Abstract
The Amber Lipid14 force field is expanded to include cholesterol parameters for all-atom cholesterol and lipid bilayer molecular dynamics simulations. The General Amber and Lipid14 force fields are used as a basis for assigning atom types and basic parameters. A new RESP charge derivation for cholesterol is presented, and tail parameters are adapted from Lipid14 alkane tails. 1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayers are simulated at a range of cholesterol contents. Experimental bilayer structural properties are compared with bilayer simulations and are found to be in good agreement. With this parameterization, another component of complex membranes is available for molecular dynamics with the Amber Lipid14 force field.
Graphical abstract
INTRODUCTION
Membranes are composed of a diverse mix of glycerophospholipids, sphingolipids, glycolipids, and sterols. Cholesterol is a common yet important component of mammalian cell membranes, and sterols may be found in mammalian membranes in molar fractions up to 50%.1,2
Cholesterol affects the phase and order of lipid bilayers. Structural studies indicate that the effects of cholesterol on lipid bilayers are dependent on lipid type.3,4 In certain bilayer types, cholesterol facilitates tighter packing of the lipid bilayer, resulting in lateral compression of the bilayer. In some lipid bilayers, increased cholesterol content introduces a liquid ordered phase, which is characterized by ordered saturated and unsaturated chain tails. The gel phase differs from the liquid ordered phase in that the liquid ordered phase lipids still diffuse through the bilayer. The liquid ordered phase has been observed with certain bilayer mixtures composed of sphingolipids, glycolipids, and cholesterol.5,6
The liquid ordered phase of lipid bilayers also may be related to raft assemblies in biological membranes. Lipidomic studies indicate membranes with rafts contain a mixture of components similar to model bilayers.2 Model lipid bilayers in the liquid ordered phase are often compared with raft assemblies in membranes;3 however, large membrane raft domains have not been directly observed in biological membranes.4 While there are experimental approaches to capture membrane dynamics, it still remains difficult to resolve structures of membranes at atomic levels of detail.7
Cholesterol not only plays a structural role in cellular membranes but also may influence membrane protein dynamics and function. Cholesterol has been identified as a ligand to certain membrane receptors and in some cases has been shown to cause conformational shifts in protein structure.8,9 One such example is the case of serotonin 1A receptors.9 This suggests that the surrounding membrane environment is important for the dynamics of membrane proteins.
It is also notoriously difficult to resolve atomic structures of membrane proteins. While the number of resolved solution protein structures continues to grow exponentially, the number of resolved membrane protein structures grows at a fraction of that rate.10 Currently, there are only 512 unique membrane protein structures available.10
Therefore, complementary methods to study the structure of these membranes and membrane proteins are valuable. One widely used computational method, molecular dynamics (MD), can be applied to bilayers and membrane environments.11–14 Previous molecular dynamics of bilayers required extensive parameterization of lipid force fields to reproduce lipid bilayer structure and dynamics.13 To accurately predict membrane bound protein dynamics it is necessary to have an accurate model of the complex surrounding membrane environment. While sterols are important membrane components, the Amber family of molecular force fields has not included an extensively tested parameter set for cholesterol in diverse membrane environments.
Many studies and parameter sets have been developed for molecular dynamics simulations of lipid bilayers and membrane proteins. Cholesterol has been included in MD simulations of lipid bilayers with parameterization at different levels of molecular detail. The MARTINI force field includes a coarse-grain model for cholesterol for use with their other lipid parameters.15 Gromacs force fields (GROMOS) include several united-atom cholesterol models.16 All-atom parameters for cholesterol have also been developed for molecular dynamics simulations. Cournia et al. published an all-atom cholesterol parameter set for molecular dynamics; however, simulations with that parameter set requires an additional constant surface tension term.17 The Charmm force fields have lipid parameter sets including parameterization for cholesterol.18 Recently, the Slipids force field was expanded to include additional lipid components and cholesterol.19
Previously, the Amber Lipid11 force field included a basic parameter set for cholesterol;12 however, with the recent release of the Lipid14 force field and changes in the parameterization strategy for lipids, a detailed investigation of cholesterol parameters and their interaction with Lipid14 bilayers was warranted.13 Because of the importance of cholesterol on biological bilayer function and structure, a cholesterol force field developed based on the parameterization strategy of Lipid14 is presented in this article. The Lipid14 parameter set is expanded to include cholesterol for Lipid14 bilayer MD simulations.
The parameterization of cholesterol presented here is based on the Lipd14 set of parameters required to independently reproduce relevant physical, thermodynamic, and quantum data. Parameterization was modular and independent of the final validation simulations. The final test set consisted of simulations of phospholipid and cholesterol bilayer mixtures. Lipid and cholesterol bilayer molecular dynamics simulations were compared to available bilayer structural data at a range of cholesterol fractions.
PARAMETERIZATION OF CHOLESTEROL
Previous all-atom cholesterol force fields included several different strategies for the refinement of force field parameters. All previous publications included atomic partial charges for the cholesterol molecule.17–19 Some cholesterol parameterizations have examined the van der Waals parameters of model molecules to fit experimental heats of vaporization and densities.18 The cholesterol tail dihedral parameters have been compared with quantum energies from torsion scans.18 Other parameterizations have investigated fitting parameters to molecular frequencies or examining the cholesterol partition coefficients.15,17
For this work, initial cholesterol parameters were drawn from the General Amber Force Field (GAFF) included in the Amber 14 release.20 The Amber 14 program Antechamber was used to automatically assign atom types from GAFF and their corresponding bond, angle, and dihedral parameters. The following sections describe further modifications to the GAFF parameters necessary for cholesterol molecular dynamics.
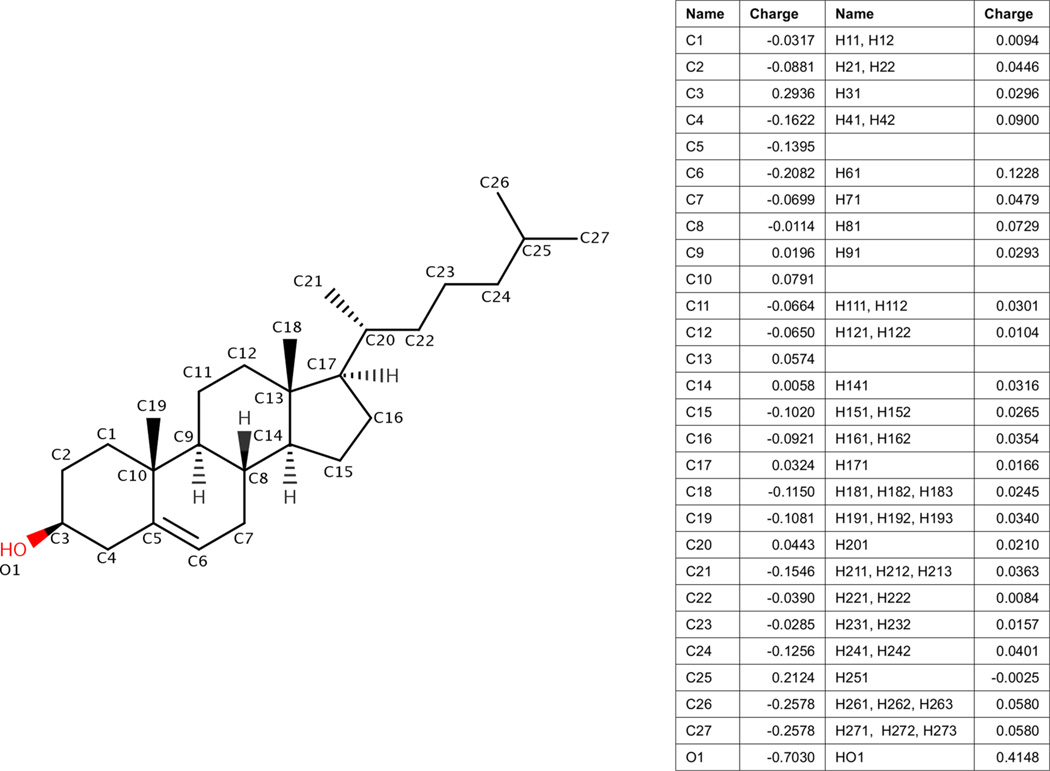
Figure 1 shows the molecular structure of cholesterol. The atom types and the partial charges of cholesterol obtained in this parameterization are listed in Figure 1. The full parameter set is included in the Supporting Information in Figure S3, Table S3, and a full printout of the parameter file. The parameters will be freely available as an update to AmberTools 15.
Figure 1.
Partial charges for cholesterol in Lipid14 bilayers. Partial charges were derived with the same method as charge derivation of units for Lipid14.13 Atom names and charges are listed in the table along with bonded hydrogen atom names and charges.
Partial Atomic Charges
Partial atomic charges are necessary for force fields using the Amber energy function. Partial charges were refined with the RESP charge derivation method as described in Lipid14.13 The charge derivation strategy of Lipid14 is compatible with the other all-atom Amber force fields. Charges were refined using a multiconformation RESP method compatible with the Amber RESP procedure used for proteins, nucleic acids, and the GAFF. An implicitly Boltzmann-weighted multiconformation RESP fit was calculated for the entire cholesterol molecule.21 No additional constraints upon the partial charges were used in the RESP charge derivation, consistent with the Lipid14 derivation.
