Abstract
Although brain tumors have been known tremendously over the past decade, there are still many problems to be solved. The etiology of brain tumors is not well understood and the treatment remains modest. There is in great need to develop a suitable brain tumor models that faithfully mirror the etiology of human brain neoplasm and subsequently get more efficient therapeutic approaches for these disorders. In this review, we described the current status of animal models of brain tumors and analyzed their advantages and disadvantages. Additionally, prokaryotic clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9), a versatile genome editing technology for investigating the functions of target genes, and its application were also introduced in our present work. We firstly proposed that brain tumor modeling could be well established via CRISPR/Cas9 techniques. And CRISPR/Cas9-mediated brain tumor modeling was likely to be more suitable for figuring out the pathogenesis of brain tumors, as CRISPR/Cas9 platform was a simple and more efficient biological toolbox for implementing mutagenesis of oncogenes or tumor suppressors that were closely linked with brain tumors.
Keywords: brain tumors, animal models, CRISPR, oncogene, tumor suppressor
CURRENT STATUS: ANIMAL MODELS OF BRAIN TUMORS
Primary brain tumors have the highest mortality rates among all the cancers worldwide [1]. From the knowledge about the subtypes of gliomas, the etiology and the molecular mechanism, especially the origins [2], we learned that there were differences between rodent model of gliomas and glioblastoma in human patients. Due to the inconvenience of interference of brain tumor patients in clinical practices, it is of desperate need to develop in vivo animal models with brain tumors that faithfully mirror human disease, finally finding an effectively therapeutic target for treating brain tumors.
Several brain tumor models have been established as shown in Table 1. For example, it was reported that the rat glioma model could be induced by implantation of cultured glioma cells (Cell-derived xenograft: CDX) [3, 4] and transplanted tumor fragments (Patient-derived xenograft: PDX) [5]. Medulloblastom was regarded as the most common malignant brain tumor in pediatrics with a poor prognosis and in vivo investigation; it was often induced by transplantation of chemically modified human medulloblastoma cells such as Daoy, ONS76 and D425 [6]. Although these animal models have the advantage of making the tumor models for a short time and are widely used for the study of brain tumors, they do not comprehensively recapitulate human neoplasms and often caused the inhibitory tumor-host immunoresponses [7, 8].
Table 1. Animal models of brain tumors.
| Tumor type | Model type | Engineered drivers | Description | Ref |
|---|---|---|---|---|
| Glioma | CDX | HOTAIR-knock down; Bmi1- deficient | Transplantation of HOTAIR shRNA; Injection of Bmi1 -deficient astrocytes | [6-7] |
| Glioma | PDX | Hedgehog-responsive | Injection of Biopsy -derived glioma cells | [5] |
| Glioma | GEMM | Platelet-derived growth factor subunit B overexpression | Tranfection of human cells by avian sarcoma- leucosis virus | [13] |
| Medulloblastom | CDX | MET kinase-driven | Injection of Daoy cells to prepare intracranial xenografts | [6] |
Abbreviations: CDX: Cell-derived xenograft; PDX: Patient-derived xenograft; GEMM: Genetically engineered mouse model
Besides, genetically engineered mouse models (GEMMs) are also extensively employed for investigating the pathogenesis of glioma. It was usually generated by introducing the mutations and genetic aberrations of both germ-line cells and somatic cells [9]. For instance, previous investigations illustrated that the introductions of mutant Ras protein, including HRas, VRas and NRas could trigger brain tumors [10, 11]. Postnatal PTEN loss or mutant epidermal growth factor receptor expression was also found to result in the generation of glioma in a transgenic mouse glioma model [12]. Similarly, platelet-derived growth factor subunit B overexpression contributed to the occurrence of brainstem glioma after human cells were injected into the brain stem of neonatal mouse [13]. The GEMMs has the advantage of resembling human glioblastoma as the tumor histology in the transgenic mouse is similar to that in human. However, the big disadvantage is that GEMM generation takes really long time and it is difficult to distinguish the primary mutation and the secondary mutation.
Collectively, those animal models as noted above possess respective shortages and it is essential for developing a new brain tumor model in order to more deeply probe molecular mechanisms of brain tumors and obtain better therapeutic approaches.
CRISPR/CAS9 SYSTEM: A POWERFUL TOOL FOR GENOME EDITING
The recently mentioned prokaryotic clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system is a powerful genetic engineering tool in which guide RNA (gRNA) targets the programmable nuclease Cas9 to a desired genomic DNA sequence and Cas9 precisely cleaves both strands of interest [14-18].
As shown in Figure 1, the genomic editing for CRISPR/Cas9 system mainly requires two biological components: Cas9 and an engineered single guide RNA (sgRNA). The sgRNA is composed of both a CRISPR RNA (crRNA) component and a trans-activating crRNA (tracrRNA). SgRNA recognizes the complementary genomic DNA sequences flanked by a protospacer adjacent motif (PAM), which is made up of NGG or NAG trinucleotide for Cas9 [19]. After Cas9 is combined with a sgRNA which is complementary to a target DNA sequence, the double-stranded break (DSB) is formed and then repaired by either non-homologous end-joining (NHEJ) or homology-directed repair (HDR) pathway [20]. Figure 2A and Figure 2B showed the crystal structure and model graph of wild type Cas9 from S. pyogenes (wt SpCas9). Additionally, the catalytically dead Cas9 mutant (dSpCas9) and other SpCas9 variants were also indicated in this Figure 2B. These SpCas9 mutants were shown to significantly minimal off-target effects [21, 22]. A small Cas9 from Staphylococcus aureus (SaCas9) was also recently obtained for eukaryotic genome engineering [23]. SaCas9 shared only 17% of identical sequence with SpCas9 [23]. And smaller size of SaCas9 makes it easier deliver to somatic tissues for genome editing, compared with SpCas9 (Figure 2B). The centromere and promoter factor 1 (Cpf1) was hypothesized to be the effector of a CRISPR locus that was different from the Cas9-containing class 2 CRISPR system [24]. Structurally, Cpf1 is lack of a second HNH endonuclease domain, which is inserted within the RuvC-like domain of Cas9, as shown in Figure 2B. It was previously reported that Cpf1was a CRISPR-associated two-component RNA programmable DNA nuclease and exhibited robust nuclease activity in human cells [24].
