Abstract
Green fluorescent protein (GFP), which was originally isolated from jellyfish, is a widely used tool in biological research, and homologs from other organisms are available. However, researchers must determine which GFP is the most suitable for a specific host. Here, we expressed GFPs from several sources in codon-optimized and non-codon-optimized forms in the yeast Saccharomyces cerevisiae, which represents an ideal eukaryotic model. Surprisingly, codon-optimized mWasabi and mNeonGreen, which are typically the brightest GFPs, emitted less green fluorescence than did the other five codon-optimized GFPs tested in S. cerevisiae. Further, commercially available GFPs that have been optimized for mammalian codon usage (e.g., EGFP, AcGFP1 and TagGFP2) unexpectedly exhibited extremely low expression levels in S. cerevisiae. In contrast, codon-optimization of the GFPs for S. cerevisiae markedly increased their expression levels, and the fluorescence intensity of the cells increased by a maximum of 101-fold. Among the tested GFPs, the codon-optimized monomeric mUkG1 from soft coral showed the highest levels of both expression and fluorescence. Finally, the expression of this protein as a fusion-tagged protein successfully improved the reporting system’s ability to sense signal transduction and protein–protein interactions in S. cerevisiae and increased the detection rates of target cells using flow cytometry.
Green fluorescent protein (GFP) was discovered more than 50 years ago1 and has fueled a new era in cell and molecular biology. GFP comprises 238 amino acid residues (approximately 27 kDa) with a β-barrel structure consisting of eleven β-strands, and it emits green fluorescence when exposed to blue light2. GFP requires no external cofactors other than oxygen to form the chromophore, unlike other proteins that require cofactors or substrates for their activities (e.g., opsin, β-glucuronidase, β-galactosidase, chloramphenicol acetyl-transferase, and luciferase)3. Therefore, GFP represents an excellent reporter gene and fusion tag for the analysis of gene regulation, protein localization, and specific organelle labeling in almost every organism4,5,6,7,8.
The budding yeast Saccharomyces cerevisiae is an ideal eukaryotic model organism for studying protein localization due to the ease and tractability of its genetic modification7. Recent studies have applied this tool widely, for example in genome-wide approaches to analyze messenger RNA (mRNA) abundance9,10, transcriptional regulation11, protein abundance or localization3,12,13, and protein–protein interactions14,15. In many cases, these studies used GFP or other chromophoric proteins for the facile tracking and monitoring of target proteins. Further, the use of flow cytometry has accelerated the screening of target cells. GFP has been widely applied to the yeast S. cerevisiae, thus rendering it a useful tool for a wide variety of biological studies.
Several varieties of GFPs were tested in this study, and their properties are listed in Table 1. The GFP that was first isolated from jellyfish Aequorea victoria forms a weakly associated dimer that exhibits a low molar extinction coefficient (ε) and two excitation maxima (395 nm and 475 nm)16,17,18. Enhanced GFP (EGFP), a mutant of the original GFP that was developed through mutagenesis studies, is among the most popular and widely used monomeric GFPs, and it exhibits a single excitation peak at 488 nm19 (Table 1). EGFP contains codons that were optimized for mammalian cell expression and S65T and F64L mutations that improve the spectral characteristics, fluorescence intensity and stability of the protein and increased protein maturation efficiency at 37 °C19,20.
Table 1. Properties of green fluorescent proteins tested.
| Protein | λex (nm) | λem (nm) | EC (ε) (M−1cm−1) | QY | RB (% of EGFP) | Association state | Reference or source |
|---|---|---|---|---|---|---|---|
| EGFP | 488 | 507 | 56,000 | 0.60 | 100 | Monomer | 18 |
| AcGFP1 | 475 | 505 | 32,500 | 0.82 | 79 | Monomer | Clontech |
| TagGFP2 | 483 | 506 | 56,500 | 0.60 | 101 | Monomer | Evrogen |
| mUkG1 | 483 | 499 | 60,000 | 0.72 | 129 | Monomer | 26 |
| ZsGreen | 493 | 505 | 43,000 | 0.91 | 116 | Tetramer | 27 |
| mWasabi | 493 | 509 | 70,000 | 0.80 | 167 | Monomer | 29 |
| mNeonGreen | 506 | 517 | 116,000 | 0.80 | 276 | Monomer | 24 |
Along with the common name for each GFP, the peak excitation (λex) and emission (λem) wavelengths, molar extinction coefficient (EC), quantum yield (QY), relative brightness (RB), association state and reference or source are listed. Brightness values were derived from the product of EC and QY, divided by the value for EGFP.
Recently, GFP variants have been discovered in a wide range of sources, including various Aequorea species21,22, copepods23, amphioxus24,25 and reef corals26,27,28,29, and these variants are now commercially available. For example, AcGFP1, a brilliant green monomeric GFP variant isolated from Aequorea coerulescens, exhibits 94% sequence homology to EGFP21 (Table 1). The monomeric and pH-stable GFP, TagGFP2 (mTagGFP), which is the improved variant of TagGFP derived from Aequorea macrodactyla, exhibits 80% sequence homology to EGFP22 (Table 1). The coral-derived GFPs mUkG126, ZsGreen27 and mWasabi29, bear surprisingly low sequence similarity to the GFPs that were originally identified in Aequorea species. Monomeric mUkG1 emits intense fluorescence and is extremely stable against high pH, owing to the introduction of five mutations to the dimeric wild-type fluorescent protein UkG1, which was isolated from the soft coral Sarcophyton sp.26 (Table 1). Tetrameric ZsGreen is a Zoanthus sp. fluorescent protein variant with a single amino acid substitution that enhances the emission characteristics; the protein has a substantially higher quantum yield (QY)27 (Table 1). The bright, monomeric green fluorescent protein mWasabi is a variant of monomeric mTFP1, which is derived from the tetrameric cyan fluorescent protein cFP484 of Clavularia sp.29 (Table 1). mNeonGreen is a monomeric yellow-green fluorescent protein that emits the brightest green or yellow fluorescence reported to date; this protein was derived from the tetrameric yellow fluorescent protein (LanYFP) of the cephalochordate Branchiostoma lanceolatum by structure-guided directed evolution24 (Table 1).
Thus, a wide range of GFPs are now available. These proteins generating green fluorescence are crucially important as versatile experimental tools, because of their compatibilities for varied fluorometric instruments. Codon optimization has reported to increase the expression levels of several fluorescent proteins in S. cerevisiae30,31,32. However, the subjects of investigation were limited to the EGFP or other major color fluorescent proteins, and the choice of a suitable GFP for expression in S. cerevisiae is less well studied among a wide variety of GFPs.
Here, we expressed seven codon-optimized GFPs and five non-codon-optimized GFPs in S. cerevisiae and compared the protein expression levels and green fluorescence intensities. Through the evaluations, we determined the nucleic acid sequences of the codon-optimized GFPs that were expressed at high levels and that emitted intense green fluorescence in S. cerevisiae. Furthermore, we tested these GFPs that were expressed at relatively high levels for use as reporter genes in fusion form for detecting signal transduction and protein–protein interactions. Finally, we demonstrated the codon-optimized mUkG1 fusion reporter successfully increased the detection rates of target cells using flow cytometry.
