Abstract
Runx2 is a transcription factor controlling skeletal development, and is also expressed in extraskeletal tissues where its function is not well understood. Existing Runx2 mutant and transgenic mouse models do not allow the necessary control of Runx2 expression to understand its functions in different tissues. We generated conditional, doxycyline-inducible, triple transgenic mice (CMV-Cre;ROSA26-neoflox/+-rtTA;Tet-O-Runx2) to investigate the effects of wide spread overexpression of Runx2. Osteoblasts isolated from CMV-Cre;ROSA26-neoflox/+-rtTA; Tet-O-Runx2 mice demonstrated a dose-dependent effect of doxycycline to stimulate Runx2 transgene expression. Doxycycline administration to CMV-Cre;ROSA26-neoflox/+-rtTA;Tet-O-Runx2 mice induced Runx2 transgene expression in all tissues tested, with the highest levels observed in kidney, ovary, and bone. Runx2 overexpression resulted in deceased body size and reduced viability. With regard to bone, Runx2 overexpressing mice paradoxically displayed profound osteopenia and diminished osteogenesis. Induced expression of Runx2 in extraskeletal tissues resulted in ectopic calcification and induction of the osteogenic program in a limited number of tissues, including lung and muscle. In addition, the triple transgenic mice showed evidence of a myeloproliferative disorder and an apparent inhibition of lymphocyte development. Thus, overexpression of Runx2 both within and outside of the skeleton can have diverse biological effects. Use of tissue specific Cre mice will allow this model to be used to conditionally and inducibly overexpress Runx2 in different tissues and provide a means to study the post-natal tissue- and cell context-dependent functions of Runx2.
Keywords: RUNX2, INDUCIBLE EXPRESSION, OSTEOPENIA, ECTOPIC CALCIFICATION, MYELOPROLIFERATIVE DISORDER
Runx2 (also called Cbfa1, Osf2, PEBP2αA, and AML3) is a transcription factor that regulates the lineage determination and differentiation of mesenchymal precursors into osteoblasts and the terminal differentiation of chondrocytes essential for normal skeletogenesis [Ducy et al., 1997, 1999; Komori et al., 1997; Otto et al., 1997; Enomoto et al., 2000; Yoshida and Komori, 2005]. The original targeted disruption of Runx2 in mice reported by Komori et al. [1997] and Otto et al. [1997] and the overexpression of dominant negative Runx2 or C-terminal truncated constructs established the importance of Runx2 in skeletogenesis [Choi et al., 2001]. Runx2 also plays a role in postnatal bone metabolism [Liu et al., 2001; Geoffroy et al., 2002; Maruyama et al., 2007], but overexpression of Runx2 postnatally paradoxically reduces bone mass. In addition, we defined the differential roles of Runx2-I and Runx2-II isoforms in bone development [Xiao et al., 2004] by creating selective Runx2-II deficient mice and compound heterozygous Runx2-II and global Runx2 deficient mice. We found that the P1 promoter regulation of Runx2-II expression is critical for endochondral bone formation and full osteogenic capacity of cells within the osteoblasts lineage, whereas P2 promoter-dependent Runx2-I expression is sufficient for early osteoblastogenesis and intramembranous bone formation [Xiao et al., 2004; Zhang et al., 2009]. These differences occurred, in spite of similar transactivation potential of Runx2-I and Runx2-II. Runx2-I and II are identical with respect to their runt domains and C-termini and have indistinguishable transactivation potential, in spite of their different biological functions [Ahn et al., 1996; Ueta et al., 2001; Wang et al., 2005].
Runx2 is also expressed in extraskeletal tissues where its function is less well understood [Ogawa et al., 1993; Satake et al., 1995; Meyers et al., 1996; Banerjee et al., 2001]. For example, Runx2 was identified from a retroviral insertion site (Til-1 locus) in the Runx2 gene as a cooperative factor in the development of T cell lymphomas [Stewart et al., 1997; Blyth et al., 2001]. Depending on the cell context, Runx2 may function as a tumor suppressor [Blyth et al., 2005] or an oncogene. Runx2 may contribute to the tumorigenesis and metastatic potential of breast [Javed et al., 2005] prostate cancer cells [van der Deen et al., 2010]. Runx2 is also expressed hematopoietic precursors [Kuo et al., 2009] and hematological malignances, including multiple myeloma [Colla et al., 2005] and myeloid leukemia [Kurokawa and Hirai, 2003; Kuo et al., 2009]. In addition, the ectopic expression of Runx2 in vascular smooth muscle induced by environmental factors such as hyperphosphatemia and oxidative stress has been implicated in ectopic bone formation and vascular calcifications [Byon et al., 2008; Speer et al., 2010]. It is possible that the mechanisms underlying the distinct biological responses of Runx2 observed in different tissues depend more on the amount of Runx2 expression and the importance of factors related to the cell-context and temporal factors (i.e., time and place), rather than intrinsic transcriptional activation potential of the different members of this family.
To fully understand the cell context-dependent functions of Runx2 in different tissues, conditional and inducible deletion and/or overexpression of this transcription factor will be required. Recent studies have used a retroviral system to deliver Runx2 under the control of the tetracycline-inducible (tet-off) promoter in vitro [Gersbach et al., 2006], but to date, no inducible and conditional system for overexpression of Runx2 have been evaluated in vivo. A conditional and inducible transgene expression system has been developed in vivo using Cre-mediated recombination to express rtTA transgene in ROSA26-neoflox/+-rtTA mice [Belteki et al., 2005]. To address the question of time and place of Runx2 expression on functions of this transcription factor, we have developed a Tet-O-driven Runx2 transgenic mouse model and used the CMV-Cre and ROSA26-neoflox/+-rtTA mice to conditionally and inducibly overexpress Runx2, thereby allowing the assessment of the context-dependent functions of Runx2 in multiple tissues.
MATERIALS AND METHODS
CREATING EXPRESSION OF DOXYCYCLINE-INDUCIBLE RUNX2 (TET-O-RUNX2) TRANSGENIC MICE
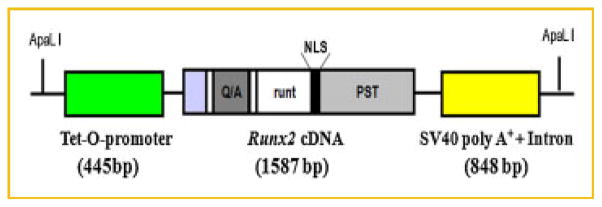
We used pW1 vector containing a multiple cloning site and an SV40 intron plus polyadenylation site and the following strategy to create inducible Tet-O-Runx2 transgene construct by doxycycline (Dox; Fig. 1). Briefly, a 445 bp of Tet-O-promoter, consisting of seven direct repeats of tetracycline (Tet) Operator sequences [(Tet-O)7] and a minimal CMV promoter, was excised from pSK-tet-Cre plasmid (a gift from Dr. Andras Nagy, University of Toronto) [Belteki et al., 2005] with XhoI and EcoRI double digest and inserted into pW1 vector digested by SalI and EcoRI, thereby generating pW1-Tet-O plasmid. The pW1_Tet-O-Runx2-II construct was generated by PCR amplification of the mouse 1,587 bp Runx2-II cDNA in pcDNA3.1 plasmids [Xiao et al., 1999] using a XbaI-containing Runx2-II forward primer 5′-GCT CTA GAG CAT GGC GTC AAA CAG CCT CTT CAG CGC AG-3′ in combination with a XbaI-containing Runx2 reverse primer 5′-GCT CTA GAG CTC AAT ATG GCC GCC AAA CAG ACT CAT CC-3′ followed by subcloning of the product into the XbaI site of pW1-Tet-O plasmid. The correct orientation of the Runx2-II in pW1-Tet-O-Runx2-II plasmid was confirmed by direct sequencing. Then pW1_Tet-O-Runx2-II was purified using the EndoFree Kit (Qiagen, Valencia, CA) to remove endotoxins. A 4.5 kb of Tet-O-Runx2-II fragment was released from the vector pW1 using ApaLI degestion, separated by agarose gel, and gel-purified twice using GENECLEAN® Turbo (MP Biomedicals, LLC, Solon, OH) according to the manufacturer’s instructions and quantified using the PicoGreen Assay Kit (Molecular Probes, Eugene, OR). Then transgenic mouse was made by the KUMC Transgenic and Gene-targeting Institutional Facility by microinjection of C57Bl/6 × SJL hybrid fertilized mouse eggs with DNA at a concentration of 2–3 ng/μl according to standard techniques. The Tet-O-Runx2-II transgenic mouse was genotyped by PCR using tail DNA and the following primers: Runx2 forward 5′-TCT TCC CAA AGC CAG AGT GG-3′ and SV40pA reverse 5′-ATC AGT TCC ATA GGT TGG AAT C-3′. We have successfully generated 3 founder lines of Tet-O-Runx2-II transgenic mice. All animal research was conducted according to guidelines provided by the National Institute of Health and the Institute of Laboratory Animal Resources, National Research Council. The University of Tennessee Health Science Center’s Animal Care and Use Committee approved all animal studies (Protocol number: 1885R2).