Figure 1. The CRISPR/Cas9 system for genome engineering. The CRISPR is composed of two major components including a CRISPR-associated endonuclease (Cas9) and a single guide RNA (sgRNA).
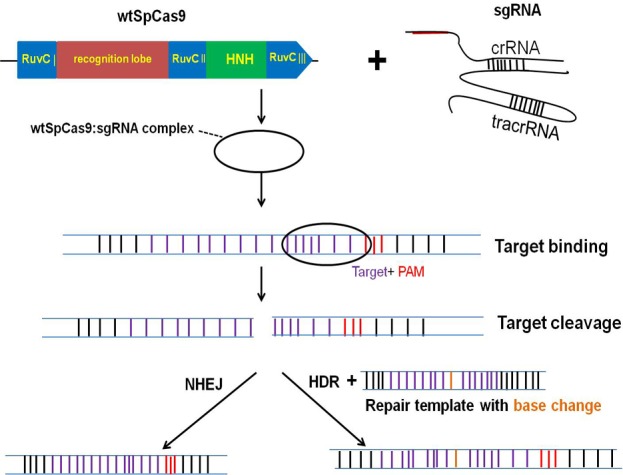
The Cas9 from S. pyogenes (wt SpCas9) is shown in this figure as it is the most widely used in genome editing nowadays. After wt SpCas9 and sgRNA form a riboprotein complex, they can bind any genomic sequence with a protospacer adjacent motif (PAM), directing DNA double-strand breaks (DSBs) at the target site. DSBs are then repaired by either non-homologous end-joining (NHEJ) or homology-directed repair (HDR) pathway.
Figure 2. Proposed applications of the CRISPR/Cas9 system for brain tumor modeling.
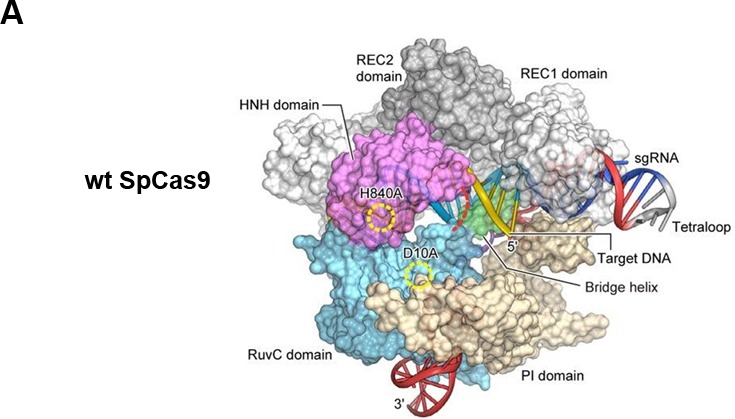
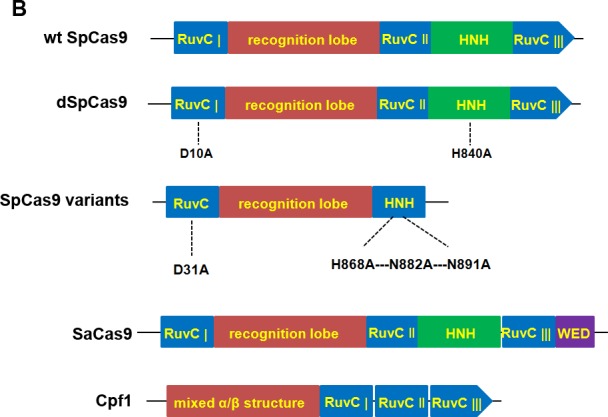
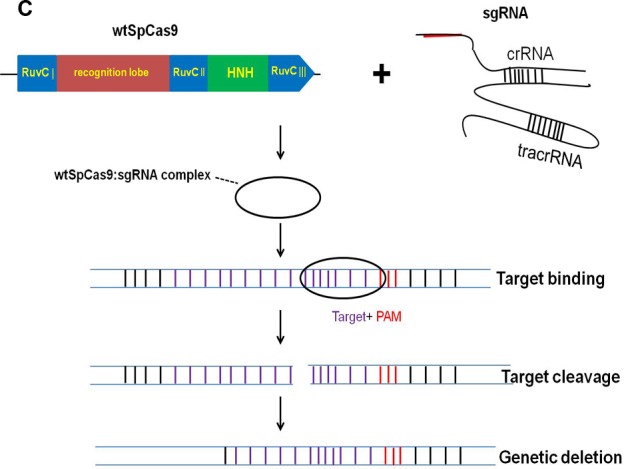
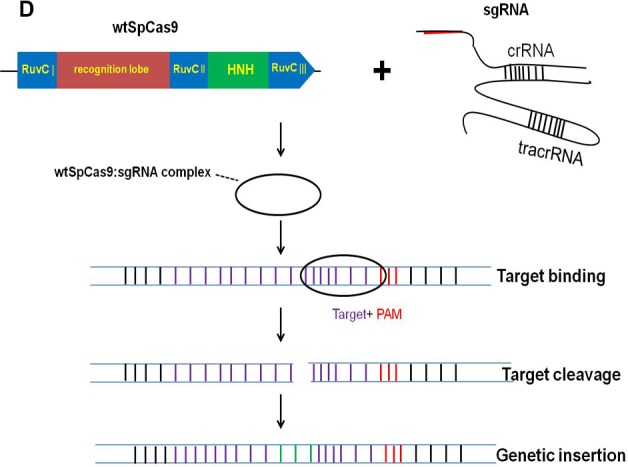
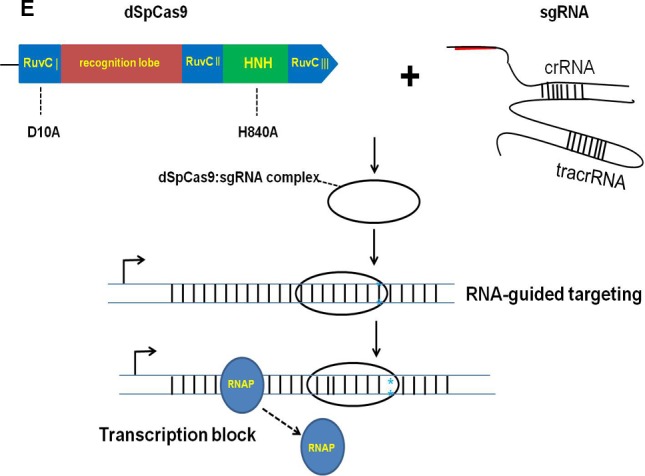
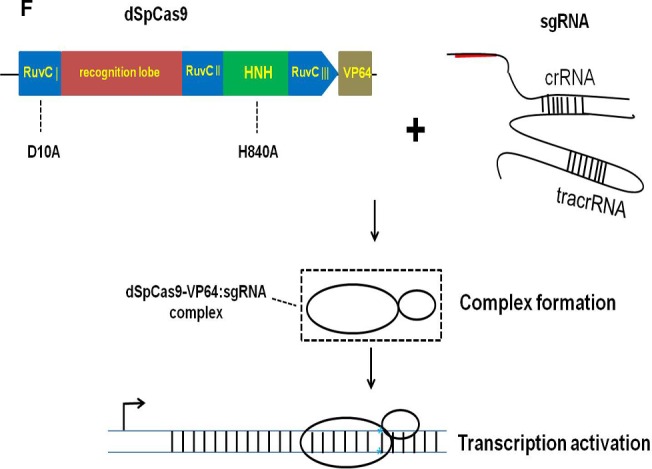
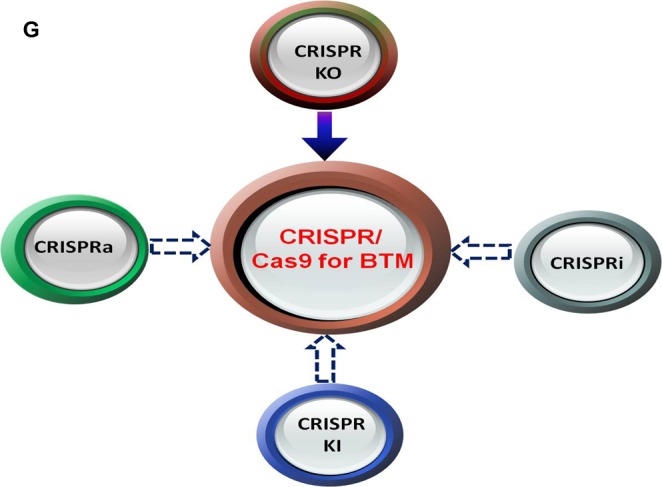
A. shows the crystal structure of wt SpCas9 referred to Zhang et al [70]; B. is the model graphs of wt SpCas9, dSpCas9, SpCas9 variants, SaCas9 and Cpf1; C. D. E. F. display the processions of four different types of CRISPR-mediated genome editing including CRISPR knock out (CRISPR KO), CRISPR knock in (CRISPR KI), CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa), respectively; G. proposes four different sorts of CRISPR/Cas9 techniques are possibly involved in brain tumor modeling (BTM). CRISPR KO is presently successfully applied for BTM (as shown by solid arrow). We propose CRISPR KI, CRISPRi and CRISPRa are used for BTM (as shown by dotted arrows).
RNA-guided nucleases Cas9 employs simpler, Watson-Crick base-pairing rules between the sgRNA and the genomic DNA sequence of interest, compared with the early method including zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), which acquire the targeting genomes via protein-DNA interactions [25-27]. Although ZFNs and TALENs are very beneficial for performing precise genome editing, their applications have been limited as a result of high cost and difficult design of this endonucleases. The RNA-guided endonucleases Cas9 from the microbial adaptive immune system CRISPR can be easily targeted to any selected genomic location by a short RNA guide.
