Abstract
The androgen receptor (AR) was found to suppress hepatocellular carcinoma (HCC) metastasis at late stages. Due to this discovery, we searched for some AR enhancers to increase the efficacy of Sorafenib chemotherapy, and identified the microRNA (miR)-367-3p, whose expression is positively correlated with AR expression in advanced HCC, as an HCC metastasis suppressor. Combining miR-367-3p with Sorafenib showed better efficacy to suppress HCC cell invasion in vitro and in vivo. Mechanism dissection revealed that miR-367-3p could increase AR expression via directly targeting the 3′UTR of MDM2 to decrease MDM2 protein expression. The resultant increase of AR expression might then promote the expression of FKBP5 and PHLPP, thus dephosphorylating and inactivating AKT and ERK, to suppress the HCC cell invasion. Interestingly, the suppression of pAKT by miR-367-3p could subsequently attenuate the phosphorylation of AR and MDM2, giving rise to additional enhancement of AR protein expression, effectively forming a positive feedback loop. Together, these results suggest that miR-367-3p may function as an AR enhancer to increase Sorafenib chemotherapy efficacy via altering the MDM2/AR/FKBP5/PHLPP/(pAKT and pERK) signals to better suppress HCC metastasis. Successful development of this newly combined chemotherapy in the future may help us to better suppress the HCC metastasis at late stages.
Keywords: Androgen receptor, Hepatocellular carcinoma, Metastasis, miR-367-3p
Highlights
-
•
As an HCC metastasis suppressor, miR-367-3p is expressed at lower levels and positively correlated with AR in advanced HCC.
-
•
The miR-367-3p enhances Sorafenib chemotherapy efficacy via MDM2-AR-(pAKT and pERK) signals and a positive feedback loop.
-
•
Successful clinical application of these findings in the future may help us to better retard HCC metastasis at late stages.
MiRNAs may be promising candidates for therapeutic targets. In this study, we show that miR-367-3p could enhance Sorafenib chemotherapy efficacy via altering MDM2-AR-(pAKT and pERK) signals and formed a positive feedback loop to better suppress the metastasis of hepatocellular carcinoma. Successful clinical application of these findings in the future may help us to better suppress the process of late-stage hepatocellular carcinoma.
1. Introduction
Hepatocellular carcinoma (HCC) is listed as the fifth and seventh most frequently diagnosed cancer in men and women, respectively, across the globe, and the most common primary liver tumor with increasing incidence in the United States and elsewhere. With a dismal outcome largely due to postsurgical recurrence and/or metastasis, HCC is the third most common cause of cancer-related death worldwide (Jemal et al., 2011, Siegel et al., 2012). Although targeted chemotherapy with special target(s) has experienced a rapid progress in the recent decade, the management of patients with advanced HCC, who become unqualified for liver transplantation or liver resection, is still far from satisfactory. Indeed, even patients who have received liver resection often have a high frequency of metastasis/recurrence (Maluccio and Covey, 2012). While clinical trials demonstrated that Sorafenib prolonged the median overall survival time for 2.3–3 months in advanced HCC patients (Cheng et al., 2009), limited efficacy with side effects demanded improvement of this therapy.
The androgen receptor (AR) is a ligand-dependent transcriptional factor that belongs to the nuclear receptor superfamily. Androgen/AR signals play important roles in normal liver function and in the development of liver diseases including HCC (Ma et al., 2014). Previous studies revealed a male predominance in HCC, which suggested that androgen/AR signals may promote hepatocarcinogenesis (Yeh and Chen, 2010). However, the results from several clinical trials using various anti-androgens to treat HCC remain controversial (Groupe d'etude et de traitement du carcinome, H., 2004, Matsuura et al., 1994, Chao et al., 1996). Interestingly, Ma et al. revealed that AR might suppress advanced-stage HCC metastasis using liver-specific deletion of AR (L-ARKO) mice model and in vitro HCC cell lines, and over-expressing AR with Sorafenib treatment could better suppress the HCC progression at late stages (Ma et al., 2012). How to identify a potential molecule to increase AR expression in order to enhance Sorafenib chemotherapy efficacy, however, remains to be further elucidated.
The microRNAs (miRNAs or miRs) are a group of small (around 22 nucleotides), non-coding RNA gene products existing in many organisms that play crucial post-transcriptional regulatory roles in mRNA translation and degradation by complementary base pairing, predominantly in the 3′-untranslated region (3′UTR) (Lau et al., 2001). The discovery of miRNAs has increased our understanding of the post-transcriptional regulation of genes and how this contributes to diverse physiological, developmental, and pathophysiological processes, including cancer initiation and progression (Wang et al., 2012). Several studies have linked deregulation of miRNA expressions to HCC carcinogenesis and metastasis in recent years (Budhu et al., 2008, Salvi et al., 2013). Thus, miRNAs may become promising candidates for therapeutic targets. For example, Miravirsen, a miR-122 inhibitor that is already used in clinical trials, was found to be safe and have no dose-limiting adverse events or escape mutations in the miR-122 binding sites of the HCV genome so far (Janssen et al., 2013).
Here we found miR-367-3p, whose expression is down regulated in advanced HCC, might function as an invasion suppressor in advanced HCC via altering the MDM2/AR/FKBP5/PHLPP/(pAKT and pERK) signals.
2. Materials and Methods
2.1. Materials
Sorafenib was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). MK2206 was purchased from ApexBio (Houston, TX). MG132 (M8699) and Cycloheximide (CHX, 227048) were purchased from Sigma-Aldrich (St. Louis, MO). MISSION® Synthetic miR-367-3p inhibitor was purchased from Sigma-Aldrich also and transfected according to the technical bulletin provided by the manufacturer.
2.2. Invasion Assay
Before invasion assays, HCC cells were seeded in 6-well plates at 5 × 105 (SKhep1, HepG2, Huh7 and SNU398) or 2 × 105 (HA22T and SNU423) cells/well and we followed the designated treatments for 48 h. Then the cells were harvested by trypsinization and 3 × 104 HA22T cells, 5 × 104 SKhep1 and SNU423 cells, 1 × 105 HepG2, Huh7 and SNU398 cells in serum free DMEM were plated into the upper chambers of transwells and 700 μl 10% FCS media was placed in the lower chambers for 24 h incubation at 37 °C in 5% (v/v) CO2 incubator. The upper chambers with 8 μm-pore-size polycarbonate membrane filters (Corning, Inc., Corning, NY) were pre-coated with diluted growth factor-reduced matrigel (1:14 serum free DMEM) (BD Biosciences). After incubation, the cells on the upper layer of membranes were scrapped out, and the cells invaded to the lower membrane were stained with 0.1% (w/v) crystal violet. The invaded cells were counted in ten randomly chosen microscopic fields (100 ×) in each experiment and averaged for quantification. Each sample was run in triplicate and multiple experiments were performed.
2.3. 3D Invasion Assay
Briefly, 5 × 104 of cells in 3 ml media containing 2.5% Matrigel and 30 ng epidermal growth factor (EGF) were plated into collagen/Matrigel mixture coated plate. The media were replenished every 3 days for 10–12 days. The cells with protrusions were regarded as invaded cells and 10 random different fields under 200 × magnification were counted for quantification.
2.4. Luciferase Assay
540 bp fragments of MDM2 3′UTR containing wild type or mutant miRNA-responsive elements (Sequences listed on Supplementary Table 1) were cloned into the psiCheck2 construct (Promaga, Madison, WI) downstream of the Renilla luciferase ORF. Cells were plated in 24-well plates and the cDNA were transfected using Lipofectamine 3000 (Invitrogen) according to the manufacturer's instructions. Luciferase activity was measured by Dual-Luciferase Assay (Promega) according to the manufacturer's manual.
2.5. Tissue Samples
The current study conformed to the principles of the Declaration of Helsinki and was approved by the Institutional Review Board of the Sir Run-Run Shaw Hospital. A total of 126 randomly selected HCC clinical samples (102 paraffin-fixed and 24 freshly frozen samples) were collected from the Department of General Surgery, Sir Run-Run Shaw Hospital, Zhejiang University starting in February 2006, with all patients signing Informed Consent for the use of their tissues for scientific research. We reviewed pathology records to identify samples with confirmed HCC. The demographic and clinical information of the patients were listed in Table 1.
2.6. In Vivo Studies
32 6–8 weeks old athymic nude male mice were purchased from NCI and randomized into 4 groups with standard housing and husbandry. SKhep1 cells were prepared as stable luciferase clones by infection with pLKO-Luciferase lentivirus and pLKO.1-shGFP or pLKO.1-miR-367-3p. Intrahepatic injections of 1 × 106 SKhep1-luc cell lines/100 μL serum-free DMEM and matrigel (1:1) were performed in mice in groups as follows: 1) SKhep1-luc-shGFP + vehicle; 2) SKhep1-luc-miR-367-3p + vehicle; 3) SKhep1-luc-shGFP + Sorafenib; and 4) SKhep1-luc-miR-367-3p + Sorafenib. After 1 month, the mice were treated with/without Sorafenib (30 mg/kg/mice; daily, oral gavage) as indicated for another month. Sorafenib (for groups 3 and 4) was suspended in an oral vehicle containing Cremophor (Sigma-Aldrich), 95% ethanol and water in a ratio of 1:1:6. All control mice (groups 1 and 2) received an equal volume of carrier solution by gavage. Tumor development and metastasis was monitored by IVIS once a week starting after drug treatment following intraperitoneal injection of 150 mg/kg D-Luciferin. Mice were sacrificed after 4 weeks of treatment and tumors were removed for studies. All animal studies were performed under the supervision and guidelines of the University of Rochester Medical Center Animal Care and Use Committee, and experiments were adjusted to minimize suffering and to use the smallest number of animals that could obtain statistical results.
2.7. Statistical Analysis
Data are expressed as mean ± SEM from at least 3 independent experiments. Statistical analyses involved Student's t-test, Log-rank (Mantel-Cox) test, Kolmogorov-Smirnow test, Mann-Whitney U Test and Spearman rank correlation with GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA). p < 0.05 was considered statistically significant. The Western blot bands were analyzed by Gel-Pro analyzer (Meyer Instruments, Inc., Houston, TX) and shown on Supplementary Fig. 5. More detailed materials and methods information please see Supplementary Materials and Methods.
