Abstract
The recent Zika virus (ZIKV) epidemic has highlighted the poor knowledge on its physiopathology. Recent studies showed that ZIKV of the Asian lineage, responsible for this international outbreak, causes neuropathology in vitro and in vivo. However, two African lineages exist and the virus is currently found circulating in Africa. The original African strain was also suggested to be neurovirulent but its laboratory usage has been criticized due to its multiple passages. In this study, we compared the French Polynesian (Asian) ZIKV strain to an African strain isolated in Central African Republic and show a difference in infectivity and cellular response between both strains in human neural stem cells and astrocytes. Consistently, this African strain led to a higher infection rate and viral production, as well as stronger cell death and anti-viral response. Our results highlight the need to better characterize the physiopathology and predict neurological impairment associated with African ZIKV.
Keywords: Zika virus, Lineages, Neural stem cells, Astrocytes
Highlights
-
•
ZIKV from two Asian and African strains infect differentially IPSc-derived NSCs
-
•
African ZIKV strain displays more infectivity and anti-viral response
-
•
African and Asian ZIKV strains infect and replicate efficiently in human astrocytes
Zika virus (ZIKV) is involved in a major epidemic in Latin America. Some neurological complications are reported following infection, in particular microcephaly and Guillain-Barré syndromes. Analyses show that one Asian and two Africans lineages exist. Although originally discovered in Africa in 1947, the actual epidemic is due to Asian ZIKV. Here, we compared the virulence of two African and Asian ZIKV strains in neural cells. We showed that in human neural stem cells, African ZIKV strain had a higher infectivity and triggered stronger cellular response. Altogether, our results highlight the need to better characterize ZIKV lineages to anticipate further epidemics.
1. Introduction
The current (2015–2016) Zika virus (ZIKV) epidemic in South America and in the Caribbean islands revealed the lack of knowledge on the pathophysiological mechanisms of a virus discovered almost 70 years ago in Uganda (Dick et al., 1952). This arbovirus is transmitted by vectors from the Aedes family, in particular Aedes aegypti and Aedes albopictus. Phylogenetic analyses revealed recently that three lineages (one Asian and two Africans) exist and explain the evolution of ZIKV over the last 70 years (Shen et al., 2016, Gong et al., 2016). The current epidemic is due to ZIKV from the Asian lineage, even though some further genetic evolution seems to have occurred (Wang et al., 2016).
While a large proportion of infected persons are asymptomatic (70–80%), others develop rather classical clinical signs of an arboviral infection namely, skin rash, headache, myalgia, joint pain, conjunctivitis, but also moderate fever. In some cases, neurological disorders have been linked to ZIKV infections, in particular Guillain-Barré syndrome (GBS), myelitis, encephalitis, neuralgia and microcephaly in newborns and infants born to women with ZIKV infection during pregnancy (Musso et al., 2016). Microcephaly is now well documented as a direct disorder triggered by ZIKV infection, as the virus can cross the placenta (Miner et al., 2016), is found in the amniotic fluid (Calvet et al., 2016) and can be detected in post-mortem newborn brains (Brasil Martines et al., 2016). In particular, infection during the first trimester of pregnancy has been proposed to lead to microcephaly in 1% of cases (Cauchemez et al., 2016). Studies on animal models (rodent) show that ZIKV can cause a similar pathology as in humans, such as trans-placental fetal transmission and neurological impairment in fetal and adult brain (Rossi et al., 2016, Dowall et al., 2016, Aliota et al., 2016, Cugola et al., 2016, Lazear et al., 2016, Miner et al., 2016). However, most of these models are based on an immunodeficient background. Recent reports clearly demonstrate that ZIKV can infect human and murine neural precursors. Ex vivo works using induced pluripotent cells (IPSc)-derived neural precursors cells (NPCs) and neural stem cells (NSCs) show that ZIKV has a preferential tropism for cells of the neuronal lineage during development (Tang et al., 2016, Qian et al., 2016, Garcez et al., 2016). Notably, some reports suggest that impairment of proliferation and cell death is associated with ZIKV infection (Tang et al., 2016, Li et al., 2016), whereas another report suggests that ZIKV poorly stimulates immune response and cytopathic effect in human neuroprecursors obtained from fetal tissue (Hanners et al., 2016). This lack of strong immune response would allow ZIKV to persist during development and is consistent with the replication seen over few weeks in this tissue. These observations are strengthening the hypothesis that infection early during brain development can have drastic effects.
While most of the focus has been directed to Asian ZIKV strains or to African MR-766, much less effort has been undertaken to monitor potential circulating ZIKV of the African lineage (Grard et al., 2014, Baraka and Kweka, 2016, Meda et al., 2016). In this context, there is an urgent need to have clear understanding of the pathophysiological mechanisms involved in infection by African ZIKV, in particular in terms of neurovirulence. In other words, to know whether the neurological effects observed with the Asian lineage are specifically associated with the Asian strain, in terms of severity and specificity, or if we can expect African strains to lead to similar disorders. So far, most of the studies that investigated African ZIKV strain used the original strain of ZIKV (MR-766, isolated in 1947). Lately, criticisms emerged concerning the pertinence of this strain, isolated from primates and extensively amplified in suckling mouse brains and ex vivo on cells (Haddow et al., 2012, Musso et al., 2016).
Here, we used IPSc-derived human NSCs to better compare the neural infectivity of an Asian strain (ZIKV AS) and an African strain (ZIKV AF) that underwent low passages. We demonstrate that this ZIKV AF strain is more infectious than the French Polynesian ZIKV AS PF-13 strain (H/PF/2013): indeed, this strain showed a higher rate of infection, viral production and cellular response (cell death and anti-viral response) than ZIKV AS. Finally, we show that ZIKV AF and AS strains also display difference in infection of human astrocytes.
2. Material and Methods
2.1. Material
Antibodies used in this study are: anti-pan-flavivirus (MAB10216, clone D1-4G2) and anti-nestin, (Millipore), anti-GFAP (Abcam), anti-PDI and anti-activated caspase 3 (Cell Signalling Technology), anti-TRA1-60 (Becton Dickinson) and anti-PAX6 (BioLegend). Carboxyfluorescein succinimidyl ester (CFSE) dye was purchased from Thermoscientific.
2.2. ZIKV Strains, Production and Cellular Infection
H/PF/2013 ZIKV of Asian lineage (French Polynesia, 2013) and ArB41644 ZIKV of African lineage (Bangui, Central African Republic, 1989, isolated from mosquitoes by Pasteur Institute of Dakkar) were produced and provided by the National Reference Center for arboviruses (NRC) and have both no > 5 passages on Vero cells. Viral stocks were prepared by infecting sub confluent Vero cells at the multiplicity of infection (MOI) of 0.01 in D-MEM medium (Thermoscientific) supplemented by 2% heat-inactivated fetal bovine serum (Sigma). Cell supernatant was collected 6 days later and viral stock harvested after centrifugation at 300g to remove cellular debris. Viral titers were determined by the 50% tissue culture infective dose (TCID50), which was calculated using the Spearman-Kärber method (Kärber, 1931) and were expressed as TCID50 per mL. Titers were calculated twice, once at the NRC and once in our laboratory. Another stock from each ZIKV strain was also produced in C6/36 cells and had similar results (data not shown).
