Abstract
The circadian clock orchestrates diverse physiological processes critical for health and disease. CREB, hepatocyte specific (CREBH) is a liver-enriched, endoplasmic reticulum (ER)–tethered transcription factor known to regulate the hepatic acute phase response and energy homeostasis under stress conditions. We demonstrate that CREBH is regulated by the circadian clock and functions as a circadian regulator of hepatic lipid metabolism. Proteolytic activation of CREBH in the liver exhibits typical circadian rhythmicity controlled by the core clock oscillator BMAL1 and AKT/glycogen synthase kinase 3β (GSK3β) signaling pathway. GSK3β-mediated phosphorylation of CREBH modulates the association between CREBH and the coat protein complex II transport vesicle and thus controls the ER-to-Golgi transport and subsequent proteolytic cleavage of CREBH in a circadian manner. Functionally, CREBH regulates circadian expression of the key genes involved in triglyceride (TG) and fatty acid (FA) metabolism and is required to maintain circadian amplitudes of blood TG and FA in mice. During the circadian cycle, CREBH rhythmically regulates and interacts with the hepatic nuclear receptors peroxisome proliferator–activated receptor α and liver X receptor α as well as with the circadian oscillation activator DBP and the repressor E4BP4 to modulate CREBH transcriptional activities. In conclusion, these studies reveal that CREBH functions as a circadian-regulated liver transcriptional regulator that integrates energy metabolism with circadian rhythm.
Introduction
Mammalian circadian rhythms are biological processes that exhibit endogenous oscillations over a 24-h light-dark cycle and are entrainable by internal and external stimuli. Circadian rhythms are generated at the level of gene transcription by a network of clock-controlled genes that form an autoregulatory feedback loop (1). The CLOCK/BMAL1 heterodimer drives circadian expression of many other transcription factors, thereby extending and enhancing other circadian regulatory functions. Peripheral organs, such as the liver, have local rhythms synchronized by master clock oscillators located in the suprachiasmatic nuclei of the anterior hypothalamus (2). Previous work demonstrated the intimate and reciprocal interaction between the circadian clock system and fundamental metabolic pathways (3,4). In the liver, nuclear receptors or transcription factors are inducible by metabolites or hormones, whereas one-half of them exhibit rhythmic expression (5). Therefore, the liver nuclear receptors or transcriptional regulators may serve as direct links between metabolic pathways and circadian regulation.
We reported that the endoplasmic reticulum (ER)–tethered, liver-enriched transcription factor CREB, hepatocyte specific (CREBH) regulates energy homeostasis under metabolic stress. The expression and activation of CREBH in the liver are regulated by a variety of inflammatory and metabolic signals, such as proinflammatory cytokines, saturated fatty acid (FA), insulin, fasting, and atherogenic high-fat diets (6,7). Activated CREBH is a multifaceted activator of transcription that induces expression of the genes involved in hepatic acute phase response, FA oxidation, lipolysis, lipogenesis, and gluconeogenesis (6–10). CREBH-null mice develop profound nonalcoholic steatohepatitis and hypertriglyceridemia when fed an atherogenic high-fat diet (6). Other studies have confirmed that patients with hypertriglyceridemia exhibit a high rate of nonsense mutations or rare genetic variant accumulation in the human CREBH gene (9,11,12).
In this study, we demonstrate that CREBH is an organ-specific circadian regulator of lipid metabolism and that CREBH activation is regulated by the circadian oscillation in the liver. CREBH plays an indispensable role in maintaining lipid homeostasis under circadian control, and dysfunction of CREBH in mice leads to impaired rhythmic profiles of triglycerides (TGs) and FAs. The finding of CREBH as a circadian metabolic regulator has important implications in the understanding of the molecular basis of circadian metabolism and the development of metabolic disorders.
Research Design and Methods
Animal Model
All animal experiments were performed with the approval of the Institutional Animal Care and Use Committee of Wayne State University. Four-month-old male wild-type (WT) and CREBH-null C57BL/6 mice (6) were housed in 12-h light-dark cycles with free access to food and water for at least 2 weeks before switching to constant darkness for 24 h to allow endogenous clocks to free run. Mice were killed with isoflurane followed by rapid cervical dislocation. Liver samples from three to five mice per time point per genotype group were collected in constant darkness every 4 h for a 24-h period.
In Vitro Circadian Synchronization of Mouse Primary Hepatocytes
Primary hepatocytes isolated from C57BL/6J mice were infected with recombinant adenovirus expressing Bmal1 short hairpin RNA (shRNA), dominant-negative or constitutively activated AKT, or dominant-negative or constitutively activated glycogen synthase kinase 3β (GSK3β) for 24 h before being subjected to serum shock (50% horse serum) for 2 h for circadian synchronization (13). After serum shock synchronization, the shock medium was replaced with serum-free medium. Cell lysates were collected at 8-h intervals between 24 h (circadian 0 h) and 72 h (circadian 48 h) post–serum shock for Western blot analysis.
Chromatin Immunoprecipitation Assays With Mouse Liver Chromatin
Mouse liver chromatin was fragmented to an average size of 500 base pairs by sonication as previously described (14). Fragmented chromatin was precleared by incubating with rabbit IgG followed by incubation with protein G agarose. CREBH-binding complexes were pulled down by using a rabbit anti-CREBH antibody (8). As controls, the precleared chromatin samples were pulled down by using a rabbit anti-HA antibody. Immunoprecipitated chromatin fragments were reverse cross linked and digested by proteinase K. Presence of CREBH in gene promoters during various circadian phases was analyzed by quantification real-time PCR (qPCR) and expressed relative to the input genomic DNA. The primer sequences for chromatin immunoprecipitation (ChIP)-quantification real-time PCR (ChIP-qPCR) are described in Supplementary Table 1.
Site-Directed Mutagenesis
Site-directed mutations were introduced into the putative GSK3β phosphorylation sites serine (S) 256 and S260 of human CREBH by using a Stratagene QuikChange II Site-Directed Mutagenesis Kit (Agilent). The S256A or S260A mutation (where S was changed to alanine [A]) was achieved by site-directed mutagenesis PCR by using a human full-length CREBH expression plasmid as the template and the primers 5′- GAACAAGCAGGCGGCGCAAGAAA-3′ and 5′- TTTCTTGCGCCGCCTGCTTGTTC-3′ for S256A or 5′- GGCGCAAGAAGCCAGGAAGAAGA-3′ and 5′- TCTTCTTCCTGGCTTCTTGCGCC-3′ for S260A. All constructs were confirmed by sequencing analysis.
Statistics
Results were analyzed by several statistical methods. Unpaired Mann-Whitney U test was used for nonparametric comparisons. One-way ANOVA test was used for parametric comparisons. Two-way ANOVA was used to distinguish the effects of genotypes from the effects of circadian time on gene expression, levels of mouse blood lipids, and quantification of food intake. When multiple testing procedures were implemented, the Bonferroni correction was used.
