Abstract
MicroRNAs (miRNAs) are a class of endogenous and non-coding single-stranded RNAs of approximately 22 nucleotides, many of which are evolutionarily conserved. Genome-wide association studies have identified a robust statistical association between the MIR137 gene and schizophrenia in Europeans, which was replicated in the Han Chinese population in a case-control study. In the previous study, we provided evidence for a significant association between the EFNB2 gene and schizophrenia in Han Chinese subjects. In the current study, we utilized computational analysis, vector construction of point mutations, luciferase reporter assays and gene expression assays including RT-qPCR and western blotting methods to investigate miR-137 directly targeting EFNB2 gene and explore the reversal effect of a genetic variant of SNP rs550067317 in the putative seed-pair region of EFNB2 3′-UTR. We also found that miR-137 could be detected in the peripheral blood of a cohort of first-onset schizophrenia patients and healthy controls, and the area under curve was 0.795 (95% confidence interval 0.700–0.890), which is a middle diagnostic value for disease, suggesting that it might be valuable for diagnosing schizophrenia. In summary, this study would improve our understanding of the role of miR-137 in schizophrenia-associated signaling pathways and identify the genetic basis of rs550067317 for schizophrenia. Furthermore, we provided new evidence for the involvement of miR-137 in the etiology and diagnosis of schizophrenia, which might contribute to the discovery of new biomarkers and therapeutic targets for the disease.
Keywords: Schizophrenia, MiR-137, EFNB2, SNP, Regulation
Highlights
-
•
We demonstrated that miR-137 inhibiting EFNB2 gene at protein level by using classical methods of dual luciferase report assay, RT-qPCR, and western blotting.
-
•
MiR-137 inhibiting EFNB2 gene was affected by a genetic variant of SNP rs550067317 tested by the method of dual luciferase reporter assay through point mutation.
-
•
MiR-137 expressed aberrantly in peripheral blood of schizophrenia patients, and the area under curve was 0.795 which is a middle diagnostic value for disease.
In this study, we provided evidence that miR-137 targets EFNB2 gene directly and a genetic variant in 3’UTR of the EFNB2 affects the regulation. MiR-137 regulated EFNB2 gene expression at protein level. We further found that miR-137 expressed abnormally in peripheral blood of schizophrenia patients and the area under curve reaches a middle diagnostic value for disease.
1. Introduction
MicroRNA (miRNAs) are a class of endogenous and non-coding single-stranded RNAs of approximately 22 nucleotides (Bartel, 2004), many of which are evolutionarily conserved (Farh et al., 2005). Nucleotides 2–7, from the 5′ end of the miRNA, are referred to as the seed sequence and are critical for hybridization to targets (Kim et al., 2010). They bind to a complementary sequence of 6 to 8 base-pairs typically located in the 3′ untranslated region (3′-UTRs) of target mRNAs. This interaction either facilitates the degradation of the mRNA or delays its translation (Kosik, 2006).
The widespread expression and activity of miRNAs in the brain implicates them in numerous neurological disorders (Cao et al., 2006). They are emerging as key regulators of neurodevelopmental and neurological processes and dysregulation of miRNAs in developing or mature brain, could cause pervasive changes to the molecular networks linked to the pathophysiology of schizophrenia (Beveridge and Cairns, 2012). MiR-137 is enriched in the dentate gyrus, a subregion of the hippocampal formation that regulates adult neurogenesis and neuronal maturation (Silber et al., 2008, Smrt et al., 2010). In a mega genome-wide association study (GWAS) performed by the Psychiatric GWAS Consortium (PGC), the second strongest link to schizophrenia was the genetic variant rs1625579. This variant is located in an intron of a putative primary transcript of the MIR137 gene, which encodes miR-137 (Ripke et al., 2011). Subsequent multi-stage and large-scale GWA studies identified MIR137 transcript and within a subset of predicted miR-137 target genes strongly associated with schizophrenia (Ripke et al., 2013, Ripke et al., 2014).
Schizophrenia is a chronic, severe and disabling mental disorder with a lifetime risk of approximately 1%; it is characterized by hallucinations, delusions and cognitive deficits, with heritability estimated at up to 80% (Cardno and Gottesman, 2000, Sullivan et al., 2003). Previously, we showed that the gene EFNB2 is associated with schizophrenia in Han Chinese samples (Zhang et al., 2010). EFNB2 is located on 13q33, a region strongly linked to schizophrenia by independent genetic linkage studies (Lin et al., 1997). It encodes a member of the protein-tyrosine kinase family (ephrin), which is implicated in mediating NMDA receptor activated regulation of development and remodeling of synaptic connections (Takasu et al., 2002). EFNB2 is also an essential component of the Reelin pathway, which controls neuronal migration during nervous system development (Senturk et al., 2011). Using TargetScan and Miranda online software and the UCSC genome browser, we found that EFNB2 is a predicted target of miR-137 and that it contains a genetic variant in the binding site located in the 3′-UTR of EFNB2.
A combination of literature searches, ingenuity pathway analysis (IPA), and freely accessible bioinformatics resources identified > 1000 genes predicted to interact with miR-137 (Wright et al., 2015, Wright et al., 2013). However, few of these schizophrenia associated target genes have been validated by biological experimentation, and even fewer showed alterations at the protein level (Kim et al., 2012, Kwon et al., 2013, Xia et al., 2015). In addition, Sullivan et al. showed significant gene expression similarities between whole blood and multiple brain tissues after a through comparison of gene expression in blood and brain (Sullivan et al., 2006). Harris et al. found that schizophrenia could be studied using blood-based biomarkers because peripheral biomarkers may mirror pathological processes in the brain (Harris et al., 2012).
Therefore, we initiated a study using computational analysis, luciferase reporter assays and gene expression assays, including mRNA and protein levels, to investigate miR-137 directly targeting EFNB2 gene and to explore the effect of a genetic variant in the putative seed-pair region of EFNB2 3′-UTR. We also tested for the expression of miR-137 in the peripheral blood of a cohort of first-onset schizophrenia patients and healthy controls to determine its potential diagnostic value.
2. Materials and Methods
2.1. Bioinformatic Analyses
TargetScan (TargetScan 6.2, http://www.targetscan.org) and Miranda (http://www.microrna.org) were used to predicate miR-137 and EFNB2 interaction according to the presence of binding sites in the seed region, the efficacy of targeting and the probability of conserved targeting (Lewis et al., 2005). The human hsa-miR-137 sequences were obtained from miRBase (miRBase, http://www.mirbase.org). EFNB2 mRNA 3′-UTR sequences, the location of the single nucleotide polymorphism (SNP) rs550067317 in EFNB2 and allele frequencies were obtained from the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgTracks) and NCBI database (http://www.ncbi.nlm.nih.gov/gquery/). RNA folding structures between miRNAs and targeting sequences and the minimum folding energy (MFE) were predicted by RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid) (Kruger and Rehmsmeier, 2006, Rehmsmeier et al., 2004).
2.2. Vector Construction and Sequence Mutagenesis
A 500-bp-long 3′-UTR fragment of the EFNB2 gene containing the putative miR-137 binding site was synthesized by GenePharm Corporation. The sequence was cloned into the pmirGLO luciferase vector (Promega, USA) downstream of the firefly luciferase coding region, between the SacI and XbaI sites. The DNA sequences of the amplified fragments were identified by restriction enzyme digestion and sequencing and were consistent with the RCh38/hg38 reference sequence from UCSC. This vector was referred to as the wild type or rs550067317 (A) vector. The mutated EFNB2 3′-UTR sequences, containing all six nucleotides in the putative seed-pair region, was synthesized according to A-T, C-G substitution criteria. PCR-directed mutation was used to generate a mutation of minor allele C of SNP rs550067317. Likewise, these two mutations were cloned into the pmirGLO vector and referred to as full mutant and rs550067317(C) vector respectively. Both of them are validated by two methods of restriction enzyme digestion and sequencing. The primer information for the gene 3′-UTR to construct different vectors are showed in Table 1. In addition, the ZNF804A 3′-UTR was synthesized, and the pmirGLO + ZNF804A was constructed as a positive control.
Table 1.