Preliminary partial atomic charges were needed for initial MD of cholesterol. Charges for preliminary MD of a single cholesterol molecule were refined using the Amber RESP procedure.21 The molecular geometry of a single cholesterol was optimized, and the ESP was calculated at the HF/6–31G* level of theory and basis set with Gaussian 2009.22 Besides the normal Amber RESP constraints, no additional capping groups or constraints were used for cholesterol partial atomic charges.
MD of cholesterol was simulated with 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) bilayers. The same protocol for MD was used as described for production MD for Lipid14 bilayers.13 POPC and POPE were chosen because they include the two main phospholipid head groups included in Lipid14 as well as saturated and unsaturated tails. Starting bilayer conformations were obtained with the CHARMM-GUI membrane builder.23 Each system was simulated for 125 ns with 0.25 molar fraction cholesterol in a bilayer with 128 total lipid and cholesterol molecules. POPC was simulated at 303 K with 32 TIP3P water molecules per bilayer molecule.24 POPE was simulated at 310 K with 32 TIP3P water molecules per bilayer molecule. Each simulation included 150 mM K+ and Cl− molecules using Joung et al.’s ion parameters.25
Random structures were extracted from the molecular dynamics trajectories of cholesterol and POPC and POPE. Fifty cholesterol conformations were extracted from the POPC simulation, and 50 conformations were extracted from POPE simulation. RESP charges were obtained at the HF/6–31G* level for each individual conformation. The mean partial atomic charge from all conformations was used for the final charge set. The partial charges of the terminal methyl groups at carbon 26 and 27 were adjusted to their mean value because they are equivalent methyl groups. Because the structures were extracted from an initial MD simulation, the mean RESP charges are assumed to be implicitly Boltzmann-weighted.
Lipid14 Alkane Parameters
A major dynamic portion of cholesterol is its alkane tail. Cholesterol is normally oriented in bilayers such that the hydroxyl group of the sterol is oriented toward the polar heads of the lipids. The cholesterol alkane tail is usually oriented toward the nonpolar tails of the lipids. Thus, the cholesterol tails normally interact with the other lipid tails of the bilayer. As presented in Lipid14, the phospholipid tail parameters are very important for overall bilayer structure and dynamics in tensionless MD simulations.13 Because of issues with the order of GAFF long alkane chain parameters, the Lipid14 tail parameters were refit.11,26 RESP charges were derived for each alkane chain with capping groups. The hydrogen van der Waals parameters of long alkane chains were adjusted such that simulations of pure alkanes matched physical and thermodynamic data. The alkane dihedral angles were also fit to torsional scan quantum energies.
Therefore, instead of using the GAFF parameters for cholesterol tails, Lipid14 alkane parameters were applied.13 All carbon and hydrogen atoms in the cholesterol tail were assigned the default saturated tail sp3 hybridized carbon and the associated hydrogen atom type from Lipid14. The cholesterol tails include all of the modifications from Lipid14 and are thus consistent with Lipid14 molecules in the interior environment of the bilayer.
Sterol Ring Parameters
Parameters were applied from the General Amber Force Field and Lipid14 according to their atom type.13,20 Given the size and rigidity of the sterol portion of the molecule, this portion of the molecule may significantly influence bilayer structure.18 Therefore, the van der Waals parameters of the sterol group hydrogens were re-examined. A ringed model molecule, trans-decalin, was simulated with MD under a range of conditions. Trans-decalin was also simulated with various mixtures of hexadecane, which models the mixing of cholesterol with phospholipid tails. Parameters for trans-decalin were taken from cholesterol and parameters for hexadecane were adapted from Lipid14.13 The full details of trans-decalin and hexadecane parameters are presented in the Supporting Information (Figure S1, Table S1).
To evaluate the van der Waals parameters, trans-decalin MD simulations were compared with experimental molar volume, density, and enthalpy of vaporization measurements.27 Molar volume was calculated directly from MD simulation system volume. Average bulk density was calculated as the simple average calculated from the volume of the system and the mass of molecular components at the measured temperature. Enthalpy of vaporization ΔHvap was calculated in the same manner as in Lipid14 (eq 1).13 Enthalpy of vaporization calculations require two sets of simulations: individual trans-decalin molecules in gas-phase and liquid-phase trans-decalin systems both at the vaporization temperature.
Gas-phase trans-decalin systems consisted of multiple simulations of a single trans-decalin molecule. Liquid-phase trans-decalin systems were assembled with 256 trans-decalin molecules using the program Packmol.28 Mixtures of 256 molecules of trans-decalin and hexadecane were also assembled at various molar fractions of trans-decalin and hexadecane.
Simulations employed SHAKE constraints for bonds with hydrogen and used a 0.002 fs time step.29 All systems were simulated with MD for 20 ns. Gas-phase trans-decalin was simulated in the constant volume ensemble at a temperature of 461.45 K using the Langevin thermostat and 1 ps−1 collision frequency.30 Pure liquid-phase trans-decalin simulations were simulated at 298.15 and 461.45 K. Liquid-phase mixtures of trans-decalin and hexadecane were simulated at 298.15 K. The simulations used the Berendsen barostat with a pressure relaxation time of 1 ps.31 Periodic boundary conditions were set with the particle mesh Ewald method for long-range electrostatics interactions with a 10 Å cutoff for nonbonded interactions.32
Table 1 summarizes the enthalpy of vaporization from the MD simulations as well as experimental constants. From simulation, the enthalpy of vaporization was calculated to be 10.03 kcal/mol, while the reported experimental value is 9.61 kcal/mol.27 MD with the current sterol van der Waals parameters slightly overestimates the enthalpy of vaporization. The predicted liquid density of trans-decalin at 298.15 K was 0.8641 g/cm3, which reproduced the experimental constant of 0.8659 g/cm2.27 The agreement between simulation and experiment for densities suggests that the van der Waals parameters for trans-decalin may be acceptable for predicting volumes of cholesterol in lipid bilayers.
Table 1.
Trans-Decalin Enthalpy of Vaporization ΔHvap and Density ρ
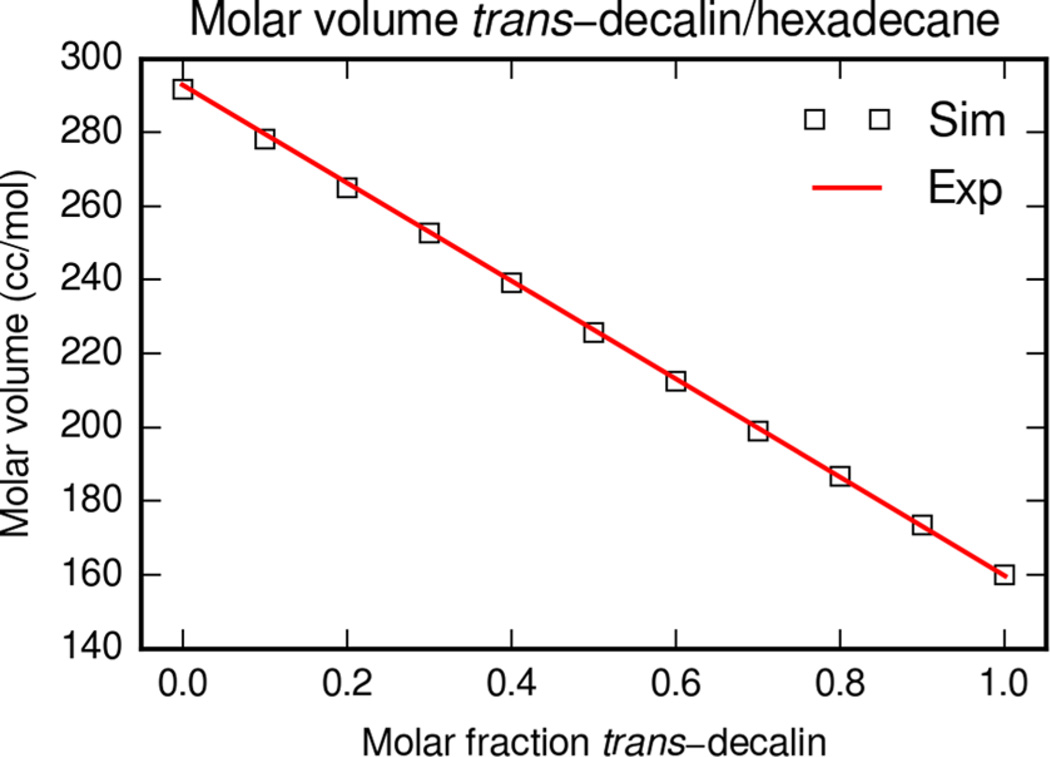
trans-Decalin was also simulated in binary mixtures of hexadecane to examine the molar volume. The mean molar volume was calculated from the equilibrated MD trajectory and compared with experimental molar volumes. The experimental molar volume of pure trans-decalin from Benson et al., the molar volume of pure hexadecane from Fuchs et al., and the excess molar volumes of binary mixtures from Letcher et al. are shown in Figure 2. The simulation and experimental molar volumes exhibit the same trend across the range of trans-decalin and hexadecane mixtures.
Figure 2.
Molar volume of trans-decalin and hexadecane mixtures at 298 K. Experimental values are calculated from Benson et al. and Fuchs et al. combined with excess molar volumes from Letcher et al.33–35 Simulation molar volumes are from MD simulations of trans-decalin with cholesterol parameters and hexadecane with the default Lipid14 parameters.
Simulations of trans-decalin indicate that the GAFF hydrogen van der Waals parameters are able to reproduce physical and thermodynamic data for this molecule. This is consistent with recent studies that have investigated GAFF parameters for a range of small organic molecules.36 Therefore, no further changes to the GAFF hydrogen van der Waals parameters were applied to the cholesterol sterol rings.