PROPOSED APPLICATIONS OF THE CRISPR/CAS9 SYSTEMS IN BRAIN TUMOR MODELING
Modeling cancer including brain tumors in mice through genetic manipulation in the germline of an organism has long been regarded as the gold criteria for seeking putative oncogenes or tumor suppressor genes (TSGs). Loss-of-function mutations in TSGs and gain-of-function mutations in a proto-oncogene were reported to be involved in the generation and progression of glioma [28, 29]. Traditional cell-type-specific knockout techniques via homologous recombination are shown to cause the loss of function for TSGs in embryonic stem cells, finally leading to the malignant transformation [30]. Nevertheless, the low efficiency of homologous recombination and the time-consuming property for the generation of GEMMs hamper its applications. As alternatively, the CRISPR/Cas9-guided endonuclease technique provides more efficient and precise modification of the DSB sites at target genomes [31]. One big advantage of CRISPR/Cas9 is that it takes short time to generate GEMM model. Nowadays, this versatile genome editing technique has been developed for producing gene knockout models of various animals including mouse [32], rat [33-35], as it can more precisely understanding human diseases than the conventional gene knockout models.
In general, CRISPR/Cas9 system contains four types of genome editing techniques including CRISPR knock out (CRISPR KO), CRISPR knock in (CRISPR KI), CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa) (Figure 2C-2F). Using the CRISPR KO-mediated target PTEN efficiently diminished PTEN expression in neurons, inducing neuronal hypertrophy and altering neuronal excitation, while targeting NF1 was shown to facilitate astrocytogenesis and combined targeting of three TSGs (including PTEN, NF1 and P53) by multiplex CRISPR/Cas9 triggered the pathogenesis of glioblastoma [36]. Besides, CRISPR KO technology was also previously found to efficiently and completely delete two well-known synaptic proteins including GluN1 subunit of the NMDA receptor and the GluA2 subunit of the AMPA receptor in neurons [37].