Primer information for genes and small RNAs.
| Gene name | Accession number | Primer | Location for the first nucleotide of the primera | Sequence (from 5′ to 3′) | Annealing temperature (°C) |
|---|---|---|---|---|---|
| EFNB2 | NM_004093 | Forward for RT-qPCR | Chr13:106,494,962 | GATCCAACAAGACGTCCAGAA | 60 |
| Reverse for RT-qPCR | Chr13:106,493,342 | CTGAAGCAATCCCTGCAAATA | |||
| 3′-UTR Forward for mutationb | Chr13:106,492,317 | CGAGCTCAGTGGGGCTGGGGAAAGGGCT | 58 | ||
| 3′-UTR Reverse for mutationb | Chr13:106,491,818 | GCTCTAGATGACCTTTGGCTTTTCAAC | |||
| rs550067317(C) Forward for mutation | Chr13:106,491,988 | TACTGGCACTAAGATACACAGCTCCGAGCT | 58 | ||
| rs550067317(C) Reverse for mutation | Chr13:106,491,972 | GTATCTTAGTGCCAGTACCACAACAGTCCT | |||
| GAPDH | NM_002046 | Forward for RT-qPCR | Chr12:6,537,347 | CCAAGGTCATCCATGACAACT | 60 |
| Reverse for RT-qPCR | Chr12:6,537,683 | CAGGGATGATGTTCTGGAGAG | |||
| Hsa-miR-137 | NR_029679 | Forward for RT-qPCR | Chr1:98,046,113 | TTATTGCTTAAGAATACGCGTAG | 60 |
| U6 snRNA | NR_004394 | Forward for RT-qPCR | Chr15:67,840,042 | CTCGCTTCGGCAGCACA | 60 |
| Reverse for RT-qPCR | Chr15:67,839,949 | AACGCTTCACGAATTTGCGT |
UCSC Browser, Dec. 2013; http://genome,ucsc.edu/cgi-bin/hgGateway.
Several nucleotides (restriction enzyme cutting site) have been added to the 5′ terminal of the primer.
2.3. Cell Culture
HEK293T and SH-SY5Y cell lines were maintained at 37 °C with a humidified atmosphere of 5% CO2. HEK293T cells were cultured in Dulbecco's modified Eagle medium and high glucose medium (DMEM/HIGH GLUCOSE) (Hyclone, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, Australia), 100 U/mL penicillin, and 100 μg/mL streptomycin (Penicillin-Streptomycin Solution) (Hyclone, USA). SH-SY5Y cells were cultured in DMEM:Nutrient Mixture F-12 containing 1.5 mM l-glutamine at a ratio of 1:1 (DMEM/F-12) supplemented with 10% FBS, 100 U/mL of penicillin, and 100 μg/mL of streptomycin.
2.4. Dual Luciferase Reporter Assay
Luciferase reporter constructs were transfected into HEK293T cell. The mature miR-137 sequence (miR-137 mimics) and negative control miRNAs (mimic NC) were ordered from GenePharm Corporation. The combination of miR-137 mimics and pmirGLO + ZNF804A was co-transfected into SH-SY5Y cells as a positive control. To test for an interaction between miR-137 and the EFNB2 3′-UTR, combinations of mimics or NC with full mutant and wild type/rs550067317(A) were co-transfected respectively and were harvested after 24 h or 48 h. To determine whether rs550067317(C) affects the inhibition, we tested combinations of miR-137 or NC with different constructs and harvested after 24 h. The activity of the luciferase reporter gene was detected using the Dual-Luciferase® Reporter Assay System (Promega, USA). Each sample was tested in triplicate and the relative luciferase activity was calculated as the ratio of firefly to Renilla luciferase activity and normalized against a blank control.
2.5. RNA Isolation and Real-Time Quantitative PCR (RT-qPCR)
SH-SY5Y cells were transfected with miR-137 mimics or with NC. For comparison, we generated a blank control using the same volume of media and a mock control using the same volume of media and transfection reagents. Samples for each group were transfected in triplicate. Twenty-four hours after transfection, total RNA was extracted with TRIzol® (Invitrogen, USA) and quantified by NanoDrop2000 (Thermo, USA). The first strand of cDNA for miRNA was synthesized using Mir-X miRNA First-Strand Synthesis Kit (Clontech, USA), and the first strand for mRNA was synthesized using RevertaidTM First Strand cDNA Synthesis kit (Fermentas, Canada). Real-time quantitative PCR (RT-qPCR) was performed with the FastStart Universal SYBR Green Master (Rox) (Roche, USA) for quantification. Gene and miRNA expression were normalized by GAPDH and U6 small nuclear RNA (snRNA), respectively. Each sample was tested in three repeats and all data were analyzed using the 2-ΔΔCt method. The primer information for genes and RNAs was shown in Table 1.
2.6. Protein Isolation and Western Blotting
Forty-eight hours after transfection with four groups, the SH-SY5Y cells were washed with cold PBS and lysed in RIPA Lysis Buffer (Beyotime, China). Protein concentrations were determined using a bicinchonininc acid (BCA) assay kit (Pierce, USA). On a 10% SDS-PAGE gel, the total protein was separated and transferred to a PVDF membrane (Millipore, USA). GAPDH on the same membrane was used as a loading control. The primary monoclonal antibodies were anti-EFNB2 (ab150411, Abcam, UK) or anti-GAPDH (TA-08, ZSGB-BIO, China) and anti-mouse (31430, Thermo, USA) or anti-rabbit IgG (31460, Thermo, USA) were used as the secondary antibodies to further detect the signal. The target proteins were determined on membranes by an enhanced chemiluminescence system (Millipore, USA). The intensities of the immunoblots were quantified with a bio imaging (Syngene, UK) coupled to a personal computer. Expression level was accomplished by obtaining the ratio of the band density of EFNB2 to that of GAPDH from the same sample.
2.7. Peripheral Blood Collection and RT-qPCR
All subjects were Han Chinese in origin and from Northwest China. All patients were independently diagnosed by at least two experienced psychiatrists according to the Diagnosis and Statistical Manual of Mental Disorders Fourth Edition (DSM-IV) criteria for schizophrenia. A total of 88 unrelated individuals were recruited from the Northwest China that included 44 patients (22 males, mean age = 28.45 ± 6.79 years; 22 females, mean age = 30.73 ± 7.67 years) with schizophrenia and 44 healthy controls (22 males, mean age = 30.22 ± 4.22 years; 22 females, mean age = 30.36 ± 7.59 years). All patients were drug naive and untreated from any other methods. Controls were drawn from unrelated volunteers and blood donors. Any individuals with current or past evidence of mental illness or a relative with mental illness were excluded from the study. The study was approved by the medical ethics committee of Xi'an Jiaotong University Health Science Center and informed consent was obtained from all subjects.
Peripheral blood samples from the participants were collected in EDTA tubes and placed on ice. Within 2 h, total RNA was harvested using TRIzol® (Invitrogen, USA) according to the manufacturer's instructions and quantified using a NanoDrop 2000 (Thermo, USA). Reverse transcription and RT-qPCR for the mRNA expression were performed as previously described for SH-SY5Y cells. Furthermore, GAPDH, ATCB and B2M were selected as multiple internal control genes in peripheral blood samples according to the complexity of blood components, and the geometric mean of the three control genes was used to normalize the expression of EFNB2 (Vandesompele et al., 2002).
2.8. Statistical Analyses
All data are presented as the mean ± standard error of the mean (SEM). Statistically significant differences between groups were calculated using an analysis of variance. The results within a group were evaluated using the student's t-test. For the data that did not fit the normal distribution and homogeneity of variance, the non-parametric test was performed. The receiver operating characteristics (ROC) curve was constructed to determine the sensitivity, specificity and area under curve (AUC) for miR-137 levels in peripheral blood. Data analyses were performed using GraphPad Prism 5 software (GraphPad Co. Ltd., San Diego, CA, USA) or SPSS software version 18.0 (SPSS, Inc., Chicago, IL, USA). Differences with P-values of < 0.05 were considered statistically significant.