3. Results
3.1. AR Suppresses HCC Cell Invasion
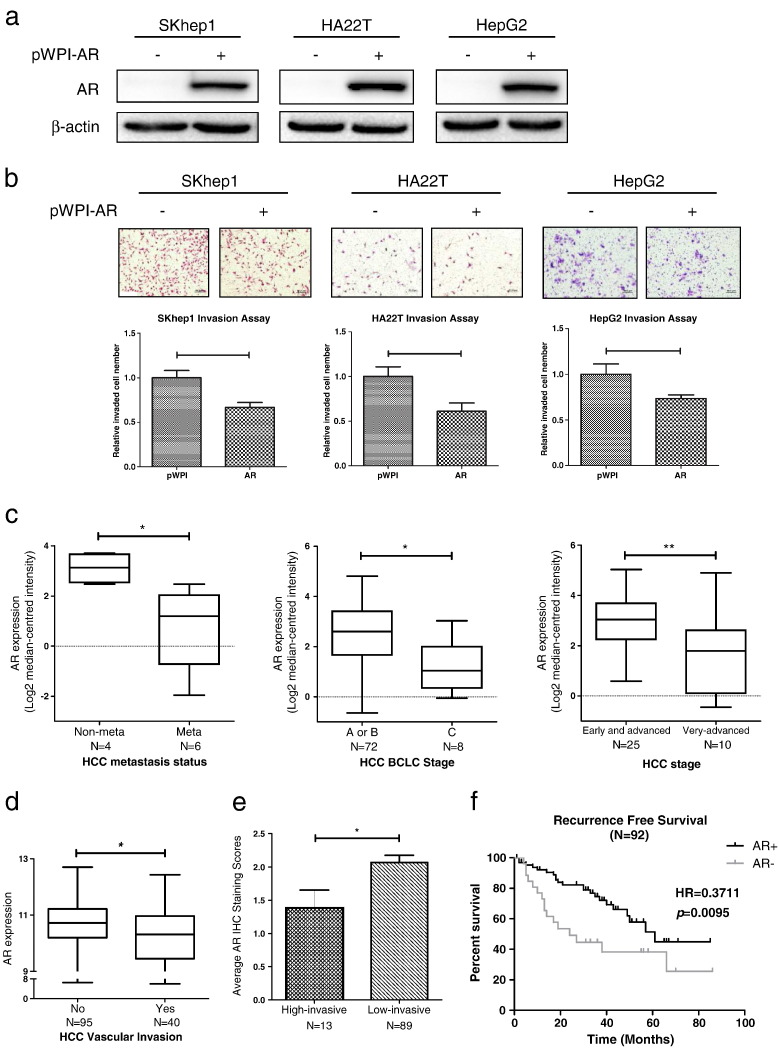
Early in vivo mice studies suggested that hepatic AR could function as a late-stage HCC metastasis suppressor (Ma et al., 2012). In the present study, we examined AR's effect on the invasion of various HCC cell lines using chamber invasion assays. Results revealed that over-expression of AR decreased the cell invasion in all 3 HCC cell lines (SKhep1, HA22T, and HepG2) (Fig. 1a-b), suggesting that AR could function as a late-stage HCC metastasis suppressor and is in agreement with the previous study (Ma et al., 2012).
Fig. 1.
AR suppresses HCC cells invasion.
(a), verification of AR over-expression by western blot assays; (b), chamber-transwell invasion assays showed that over-expression of AR decreased the cell invasion in all 3 HCC cell lines (SKhep1, HA22T, and HepG2). Upper panel, representative images of the chamber-transwell invasion assays; Lower panel, quantification of the invaded cells. The invaded cells were counted in 10 randomly chosen microscopic fields (100 ×) of each experiment and pooled. Each sample was run in triplicate and multiple experiments were performed. (c-d), 4 individual research databases clearly showed that AR levels were lower in advanced stages compared with early stages, implicating that AR played a vital role in late-stage HCC and was negative correlated with HCC metastasis. Data were extracted from Oncomine® Platform (c) and Gene Expression Omnibus Datasets (d). (e), the average AR IHC staining scores of patients were evaluated by experienced pathologists both in high-invasive group (N = 13) and low-invasive group (N = 89). Lower AR IHC staining scores were found in high-invasive group as compared to those found in low-invasive group. (f), recurrence-free survival curve of HCC patients who received surgery (N = 92) indicated that patients with HCC (AR+) (defined by IHC staining) had significant higher recurrence-free survival (HR = 0.3711) than patients with HCC (AR-). p < 0.05 was considered statistically significant. * p < 0.05 and ** p < 0.01.
Importantly, multiple HCC transcriptome analyses (Fig. 1c–d) demonstrated that AR expression was lower in advanced stages compared with early stages of HCC (Liao et al., 2008, Minguez et al., 2011, Wurmbach et al., 2007, Chiang et al., 2008), implicating that AR played a vital role in late-stage HCC and was negatively related with HCC metastasis.
To further confirm the conclusion above, we performed IHC staining of AR on the clinical samples collected from HCC patients who received surgery. We used multiple tumor foci, satellite nodules, micro-vascular invasion and portal vein tumor thrombosis, to define/classify these patients into 2 groups: high-invasive (patients with either multiple tumor foci, satellite nodules, micro-vascular invasion, portal vein tumor thrombosis, or combinations of these) and low-invasive (patients with neither multiple tumor foci, satellite nodules, micro-vascular invasion nor portal vein tumor thrombosis). The results indicated that lower AR IHC staining scores were found in high-invasive group as compared to those found in low-invasive group (Fig. 1e), suggesting that AR might relate negatively with HCC metastasis at late stages. Importantly, it was also found that patients with positive AR (AR+) HCC (as defined by IHC staining) had significant higher recurrence-free survival (HR = 0.3711) than patients with negative AR (AR-) HCC (Fig. 1f).
Together, results from Fig. 1 suggest that AR may act as a late-stage HCC metastasis suppressor.
3.2. The miR-367-3p Increases AR Protein Expression in Late-stage HCC
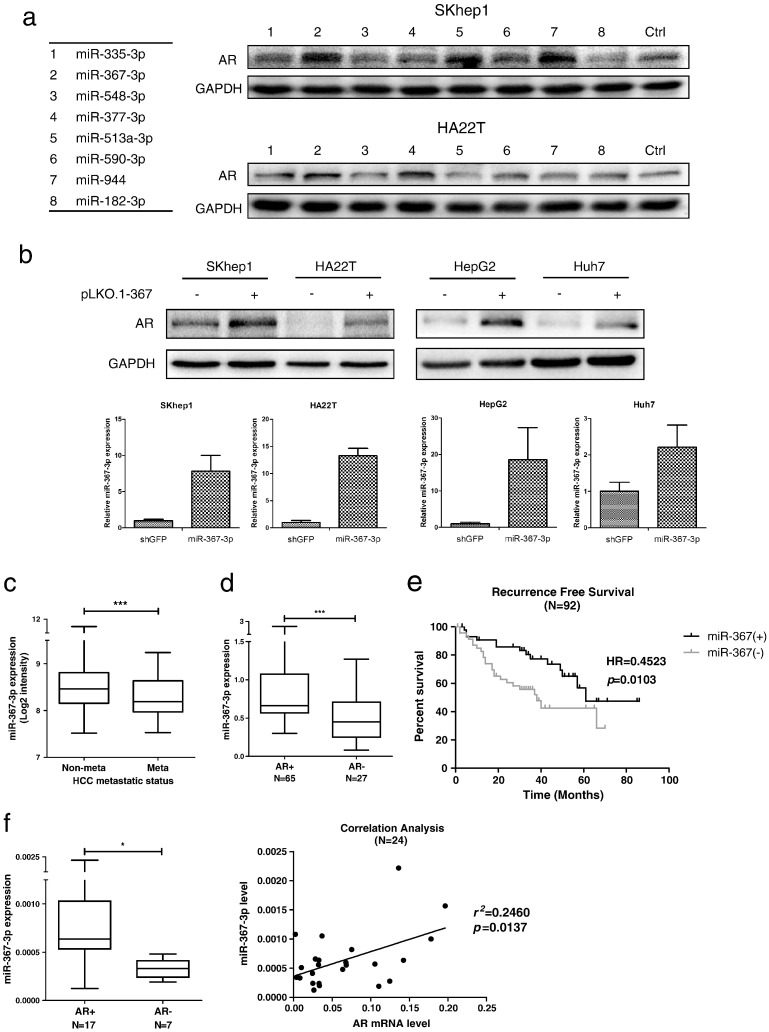
Since AR serves as an HCC suppressor at late stages, we were interested in targeting AR via some potential molecules that could increase or stabilize AR expression in order to provide potential therapies to better fight HCC metastasis. We focused on those miRNAs related to the E3 ligases that might alter the AR degradation. We first developed a specific screening strategy to seek for AR expression enhancers. According to recent literature (Lin et al., 2002, Dirac and Bernards, 2010, Li et al., 2008, Rees et al., 2006, An et al., 2014, Nishida and Yasuda, 2002), we picked 7 E3 ligases that could target AR and enhance AR degradation. From databases (TargetScan, miRDB and MicroCosm Targets) analyses, we found limited potential candidates and focused on 8 miRNAs that might possibly increase AR expression according to the databases prediction (Supplementary Fig. 1a). We then over-expressed these miRNAs in SKhep1 and HA22T cell lines to examine their effects on AR expression. As shown in Fig. 2a, the presence of miR-367-3p, miR-513a-3p, and miR-944 significantly increased the AR expression in SKhep1 cells; however, AR expression could only be elevated by miR-367-3p and miR-377-3p in HA22T cells.
Fig. 2.
The miR-367-3p increases AR protein levels and is expressed at lower levels in late-stage HCC.
