Abstract
Human immunodeficiency virus type 1 (HIV-1) has evolved a sophisticated strategy to conceal conserved epitopes of its envelope glycoproteins (Env) recognized by antibody-dependent cellular cytotoxicity (ADCC)-mediating antibodies. These antibodies, which are present in the sera of most HIV-1-infected individuals, preferentially recognize Env in its CD4-bound conformation. Accordingly, recent studies showed that small CD4-mimetics (CD4mc) able to “push” Env into this conformation sensitize HIV-1-infected cells to ADCC mediated by HIV + sera. Here we test whether CD4mc also expose epitopes recognized by anti-cluster A monoclonal antibodies such as A32, thought to be responsible for the majority of ADCC activity present in HIV + sera and linked to decreased HIV-1 transmission in the RV144 trial. We made the surprising observation that CD4mc are unable to enhance recognition of HIV-1-infected cells by this family of antibodies in the absence of antibodies such as 17b, which binds a highly conserved CD4-induced epitope overlapping the co-receptor binding site (CoRBS). Our results indicate that CD4mc initially open the trimeric Env enough to allow the binding of CoRBS antibodies but not anti-cluster A antibodies. CoRBS antibody binding further opens the trimeric Env, allowing anti-cluster A antibody interaction and sensitization of infected cells to ADCC. Therefore, ADCC responses mediated by cluster A antibodies in HIV-positive sera involve a sequential opening of the Env trimer on the surface of HIV-1-infected cells. The understanding of the conformational changes required to expose these vulnerable Env epitopes might be important in the design of new strategies aimed at fighting HIV-1.
Keywords: HIV-1, Envelope glycoproteins, CD4, Non-neutralizing antibodies, ADCC, CD4-mimetics
Highlights
-
•
CD4-mimetics fail to enhance recognition of infected cells by anti-cluster A antibodies (Abs).
-
•
Co-receptor binding site Abs in conjunction with CD4-mimetics allow binding of Env by anti-cluster A Abs.
-
•
Co-receptor binding site Abs help CD4-mimetics sensitize HIV-1-infected cells to ADCC.
HIV-1 developed sophisticated strategies to conceal vulnerable epitopes of its envelope glycoproteins (Env) recognized by antibody-dependent cellular cytotoxicity (ADCC)-mediating antibodies. CD4-mimetics (CD4mc) were shown to sensitize HIV-1-infected cells to ADCC induced by HIV + sera. Here we show that this response requires a sequential opening of Env at the surface of HIV-1-infected cells. Co-receptor binding site antibodies, also present in HIV + sera, are required to expose ADCC-mediating epitopes recognized by anti-cluster A antibodies upon CD4mc addition. The understanding of the conformational changes required to expose anti-cluster A epitopes might be important in the design of new strategies aimed at fighting HIV-1.
Graphical Abstract
1. Introduction
Several lines of evidence support a role for antibody-dependent cellular cytotoxicity (ADCC) in controlling human immunodeficiency virus type 1 (HIV-1) infection and replication (Alpert et al., 2012, Banks et al., 2002, Baum et al., 1996, Chung et al., 2011, Forthal et al., 1999, Mabuka et al., 2012, Sun et al., 2011, Williams et al., 2015). Analysis of the correlates of protection in the RV144 vaccine trial suggested that decreased HIV-1 acquisition was linked to increased ADCC activity in protected vaccinees (Haynes et al., 2012). Accordingly, potent ADCC-mediating antibodies (Abs) targeting anti-cluster A epitopes were isolated from some RV144 vaccinees (Bonsignori et al., 2012) and were shown to preferentially recognize the HIV-1 envelope glycoproteins (Env) in their CD4-bound conformation (Veillette et al., 2014b). The CD4-bound Env conformation is also recognized by non-neutralizing CD4-induced (CD4i) ADCC-mediating antibodies present in sera (Veillette et al., 2015, Richard et al., 2015, Richard et al., 2016, Williams et al., 2015), breast milk (Richard et al., 2015) and cervicovaginal lavages (Batraville et al., 2014, Richard et al., 2015) of HIV-1-infected individuals. These antibodies represent a significant portion of the anti-Env Abs elicited during natural HIV-1 infection (Veillette et al., 2015, Decker et al., 2005). However, to limit the exposure of CD4-bound Env on the surface of infected cells, HIV-1 evolved sophisticated mechanisms to efficiently internalize Env (Von Bredow et al., 2015), to counteract the host restriction factor BST-2 with the viral Vpu protein (Arias et al., 2014, Alvarez et al., 2014, Veillette et al., 2014b), and to downregulate CD4 by Nef and Vpu (Veillette et al., 2015, Veillette et al., 2014b). The requirement to evade ADCC provides one explanation why the vast majority of circulating HIV-1 strains worldwide express functional Nef and Vpu proteins, which limit the exposure of CD4i Env epitopes on the surface of infected cells.
In agreement with the requirement for HIV-1 to avoid exposing the CD4-bound conformation of Env, we recently showed that forcing Env to adopt this conformation with small CD4-mimetics (CD4mc) sensitizes HIV-1-infected cells to ADCC mediated by non-neutralizing Abs present in sera, breast-milk and cervicovaginal fluids from HIV-1-infected subjects (Richard et al., 2015). These non-neutralizing ADCC-mediating Abs target gp120 epitopes that overlap the epitope recognized by the anti-cluster A A32 Ab (Ferrari et al., 2011, Guan et al., 2013, Veillette et al., 2015, Veillette et al., 2014b, Ding et al., 2016). But whether these anti-cluster A Abs respond to CD4mc is not yet known. Here we tested whether CD4mc expose epitopes recognized by anti-cluster A Abs and enhance the susceptibility of HIV-1-infected cells to ADCC mediated by this class of antibodies.
2. Materials and Methods
2.1. Cell Lines and Isolation of Primary Cells
293T human embryonic kidney cells (obtained from ATCC, Cat# CRL-3216, RRID:CVCL_0063), CEM.NKr cells (obtained from Dr. David Evans, Harvard Medical School) and primary cells were grown as previously described (Richard et al., 2010, Veillette et al., 2014b). CD4 T lymphocytes were purified from resting PBMCs by negative selection and activated as previously described (Richard et al., 2015).
2.2. Viral Production, Infections, Ex Vivo Amplification and Detection of Infected Cells
Vesicular stomatitis viruses G (VSVG)-pseudotyped viruses were produced and titrated as described (Veillette et al., 2015). Viruses were used to infect CEM.NKr cells or primary CD4 T cells from healthy donors by spin infection at 800 g for 1 h in 96-well plates at 25 °C. In order to expand endogenously-infected CD4 T cells, primary CD4 T cells were isolated from PBMCs obtained from viremic HIV-1-infected individuals. Purified CD4 + T cells were activated with PHA-L at 10 μg/ml for 36 h and then cultured for 7 days in RPMI-1640 complete medium supplemented with rIL-2 (100 U/ml) (obtained from the NIH AIDS Reagent Program, Cat# 136).
2.3. CD4-mimetics
The mini-protein M48U1 was produced and purified as previously described (Martin et al., 2003). The CD4-mimetic small molecules JP-III-48 and BNM-III-170 were synthesized as described (Melillo et al., 2016). The compounds were dissolved in dimethyl sulfoxide (DMSO) at a stock concentration of 10 mM, aliquoted, and stored at − 20 °C. Each compound was then diluted to 50–100 μM in PBS for cell-surface staining or in RPMI-1640 complete medium for ADCC assays.
2.4. Antibodies and Sera
The anti-cluster A mAbs A32, C11 and N5-i5 were conjugated with Alexa-Flour 647 probe (Thermo Fisher Scientific, Cat# A20186) as per the manufacturer instructions and used for cell-surface staining of HIV-1-infected primary CD4 + T cells at 0.35 μg/ml. The following anti-Env mAbs were used in combination of anti-cluster A mAbs during cell-surface staining: 48d, 412d, 19b (kindly provided by Dr. J. Robinson), VRC01, b12 (NIH AIDS Reagent, Cat# 12033 and 2640), CH58 (kindly provided by Dr. B. Haynes), C2 and the Fab fragments, Fab′2 fragments or full 17b and N12-i2 mAbs. Goat anti-human Alexa Fluor-647 mAbs (Thermo Fisher Scientific, Cat# A-21445 RRID:AB_2535862) were used as secondary Abs for sera binding and AquaVivid (Thermo Fisher Scientific, Cat# L43957) as a viability dye.
Sera from HIV-infected (Supplemental Table 1) and healthy donors were collected, heat-inactivated and conserved as previously described. Written informed consent was obtained from all study participants [the Montreal Primary HIV Infection Cohort (Fontaine et al., 2011, Fontaine et al., 2009) and the Canadian Cohort of HIV Infected Slow Progressors (Peretz et al., 2007, Kamya et al., 2011, International et al., 2010)], and research adhered to the ethical guidelines of CRCHUM and was reviewed and approved by the CRCHUM institutional review board (ethics committee). Research adhered to the standards indicated by the Declaration of Helsinki. All sera were heat-inactivated for 30 min at 56 °C and stored at 4 °C until ready to use in subsequent experiments. A random number generator (GraphPad, QuickCalcs) was used to randomly select a number of sera for each experiment.
2.5. Plasmids
pNL43-ADA(Env)-GFP.IRES.Nef proviral vector was previously described (Veillette et al., 2015). The plasmid encoding the HIV-1 transmitted founder (T/F) IMC CH58 was previously described (Ochsenbauer et al., 2012, Bar et al., 2012, Parrish et al., 2013, Fenton-May et al., 2013, Richard et al., 2015).