Results and Discussion
Expression of GFPs in S. cerevisiae
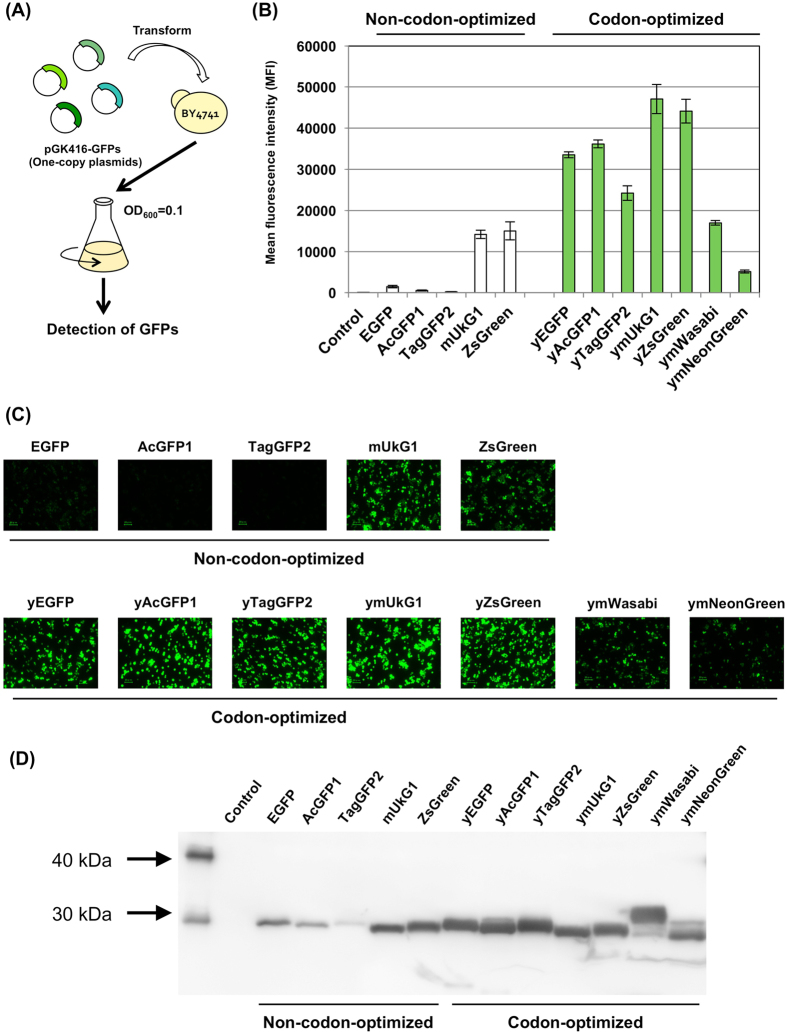
We compared the expression of seven GFPs from various sources in S. cerevisiae: EGFP, AcGFP1, TagGFP2, mUkG1, ZsGreen, mWasabi and mNeonGreen (Table 1). The genes encoding the seven GFPs were optimized for expression in the yeast S. cerevisiae (yEGFP, yAcGFP1, yTagGFP2, ymUkG1, yZsGreen, ymWasabi and ymNeonGreen) (Supplementary Table S1) and inserted into the multiple-cloning site of a pGK416 single-copy autonomous replicating plasmid for constitutive expression under the control of the PGK1 promoter33 (Fig. 1A and Table 2). The yeast BY4741 strain was transformed with the constructed plasmids (Tables 2 and 3 and Supplementary Table S2), and their green fluorescence was evaluated using flow cytometry and fluorescence microscopy (Fig. 1 and Supplementary Table S3).
Figure 1. GFP expression in S. cerevisiae.
(A) Flow diagram of GFP detection assays. GFP expression plasmids were transformed into the BY4741 yeast strain; the cells were then grown, and GFP expression was analyzed. (B) Mean GFP fluorescence intensities (MFIs). The MFIs of 10,000 cells were measured by flow cytometry. (C) Green fluorescence images of the cells. The images were acquired with a fluorescence microscope equipped with a 60× objective lens. Scale bar: 20 μm. The exposure time was 1/15 s. (D) Western blot analysis. Western blot analysis was performed using as the primary antibody monoclonal anti-frag M2 antibody for the GFPs fused frag tag. Alkaline phosphatase-conjugated anti-mouse IgG was used as the secondary antibody, and colorimetric detection of alkaline phosphatase activity was performed using CDP-Star detection reagent. ‘Control’ indicates the BY4741 yeast strain harboring a mock pGK416 plasmid.
Table 2. List of plasmids used in this study.
| Plasmids | Genotype | Reference or source |
|---|---|---|
| pGK416 | Expression vector containing PGK1 promoter, CEN/ARS single-copy origin and URA3 marker | 33 |
| pGK416-EGFP | EGFP expression, in pGK416 | 33 |
| pGK416-AcGFP1 | AcGFP1 expression, in pGK416 | This study |
| pGK416-TagGFP2 | TagGFP2 expression, in pGK416 | This study |
| pGK416-mUkG1 | mUkG1 expression, in pGK416 | This study |
| pGK416-ZsGreen | ZsGreen expression, in pGK416 | 34 |
| pGK416-yEGFP | yEGFP (codon-optimized EGFP) expression, in pGK416 | This study |
| pGK416-yAcGFP1 | yAcGFP1 (codon-optimized AcGFP1) expression, in pGK416 | This study |
| pGK416-yTagGFP2 | yTagGFP2 (codon-optimized TagGFP2) expression, in pGK416 | This study |
| pGK416-ymUkG1 | ymUkG1 (codon-optimized mUkG1) expression, in pGK416 | This study |
| pGK416-yZsGreen | yZsGreen (codon-optimized ZsGreen) expression, in pGK416 | This study |
| pGK416-ymWasabi | ymWasabi (codon-optimized mWasabi) expression, in pGK416 | This study |
| pGK416-ymNeonGreen | ymNeonGreen (codon-optimized mNeonGreen) expression, in pGK416 | This study |
| pGK416-EGFP-F | EGFP-FLAG expression, in pGK416 | This study |
| pGK416-AcGFP1-F | AcGFP1-FLAG expression, in pGK416 | This study |
| pGK416-TagGFP2-F | TagGFP2-FLAG expression, in pGK416 | This study |
| pGK416-mUkG1-F | mUkG1-FLAG expression, in pGK416 | This study |
| pGK416-ZsGreen-F | ZsGreen-FLAG expression, in pGK416 | This study |
| pGK416-yEGFP-F | yEGFP (codon-optimized EGFP) -FLAG expression, in pGK416 | This study |