(a), western blot assays were applied to check the AR protein levels after over-expressing the 8 miRNA candidates according to the screening strategy in SKhep1 and HA22T cell lines. (b), upper panel: AR levels in 4 different HCC cell lines (SKhep1, HA22T, HepG2, and Huh7) with overexpressed miR-367-3p were checked by western blot assays and were found consistently and significantly increased; lower panel: verification of miR-367-3p over-expression by qRT-PCR. (c), miR-367-3p was expressed at lower levels in metastatic patients than non-metastatic patients according to the non-coding RNA microarray data submitted on public Gene Expression Omnibus Datasets. (d), paraffin-fixed clinical sample surveys disclosed a significantly higher miR-367-3p expression in patients with HCC (AR+, N = 65) than in those with HCC (AR-, N = 27). HCC (AR+) and HCC (AR-) were determined by IHC staining. (e), recurrence-free survival curve of HCC patients who received surgery (N = 92) indicated that patients with higher miR-367-3p expression had significantly higher recurrence-free survival (HR = 0.4523) than patients with lower miR-367-3p expression. (f), left panel: freshly frozen clinical samples showed a significantly higher miR-367-3p expression in patients with HCC (AR+, N = 17) than in those with HCC (AR-, N = 7). HCC (AR+) and HCC (AR-) were determined by IHC staining; right panel: correlation analysis of miR-367-3p and AR mRNA levels from 24 HCC samples by qRT-PCR indicated a significant positive correlation between miR-367-3p and AR mRNA levels. p < 0.05 was considered statistically significant. * p < 0.05 and *** p < 0.001.
We then focused on miR-367-3p as it could up-regulate AR expression in both HCC cell lines, and further confirmed these findings in 4 HCC cell lines (SKhep1, HA22T, HepG2, and Huh7), showing a consistent increase of AR expression upon over-expressing miR-367-3p (Fig. 2b). Interestingly, based on public Gene Expression Omnibus Datasets submitted by Budhu A et al. (Budhu et al., 2008) that contained the largest sample size related to HCC metastasis, we found that miR-367-3p was expressed lower in metastatic HCC patients as compared to those in non-metastatic HCC patients (Fig. 2c). Similarly, miR-302a/b/c/d, members of the miR-302/367 cluster, which are located in the same genomic region and share the same promoter (Sewer et al., 2005), also had lower expressions in metastatic HCC patients (Supplementary Fig. 1b), indicating this down-regulation in the advanced stage of HCC might be controlled or modified by the shared promoter of the miR-302/367 cluster. Moreover, 6 HCC cell lines were divided into 2 groups based on their invasive abilities (Supplementary Fig. 1c-d, Group I: high invasive; Group II: low invasive), and we found that the expression of miR-367-3p was significantly lower in Group I compared to Group II, which further supported the role of miR-367-3p as an HCC metastasis suppressor (Supplementary Fig. 1e).
Paraffin-fixed clinical sample surveys disclosed a significantly higher miR-367-3p expression in patients with HCC (AR+) than in those with HCC (AR-) (Fig. 2d). Importantly, results showed that patients with higher miR-367-3p expression had significant higher recurrence-free survival (HR = 0.4523) than patients with lower miR-367-3p expression (Fig. 2e). Moreover, frozen clinical samples also showed significantly higher miR-367-3p expression in patients with HCC (AR+) than in those with HCC (AR-) (Fig. 2f, left panel). In addition, we also found a significant positive correlation between miR-367-3p and AR mRNA expression, which strongly supported our findings in 4 different HCC cell lines (Fig. 2f, right panel).
Together, results from Fig. 2 and Supplementary Fig. 1 suggest that miR-367-3p, which is lower expressed and positively correlates with AR expression in late-stage HCC, may increase AR protein levels in HCC.
3.3. The miR-367-3p Suppresses HCC Cell Invasion via Altering the AR-FKBP5-PHLPP-(pAKT and pERK) Signals
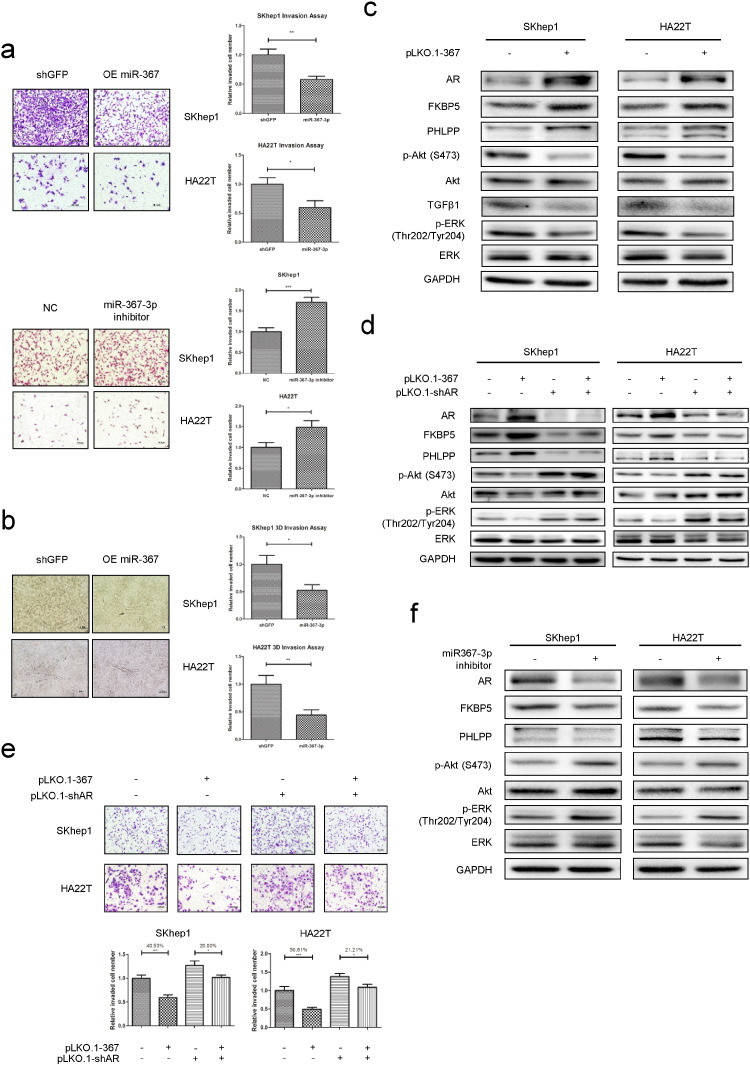
To further verify the positive correlation between miR-367-3p and AR expression and their impacts on HCC progression at late stages, we performed the chamber-transwell invasion assays on 2 invasive HCC cell lines (SKhep1 and HA22T) and the results showed that over-expression of miR-367-3p could significantly decrease (Fig. 3a, upper panel), while miR-367-3p inhibitor could significantly increase the invasion ability of both HCC cell lines (Fig. 3a, lower panel). Similar results were also obtained when we replaced the chamber-transwell invasion assay with the 3D invasion assay in both SKhep1 and HA22T cells (Fig. 3b).
Fig. 3.
The miR-367-3p suppresses HCC cell invasion via altering the AR/FKBP5/PHLPP/(pAKT and pERK) signals pathway.
(a), chamber-transwell invasion assays on SKhep1 and HA22T cell lines showed that over-expression of miR-367-3p could significantly decrease, while miR-367-3p inhibitor could significantly increase the invasion ability. The invaded cells were counted in 10 randomly chosen microscopic fields (100 ×) of each experiment and pooled for quantification. (b), 3D-invasion assays on SKhep1 and HA22T cell lines also showed that over-expression of miR-367-3p could significantly decrease the invasion ability. The cells with protrusions were regarded as invaded cells and 10 random different fields at 200 × magnification were counted for quantification; (c), we used western blot assays to test downstream altered molecules upon over-expressing miR-367-3p in SKhep1 and HA22T cells. (d, e), rescue assays by knocking down AR showed partially reversed FKBP5-PHLPP-(pAKT/pERK) levels (by western blot assays) and cell invasion ability (by chamber-transwell invasion assays). (f), we used western blot assays to test downstream altered molecules upon transfecting miR-367-3p inhibitor (50 nM) in SKhep1 and HA22T cells.
To dissect the potential molecular mechanism, we first hypothesized that miR-367-3p might regulate some AR-modulated metastasis-related genes, such as pAKT, TGFβ1, COX-2, NF-κB and p27, as recent studies indicated that these AR-modulated genes might be able to mediate the AR-suppressed cell invasion in prostate cancer (Niu et al., 2008). In a pilot study, we found miR-367-3p could significantly decrease TGFβ1 expression and decrease AKT activity (by inhibiting phosphorylation of Akt in Ser-473) in both SKhep1 and HA22T cells (Fig. 3c), but not for the other genes mentioned above (Supplementary Fig. 2a). However, TGFβ1 expression changed inconsistently in SKhep1 and HA22T cells with AR knocked down, suggesting the roles of TGFβ1 in these cells might be limited (Supplementary Fig. 2b).
We then examined other potential metastasis-related genes and found that the expression of PHLPP (PH domain and Leucine rich repeat Protein Phosphatases), which could inhibit the phosphorylation of AKT through different signaling components (Brognard et al., 2007), was increased (Fig. 3c), whereas the expression of other metastasis-related genes remained unchanged or inconsistently changed (Supplementary Fig. 2a). These results suggested that miR-367-3p might be able to function through increasing PHLPP expression to dephosphorylate AKT at Ser-473 to decrease AKT activity. Furthermore, a previous study suggested that p-ERK could also be regulated by PHLPP (Li et al., 2014), and we indeed found p-ERK (Thr202/Tyr204) significantly decreased when over-expressing miR-367-3p (Fig. 3c). The enhanced de-phosphorylation by PHLPP is likely facilitated by FKBP5 that functions as a scaffold to enhance the PHLPP-AKT interaction and PHLPP-mediated de-phosphorylation (Brognard and Newton, 2008, Pei et al., 2009). In our study, FKBP5 was significantly increased upon AR elevation by miR-367-3p as it is known that FKBP5 is a transcription target of AR. (Fig. 3c).
An interruption approach via knocking down AR revealed that a reduction of AR partially reversed the expression of FKBP5, PHLPP, pAKT (S473) and p-ERK (Thr202/Tyr204), and miR-367-3p could no longer significantly inhibit pAKT (S473) and p-ERK (Thr202/Tyr204) in AR knocked-down HCC cells (Fig. 3d). The rescue assay for invasion showed the inhibition of invasion by miR-367-3p in SKhep1 cells was diminished in AR knocked-down group (Fig. 3e). Similar results were also obtained when we replaced SKhep1 cells with HA22T cells, indicating this newly identified signal is not unique to a particular cell line (Fig. 3e).