Additionally, knockin animal models has been successfully prepared using CRISPR/Cas9 system (which referred to CRISPR KI) [38-42]. It was previously found that Cre-dependent Cas9 konckin mice exhibited indel formation near predicted cleavage site on NeuN (a neuron-specific marker) locus indels in the brain via virus-mediated sgRNA expression as reported by J. Platt [38]. Delivery of a single adeno-associated virus (AAV) vector in the lung caused the genetic mutations of p53, LKB1 and KRAS using the Cas9 knockin mouse, resulting in the macroscopic tumors of adeno-carcinoma pathology [38]. The feasibility of direct mutations of TSGs and oncogenes was also reported in the liver by the CRISPR/Cas system [43]. What's more important, recent investigations illustrated that the somatic gene transfer of CRISPR plasmids encoding Cas9 and gRNAs was suitable to induce distinct brain tumors including sonic hedgehog medulloblastoma and glioblastoma [44].
CRISPR/Cas9-mediated control of gene suppression (which referred to CRISPRi) is also an important application for this genetic engineering technology. The CRISPRi system, derived from the Streptococcus pyogenes CRISPR pathway, is established by the coexpression of catalytically incactive Cas9 (dCas9) fusion proteins and a customizable sgRNA [15, 45-47]. As dCas9 is lack of endonuclease activity and generates a targeted protein-RNA complex when coexpressed with a guide RNA. This Cas9-sgRNA complex binds to DNA sequences complementary to the sgRNA and specifically blocks transcription elongation within protein-coding regions, RNA polymerase binding, or transcription factor binding. It was previously found that usage of CRISPRi could remarkably suppress expressions of any target gene of interest in Escherichia coli, with no obvious off-target effects [46]. An additional investigation revealed more efficient gene suppression in eukaryotes by dCas9 fused with a transcription repression domain or exogenous transgene activation [45]. The multiple target genes were also evidently repressed in human cells via this system. In fact, other targeted gene modulation methods have been widely established in the past few years including RNA interference (RNAi) [48] and polydactyl zinc-finger proteins [49-51] and sequence-specific transcription activator-like effector (TALE) [52-54]. Among these traditional methods, RNAi is a classical technology for perturbing target genes on the mRNA level by designing complementary RNAs [28], but it is limited by evident off-target effects, low efficiency, serious toxicity and small-scale use in particular organisms [55]. The CRISPRi technology is more efficient and has the minimal off-target effects due to its simple and precise design. Engineered DNA-binding proteins such as custom zinc-finger or TALE proteins provide a very useful platform for achieving diverse targeted regulatory functions when combined with effector domains. Nevertheless, this method is a time-consuming process and the construct development requires high cost. As a result, it is likely to be very difficult to build a comprehensive protein library to simultaneously perturb multiple target genes [56]. In contrast, the CRISPRi method is very convenient and cheap for suppressing target genes due to use of sgRNA with a specific 20-nt-long complementary region. And with the faster and cheaper synthesis of wide-range DNA oligonucleotide, it allows for targeting large amounts of genes to probe gene function using CRISPRi system.
Furthermore, dCas9 could also be fused to transcription activation domains to form RNA-guided transcriptional activator system (which referred as CRISPRa), finally regulating gene expression via targeting the promoter region of endogenous genes [57, 58]. Usage of dCas9-based transcription activators was observed to result in the up-regulation of several endogenous loci [25]. In human cells, it was also illustrated that the dCas9-VP64 fusion protein was found to be directed by single or multiple gRNAs and produce robust transcriptional activations of endogenous human genes such as VEGFA [58]. This CRISPRa system was also reported to powerfully up-regulate multiple exogenous reporter genes in both human and mouse transformed cells as well as in embryonic stem cell cells in a tunable manner [59]. Endogenous IL1RN, SOX2, and OCT4 genes were also observed to be activated in human embryonic kidney 293T (HEK293T) cells via CRISPRa technology [59]. It implicates that CRISPRa is likely to be used to target heterologous effector domains in human cells. In a word, CRISPR KO and CRISPR KI are more effective for making genetic mutations while CRISPRi and CRISPRa for genetic expressions. Besides, the latter two techniques seem to interference lncRNA more efficiently.
According to these findings as mentioned, we propose different types of brain tumors modeling can be established using CRISPR/Cas9, as displayed in Figure 2G. CRISPR/Cas9-mediated brain tumor modeling is likely to be more suitable for exploring the pathogenesis of brain tumors, as CRISPR/Cas9 platform is a simple and more efficient biological toolbox for implementing mutagenesis of oncogenes or tumor suppressors that are closely linked with brain tumors. Indeed, prior investigations supported the notion that the preparation of brain tumor modeling could be carried out via CRISPR knock out (CRISPR KO) technology [36, 44, 60-62], which were partly in line with our hypothesis that CRISPR technology was served to establish efficient knock-out brain tumor models. Recently, Zhang et al also emphasized that CRISPR/Cas9-mediated precise and efficient genome editing could help to deeply figure out the logic of neural circuits and disclose the mysteries of diverse neurological diseases including brain tumors [63].
FURTHER REQUIRED CLARIFICATION FOR CRISPR/CAS9 TECHNIQUE
Despite great progress has been made in the past two years with the CRISPR/Cas9 system and the establishment of diverse cancer models, there are still some issues that require further clarification.
Firstly, the delivery of Cas9 for establishing somatic mutation of mouse models should be further improved. It was previously demonstrated that efficient delivery of genome-editing proteins could significantly enhance Cas9-mediated genetic engineering of target sequences [64, 65]. Split Cas9 was also reported to be beneficial for in vivo genome editing in HEK293FT cells [66, 67]. Finding smaller Cas9 orthologs could also improve CRISPR delivery as a result of facilitating viral vector package [23, 68, 69]. However, it is still essential to optimize the methods for efficient delivery and expression of CRISPR-Cas9 system in order to be suitable for each cell-type or organism. Since some cell types or tissues are resistant to transfection or infection by viral vectors, researchers should develop methods that alter the expressions of Cas9 endonuclease or gRNAs that is specific to a tissue or cell type.
Secondly, the specificity of genetic modification is also an important issue to be considered. As targeting by CRISPR/Cas9 is dependent on nearly 23 base pair matches [70], it may generate many nonspecific mutations in the genome editing. Great efforts should be made to improve the ratio of on- and off-target effects. Several research groups have developed new technologies to maximally diminished off-target genome editing such as truncated sgRNA [71, 72] and dCas9-Fok I fusions [73]. Additionally, employment of bioinformatic screening and paired Cas9 nickases were also found to remarkably enhance genome editing specificity [74, 75]. Cas9 off-target sites have been reported by Genome-wide analysis [76-78]. Evaluation of off-target effects is a critical step of developing this method. Although reduced off-target effects are observed by tru-gRNAs and paired nickases, further improvements will be required, especially for therapeutic interferences.