2.6. Flow Cytometry Analysis of Cell-surface Staining and ADCC Responses
Cell-surface staining was performed as previously described and MFI histograms shows signal on live infected populations (Richard et al., 2015, Veillette et al., 2015). Binding of HIV-1-infected cells by HIV + sera (1:1000 dilution) or Alexa-Fluor 647-conjugated anti-cluster A Abs A32, C11 and N5-i5 was performed 48 h after in vitro infection or 7 days post-activation for endogenously-infected (clade B) ex-vivo-amplified cells, in presence of CD4mc JP-III-48 (50 μM), BNM-III-170 (50uM), M48U1 (100 nM) or with equivalent volume of vehicle (DMSO). Binding of HIV-1-infected cells with Alexa-Fluor 647-conjugated anti-cluster A Abs A32, C11 and N5-i5 was performed alone or in combination with different mAbs or Fab fragments (5 μg/ml), or in combination with HIV- or HIV + sera (1:1000 dilution), with or without Fab fragments of 17b or N12-i2 (5 μg/ml). Detection of GFP + or p24 + infected cells was performed as described (Richard et al., 2015). The percentage of infected cells (GFP + or p24 + cells) was determined by gating the living cell population based on the viability dye staining (Aqua Vivid, Thermo Fisher Scientific, Cat# L43957). Samples were analyzed on a LSRII cytometer (BD Biosciences, Mississauga, ON, Canada) and data analysis was performed using FlowJo vX.0.7 (Tree Star, Ashland, OR, USA).
Measurement of ADCC-mediated killing was performed with a previously described assay (Richard et al., 2015). Briefly, primary CD4 + T cells infected for 48 h with the CH58 T/F virus were incubated with autologous PBMC (Effector: Target ratio of 10:1) in presence of A32 (0.3125, 0,625, 1,25 or 2,5 μg/ml, kindly provided by Dr. G. Ferrari) and 17b (5 μg/ml) or17b Fab fragments alone or in combination, or with HIV + sera (1:1000), in presence of CD4mc JP-III-48, BNM-III-170 or with equivalent volume of vehicle (DMSO). The percentage of cytotoxicity was calculated as described (Richard et al., 2015).
2.7. Statistical Analyses
Statistics were analyzed using GraphPad Prism version 6.01 (GraphPad, San Diego, CA,USA). Every data set was tested for statistical normality and this information was used to apply the appropriate (parametric or nonparametric) statistical test. P values < 0.05 were considered significant; significance values are indicated as *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
3. Results
3.1. CD4mc Fail to Enhance Recognition of HIV-1-infected Cells by ADCC-mediating Anti-cluster A Abs
We recently reported that small CD4mc enhance the recognition of HIV-1-infected cells by autologous and heterologous sera in both in vitro and ex vivo experiments (Richard et al., 2015, Lee et al., 2015). Accordingly, primary CD4 + T cells infected with a pNL4.3 full-length HIV-1 molecular clone coding for the primary ADA Env and a GFP reporter gene (NL4.3 ADA GFP) were better recognized by HIV + sera after the addition of rationally-designed small CD4mc compounds (JP-III-48, BNM-III-170) (Melillo et al., 2016) or CD4-mimetic peptides (M48U1) (Van Herrewege et al., 2008) (Fig. 1A). These molecules engage gp120 within the Phe43 cavity (Madani et al., 2008, Melillo et al., 2016) and can act as CD4 agonists, inducing thermodynamic changes in the Env trimer similar to those observed upon membrane CD4 binding (Schon et al., 2006, Lalonde et al., 2012). As previously reported (Ding et al., 2016, Richard et al., 2015, Veillette et al., 2015), Env present on the surface of cells infected with a wild-type (wt) virus is poorly recognized by HIV-1 + sera (Fig. 1A, C). This is due to efficient CD4 downregulation by the virus; Env cannot engage with CD4 and therefore remains in the unbound conformation, preventing CD4i epitope exposure (Veillette et al., 2015, Veillette et al., 2014a, Veillette et al., 2014b). However, CD4mc promote the exposure of Env CD4i epitopes, resulting in enhanced recognition of Env on the surface of HIV-1-infected cells by HIV-1 + sera known to contain high levels of CD4i antibodies (Fig. 1A, C) (Decker et al., 2005, Veillette et al., 2015).
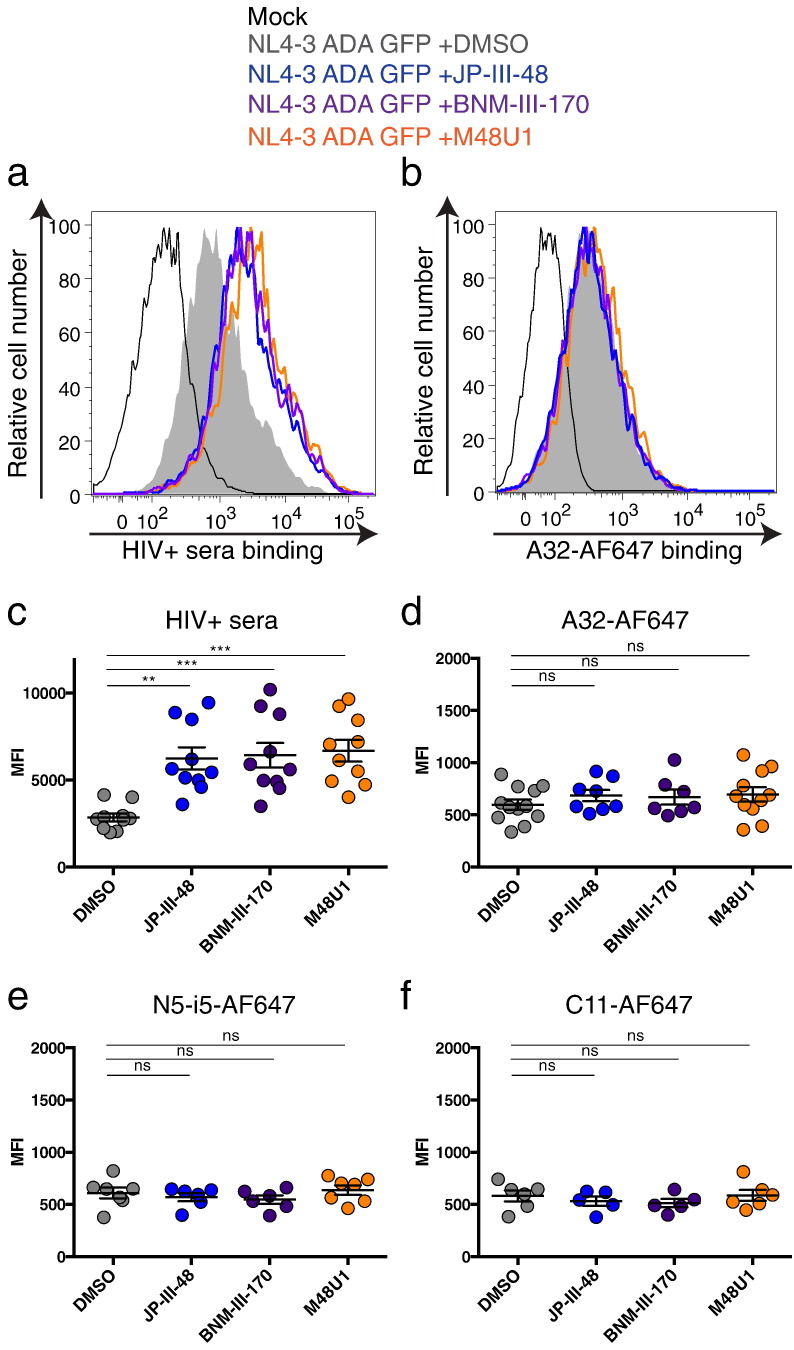
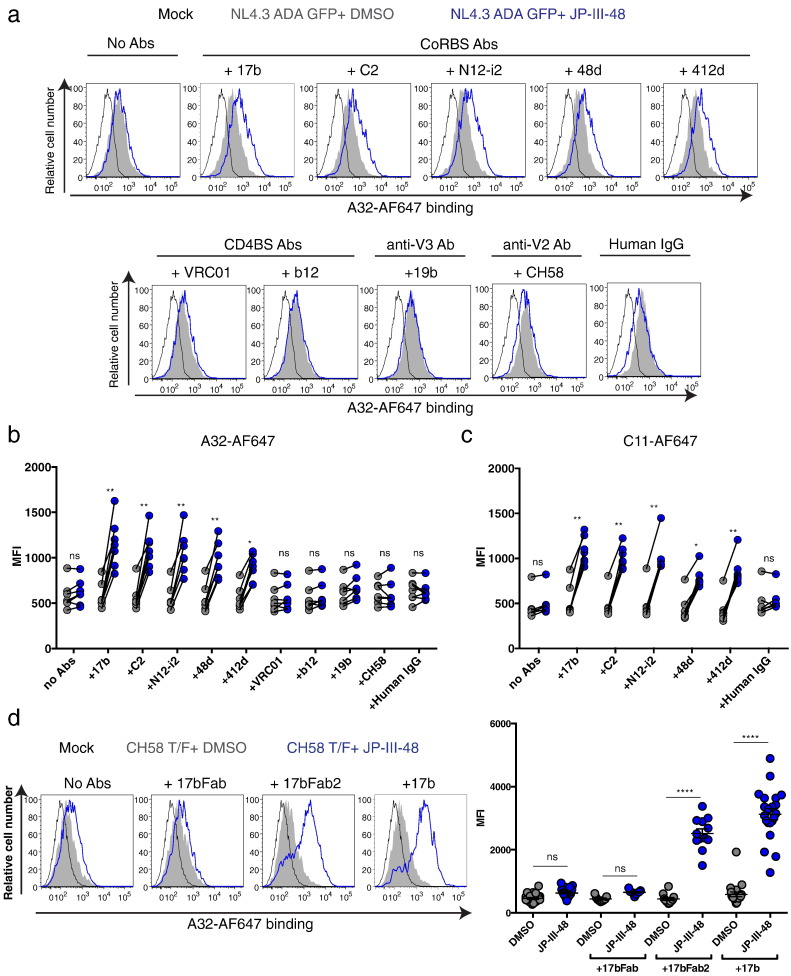
Fig. 1.