Importantly, expected changes in the signaling components were demonstrated in SKhep1 and HA22T cells in response to the inhibitor of miR-367-3p (Fig. 3f), which further proved miR-367-3p could regulate AR-FKBP5-PHLPP-(pAKT and pERK) signaling pathway to inhibit the cell invasion of HCC. Consistent with the in vitro findings, a clinical sample survey also showed a negative correlation of pAKT/pERK and AR in HCC patients (Table 2; Supplementary Fig. 2c). (See Table 1.)
Table 2.
IHC grades of AR and pERK/pAKT from 102 clinical HCC samples.
| IHC grade | AR |
||||
|---|---|---|---|---|---|
| − | + | ++ | +++ | ||
| pERK⁎ | − | 3 | 2 | 10 | 23 |
| + | 4 | 8 | 6 | 3 | |
| ++ | 2 | 3 | 15 | 11 | |
| +++ | 3 | 5 | 1 | 3 | |
| pAKT⁎⁎ | − | 3 | 2 | 10 | 23 |
| + | 4 | 8 | 6 | 3 | |
| ++ | 2 | 3 | 15 | 11 | |
| +++ | 3 | 5 | 1 | 3 | |
Each IHC slide was checked and the IHC grades were determined by a specialized pathologist.
Spearman rank correlation: r = − 0.2613, p = 0.0080.
Spearman rank correlation: r = − 0.2047, p = 0.0390.
Table 1.
Demographic and clinical information of the patients.
| Paraffin samples | Frozen samples | ||
|---|---|---|---|
| Total | 102 | 24 | |
| Gender | Male | 84 | 19 |
| Female | 18 | 5 | |
| Age | < 50 | 35 | 3 |
| ≥ 50 | 67 | 21 | |
| Multinodular | Yes | 5 | 2 |
| No | 97 | 22 | |
| Tumor size (cm) | < 5 | 61 | 15 |
| ≥ 5 | 41 | 9 | |
| Cirrhosis | Yes | 62 | 14 |
| No | 40 | 10 | |
| Metastasis | Yes | 13 | 9 |
| No | 89 | 15 | |
| ALT (IU/L) | Normal⁎ | 75 | 12 |
| Abnormal⁎ | 37 | 12 | |
| AFP (μg/L) | Normal⁎⁎ | 48 | 13 |
| Abnormal⁎⁎ | 56 | 11 | |
| Child-Pugh Class | A | 97 | 21 |
| B | 5 | 3 | |
| BCLC stage | 0 | 15 | 0 |
| A | 73 | 13 | |
| B | 1 | 2 | |
| C | 13 | 9 | |
| HBV | Yes | 85 | 19 |
| No | 17 | 5 | |
| HCV | Yes | 0 | 0 |
| No | 102 | 24 | |
| Schistosomiasis | Yes | 3 | 1 |
| No | 99 | 23 | |
| Fatty liver | Yes | 5 | 3 |
| No | 97 | 21 | |
All the information were collected before the surgery.
Normal: ALT ≤ 50 IU/L; Abnormal: ALT > 50 IU/L.
Normal: AFP ≤ 25 μg/L; Abnormal: AFP > 25 μg/L.
Together, results from Table 1, Fig. 3 and Supplementary Fig. 2 suggest that miR-367-3p may be able to suppress HCC cell invasion via increasing AR expression that results in modulating the AR-FKBP5-PHLPP-(pAKT and pERK) signaling pathway.
3.4. Mechanism Dissection How miR-367-3p Increases AR Expression
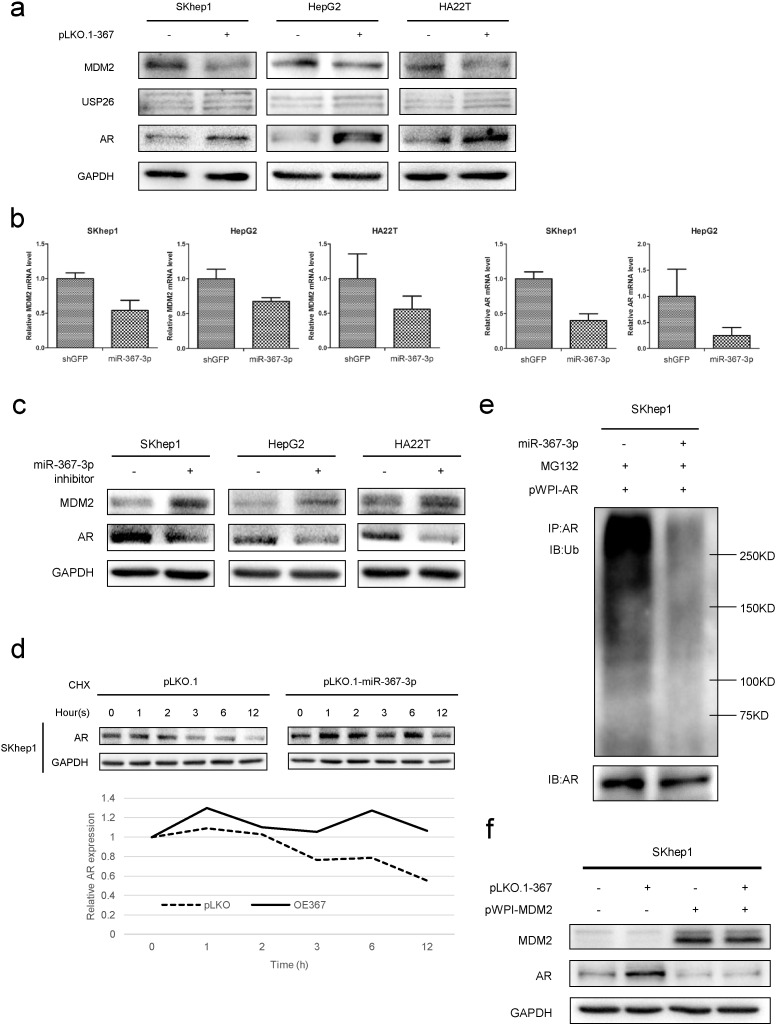
To further dissect the molecular mechanism(s) how miR-367-3p increases AR expression, according to the screening strategy and bioinformatics analysis, we focused on the 2 miR-367-3p predicted targets, MDM2 and USP26 (Supplementary Fig. 1a), that were reported to modulate AR protein expression. We found MDM2 was consistently decreased in these 3 HCC cell lines (SKhep1, HepG2 and HA22T), whereas USP26 showed a slight increase after over-expression of miR-367-3p (Fig. 4a). Moreover, the transcript levels of MDM2 were consistently decreased after 7 days expression of exogenous miR-367-3p (Fig. 4b left panels). Interestingly, we also found AR mRNA levels decreased after over-expression of miR-367-3p (Fig. 4b right panels), thus were incompatible with increased AR protein levels while supporting a post-transcriptional/translational upregulation of AR in response to miR-367-3p. This mechanism is entirely consistent with miR-367-3p-mediated suppression of MDM2, which degrades AR in a proteasome-dependent manner. With regard to the decrease of AR mRNA levels upon over-expressing miR-367-3p, we speculated that miR-367-3p might have several other targets, which resulted in alteration of AR mRNA transcription. In addition, the post-translational regulation of AR by MDM2 resulted in consistent increases in AR protein levels. Indeed, in the 3 HCC cell lines, we found the miR-367-3p inhibitor could significantly increase MDM2 and decrease AR at a protein level (Fig. 4c). Furthermore, we found the expression of miR-367-3p could increase AR protein stability when treated with cyclohexane (CHX) (Fig. 4d) and decrease AR ubiquitination when treated with MG132, a specific potent, reversible, and cell-permeable proteasome inhibitor that reduces the degradation of ubiquitin-conjugated proteins in mammalian cells (Fig. 4e). Importantly, we applied the interruption approach via over-expressing MDM2, and results revealed that increased MDM2 could significantly decrease AR and could partially abolish the miR-367-3p-increased AR protein expression (Fig. 4f), indicating that miR-367-3p could increase AR protein levels via altering the MDM2-mediated AR ubiquitination and degradation.
Fig. 4.
The miR-367-3p mediates AR protein stability and ubiquitination via targeting MDM2.
(a), we used western blot assays to test MDM2, USP26 and AR protein levels upon over-expressing miR-367-3p and found MDM2 was consistently decreased in these 3 HCC cell lines (SKhep1, HepG2 and HA22T), whereas USP26 increased slightly after over-expression of miR-367-3p. (b), left panels, transcript levels of MDM2 were consistently decreased after 7 days exogenous expression of miR-367-3p. Right panels, AR mRNA levels decreased after over-expression of miR-367-3p. MDM2 and AR mRNA levels were determined by qRT-PCR. (c), miR-367-3p (50 nM) inhibitor could significantly increase MDM2 and decrease AR at protein levels as shown by western blot assays. (d), the expression of miR-367-3p could increase AR protein stability as determined by western blot assays after cycloheximide (CHX) treatment for 0, 1, 2, 3, 6, and 12 h. (e), in AR over-expressed SKhep1 cells, the expression of miR-367-3p could decrease AR ubiquitination as determined by immunoprecipitation (AR) and western blot (ubiquitin) after 3 h 20 μM MG132 treatment. (f), interruption approach via over-expressing MDM2 revealed that increased MDM2 could significantly decrease AR expression and could partially abolish miR-367-3p-increased AR protein expression.