Molecular Dynamics of Lipid Bilayers with Cholesterol
For further testing and validation of the cholesterol parameters, lipid bilayer types with available structural experimental data were simulated. 1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) are three common bilayer types used in cholesterol structural studies.
A summary of the validation simulation system parameters is included in Table 2. This Table lists the molar fraction of cholesterol in the bilayer, Xc, and the respective number of lipids and cholesterol, Nlip and Nchl. The number of water molecules per bilayer molecule (lipid and cholesterol), nw, is also listed.37,38 Molar fractions of cholesterol were considered in the range from 0.0 to 0.5, a common physiological range of cholesterol content.1 Each bilayer was simulated in triplicate for 200 ns resulting in total sampling of 10.8 µs.
Table 2.
Summary of Cholesterol and Lipid Bilayer Simulations at Varying Cholesterol Molar Fractionsa
| Xc | Nlip | Nchl | sim. time (ns) | T (K) | nw | |
|---|---|---|---|---|---|---|
| DMPC | 0.0 | 128 | 0 | 3 × 200 | 303 | 25.6 |
| DMPC | 0.1 | 116 | 12 | 3 × 200 | 303 | 25.6 |
| DMPC | 0.2 | 102 | 26 | 3 × 200 | 303 | 25.6 |
| DMPC | 0.3 | 90 | 38 | 3 × 200 | 303 | 25.6 |
| DMPC | 0.4 | 76 | 52 | 3 × 200 | 303 | 25.6 |
| DMPC | 0.5 | 64 | 64 | 3 × 200 | 303 | 25.6 |
| DOPC | 0.0 | 128 | 0 | 3 × 200 | 303 | 32.8 |
| DOPC | 0.1 | 116 | 12 | 3 × 200 | 303 | 32.8 |
| DOPC | 0.2 | 102 | 26 | 3 × 200 | 303 | 32.8 |
| DOPC | 0.3 | 90 | 38 | 3 × 200 | 303 | 32.8 |
| DOPC | 0.4 | 76 | 52 | 3 × 200 | 303 | 32.8 |
| DOPC | 0.5 | 64 | 64 | 3 × 200 | 303 | 32.8 |
| POPC | 0.0 | 128 | 0 | 3 × 200 | 303 | 31.0 |
| POPC | 0.07 | 120 | 8 | 3 × 200 | 303 | 31.0 |
| POPC | 0.15 | 108 | 20 | 3 × 200 | 303 | 31.0 |
| POPC | 0.34 | 84 | 44 | 3 × 200 | 303 | 31.0 |
| POPC | 0.5 | 64 | 64 | 3 × 200 | 303 | 31.0 |
| POPC | 0.6 | 52 | 76 | 3 × 200 | 303 | 31.0 |
In the initial input structures, cholesterol molecules were evenly distributed between the two bilayer leaflets. Initial structures at varying cholesterol contents were obtained using the CHARMM-GUI web server.23 Structures were then converted to the Amber Lipid14 naming convention with the charmmlipid2amber.py script, available with AmberTools v14.39 Formatted structure files were loaded into the program Leap, and parameters and topology were assigned. Glycerophospholipid parameters from Lipid14 were used for the lipids.13 Each structure was solvated with TIP3P water molecules and contained a monovalent ionic concentration of 150 mM K+ and Cl− with parameters from Joung et al.24,25
The simulation protocol used for validation of the parameters was consistent with the Lipid14 validation protocol.13 The protocol was designed with several stages including linear heating and equilibration of the system prior to production MD.
The initial structures were first minimized using 5000 steps of steepest descent followed by 5000 steps of conjugate gradient minimization. Each system was then heated in two stages. In the first heating stage the system target temperature was slowly heated from 0 to 100 K over 5 ps with a constant volume MD simulation. The Langevin thermostat was used with a collision frequency of 1.0 ps−1.30 The second heating stage then increased the target temperature to production temperature over 1.0 ns with a constant pressure MD simulation. The Langevin thermostat was used with the Berendsen barostat with a target pressure of 1.0 bar and a relaxation time of 1.0 ps.30,31 During each heating simulation, all atoms of the bilayer were restrained with a weak harmonic restraint to the initial structure using a force constant of 10 kcal mol−1 Å−2 to prevent large structural deviations during heating.
After heating, production dynamics was simulated with constant temperature and pressure with the Langevin thermostat and the anisotropic Berendsen barostat.30,31 The thermostat target temperature used was specific to each bilayer type (Table 2) and a collision frequency of 1.0 ps−1 was set. The barostat reference pressure was 1.0 bar, and a relaxation time of 1.0 ps was used. In all simulations, the SHAKE algorithm was used to constrain bonds involving hydrogen with a relative tolerance of 1 × 10−7.29 A 0.002 ps time step was used in all stages of dynamics. The particle mesh Ewald summation method was used for long-range electrostatics with a real-space cutoff of 10.0 Å for the electrostatics.32 The same cutoff was used for van der Waals interactions. A long-range dispersion correction was applied to the energy and pressure beyond the cutoff.40 Lipid bilayer equilibration was monitored via system volume and density. Equilibration times were between 25 and 50 ns for all systems, and there was no correlation between bilayer composition and equilibration times.
All simulations used the GPU-accelerated version of the MD program Pmemd using the SPFP precision model of Le Grand and Walker.39,41–44 Each bilayer simulation consisted of three independent runs, begun from different initial random seeds for a duration of 200 ns each. Analysis was conducted using the AmberTools programs Ptraj and Cpptraj on the equilibrated portion of the trajectories.39,45
VALIDATION
There is limited experimental data for lipids containing cholesterol, but some data can be directly compared with simulation bilayer structural properties. Bilayers have been studied experimentally at a range of cholesterol contents and conditions.46 Along with estimates of area per molecule and volume per molecule of the bilayer, there are several other data sets available for comparison with experiment. Simulation results may be compared with X-ray and neutron scattering form factor data and electron density profiles of these bilayers. NMR studies provide important information regarding cholesterol deuterium order parameters and lipid acyl chain order. In the following sections, the cholesterol and lipid bilayers are compared with experimental structural properties as well as other all-atom force fields.
Volume per Molecule
The volumes of a given bilayer under various conditions may be obtained from experiments by fitting X-ray and neutron scattering results to bilayer models and measurements of volume per bilayer molecule are precise.47 In the simulation of mixed bilayers, the average volume per molecule for both lipid and cholesterol molecules was calculated using simulation periodic box dimensions and then subtracting the volume of water molecules. The volume per molecule of the bilayer is defined as the average volume per molecule divided by the number of molecules (including cholesterol and lipids) in the bilayer. The volume of water was estimated to be 30.51 Å3 from MD simulations of neat TIP3P water molecules under identical production simulation conditions.
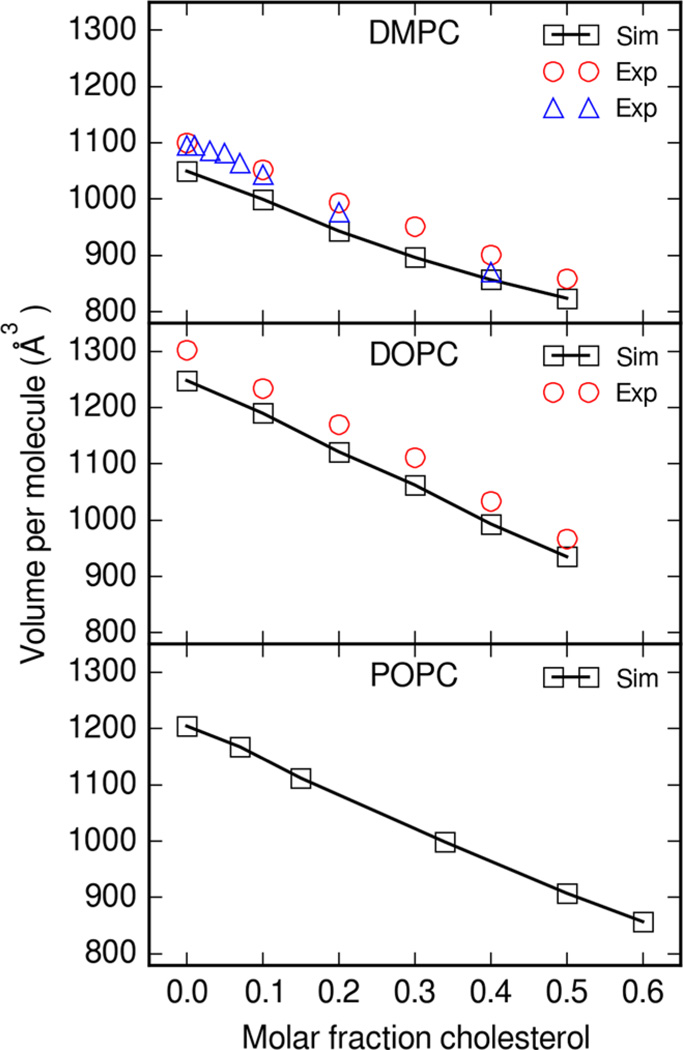
Figure 3 shows the change in average volume per molecule while varying the cholesterol content in bilayers. Averages for all simulation properties were calculated as the mean value across all three 200 ns simulations. As the fraction of cholesterol increases, the average volume per molecule decreases. The change in the volume per molecule with change in the molar fraction of cholesterol is reproduced between simulation and experiment. While the simulation volume per lipid is systematically underestimated in both DMPC and DOPC simulations, the change in volume follows the same trend as experiment. Some variability also exists between reported experimental values at different temperatures. Volume per molecule in pure Lipid14 systems were also systematically underestimated.13 As discussed in the Lipid14 article, this may be due to an underestimation of phospholipid headgroup van der Waals parameters.