CD4mc fail to enhance recognition of HIV-1-infected cells by anti-cluster A antibodies.
Primary CD4 + T cells, which were either mock-infected or infected with NL4.3 GFP expressing the primary R5 ADA Env (NL4.3 ADA GFP), were stained with sera from 10 HIV-1-infected individuals followed by an anti-human Alexa-Fluor 647 (AF647)-conjugated secondary antibody (a, c) or with (b, d) AF647-conjugated anti-cluster A (b, d) A32, (e) N5-i5 or (f) C11, in the presence of CD4mc JP-III-48, BNM-III-170, M48U1 or DMSO (vehicle). Shown are histograms depicting representative staining obtained with (a) HIV + sera or (b) A32-AF647 and (c-f) the mean fluorescence intensities (MFI) obtained from at least three experiments. Error bars indicate mean ± SEM. Statistical significance was tested using a Kruskal-Wallis with a Dunn's post-test (**p < 0.01, ***p < 0.001, ns: non-significant).
It has been previously shown that ADCC-mediating anti-cluster A Abs such as A32 recognize a highly-conserved region in gp120 that is buried inside the Env trimer on the surface of HIV-1-infected (p24 +) cells and is not readily accessible in the ligand-free closed state (Veillette et al., 2014b, Ding et al., 2016, Acharya et al., 2014, Von Bredow et al., 2016, Tolbert et al., 2016). This explains why the A32 antibody recognizes HIV-1-infected cells poorly in the absence of CD4mc (Fig. 1B, D), yet efficiently recognizes shed gp120 bound to CD4 on the surface of CD4 + uninfected bystander cells present in cultures of HIV-1-infected cells (Richard et al., 2016). We therefore explored the capacity of different CD4mc to promote the CD4-bound conformation of Env and thereby enhance Env recognition on the surface of HIV-1-infected cells by the anti-cluster A class of antibodies. Surprisingly, none of the CD4mc tested (JP-III-48, BNM-III-170, M48U1) were able to enhance recognition of wild-type HIV-1-infected cells by anti-Cluster A Abs (A32, N5-i5, C11) (Fig. 1 B, D–F) at concentrations that were sufficient to enhance recognition by HIV + sera (Fig. 1A, C). This is in contrast with the effects of membrane-bound CD4, which has been shown to enable A32 recognition (Veillette et al., 2014b). Apparently, the conformational changes in Env induced by CD4mc differ qualitatively or quantitatively from those induced by membrane-bound CD4.
3.2. CD4mc Enhance Recognition of HIV-1-infected Cells by Anti-cluster A Abs in the Presence of HIV + Sera
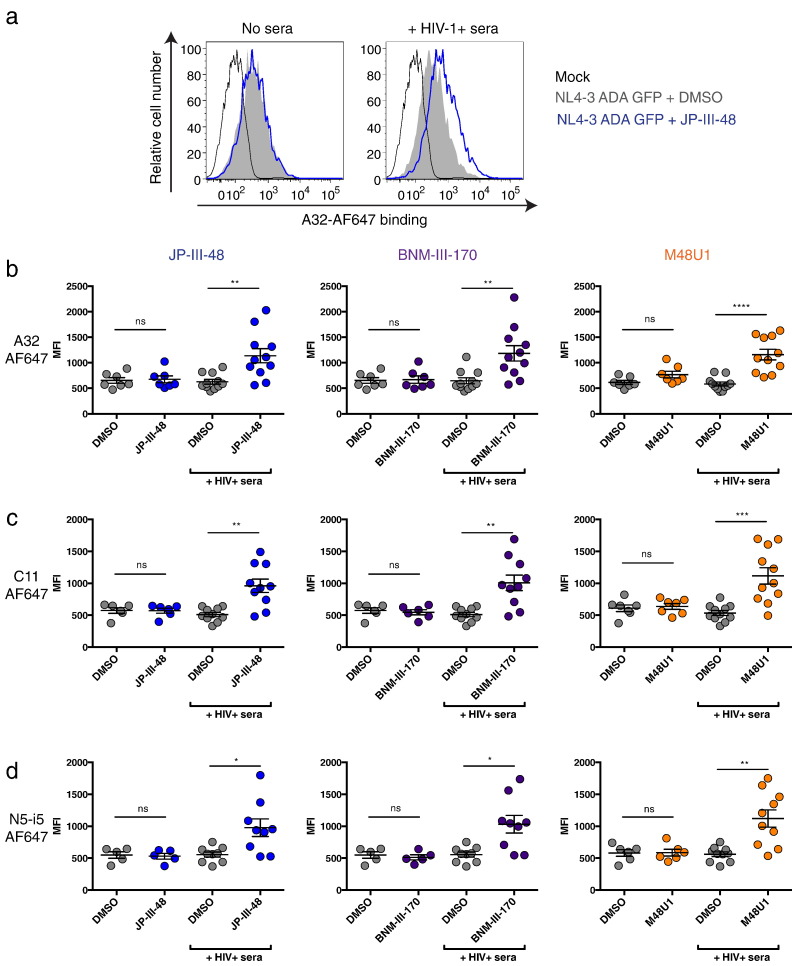
It was previously shown that the ADCC activity present in sera from chronically HIV-1-infected individuals could be blocked by an A32 Fab fragment (Ferrari et al., 2011). Subsequent studies showed that the majority of ADCC responses were targeted against the conserved gp120 core but not the gp120 variable regions V1, V2, V3 and V5 (Veillette et al., 2015). Some ADCC responses depended on the highly-conserved tryptophan 69 (W69) of the gp120 inner domain, required for some anti-cluster A antibody binding (Ding et al., 2016, Tolbert et al., 2016). Therefore, if the ADCC activity present in HIV + sera is mediated by anti-cluster A Abs and CD4mc enhance this activity, why then do CD4mc fail to augment recognition of Env by anti-cluster A antibodies? We hypothesized that a different class of antibodies present in the sera of HIV-1-infected individuals enabled anti-cluster A antibodies to recognize Env in the presence of CD4mc. To test this possibility, we infected primary CD4 + T cells with the NL4.3 ADA GFP virus and evaluated the ability of HIV + sera to enhance the recognition of these HIV-1-infected cells by anti-cluster A antibodies upon CD4mc addition. Monoclonal A32 and N5-i5 antibodies recognize an epitope located at the interface of the gp120 inner domain Layers 1 and 2 (Veillette et al., 2014a, Finzi et al., 2010, Acharya et al., 2014, Tolbert et al., 2016), whereas C11 recognizes the N- and C-termini of gp120 (Finzi et al., 2010, Moore et al., 1994, Gohain et al., 2015). To unequivocally measure the binding capacity of these antibodies to HIV-1-infected cells in the presence of CD4mc, with and without co-incubation with HIV + sera, A32, N5-i5 and C11 were conjugated to the Alexa-Fluor 647 probe. Remarkably, the addition of a 1/1000 dilution of sera from 10 HIV-1-infected individuals but not from healthy donors enhanced recognition of infected cells by these antibodies in the presence of all three CD4mc (Fig. 2 and Supplemental Fig. 1). Importantly, similar results were obtained with the unmodified clinically-relevant primary transmitted/founder (T/F) virus CH58 (CH58 T/F) and endogenously-infected ex-vivo-amplified primary CD4 + T cells from HIV-1-infected individuals (Fig. 3), indicating that the phenomenon is not restricted to laboratory-adapted viruses. These results suggest that some antibodies present in the sera of HIV-1-infected individuals facilitate the exposure of anti-cluster A epitopes upon CD4mc addition.
Fig. 2.
Sera from HIV-1-infected individuals enable recognition of HIV-1-infected cells by anti-cluster A antibodies upon CD4mc addition.
Cell-surface staining of primary CD4 + T cells either mock-infected or infected with NL4.3 GFP expressing the primary R5 ADA Env with AF647-conjugated anti-cluster A antibodies (a–b) A32, (c) C11 or (d) N5-i5, in the presence or absence of HIV + sera and CD4mc JP-III-48, BNM-III-170, M48U1 or DMSO. Shown in (a) are histograms depicting representative staining obtained with A32-AF647 in the presence the CD4mc JP-III-48 or DMSO. Shown in (b–d) are the mean fluorescence intensities (MFI) obtained for staining obtained in at least 5 experiments with (b) A32-AF647, (c) C11-AF647 and (d) N5-i5-AF647 in the presence or absence of 10 different HIV + sera. Error bars indicate mean ± SEM. Statistical significance was tested using a Kruskal-Wallis with a Dunn's post-test (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns: non-significant). See also Fig. S1.