Results
CREBH Is a Clock-Regulated Diurnal Regulator in the Liver
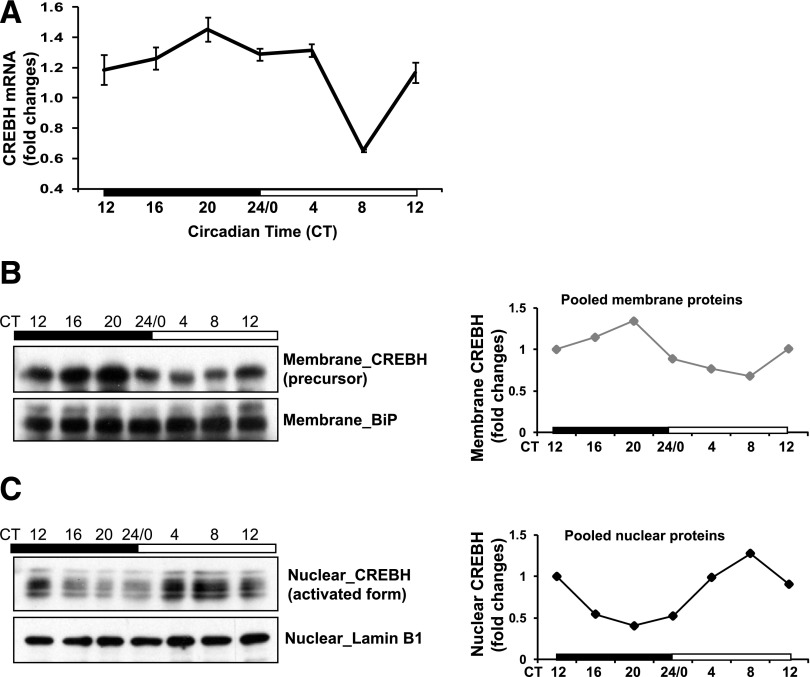
To test whether circadian-dependent regulation for CREBH is present in the liver, we examined the expression and proteolytic activation profiles of CREBH in the livers of WT mice during a 24-h circadian cycle. Expression of CrebH mRNA in mouse liver was increased during the late phase of daytime and decreased during the late phase of nighttime but did not exhibit typical circadian rhythmicity (Fig. 1A). We next examined levels of precursor and activated forms of CREBH proteins in mouse liver throughout the day-night cycle. Production of the activated CREBH protein involves translocation of CREBH precursor from the ER to the Golgi apparatus where it is cleaved by S1P and S2P proteases (7); therefore, levels of the activated CREBH can be evaluated by examining cleaved CREBH proteins. Western blot analyses with the membrane and nuclear protein fractions prepared from the pooled mouse liver tissues taken during the circadian circle showed that levels of the membrane-bound CREBH precursor protein during the daytime phases were lower than those of the nighttime phases, which displayed a peak at circadian time point (CT) 20 and a trough at CT8 (Fig. 1B). In an opposite manner, levels of the activated nuclear CREBH protein reached a trough at CT20 and peak at CT8, 12 h after the peak production of CREBH precursor protein (Fig. 1C). The decreases in the levels of CREBH precursor were coincident with the increase in the levels of the cleaved CREBH, which exhibited typical circadian rhythmicity. The rhythmicity in the production of CREBH precursor and cleaved proteins was confirmed by Western blot analysis with total liver protein lysates from WT and CREBH-null animals during the circadian cycle (Supplementary Fig. 1A). Taken together, these results indicate that the proteolytic cleavage and activation processes of CREBH in the liver are rhythmically regulated by the circadian clock during the day-night cycle.
Figure 1.
Activation of CREBH in the liver is regulated by circadian rhythm. A: Circadian oscillations of CrebH mRNA expression levels in WT mouse liver tissue collected every 4 h over a 24-h period in constant darkness, determined by qPCR. Fold changes of mRNA levels were compared with the nadir mRNA levels at CT8. Data are mean ± SEM (n = 3 mice/time point). B and C: Western blot analysis of levels of membrane-bound CREBH precursor and nuclear forms in mouse livers during the circadian cycle. Cellular membrane and nuclear protein fractions were prepared from pooled liver tissues of WT mice collected every 4 h over a 24-h circadian cycle (n = 3 mice/time point). Levels of the ER chaperone BiP or the nuclear protein lamin B1 were determined as controls. The graphs beside the images show the quantifications of CREBH precursor and nuclear forms in the mouse livers under the circadian clock. CREBH protein signals in the pooled liver membrane and nuclear protein fractions, determined by Western blot densitometry, were normalized to that of the ER membrane protein control BiP or the nuclear protein control lamin B1. Fold changes of protein levels are compared with that at CT12.
Circadian Rhythmic Activation of CREBH Is Regulated by the BMAL1-AKT-GSK3β Signaling Pathway
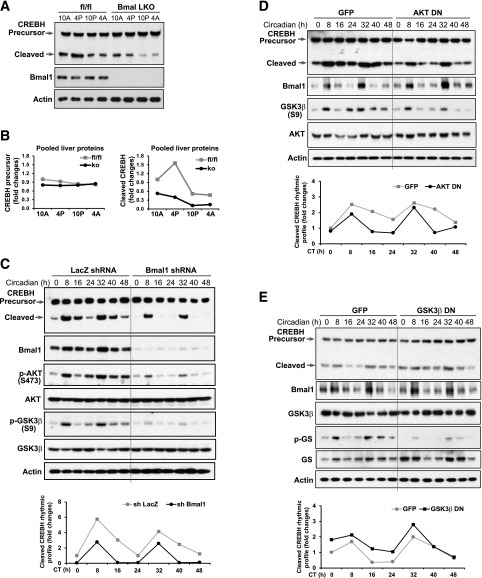
BMAL1, the core circadian oscillator, plays a central role in regulating expression or activities of circadian output regulators (1). To determine whether BMAL1 regulates circadian rhythmic activation of CREBH, we examined the expression and proteolytic cleavage of CREBH protein in liver-specific Bmal1 conditional knockout (Bmal1 LKO) and control mouse livers collected every 6 h during a 24-h circadian period (15). Western blot analysis showed that the levels of the cleaved/activated CREBH protein, but not the CREBH precursor, were reduced in the livers of Bmal1 LKO mice across the day-night periods compared with those in the control fl/fl mouse livers (Fig. 2A and B). Additionally, we examined activation of CREBH in livers of Bmal1 LKO and control mice under the normal feeding condition or after 16 h of fasting. The levels of the activated CREBH protein was significantly decreased in the livers of Bmal1 LKO mice under the feeding condition but not the fasting condition (Supplementary Fig. 1B), confirming that BMAL1 plays a major role in regulating CREBH activation under normal physiological conditions. However, under metabolic stress (fasting), CREBH is likely activated through alternative regulatory mechanisms independent of BMAL1, an interesting question to be studied in the future.
Figure 2.
Circadian activation of CREBH is regulated by the BMAL1-AKT-GSK3β signaling pathway. A: Western blot analysis of CREBH precursor and activated form in the livers of Bmal1 LKO and fl/fl control mice during various circadian phases. The liver protein lysates were prepared from pooled liver tissues of Bmal1 LKO and fl/fl mice collected every 6 h during a 24-h circadian period (n = 3–4 mice/genotype/time point) (15). Levels of BMAL1 and β-actin were determined as controls. B: Rhythmic fold changes of CREBH precursor and activated proteins in the livers of Bmal1 LKO and fl/fl mice during the circadian cycle. The CREBH protein signals, determined by Western blot densitometry, were normalized to that of β-actin. Fold change of the normalized CREBH precursor or activated protein levels was determined by comparing with that at 10 a.m. Western blot analysis of CREBH precursor and activated protein in Bmal1 knockdown (C), AKT dominant-negative (DN) (D), GSK3β-DN (E), and control primary hepatocytes during a 48-h circadian cycle. Mouse primary hepatocytes were infected with adenovirus expressing Bmal1 shRNA, AKT-DN, GSK3β-DN, control LacZ shRNA, or control GFP for 24 h before being subjected to horse serum shock for circadian synchronization. Cell lysates were collected at 8-h intervals during a 48-h circadian cycle for Western blot analysis to determine levels of CREBH, BMAL1, phosphorylated AKT2 (S473), total AKT2, phosphorylated GSK3β (S9), total GSK3β, phosphorylated glycogen synthase (GS) (S641), total GS, or β-actin. The graphs below the images show the rhythmic fold changes of cleaved CREBH proteins in Bmal1 knockdown, AKT-DN, GSK3β-DN, and control primary hepatocytes during the circadian cycle. CREBH protein signals were normalized to that of β-actin. Fold change of the normalized CREBH protein levels at each circadian time point was compared with that of the starting circadian time. A, a.m.; ko, knockout; P, p.m.