Figure 3.
Average volume per molecule for lipid and cholesterol bilayers. Boxes are the mean simulation volume per molecule across three simulations. Circles are experimental volume per average molecule at 30 °C from Greenwood et al.,47 and triangles are values at 35 °C from Hodzic et al.48
Bilayer Thickness
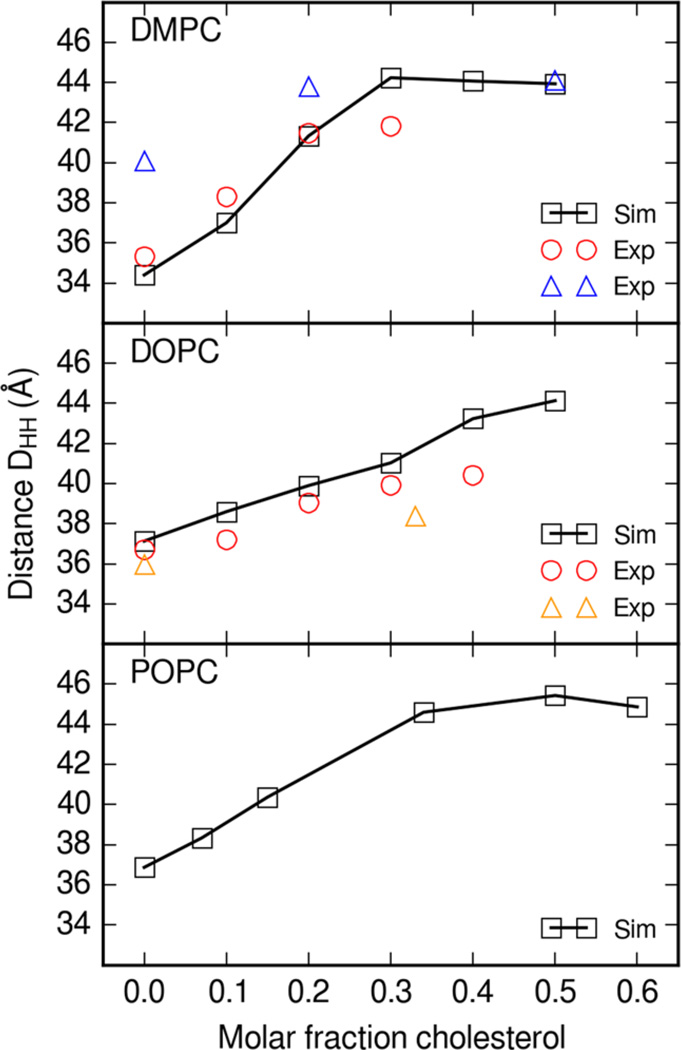
Another available structural measurement is that of bilayer thickness. Bilayer head-to-head (DHH) thickness was calculated from electron density profiles of equilibrated production simulation data. Bilayer thicknesses from each simulation were averaged and the mean values are plotted in Figure 4. Simulation electron densities were obtained with Hannes Loeffler’s Ptraj and Cpptraj program modifications available in AmberTools 14.39,45 Head-to-head distance was calculated as the distance between the two maxima of electron density corresponding to the head groups.
Figure 4.
Head-to-head thickness DHH of lipid and cholesterol bilayers. Thickness is calculated from the lipid and cholesterol electron density profile. In DMPC, circles are experimental thicknesses at 30 °C from Pan et al.46 and upward triangles are thicknesses at 30 °C from Pencer et al.49 In DOPC, upward triangles are at 20 °C from Gandhavadi et al.50
Figure 4 shows the dependence of bilayer head-to-head thickness on cholesterol content. As the molar fraction of cholesterol increases, the thickness of the bilayer increases. The simulation DMPC thickness falls within the range of experimental values from Pan et al. and Pencer et al.46,49 Simulation DOPC thickness follows the trend of experimental results from Pan et al.46
Bilayer thicknesses were previously calculated in comparable simulations with other force fields. The Charmm C36c force field presented by Lim et al. includes mixed lipid bilayer simulations from which electron density profiles.18 Simulations of phospholipid and cholesterol bilayers with the C36c force field yielded head-to-head bilayer thicknesses of ~38 Å and ~42 Å DMPC with 10% cholesterol and 30% cholesterol, respectively.18 Bilayers simulated with Lipid14 DMPC and cholesterol resulted in bilayer thicknesses of ~37.5 and ~44 Å for 10 and 30% cholesterol, respectively. The estimated thickness from simulation of Charmm C36c DOPC bilayers was ~40 Å with 10% cholesterol.18 Simulations of Lipid14 DOPC and 10% cholesterol bilayers yielded an estimated thickness of ~37.5 Å.
The Slipids force field by Jämbeck et al. also includes bilayer thicknesses for cholesterol and lipid simulations.19 For simulations of DMPC bilayers with Slipids parameters, the head-to-head thickness is slightly lower overall compared with the Lipid14 simulations by ~2 Å. For DOPC, Lipid14 simulations follow the trend of Slipids simulations with DOPC bilayer thickness with thicknesses within ~1 Å of each other. Simulations of membranes with Lipid14 parameters yield bilayer thickness that falls within the range of thickness reported from Charmm and Slipids simulations.
DMPC and DOPC Order Parameters
NMR order parameters provide important information about glycerophospholipid acyl tail configurations. Experiments with 2H NMR or 1H–13C NMR measure the splitting or coupling that is proportional to an order parameter value. The deuterium order parameter |SCD| is defined by the equation
| (1) |
The angle between the C–D vector and the bilayer normal is defined as θ. Simulation deuterium order parameters were calculated from equilibrated production trajectories with Hannes Loeffler’s Ptraj program modifications.39,45
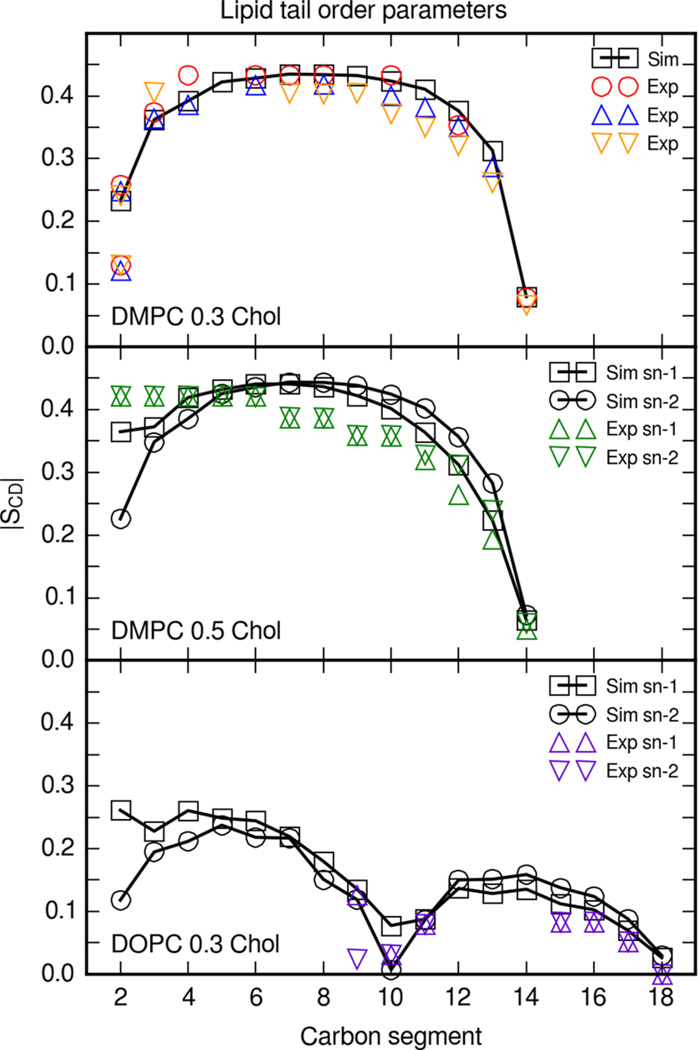
In Figure 5, the deuterium order parameter |SCD| is shown as a function of each carbon atom along the acyl chain. Order parameters were calculated for each lipid tail at every simulated molar fraction of cholesterol; however, experimental order profiles only exist for the cholesterol and DMPC and DOPC bilayer compositions shown in Figure 5. Simulation data are shown for DMPC at 0.3 molar fraction cholesterol. Order parameters from these simulations fall within the range of experimental order parameters.
Figure 5.
