ABSTRACT
Alphavirus vectors are promising tools for cancer treatment. However, relevant entry mechanisms and interactions with host cells are still not clearly understood. The first step toward a more effective therapy is the identification of novel intracellular alterations that could be associated with cancer aggressiveness and could affect the therapeutic potential of these vectors. In this study, we observed that alphaviruses efficiently infected B16 mouse melanoma tumors/tumor cells in vivo, whereas their transduction efficiency in B16 cells under in vitro conditions was blocked. Therefore, we further aimed to understand the mechanisms pertaining to the differential transduction efficacy of alphaviruses in B16 tumor cells under varying growth conditions. We hypothesized that the tumor microenvironment might alter gene expression in B16 cells, leading to an up-regulation of the expression of virus-binding receptors or factors associated with virus entry and replication. To test our hypothesis, we performed a proteomics analysis of B16 cells cultured in vitro and of B16 cells isolated from tumors, and we identified 277 differentially regulated proteins. A further in-depth analysis to identify the biological and molecular functions of the detected proteins revealed a set of candidate genes that could affect virus infectivity. Importantly, we observed a decrease in the expression of interferon α (IFN-α) in tumor-isolated cells that resulted in the suppression of several IFN-regulated genes, thereby abrogating host cell antiviral defense. Additionally, differences in the expression of genes that regulate cytoskeletal organization caused significant alterations in cell membrane elasticity. Taken together, our findings demonstrated favorable intracellular conditions for alphavirus transduction/replication that occurred during tumor transformation. These results pave the way for optimizing the development of strategies for the application of alphaviral vectors as a potent cancer therapy.
KEYWORDS: Alphavirus, B16 melanoma, cancer, intracellular alterations, tumor microenvironment
Introduction
The application of recombinant viral vectors has become one of the most intensively developed strategies in cancer gene therapy. Such therapy is based on the ability of the viruses to preferentially infect and kill cancer cells. Alphaviral vectors function as efficient mammalian expression systems because of their high-level transgenic expression and induction of p53-independent apoptosis in infected cells.1,2 These vectors also have a broad range of hosts and induce a weak immune response against the vector.3 Alphaviruses are small, enveloped, positive-stranded RNA viruses that belong to the Togaviridae family. The classical Semliki Forest virus (SFV) replicon vector is generated by replacing the structural genes under the control of the 26S viral subgenomic promoter with a heterologous insert of interest.4 The vector RNA can be packaged into recombinant viral particles during co-transfection of the host cells with a helper RNA that encodes structural genes, i.e., capsid and envelope proteins. SFV RNA replicates actively during infection, and the heterologous gene is expressed at a high level. However, the vector cannot propagate because it lacks genes encoding the viral structural proteins.
The expression efficacy of all viral vectors relies on the virus transduction, replication and distribution ability. Alphaviruses are able to infect a broad range of cancer cell lines with widely divergent biochemical and genetic environments both in vitro and in vivo.5-7 However, the tumor microenvironments are capable of forming a barrier that is highly impermeable to the virus.8 Viral penetration, persistence and spreading may be impeded alone or in combination, thus causing a wide variation in viral transduction/replication capacity even within a single cell line under in vitro and in vivo conditions. We and other authors have found that alphaviruses can efficiently infect B16 mouse melanoma tumors in vivo, whereas the infection in vitro is blocked for unknown reasons.6 This observation has encouraged us to perform an in-depth analysis of intracellular factors that could vary in the same cells before and after administration in mice. Melanoma is a complex multi-step heterogeneous disease in which most of the steps in the tumor transformation process, such as proliferation, invasion, angiogenesis and metastasis, are modulated by microenvironmental factors such as growth factors and proteolytic enzymes produced by stromal cells.9 However, the ability of these factors to affect viral infectivity has yet to be explored. Taking into consideration that the tumor microenvironment is able to influence gene expression in cancer cells, we hypothesized that it might also play a role in the upregulation of virus-binding receptors or other factors, which in turn affect viral entry and replication. To date, only Sindbis virus has demonstrated tumor tropism in vivo.10-11 We recently demonstrated that SFV is capable of predominant tumor infection upon systemic vector administration at an optimized dose.12 However, the interaction/entry mechanisms have not been investigated in detail.
Various studies have reported that alphaviruses enter the cell via receptor-mediated endocytosis, involving multiple proteins implicated in virus absorption/transduction, such as heparan sulfate, laminin receptor, the major histocompatibility complex (MHC), DC-SIGN, L-SIGN, heat shock 70 protein, and α1β1 integrin, among others.13-18 However, it is most likely that alphaviruses may utilize multiple surface proteins as receptors or alternative entry pathways in different cells. In addition to receptors, cytoskeletal organization plays an essential role in the interaction of viruses with the host cell, affecting penetration through the membrane and further development of infection and viral expression.19,20 The microtubule network regulates several processes, including intracellular transport, transcription, replication and secretion of progeny virions, as well as assembly and cell-to-cell spread.21 Recent studies indicate that the cell membrane interacts with the attached cytoskeleton/associated motor proteins, thus controlling endocytosis/exocytosis and modulating physical features of the cell such as its shape, motility and membrane elasticity.21 All these processes could potentially affect viral activity;22 however, the mechanism(s) associated with the alphaviruses remain to be elucidated.
Efficient viral replication is one the most important prerequisites for successful gene therapy. Alphaviral replication is extremely high under favorable intracellular conditions, resulting in expression levels of nearly 20% of the total cell protein.23 However, infection triggers cellular defenses induced by a combination of different factors. Thus, the up-regulation of several immune system proteins or stress factors, such as interferon-stimulated genes (ISGs),24 zinc-finger antiviral proteins25 or eukaryotic translation initiation factors,26 can significantly suppress alphavirus-driven gene expression. Interferons were shown to be the major players in the modulation of the pervasive antiviral environment, leading to the prevention of viral spread and replication. Several studies have shown that enhanced alphavirus infectivity is mediated by a decrease in the intracellular level of type I IFNs, which is true both in vitro and in vivo.27,28 Interesting results, which are consistent with the current research, have been shown for Vesicular Stomatitis Indiana virus (VSV). Ovarian and breast cancer cells were highly susceptible to VSV infection in vitro, but the infection of the developed tumors in vivo was inefficient due to activation of the JAK/STAT pathways and overexpression of interferon-stimulated genes induced by tumor-infiltrating macrophages.29 This study confirms our hypothesis that the tumor microenvironment is able to induce intracellular changes in cancer cells, thus leading to variation in viral activity in vivo.
In this current study, we performed a proteomics analysis of B16 cells cultivated in vitro and of tumor-isolated B16 cells and the results were compared. Our goal was to identify genes that are differentially expressed in mouse melanoma cells before and after their inoculation in mice. Based on a quantitative analysis of the detected proteins, we report a list of gene candidates (S100b, Pycard, CD97, Pdcd4, Gpx1, Csnk2b, Gstz1, Gsto1, Scrib, Hsp90aa1, Hspb1, Cryab, Csf1, Adh7, Sgtb, Aif1l, Crem, Etv6, Nfe2l2, Stat1, IFI35, S100a11, Txnl1) that may be involved in the antiviral response. We also identified cytoskeletal organization gene candidates (Tmod, Dst, Dynll2, Opa1, Mlph, Actc1, Myo18a, Dynlt1, Myo5a, Map7, Csnk2b, Lmna, Dync1h1, Timm10b, Kif1a, Gfap, Capg, Flnc, Aspm, Hist1h1a, Ss18, Hist1h1t, Cfl1, Cald1, Rps21, Tubb6), which might provide optimal intracellular conditions for the infection and further expression of transgenes. Our findings could extend what is currently known about the alterations in the melanoma microenvironment during tumor development. In turn, this information could provide a significant contribution toward the modulation of more efficient strategies for cancer gene therapy.
Results
Comparison of SFV infectivity in B16 cells and B16 tumors
To compare the efficiency of SFV-driven transgene expression in mouse melanoma cells in vitro and in vivo, cultured B16 cells were infected (in vitro) with the SFV/Enh.Luc vector at a multiplicity of infection (MOI) of 10. To examine SFV infectivity in vivo, immunocompetent B16 tumor-bearing mice were inoculated intratumorally (i.t.) with 108 SFV/Enh.Luc particles. Luciferase expression was analyzed in the cell lysates and tumor homogenates at 24 h post-infection.
In vitro, very low expression of the transgene was detected in B16 cells post-infection, revealing the transduction/replication inefficiency of the SFV vector under such conditions. By contrast, intratumoral administration of the same recombinant virus resulted in high expression of the transgene in B16 melanoma tumors (Fig. 1A). These data indicate that there is variation in the genetic background between in vitro-cultivated B16 cells and B16 cells in tumor-bearing mice, which facilitates transduction or replication of the SFV vector.
Figure 1.
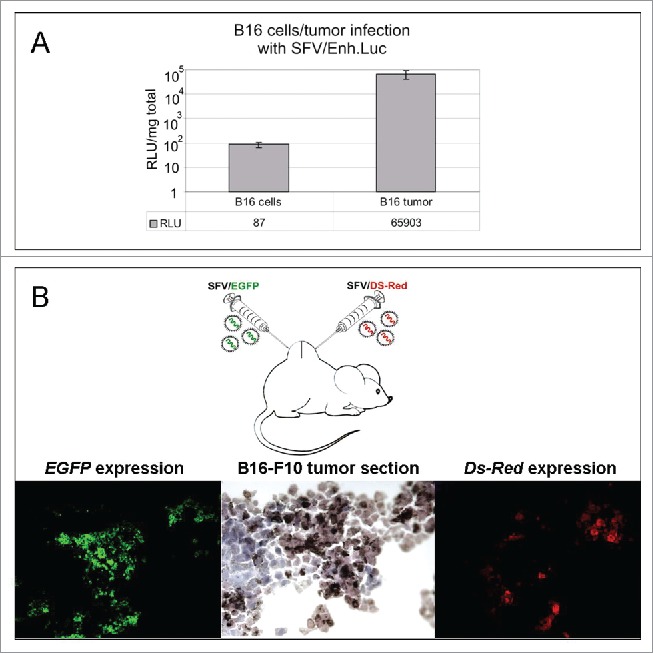
SFV expression and intratumoral spread in a melanoma mouse model. (A) Infection of B16 melanoma cells in vitro and B16 tumor cells in vivo with SFV/Enh.Luc vector. The B16 cells were infected with SFV at an MOI of 10 in vitro. For the in vivo experiment, B16 tumor-bearing mice were i.t. inoculated with 108 SFV v.p. The luciferase expression analysis in cell lysates and tumor homogenates was performed 24 h post-infection by luminometry. The bar graph presents the RLUs per 1 mg protein in the cell lysate/tumor homogenate. The results represent the mean ± s.e. RLU - relative light unit. (B) Administration strategy of SFV vectors and fluorescence microscopy of B16 tumor cryosections, demonstrating SFV/FGFP and SFV/Ds-Red virus spread in the tumor. A total of 106 v.p. of SFV/EGFP and SFV/Ds-Red were injected in different tumor sides by direct intratumoral injections. The tumors were cryosectioned and analyzed 24 h after SFV vector administration.
To determine the vector distribution within a subcutaneous melanoma tumor nodule, 2 SFV vectors expressing green fluorescence protein (SFV/EGFP) and red fluorescence protein (SFV/DS-Red) were inoculated into different points of a B16 tumor nodule (Fig. 1B). The analysis of tumor cryosections revealed only local expression of the corresponding fluorescent protein at the location of the intratumoral injections, with no broad intratumoral dissemination of the virus due to the absence of SFV/EGFP expression at locations of SFV/DS-Red injection and vice versa. Therefore, it is possible to conclude that although the SFV vector is capable of a high level of expression of the transgenes in B16 tumors, the efficacy of the alphavirus-based therapy could be increased by enhancing the tumor permeability, thus promoting the wide intratumoral spread of the vector.