Fig. 1.
Schematic diagram of inducible Tet-O-Runx2 transgene construct by doxycycline. A 445 bp of Tet-O-promoter, consisting of seven direct repeats of Tet Operator sequences [(Tet-O)7] and a minimal CMV promoter, followed by a 1,587 bp mouse Runx2-II cDNA and a 848 bp SV40 intron plus poly A+ site was subcloned pW1 plasmid and linearized by ApaLI restriction enzyme.
GENERATION OF CMV-CRE-MEDIATED AND DOXYCYCLINE-INDUCED OVEREXPRESSION OF RUNX2 MOUSE MODELS IN A TRIPLE TRANSGENIC SYSTEM
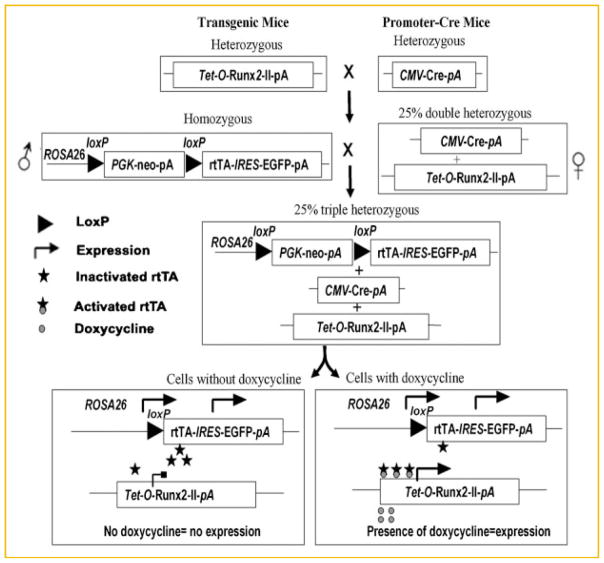
We have successfully developed an inducible transgenic mouse models to overexpress Runx2-II and Runx2-I isoform. We used a triple transgenic system: (1) a ROSA26-neoflox/flox-rtTA mouse, which carries floxed PGK-neo cassette between the ROSA26 locus and the rtTA-IRES-EGRP transgene, (2) a CMV promoter driven-Cre mouse obtained from The Jackson Laboratory, which expresses Cre in all the cells of the early embryo, and (3) a transgenic mouse expressing Runx2-II or Runx2-I under the control of the Tet-O promoter. We used a two-step breeding strategy (Fig. 2) to create double transgenic CMV-Cre;Tet-O-Runx2-II or CMV-Cre;Tet-O-Runx2-I mice, and then crossing CMV-Cre;Tet-O-Runx2-II or CMV-Cre;Tet-O-Runx2-I with homozygous ROSA26-neoflox/flox-rtTA mouse to generate the following four genotypes: ROSA26-neoflox/+-rtTA, CMV-Cre; ROSA26-neoflox/+-rtTA, Tet-O-Runx2-II or CMV-Cre;Tet-O-Runx2-I, and CMV-Cre; ROSA26-neoflox/+-rtTA; Tet-O-Runx2-II or CMV-Cre;ROSA26-neoflox/+-rtTA;Tet-O-Runx2-I (designated TgCMV-rtTA-Tet-O-Runx2-II or TgCMV-rtTA-Tet-O-Runx2-I or triple transgenic mouse; Fig. 2). For these initial studies we used the CMV-Cre-recombinase to remove the floxed PGK-neo cassette, thereby allowing the broad tissue expression of rtTA and GFP from the rtTA-IRES-EGRP cassette. The administration of doxycyline will result in over-expression of Runx2-II or Runx2-I in all the cells where the CMV promoter is active, thereby allowing us to explore in the same animal the cell context dependent effects of Runx2-II or Runx2-I.
Fig. 2.
Breeding strategy for CMV-Cre-mediated and doxycycline-induced overexpression Runx2 mouse model in a triple transgenic system. A triple Cre-loxP system was used to generate global inducible Tet-O-Runx2-II transgenic mice by conditional deletion of the floxed PGK-neo cassette in all tissues using CMV-Cre through two-round crossing Tet-O-Runx2-II, CMV-Cre, and ROSA26-neoflox/flox-rtTA mice. For the first round, Tet-O-Runx2-II mice were crossed with CMV-Cre to generate CMV-Cre;Tet-O-Runx2-II mice. For the second round, CMV-Cre;Tet-O-Runx2-II mice were mated with ROSA26-neoflox/flox-rtTA mice to generate CMV-Cre; ROSA26-neoflox/+-rtTA;Tet-O-Runx2-II mice (triple transgenic mice).
DOXYCYCLINE-INDUCED RUNX2-II OR RUNX2-I EXPRESSION IN VITRO
Primary osteoblasts were isolated from newborn control and TgCMV-rtTA-Tet-O-Runx2-II or TgCMV-rtTA-Tet-O-Runx2-I triple transgenic mice. The cells were incubated with various dose of doxycycline (10,100, 500, 1,000, and 2,000 μg/ml) in the culture media for 48 h, then total RNAs were extracted from the cells and a real-time RT-PCR was performed to examine the expression of the Runx2-II or Runx2-I transgene and its downstream genes.
DOXYCYCLINE-INDUCED RUNX2-II OR RUNX2-I EXPRESSION IN VIVO
We examined the tissue expression of the Runx2-II or Runx2-I transgene after administration of doxycyline for 1 week to TgCMV-rtTA-Tet-O-Runx2-II or TgCMV-rtTA-Tet-O-Runx2-I 3-week-old mice. For these studies, we administered doxycycline (Dox) to achieve activation of the Tet-O-Runx2-II or Tet-O-Runx2-I (Tet-ON) in the triple transgenic mice. Dox (2 mg/ml in 5% sucrose solution) was added to the drinking water to 3-week-old mice. At the end of a week, various tissues were harvested and the levels of Runx2-II or Runx2-I transgene expression were quantified using real-time RT-PCR.
REAL-TIME RT-PCR
For quantitative real-time RT-PCR, 2.0 μg of total RNA isolated from either the long bone of 4-week-old mice or 3 days cultured primary osteoblasts in inducible conditions was reverse transcribed as previously described [Xiao et al., 2004]. PCR reactions contained 100 μg template (cDNA or RNA), 300 μM each forward and reverse primers, and 1 × iQ™ SYBR® Green Supermix (Bio-Rad, Hercules, CA) in 50 μl. The threshold cycle (Ct) of tested-gene product from the indicated genotype was normalized to the Ct for cyclophilin A.
ASSESSMENT OF SOFT TISSUE CALCIFICATION
The whole organ specimens were stained for overnight in a 1% aqueous Alizarin Red S solution (Sigma Cat. No. A5533), followed by exhaustively rinsed with tap water for 10 min. Then the dye was extracted with 10% cetylpyridinium chloride and quantified at 562 nm as previously described [Stubbs et al., 2007].
SERUM BIOCHEMISTRY
Serum osteocalcin levels were measured using a mouse osteocalcin EIA kit (Biomedical Technologies Inc., Stoughton, MA). Serum urea nitrogen (BUN) was determined using a BUN diagnostic kit from Pointe Scientific, Inc. Serum calcium (Ca) was measured by the colorimetric cresolphthalein binding method, and phosphorus (P) was measured by the phosphomolybdate–ascorbic acid method (Stanbio Laboratory, TX). Serum TRAP was assayed with the ELISA-based SBA Sciences mouseTRAP™ assay (Immunodiagnostic Systems, Fountain Hills, AZ). Serum PTH levels were quantitated with the use of an enzyme-linked immunosorbant assay from Immunotopics according to the directions of the manufacturer.