IPSc-derived NSCs and human astrocytes (see below) at 60–70% confluence were rinsed once with phosphate-buffered saline (PBS), and ZIKV diluted to the required MOI (0.01, 0.1 or 1) was added to the cells in a low medium volume. Cells were incubated for 2 h at 37 °C with permanent gentle agitation and then the inoculum was removed and cells washed with PBS. Culture medium was added to each well, and cells were incubated at 37 °C and 5% CO2. As control, cells were incubated with the culture supernatant from Vero cells (mock condition).
2.3. NSC Generation and Maintenance
NSCs were obtained from the SAFE-IPSc platform at IRMB. Briefly, iPSCs generated from healthy patient using Lentivirus-derived vectors were individualized with Gentle Cell Dissociation Reagent (Stemcell, 07174). They were rinsed out with Dulbecco's modified Eagle's medium/Ham's F12 (DMEM/F-12, Gibco, 31330038) and centrifuged at 300g for 5 min. Dissociated cells were plated on matrigel at a density of 20,000–40,000 cells/cm2 and cultured in neural induction medium (Stemcell, 05835) supplemented with 10 μM ROCK-inhibitor (Y-27632). Cells were allowed to reach 80–90% confluence over 6 days. Medium was changed daily with neural induction medium without Y-27632. iPSC-derived NSCs were passaged by incubation with Trypsin at 0.005% to allow dissociation, and then seeded on poly-D-ornithine/laminin coated plates at 20,000 cells/cm2 in 50% DMEM/F-12 and 50% Neurobasal medium (Thermoscientific) supplemented with 1 × N2 (Thermoscientific), 1 × B27 (Thermoscientific), Glutamax (Thermoscientific) and β-FGF plus EGF (Peprotech, 20 ng/mL each). The medium was changed every two days. Cells were used between passages 5 and 8.
2.4. Astrocyte Cultures
Astrocytes were purchased from ScienCell™ and cultured according to the manufacturer's instruction. Cells were cultured on Poly-lysine coated plates and were used between passages 3 and 7.
2.5. Immunofluorescence Assays
NSCs plated on poly-D-ornithine/laminin coverslips and astrocytes plated on poly-D-lysin coverslips were imaged using Zeiss SP8 confocal microscope, with 40 × or 63 × 1.4 NA Plan Apochromat oil-immersion objectives. For indirect immunofluorescence, cells were fixed with 4% PFA and permeabilized with 0.1% Triton X-100/PBS for 5 min at room temperature (RT), followed by a blocking step with 2% bovine serum albumin (BSA) and 10% horse serum for 30 min to 1 h at RT. Primary and secondary antibodies were diluted in blocking solution and incubated sequentially for 1 h at RT. Samples were then mounted with fluorescent mounting medium (Prolongold, Thermofischer) with DAPI (Sigma) and imaged by confocal microscopy using the Zeiss SP8 confocal microscope, with 40 × or 63 × 1.4 NA Plan Apochromat oil-immersion objectives. Nuclei and ZIKV infected cells were counted using a plugin of the Image J software.
2.6. Flow Cytometry and CFSE Assays
NSCs were incubated with CFSE according to the manufacturer (Thermoscientific) for 3 min at RT after enzymatic dissociation and replated after the dye was quenched and cells were washed with PBS. NSCs infected with ZIKV strains were collected in PBS at various days, washed with PBS, fixed with 2% PFA and analyzed with FACSCalibur (BD Biosciences). Data analysis was performed using the FlowJo software. Decrease in fluorescence intensity is consistent with cell division.
2.7. RT-qPCR
RT-PCR on ZIKV-infected NSC supernatants was performed after mRNA extraction with the Altona Diagnostics kit RealStar® Zika Virus RT-PCR Kit 1.0, according to the manufacturer's instructions. Experiments done with similar MOI for ZIKV AF and AS showed equal amplification for both strains (data not shown). For PCR arrays, NSCs infected with ZIKV AF or AS or mock-treated cells were harvested in RLT buffer (Qiagen). Total RNA was extracted using RNeasy® mini-kit (Qiagen). Complementary DNA was synthesized using Omniscript® reverse transcriptase (Life Technologies). RT2 Profiler PCR arrays for Human Antiviral Response (96 well format, Qiagen) were used for real-time quantitative PCR analysis, with the use of the LC480 real time PCR instrument (Roche) and the Light Cycler 480 SYBR Green I master Mix (Roche). Volumes of mix, cDNA, RNAse-free water, and cycling conditions were determined according to manufacturer's instructions. Data on gene expression were normalized according to data from the HPRT housekeeping gene. Genes with uninterpretable amplifying curves were excluded from the analysis.
2.8. Phylogenetic Analysis
The phylogenetic analysis was performed through a consensus flavivirus RT-PCR fragment that targeted a conserved NS5 gene region as previously described by Scaramozzino et al. (2001). Briefly, The NS5 PCR product was purified with the QIAquick PCR purification kit (Qiagen) and sequenced on an ABI 310 sequencer with a fluorescent dye terminator kit (Applied Biosystem). Sequencing primers were: GTGTCCCAGCCGGCGGTGTCATCAGC and AACATGATGGGRAARAGRGARAA and the PCR cycles were with an initial amplification of 25 cycles of incubation at 94 °C, 53 °C, and 72 °C for 1 min each. The resulting sequences (ZIKV AF and ZIKV AS) and a set of sequences from both Asian and African ZIKV isolates (Accession numbers: KF268950, HQ234500, KF383116, LC002520, KF383119, KF383115, KF383118, KF383117, KF993678, HQ234499, JN860885, KF258813, EU545988, KJ776791, KU955595) were submitted to the French phylogeny website (http://www.phylogeny.fr) using the “one click version” for analysis. The nucleotide sequences are aligned using the MUSCLE software, cured with Gblocks and subsequent phylogeny performed with the PhyML software. One hundred bootstrap data sets with random sequence addition were computed to generate a consensus tree, rooted with a West Nile isolate sequence (AY532665), through TreeDyn.
2.9. Statistical Analyses
For all quantitative analyses, a minimum of three independent experiments were performed. Student's t-test or Mann-Whitney test were performed to analyze unpaired data. A p < 0.05 is reported as significant.
2.10. Ethic Statement
All experiments for IPSc generation were performed with the approval of local biomedical ethic committee.