Deuterium order parameters |SCD| for DMPC and DOPC tails in bilayers with cholesterol. Simulation profiles were calculated directly from the trajectory C–H vectors and averaged across the three simulations. DMPC and 0.3 molar fraction cholesterol order parameters shown are for the sn-2 tail. DMPC and 0.3 molar fraction cholesterol: Circles are experimental order parameters at 25 °C from Douliez et al.,51 upward triangles are at 25 °C from Urbina et al.,52 and orange downward triangles are with 0.33 molar fraction cholesterol at 30 °C from Vermeer et al.53 DMPC and 0.5 molar fraction cholesterol: Upward triangles and downward triangles are sn-1 and sn-2 perdeuterated order parameters at 40 °C from Trouard et al.54 DOPC and 0.3 molar fraction cholesterol: Upward triangles and downward triangles are sn-1 and sn-2 order parameters at 37 °C from Warschawski et al.55
Cholesterol order parameters have been calculated from simulations of bilayers with other cholesterol force fields over a range of cholesterol contents.18,19 Order parameters for Charmm C36c simulations of DMPC and 0.3 molar fraction cholesterol have a similar order profile;18 however, simulations of bilayers with the C36c force field have a slightly lower order plateau (~0.38). The order profile of the sn-2 8–13 carbons from Slipids simulations resembles the order parameters from simulations with Lipid14 parameters, although lipids from Lipid14 simulations were more ordered.19 The order parameters presented in this article lie within the range of experimental order parameters at 25 and 30 °C and follow a similar trend as for order parameters calculated from simulations with the Charmm C36c and Slipids force fields.
Simulation deuterium order parameters for DMPC at 0.5 molar fraction cholesterol are compared with order parameters from diperdeuterated DMPC experiments from Trouard et al., in which every carbon along the hydrocarbon tails is deuterated.54 Note that perdeuterated experimental order parameters are usually assumed to monotonically decrease along the carbon chain.56 Myristoyl order parameters calculated from bilayer simulations with the Slipids force field are comparable with Lipid14 myristoyl order parameters, although with lower order in the plateau region for the sn-1 tail.19
DOPC order parameters with 0.3 molar fraction cholesterol were reported by Warschawski.55 DOPC and cholesterol bilayers simulated under the same conditions revealed that the order parameters for carbons 9–11 and 15–18 resemble but slightly overestimated the experimental values.
Order parameters for Slipids DOPC bilayer simulations with cholesterol showed that the oleoyl order is higher than experiment.19 Lipid14 bilayer simulations, in comparison, have oleoyl order parameters closer to the experimental results presented by Warschawski.
POPC Order Parameters
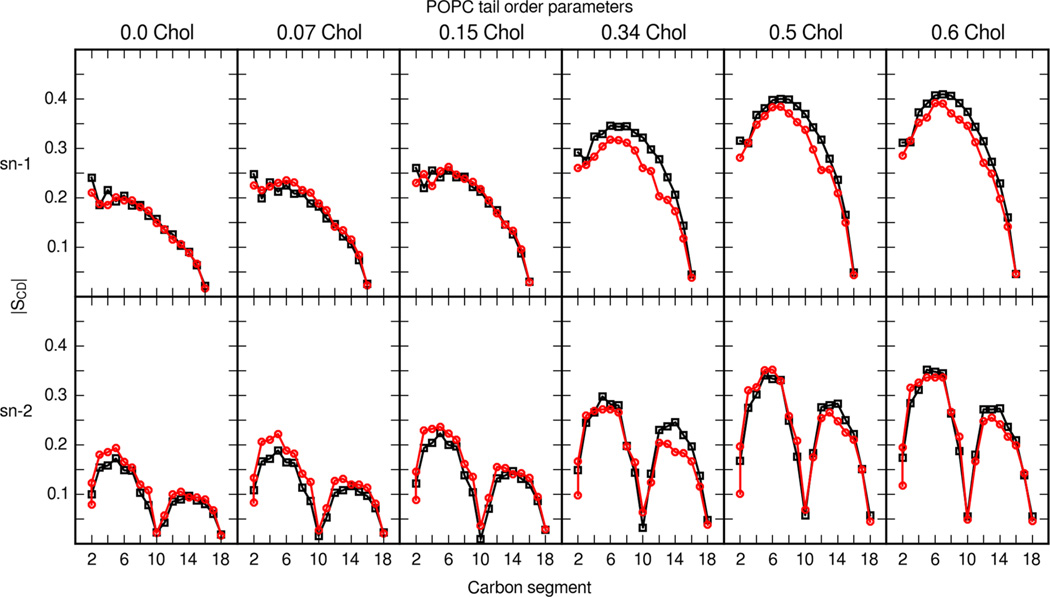
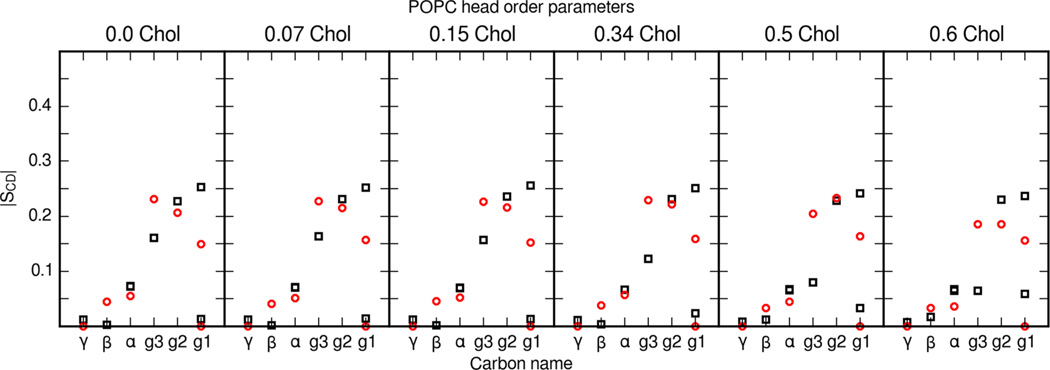
Ferreira et al. present an extensive data set on POPC phospholipid and cholesterol order parameters in bilayers with varying cholesterol content. Figures 6 and 7 present a detailed comparison of POPC tail and head order parameters from MD simulation and experiment. Simulations with the updated cholesterol parameter set and Lipid14 are able to reproduce the change in lipid order throughout a range of cholesterol contents. Ferreira et al. also compared experimental order parameters with MD simulations using the Berger parameter set for phospholipids and a GROMOS-based parameter set for cholesterol.57 The tail and head order parameters presented here match Ferreira et al.’s experimental results well.
Figure 6.
POPC tail-order parameters in mixed POPC and cholesterol bilayers. The top six panels show the order parameters of the sn-1 tail of POPC (palmitoyl), and the bottom six panels show the sn-2 tail of POPC (oleoyl). Black squares are order parameters calculated from MD simulations with the updated cholesterol parameter set. Red circles are experimental values from Ferreira et al.57
Figure 7.
POPC head segment order parameters in mixed POPC and cholesterol bilayers. POPC Head group segments are labeled as in Ferreira et al.57 Black squares are calculated values from the present MD simulations and red circles are values reported by Ferreira et al.57
Cholesterol Order Parameters in DMPC and POPC
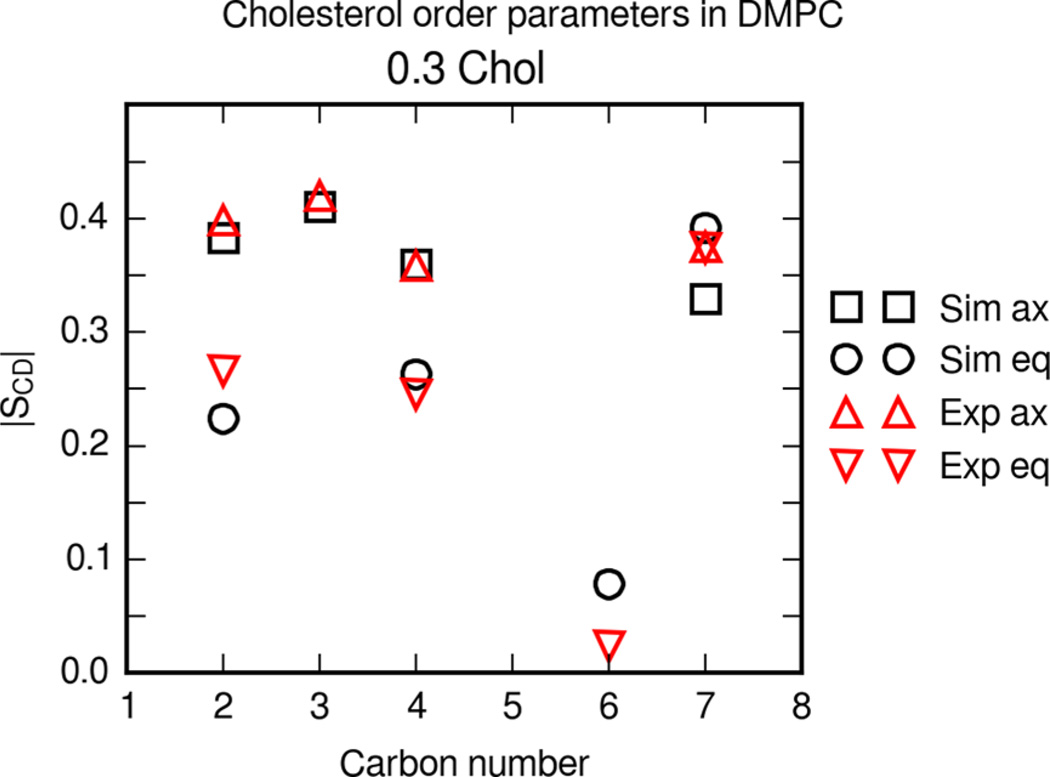
Vermeer et al. report order parameters for axial and equatorial C–D bonds in cholesterol.53 From these data it is possible to examine the effect of cholesterol content on the orientation of cholesterol within the lipid bilayer.