Comparison of SFV infectivity of in vitro B16 cells and ex vivo B16 cells
To investigate whether B16 cells were modified by the tumor microenvironment, rendering them susceptible to SFV vector infection, we conducted a series of SFV transduction experiments using 2 types of melanoma cells: i) B16 in vitro cells – a standard B16-F10 cell line cultured under ordinary cell culturing conditions in vitro, and ii) B16 ex vivo cells - melanoma cells isolated from B16-F10 tumors. The susceptibility of B16 ex vivo cells to SFV infection was analyzed over time at different cell passages. In vitro and ex vivo B16 cells were infected with SFV/Ds-Red at an MOI of 10. Ds-Red gene expression was analyzed 24 h post-infection by fluorescence microscopy.
Our results demonstrated extremely high SFV-mediated transgene expression during the first passage of B16 ex vivo cells in contrast to control B16 cells in vitro (Fig. 2). Surprisingly, further cultivation of the ex vivo cells led to a dramatic inhibition of SFV vector infection. We hypothesize that the tumor microenvironment can induce alterations at the level of gene expression, which affect the morphology and physical parameters of cells, such as cell shape, motility and membrane elasticity. In turn, these changes provide conditions favorable for SFV vector transduction/replication. Notably, although the ex vivo B16 cells displayed these features for a short period, these cells subsequently began to lose susceptibility to SFV infection by the second passage upon splitting. Remarkably, the replacement of fetal bovine serum to freshly prepared autologous mouse serum for cultivation of ex vivo and in vitro B16 cells had no effect on SFV infectivity. We did not observe any significant increase in SFV/Ds-Red activity in B16 cells cultivated in cell medium supplemented with mouse serum (not shown).
Figure 2.
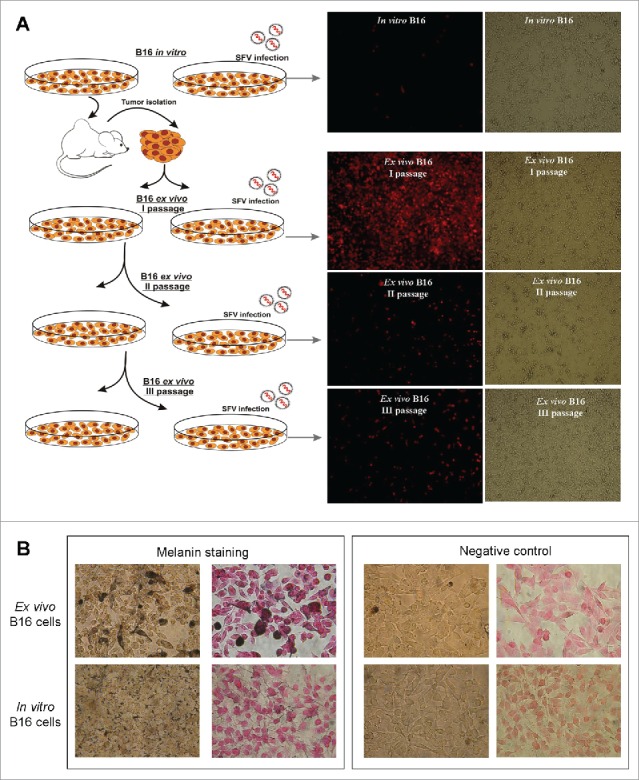
Comparison of SFV infection susceptibility and melanin expression of in vitro and ex vivo B16 cells. (A) Schematic illustration of the isolation, propagation and infection of ex vivo B16 cells with SFV vector compared with the control in vitro B16 cells. The control in vitro B16 cultured cells were subcutaneously injected into immunocompetent C57BL/6 mice. Ten days after cell inoculation, B16 tumor nodules were isolated, homogenized and plated as ex vivo B16 cells in 2 plates as the first cell passage. When the cell monolayer reached 80% confluency, the 1st plate of cells was split for further cultivation (second cell passage), whereas the 2nd plate of cells was infected with SFV/Ds-Red vector for transgene expression analysis, which is presented as fluorescence and phase contrast microscopy images. (B) Melanin staining of in vitro and ex vivo B16 cells. The cells were treated with Fontana-Masson silver stain to detect melanin (dark brown, black color). Negative control – non-stained cells. The nuclei were counterstained with nuclear fast red.
To demonstrate that the ex vivo-isolated cells were B16 melanoma cells, we performed melanin staining using the classical Fontana-Manson method (Fig. 2B). The results revealed melanin production in the isolated cells. Moreover, in contrast to in vitro B16 cells, the level of expression of melanin in ex vivo cells was significantly increased. Further cultivation of ex vivo cells led to a decrease in melanin synthesis (data not shown).
Protein profile analysis of ex vivo and in vitro B16 cells
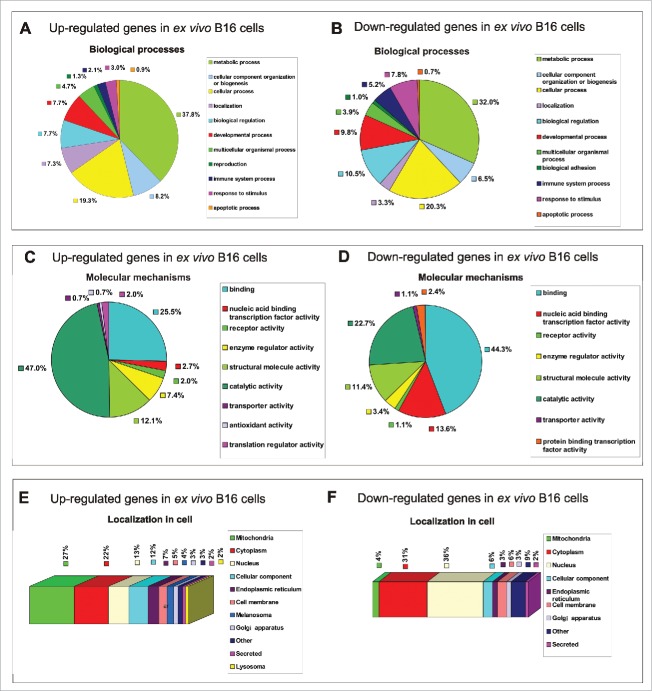
To determine the intracellular conditions that could enhance SFV infection, we performed a comparative proteomics analysis of ex vivo B16 (first passage) and in vitro B16 cells using liquid chromatography-mass spectrometry (LC-MS). To characterize the in vitro and ex vivo B16 cells, we identified individual proteomes of 3 in vitro B16 cell samples and 3 ex vivo B16 samples (isolated from 3 different mice). A total of 4980 proteins were identified using the UniProt database, among which 277 proteins were differentially regulated (165 up-regulated and 112 down-regulated), with a fold change >1.4 in ex vivo B16 cells in comparison to the in vitro B16 cells (p < 0.01; Table S1 and Table S2). All these proteins were identified in triplicate for each sample. The quantified proteins were functionally annotated using the PANTHER bioinformatics resource (version PANTHER 9.0; http://www.pantherdb.org/) and further classified according to their functions in biological processes (Fig. 3A-B) and molecular mechanisms (Fig. 3C-D).30 The cellular localization of the detected proteins were determined by manual classification using the UniProt database (http://www.uniprot.org/) (Fig. 3E-F).
Figure 3.
Functional classification of 277 differentially regulated proteins in ex vivo B16 cells identified in total protein extracts by LC-MS. Protein profile analysis of both in vitro and ex vivo B16 cells was performed by LC-MS, and in vitro B16 cells were used as reference cells. The pie charts demonstrate the distribution of 165 upregulated (left panel) and 112 downregulated (right panel) proteins in ex vivo B16 cells according to their biological processes (A-B) and molecular mechanisms (C-D). Categorizations were based on information provided by the PANTHER classification system (see methods). The subcellular localization prediction of up-regulated (E) and down-regulated (F) proteins in B16 ex vivo cells was annotated manually using the UniProt database. The percentages shown in the pie and bar charts represent the percentage of genes belonging to each group.
We identified 165 proteins that were upregulated in the ex vivo B16 cells during tumor development in comparison to the in vitro B16 cells. According to the analysis of the biological functions, the majority of the identified proteins belonged to different metabolic processes (37.8%). Smaller groups comprised cellular process proteins (19.3%), cell component organization and biogenesis proteins (8.2%), biological regulation proteins (7.7%), developmental process proteins (7.7%) and localization proteins (7.3%). Several other identified proteins were classified as multicellular organismal process (4.7%), response to stimulus (3%), immune system process (2.1%), reproduction (1.3%) and apoptotic process (0.9%) proteins (Fig. 3A).
Regarding molecular functions, more that 70% of all up-regulated proteins were grouped into 2 leading categories: catalytic activity proteins (47%) and binding proteins (25.5%). The other overexpressed gene products were functionally distributed as structural molecule activity proteins (12.1%), enzyme regulator activity proteins (7.4%), receptor activity (2%), translation regulators (2%), nucleic acid binding/transcription factor activity (2.7%), transporters (0.7%) and antioxidant activity (0.7%) proteins (Fig. 3C).
Proteins perform their functions in specific cellular locations. Careful analysis of their subcellular localization revealed that the majority of the upregulated proteins were localized in the mitochondria (27%) and the cytoplasm (21%) (Fig. 3E). Importantly, a large portion of the mitochondrial proteins were NADH dehydrogenases, which are fundamentally crucial for growth signaling and transcription in a broad array of melanoma tumors,31 indicating enhanced aggressiveness of the ex vivo B16 cells in contrast to the control in vitro cells. The intracellular distribution of the other proteins was predicted to be localized in the nucleus (13%), cellular components (12%), endoplasmic reticulum (7%), cell membrane (5%), Golgi apparatus (3%) and both lysosomal and secreted (2%). More stringent analysis was performed for the proteins localized in melanosomes (4%), which are unique organelles in melanoma cells. The ex vivo B16 cells overexpressed genes such us Typr1, Tyr, Dct, Mlph and Myo5a, which are involved in melanin synthesis, explaining the enhanced pigmentation of the ex vivo cells, which resulted in the histological difference between the ex vivo and in vitro B16 cells (Fig. 2B).
LC-MS analysis identified 112 downregulated proteins in the ex vivo B16 cells. Regarding their involvement in biological processes, the majority of the down-regulated proteins were also categorized into different metabolic and cellular processes (32% and 20.3%, respectively) in a similar manner as the up-regulated proteins (see above). The other proteins were involved in biological regulation (10.5%), developmental process (9.8%), response to stimulus (7.8%), biogenesis (6.5%), immune system process (5.2%), multicellular organismal process (3.9%), localization (3.3%), biological adhesion (1%) and apoptotic process (0.7%) (Fig. 3B).
Biological classification of the downregulated proteins in terms of their molecular mechanisms revealed that most of them had binding (44.3%) and catalytic activity (22.7%) functions. A significantly smaller number of proteins were related to nucleic acid binding transcription factor activity (13.6%) and structural molecule activity (11.4%) proteins, respectively. The smallest number of proteins were categorized as enzyme regulator (3.4%), protein binding transcription factor (2.3%), and receptor and transporter (1.1%) activity proteins (Fig. 3D).
Most of the down-regulated proteins were predicted to be localized in the nucleus (36%) and the cytoplasm (31%) (Fig. 3F). A total of 6% were classified as cell membrane and cellular component proteins. Interestingly, only 4% of the downregulated proteins exhibited mitochondrial localization, which is in contrast to 27% of the upregulated proteins (see above), indicating decreased mitochondrial activity in the in vitro B16 cells. A total of 3% of the proteins were localized in the endoplasmic reticulum and Golgi apparatus, and 2% of the proteins were identified as secreted proteins.