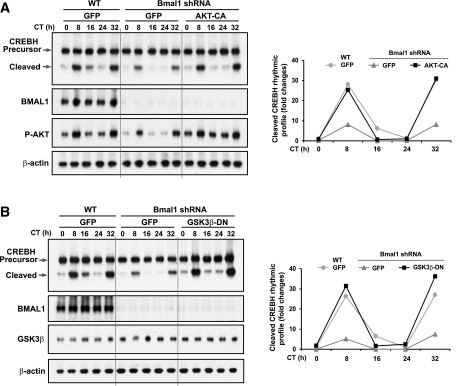
To gain further insights into the regulatory mechanism underlying circadian activation of CREBH, we explored the signal transduction pathway through which BMAL1 regulates CREBH activation in primary hepatocytes during circadian oscillation. Bmal1 was knocked down in mouse primary hepatocytes using adenoviral-based expression of Bmal1 shRNA (16) and then subjected to horse serum shock for circadian synchronization (13). Western blot analysis showed that the amplitudes of CREBH cleavage in hepatocytes across a 48-h circadian period were significantly repressed by Bmal1 knockdown (Fig. 2C). Indeed, the cleaved/activated form of CREBH was diminished in Bmal1 knockdown hepatocytes during the circadian cycle, except CT8 or CT32. AKT, a serine/threonine protein kinase, played a key role in regulating energy homeostasis (17). Studies have shown that circadian oscillation of AKT activities is impaired in Bmal1 knockout mice (18,19) and that BMAL1 regulates hepatic lipogenesis through the AKT2 signaling pathway (16). Furthermore, GSK3β, a constitutively active serine/threonine kinase that is negatively regulated by AKT2-mediated phosphorylation (17), can regulate energy homeostasis and circadian function through phosphorylation-mediated suppression of its downstream substrates (20). Through bioinformatics analysis, we identified that mammalian CREBH proteins possess conserved GSK3β phosphorylation sites within their basic leucine zipper (bZIP) domains (Supplementary Fig. 1C). This information prompted us to speculate that BMAL1 may regulate CREBH circadian activation through the AKT-GSK3β signaling pathway. To test this hypothesis, we examined hepatic AKT and GSK3β phosphorylation states in Bmal1 knockdown primary hepatocytes under the circadian clock. Phosphorylation at S473 of AKT2, an indication of AKT activity in insulin signaling and circadian regulation (17,19), was significantly reduced in Bmal1 knockdown hepatocytes (Fig. 2C). Bmal1 knockdown largely repressed phosphorylation of GSK3β at S9, the inhibitory phosphorylation site regulated by AKT (17). Of note, the circadian time points where the cleaved CREBH protein was elevated were coincidental with the time points when phosphorylation of AKT or GSK3β was upregulated (Fig. 2C), indicating that circadian-regulated cleavage of CREBH may be regulated through a BMAL1-AKT-GSK3β regulatory axis. To validate this possibility, we examined proteolytic activation of CREBH in AKT or GSK3β dominant-negative primary hepatocytes across the circadian cycle. The levels of cleaved CREBH in AKT dominant-negative hepatocytes during a 48-h circadian cycle were reduced ∼20–70% compared with control hepatocytes (Fig. 2D). In contrast, overexpression of a dominant-negative GSK3β led to increased proteolytic cleavage of CREBH in hepatocytes during the circadian cycle (Fig. 2E), thus supporting a regulatory role of AKT-GSK3β signaling in BMAL1-controlled CREBH cleavage. To validate that BMAL1 regulates CREBH cleavage through AKT/GSK3β signaling under the circadian clock, we performed the reconstitution experiments by overexpressing a constitutively active AKT (16) or dominant-negative GSK3β in Bmal1 knockdown primary hepatocytes. Expression of constitutively active AKT increased the levels of cleaved CREBH in the Bmal1 knockdown hepatocytes compared with expression of green fluorescent protein (GFP) control (Fig. 3A). Similarly, expression of dominant-negative GSK3β can rescue the decreased CREBH cleavage activity in the Bmal1 knockdown hepatocytes under the circadian clock (Fig. 3B), thus confirming that BMAL1 controls circadian-regulated CREBH cleavage/activation through AKT/GSK3β signaling in hepatocytes.
Figure 3.
Expression of a constitutively active AKT or a dominant-negative GSK3β can rescue decreased CREBH cleavage activities in Bmal1 knockdown primary hepatocytes. Western blot analyses of CREBH precursor and cleaved protein in Bmal1 knockdown and WT control mouse primary hepatocytes expressing the constitutively active AKT (AKT-CA) (A) or the dominant-negative GSK3β (GSK3β-DN) (B) under the circadian clock. Mouse primary hepatocytes were infected with adenovirus expressing Bmal1 shRNA, AKT-CA, GSK3β-DN, or GFP control before being subjected to horse serum shock for circadian synchronization. Cell lysates were collected at 8-h intervals for Western blot analysis to determine levels of CREBH, BMAL1, phosphorylated AKT, or β-actin. The graphs next to each blot show the rhythmic fold changes of cleaved CREBH proteins in Bmal1 knockdown and WT control hepatocytes expressing AKT-CA, GSK3β-DN, or GFP control. CREBH protein signals were normalized to that of β-actin. Fold change of the normalized CREBH protein levels at each circadian time point was compared with that of the starting circadian time.
GSK3β-Mediated Phosphorylation Modulates CREBH Association With COPII Vesicle Proteins and Subsequent CREBH Cleavage Under the Circadian Clock
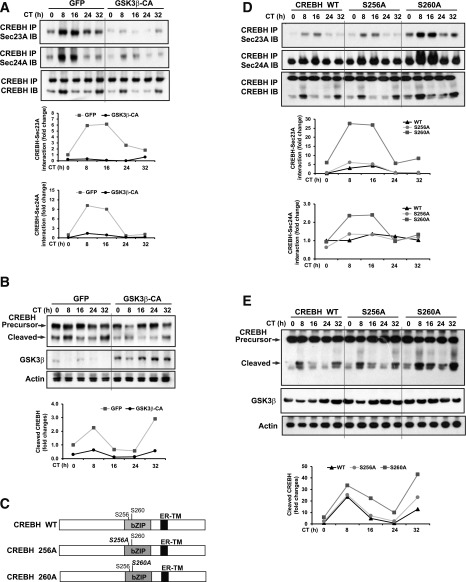
The CREBH cleavage process involves transport of the ER-bound CREBH from the ER to the Golgi apparatus where it is cleaved by S1P and S2P proteases (7). To undergo the sequential proteolysis at the Golgi, ER-bound nascent proteins need to be transported to the Golgi through COPII transport vesicles (21,22). Formation of COPII vesicles is initiated by the small GTPase Sar1, which sequentially recruits the heterodimeric protein complexes Sec23/24 and Sec13/31 to the ER membrane to facilitate vesicle formation (21). We reasoned that GSK3β-mediated phosphorylation of CREBH may be a control point for circadian-regulated transport of CREBH from the ER to the Golgi and subsequent proteolytic cleavage of CREBH. To test this possibility, we first overexpressed a constitutively active GSK3β in which the regulatory phosphorylation site S9 was mutated to an alanine and thus was resistant to inactivation mediated by Akt phosphorylation (23) or GFP control in mouse primary hepatocytes. After circadian synchronization by serum shock, we examined the interaction between CREBH and Sec23/24, the core component proteins of COPII transport vesicles, in the hepatocytes. CREBH complexed with Sec23A/Sec24A in the primary hepatocytes expressing GFP control in a circadian-dependent manner (Fig. 4A). However, the circadian-regulated association between CREBH and Sec23A/Sec24A in the hepatocytes expressing constitutively active GSK3β was significantly repressed. Moreover, the CREBH-Sec23/24 interactive activities complied with the levels of cleaved CREBH protein in the hepatocytes expressing GFP or constitutively active GSK3β under the circadian clock (Fig. 4B). These results indicate that CREBH is associated with the COPII vesicle proteins in a circadian-dependent manner and that GSK3β modulates circadian-regulated association of CREBH with the COPII vesicle.