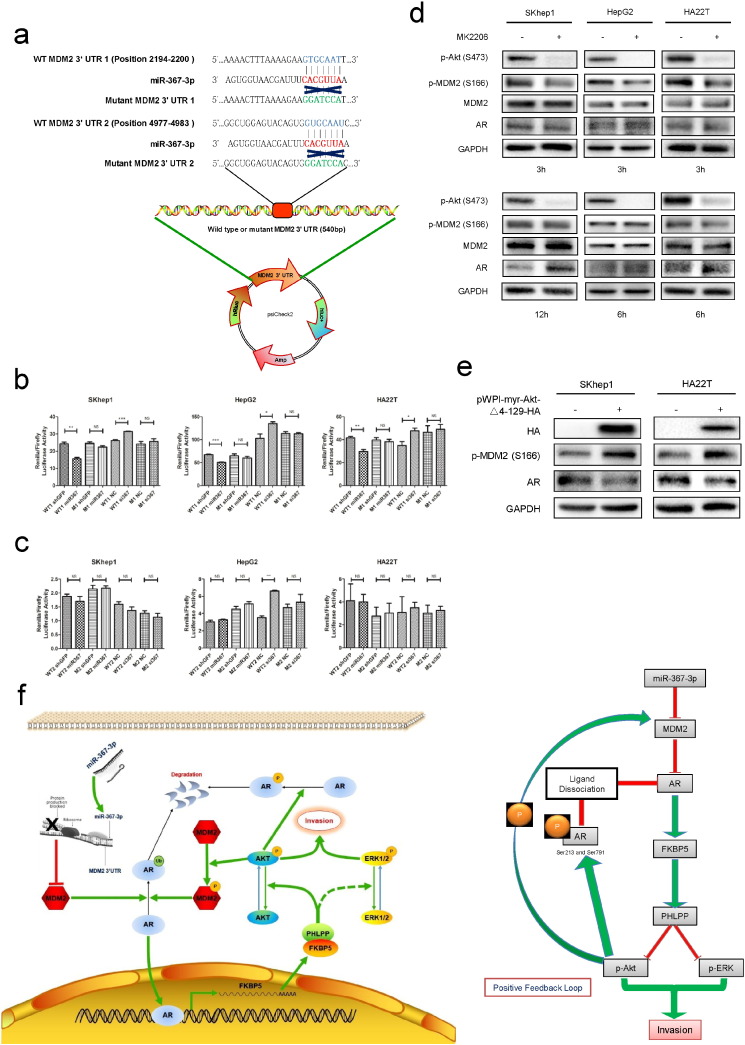
To further confirm that the MDM2 gene is a direct target of miR-367-3p to mediate its downstream events, we constructed two luciferase reporter constructs that bear the miR-367-3p target site in the 3′UTR of MDM2 gene as well as the attendant construct carrying the mutation in the target sequence (Fig. 5a). As expected, the luciferase assay results indicated that miR-367-3p could decrease MDM2 3′UTR #1 constructs containing the wild type (WT) but not the mutated (M) site (Fig. 5b), suggesting miR-367-3p might directly target the MDM2 3′UTR to induce its mRNA degradation (Djuranovic et al., 2012). On the other hand, miR-367-3p could not regulate MDM2 3′UTR #2 construct either with WT or M target sequences, suggesting that the surrounding sequence context might contribute to the efficacy of microRNA (Fig. 5c).
Fig. 5.
The miR-367-3p directly targets the 3′UTR of MDM2 to mediate AR levels and subsequently generates a positive feedback loop.
(a), MDM2 3′UTR containing wild type (WT) or mutant (M) miRNA-response elements were cloned into the psiCheck2 construct downstream of the Renilla luciferase ORF, (b), co-transfection of MDM2 3′UTR constructs containing WT seed regions with pLKO.1-miR-367-3p decreased the luciferase activity and with miR-367-3p inhibitor increased the luciferase activity, whereas the reporter with a M seed region in MDM2 3′UTR did not, in the first predicted miR-367-3p seed regions. (c), co-transfection of MDM2 3′UTR constructs containing WT seed regions with pLKO.1-miR-367-3p or with miR-367-3p inhibitor (50 nM) could not alter the luciferase activity, neither did the reporter with a M seed region in MDM2 3′UTR, in the second predicted miR-367-3p seed regions. (d), western blot assays showed that MK2206 (1 μM in SKhep1 and HA22T cells, 2.5 μM in HepG2 cells) suppressed MDM2 phosphorylation and increased the AR protein expression at various time points. (e), over-expressing pWPI-myr-Akt-Δ4-129-HA in HCC cells showed significant increases of pMDM2 (S166) and decreases of AR. (f), the schematic depiction showing the suppression of pAkt by miR-367-3p could subsequently attenuate the phosphorylation of AR and MDM2, giving rise to additionally increased AR protein levels, which formed a positive feed-back circuitry. p < 0.05 was considered statistically significant. * p < 0.05, ** p < 0.01, and *** p < 0.001.
Early studies suggested that MDM2-mediated and phosphorylation-dependent AR ubiquitination plays key roles for AR to be degraded by the proteasome (Lin et al., 2001, Lin et al., 2002). Thus, we applied the MK2206, a highly selective inhibitor of AKT1/2/3, which inhibits the phosphorylation of AKT (Ser473) that could then influence MDM2 protein expression via suppressing MDM2 phosphorylation (S166) (Lin et al., 2002), and examined its effects on AR protein expression in response to MDM2 expression. As expected, MK2206 suppressed MDM2 phosphorylation and increased the AR protein expression in 3 HCC cell lines (Fig. 5d), suggesting a repression of AKT activity could result in increased AR protein expression, mimicking the decreased phosphorylation of AKT by a higher expression of miR-367-3p. Moreover, we constructed pWPI-myr-Akt-Δ4-129-HA to over-express active AKT in HCC cells and found significant increases of pMDM2 (S166) and decreases of AR (Fig. 5e). Hence, the suppression of pAKT by miR-367-3p could subsequently attenuate the phosphorylation of AR and MDM2, giving rise to additional enhancement of AR protein levels, effectively forming a positive feedback loop (Fig. 5f).
As an early study suggested, it was possible that miR-367-3p might function through p53 because of the well-known connection between MDM2 and p53. In this study, we also checked the p53 levels upon miR-367-3p over-expression and results showed that the p53 decreased in SKhep1 and had little change in HA22T cells (Supplementary Fig. 3a), which indicated that p53 might unlikely to play a role in the case of invasion in terms of HCC.
Together, results from Fig. 4, Fig. 5 and Supplementary Fig. 3a suggest that miR-367-3p could directly target the 3′UTR of MDM2 to decrease MDM2 expression that results in increased AR expression and subsequently generates a positive feedback loop resulting in further augmentation of AR expression.
3.5. The miR-367-3p Increases Sorafenib Efficacy to Better Suppress HCC Metastasis Both In Vitro and In Vivo
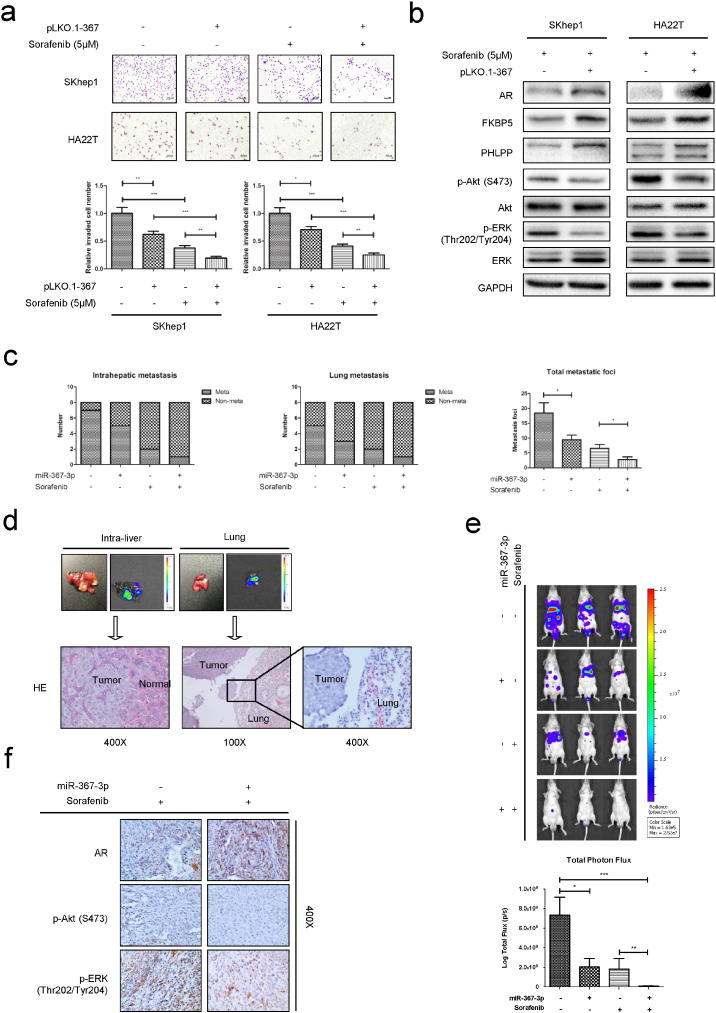
While the current HCC therapy with Sorafenib showed survival benefits in Phase III clinical studies (Llovet et al., 2008, Cheng et al., 2009), many HCC patients still failed to respond, or suffered from side effects or developed resistance after being treated for several months (Villanueva and Llovet, 2011, Berk et al., 2013). Interestingly, a recent study indicated that a moderate dose of Sorafenib (5 μM) could be sensitized with exogenous expression of AR that resulted in better efficacy to suppress HCC progression at late stages (Ma et al., 2012). However, using potential molecules to increase AR expression to enhance the Sorafenib chemotherapy efficacy remains elusive. We hypothesized that introducing miR-367-3p to increase AR expression might be able to sensitize HCC to Sorafenib chemotherapy. We first demonstrated that over-expression of miR-367-3p enhanced Sorafenib efficacy to suppress SKhep1 and HA22T cell invasion compared to Sorafenib or miR-367-3p alone (Fig. 6a), indicating that the combination of Sorafenib and miR-367-3p might provide a new robust approach to better treat advanced-stage HCC patients. Interestingly, we noticed that under moderate Sorafenib chemotherapy, miR-367-3p could significantly increase AR, FKBP5 and PHLPP expressions, therefore inhibit p-Akt (S473) and p-ERK (Thr202/Tyr204) in both cell lines, suggesting a potential positive feed-back mechanism to further increase the combined (Sorafenib + miR-367-3p) therapy efficacy (Fig. 6b).
Fig. 6.
Sorafenib and miR-367-3p serve as a better combination therapy to suppress HCC metastasis in vitro and in vivo.