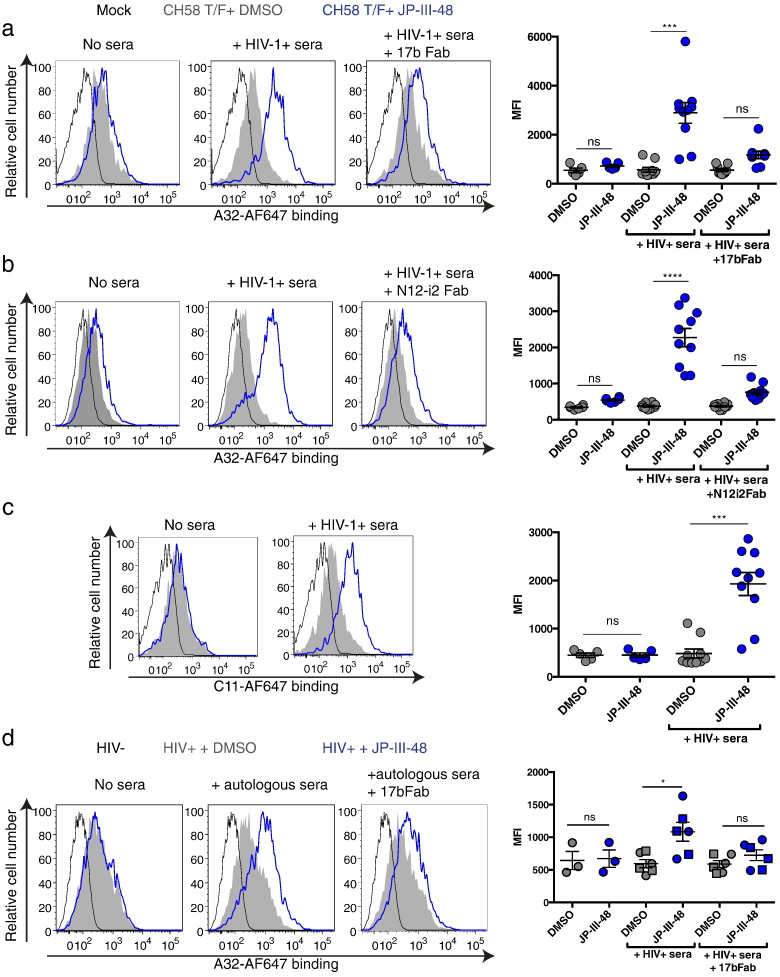
Fig. 3.
Co-receptor binding site Fab fragments reduce HIV + sera ability to enhance recognition of infected cells in the presence of CD4mc.
(a–c) Cell-surface staining of primary CD4 + T cells either mock-infected or infected with CH58 T/F with AF647 anti-cluster A antibodies (a–b) A32 or (c) C11, in the presence or not of HIV + sera, with or without the Fab fragment of (a) 17b or (b) N12-i2 and CD4mc JP-III-48 or DMSO (vehicle). Shown in (a–c) are (left) histograms depicting representative staining and (right) the mean fluorescence intensities (MFI) obtained for staining obtained in at least 5 experiments in the presence of 10 different HIV + sera. (d) Cell-surface staining of primary CD4 + T cells isolated from 3 HIV-1-infected individuals after ex-vivo expansion with AF647-A32 in the presence or not of autologous (circle), an HIV + heterologous (square) sera, with or without the 17b Fab fragment and CD4mc JP-III-48 or DMSO. Left panels show histograms depicting representative staining with autologous sera. Right panels present the mean fluorescence intensities (MFI) obtained for multiple staining. Error bars indicate mean ± SEM. Statistical significance was tested using a Kruskal-Wallis with a Dunn's post-test (*p < 0.05, ***p < 0.001, ****p < 0.0001, ns: non-significant).
3.3. CoRBS Abs Allow Recognition of HIV-1-infected Cells by Anti-cluster A Abs
In the same report describing the ADCC blocking capacity of the A32 Fab (Ferrari et al., 2011), the Fab fragment from the 17b antibody, which belongs to the co-receptor binding site (CoRBS) family of anti-Env antibodies (Wyatt et al., 1995), was able to block the ADCC activity in approximately 60% of the HIV + sera tested. In some cases, the blocking effect was comparable to that obtained with the A32 Fab (Ferrari et al., 2011). To explore the possibility that CoRBS antibodies present in sera from HIV-1-infected individuals facilitate Env recognition by anti-cluster A antibodies in the presence of CD4mc, we added the Fab fragments of 17b or another CoRBS antibody, N12-i2 (Guan et al., 2013), to 10 HIV + sera. In all the HIV + sera tested, addition of CoRBS Fab fragments decreased the recognition of cells infected with CH58 T/F viruses (Fig. 3 A–B) and endogenously-infected ex-vivo-amplified primary CD4 + T cells from three HIV-1-infected individuals (Fig. 3D) by Alexa-Fluor 647-conjugated A32 Abs (A32-AF647). These observations suggest that CoRBS antibodies facilitate the access of anti-cluster A antibodies to trimeric Env in the presence of CD4mc.
To test this conclusion directly, we co-incubated primary CD4 + T cells infected with NL4.3 ADA GFP or CH58 T/F viruses in the presence or absence of CD4mc with the full 17b antibody and evaluated A32-AF647 and C11-AF647 recognition of infected cells. Incubation with the 17b antibody was sufficient to allow recognition of HIV-1-infected cells by these two anti-cluster A antibodies (Fig. 4 A–C). Importantly, while this was recapitulated by the 17b Fab′2 fragment, the 17b Fab fragment was unable to do so (Fig. 4D). This was reiterated with a different CoRBS (N12-i2) antibody and Fab fragments (Supplemental Fig. 2), indicating that the bivalent recognition of Env by CoRBS antibodies may be required to facilitate anti-cluster A antibody binding in the presence of CD4mc. Of note, similar to the CD4mc JP-III-48, soluble CD4 (sCD4) alone was unable to facilitate A32 recognition of CH58 T/F-infected primary CD4 + T cells (Supplemental Fig. 3A). Interestingly, the 17b CoRBS antibody allowed recognition of CH58 T/F-infected primary CD4 + T cells by the A32 antibody in the presence of JP-III-48, but not in the presence of sCD4 (Supplemental Fig. 3B). This is in agreement with our previous observations indicating that CD4mc but not sCD4 sensitize primary CD4 + T cells infected with primary HIV-1 viruses to ADCC (Richard et al., 2015).
Fig. 4.
Co-receptor binding site antibodies facilitate anti-cluster A antibody recognition of HIV-1-infected cells in the presence of CD4mc.
(a–c) Cell-surface staining of primary CD4 + T cells either mock-infected or infected with NL4.3 GFP expressing the primary R5 ADA Env with AF647-conjugated (a–b) A32 or (c) C11, in the presence or not of co-receptor binding site (CoRBS) mAbs 17b, C2, N12-i2, 48d or 412d, CD4 binding site (CD4BS) mAbs VRC01 or b12, anti-V3 mAbs 19b, anti-V2 mAbs CH58 or human IgG, with CD4mc JP-III-48 or DMSO. Shown in (a) are histograms depicting representative A32-AF647 staining and (b–c) the mean fluorescence intensities (MFI) obtained for at least 7 different staining. (d) Cell-surface staining of primary CD4 + T cells either mock-infected or infected with CH58 T/F with AF647-conjugated A32 in the presence or not of the Fab fragment, the Fab′2 fragment or the full 17b with CD4mc JP-III-48 or DMSO. Shown in (d) are (left) histograms depicting representative staining and (right) the MFI for at least 9 different staining. Error bars indicate mean ± SEM. Statistical significance was tested using a Kruskal-Wallis with a Dunn's post-test (*p < 0.05, **p < 0.01, ****p < 0.0001, ns: non-significant). See also Fig. S2, S3 and S4.
To extend these observations to additional CoRBS antibodies, we evaluated the ability of well-established CoRBS antibodies (48d, 412d) or a newly-isolated CoRBS antibody from a clade-B HIV-1-infected individual (C2, Supplemental Fig. 4) to allow A32-AF647 or C11-AF647 binding. All tested CoRBS antibodies facilitated A32-AF647 and C11-AF647 binding upon CD4mc addition (Fig. 4 A–C). We also evaluated whether this activity was shared by antibodies with different Env specificities. We tested anti-V3 (19b), anti-V2 (CH58) and anti-CD4 binding site (VRC01, b12) antibodies. These antibodies did not facilitate A32 recognition of infected cells in the presence of CD4mc (Fig. 4 A–B). These observations suggest that antibodies that recognize CD4-induced epitopes near the CoRBS specifically enhance the recognition of the HIV-1 Env trimer by anti-cluster A antibodies in the presence of CD4mc. It remains possible that antibodies with different Env specificities, present in HIV + sera, might also contribute to the exposure of epitopes recognized by anti-cluster A antibodies.