3. Results
3.1. Bioinformatic Prediction of EFNB2 as a miR-137 Target, and Different Allele of SNP rs550067317 within EFNB2 Affects the MFE
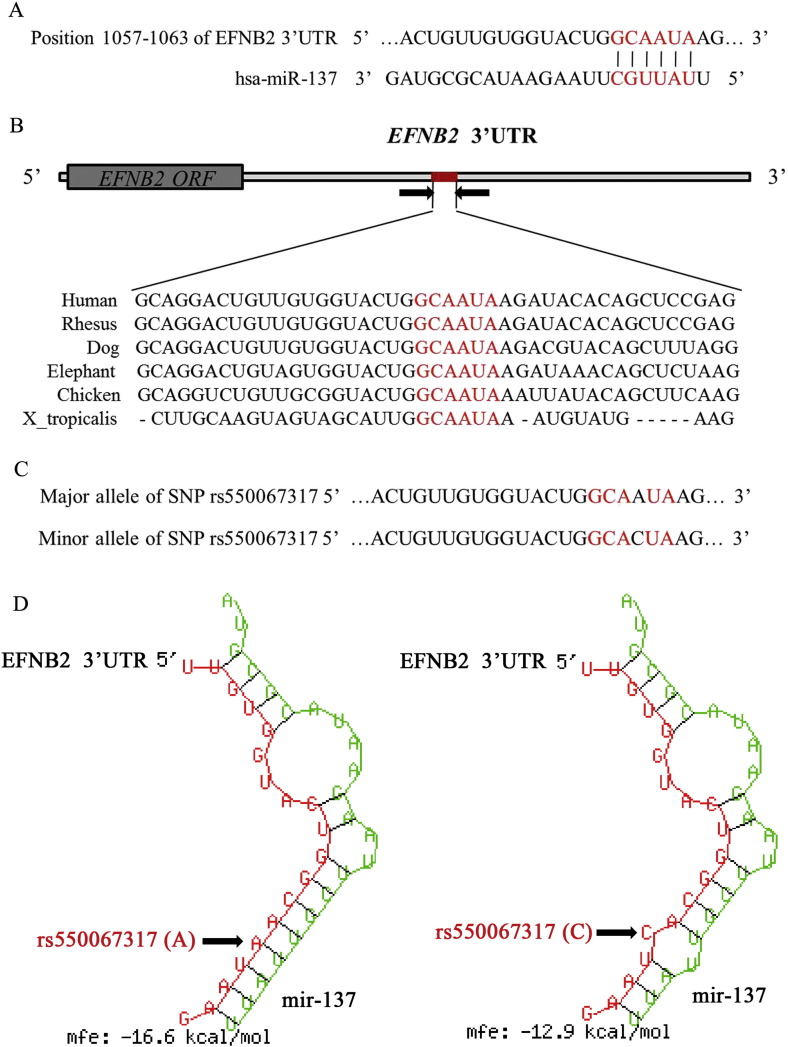
The TargetScan and Miranda online software predicted that hsa-miR-137 was a candidate miRNA for targeting the EFNB2 gene at nucleotides 1057–1063 of its 3′-UTR. The complementary sequence was shown in Fig. 1A, with the miR-137 seed sequence shown in red. The sequence in the binding site of EFNB2 3′-UTR was broadly conserved among vertebrates, including human, rhesus, dog, elephant and chicken, suggesting its potential important function for the gene (Fig. 1B).
Fig. 1.
EFNB2 as target of miR-137, and different allele of SNP rs550067317 affects the MFE. A. Schematic diagram of the pairing relationship between miR-137 and EFNB2 mRNA 3′-UTR analyzed by TargetScan and Miranda online software. The putative seed-pair region was in red. B. Broad conservation of the target sites in EFNB2 3′-UTR among vertebrates. C. The location of SNP rs550067317 in EFNB2 3′-UTR binding site. D. Location of the SNP rs550067317 alleles.
The SNP rs550067317 is in the EFNB2 3′-UTR binding site for miR-137 and the minor allele is the nucleotide C (Fig. 1C). With the major allele A at rs550067317, base-pairing between EFNB2 3′-UTR mRNA and mature miR-137 results in a typical stem-loop structure essential for miRNA-mediated expression repression. When the minor allele of C is present at rs550067317, it disrupts the formation of the stem in this essential structure. With the minor allele, the MFE increases to − 12.9 kcal/mol, compared with an MFE of − 16.6 kcal/mol for the major allele A, indicating decreased stability in the secondary RNA folding structure (Fig. 1D).
3.2. Full Mutant and rs550067317(C) Vector in the Binding Site of EFNB2 3′-UTR Independently Reverses the Inhibition of miR-137 in Luciferase Activity Assays
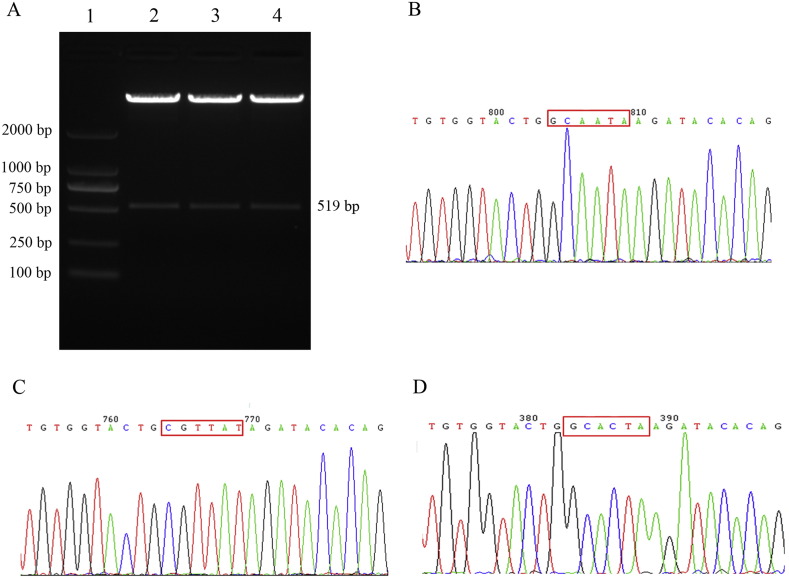
All constructed vectors were validated by restriction enzyme digestion and sequencing methods (Fig. 2). Data for pmirGLO + ZNF804A were not shown. The constructs of full mutant and rs550067317 (C) vector were shown on Fig. 3A.
Fig. 2.
The results of restriction enzyme digestion and sequencing. A. The results of restriction enzyme digestion. Lane 1 shows the marker of D2000, lanes 2–4 shows the restriction enzyme digestion result of wild type/rs550067317(A), full mutant, rs550067317(C) construct, respectively. B, C, D. The sequencing result of wild type/rs550067317(A), full mutant, rs550067317(C) construct, respectively.
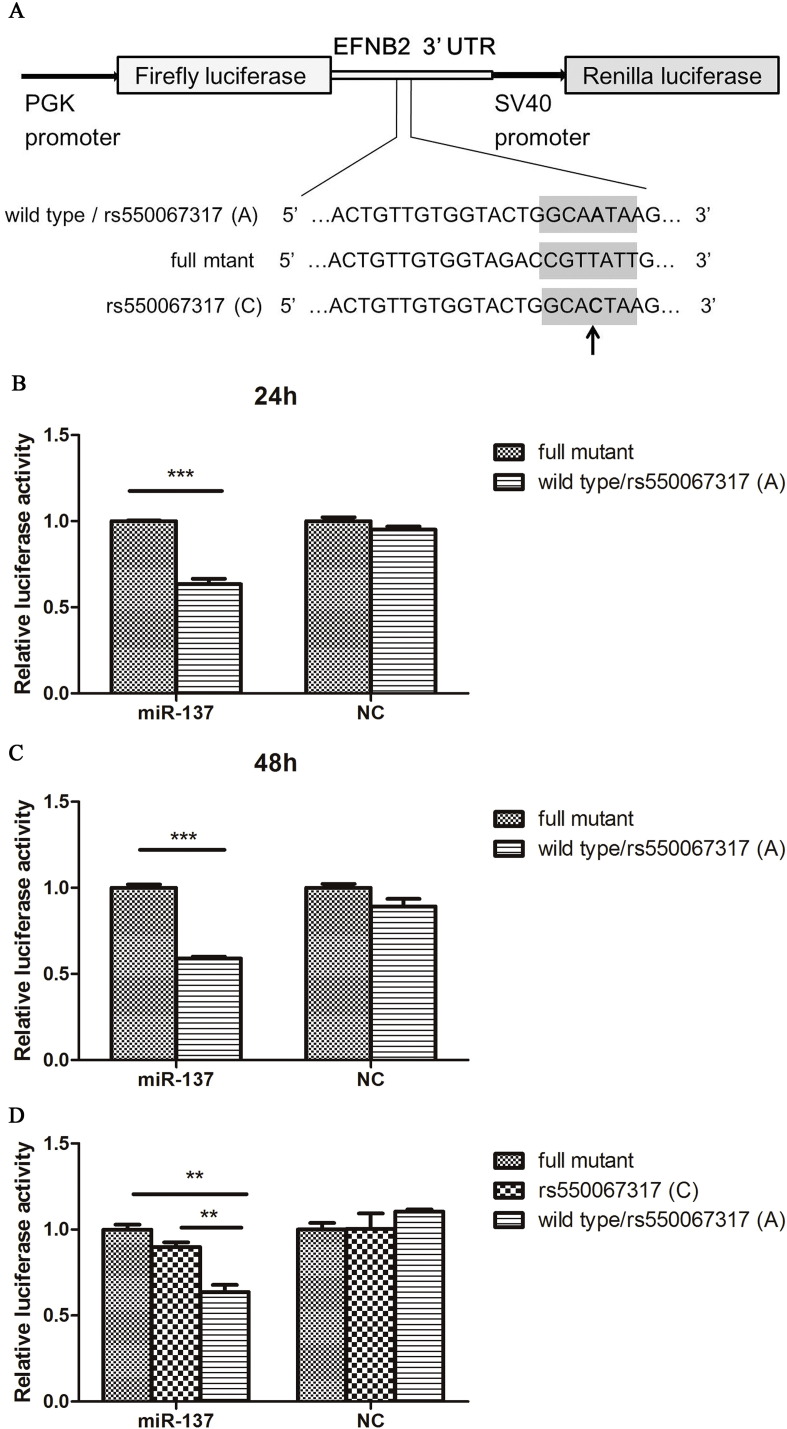
Fig. 3.