Another issue to be concerned is the safety of the CRISPR/Cas9 platform [79]. As this genome engineering technology has the unparalleled potential for modifying human and nonhuman genomes, it is likely to confound unknown risks to human health. Research is required to understand and avoid risks when using the CRISPR/Cas9 technique.
CONCLUSION REMARKS AND FUTURE PERSPECTIVES
Although brain tumors have been known for more than 50 years, there are still many problems to be solved. The etiology of brain tumors is elusive and the treatment is not satisfactory [80]. There is in great need to develop suitable brain tumor models that faithfully reflect the etiology of human brain neoplasms.
Traditional animal models of brain tumor possess various faults. For example, the cells transplanted in the CDX model are mainly high-grade origin and this model can not comprehensively mimic different grades of brain tumors. In terms of the GEMM model, the generation is a low-efficient and time-consuming process. The CRISPR/Cas9 system can be served as a novel tool to overcome the shortages of the existing methods. It can precisely and efficiently obtain the genetic mutations of the target genome at a selected DNA site. In central nervous system, previous investigations illustrated that animal models of sonic hedgehog medulloblastoma and glioblastoma have been successfully established using CRISPR KO [81, 82]. Table 2 listed several regulatory factors (p53, Nf1, Pten, Ptch1, Bmi1, Met, Notch1, CDK6, miR-10b, TERT, LSD1, miR-218 and miR-128) which were closely related to the etiology of brain tumors [28, 43, 44, 83-92]. It was found that Cas9-deletion of multiple TSGs including p53, Nf1, Pten and Ptch1 could induce the formation of brain tumors [44]. We propose that other regulators that are associated with the generation of brain tumors can be manipulated by CRISPR/Cas9 system.
Table 2. Representative factors that can be manipulated by different types of CRISPR genome editing technologies for brain tumor modeling.
| Factors | Role in tumorigenesis | Proposed CRISPR methods | Previous methods | Ref |
|---|---|---|---|---|
| p53 (a) (b) | tumor suppression | CRISPR KO | CRISPR KO | [43, 44] |
| Nf1 (a) | tumor suppression | CRISPR KO | CRISPR KO | [44] |
| Pten (a) (b) | tumor suppression | CRISPR KO | CRISPR KO | [43, 44] |
| Ptch1 (c) | tumor suppression | CRISPR KO | CRISPR KO | [44] |
| Bmi1 (a) (d) | tumor facilitation | CRISPR KI | Bmi1 shRNA | [83, 84] |
| Met (e) | tumor facilitation | CRISPR KI | CRISPR KO | [85] |
| Notch1(a) | tumor facilitation | CRISPR KI | Notch1 siRNA | [86] |
| CDK6(a) | tumor facilitation | CRISPR KI | CDK6 shRNA | [87] |
| miR-10b(a) | tumor facilitation | CRISPRa | miR-10b mimics | [88] |
| TERT(e) | tumor facilitation | CRISPRa | CRISPRa | [90] |
| LSD1(f) | tumor facilitation | CRISPRa | CRISPRa | [90] |
| miR-218(a) | tumor suppression | CRISPRi | anti- miR-218 | [28] |
| miR-128(a) | tumor suppression | CRISPRi | anti-miR-128 | [91] |
Abbreviations: CRISPR KO: CRISPR knock out; CRISPR KI: CRISPR knock in; CRISPRa: CRISPR activation; CRISPRi: CRISPR interference.
Note: (a) glioma; (b) liver; (c) medulloblastoma; (d) breast cancer with brain metastases; (e) HEK293 cell line; (f) embryonic stem cell
Furthermore, the CRISPR/Cas9 technology has been widely used for genome engineering in many stem cell organoids including stomach [93, 94], intestine [95-97] and pancreas [38, 98, 99]. In details, the locus of the cystic fibrosis transmembrane conductor receptor in cultured intestinal stem cells derived from cystic fibrosis patients was accurately repaired using CRISPR/Cas9 genome editing system via homologous recombination [97]. Efficient gene transfer was also successfully established in human intestinal organoids by this platform [96]. Human pluripotent stem cells (hPSCs) such as induced pluripotent stem cells (iPSCs) and embryonic stem cells (ESCs) are considered as very useful tools for elucidating regulatory processes during early development and the pathogenesis of genetic disorders [100, 101]. Since the human iPSCs retain the individual genetic information, combination with this cell and CRISPR-mediated genetic editing may be critical for exploring this phenotype that appears during cell differentiation [63]. Although traditional knockout of genes related to hPSCs self-renewal or survival may block cell propagation and survival, a recent investigation reported that successful knockout of multiple genes including SOX2, PAX6, OTX2 and AGO2 was established in human iPSCs and ESCs using CRISPR system [102]. In the future, CRISPR/Cas9 system is likely to apply for studying human brain tumors in human iPSCs derived from brain tumor patients. The large genomic mutation databases generated from sequencing of the GBM patient indicate a lot new gene mutations can potentially response for glioma formation. With the CRISPR/Cas9 tool, we can also easily generate a new model based on the information we learned from this database in a more effective way.
Although the development of CRISPR/Cas9 technology for genome editing raises some social challenges to some extent and possibly brings out a series of uncertainty and fear of catastrophic misuse, it can never cease to inspire us for investigating the precise molecular mechanisms of brain tumors using this biological tool.