In summary, a functional analysis of the biological processes of differentially regulated proteins revealed a similar distribution, in which more than 50% of all up/down-regulated proteins participated in different metabolic processes (mostly in primary metabolic processes such as protein, nucleobase-containing compound, lipid, carbohydrate and tricarboxylic acid cycle metabolic processes) and cellular processes, which included cell communication, cell cycle, movement of cellular components, chromosome segregation and cytokinesis (PANTHER annotation). According to the molecular mechanisms, most of the detected up-regulated proteins were related to catalytic activity proteins, indicating enzyme regulator, hydrolase, isomerase, ligase, oxidoreductase and transferase activity. By contrast, the majority of the downregulated proteins were responsible for binding calcium ions, calcium-dependent phospholipids, chromatin, nucleic acids and other proteins. The difference in molecular functions between the up- and down-regulated proteins was the reason for the different predominant intracellular localization of these proteins.
Classical alphavirus-recognizing receptors, such as membrane heparan sulfate, laminin receptor, major histocompatibility complex (MHC), DC-sign, L-sign, heat shock 70 protein, and α1β1 integrin,13-18 which are implicated in virus absorption/transduction and are described in several studies, were not identified by LC-MS analysis in our experiment or were not found to be significantly up/downregulated. In addition, real-time PCR analysis did not demonstrate any significant differences in the gene expression of some of these candidates (data not shown).
Antiviral response genes
The main biological processes that initiate intracellular responses to viral infection and could potentially block replication primarily comprise the response to a stimulus, immune system processes and apoptosis.32 The category of responses to environmental stimuli (such as stress, endogenous or external stimuli) displays considerable overlap with the immune response category, usually regulating the expression of the same genes. The induction of apoptosis is often a consequence of such processes because all these features largely overlap. Some of the genes could be classified into 2 or 3 biological processes by the PANTHER classification system. Based on our proteomics classification results (as mentioned above), we performed a detailed analysis of the up/down-regulated genes that participated in 3 biological processes, response to stimulus, immune system response and apoptosis, summarizing these genes in one “antiviral response genes” category (Fig. 4A). In total, we identified 10 upregulated antiviral response genes in the ex vivo B16 cells: S100b (6.2-fold increase), Pycard (4.9-fold increase), CD97 (4-fold increase), Pdcd4 (3.1-fold increase), Gpx1 (2.3-fold increase), Csnk2b (2.2-fold increase), Gstz1 (2.2-fold increase), Gsto1 (1.9-fold increase), Scrib (1.7-fold increase) and Hsp90aa1 (1.6-fold increase).
Figure 4.
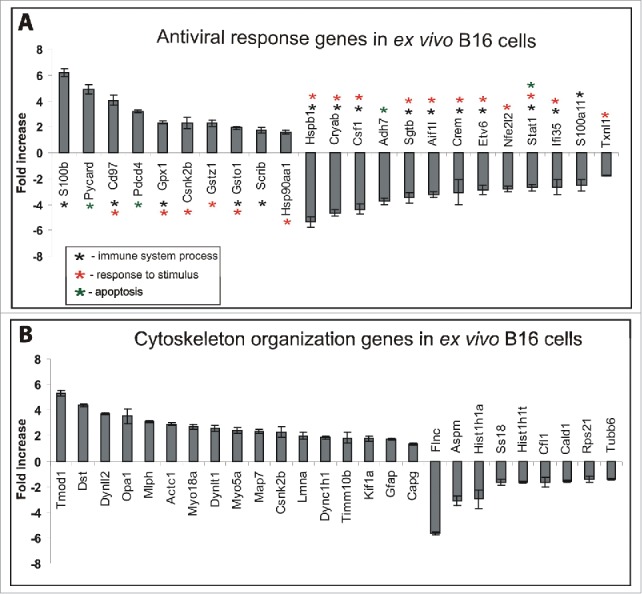
Up/downregulated genes in ex vivo B16 cells classified as antiviral response genes and cytoskeletal organization genes. (A) Based on the LC-MS data classification by PANTHER, genes that participated in apoptosis, response to stimulus and immune system processes were summarized in the antiviral response genes group. (B) Genes responsible for cellular component organization. The graph shows the fold-change in the expression of corresponding genes in ex vivo B16 cells compared with in vitro B16 cells. The results represent the mean ± s.e.
Overexpression of casein kinase II Csnk2b, scribble protein Scrib and heat shock protein Hsp90aa1 has been reported to potentially improve alphavirus-host cell binding and transduction (see Discussion). Additionally, all these proteins are correlated with high tumor aggressiveness and poor prognosis.
Overexpression of S100b (S100b) in the ex vivo B16 cells could be explained by active B16 tumor development because S100b is generally synthesized only under pathological circumstances and not under normal physiological conditions.33 Moreover, a high level of the S100B protein serves as a reliable prognostic biomarker in patients with malignant melanoma.34
The overexpressed glutathione peroxidase 1 Gpx1, maleylacetoacetate isomerase Gstz1 and glutathione S-transferase omega-1 Gsto were sub-classified as toxic substance responsible genes (PANTHER classification). Because there is limited information available regarding these multifunctional enzyme activities in cancer models, the upregulation of the genes could be explained as a consequence of a stress response caused by B16 cells upon transfer from the mice (in vivo conditions) to the plates (ex vivo conditions).
Although the up-regulation of CD97 antigen in melanoma cells has not been previously described, it was overexpressed in advanced stages of different cancers. The protein is mostly located in invasive tumors with higher cell motility than the cells in solid tumors.35-37
The up-regulated apoptotic genes, Pycard (apoptosis-associated speck-like protein containing a CARD) and Pdcd4 (programmed cell death protein 4), in the ex vivo B16 cells could act as key mediators in apoptosis and inflammation; however, no studies have described their roles in melanoma or their impacts on viral infectivity and replication.
In addition to the 10 upregulated genes described above, 13 downregulated genes involved in apoptosis, response to stimuli and immune system processes have been identified: Hspb1 (5.3-fold decrease), Cryab (4.6-fold decrease), Csf1 (4.3-fold decrease), Adh7 (3.7-fold decrease), Sgtb (3.4-fold decrease), Aif1l (3.2-fold decrease), Crem (3-fold decrease), Etv6 (2.9-fold decrease), Nfe2l2 (2.8-fold decrease), Stat1 (2.6-fold decrease), IFI35 (2.6-fold decrease), S100a11 (2.5-fold decrease) and Txnl1 (1.7-fold decrease) (Fig. 4A).
The roles of genes such as alcohol dehydrogenase 7 (Adh7, also known as Adh3) and small glutamine-rich tetratricopeptide repeat-containing protein Sgtb have not been described for alphavirus infection/replication. However, reduced Adh7 expression has been implicated in oncogenesis and might influence viral life cycles through increased S-nitrosylation and formaldehyde-induced changes in cellular redox.38,39 By contrast, the down-regulation of SGT proteins remarkably enhances the activity of HIV-1 virus.40
An in-depth analysis of other down-regulated genes showed that most of them participated in 3 IFN-regulated pathways: JAK-STAT, p38 MAPK and PI3K. Specifically, those genes were the heat shock protein Hspb1, α-B crystallin Cryab, macrophage colony-stimulating factor 1 Csf1, cAMP-responsive element modulator Crem, allograft inflammatory factor 1 Aif1l, transcription factor Etv6, nuclear factor erythroid 2-related factor 2 Nfe2l2, signal transducer and activator of transcription 1 Stat1, protein S100-A11 S100a11 and thioredoxin-like protein 1 Txnl1 (Table S3). These findings indicated that the described signaling pathways were suppressed in the ex vivo B16 cells. The expression of interferon-inducible 35 kDa protein (IFI35) was also significantly inhibited. These results present a network of downregulated genes modulated by IFN that could cooperatively provide favorable conditions for SFV transduction/replication.
Cytoskeletal organization genes
The cytoskeleton plays an important role in the life cycle of every virus during attachment, internalization, endocytosis, nuclear targeting, transcription, replication, transport of progeny subviral particles, assembly, exocytosis or cell to-cell spread.21 To investigate the genes regulating cytoskeletal organization that could facilitate alphavirus activity in ex vivo B16 cells, we performed a detailed analysis of the up/down-regulated proteins that participated in cellular component organization (or biogenesis) categorized by the PANTHER classification system (Fig. 4B). We identified 17 up-regulated genes: Tmod (5.3-fold increase), Dst (4.3-fold increase), Dynll2 (3.7-fold increase), Opa1 (3.5-fold increase), Mlph (3.1-fold increase), Actc1 (2.9-fold increase), Myo18a (2.7-fold increase), Dynlt1 (2.5-fold increase), Myo5a (2.4-fold increase), Map7 (2.3-fold increase), Csnk2b (2.2-fold increase), Lmna (1.9-fold increase), Dync1h1 (1.8-fold increase), Timm10b (1.8-fold increase), Kif1a (1.7-fold increase), Gfap (1.7-fold increase), and Capg (1.7-fold increase).
The cytoskeleton has 3 major types of filaments: actin, intermediate filaments and microtubules. Each type of filament has its own specific stabilization and motor proteins. In this experiment, we identified the upregulation of the filamentous protein α actin, Actc1. Moreover, both tropomodulin 1 (Tmod1) and macrophage-capping protein (Capg), which serve as actin filament stabilization proteins,41,42 were also found to be overexpressed in B16 ex vivo cells. The up-regulation of the intermediate filament protein Laminin (Lmna) was shown to increase the stiffness matrix, to confer nuclear mechanical properties, and to influence the differentiation of mesenchymal stem cells.43 Conversely, tubulin, which is the main component of microtubules, was found to be 1.4-fold downregulated in the ex vivo B16 cells (see below). However, the microtubule-stabilizing protein, Map7, which might play an important role during the reorganization of microtubules, was found to be 2.3-fold overexpressed.
Interestingly, in the ex vivo B16 cells, all the major motor proteins involved in movement on cytoskeletal filaments were upregulated. The myosins, Myo18a and Myo5a, are the only motor proteins that are able to bind to actin. Myosin acts upon actin filaments to generate cell surface contractions and other morphological changes, including vesicle motility and cytoplasmic streaming. Additionally, Myo5a has been considered to regulate melanosome transport in cooperation with the up-regulated Mlph.44 Both the upregulated kinesin, Kif1a, and the dyneins, Dynll2, Dynlt1, and Dync1h1, act as the main microtubule motor proteins, providing the intracellular retrograde motility of vesicles and organelles along microtubules.45 There are no known motor proteins for intermediate filaments.
