP3 ΔTM impairs its interaction with PINK1. HEK293 cells were co-transfected PINK1-FLAG with an empty plasmid (−), Myc-tagged BNIP3 (WT), or Myc-tagged BNIP3 ΔTM followed by CCCP treatment. Lysates were immunoprecipitated with anti-FLAG followed by immunoblotting with BNIP3 or a FLAG tag. Input controls are shown on the left panel. F, quantitation of PINK1 variants regulated by BNIP3. The relative levels of 64- and 55-kDa PINK1 variants and the ratio of 64-kDa/55-kDa PINK1 with/without CCCP treatment were quantified, calculated, and analyzed by one-way ANOVA and Dunnett's test or Tukey's test. Error bars, S.E.; n = 4. *, p < 0.05; **, p < 0.01; ***, p < 0.001. G, PINK1 cleavage affected by BNIP3 with MG132 treatment. HEK293 cells expressing PINK1-FLAG or/and Myc-BNIP3 were treated with MG132. Lysates were immunoblotted with anti-PINK1 (top panel) or anti-Myc (middle panel). β-Tubulin (bottom panel) was the loading control. H and I, stability of full-length PINK1 increased by BNIP3. HEK293 cells expressing PINK1-FLAG or/and Myc-BNIP3 were treated with CHX followed by immunoblotting with either anti-PINK1 (top panel) anti-Myc (middle panel). β-Actin (bottom panel) was the loading control (H). Quantification of PINK1 variants affected by BNIP3 is shown. Error bars, S.E.; n = 3 (I).
PINK1 is proteolytically processed to a 55-kDa fragment lacking the N-terminal mitochondrial localization sequence (4, 5). To our surprise, expression of BNIP3 resulted in increased detection of the 64-kDa full-length PINK1 and reduced detection of the 55-kDa PINK1 proteolytic fragment. Consistent with previous reports that full-length PINK1 is increased with mitochondrial depolarization (6, 15, 16), the accumulation of full-length PINK1 was further potentiated with CCCP treatment (Fig. 1A). Quantitative analyses verified a significant increase in 64-kDa full-length PINK1 with expression of BNIP3 (Fig. 1F). Immunofluorescent staining confirmed that the C-terminal FLAG-tagged PINK1 (PINK1-FLAG) was detected mostly in the cytoplasm. It became localized to the mitochondria following mitochondrial depolarization with CCCP treatment (supplemental Fig. 2). The results suggest that PINK1-FLAG resembles wild-type PINK1 in cellular localization and that BNIP3 promotes accumulation of 64 kDa full-length PINK1.
To determine whether full-length PINK1 accumulation following BNIP3 overexpression was due to inhibition of PINK1 proteasomal degradation, we treated cells with MG132, a proteasome inhibitor. The results showed that the 55-kDa PINK1 fragment accumulated following MG132 treatment as described previously (5, 16). Nevertheless, the levels of 55-kDa PINK1 in cells overexpressing BNIP3 remained markedly lower than those in control cells even after MG132 treatment (Fig. 1G). Cycloheximide (CHX)-mediated chase analysis revealed that half-life of full-length PINK1 was about 1.5 h in cells overexpressing BNIP3, whereas it was less than 0.5 h in control cells. In contrast, the half-life of the 55-kDa PINK1 proteolytic fragment remained similar in cells overexpressing BNIP3 and in cells with control transfection (Fig. 1, H and I). Together, these data show that BNIP3 suppresses the proteolytic cleavage of PINK1.
Mitochondrial Localization of BNIP3 Is Critical for Its Inhibition of PINK1 Cleavage
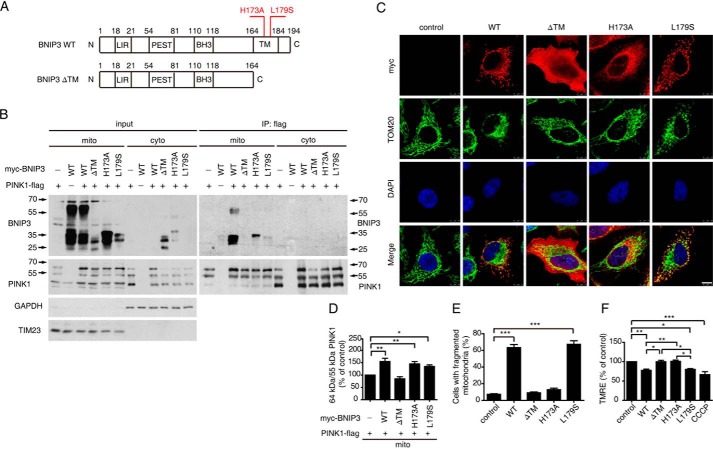
To investigate the mechanism of BNIP3 regulation of PINK1 cleavage, we employed three BNIP3 mutants involving its transmembrane domain, including a transmembrane domain deletion mutant (BNIP3 ΔTM) and two previously reported dimerization defect point mutants, BNIP3 H173A and BNIP3 L179S (30–33) (Fig. 2A). Both point mutants localize to the MOM. However, BNIP3 H173A does not change the mitochondrial membrane potential, whereas BNIP3 L179S does (30, 33). Consistent with previous reports, all three mutants are detected as a 30-kDa band and lack the 60-kDa dimer (Fig. 2B). Immunoprecipitation analysis revealed that BINP3 H173A and BNIP3 L179S interacted with PINK1 on mitochondria as its wild-type counterpart did, whereas BINP3 ΔTM did not (Fig. 2B). The ratio of 64-kDa/55-kDa PINK1 in cells expressing BNIP3 WT, H173A, and L179S was higher than in cells expressing BNIP3 ΔTM (Fig. 2, B and D). Immunofluorescent staining showed that BNIP3 WT, L179S, and H173A had prominent mitochondrial localization. In contrast, BNIP3 ΔTM was diffusely distributed in the cytoplasm (Fig. 2C). Together, the results suggest that mitochondrial membrane-associated BNIP3 interacts with PINK1 and suppresses full-length PINK1 cleavage.
FIGURE 2.
Mitochondrial localization is essential for BNIP3 to suppress PINK1 cleavage. A, illustration of BNIP3 variants. BNIP3 ΔTM, deletion of the C-terminal transmembrane domain; BNIP3 H173A, dimerization mutant; BNIP3 L179S, depolarization mutant; LIR, LC3-interacting region; PEST, proline, glutamic acid, serine, and threonine rich sequences. B, interaction of PINK1 and BNIP3 variants. HEK293 cells expressing PINK1-FLAG and Myc-tagged BNIP3 variants (WT, ΔTM, H173A, and L179S) in various combinations were fractionated to separate cytoplasm (cyto) and mitochondria (mito) followed by immunoprecipitation (IP) with anti-FLAG tag (to detect PINK1) and immunoblotting with anti-BNIP3 or anti-PINK1 (right panel). Input controls are shown on the left panel. TIM23 and GAPDH, a mitochondrial marker and a cytosolic marker, respectively. C, mitochondrial morphology in cells expressing BNIP3 variants. HeLa cells expressing Myc-tagged BNIP3 variants (WT, ΔTM, H173A, and L179S) were immunostained with anti-Myc tag (red) or anti-TOM20 (green). Cell nuclei were labeled with DAPI (blue). Colocalization is shown on the bottom panel (Merge). Note that significant mitochondrial fragmentation is observed in cells expressing WT and L179S. Bar = 10 μm. D, 64-kDa/55-kDa PINK1 ratios affected by BNIP3 variants. Relative protein levels from the mitochondrial fraction were measured using ImageJ and analyzed by one-way ANOVA and Dunnett's test. Error bars, S.E.; n = 4. *, p < 0.05; **, p < 0.01. E, quantitation of mitochondrial fragmentation. >300 transfected cells were counted for each experiment. The results were analyzed by one-way ANOVA and Dunnett's test. Error bars, S.E.; n = 3. ***, p < 0.001. F, mitochondrial membrane potential in cells expressing BNIP3 variants. Mitochondrial membrane potential was detected by TMRE staining followed by FACS analysis using HEK293 cells expressing Myc-tagged BNIP3 variants (WT, ΔTM, H173A, and L179S). HEK293 cells treated with CCCP (10 μm) for 2 h served as a positive control. Results were analyzed by one-way ANOVA and Dunnett's test. Error bars, S.E.; n = 4. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
We next analyzed the mitochondrial fragmentation and membrane potential of cells expressing BNIP3 variants. Expression of both BNIP3 WT and L179S, but not BNIP3 ΔTM and H173A, resulted in mitochondrial fragmentation (Fig. 2, C and E). Meanwhile, BNIP3 WT and L179S induced depolarization of mitochondria similar to the level seen with CCCP treatment. However, BNIP3 ΔTM and H173A had little effect on mitochondrial depolarization (Fig. 2F). Therefore, mitochondrial fragmentation and depolarization are unlikely to be the cause of BNIP3 suppression of PINK1 cleavage. The effects of BNIP3 variants on mitochondria and PINK1 cleavage are summarized in Table 1. These results suggest that the mitochondrial localization of BNIP3 plays an essential role in its suppression of PINK1 cleavage. Moreover, BNIP3-induced mitochondrial fragmentation and depolarization are unlikely the sole mechanisms for inhibition of PINK1 cleavage inhibition by BNIP3.