3. Results
3.1. Two African and Asian ZIKV Strains Show Different Infectivity in IPS-derived NSCs
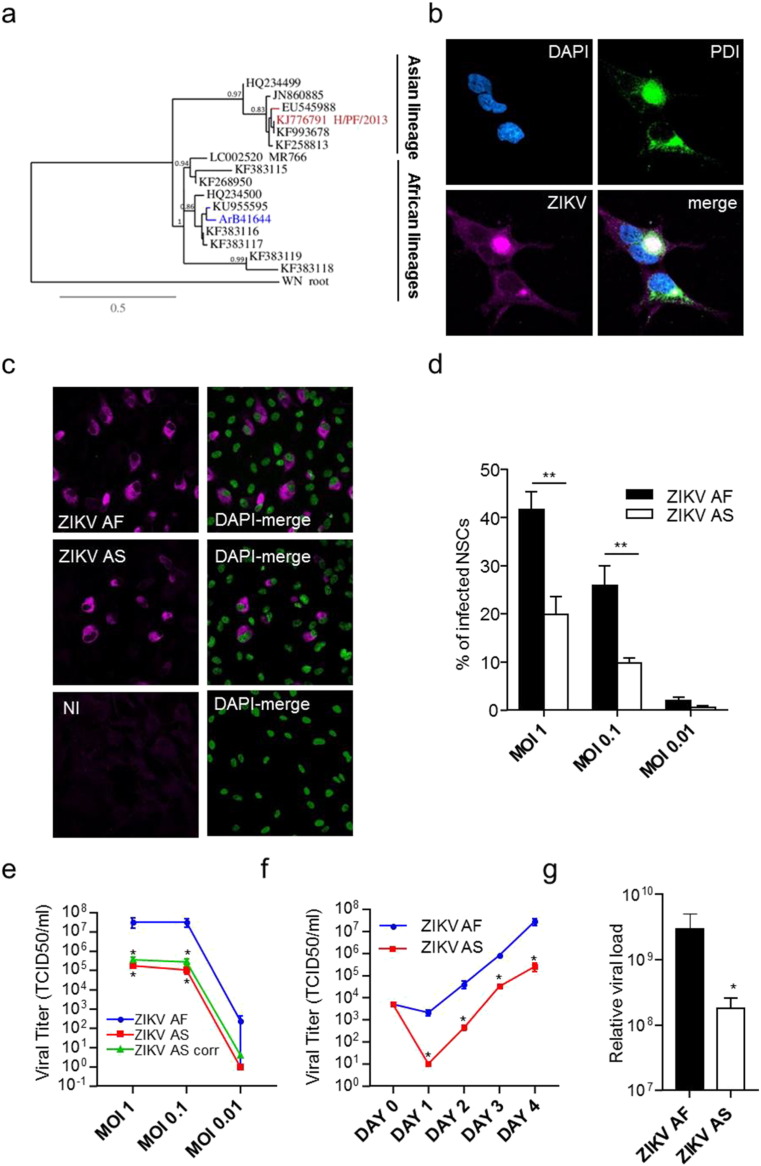
There is no consistency regarding ZIKV strain usage and relevance, which make difficult any true comparison of the neurovirulence between African and Asian strains. To have a better understanding on how ZIKV infect the central nervous system (CNS) and potential differences between lineages, we decided to use two low-passage ZIKV strains as it is known that RNA viruses can easily derive upon culture. These two strains were provided by the French National Reference Center and were isolated from the Asian lineage (PF-13, isolated in French Polynesia in 2013, hereafter named ZIKV AS) and an African strain (ArB41644, isolated in Central African Republic in 1989, hereafter named ZIKV AF), which partial sequencing showed that it has strong homology to other strains of the African lineage I (Fig. 1A) (Shen et al., 2016). IPS-derived NSCs were obtained according to standard methods (see Material and Methods section and Supplemental Figure 1A). Consistent with previous report, in NSCs infected with ZIKV AS at multiplicity of infection (MOI) of 1, we detected efficient ZIKV replication after 4 days by an immunofluorescence (IF) assay using a pan-flavivirus antibody (Fig. 1B). This staining was in close proximity to the endoplasmic reticulum, labeled with anti-PDI, a common replication site for Flaviviruses (Fig. 1B). When we performed comparative analyses of ZIKV AF and AS infection at different MOIs (1, 0.1 and 0.01), we reproducibly detected a higher infection rate for ZIKV AF than for ZIKV AS (Fig. 1C and D). Interestingly, even with a log difference in MOI, ZIKV AF led to more infected NSCs than ZIKV AS (Fig. 1D). Notably, percentage of infected cells at stronger MOI for ZIKV AS was not significantly higher (data not shown). Importantly, we used other NSCs clones and other ZIKV AF and AS stocks from C6/36 cells (Aedes albopictus cells) and showed similar differences between the strains (data not shown). CPE (cytopathic effect) analyses in supernatants from NSCs at day 4 post-infection showed again a significant difference in viral titers quantified by TCID (tissue culture infective dose) 50 between ZIKV AF and ZIKV AS (Fig. 1E). Kinetics of viral production showed early dichotomy in viral titers as differences were already seen from day 1 post-infection (Fig. 1F). Finally, real-time RT-PCR from supernatants harvested at day 4 post-infection (MOI 1) showed also a higher relative viral charge for ZIKV AF (Fig. 1G).
Fig. 1.
Differential infectivity of ZIKV AF and AS in human IPSc-derived NSCs.
(a) Phylogenetic analysis of ZIKV strains used in this study was performed through a consensus flavivirus RT-PCR fragment that targeted a conserved NS5 gene region. Bootstrap values are indicated at main nodes.
(b) Indirect immunofluorescence (IF) of ZIKV AS-infected NSCs (MOI 1, day 4 post-infection) where the nucleus is stained using DAPI (cyan), the endoplasmic reticulum is stained using an anti-PDI (green) and the virus is labeled using a pan-flavivirus antibody (magenta). Scale bar = 10 μm.
(c) IF of non-infected (NI), ZIKV AF- and ZIKV AS-infected NSCs (MOI 1, day 4 post-infection). The virus is labeled using a pan-flavivirus antibody (magenta) and nuclei by DAPI (false colored in green). Scale bar = 10 μm.
(d) Quantitative analyses show differences in rate of infection between ZIKV AF and ZIKV AS that is consistently seen using different MOIs. 4 independent experiments were quantified with 200–600 cells per conditions. Results are expressed as means ± SEM and analyzed using an unpaired t-test **p < 0.01.
(e) Viral titers were determined by TCID50 on Vero cells. Supernatants from NSCs infected at different MOIs and collected day 4 post-infection show differential titers between ZIKV AF and AS, even when % of infected cells were taken in account (green curve, ZIKV AS corrected, titer normalized assuming same infection rate than ZIKV AF (i.e. 41%). Results are expressed as means ± SEM, for three independent experiments and analyzed using Mann-Whitney test *p < 0.05.
(f) Kinetics of viral production show early dichotomy in viral replication between ZIKV AF and ZIKV AS. Day 0 corresponds to the input inoculum. Results are expressed as means ± SEM, for three independent experiments and analyzed using Mann-Whitney test *p < 0.05.
(g) Quantitative RT-PCR done on extracts of supernatants from NSCs infected at MOI 1 by the different ZIKV strains and collected at day 4 post-infection shows also a difference in the relative physical viral load between ZIKV AF and AS. CTs from amplifying curves were arbitrarily converted using internal control (see Material and Methods section). Results of three independent experiments are expressed as means of relative charges ± SEM and analyzed using Mann-Whitney test. *p < 0.05.
Together, these data suggest that both lineages are efficiently targeting NSCs and that neurovirulence and neuropathologies are likely to be associated with this ZIKV AF strain as it infects and replicates more efficiently than ZIKV AS in NSCs.