BONE DENSITOMETRY, HISTOMORPHOMETRIC, AND MICRO-CT ANALYSIS
Bone mineral density (BMD) of femurs was assessed before and after doxycycline administration for 2 weeks at 3 and 5 weeks of age using a LUNARPIXIMUS bone densitometer (Lunar Corp, Madison, WI). Alizarin Red (Sigma, St. Louis, MO) double labeling of bone and histomorphometric analyses of periosteal mineral apposition rate (MAR) in tibias were performed using the osteomeasure analysis system (Osteometrics). Goldner and Von Kossa staining were performed according to standard protocols [Glass et al., 2005; Xiao et al., 2005]. The distal femoral metaphyses were also scanned using a Scanco μCT 40 (Scanco Medical AG, Brüttisellen, Switzerland). A 3D images analysis was done to determine bone volume (BV/TV) and cortical thickness (Ct.Th) as previously described [Xiao et al., 2005, 2006].
HISTOLOGY AND CYTOLOGY ANALYSES
Tissue samples (spleen, liver, and thymus) collected from control and triple transgenic mice were fixed in 10% buffered formalin, embedded in paraffin, and sectioned specimens were stained with hematoxylin and eosin (H&E). The bone marrow (BM) cells were isolated from femurs and tibias via cytocentrifuged preparations. BM smear were prepared and stained with Wright-Giemsa (Fisher Scientific, Pittsburgh, PA) for cytologic and morphologic changes according to the manufacturer’s instructions. Images were taken under a microscope with magnifications of 20× using a Nikon E800 and an optronics CCD camera driven by the AnalySIS software (Olympus Soft Imaging Solutions Corp, Lakewood, CO).
IMMUNOFLUORESCENCE
Freshly dissected tissues were covered with cryoembedding media, and then rapidly frozen in liquid nitrogen. The 10 μm thick frozen sections were fixed in 4% PFA for 10 min at room temperature and rinsed 3 times in 1 × PBS containing 0.3% Triton X-100 (PBST; Sigma; pH 7.4). The sections were blocked with normal serum (from same host species as secondary antibody) plus BSA (Sigma) at room temperature for 1 h. The sections were then incubated overnight at room temperature with antiserum against Runx2 (1:200; rabbit polyclonal IgG; SC-10758, Santa Cruz). Primary antibody binding was visualized with Cy3 goat anti-rabbit IgG (1:200; Jackson, West Grove, PA) incubation for 1 h at room temperature. All incubations were carried out in a humidified chamber and slides had coverslips placed on them with Fluoromount G (Southern Biotechnology Associates, Inc., Birmingham, AL). Images were captured with Nikon Eclipse 80i microscope system.
FLOW CYTOMETRY AND PHENOTYPE ANALYSIS
Mice were killed at the indicated time points to collect peripheral blood, spleen, thymus, and bone marrow (BM). Blood was counted by Hemav (Drew Scientific Ltd., Waterbury, CT) and the other sample cells were counted by Cell Lab Quanta SC(Beckman Coulter Inc., Fullerton, CA). After lyses of red blood cells, total nucleated cells from peripheral blood, spleen, thymus, and BM were stained with cell surface markers for phenotype analysis by using flow cytometry as described previously [Zhang et al., 2006].
STATISTICAL ANALYSIS
We evaluated differences between groups by one-way analysis of variance. All values are expressed as means ± SD. All computations were performed using the GraphPad Prism5 (GraphPad Software Inc., La Jolla, CA).
RESULTS
INDUCIBLE EXPRESSION OF RUNX2 IN CMV-CRE;ROSA26-NEOFLOX/+-RTTA;TET-O-RUNX2-II MICE IS LETHAL
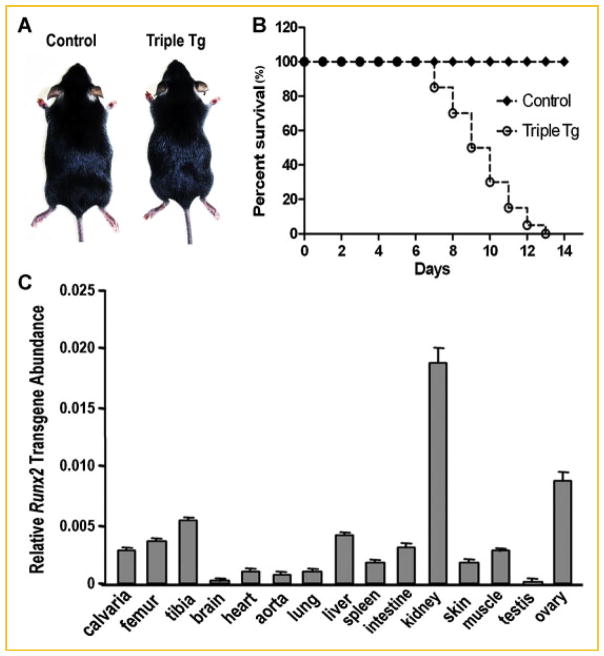
CMV-Cre;ROSA26-neoflox/+-rtTA;Tet-O-Runx2-II mice were born with the expected Mendelian frequency, developed normally, and where identical to wild-type littermates in the absence of rtTA-inducer doxycycline (DOX; Fig. 3A). Dox treatment also had no demonstrable effects on WT mice. In contrast, CMV-Cre;ROSA26-neoflox/+-rtTA;Tet-O-Runx2-II treated with DOX from the time of weaning (3-weeks-of-age) displayed growth retardation, developed a sickly appearance characterized by patchy hair loss and the inability to open their eyelids (Fig. 3A). While vehicle treated and non-treated CMV-Cre; ROSA26-neoflox/+-rtTA;Tet-O-Runx2-II mice had normal survival identical to non-transgenic wild-type mice, CMV-Cre;ROSA26-neoflox/+-rtTA;Tet-O-Runx2-II mice began to die 1 week after Dox treatment and no mice survived beyond 2 weeks of treatment (Fig. 3B). In contrast, Dox administration to control mice had no effect on survival. We lowered the dose of Dox to 0.25 and 1 mg/ml, but this did not improve the survival of CMV-Cre;ROSA26-neoflox/+-rtTA;Tet-O-Runx2-II mice. We also found that CMV-mediated expression of rtTA resulted in wide expression of the Runx2 transgene in response to Dox treatment. Indeed, the Runx2-II transgene was induced in all tissues tested, with the highest levels observed in kidney and ovary (Fig. 3C).
Fig. 3.
Gross appearance and Runx2 expression in Dox treated CMV-Cre; ROSA26-neoflox/+-rtTA; Tet-O-Runx2-II (Triple Tg) and control mice. Dox (2 mg/ml in 5% sucrose solution) was added to the drinking water to 3-week-old mice for 2 weeks. A: Gross appearance of 5-week-old mice after treated with Dox for 2 weeks. B: Kaplan–Meier survival curve of CMV-Cre;ROSA26-neoflox/+-rtTA;Tet-O-Runx2-II (Triple Tg) and control mice. Dox treatment was started at day 0 in 3-week-old mice. C: Runx2-II transgene expression in various tissues after Dox treatment of CMV-Cre; ROSA26-neoflox/+-rtTA; Tet-O-Runx2-II mice by quantitative RT-PCR. Data represent the mean ± SD from four individual samples.
To gain insights into the tissue and cellular context dependent functions of Runx2 and to understand the potential mechanisms underlying the apparent toxic effects of widespread Runx2 overexpression, we examined the multiple tissues for abnormalities.