3.4. CoRBS Abs Are Required for CD4mc to Sensitize Infected Cells to ADCC Mediated by Anti-cluster A Abs
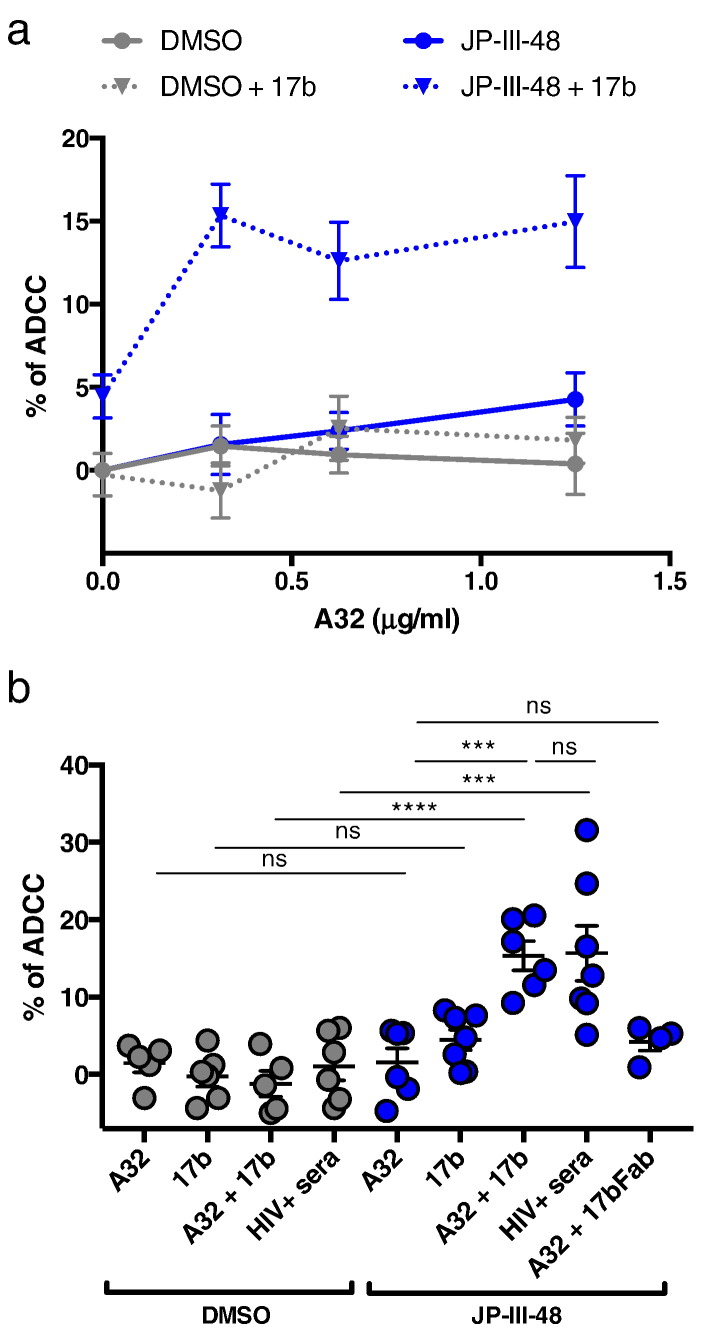
To evaluate whether the enhanced recognition of HIV-1-infected cells by anti-cluster A antibodies in presence of CD4mc and CoRBS mAbs resulted in ADCC killing, we infected primary CD4 + T cells with CH58 T/F virus and evaluated their susceptibility to ADCC mediated by autologous PBMCs using a previously-described FACS-based assay (Richard et al., 2014, Richard et al., 2015). As expected, A32 and 17b alone or in combination were unable to mediate ADCC (Fig. 5). Recognition of HIV-1-infected cells by the CoRBS 17b antibody upon CD4mc addition (Supplemental Fig. 5) did not translate into enhanced ADCC killing (Fig. 5). This is in agreement with recent reports indicating that CoRBS antibodies do not mediate efficient ADCC, despite good recognition of target cells when Env samples the CD4-bound conformation (Ding et al., 2016, Guan et al., 2013). However, when the anti-cluster A A32 and CoRBS 17b antibodies were added, at different concentrations, in conjunction with CD4mc JP-III-48 or BNM-III-170, we observed a marked increase in the killing of HIV-1-infected cells (Fig. 5 and Supplemental Fig. 6), reminiscent of the CD4mc sensitization of infected cells to ADCC mediated by HIV + sera (Richard et al., 2015) (Fig. 5B). Importantly, this response was only observed when the full 17b antibody but not its Fab fragment was added in combination with A32 in the presence of CD4mc (Fig. 5B). Thus, CD4mc and CoRBS antibodies participate in the formation of a suitable target for ADCC responses mediated by anti-cluster A antibodies.
Fig. 5.
CD4-mimetic sensitize HIV-1-infected cells to ADCC-mediated by anti-cluster A antibodies in the presence of CoRBS antibodies.
Primary CD4 T cells infected with CH58 T/F were used as target cells and autologous PBMC as effector cells in our FACS-based ADCC assay. Shown in (a) are the percentages of ADCC-mediated killing obtained with serial dilution of A32 (0.3125, 0.625, and 1.25 μg/ml) alone or in combination with 17b (5 μg/ml), in the presence of CD4mc JP-III-48 or equivalent volume of DMSO for 7 different experiments. Shown in (b) are the percentages of ADCC-mediated killing obtained with A32 (0.3125 μg/ml), 17b, 17b Fab, alone or in combination, or with sera from an HIV + individual, in the presence of JP-III-48 or equivalent volume of DMSO. Error bars indicate mean ± SEM. Statistical significance was tested using an Ordinary one-way ANOVA with a Holm-Sidak's post-test (***p < 0.001, ****p < 0.0001, ns: non-significant), See also Fig. S5 and S6.
4. Discussion
HIV-1 has evolved several mechanisms to avoid the robust humoral response elicited against its envelope glycoproteins, including sequence-variable loops, extensive glycosylation, and conformational masking of vulnerable epitopes (Kwong et al., 2002, Wyatt et al., 1998, Wyatt and Sodroski, 1998). While the majority of the antibodies elicited during natural infection are strain-specific neutralizing or non-neutralizing antibodies (reviewed in Kwong and Mascola, 2012) and thought to play only a minimal role in controlling viral replication, recent studies showed that these antibodies can exert a constant selection pressure through their weakly neutralizing activities and thus drive viral evolution (Moody et al., 2015). In addition to their non- or weakly-neutralizing activities, these antibodies possess Fc-mediated effector functions with the potential to eliminate HIV-1-infected cells through several immune responses, including ADCC. The analysis of the correlates of protection in the RV144 vaccine trial suggesting that decreased HIV-1 acquisition was linked to increased ADCC activity in protected vaccinees (Haynes et al., 2012) spurred a renewed interest in these Fc-mediated functions (Chung et al., 2015, Ackerman et al., 2016, Williams et al., 2015, Veillette et al., 2014b, Ding et al., 2016, Richard et al., 2015, Veillette et al., 2015, Richard et al., 2016, Ferrari et al., 2011, Von Bredow et al., 2016). These ADCC-mediating antibodies appear to preferentially target Env in its CD4-bound conformation (Ding et al., 2016, Veillette et al., 2015). In other words, these antibodies primarily recognize Env epitopes exposed upon interaction with the viral receptor CD4. To avoid exposing these epitopes at the surface of infected cells, HIV-1 limits the amount of Env and CD4 present at the cell surface (reviewed in Veillette et al., 2016).
Small CD4mc with the capacity to “push” Env to the CD4-bound conformation were recently shown to enhance the recognition of HIV-1-infected cells by sera, cervicovaginal lavages and breast milk from HIV-1-infected individuals (Richard et al., 2015). However, the specificity of the antibodies mediating this response in these complex biological fluids was not known. Previous reports indicated that the ADCC activity present in HIV + sera is mediated by anti-cluster A antibodies (Guan et al., 2013, Ferrari et al., 2011, Ding et al., 2016). Accordingly, competition experiments using a gp120 inner domain recombinant protein exposing only A32-like epitopes showed the importance of A32-like antibodies in ADCC responses mediated by HIV + sera (Tolbert et al., 2016). We therefore evaluated whether CD4mc were able to sensitize HIV-1-infected cells to ADCC by this family of antibodies. Surprisingly, all CD4mc tested (M48U1, JP-III-48, BNM-III-170) failed to enhance recognition of HIV-1 Env present on the surface of infected cells by anti-cluster A antibodies. Interestingly, we found that HIV + sera enabled recognition of Env by anti-cluster A antibodies in the presence of CD4mc. Importantly, this depended on the presence of CoRBS antibodies. Indeed, 17b and N12-i2 Fab fragments were able to negate the effect of HIV + sera on Env recognition by these antibodies. Moreover, several CoRBS antibodies but not anti-V2, anti-V3 or anti-CD4bs antibodies were able to recapitulate the effect of HIV + plasma in the presence of CD4mc; and the CoRBS antibodies facilitated ADCC-mediated killing of HIV-1-infected cells by the anti-cluster A family of antibodies. Interestingly, the Fab′2 but not Fab fragments of CoRBS antibodies were able to recapitulate the effect of full antibodies, indicating that the bivalent recognition of Env by these antibodies is required to facilitate anti-cluster A antibody binding in the presence of CD4mc.
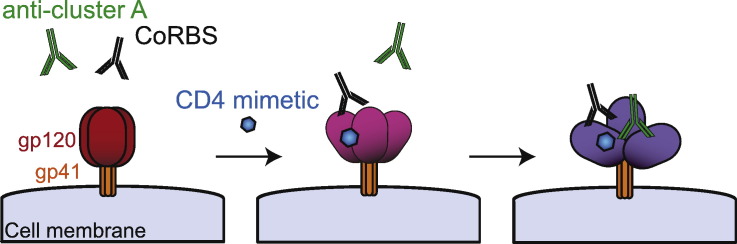
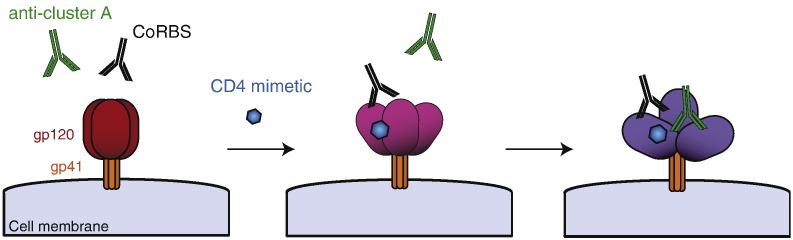
The unexpected requirement for CoRBS antibodies to allow Env recognition by anti-cluster A antibodies in the presence of CD4mc suggests a model for the sequential opening of trimeric Env. In this model (Fig. 6), CD4mc engagement in the Phe43 cavity induces a partial opening of trimeric Env, which is sufficient to expose the CoRBS but not enough to expose inner domain cluster-A epitopes. Interaction of CoRBS antibodies with two gp120 subunits within this CD4mc-sensitized trimer further opens the trimeric Env and exposes highly-conserved epitopes recognized by anti-cluster A antibodies. It is unclear why a bivalent recognition of Env by CoRBS is required to expose anti-cluster A antibodies in the presence of CD4mc. This could be related to a potential asymmetric transition of Env upon CD4mc engagement towards downstream conformations, as suggested for CD4 binding (Personal communication, Ma, Mothes, Munro). While in theory CD4mc should induce conformational changes in all Envs with an available Phe 43 cavity (the majority of HIV-1 strains), the results presented here have been generated using clade B HIV-1 strains only. How these findings extend to additional HIV-1 strains remains to be evaluated.