Figure 4.
GSK3β-mediated phosphorylation modulates CREBH association with the COPII vesicle proteins Sec23/24 and subsequent CREBH cleavage under the circadian clock. A: Immunoprecipitation (IP)-Western blot analysis of interactions among CREBH, Sec23A, and Sec24A in mouse primary hepatocytes expressing the constitutively active GSK3β (GSK3β-CA) or GFP control. Mouse primary hepatocytes were infected with adenovirus expressing GSK3β-CA or GFP followed by horse serum shock for circadian synchronization. Cell lysates collected after serum shock were pulled down by the anti-CREBH antibody and probed with the Sec23A or Sec24A antibody. As control, the CREBH pulldown lysates were probed with the CREBH antibody. The graphs show the rhythmic fold changes of CREBH-associated Sec23A or Sec24A in hepatocytes expressing GSK3β-CA or GFP control. The immunoprecipitated Sec23A and Sec24A protein signals were normalized to that of CREBH. Fold change of the normalized Sec23A and Sec24A protein levels at each circadian time point was compared with that of the starting circadian time. B: Western blot analysis of CREBH, GSK3β, and β-actin in the mouse primary hepatocytes expressing GSK3β-CA or GFP control as described in A. The graph shows the rhythmic fold changes of cleaved CREBH protein in the hepatocytes expressing GSK3β-CA or GFP control. C: Illustration of WT CREBH protein and two CREBH mutants, S256A and S260A, in which the GSK3β phosphorylation residue S256 or S260 was mutated to A. D: IP-Western blot analysis of interactions among CREBH, Sec23A, and Sec24A in Hepa1-6 cells expressing CREBH-WT, S256A, or S260A under the circadian clock. Hepa1-6 cells were transfected with plasmid vector expressing CREBH-WT, S256A, or S260A, followed by horse serum shock for circadian synchronization. Cell lysates collected after serum shock were pulled down by the anti-CREBH antibody and then probed with the Sec23A, Sec24A, or CREBH antibody. The graphs show the rhythmic fold changes of CREBH-associated Sec23A or Sec24A in Hepa1-6 cells expressing CREBH-WT, S256A, or S260A. The protein signal normalization and fold change calculation methods were as described in A. E: Western blot analysis of CREBH, GSK3β, and β-actin in the Hepa1-6 cells expressing CREBH-WT, S256A, or S260A as described in D. The graphs show the rhythmic fold changes of cleaved CREBH in the Hepa1-6 cells expressing WT CREBH, S256A, or S26A. IB, immunoblot; TM, transmembrane.
To further delineate the regulation of the CREBH-COPII interaction by GSK3β, we generated two CREBH mutants in which the putative GSK3β phosphorylation sites within the bZIP domain, S256 and S260, were changed to S256A and S260A, respectively (Fig. 4C and Supplementary Fig. 1B). The vectors expressing the WT CREBH, CREBH mutant S256A, or CREBH mutant S260A were transferred into the mouse hepatoma cell line Hepa1-6 followed by serum shock for circadian synchronization. Confirming the circadian-regulated association of CREBH with the COPII vesicle proteins, WT CREBH was rhythmically associated with Sec23A and Sec24A in a circadian-dependent manner (Fig. 4D). In comparison, the CREBH mutant S260A exhibited an increased affinity to form a complex with Sec23A and Sec24A, indicating that GSK3β-mediated phosphorylation of CREBH at S260 represses the association of CREBH with Sec23/24 (Fig. 4D). Of note, the site mutation at S256 (S256A) only slightly increased the affinity of CREBH to form a complex with Sec23A or Sec24A, suggesting that S260 but not S256 is the primary GSK3β phosphorylation site within CREBH that affects the interaction between CREBH and Sec23/24. Furthermore, we examined the levels of cleavage of CREBH WT, S256A, and S260A in hepatocytes under the circadian clock. Consistent the circadian-regulated associations between CREBH isoforms and Sec23/24, the CREBH mutant S260A displayed increased cleavage activities compared with WT CREBH under the circadian clock (Fig. 4E). Together, these results indicate that GSK3β-mediated phosphorylation of CREBH at S260 alters the affinity of CREBH to complex with the COPII vesicular proteins and thus modulates the ER-to-Golgi transport and subsequent proteolytic cleavage of CREBH under the circadian clock.
CREBH Regulates Circadian Rhythmic Levels of TG and FA by Activating the Genes Encoding Functions in Lipolysis, FA Oxidation, and Lipogenesis
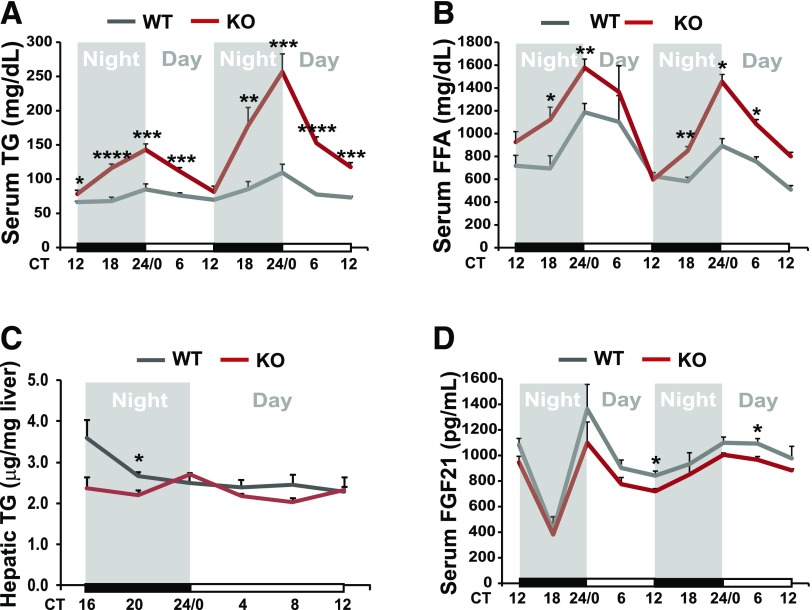
To elucidate whether CREBH regulates energy homeostasis during the circadian cycle, we characterized rhythmic profiles of circulating lipids in CREBH-null mice. Compared with WT mice, CREBH-null mice exhibited significantly higher serum TG and FA, but not cholesterol, levels during a 48-h circadian period (Fig. 5A and B and Supplementary Fig. 1D). However, hepatic TG levels of CREBH-null mice were reduced compared with that of WT mice at CT20 (equal to nighttime 2:00 a.m.) when mice usually take most of their meals of the day (Fig. 5C). Furthermore, serum levels of the metabolic hormone FGF21, a known target of CREBH under metabolic stress (8), were reduced in CREBH-null mice during the circadian period (Fig. 5D). Additionally, we examined body weights, body composition, blood glucose levels, and food consumption of CREBH-null and WT control mice. CREBH-null mice displayed a modest reduction in body fat mass while they consumed modestly increased amounts of food compared with WT mice (Supplementary Fig. 2). These metabolic phenotypes may be partially attributed to the defects in lipolysis and FA oxidation in CREBH-null mice as we previously demonstrated (6,8).