MiR-137 directly targets EFNB2 gene and SNP rs550067317(C) independently reverses the inhibition. A. Schematic diagram of three different constructs. The arrow marked the minor allele C in the construct. B, C. Dual luciferase reporter assay of miR-137 mimics/negative co-transfected with full mutant and wild type construct in HEK293T cells after 24 h and 48 h. D. Dual luciferase reporter assay of miR-137 mimics/negative co-transfected with full mutant, rs55006731(C), and wild type construct in HEK293T cells. Bars represent SEM from three experiments.
NC, negative control. ** indicates P < 0.01, *** indicates P < 0.001.
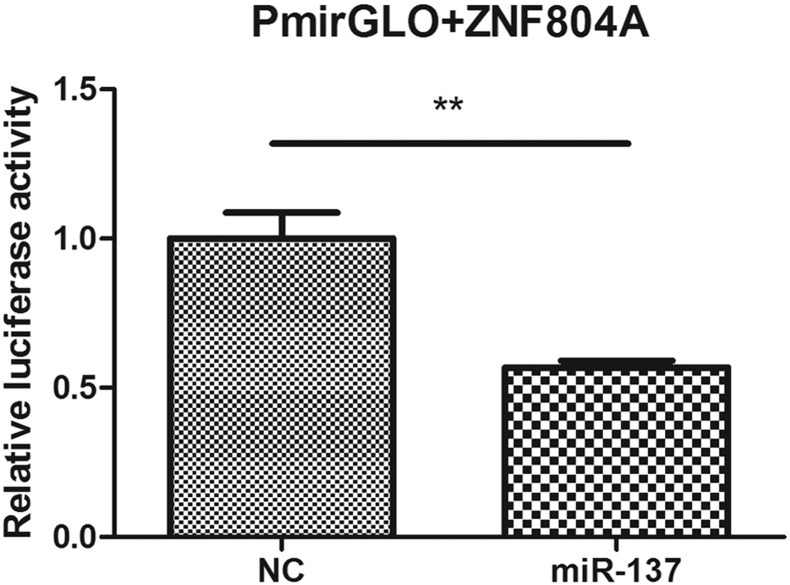
As shown in Fig. 4, luciferase activity in HEK293T cells co-transfected with miR-137 mimics and pmirGLO + ZNF804A was significantly decreased compared with NC and the reporter gene construct, which was consistent with the previous reports (Kim et al., 2012, Xia et al., 2015). Fig. 3B and C showed that the luciferase activity in cells co-transfected with miR-137 mimics and wild type target sequence was significantly decreased after 24 h and 48 h compared with mimics and full mutated vector. In the group of NC, there was no significant difference in luciferase activity, suggesting that miR-137 directly binds to the EFNB2 3′-UTR and inhibits its expression. In Fig. 3D, the luciferase activity decreased significantly in the group of co-transfection of miR-137 mimics and wild type, causing 36% reduction compared with the full mutant, and 29% reduction compared with the rs550067317 (C) vector, while the luciferase activity did not differ significantly with the combinations of NC and the different constructs, suggesting that SNP rs550067317 (C) also disrupted the miR-137-mediated repression. Luciferase activity in cells co-transfected with a combination of miR-137 with the full mutant and the rs550067317 (C) vector did not show significant differences, suggesting that the minor allele of SNP rs550067317 reverses the inhibition independently. The decreased effect of rs550067317(C) was consistent with in silico analyses of the SNP effect on RNA-folding structures formed by miR-137 and its target sequence in EFNB2 (Fig. 1D).
Fig. 4.
The dual luciferase reporter assay of miR-137 mimics/negative co-transfecting with PmirGLO + ZNF804A in HEK293T cells. 24 h after transfection, the cells were assayed for firefly luciferase activity and Renilla luciferase activity test and the ratio of firefly/Renilla is obtained. Bars represent SEM from three experiments.
NC, negative control. ** indicates P < 0.01.
3.3. MiR-137 Inhibits Endogenous Expression of EFNB2 at the Protein Level in SH-SY5Y Cells
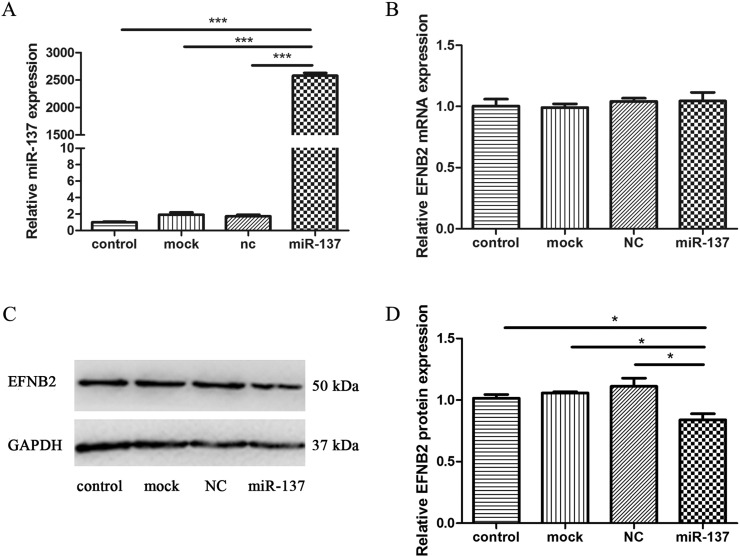
Reporter gene assays were used to confirm the direct targeting of the miR-137 to the 3′-UTR of EFNB2. To determine whether endogenously expressed EFNB2 is subject to regulation by miR-137, miR-137 mimics and the negative control were transfected into SH-SY5Y cells, a neuroblastoma cell line commonly used in the study of schizophrenia. The RT-qPCR results of miRNA expression showed that miR-137 mimics caused mature miR-137 levels to increase by > 2000-folds compared with NC, mock or control, confirming the transfection efficiency (Fig. 5A). Analyses of mRNA expression show that miR-137 has no significant effect on the stability or expression level of EFNB2 mRNA (Fig. 5B). Western blots indicated that miR-137 mimics were able to decrease the EFNB2 protein level compared with the NC, mock or control (Fig. 5C). Three independent western blotting experiments were performed and the relative expression of EFNB2 was significantly decreased (Fig. 5D). These data suggest that endogenous EFNB2 expression is down-regulated by miR-137 at the protein level.
Fig. 5.
MiR-137 inhibits endogenous expression of EFNB2 at protein level. A. RT-qPCR results of miR-137 relative expression in SH-SY5Y cells. B. RT-qPCR results of EFNB2 mRNA relative expression SH-SY5Y cells. C. Western blotting results of EFNB2 and GAPDH as an internal control. The band size of EFNB2 is 50 kDa which is consistent with the specification of the antibody kit. D. Statistical analysis of Western blotting results from three independent western blotting experiments.
NC, negative control. * indicates P < 0.05, ***indicates P < 0.001.
