Abstract
Self-assembly of short peptides into nanostructures has become an important strategy for the bottom-up fabrication of nanomaterials. Significant interest to such peptide-based building blocks is due to the opportunity to control the structure and properties of well-structured nanotubes, nanofibrils, and hydrogels. X-ray crystallography and solution NMR, two major tools of structural biology, have significant limitations when applied to peptide nanotubes because of their non-crystalline structure and large weight. Polarized Raman spectroscopy was utilized for structural characterization of well-aligned D-Diphenylalanine nanotubes. The orientation of selected chemical groups relative to the main axis of the nanotube was determined. Specifically, the C-N bond of CNH3+groups is oriented parallel to the nanotube axis, the peptides’ carbonyl groups are tilted at approximately 54° from the axis and the COO− groups run perpendicular to the axis. The determined orientation of chemical groups allowed the understanding of the orientation of D-diphenylalanine molecule that is consistent with its equilibrium conformation. The obtained data indicate that there is only one orientation of D-diphenylalanine molecules with respect to the nanotube main axis.
Keywords: Peptide nanotube, polarized Raman spectroscopy, nanotube structure, orientation distribution function, diphenylalanine
Introduction
The self-assembly of short hydrophobic dipeptides into microporous materials has become an important strategy for bottom-up fabrications of nanostructures. Such peptide-based molecular building blocks can be used to design highly organized nanoscale structures. Wide variety of hydrophobic dipeptides have showed a great potential for application in areas such as gas storage and selective adsorption.[1] Due to the ability to form unique structures with hydrophilic orifices, they are attractive candidates as models for membrane channels through which transportation of water, ions, and larger molecules can theoretically take place.[2] To address specific applications, nanotubes with different morphologies and aspect ratios can be produced by varying preparation conditions, including copolymerization with organic monomers or using peptides with different functional groups.[3, 4] Varying the processing conditions allows for control of the surface hydrophobicity that can then serve as the basis for designing enhanced biological interfaces, which could further lead to potential biosensors.[3, 5]
One of the most commonly used building blocks is diphenylalanine (FF), a short dipeptide which has been identified as playing a key role in the core-recognition motif of Alzheimer’s disease, the β-amyloid protein (Aβ).[6, 7] Diphenylalanine nanotubes exhibit unique functional properties such as crystallinity, high aspect ratios and high thermal stability[8] and their applications include drug delivery, sensing, and nanoelectronics.[9–11] Several groups have demonstrated the rich polymorphism of self-assembled diphenylalanine and their ability to form nanotubes, nanofibrils, nanospheres and hydrogels.[12]
Most of the previously reported nanotubes were self-assembled in either aqueous or organic solutions. In this study, we have characterized di-D-diphenylalanine nanotubes grown using a hybrid vapor deposition procedure, plasma enhanced chemical vapor deposition (PECVD).[3, 13] PECVD-based self-assembly of peptide nanotubes has produced highly ordered and uniform three-dimensional nanostructures.
X-ray crystallography and solution NMR, two major tools of structural biology, have significant limitations when applied to peptide nanotubes due to their non-crystalline and insoluble nature as well as their large molecular weight. However, several structural models were proposed based on single-crystal X-ray diffraction measurements carried out on microcrystals grown at different conditions.[1, 2, 14] Peptide nanotubes have been intensively investigated by scanning electron microscopy and atomic-force microscopy, focusing mainly on physical properties, such as thermal, chemical and conformational stability,[15] rather than structural organization.
Raman spectroscopy is a powerful nondestructive technique used for studying the structure and conformation of molecular species. It has been applied to study many types of materials, including inorganic[16, 17] and organic compounds, [18, 19] polymers,[20] proteins[21] and peptides.[22] Raman spectroscopy has the ability to probe a protein’s secondary and tertiary structure, determine the conformation of side chains, and determine the local environment of aromatic amino acid residues.[21, 23, 24] In particular, Asher with co-workers[25] have developed a semiempirical approach for obtaining the distribution of a dihedral Ψ angle that determines the conformation of the polypeptide backbone.[26] Ultraviolet resonance Raman spectroscopy is a powerful tool for structural characterization of protein aggregates, including the core of amyloid fibrils.[27–29] The structure and composition of the surface of amyloid fibrils can be probed using a novel technique called tip enhanced Raman spectroscopy.[30, 31]
Polarized vibrational spectroscopy is a powerful tool for probing the structure of elongated species including synthetic and natural fibers,[32, 33] amyloid fibrils[34–36] and nanotubes.[37, 38] Polarized Raman spectroscopy can provide information about the orientation of chemical groups in an anisotropic sample of aligned species. Strempel and Kiefer used polarized Raman scattering for accurate determination of repulsive states.[17] The polarization characteristics of Raman scattering are related to the polarizability tensor and carry symmetry information about the corresponding chemical group. If the Raman tensor is known, Raman band anisotropy measurements allow for the retrieval of this information.[39–48] In particular, this method has been utilized for studying diphenylalanine nanotubes,[38] single-walled carbon nanotubes,[37, 49] polymers[20, 32, 50] and small molecules,[47] silk[51] and amyloid fibrils.[34, 52]
The theoretical background of orientation measurements by Raman spectroscopy has been extensively described in literature devoted to polarized Raman spectroscopic measurements.[42, 43] A quantitative interpretation of polarized Raman spectra requires the knowledge of a Raman tensor of a particular vibrational mode that can be described in terms of three non-zero components.[39, 53, 54] The orientation determination is based on the measurement of four polarized Raman spectra in the backscattering configuration.[33, 55] This approach avoids the displacement or rotation of the sample, thus ensuring that all four polarized spectra are measured from the same spot on the sample.[46] The calculation of two depolarization ratios allows for the determination of the second and fourth coefficients, or order parameters, of the orientation distribution function.[44, 51, 56]
In this study, polarized Raman spectroscopy was used for determining the orientation of di-D-phenylalanine (D-FF) molecules within a nanotube. Raman spectra were acquired for four different polarization configurations for a highly aligned bundle of the nanotubes. This data, in conjunction with a high orientation order of the sample and known local Raman tensors for specific vibrational modes, was used to obtain both qualitative and quantitative information about the orientation of specific chemical groups with respect to the main axis of the nanotube. An orientation of D-FF molecules with regard to the main axis of the nanotube was also predicted.