MUSCULOSKELETAL PHENOTYPE IN DOX-TREATED CMV-CRE; ROSA26-NEOFLOX/+-rTTA;TET-O-RUNX2-II MICE
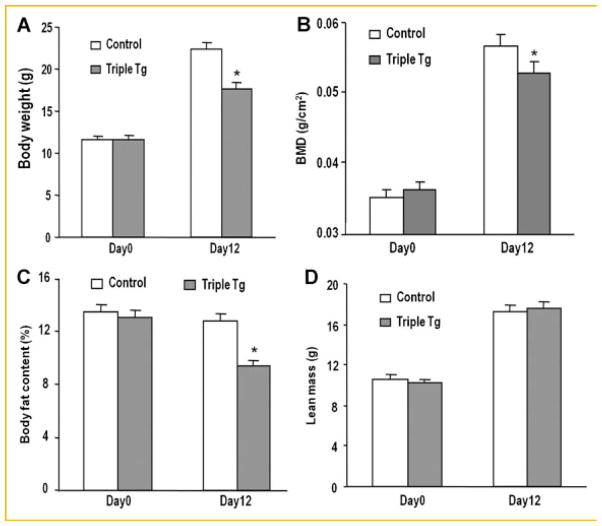
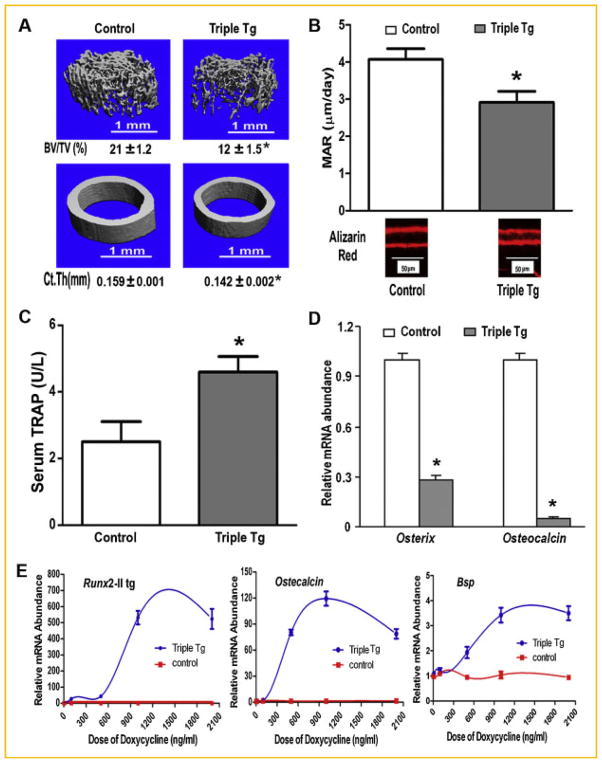
Although the triple transgenic mice overexpressing Runx2-II survived for only 2 weeks, the inducible overexpression of Runx2-II had profound effects on the musculoskeletal system. Dox-treated CMV-Cre; ROSA26-neoflox/+-rtTA;Tet-O-Runx2-II mice exhibited lower body weights (Fig. 4A), reductions in BMD (Fig. 4B), and body fat (Fig. 4C), but no change in lean body mass (Fig. 4D) by DEXA analysis. The lower bone density in Dox-treated CMV-Cre; ROSA26-neoflox/+-rtTA; Tet-O-Runx2-II mice was due to loss of trabecular bone volume and cortical thickness measured by μCT analysis of femoral metaphyseal bone (Fig. 5A) due to suppression of osteoblast-mediated bone formation and increased bone resorption. In this regard, we respectively found a significant decrease in periosteal and endosteal mineral apposition rate (MAR; Fig. 5B) and an increase in circulating TRAP levels (Fig. 5C) in Dox-treated Runx2-II triple transgenic mice. Consistent with suppressed osteoblastic function, Osx and Oc were markedly suppressed in the femur (Fig. 5D). To quantify DOX-induced Runx2 expression, we examined Runx2-II transgene expression in primary osteoblast cultures from CMV-rtTA-Tet-O-Runx2-II mice in which doxycycline was added to the culture medium in vitro (Fig. 5E). Doxycycline induced Runx2-II transgene expressions in a dose-dependent manner in triple transgenic osteoblasts, but not in the control cells, achieving a maximal induction at doxycycline concentrations of ~1,200 ng/ml. Similarly, Runx2-dependent osteogenic genes, including Osteocalcin and Bone sialoprotein (Bsp) were increased in a dose-dependent fashion by Dox (Fig. 5E), indicating that the Runx2-II transgene was highly expressed and functional in osteoblasts and that the effect was different between ex vivo and in vivo induction of Runx2 expression.
Fig. 4.
Musculoskeletal phenotype in Dox treated CMV-Cre; ROSA26-neoflox/+-rtTA; Tet-O-Runx2-II (Triple Tg) and control mice. A: Body weight. B–D: DEXA analysis of bone density (BMD), fat mass and lean body mass. Data represent the mean ± SD from a minimum of four samples. Asterisk (*) indicates significant difference from control mice at P <0.05.
Fig. 5.
Osteoblast-mediated bone formation and bone resorption in Dox-induced Runx2-II transgenic mice. A: μCT analysis of femoral trabecular and cortical bone; B: Bone mineral apposition rate (MAR) by bone histomorphometric analyses. C: The levels of serum TRAP. D: Osterix and Osteocalcin messages of tibias by real time RT-PCR. E: Inducible regulation of Runx2-II and its downstream gene expression [Osteocalcin, Bone sialoprotein (Bsp)] mRNA abundance by quantitative RT-PCR in cultured osteoblasts. Data represent the mean ± SD from a minimum of four samples. Asterisk (*) indicates significant difference from control mice at P <0.05.
RUNX2 MEDIATED “OSTEOGENIC PROGRAM” AND ECTOPIC CALCIFICATION IN EXTRASKELETAL SITES ARE CELL CONTEXT DEPENDENT
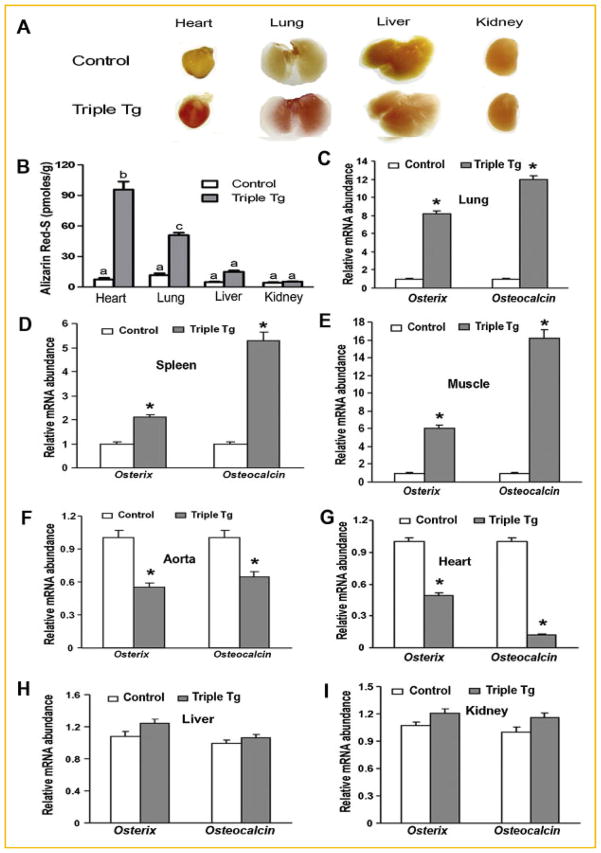
We examined the effects of Runx2 overexpression in different tissues to induce an “osteogenic program” as measured by the expression of Osterix (Osx) and Osteocalcin (Oc), early and late marker of the osteogenic lineage, respectively and by the presence of ectopic calcifications, as measured by Alizarin Red S staining. Of the multiple tissues overexpressing Runx2, extensive calcification of the heart and lung as evidenced by both the greater intensity of staining and amount of Alizarin Red S eluted from these tissues in Runx2-II triple transgenic mice compared to control mice (Fig. 6A,B). In contrast, the liver and kidney had spotty Alizarin-red staining and lower amounts of Alizarin red elution. Other tissues, such as the skin, spleen, and muscle, displayed no evidence of calcifications (data not shown).
Fig. 6.
Variable effects of overexpressing Runx2-II to stimulate an osteogenic program in different tissues in Dox treated CMV-Cre;ROSA26-neoflox/+-rtTA;Tet-O-Runx2-II mice. A: Multiple organs calcification with Alizarin red staining. B: Quantification of Alizarin red staining. C–I: Expression of Runx2 downstream targeting genes such as Osterix and Osteocalcin messages by real time RT-PCR in lung, spleen, muscle, aorta, heart, liver, and kidney samples. Data represent the mean ± SD (n = 4). Asterisk (*) indicates significant difference from control mice at P <0.05.
There was also tissue heterogeneity in the ability of Runx2 overexpression to induce an osteogenic program. In this regard, overexpression of Runx2 induced expression of Osx and Oc in the lung, spleen, and muscle (Fig. 6C–E), but not the aorta, heart, liver or kidney (Fig. 6F–I). We also found discordance between Runx2-mediated up-regulation of Osx and Oc and organ calcification.
The degree of calcification did not correlate with the levels of ectopic Runx2 expression or the induction of the osteogenic program. For example, heart calcifications were associated with reductions in Osx and Oc expression, lung calcifications were associated with the upregulation of Osx and Oc and kidney calcifications were associated with no changes in expression of these osteoblastic markers (Fig. 6C–I). In non-calcifying tissues in Runx2 overexpressing mice, the spleen and muscle exhibited up-regulation of Osx and Oc, whereas these osteogenic markers were decreased in the heart and aorta of Runx2 overexpressing mice (Fig. 6C–I). Differences in serum phosphate did not explain the soft tissue calcifications, since serum phosphate in the triple transgenic mice was not different from controls (8.8 ± 3.7 mg/dl vs. 7.9 ± 2.3 mg/dl, respectively).