3.2. Cellular and Anti-viral Responses in ZIKV-infected NSCs
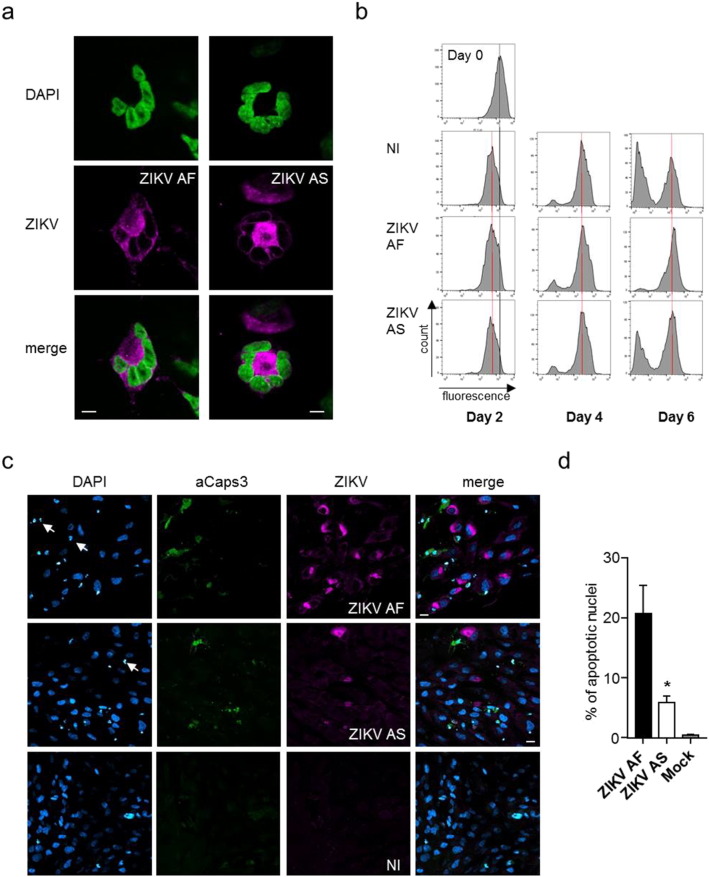
In a recent study, minimum cell death and cell cycle impairment in neuronal precursors upon infection with ZIKV from the Asian lineage was reported, suggesting that the virus could establish persistence in this cell type (Hanners et al., 2016). We therefore next sought to characterize the cellular response in ZIKV AF- and ZIKV AS-infected NSCs by undertaking comparative analyses. Few multi-nucleated cells could be observed at day 4 post-infection with both lineages, possibly resulting from endoreplication (Fig. 2A), which is not observed in non-infected cells and suggests defect in the cell cycle. We therefore monitored cell division in ZIKV-infected NSCs by flow cytometry analyzes. Using the CFSE dye, which is cell-permeable incorporated to cellular proteins and which fluorescence is serially lost upon cell division, we analyzed cell cycle progression in non-infected, ZIKV AF- and ZIKV AS-infected (MOI 1) NSCs. Fig. 2B shows representative graphs after 2, 4, and 6 days post-infection. No significant differences were seen in cell cycle progression 2 and 4 days after infection, whereas NSCs infected with ZIKV AF showed an important impairment in cell division after 6 days post-infection (Fig. 2B). ZIKV AS-infected cells also displayed delayed division, albeit much less than ZIKV AF-infected NSCs (Fig. 2B). Similar that what was reported by Hanners et al. (2016) with another Asian strain (PRVABC59), we found low cell death associated with infection at day 4 as seen by the presence of few fragmented nuclei and activated caspase 3, a marker of apoptosis, for both lineages, even though ZIKV AF infection led to more apoptotic nuclei than ZIKV AS (Fig. 2C and D). Surprisingly, in ZIKV-infected conditions, we also observed non-infected cells positive for activated-caspase 3 that could suggest bystander cell death, possibly related to cytokine production following infection. Using bright light, we also scarcely detected “rounded” cells, which suggest a poor CPE at early time post-infection (day 4) (Supplemental Figure 1B).
Fig. 2.
Effect of ZIKV AF and AS on NSC cell cycle and survival.
(a) Nuclear fragmentation or endoreplication in ZIKV-infected cells. IF of ZIKV-infected NSCs (ZIKV AF or AS MOI 1 day 4 post-infection). The nucleus is labeled with DAPI (false colored in green) and viruses with a pan-flavivirus antibody (magenta). Scale bar = 5 μm.
(b) Flow cytometry analyses of NSCs incubated with CFSE at day 0 and infected with ZIKV AF and AS for 2 to 6 days. The shift in fluorescence is consistent with cell division.
(c) IF of non-infected (NI), ZIKV AF and ZIKV AS-infected NSCs (MOI 1, day 4 post-infection) show the presence of apoptotic cells in infected cells (white arrows show condensed nuclei and anti-activated caspase 3 is in green). Scale bar = 10 μm.
(d) Apoptotic nuclei were quantified in ZIKV AF and AS-infected NSCs at day 4 post-infection. Three independent experiments were quantified with 200–400 nuclei per condition. Results are expressed as means ± SEM and analyzed using an unpaired t-test *p < 0.05.
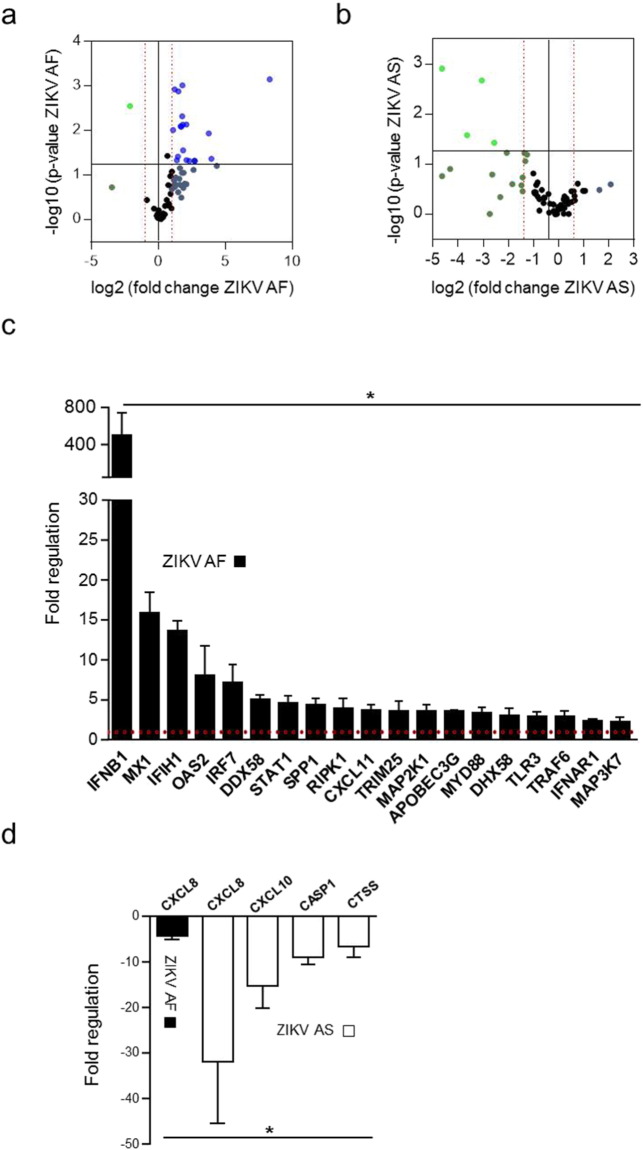
To better characterize and compare the cellular response to ZIKV AF- and AS-NSC infection, we performed a PCR array consisting of 79 genes involved in anti-viral responses (see Material and Methods section and Supplemental Table 1). NSCs were infected with ZIKV AF or ZIKV AS at a MOI of 1 and mRNA collected at day 4 post-infection. Twenty genes were significantly modulated in NSCs upon ZIKV AF infection (Fig. 3A and C). In particular, 19 genes were upregulated, among them the cellular pattern recognition receptor (PRR) genes DDX58 (RIG-I), IFH1 (MDA5) and TLR3, and accompanied by an increase > 400 times in IFNB1 expression, whereas no IFNA variation was found, as recently described (Hanners et al., 2016). Only one gene was found to be significantly downregulated, namely the chemokine CXCL8, an important mediator of the inflammatory responses to many viruses and bacteria (Fig. 3D). The antiviral response to ZIKV AS in NSCs was quite different as no genes were significantly upregulated (Fig. 3B). Interestingly, four genes were down regulated: CXCL8, similar to ZIKV AF, CXCL10, CASP1 and CTSS (Fig. 3D). As mentioned previously, the percentage of infected cells by the Asiatic strain at stronger MOI is not significantly higher, suggesting that the cytokine response remains weak in NSCs ZIKV AS-infected cells.