TABLE 1.
Role of BNIP3 variants in mitochondrial function and PINK1 cleavage
+, with the indicated functions or cleavage; −, without indicated functions or cleavage.
| BNIP3 |
||||
|---|---|---|---|---|
| WT | ΔTM | H173A | L179S | |
| BNIP3 dimerization | + | − | − | − |
| Mitochondrial fragmentation | + | − | − | + |
| Mitochondrial depolarization | + | − | − | + |
| Mitochondrial location | + | − | + | + |
| Interaction with PINK1 | + | − | + | + |
| Inhibition of PINK1 cleavage | + | − | + | + |
BNIP3 Interacts with Full-length PINK1 on MOM and Facilitates Parkin Recruitment to Mitochondria as well as Mitophagy
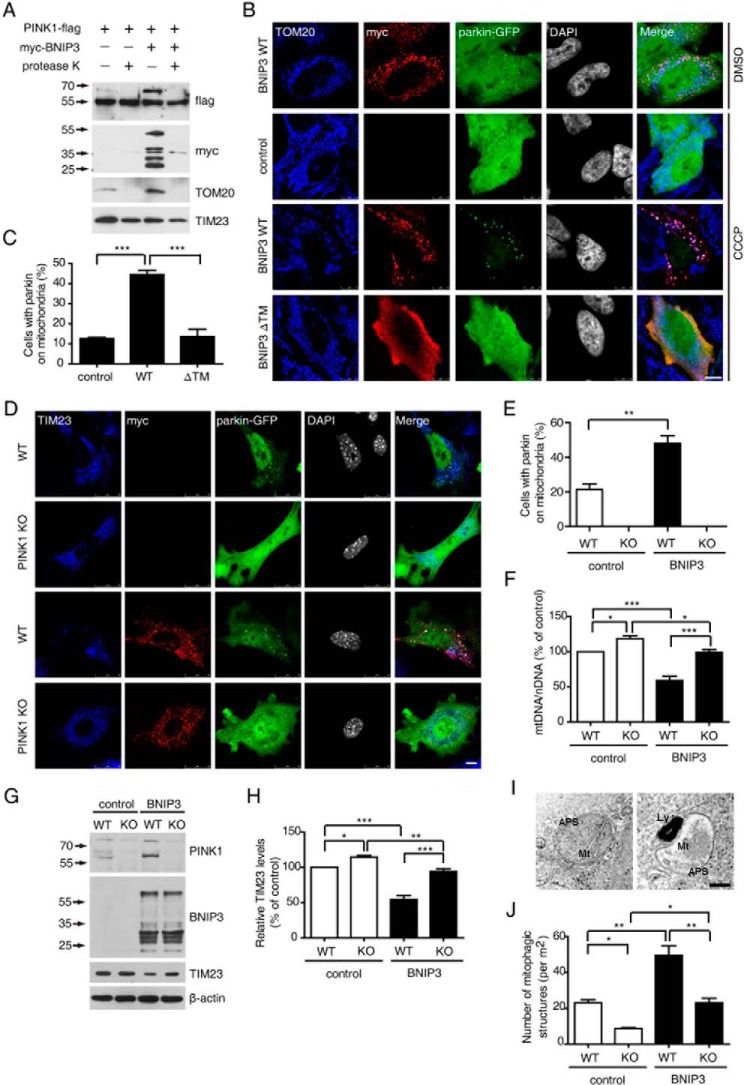
To further analyze the subcellular localization of BNIP3-PINK1 interaction, we conducted a proteinase K protection assay. Mitochondria isolated from HEK293 cells overexpressing PINK1 and BNIP3 were incubated with 5 μg/ml proteinase K. The full-length PINK1, together with the MOM protein TOM20 and BNIP3, was rapidly degraded by proteinase K treatment. In contrast, the inner membrane protein, TIM23, remained intact (Fig. 3A). The results suggest that the BNIP3 interacts with full-length PINK1 on the outer membrane of mitochondria with the PINK1 kinase domain likely facing the cytosol.
FIGURE 3.
BNIP3 facilitates PINK1-depandant parkin recruitment to mitochondria. A, BNIP3-induced full-length PINK1 is on MOM. Mitochondria from HEK293 cells expressing PINK1-FLAG and Myc-BNIP3 in various combinations were treated with proteinase K followed by immunoblotting with antibodies against FLAG tag (PINK1), Myc tag (BNIP3), MOM protein TOM20, or IMM protein TIM23. B, BNIP3 facilitates parkin recruitment to depolarized mitochondria. HeLa cells co-expressing either parkin-GFP and Myc-BNIP3 WT or parkin-GFP and Myc-BNIP3 ΔTM with/without CCCP treatment were immunostained with anti-TOM20 (blue) or anti-Myc tag (red). Controls were cells expressing parkin-GFP alone. Cell nuclei are shown in gray. Merge panel, colocalization of TOM20, Myc tag, and GFP. Bar = 10 μm. C, quantification of parkin recruitment to mitochondria. >200 cells in each group were counted for parkin on mitochondria from the CCCP treatment experiment shown in B. Results were analyzed by one-way ANOVA and Tukey's test. Error bars, S.E.; n = 3. ***, p < 0.001. D, MEFs from PINK1 KO mice or WT control littermates were co-transfected with Parkin-GFP and Myc-BNIP3 WT followed by CCCP (20 μm) treatment. Cells were immunostained with anti-TOM20 (blue) or anti-Myc tag (red). Cell nuclei are shown in gray. Merge panel, colocalization of TOM20, Myc tag, and GFP. Bar = 10 μm. E, quantification of MEFs with parkin on mitochondria. >100 cells in each group were counted for parkin on mitochondria from the experiment shown in D. Results were analyzed by two-tailed Student's t test. Error bars, S.E.; n = 3. **, p = 0.0071. F, mitochondrial DNA content. WT and PINK1-deficient (KO) HEK293 cells transfected with either an empty plasmid (control) or Myc-BNIP3 were treated with CCCP (5 μm). Relative mitochondrial DNA content (mtDNA/nDNA) was detected and analyzed by one-way ANOVA and Tukey's test. Error bars, S.E.; n = 4. *, p < 0.05; ***, p < 0.001. G, BNIP3 promotes PINK1-mediated TIM23 degradation. Cells treated as described in F were lysed and immunoblotted with antibodies against PINK1, BNIP3, or TIM23. β-Actin (bottom panel) was the loading control. H, quantification of TIM23 levels in experiments shown in G. Relative levels of endogenous TIM23 affected by BNIP3 were quantified and analyzed by one-way ANOVA and Tukey's test. Error bars, S.E., n = 4. *, p < 0.05; **, p < 0.01; ***, p < 0.001. I, representative TEM images of autophagosomes engulfing mitochondria. Wild-type control (left panel) and PINK1-deficient (right panel) HEK293 cells were transfected with Myc-BNIP3 and treated with 5 μm CCCP followed by TEM analysis. Note that an autophagosome containing mitochondrial debris-like structures is shown in each image. Ly, lysosome; APS, autophagosome; Mt, mitochondrial debris. Bar = 100 nm. J, quantification of mitophagy-like structures in experiments shown in I. >0.72-mm2 cell areas in each group were randomly selected and counted for double membrane engulfed mitochondrion-like structures. The results were analyzed by one-way ANOVA and Tukey's test. Error bars, S.E.; n = 3. *, p < 0.05; **, p < 0.01.
PINK1 recruits parkin to the mitochondria to promote mitophagy of damaged mitochondria (14–16). We hypothesized that the stabilization of full-length PINK1 by BNIP3 would facilitate parkin recruitment to mitochondria. Expression of BNIP3 WT resulted in the colocalization of parkin to mitochondria in ∼6.7% cells (Fig. 3B, top panel). Upon low concentration CCCP (5 μm) induction for 2.5 h, <15% of cells transfected with either control vector alone or BNIP3 ΔTM were detected to have parkin co-localized to mitochondria (Fig. 3, B and C). In contrast, >40% of cells expressing BNIP3 WT showed parkin recruitment to mitochondria (Fig. 3B, 3rd panel from top). These results suggest that BNIP3 promotes parkin recruitment to mitochondria.