Abbreviations
- CDX
cell-derived xenograft
- PDX
patient-derived xenograft
- GEMM
genetically engineered mouse model
- CRISPR
clustered regularly interspaced short palindromic repeat
- Cas9
CRISPR-associated protein 9
- gRNA
guide RNA
- sgRNA
single guide RNA
- crRNA
CRISPR RNA
- tracrRNA
trans-activating crRNA
- PAM
protospacer adjacent motif
- DSB
double-stranded break
- NHEJ
non-homologous end-joining
- HDR
homology-directed repair
- dSpCas9
dead Cas9 mutant
- SaCas9
Staphylococcus aureus Cas9
- Cpf1
centromere and promoter factor 1
- ZFN
zinc finger nuclease
- TALEN
transcription activator-like effector nuclease
- TSG
tumor suppressor gene
- CRISPR KO
CRISPR knock out
- CRISPR KI
CRISPR knock in
- CRISPRi
CRISPR interference
- CRISPRa
CRISPR activation
- RNAi
RNA interference
- TALE
transcription activator-like effector
- hPSC
human pluripotent stem cells
- iPSC
induced pluripotent stem cell
- ESC
embryonic stem cell
Footnotes
CONFLICTS OF INTEREST
The authors declared no potential financial conflicts of interest.
GRANT SUPPORT
This work was in part supported by National Natural Science Foundation of China (Nos. 81272801, 81302750 and 81373490).
REFERENCES
- 1.Segal G. Re: Brain and other central nervous system cancers: recent trends in incidence and mortality. J Natl Cancer Inst. 2000;92:77–78. doi: 10.1093/jnci/92.1.77. [DOI] [PubMed] [Google Scholar]
- 2.Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pastori C, Kapranov P, Penas C, Peschansky V, Volmar CH, Sarkaria JN, Bregy A, Komotar R, St Laurent G, Ayad NG, Wahlestedt C. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc Natl Acad Sci U S A. 2015;112:8326–8331. doi: 10.1073/pnas.1424220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benedetti E, Antonosante A, d'Angelo M, Cristiano L, Galzio R, Destouches D, Florio TM, Dhez AC, Astarita C, Cinque B, Fidoamore A, Rosati F, Cifone MG, Ippoliti R, Giordano A, Courty J, Cimini A. Nucleolin antagonist triggers autophagic cell death in human glioblastoma primary cells and decreased in vivo tumor growth in orthotopic brain tumor model. Oncotarget. 2015;6:42091–104. doi: 10.18632/oncotarget.5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monje M, Mitra SS, Freret ME, Raveh TB, Kim J, Masek M, Attema JL, Li G, Haddix T, Edwards MS, Fisher PG, Weissman IL, Rowitch DH, Vogel H, Wong AJ, Beachy PA. Hedgehog-responsive candidate cell of origin for diffuse intrinsic pontine glioma. Proc Natl Acad Sci U S A. 2011;108:4453–4458. doi: 10.1073/pnas.1101657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faria CC, Golbourn BJ, Dubuc AM, Remke M, Diaz RJ, Agnihotri S, Luck A, Sabha N, Olsen S, Wu X, Garzia L, Ramaswamy V, Mack SC, Wang X, Leadley M, Reynaud D, Ermini L, Post M, Northcott PA, Pfister SM, Croul SE, Kool M, Korshunov A, Smith CA, Taylor MD, Rutka JT. Foretinib is effective therapy for metastatic sonic hedgehog medulloblastoma. Cancer Res. 2015;75:134–146. doi: 10.1158/0008-5472.CAN-13-3629. [DOI] [PubMed] [Google Scholar]
- 7.Jones TS, Holland EC. Animal models for glioma drug discovery. Expert Opin Drug Discov. 2011;6:1271–1283. doi: 10.1517/17460441.2011.632628. [DOI] [PubMed] [Google Scholar]
- 8.Candolfi M, Curtin JF, Nichols WS, Muhammad AG, King GD, Pluhar GE, McNiel EA, Ohlfest JR, Freese AB, Moore PF, Lerner J, Lowenstein PR, Castro MG. Intracranial glioblastoma models in preclinical neuro-oncology: neuropathological characterization and tumor progression. J Neurooncol. 2007;85:133–148. doi: 10.1007/s11060-007-9400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swartling FJ, Hede SM, Weiss WA. What underlies the diversity of brain tumors? Cancer Metastasis Rev. 2013;32:5–24. doi: 10.1007/s10555-012-9407-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmen SL, Williams BO. Essential role for Ras signaling in glioblastoma maintenance. Cancer Res. 2005;65:8250–8255. doi: 10.1158/0008-5472.CAN-05-1173. [DOI] [PubMed] [Google Scholar]
- 11.Uhrbom L, Dai C, Celestino JC, Rosenblum MK, Fuller GN, Holland EC. Ink4a-Arf loss cooperates with KRas activation in astrocytes and neural progenitors to generate glioblastomas of various morphologies depending on activated Akt. Cancer Res. 2002;62:5551–5558. [PubMed] [Google Scholar]
- 12.Wei Q, Clarke L, Scheidenhelm DK, Qian B, Tong A, Sabha N, Karim Z, Bock NA, Reti R, Swoboda R, Purev E, Lavoie JF, Bajenaru ML, Shannon P, Herlyn D, Kaplan D, Henkelman RM, Gutmann DH, Guha A. High-grade glioma formation results from postnatal pten loss or mutant epidermal growth factor receptor expression in a transgenic mouse glioma model. Cancer Res. 2006;66:7429–7437. doi: 10.1158/0008-5472.CAN-06-0712. [DOI] [PubMed] [Google Scholar]
- 13.Becher OJ, Hambardzumyan D, Walker TR, Helmy K, Nazarian J, Albrecht S, Hiner RL, Gall S, Huse JT, Jabado N, MacDonald TJ, Holland EC. Preclinical evaluation of radiation and perifosine in a genetically and histologically accurate model of brainstem glioma. Cancer Res. 2010;70:2548–2557. doi: 10.1158/0008-5472.CAN-09-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tu Z, Yang W, Yan S, Guo X, Li XJ. CRISPR/Cas9: a powerful genetic engineering tool for establishing large animal models of neurodegenerative diseases. Mol Neurodegener. 2015;10:35. doi: 10.1186/s13024-015-0031-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boettcher M, McManus MT. Choosing the Right Tool for the Job: RNAi, TALEN, or CRISPR. Mol Cell. 2015;58:575–585. doi: 10.1016/j.molcel.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, Katsanos D, Gribble M, Baker D, Marois E, Russell S, Burt A, Windbichler N, Crisanti A, Nolan T. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol. 2015 doi: 10.1038/nbt.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malina A, Cameron CJ, Robert F, Blanchette M, Dostie J, Pelletier J. PAM multiplicity marks genomic target sites as inhibitory to CRISPR-Cas9 editing. Nat Commun. 2015;6:10124. doi: 10.1038/ncomms10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 27.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 28.Tu Y, Gao X, Li G, Fu H, Cui D, Liu H, Jin W, Zhang Y. MicroRNA-218 inhibits glioma invasion, migration, proliferation, and cancer stem-like cell self-renewal by targeting the polycomb group gene Bmi1. Cancer Res. 2013;73:6046–6055. doi: 10.1158/0008-5472.CAN-13-0358. [DOI] [PubMed] [Google Scholar]