3.4. MiR-137 Is aberrantly Expressed in Peripheral Blood of Schizophrenia Patients and Has some Diagnostic Value for the Disease
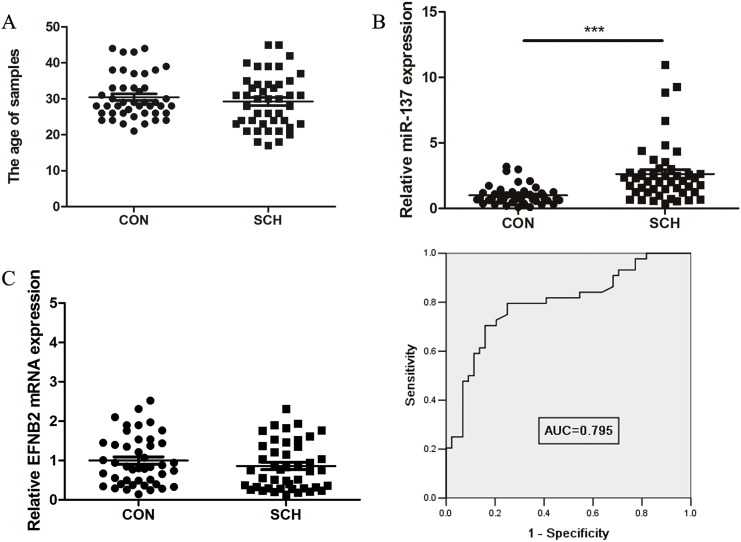
The age distribution of samples was shown in Fig. 6A. In the peripheral blood of schizophrenia patients, there was a significant increase of 2.63-fold in the expression of miR-137 compared with healthy controls (Fig. 6B). Expression of the target gene, EFNB2, was not significantly altered (Fig. 6C). The relationship between miR-137 and EFNB2 mRNA expression levels was consistent with data obtained using SH-SY5Y cells. Because miR-137 was aberrantly expressed in the peripheral blood of schizophrenia patients, we performed a receiver operating characteristics (ROC) curve analysis to evaluate the diagnostic value of miR-137 in differentiating between patients with schizophrenia and healthy controls. As shown in Fig. 6D, the area under the curve (AUC) of miR-137 was 0.795 (95% confidence interval 0.700–0.890), the sensitivity of the cut-off point was 70.5%, and the specificity was 84.1%. These results suggested that miR-137 in peripheral blood has certain diagnostic value for schizophrenia.
Fig. 6.
MiR-137 is aberrantly expressed in peripheral blood of schizophrenia patients. A. The distribution of sample ages. B. The expression of miR-137 in the peripheral blood of schizophrenia patients and healthy controls. C. The expression of EFNB2 in the peripheral blood of schizophrenia patients and normal controls. D. Receiver operating characteristic (ROC) curve analysis of the diagnostic value of miR-137 expression for schizophrenia.
CON, normal control; SCH, schizophrenia patients; AUC, area under curve. ***indicates P < 0.001.
4. Discussion
Although genome-wide association studies provide a systematic, powerful, and unbiased method for studying the common disease/common variant (CDCV) hypothesis of complex disorders such as schizophrenia, the identification of susceptibility loci for schizophrenia was not replicated across studies because of the genetic heterogeneity of schizophrenia among different populations (Shi et al., 2011, Yue et al., 2011). Nonetheless, a robust statistical association between the MIR137 gene and schizophrenia identified in Europeans through genome-wide association studies was replicated in the Han Chinese population in a case-control study (Guan et al., 2014, Ripke et al., 2011, Ripke et al., 2013, Ripke et al., 2014). The type of miR-137 seed sequence was canonical and the sequence of binding site within EFNB2 were evolutionary conserved (Bartel, 2009). Using a computational prediction analysis and report gene assays, EFNB2 gene was directly targeted by miR-137. Furthermore, in our previous study of EFNB2 associated with schizophrenia, all the samples were recruited from Northwest China (Zhang et al., 2010). The comparative genetic homogeneity is helpful to find some susceptibility loci, especially that might increase susceptibility to the disease in a subpopulation.
Both EFNB2 and DAOA genes involved in NMDA receptor signaling-mediated susceptibility to schizophrenia have been identified in a Han Chinese population in our previous studies (Ma et al., 2006, Zhang et al., 2010). Interestingly, a recent genome-wide association analysis identified 13 new risk loci for schizophrenia and suggested the involvement of glutamatergic neurotransmission signaling in the etiology of schizophrenia, providing robust support for the association of NMDA receptor signaling with schizophrenia (Ripke et al., 2014). Sentürk et al. reported that the activation of ephrin Bs, including EFNB2, was sufficient to rescue the absence of Reelin (Senturk et al., 2011). Reelin signaling was required for the nervous system to function properly, and disruption of this pathway results in lissencephaly, a severe developmental disorder associated with schizophrenia (Costa et al., 2002, Herz and Chen, 2006, Hong et al., 2000). Previous studies have demonstrated that miR-137 regulates adult neurogenesis, neuronal maturation, and presynaptic plasticity (Siegert et al., 2015, Silber et al., 2008, Smrt et al., 2010, Szulwach et al., 2010). Siegert et al. showed that overexpression of miR-137 caused impairment in synaptic vesicle trafficking and alterations in synaptic plasticity (Siegert et al., 2015). Using meta gene set enrichment analyses, Wright et al. found that potential target gene sets of miR-137 were enriched with variants associated with Ephrin receptor signaling and some other signal pathways (Wright et al., 2015). In current study, we provided the molecular experimental evidence to validate EFNB2 as a direct target of miR-137, including dual luciferase reporter gene assays and western blotting methods performed in two cell lines, suggesting that miR-137 as a risk factor in schizophrenia, might be implicated in NMDA receptor signaling and/or Reelin signaling pathway.
It was more intriguing that the SNP rs550067317 lies in the binding site in EFNB2 3′-UTR and rs550067317(C) is a genetic variant. The current study confirmed that the rs550067317(C) vector reversed the repression of luciferase activity caused by miR-137, validating this SNP is a functional genetic variant. The functional genetic variant may also have implications in understanding the genetic architecture of schizophrenia. The predominant hypothesis in recent years has been that as in other common diseases, the genetic architecture of schizophrenia involves the action of numerous common alleles with small to moderate effects and possibly some rare alleles with much larger effects (Wang et al., 2005, Williams et al., 2009). It is not contradictory to have both common and rare variants as the genetic basis of a complex trait such as schizophrenia; the effects of both classes can be readily reconciled (Gibson, 2012, McCarthy et al., 2008, Owen et al., 2010). In studies of schizophrenia, people are categorized as cases or controls. However, the genetic contributions are complex, and the schizophrenia generally displays a threshold-dependent response superimposed on a continuous liability composed of many variants. A rare variant can either increase or decrease function, and its association with the disease will be conditional on the patient's background liability (Gibson, 2012). The SNP rs550067317 is a rare variant. The minor allele C is only exist in Northern Han Chinese population (Beijing, CHB) and the frequency is 0.97% according to the present data base (http://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/?q=rs550067317). The current study demonstrated that the minor allele of rs550067317 reversed the inhibition of miR-137 similar to the full mutation in the binding sites of EFNB2 3’UTR. It might strengthen EFNB2 function by impairing the direct interaction of miR-137 with EFNB2 in conditions where miR-137 is aberrantly overexpressing.
To determine if the overexpression of miR-137 negatively regulate EFNB2 expression, human neuroblastoma SH-SY5Y cells were transfected with mature miR-137 sequences or negative control miRNAs. The results showed that the overexpression of miR-137 did not affect the mRNA expression of EFNB2 but significantly repressed the accumulation of EFNB2 protein. This post-transcriptional function was consistent with initial discovery of animal miRNA, which inhibited protein synthesis and preserved the stability of the mRNA target (Ambros, 2004, Olsen and Ambros, 1999, Seggerson et al., 2002). Similarly, we demonstrated that in the peripheral blood of first-onset schizophrenia patients compared with healthy controls, the expression of miR-137 was significantly increased, and the mRNA levels of EFNB2 were not significantly different.
We used the conventional ROC curve of miR-137 in peripheral blood to calculate the area under the curve (AUC) with 95% confidence intervals. The AUC reached 0.795, indicating that miR-137 has certain value for diagnosing schizophrenia. Schizophrenia is a chronic, frequently disabling mental disorder with neurodevelopmental impairment (Insel, 2010). In both the neurodevelopment and the etiology of psychiatric disease such as schizophrenia, miRNA plays a pivotal role (Miller and Wahlestedt, 2010, Welberg, 2010, Xu et al., 2010). Because brain tissue is not readily accessible, blood-based expression profiling of miRNAs is increasingly common for the discovery of pathophysiology and identification of potential biomarkers for schizophrenia (Lai et al., 2011, Yu et al., 2015, Zhang et al., 2014). A major advantage of blood sample is avoiding potential changes in miRNA levels by different processing conditions (Cheng et al., 2013). The current study also showed that miR-137 in peripheral blood is a potential biomarker for differentiating between schizophrenia patients and healthy controls.
It is important to note that although the rare SNP rs550067317 in EFNB2 gene was not genotyped in a large population and the frequency of the minor allele C in the patients of schizophrenia and healthy controls was unknown, but we uncovered that it is a functional genetic variant through point mutation and dual luciferase activity assay. The other limitation of the current study is that TaqMan method for quantifying gene expression might be more sensitive than SYBR. However, the latter is widely accepted and applied to many studies.