Figure 5.
CREBH is required to maintain rhythmic levels of lipids and FGF21. Levels of serum TG (A), serum free FA (B), hepatic TG (C), and serum FGF21 (D) in CREBH-null and WT mice under the circadian clock. Blood samples were collected every 6 h for 48 h in constant darkness to measure TG, FA, and FGF21 levels. Liver tissue samples of CREBH-null and WT control mice were collected every 4 h for 24 h in constant darkness to measure hepatic TG content. Data are mean ± SEM (n = 8 mice/time point). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. FFA, free fatty acid; KO, knockout.
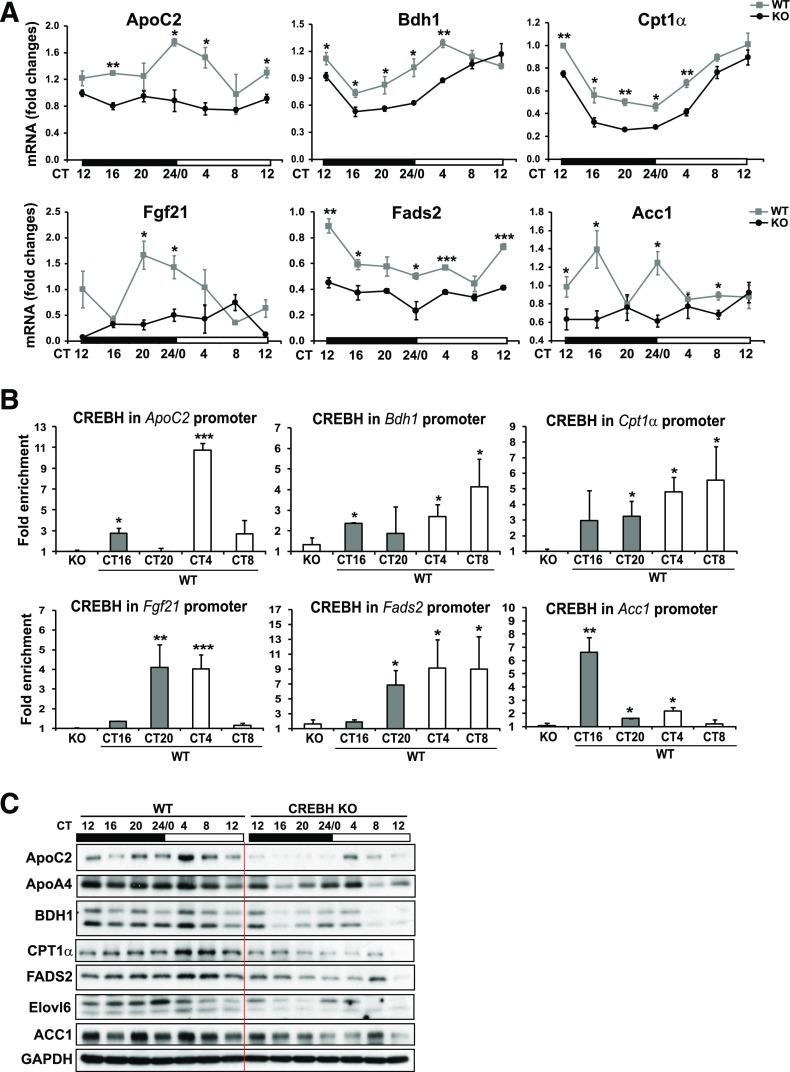
To understand the molecular basis underlying the lipid-associated phenotype of CREBH-null mice under the circadian clock, we examined whether CREBH as a transcription factor rhythmically regulates expression of the genes involved in hepatic lipid metabolism. qPCR indicated that rhythmic expression of the following genes was significantly repressed in CREBH-null mice (Fig. 6A and Supplementary Fig. 3): 1) the genes encoding the coactivators or enzymes of lipolysis, including apolipoprotein C2 (ApoC2), ApoA4, and FA desaturase 2 (Fads2); 2) the genes encoding the enzymes or regulators in FA oxidation, including carnitine palmitoyltransferase 1A (CPT1α), 3-hydroxybutyrate dehydrogenase 1 (BDH1), and fibroblast growth factor 21 (FGF21); and 3) the genes encoding the enzymes in lipogenesis, including acetyl-CoA carboxylase 1 (ACC1) and FA elongase 6 (Elvol6). Consistent with the mRNA expression profiles, levels of ApoC2, ApoA4, CPT1α, BDH1, Fads2, Elovl6, and ACC1 proteins were decreased in the livers of CREBH-null mice (Fig. 6C and Supplementary Fig. 4). Additionally, rhythmic expression of other key metabolic genes involved in lipolysis, FA oxidation, and lipogenesis, including Dhcr24, Lcat, Elvol2, Acot4, Hmgcl, and Dgat2, was repressed in CREBH-null mouse livers (Supplementary Fig. 3). These results support that CREBH functions as a critical regulator of circadian TG and FA metabolism by regulating expression of the key enzymes or regulators involved in lipolysis, FA oxidation, and de novo lipogenesis.
Figure 6.
CREBH regulates expression of the genes involved in lipolysis, FA oxidation, and lipogenesis. A: Rhythmic expression of CREBH target genes involved in lipolysis, FA oxidation, and lipogenesis in CREBH-null and WT mouse livers under the circadian clock. Levels of mRNA were determined by qPCR. Fold changes of mRNA levels were compared with that of one WT mouse at the starting circadian time point. Data are mean ± SEM (n = 3–5 mice/time point). B: CREBH enrichment in CREBH target gene promoters in WT mouse livers during various circadian phases determined by ChIP-qPCR. CREBH-null liver nuclei were used as negative control for the endogenous CREBH ChIP assays. Quantification of CREBH enrichment in the gene promoters at various circadian phases was determined by comparing ChIP-qPCR signals from the samples pulled down by the anti-CREBH antibody with that pulled down by a rabbit anti-IgG antibody. Data are mean ± SEM (n = 3 mice/time point). C: Circadian rhythmic levels of the proteins encoded by the CREBH target genes, including ApoC2, ApoA4, BDH1, CPT1α, FADS2, Elovl6, and ACC1, in the livers of CREBH-null and WT control mice. The liver tissue samples from CREBH-null and WT control mice were collected every 4 h in a 24-h circadian period. Pooled liver protein lysates from three to five mice per genotype group per time point were used for the Western blot analyses. Levels of GAPDH were determined as loading controls. The quantifications of the rhythmic fold changes of these proteins are shown in Supplementary Fig. 4. *P < 0.05, **P < 0.01, ***P < 0.001. KO, knockout.