Up to 4.3-fold upregulation of dystonin (Dst), which acts as an integrator of intermediate filaments and actin and microtubule cytoskeleton networks, facilitates intracellular transport by regulating organelle organization.46 The overexpression of another cytoskeletal organization factor, glial fibrillary acidic protein (Gfap), could be associated with cell mechanical strength and shape, but its exact function remains poorly understood.47
Casein kinase II subunit β (Csnk2b) is the only common protein that was detected in both categories of antiviral response genes and cellular component organization genes (Fig. 3A-B). Several studies have shown that casein kinase II is able to phosphorylate cytoskeletal proteins including tubulin48 or microtubule-associated protein 1B,49 thus contributing to morphological changes that occur during mitosis and cytokinesis in differentiating cells. Additionally, we identified the up-regulation of 2 mitochondrial membrane organization proteins, Opa1 and Timm10b, which are responsible for multi-pass transmembrane protein transport.50,51
Nine down-regulated genes responsible for cytoskeletal organization have been identified in the ex vivo B16 cells: Flnc (5.6-fold decrease), Aspm (3.1-fold decrease), Hist1h1a (2.9-fold decrease), Ss18 (1.6-fold decrease), Hist1h1t (1.6-fold decrease), Cfl1 (1.6-fold decrease), Cald1 (1.5-fold decrease), Rps21 (1.4-fold decrease) and Tubb6 (1.4-fold decrease). In this gene category, the muscle-specific filamin-C (Flnc) was detected as one of the most downregulated genes in B16 ex vivo cells that could potentially affect melanoma cell structure. The reduced expression of Flnc in myoblast cells has been shown to lead to defects in cell differentiation and fusion ability. These cells form multinucleated “myoballs” rather than maintaining an elongated morphology.52 Interestingly, the elongation of B16 ex vivo cells could also be impeded by a low level of the protein SSXT (Ss18), which has been described as a cytoskeletal phenotype-associated protein that plays a role in the elongation of the cell body via the induction of detyrosinated Glu tubulin.53 Our results demonstrate the downregulation of abnormal spindle-like microcephaly-associated (Aspm) protein in B16 ex vivo cells. Some studies have suggested that mutations or deregulation of Aspm could cause microcephaly due to the dysregulation of mitotic spindle activity, which increases the probability of asymmetric cell division.54
Caldesmon (Cald1) and cofilin (Cfl1), which are important for cytoskeletal organization and dynamics, were also found to be down-regulated up to 1.5- and 1.6-fold, respectively. Caldesmon is a multifunctional ubiquitous regulator of the actin cytoskeleton that when expressed at low levels, determines the bipolar shape and linear migration of cells.55 By contrast, defects in cofilin expression alter the morphology of actin networks in vivo and reduce the rate of actin flux through actin networks. The consequences of decreasing actin flux are manifested by decreased, but not blocked, endocytic internalization at the plasma membrane.56 Additionally, inhibition of the production of the 40S ribosomal protein S21 (Rps21) was also detected in B16 ex vivo cells.
Finally, suppression of H1 histone expression was detected in B16 ex vivo cells (2.9-fold decrease in Hist and 1.6-fold decrease in Hist1h1t), which could significantly impair the cell differentiation capacity. H1 histones have been shown to contribute to efficient repression of the expression of pluripotency factors and to participate in the establishment and maintenance of the epigenetic marks necessary for silencing pluripotency genes during embryogenesis and stem cell differentiation.57
Comparison of membrane elasticity between in vitro and ex vivo B16 cells
The transition of a tumor cell from fluid to adhesive conditions involves an early polarization event and major rearrangements of the submembrane cytoskeleton. This process can change the mechanical properties of the membrane, for example by increasing its elastic properties, which could affect cell endocytosis,58 virus-cell interactions and transduction.22 In our study, we used atomic force microscopy (AFM) to compare the cell membrane elasticity (or hardness)59 of the in vitro B16 cells and the first passage of the ex vivo B16 cells.
Several AFM force measurements were performed in which equal contact points of the AFM tip and cell surface were selected (the middle point between the cell nucleus and the elongated cell body) (Fig. 5A). After tip-cell contact, the pushing force was slowly increased until the membrane was punctured. The elasticity (or hardness) defined by the resistance of the cell to the changes was measured. The greater the resistance, the greater was the elasticity of the material and the faster it regained its original shape or configuration upon withdrawal of the deforming force.
Figure 5.
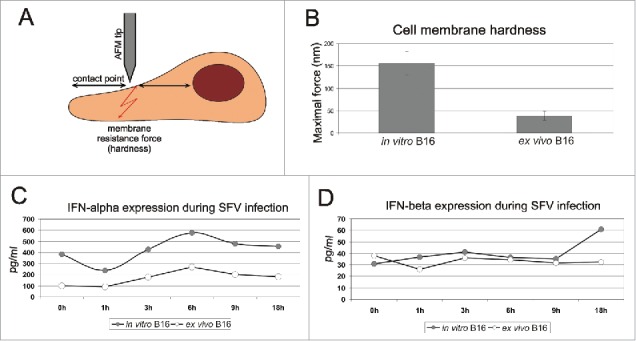
Comparison of cell membrane elasticity and IFN-α/β expression between in vitro and first-passage ex vivo B16 cells. The principle of AFM measurement is schematically demonstrated in panel A. The middle point between the cell nucleus and the elongated cell body was selected as the contact point of the AFM tip and the cell surface. The pushing force was increased slowly until the membrane was punctured. The acquired maximal forces essential for membrane puncturing of in vitro and ex vivo B16 cells are shown in panel B. The membrane resistance force indicates the elasticity of the sample surface; higher maximal force indicates higher membrane hardness. The results represent the mean ± s.e. Expression levels of IFN-α (C) and IFN-β (D) were determined in in vitro and first-passage ex vivo B16 cell lysates before SFV infection (0 h) and at 1 h, 3 h, 6 h, 9 h and 18 h post-infection. The results represent the concentration of IFN protein per 1 ml of cell lysate (pg/ml).
In this experiment, the membrane resistance force indicated the elasticity of the sample surface. A higher maximal force indicated a greater membrane hardness. The AFM data demonstrated that the ex vivo B16 cells had significantly lower cell membrane elasticity (Fig. 5B), with an average maximal force of 38 nN compared with the control cells with maximal forces within the range of 120–180 nN. The low level of membrane hardness in the case of the ex vivo B16 cells could be one of the reasons for the improved alphavirus-cell interaction and transduction by endocytosis.
IFN-α/β analysis
Type I interferon (IFN) is an extremely powerful antiviral response factor that is capable of affecting alphavirus infections.60 We compared the expression levels of IFN-α and IFN-β in the in vitro B16 cells and the first passage of the ex vivo B16 cells before infection (0 h) and at different time points during infection and replication (1 h, 3 h, 6 h, 9 h, and 18 h) (Fig. 5C-D). Our data demonstrated a significant down-regulation of IFN-α expression (4-fold decrease) in the ex vivo B16 cells before infection compared with the control in vitro cells. Melanoma cells do not secrete IFN under normal conditions, which results in the accumulation of IFN within the cells. In our experiment, we observed an equal tendency of IFN-α accumulation in both in vitro and ex vivo B16 cells starting from 1 h after infection (Fig. 5C). However, the expression level of IFN-α in in vitro cells was remarkably higher 1 h post-infection and increased over time to reach a maximum of 576 pg/ml at 6 h post-infection. By contrast, the maximum expression in ex vivo cells reached only 269 pg/ml at the same time point. The inhibited expression of IFN-α could be the main cause of the enhanced alphavirus replication in the ex vivo melanoma cells.
There were no significant differences in the expression and accumulation of IFN-β in either the in vitro or the ex vivo B16 cells, thus providing evidence that IFN-β might play a less important role in host cell protection against alphaviral infection.
Discussion
Although alphaviruses are successfully used as tumor-targeting agents in cancer gene therapy, the key factors providing intracellular conditions that are favorable for more efficient viral activity as well as the virus-host interaction dynamics are not well understood. The tumor microenvironment and innate immune responses could cause a wide range of variations in viral activity, blocking or facilitating the replication and spread of the viral particles. We obtained interesting results demonstrating an extremely high infectiousness of alphaviruses in B16 melanoma tumors in vivo, whereas delivery of the marker transgenes was inefficient for the same cells in vitro (Fig. 1A). These data corresponded well with the results of other authors.6 We hypothesized that this difference resulted from the influence of the tumor microenvironment on the total cellular proteome.
Alphavirus infectivity and spread in vivo
Although a high level of alphavirus-driven transgene expression was determined, the SFV vector displayed a limited distribution in B16 tumors, demonstrating patchy expression of the virus only in local injection areas (Fig. 1B). Several factors affect the capacity of the virus to spread within the tumor, such as virus neutralization by blood components or rapid generation of an antiviral immune response; however, the main factor is the intratumoral stromal barriers. For alphaviruses, it is possible to increase the tissue permeability and promote their intratumoral distribution, simultaneously using synergic chemotherapeutic drugs.27,61 However, the slow kinetics of viral spread after intratumoral injections is a common disadvantage that has also been reported for other vectors, e.g., adenoviruses62 and retroviruses.63 The oncolytic properties of alphaviruses make them a promising tool with the potential to significantly improve the field of cancer treatment. Thus, it is clearly important to investigate the details of the processes that determine the efficiency of virus spread and infection within tumor microenvironments.
Alphavirus infectivity in vitro
The ex vivo B16 tumor cells clearly demonstrated a high efficacy of SFV infection in the first passage after isolation, whereas the control B16 cells cultivated in vitro did not support the viral activity. However, the ex vivo cells were not able to retain these properties for a long time during the cultivation in vitro (Fig. 2A). This observation indicated that the B16 tumor microenvironment was able to provide favorable conditions for alphaviral transduction and replication that were not present in the in vitro culture.
The ability to support virus entry and subsequent expression of the marker transgene is most likely determined by differences between the protein profile of the cells residing in the tumor microenvironment and the cells cultivated in a monolayer. The alterations of cellular protein levels might be reflected in numerous ways, from changes in metabolic activity to the physical parameters of the cell membranes, which facilitate endocytosis. Moreover, it is likely that a combination of several appropriate properties is required simultaneously, which is why we focused our efforts on the systematic top-down analysis of the proteomic differences between ex vivo and in vitro cells.
Role of antiviral response genes in viral activity
The comparative analysis of the protein profile of B16 ex vivo cells yielded the list of up/downregulated gene candidates, which were classified into several groups based on their functions and roles in biological processes and molecular mechanisms (Fig. 3A-D). The statistical analysis showed that none of the designated subgroups representing specific biological processes and molecular mechanisms was significantly up- or down-regulated in the ex vivo cells in general. This result was expected because although the ex vivo and in vivo cells demonstrate altered properties, they still exhibit similar phenotypes and thus similar expression patterns.
Because the main biological processes capable of dramatically affecting viral activity are the response to stimuli, immune system processes and apoptosis,32 it is very likely that alterations of the expression and translation levels of these specific proteins could ensure favorable conditions for viral activity (Fig. 4A). The most interesting candidate genes that were found to be up-regulated were Csnk2b ((CK2) family member), Scrib and Hsp90aa1. Overexpression of all these genes not only serves as an indicator of the increased aggressiveness of the ex vivo B16 cells by facilitating malignant transformation64,65 but also potentially facilitates viral vector activity. Recent studies have shown that CK2 is essential for the infectious cycle of alphaviruses and other different DNA/RNA viruses.66,67 The inhibition of Hsp90 reduces chikungunya virus infection and inflammation in vivo.68 Scrib acts as a binding protein and plays a role in virus-host cell interactions in the case of several viral infections.69 Moreover, Scrib is important for cell polarity, acting as a crucial factor in cell membrane architecture and physiology.70,71 Therefore, upregulation of Scrib could play a significant role in the increased membrane elasticity of ex vivo cells, which could in turn impact the process of viral entry.