5. Conclusion
We provided strong experimental evidence that miR-137 directly down-regulates the expression of EFNB2, and a genetic variant in the RNA binding site in EFNB2 gene affects the expression regulation. MiR-137, as a risk factor for schizophrenia, might be implicated in NMDA receptor signaling and/or Reelin signaling pathway. Further clinical experiment in peripheral blood verified that miR-137 is aberrantly expressed in the first-onset patients of schizophrenia compared healthy controls and it has certain diagnostic value for the disease. This study not only improved the understanding of the roles of EFNB2 and the genetic basis of rs550067317 for schizophrenia, but also provided new evidences for miR-137 involved in schizophrenia etiology and diagnosis, which might contribute to discover new biomarkers and therapeutic targets for the disease.
Author Contribution
J.M., Y.S., and W.L. were responsible for conceiving and designing the original protocol. S.W., R.Z., F.N., X.W., and M.L. performed the experiments. R.Z., C.J., R.K.V., and J.M. analyzed the data. X.W., C.J., and M.L. contributed reagents/materials/analysis tools. R.Z., and R.K.V. wrote the first draft of the manuscript. All authors contributed to and approved the final manuscript.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Acknowledgments
We are indebted to all of the individuals who have participated in, or helped with, our research. This study was supported by the National Natural Science Foundation of China (No. 81301151; 31371298), the China Postdoctoral Science Foundation (No. 2013M542337; 2015T81016), and the Program for New Century Excellent Talents in University (NCET-13-0452).
Study sponsors had no role in study design, data collection, data analysis, interpretation, writing of the report, and in the decision to submit the paper for publication.
Contributor Information
Rui Zhang, Email: zhangruity12@163.com.
Jie Ma, Email: majie_article@163.com.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge N.J., Cairns M.J. MicroRNA dysregulation in schizophrenia. Neurobiol. Dis. 2012;46(2):263–271. doi: 10.1016/j.nbd.2011.12.029. [DOI] [PubMed] [Google Scholar]
- Cao X., Yeo G., Muotri A.R., Kuwabara T., Gage F.H. Noncoding RNAs in the mammalian central nervous system. Annu. Rev. Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Cardno A.G., Gottesman I.I. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am. J. Med. Genet. 2000;97(1):12–17. [PubMed] [Google Scholar]
- Cheng H.H., Yi H.S., Kim Y., Kroh E.M., Chien J.W., Eaton K.D., Goodman M.T., Tait J.F., Tewari M., Pritchard C.C. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0064795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa E., Chen Y., Davis J., Dong E., Noh J.S., Tremolizzo L., Veldic M., Grayson D.R., Guidotti A. REELIN and schizophrenia: a disease at the interface of the genome and the epigenome. Mol. Interv. 2002;2(1):47–57. doi: 10.1124/mi.2.1.47. [DOI] [PubMed] [Google Scholar]
- Farh K.K., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310(5755):1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Gibson G. Rare and common variants: twenty arguments. Nat. Rev. Genet. 2012;13(2):135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan F., Zhang B., Yan T., Li L., Liu F., Li T., Feng Z., Liu X., Li S. MIR137 gene and target gene CACNA1C of miR-137 contribute to schizophrenia susceptibility in Han Chinese. Schizophr. Res. 2014;152(1):97–104. doi: 10.1016/j.schres.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Harris L.W., Pietsch S., Cheng T.M., Schwarz E., Guest P.C., Bahn S. Comparison of peripheral and central schizophrenia biomarker profiles. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0046368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz J., Chen Y. Reelin, lipoprotein receptors and synaptic plasticity. Nat. Rev. Neurosci. 2006;7(11):850–859. doi: 10.1038/nrn2009. [DOI] [PubMed] [Google Scholar]
- Hong S.E., Shugart Y.Y., Huang D.T., Shahwan S.A., Grant P.E., Hourihane J.O., Martin N.D., Walsh C.A. Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with human RELN mutations. Nat. Genet. 2000;26(1):93–96. doi: 10.1038/79246. [DOI] [PubMed] [Google Scholar]
- Insel T.R. Rethinking schizophrenia. Nature. 2010;468(7321):187–193. doi: 10.1038/nature09552. [DOI] [PubMed] [Google Scholar]
- Kim Y.K., Heo I., Kim V.N. Modifications of small RNAs and their associated proteins. Cell. 2010;143(5):703–709. doi: 10.1016/j.cell.2010.11.018. [DOI] [PubMed] [Google Scholar]
- Kim A.H., Parker E.K., Williamson V., McMichael G.O., Fanous A.H., Vladimirov V.I. Experimental validation of candidate schizophrenia gene ZNF804A as target for hsa-miR-137. Schizophr. Res. 2012;141(1):60–64. doi: 10.1016/j.schres.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik K.S. The neuronal microRNA system. Nat. Rev. Neurosci. 2006;7(12):911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Kruger J., Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(Web Server issue):W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon E., Wang W., Tsai L.H. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Mol. Psychiatry. 2013;18(1):11–12. doi: 10.1038/mp.2011.170. [DOI] [PubMed] [Google Scholar]
- Lai C.Y., Yu S.L., Hsieh M.H., Chen C.H., Chen H.Y., Wen C.C., Huang Y.H., Hsiao P.C., Hsiao C.K., Liu C.M., Yang P.C., Hwu H.G., Chen W.J. MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS One. 2011;6(6) doi: 10.1371/journal.pone.0021635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lin M.W., Sham P., Hwu H.G., Collier D., Murray R., Powell J.F. Suggestive evidence for linkage of schizophrenia to markers on chromosome 13 in Caucasian but not Oriental populations. Hum. Genet. 1997;99:417–420. doi: 10.1007/s004390050382. [DOI] [PubMed] [Google Scholar]
- Ma J., Qin W., Wang X.Y., Guo T.W., Bian L., Duan S.W., Li X.W., Zou F.G., Fang Y.R., Fang J.X., Feng G.Y., Gu N.F., St Clair D., He L. Further evidence for the association between G72/G30 genes and schizophrenia in two ethnically distinct populations. Mol. Psychiatry. 2006;11(5):479–487. doi: 10.1038/sj.mp.4001788. [DOI] [PubMed] [Google Scholar]