EFFECTS OF DOXYCYCLINE-INDUCIBLE RUNX2-II EXPRESSION ON SPLEEN, THYMUS, LIVER AND BONE MARROW
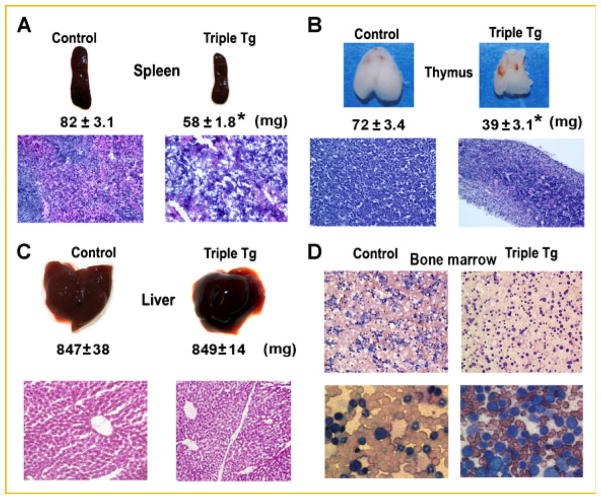
The spleen and thymus were significantly smaller in Dox-treated CMV-Cre; ROSA26-neoflox/+-rtTA;Tet-O-Runx2-II compared to controls (Fig. 7A,B). The control spleen consists of the lymphoid white pulp (composed of the periarteriolar lymphoid sheath (T-cell area), the adjacent follicles (B-cell area), and marginal zone (B-cell area), and the hematogenous red pulp (which is predominantly composed of red blood cells, but also contains lymphocytes and macrophages; Fig. 7A, left panel). Both the white and red pulp are disorganized and depleted of cells in Runx2-II overexpressing mice (Fig. 7A, right panel). With regard to the thymus, control mice show densely packed T-lymphocytes (Fig. 7B, left panel), whereas the Runx2 overexpressing mice show significantly smaller thymus glands that on histological examination demonstrate lighter areas consisting of what appear to be less densely arranged T-lymphocytes and the presence of a greater number of reticular cells that have fused into Hassall’s corpuscle, that typically appear in involved thymus glands (Fig. 7B, right panel). We did not observe any significant changes of B220+ B cells in peripheral blood, BM and spleen in Runx2 transgenic mice. The absolute number of thymus mature and immature T cells decreased in Runx2 transgenic mice by fluorescence activated cell sorting (FACS) analysis (data not shown). In contrast, there were no abnormalities of liver size or histology (Fig. 7C), in spite of higher levels of the Runx2 transgene expression in liver compared to spleen.
Fig. 7.
Hematological abnormalities in Dox treated CMV-Cre; ROSA26-neoflox/+-rtTA; Tet-O-Runx2-II (Triple Tg) and control mice. Gross appearance, weight and H&E stained histological section of the spleen (A), thymus (B), and liver (C). D: Bone marrow smear (Wright-Giemsa stain) showing altered myeloid-erythroid ratio in Runx2 overexpressing mice (low power, upper panel) and uniform appearing immature “blast” like cells in Runx2-II expressing mice (high power, lower panel). Data represent the mean ± SD (n = 4). Asterisk (*) indicates significant difference from control mice at P <0.05.
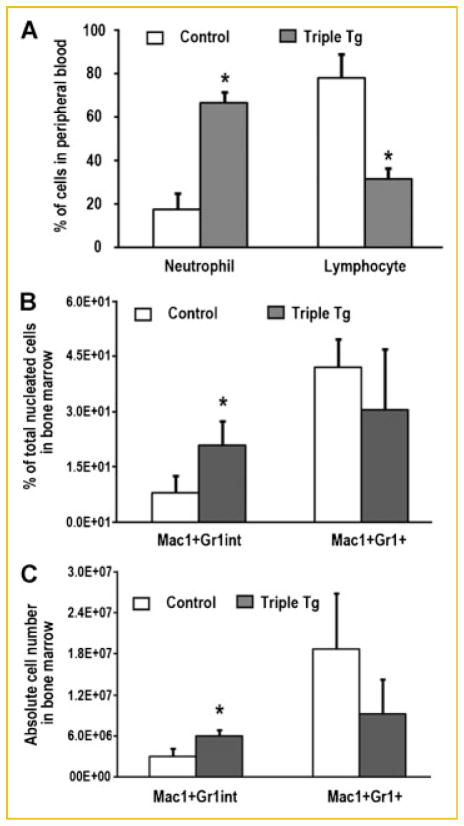
Analysis of doxycycline-inducible Runx2-II in the peripheral blood and bone marrow suggests the potential presence of a myeloid proliferative disorder (MPD). The Wright-Giemsa stained bone marrow smear from Runx2-II overexpressing mice reveals an altered myeloid-erythroid ratio and a preponderance of immature appearing cells of uniform appearance (Fig. 7D). We assessed cell types in peripheral blood and bone marrow (BM) by fluorescence activated cell sorting (FACS) analysis in these mice. We have found that the percentage of neutrophil was 3.8 folds increase in peripheral blood while the percentage of lymphocyte was 2.5 folds decrease in Runx2 transgenic mice (Fig. 8A). Consistent with the peripheral blood count, the percentage and absolute number of myeloid and monocyte precursors (Mac1+Gr1int) were 2.6 and 2.0 folds increase in bone marrow (Fig. 8B,C) from Runx2-II transgenic mice, but there was no significant change in Mac1+Gr1+ myeloid cells in the BM from Runx2-II transgenic mice. These data suggested Runx2-II transgenic mice might develop MPD.
Fig. 8.
Increase in neutrophil and myeloid and monocyte precursors (Mac1+−Gr1int) in Dox-induced Runx2-II triple transgenic mice. A: Peripheral blood count (n = 3) showed that the percentage of neutrophil and lymphocyte was upside down in Runx2 transgenic (Triple Tg) mice compared with control mice treated with Dox for 1 week. B,C: Flow cytometry analyses of bone marrow (BM) cells from control and Runx2-II transgenic (Tg) mice treated with Dox for 1 week. The data showed that the percentage and absolute number of bone marrow myeloid and monocyte precursors (Mac1+Gr1int) in Runx2 transgenic mice significantly increased when compared with control BM. Data are expressed as the mean ± SD (n = 3). Asterisk (*) indicates significant difference from control mice at P <0.05.
DISCUSSION
Runx2 is a widely expressed transcription factor but its physiological function is only well understood in bone development, where Runx2 is an essential nodal point for the commitment of undifferentiated mesenchymal cells to the osteoblast lineage and for the terminal differentiation of chondrocytes [Komori et al., 1997; Otto et al., 1997; Choi et al., 2001]. Since Runx2 null mice are embryonic lethal, the extraskeletal effects of Runx2 remain largely undefined, principally due to the lack tools to conditionally and inducibly manipulate the expression of Runx2. In the current study we tested the hypothesis that Runx2 biological effects are cell-context dependent using a newly developed conditional and inducible in vivo mouse model for overexpression of Runx2 under control of a heterologous promoter. Using CMV-Cre to release the inhibition of rtTA expression and doxycyline administration to stimulate Runx2 cDNA expression from a Tet-On-promoter in combined CMV-Cre; ROSA26-neoflox/+-rtTA;Tet-O-Runx2-II mice, we were successful in increasing the expression of Runx2 in multiple tissues, postnatally. We found that the wide spread extraskeletal expression of Runx2 in adult mice was lethal, with most of the mice dying within 2 weeks of inducing Runx2 expression. The precise mechanism of death could not be identified, in part because abnormalities were identified in multiple organs. Indeed, the lethal phenotype in resulting from CMV-Cre mediated and doxycycline induced overexpression of Runx2 was associated with paradoxical osteopenia, a myleodysplastic syndrome, and extensive soft-tissue calcifications.