Materials and Methods
Sample preparation
Materials
The peptide di-D-phenylalanine was purchased from Bachem (Bubendorf, Switzerland). The di-D-phenylalanine nanotubes were formed at high concentrations in water. Their self-assembly has been observed both in aqueous and organic solutions and the nanotubes used in these experiments were formed via PECVD process. A home-built reactor was used for the PECVD deposition of peptide nanotubes on rigid and flexible substrates such as silicon and polydimethylsiloxane (PDMS). Briefly, the D-diphenylalanine was heated under vacuum and deposited on a substrate at room temperature. The sample set-up consists of a sample boat containing peptides, (sublime at ~220 °C) and the substrate, which was placed downstream from the plasma zone to avoid plasma bombardment effects and to allow it to be maintained at lower temperatures. Deposition was carried out under low vacuum conditions, with low chamber pressure (0.03 Torr), low argon flow rate (99.99% purity, flow rate of 10 cm3/min) and low RF power (30 W).[3] The RF power was pulsed with frequency ranging between 100 Hz to 1000 Hz and duty cycles ranging from 25% to 100%, since previous studies have reported that using a low power pulsed plasma helps the activation as well as the retention of chemical activity of the deposited molecule.[57, 58] The morphology and PECVD deposited di-D-phenylalanine nanotubes are shown in Fig. S1. Suspension of nanotubes were prepared by adding dry nanotubes to deionized water. The prepared suspension was sonicated for 5 min and then aliquots (5–20 μL) of the nanotube suspensions were dropped onto aluminum foil and air-dried.
Polarized Raman spectroscopy
Spectra were recorded in the backscattering geometry with a LabRam HR Evolution Raman microscope (Horiba Jobin Yvon). A 785 nm diode laser was focused on the sample with a 100x objective (0.9 NA-Olympus) and the power adjusted to approximately 10 mW. No damage or spectral modifications were observed in samples under these conditions. A half wave plate was used to select the polarization of the incident laser beam and a polarizer was used to select the X or Z component of the scattered beam. An optical scrambler was installed before the spectrometer entrance slit to eliminate the polarization dependence of the grating. Polarized Raman spectra were recorded for various polarization geometries, XX, XZ, ZX and ZZ, where the Z direction corresponds to the long axis of the nanotubes and the Y-axis corresponds to the propagation direction of the incident and scattered light.
Data Analysis
Data acquisition and processing were performed using LabSpec6 software (Horiba Jobin Yvon). The spectra were baseline corrected over the 300–1800 cm−1 spectral range using a polynomial baseline, followed by 7–11 points smoothing. From the intensity ratios, RZ and RX, the orientation order parameters, 〈P2〉 and 〈P4〉, were calculated by applying methods previously described.[38, 46] All calculations were performed in MATLAB (version R2105a) and Maxima, a computer algebraic system.[59]
Determining the orientation of chemical groups
The orientation of chemical groups in an anisotropic sample can result in an anisotropic orientation of the corresponding Raman tensors. For a uniaxial system with cylindrical symmetry, the molecular orientation can be described by an orientation distribution function, N(θ), where θ is the angle between the unique laboratory axis of the system and the principal axis of the polarizability tensor corresponding to the vibrational mode.[32, 38] This distribution can be expanded in a series of even Legendre polynomials.[44, 46] Polarized Raman spectroscopy provides information to obtain the second and fourth coefficients, 〈P2〉 and 〈P4〉, where 〈Pi〉 is the Legendre polynomial of degree i, whose average value is defined as the orientation order parameter. It has been shown for systems possessing cylindrical symmetry that the order parameters 〈P2〉 and 〈P4〉 can be determined using intensity ratios RZ and RX. The former parameters allow for obtaining the most probable orientation distribution Nmp(θ) using methodology described elsewhere.[38, 44, 46, 51, 56, 60] Briefly, the method is based on obtaining four polarized Raman spectra (ZZ, XX, ZX, and XZ) in the backscattering configuration, where the letters correspond to the incident and scattered light polarizations, respectively. The intensity ratios RZ and RX for individual Raman bands (IZZ, IZX, IXX, and IXZ) are related to parameters A and B, which take into account the depolarization of the incident and scattered beams (see below for more details)
| (1) |
| (2) |
Components of the tensor, 〈(αij)2〉, can be related to the components in the molecular frame and the orientation parameters 〈P2〉 and 〈P4〉[38, 42, 46]
| (3) |
| (4) |
| (5) |
| (6) |
where
| (7) |
| (8) |
| (9) |
For a Raman tensor with cylindrical symmetry
where a = α1/α3 = α2/α3.
The parameter a of the Raman tensor can be determined from the depolarization ratio, Riso, acquired for an isotropic sample from Eqns (1, 2) (for which 〈P2〉 = 〈P4〉 = 0).
| (10) |
The calculations (Eqns 1, 2 and 10) also require the use of additional correction parameters,[61, 62] A = 4.088 and B = 1.0624, which come from integrations of the squares of the electric vector components (Eqn 11) over the solid angle of light collection, Ω
| (11) |
where V is the total volume, Ee and Es are the electric vectors of the excitation and scattered radiation respectively, and α is the polarizability tensor.[46, 62]
The constants A and B take into account the depolarization of the incident and scattered beams in the focal plane associated with the use of a high numerical aperture objective (NA = 0.9), objective semi-angular aperture (θm), and a sample refractive index (n = 1.4)[38] according to Eqns (12–14).