Among the downregulated proteins, one of the most attractive candidates was IFN-α because it is one of the main negative regulators of tumor growth. Inhibition of IFN-α expression has been shown to cause transcriptional downregulation of several anti-proliferative IFN-α-inducible signaling pathways, namely, PI3K, p38 MAPK and JAK-STAT,72,73 which was correlated with extremely high alphavirus activity.74,75 A detailed analysis of down-regulated antiviral response genes in B16 ex vivo cells showed that most of them participate in the PI3K, p38 MAPK and JAK-STAT pathways (Table S3), but the expression of genes such as Hspb1 (Hsp27), Cryab, Csf1, Stat1 and S100a11 is mediated by all 3 pathways. The PI3K pathway has been shown to be essential for the replication of different viruses. However, inhibition of the activity of PI3K in B16 ex vivo cells apparently did not function as a key factor affecting SFV vector-driven expression. Alphaviruses are able to auto-activate and auto-regulate PI3K signaling during infection, providing essential conditions for cap-dependent translation of viral proteins.76 By contrast, down-regulation of p38 MAPK and JAK-STAT signaling has a remarkable effect on the virulence potential of alphaviruses. Both of these pathways can promote apoptosis, thereby enhancing the cytopathic properties of the virus during infection.74,75 The inhibited activity of both the p38 MAPK and JAK-STAT pathways in B16 ex vivo cells facilitated alphavirus replication and transgene expression, possibly by delaying the onset of apoptosis during infection. The downregulation of several genes coordinated by the IFN-inducible pathways shown in Table S3 such us Nfe2l,77-79 Ifi35,80 S100A1181 has been shown to promote different levels of virus infectivity.
Role of genes regulating cytoskeletal organization during virus entry
An additional group, which might be responsible for the improved viral activity, comprises the cytoskeleton-related proteins. Genes regulating cytoskeletal organization are essential during the virus life cycle. Up-regulation of several cytoskeleton motor proteins such us dynein and kinesin in ex vivo B16 cells facilitates the activity of various virus vectors. Generally, invading viruses use dynein to reach the nucleus for replication. In turn, kinesin is used to reach the cell membrane where viral budding and exit occur.82 Dynein has been reported to interact with purified adenovirus,83 adeno-associated virus,84 parvovirus,85 herpes virus86 and HIV-1.87 Kinesin appears to have several important roles during the replication cycle of vaccinia virus88 and to provide cellular transport for herpes virus (HSV)89 along microtubules.90 Interestingly, the release of enveloped HSV virions at the plasma membrane was promoted by Myosin 5A (Myo5a), which is another overexpressed gene candidate detected in ex vivo B16 cells.91 Additionally, a delay of HSV replication and inhibition of capsid movement in the cytoplasm during egress was observed under the condition of dystonin (Dst) depletion, suggesting that this non-motor protein is an important part of the virus transport machinery.92 Furthermore, changes in the expression of cellular component organization genes could also affect virus entry by altering the physical features of the cell membrane, such as the elasticity and the rigidness. During the endocytosis process, the cell membrane must deform and accommodate a high degree of curvature that requires low elastic features. We presume that the reduced elasticity of the ex vivo B16 cells observed by AFM analysis (Fig. 5B) was responsible for the improved alphavirus transduction, whereas the re-organized cytoskeleton facilitated its replication and spread.
In summary, we presented candidate genes (Fig. 4) that provided favorable conditions in combination for increased alphavirus infectivity, thereby providing new possibilities for the enhanced efficacy of alphavirus-based cancer gene therapy. Based on the results of this study, we emphasize the role of combined alterations in gene expression. Individual functional experiments of the identified genes were not a focus of this study because such experiments fail to reproduce the full network of different factors that occur during malignant tumor development and affect alphavirus infection. However, continuous progress in these investigations in the future might impact our understanding of the alphavirus transduction/replication process.
Materials and methods
Cell lines and animals
The BHK-21 (baby hamster kidney cells) and B16-F10 (metastatic mouse melanoma) cells were obtained from the American Type Culture Collection (ATCC/LGC Prochem). The BHK-21 cells were propagated in BHK - Glasgow MEM (GIBCO) supplemented with 5% fetal bovine serum (FBS), 10% tryptose phosphate broth, 2 mM L-glutamine, 20 mM HEPES and antibiotics (streptomycin 100 mg ml−1 and penicillin 100 U ml−1). The B16-F10 cell line was cultured in Dulbecco's GlutaMAX medium (GIBCO) supplemented with 10% FBS and antibiotics streptomycin 100 mg ml−1 and 40 µg ml−1 gentamicin. Specific pathogen-free 4- to 6-week-old female C57BL/6 mice were obtained from Latvian Experimental Animal Laboratory of Riga Stradins University and maintained under pathogen-free conditions in the accordance with the principles of the Latvian and European Community laws. All experiments were approved by the local Animal Protection Ethical Committee and the Latvian Food and Veterinary Service (permission for animal experiments no. 32/23.12.2010).
Production of SFV (SFV/EGFP, SFV/DS-Red and SFV/Enh.Luc) recombinant virus particles
The pSFV193 vector was used in this study. The pSFV/EGFP, pSFV/Ds-Red and pSFV/Enh.Luc vectors were generated as described previously.12,27 The resulting plasmids were used to produce SFV/EGFP, SFV/DS-Red and SFV/Enh.Luc virus particles. Briefly, the pSFV/EGFP, pSFV/DS-Red, pSFV/Enh.Luc and pSFV-Helper93 plasmids were linearized using the SpeI restriction enzyme. In vitro RNA transcription was performed using 1–2 μg of linearized DNA and 40 U of SP6 RNA polymerase (Thermo Scientific) in a 50 μl reaction mixture, as described by the manufacturer. The RNA transcripts were capped during the transcription reaction by adding 1 mM of the 5′(ppp)5′G cap-analog (New England Biolabs). The DNA template was removed by digestion with RNase-free DNase (Thermo Scientific).
For packaging, the corresponding in vitro transcribed recombinant RNA (20 μg each) were co-electroporated with the helper RNA into 1 × 107 BHK-21 cells (850 V, 25 μF, 2 pulses) using a Bio-Rad Gene Pulser apparatus without the pulse controller unit. The electroporated cells were resuspended in 15 ml of complete BHK medium, transferred into tissue culture flasks (75 cm2) and incubated at 37°C (5% CO2). After 24 h, the recombinant SFV particle-containing medium was harvested, rapidly frozen and subsequently used as the virus stock for cell culture infection.
The virus titer (infectious units per ml, iu ml−1) was quantified by infection of BHK-21 cells with serial dilutions of viral stock and analysis of EGFP or DS-Red expression by fluorescence microscopy on a Leica DM IL microscope (Leica Microsystems Wetzlar GmbH). SFV/Enh.Luc virus titer was quantified by Real-time PCR as previously described.12
Infection of cell lines with recombinant virus particles
The BHK-21 and B16-F10 cells were cultivated in 24-well plates at a density of 2 × 105 cells per well in a humidified 5% CO2 incubator at 37°C. For transduction, the cells were washed twice with PBS containing Mg2+ and Ca2+ (Invitrogen). Then, 0.3 ml of the medium containing the virus particles was added. The SFV/DS-Red and SFV/Enh.Luc virus particles were diluted in PBS (containing Mg2+ and Ca2+) to achieve a multiplicity of infection (MOI) of 10. The cells were incubated for 1 h in a humidified 5% CO2 incubator at 37°C. The control cells (uninfected) were incubated with PBS (containing Mg2+ and Ca2+). After incubation, the solution containing virus was replaced with 0.5 ml of growth medium.
Induction of tumor nodules
The B16-F10 cells were trypsinized, washed with PBS and resuspended in PBS at a final concentration of 3 × 105 cells ml−1. Two hundred microliters of the B16-F10 cell suspension was subcutaneously injected above the right shoulder blade of the mice. After 8–12 days, when tumor volumes reached 1000 mm3, the B16 nodules were i.t. injected with SFV particles or isolated for further tumor cell cultivation.
SFV/Enh.Luc administration and analysis of luciferase gene expression in tumors
B16 tumor-bearing mice (n = 3) were i.t. inoculated with 0.25 ml of SFV/Enh.Luc particle-containing stocks (4 × 108 i.u. per ml). The Luc gene expression level was estimated by measuring luciferase enzymatic activity in tumor homogenates 24h after SFV/EnhLuc virus administration. The tumors were excised and manually homogenized in a 1x concentration of ice-cold lysis buffer (Cell Culture Lysis buffer, Promega) containing a protease inhibitor cocktail (10 μl per 1 ml of lysis buffer) (Sigma). After homogenization, the samples were centrifuged for 10 min at 9000 × g, and the protein concentration was determined in tissue lysates using the BCA Protein Assay Kit (Thermo Scientific). Luciferase activity was measured by adding 100 μl of freshly reconstituted luciferase assay buffer to 20 μl of the tissue homogenate (Luciferase Assay System, Promega) and then was quantified as relative light units (RLUs) using a luminometer (Luminoskan Ascent, Thermo Scientific). The RLU values were expressed per mg of protein in the lysates. As a negative control, B16 tumor-bearing mice were inoculated with PBS, and the maximal negative value was subtracted from the presented results.
SFV/EGFP, SFV/Ds-Red administration and analysis of vector intratumoral spread
B16 tumor-bearing mice were i.t. inoculated with 106 i.u. of SFV/EGFP and SFV/Ds-Red in different tumor sides (SFV/EGFP was injected into right side of tumor, where SFV/Ds-Red was injected into tumor left side). 24h after vectors administrations the tumors were isolated and frozen in OCT compound (Sigma). The cryosections (5–10 μm) were prepared and EGFP and Ds-Red expression was visualized by fluorescent microscopy.
Isolation and cultivation of ex vivo B16 cells
Freshly isolated B16-F10 tumors were manually homogenized in PBS and filtered through 40-µm diameter filters. The obtained cells were washed twice with PBS and seeded in 24-well plates at a density of approximately 5 × 105 cells per well in Dulbecco's GlutaMAX medium (GIBCO/Invitrogen) supplemented with 10% FBS, 100 mg ml−1 streptomycin, and 40 µg ml−1 gentamicin. Two days later, the cells were washed with PBS to remove unattached cells and cultivated in Dulbecco's GlutaMAX medium containing 10% FBS, 100 mg ml−1 streptomycin, and 40 µg ml−1 gentamicin for 5–7 d until the cell monolayer reached 80% confluency. The medium was replaced every 2 d during cultivation. Next, the 80% monolayer 1st passage of ex vivo cells was trypsinized and plated in 24-well plates at a low density of approximately 4 × 103 cells per well (for infection or melanin staining) and in 10-cm Petri dishes at a density of 2 × 104 cells for further ex vivo B16 cell passaging (2nd passage, 3rd passage, etc.).
Melanin staining of B16 cells was performed using a Fontana-Masson Stain kit according to the protocol provided by the manufacturer (Abcam). Briefly, 80–100% monolayered cells were washed with PBS and fixed with 3% paraformaldehyde for 10 min. The cells were then washed with PBS and treated according to the Fontana-Masson staining protocol. The cell nuclei were counterstained with nuclear fast red.
Sample preparation for label-free LC-MS analysis
Ex vivo B16 cells were isolated from 3 B16-F10 tumor-bearing mice and cultivated for 5–7 d until the monolayer of the first passage reached 80% confluency (see above). The control in vitro B16-F10 cells were cultivated as described above until the monolayer reached 80% confluency. Both in vitro and ex vivo B16 cells were lysed with 0.1% ProteaseMax in 50 mM ammonium bicarbonate buffer (Promega) and sonicated for 15 min at 35 kHz. The lysate protein concentration was measured using Direct Detect® Assay-free Cards (Merck Millipore). For LC-MS analysis, samples were prepared using the FASP protocol.94 Briefly, 80 μg of proteins from each sample were mixed with DTT at a final concentration of 10 mM and incubated for 15 min at 56°C. The cell lysate was then incubated with 6 M urea and filtered through a YM-30 spin column (Millipore). The sample-containing columns were washed twice with 100 mM Tris pH 8.5 and then digested by addition of lysyl endopeptidase (Lys-C) (Wako) solution or trypsin to the column overnight at 37°C. The resulting Lys-C and trypsin fractions were eluted with H2O by centrifugation and mixed with 0.1% trifluoroacetic acid (TFA). All samples were purified using microcolumns prepared by placing a C18 Empore Extraction Disk (Varian, St. Paul, MN) into 200-μl pipet tips. Peptides were eluted by applying 80 μl of 80% acetonitrile (ACN) and 0.1% formic acid in water. ACN was evaporated in a vacuum drier, and the samples were diluted in 0.1% formic acid in water.