Fig. 6.
Sequential opening of Env required for anti-cluster A antibody binding.
CD4mc induce a partial opening of trimeric Env allowing subsequent exposure of the co-receptor binding site. Binding of Env by CoRBS antibodies induce additional conformational changes in Env resulting in the exposure of highly-conserved epitopes recognized by anti-cluster A antibodies. Env exposing anti-cluster A epitopes become a suitable target for ADCC.
We previously reported that HIV-1 avoids exposing Env epitopes of anti-cluster A antibodies through CD4 downregulation by Nef and Vpu accessory proteins (Veillette et al., 2014b, Ding et al., 2016, Veillette et al., 2015, Alsahafi et al., 2016). Our findings herein indicate that several conformational transitions are required to expose these epitopes and suggest that trimeric HIV-1 Env has evolved to minimize their exposure. This represents another example of conformational masking of vulnerable epitopes and helps to explain why the A32 antibody failed to provide sterilizing protection in non-human primates (Santra et al., 2015). Why has HIV-1 evolved multiple mechanisms to avoid anti-cluster A epitope exposure? This highly-conserved region contributes to Env stability (Finzi et al., 2010), so its buried nature may be critical for preserving the global architecture of the HIV-1 Env trimer. This architecture has evolved to minimize the exposure of this and other elements that represent suitable targets for ADCC.
Altogether, our data highlights the difficulty of exposing anti-cluster A epitopes at the surface of infected cells but also indicates that sera from HIV-1-infected individuals have all the components required to do so, provided that the initial opening of the trimer by CD4mc occurs. While it is unknown at the moment whether CD4mc will ultimately be used therapeutically to boost ADCC activity in vivo, the understanding of the conformational changes required to expose anti-cluster A epitopes might expedite the design and application of new strategies aimed at fighting HIV-1.
The following are the supplementary data related to this article.
Characteristics of HIV-infected sera donors.
Supplementary figures.
Funding Sources
This work was supported by a Canada Foundation for Innovation Program Leader grant #29866, by CIHR foundation grant #352417, by an amfAR Innovation Grant #109343-59-RGRL to A.F. and by the FRQS AIDS and Infectious Diseases Network. A.F. is the recipient of a Canada Research Chair on Retroviral Entry. M.V. was supported by a CIHR Doctoral Research Award #291485. J.R. is the recipient of a CIHR Fellowship Award #135349. N.A. is the recipient of a King Abdullah scholarship for higher education from the Saudi Government. D.E.K. and C.T. are supported by a Research Scholar Career Award of the Quebec Health Research Fund (FRQS). This study was also supported by CIHR MOP93770, NIH AI100645 and AI100663 Center for HIV/AIDS Vaccine Immunology and Immunogen Design (CHAVI-ID), by the National Institutes of Health grant #GM56550, the late William F. McCarty-Cooper, NIH R01AI116274 and the Bill and Melinda Gates Foundation #OPP1033109. Our funding sources had no role in data collection, analysis or interpretation, and were not involved in the writing of this manuscript.
Conflicts of Interest
The authors have no conflicts of interest to report.
Author Contributions
J.R. and A.F. conceived and designed the experiments; J.R., B.P., N.G., M.V., S.D., N.A., W.D.T., J.P., J.P.C., M.C., M.J. and N.B. performed the experiments; J.R., B.P., N.G., M.V., S.D., N.A., W.D.T., J.P., J.P.C., M.C., M.J., J.P., J.R.C., B.M., L.M., C.T., B.H.H., D.E.K., X.W., A.B.S, J.S., M.P. and A.F. contributed unique reagents and analyzed the data. J.R. J.S. and A.F. wrote the paper.
Acknowledgments
We thank Elizabeth Carpelan for help with manuscript preparation. The authors thank Dominique Gauchat from the CRCHUM Flow Cytometry Platform for technical assistance, Mario Legault for cohort coordination and clinical samples, Walther Mothes and James Munro for helpful discussions, Barton Haynes for CH58 and James Robinson for A32, 48d, 412d and 19b antibodies.
Contributor Information
Jonathan Richard, Email: jonathan.richard.1@umontreal.ca.
Andrés Finzi, Email: andres.finzi@umontreal.ca.
References
- Acharya P., Tolbert W.D., Gohain N., Wu X., Yu L., Liu T., Huang W., Huang C.C., Kwon Y.D., Louder R.K., Luongo T.S., Mclellan J.S., Pancera M., Yang Y., Zhang B., Flinko R., Foulke J.S., Jr., Sajadi M.M., Kamin-Lewis R., Robinson J.E., Martin L., Kwong P.D., Guan Y., Devico A.L., Lewis G.K., Pazgier M. Structural definition of an antibody-dependent cellular cytotoxicity response implicated in reduced risk for HIV-1 infection. J. Virol. 2014;88:12895–12906. doi: 10.1128/JVI.02194-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman M.E., Mikhailova A., Brown E.P., Dowell K.G., Walker B.D., Bailey-Kellogg C., Suscovich T.J., Alter G. Polyfunctional HIV-specific antibody responses are associated with spontaneous HIV control. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpert M.D., Harvey J.D., Lauer W.A., Reeves R.K., Piatak M., Jr., Carville A., Mansfield K.G., Lifson J.D., Li W., Desrosiers R.C., Johnson R.P., Evans D.T. ADCC develops over time during persistent infection with live-attenuated SIV and is associated with complete protection against SIV(mac)251 challenge. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsahafi N., Ding S., Richard J., Markle T., Brassard N., Walker B., Lewis G.K., Kaufmann D.E., Brockman M.A., Finzi A. Nef proteins from HIV-1 elite controllers are inefficient at preventing antibody-dependent cellular cytotoxicity. J. Virol. 2016;90:2993–3002. doi: 10.1128/JVI.02973-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez R.A., Hamlin R.E., Monroe A., Moldt B., Hotta M.T., Rodriguez Caprio G., Fierer D.S., Simon V., Chen B.K. HIV-1 Vpu antagonism of tetherin inhibits antibody-dependent cellular cytotoxic responses by natural killer cells. J. Virol. 2014;88:6031–6046. doi: 10.1128/JVI.00449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias J.F., Heyer L.N., Von Bredow B., Weisgrau K.L., Moldt B., Burton D.R., Rakasz E.G., Evans D.T. Tetherin antagonism by Vpu protects HIV-infected cells from antibody-dependent cell-mediated cytotoxicity. Proc. Natl. Acad. Sci. U. S. A. 2014;111:6425–6430. doi: 10.1073/pnas.1321507111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks N.D., Kinsey N., Clements J., Hildreth J.E. Sustained antibody-dependent cell-mediated cytotoxicity (ADCC) in SIV-infected macaques correlates with delayed progression to AIDS. AIDS Res. Hum. Retrovir. 2002;18:1197–1205. doi: 10.1089/08892220260387940. [DOI] [PubMed] [Google Scholar]
- Bar K.J., Tsao C.Y., Iyer S.S., Decker J.M., Yang Y., Bonsignori M., Chen X., Hwang K.K., Montefiori D.C., Liao H.X., Hraber P., Fischer W., Li H., Wang S., Sterrett S., Keele B.F., Ganusov V.V., Perelson A.S., Korber B.T., Georgiev I., Mclellan J.S., Pavlicek J.W., Gao F., Haynes B.F., Hahn B.H., Kwong P.D., Shaw G.M. Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batraville L.A., Richard J., Veillette M., Labbe A.C., Alary M., Guedou F., Kaufmann D.E., Poudrier J., Finzi A., Roger M. Short communication: anti-HIV-1 envelope immunoglobulin Gs in blood and cervicovaginal samples of Beninese commercial sex workers. AIDS Res. Hum. Retrovir. 2014;30:1145–1149. doi: 10.1089/aid.2014.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum L.L., Cassutt K.J., Knigge K., Khattri R., Margolick J., Rinaldo C., Kleeberger C.A., Nishanian P., Henrard D.R., Phair J. HIV-1 gp120-specific antibody-dependent cell-mediated cytotoxicity correlates with rate of disease progression. J. Immunol. 1996;157:2168–2173. [PubMed] [Google Scholar]