| (12) |
| (13) |
| (14) |
Results and Discussion
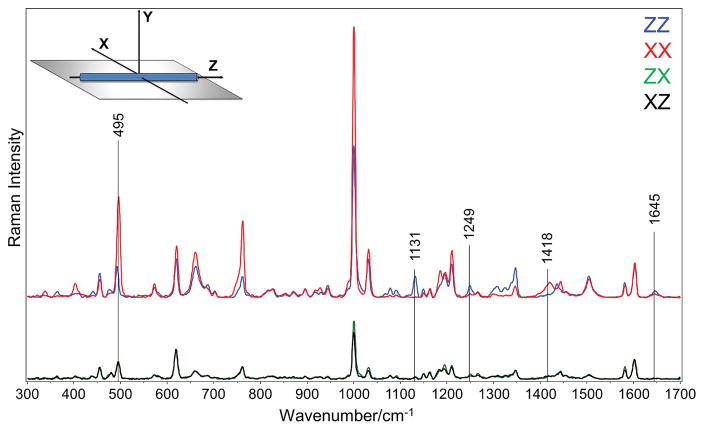
Based on SEM data and AFM analysis, the PECVD deposited di-D-diphenylalanine nanotubes had a tube-like morphology with a diameter of about 100 nm and a length between 10 and 20μm (Fig. S1).[3] Raman spectra collected from an anisotropic sample at various polarization configurations is expected to show substantial differences in peak intensities when the orientation of chemical groups causes polarization anisotropy. A set of Raman spectra acquired using four polarization geometries (XX, ZZ, XZ, and ZX), is typically sufficient for characterizing the orientation of chemical groups, providing that the polarizability tensors are known. Polarized Raman spectroscopy was used for characterization of the orientation of chemical groups in D-FF nanotubes. Polarized Raman spectra XX, ZZ, XZ, and ZX were measured for a bundle of well-aligned nanotubes, the optical image of which is shown in supplementary Fig. S2. The laboratory coordinate system (XYZ) is defined in such a way that the Z-axis corresponds to the main axis of the aligned nanotubes and the Y-axis corresponds to the propagation direction of the incident and scattered, or collected, light (Fig. 1).
Figure 1.
Normalized polarized Raman spectra of oriented di-D-phenylalanine nanotubes.
Significant differences in the intensities were found between the ZZ and XX polarized Raman spectra for many bands (Fig. 1). For example, the band at 1131 cm−1, which can be assigned to the NH3+ rocking vibration mode,[63, 64] ρ(NH3+), exhibited major variations. The position of this band in the Raman spectrum indicates that amine group in the D-FF nanotube is protonated. Moreover, the appearance of the band is in agreement with another band at about 495 cm−1, which is assigned to the torsional vibration of NH3+ group, τ(NH3+).[64–66]
It is important to make sure that the nanotubes are well aligned in the sample. To estimate the degree of alignment, we calculated the orientation parameter f, also known as the pseudo-order parameter.[32]
where IXX and IZZ are the Raman intensities of the peak of interest in the XX and ZZ spectra, respectively. For an isotropic sample, f equals zero; the value of f increases as the molecular orientation becomes increasingly more aligned until it reaches 1, which signifies a perfect parallel orientation. We obtained a value for f of 0.98 for the 1131 cm−1 band, which indicates excellent alignment of the nanotube.
The lower portion of Fig. 1 shows the Raman spectra from the two cross-polarizations, XZ and ZX. Because these crossed-polarized spectra overlap within experimental error confirms that no displacements or rotations of the sample occurred during the measurements.[46] Most importantly, this result experimentally verifies the accuracy of the polarization Raman measurements and also shows that the conditions for the application of the uniaxial model for polarized Raman spectral data analysis are satisfied.[38, 67, 68] For systems with uniaxial symmetry, the polarized Raman spectroscopy allows only the second and fourth order parameters, 〈P2〉 and 〈P4〉, to be obtained. As previously mentioned, these are the first two coefficients of the Legendre polynomial expansion of the orientation distribution function.[45, 46, 55, 69]
Quantitative interpretation of polarized Raman spectra (Fig. 1) requires the knowledge of a Raman tensor for a specific vibrational mode of interest. In general, a Raman tensor relates the polarization direction of the exciting light to the polarization direction of the scattered light.[39, 53] A Raman tensor corresponding to a particular vibrational mode can be described in terms of three non-zero components (principal axes) that can be determined using polarized Raman spectroscopic measurements.[54] The procedure for determining the orientation of chemical groups based on polarized Raman spectroscopy has been previously reported.[20, 38, 46, 60]
Orientation of chemical groups
Amide I and Amide III Raman bands
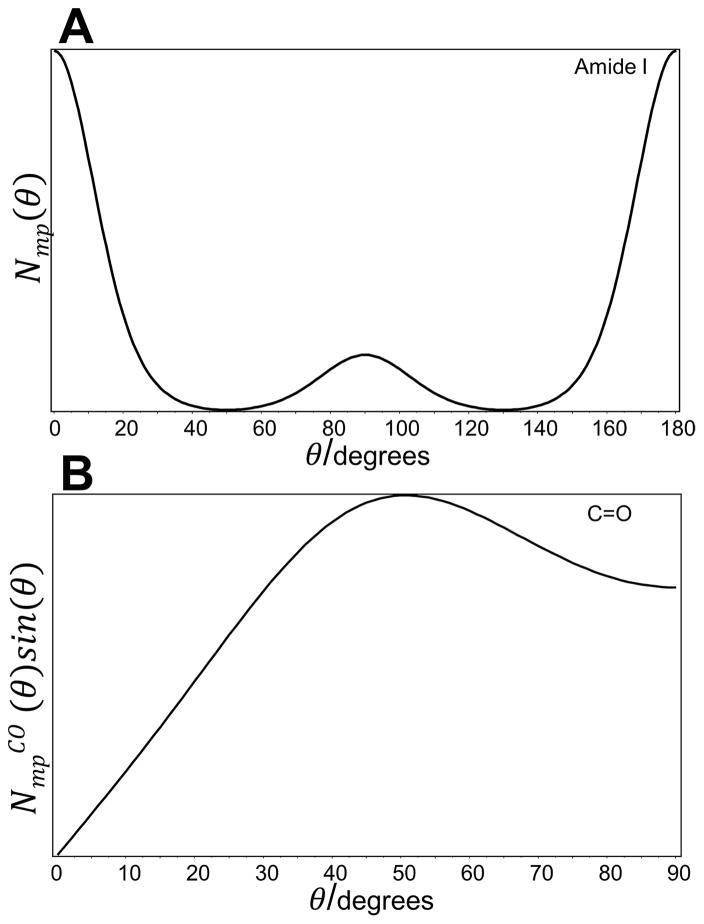
The intensity of the amide I band was used to determine the orientation of the carbonyl groups in D-FF nanotubes, with respect to the nanotube axis. The amide I Raman band is mainly associated with the C=O stretching vibrational mode mixed with C-N stretching and Cα-C-N deformation.[70–73] For nanotubes, the amide I band is located at 1645 cm−1, as seen in Fig. 2. For the aligned D-FF nanotubes, calculated intensity ratios RZ and RX for this peak were 0.1±0.02 and 0.2±0.03. To evaluate the orientation of the principal axis of the Raman tensor (PART) with respect to the main axis of D-FF nanotubes, the Raman tensor of the amide I and amide III modes reported by Lekprasert et al was used.[38] Tensor elements of the amide I and the amide III vibrational modes are represented in the form of ratios r1=α1/α3 and r2=α2/α3. For the amide I, r1 and r2 were −0.016and 0.274 and for the amide III, r1 and r2 were −0.085 and0.356.[38]
Figure 2.