(a), chamber-transwell invasion assays showed the combination of Sorafenib and miR-367-3p exhibited better efficacy to suppress SKhep1 and HA22T cells invasion compared to Sorafenib or miR-367-3p alone. The invaded cells were counted in 10 randomly chosen microscopic fields (100 ×) of each experiment and pooled for quantification. (b), western blot assays showed that under moderate Sorafenib chemotherapy, miR-367-3p could significantly increase AR, FKBP5 and PHLPP expressions, therefore inhibit p-Akt (S473) and p-ERK (Thr202/Tyr204) in SKhep1 and HA22T cells. (c), left and middle panel, IVIS imaging in mice was used to determine the intrahepatic metastasis and lung metastasis, and lower intra-hepatic and lung metastasis occurrence in miR-367-3p-expressing group than control group with or without Sorafenib treatment were found. Right panel, total metastatic foci were counted after sacrifice, and less total metastatic foci in miR-367-3p-expressing group than control group with or without Sorafenib treatment were found. (d), representative bioluminescent images of intrahepatic metastasis and lung metastasis (upper panel). HE staining confirmed the tumor tissue in liver (left lower panel) and lung (right and middle lower panels). (e), representative bioluminescent images in different groups after 2 months of orthotopic HCC xenograft (upper panel), and the total photon flux were measured and analyzed using Living Image® software (PerkinElmer, lower panel), showing less total tumor burden in miR-367-3p-expressing group than control group with or without Sorafenib treatment. (f), representative images of IHC staining for AR, p-Akt (S473) and p-ERK (Thr202/Tyr204) in different treatment groups. p < 0.05 was considered statistically significant. * p < 0.05, ** p < 0.01, and *** p < 0.001.
To verify the efficacy in vivo of the combined therapy characterized in HCC cell lines in vitro, we applied orthotopic HCC xenografts in a mouse model. We found lower intra-hepatic and lung metastasis occurrence in miR-367-3p-expressing group than control group with or without Sorafenib treatment (Fig. 6c left and middle panel, Fig. 6d and Supplementary Fig. 4a-b). Significantly, we also demonstrated less total metastatic foci and total tumor burden (determined by total photon flux) in miR-367-3p-expressing group than control group with or without Sorafenib treatment (Fig. 6c right panel, Fig. 6e). Consistent with the findings in vitro, through IHC staining of AR, pAKT and pERK in the tumors formed in these mice, we found a similar signaling transduction regulated by miR-367-3p expression (Fig. 6f).
4. Discussion
HCC represents the most common histological subtype among primary liver cancers and the leading cause of death among patients with cirrhosis, accounting for 70% to 85% of the total liver cancer burden worldwide (Perz et al., 2006, Alazawi et al., 2010). Increasing incidence of HCC around the world, especially in the United States and Central Europe, allows a more profound understanding of the cancer progression and propelled a rapid development of life-prolonging therapies. With advances in liver surgery and transplantation in the past few decades, curative treatment can now be offered to HCC patients with liver resection and regeneration if diagnosed early enough or those who meet the transplant criteria, yielding over 50% 5-year survival rate through the two surgical treatments (Asham et al., 2013).
However, for late-stage HCC patients with poor liver functions who cannot tolerate liver resection or transplantation, several local-regional therapies such as tumor ablation, TACE and radiation therapies are available. Moreover, systemic chemotherapy and molecularly targeted therapies also yield promising results. Among these, Sorafenib, the first effective drug approved by the FDA, represented a milestone in the therapy for HCC, especially for the patients at late stages. Nonetheless, irresponsiveness and resistance along with undesired side effects of Sorafenib suggest the need for more and better efficacious drugs with fewer side effects.
In the past years, efforts have been made to dissect mechanisms by which targeting AR could inhibit HBV- and carcinogen-induced early-stage HCC development (Wu et al., 2010). Nevertheless, clinical trials using various anti-androgens in the attempt to treat HCC have inconsistent conclusions (Groupe d'etude et de traitement du carcinome, H., 2004, Matsuura et al., 1994, Chao et al., 1996). Several hypotheses have been proposed to elucidate this riddle, such as the fact that most of the anti-androgen therapies were developed to reduce/antagonize androgens from binding to AR, but not to target AR itself. Recently, the dual roles of AR in HCC were demonstrated showing AR might function as a stimulator to promote HCC initiation and development at early stages, yet might function as a metastasis suppressor in the advanced stages of HCC, implying that targeting AR should be stage-dependent (Ma et al., 2012). Importantly, results from in vivo mouse model studies also indicated that increased AR expression with a moderate dose (5 μM) of Sorafenib might result in higher efficacy of HCC suppression at late stages. Furthermore, supported by data from cell line studies in vitro, mouse model studies in vivo, bioinformatic analyses and human clinical evidences, the present study provides clearer evidences that AR indeed acts as a metastasis suppressor in late-stage HCC. It remains to be seen whether activation of AR through hormone stimulation is equal to lentiviral induction of AR expression, thus a clinical application of these findings could be beneficial for patients with late stages of liver cancer.
Alternatively, identifying potential molecules to increase AR expression in order to enhance Sorafenib chemotherapy efficacy could be one choice and remains to be explored. Growing evidences indicate that the deregulation of miRNAs plays a vital role in HCC onset as well as progression. The diagnostic, prognostic, and even therapeutic values of HCC related miRNAs have also been widely discussed. At advanced stages, several miRNAs associated with HCC venous metastasis were reported to be up-regulated or down-regulated (Budhu et al., 2008). Several studies further developed miRNA-based classification of HCC subclasses (Toffanin et al., 2011), miRNA-based risk assessment of HCC recurrence after liver resection (Sato et al., 2011), and miRNA-based prediction and alteration of sensitivity to chemotherapeutics and molecularly targeted therapies (Bai et al., 2009, Zhou et al., 2011). Furthermore, accumulating studies focusing on miRNA therapies in HCC indicated that, either introducing the oncosuppressor miRNAs or inhibiting oncomiRs resulted in impaired growth or invasion ability in vitro by HCC cell lines, in vivo in animal xenografts models, and even in clinical trials (Kota et al., 2009, Lanford et al., 2010, Callegari et al., 2012). Miravirsen (SPC3649), the first anti-miR drug candidate, is currently in clinical testing for treatment of HCV infections. Targeting miR-122, a liver-specific miRNA with an important role in the life cycle of hepatitis C virus (HCV), Miravirsen monotherapy provides long-lasting suppression of viremia, has a high barrier to viral resistance, and is well tolerated in patients with chronic HCV infection (Lindow and Kauppinen, 2012). This break-through encouraged us to seek for new miRNA targets as novel therapeutics, especially combining with Sorafenib, to better control HCC progression.
In this study, we identified miR-367-3p, whose expression is positively correlated with AR expression and expressed at lower levels in advanced human HCC, as an HCC metastasis suppressor. These findings are supported by solid clinical data and had established a foundation for a potential use of miRNA for late-stage HCC. Importantly, we identified that miR-367-3p could directly target the 3′UTR #1 of MDM2 to decrease MDM2 protein levels. Interestingly, an early study showed that MDM2 was a direct target of miR-25 and miR-32 (from the same microRNA family as miR-367-3p) in glioblastoma multiforms (Suh et al., 2012), with exactly the same sites predicted in that study as those in the present study. However, we only identified the 1st predicted site as the microRNA response element (MRE) mediating the silencing effect, but not the 2nd site, with 3 different HCC cell lines being tested. Different disease models, different sources of tissue, different ways of cloning and different specific microRNAs are all possible reasons for the varied results. Nevertheless, our data demonstrated that miR-367-3p could directly target the 3′UTR of MDM2 to decrease MDM2 protein levels, which in turn increases AR protein expression. MDM2 is a RING finger-containing E3 ligase, which serves as the rate-limiting step of protein ubiquitin modification. An early study indicated that the E3 ligase activity of MDM2 and phosphorylation of MDM2 by AKT were essential for MDM2 to affect AR ubiquitination and degradation (Lin et al., 2002). Indeed, in the present study, the MDM2 mediated ubiquitination and degradation of AR were also observed in HCC.
The consequences of such increased AR protein levels may then function through altering the downstream FKBP5/PHLPP signals, thus dephosphorylating and deactivating AKT and ERK to inhibit the HCC cell invasion. Although it is possible that miR-367-3p may suppress HCC invasion through MDM2/p53 signals, our results indicate that p53 had little change or even decreased in HCC cells upon over-expressing miR-367-3p. For MDM2 to degrade p53, there may be other factors involved or required, but those factors are not there in this case. On the other hand, miR-367-3p may have other effects beside MDM2 that ultimately may influence p53 levels. In brief, p53 is unlikely to play a role in miR-367-3p regulated HCC invasion.
Although it was clear in the present study that miR-367-3p inhibits HCC invasion, potential targets of miR-367-3p other than AR might also play roles in regulating cell invasion and metastasis. For example, miR-367-3p was recently reported to target Rab23, a member of the RAS oncogene family, to inhibit the invasion and metastasis of gastric cancer (Bin et al., 2015). Moreover, this miRNA can also potentially target oncoproteins like MAP2K4, SOX4 and FGF2, among many other proteins, to negatively regulate HCC metastasis (Dweep and Gretz, 2015). Nevertheless, our studies provided compelling evidence that MDM2/AR signaling played a clear role in mediating HCC invasion. This is consistent with the central role of AR in regulating HCC initiation and development as HCC is one of several human malignancies exhibiting a clear gender disparity (Clocchiatti et al., 2016).
We noted that the interaction and cross-talk among AKT, MDM2 and AR were investigated in different contexts in previous studies (Lin et al., 2002, Lee et al., 2015, Mcclurg et al., 2014), and here we provided the evidence of a positive feedback loop linking all three components in HCC cells (Fig. 5F). Furthermore, such a positive feedback loop also likely contributes to an extensive and consistent elevation of AR in the context of introducing miR-367-3p. Importantly, the combination of miR-367-3p (to increase AR) and Sorafenib could better suppress HCC cell invasion. Successful clinical application of these findings in the future may help us to better retard HCC metastasis at late stages.