We found that the ability of Runx2 to induce an osteogenic program, which is the principal known function of this transcription factor [Choi et al., 2001], was highly influenced by the tissue and cell context. Interestingly, CMV-directed increased expression of Runx2 in mature bone resulted in a reduction in bone mass. The reduction in bone mass was caused by both a reduction in osteoblastic markers consistent with decreased bone formation as well as a nearly twofold increase in serum TRAP activity, consistent with increased bone resorption in Runx2 transgenic mice. There are several studies consistent with our findings that Runx2 overexpression in mature osteoblasts inhibits bone formation and stimulates resorption [Liu et al., 2001; Ueta et al., 2001; Geoffroy et al., 2002; Hassan et al., 2006; Kanatani et al., 2006]. This paradoxical effects of Runx2 to either stimulate skeletogenesis or cause net skeletal catabolism has been explained by an effect of Runx2 antagonizing the final steps of osteoblast maturation and/or the bone-forming activity of fully differentiated osteoblasts [Liu et al., 2001; Geoffroy et al., 2002; Maruyama et al., 2007; Merciris et al., 2007]. These catabolic effects of Runx2 differ from other observations showing a positive association between increased Runx2 and bone mass. For example, a human mutation in the P2 promoter of Runx2 is associated with increased BMD [Doecke et al., 2006], a high bone density mouse model is caused by inhibition of Runx2 degradation [Jones et al., 2006], and Mmp13-promoter directed Runx2 overexpression stimulates net increases in bone formation in adult mice [Selvamurugan et al., 2006]. The disparities between the catabolic and osteoinductive effects of Runx2 overexperssion may be explained by the amount of Runx2 and the timing of expression controlled by the different promoters driving its expression.
The more interesting aspect of our studies is the finding that the ectopic induction of the osteogenic program was limited to certain tissues. Whereas in some tissues, such as muscle, lung and spleen, overexpression of Runx2 induced an osteogenic program leading to vascular calcifications, in other tissues, such as liver, heart, aortal and kidney, Runx2 overexpression did not induce markers of osteoblast differentiation. The ability of Runx2 overexpression to induce an osteogenic program in lung, is consistent with clinical observations of pathological calcifications of lung tissue in both hereditary disorders, such as pulmonary alveolar microlithiasis, and in acquired disorders, such as chronic kidney disease [Bendayan et al., 2000]. Runx2 overexpression also induced an osteogenic program in skeletal muscle, which also undergoes ossification in a variety of hereditary and acquired disorders [Shore and Kaplan, 2008]. In contrast, the cardiovascular system, which has a special susceptibility to calcifications in chronic kidney disease, diabetes and aging [Chillon et al., 2009; Speer et al., 2009], did not exhibit an osteogenic program in response to Runx2 overexpression. Our findings add complexity to the idea that ectopic Runx2 expression is always sufficient to initiate an osteogenic program leading to soft-tissue and vascular calcifications in various disease states, by suggesting that the initiation of the Runx2 osteogenic development cascades requires other co-factors in some but not all tissues. There is growing evidence that environmental signals, such as hyperphosphatemia or oxidative stress can promote a phenotypic switch of vascular smooth muscle cells from contractile to osteogenic phenotype that is associated with an increase in Runx2 expression and transactivity [Byon et al., 2008; Mathew et al., 2008; Speer et al., 2009]. In addition, Runx2 siRNA blocks VSMC calcification, and adenovirus-mediated overexpression of Runx2 induces VSMC calcification in vitro. Also, we found that the presence of soft-tissue calcifications did not correlate with the induction of the osteogenic program. For example, increased alizarin red staining was observed in the heart and lung, but the osteogenic program was induced only in the lung. The rapid loss of bone mass in Runx2 transgenic mice, and the consequent release of calcium and phosphate from bone, might explain the widespread organ calcification, similar to the proposed mechanism for soft tissue calcifications caused by the high turnover bone loss in OPG null mice [Bucay et al., 1998].
We also found that overexpression of Runx2 under control of the CMV-promoter and doxcycyline production rapidly induced abnormalities in hematopoietic stem cell development. Ectopic expression of Runx2, either by retroviral insertion into Runx2 gene or by the generation of transgenic mice that overexpress a full-length Runx2 cDNA in the T-cell compartment (CD2-Runx2) is reported to cause lymphomas [Vaillant et al., 1999; Blyth et al., 2001, 2006]. We found, however, that overexpression of Runx2 resulted in impaired development of the B- and T-cells. While it is well established that Runx1, a related family member, controls hematopoiesis, conditional knockout of Runx1 in hematopoietic compartments after birth, results in greater impairment of the maturation of megakaryocytes and development of both T and B lymphocytes than myeloid and erythroid development [Wang et al., 1996; Ichikawa et al., 2004a,b]. Recently, the p2-Runx2-I isoform has been shown to be expressed in hematopoietic stem cells, where sustained expression cooperates with abnormal Cbfβ in mice expressing the fusion oncogene Cbfβ-MYH11 to cause acute myeloid leukemia [Blyth et al., 2001, 2006; Kuo et al., 2009]. Consistent with this later finding, we found that overexpression of Runx2 resulted in a myeloproliferative-like disorder in doxycycline treated CMV-Cre;ROSA26-neoflox/+-rtTA;Tet-O-Runx2-II mice. Our findings of differential effects on myeloid and lymphocyte compartments is consistent with the finding that depending on the cell context, Runx2 may function as a tumor suppressor [Blyth et al., 2005] or an oncogene. This cell-context proliferative potential of Runx2 is not limited to cancer, since Runx2 also stimulates proliferation in endothelial cells [Sun et al., 2004; Qiao et al., 2006] and may be involved in endothelial-mesenchymal transformation during cardiac valve formation [Mercado-Pimentel et al., 2007]. Cardiac valves of doxycycline treated CMV-Cre; ROSA26-neoflox/+-rtTA;Tet-O-Runx2-II mice also showed abnormalities of proliferation of endocardial cells lining the heart valves (Supplemental Fig. 1).
There are several limitations to our model. First, we cannot distinguish whether the extraskeletal effects represent a biological function of Runx2 per se or whether it is mimicking the function of a paralogue normally expressed in that tissue. Runx2 isoforms and Runx paralogues have shared gene structures and functional domains encoded by exons 2–8 and overlapping transactivation potential [Lindenmuth et al., 1997; Imai et al., 1998; Thirunavukkarasu et al., 1998; Quack et al., 1999; Javed et al., 2000; Zaidi et al., 2001]. Second, we also did not test the differential effects of the Runx2-I and Runx2-II isoforms, which have been shown to have similar transactivation in vitro [Xiao et al., 2004]. Finally, since we used the broadly expressed CMV promoter to overexpress Runx2, we have not fully utilized the advantage of our triple transgenic system to target Runx2 overexpression to specific tissues.
In conclusion, we have addressed the differential functions of Runx2 in different tissues. We found that the ectopic overexpression of Runx2 recapitulated phenotypes in the mouse that have been previously associated with the pathological unregulated expression of Runx2 in various disease states. Our results emphasize the importance that the cell context plays, rather than intrinsic transcriptional activation potential of the transcription factor, in determining whether Runx2 promotes osteogenic differentiation, suppresses cell proliferation or functions as an oncogene. There appears to be a physiological requirement for stringent control of Runx2 expression, such that overexpression both within and outside of the skeleton can have adverse consequences and contribute to a variety of disease states. Overall, our findings support the idea that differential regulation of Runx2 promoters that control the time, place and amount of gene expression, the presence of different cofactors that bind to Runx proteins, and concurrent transcriptional programs that cross-talk with Runx all act in concert to determine the separate functions of Runx 2 isoforms. By developing a conditional and inducible mouse model to overexpress Runx2, and which can also modified to knockdown Runx2 isoforms by developing Tet-O-siRNA for Runx2-I and II, we have tools to investigate the cell context-dependent functions of Runx2 isoforms. Understanding the molecular basis for the cell context dependent actions of Runx2 may allow us to exploit this knowledge to understand a wide range of disorders ranging from bone anabolism and catabolism to the oncogenic and tumor suppressor potential of Runx2.
Acknowledgments
Grant sponsor: National Institutes of Health; Grant number: RO1-AR049712.
Footnotes
Additional supporting information may be found in the online version of this article.