- 29.Ding MH, Wang Z, Jiang L, Fu HL, Gao J, Lin XB, Zhang CL, Liu ZY, Shi YF, Qiu GZ, Ma Y, Cui DX, Hu GH, Jin WL. The transducible TAT-RIZ1-PR protein exerts histone methyltransferase activity and tumor-suppressive functions in human malignant meningiomas. Biomaterials. 2015;56:165–178. doi: 10.1016/j.biomaterials.2015.03.058. [DOI] [PubMed] [Google Scholar]
- 30.Capecchi MR. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 31.Wyman C, Kanaar R. DNA double-strand break repair: all's well that ends well. Annu Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 32.Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23:720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W, Teng F, Li T, Zhou Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol. 2013;31:684–686. doi: 10.1038/nbt.2652. [DOI] [PubMed] [Google Scholar]
- 34.Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, Zhao Y. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol. 2013;31:681–683. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- 35.Ma Y, Shen B, Zhang X, Lu Y, Chen W, Ma J, Huang X, Zhang L. Heritable multiplex genetic engineering in rats using CRISPR/Cas9. PLoS One. 2014;9:e89413. doi: 10.1371/journal.pone.0089413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen F, Rosiene J, Che A, Becker A, LoTurco J. Tracking and transforming neocortical progenitors by CRISPR/Cas9 gene targeting and piggyBac transposase lineage labeling. Development. 2015;142:3601–3611. doi: 10.1242/dev.118836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Incontro S, Asensio CS, Edwards RH, Nicoll RA. Efficient, complete deletion of synaptic proteins using CRISPR. Neuron. 2014;83:1051–1057. doi: 10.1016/j.neuron.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng J, Wang Y, Jiang J, Zhou X, Song L, Wang L, Ding C, Qin J, Liu L, Wang W, Liu J, Huang X, Wei H, Zhang P. Production of Human Albumin in Pigs Through CRISPR/Cas9-Mediated Knockin of Human cDNA into Swine Albumin Locus in the Zygotes. Sci Rep. 2015;5:16705. doi: 10.1038/srep16705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma Y, Ma J, Zhang X, Chen W, Yu L, Lu Y, Bai L, Shen B, Huang X, Zhang L. Generation of eGFP and Cre knockin rats by CRISPR/Cas9. FEBS J. 2014;281:3779–3790. doi: 10.1111/febs.12935. [DOI] [PubMed] [Google Scholar]
- 41.Li S, Xue H, Wu J, Rao MS, Kim DH, Deng W, Liu Y. Human Induced Pluripotent Stem Cell NEUROG2 Dual Knockin Reporter Lines Generated by the CRISPR/Cas9 System. Stem Cells Dev. 2015;24:2925–2942. doi: 10.1089/scd.2015.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu Z, Verma N, Gonzalez F, Shi ZD, Huangfu D. A CRISPR/Cas-Mediated Selection-free Knockin Strategy in Human Embryonic Stem Cells. Stem Cell Reports. 2015;4:1103–1111. doi: 10.1016/j.stemcr.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai W, Yang G, Bronson R, Crowley DG, Zhang F, Anderson DG, Sharp PA, Jacks T. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, Zapatka M, Northcott PA, Schramm K, Belic J, Jones DT, Tschida B, Moriarity B, Largaespada D, Roussel MF, Korshunov A, Reifenberger G, Pfister SM, Lichter P, Kawauchi D, Gronych J. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun. 2015;6:7391. doi: 10.1038/ncomms8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 49.Segal DJ, Barbas CF., 3rd Custom DNA-binding proteins come of age: polydactyl zinc-finger proteins. Curr Opin Biotechnol. 2001;12:632–637. doi: 10.1016/s0958-1669(01)00272-5. [DOI] [PubMed] [Google Scholar]
- 50.Beerli RR, Barbas CF., 3rd Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 51.Liu Q, Segal DJ, Ghiara JB, Barbas CF., 3rd Design of polydactyl zinc-finger proteins for unique addressing within complex genomes. Proc Natl Acad Sci U S A. 1997;94:5525–5530. doi: 10.1073/pnas.94.11.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garg A, Lohmueller JJ, Silver PA, Armel TZ. Engineering synthetic TAL effectors with orthogonal target sites. Nucleic Acids Res. 2012;40:7584–7595. doi: 10.1093/nar/gks404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim D, Rossi J. RNAi mechanisms and applications. Biotechniques. 2008;44:613–616. doi: 10.2144/000112792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 57.Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2015;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toledo Chad M, Ding Y, Hoellerbauer P, Davis Ryan J, Basom R, Girard Emily J, Lee E, Corrin P, Hart T, Bolouri H, Davison J, Zhang Q, Hardcastle J, Aronow Bruce J, Plaisier Christopher L, Baliga Nitin S, Moffat J, Lin Q, Li X-N, Nam D-H, Lee J, Pollard Steven M, Zhu J, Delrow Jeffery J, Clurman Bruce E, Olson James M, Paddison Patrick J. Genome-wide CRISPR-Cas9 Screens Reveal Loss of Redundancy between PKMYT1 and WEE1 in Glioblastoma Stem-like Cells. Cell Reports. 2015;13:2425–2439. doi: 10.1016/j.celrep.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shinmyo Y, Tanaka S, Tsunoda S, Hosomichi K, Tajima A, Kawasaki H. CRISPR/Cas9-mediated gene knockout in the mouse brain using in utero electroporation. Scientific Reports. 