LC-MS
All experiments were performed on an Easy nLC1000 nano-LC system connected to a quadrupole – Orbitrap (QExactive) mass spectrometer (ThermoElectron) equipped with a nanoelectrospray ion source (EasySpray/Thermo). For liquid chromatography separation, we used an EasySpray column (C18, 2-µm beads, 100 Å, 75-μm inner diameter) (Thermo) capillary with a 25-cm bed length. The flow rate was 300 nl/min, and the solvent gradient was 2% B to 5% B in 10 min followed by 5% to 26% B in 230 min, and then 90% B wash in 20 min. Solvent A was aqueous 0.1% formic acid, whereas solvent B was 100% acetonitrile in 0.1% formic acid. The column temperature was kept at 60°C.
The mass spectrometer was operated in data-dependent mode to automatically switch between MS and MS/MS acquisition. Survey full-scan MS spectra (from m/z 400 to 1,200) were acquired in the Orbitrap with resolution R = 70,000 at m/z 200 (after accumulation to a target of 3,000,000 ions in the quadruple). The method used allowed for the sequential isolation of the most intense multiply charged ions, consisting of up to 10 depending on the signal intensity, for fragmentation on the HCD cell using high-energy collision dissociation at a target value of 100,000 charges or a maximum acquisition time of 100 ms. MS/MS scans were collected at a resolution of 17,500 at the Orbitrap cell. Target ions already selected for MS/MS were dynamically excluded for 30 seconds. General mass spectrometry conditions were as follows: electrospray voltage of 2.1 kV, no sheath and auxiliary gas flow, heated capillary temperature of 250°C, and normalized HCD collision energy of 25%. The ion selection threshold was set to 1e4 counts. An isolation width of 3.0 Da was used.
Protein identification and label-free quantitation
MS raw files were submitted to MaxQuant software version 1.4.0.5 for protein identification.95 Parameters were set as follows: protein N-acetylation, methionine oxidation and pyroglutamate conversion of Glu and Gln as variable modifications. First, we used a search error window of 20 ppm and a main search error of 6 ppm. The Lys-C or Trypsin enzyme option, both without proline restriction, was used depending on the sample, with 2 allowed miscleavages. Minimal unique peptides were set to 1, and the FDR allowed was 0.01 (1%) for peptide and protein identification. Label-free quantitation was set with a retention time alignment window of 3 min. The UniProt Reference Proteome mouse database was used (download from April 2014). Generation of reversed sequences was selected to assign FDR rates. All quantitative analyses were performed using the Perseus suit from MaxQuant. Briefly, MaxLFQ values were loaded and log-transformed, and 0 values were replaced by noise detection values using an imputation approach based on the normal distribution of the whole data. Differential proteins were assigned by a t-test analysis using S0 = 0.5, a p-value threshold of 0.01 and a permutation-based FDR correction.
Bioinformatics analysis
A total of 277 differentially expressed genes in ex vivo and in vitro B16 cells were selected by filtering with confidence at p < 0.01 from a total of 4980 proteins with a difference in expression of at least 1.4-fold. The biological classification of associated genes in terms of their biologic processes and molecular functions was obtained by Gene Ontology (GO) analysis using the Protein ANalysis THrough Evolutionary Relationships (PANTHER) classification system (version: PANTHER 9.0; http://www.pantherdb.org).30,96 Analysis of the cellular localization of selected genes was conducted using the UniProt Knowledgebase (http://www.uniprot.org/).
Analysis of IFN-α and IFN-β in ex vivo and in vitro B16 cells
Control in vitro B16 cells and freshly isolated ex vivo B16 cells were seeded in 24-well plates and cultivated until the cell monolayer reached 80% confluency as described above. Expression levels of the IFN-α and IFN-β were determined in in vitro and first-passage ex vivo B16 cell lysates before SFV infection (0 h) and at 1 h, 3 h, 6 h, 9 h and 18 h after infection with SFV/Ds-Red at an MOI of 10. The cells were trypsinized, washed with PBS and resuspended in 100 µl of PBS. For the lysates, 3 freeze-thaw cycles of the cell suspensions were performed. The cell lysates were centrifuged for 10 min at 5000 × g, and the protein concentration was equalized in all samples using the BCA Protein Assay Kit (Pierce™ BCA Protein Assay Kit, Thermo Scientific). The expression levels of IFN-α and IFN-β in the cell lysates were determined using the Interferon Alpha ELISA Kit (Uscn Life Science Inc.) and Interferon Beta ELISA Kit (Cusabio Biotech) according to the manufacturers' protocols.
Analysis of cell membrane elasticity by AFM
Control in vitro B16 cells and freshly isolated ex vivo B16 cells were seeded in 8-well plastic chambers and cultivated until the cell monolayer reached 80% confluency as described above. To measure cell membrane hardness, an uncoated atomic force microscope cantilever (Olympus AC240TS) with spring constant C = 2 N/m and resonant frequency F = 70 kHz was used. Cantilever calibration was conducted by standard operations using an MFP-3D atomic force microscope (Asylum Research) and Igor Pro 6.34A software. The AFM tip with a radius of 10 nm was manually positioned at the middle point between the cell nucleus and the elongated cell body (Fig. 5A) using an OLYMPUS IX71 inverted optical microscope. After positioning, the tip was engaged without scanning the surface of the cell to maintain viability, and single force curves were acquired. Single force curve data were exported to Microsoft Excel. For each cell, a particular force curve was acquired using only trace data, where x was the distance to the cell and y was the applied force. The maximum force that could be endured by a cell was calculated by identifying the intersection point of 2 linear trendlines: one describing the tip approach region and the second describing the cell perforation region. The trendlines fit the experimental data with a coefficient of determination of no less than 95%.
Statistical analysis
The RLU results of the in vivo and in vitro experiments presented in Fig. 1A are presented as the mean ± s.e. of replicate analyses and are representative of 2 independent experiments. The data were transformed to the logarithmic scale. All error terms shown in Figs. 4 and 5B are expressed as the standard error of the mean from at least 3 different samples. Statistical analyses of the results were performed using Microsoft Excel and Statistica7 (StatSoft, Tulsa). Statistically significant differences were determined using Student's t-test (p < 0.05).
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Professor P. Pumpens and his lab members for helpful discussions and excellent technical assistance.
Funding
This work was supported by Norway Grants Program 2009–2014 under project contract NFI/R/2014/051.
References
- 1.Glasgow GM, McGee MM, Tarbatt CJ, Mooney DA, Sheahan BJ, Atkins GJ. The Semliki Forest virus vector induces p53-independent apoptosis. J Gen Virol 1998; 79(Pt 10):2405-10; PMID:9780045; http://dx.doi.org/ 10.1099/0022-1317-79-10-2405 [DOI] [PubMed] [Google Scholar]
- 2.Venticinque L, Meruelo D. Sindbis viral vector induced apoptosis requires translational inhibition and signaling through Mcl-1 and Bak. Mol Cancer 2010; 9:37; PMID:20152035; http://dx.doi.org/ 10.1186/1476-4598-9-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riezebos-Brilman A, de Mare A, Bungener L, Huckriede A. Recombinant alphaviruses as vectors for anti-tumour and anti-microbial immunotherapy. J Clin Virol 2006; 35:233-243; PMID:16448844; http://dx.doi.org/ 10.1016/j.jcv.2005.12.001 [DOI] [PubMed] [Google Scholar]
- 4.Liljestrom P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology (NY) 1991; 9:1356-61; http://dx.doi.org/ 10.1038/nbt1291-1356 [DOI] [PubMed] [Google Scholar]
- 5.Wahlfors JJ, Zullo SA, Loimas S, Nelson DM, Morgan RA. Evaluation of recombinant alphaviruses as vectors in gene therapy. Gene Ther 2000; 7:472-80; PMID:10757020; http://dx.doi.org/ 10.1038/sj.gt.3301122 [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Madoz JR, Prieto J, Smerdou C. Biodistribution and tumor infectivity of semliki forest virus vectors in mice: effects of re-administration. Mol Ther 2007; 15:2164-71; PMID:17667947; http://dx.doi.org/ 10.1038/sj.mt.6300274 [DOI] [PubMed] [Google Scholar]
- 7.Kononchik JP Jr, Hernandez R, Brown DT. An alternative pathway for alphavirus entry. Virol J 2011; 8:304; PMID:21676248; http://dx.doi.org/ 10.1186/1743-422X-8-304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wojton J, Kaur B. Impact of tumor microenvironment on oncolytic viral therapy. Cytokine Growth Factor Rev 2010; 21(2–3):127-34; PMID:20399700; http://dx.doi.org/ 10.1016/j.cytogfr.2010.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villanueva J, Herlyn M. Melanoma and the tumor microenvironment. Curr Oncol Rep 2008; 5:439-46; http://dx.doi.org/ 10.1007/s11912-008-0067-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meruelo D. Systemic gene therapy by Sindbis vectors: A potentially safe and effective targeted therapy for identifying and killing tumor cells in vivo. Discov Med 2004; 20:54-7; http://dx.doi.org/ 10.1002/0471234303.ch17 [DOI] [PubMed] [Google Scholar]
- 11.Tseng JC, Granot T, DiGiacomo V, Levin B, Meruelo D. Enhanced specific delivery and targeting of oncolytic Sindbis viral vectors by modulating vascular leakiness in tumor. Cancer Gene Ther 2010; 4:244-55; http://dx.doi.org/ 10.1038/cgt.2009.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasilevska J, Skrastina D, Spunde K, Garoff H, Kozlovska T, Zajakina A. Semliki Forest virus biodistribution in tumor-free and 4T1 mammary tumor-bearing mice: a comparison of transgene delivery by recombinant virus particles and naked RNA replicon. Cancer Gene Ther 2012; 8:579-87; http://dx.doi.org/ 10.1038/cgt.2012.37 [DOI] [PubMed] [Google Scholar]
- 13.Smit JM, Waarts BL, Kimata K, Klimstra WB, Bittman R, Wilschut J. Adaptation of alphaviruses to heparan sulfate: interaction of Sindbis and Semliki forest viruses with liposomes containing lipid-conjugated heparin. J Virol 2002; 76:10128-37; PMID:12239287; http://dx.doi.org/ 10.1128/JVI.76.20.10128-10137.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang KS, Kuhn RJ, Strauss EG, Ou S, Strauss JH. High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J Virol 1992; 66:4992-5001; PMID:1385835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helenius A, Morein B, Fries E, Simons K, Robinson P, Schirrmacher V, Terhorst C, Strominger JL. Human (HLA-A and HLA-B) and murine (H-2K and H-2D) histocompatibility antigens are cell surface receptors for Semliki Forest virus. Proc Natl Acad Sci USA 1978; 75:3846-50; PMID:278998; http://dx.doi.org/ 10.1073/pnas.75.8.3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klimstra WB, Nangle EM, Smith MS, Yurochko AD, Ryman KD. DC-SIGN and L-SIGN can act as attachment receptors for alphaviruses and distinguish between mosquito cell- and mammalian cell-derived viruses. J Virol 2003; 77:12022-32; PMID:14581539; http://dx.doi.org/ 10.1128/JVI.77.22.12022-12032.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan C, Stevens TH, Schlesinger MJ. Inhibitory effects of HSP70 chaperones on nascent polypeptides. Protein Sci 1992; 1:980-85; PMID:1304386; http://dx.doi.org/ 10.1002/pro.5560010803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.La Linn M, Eble JA, Lubken C, Slade RW, Heino J, Davies J, Suhrbier A. An arthritogenic alphavirus uses the alpha1beta1 integrin collagen receptor. Virol 2005; 336:229-39; http://dx.doi.org/ 10.1016/j.virol.2005.03.015 [DOI] [PubMed] [Google Scholar]