References
- Ahn MY, Bae SC, Maruyama M, Ito Y. Comparison of the human genomic structure of the Runt domain-encoding PEBP2/CBFalpha gene family. Gene. 1996;168:279–280. doi: 10.1016/0378-1119(95)00751-2. [DOI] [PubMed] [Google Scholar]
- Banerjee C, Javed A, Choi JY, Green J, Rosen V, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Differential regulation of the two principal Runx2/Cbfa1 n-terminal isoforms in response to bone morphogenetic protein-2 during development of the osteoblast phenotype. Endocrinology. 2001;142:4026–4039. doi: 10.1210/endo.142.9.8367. [DOI] [PubMed] [Google Scholar]
- Belteki G, Haigh J, Kabacs N, Haigh K, Sison K, Costantini F, Whitsett J, Quaggin SE, Nagy A. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 2005;33:e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendayan D, Barziv Y, Kramer MR. Pulmonary calcifications: A review. Respir Med. 2000;94:190–193. doi: 10.1053/rmed.1999.0716. [DOI] [PubMed] [Google Scholar]
- Blyth K, Terry A, Mackay N, Vaillant F, Bell M, Cameron ER, Neil JC, Stewart M. Runx2: A novel oncogenic effector revealed by in vivo complementation and retroviral tagging. Oncogene. 2001;20:295–302. doi: 10.1038/sj.onc.1204090. [DOI] [PubMed] [Google Scholar]
- Blyth K, Cameron ER, Neil JC. The RUNX genes: Gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- Blyth K, Vaillant F, Hanlon L, Mackay N, Bell M, Jenkins A, Neil JC, Cameron ER. Runx2 and MYC collaborate in lymphoma development by suppressing apoptotic and growth arrest pathways in vivo. Cancer Res. 2006;66:2195–2201. doi: 10.1158/0008-5472.CAN-05-3558. [DOI] [PubMed] [Google Scholar]
- Bucay N, Sarosi I, Dunstan CR, Morony S, Tarpley J, Capparelli C, Scully S, Tan HL, Xu W, Lacey DL, Boyle WJ, Simonet WS. Osteoprotegerin-deficient mice develop early onset osteoporosis and arterial calcification. Genes Dev. 1998;12:1260–1268. doi: 10.1101/gad.12.9.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byon CH, Javed A, Dai Q, Kappes JC, Clemens TL, Darley-Usmar VM, McDonald JM, Chen Y. Oxidative stress induces vascular calcification through modulation of the osteogenic transcription factor Runx2 by AKT signaling. J Biol Chem. 2008;283:15319–15327. doi: 10.1074/jbc.M800021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillon JM, Mozar A, Six I, Maizel J, Bugnicourt JM, Kamel S, Slama M, Brazier M, Massy ZA. Pathophysiological mechanisms and consequences of cardiovascular calcifications: Role of uremic toxicity. Ann Pharm Fr. 2009;67:234–240. doi: 10.1016/j.pharma.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Choi JY, Pratap J, Javed A, Zaidi SK, Xing L, Balint E, Dalamangas S, Boyce B, van Wijnen AJ, Lian JB, Stein JL, Jones SN, Stein GS. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc Natl Acad Sci USA. 2001;98:8650–8655. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colla S, Morandi F, Lazzaretti M, Rizzato R, Lunghi P, Bonomini S, Mancini C, Pedrazzoni M, Crugnola M, Rizzoli V, Giuliani N. Human myeloma cells express the bone regulating gene Runx2/Cbfa1 and produce osteopontin that is involved in angiogenesis in multiple myeloma patients. Leukemia. 2005;19:2166–2176. doi: 10.1038/sj.leu.2403976. [DOI] [PubMed] [Google Scholar]
- Doecke JD, Day CJ, Stephens AS, Carter SL, van Daal A, Kotowicz MA, Nicholson GC, Morrison NA. Association of functionally different RUNX2 P2 promoter alleles with BMD. J Bone Miner Res. 2006;21:265–273. doi: 10.1359/JBMR.051013. [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Ducy P, Starbuck M, Priemel M, Shen J, Pinero G, Geoffroy V, Amling M, Karsenty G. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 1999;13:1025–1036. doi: 10.1101/gad.13.8.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto H, Enomoto-Iwamoto M, Iwamoto M, Nomura S, Himeno M, Kitamura Y, Kishimoto T, Komori T. Cbfa1 is a positive regulatory factor in chondrocyte maturation. J Biol Chem. 2000;275:8695–8702. doi: 10.1074/jbc.275.12.8695. [DOI] [PubMed] [Google Scholar]
- Geoffroy V, Kneissel M, Fournier B, Boyde A, Matthias P. High bone resorption in adult aging transgenic mice overexpressing cbfa1/runx2 in cells of the osteoblastic lineage. Mol Cell Biol. 2002;22:6222–6233. doi: 10.1128/MCB.22.17.6222-6233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersbach CA, Le Doux JM, Guldberg RE, Garcia AJ. Inducible regulation of Runx2-stimulated osteogenesis. Gene Ther. 2006;13:873–882. doi: 10.1038/sj.gt.3302725. [DOI] [PubMed] [Google Scholar]
- Glass DA, II, Bialek P, Ahn JD, Starbuck M, Patel MS, Clevers H, Taketo MM, Long F, McMahon AP, Lang RA, Karsenty G. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell. 2005;8:751–764. doi: 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Hassan MQ, Tare RS, Lee SH, Mandeville M, Morasso MI, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB. BMP2 commitment to the osteogenic lineage involves activation of Runx2 by DLX3 and a homeodomain transcriptional network. J Biol Chem. 2006;281:40515–40526. doi: 10.1074/jbc.M604508200. [DOI] [PubMed] [Google Scholar]
- Ichikawa M, Asai T, Chiba S, Kurokawa M, Ogawa S. Runx1/AML-1 ranks as a master regulator of adult hematopoiesis. Cell Cycle. 2004a;3:722–724. [PubMed] [Google Scholar]
- Ichikawa M, Asai T, Saito T, Seo S, Yamazaki I, Yamagata T, Mitani K, Chiba S, Ogawa S, Kurokawa M, Hirai H. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004b;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- Imai Y, Kurokawa M, Tanaka K, Friedman AD, Ogawa S, Mitani K, Yazaki Y, Hirai H. TLE, the human homolog of groucho, interacts with AML1 and acts as a repressor of AML1-induced transactivation. Biochem Biophys Res Commun. 1998;252:582–589. doi: 10.1006/bbrc.1998.9705. [DOI] [PubMed] [Google Scholar]
- Javed A, Guo B, Hiebert S, Choi JY, Green J, Zhao SC, Osborne MA, Stifani S, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Groucho/TLE/R-esp proteins associate with the nuclear matrix and repress RUNX (CBF(alpha)/AML/PEBP2(alpha)) dependent activation of tissue-specific gene transcription. J Cell Sci. 2000;113(Pt 12):2221–2231. doi: 10.1242/jcs.113.12.2221. [DOI] [PubMed] [Google Scholar]
- Javed A, Barnes GL, Pratap J, Antkowiak T, Gerstenfeld LC, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc Natl Acad Sci USA. 2005;102:1454–1459. doi: 10.1073/pnas.0409121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DC, Wein MN, Oukka M, Hofstaetter JG, Glimcher MJ, Glimcher LH. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science. 2006;312:1223–1227. doi: 10.1126/science.1126313. [DOI] [PubMed] [Google Scholar]
- Kanatani N, Fujita T, Fukuyama R, Liu W, Yoshida CA, Moriishi T, Yamana K, Miyazaki T, Toyosawa S, Komori T. Cbf beta regulates Runx2 function isoform-dependently in postnatal bone development. Dev Biol. 2006;296:48–61. doi: 10.1016/j.ydbio.2006.03.039. [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Kuo YH, Zaidi SK, Gornostaeva S, Komori T, Stein GS, Castilla LH. Runx2 induces acute myeloid leukemia in cooperation with Cbfbeta-SMMHC in mice. Blood. 2009;113:3323–3332. doi: 10.1182/blood-2008-06-162248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa M, Hirai H. Role of AML1/Runx1 in the pathogenesis of hematological malignancies. Cancer Sci. 2003;94:841–846. doi: 10.1111/j.1349-7006.2003.tb01364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenmuth DM, van Wijnen AJ, Hiebert S, Stein JL, Lian JB, Stein GS. Subcellular partitioning of transcription factors during osteoblast differentiation: Developmental association of the AML/CBF alpha/PEBP2 alpha-related transcription factor-NMP-2 with the nuclear matrix. J Cell Biochem. 1997;66:123–132. [PubMed] [Google Scholar]