- McCarthy M.I., Abecasis G.R., Cardon L.R., Goldstein D.B., Little J., Ioannidis J.P., Hirschhorn J.N. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9(5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- Miller B.H., Wahlestedt C. MicroRNA dysregulation in psychiatric disease. Brain Res. 2010;1338:89–99. doi: 10.1016/j.brainres.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen P.H., Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 1999;216(2):671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Owen M.J., Craddock N., O'Donovan M.C. Suggestion of roles for both common and rare risk variants in genome-wide studies of schizophrenia. Arch. Gen. Psychiatry. 2010;67(7):667–673. doi: 10.1001/archgenpsychiatry.2010.69. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10(10):1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S., Sanders A.R., Kendler K.S., Levinson D.F., Sklar P., Holmans P.A., Lin D.Y., Duan J., Ophoff R.A., Andreassen O.A., Scolnick E., Cichon S., Clair D., Corvin A., Gurling H., Werge T., Rujescu D., Blackwood D.H., Pato C.N., Malhotra A.K., Purcell S., Dudbridge F., Neale B.M., Rossin L., Visscher P.M., Posthuma D., Ruderfer D.M., Fanous A., Stefansson H., Steinberg S., Mowry B.J., Golimbet V., De Hert M., Jönsson E.G., Bitter I., Pietiläinen O.P., Collier D.A., Tosato S., Agartz I., Albus M., Alexander M., Amdur R.L., Amin F., Bass N., Bergen S.E., Black D.W., Børglum A.D., Brown M.A., Bruggeman R., Buccola N.G., Byerley W.F., Cahn W., Cantor R.M., Carr V.J., Catts S.V., Choudhury K., Cloninger C.R., Cormican P., Craddock N., Danoy P.A., Datta S., de Hann L., Demontis D., Dikeos D., Djurovic S., Donnelly P., Donohoe G., Duong L., Dwyer S., Fink-Jensen A., Freedman R., Freimer N.B., Friedl M., Georgieva L., Giegling I., Gill M., Glenthøj B., Godard S., Hamshere M., Hansen M., Hansen T., Hartmann A.M., Henskens F.A., Hougaard D.M., Hultman C.M., Ingason A., Jablensky A.V., Jakobsen K.D., Jay M., Jürgens G., Kahn R.S., Keller M.C., Kenis G., Kenny E., Kim Y., Kirov G.K., Konnerth H., Konte B., Krabbendam L., Krausucki R., Lasseter V.K., Laurent C., Lawrence J., Lencz T., Lerer F.B., Liang K.Y., Lichtenstein P., Lieberman J.A., Linszen D.H., Lönnqvist J., Loughland C.M., Maclean A.W., Maher B.S., Maier W., Mallet J., Malloy P., Mattheisen M., Mattinsgsdal M., McGhee K.A., McGrath J.J., McIntosh A., McLean D.E., McQuillin A., Melle I., Michie P.T., Milanova V., Morris D.W., Mors O., Mortensen P.B., Moskvina V., Muglia P., Myin-Germeys I., Nertney D.A., Nestadt G., Nielsen J., Nikolov I., Nordentroft M., Norton N., Nöthen M.M., O'Dushlaine C.T., Olincy A., Olsen L., O'Neill F.A., Ørntoft T., Owen M.J., Pantelis C., Papadimitriou G., Pato M.T., Peltonen L., Petursson H., Pickard B., Pimm J., Pulver A.E., Puri V., Quested D., Quinn E.M., Rasmussen H.B., Réthelyi J.M., Ribble R., Rietschel M., Riley B.P., Ruggeri M., Schall U., Schulze T.G., Schwab S.G., Scott R.J., Shi J., Sigurdsson E., Silverman J.M., Spencer C.C., Stefansson K., Strange A., Strengman E., Stroup T.S., Suvisaari J., Tereniuis L., Thirumalai S., Thygesen J.H., Timm S., Toncheva D., van den Oord E., van Os J., van Winkel R., Veldink J., Walsh D., Wang A.G., Wiersma D., Wildenauer D.B., Williams H.J., Williams N.M., Wormley B., Zammit S., Sullivan P.F., O'Donovan M.C., Daly M.J., Gejman P.V. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43(10):969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S., O'Dushlaine C., Chambert K., Moran J.L., Kahler A.K., Akterin S., Bergen S.E., Collins A.L., Crowley J.J., Fromer M., Kim Y., Lee S.H., Magnusson P.K., Sanchez N., Stahl E.A., Williams S., Wray N.R., Xia K., Bettella F., Borglum A.D., Bulik-Sullivan B.K., Cormican P., Craddock N., de Leeuw C., Durmishi N., Gill M., Golimbet V., Hamshere M.L., Holmans P., Hougaard D.M., Kendler K.S., Lin K., Morris D.W., Mors O., Mortensen P.B., Neale B.M., O'Neill F.A., Owen M.J., Milovancevic M.P., Posthuma D., Powell J., Richards A.L., Riley B.P., Ruderfer D., Rujescu D., Sigurdsson E., Silagadze T., Smit A.B., Stefansson H., Steinberg S., Suvisaari J., Tosato S., Verhage M., Walters J.T., Levinson D.F., Gejman P.V., Laurent C., Mowry B.J., O'Donovan M.C., Pulver A.E., Schwab S.G., Wildenauer D.B., Dudbridge F., Shi J., Albus M., Alexander M., Campion D., Cohen D., Dikeos D., Duan J., Eichhammer P., Godard S., Hansen M., Lerer F.B., Liang K.Y., Maier W., Mallet J., Nertney D.A., Nestadt G., Norton N., Papadimitriou G.N., Ribble R., Sanders A.R., Silverman J.M., Walsh D., Williams N.M., Wormley B., Arranz M.J., Bakker S., Bender S., Bramon E., Collier D., Crespo-Facorro B., Hall J., Iyegbe C., Jablensky A., Kahn R.S., Kalaydjieva L., Lawrie S., Lewis C.M., Linszen D.H., Mata I., McIntosh A., Murray R.M., Ophoff R.A., Van Os J., Walshe M., Weisbrod M., Wiersma D., Donnelly P., Barroso I., Blackwell J.M., Brown M.A., Casas J.P., Corvin A.P., Deloukas P., Duncanson A., Jankowski J., Markus H.S., Mathew C.G., Palmer C.N., Plomin R., Rautanen A., Sawcer S.J., Trembath R.C., Viswanathan A.C., Wood N.W., Spencer C.C., Band G., Bellenguez C., Freeman C., Hellenthal G., Giannoulatou E., Pirinen M., Pearson R.D., Strange A., Su Z., Vukcevic D., Langford C., Hunt S.E., Edkins S., Gwilliam R., Blackburn H., Bumpstead S.J., Dronov S., Gillman M., Gray E., Hammond N., Jayakumar A., McCann O.T., Liddle J., Potter S.C., Ravindrarajah R., Ricketts M., Tashakkori-Ghanbaria A., Waller M.J., Weston P., Widaa S., Whittaker P., McCarthy M.I., Stefansson K., Scolnick E., Purcell S., McCarroll S.A., Sklar P., Hultman C.M., Sullivan P.F. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 2013;45(10):1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S., Neale B.M., Corvin A., Walters J.T., Farh K.H., Holmans P.A., Lee P., Bulik-Sullivan B., Collier D.A., Huang H., Pers T.H., Agartz I., Agerbo E., Albus M., Alexander M., Amin F., Bacanu S.A., Begemann M., Belliveau R.A., Jr., Bene J., Bergen S.E., Bevilacqua E., Bigdeli T.B., Black D.W., Bruggeman R., Buccola N.G., Buckner R.L., Byerley W., Cahn W., Cai G., Campion D., Cantor R.M., Carr V.J., Carrera N., Catts S.V., Chambert K.D., Chan R.C., Chen R.Y., Chen E.Y., Cheng W., Cheung E.F., Chong S.A., Cloninger C.R., Cohen D., Cohen N., Cormican P., Craddock N., Crowley J.J., Curtis D., Davidson M., Davis K.L., Degenhardt F., Del Favero J., Demontis D., Dikeos D., Dinan T., Djurovic S., Donohoe G., Drapeau E., Duan J., Dudbridge F., Durmishi N., Eichhammer P., Eriksson J., Escott-Price V., Essioux L., Fanous A.H., Farrell M.S., Frank J., Franke L., Freedman R., Freimer N.B., Friedl M., Friedman J.I., Fromer M., Genovese G., Georgieva L., Giegling I., Giusti-Rodríguez P., Godard S., Goldstein J.I., Golimbet V., Gopal S., Gratten J., de Haan L., Hammer C., Hamshere M.L., Hansen M., Hansen T., Haroutunian V., Hartmann A.M., Henskens F.A., Herms S., Hirschhorn J.N., Hoffmann P., Hofman A., Hollegaard M.V., Hougaard D.M., Ikeda M., Joa I., Julià A., Kahn R.S., Kalaydjieva L., Karachanak-Yankova S., Karjalainen J., Kavanagh D., Keller M.C., Kennedy J.L., Khrunin A., Kim Y., Klovins J., Knowles J.A., Konte B., Kucinskas V., Ausrele Kucinskiene Z., Kuzelova-Ptackova H., Kähler A.K., Laurent C., Keong J.L., Lee S.H., Legge S.E., Lerer B., Li M., Li T., Liang K.Y., Lieberman J., Limborska S., Loughland C.M., Lubinski J., Lönnqvist J., Macek M., Jr., Magnusson P.K., Maher B.S., Maier W., Mallet J., Marsal S., Mattheisen M., Mattingsdal M., McCarley R.W., McDonald C., McIntosh A.M., Meier S., Meijer C.J., Melegh B., Melle I., Mesholam-Gately R.I., Metspalu A., Michie P.T., Milani L., Milanova V., Mokrab Y., Morris D.W., Mors O., Murphy K.C., Murray R.M., Myin-Germeys I., Müller-Myhsok B., Nelis M., Nenadic I., Nertney D.A., Nestadt G., Nicodemus K.K., Nikitina-Zake L., Nisenbaum L., Nordin A., O'Callaghan E., O'Dushlaine C., O'Neill F.A., Oh S.Y., Olincy A., Olsen L., Van Os J., Pantelis C., Papadimitriou G.N., Papiol S., Parkhomenko E., Pato M.T., Paunio T., Pejovic-Milovancevic M., Perkins D.O., Pietiläinen O., Pimm J., Pocklington A.J., Powell J., Price A., Pulver A.E., Purcell S.M., Quested D., Rasmussen H.B., Reichenberg A., Reimers M.A., Richards A.L., Roffman J.L., Roussos P., Ruderfer D.M., Salomaa V., Sanders A.R., Schall U., Schubert C.R., Schulze T.G., Schwab S.G., Scolnick