- Bonsignori M., Pollara J., Moody M.A., Alpert M.D., Chen X., Hwang K.K., Gilbert P.B., Huang Y., Gurley T.C., Kozink D.M., Marshall D.J., Whitesides J.F., Tsao C.Y., Kaewkungwal J., Nitayaphan S., Pitisuttithum P., Rerks-Ngarm S., Kim J.H., Michael N.L., Tomaras G.D., Montefiori D.C., Lewis G.K., Devico A., Evans D.T., Ferrari G., Liao H.X., Haynes B.F. Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J. Virol. 2012;86:11521–11532. doi: 10.1128/JVI.01023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung A.W., Isitman G., Navis M., Kramski M., Center R.J., Kent S.J., Stratov I. Immune escape from HIV-specific antibody-dependent cellular cytotoxicity (ADCC) pressure. Proc. Natl. Acad. Sci. U. S. A. 2011;108:7505–7510. doi: 10.1073/pnas.1016048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung A.W., Kumar M.P., Arnold K.B., Yu W.H., Schoen M.K., Dunphy L.J., Suscovich T.J., Frahm N., Linde C., Mahan A.E., Hoffner M., Streeck H., Ackerman M.E., Mcelrath M.J., Schuitemaker H., Pau M.G., Baden L.R., Kim J.H., Michael N.L., Barouch D.H., Lauffenburger D.A., Alter G. Dissecting polyclonal vaccine-induced humoral immunity against HIV using systems serology. Cell. 2015;163:988–998. doi: 10.1016/j.cell.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker J.M., Bibollet-Ruche F., Wei X., Wang S., Levy D.N., Wang W., Delaporte E., Peeters M., Derdeyn C.A., Allen S., Hunter E., Saag M.S., Hoxie J.A., Hahn B.H., Kwong P.D., Robinson J.E., Shaw G.M. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 2005;201:1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Veillette M., Coutu M., Prevost J., Scharf L., Bjorkman P.J., Ferrari G., Robinson J.E., Sturzel C., Hahn B.H., Sauter D., Kirchhoff F., Lewis G.K., Pazgier M., Finzi A. A highly conserved residue of the HIV-1 gp120 inner domain is important for antibody-dependent cellular cytotoxicity responses mediated by anti-cluster A antibodies. J. Virol. 2016;90:2127–2134. doi: 10.1128/JVI.02779-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton-May A.E., Dibben O., Emmerich T., Ding H., Pfafferott K., Aasa-Chapman M.M., Pellegrino P., Williams I., Cohen M.S., Gao F., Shaw G.M., Hahn B.H., Ochsenbauer C., Kappes J.C., Borrow P. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology. 2013;10:146. doi: 10.1186/1742-4690-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari G., Pollara J., Kozink D., Harms T., Drinker M., Freel S., Moody M.A., Alam S.M., Tomaras G.D., Ochsenbauer C., Kappes J.C., Shaw G.M., Hoxie J.A., Robinson J.E., Haynes B.F. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J. Virol. 2011;85:7029–7036. doi: 10.1128/JVI.00171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi A., Xiang S.H., Pacheco B., Wang L., Haight J., Kassa A., Danek B., Pancera M., Kwong P.D., Sodroski J. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol. Cell. 2010;37:656–667. doi: 10.1016/j.molcel.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine J., Coutlee F., Tremblay C., Routy J.P., Poudrier J., Roger M., Montreal Primary, H. I. V. I., Long-Term Nonprogressor Study, G. HIV infection affects blood myeloid dendritic cells after successful therapy and despite nonprogressing clinical disease. J. Infect. Dis. 2009;199:1007–1018. doi: 10.1086/597278. [DOI] [PubMed] [Google Scholar]
- Fontaine J., Chagnon-Choquet J., Valcke H.S., Poudrier J., Roger M., Montreal Primary, H. I. V. I., Long-Term Non-Progressor Study, G. High expression levels of B lymphocyte stimulator (BLyS) by dendritic cells correlate with HIV-related B-cell disease progression in humans. Blood. 2011;117:145–155. doi: 10.1182/blood-2010-08-301887. [DOI] [PubMed] [Google Scholar]
- Forthal D.N., Landucci G., Haubrich R., Keenan B., Kuppermann B.D., Tilles J.G., Kaplan J. Antibody-dependent cellular cytotoxicity independently predicts survival in severely immunocompromised human immunodeficiency virus-infected patients. J. Infect. Dis. 1999;180:1338–1341. doi: 10.1086/314988. [DOI] [PubMed] [Google Scholar]
- Gohain N., Tolbert W.D., Acharya P., Yu L., Liu T., Zhao P., Orlandi C., Visciano M.L., Kamin-Lewis R., Sajadi M.M., Martin L., Robinson J.E., Kwong P.D., Devico A.L., Ray K., Lewis G.K., Pazgier M. Cocrystal structures of antibody N60-i3 and antibody JR4 in complex with gp120 define more cluster a epitopes involved in effective antibody-dependent effector function against HIV-1. J. Virol. 2015;89:8840–8854. doi: 10.1128/JVI.01232-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Pazgier M., Sajadi M.M., Kamin-Lewis R., Al-Darmarki S., Flinko R., Lovo E., Wu X., Robinson J.E., Seaman M.S., Fouts T.R., Gallo R.C., Devico A.L., Lewis G.K. Diverse specificity and effector function among human antibodies to HIV-1 envelope glycoprotein epitopes exposed by CD4 binding. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E69–E78. doi: 10.1073/pnas.1217609110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes B.F., Gilbert P.B., Mcelrath M.J., Zolla-Pazner S., Tomaras G.D., Alam S.M., Evans D.T., Montefiori D.C., Karnasuta C., Sutthent R., Liao H.X., Devico A.L., Lewis G.K., Williams C., Pinter A., Fong Y., Janes H., Decamp A., Huang Y., Rao M., Billings E., Karasavvas N., Robb M.L., Ngauy V., De Souza M.S., Paris R., Ferrari G., Bailer R.T., Soderberg K.A., Andrews C., Berman P.W., Frahm N., De Rosa S.C., Alpert M.D., Yates N.L., Shen X., Koup R.A., Pitisuttithum P., Kaewkungwal J., Nitayaphan S., Rerks-Ngarm S., Michael N.L., Kim J.H. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International H.I.V.C.S., Pereyra F., Jia X., Mclaren P.J., Telenti A., De Bakker P.I., Walker B.D., Ripke S., Brumme C.J., Pulit S.L., Carrington M., Kadie C.M., Carlson J.M., Heckerman D., Graham R.R., Plenge R.M., Deeks S.G., Gianniny L., Crawford G., Sullivan J., Gonzalez E., Davies L., Camargo A., Moore J.M., Beattie N., Gupta S., Crenshaw A., Burtt N.P., Guiducci C., Gupta N., Gao X., Qi Y., Yuki Y., Piechocka-Trocha A., Cutrell E., Rosenberg R., Moss K.L., Lemay P., O'leary J., Schaefer T., Verma P., Toth I., Block B., Baker B., Rothchild A., Lian J., Proudfoot J., Alvino D.M., Vine S., Addo M.M., Allen T.M., Altfeld M., Henn M.R., Le Gall S., Streeck H., Haas D.W., Kuritzkes D.R., Robbins G.K., Shafer R.W., Gulick R.M., Shikuma C.M., Haubrich R., Riddler S., Sax P.E., Daar E.S., Ribaudo H.J., Agan B., Agarwal S., Ahern R.L., Allen B.L., Altidor S., Altschuler E.L., Ambardar S., Anastos K., Anderson B., Anderson V., Andrady U., Antoniskis D., Bangsberg D., Barbaro D., Barrie W., Bartczak J., Barton S., Basden P., Basgoz N., Bazner S., Bellos N.C., Benson A.M., Berger J., Bernard N.F., Bernard A.M., Birch C., Bodner S.J., Bolan R.K., Boudreaux E.T., Bradley M., Braun J.F., Brndjar J.E., Brown S.J., Brown K. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamya P., Boulet S., Tsoukas C.M., Routy J.P., Thomas R., Cote P., Boulassel M.R., Baril J.G., Kovacs C., Migueles S.A., Connors M., Suscovich T.J., Brander C., Tremblay C.L., Bernard N., Canadian Cohort Of, H. I. V. I. S. P. Receptor-ligand requirements for increased NK cell polyfunctional potential in slow progressors infected with HIV-1 coexpressing KIR3DL1*h/*y and HLA-B*57. J. Virol. 2011;85:5949–5960. doi: 10.1128/JVI.02652-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D., Mascola J.R. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D., Doyle M.L., Casper D.J., Cicala C., Leavitt S.A., Majeed S., Steenbeke T.D., Venturi M., Chaiken I., Fung M., Katinger H., Parren P.W., Robinson J., Van Ryk D., Wang L., Burton D.R., Freire E., Wyatt R., Sodroski J., Hendrickson W.A., Arthos J. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- Lalonde J.M., Kwon Y.D., Jones D.M., Sun A.W., Courter J.R., Soeta T., Kobayashi T., Princiotto A.M., Wu X., Schon A., Freire E., Kwong P.D., Mascola J.R., Sodroski J., Madani N., Smith A.B., 3rd Structure-based design, synthesis, and characterization of dual hotspot small-molecule HIV-1 entry inhibitors. J. Med. Chem. 2012;55:4382–4396. doi: 10.1021/jm300265j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.S., Richard J., Lichtfuss M., Smith A.B., 3rd, Park J., Courter J.R., Melillo B.N., Sodroski J.G., Kaufmann D.E., Finzi A., Parsons M.S., Kent S.J. Antibody-dependent cellular cytotoxicity against reactivated HIV-1-infected cells. J. Virol. 2015;90:2021–2030. doi: 10.1128/JVI.02717-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuka J., Nduati R., Odem-Davis K., Peterson D., Overbaugh J. HIV-specific antibodies capable of ADCC are common in breastmilk and are associated with reduced risk of transmission in women with high viral loads. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madani N., Schon A., Princiotto A.M., Lalonde J.M., Courter J.R., Soeta T., Ng D., Wang L., Brower E.T., Xiang S.H., Do Kwon Y., Huang C.C., Wyatt R., Kwong P.D., Freire E., Smith A.B., 3rd, Sodroski J. Small-molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure. 2008;16:1689–1701. doi: 10.1016/j.str.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L., Stricher F., Misse D., Sironi F., Pugniere M., Barthe P., Prado-Gotor R., Freulon I., Magne X., Roumestand C., Menez A., Lusso P., Veas F., Vita C. Rational design of a CD4 mimic that inhibits HIV-1 entry and exposes cryptic neutralization epitopes. Nat. Biotechnol. 2003;21:71–76. doi: 10.1038/nbt768. [DOI] [PubMed] [Google Scholar]