In PINK1-deficient mouse embryonic fibroblasts (MEFs), parkin was distributed in the cytoplasm with or without BNIP3 overexpression even with 20 μm CCCP exposure (Fig. 3, D and E). In contrast, CCCP treatment induced parkin recruitment to mitochondria in about 20% of wild-type MEFs. Overexpression of BNIP3 further increased mitochondrial localization of parkin to >50% of wild-type MEFs (Fig. 3, D and E). Taken together, these findings show that BNIP3-induced parkin recruitment to mitochondria is PINK1-dependent. We next examined whether BNIP3-induced parkin recruitment to mitochondria facilitated mitophagy. Mitophagy was first determined by measuring the relative mitochondrial DNA content, a ratio of mitochondrial DNA (mtDNA) to nuclear DNA (nDNA), in PINK1-deficient HEK293 cells. Consistent with previous reports, mitophagy was impaired in PINK1-deficient cells as shown by an increase of the mtDNA/nDNA ratio. Moreover, upon 5 μm CCCP induction for 24 h, the mtDNA/nDNA ratio in HEK293 cells expressing BNIP3 was significantly lower than that in control HEK293 cells, suggesting that overexpressing BNIP3 promotes mitophagy in HEK293 cells. Inactivation of PINK1 restored the effect on the BNIP3 mtDNA/nDNA ratio, indicating that BNIP3-promoted mitophagy was mostly PINK1-mediated (Fig. 3F). We also analyzed the relative mitochondrial protein content in cells. In agreement with the results of the mtDNA/nDNA analysis, the level of the mitochondrial protein TIM23 was lower in control HEK293 cells expressing BNIP3 than in PINK1 KO cells expressing BNIP3 after 5 μm CCCP treatment for 24 h (Fig. 3, G and H). The effect of BNIP3 on promoting PINK1-mediated mitophagy was further verified using transmission electronic microscopy (TEM) (Fig. 3I). After 5 μm CCCP induction for 24 h, more double membrane vacuoles containing mitochondrion-like structures were observed in control HEK293 cells expressing BNIP3 than in PINK1-deficeint HEK293 cells (Fig. 3, I and J). Therefore, the expression of BNIP3 promotes PINK1/parkin-mediated mitophagy. It was notable that PINK1 knock-out did not completely abolish BNIP3-induced mitophagy (Fig. 3, F–J). One possible explanation is that BNIP3 induces mitophagy directly by interacting with LC3 (26).
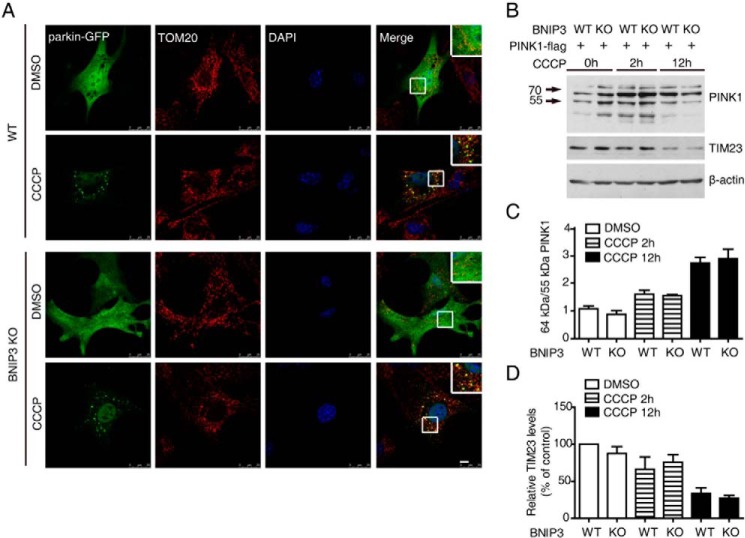
BNIP3 Is Not Required for CCCP-induced Full-length PINK1 Accumulation and Mitophagy
Previous studies suggest that CCCP treatment induces the accumulation of full-length PINK1 (6, 15, 16). To determine the roles of BNIP3 in CCCP-induced full-length PINK1 accumulation, we first examined parkin recruitment to mitochondria in MEFs generated from BNIP3 knock-out mice and their wild-type control littermates. Upon 20 μm CCCP induction, remarkable colocalization of parkin and the mitochondrial protein TOM20 was detected in both wild-type and BNIP3 KO MEFs (Fig. 4A). Likewise, the accumulation of full-length PINK1 was increased, whereas the mitochondrial protein TIM23 was reduced after CCCP treatment in both wild-type and BNIP3 KO MEFs (Fig. 4, B–D). These results suggest that BNIP3 is not essential for CCCP treatment-induced full-length PINK1 accumulation and for parkin recruitment to mitochondria or mitophagy.
FIGURE 4.
Full-length PINK1 accumulation, parkin recruitment to mitochondria, and mitophagy in BNIP3 KO MEFs. A, CCCP induces parkin recruitment to mitochondria in BNIP3 KO MEFs. BNIP3 KO and WT control MEFs were transfected with parkin-GFP followed by treatment with either control solvent (DMSO) or 20 μm CCCP for 6 h. Cells were immunostained with antibodies against TOM20 (red). Nuclei were stained with DAPI (blue). Colocalization of parkin (green) and TOM20 is shown (Merge panel). The insets in the Merge panel show amplified images of colocalization. Bar = 10 μm. B, full-length PINK1 in BNIP3 KO MEFs. BNIP3 KO and WT control MEFs were transfected with PINK1-FLAG followed by treatment with 20 μm CCCP for 0, 2, and 12 h. The cell lysates were immunoblotted with antibodies against PINK1, TIM23, or β-actin. C, 64-kDa/55-kDa PINK1 ratio in CCCP-treated BNIP3 KO MEFs. The results are an average of three independent experiments as shown in B. Error bar, represents S.E. D, level of TIM23 in CCCP-treated BNIP3 KO MEFs. The results are an average of three independent experiments as shown in B. Error bar, represents S.E.
Endogenous BNIP3 Regulates PINK1 Cleavage and Mitophagy
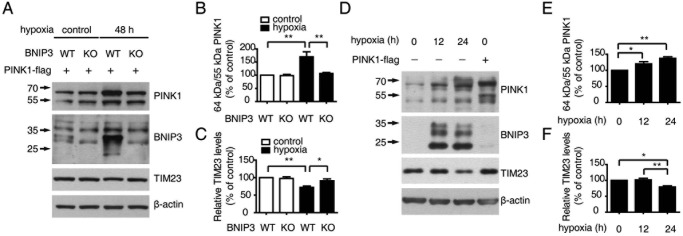
BNIP3 is transcriptionally regulated by HIF-1 (34). To investigate the roles of endogenous BNIP3 in regulating PINK1, we first employed BNIP3-deficient MEFs expressing PINK1-FLAG. After 48 h of hypoxia, BNIP3 expression was increased in wild-type control MEFs but not in BNIP3-deficient MEFs (Fig. 5A). Accordingly, the accumulation of full-length PINK1 and the decrease in the mitochondrial protein TIM23 were observed consistently in wild-type MEFs but neither in BNIP3-deficient MEFS nor in wild-type MEFs without hypoxia treatment (Fig. 5, A–C). In HEK293 cells, endogenous full-length PINK1 was increased with the induction of endogenous BNIP3 upon hypoxia treatment for 12 and 24 h. Concomitantly, TIM23 was reduced compared with untreated control cells (Fig. 5, D–F). These results provide strong evidence that endogenous BNIP3 regulates PINK1 cleavage and mitophagy under physiological conditions such as hypoxia.
FIGURE 5.
Endogenous BNIP3 regulates PINK1 cleavage and mitophagy. A, endogenous BNIP3 inhibits PINK1 cleavage and promotes mitophagy. MEFs from BNIP3 KO mice and WT control littermates were transfected with PINK1-FLAG followed by incubation under either normoxia (21% O2, control) or hypoxia (5% O2, 48 h) to up-regulate BNIP3. The cell lysates were immunoblotted with antibodies against PINK1, BNIP3, or TIM23. β-Actin (bottom panel) was the loading control. B, ratio of PINK1 64-kDa/55-kDa fragments affected by endogenous BNIP3. The relative levels of PINK1 64- and 55-kDa fragments in the experiments described in A were quantified and analyzed by one-way ANOVA and Tukey's test. Error bars, S.E.; n = 4. **, p < 0.01. C, quantification of TIM23 affected by endogenous BNIP3. The relative levels of TIM23 in experiments described in A were quantified and analyzed by one-way ANOVA and Tukey's test. Error bars, S.E.; n = 4. *, p < 0.05; **, p < 0.01. D, endogenous BNIP3 inhibits endogenous PINK1 cleavage and promotes mitophagy. HEK293 cells were incubated under hypoxia (5% O2) conditions for 0, 12, and 24 h followed by immunoblotting with antibodies against PINK1, BNIP3, or TIM23. β-Actin (bottom panel) was the loading control. Cells transfected with PINK1-FLAG served as a positive control. E, quantification of endogenous PINK1 64-kDa/55-kDa ratio affected by endogenous BNIP3. Relative levels of endogenous PINK1 64- and 55-kDa fragments affected by hypoxia-induced BNIP3 were measured by ImageJ and analyzed by one-way ANOVA and Dunnett's test. Error bars, S.E.; n = 3. *, p < 0.05; **, p < 0.01. F, quantification of TIM23 affected by endogenous BNIP3. The relative levels of IMM protein TIM23 affected by hypoxia-induced BNIP3 were measured by ImageJ and analyzed by one-way ANOVA and Tukey's test. Error bars, S.E.; n = 3. *, p < 0.05; **, p < 0.01.