In conclusion, miR-367-3p might regulate the MDM2/AR-FKBP5/PHLPP/(pAKT and pERK) signaling pathway, and miR-367-3p might serve as a metastasis biomarker and possible novel candidate for therapeutic intervention for late-stage HCC, especially combined with a moderate dose of Sorafenib.
Funding Sources
This work was supported by NIH grants (CA155477 and CA156700), George Whipple Professorship Endowment and Taiwan Department of Health Clinical Trial, Research Center of Excellence (DOH99-TD-B-111-004 to China Medical University, Taichung, Taiwan), International scientific and technological cooperation projects (2012DFA30410), Natural Science Foundation of Zhejiang Province (LZ14H160002), National Natural Science Foundation of China (81201942), The National Science-technology Support Plan Projects (2012BAI14B06). No one was paid to write this article by a pharmaceutical company or other agency.
Conflict of Interest Statement
None.
Authors' contributions
Cai X and Chang C are the principal investigators for the study. Xu J, Lin H and Li G conceived of the present study and carried out the major part of this project. Xu J also collected the data and wrote the manuscript. Sun Y, Chen J and Shi L participated in this project and collected and analyzed the data. Cai X and Chang C contributed to the interpretation of the data and to critical review of the manuscript. Lin H, Cai X and Chang C provided the funding for this study.
Acknowledgements
We acknowledge Karen Wolf for checking the text thoroughly for accuracy and grammar.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.07.013.
Contributor Information
Xiujun Cai, Email: cxjzu@hotmail.com.
Chawnshang Chang, Email: chang@urmc.rochester.edu.
Appendix A. Supplementary Data
Supplementary Fig. 1 (a), a specific screening strategy to determine AR expression enhancers. According to recent literature, we picked 7 E3 ligases that could target AR and enhance AR degradation. Then we predicted the microRNAs which could target these E3 ligases by well-known online prediction databases (TargetScan, miRDB and MicroCosm Targets). Those targeting > 3 out of 7 (left) or no < 2 out of 5 (right) were the candidates (we put more weight on USP26 because of the less predicted candidate number). Finally, we focused on 8 miRNAs that could possibly increase AR expression according to the prediction, and over-expressed these miRNAs in SKhep1 and HA22T cell lines by lentivirus transduction. (b), miR-302a/b/c/d were expressed lower in metastatic patients than non-metastatic patients according to the non-coding RNA microarray data submitted on public Gene Expression Omnibus Datasets, which contained the largest sample size related to HCC metastasis. (c), chamber-transwell invasion assays were performed using SKhep1, HA22T, HepG2, Huh7, SNU398, SNU423 cells. (d), the invaded cells were counted in 10 randomly chosen microscopic fields (100 ×) of each experiment and pooled for quantification. (e), according to invasion ability, these 6 HCC cell lines were classified into 2 groups (group I: high-invasive; group II: low-invasive). The expression of miR-367-3p was significantly lower in Group I compared to Group II. * p < 0.05 and ** p < 0.01.
Supplementary Fig. 2. (a), PTEN, Cox-2, NF-κB, and p27 alteration after over-expressing miR-367-3p in SKhep1 and HA22T cells were determined by western blot assays, but were not consistent between 2 cell lines or changed little. (b), TGFβ1 alteration after AR knocked down in SKhep1 and HA22T cells as determined by western blot assays showing that TGFβ1 changed inconsistently in SKhep1 and HA22T cells. (c), representative images of IHC staining grades (∓/++/+++) of pAKT, pERK and AR in human HCC samples.
Supplementary Fig. 3. (a), p53 alteration after over-expressing miR-367-3p in SKhep1 and HA22T cells as determined by western blot assays showing that the p53 decreased in SKhep1 and had little change in HA22T cells
Supplementary Fig. 4. (a), representative bioluminescent images of metastatic foci in kidney, diaphragm, stomach and spleen. (b), representative macroscopic images of metastatic foci in diaphragm, intestine, spleen, kidney, intra-liver, mesentery, abdominal wall and stomach. Green arrows indicate metastatic foci.
Supplementary Fig. 5. Quantification of the western blots from Figs. 3c, d, f, 4a, c, f, 5d, e, 6b.
Supplementary material.
References
- Alazawi W., Cunningham M., Dearden J., Foster G.R. Systematic review: outcome of compensated cirrhosis due to chronic hepatitis C infection. Aliment. Pharmacol. Ther. 2010;32:344–355. doi: 10.1111/j.1365-2036.2010.04370.x. [DOI] [PubMed] [Google Scholar]
- An J., Wang C., Deng Y., Yu L., Huang H. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 2014;6:657–669. doi: 10.1016/j.celrep.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asham E.H., Kaseb A., Ghobrial R.M. Management of hepatocellular carcinoma. Surg. Clin. North Am. 2013;93:1423–1450. doi: 10.1016/j.suc.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Bai S., Nasser M.W., Wang B., Hsu S.H., Datta J., Kutay H., Yadav A., Nuovo G., Kumar P., Ghoshal K. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J. Biol. Chem. 2009;284:32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk V., Kaplan M.A., Tonyali O., Buyukberber S., Balakan O., Ozkan M., Demirci U., Ozturk T., Bilici A., Tastekin D., Ozdemir N., Unal O.U., Oflazoglu U., Turkmen E., Erdogan B., Uyeturk U., Oksuzoglu B., Cinkir H.Y., Yasar N., Gumus M. Efficiency and side effects of sorafenib therapy for advanced hepatocellular carcinoma: a retrospective study by the anatolian society of medical oncology. Asian Pac. J. Cancer Prev. 2013;14:7367–7369. doi: 10.7314/apjcp.2013.14.12.7367. [DOI] [PubMed] [Google Scholar]
- Bin Z., Dedong H., Xiangjie F., Hongwei X., Qinghui Y. The microRNA-367 inhibits the invasion and metastasis of gastric cancer by directly repressing Rab23. Genet Test Mol Biomarkers. 2015;19:69–74. doi: 10.1089/gtmb.2014.0210. [DOI] [PubMed] [Google Scholar]
- Brognard J., Newton A.C. PHLiPPing the switch on Akt and protein kinase C signaling. Trends Endocrinol. Metab. 2008;19:223–230. doi: 10.1016/j.tem.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brognard J., Sierecki E., Gao T., Newton A.C. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol. Cell. 2007;25:917–931. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Budhu A., Jia H.L., Forgues M., Liu C.G., Goldstein D., Lam A., Zanetti K.A., Ye Q.H., Qin L.X., Croce C.M., Tang Z.Y., Wang X.W. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- Callegari E., Elamin B.K., Giannone F., Milazzo M., Altavilla G., Fornari F., Giacomelli L., D'abundo L., Ferracin M., Bassi C., Zagatti B., Corra F., Miotto E., Lupini L., Bolondi L., Gramantieri L., Croce C.M., Sabbioni S., Negrini M. Liver tumorigenicity promoted by microRNA-221 in a mouse transgenic model. Hepatology. 2012;56:1025–1033. doi: 10.1002/hep.25747. [DOI] [PubMed] [Google Scholar]
- Chao Y., Chan W.K., Huang Y.S., Teng H.C., Wang S.S., Lui W.Y., Whang-Peng J., Lee S.D. Phase II study of flutamide in the treatment of hepatocellular carcinoma. Cancer. 1996;77:635–639. [PubMed] [Google Scholar]
- Cheng A.L., Kang Y.K., Chen Z., Tsao C.J., Qin S., Kim J.S., Luo R., Feng J., Ye S., Yang T.S., Xu J., Sun Y., Liang H., Liu J., Wang J., Tak W.Y., Pan H., Burock K., Zou J., Voliotis D., Guan Z. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- Chiang D.Y., Villanueva A., Hoshida Y., Peix J., Newell P., Minguez B., Leblanc A.C., Donovan D.J., Thung S.N., Sole M., Tovar V., Alsinet C., Ramos A.H., Barretina J., Roayaie S., Schwartz M., Waxman S., Bruix J., Mazzaferro V., Ligon A.H., Najfeld V., Friedman S.L., Sellers W.R., Meyerson M., Llovet J.M. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–6788. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clocchiatti A., Cora E., Zhang Y., Dotto G.P. Sexual dimorphism in cancer. Nat. Rev. Cancer. 2016;16:330–339. doi: 10.1038/nrc.2016.30. [DOI] [PubMed] [Google Scholar]
- Dirac A.M., Bernards R. The deubiquitinating enzyme USP26 is a regulator of androgen receptor signaling. Mol. Cancer Res. 2010;8:844–854. doi: 10.1158/1541-7786.MCR-09-0424. [DOI] [PubMed] [Google Scholar]
- Djuranovic S., Nahvi A., Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweep H., Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat. Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- Groupe d'etude et de traitement du carcinome, H. Randomized trial of leuprorelin and flutamide in male patients with hepatocellular carcinoma treated with tamoxifen. Hepatology. 2004;40:1361–1369. doi: 10.1002/hep.20474. [DOI] [PubMed] [Google Scholar]