To determine whether CREBH directly regulates its target genes involved in lipid metabolism during the circadian cycle, we performed ChIP-qPCR analysis to determine CREBH enrichment in the promoter regions of metabolic genes whose rhythmic expression profiles were altered in CREBH-null mouse livers. ChIP-qPCR analyses with WT mouse livers collected at various circadian phases indicated that CREBH binds in a circadian phase–dependent manner to the ApoC2, Bdh1, Cpt1a, Fgf21, Fads2, or Acc1 gene promoter that possesses one or multiple CRE-binding elements (Fig. 6B and Supplementary Fig. 5). Increased enrichment of CREBH in the ApoC2 gene promoter was detectable at CT16 and peaked at CT4, which is consistent with the rhythmic expression profile of the ApoC2 mRNA in the liver. Similarly, consistent with the mRNA expression profiles, the enrichment of CREBH in the promoters of the genes encoding functions in lipogenesis, including Fads2 and Acc1, reached peak levels at CT8 and CT16, respectively (Fig. 6B). Taken together, these results suggest that CREBH activates expression of genes involved in the metabolic pathways of both energy utilization (lipolysis and FA oxidation) and storage (lipogenesis), depending on the circadian periods.
CREBH Rhythmically Regulates and Interacts With the Nuclear Receptors Peroxisome Proliferator–Activated Receptor α and Liver X Receptor α Across the Circadian Cycle
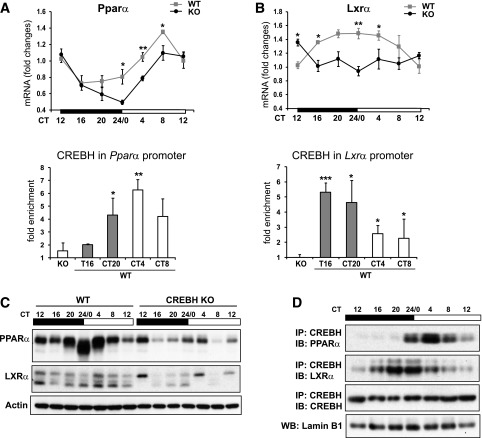
To elucidate whether CREBH regulates and/or interacts with other local circadian regulators, we first examined the expression of the genes involved in hepatic lipid metabolism in the livers of CREBH-null and WT control mice. Among others, peroxisome proliferator–activated receptor α (PPARα) is a liver-enriched, clock-regulated nuclear receptor that plays key roles in regulating lipid utilization pathways during the starvation phase (24). Liver X receptor α (LXRα) is a nuclear receptor that regulates de novo lipogenesis and lipid uptake (25). Rhythmic levels of the Pparα and Lxrα mRNA were significantly reduced in CREBH-null livers compared with those in WT control mice (Fig. 7A and B). ChIP-qPCR analysis indicated that CREBH binds in a day-night–dependent manner to the Pparα or Lxrα gene promoters in mouse livers (Fig. 7A and B), suggesting that Pparα and Lxrα are circadian-dependent targets of CREBH. Furthermore, Western blot analysis indicated that rhythmic levels of PPARα and LXRα proteins were decreased in CREBH-null livers (Fig. 7C and Supplementary Fig. 6A).
Figure 7.
CREBH rhythmically regulates and interacts with circadian transcription regulators involved in hepatic lipid metabolism. A and B: Rhythmic expression levels and CREBH enrichment in the promoters of the Pparα and Lxrα genes in the CREBH-null and WT mouse livers under the circadian clock. Fold changes of mRNA levels were determined by qPCR, and rhythmic enrichment of endogenous CREBH in the target gene promoters in WT mouse livers during various circadian phases were determined by ChIP-qPCR. Data are mean ± SEM (n = 3 mice/time point). C: Western blot analysis of rhythmic levels of PPARα and LXRα proteins in CREBH-null and WT mouse livers collected every 4 h in a 24-h circadian period. Pooled liver protein lysates from three to five mice per time point per genotype group were used. D: Immunoprecipitation (IP)-Western blot analysis of interactions between CREBH and PPARα or LXRα in the liver nuclear fractions of WT mice during various circadian phases of a 24-h circadian period. Liver nuclear proteins pooled from three WT mice per time point was pulled down by the anti-CREBH antibody and then probed with the PPARα, LXRα, or CREBH antibody. Levels of lamin B1 were determined as controls. The quantifications of protein rhythmic fold changes for C and D are shown in Supplementary Fig. 6. *P < 0.05, **P < 0.01, ***P < 0.001. IB, immunoblot; KO, knockout; WB, Western blot.
Circadian transcriptional regulators may interact with local transcription factors or nuclear receptors to control the circadian output of gene expression (26). Immunoprecipitation-Western blot analyses with the mouse livers collected across the day-night cycle indicated that CREBH interacts with PPARα and LXRα at the various circadian phases (Fig. 7D and Supplementary Fig. 6B). The interaction between CREBH and PPARα was detected from CT24/0 to CT12 and peaked at CT4, the day-time phases when lipolysis and FA oxidation are activated in mice upon energy demands. CREBH also interacted with LXRα, a nuclear receptor involved in lipogenesis and lipid uptake, during the circadian cycle (Fig. 7D and Supplementary Fig. 6B). The interaction between CREBH and LXRα peaked from CT20 to CT24/0, a nighttime period when lipogenesis is more active in mice. Apparently, the phases of the interactions among CREBH, PPARα, and LXRα correlate with the time of feeding in mice and support the dual functions of CREBH in energy utilization (lipolysis and FA oxidation) and storage (lipogenesis) during the day-night cycle.
The Circadian Output Activator DBP and the Repressor E4BP4 Interact With CREBH to Modulate CREBH Transcriptional Activities
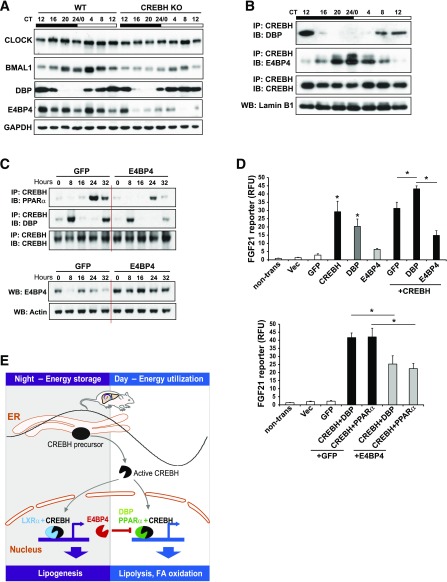
We next examined whether CREBH regulates or interacts with the core or output circadian oscillation regulators to modulate its transcriptional activity. CREBH deficiency resulted in marginally altered rhythmic expression of the genes encoding the core circadian oscillators, including Bmal1, Clock, Per2, and Rev-erbα, in mouse livers (Supplementary Fig. 7A). DBP, a direct target of CLOCK/BMAL1, functions as a circadian output transcriptional activator to activate expression of genes involved in energy metabolism (3,27). E4BP4 is a bZIP transcription factor that functions as a repressor of circadian oscillations (28). CREBH-null mice exhibited decreased expression of Dbp mRNA at CT8, whereas CREBH deficiency resulted in a phase-inversed expression pattern of E4bp4 mRNA in the liver (Supplementary Fig. 7B). Western blot analysis showed that the rhythmic levels of the circadian oscillator BMAL1 but not CLOCK in CREBH-null livers were modestly decreased compared with those in the WT livers (Fig. 8A and Supplementary Fig. 8A). Rhythmic expression of E4BP4 protein was repressed in CREBH-null livers compared with that in WT livers. Expression levels of DBP was not affected by CREBH deletion, but CREBH-null mice exhibited a phase shift in DBP expression during the circadian period from CT16 to CT4 (Fig. 8A).
Figure 8.