BNIP3 Suppresses Phenotypes of PINK1 Mutant Drosophila
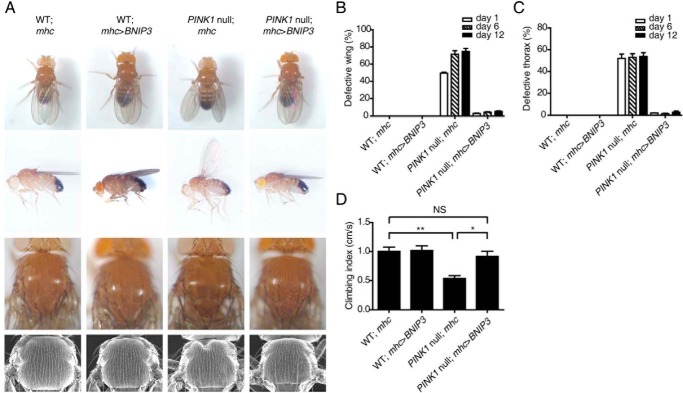
To further explore the functional interaction of PINK1 and BNIP3 in vivo, we examined whether BNIP3 compensates for loss of PINK1 function using Drosophila. PINK1 inactivation in Drosophila results in multiple phenotypes, including abnormally positioned wings and crushed thorax (35–37). The ubiquitous expression of BNIP3 resulted in embryonic lethality of Drosophila. Muscle-specific expression of BNIP3 driven by mhc-gal4 led to a near complete reversal of the abnormal wing posture and crushed thorax in PINK1 null flies (PINK1B9) (Fig. 6, A–C). Consistently, BNIP3 also successfully rescued the abnormal wing and thorax phenotypes of Drosophila with muscle-specific PINK1 RNAi treatment (38) (not shown). Likewise, the climbing defect of PINK1 null flies was also evidently rescued by expressing BNIP3 in muscles (Fig. 6D). As controls, UAS-BNIP3 transgene or expression of mhc-gal4 in muscles showed little effect on the wing posture or thorax development of Drosophila (Fig. 6, A–C). The results suggest that BNIP3 interact functionally with PINK1 in vivo and that BNIP3 and PINK1 function in the same pathway.
FIGURE 6.
BNIP3 suppresses abnormal phenotypes of PINK1 null Drosophila. A, the wing posture (upper two panels) and thorax (lower two panels) of WT and PINK1 null mutant Drosophila expressing either mhc-gal4 (mhc) or mhc-gal4-driven BNIP3 (mhc>BNIP3) are shown. The back view (top panel) and side view (upper middle panel) of the wing posture and light microscopic images (lower middle panel) and scan electronic microscopic images (bottom panel) of the thorax are shown. Note that BNIP3 expression rescues the wing posture abnormality and thorax defect in PINK1 null flies. B, quantitation of PINK1 null-induced wing posture rescued by BNIP3. >300 flies were analyzed for each experiment. Error bars, S.E.; n = 3. C, quantitation of PINK1 null-induced thorax defects rescued by BNIP3. >300 flies were analyzed for each experiment. Error bars, S.E.; n = 3. D, rescue of PINK1 null-induced climbing ability by BNIP3. Climbing index (cm/s) of WT and PINK1 null mutant Drosophila expressing either mhc-gal4 (mhc) or mhc-gal4-driven BNIP3 (mhc>BNIP3) are shown. The climbing ability of >100 flies was determined for each tested genotype per experiment and analyzed by one-way ANOVA and Tukey's test. Error bars, S.E.; n = 5. NS, not statistically significant; *, p < 0.05; **, p < 0.01.
BNIP3 Restores Mitochondrial Morphology and ATP Production in Drosophila with Mutated PINK1
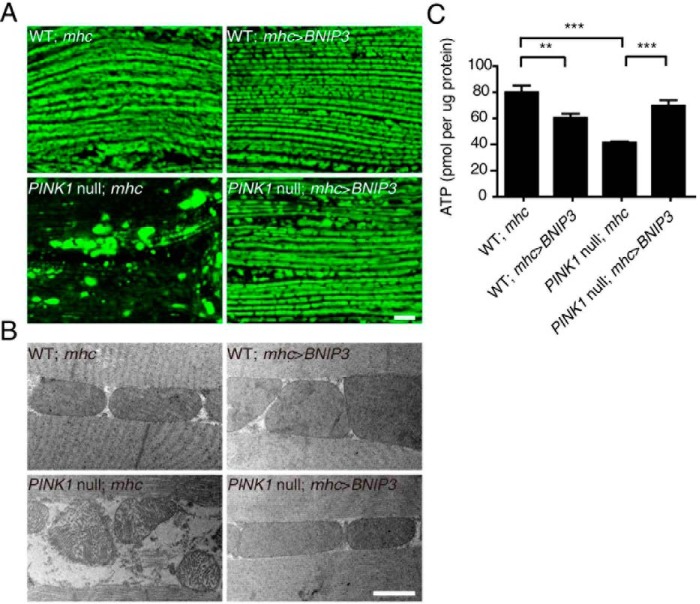
PINK1 inactivation in Drosophila results in mitochondrial abnormality leading to indirect flight muscle (IFM) degeneration (35–37). We next examined whether BNIP3 rescues mitochondrial impairment in PINK1 null flies. Mitochondria were labeled by expressing a mitochondria-targeting green fluorescent protein (mitoGFP). In control wild-type flies, co-expression of either mhc-gal4 and mitoGFP or BNIP3 and mitoGFP leads to well organized mitochondria in IFM along the muscle fibers (Fig. 7A). Consistent with previous findings (36, 38, 39), mitochondria were aggregated in IFM of PINK1B9 flies expressing mitoGFP alone. PINK1B9 flies expressing BNIP3 in IFM exhibited the normal morphology and organization as seen in the wild-type control flies (Fig. 7A). The effect of BNIP3 on mitochondria of PINK1B9 flies was further verified using TEM (Fig. 7B). Previous studies report that PINK1 inactivation promotes mitochondrial aggregation in dopaminergic neurons (36, 37). Dopaminergic neuron-specific expression of BNIP3 and mitoGFP in PINK1B9 flies driven by th-gal4 resulted in a remarkable decrease of mitochondrial aggregation and increased fragmentation (supplemental Fig. 3). Thus, expression of BNIP3 rescues abnormal mitochondrial morphology caused by PINK1 deficiency in vivo.
FIGURE 7.
BNIP3 restores mitochondrial morphology and ATP production of PINK1 null Drosophila. A, mitoGFP (green)-labeled mitochondria in IFM of WT and PINK1 null mutant Drosophila expressing either mhc-gal4 (mhc) or mhc-gal4-driven BNIP3 (mhc>BNIP3) are shown. Note that mitochondrial aggregates are seen in PINK1 null flies and suppressed by muscle-specific expression of BNIP3 (PINK1 null; mhc>BNIP3). Bar = 10 μm. B, TEM images of mitochondria in IFM. Representative images of WT and PINK1 null mutant Drosophila expressing either mhc-gal4 or mhc-gal4-driven BNIP3 (mhc>BNIP3) are shown. Bar = 1 μm. C, ATP production of thoraces. ATP levels in thoraces of WT and PINK1 null mutant Drosophila expressing either mhc-gal4 or mhc-gal4 driven BNIP3 (mhc>BNIP3) are measured and analyzed by one-way ANOVA and Tukey's test. Error bars, represent S.E.; n = 5. **, p < 0.01; ***, p < 0.001. Note that PINK1 null expression results in decreased ATP production, which is restored by muscle-specific expression of BNIP3.
To determine the role of BNIP3-PINK1 interaction in mitochondrial function, we analyzed ATP production in IFM of genetically engineered flies. The ATP level in the IFM of PINK1B9 flies (41.4 pmol/μg protein) is approximately half of that in the IFM of their wild-type control flies (79.2 pmol/μg protein). Overexpression of BNIP3 led to a significant increase of ATP production in the IFM of PINK1B9 flies (69.7 pmol/μg protein) (Fig. 7C). Thus, BNIP3 compensates functionally for the mitochondrial abnormality caused by PINK1 deficiency in vivo.