| pGK416-yAcGFP1-F | yAcGFP1 (codon-optimized AcGFP1) -FLAG expression, in pGK416 | This study |
| pGK416-yTagGFP2-F | yTagGFP2 (codon-optimized TagGFP2) -FLAG expression, in pGK416 | This study |
| pGK416-ymUkG1-F | ymUkG1 (codon-optimized mUkG1) -FLAG expression, in pGK416 | This study |
| pGK416-yZsGreen-F | yZsGreen (codon-optimized ZsGreen)-FLAG expression, in pGK416 | This study |
| pGK416-ymWasabi-F | ymWasabi (codon-optimized mWasabi)-FLAG expression, in pGK416 | This study |
| pGK416-ymNeonGreen-F | ymNeonGreen (codon-optimized mNeonGreen)-FLAG expression, in pGK416 | This study |
| pBlueScript II KS(+) | Cloning vector | Agilent Technologies |
| pBlue-FIG1pt-ZsGreen | PFIG1 (300 bp)-URA3-ZsGreen-TFIG1 (200 bp) in pBlueScript II KS(+) | 34 |
| pBlue-FIG1t | TFIG1 (200 bp) in pBlueScript II KS(+) | This study |
| pBlue-UFt-AcGFP1 | C-terminus of FIG1 (50 bp)-AcGFP1-URA3-TFIG1 (200 bp) in pBlueScript II KS(+) | This study |
| pBlue-UFt-TagGFP2 | C-terminus of FIG1 (50 bp)-TagGFP2-URA3-TFIG1 (200 bp) in pBlueScript II KS(+) | This study |
| pBlue-UFt-mUkG1 | C-terminus of FIG1 (50 bp)-mUkG1-URA3-TFIG1 (200 bp) in pBlueScript II KS(+) | This study |
| pBlue-UFt-ZsGreen | C-terminus of FIG1 (50 bp)-ZsGreen-URA3-TFIG1 (200 bp) in pBlueScript II KS(+) | This study |
| pBlue-UFt-yEGFP | C-terminus of FIG1 (50 bp)-yEGFP-URA3-TFIG1 (200 bp) in pBlueScript II KS(+) | This study |
| pBlue-UFt-yAcGFP1 | C-terminus of FIG1 (50 bp)-yAcGFP1-URA3-TFIG1 (200 bp) in pBlueScript II KS(+) | This study |
| pBlue-UFt-yTagGFP2 | C-terminus of FIG1 (50 bp)-yTagGFP2-URA3-TFIG1 (200 bp) in pBlueScript II KS(+) | This study |
| pBlue-UFt-ymUkG1 | C-terminus of FIG1 (50 bp)-ymUkG1-URA3-TFIG1 (200 bp) in pBlueScript II KS(+) | This study |
| pBlue-UFt-yZsGreen | C-terminus of FIG1 (50 bp)-yZsGreen-URA3-TFIG1 (200 bp) in pBlueScript II KS(+) | This study |
| pGK426 | Expression vector containing PGK1 promoter, 2 μ origin and URA3 marker | 33 |
| pGK426-GPTK | URA3-PSTE18-kanMX4-TSTE18 in pGK426 | 42 |
| pUSTE18p-Gγcyto-HIS3t | URA3-PSTE18-Gγcyto-TPGK1-THIS3 in pGK426 | 45 |
| pUMGP-GγMFcH | URA3-PSTE18-Gγcyto-Fc-TPGK1 in pUSTE18p-Gγcyto-HIS3t | 42 |
| pGK413 | Expression vector containing PGK1 promoter, CEN/ARS single-copy origin and HIS3 marker | 33 |
| pGK413-ZWTmem | ZWT and C-terminus of Ste18 (9 a.a.) fusion expression, in pGK413 | 40 |
| pGK413-ZK35Amem | ZK35A and C-terminus of Ste18 (9 a.a.) fusion expression, in pGK413 | 40 |
| pGK413-ZI31Amem | ZI31A and C-terminus of Ste18 (9 a.a.) fusion expression, in pGK413 | 40 |
| pGK413-Z955mem | Z955 and C-terminus of Ste18 (9 a.a.) fusion expression, in pGK413 | 40 |
| pGK426-GPTK | URA3-PSTE18-kanMX4-TSTE18 in pGK426 | 42 |
| pUMGPTK-Gpa1N-Fc | URA3-PSTE18-PPGK1-Gpa1N (9 a.a.)-Fc-TPGK1-kanMX4-TSTE18 in pGK426-GPTK | 45 |
| pUMGPTK-Fc-Ste18C | URA3-PSTE18-PPGK1-Fc-Ste18C (9 a.a.)-TPGK1-kanMX4-TSTE18 in pGK426-GPTK | 45 |
| pUSTE18p-Gγcyto-HIS3t | URA3-PSTE18-Gγcyto-TPGK1-THIS3 in pGK426 | 45 |
| pUSTE18p-Gγcyto-ZWT-H | URA3-PSTE18-Gγcyto-ZWT-TPGK1 in pUSTE18p-Gγcyto-HIS3t | 45 |
| pUSTE18p-Gγcyto-ZK35A-H | URA3-PSTE18-Gγcyto-ZK35A-TPGK1 in pUSTE18p-Gγcyto-HIS3t | 45 |
| pUSTE18p-Gγcyto-ZI31A-H | URA3-PSTE18-Gγcyto-ZI31A-TPGK1 in pUSTE18p-Gγcyto-HIS3t | 45 |
| pUSTE18p-Gγcyto-Z955-H | URA3-PSTE18-Gγcyto-Z955-TPGK1 in pUSTE18p-Gγcyto-HIS3t | 45 |
Table 3. Yeast strains used in this study.