Figure 8 shows the deuterium order parameters |SCD| for axial and equatorial C–D vectors in the cholesterol rings near the hydroxyl group. Experimental data for DMPC with 30% cholesterol are shown from Vermeer et al.53 The simulation and experimental cholesterol order parameters are in good agreement. This suggests that on average the cholesterol in the simulations is oriented in a similar configuration as experimental bilayers.
Figure 8.
Deuterium order parameters |SCD| for cholesterol in a DMPC bilayer. The bilayer contains 0.3 molar fraction cholesterol. Boxes and circles are axial and equatorial simulation order parameters, respectively. Upward triangles are axial and downward triangles are equatorial order parameters at 30 °C from Vermeer et al.53
Jämbeck et al. calculate order parameters for cholesterol in DMPC bilayer simulations with the Slipids force field under the same conditions.19 Overall, bilayer simulations with Slipids yield average order parameters that are close to available experimental values from Vermeer et al.53 The order parameters of cholesterol gives insight into the orientation and configuration of cholesterol molecules in bilayers.
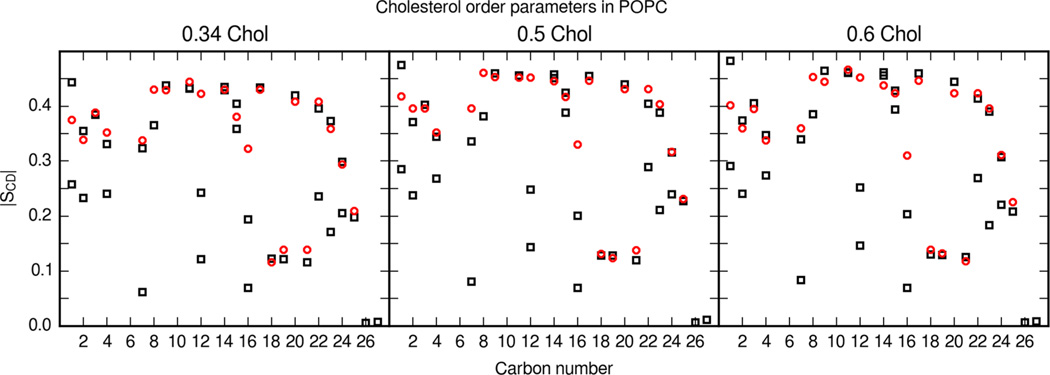
Ferreira et al. also include detailed cholesterol order parameters in their study of mixed POPC and cholesterol bilayers.57 Remarkably, this data set includes detailed order parameters for all cholesterol C–H vectors as well. Order parameters were calculated from the entire set of POPC and cholesterol MD simulations and compared with experimental values in Figure 9. Ferreira et al. note that NMR experiments usually only produce the larger of the two order parameters.57 Considering this fact, there is agreement between the larger cholesterol order parameters calculated from MD simulations of the updated cholesterol parameter set and Ferreira et al.’s NMR order parameters.
Figure 9.
Cholesterol order parameters in POPC bilayers. Molar fraction of cholesterol is labeled in each panel. Black boxes are calculated values from MD simulations and red circles are experimental values reported by Ferreira et al.57 Simulations show one order parameter value for single hydrogens and methyl groups. Otherwise, an order parameter value is shown for each hydrogen. Carbon segments follow the cholesterol carbon numbering of Figure 1.
Scattering Form Factors
X-ray and neutron scattering form factors are an important direct comparison for bilayer structures. In particular, experimental form factors may be modeled to calculate electron density profiles of bilayers. From the electron density profile, it is possible to determine the thickness and phase of the bilayer.
Conversely, simulation density profiles can be converted to scattering form factors. The SIMtoEXP program allows for direct comparison of simulation form factors with experimental form factors without requiring additional modeling of the experimental data.58 Number density profiles were generated from equilibrated production trajectories using Hannes Loeffler’s Ptraj program modifications.39,45 These number densities, in turn, were loaded and processed with SIMtoEXP. Default SIMtoEXP settings were used during analysis, and the magnitude of experimental form factors was automatically scaled to minimize the mean square of deviation between form factors.
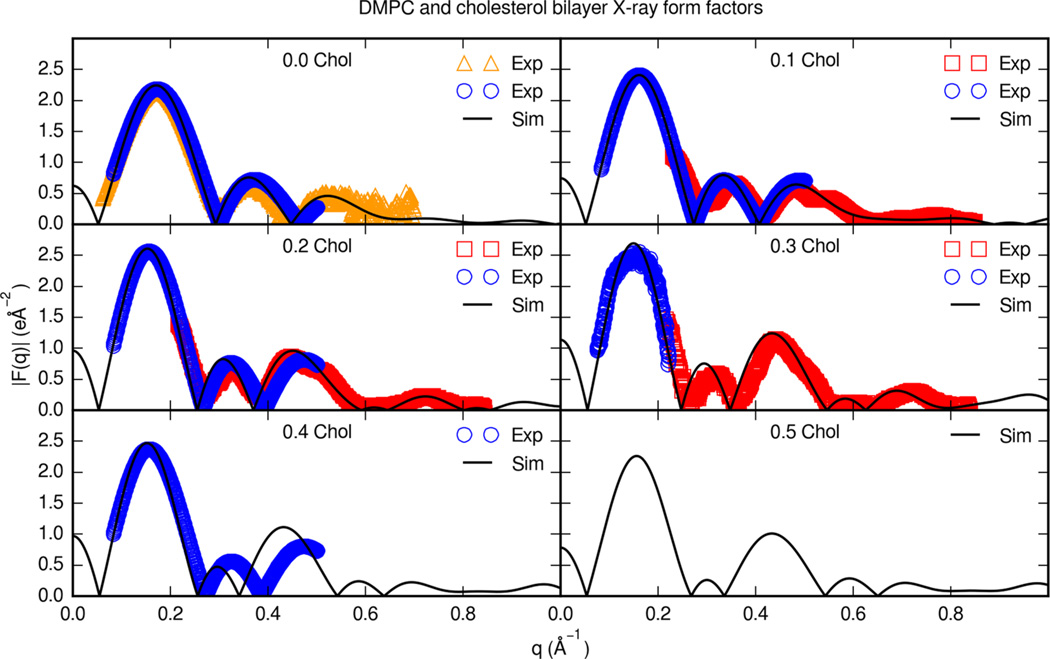
Figure 10 shows the X-ray form factors for cholesterol and DMPC bilayers in comparison with experimental form factors. X-ray scattering form factors are available for DMPC and cholesterol bilayers at a range of cholesterol contents.46,48 For a DMPC bilayer with no cholesterol, form factors were consistent with experimental data and previous Lipid14 simulations.46,59 Form factor maxima and minima are reproduced in the simulation form factors. At cholesterol molar fractions of 0.2 and above, the maxima and minima for some form factors appear to be slightly shifted toward lower q values. The simulations with Lipid14 DMPC cholesterol parameters compare favorably with this set of experimental scattering profiles up to 0.3 molar fraction cholesterol.
Figure 10.
Experimental X-ray form factors for cholesterol and DMPC bilayers. Each panel is a different cholesterol molar fraction. Solid lines are simulation form factors. Boxes are oriented (ORI) and unilamellar vesicle (ULV) form factors at 30 °C from Pan et al.,46 circles are form factors at 35 °C from Hodzic et al.,48 and triangles are ULV form factors at 30 °C from Kučerka et al.59
Cholesterol bilayer densities calculated from simulations with other force fields have been compared with experimental scattering profiles previously.18,19 The form factors for DMPC and cholesterol simulated with the Slipids force field closely resemble scattering form factors calculated from Lipid14 simulations.19 A difference in the form factors from Slipids simulations is a shift of maxima and minima to smaller q values relative to the form factors from Lipid14 simulations. Lim et al. calculate a scattering form factor from simulations of cholesterol and 0.1 fraction cholesterol with the Charmm C36c form field.18 The scattering form factor from Charmm C36c bilayer simulations resembles the form factor reported with Lipid14 parameters, however, with differences noticeable at higher q values. The Lipid14 cholesterol parameters reproduce several small nodes at high q values.
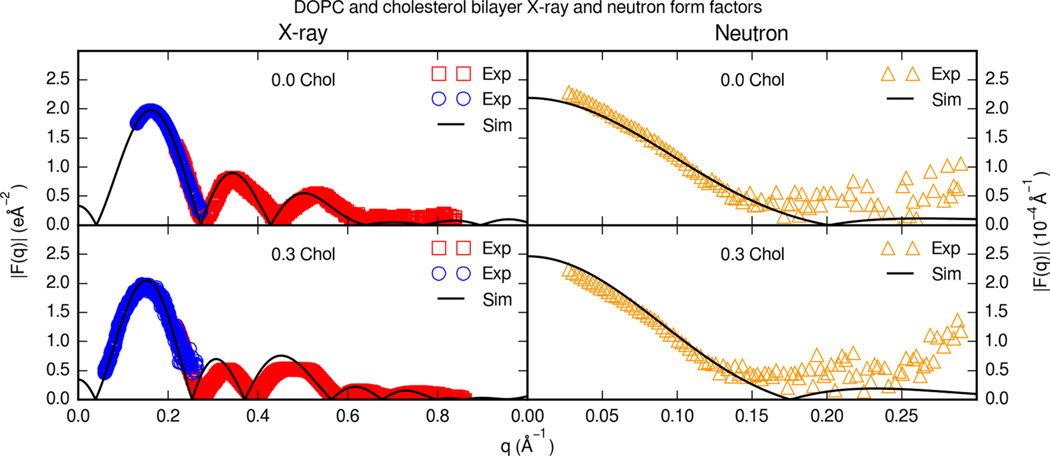
Figure 11 displays the X-ray and neutron form factors for cholesterol and DOPC bilayers. For DOPC and cholesterol bilayers, experimental X-ray and neutron form factors are only available for a limited set of cholesterol contents.46,60 At 0.3 molar fraction cholesterol, simulation X-ray form factors reproduce experimental form factors. Scaled simulation neutron form factors fit experimental form factors in 100% D2O at 0.3 molar fraction cholesterol. It should be noted that membranes were simulated with a normal water model and compared with neutron form factors in heavy water. Previous united-atom simulations with heavy water isotope models suggest that bilayer dynamics may be affected to some degree;61,62 however, alternate isotope solvation models have yet to be extensively investigated in all-atom MD of membrane systems.