Discussion
The major finding of this study is the identification of a physiological regulatory mechanism of PINK1/parkin-mediated mitophagy by BNIP3. Our study demonstrates that the interaction of BNIP3 and PINK1 results in the inhibition of proteolytic cleavage of PINK1, leading to an increased accumulation of full-length PINK1 on the MOM, which promotes parkin recruitment to mitochondria and PINK1/parkin-mediated mitophagy. In Drosophila, BNIP3 compensates for PINK1 deficiency to restore mitochondrial morphology and function. In mammalian cells, BNIP3 potentiates PINK1-mediated mitophagy induced by hypoxia. Thus, BNIP3-PINK1 interactions play an important role in mitochondrial quality control under physiological conditions. In addition, the study identifies a novel function for BNIP3 in PINK1/parkin-mediated mitophagy via a mechanism other than BNIP3 directly serving as an autophagy receptor. These observations could have implications for the therapeutic modulation of BNIP3 levels in diseases such as PD.
Previous studies have detected two forms of PINK1 proteins in the cell, a full-length form and a 55-kDa proteolytic fragment lacking the N-terminal mitochondrial localization sequence (4–6). The full-length form, PINK1, is found mainly on mitochondria and likely functions in regulating mitophagy (12–16). In contrast, the 55-kDa proteolytic fragments are detected largely in the cytosol to activate parkin E3 ligase activity and to promote the degradation of un-/misfolded proteins (11, 40). Several mitochondrial proteases are implicated in PINK1 cleavage (41, 42). It is suggested that PINK1 cleavage is coupled with its import into the mitochondrial inner membrane (IMM). We show in this study that BNIP3 interacts with PINK1 on MOM and inhibits PINK1 cleavage. We therefore propose a model in which BNIP3 inhibits PINK1 from cleavage by functioning as an anchoring force to prevent PINK1 import into mitochondrial IMM (Fig. 8). It has been reported that mitochondrial depolarization by CCCP treatment results in the accumulation of full-length PINK1 in the cell (6, 15, 16). In this study, BNIP3 H173A, a mutant lacking the ability to induce mitochondrial depolarization, showed the ability to suppress PINK1 cleavage, suggesting that BNIP3-induced accumulation of full-length PINK1 is unlikely to be the result of mitochondrial depolarization or damage. Consistently, BNIP3 was not required for CCCP-induced full-length PINK1 accumulation or for parkin recruitment to mitochondria or mitophagy. We postulate that BNIP3 stabilizes PINK1 on MOM during physiological responses, whereas mitochondrial depolarization causes PINK1 accumulation on MOM with mitochondrial damage.
FIGURE 8.
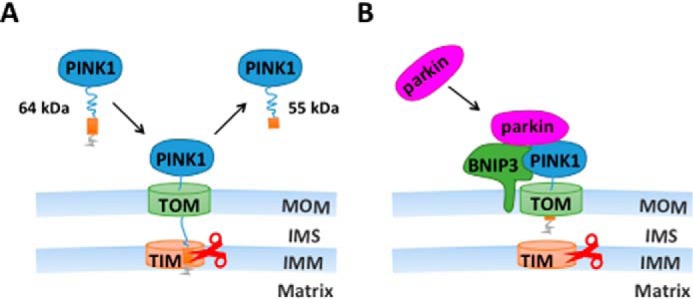
Schematic illustration of BNIP3-PINK1 interaction. A, under normal conditions, full-length PINK1 (64 kDa) is synthesized and transported to the mitochondria. Its mitochondrial targeting sequence reaches the IMM followed by enzymatic cleavage resulting in a 55-kDa fragment. The 55-kDa fragment is released to and eventually degraded in the cytoplasm. B, with stress, BNIP3 is up-regulated and interacts with PINK1 to anchor the full-length PINK1 (65 kDa) on the MOM. The PINK1 on MOM recruits parkin from the cytoplasm to activate mitophagy. TOM, translocase of the MOM complex; TIM, translocase of the IMM complex; IMS, mitochondrial intermembrane space.
BNIP3 and its homologous protein BNIP3L/NIX are both shown to regulate mitophagy (12–16, 23, 26, 44–46). BNIP3/BNIP3L disrupts the BCL-2·Beclin-1 complex, releasing Beclin-1 from the complex to induce mitophagy. BNIP3/BNIP3L complexes with Bcl-2 to promote cell survival (24). BNIP3L plays also an important role in the autophagic recognition of mitochondria. It interacts with LC3 and GABARAP and likely is involved in the recruitment of damaged mitochondria to autophagosome (28, 45, 46). The Results from this study reveal a functional interaction of PINK1 and BNIP3, two proteins previously shown to participate independently in mitophagy. Although it remains unclear how PINK1 affects BNIP3-mediated mitophagy, three lines of evidences from this study support the notion that BNIP3 potentiates PINK1/parkin-mediated mitophagy. First, mtDNA is significantly reduced in cells expressing BNIP3 but not in PINK1-deficient cells expressing BNIP3. Likewise, TOM20 is significantly reduced in cells expressing BNIP3 compared with PINK1-deficient cells expressing BNIP3. Finally, expression of BNIP3 results in increased detection of autophagosomes containing mitochondrion-like structures in wild-type HEK293 cells but not in PINK1-deficient HEK293 cells. Mechanistically, BNIP3 likely increases the presence of PINK1 on MOM to promote parkin recruitment leading to enhanced mitophagy. This is consistent with a previous report that BNIP3 promotes the recruitment of parkin in cardiac myocytes (44). In Drosophila, mitochondrial fusion contributes to PINK1 deficiency-associated mitochondrial abnormalities (38, 47). The fact that BNIP3 restores the mitochondrial morphology and function of PINK1 null flies suggests that BNIP3 may inhibit PINK1 deficiency-caused mitochondrial fusion. In this study, overexpression of BNIP3 causes mitochondrial fragmentation in both cultured cells and neurons in Drosophila. Consistent with such a notion, BNIP3 is shown to induce the mitochondrial translocation of dynamin-related protein 1 (Drp1), a protein implicated in mitochondrial fission in adult myocytes (44). Overexpression of Drp1 suppresses the mutant phenotypes caused by PINK1 deficiency in Drosophila by promoting mitochondrial fission (38, 47–49). However, although it is sufficient to compensate the mitochondrial abnormalities of PINK1 null flies, BNIP3 can induce mitophagy in the absence of PINK1, suggesting that BNIP3 likely induces mitophagy via additional mechanisms. Thus, one cannot exclude the possibility that BNIP3 simply promotes mitophagy in PINK1 null flies. The exact mechanism by which BNIP3 rescues the mitochondrial abnormality of PINK1 null flies remains unknown. Nevertheless, parkin is able to restore the mitochondrial morphology of PINK1 null flies despite the fact that PINK1 plays an essential role in parkin recruitment to mitochondria and PINK1/parkin-mediated mitophagy in mammalian cells (35–37). It is possible that BNIP3 and parkin share a similar mechanism in rescuing the mitochondrial abnormalities of PINK1 null flies.
The functional significance of autophagy in a physiological context is poorly understood. This study demonstrates the up-regulation of endogenous BNIP3, resulting in increased full-length PINK1 and enhanced mitophagy. BNIP3 is both proapoptotic and neuroprotective against oxidative stress (50). Autophagy is proposed to play either a protective or a pathogenic role under different conditions. In response to hypoxia, mitophagy is shown as an adaptive reaction to maintaining cell viability (51). It is possible that a fine-tuned regulation of BNIP3 expression of BNIP3 is beneficial for neuronal survival. The results from this study may open a new avenue to the design of novel treatments for PD.