| Strain | Relevant feature | Reference |
|---|---|---|
| BY4741 | MATa his3Δ1 ura3Δ0 leu2Δ0 met15Δ0 | 51 |
| BY4742 | MATα his3Δ1 ura3Δ0 leu2Δ0 lys2Δ0 | 51 |
| MC-F1 | BY4741 fig1::FIG1-EGFP | 44 |
| BYFAG1 | BY4741 fig1::FIG1-AcGFP1 | This study |
| BYFTG1 | BY4741 fig1::FIG1-TagGFP2 | This study |
| BYFUG1 | BY4741 fig1::FIG1-mUkG1 | This study |
| BYFZG1 | BY4741 fig1::FIG1-ZsGreen | This study |
| BYFEG2 | BY4741 fig1::FIG1-yEGFP (codon optimized EGFP) | This study |
| BYFAG2 | BY4741 fig1::FIG1-yAcGFP1 (codon optimized AcGFP1) | This study |
| BYFTG2 | BY4741 fig1::FIG1-yTagGFP2 (codon optimized TagGFP2) | This study |
| BYFUG2 | BY4741 fig1::FIG1-ymUkG1 (codon optimized mUkG1) | This study |
| BYFZG2 | BY4741 fig1::FIG1-yZsGreen (codon optimized ZsGreen) | This study |
| BFG2118 | MC-F1 ste18Δ::kanMX4 his3Δ::URA3-PSTE18-Gγcyto-Fc | 42 |
| UGW2 | BYFUG2 ste18Δ::kanMX4 | This study |
| UGFG2 | BYFUG2 ste18Δ::kanMX4 his3Δ::URA3-PSTE18-Gγcyto-Fc | This study |
| MC-FC | MC-F1 ste18Δ::kanMX4-PPGK1-Fc-Ste18C | 45 |
| MC-FN | MC-F1 ste18Δ::kanMX4-PPGK1-Gpa1N-Fc | 45 |
| FC-GW | MC-F1 ste18Δ::kanMX4-PPGK1-Fc-Ste18C his3Δ::URA3-PSTE18-Gγcyto-ZWT | 45 |
| FC-GK | MC-F1 ste18Δ::kanMX4-PPGK1-Fc-Ste18C his3Δ::URA3-PSTE18-Gγcyto-ZK35A | 45 |
| FC-GI | MC-F1 ste18Δ::kanMX4-PPGK1-Fc-Ste18C his3Δ::URA3-PSTE18-Gγcyto-ZI31A | 45 |
| FC-G9 | MC-F1 ste18Δ::kanMX4-PPGK1-Fc-Ste18C his3Δ::URA3-PSTE18-Gγcyto-Z955 | 45 |
| FC-G0 | MC-F1 ste18Δ::kanMX4-PPGK1-Fc-Ste18C his3Δ::URA3 | 45 |
| FN-GW | MC-F1 ste18Δ::kanMX4-PPGK1-Gpa1N-Fc his3Δ::URA3-PSTE18-Gγcyto-ZWT | 45 |
| FN-GK | MC-F1 ste18Δ::kanMX4-PPGK1-Gpa1N-Fc his3Δ::URA3-PSTE18-Gγcyto-ZK35A | 45 |
| FN-GI | MC-F1 ste18Δ::kanMX4-PPGK1-Gpa1N-Fc his3Δ::URA3-PSTE18-Gγcyto-ZI31A | 45 |
| FN-G9 | MC-F1 ste18Δ::kanMX4-PPGK1-Gpa1N-Fc his3Δ::URA3-PSTE18-Gγcyto-Z955 | 45 |
| FN-G0 | MC-F1 ste18Δ::kanMX4-PPGK1-Gpa1N-Fc his3Δ::URA3 | 45 |
| BYFUG2-FC | BYFUG2 ste18Δ::kanMX4-PPGK1-Fc-Ste18C | This study |
| BYFUG2-FN | BYFUG2 ste18Δ::kanMX4-PPGK1-Gpa1N-Fc | This study |
| UG2-FCGW | BYFUG2 ste18Δ::kanMX4-PPGK1-Fc-Ste18C his3Δ::URA3-PSTE18-Gγcyto-ZWT | This study |
| UG2-FCGK | BYFUG2 ste18Δ::kanMX4-PPGK1-Fc-Ste18C his3Δ::URA3-PSTE18-Gγcyto-ZK35A | This study |
| UG2-FCGI | BYFUG2 ste18Δ::kanMX4-PPGK1-Fc-Ste18C his3Δ::URA3-PSTE18-Gγcyto-ZI31A | This study |
| UG2-FCG9 | BYFUG2 ste18Δ::kanMX4-PPGK1-Fc-Ste18C his3Δ::URA3-PSTE18-Gγcyto-Z955 | This study |
| UG2-FCG0 | BYFUG2 ste18Δ::kanMX4-PPGK1-Fc-Ste18C his3Δ::URA3 | This study |
| UG2-FNGW | BYFUG2 ste18Δ::kanMX4-PPGK1-Gpa1N-Fc his3Δ::URA3-PSTE18-Gγcyto-ZWT | This study |
| UG2-FNGK | BYFUG2 ste18Δ::kanMX4-PPGK1-Gpa1N-Fc his3Δ::URA3-PSTE18-Gγcyto-ZK35A | This study |
| UG2-FNGI | BYFUG2 ste18Δ::kanMX4-PPGK1-Gpa1N-Fc his3Δ::URA3-PSTE18-Gγcyto-ZI31A | This study |
| UG2-FNG9 | BYFUG2 ste18Δ::kanMX4-PPGK1-Gpa1N-Fc his3Δ::URA3-PSTE18-Gγcyto-Z955 | This study |
| UG2-FNG0 | BYFUG2 ste18Δ::kanMX4-PPGK1-Gpa1N-Fc his3Δ::URA3 | This study |
The green fluorescence intensities of 10,000 cells were quantitatively measured using flow cytometry, and the values were recorded as the mean fluorescence intensities (MFIs). Monomeric ymUkG1 and tetrameric yZsGreen exhibited the highest fluorescence intensities over 40,000–45,000 MFIs (Fig. 1B and Supplementary Table S3). In contrast, ymWasabi and ymNeonGreen exhibited remarkably low fluorescence intensities (less than 20,000 MFIs) (Fig. 1B and Supplementary Table S3), even though both GFPs are the brightest green fluorescent proteins (mWasabi, ε = 70,000 M−1cm−1, QY = 0.80; mNeonGreen, ε = 116,000 M−1cm−1, QY = 0.80)24,29 (Table 1). Fluorescence microscopy observations also showed that the green fluorescence of these two proteins was less intense than that of the other five GFPs (Fig. 1C). Therefore, we decided to use GFPs other than mWasabi and mNeonGreen (EGFP, AcGFP1, TagGFP2, mUkG1 and ZsGreen) in the later experiments.
Next, we compared the expression of the codon-optimized sequences and the commercially available (non-optimized for S. cerevisiae) sequences of five GFPs (Table 2 and Supplementary Table S2) by flow cytometry, fluorescence microscopy and western blot analysis (Fig. 1B–D). Of the commercially available GFP sequences tested, EGFP, AcGFP1 and TagGFP2 include codons that were optimized for mammalian cell expression, and ZsGreen and mUkG1 used the codons from the isolated, wild type organism. The flow cytometry and fluorescence microscopy observations for codon-optimized GFPs are shown in Fig. 1B,C. mUkG1 and ZsGreen (non-codon-optimized), including the natural codons of the soft coral Sarcophyton sp. and the button polyp coral Zoanthus sp., exhibited good fluorescence intensities (mUkG1, MFI = 14,194; ZsGreen, MFI = 15,019) (Fig. 1B and Supplementary Table S3). The MFIs of codon-optimized GFPs (ymUkG1, MFI = 47,088; yZsGreen, MFI = 44,154) were almost three-fold greater than those of non-codon-optimized sequences (Fig. 1B, Supplementary Table S3 and Fig. S1). In contrast, EGFP, AcGFP1 and TagGFP2, which were not codon-optimized for S. cerevisiae (these were optimized for mammalian cells) unexpectedly exhibited considerably lower fluorescence intensities (EGFP, MFI = 1,490; AcGFP1, MFI = 542; TagGFP2, MFI = 240) (Fig. 1B, Supplementary Table S3 and Fig. S1). The MFIs of these GFPs, when codon-optimized for S. cerevisiae (yEGFP, MFI = 33,351; yAcGFP1, MFI = 36,177; yTagGFP2, MFI = 24,250) were surprisingly 22-, 67- and 101-fold higher, respectively, than those of corresponding non-codon-optimized GFPs (Fig. 1B, Supplementary Table S3 and Fig. S1). Fluorescence microscopy revealed clear differences in the green fluorescence intensity of the codon-optimized and non-codon-optimized GFPs (Fig. 1C). Using the FLAG tag fused GFPs (Table 2 and Supplementary Table S2), the western blot analysis showed increased expression levels of the codon-optimized GFPs (approx. 27 kDa) in S. cerevisiae (Fig. 1D). Moreover, intriguingly, all yeast cells expressing codon-optimized GFPs generated light green colors under natural light, unlike non-codon-optimized EGFP, AcGFP1 and TagGFP2, which were never green under natural light (Supplementary Fig. S2).