Figure 11.
Experimental X-ray and neutron form factors for cholesterol and DOPC bilayers. Each panel is a different cholesterol content. Solid lines are simulation form factors. Boxes are oriented form factors and blue circles are ULV form factors at 30 C from Pan et al.46 Triangles are neutron form factors in 100% D2O at 30 °C from Kučerka et al.60
Jämbeck et al. also reports DOPC X-ray form factors.19 At 0.3 molar fraction cholesterol, the calculated form factors from Slipids simulations resemble the form factors from simulations with Lipid14 parameters closely.
CONCLUSIONS
Cholesterol force-field refinement began with atom types and parameters selected from the General Amber Force Field and Lipid14 force field.13 The charge derivation included a RESP fit on multiple cholesterol conformations extracted from previous cholesterol molecular dynamics on representative bilayers. Sterol van der Waals and tail dihedral parameters were reexamined. This is a parameterization approach that ultimately included fitting based on Lipid14 parameters to physical and thermodynamic data, quantum torsion scans, and charges fit to ESP calculations for cholesterol. Then, bilayer simulations were compared with available experimental structural information and were found to be in good agreement with reported values.
The change in the bilayer structure is manifested in lower average volume per bilayer molecule and an increase in the thickness of the bilayer. Consequently, the phospholipid tails are more constrained and exhibit higher deuterium order parameters. This is especially clear in POPC and cholesterol bilayer MD simulations, which reproduce experimental order parameters from Ferreira et al. These structural changes are consistent with the shift in phase from liquid disordered toward liquid ordered. This shift of bilayer phase is also visible in the X-ray and deuterium scattering form factors. Cholesterol order parameters match experimental values in a wide range of cholesterol contents. This suggests that cholesterol is oriented appropriately within DMPC throughout the MD simulation. Similarly, simulation scattering profiles fit experimental scattering profiles within this range of cholesterol contents.
The development of the Lipid14 force field continues with the refinement of parameters for mixed membrane simulations. The parameters presented here for cholesterol are fully compatible with Lipid14 phospholipids and are available for tensionless anisotropic bilayer simulations. Cholesterol is a common yet crucial component in bilayers and is essential for future simulations of complex membranes and membrane proteins. Future simulations will investigate the behavior of more complex membranes including other common membrane components. This ultimately allows for diverse membrane environments to be combined with other Amber force fields.
Supplementary Material
Acknowledgments
This work was supported by NSF SI2-SSE grants (NSF-1047875 and 1148276) to R.C.W. R.C.W. also acknowledges funding through fellowships from Intel Corp. and NVIDIA, Inc. B.D.M. acknowledges funding for this work provided by the NIH Molecular Biophysics Training Grant (T32 GM008326) and the NVIDIA Graduate Fellowship Program. I.R.G. also acknowledges funding from the EU in the form of the project “HeCaToS - Hepatic and Cardiac Toxicity Systems modeling” FP7-HEALTH-2013-INNOVATION-1 (Project number 602156).
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Details of parameterization and the full parameter print out for cholesterol. (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
REFERENCES
- 1.van Meer G, Voelker DR, Feigenson GW. Membrane Lipids: Where They Are and How They Behave. Nat. Rev. Mol. Cell Biol. 2008;9(2):112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wenk MR. The Emerging Field of Lipidomics. Nat. Rev. Drug Discovery. 2005;4(7):594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 3.Edidin M. The State of Lipid Rafts: From Model Membranes to Cells. Annu. Rev. Biophys. Biomol. Struct. 2003;32(1):257–283. doi: 10.1146/annurev.biophys.32.110601.142439. [DOI] [PubMed] [Google Scholar]
- 4.Simons K, Vaz WLC. Model Systems, Lipid Rafts, and Cell Membranes. Annu. Rev. Biophys. Biomol. Struct. 2004;33(1):269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 5.de Almeida RFM, Fedorov A, Prieto M. Sphingomyelin/Phosphatidylcholine/Cholesterol Phase Diagram: Boundaries and Composition of Lipid Rafts. Biophys. J. 2003;85(4):2406–2416. doi: 10.1016/s0006-3495(03)74664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leonenko ZV, Finot E, Ma H, Dahms TES, Cramb DT. Investigation of Temperature-Induced Phase Transitions in DOPC and DPPC Phospholipid Bilayers Using Temperature-Controlled Scanning Force Microscopy. Biophys. J. 2004;86(6):3783–3793. doi: 10.1529/biophysj.103.036681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simons K, Gerl MJ. Revitalizing Membrane Rafts: New Tools and Insights. Nat. Rev. Mol. Cell Biol. 2010;11(10):688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 8.Boesze-Battaglia K, Schimmel R. Cell Membrane Lipid Composition and Distribution: Implications for Cell Function and Lessons Learned from Photoreceptors and Platelets. J. Exp. Biol. 1997;200(23):2927–2936. doi: 10.1242/jeb.200.23.2927. [DOI] [PubMed] [Google Scholar]
- 9.Pucadyil TJ, Chattopadhyay A. Cholesterol Modulates Ligand Binding and G-Protein Coupling to Serotonin 1A Receptors from Bovine Hippocampus. Biochim. Biophys. Acta, Biomembr. 2004;1663(1–2):188–200. doi: 10.1016/j.bbamem.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 10.White S. [accessed Dec 14, 2014];Membrane Proteins of Known 3D Structure. http://blanco.biomol.uci.edu/mpstruc/ [Google Scholar]
- 11.Dickson CJ, Rosso L, Betz RM, Walker RC, Gould IR. GAFFlipid: A General Amber Force Field for the Accurate Molecular Dynamics Simulation of Phospholipid. Soft Matter. 2012;8(37):9617. [Google Scholar]
- 12.Skjevik ÅA, Madej BD, Walker RC, Teigen K. LIPID11: A Modular Framework for Lipid Simulations Using Amber. J. Phys. Chem. B. 2012;116(36):11124–11136. doi: 10.1021/jp3059992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickson CJ, Madej BD, Skjevik ÅA, Betz RM, Teigen K, Gould IR, Walker RC. Lipid14: The Amber Lipid Force Field. J. Chem. Theory Comput. 2014;10(2):865–879. doi: 10.1021/ct4010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skjevik ÅA, Madej BD, Dickson CJ, Teigen K, Walker RC, Gould IR. All-Atom Lipid Bilayer Self-Assembly with the AMBER and CHARMM Lipid Force Fields. Chem. Commun. 2015;51(21):4402–4405. doi: 10.1039/c4cc09584g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J. Phys. Chem. B. 2007;111(27):7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 16.Chiu S-W, Pandit SA, Scott HL, Jakobsson E. An Improved United Atom Force Field for Simulation of Mixed Lipid Bilayers. J. Phys. Chem. B. 2009;113(9):2748–2763. doi: 10.1021/jp807056c. [DOI] [PubMed] [Google Scholar]
- 17.Cournia Z, Ullmann GM, Smith JC. Differential Effects of Cholesterol, Ergosterol and Lanosterol on a Dipalmitoyl Phosphatidylcholine Membrane: A Molecular Dynamics Simulation Study. J. Phys. Chem. B. 2007;111(7):1786–1801. doi: 10.1021/jp065172i. [DOI] [PubMed] [Google Scholar]
- 18.Lim JB, Rogaski B, Klauda JB. Update of the Cholesterol Force Field Parameters in CHARMM. J. Phys. Chem. B. 2012;116(1):203–210. doi: 10.1021/jp207925m. [DOI] [PubMed] [Google Scholar]
- 19.Jämbeck JPM, Lyubartsev AP. Another Piece of the Membrane Puzzle: Extending Slipids Further. J. Chem. Theory Comput. 2013;9(1):774–784. doi: 10.1021/ct300777p. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 21.Bayly CI, Cieplak P, Cornell W, Kollman PA. A Well-Behaved Electrostatic Potential Based Method Using Charge Restraints for Deriving Atomic Charges: The RESP Model. J. Phys. Chem. 1993;97(40):10269–10280. [Google Scholar]
- 22.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, et al. Gaussian 09. Wallingford, CT: Gaussian, Inc.; 2009. [Google Scholar]
- 23.Jo S, Lim JB, Klauda JB, Im W. CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophys. J. 2009;97(1):50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983;79(2):926–935. [Google Scholar]