E.M., Scott R.J., Seidman L.J., Shi J., Sigurdsson E., Silagadze T., Silverman J.M., Sim K., Slominsky P., Smoller J.W., So H.C., Spencer C.A., Stahl E.A., Stefansson H., Steinberg S., Stogmann E., Straub R.E., Strengman E., Strohmaier J., Stroup T.S., Subramaniam M., Suvisaari J., Svrakic D.M., Szatkiewicz J.P., Söderman E., Thirumalai S., Toncheva D., Tosato S., Veijola J., Waddington J., Walsh D., Wang D., Wang Q., Webb B.T., Weiser M., Wildenauer D.B., Williams N.M., Williams S., Witt S.H., Wolen A.R., Wong E.H., Wormley B.K., Xi H.S., Zai C.C., Zheng X., Zimprich F., Wray N.R., Stefansson K., Visscher P.M., Adolfsson R., Andreassen O.A., Blackwood D.H., Bramon E., Buxbaum J.D., Børglum A.D., Cichon S., Darvasi A., Domenici E., Ehrenreich H., Esko T., Gejman P.V., Gill M., Gurling H., Hultman C.M., Iwata N., Jablensky A.V., Jönsson E.G., Kendler K.S., Kirov G., Knight J., Lencz T., Levinson D.F., Li Q.S., Liu J., Malhotra A.K., McCarroll S.A., McQuillin A., Moran J.L., Mortensen P.B., Mowry B.J., Nöthen M.M., Ophoff R.A., Owen M.J., Palotie A., Pato C.N., Petryshen T.L., Posthuma D., Rietschel M., Riley B.P., Rujescu D., Sham P.C., Sklar P., St Clair D., Weinberger D.R., Wendland J.R., Werge T., Daly M.J., Sullivan P.F., O'Donovan M.C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seggerson K., Tang L., Moss E.G. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 2002;243(2):215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- Senturk A., Pfennig S., Weiss A., Burk K., Acker-Palmer A. Ephrin Bs are essential components of the Reelin pathway to regulate neuronal migration. Nature. 2011;472(7343):356–360. doi: 10.1038/nature09874. [DOI] [PubMed] [Google Scholar]
- Shi Y., Li Z., Xu Q., Wang T., Li T., Shen J., Zhang F., Chen J., Zhou G., Ji W., Li B., Xu Y., Liu D., Wang P., Yang P., Liu B., Sun W., Wan C., Qin S., He G., Steinberg S., Cichon S., Werge T., Sigurdsson E., Tosato S., Palotie A., Nothen M.M., Rietschel M., Ophoff R.A., Collier D.A., Rujescu D., Clair D.S., Stefansson H., Stefansson K., Ji J., Wang Q., Li W., Zheng L., Zhang H., Feng G., He L. Common variants on 8p12 and 1q24.2 confer risk of schizophrenia. Nat. Genet. 2011;43(12):1224–1227. doi: 10.1038/ng.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert S., Seo J., Kwon E.J., Rudenko A., Cho S., Wang W., Flood Z., Martorell A.J., Ericsson M., Mungenast A.E., Tsai L.H. The schizophrenia risk gene product miR-137 alters presynaptic plasticity. Nat. Neurosci. 2015;18(7):1008–1016. doi: 10.1038/nn.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber J., Lim D.A., Petritsch C., Persson A.I., Maunakea A.K., Yu M., Vandenberg S.R., Ginzinger D.G., James C.D., Costello J.F., Bergers G., Weiss W.A., Alvarez-Buylla A., Hodgson J.G. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrt R.D., Szulwach K.E., Pfeiffer R.L., Li X., Guo W., Pathania M., Teng Z.Q., Luo Y., Peng J., Bordey A., Jin P., Zhao X. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28(6):1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan P.F., Kendler K.S., Neale M.C. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry. 2003;60(12):1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- Sullivan P.F., Fan C., Perou C.M. Evaluating the comparability of gene expression in blood and brain. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006;141B(3):261–268. doi: 10.1002/ajmg.b.30272. [DOI] [PubMed] [Google Scholar]
- Szulwach K.E., Li X., Smrt R.D., Li Y., Luo Y., Lin L., Santistevan N.J., Li W., Zhao X., Jin P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 2010;189(1):127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasu M.A., Dalva M.B., Zigmond R.E., Greenberg M.E. Modulation of NMDA receptor-dependent calcium influx and gene expression through EphB receptors. Science. 2002;295(5554):491–495. doi: 10.1126/science.1065983. [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:1–12. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.Y., Barratt B.J., Clayton D.G., Todd J.A. Genome-wide association studies: theoretical and practical concerns. Nat. Rev. Genet. 2005;6(2):109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- Welberg L. Neurodegeneration: export disrupts transport. Nat. Rev. Neurosci. 2010;11(2):74. doi: 10.1038/nrn2796. [DOI] [PubMed] [Google Scholar]
- Williams H.J., Owen M.J., O'Donovan M.C. Schizophrenia genetics: new insights from new approaches. Br. Med. Bull. 2009;91:61–74. doi: 10.1093/bmb/ldp017. [DOI] [PubMed] [Google Scholar]
- Wright C., Turner J.A., Calhoun V.D., Perrone-Bizzozero N. Potential impact of miR-137 and its targets in schizophrenia. Front. Genet. 2013;4:58. doi: 10.3389/fgene.2013.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C., Calhoun V.D., Ehrlich S., Wang L., Turner J.A., Bizzozero N.I. Meta gene set enrichment analyses link miR-137-regulated pathways with schizophrenia risk. Front. Genet. 2015;6:147. doi: 10.3389/fgene.2015.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhou X., Wang T., Zhang Q., Li Q., Liu Y., Xing Q., Wang L., He L., Zhao X. Experimental validation of candidate schizophrenia gene CALN1 as a target for microRNA-137. Neurosci. Lett. 2015;602:110–114. doi: 10.1016/j.neulet.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Xu B., Karayiorgou M., Gogos J.A. MicroRNAs in psychiatric and neurodevelopmental disorders. Brain Res. 2010;1338:78–88. doi: 10.1016/j.brainres.2010.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.C., Wu J., Zhang H.X., Zhang G.L., Sui J., Tong W.W., Zhang X.Y., Nie L.L., Duan J.H., Zhang L.R., Lv L.X. Alterations of miR-132 are novel diagnostic biomarkers in peripheral blood of schizophrenia patients. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2015;63:23–29. doi: 10.1016/j.pnpbp.2015.05.007. [DOI] [PubMed] [Google Scholar]
- Yue W.H., Wang H.F., Sun L.D., Tang F.L., Liu Z.H., Zhang H.X., Li W.Q., Zhang Y.L., Zhang Y., Ma C.C., Du B., Wang L.F., Ren Y.Q., Yang Y.F., Hu X.F., Wang Y., Deng W., Tan L.W., Tan Y.L., Chen Q., Xu G.M., Yang G.G., Zuo X.B., Yan H., Ruan Y.Y., Lu T.L., Han X., Ma X.H., Cai L.W., Jin C., Zhang H.Y., Yan J., Mi W.F., Yin X.Y., Ma W.B., Liu Q., Kang L., Sun W., Pan C.Y., Shuang M., Yang F.D., Wang C.Y., Yang J.L., Li K.Q., Ma X., Li L.J., Yu X., Li Q.Z., Huang X., Lv L.X., Li T., Zhao G.P., Huang W., Zhang X.J., Zhang D. Genome-wide association study identifies a susceptibility locus for schizophrenia in Han Chinese at 11p11.2. Nat. Genet. 2011;43(12):1228–1231. doi: 10.1038/ng.979. [DOI] [PubMed] [Google Scholar]
- Zhang R., Zhong N.N., Liu X.G., Yan H., Qiu C., Han Y., Wang W., Hou W.K., Liu Y., Gao C.G., Guo T.W., Lu S.M., Deng H.W., Ma J. Is the EFNB2 locus associated with schizophrenia? Single nucleotide polymorphisms and haplotypes analysis. Psychiatry Res. 2010;180(1):5–9. doi: 10.1016/j.psychres.2010.04.037. [DOI] [PubMed] [Google Scholar]
- Zhang F., Xu Y., Shugart Y.Y., Yue W., Qi G., Yuan G., Cheng Z., Yao J., Wang J., Wang G., Cao H., Guo W., Zhou Z., Wang Z., Tian L., Jin C., Yuan J., Liu C., Zhang D. Converging evidence implicates the abnormal microRNA system in schizophrenia. Schizophr. Bull. 2014;41(3):728–735. doi: 10.1093/schbul/sbu148. [DOI] [PMC free article] [PubMed] [Google Scholar]