Experimental Procedures
Cells, Antibodies, Reagents, and Drosophila Lines
HEK293 and HeLa cells were obtained from American Type Culture Collection. MEFs were generated from PINK1 and BNIP3 KO mouse embryos along with their WT control littermates as described (11). All cells were cultured in DMEM (HyClone, SH30022) containing 2 mm/liter l-glutamine (Life Technologies, catalog No. 25030) and 1 mm/liter sodium pyruvate (Life Technologies, catalog No. 11360) supplemented with 10% fetal bovine serum (HyClone, SH30084.03). Plasmid encoding C-terminal FLAG-tagged PINK1 WT and PINK1 G309D were described previously (11). pDsRed1-Mito plasmid was from Clontech (catalog No. 6928-1). cDNA encoding Myc-tagged BNIP3 were generated by PCR and subcloned into pcDNA3.1(A-) (Invitrogen, V795-20). cDNA encoding parkin was PCR-generated and subcloned into pEGFPN3 to express parkin-GFP (Clontech, catalog No. 6080-1). cDNA encoding parkin was PCR-generated and subcloned into pcDNA3.1(B-) (Invitrogen, V795-20) to express parkin-FLAG. cDNA encoding LC3 was PCR-generated and subcloned into pEGFPN1 (Clontech, catalog No. 6085-1) to express LC3-GFP. PINK1 ΔN (1–110-amino acid deletion), PINK1 D384N, BNIP3 ΔTM (164–194-amino acid deletion), BNIP3 H173A, and BNIP3 L179S were generated by PCR. The primers were: BNIP3 WT (forward, 5′-CAGAATTCATGGAGCAGAAACTCATCTCTGAAGAGGATCTGATGTCGCAGAACGGAGCG-3′; reverse, 5′-TAGGATCCTCAAAAGGTGCTGGTGGAGG-3′), parkin-GFP (forward, 5′-GCGAATTCGCCACCATGATAGTGTTTGTCAGGTTCAAC-3′; reverse, 5′-GCGGATCCCACGTCGAACCAGTGGTCCCCCAT-3′), parkin-FLAG (forward, 5′-CGGCTAGCGCCACCATGATAGTGTTTGTCAGGTTCAAC-3′; reverse, 5′-CGGGATCCCTAGATCTTATCGTCGTCATCCTTGTAATCCACGTCGAACCAGTGGTCCCCCAT-3′), LC3 (forward, 5′-CGGGATCCATGATCCCTAACCCTCTCCTCGGTCTCGATTCTACGATGCCGTCGGAGAAGACCTT-3′; reverse, 5′-CGGAATTCTTACACTGACAATTTCATCCCGAACG-3′), PINK1 ΔN (forward, 5′-CGGAATTCGCCACCATGGAGGAAAAACAGGC-3′; reverse, 5′-CGGGATCCTCAGATCTTATCGTCGT-3′), PINK1 D384N (forward, 5′-CTGGTGATCGCAAATTTTGGCTGCTGC-3′; reverse, 5′-GCAGCCAAAATTTGCGATCACCAGCCA-3′), BNIP3 ΔTM (forward, 5′-CGGAATTCGCCACCATGGAGCAGAAACTCATCTC-3′; reverse, 5′-CGGGATCCTCATTTCAGAAATT-3′), BNIP3 H173A (forward, 5′-TCTCTGCTGCTCTCTGCTTTGCTGGCCATCGGATTG-3′; reverse, 5′-TCCGATGGCCAGCAAAGCAGAGAGCAGCAGAGATGG-3′), and BNIP3 L179S (forward, 5′-TTGCTGGCCATCGGATCGGGGATCTATATTGGAAGG-3′; reverse, 5′-TCCAATATAGATCCCCGATCCGATGGCCAGCAAATG-3′). All plasmids were confirmed by sequencing.
Rabbit anti-BNIP3 (catalog No. 3485-1) monoclonal antibody was from Epitomics. Mouse anti-TIM23 (catalog No. 611222) monoclonal antibody was from BD Biosciences. Anti-PINK1 antibody (BC100-494) was from Novus Biologicals. Anti-TOM20 (sc-17764) monoclonal antibody was from Santa Cruz Biotechnology. Rabbit anti-PINK1 antibody (6946S), rabbit anti-Myc monoclonal antibody (2272S), and mouse anti-Myc monoclonal antibody (2276S) were from Cell Signaling Technology. Rabbit anti-FLAG polyclonal antibody (F7425), mouse anti-β-actin monoclonal antibody (A5441), mouse anti-β-tubulin monoclonal antibody (T8328), rabbit anti-GAPDH polyclonal antibody (SAB1405848), mouse anti-FLAG (M2) affinity gel (A2220), mouse anti-c-Myc-agarose affinity gel antibody (A7470), CHX (C7698), MG132 (C2211), CCCP (C2759), and DMSO (D2650) were from Sigma-Aldrich. Mouse anti-BNIP3 (ab10433) monoclonal antibody was from Abcam. Protein G-agarose was from KPL Inc. (catalog No. 223-51-01). Mouse IgG, rabbit IgG, and all of the secondary antibodies were from Jackson ImmunoResearch Laboratories. Protease K was from EMD Chemicals (catalog No. 539430). Tetramethylrhodamine ethyl ester (TMRE) was from Life Technologies (T-669), and the ENLITEN® ATP assay system was from Promega (FF2000).
Transgenic flies were generated by embryo injection. cDNAs encoding human BNIP3 with a C-terminal HA tag were cloned into the pUAST (Addgene, No. 31223). Primers were: forward, 5′-CGGAATTCGCCACCATGTACCCATACGACGTCCCAGACTACGCTATGTCGCAGAACGGAGCG-3′; and reverse, 5′-CCGCTCGAGTCAAAAGGTGCTGGTGGAGG-3′. The construct was confirmed by sequencing and injected into w1118 embryos. The PINK1 mutant fly (PINK1B9) was kindly provided by Dr. Jongkyeong Chung (36). Flies harboring th-gal4, mhc-gal4, and uas-mitoGFP were obtained from the Bloomington Drosophila Stock Center. Fly strains were grown on standard cornmeal media at 25 °C unless otherwise specified.
Generation of PINK1-deficient HEK293 Cells Using CRISPR/Cas9 System
HEK293 cells with PINK1 knock-out were generated using the CRISPR/Cas9 system as described previously (52). The targeting single-guide RNA sequence, 5′-CGCCACCATGGCGGTGCGAC-3′, was selected by an online CRISPR design tool followed by cloning into plentiCRISPR v2 (Addgene, catalog No. 49535). The recombinant lentiCRISPR plasmid was co-transfected with pVSVg (Addgene, No. 8454) and psPAX2 (Addgene, catalog No. 12260) into HEK293FT cells to package infectious lentivirus. HEK293 cells were infected followed by selection with 1 μg/ml puromycin (Thermo Fisher Scientific, A1113803). The puromycin-resistant cell clones were genotyped by sequencing. Three PINK1 mutant cell lines were isolated and confirmed for PINK1 deficiency using a PINK1 antibody.
Mitochondrial Fractionation
Cells were grown on 100-mm plates until 80–90% confluency, washed twice with ice-cold PBS, and then scraped into ice-cold PBS followed by centrifugation at 1000 × g for 5 min at 4 °C. Cell pellets were resuspended in mitochondrial isolation buffer (5 mm Hepes (pH 7.4), 3 mm MgCl2, 1 mm EGTA, and 250 mm sucrose) containing protease and phosphatase inhibitors (10 μg/ml aprotinin, 10 μg/ml pepstatin A, 10 μg/ml leupeptin, 1 mm PMSF, 2 mm sodium orthovanadate, and 5 mm sodium fluoride). Lysates were passed through a 5/8-inch 25-gauge needle 20 times using a 1-ml syringe and centrifuged at 1000 × g for 20 min. Supernatants were collected, and cytosolic extracts were recovered by centrifugation at 10,000 × g for 15 min to obtain a crude mitochondrial pellet. Protein concentration was determined using the BCA kit (Pierce, catalog No. 23228) according to the manufacturer's instructions. For immunoprecipitation, 300 μg of mitochondrial and cytosolic proteins was used (53, 54).
Immunoblotting
Cells were washed twice with cold PBS and lysed with SDS sample buffer (63 mm Tris-HCl, 10% glycerol, and 2% SDS) containing protease and phosphatase inhibitors. 20 μg of proteins was separated on an SDS-polyacrylamide gel and immunoblotted with corresponding antibodies. Proteins for input controls were 1–2% of the total proteins used for immunoprecipitation. The band intensity was quantified by ImageJ.
Immunoprecipitation
Immunoprecipitation was done essentially as described previously (55). Briefly, cells were lysed in Triton X-100 lysis buffer (1% Triton X-100, 10 mm HEPES (pH 7.5), 142.5 mm KCl, 5 mm MgCl2, and 1 mm EGTA) supplemented with protease and phosphatase inhibitors followed by incubating for 1 h at 4 °C with gentle rotation. Lysates were centrifuged at 14,000 × g for 30 min. Supernatants were collected and measured for protein concentration. Immunoprecipitation was performed by incubation with an anti-FLAG M2 affinity gel, an anti-Myc affinity gel, or corresponding antibodies with protein G at 4 °C for 12 h. The beads were washed with 1% Triton X-100 buffer three times followed by boiling for 5 min in reducing SDS sample buffer. The immunoprecipitated proteins were separated on an SDS-polyacrylamide gel and immunoblotted with corresponding antibodies. The band intensity was quantified by ImageJ.
Immunofluorescent Staining
Cultured cells were washed twice with PBS followed by fixation with 3.7% paraformaldehyde for 10 min and permeabilization with 0.1% Triton X-100. After blocking with 5% BSA for 30 min, cells were incubated with primary antibodies and detected with Alexa-conjugated secondary antibodies.
Whole-mount immunostaining of fly brains was performed essentially as described (29). Briefly, fly heads were fixed overnight with 4% paraformaldehyde containing 0.2% Triton X-100 and washed with 0.1% Triton X-100 three times. Brains were dissected in blocking buffer (5% goat serum in 0.5% PBST) followed by blocking at room temperature for 1 h. Brains were immunostained with the corresponding primary antibodies at 4 °C overnight followed by respective Alexa-conjugated secondary antibodies at room temperature for 3 h.