- Janssen H.L., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-torres M., Patel K., Van der meer A.J., Patick A.K., Chen A., Zhou Y., Persson R., King B.D., Kauppinen S., Levin A.A., Hodges M.R. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Kota J., Chivukula R.R., O'donnell K.A., Wentzel E.A., Montgomery C.L., Hwang H.W., Chang T.C., Vivekanandan P., Torbenson M., Clark K.R., Mendell J.R., Mendell J.T. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford R.E., Hildebrandt-Eriksen E.S., Petri A., Persson R., Lindow M., Munk M.E., Kauppinen S., Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee S.H., Johnson D., Luong R., Sun Z. Crosstalking between androgen and PI3K/AKT signaling pathways in prostate cancer cells. J. Biol. Chem. 2015;290:2759–2768. doi: 10.1074/jbc.M114.607846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Xu L.L., Masuda K., Raymundo E., Mcleod D.G., Dobi A., Srivastava S. A feedback loop between the androgen receptor and a NEDD4-binding protein, PMEPA1, in prostate cancer cells. J. Biol. Chem. 2008;283:28988–28995. doi: 10.1074/jbc.M710528200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Stevens P.D., Liu J., Yang H., Wang W., Wang C., Zeng Z., Schmidt M.D., Yang M., Lee E.Y., Gao T. PHLPP is a negative regulator of RAF1, which reduces colorectal cancer cell motility and prevents tumor progression in mice. Gastroenterology. 2014;146:1301. doi: 10.1053/j.gastro.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y.L., Sun Y.M., Chau G.Y., Chau Y.P., Lai T.C., Wang J.L., Horng J.T., Hsiao M., Tsou A.P. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27:5578–5589. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- Lin H.K., Wang L., Hu Y.C., Altuwaijri S., Chang C. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 2002;21:4037–4048. doi: 10.1093/emboj/cdf406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.K., Yeh S., Kang H.Y., Chang C. Akt suppresses androgen-induced apoptosis by phosphorylating and inhibiting androgen receptor. Proc. Natl. Acad. Sci. U. S. A. 2001;98:7200–7205. doi: 10.1073/pnas.121173298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow M., Kauppinen S. Discovering the first microRNA-targeted drug. J. Cell Biol. 2012;199:407–412. doi: 10.1083/jcb.201208082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet J.M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J.F., De Oliveira A.C., Santoro A., Raoul J.L., Forner A., Schwartz M., Porta C., Zeuzem S., Bolondi L., Greten T.F., Galle P.R., Seitz J.F., Borbath I., Haussinger D., Giannaris T., Shan M., Moscovici M., Voliotis D., Bruix J. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- Ma W.L., Hsu C.L., Yeh C.C., Wu M.H., Huang C.K., Jeng L.B., Hung Y.C., Lin T.Y., Yeh S., Chang C. Hepatic androgen receptor suppresses hepatocellular carcinoma metastasis through modulation of cell migration and anoikis. Hepatology. 2012;56:176–185. doi: 10.1002/hep.25644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W.L., Lai H.C., Yeh S., Cai X., Chang C. Androgen receptor roles in hepatocellular carcinoma, fatty liver, cirrhosis and hepatitis. Endocr Relat Cancer. 2014;21:R165–R182. doi: 10.1530/ERC-13-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluccio M., Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J. Clin. 2012;62:394–399. doi: 10.3322/caac.21161. [DOI] [PubMed] [Google Scholar]
- Matsuura B., Taniguchi Y., Ohta Y. Effect of antiandrogen treatment on chemical hepatocarcinogenesis in rats. J. Hepatol. 1994;21:187–193. doi: 10.1016/s0168-8278(05)80393-9. [DOI] [PubMed] [Google Scholar]
- Mcclurg U.L., Summerscales E.E., Harle V.J., Gaughan L., Robson C.N. Deubiquitinating enzyme Usp12 regulates the interaction between the androgen receptor and the Akt pathway. Oncotarget. 2014;5:7081–7092. doi: 10.18632/oncotarget.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minguez B., Hoshida Y., Villanueva A., Toffanin S., Cabellos L., Thung S., Mandeli J., Sia D., April C., Fan J.B., Lachenmayer A., Savic R., Roayaie S., Mazzaferro V., Bruix J., Schwartz M., Friedman S.L., Llovet J.M. Gene-expression signature of vascular invasion in hepatocellular carcinoma. J. Hepatol. 2011;55:1325–1331. doi: 10.1016/j.jhep.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida T., Yasuda H. PIAS1 and PIASxalpha function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J. Biol. Chem. 2002;277:41311–41317. doi: 10.1074/jbc.M206741200. [DOI] [PubMed] [Google Scholar]
- Niu Y., Altuwaijri S., Lai K.P., Wu C.T., Ricke W.A., Messing E.M., Yao J., Yeh S., Chang C. Androgen receptor is a tumor suppressor and proliferator in prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12182–12187. doi: 10.1073/pnas.0804700105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H., Li L., Fridley B.L., Jenkins G.D., Kalari K.R., Lingle W., Petersen G., Lou Z., Wang L. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16:259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perz J.F., Armstrong G.L., Farrington L.A., Hutin Y.J., BELL B.P. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J. Hepatol. 2006;45:529–538. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Rees I., Lee S., Kim H., Tsai F.T. The E3 ubiquitin ligase CHIP binds the androgen receptor in a phosphorylation-dependent manner. Biochim. Biophys. Acta. 2006;1764:1073–1079. doi: 10.1016/j.bbapap.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Salvi A., Abeni E., Portolani N., Barlati S., De Petro G. Human hepatocellular carcinoma cell-specific miRNAs reveal the differential expression of miR-24 and miR-27a in cirrhotic/non-cirrhotic HCC. Int. J. Oncol. 2013;42:391–402. doi: 10.3892/ijo.2012.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato F., Hatano E., Kitamura K., Myomoto A., Fujiwara T., Takizawa S., Tsuchiya S., Tsujimoto G., Uemoto S., Shimizu K. MicroRNA profile predicts recurrence after resection in patients with hepatocellular carcinoma within the Milan Criteria. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewer A., Paul N., Landgraf P., Aravin A., Pfeffer S., Brownstein M.J., Tuschl T., Van Nimwegen E., Zavolan M. Identification of clustered microRNAs using an ab initio prediction method. BMC Biochem. 2005;6:267. doi: 10.1186/1471-2105-6-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R., Naishadham D., Jemal A. Cancer statistics, 2012. CA Cancer J. Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Suh S.S., Yoo J.Y., Nuovo G.J., Jeon Y.J., Kim S., Lee T.J., Kim T., Bakacs A., Alder H., Kaur B., Aqeilan R.I., Pichiorri F., Croce C.M. MicroRNAs/TP53 feedback circuitry in glioblastoma multiforme. Proc. Natl. Acad. Sci. U. S. A. 2012;109:5316–5321. doi: 10.1073/pnas.1202465109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffanin S., Hoshida Y., Lachenmayer A., Villanueva A., Cabellos L., Minguez B., Savic R., Ward S.C., Thung S., Chiang D.Y., Alsinet C., Tovar V., Roayaie S., Schwartz M., Bruix J., Waxman S., Friedman S.L., Golub T., Mazzaferro V., Llovet J.M. MicroRNA-based classification of hepatocellular carcinoma and oncogenic role of miR-517a. Gastroenterology. 2011;140(1618–28) doi: 10.1053/j.gastro.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva A., Llovet J.M. Targeted therapies for hepatocellular carcinoma. Gastroenterology. 2011;140:1410–1426. doi: 10.1053/j.gastro.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Heegaard N.H., Orum H. MicroRNAs in liver disease. Gastroenterology. 2012;142:1431–1443. doi: 10.1053/j.gastro.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.H., Ma W.L., Hsu C.L., Chen Y.L., Ou J.H., Ryan C.K., Hung Y.C., Yeh S., Chang C. Androgen receptor promotes hepatitis B virus-induced hepatocarcinogenesis through modulation of hepatitis B virus RNA transcription. Sci. Transl. Med. 2010;2 doi: 10.1126/scitranslmed.3001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmbach E., Chen Y.B., Khitrov G., Zhang W., Roayaie S., Schwartz M., Fiel I., Thung S., Mazzaferro V., Bruix J., Bottinger E., Friedman S., Waxman S., Llovet J.M. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- Yeh S.H., Chen P.J. Gender disparity of hepatocellular carcinoma: the roles of sex hormones. Oncology. 2010;78(Suppl. 1):172–179. doi: 10.1159/000315247. [DOI] [PubMed] [Google Scholar]
- Zhou C., Liu J., Li Y., Liu L., Zhang X., Ma C.Y., Hua S.C., Yang M., Yuan Q. microRNA-1274a, a modulator of sorafenib induced a disintegrin and metalloproteinase 9 (ADAM9) down-regulation in hepatocellular carcinoma. FEBS Lett. 2011;585:1828–1834. doi: 10.1016/j.febslet.2011.04.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 (a), a specific screening strategy to determine AR expression enhancers. According to recent literature, we picked 7 E3 ligases that could target AR and enhance AR degradation. Then we predicted the microRNAs which could target these E3 ligases by well-known online prediction databases (TargetScan, miRDB and MicroCosm Targets). Those targeting > 3 out of 7 (left) or no < 2 out of 5 (right) were the candidates (we put more weight on USP26 because of the less predicted candidate number). Finally, we focused on 8 miRNAs that could possibly increase AR expression according to the prediction, and over-expressed these miRNAs in SKhep1 and HA22T cell lines by lentivirus transduction. (b), miR-302a/b/c/d were expressed lower in metastatic patients than non-metastatic patients according to the non-coding RNA microarray data submitted on public Gene Expression Omnibus Datasets, which contained the largest sample size related to HCC metastasis. (c), chamber-transwell invasion assays were performed using SKhep1, HA22T, HepG2, Huh7, SNU398, SNU423 cells. (d), the invaded cells were counted in 10 randomly chosen microscopic fields (100 ×) of each experiment and pooled for quantification. (e), according to invasion ability, these 6 HCC cell lines were classified into 2 groups (group I: high-invasive; group II: low-invasive). The expression of miR-367-3p was significantly lower in Group I compared to Group II. * p < 0.05 and ** p < 0.01.
Supplementary Fig. 2. (a), PTEN, Cox-2, NF-κB, and p27 alteration after over-expressing miR-367-3p in SKhep1 and HA22T cells were determined by western blot assays, but were not consistent between 2 cell lines or changed little. (b), TGFβ1 alteration after AR knocked down in SKhep1 and HA22T cells as determined by western blot assays showing that TGFβ1 changed inconsistently in SKhep1 and HA22T cells. (c), representative images of IHC staining grades (∓/++/+++) of pAKT, pERK and AR in human HCC samples.
Supplementary Fig. 3. (a), p53 alteration after over-expressing miR-367-3p in SKhep1 and HA22T cells as determined by western blot assays showing that the p53 decreased in SKhep1 and had little change in HA22T cells
Supplementary Fig. 4. (a), representative bioluminescent images of metastatic foci in kidney, diaphragm, stomach and spleen. (b), representative macroscopic images of metastatic foci in diaphragm, intestine, spleen, kidney, intra-liver, mesentery, abdominal wall and stomach. Green arrows indicate metastatic foci.
Supplementary Fig. 5. Quantification of the western blots from Figs. 3c, d, f, 4a, c, f, 5d, e, 6b.
Supplementary material.