Fig. 3.
Anti-viral response in NSCs infected by ZIKV AF or AS.
(a) and (b) Volcano plots of genes modulated upon ZIKV infection in NSCs. mRNA from supernatants of NSCs infected with ZIKV AF (a) or ZIKV AS (b) at MOI 1 for 4 days were subjected to qRT-PCR analyses. The experiment was performed in triplicates and each point represents the mean. Statistically significant changes in fold regulation appear in the top-right cadran (genes upregulated) and top-left cadran (genes downregulated).
(c) and (d) Fold regulation of statistically significant genes modulated (upregulated (c) or downregulated (d)) upon ZIKV AF and AS-infection shown in (a) and (b). Results are expressed as means ± SD and analyzed using an unpaired t-test *p < 0.05.
This set of data suggest that these two ZIKV AF and ZIKV AS strains differentially affect cellular responses, whether during the cell cycle or for the anti-viral response, and could potentially lead to pathophysiological differences in vivo depending of the strain involved.
3.3. Human Astrocytes Are Permissive for ZIKV Infection
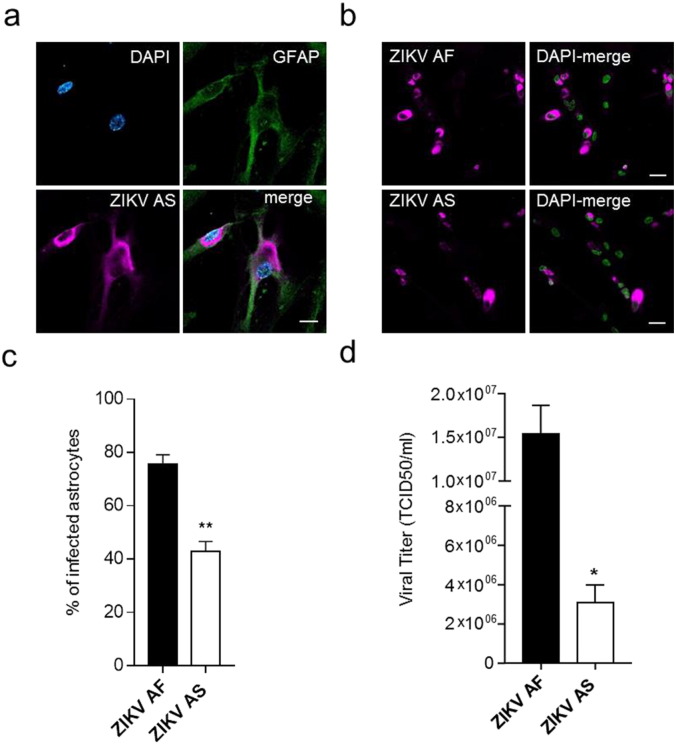
The putative ZIKV attachment factor AXL is highly expressed in astrocytes (Nowakowski et al., 2016). Moreover, astrocyte hypertrophy has been seen in animal models of ZIKV infection (Bell et al., 1971) and astrocytic activation is often described during Flavivirus infections (King et al., 2007). In this context, we characterized ZIKV AF and ZIKV AS susceptibility in this cell type. Using primary human astrocytes, we could detect efficient ZIKV infection (Fig. 4A). When we performed quantitative comparative analyses of the infection rate, we could show that (i) here also ZIKV AF led to a higher cellular infection rate than ZIKV AS and (ii) ZIKV AF and AS were potentially more efficient in terms of percentage of infection in astrocytes that in NSCs (75% vs 25% for ZIKV AF at MOI 0.1 and 43% vs 9.7% for ZIKV AS at MOI 0.1) (Fig. 4B and C). Consistently, viral titers determined by TCID50 showed a significant difference between strains, albeit less marked than in NSCs, suggesting that this feature could be found in several cell types (Fig. 4D). However, the difference between ZIKV AF and ZIKV AS production were less important than in NSCs.
Fig. 4.
Human astrocytes are permissive to ZIKV infection.
(a) IF of ZIKV AS-infected astrocytes (MOI 1, day 4 post-infection). The astrocytic marker GFAP is detected using a specific antibody (green) and the virus is labeled with a pan-flavivirus antibody (magenta) and nuclei by DAPI (cyan). Scale bar = 10 μm.
(b) IF of ZIKV AF and ZIKV AS-infected astrocytes (MOI 0.1, day 4 post-infection). The virus is labeled using a pan-flavivirus antibody (magenta) and nuclei by DAPI (false colored in green). Scale bar = 20 μm.
(c) Quantitative analyses show differences in rate of infection between ZIKV AF and ZIKV AS (MOI 0.1, day 4 post-infection). Three independent experiments were quantified with 150–200 cells per condition. Results are expressed as means ± SEM and analyzed using an unpaired t-test **p < 0.01.
(d) Viral titers from supernatant of ZIKV AF and AS-infected astrocytes (MOI 0.1, day 4 post-infection) were determined by TCID50 on Vero cells. Three independent experiments were quantified. Results are expressed as means ± SEM and analyzed using an unpaired t-test *p < 0.05.
These data open new questions regarding the cellular response in the brain after ZIKV infection as astrocytes play crucial roles in immune response, synaptogenesis and blood brain barrier (BBB) integrity among other processes.
4. Discussion
In this study, we show significant differences in the infectivity and cellular response between two Asian and African ZIKV strains. Using a combination of cell and molecular biology approaches, we observed in two human neural cell types (NSCs and astrocytes) a higher infectivity for the African strain. Similarly to the accumulating evidences reporting infection of neural stem or precursor cells by ZIKV, we report that both ZIKV AF and AS can infect IPSc-derived NSCs. However, ZIKV AF led to a stronger modulation of cellular homeostasis, in term of effects on cell cycle progression and anti-viral response. We also report that both strains of ZIKV can infect human astrocytes, opening new questions on the pathogenesis of this Flavivirus in the developing and adult brain as this most abundant cell type in the brain is responsible for multiple functions such as BBB maintenance, synaptogenesis and immune regulation among others (Molofsky and Deneen, 2015).