A) The most probable orientation distribution function Nmp(θ) and (B) the calculated orientation distribution function NmpCO (θ)sin(θ) of C=O groups relative to the main axis of D-Diphenylalanine nanotubes.
Based on the above, values of 〈P2〉 and 〈P4〉 were calculated to be approximately 0.18 and 0.31, respectively for the amide I band. The 〈P2〉 order parameter determines the molecular orientation; values −0.5 and 1 correspond to a perfect orientation at 90° and 0°, respectively, from the nanotube axis. It also has been shown that parameters 〈P2〉 and 〈P4〉 are not totally independent, and values of 〈P4〉 associated with a given 〈P2〉 value are limited by:[44]
| (15) |
The maximum value of 〈P4〉 results in a bimodal distribution of the Raman tensor centered at 0° and 90°. If 〈P4〉 = 〈P4〉min the orientation distribution function is unimodal and is given by the delta function centered at an angle θ0.[44, 74, 75] This angle also corresponds to the mean value of the orientation distribution and can be calculated when only 〈P2〉 is known.[38, 46]
| (16) |
For all other 〈P2〉 values, the orientation distribution can be obtained from the most probable orientation distribution function, Nmp(θ).[43] Based on the experimentally determined 〈P2〉 and 〈P4〉 values and numerically calculated Lagrangian multipliers, the function Nmp(θ) has been calculated using a well-developed approach.[44, 51, 56, 75]
Figure 2A shows that Nmp(θ) of the PART for the amide I band is bimodal with a main Gaussian peak centered at 0° with a width at half height of 15°, and a second much smaller contribution centered at 90°. In this case, it is indicated that only a fraction of the PART for the amide I band is oriented perpendicularly to the nanotubes axis, while the remaining fraction is oriented parallel to the main axis. Although information about the orientation of the PART was obtained, the orientation of carbonyl groups within the aligned nanotubes was calculated instead. To evaluate the orientation of C=O groups, the angle (~ 33°)[39, 68] between the PART and carbonyl bond has to be taken into consideration. The orientation order parameters for the C=O bonds, 〈P2〉CO and 〈P4〉CO, were then calculated from the experimentally determined 〈P2〉 and 〈P4〉 values for PART of the amide I Raman tensor using the Legendre addition theorem[76] followed by calculation of the most probable orientation distribution function NmpCO (θ). It should be noted that due to a uniaxial cylindrical symmetry, multiplication of the most probable orientation distribution function by sin(θ) allows for obtaining the preferred orientation with respect to the nanotube axis, which corresponds to the maximum NmpCO (θ)sin(θ) function (Fig. 2B). The mean of this distribution function characterizes the average orientation angle.[77, 78] Figure 3B clearly shows that in the case of carbonyl groups, the NmpCO (θ)sin(θ) function was much broader, however, it reached a maximum at θ = 50° with an average orientation angle of approximately 52° relative to the main Z-axis (Fig. 2B).
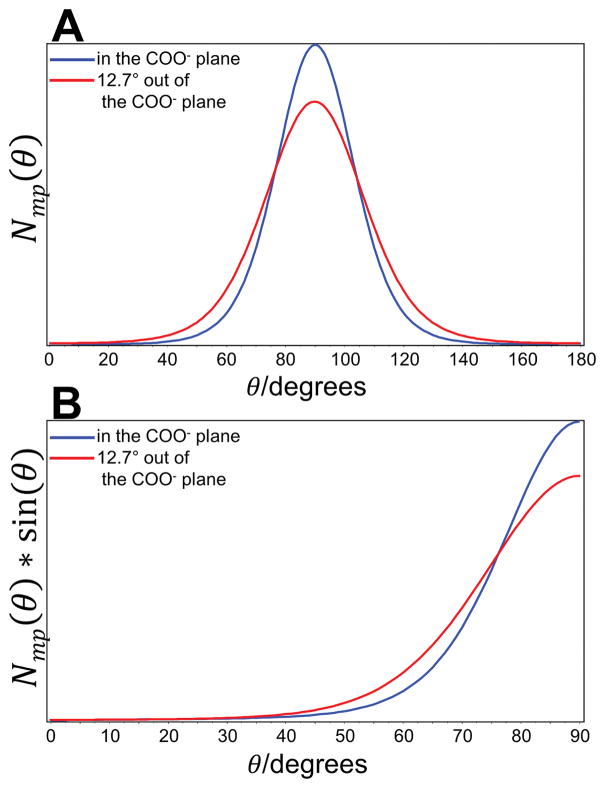
Figure 3.
(A) The most probable orientation distribution function Nmp(θ) and (B) the calculated orientation distribution function Nmp(θ)sin(θ) of the major axis of the Raman tensor of the symmetric stretching vibration νsCOO− with respect to the oriented di-D-phenylalanine nanotubes.