- Melillo B., Liang S., Park J., Schon A., Courter J.R., Lalonde J.M., Wendler D.J., Princiotto A.M., Seaman M.S., Freire E., Sodroski J., Madani N., Hendrickson W.A., Smith A.B., 3rd Small-molecule CD4-mimics: structure-based optimization of HIV-1 entry inhibition. ACS Med. Chem. Lett. 2016;7:330–334. doi: 10.1021/acsmedchemlett.5b00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody M.A., Gao F., Gurley T.C., Amos J.D., Kumar A., Hora B., Marshall D.J., Whitesides J.F., Xia S.M., Parks R., Lloyd K.E., Hwang K.K., Lu X., Bonsignori M., Finzi A., Vandergrift N.A., Alam S.M., Ferrari G., Shen X., Tomaras G.D., Kamanga G., Cohen M.S., Sam N.E., Kapiga S., Gray E.S., Tumba N.L., Morris L., Zolla-Pazner S., Gorny M.K., Mascola J.R., Hahn B.H., Shaw G.M., Sodroski J.G., Liao H.X., Montefiori D.C., Hraber P.T., Korber B.T., Haynes B.F. Strain-specific V3 and CD4 binding site autologous HIV-1 neutralizing antibodies select neutralization-resistant viruses. Cell Host Microbe. 2015;18:354–362. doi: 10.1016/j.chom.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.P., Sattentau Q.J., Wyatt R., Sodroski J. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J. Virol. 1994;68:469–484. doi: 10.1128/jvi.68.1.469-484.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenbauer C., Edmonds T.G., Ding H., Keele B.F., Decker J., Salazar M.G., Salazar-Gonzalez J.F., Shattock R., Haynes B.F., Shaw G.M., Hahn B.H., Kappes J.C. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J. Virol. 2012;86:2715–2728. doi: 10.1128/JVI.06157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish N.F., Gao F., Li H., Giorgi E.E., Barbian H.J., Parrish E.H., Zajic L., Iyer S.S., Decker J.M., Kumar A., Hora B., Berg A., Cai F., Hopper J., Denny T.N., Ding H., Ochsenbauer C., Kappes J.C., Galimidi R.P., West A.P., Jr., Bjorkman P.J., Wilen C.B., Doms R.W., O'brien M., Bhardwaj N., Borrow P., Haynes B.F., Muldoon M., Theiler J.P., Korber B., Shaw G.M., Hahn B.H. Phenotypic properties of transmitted founder HIV-1. Proc. Natl. Acad. Sci. U. S. A. 2013;110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretz Y., Ndongala M.L., Boulet S., Boulassel M.R., Rouleau D., Cote P., Longpre D., Routy J.P., Falutz J., Tremblay C., Tsoukas C.M., Sekaly R.P., Bernard N.F. Functional T cell subsets contribute differentially to HIV peptide-specific responses within infected individuals: correlation of these functional T cell subsets with markers of disease progression. Clin. Immunol. 2007;124:57–68. doi: 10.1016/j.clim.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Richard J., Sindhu S., Pham T.N., Belzile J.P., Cohen E.A. HIV-1 Vpr up-regulates expression of ligands for the activating NKG2D receptor and promotes NK cell-mediated killing. Blood. 2010;115:1354–1363. doi: 10.1182/blood-2009-08-237370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard J., Veillette M., Batraville L.A., Coutu M., Chapleau J.P., Bonsignori M., Bernard N., Tremblay C., Roger M., Kaufmann D.E., Finzi A. Flow cytometry-based assay to study HIV-1 gp120 specific antibody-dependent cellular cytotoxicity responses. J. Virol. Methods. 2014;208:107–114. doi: 10.1016/j.jviromet.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Richard J., Veillette M., Brassard N., Iyer S.S., Roger M., Martin L., Pazgier M., Schon A., Freire E., Routy J.P., Smith A.B., 3rd, Park J., Jones D.M., Courter J.R., Melillo B.N., Kaufmann D.E., Hahn B.H., Permar S.R., Haynes B.F., Madani N., Sodroski J.G., Finzi A. CD4 mimetics sensitize HIV-1-infected cells to ADCC. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E2687–E2694. doi: 10.1073/pnas.1506755112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard J., Veillette M., Ding S., Zoubchenok D., Alsahafi N., Coutu M., Brassard N., Park J., Courter J.R., Melillo B., Smith A.B., 3rd, Shaw G.M., Hahn B.H., Sodroski J., Kaufmann D.E., Finzi A. Small CD4 mimetics prevent HIV-1 uninfected bystander CD4 + T cell killing mediated by antibody-dependent cell-mediated cytotoxicity. EBioMedicine. 2016;3:122–134. doi: 10.1016/j.ebiom.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra S., Tomaras G.D., Warrier R., Nicely N.I., Liao H.X., Pollara J., Liu P., Alam S.M., Zhang R., Cocklin S.L., Shen X., Duffy R., Xia S.M., Schutte R.J., Pemble Iv C.W., Dennison S.M., Li H., Chao A., Vidnovic K., Evans A., Klein K., Kumar A., Robinson J., Landucci G., Forthal D.N., Montefiori D.C., Kaewkungwal J., Nitayaphan S., Pitisuttithum P., Rerks-Ngarm S., Robb M.L., Michael N.L., Kim J.H., Soderberg K.A., Giorgi E.E., Blair L., Korber B.T., Moog C., Shattock R.J., Letvin N.L., Schmitz J.E., Moody M.A., Gao F., Ferrari G., Shaw G.M., Haynes B.F. Human non-neutralizing HIV-1 envelope monoclonal antibodies limit the number of founder viruses during SHIV mucosal infection in rhesus macaques. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon A., Madani N., Klein J.C., Hubicki A., Ng D., Yang X., Smith A.B., 3rd, Sodroski J., Freire E. Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry. 2006;45:10973–10980. doi: 10.1021/bi061193r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Asmal M., Lane S., Permar S.R., Schmidt S.D., Mascola J.R., Letvin N.L. Antibody-dependent cell-mediated cytotoxicity in simian immunodeficiency virus-infected rhesus monkeys. J. Virol. 2011;85:6906–6912. doi: 10.1128/JVI.00326-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert W.D., Gohain N., Veillette M., Chapleau J.P., Orlandi C., Visciano M.L., Ebadi M., Devico A.L., Fouts T.R., Finzi A., Lewis G.K., Pazgier M. Paring down HIV Env: design and crystal structure of a stabilized inner domain of HIV-1 gp120 displaying a major ADCC target of the A32 region. Structure. 2016;24:697–709. doi: 10.1016/j.str.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Herrewege Y., Morellato L., Descours A., Aerts L., Michiels J., Heyndrickx L., Martin L., Vanham G. CD4 mimetic miniproteins: potent anti-HIV compounds with promising activity as microbicides. J. Antimicrob. Chemother. 2008;61:818–826. doi: 10.1093/jac/dkn042. [DOI] [PubMed] [Google Scholar]
- Veillette M., Coutu M., Richard J., Batraville L.A., Desormeaux A., Roger M., Finzi A. Conformational evaluation of HIV-1 trimeric envelope glycoproteins using a cell-based ELISA assay. J. Vis. Exp. 2014 doi: 10.3791/51995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillette M., Desormeaux A., Medjahed H., Gharsallah N.E., Coutu M., Baalwa J., Guan Y., Lewis G., Ferrari G., Hahn B.H., Haynes B.F., Robinson J.E., Kaufmann D.E., Bonsignori M., Sodroski J., Finzi A. Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity. J. Virol. 2014;88:2633–2644. doi: 10.1128/JVI.03230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillette M., Coutu M., Richard J., Batraville L.A., Dagher O., Bernard N., Tremblay C., Kaufmann D.E., Roger M., Finzi A. The HIV-1 gp120 CD4-bound conformation is preferentially targeted by antibody-dependent cellular cytotoxicity-mediating antibodies in sera from HIV-1-infected individuals. J. Virol. 2015;89:545–551. doi: 10.1128/JVI.02868-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillette M., Richard J., Pazgier M., Lewis G.K., Parsons M.S., Finzi A. Role of HIV-1 envelope glycoproteins conformation and accessory proteins on ADCC responses. Curr. HIV Res. 2016;14:9–23. doi: 10.2174/1570162x13666150827093449. [DOI] [PubMed] [Google Scholar]
- Von Bredow B., Arias J.F., Heyer L.N., Gardner M.R., Farzan M., Rakasz E.G., Evans D.T. Envelope glycoprotein internalization protects human and simian immunodeficiency virus infected cells from antibody-dependent cell-mediated cytotoxicity. J. Virol. 2015 doi: 10.1128/JVI.01911-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Bredow B., Arias J.F., Heyer L.N., Moldt B., Le K., Robinson J.E., Zolla-Pazner S., Burton D.R., Evans D.T. Comparison of antibody-dependent cell-mediated cytotoxicity and virus neutralization by HIV-1 Env-specific monoclonal antibodies. J. Virol. 2016;90:6127–6139. doi: 10.1128/JVI.00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K.L., Cortez V., Dingens A.S., Gach J.S., Rainwater S., Weis J.F., Chen X., Spearman P., Forthal D.N., Overbaugh J. HIV-specific CD4-induced antibodies mediate broad and potent antibody-dependent cellular cytotoxicity activity and are commonly detected in plasma from HIV-infected humans. EBioMedicine. 2015;2:1464–1477. doi: 10.1016/j.ebiom.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R., Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Wyatt R., Moore J., Accola M., Desjardin E., Robinson J., Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R., Kwong P.D., Desjardins E., Sweet R.W., Robinson J., Hendrickson W.A., Sodroski J.G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of HIV-infected sera donors.
Supplementary figures.