2016;6:20611. doi: 10.1038/srep20611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heidenreich M, Zhang F. Applications of CRISPR-Cas systems in neuroscience. Nat Rev Neurosci. 2015 doi: 10.1038/nrn.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zetsche B, Volz SE, Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wright AV, Sternberg SH, Taylor DW, Staahl BT, Bardales JA, Kornfeld JE, Doudna JA. Rational design of a split-Cas9 enzyme complex. Proc Natl Acad Sci U S A. 2015;112:2984–2989. doi: 10.1073/pnas.1501698112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, Joung JK. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, Jaenisch R, Zhang F, Sharp PA. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baltimore D, Berg P, Botchan M, Carroll D, Charo RA, Church G, Corn JE, Daley GQ, Doudna JA, Fenner M, Greely HT, Jinek M, Martin GS, Penhoet E, Puck J, Sternberg SH, Weissman JS, Yamamoto KR. Biotechnology. A prudent path forward for genomic engineering and germline gene modification. Science. 2015;348:36–38. doi: 10.1126/science.aab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frosina G. DNA repair and resistance of gliomas to chemotherapy and radiotherapy. Mol Cancer Res. 2009;7:989–999. doi: 10.1158/1541-7786.MCR-09-0030. [DOI] [PubMed] [Google Scholar]
- 81.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A, Ellison DW, Lichter P, Gilbertson RJ, Pomeroy SL, Kool M, Pfister SM. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:829–848. doi: 10.1007/s00401-015-1432-1. [DOI] [PubMed] [Google Scholar]
- 83.Hoenerhoff MJ, Chu I, Barkan D, Liu ZY, Datta S, Dimri GP, Green JE. BMI1 cooperates with H-RAS to induce an aggressive breast cancer phenotype with brain metastases. Oncogene. 2009;28:3022–3032. doi: 10.1038/onc.2009.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bruggeman SW, Hulsman D, Tanger E, Buckle T, Blom M, Zevenhoven J, van Tellingen O, van Lohuizen M. Bmi1 controls tumor development in an Ink4a/Arf-independent manner in a mouse model for glioma. Cancer Cell. 2007;12:328–341. doi: 10.1016/j.ccr.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 85.Togashi Y, Mizuuchi H, Tomida S, Terashima M, Hayashi H, Nishio K, Mitsudomi T. MET gene exon 14 deletion created using the CRISPR/Cas9 system enhances cellular growth and sensitivity to a MET inhibitor. Lung Cancer. 2015 doi: 10.1016/j.lungcan.2015.10.020. [DOI] [PubMed] [Google Scholar]
- 86.Zhang X, Chen T, Zhang J, Mao Q, Li S, Xiong W, Qiu Y, Xie Q, Ge J. Notch1 promotes glioma cell migration and invasion by stimulating beta-catenin and NF-kappaB signaling via AKT activation. Cancer Sci. 2012;103:181–190. doi: 10.1111/j.1349-7006.2011.02154.x. [DOI] [PubMed] [Google Scholar]
- 87.Li B, He H, Tao BB, Zhao ZY, Hu GH, Luo C, Chen JX, Ding XH, Sheng P, Dong Y, Zhang L, Lu YC. Knockdown of CDK6 enhances glioma sensitivity to chemotherapy. Oncol Rep. 2012;28:909–914. doi: 10.3892/or.2012.1884. [DOI] [PubMed] [Google Scholar]
- 88.Sun L, Yan W, Wang Y, Sun G, Luo H, Zhang J, Wang X, You Y, Yang Z, Liu N. MicroRNA-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. Brain Res. 2011;1389:9–18. doi: 10.1016/j.brainres.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 89.Xi L, Schmidt JC, Zaug AJ, Ascarrunz DR, Cech TR. A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression. Genome Biol. 2015;16:231. doi: 10.1186/s13059-015-0791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12:401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang Y, Chao T, Li R, Liu W, Chen Y, Yan X, Gong Y, Yin B, Qiang B, Zhao J, Yuan J, Peng X. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J Mol Med (Berl) 2009;87:43–51. doi: 10.1007/s00109-008-0403-6. [DOI] [PubMed] [Google Scholar]
- 92.Gao X, Jin W. The emerging role of tumor-suppressive microRNA-218 in targeting glioblastoma stemness. Cancer Lett. 2014;353:25–31. doi: 10.1016/j.canlet.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 93.McCracken KW, Cata EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai YH, Mayhew CN, Spence JR, Zavros Y, Wells JM. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gannon HS, Jr, Kaplan N, Tsherniak A, Vazquez F, Weir BA, Hahn WC, Meyerson M. Identification of an “Exceptional Responder” Cell Line to MEK1 Inhibition: Clinical Implications for MEK-targeted Therapy. Mol Cancer Res. 2015 doi: 10.1158/1541-7786.MCR-15-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H, Korving J, van de Wetering M, Schwank G, Logtenberg M, Cuppen E, Snippert HJ, Medema JP, Kops GJ, Clevers H. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 96.Fujii M, Matano M, Nanki K, Sato T. Efficient genetic engineering of human intestinal organoids using electroporation. Nat Protoc. 2015;10:1474–1485. doi: 10.1038/nprot.2015.088. [DOI] [PubMed] [Google Scholar]
- 97.Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK, Nieuwenhuis EE, Beekman JM, Clevers H. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 98.Chiou SH, Winters IP, Wang J, Naranjo S, Dudgeon C, Tamburini FB, Brady JJ, Yang D, Gruner BM, Chuang CH, Caswell DR, Zeng H, Chu P, Kim GE, Carpizo DR, Kim SK, Winslow MM. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev. 2015;29:1576–1585. doi: 10.1101/gad.264861.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, Lowe SW, Ventura A. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 101.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 102.Chen Y, Cao J, Xiong M, Petersen AJ, Dong Y, Tao Y, Huang CT, Du Z, Zhang SC. Engineering Human Stem Cell Lines with Inducible Gene Knockout using CRISPR/Cas9. Cell Stem Cell. 2015;17:233–244. doi: 10.1016/j.stem.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