- 19.Naghavi MH1, Goff SP. Retroviral proteins that interact with the host cell cytoskeleton. Curr Opin Immunol 2007; 4:402-7; http://dx.doi.org/ 10.1016/j.coi.2007.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radtke K, Döhner K, Sodeik B. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell Microbiol 2006; 3:387-400; PMID:NOT_FOUND; http://dx.doi.org/ 10.1111/j.1462-5822.2005.00679.x [DOI] [PubMed] [Google Scholar]
- 21.Ward BM. The taking of the cytoskeleton one two three: how viruses utilize the cytoskeleton during egress. Virology 2011; 2:244-50; http://dx.doi.org/ 10.1016/j.virol.2010.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun SX, Wirtz D. Biophys J. Mechanics of enveloped virus entry into host cells. Biophys J 2006; 1:L10-2; http://dx.doi.org/ 10.1529/biophysj.105.074203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pushko P, Parker M, Ludwig GV, Davis NL, Johnston RE, Smith JF. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology 1997; 239:389-401; PMID:9434729; http://dx.doi.org/ 10.1006/viro.1997.8878 [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Burke CW, Ryman KD, Klimstra WB. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J Virol 2007; 20:11246-55; http://dx.doi.org/ 10.1128/JVI.01282-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bick MJ, Carroll JW, Gao G, Goff SP, Rice CM, MacDonald MR. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol 2003; 21:11555-62; http://dx.doi.org/ 10.1128/JVI.77.21.11555-11562.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljeström P. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol Biol Cell 2005; 8:3753-63; http://dx.doi.org/ 10.1091/mbc.E05-02-0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zajakina A, Vasilevska J, Zhulenkovs D, Skrastina D, Spaks A, Plotniece A, Kozlovska T. High efficiency of alphaviral gene transfer in combination with 5-fluorouracil in a mouse mammary tumor model. BMC Cancer 2014; 14:460; PMID:24950740; http://dx.doi.org/ 10.1186/1471-2407-14-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang PY, Guo JH, Hwang LH. Oncolytic Sindbis virus targets tumors defective in the interferon response and induces significant bystander antitumor immunity in vivo. Mol Ther 2012; 2:298-305; http://dx.doi.org/ 10.1038/mt.2011.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu YP, Suksanpaisan L, Steele MB, Russell SJ, Peng KW. Induction of antiviral genes by the tumor microenvironment confers resistance to virotherapy. Sci Rep 2013; 3:2375; PMID:23921465; http://dx.doi.org/doi.org/ 10.1038/srep02375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 2003; 9:2129-41; http://dx.doi.org/ 10.1101/gr.772403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brar SS, Kennedy TP, Sturrock AB, Huecksteadt TP, Quinn MT, Whorton AR, Hoidal JR. An NAD(P)H oxidase regulates growth and transcription in melanoma cells. Am J Physiol Cell Physiol 2002; 6:C1212-24; http://dx.doi.org/ 10.1152/ajpcell.00496.2001 [DOI] [PubMed] [Google Scholar]
- 32.Barber GN. Host defense, viruses and apoptosis. Cell Death Differ 2001; 2:113-26; http://dx.doi.org/ 10.1038/sj.cdd.4400823 [DOI] [PubMed] [Google Scholar]
- 33.Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, Geczy CL. Functions of S100 proteins. Curr Mol Med 2013; 1:24-57; http://dx.doi.org/ 10.2174/156652413804486214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leclerc E, Heizmann CW, Vetter SW. RAGE and S100 protein transcription levels are highly variable in human melanoma tumors and cells. Gen Physiol Biophy 2009; Spec No Focus:F65-75; PMID:20093728; http://dx.doi.org/ 10.5167/uzh-26499 [DOI] [PubMed] [Google Scholar]
- 35.Steinert M, Wobus M, Boltze C, Schütz A, Wahlbuhl M, Hamann J, Aust G. Expression and regulation of CD97 in colorectal carcinoma cell lines and tumor tissues. Am J Pathol 2002; 5:1657-67; http://dx.doi.org/ 10.1016/S0002-9440(10)64443-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoang-Vu C1, Bull K, Schwarz I, Krause G, Schmutzler C, Aust G, Köhrle J, Dralle H. Regulation of CD97 protein in thyroid carcinoma. J Clin Endocrinol Metab 1999; 3:1104-9; PMID:10084602; http://dx.doi.org/ 10.1210/jc.84.3.1104 [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Chen L, Peng SY, Chen ZX, Hoang-Vu C. Role of CD97(stalk) and CD55 as molecular markers for prognosis and therapy of gastric carcinoma patients. J Zhejiang Univ Sci B 2005; 6:913-18; PMID:16130195; http://dx.doi.org/ 10.1631/jzus.2005.B0913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei W, Li B, Hanes MA, Kakar S, Chen X, Liu L. Snitrosylation from GSNOR deficiency impairs DNA repair and promotes hepatocarcinogenesis. Sci Transl Med 2010; 19:19ra13; PMID:20371487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson CM, Grafström RC. Commentary: mechanistic considerations for associations between formaldehyde exposure and nasopharyngeal carcinoma. Environ Health 2009; 8:53; PMID:19939253; http://dx.doi.org/ 10.1186/1476-069X-8-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dutta S, Tan YJ. Structural and functional characterization of human SGT and its interaction with Vpu of the humanimmunodeficiency virus type 1. Biochemistry 2008; 38:10123-31; PMID:18759457; http://dx.doi.org/ 10.1021/bi800758a [DOI] [PubMed] [Google Scholar]
- 41.Nowak RB, Fischer RS, Zoltoski RK, Kuszak JR, Fowler VM. Tropomodulin1 is required for membrane skeleton organization and hexagonal geometry of fiber cells in the mouse lens. J Cell Biol 2009; 6:915-28; PMID:19752024; http://dx.doi.org/ 10.1083/jcb.200905065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu FX, Johnston PA, Südhof TC, Yin HL. gCap39, a calcium ion- and polyphosphoinositide-regulated actin capping protein. Science 1990; 250(4986):1413-15; PMID:2255912; http://dx.doi.org/ 10.1126/science.2255912 [DOI] [PubMed] [Google Scholar]
- 43.Buxboim A, Swift J, Irianto J, Spinler KR, Dingal PC, Athirasala A, Kao YR, Cho S, Harada T, Shin JW, Discher DE. Matrix elasticity regulates lamin- A,C phosphorylation and turnover with feedback to actomyosin. Curr Biol 2014; 16:1909-17; PMID:25127216; http://dx.doi.org/ 10.1016/j.cub.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fukuda M, Kuroda TS, Mikoshiba K. Slac2-a/Melanophilin, the Missing Link between Rab27 and Myosin Va. Implications of a tripartite protein complex for melanosome transport. J Biol. Chem 2002; 277:12432-6; PMID:11856727; http://dx.doi.org/ 10.1074/jbc.C200005200 [DOI] [PubMed] [Google Scholar]
- 45.Goldstein LS, Yang Z. Microtubule-based transport systems in neurons: the roles of kinesins and dyneins. Annu Rev Neurosci 2000; 23:39-71; PMID:10845058; http://dx.doi.org/ 10.1146/annurev.neuro.23.1.39 [DOI] [PubMed] [Google Scholar]
- 46.Dalpé G, Leclerc N, Vallée A, Messer A, Mathieu M, De Repentigny Y, Kothary R. Dystonin is essential for maintaining neuronal cytoskeleton organization. Mol Cell Neurosci 1998; 5–6:243-57; http://dx.doi.org/ 10.1006/mcne.1997.0660 [DOI] [PubMed] [Google Scholar]
- 47.Cullen DK, Simon CM, LaPlaca MC. Strain rate-dependent induction of reactive astrogliosis and cell death in three-dimensional neuronal-astrocytic co-cultures. Brain Res 2007; 1158:103-15; PMID:17555726; http://dx.doi.org/ 10.1016/j.brainres.2007.04.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serrano L, Díaz-Nido J, Wandosell F, Avila J. Tubulin phosphorylation by casein kinase II is similar to that found in vivo. J Cell Biol 1987; 105:1731-39; PMID:3478337; http://dx.doi.org/ 10.1083/jcb.105.4.1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ulloa L, Díaz-Nido J, Avila J. Depletion of casein kinase II by antisense oligonucleotide prevents neuritogenesis in neuroblastoma cells. EMBO J 1993; 12:1633-40; PMID:8467810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boissan M, Montagnac G, Shen Q, Griparic L, Guitton J, Romao M, Sauvonnet N, Lagache T, Lascu I, Raposo G, et al.. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membraneremodeling. Science 2014; 6191:1510-5; PMID:24970086; http://dx.doi.org/ 10.1126/science.1253768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin H, Kendall E, Freeman TC, Roberts RG, Vetrie DL. The human family of Deafness/Dystonia peptide (DDP) related mitochondrial import proteins. Genomics 1999; 3:259-67; PMID:10552927; http://dx.doi.org/ 10.1006/geno.1999.5966 [DOI] [PubMed] [Google Scholar]
- 52.Dalkilic I, Schienda J, Thompson TG, Kunkel LM. Loss of FilaminC (FLNc) results in severe defects in myogenesis and myotube structure. Mol Cell Biol 2006; 17:6522-34; PMID:16914736; http://dx.doi.org/ 10.1128/MCB.00243-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barco R, Hunt LB, Frump AL, Garcia CB, Benesh A, Caldwell RL, Eid JE. The synovial sarcoma SYT-SSX2 oncogene remodels the cytoskeleton through activation of the ephrin pathway. Mol Biol Cell 2007; 10:4003-12; PMID:17686994; http://dx.doi.org/ 10.1091/mbc.E07-05-0496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fish JL, Kosodo Y, Enard W, Pääbo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci U S A 2006; 27:10438-43; PMID:16798874; http://dx.doi.org/ 10.1073/pnas.0604066103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mayanagi T, Sobue K. Diversification of caldesmon-linked actin cytoskeleton in cell motility. Cell Adh Migr 2011; 2:150-9; PMID:21350330; http://dx.doi.org/ 10.4161/cam.5.2.14398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okreglak V, Drubin DG. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J Cell Biol 2007; 7:1251-64; PMID:17875745; http://dx.doi.org/ 10.1083/jcb.200703092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, Cooke M, Panjwani S, Cao K, Krauth B, Ho PY, Medrzycki M, Berhe DT, Pan C, McDevitt TC, et all.. Histone h1 depletion impairs embryonic stem cell differentiation. PLoS Genet 2012; 5:e1002691; PMID:22589736; http://dx.doi.org/ 10.1371/journal.pgen.1002691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farge E. Increased vesicle endocytosis due to an increase in the plasma membrane phosphatidylserine concentration. Biophys J 1995; 6:2501-6; PMID:8599656; http://dx.doi.org/ 10.1016/S0006-3495(95)80120-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kwon S, Yang W, Choi YK, Park JK. Force spectroscopy of membrane hardness of SH-SY5Y neuroblastoma cells before and after differentiation. JKPS 2014; 10:1595-99; http://dx.doi.org/ 10.3938/jkps.64.1595 [DOI] [Google Scholar]