As shown in Table 4, codon-optimized GFPs for S. cerevisiae presented high codon adaptation indexes (CAIs; 0.87~0.90) and moderate GC contents (39.7~40.4%). In contrast, commercially available, non-codon-optimized EGFP, AcGFP1 and TagGFP2 sequences unexpectedly exhibited much lower CAIs (0.50~0.56) and higher GC contents (59.0~63.6%) (Table 4), although the codon usage had been optimized for mammalian expression. This may have misled us into thinking that the mammalian cells had reasonably similar codon usages to the eukaryotic organism S. cerevisiae. For mUkG1 and ZsGreen, the wild type codons from the soft coral Sarcophyton sp. and the button polyp coral Zoanthus sp. were used, unexpectedly resulting in a relatively similar codon usage to that in yeast (CAIs = 0.69~0.70 and GC contents = 44.9~45.0%) (Table 4). These support our previous result that ZsGreen (original codons) exhibited more intense fluorescence than EGFP (non-codon-optimized) when used as a fluorescent reporter protein34. Among the codon-optimized GFPs tested in this study, monomeric ymUkG1 and tetrameric yZsGreen exhibited the most intense green fluorescence in S. cerevisiae (Fig. 1B).
Table 4. Codon adaptation index (CAI) and GC content of varied GFPs tested.
| Protein | CAI | GC content (%) |
|---|---|---|
| EGFP | 0.52 | 61.8 |
| AcGFP1 | 0.56 | 59.0 |
| TagGFP2 | 0.50 | 63.6 |
| mUkG1 | 0.69 | 45.0 |
| ZsGreen | 0.70 | 44.9 |
| yEGFP | 0.87 | 40.4 |
| yAcGFP1 | 0.87 | 40.2 |
| yTagGFP2 | 0.87 | 40.2 |
| ymUkG1 | 0.88 | 40.1 |
| yZsGreen | 0.90 | 40.0 |
| ymWasabi | 0.87 | 39.7 |
| ymNeonGreen | 0.90 | 40.0 |
GFP expression when used as fusion-tagged proteins to report the activation of signal transduction in S. cerevisiae
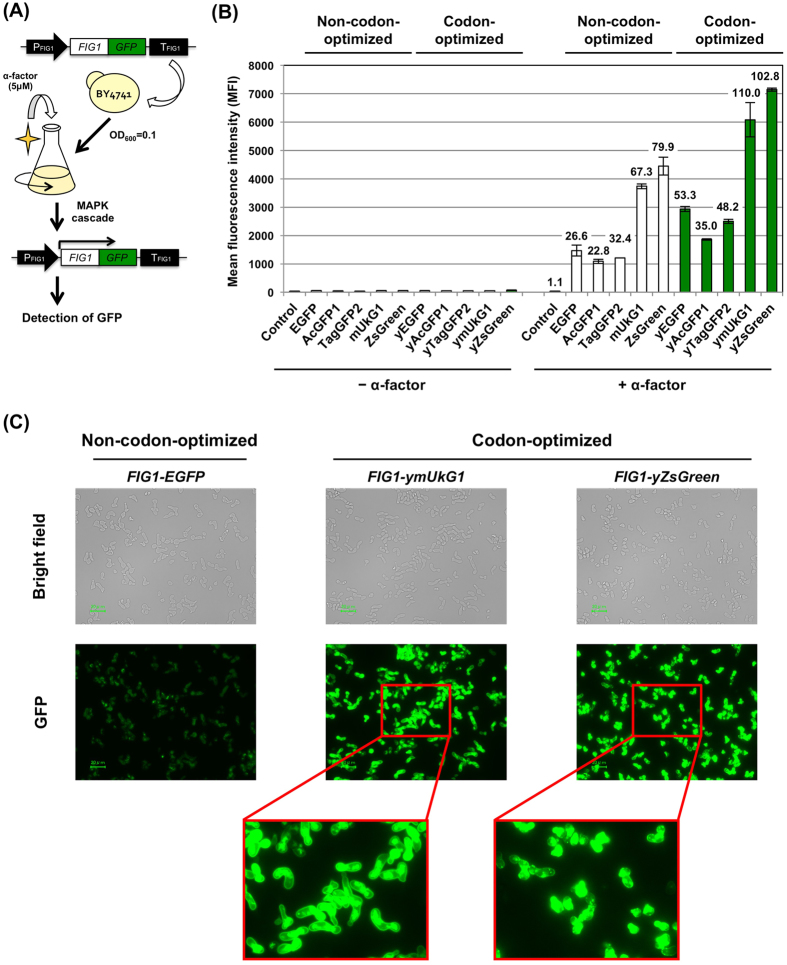
GFP is most widely applied as a protein tag or as a reporter gene4,7,32,35,36. To determine the applicability of the GFPs tested above, we decided to test the pheromone-responsive FIG1 gene, which is involved in conjugation and cellular fusion on plasma membrane during the mating process37. FIG1 gene expression is induced in response to activation of the yeast mating signaling pathway; therefore, this gene has been often used to sense the signal promoted in the mating pathway (Fig. 2A) or the agonist response to heterologous GPCRs using the non-codon-optimized EGFP as the reporter gene38,39,40,41.
Figure 2. Expression of GFPs as fusion-tagged proteins to report the activation of signal transduction in S. cerevisiae.
(A) Flow diagram of GFP transcription assays used to detect signal transduction. FIG1-GFP fusion genes were substituted for the FIG1 gene in the yeast genome. The cells were grown in media with and without 5 μM α-factor, and GFP expression was measured. (B) Mean GFP fluorescence intensities (MFIs). The MFIs of 10,000 cells were measured by flow cytometry. (C) Green fluorescence images of the cells. The images were acquired with a fluorescence microscope equipped with a 60× objective lens. Scale bar: 20 μm. The exposure time was 1/4 s. Codon-optimized and non-codon-optimized GFPs were evaluated. ‘Control’ indicates the BY4741 wild-type yeast strain.
The five codon-optimized and non-codon-optimized GFP genes were integrated into the chromosome of BY4741 haploid yeast a-cells to fuse to the C-terminus of the genomic FIG1 gene (Table 3 and Supplementary Table S4). To induce the transcription of FIG1-GFPs, the cells were incubated for 6 hours in YPD medium containing the a-cell-specific mating pheromone (α-factor) (Fig. 2A). The cultured cells were then analyzed by flow cytometry and observed under a fluorescence microscope (Fig. 2B,C).