- 25.Joung IS, Cheatham TE. Determination of Alkali and Halide Monovalent Ion Parameters for Use in Explicitly Solvated Biomolecular Simulations. J. Phys. Chem. B. 2008;112(30):9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jójárt B, Martinek TA. Performance of the General Amber Force Field in Modeling Aqueous POPC Membrane Bilayers. J. Comput. Chem. 2007;28(12):2051–2058. doi: 10.1002/jcc.20748. [DOI] [PubMed] [Google Scholar]
- 27.Haynes WM. CRC Handbook of Chemistry and Physics. Boca Raton, FL: CRC Press; 2013. [Google Scholar]
- 28.Martínez L, Andrade R, Birgin EG, Martínez JM. PACKMOL: A Package for Building Initial Configurations for Molecular Dynamics Simulations. J. Comput. Chem. 2009;30(13):2157–2164. doi: 10.1002/jcc.21224. [DOI] [PubMed] [Google Scholar]
- 29.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of N-Alkanes. J. Comput. Phys. 1977;23(3):327–341. [Google Scholar]
- 30.Izaguirre JA, Catarello DP, Wozniak JM, Skeel RD. Langevin Stabilization of Molecular Dynamics. J. Chem. Phys. 2001;114(5):2090–2098. [Google Scholar]
- 31.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984;81(8):3684–3690. [Google Scholar]
- 32.Darden T, York D, Pedersen L. Particle Mesh Ewald: An N log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993;98(12):10089–10092. [Google Scholar]
- 33.Benson GC, Murakami S, Lam VT, Singh J. Molar Excess Enthalpies and Volumes of Cyclohexane – Isomeric Decalin Systems at 25 °C. Can. J. Chem. 1970;48(2):211–218. [Google Scholar]
- 34.Fuchs R, Chambers EJ, Stephenson WK. Enthalpies of Interaction of Nonpolar Solutes with Nonpolar Solvents. The Role of Solute Polarizability and Molar Volume in Solvation. Can. J. Chem. 1987;65(11):2624–2627. [Google Scholar]
- 35.Letcher TM, Lucas A. Excess Volumes of Decahydronaphthalene in N-Alkanes at 10 and 25°C. J. Solution Chem. 1981;10(12):863–870. [Google Scholar]
- 36.Wang J, Hou T. Application of Molecular Dynamics Simulations in Molecular Property Prediction. 1. Density and Heat of Vaporization. J. Chem. Theory Comput. 2011;7(7):2151–2165. doi: 10.1021/ct200142z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagle JF, Tristram-Nagle S. Structure of Lipid Bilayers. Biochim. Biophys. Acta, Rev. Biomembr. 2000;1469(3):159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kučerka N, Tristram-Nagle S, Nagle JF. Structure of Fully Hydrated Fluid Phase Lipid Bilayers with Monounsaturated Chains. J. Membr. Biol. 2006;208(3):193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 39.Case D, Babin V, Berryman J, Betz R, Cai Q, Cerutti D, Cheatham T, III, Darden T, Duke R, Gohlke H, et al. Amber 14. San Francisco: University of California; 2014. [Google Scholar]
- 40.Allen M, Tildesley D. Computer Simulation of Liquids. Oxford, UK: Oxford University Press; 1988. [Google Scholar]
- 41.Salomon-Ferrer R, Case DA, Walker RC. An Overview of the Amber Biomolecular Simulation Package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013;3(2):198–210. [Google Scholar]
- 42.Götz AW, Williamson MJ, Xu D, Poole D, Le Grand S, Walker RC. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 2012;8(5):1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, Walker RC. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013;9(9):3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 44.Le Grand S, Götz AW, Walker RC. SPFP: Speed Without Compromise—A Mixed Precision Model for GPU Accelerated Molecular Dynamics Simulations. Comput. Phys. Commun. 2013;184(2):374–380. [Google Scholar]
- 45.Loeffler H. [accessed Dec 14, 2014];Handy Routines for Ptraj/Cpptraj. http://www.hecbiosim.ac.uk/ptraj-routines. [Google Scholar]
- 46.Pan J, Tristram-Nagle S, Nagle JF. Effect of Cholesterol on Structural and Mechanical Properties of Membranes Depends on Lipid Chain Saturation. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2009;80(2 Pt 1):021931. doi: 10.1103/PhysRevE.80.021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greenwood AI, Tristram-Nagle S, Nagle JF. Partial Molecular Volumes of Lipids and Cholesterol. Chem. Phys. Lipids. 2006;143(1–2):1–10. doi: 10.1016/j.chemphyslip.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hodzic A, Rappolt M, Amenitsch H, Laggner P, Pabst G. Differential Modulation of Membrane Structure and Fluctuations by Plant Sterols and Cholesterol. Biophys. J. 2008;94(10):3935–3944. doi: 10.1529/biophysj.107.123224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pencer J, Nieh M-P, Harroun TA, Krueger S, Adams C, Katsaras J. Bilayer Thickness and Thermal Response of Dimyristoylphosphatidylcholine Unilamellar Vesicles Containing Cholesterol, Ergosterol and Lanosterol: A Small-Angle Neutron Scattering Study. Biochim. Biophys. Acta, Biomembr. 2005;1720(1–2):84–91. doi: 10.1016/j.bbamem.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 50.Gandhavadi M, Allende D, Vidal A, Simon SA, McIntosh TJ. Structure, Composition, and Peptide Binding Properties of Detergent Soluble Bilayers and Detergent Resistant Rafts. Biophys. J. 2002;82(3):1469–1482. doi: 10.1016/S0006-3495(02)75501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Douliez JP, Léonard A, Dufourc EJ. Restatement of Order Parameters in Biomembranes: Calculation of C-C Bond Order Parameters from C-D Quadrupolar Splittings. Biophys. J. 1995;68(5):1727–1739. doi: 10.1016/S0006-3495(95)80350-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Urbina JA, Pekerar S, Le H, Patterson J, Montez B, Oldfield E. Molecular Order and Dynamics of Phosphatidylcholine Bilayer Membranes in the Presence of Cholesterol, Ergosterol and Lanosterol: A Comparative Study Using 2H-, 13C- and 31P-NMR Spectroscopy. Biochim. Biophys. Acta, Biomembr. 1995;1238(2):163–176. doi: 10.1016/0005-2736(95)00117-l. [DOI] [PubMed] [Google Scholar]
- 53.Vermeer LS, de Groot BL, Réat V, Milon A, Czaplicki J. Acyl Chain Order Parameter Profiles in Phospholipid Bilayers: Computation from Molecular Dynamics Simulations and Comparison with 2H NMR Experiments. Eur. Biophys. J. 2007;36(8):919–931. doi: 10.1007/s00249-007-0192-9. [DOI] [PubMed] [Google Scholar]
- 54.Trouard TP, Nevzorov AA, Alam TM, Job C, Zajicek J, Brown MF. Influence of Cholesterol on Dynamics of Dimyristoylphosphatidylcholine Bilayers as Studied by Deuterium NMR Relaxation. J. Chem. Phys. 1999;110(17):8802–8818. [Google Scholar]
- 55.Warschawski DE, Devaux PF. Order Parameters of Unsaturated Phospholipids in Membranes and the Effect of Cholesterol: A 1H–13C Solid-State NMR Study at Natural Abundance. Eur. Biophys. J. 2005;34(8):987–996. doi: 10.1007/s00249-005-0482-z. [DOI] [PubMed] [Google Scholar]
- 56.Lafleur M, Fine B, Sternin E, Cullis PR, Bloom M. Smoothed Orientational Order Profile of Lipid Bilayers by 2H-Nuclear Magnetic Resonance. Biophys. J. 1989;56(5):1037–1041. doi: 10.1016/S0006-3495(89)82749-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferreira TM, Coreta-Gomes F, Ollila OHS, Moreno MJ, Vaz WLC, Topgaard D. Cholesterol and POPC Segmental Order Parameters in Lipid Membranes: Solid State 1H–13C NMR and MD Simulation Studies. Phys. Chem. Chem. Phys. 2013;15(6):1976–1989. doi: 10.1039/c2cp42738a. [DOI] [PubMed] [Google Scholar]
- 58.Kučerka N, Katsaras J, Nagle JF. Comparing Membrane Simulations to Scattering Experiments: Introducing the SIMtoEXP Software. J. Membr. Biol. 2010;235(1):43–50. doi: 10.1007/s00232-010-9254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kučerka N, Nieh M-P, Katsaras J. Fluid Phase Lipid Areas and Bilayer Thicknesses of Commonly Used Phosphatidylcholines as a Function of Temperature. Biochim. Biophys. Acta, Biomembr. 2011;1808(11):2761–2771. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 60.Kučerka N, Pencer J, Nieh M-P, Katsaras J. Influence of Cholesterol on the Bilayer Properties of Monounsaturated Phosphatidylcholine Unilamellar Vesicles. Eur. Phys. J. E: Soft Matter Biol. Phys. 2007;23(3):247–254. doi: 10.1140/epje/i2007-10202-8. [DOI] [PubMed] [Google Scholar]
- 61.Róg T, Murzyn K, Milhaud J, Karttunen M, Pasenkiewicz-Gierula M. Water Isotope Effect on the Phosphatidylcholine Bilayer Properties: A Molecular Dynamics Simulation Study. J. Phys. Chem. B. 2009;113(8):2378–2387. doi: 10.1021/jp8048235. [DOI] [PubMed] [Google Scholar]
- 62.Beranová L, Humpolíčková J, Sýkora J, Benda A, Cwiklik L, Jurkiewicz P, Gröbner G, Hof M. Effect of Heavy Water on Phospholipid Membranes: Experimental Confirmation of Molecular Dynamics Simulations. Phys. Chem. Chem. Phys. 2012;14(42):14516–14522. doi: 10.1039/c2cp41275f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.