- 60.Zhang Y, Burke CW, Ryman KD, Klimstra WB. Identification and characterization 995 of interferon-induced proteins that inhibit alphavirus replication. J Virol 2007; 81:11246-55; PMID:17686841; http://dx.doi.org/ 10.1128/JVI.01282-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tseng JC, Granot T, DiGiacomo V, Levin B, Meruelo D. Enhanced specific delivery and targeting of oncolytic Sindbis viral vectors by modulating vascular leakiness in tumor. Cancer Gene Ther 2010; 4:244-55; PMID:17686841; http://dx.doi.org/ 10.1038/cgt.2009.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sauthoff H, Hu J, Maca C, Goldman M, Heitner S, Yee H, Pipiya T, Rom WN, Hay JG. Intratumoral spread of wild-type adenovirus is limited after local injection of human xenograft tumors: viruspersists and spreads systemically at late time points. Hum Gene Ther 2003; 5:425-33; PMID:12691608; http://dx.doi.org/ 10.1089/104303403321467199 [DOI] [PubMed] [Google Scholar]
- 63.Currier MA, Adams LC, Mahller YY, Cripe TP. Widespread intratumoral virus distribution with fractionated injection enables local control of large human rhabdomyosarcoma xenografts by oncolytic herpes simplex viruses. Cancer Gene Ther 2005; 4:407-16; PMID:15665822; http://dx.doi.org/ 10.1038/sj.cgt.7700799 [DOI] [PubMed] [Google Scholar]
- 64.Faust RA, Niehans G, Gapany M, Hoistad D, Knapp D, Cherwitz D, Davis A, Adams GL, Ahmed K. Subcellular immunolocalization of protein kinase CK2 in normal and carcinoma cells. Int J Biochem Cell Biol 1999; 31:941-9; PMID:10533285; http://dx.doi.org/ 10.1016/S1357-2725(99)00050-3 [DOI] [PubMed] [Google Scholar]
- 65.Sankhala KK, Mita MM, Mita AC, Takimoto CH. Heat shock proteins: a potential anticancer target. Curr Drug Targets 2011; 14:2001-8; PMID:21777196; http://dx.doi.org/ 10.2174/138945011798829339 [DOI] [PubMed] [Google Scholar]
- 66.Barroso MM, Lima CS, Silva-Neto MA, Da Poian AT. Mayaro virus infection cycle relies on casein kinase 2 activity. Biochem Biophys Res Commun 2002; 5:1334-9; PMID:12207921; http://dx.doi.org/ 10.1016/S0006-291X(02)02093-4 [DOI] [PubMed] [Google Scholar]
- 67.Alvarez DE, Agaisse H. Casein kinase 2 regulates vaccinia virus actin tail formation. Virology 2012; 2:143-51; PMID:22209233; http://dx.doi.org/ 10.1016/j.virol.2011.12.003 [DOI] [PubMed] [Google Scholar]
- 68.Rathore AP, Haystead T, Das PK, Merits A, Ng ML, Vasudevan SG. Chikungunya virus nsP3 & nsP4 interacts with HSP-90 to promote virus replication: HSP-90 inhibitors reduce CHIKV infection and inflammation in vivo. Antiviral Res 2014; 103:7-16; PMID:24388965; http://dx.doi.org/ 10.1016/j.antiviral.2013.12.010 [DOI] [PubMed] [Google Scholar]
- 69.Javier RT, Rice AP. Emerging theme: cellular PDZ proteins as common targets of pathogenic viruses. J Virol 2011; 22:11544-56; PMID:21775458; http://dx.doi.org/ 10.1128/JVI.05410-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roche JP, Packard MC, Moeckel-Cole S, Budnik V. Regulation of synaptic plasticity and synaptic vesicle dynamics by the PDZ protein Scribble. J Neurosci 2002; 15:6471-9; PMID:12151526[ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pieczynski J, Margolis B. Protein complexes that control renal epithelial polarity. Am J Physiol Renal Physiol 2011; 300:F589-F601; PMID:21228104; http://dx.doi.org/ 10.1152/ajprenal.00615.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Inamura K, Matsuzaki Y, Uematsu N, Honda A, Tanaka N, Uchida K. Rapid inhibition of MAPK signaling and anti- proliferation effect via JAK/STAT signaling by interferon-alpha inhepatocellular carcinoma cell lines. Biochim Biophys Acta 2005; 3:401-10; PMID:16054712; http://dx.doi.org/ 10.1016/j.bbamcr.2005.06.003 [DOI] [PubMed] [Google Scholar]
- 73.Brierley MM, Fish EN. Review: IFN- alpha/beta receptor interactions to biologic outcomes: understanding the circuitry. J Interferon Cytokine Res 2002; 8:835-45; PMID:12396722; http://dx.doi.org/ 10.1089/107999002760274845 [DOI] [PubMed] [Google Scholar]
- 74.Nakatsue T, Katoh I, Nakamura S, Takahashi Y, Ikawa Y, Yoshinaka Y. Acute infection of Sindbis virus induces phosphorylation and intracellular translocation of small heat shockprotein HSP27 and activation of p38 MAP kinase signaling pathway. Biochem Biophys Res Commun 1998; 1:59-64; PMID:9875220; http://dx.doi.org/ 10.1006/bbrc.1998.9724 [DOI] [PubMed] [Google Scholar]
- 75.Simmons JD, Wollish AC, Heise MT. A determinant of Sindbis virus neurovirulence enables efficient disruption of Jak/STAT signaling. J Virol 2010; 21:11429-39; PMID:20739538; http://dx.doi.org/ 10.1128/JVI.00577-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Patel RK, Hardy RW. Role for the phosphatidylinositol 3-kinase-Akt-TOR pathway during sindbis virus replication in arthropods. J Virol 2012; 7:3595-604; PMID:22258238[; http://dx.doi.org/ 10.1128/JVI.06625-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kesic MJ, Simmons SO, Bauer R, Jaspers I. Nrf2 expression modifies influenza A entry and replication in nasal epithelial cells. Free Radic Biol Med 2011; 2:444-53; PMID:21549835; http://dx.doi.org/ 10.1016/j.freeradbiomed.2011.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen WC, Wang SY, Chiu CC, Tseng CK, Lin CK, Wang HC, Lee JC. Lucidone suppresses hepatitis C virus replication by Nrf2-mediated heme oxygenase-1 induction. Antimicrob Agents Chemother 2013; 3:1180-91; PMID:23254429; http://dx.doi.org/ 10.1128/AAC.02053-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang HS, Li HY, Zhou Y, Wu MR, Zhou HS. Nrf2 is involved in inhibiting Tat-induced HIV-1 long terminal repeat transactivation. Free Radic Biol Med 2009; 3:261-8; PMID:19409485; http://dx.doi.org/ 10.1016/j.freeradbiomed.2009.04.028 [DOI] [PubMed] [Google Scholar]
- 80.Tan J, Qiao W, Wang J, Xu F, Li Y, Zhou J, Chen Q, Geng Y. IFP35 is involved in the antiviral function of interferon by association with the viral tas transactivator of bovine foamy virus. J Virol 2008; 9:4275-83; PMID:18305040; http://dx.doi.org/ 10.1128/JVI.02249-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bin L, Howell MD, Kim BE, Hall CF, Streib JE, Leung DY. Inhibition of S100A11 gene expression impairs keratinocyte response against vaccinia virus through downregulation of the IL-10 receptor 2 chain. J Allergy Clin Immunol 2009; 2:270-7; PMID:19577285; http://dx.doi.org/ 10.1016/j.jaci.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Döhner K, Nagel CH, Sodeik B. Viral stop-and-go along microtubules: taking a ride with dynein and kinesins. Trends Microbiol 2005; 7:320-7; PMID:15950476; http://dx.doi.org/ 10.1016/j.tim.2005.05.010 [DOI] [PubMed] [Google Scholar]
- 83.Bremner KH, Scherer J, Yi J, Vershinin M, Gross SP, Vallee RB. Adenovirus transport via direct interaction of cytoplasmic dynein with the viral capsid hexon subunit. Cell Host Microbe 2009; 6:523-35; PMID:20006841; http://dx.doi.org/ 10.1016/j.chom.2009.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kelkar S, De BP, Gao G, Wilson JM, Crystal RG, Leopold PL. A common mechanism for cytoplasmic dynein-dependent microtubule binding shared among adeno-associated virus and adenovirus serotypes. J Virol 2006; 15:7781-5; PMID:16840360; http://dx.doi.org/ 10.1128/JVI.00481-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Suikkanen S1, Aaltonen T, Nevalainen M, Välilehto O, Lindholm L, Vuento M, Vihinen-Ranta M. Exploitation of microtubule cytoskeleton and dynein during parvoviral traffic toward the nucleus. J Virol 2003; 19:10270-9; PMID:12970411; http://dx.doi.org/ 10.1128/jvi.77.19.10270-10279.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Douglas MW, Diefenbach RJ, Homa FL, Miranda-Saksena M, Rixon FJ, Vittone V, Byth K, Cunningham AL. Herpes simplex virus type 1 capsid protein VP26 interacts with dynein light chains RP3 and Tctex1 and plays arole in retrograde cellular transport. J Biol Chem 2004; 27:28522-30; PMID:15117959; http://dx.doi.org/ 10.1074/jbc.M311671200 [DOI] [PubMed] [Google Scholar]
- 87.McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol 2002; 3:441-52; PMID:12417576; http://dx.doi.org/ 10.1083/jcb.200203150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schepis A, Stauber T, Krijnse Locker J. Kinesin-1 plays multiple roles during the vaccinia virus life cycle. Cell Microbiol 2007; 8:1960-73; PMID:17394562; http://dx.doi.org/ 10.1111/j.1462-5822.2007.00927.x [DOI] [PubMed] [Google Scholar]
- 89.Diefenbach RJ, Miranda-Saksena M, Diefenbach E, Holland DJ, Boadle RA, Armati PJ, Cunningham AL. Herpes simplex virus tegument protein US11 interacts with conventional kinesin heavy chain. J Virol 2002; 7:3282-91; PMID:11884553; http://dx.doi.org/ 10.1128/JVI.76.7.3282-3291.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dodding MP, Way M. Coupling viruses to dynein and kinesin-1. EMBO J 2011; 17:3527-39; PMID:21878994; http://dx.doi.org/ 10.1038/emboj.2011.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roberts KL, Baines JD. Myosin Va enhances secretion of herpes simplex virus 1 virions and cell surface expression of viral glycoproteins. J Virol 2010; 19:9889-96; PMID:20631136; http://dx.doi.org/ 10.1128/JVI.00732-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pasdeloup D, McElwee M, Beilstein F, Labetoulle M, Rixon FJ. Herpesvirus tegument protein pUL37 interacts with dystonin/BPAG1 to promote capsid transport on microtubulesduring egress. J Virol 2013; 5:2857-67; PMID:23269794; http://dx.doi.org/ 10.1128/JVI.02676-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liljestrom P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology (N Y) 1991; 9:1356-61; PMID:1370252; http://dx.doi.org/ 10.1038/nbt1291-1356 [DOI] [PubMed] [Google Scholar]
- 94.Wiśniewski JR, Mann M. Consecutive proteolytic digestion in an enzyme reactor increases depth of proteomic and phosphoproteomic analysis. Anal Chem 2012; 6:2631-7; PMID:22324799; http://dx.doi.org/ 10.1021/ac300006b [DOI] [PubMed] [Google Scholar]
- 95.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008; 12:1367-72; PMID:19029910; http://dx.doi.org/ 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 96.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 2013; 8:1551-66; PMID:23868073; http://dx.doi.org/ 10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yokota S, Yokosawa N, Kubota T, Okabayashi T, Arata S, Fujii N. Suppression of thermotolerance in mumps virus-infected cells is caused by lack of HSP27 induction contributed by STAT-1. J Biol Chem 2003; 43:41654-60; PMID:12917439; http://dx.doi.org/ 10.1074/jbc.M305701200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.