The amide III band spans the range of 1220–1250 cm−1 and is primarily due to the in-phase combination of N-H in-plane-bending and C-N stretching vibrations.[79] For D-FF, the amide III band was located at 1245 cm−1. Applying the same approach, the order parameters 〈P2〉 and 〈P4〉 for the amide III Raman tensor were 0.35 and −0.55. Surprisingly, the value of 〈P4〉 falls in the allowed region determined by the condition of 〈P4〉= 〈P4〉min. As already mentioned, fulfilling the requirements where 〈P4〉= 〈P4〉min, the orientation angle was obtained using Eqn (16), showing that the amide III Raman tensor makes 41° with the nanotube axis. Moreover, applying the Legendre addition theorem,[76] the orientation of C=O groups was found to be at 56° from the nanotube’s main axis. Excellent agreement between two estimations based on two independent vibrational modes indicates a high reliability of the method. It should be noted that in case of the amide I band, the value of 〈P4〉 may fall higher than the 〈P4〉= 〈P4〉min level due to possible limited applicability of the known localized polarizability tensors used for calculations, and/or experimental errors. Interestingly, Lekpraset at al. have shown that for nanotubes prepared from L-diphenylalanine the orientation of the PART corresponding to the amide I and amide III were 59±5° and 41±4°.[38] However, the predominant orientation of C=O bond was 0±5° to the nanotube axis.[38]
1418 cm−1 Raman band
The band at 1418 cm−1, which was assigned to the symmetric stretching vibration νsCOO−,[64, 80] also showed a strong polarization effect (Fig. 2). The intensity ratios RZ= IZX/IZZ=1.1 and RX= IXZ/IXX=0.2 were measured for this band. The orientation of the local Raman tensor axes and r1=1.072 and r2=4.896 ratios of νsCOO− vibrational mode have been calculated for the glycine zwitterions in the α-glycine crystal.[64] However, different tensor values have been reported by Pajcini et al. where r1=α1/α3 and r2=α2/α3 for the symmetric stretching vibration were 3.03 and −1.62, respectively.[68] The main difference is in the absolute value of α1 and α2 elements of corresponding Raman tensors. To overcome possible mistakes in the orientation determination an alternative approach was applied. 〈P2〉 and 〈P4〉 values and the most probable distribution function were determined following the procedures described elsewhere.[20, 46, 60, 77] The form of the Raman tensor is described by parameter a, which can be obtained from the depolarization ratio (Riso) of a perfectly isotropic sample.[81].The calculated isotropic depolarization ratio of 0.52 was previously reported for νsCOO− vibrational mode.[68]
Values of 〈P2〉 and 〈P4〉 obtained for the band at 1418 cm−1 were approximately −0.4 and 0.2. The function Nmp(θ) is unimodal with a Gaussian peak centered at 90° with a width at half height of 15 (Fig. 4). Interestingly, the Nmp(θ)sin(θ) function peaks at 90° with the average orientation angle of 78°. As already mentioned, the orientation of the PART of the corresponding carboxylate symmetric stretching band at 1418 cm−1 has been calculated. In addition to the difference in tensor values, the orientation of the local Raman tensor axes has been shown to be slightly different. In particular, for the COO− stretching vibration, the major axis of the Raman tensor lies within the carboxylate plane[64] (Fig. 3, blue lines) and 12.7° out of the COO− plane.[68] Parameters 〈P2〉COO− and 〈P4〉COO− and the most probable orientation distribution function for the case when the major axis of the Raman tensor is 12.7° out of COO− plane were calculated using the Legendre addition theorem[76] (Fig. 3, red lines). The functions represent the range of distributions present within the nanotubes. Obtained results clearly show that the majority of COO− groups were perpendicular to the nanotube axis although the distribution was broader when an angle at 12.7° out of COO− plane was considered.
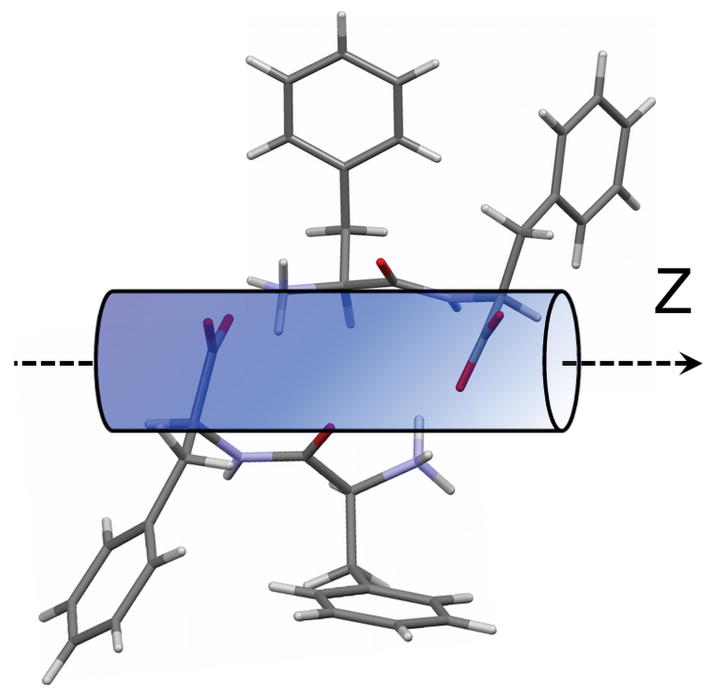
Figure 4.
Schematic diagram for the di-D-phenylalanine molecule with respect to the Z-axis based on obtained qualitative and quantitative data. Orientation of phenyl rings is arbitrary.
1000 cm−1 Raman band
The intense band located at 1000 cm−1, seen in Fig. 1, corresponds to the in-plane breathing mode of the phenylalanine benzene rings.[82–85] Due to the lack of the information on Raman tensors for this vibrational mode, the orientation cannot be obtained. However, qualitative information was still obtained due to the difference in the intensity of this band between the perpendicular and parallel polarizations, (Fig. 1) and by assuming that the largest polarizability change occurs within the plane of the phenyl ring. The band has higher intensity in the case of XX polarization, and thus, the plane normal to the phenylalanine ring planes is closer to being perpendicular than to being parallel to the axis of the nanotube. Therefore, the phenylalanine ring planes should be oriented parallel to rather than perpendicular to the nanotube axis.