Samples were imaged using a confocal microscope (TCS SP5, Leica) with a Plan-Apochromat 63× NA 1.4 oil differential interference contrast objective lens. The images were shown as a montage of 3 × 0.5-μm sections.
Protease K Protection Assay
For protease K protection assay, fresh isolate mitochondria were resuspended in mitochondrial isolation buffer and incubated with 5 μg/ml protease K for 30 min on ice. Digestion was terminated with protease inhibitors. Mitochondrial proteins were detected by immunoblotting, and PINK1 was detected by immunoprecipitation.
Cycloheximide Chase Experiments
To determine the half-life of the PINK1 protein, HEK293 cells were co-transfected with PINK1-FLAG and Myc-BNIP3 followed by cycloheximide treatment (10 μg/ml) for 0, 0.5, 1, and 1.5 h. PINK1 protein levels were detected by immunoprecipitation. Cells co-transfected with PINK1 and an empty vector were included as controls.
Mitochondrial Membrane Potential Assay
To access mitochondrial membrane potential, HeLa cells were transfected with BNIP3 WT or BNIP3 mutants followed by staining with TMRE (30 nm) for 20 min. Post-treatment, cells were immediately analyzed using a FACScan flow cytometer (BD Biosciences) with excitation at 488 nm and emission at 610 nm. 20,000 cells were analyzed for each treatment.
Quantification of Mitochondrial DNA Content in Cells
WT control and PINK1 KO HEK293 cells were transfected with either empty control plasmid or Myc-BNIP3 WT (BNIP3) followed by CCCP (5 μm) treatment for 24 h. Total cellular DNA was extracted using the DNeasy blood and tissue kit (Qiagen, catalog No. 69506) according to the manufacturer's instruction. The relative content of mitochondrial DNA (mtDNA) in cells was measured as described previously (43). Briefly, both mtDNA (measured by mitochondrial 16S rRNA gene) and nuclear DNA (nDNA, measured by the β2-microglobulin gene) under the same experimental condition were quantified by quantitative PCR using a CFX96 real-time PCR detection system (Bio-Rad) with SyBR GreenER qPCR SuperMix (Invitrogen, catalog No. 11762-500). The primers used were: 16S rRNA gene (forward, 5′-GCCTTCCCCCGTAAATGATA-3′; reverse, 5′-TTATGCGATTACCGGGCTCT-3′) and β2-microglobulin (forward, 5′-TGCTGTCTCCATGTTTGATGTATCT-3′; and reverse, 5′-TCTCTGCTCCCCACCTCTAAGT-3′). The CT values were obtained automatically. The relative content of mtDNA in cells was shown as a ratio of mtDNA to nDNA, calculated by 2 × 2(ΔCT). ΔCT is the difference of CT values between the β2-microglobulin gene and the 16S rRNA gene.
Hypoxia Induction
Hypoxia culture conditions (5%) were achieved by culturing cells in a multi-gas cell culture incubator (Thermo Scientific HERAcell 150i).
Climbing Assay
A climbing assay was performed as described previously, with modification (36). Briefly, groups of 10-day-old male flies were anesthetized by CO2 and then transferred into transparent plastic vials 25 cm in length and 1.5 cm in diameter. Flies were incubated for 30 min for environmental acclimatization. Flies were tapped down to the bottom of the vials. The time that more than five flies required to climb over the 10-cm finish line was recorded. The climbing index equaled the speed of the flies (cm/s). The climbing assay was repeated three times for each group at 5-min intervals. At least 100 flies for each genotype were tested. The average climbing time for each genotype was calculated.
Quantitation of Wing and Thorax Phenotypes
The percentage of male flies with abnormally positioned wings and crushed thorax was determined. >300 flies were analyzed for each genotype/experiment, and 3 experiments were done.
ATP Assay
The ATP level was quantified as described previously (36). Briefly, lysates from five thoraces of 3-day-old flies were prepared for each experiment. Samples were mixed with luminescent solution. The luminescence was measured by an illuminometer (Berthold Technologies). Values were normalized to protein content measured by the BCA protein assay reagent.
TEM
Dissected thoraces from 3-day-old male flies or HEK293 cells were fixed in paraformaldehyde (Sigma-Aldrich, catalog No. 158127)/glutaraldehyde (Ted Pella Inc., catalog No. 18462), postfixed in osmium tetraoxide (Electron Microscopy Sciences, catalog No. 19150), dehydrated in ethanol (Sigma-Aldrich, catalog No. 46139), and embedded in Epon (Sigma-Aldrich, catalog No. 45345). After polymerization of the Epon, blocks were sectioned to generate 70-nm-thick sections using a diamond knife on a microtome (Leica, Wetzlar, Germany). The sections were stained with uranyl acetate (Ted Pella Inc., catalog No. 19481) and lead citrate (Sigma-Aldrich, catalog No. 15326). Digital images were obtained on a Tecnai G2 Spirit by FEI equipped with an Eagle 4k HS digital camera with >6 thoraces examined for each sample.
Statistical Analysis
Statistical analysis was performed using Prism 5 software (GraphPad). Two-tailed Student's t test was used to determine the significance of difference between two groups. Statistical significance between multiple groups was derived using one-way ANOVA followed by Tukey's test. One-way ANOVA with Dunnett's tests were used to assess the difference between treatment groups against their controls. All error bars indicate S.E. The quantitation was performed double-blinded.
Author Contributions
T. Z. conceived and performed the experiments. L. X., L. L., C. T., Z. W., R. W., J. T., Y. T., H. H., and R. T. performed the experiments. W. A. T., T. R. B., and Z. Z. conceived, supervised, and wrote the manuscript.
Supplementary Material
This work was supported by Chinese National 973 Projects 2011CB510000 (to Z. Z.), Natural Science Foundation of China Grants 313300257, 81429002, and 81161120498 (to Z. Z.), and the Discipline Innovative Engineering Plan (111 Program) of China (to Z. Z.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. 1–3.
- PD
- Parkinson's disease
- MOM
- mitochondrial outer membrane
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- CHX
- cycloheximide
- MEF
- mouse embryonic fibroblast
- TEM
- transmission electronic microscopy
- IFM
- indirect flight muscle
- IMM
- mitochondrial inner membrane
- TMRE
- tetramethylrhodamine ethyl ester
- ANOVA
- analysis of variance.