The reports of the first suspected cases of microcephaly in Brazil in 2015 (Schuler-Faccini et al., 2016, ECDC, 2015, COES, 2016) led to an unprecedented effort of the scientific community to understand the pathophysiological mechanisms of a virus barely characterized, despite its discovery almost 70 years ago. Numerous studies were subsequently published describing the placental transcytosis of ZIKV (Bayer et al., 2016, Miner et al., 2016) and highlighting the strong tropism for neural precursor cells (Tang et al., 2016, Qian et al., 2016, Lazear et al., 2016, Dang et al., 2016, Cugola et al., 2016). These observations, together with cohort studies and the demonstration of viral shedding in the amniotic fluid and in fetal tissue (brain included), provide strong conclusions regarding the link between ZIKV infection and microcephaly. Retrospective studies in French Polynesia also reported neurodevelopmental disorders associated with ZIKV infection (Besnard et al., 2016), suggesting that this feature of ZIKV was not specific to the Brazilian epidemic. Regarding Africa, due to the poor surveillance and traceability in the human health system, it is difficult to apprehend and predict the circulation of this virus (Meda et al., 2016). However, Cape Verde has reported an outbreak and several cases were recently found in Guinea-Bissau (both due to ZIKV of the Asian lineage) in 2015–2016. Occasional studies or reports show that ZIKV AF strains are currently circulating and support the need to closely monitor ZIKV AF (Baraka and Kweka, 2016, Grard et al., 2014, Van Esbroeck et al., 2016).
Surprisingly, we repeatedly detected a stronger infectivity, viral production and cellular response of the ZIKV AF strain ArB41644 vs ZIKV AS strain H/PF/2013 ex vivo in neural cells. Because there is strong probability that these strains have not derived due to extensive passages (no > 5 passages for both strains), our observations could suggest a potential difference in the physiopathology between the two strains. A rather common trait among Flaviviruses is their ability to access the CNS and cause neuronal impairment (Neal, 2014). However, some Flaviviruses can display strain-dependent neurovirulence. For instance, the neuroinvasive potential of West Nile virus (WNV) is strain-dependent but mostly because of difference in their ability to reach the CNS (Beasley et al., 2002). However, in some case this difference could also be due to different replication abilities in cells of the BBB or directly in neural cells, as was shown for WNV (Hussmann et al., 2013) and the Alphavirus Semliki Forest virus (Fazakerley et al., 2002). One mechanism responsible for difference in neurovirulence is the glycosylation status of the envelope protein (Kawano et al., 1993, Shirato et al., 2004). Crystal structure of ZIKV demonstrated that its envelop protein contains a glycosylation site at Asn154 (Dai et al., 2016, Kostyuchenko et al., 2016), similarly to the site responsible for neuroinvasiveness of WNV (Beasley et al., 2005). Moreover, passage history likely influenced glycosylation sites in the (original) MR-766 strain (Haddow et al., 2012). This observation, among other critics such as number and way of passages, are suggesting that MR-766 may not be the ideal African reference strain. In this context, a study described a stronger replication in a Brazilian ZIKV strain in NPCs as compared to the MR-766 strain, which therefore appears to have less capacity of replication than the low passage African strain used in our study (Cugola et al., 2016).
Regarding the anti-viral response occurring during ZIKV infections, few reports using different cell types led to different observations. ZIKV AS (H/PF/2013) has been shown to induce an innate antiviral response in primary human skin fibroblasts characterized by the upregulation of TLR3, DDX58 (RIG-I) and IFIH1 (MDA5) mRNA expression, three PRRs known to be activated by viral dsRNA (Hamel et al., 2015). This upregulation is associated with an increase of Type-1 IFN and several interferon-stimulated genes (ISGs) including OAS2, ISG15, and MX1. Several chemokines as CXCL10, or the inflammatory antiviral chemokine CCL5, were also induced by ZIKV AS in this cellular context. H/PF/2013 also stimulated the production of Type-I IFNs, ISGs, and pro-inflammatory cytokines in the human lung epithelial cell line A549 (Frumence et al., 2016). Human choriocarcinoma JEG-3 cells can be infected both by ZIKV AS (FSS13025 of Cambodian origin) and ZIKV AF (MR-766) leading to release of antiviral type III IFN as well as the ISG 2′-5′-oligoadenylate synthetase 1 (OAS1), whereas placental trophoblasts (PHT) appear to be refractory to ZIKV infection probably due to the fact that PHT cells constitutively release the anti-viral type III interferon IFNλ1 (Bayer et al., 2016). In the neuronal context, and similarly to what we detected for the ZIKV AF strain in NSCs, TLR3 was shown to be upregulated in cerebral organoids and human neurospheres after ZIKV MR-766 strain infection by RT-qPCR analysis (Dang et al., 2016). However, more recently, Hanners et al. demonstrated that ZIKV AS (PRVABC59, Puerto Rico, 2015) did not stimulate cytokine secretion in human neuroprecursors (hNPs) and in the THP-1 human monocytic cells (Hanners et al., 2016). Although a viral RNA mimic (pIC) can induce cytokine responses in hNPs, these neural progenitors appears therefore refractory to the induction of neuroinflammatory or neuroprotective cytokines in response to this ZIKV AS strain (Hanners et al., 2016). This is consistent with our observation in NSCs where H/PF/2013, contrary to ZIKV AF, did not lead to any upregulation of genes involved in anti-viral response and triggered the down-regulation of CXCL8, CXCL10, CASP1 and CTSS. Similarly to what was proposed by Hanners et al., our data strongly suggest that ZIKV AS could persist and replicate in a proportion of infected NSCs, albeit we cannot nevertheless exclude that ZIKV AS could infect more cells at longer time and lead to a stronger cellular impairment. This could however explain why ZIKV AS is found in newborns who were infected during the first trimester of pregnancy (Johansson et al., 2016). It is tempting to speculate that ZIKV AS being poorly cytopathic and immunogenic could establish cellular reservoir(s). This could lead to impairment of neuronal development through subtle mechanisms such as cell cycle modification and low noise apoptosis.
Finally, in the mature and developing brain, astrocytes are one of the first cell type to respond to viral infection (Furr and Marriott, 2012). Astrocyte hypertrophy has been detected in a mouse model of ZIKV infection (Bell et al., 1971) and is occurring for other Flavivirus CNS infection such as WNV (Hussmann et al., 2013), DENV (Lee et al., 2016) and the Alphavirus CHIKV (Inglis et al., 2016). Interestingly, AXL, a molecule suggested as a putative attachment receptor for ZIKV (Hamel et al., 2015) is highly expressed in NSCs but also in astrocytes (Nowakowski et al., 2016). Indeed, using single-cell RNA-seq approaches and confirming with immunohistochemistry, the authors showed that NSCs, radial glial cells, astrocytes and also microglia are expressing AXL and could therefore represent a target of choice for ZIKV (Nowakowski et al., 2016). Moreover recent studies described permissiveness of astrocytes to AS strains of ZIKV and that AXL receptor decreased ZIKV infection in human astrocytes ex vivo (Xu et al., 2016, Retallack et al., 2016). Recent histopathological studies in ZIKV-infected newborns showed the presence of viral antigens in glial cells (Martines et al., 2016). These sets of observations prompted us to monitor the permissibility of astrocytes to ZIKV AS and AF infection. Surprisingly, we found a higher infection rate in astrocytes than in NSCs for both lineages. One possible explanation would be a differential use of receptors between NSCs and astrocytes or a difference in AXL expression between these cell types. Interestingly, there was also a difference in infectivity between ZIKV AF and AS, albeit less pronounced than in NSCs. In this context, the effect of ZIKV infection on astrocyte homeostasis, in particular immune and anti-viral response will be particularly pertinent to study. Finally, astrocytes are constantly involved in cell-to-cell contact and could potentially favor viral spread to other cell types.