CREBH rhythmically interacts with the circadian activator DBP and repressor E4BP4 to modulate CREBH transcriptional activity. A: Western blot analysis of rhythmic levels of CLOCK, BMAL1, DBP, and E4BP4 proteins in CREBH-null and WT mouse livers collected every 4 h during a 24-h circadian period. Pooled liver protein lysates from three to five mice per time point per genotype were used. Levels of GAPDH were included as loading controls. B: Immunoprecipitation (IP)-Western blot analysis of interactions between CREBH and DBP or E4BP4 in the liver nuclear fractions of WT mice during various phases of a 24-h circadian period. Liver nuclear proteins pooled from three WT mice per time point were pulled down by the anti-CREBH antibody and then probed with the DBP or E4BP4 antibody. Levels of lamin B1 were determined as controls. C: IP-Western blot analysis of interactions among CREBH, PPARα, and DBP in mouse primary hepatocytes expressing E4BP4 or GFP control. Mouse primary hepatocytes were infected with adenovirus expressing E4BP4 or GFP followed by serum shock for circadian synchronization. Cell lysates collected after serum shock were pulled down by the anti-CREBH antibody and then probed with the PPARα, DBP, or CREBH antibody. The same protein samples were subjected to Western blot analyses of E4BP4 and β-actin levels. In A–C, the quantifications of protein rhythmic fold changes are shown in Supplementary Fig. 8. D: Luciferase reporter analyses of transcriptional activation of the human FGF21 gene promoter by CREBH alone or in combination with DBP, PPARα, and/or E4BP4. Hepa1-6 cells were transduced with the Fgf21 reporter vector or vehicle. After 24 h, the transfected cells were infected with adenovirus expressing GFP (control), CREBH, DBP, PPARα, and/or E4BP4. Renilla reporter plasmid was included in the cotransfection for normalization of luciferase reporter activities. The same amounts of adenovirus titers were used for individual infections. Data are mean ± SEM. *P < 0.05. E: A working model for CREBH function as a circadian metabolic regulator. IB, immunoblot; KO, knockout; non-trans, nontransfected; RFU, relative fluorescence unit; Vec, vector; WB, Western blot.
Furthermore, we examined the interactions among CREBH, DBP, and E4BP4 in mouse liver during the circadian cycle. The interaction between CREBH and DBP was detected at CT8 and CT12, whereas the interaction between CREBH and E4BP4 was detected from CT16 to CT8 (Fig. 8B and Supplementary Fig. 8B). Of note, robust interactions between CREBH and E4BP4 were detected at CT20 to CT4 when the interaction between CREBH and DBP was diminished (Fig. 8B). Overall, the phase and amplitudes of the CREBH-DBP interaction roughly oppose that of the CREBH-E4BP4 interaction, implying that DBP and E4BP4 may compete to interact with CREBH in a circadian phase–dependent manner. To verify the suppressive effects of E4BP4 on CREBH interaction with its partners, mouse primary hepatocytes were infected with the adenovirus expressing E4BP4 or GFP control followed by serum shock for circadian synchronization. Upon E4BP4 overexpression, the circadian-regulated interactions between CREBH and PPARα or DBP were repressed in the hepatocytes (Fig. 8C and Supplementary Fig. 8C), thus confirming that E4BP4, as a circadian output repressor, can suppress CREBH interactions with its partners under the circadian clock.
To explore the functional significance of the interactions between CREBH and DBP or E4BP4, we performed a reporter analysis with the Fgf21 gene promoter, a common target of CREBH and its interaction partner PPARα (8). Although overexpression of the activated CREBH or DBP alone can significantly increase Fgf21 gene promoter activity, coexpression of CREBH with DBP or PPARα further augmented the reporter activity (Fig. 8D). In contrast, coexpression of CREBH with E4BP4 significantly decreased Fgf21 promoter activity compared with expression of CREBH alone or coexpression of CREBH with GFP. These results suggest that DBP and PPARα function as coactivators of CREBH in driving Fgf21 gene transcription, whereas E4BP4 acts as a repressor of CREBH-dependent Fgf21 gene expression. Moreover, we observed that coexpression of E4BP4 with the combination of CREBH and DBP or PPARα repressed expression of the Fgf21 gene reporter compared with coexpression of CREBH with DBP or PPARα (Fig. 8D), thus supporting the suppressive effect of E4BP4 on CREBH transcriptional activity through competition with the coactivator DBP or PPARα. Given that the rhythmic expression of E4BP4 is regulated by CREBH (Fig. 8A), the repressive effect of E4BP4 on CREBH activity may represent a negative feedback regulation of CREBH activity under circadian constraint.
Discussion
In this study, we demonstrated that CREBH is a liver circadian oscillator that plays key roles in integrating energy metabolism with circadian rhythm (Fig. 8E). This study revealed that proteolytic cleavage of CREBH, but not CrebH mRNA transcription, exhibits typical circadian rhythmicity (Fig. 1), suggesting that CREBH is an output circadian factor that is regulated by the clock at the posttranslational level. This finding is consistent with other rhythmic proteome studies showing that approximately one-half of rhythmic proteins are under significant translational or posttranslational diurnal control and have no corresponding rhythmic mRNAs (29,30). The current finding that BMAL1, the core circadian oscillator, regulates cleavage/activation of CREBH through AKT-GSK3β signaling implicated a major molecular network through which BMAL1 regulates circadian energy metabolism in the liver. Importantly, GSK3β-mediated phosphorylation of ER-bound CREBH, which is under the control of the Bmal1-AKT regulatory axis, is a critical regulatory event for the ER-to-Golgi transport and subsequent proteolytic cleavage of CREBH (Figs. 3 and 4). Upon GSK3β-mediated phosphorylation, CREBH protein exhibits a decreased affinity to complex with Sec23/24, the core protein components of the ER-to-Golgi transport vesicle COPII, under the circadian clock. It is possible that phosphorylation of CREBH by GSK3β leads to altered CREBH conformation with a resulting decreased affinity toward the COPII-coated transport complex. To assemble the COPII complex, Sec23 and Sec24 form a heterodimer where Sec24 is mainly responsible for cargo recognition and Sec23 binds Sar1-GTP (21). CREBH may indirectly interact with Sec24 through a potential scaffold protein like the SREBP escort protein SCAP (31), an interesting question to be studied in the future.
The current study demonstrates that CREBH as a hepatic transcription factor is required to maintain homeostasis of circulating TG and FA levels by regulating expression of the key enzymes or regulators of lipolysis, FA oxidation, and de novo lipogenesis in a circadian-dependent fashion. Because CREBH is required for expression of ApoC2, ApoA4, Fads2, Dhcr24, and Lcat (Fig. 6 and Supplementary Fig. 3), all of which play important roles in TG lipolysis, CREBH-null mice have the defect in the clearance of TG-rich lipoproteins from the circulation and therefore display hypertriglyceridemia. Moreover, reduced expression of the regulators or enzymes in FA metabolism, including FGF21, CPT1α, BDH1, Fads, Elovls, and PPARα, could be partially responsible for elevated serum FA levels in CREBH-null mice. Of note, during the nighttime, CREBH regulates expression of the genes encoding the functions in hepatic de novo lipogenesis, including Acc1, Elovl2, Elovl6, Lxrα, Hmgc1, and Dgat (Figs. 6 and 7 and Supplementary Fig. 3). This is consistent with the observation that CREBH-null mice preserved reduced levels of hepatic lipids in the nighttime phase when they eat most of the meal of the day (Fig. 5C). The circadian-dependent CREBH-regulated gene expression profiles suggest that CREBH regulates multiple metabolic pathways involved in both energy utilization and storage across the circadian cycle.