- Liu W, Toyosawa S, Furuichi T, Kanatani N, Yoshida C, Liu Y, Himeno M, Narai S, Yamaguchi A, Komori T. Overexpression of Cbfa1 in osteoblasts inhibits osteoblast maturation and causes osteopenia with multiple fractures. J Cell Biol. 2001;155:157–166. doi: 10.1083/jcb.200105052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama Z, Yoshida CA, Furuichi T, Amizuka N, Ito M, Fukuyama R, Miyazaki T, Kitaura H, Nakamura K, Fujita T, Kanatani N, Moriishi T, Yamana K, Liu W, Kawaguchi H, Nakamura K, Komori T. Runx2 determines bone maturity and turnover rate in postnatal bone development and is involved in bone loss in estrogen deficiency. Dev Dyn. 2007;236:1876–1890. doi: 10.1002/dvdy.21187. [DOI] [PubMed] [Google Scholar]
- Mathew S, Tustison KS, Sugatani T, Chaudhary LR, Rifas L, Hruska KA. The mechanism of phosphorus as a cardiovascular risk factor in CKD. J Am Soc Nephrol. 2008;19:1092–1105. doi: 10.1681/ASN.2007070760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado-Pimentel ME, Hubbard AD, Runyan RB. Endoglin and Alk5 regulate epithelial-mesenchymal transformation during cardiac valve formation. Dev Biol. 2007;304:420–432. doi: 10.1016/j.ydbio.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merciris D, Marty C, Collet C, de Vernejoul MC, Geoffroy V. Overexpression of the transcriptional factor Runx2 in osteoblasts abolishes the anabolic effect of parathyroid hormone in vivo. Am J Pathol. 2007;170:1676–1685. doi: 10.2353/ajpath.2007.061069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers S, Lenny N, Sun W, Hiebert SW. AML-2 is a potential target for transcriptional regulation by the t(8;21) and t(12;21) fusion proteins in acute leukemia. Oncogene. 1996;13:303–312. [PubMed] [Google Scholar]
- Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci USA. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Qiao M, Shapiro P, Fosbrink M, Rus H, Kumar R, Passaniti A. Cell cycle-dependent phosphorylation of the RUNX2 transcription factor by cdc2 regulates endothelial cell proliferation. J Biol Chem. 2006;281:7118–7128. doi: 10.1074/jbc.M508162200. [DOI] [PubMed] [Google Scholar]
- Quack I, Vonderstrass B, Stock M, Aylsworth AS, Becker A, Brueton L, Lee PJ, Majewski F, Mulliken JB, Suri M, Zenker M, Mundlos S, Otto F. Mutation analysis of core binding factor A1 in patients with cleidocranial dysplasia. Am J Hum Genet. 1999;65:1268–1278. doi: 10.1086/302622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satake M, Nomura S, Yamaguchi-Iwai Y, Takahama Y, Hashimoto Y, Niki M, Kitamura Y, Ito Y. Expression of the Runt domain-encoding PEBP2 alpha genes in T cells during thymic development. Mol Cell Biol. 1995;15:1662–1670. doi: 10.1128/mcb.15.3.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvamurugan N, Jefcoat SC, Kwok S, Kowalewski R, Tamasi JA, Partridge NC. Overexpression of Runx2 directed by the matrix metalloproteinase-13 promoter containing the AP-1 and Runx/RD/Cbfa sites alters bone remodeling in vivo. J Cell Biochem. 2006;99:545–557. doi: 10.1002/jcb.20878. [DOI] [PubMed] [Google Scholar]
- Shore EM, Kaplan FS. Insights from a rare genetic disorder of extraskeletal bone formation, fibrodysplasia ossificans progressiva (FOP) Bone. 2008;43:427–433. doi: 10.1016/j.bone.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer MY, Yang HY, Brabb T, Leaf E, Look A, Lin WL, Frutkin A, Dichek D, Giachelli CM. Smooth muscle cells give rise to osteochondrogenic precursors and chondrocytes in calcifying arteries. Circ Res. 2009;104:733–741. doi: 10.1161/CIRCRESAHA.108.183053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer MY, Li X, Hiremath PG, Giachelli CM. Runx2/Cbfa1, but not loss of myocardin, is required for smooth muscle cell lineage reprogramming toward osteochondrogenesis. J Cell Biochem. 2010;110:935–947. doi: 10.1002/jcb.22607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M, Terry A, Hu M, O’Hara M, Blyth K, Baxter E, Cameron E, Onions DE, Neil JC. Proviral insertions induce the expression of bone-specific isoforms of PEBP2alphaA (CBFA1): Evidence for a new myc collaborating oncogene. Proc Natl Acad Sci USA. 1997;94:8646–8651. doi: 10.1073/pnas.94.16.8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JR, Liu S, Tang W, Zhou J, Wang Y, Yao X, Quarles LD. Role of hyperphosphatemia and 1,25-dihydroxyvitamin D in vascular calcification and mortality in fibroblastic growth factor 23 null mice. J Am Soc Nephrol. 2007;18:2116–2124. doi: 10.1681/ASN.2006121385. [DOI] [PubMed] [Google Scholar]
- Sun L, Vitolo MI, Qiao M, Anglin IE, Passaniti A. Regulation of TGFbeta1-mediated growth inhibition and apoptosis by RUNX2 isoforms in endothelial cells. Oncogene. 2004;23:4722–4734. doi: 10.1038/sj.onc.1207589. [DOI] [PubMed] [Google Scholar]
- Thirunavukkarasu K, Mahajan M, McLarren KW, Stifani S, Karsenty G. Two domains unique to osteoblast-specific transcription factor Osf2/Cbfa1 contribute to its transactivation function and its inability to heterodimerize with Cbfbeta. Mol Cell Biol. 1998;18:4197–4208. doi: 10.1128/mcb.18.7.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta C, Iwamoto M, Kanatani N, Yoshida C, Liu Y, Enomoto-Iwamoto M, Ohmori T, Enomoto H, Nakata K, Takada K, Kurisu K, Komori T. Skeletal malformations caused by overexpression of Cbfa1 or its dominant negative form in chondrocytes. J Cell Biol. 2001;153:87–100. doi: 10.1083/jcb.153.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant F, Blyth K, Terry A, Bell M, Cameron ER, Neil J, Stewart M. A full-length Cbfa1 gene product perturbs T-cell development and promotes lymphomagenesis in synergy with myc. Oncogene. 1999;18:7124–7134. doi: 10.1038/sj.onc.1203202. [DOI] [PubMed] [Google Scholar]
- van der Deen M, Akech J, Wang T, FitzGerald TJ, Altieri DC, Languino LR, Lian JB, van Wijnen AJ, Stein JL, Stein GS. The cancer-related Runx2 protein enhances cell growth and responses to androgen and TGFbeta in prostate cancer cells. J Cell Biochem. 2010;109:828–837. doi: 10.1002/jcb.22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Belflower RM, Dong YF, Schwarz EM, O’Keefe RJ, Drissi H. Runx1/AML1/Cbfa2 mediates onset of mesenchymal cell differentiation toward chondrogenesis. J Bone Miner Res. 2005;20:1624–1636. doi: 10.1359/JBMR.050516. [DOI] [PubMed] [Google Scholar]
- Xiao ZS, Hinson TK, Quarles LD. Cbfa1 isoform overexpression upregulates osteocalcin gene expression in non-osteoblastic and pre-osteoblastic cells. J Cell Biochem. 1999;74:596–605. [PubMed] [Google Scholar]
- Xiao ZS, Hjelmeland AB, Quarles LD. Selective deficiency of the “bone-related” Runx2-II unexpectedly preserves osteoblast-mediated skeletogenesis. J Biol Chem. 2004;279:20307–20313. doi: 10.1074/jbc.M401109200. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Awad HA, Liu S, Mahlios J, Zhang S, Guilak F, Mayo MS, Quarles LD. Selective Runx2-II deficiency leads to low-turnover osteopenia in adult mice. Dev Biol. 2005;283:345–356. doi: 10.1016/j.ydbio.2005.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Zhang S, Mahlios J, Zhou G, Magenheimer BS, Guo D, Dallas SL, Maser R, Calvet JP, Bonewald L, Quarles LD. Cilia-like structures and polycystin-1 in osteoblasts/osteocytes and associated abnormalities in skeletogenesis and Runx2 expression. J Biol Chem. 2006;281:30884–30895. doi: 10.1074/jbc.M604772200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida CA, Komori T. Role of Runx proteins in chondrogenesis. Crit Rev Eukaryot Gene Expr. 2005;15:243–254. doi: 10.1615/critreveukargeneexpr.v15.i3.60. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Javed A, Choi JY, van Wijnen AJ, Stein JL, Lian JB, Stein GS. A specific targeting signal directs Runx2/Cbfa1 to subnuclear domains and contributes to transactivation of the osteocalcin gene. J Cell Sci. 2001;114:3093–3102. doi: 10.1242/jcs.114.17.3093. [DOI] [PubMed] [Google Scholar]
- Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, Wiedemann LM, Wu H, Li L. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- Zhang S, Xiao Z, Luo J, He N, Mahlios J, Quarles LD. Dose-dependent effects of Runx2 on bone development. J Bone Miner Res. 2009;24:1889–1904. doi: 10.1359/JBMR.090502. [DOI] [PMC free article] [PubMed] [Google Scholar]