The ZIKV field is still developing, and critics regarding the use of the original ZIKV strain are starting to emerge. There is currently no appropriate African and Asian reference strains to allow proper comparison of results between laboratories. At this stage, we cannot disregard that the differences observed in our study are due to intrinsic abilities or differences in infectivity of these two ZIKV strains. Whether this difference could be explained by envelope variants and/or receptor usage or later in the replication cycle will be the aim of future studies. One caveat of our study is the few number of African and Asian strains used that did not allow us to address a general conclusion regarding differences between ZIKV lineages. However, our work address the important concern that circulating African ZIKV strains must be closely followed and that ZIKV strains could have different molecular patterns. Studies need to investigate the neurotropism and neurovirulence of more circulating ZIKV AF strains versus AS strains in order to establish their replication and cellular response abilities. It is not unlikely that ZIKV outbreaks occurred recently in Africa and remained undiscovered due to the very low capacities for detection of emerging or re-emerging pathogens in most of the continent (Meda et al., 2016). Understanding the basic mechanisms of ZIKV AF infection is therefore of crucial importance.
Funding Resources
This work was funded by REACTing through the ITMOs I3M and Neurosciences and La Région Languedoc-Roussillon (LR 0001146).
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
Conceived and designed the experiments, YS and SS; performed the experiments, YS, FL, CD, VF, OC, FL, AG, MM and SS; analyzed the data, YS, CD, VF, CFW, JPM, JML, PVDP, NN and SS; contributed reagents/materials/analysis tools, LB and ILG; contributed to the writing of the manuscript, YS and SS.
Acknowledgments
We thank Jean-François Delfraissy for the REACTing opportunity, the Montpellier Rio Imaging facility for microscope imaging and flow cytometry analyses, Eric J Kremer for the CFSE reagent, Neus Bayo-Puxan and Antonella Consiglio (Universidad of Barcelona) for human astrocytes and Anne-Sophie Gosselin for the C636 cells. JPM and SS are Inserm fellows.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.09.020.
Contributor Information
Yannick Simonin, Email: yannick.simonin@umontpellier.fr.
Sara Salinas, Email: sara.salinas@inserm.fr.
Appendix A. Supplementary Data
a. IF studies showing iPSCs expressing pluripotency markers TRA1-60 and iPSC-derived NSCs expressing neural progenitor markers Nestin and Pax6. Scale bar 100 µm. b. Transmitted light image of ZIKV AF and AS-infected NSCs at day 4 post infection show scarcely ECPs (zoom panels, black box are shown in the right). The virus is labeled using a pan-flavivirus antibody (magenta) and nuclei by DAPI (false colored in green). Scale bar = 10 μm.
References
- Aliota M.T. Characterization of lethal Zika virus infection in AG129 mice. PLoS Negl. Trop. Dis. 2016;10(4) doi: 10.1371/journal.pntd.0004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraka V., Kweka E.J. The threat of Zika virus in Sub-Saharan Africa — the need to remain vigilant. Front. Public Health. 2016;4(May):1–3. doi: 10.3389/fpubh.2016.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer A. Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection. Cell Host Microbe. 2016:1–8. doi: 10.1016/j.chom.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley D.W.C. Mouse neuroinvasive phenotype of West Nile virus strains varies depending upon virus genotype. Virology. 2002;296(1):17–23. doi: 10.1006/viro.2002.1372. [DOI] [PubMed] [Google Scholar]
- Beasley D.W.C. Envelope protein glycosylation status influences mouse neuroinvasion phenotype of genetic lineage 1 West Nile virus strains. J. Virol. 2005;79(13):8339–8347. doi: 10.1128/JVI.79.13.8339-8347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell T.M., Field E.J., Narang H.K. Zika virus infection of the central nervous system of mice by intracerebral inoculation of newborn and 5-week-old mice with Zika virus. Arch. Gesamte Virusforsch. 1971;193 doi: 10.1007/BF01249709. [DOI] [PubMed] [Google Scholar]
- Besnard M. Congenital cerebral malformations and dysfunction in fetuses and newborns following the 2013 to 2014 Zika virus epidemic in French Polynesia. Euro Surveill. 2016;21(13) doi: 10.2807/1560-7917.ES.2016.21.13.30181. [DOI] [PubMed] [Google Scholar]
- Brasil Martines R. Notes from the field: evidence of Zika virus infection in brain and placental tissues from two congenitally infected newborns and two fetal losses — Brazil, 2015. MMWR Morb. Mortal. Wkly Rep. 2016;65:1–2. doi: 10.15585/mmwr.mm6506e1. Early Release. [DOI] [PubMed] [Google Scholar]
- Calvet G. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect. Dis. 2016:653–660. doi: 10.1016/S1473-3099(16)00095-5. [DOI] [PubMed] [Google Scholar]
- Cauchemez S. Association between Zika virus and microcephaly in French Polynesia, 2013–15: a retrospective study. Lancet. 2016;6736(16):1–8. doi: 10.1016/S0140-6736(16)00651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COES INFORME EPIDEMIOLÓGICO No 10 – SEMANA EPIDEMIOLÓGICA (SE) 03/2016 (17 A 23/01/2016) MONITORAMENTO DOS CASOS DE MICROCEFALIA NO BRASIL. COES microcephalia. 2016. http://www.salud.gob.ec/wp-content/uploads/2015/12/COES-Microcefalias-Informe-Epidemiol-gico-10-SE-03-2016-27jan2016-12h04.pdf Available at. Accessed February 9, 2016.
- Cugola F.R. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534(7606):267. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L. Structures of the Zika virus envelope protein and its complex with a flavivirus broadly protective antibody. Cell Host Microbe. 2016;19(5):696–704. doi: 10.1016/j.chom.2016.04.013. [DOI] [PubMed] [Google Scholar]
- Dang J. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell. 2016:1–8. doi: 10.1016/j.stem.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick G.W.A., Kitchen S.F., Haddow A.J. Zika virus. I. Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952;46(5):509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- Dowall S.D. A susceptible mouse model for Zika virus infection. PLoS Negl. Trop. Dis. 2016;10(5) doi: 10.1371/journal.pntd.0004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC Zika Virus Epidemic in the Americas: Potential Association with Microcephaly and Guillain-Barré Syndrome. Rapid Risk Assessment. 2015. http://ecdc.europa.eu/en/publications/Publications/zika-virus-americas-association-with-microcephaly-rapid-risk-assessment.pdf Available at. Accessed February 9, 2016.