The GFPs tested as fusion reporters exhibited marked increases in green fluorescence in response to the addition of α-factor pheromone (Fig. 2B). Although these fusion proteins showed lower fluorescence intensities than those of the solely expressed untagged GFPs, the MFIs of the fusion-codon-optimized GFPs were greater than those of all five non-codon-optimized GFPs (Fig. 2B). The pheromone-stimulated cells induced 22.8~110.0-fold changes in the MFIs compared to the corresponding unstimulated cells (Fig. 2B). The MFIs of Fig1-GFPs were increased approximately 1.6~2.0-fold due to codon optimization. Codon-optimized ymUkG1 and yZsGreen exhibited higher fluorescence intensities than the most commonly used non-codon-optimized EGFP when expressed as fusion genes with the FIG1 reporter gene (FIG1-EGFP, MFI = 1,469; FIG1-ymUkG1, MFI = 6,184; FIG1-yZsGreen, MFI = 7,143) and exhibited greater than 4.2- and 4.9-fold increases (Fig. 2B). Fluorescence microscope observation revealed brighter green fluorescence in pheromone-stimulated yeast cells encoding Fig1-ymUkG1 and Fig1-yZsGreen than in cells encoding Fig1-EGFP (the cell shapes were elongated by the excessive pheromone stimulation) (Fig. 2C). In addition, Fig1-ymUkG1 fusion protein (monomeric GFP fusion) especially showed correct localization on the plasma membrane, although it has been also observed in other organelle caused by the extensive overexpression (Fig. 2C).
As shown above, similarly to the results obtained for the solely expressed untagged GFPs, codon-optimized ymUkG1 and yZsGreen exhibited the brightest fluorescence, even when expressed as fusions with the FIG1 reporter gene. Tetrameric fluorescent proteins are generally regarded as unfavorable for use as fusion tags because they interfere with the normal function of the target proteins4; however, tetrameric yZsGreen could function as the quantitative fusion reporter, although the fusion with the Fig1 protein rarely localized on the plasma membrane. In further experiments, we studied monomeric ymUkG1, which is apparently less constraining in many applications but exhibits significantly more intense fluorescence than other monomeric GFPs in S. cerevisiae.
Application of codon-optimized ymUkG1 in Gγ recruitment systems that report protein–protein interactions in S. cerevisiae
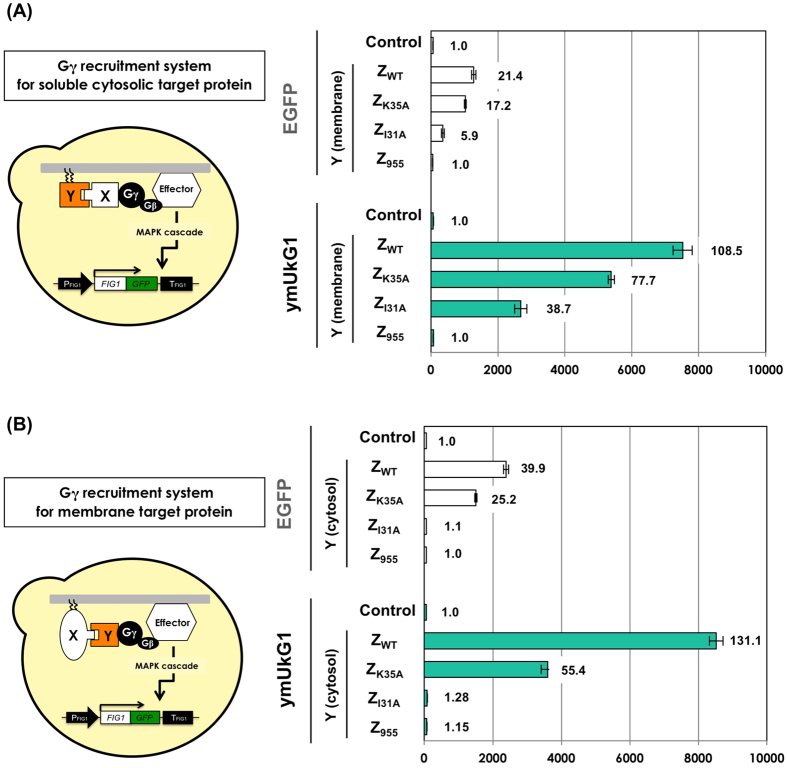
We developed the Gγ recruitment system to detect protein–protein interactions (PPIs)40,42,43,44,45 and to screen for protein variants presenting desirable affinities40. The detection method of the Gγ recruitment system is based on the fundamental principle that yeast pheromone (mating) signaling requires the localization of a complex between guanine nucleotide binding protein (G-protein) β- and γ-subunits (Gβγ) and the inner leaflet of the plasma membrane46. In brief, for the machinery to detect PPIs, an engineered Gγ mutant (termed Gγcyto) is used that lacks a membrane localization sequence (lipidation motif); this localization signal is normally expressed in the cytosol. When the target protein (X) is a soluble cytosolic protein, Gγcyto is prepared as a fusion protein with the target protein (X) (Gγcyto-X), and library proteins (Y) are attached to the artificial lipidation site to localize the proteins to the membrane (Supplementary Fig. S3A)40,42. In contrast, when the target protein (X) is a membrane-bound protein, Gγcyto is prepared as a fused protein with the library proteins (Y) (Gγcyto-Y) (Supplementary Fig. S3B)43,45. When target “X” and candidate “Y” interact, the Gγcyto-X (Supplementary Fig. S3A) or Gγcyto-Y (Supplementary Fig. S3B) fusion protein targets the Gβ to the membrane and subsequently induces pheromone-signaling pathway activation. Signaling can be monitored by a fluorescent reporter assay or a mating growth assay; therefore, the PPIs are easily detected (Supplementary Fig. S3C).
Here, FIG1–EGFP (non-optimized) was used as a fluorescent reporter gene in the Gγ recruitment system. If the flow cytometer (cell sorter) that can analyze more than several thousand cells per second is available, the fluorescent reporter assay extremely speeds up and is helpful for screening PPI protein variants. We tested whether FIG1-ymUkG1 could improve the fluorescence reporting ability of the Gγ recruitment system. As for the previously described system, the Fc protein of human immunoglobulin G (IgG) and the Z domain (derived from Staphylococcus aureus protein A (ZWT))47 were used as the PPI models. Several Z variants (ZK35A, ZI31A and Z955) with differing affinities for the Fc protein were also used as the PPI models (ZWT, 5.9 × 107 M−1; ZK35A, 4.6 × 106 M−1; ZI31A, 8.0 × 103 M−1; Z955, none)48,49.