495 and 1131 cm−1
As previously mentioned, the bands at 495 and 1131 cm−1 were assigned to torsional and rocking vibrations of the NH3+ functional group, respectively.[63, 86] Despite the fact that Raman tensors corresponding to NH3+ vibrational modes are not available, a qualitative estimation of orientation can still be made. First, the almost maximum possible polarization dependence observed for the band at 1131 cm−1 indicates that the corresponding Raman tensor is anisotropic. Furthermore, the perfect orientation parallel to the nanotube axis unambiguously implies that the PART should be oriented parallel, or along, the chemical bond. Secondly, our assumption is based on the reported polarized Raman data of L-valine crystals.[66] L-valine crystallizes in a monoclinic structure, space group P21, extending parallel to the (001) plane.[66] Authors defined the y-axis as the axis along the longest dimension of the crystal; the z-axis and x-axis were mutually perpendicular. Raman spectra have been collected with three different scattering geometries, Z(yy)Z, Z(xx)Z and Z(yx)Z, where the first and the last letter represents the incident and scattered light propagation direction. Interestingly, it has been shown that the relative intensity of the ρ(NH3+) vibration mode in case of Z(xx)Z polarization geometry was almost three times higher comparedto the intensity for Z(yy)Z geometry. Moreover, the relative intensity of the torsional vibration of NH3+ was more than four times lower than the intensity for the rocking vibrations of NH3+ group for Z(yy)Z geometry.[66] Based on the reported data and known crystal structure (Fig. S3), the orientation of the corresponding Raman tensors relative to the C-NH3+ bond could be qualitatively estimated. It is assumed that the largest polarizability oscillations occur along the line closest to being parallel and perpendicular to the C-N bond for rocking and torsional vibrations, respectively. The relative orientation of polarizability ellipsoids (close to being mutually perpendicular) of torsional and rocking vibrations of NH3+ group is consistent with the reported data[66] and our measurements.
Figure 1 clearly shows that the 1131-cm−1 Raman band is strong in the case of the ZZ spectrum and vanishes almost completely in the case of XX polarization geometry. This indicates almost perfect orientation of C-NH3+ bonds as being parallel to the main axis of the nanotube. At the same time, the higher Raman intensity for the XX geometry of the band at 495 cm−1 of the torsional vibration τ(NH3+) also implies a parallel orientation of C-NH3+ groups relative to the nanotubes’ main axis.
Orientation of a dipeptide molecule
The proposed orientation of D-FF molecules with respect to the Z-axis of the nanotube can be seen in Fig. 4. Dipeptide molecules are organized in such a way that the COO− groups are oriented perpendicular to the nanotube axis (Z) with NH3+ groups (C-N bonds) running along the axis. At the same time, the suggested orientation of the carbonyls is about 54° in the direction of the nanotube axis. Despite the fact that no quantitative information regarding the orientation of phenyl ring was obtained, the orientation of the rings’ plane was qualitatively estimated as being parallel to the nanotube axis.
At this point it should be noted that a relatively broad distribution in the case of peptide carbonyls and COO− groups (Fig. 2 and 3) can be explained by the possible existence of more than one specific orientation of dipeptide molecules or/and chemical groups. However, given that there is only one distinct orientation of NH3+ groups relative to the nanotube main axis, the existence of multiple orientations of dipeptide molecules seems unlikely. In addition, the appearance of Raman bands, which were unambiguously assigned to carboxyl and NH3+ groups, indicates water presence within the core of the nanotube, which is in good agreement with the literature data.[87]
Conclusions
Polarized Raman spectroscopy was used to determine the orientation of several chemical groups in a di-D-phenylalanine nanotube. This information provides significant constraints for the possible packing and orientation of di-D-phenylalanine molecules, and for building a structural model of the nanotube. Analysis of peak intensities between four different polarization combinations (ZZ, XX, ZX, and XZ) confirmed that the nanotube had cylindrical symmetry as well as multiple highly oriented functional groups. It was determined that the orientation angle of the carbonyl bond was 54±3° relative to the nanotube main axis. The band indicative of the NH3+ rocking mode shows that this group was oriented parallel to the nanotube axis. Finally, the COO− groups were confirmed to be oriented at 90° relative to the axis of the nanotube. Based on the obtained quantitative and qualitative data, a tentative overall orientation of the di-D-phenylalanine molecule within a nanotube was proposed.
Supplementary Material
Acknowledgments
We are grateful to Prof. Laurence Nafie for stimulating discussions and valuable advice. This work was supported by the National Institute on Aging, National Institutes of Health, Grant R01AG033719 (I.K.L.).
References
- 1.Görbitz CH. Chem Eur J. 2007;13:1022. doi: 10.1002/chem.200601427. [DOI] [PubMed] [Google Scholar]