- Fazakerley J.K. A single amino acid change in the nuclear localization sequence of the nsP2 protein affects the neurovirulence of Semliki Forest virus. J. Virol. 2002;76(1):392–396. doi: 10.1128/JVI.76.1.392-396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frumence E. The South Pacific epidemic strain of Zika virus replicates efficiently in human epithelial A549 cells leading to IFN-β production and apoptosis induction. Virology. 2016;493:217–226. doi: 10.1016/j.virol.2016.03.006. [DOI] [PubMed] [Google Scholar]
- Furr S.R., Marriott I. Viral CNS infections: role of glial pattern recognition receptors in neuroinflammation. Front. Microbiol. 2012;3:1–12. doi: 10.3389/fmicb.2012.00201. JUN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcez P.P. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352(6287):816–818. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- Gong Z., Gao Y., Han G.-Z. Zika virus: two or three lineages? Trends Microbiol. 2016;xx:1–2. doi: 10.1016/j.tim.2016.05.002. August. [DOI] [PubMed] [Google Scholar]
- Grard G. Zika virus in Gabon (Central Africa) — 2007: a new threat from Aedes albopictus? PLoS Negl. Trop. Dis. 2014;8(2):1–6. doi: 10.1371/journal.pntd.0002681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddow A.D. Genetic characterization of Zika virus strains: geographic expansion of the Asian lineage. PLoS Negl. Trop. Dis. 2012;6(2) doi: 10.1371/journal.pntd.0001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel R. Biology of Zika virus infection in human skin cells. J. Virol. 2015;89(17) doi: 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanners N.W. Western Zika virus in human fetal neural progenitors persists long term with partial cytopathic and limited immunogenic effects. Cell Rep. 2016;15(11):2315–2322. doi: 10.1016/j.celrep.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussmann K.L. Differential replication of pathogenic and nonpathogenic strains of West Nile virus within astrocytes. J. Virol. 2013;87(5):2814–2822. doi: 10.1128/JVI.02577-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis F.M. Neuropathogenesis of Chikungunya infection: astrogliosis and innate immune activation. J. Neurovirol. 2016;22(2):140–148. doi: 10.1007/s13365-015-0378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.A. Zika and the risk of microcephaly. N. Engl. J. Med. 2016 doi: 10.1056/NEJMp1605367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärber G. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Arch. Exp. Pathol. Pharmakol. 1931;162(4):480–483. [Google Scholar]
- Kawano H. Genetic determinants of dengue type 4 virus neurovirulence for mice. J. Virol. 1993:6567–6575. doi: 10.1128/jvi.67.11.6567-6575.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N.J.C. Immunopathology of flavivirus infections. Immunol. Cell Biol. 2007;85(1):33–42. doi: 10.1038/sj.icb.7100012. [DOI] [PubMed] [Google Scholar]
- Kostyuchenko V.A. Structure of the thermally stable Zika virus. Nature. 2016 doi: 10.1038/nature17994. [DOI] [PubMed] [Google Scholar]
- Lazear H.M. A mouse model of Zika virus pathogenesis. Cell Host Microbe. 2016;19(5):720–730. doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.M. The flavivirus dengue induces hypertrophy of white matter astrocytes. J. Neurovirol. 2016:1–9. doi: 10.1007/s13365-016-0461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Zika virus infects neural progenitors in the adult mouse brain and alters proliferation brief report Zika virus infects neural progenitors in the adult mouse brain and alters proliferation. Cell Stem Cell. 2016:1–6. doi: 10.1016/j.stem.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martines R.B. Pathology of congenital Zika syndrome in Brazil: a case series. Lancet. 2016;6736(16):1–7. doi: 10.1016/S0140-6736(16)30883-2. [DOI] [PubMed] [Google Scholar]
- Meda N. Zika epidemics: why should the African continent be spared? Lancet. 2016;388(10042):337–338. doi: 10.1016/S0140-6736(16)31103-5. [DOI] [PubMed] [Google Scholar]
- Miner J.J. Zika virus infection during pregnancy in mice causes placental damage and fetal demise. Cell. 2016:1–11. doi: 10.1016/j.cell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky A.V., Deneen B. Astrocyte development: a guide for the perplexed. Glia. 2015;63(8):1320–1329. doi: 10.1002/glia.22836. [DOI] [PubMed] [Google Scholar]
- Musso D., Baud D., Gubler D.J. Zika virus: what do we know? Clin. Microbiol. Infect. 2016;22(6):494–496. doi: 10.1016/j.cmi.2016.03.032. [DOI] [PubMed] [Google Scholar]
- Neal J.W. Flaviviruses are neurotropic, but how do they invade the CNS? J. Infect. 2014;69(3):203–215. doi: 10.1016/j.jinf.2014.05.010. [DOI] [PubMed] [Google Scholar]
- Nowakowski T.J. Expression analysis highlights AXL as a candidate Zika virus entry receptor in neural stem cells. Cell Stem Cell. 2016:1–6. doi: 10.1016/j.stem.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165(5):1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retallack H. BioRxiv. 2016. Zika virus in the human placenta and developing brain: cell tropism and drug inhibition. Preprint. [Google Scholar]
- Rossi S.L. Characterization of a novel murine model to study Zika virus. Am. J. Trop. Med. Hyg. 2016;94(6) doi: 10.4269/ajtmh.16-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaramozzino N. Comparison of flavivirus universal primer pairs and development of a rapid, highly sensitive heminested reverse transcription-PCR assay for detection of flaviviruses targeted to a conserved region of the NS5 gene sequences. J. Clin. Microbiol. 2001;39(5):1922–1927. doi: 10.1128/JCM.39.5.1922-1927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler-Faccini L. Possible association between Zika virus infection and microcephaly — Brazil, 2015. MMWR Morb. Mortal. Wkly Rep. 2016;65(3):59–62. doi: 10.15585/mmwr.mm6503e2. [DOI] [PubMed] [Google Scholar]
- Shen S. Phylogenetic analysis revealed the central roles of two African countries in the evolution and worldwide spread of Zika virus. Virol. Sin. 2016;31(2):118–130. doi: 10.1007/s12250-016-3774-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K. Viral envelope protein glycosylation is a molecular determinant of the neuroinvasiveness of the New York strain of West Nile virus. J. Gen. Virol. 2004;85:3637–3645. doi: 10.1099/vir.0.80247-0. [DOI] [PubMed] [Google Scholar]
- Tang H. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell. 2016:1–4. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Esbroeck M. Letter to the editor: Specificity of Zika virus ELISA: interference with malaria. Euro Surveill. 2016;21(21) doi: 10.2807/1560-7917.ES.2016.21.21.30237. [DOI] [PubMed] [Google Scholar]
- Wang L. From mosquitos to humans: genetic evolution of Zika virus. Cell Host Microbe. 2016;19(5):561–565. doi: 10.1016/j.chom.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med. 2016 doi: 10.1038/nm.4184. Epub ahead. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
a. IF studies showing iPSCs expressing pluripotency markers TRA1-60 and iPSC-derived NSCs expressing neural progenitor markers Nestin and Pax6. Scale bar 100 µm. b. Transmitted light image of ZIKV AF and AS-infected NSCs at day 4 post infection show scarcely ECPs (zoom panels, black box are shown in the right). The virus is labeled using a pan-flavivirus antibody (magenta) and nuclei by DAPI (false colored in green). Scale bar = 10 μm.