We first tested the use of soluble cytosolic target proteins (Supplementary Fig. S3A). The Fc protein was used as the cytosolic target protein ‘X’ and was fused with Gγcyto (Gγcyto-Fc). Four Z domain variants were attached to the artificial lipidation motifs (derived from Ste18 p) as the candidate ‘Y’ proteins, which were localized on the membrane (Tables 2 and 3 and Supplementary Table S5). After the addition of α-factor pheromone, the cells were cultured, and the GFP signal was measured using flow cytometry (Supplementary Fig. S3C). Both reporters (Fig1-EGFP and Fig1-ymUkG1) successfully detected PPIs between Fc and several Z variants (ZWT, ZK35A and ZI31A) (Fig. 3A). However, the fluorescence intensities of the two reporters differed greatly. When comparing the systems in ZWT, the Fig1-EGFP and Fig1-ymUkG1 reporters exhibited 21- and 109-fold more intense fluorescence than the respective negative control cells. The fluorescence intensity of Fig1-EGFP (MFI = 1,276) was 5.9-fold less than that of Fig1-ymUkG1 (MFI = 7,524). The differences in fluorescence intensities between the Fig1-EGFP and Fig1-ymUkG1 reporters were 5.2- and 7.6-fold for the ZK35A and the ZI31A variants, respectively. When using the Z variant that lacks affinity for Fc (Z955), green fluorescence was never observed. The ymUkG1 reporter exhibited an intense fluorescence signal (MFI = 2,682) even when detecting the weak PPI between Fc and ZI31A (8.0 × 103 M−1); in contrast, Fig1-EGFP exhibited a faint fluorescence signal (MFI = 350).
Figure 3. Expression of non-codon-optimized EGFP and codon-optimized ymUkG1 as fusion-tagged proteins to report PPIs using Gγ recruitment systems.
(A) Flow cytometry analyses using the Gγ recruitment system for cytosolic target proteins. The Fc protein was used as the cytosolic target protein ‘X’ and was expressed as a fusion protein with Gγcyto (Gγcyto-Fc). Membrane-anchored Z variants (ZWT, ZK35A,, ZI31A and Z955) were expressed as ‘Y’ library candidate proteins. ‘Control’ indicates BFG2118 and UGFG2 yeast strains harboring the pGK413 mock plasmid (without the expression of ‘Y’). (B) Flow cytometry analyses using the Gγ recruitment system for membrane protein targets. The Fc protein was used as the membrane target protein ‘X’ and was expressed as a membrane-associated protein with the C-terminal lipid anchor (derived from Ste18p). Four Z variants (ZWT, ZK35A,, ZI31A and Z955) were used as cytosolic candidate ‘Y’ proteins and were expressed as fusion proteins with Gγcyto (Gγcyto-Z variants). ‘Control’ indicates the FC-G0 and UG2-FCG0 yeast strains (without the expression of ‘Gγcyto-Y’). The engineered strains were grown in media containing 5 μM α-factor, and mean fluorescence intensities (MFIs) were analyzed. The MFIs of 10,000 cells were measured by flow cytometry.
Next, we tested the use of membrane target proteins (Supplementary Fig. S3B). The Fc protein was used as the membrane target protein ‘X’. To determine the positions at which to fuse the lipidation motifs, both C-terminal and N-terminal membrane-associated Fc proteins were prepared (a C-terminal lipid anchor, derived from Ste18 p; and a N-terminal lipid anchor, derived from Gpa1p). Four Z domain variants were used as cytosolic candidate ‘Y’ proteins and fused with Gγcyto (Gγcyto-Z variants) (Table 3 and Supplementary Table S6). The cells were assayed similarly to the previous experiments. For the C-terminal membrane-associated Fc proteins, both the Fig1-EGFP and Fig1-ymUkG1 reporters successfully detected the PPIs with two Z variants (ZWT and ZK35A), although they did not detect the PPI with ZI31A (Fig. 3B). This result was consistent with previous results that failed to detect a PPI between membrane-associated Fc and Gγcyto-fused ZI31A. The weak PPI (8.0 × 103 M−1) is likely below the detection limitation in the Gγ recruitment system when using Fc as the membrane target protein45. Similarly to the results of the test for the soluble cytosolic target Fc protein (Fig. 3A), a large difference was observed in the fluorescence intensities between the Fig1-EGFP and Fig1-ymUkG1 reporters (Fig. 3B). When comparing the variants in ZWT, the Fig1-EGFP and Fig1-ymUkG1 reporters emitted fluorescent signals that were 40- and 131-times more intense than those of the respective negative control cells. The fluorescence intensities were 3.6-fold different between Fig1-EGFP (MFI = 2,380) and Fig1-ymUkG1 (MFI = 8,519). When comparing the variants in ZK35A, the difference in fluorescence intensity between the Fig1-EGFP and Fig1-ymUkG1 reporters was 2.4-fold. When using the N-terminal membrane-associated Fc proteins, similar tendencies were observed (Supplementary Fig. S4). When using ZWT and ZK35A, the differences in fluorescence intensity between the Fig1-EGFP and Fig1-ymUkG1 reporters were 3.6- and 2.9-fold, respectively. Thus, when used as a fluorescence reporter, codon-optimized ymUkG1 yielded brighter fluorescence than the previous non-codon-optimized (for S. cerevisiae) EGFP, even in the Gγ recruitment systems, which detect PPIs for both soluble cytosolic target proteins and membrane target proteins.
FACS analysis of target cells in cell mixtures containing positive and negative cells
Fluorescence activated cell sorting (FACS) is a specialized instrument that enables analysis and sorting of rare target cells in a larger cell population. To evaluate the performance of ymUkG1 as the reporter for FACS analysis, the Gγ recruitment system for soluble cytosolic target proteins was used (Supplementary Fig. S3A and Fig. 3A).
To determine the gate area that dominantly included positive cells (GFP+), we analyzed the data of yeast strains expressing Gγcyto-Fc and membrane-anchored Z variants (ZWT, ZK35A, ZI31A and Z955), whose fluorescence was acquired using a flow cytometer in Fig. 3A. We set the gate area (GFP+) that completely excluded negative cells (expressing Z955 with Gγcyto-Fc) and determined the positive cells by making the dot plots (forward scatter, FSC-A vs green fluorescence, GFP-A) (Supplementary Fig. S5). The proportions of positive cells included in the 10,000 cells (expressing ZWT, ZK35A and ZI31A with Gγcyto-Fc) using the Fig1-ymUkG1 reporter were respectively 98.3%, 95.4% and 80.3%, while those using the Fig1-EGFP reporter were 58.8%, 13.9% and 2.5% (Supplementary Fig. S5 and Table S7). This indicates the Fig1-ymUkG1 reporter clearly displayed higher percentages of the cells that were defined as positive cells (GFP+) than the Fig1-EGFP reporter, even though both cells expressing the same interaction pairs could be presumed to coincide in the promoted signaling and GFP transcription levels.