- 2.Görbitz CH. Chem Eur J. 2001;7:5153. doi: 10.1002/1521-3765(20011203)7:23<5153::aid-chem5153>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 3.Vasudev MC, Koerner H, Singh KM, Partlow BP, Kaplan DL, Gazit E, Bunning TJ, Naik RR. Biomacromolecules. 2014;15:533. doi: 10.1021/bm401491k. [DOI] [PubMed] [Google Scholar]
- 4.Chen C, Liu K, Li J, Yan X. Adv Colloid Interface Sci. 2015;225:177. doi: 10.1016/j.cis.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Yemini M, Reches M, Gazit E, Rishpon J. Anal Chem. 2005;77:5155. doi: 10.1021/ac050414g. [DOI] [PubMed] [Google Scholar]
- 6.Shklovsky J, Beker P, Amdursky N, Gazit E, Rosenman G. Mater Sci Eng B. 2010;169:62. [Google Scholar]
- 7.Reches M, Gazit E. Science. 2003;300:625. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- 8.Adler-Abramovich L, Reches M, Sedman VL, Allen S, Tendler SJB, Gazit E. Langmuir. 2006;22:1313. doi: 10.1021/la052409d. [DOI] [PubMed] [Google Scholar]
- 9.Sopher NB, Abrams ZR, Reches M, Gazit E, Hanein Y. J Micromech Microeng. 2007;17:2360. [Google Scholar]
- 10.Yemini M, Reches M, Rishpon J, Gazit E. Nano Lett. 2004;5:183. doi: 10.1021/nl0484189. [DOI] [PubMed] [Google Scholar]
- 11.Meng Q, Kou Y, Ma X, Liang Y, Guo L, Ni C, Liu K. Langmuir. 2012;28:5017. doi: 10.1021/la3003355. [DOI] [PubMed] [Google Scholar]
- 12.Li Q, Jia Y, Dai L, Yang Y, Li J. ACS Nano. 2015;9:2689. doi: 10.1021/acsnano.5b00623. [DOI] [PubMed] [Google Scholar]
- 13.Adler-Abramovich L, Aronov D, Beker P, Yevnin M, Stempler S, Buzhansky L, Rosenman G, Gazit E. Nat Nanotechnol. 2009;4:849. doi: 10.1038/nnano.2009.298. [DOI] [PubMed] [Google Scholar]
- 14.Mason TO, Chirgadze DY, Levin A, Adler-Abramovich L, Gazit E, Knowles TPJ, Buell AK. ACS Nano. 2014;8:1243. doi: 10.1021/nn404237f. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Qi W, Huang R, Yang X, Wang M, Su R, He Z. J Am Chem Soc. 2015;137:7869. doi: 10.1021/jacs.5b03925. [DOI] [PubMed] [Google Scholar]
- 16.Kostova I, Peica N, Kiefer W. J Raman Spectrosc. 2007;38:1. [Google Scholar]
- 17.Strempel J, Kiefer W. Can J Chem. 1991;69:1732. [Google Scholar]
- 18.Georgieva I, Trendafilova N, Kiefer W, Rastogi VK, Kostova I. Vib Spectrosc. 2007;44:78. [Google Scholar]
- 19.Frosch T, Schmitt M, Bringmann G, Kiefer W, Popp J. J Phys Chem B. 2007;111:1815. doi: 10.1021/jp065136j. [DOI] [PubMed] [Google Scholar]
- 20.Richard-Lacroix M, Pellerin C. Macromolecules. 2012;45:1946. [Google Scholar]
- 21.Carey P. TrAC, Trends Anal Chem. 1983;2:275. [Google Scholar]
- 22.Measey T, Schweitzer-Stenner R. J Raman Spectrosc. 2006;37:248. [Google Scholar]
- 23.Oladepo SA, Xiong K, Hong Z, Asher SA, Handen J, Lednev IK. Chem Rev. 2012;112:2604. doi: 10.1021/cr200198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong K, Punihaole D, Asher SA. Biochemistry. 2012;51:5822. doi: 10.1021/bi300551b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chi Z, Chen XG, Holtz JSW, Asher SA. Biochemistry. 1998;37:2854. doi: 10.1021/bi971160z. [DOI] [PubMed] [Google Scholar]
- 26.Mikhonin AV, Bykov SV, Myshakina NS, Asher SA. J Phys Chem B. 2006;110:1928. doi: 10.1021/jp054593h. [DOI] [PubMed] [Google Scholar]
- 27.Xu M, Shashilov V, Lednev IK. J Am Chem Soc. 2007;129:11002. doi: 10.1021/ja073798w. [DOI] [PubMed] [Google Scholar]
- 28.Lednev IK. In: Protein Structures, Methods in Protein Structures and Stability Analysis. Part B. Vibrational Spectroscopy. Uversky VN, Permyakov EA, editors. Nova Science Publishers; New York: 2007. pp. 1–26. [Google Scholar]
- 29.Handen JD, Lednev IK. In: Protein Amyloid Aggregation. Methods and Protocols. Eliezer D, editor. Springer Science and Business Media; New York: 2016. pp. 89–100. [Google Scholar]
- 30.Kurouski D, Deckert-Gaudig T, Deckert V, Lednev IK. J Am Chem Soc. 2012;134:13323. doi: 10.1021/ja303263y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurouski D, Deckert-Gaudig T, Deckert V, Lednev Igor K. Biophys J. 2014;106:263. doi: 10.1016/j.bpj.2013.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frisk S, Ikeda RM, Chase DB, Rabolt JF. Appl Spectrosc. 2004;58:279. doi: 10.1366/000370204322886618. [DOI] [PubMed] [Google Scholar]
- 33.Rousseau ME, Beaulieu L, Lefèvre T, Paradis J, Asakura T, Pézolet M. Biomacromolecules. 2006;7:2512. doi: 10.1021/bm060280w. [DOI] [PubMed] [Google Scholar]
- 34.Sereda V, Sawaya MR, Lednev IK. J Am Chem Soc. 2015;137:11312. doi: 10.1021/jacs.5b07535. [DOI] [PubMed] [Google Scholar]
- 35.Rodríguez-Pérez JC, Hamley IW, Squires AM. Biomacromolecules. 2011;12:1810. doi: 10.1021/bm200167n. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez-Perez JC, Hamley IW, Squires AM. Phys Chem Chem Phys. 2013;15:13940. doi: 10.1039/c3cp52595c. [DOI] [PubMed] [Google Scholar]
- 37.Liu T, Kumar S. Chem Phys Lett. 2003;378:257. [Google Scholar]
- 38.Lekprasert B, Korolkov V, Falamas A, Chis V, Roberts CJ, Tendler SJB, Notingher I. Biomacromolecules. 2012;13:2181. doi: 10.1021/bm3005929. [DOI] [PubMed] [Google Scholar]
- 39.Tsuboi M, Kubo Y, Akahane K, Benevides JM, Thomas GJ. J Raman Spectrosc. 2006;37:240. [Google Scholar]
- 40.Tsuboi M, Kubo Y, Ikeda T, Overman SA, Osman O, Thomas GJ. Biochemistry. 2003;42:940. doi: 10.1021/bi020566v. [DOI] [PubMed] [Google Scholar]