References
- 1. Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., and Shimizu N. (1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 [DOI] [PubMed] [Google Scholar]
- 2. Farrer M. J. (2006) Genetics of Parkinson disease: paradigm shifts and future prospects. Nat. Rev. Genet. 7, 306–318 [DOI] [PubMed] [Google Scholar]
- 3. Valente E. M., Abou-Sleiman P. M., Caputo V., Muqit M. M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A. R., Healy D. G., Albanese A., Nussbaum R., González-Maldonado R., Deller T., Salvi S., et al. (2004) Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304, 1158–1160 [DOI] [PubMed] [Google Scholar]
- 4. Muqit M. M., Abou-Sleiman P. M., Saurin A. T., Harvey K., Gandhi S., Deas E., Eaton S., Payne Smith M. D., Venner K., Matilla A., Healy D. G., Gilks W. P., Lees A. J., Holton J., Revesz T., et al. (2006) Altered cleavage and localization of PINK1 to aggresomes in the presence of proteasomal stress. J. Neurochem. 98, 156–169 [DOI] [PubMed] [Google Scholar]
- 5. Lin W., and Kang U. J. (2008) Characterization of PINK1 processing, stability, and subcellular localization. J. Neurochem. 106, 464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jin S. M., Lazarou M., Wang C., Kane L. A., Narendra D. P., and Youle R. J. (2010) Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 191, 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Imai Y., Soda M., and Takahashi R. (2000) Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J. Biol. Chem. 275, 35661–35664 [DOI] [PubMed] [Google Scholar]
- 8. Zhang Y., Gao J., Chung K. K., Huang H., Dawson V. L., and Dawson T. M. (2000) Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc. Natl. Acad. Sci. U.S.A. 97, 13354–13359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shimura H., Hattori N., Kubo S., Mizuno Y., Asakawa S., Minoshima S., Shimizu N., Iwai K., Chiba T., Tanaka K., and Suzuki T. (2000) Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25, 302–305 [DOI] [PubMed] [Google Scholar]
- 10. Wenzel D. M., Lissounov A., Brzovic P. S., and Klevit R. E. (2011) UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xiong H., Wang D., Chen L., Choo Y. S., Ma H., Tang C., Xia K., Jiang W., Ronai Z., Zhuang X., and Zhang Z. (2009) Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J. Clin. Invest. 119, 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Geisler S., Holmström K. M., Treis A., Skujat D., Weber S. S., Fiesel F. C., Kahle P. J., and Springer W. (2010) The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy 6, 871–878 [DOI] [PubMed] [Google Scholar]
- 13. Narendra D., Tanaka A., Suen D. F., and Youle R. J. (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R. L., Kim J., May J., Tocilescu M. A., Liu W., Ko H. S., Magrané J., Moore D. J., Dawson V. L., Grailhe R., Dawson T. M., et al. (2010) PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U.S.A. 107, 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C. A., Sou Y. S., Saiki S., Kawajiri S., Sato F., Kimura M., Komatsu M., Hattori N., and Tanaka K. (2010) PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Narendra D. P., Jin S. M., Tanaka A., Suen D. F., Gautier C. A., Shen J., Cookson M. R., and Youle R. J. (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gegg M. E., Cooper J. M., Chau K. Y., Rojo M., Schapira A. H., and Taanman J. W. (2010) Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 19, 4861–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poole A. C., Thomas R. E., Yu S., Vincow E. S., and Pallanck L. (2010) The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PloS One 5, e10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kubli D. A., Ycaza J. E., and Gustafsson A. B. (2007) Bnip3 mediates mitochondrial dysfunction and cell death through Bax and Bak. Biochem. J. 405, 407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Regula K. M., Ens K., and Kirshenbaum L. A. (2002) Inducible expression of BNIP3 provokes mitochondrial defects and hypoxia-mediated cell death of ventricular myocytes. Circ. Res. 91, 226–231 [DOI] [PubMed] [Google Scholar]
- 21. Vande Velde C., Cizeau J., Dubik D., Alimonti J., Brown T., Israels S., Hakem R., and Greenberg A. H. (2000) BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol. Cell. Biol. 20, 5454–5468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Y., Lewis W., Diwan A., Cheng E. H., Matkovich S. J., and Dorn G. W. 2nd (2010) Dual autonomous mitochondrial cell death pathways are activated by Nix/BNip3L and induce cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 107, 9035–9042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rikka S., Quinsay M. N., Thomas R. L., Kubli D. A., Zhang X., Murphy A. N., and Gustafsson Å. B. (2011) Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 18, 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bellot G., Garcia-Medina R., Gounon P., Chiche J., Roux D., Pouysségur J., and Mazure N. M. (2009) Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol. Cell. Biol. 29, 2570–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dorn G. W., 2nd.(2010) Mitochondrial pruning by Nix and BNip3: an essential function for cardiac-expressed death factors. J. Cardiovasc. Transl. Res. 3, 374–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hanna R. A., Quinsay M. N., Orogo A. M., Giang K., Rikka S., and Gustafsson Å. B. (2012) Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J. Biol. Chem. 287, 19094–19104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu Y., Massen S., Terenzio M., Lang V., Chen-Lindner S., Eils R., Novak I., Dikic I., Hamacher-Brady A., and Brady N. R. (2013) Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. J. Biol. Chem. 288, 1099–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ding W. X., Ni H. M., Li M., Liao Y., Chen X., Stolz D. B., Dorn G. W. 2nd, and Yin X. M. (2010) Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J. Biol. Chem. 285, 27879–27890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Whitworth A. J., Theodore D. A., Greene J. C., Benes H., Wes P. D., and Pallanck L. J. (2005) Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson's disease. Proc. Natl. Acad. Sci. U.S.A. 102, 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kubli D. A., Quinsay M. N., Huang C., Lee Y., and Gustafsson A. B. (2008) Bnip3 functions as a mitochondrial sensor of oxidative stress during myocardial ischemia and reperfusion. Am. J. Physiol. Heart Circ. Physiol. 295, H2025–H2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sulistijo E. S., and MacKenzie K. R. (2006) Sequence dependence of BNIP3 transmembrane domain dimerization implicates side-chain hydrogen bonding and a tandem GxxxG motif in specific helix-helix interactions. J. Mol. Biol. 364, 974–990 [DOI] [PubMed] [Google Scholar]
- 32. Vasagiri N., and Kutala V. K. (2014) Structure, function, and epigenetic regulation of BNIP3: a pathophysiological relevance. Mol. Biol. Rep. 41, 7705–7714 [DOI] [PubMed] [Google Scholar]
- 33. Ray R., Chen G., Vande Velde C., Cizeau J., Park J. H., Reed J. C., Gietz R. D., and Greenberg A. H. (2000) BNIP3 heterodimerizes with Bcl-2/Bcl-X(L) and induces cell death independent of a Bcl-2 homology 3 (BH3) domain at both mitochondrial and nonmitochondrial sites. J. Biol. Chem. 275, 1439–1448 [DOI] [PubMed] [Google Scholar]
- 34. Bruick R. K. (2000) Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc. Natl. Acad. Sci. U.S.A. 97, 9082–9087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clark I. E., Dodson M. W., Jiang C., Cao J. H., Huh J. R., Seol J. H., Yoo S. J., Hay B. A., and Guo M. (2006) Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166 [DOI] [PubMed] [Google Scholar]
- 36. Park J., Lee S. B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J. M., and Chung J. (2006) Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161 [DOI] [PubMed] [Google Scholar]
- 37. Yang Y., Gehrke S., Imai Y., Huang Z., Ouyang Y., Wang J. W., Yang L., Beal M. F., Vogel H., and Lu B. (2006) Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl. Acad. Sci. U.S.A. 103, 10793–10798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deng H., Dodson M. W., Huang H., and Guo M. (2008) The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 105, 14503–14508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yun J., Puri R., Yang H., Lizzio M. A., Wu C., Sheng Z. H., and Guo M. (2014) MUL1 acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. eLife 3, e01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Choo Y. S., Vogler G., Wang D., Kalvakuri S., Iliuk A., Tao W. A., Bodmer R., and Zhang Z. (2012) Regulation of parkin and PINK1 by neddylation. Hum. Mol. Genet. 21, 2514–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deas E., Plun-Favreau H., Gandhi S., Desmond H., Kjaer S., Loh S. H., Renton A. E., Harvey R. J., Whitworth A. J., Martins L. M., Abramov A. Y., and Wood N. W. (2011) PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum. Mol. Genet. 20, 867–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Greene A. W., Grenier K., Aguileta M. A., Muise S., Farazifard R., Haque M. E., McBride H. M., Park D. S., and Fon E. A. (2012) Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 13, 378–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Venegas V., Wang J., Dimmock D., and Wong L. J. (2011) Real-time quantitative PCR analysis of mitochondrial DNA content. Curr. Protoc. Hum. Genet. 68, 19.7.1–19.7.12 [DOI] [PubMed] [Google Scholar]
- 44. Lee Y., Lee H. Y., Hanna R. A., and Gustafsson Å. B. (2011) Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 301, H1924–H1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ding W. X., and Yin X. M. (2012) Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol. Chem. 393, 547–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wei H., Liu L., and Chen Q. (2015) Selective removal of mitochondria via mitophagy: distinct pathways for different mitochondrial stresses. Biochim. Biophys. Acta 1853, 2784–2790 [DOI] [PubMed] [Google Scholar]
- 47. Yang Y., Ouyang Y., Yang L., Beal M. F., McQuibban A., Vogel H., and Lu B. (2008) Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc. Natl. Acad. Sci. U.S.A. 105, 7070–7075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu W., Acín-Peréz R., Geghman K. D., Manfredi G., Lu B., and Li C. (2011) Pink1 regulates the oxidative phosphorylation machinery via mitochondrial fission. Proc. Natl. Acad. Sci. U.S.A. 108, 12920–12924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Poole A. C., Thomas R. E., Andrews L. A., McBride H. M., Whitworth A. J., and Pallanck L. J. (2008) The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl. Acad. Sci. U.S.A. 105, 1638–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Awan M. U., Hasan M., Iqbal J., Lei R., Lee W. F., Hong M., Qing H., and Deng Y. (2014) Neuroprotective role of BNIP3 under oxidative stress through autophagy in neuroblastoma cells. Mol. Biol. Rep. 41, 5729–5734 [DOI] [PubMed] [Google Scholar]
- 51. Zhang H., Bosch-Marce M., Shimoda L. A., Tan Y. S., Baek J. H., Wesley J. B., Gonzalez F. J., and Semenza G. L. (2008) Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 283, 10892–10903 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., and Zhang F. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Choo Y. S., Johnson G. V., MacDonald M., Detloff P. J., and Lesort M. (2004) Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum. Mol. Genet. 13, 1407–1420 [DOI] [PubMed] [Google Scholar]
- 54. Tan J., Zhang T., Jiang L., Chi J., Hu D., Pan Q., Wang D., and Zhang Z. (2011) Regulation of intracellular manganese homeostasis by Kufor-Rakeb syndrome-associated ATP13A2 protein. J. Biol. Chem. 286, 29654–29662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang Z., Hartmann H., Do V. M., Abramowski D., Sturchler-Pierrat C., Staufenbiel M., Sommer B., van de Wetering M., Clevers H., Saftig P., De Strooper B., He X., and Yankner B. A. (1998) Destabilization of β-catenin by mutations in presenilin-1 potentiates neuronal apoptosis. Nature 395, 698–702 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.