CREBH has reciprocal interactions with the circadian transcriptional regulators PPARα and LXRα as well as the circadian oscillation activator DBP and repressor E4BP4 (Figs. 7 and 8). CREBH regulates and interacts with PPARα or LXRα to enhance CREBH transcriptional activity, which oscillates in phase with expression of the CREBH target genes involved in lipolysis, FA oxidation, and lipogenesis. On the other hand, CREBH interacts with DBP or E4BP4 to modulate CREBH transcriptional activity during the night-to-day transition period. Of note, the phase of CREBH-DBP interaction is complementary to that of CREBH-E4BP4 interaction, suggesting that DBP and E4BP4 may compete to interact with CREBH and thereby modulate CREBH activities during various circadian phases. As a coactivator of CREBH, PPARα interacts with CREBH in the circadian phase that partially overlaps with the CREBH-LXRα interaction (Fig. 7D). The interactions among CREBH, PPARα, and LXRα may represent enhancing mechanisms that facilitate CREBH peak activity.
In summary, the core circadian oscillation regulates CREBH activity through two layers (Fig. 8E): 1) The core circadian oscillator BMAL1 regulates proteolytic activation of CREBH, and 2) the output circadian modulators E4BP4 or DBP interact with the activated CREBH protein to exert a suppressive or synergizing effect on CREBH activity. Conversely, CREBH also regulates expression of the core or output circadian components, including BMAL1, DBP, and E4BP4. The reciprocal regulation between CREBH and the key circadian regulators may provide an avenue through which local and central circadian regulators are integrated to influence whole-body physiology. In the future, testing whether modulation of CREBH activity is an effective approach to metabolic disorder intervention will be important.
Article Information
Acknowledgments. The authors thank Tianqing Peng of University of Western Ontario, Canada, for providing the recombinant adenovirus expressing GSK3β and Xiao-Wei Chen and David Ginsburg of the University of Michigan for providing the anti-Sec24A antibody. The authors also thank Todd Leff of Wayne State University and Kenji Fukudome of Saga University, Japan, for critical comments on this work.
Funding. Portions of this work were supported by National Institute of Diabetes and Digestive and Kidney Diseases grant DK-090313, National Institute of Environmental Health Sciences grant ES-017829 (to K.Z.), and American Heart Association grants 0635423Z and 09GRNT2280479 (to K.Z.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. Z.Z. contributed to the study concept and design, data acquisition, data analysis and interpretation, and writing and revision of the manuscript. H.K. contributed to the study concept and design, data acquisition, and data analysis and interpretation. Y.Q., R.M., A.D., X.Z., and C.Z. contributed to the data acquisition, analysis, and interpretation. X.C. contributed technical or material support. A.C.L., L.Y., J.D.L., and P.D.W. contributed technical or material support and to the data acquisition, analysis, and interpretation. G.K. contributed technical or material support and to the data acquisition, data analysis and interpretation, and writing and revision of the manuscript. K.Z. contributed to the study concept and design, obtaining of funding, technical or material support, study supervision, data acquisition, data analysis and interpretation, and writing and revision of the manuscript. K.Z. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db16-0298/-/DC1.
References
- 1.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet 2004;5:407–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol 2001;63:647–676 [DOI] [PubMed] [Google Scholar]
- 3.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science 2010;330:1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatori M, Vollmers C, Zarrinpar A, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab 2012;15:848–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang X, Downes M, Yu RT, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell 2006;126:801–810 [DOI] [PubMed] [Google Scholar]
- 6.Zhang C, Wang G, Zheng Z, et al. Endoplasmic reticulum-tethered transcription factor cAMP responsive element-binding protein, hepatocyte specific, regulates hepatic lipogenesis, fatty acid oxidation, and lipolysis upon metabolic stress in mice. Hepatology 2012;55:1070–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang K, Shen X, Wu J, et al. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 2006;124:587–599 [DOI] [PubMed] [Google Scholar]
- 8.Kim H, Mendez R, Zheng Z, et al. Liver-enriched transcription factor CREBH interacts with peroxisome proliferator-activated receptor α to regulate metabolic hormone FGF21. Endocrinology 2014;155:769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JH, Giannikopoulos P, Duncan SA, et al. The transcription factor cyclic AMP-responsive element-binding protein H regulates triglyceride metabolism. Nat Med 2011;17:812–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee MW, Chanda D, Yang J, et al. Regulation of hepatic gluconeogenesis by an ER-bound transcription factor, CREBH. Cell Metab 2010;11:331–339 [DOI] [PubMed] [Google Scholar]
- 11.Johansen CT, Wang J, McIntyre AD, et al. Excess of rare variants in non-genome-wide association study candidate genes in patients with hypertriglyceridemia. Circ Cardiovasc Genet 2012;5:66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cefalù AB, Spina R, Noto D, et al. Novel CREB3L3 nonsense mutation in a family with dominant hypertriglyceridemia. Arterioscler Thromb Vasc Biol 2015;35:2694–2699 [DOI] [PubMed] [Google Scholar]
- 13.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 1998;93:929–937 [DOI] [PubMed] [Google Scholar]
- 14.Kapatos G, Vunnava P, Wu Y. Protein kinase A-dependent recruitment of RNA polymerase II, C/EBP beta and NF-Y to the rat GTP cyclohydrolase I proximal promoter occurs without alterations in histone acetylation. J Neurochem 2007;101:1119–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molusky MM, Ma D, Buelow K, Yin L, Lin JD. Peroxisomal localization and circadian regulation of ubiquitin-specific protease 2. PLoS One 2012;7:e47970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang D, Tong X, Arthurs B, et al. Liver clock protein BMAL1 promotes de novo lipogenesis through insulin-mTORC2-AKT signaling. J Biol Chem 2014;289:25925–25935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001;414:799–806 [DOI] [PubMed] [Google Scholar]
- 18.Rudic RD, McNamara P, Curtis AM, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol 2004;2:e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi SQ, Ansari TS, McGuinness OP, Wasserman DH, Johnson CH. Circadian disruption leads to insulin resistance and obesity. Curr Biol 2013;23:372–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahar S, Zocchi L, Kinoshita C, Borrelli E, Sassone-Corsi P. Regulation of BMAL1 protein stability and circadian function by GSK3beta-mediated phosphorylation. PLoS One 2010;5:e8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee MC, Miller EA. Molecular mechanisms of COPII vesicle formation. Semin Cell Dev Biol 2007;18:424–434 [DOI] [PubMed] [Google Scholar]
- 22.Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. COPII and the regulation of protein sorting in mammals. Nat Cell Biol 2011;14:20–28 [DOI] [PubMed] [Google Scholar]
- 23.Eldar-Finkelman H, Argast GM, Foord O, Fischer EH, Krebs EG. Expression and characterization of glycogen synthase kinase-3 mutants and their effect on glycogen synthase activity in intact cells. Proc Natl Acad Sci U S A 1996;93:10228–10233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oishi K, Shirai H, Ishida N. CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor alpha (PPARalpha) in mice. Biochem J 2005;386:575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev 1995;9:1033–1045 [DOI] [PubMed] [Google Scholar]
- 26.Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev 2010;24:345–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamaguchi S, Mitsui S, Yan L, Yagita K, Miyake S, Okamura H. Role of DBP in the circadian oscillatory mechanism. Mol Cell Biol 2000;20:4773–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clayton JD, Kyriacou CP, Reppert SM. Keeping time with the human genome. Nature 2001;409:829–831 [DOI] [PubMed] [Google Scholar]
- 29.Reddy AB, Karp NA, Maywood ES, et al. Circadian orchestration of the hepatic proteome. Curr Biol 2006;16:1107–1115 [DOI] [PubMed] [Google Scholar]
- 30.Mauvoisin D, Wang J, Jouffe C, et al. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc Natl Acad Sci U S A 2014;111:167–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun LP, Seemann J, Goldstein JL, Brown MS. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc Natl Acad Sci U S A 2007;104:6519–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]