- 41.Overman SA, Tsuboi M, Thomas GJ., Jr J Mol Biol. 1996;259:331. doi: 10.1006/jmbi.1996.0323. [DOI] [PubMed] [Google Scholar]
- 42.Bower DI. J Polym Sci Part B Polym Phys. 1972;10:2135. [Google Scholar]
- 43.Sourisseau C. Chem Rev. 2004;104:3851. doi: 10.1021/cr030042g. [DOI] [PubMed] [Google Scholar]
- 44.Labarthet FL, Buffeteau T, Sourisseau C. Appl Spectrosc. 2000;54:699. [Google Scholar]
- 45.Labarthet FL, Bruneel JL, Buffeteau T, Sourisseau C, Huber MR, Zilker SJ, Bieringer T. Phys Chem Chem Phys. 2000;2:5154. [Google Scholar]
- 46.Rousseau ME, Lefèvre T, Beaulieu L, Asakura T, Pézolet M. Biomacromolecules. 2004;5:2247. doi: 10.1021/bm049717v. [DOI] [PubMed] [Google Scholar]
- 47.Kowalska P, Cheeseman JR, Razmkhah K, Green B, Nafie LA, Rodger A. Anal Chem. 2011;84:1394. doi: 10.1021/ac202432e. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka M, Young RJ. J Mater Sci. 2006;41:963. [Google Scholar]
- 49.Gommans HH, Alldredge JW, Tashiro H, Park J, Magnuson J, Rinzler AG. J Appl Phys. 2000;88:2509. [Google Scholar]
- 50.Fanconi B, Tomlinson B, Nafie LA, Small W, Peticolas WL. J Chem Phys. 1969;51:3993. doi: 10.1063/1.1672620. [DOI] [PubMed] [Google Scholar]
- 51.Lefèvre T, Rousseau ME, Pézolet M. Biophys J. 2007;92:2885. doi: 10.1529/biophysj.106.100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sereda V, Lednev IK. J Raman Spectrosc. 2014;45:665. doi: 10.1002/jrs.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsuboi M, Ueda T, Ushizawa K. J Mol Struct. 1995;352–353:509. [Google Scholar]
- 54.Tsuboi M, Benevides JM, Thomas JGJ. Proc Jpn Acad, Ser B. 2009;85:83. doi: 10.2183/pjab/85.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rousseau ME, Lefèvre T, Pézolet M. Biomacromolecules. 2009;10:2945. doi: 10.1021/bm9007919. [DOI] [PubMed] [Google Scholar]
- 56.Lefèvre T, Paquet-Mercier F, Rioux-Dubé JF, Pézolet M. Biopolymers. 2012;97:322. doi: 10.1002/bip.21712. [DOI] [PubMed] [Google Scholar]
- 57.Vasudev MC, Anderson KD, Bunning TJ, Tsukruk VV, Naik RR. ACS Appl Mater Interface. 2013;5:3983. doi: 10.1021/am302989x. [DOI] [PubMed] [Google Scholar]
- 58.Pryce Lewis HG, Edell DJ, Gleason KK. Chem Mater. 2000;12:3488. [Google Scholar]
- 59.Maxima. 2014 http://maxima.sourceforge.net/
- 60.Church JS, Poole AJ, Woodhead AL. Vib Spectrosc. 2010;53:107. [Google Scholar]
- 61.Turrell G. J Raman Spectrosc. 1984;15:103. [Google Scholar]
- 62.Brémard C, Laureyns J, Merlin JC, Turrell G. J Raman Spectrosc. 1987;18:305. [Google Scholar]
- 63.Façanha Filho PF, Freire PTC, Lima KCV, Mendes Filho J, Melo FEA, Pizani PS. Braz J Phys. 2008;38:131. [Google Scholar]
- 64.Baran J, Ratajczak H. Vib Spectrosc. 2007;43:125. [Google Scholar]
- 65.Almeida FM, Freire PTC, Lima RJC, Remédios CMR, Mendes Filho J, Melo FEA. J Raman Spectrosc. 2006;37:1296. [Google Scholar]
- 66.Lima JA, Freire PTC, Lima RJC, Moreno AJD, Mendes Filho J, Melo FEA. J Raman Spectrosc. 2005;36:1076. [Google Scholar]
- 67.Richard-Lacroix M, Pellerin C. Appl Spectrosc y. 2013;67:409. doi: 10.1366/12-06879. [DOI] [PubMed] [Google Scholar]
- 68.Pajcini V, Chen XG, Bormett RW, Geib SJ, Li P, Asher SA, Lidiak EG. J Am Chem Soc. 1996;118:9716. [Google Scholar]
- 69.Perez R, Banda S, Ounaies Z. J Appl Phys. 2008;103:074302. [Google Scholar]
- 70.Mirkin NG, Krimm S. J Am Chem Soc. 1991;113:9742. [Google Scholar]
- 71.Chen XG, Schweitzer-Stenner R, Krimm S, Mirkin NG, Asher SA. J Am Chem Soc. 1994;116:11141. [Google Scholar]
- 72.Chen XG, Schweitzer-Stenner R, Asher SA, Mirkin NG, Krimm S. J Phys Chem. 1995;99:3074. [Google Scholar]
- 73.Schweitzer-Stenner R, Sieler G, Mirkin NG, Krimm S. J Phys Chem A. 1998;102:118. [Google Scholar]
- 74.Edwards HGM, Farwell DW. J Raman Spectrosc. 1995;26:901. [Google Scholar]
- 75.Pottel H, Herreman W, van der Meer BW, Ameloot M. Chem Phys. 1986;102:37. [Google Scholar]
- 76.Ikeda R, Chase B, Everall N. In: Vibrational spectroscopy of polymers: principles and practice. Everall NJ, Chalmers JM, Griffiths PR, editors. John Wiley & Sons, Ltd; Chichester: 2007. pp. 283–303. [Google Scholar]
- 77.Raghavan M, Sahar ND, Wilson RH, Mycek MA, Pleshko N, Kohn DH, Morris MD. J Biomed Opt. 2010;15:037001. doi: 10.1117/1.3426310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fratzl P, Paris O, Klaushofer K, Landis WJ. J Clin Invest. 1996;97:396. doi: 10.1172/JCI118428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cai S, Singh BR. Biophys Chem. 1999;80:7. doi: 10.1016/s0301-4622(99)00060-5. [DOI] [PubMed] [Google Scholar]
- 80.Movasaghi Z, Rehman S, Rehman IU. Appl Spectrosc Rev. 2007;42:493. [Google Scholar]
- 81.Richard-Lacroix M, Pellerin C. Macromolecules. 2013;46:5561. [Google Scholar]
- 82.Stewart S, Fredericks PM. Spectrochim Acta, Part A. 1999;55:1641. [Google Scholar]
- 83.Asher SA, Ludwig M, Johnson CR. J Am Chem Soc. 1986;108:3186. [Google Scholar]
- 84.Rava RP, Spiro TG. J Phys Chem. 1985;89:1856. [Google Scholar]
- 85.Rajkumar BJM, Ramakrishnan V. Spectrochim Acta, Part A. 2002;58:1923. doi: 10.1016/s1386-1425(01)00641-2. [DOI] [PubMed] [Google Scholar]
- 86.Mary MB, Sasirekha V, Ramakrishnan V. Spectrochim Acta, Part A. 2005;62:446. doi: 10.1016/j.saa.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 87.Wu X, Xiong S, Wang M, Shen J, Chu PK. Opt Express. 2012;20:5119. doi: 10.1364/OE.20.005119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.