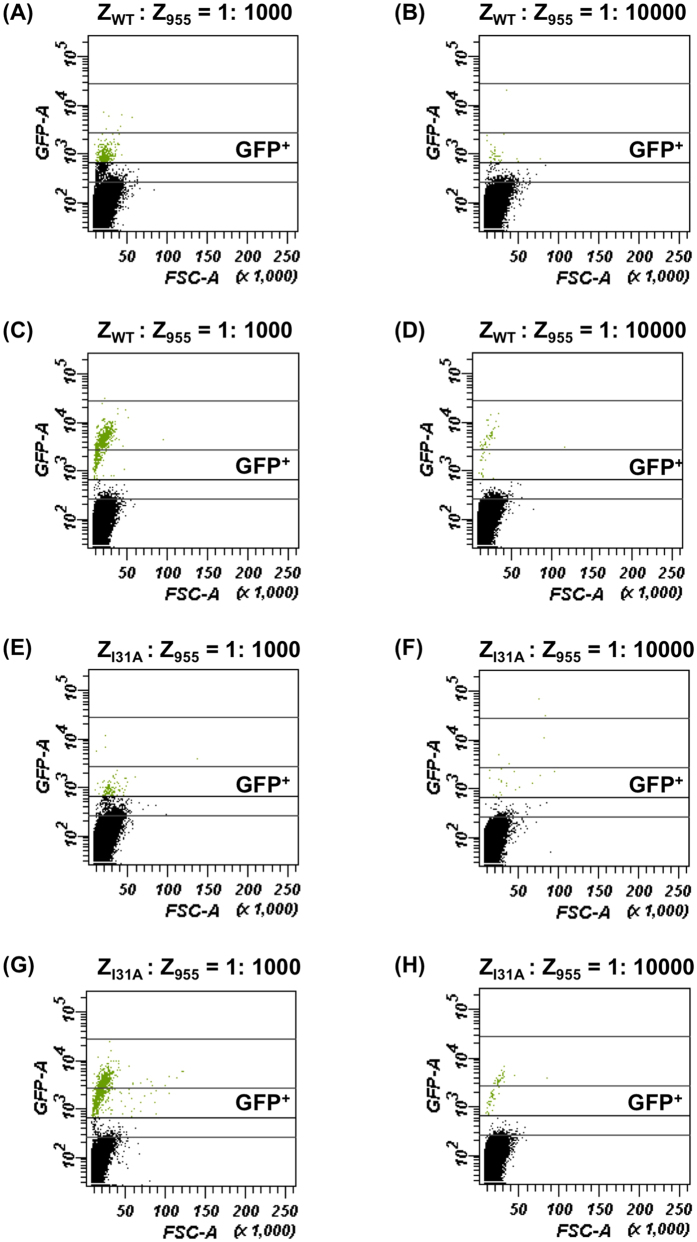
To further demonstrate the ability of the ymUkG1 reporter, we prepared the cell mixtures composed of positive cells (expressing ZWT with Gγcyto-Fc) and negative cells (expressing Z955 with Gγcyto-Fc) with two different mixing rates (0.1% and 0.01% positive cells). After cultivation of the cell mixtures in the presence of α-factor for 6 hours, 106 cells were analyzed using flow cytometer and the content rates of positive (target) cells contained in the gate area (GFP+) were evaluated (Fig. 4 and Table 5). Both Fig1-EGFP and Fig1-ymUkG1 reporters exposed the target cells contained in the GFP+ area, whereas they showed the obvious distinctions in the detection rates of the target cells (Fig. 4 and Table 5). When using the cell mixtures expressing ZWT and Z955 with Gγcyto-Fc (initially contained 0.1% and 0.01% positive cells), Fig1-ymUkG1 reporter detected 58% and 56% of the target cells (percentages of detected target cell numbers in GFP+ area against initial cell numbers of positive cells), while Fig1-EGFP reporter detected 33% and 34% of the target cells, respectively (Fig. 4A–D and Table 5). When using the cell mixtures expressing ZI31A and Z955 with Gγcyto-Fc (in the case of weak interaction), more clear differences were found between Fig1-EGFP and Fig1-ymUkG1 reporters (Fig. 4E–H and Table 5). Briefly, Fig1-ymUkG1 reporter detected 74% and 65% of the target cells, while Fig1-EGFP reporter detected 12% and 24% of the target cells (for the cell mixtures containing 0.1% and 0.01% positive cells, respectively) (Fig. 4E–H and Table 5). The reason why the use of ZI31A displayed higher percentages of the detectable target cells than that of ZWT might have arisen from the growth biases during the cultivation of cell mixtures. It is well known that the pheromone signaling (in response to the PPIs) provokes the cell-cycle arrest in G1 phase to make arrangements for the mating50. Because the percentages of the detectable positive cells to the initially contained positive cell numbers directly affect the recovery rates of the hopeful target cells, they are crucially important factors on FACS sorting. Thus, we have successfully demonstrated the superiority of the codon-optimized ymUkG1 reporter compared to the non-codon-optimized EGFP reporter in S. cerevisiae.
Figure 4. FACS detections of target cells in cell mixtures containing positive and negative cells.
Data were presented as dot plots (forward scatter, FSC-A vs green fluorescence, GFP-A). Y-axis is an indication of fluorescence and X-axis is an approximation of relative cell size. Cell mixtures were prepared by mixing positive cells (expressing membrane-anchored ZWT or ZI31A with Gγcyto-Fc) and negative cells (expressing membrane-anchored Z955 with Gγcyto-Fc) with two different mixing rates (0.1% and 0.01% positive cells). After cultivation of the cell mixtures in the presence of α-factor for 6 hours, 106 cells were analyzed using flow cytometer and the numbers of the positive (target) cells contained in the gate area (GFP+) were evaluated. (A,B) Cell mixtures containing 0.1% and 0.01% target cells expressing ZWT in BFG2118 yeast strain (EGFP reporter). (C,D) Cell mixtures containing 0.1% and 0.01% target cells expressing ZWT in UGFG2 yeast strain (ymUkG1 reporter). (E,F) Cell mixtures containing 0.1% and 0.01% target cells expressing ZI31A in BFG2118 yeast strain (EGFP reporter). (G,H) Cell mixtures containing 0.1% and 0.01% target cells expressing ZI31A in UGFG2 (ymUkG1 reporter).
Table 5. Summary of FACS analysis of cell mixtures containing positive and negative cells.
| Reporter | Interaction target | Initial ratio of target cells in mixture | Rate of detected target cell number against initial target cell number |
|---|---|---|---|
| EGFP | ZWT | 0.1% | 33% |
| EGFP | ZWT | 0.01% | 34% |
| ymUkG1 | ZWT | 0.1% | 58% |
| ymUkG1 | ZWT | 0.01% | 56% |
| EGFP | ZI31A | 0.1% | 12% |
| EGFP | ZI31A | 0.01% | 24% |
| ymUkG1 | ZI31A | 0.1% | 74% |
| ymUkG1 | ZI31A | 0.01% | 65% |
In summary, we compared the expression of seven codon-optimized GFPs and five non-codon-optimized GFPs in S. cerevisiae. Codon optimization for S. cerevisiae improved all five non-codon-optimized GFPs and surprisingly increased the fluorescence intensities up to 101-fold compared with commercially available GFPs (which were optimized for mammalian codon usage). CAI and GC content affected the expression of GFPs in S. cerevisiae more than expected. Among the codon-optimized GFPs tested, monomeric ymUkG1 and tetrameric yZsGreen exhibited the most intense fluorescence in S. cerevisiae, and green color was observed visually under natural light conditions in cells expressing these proteins. The expression of monomeric ymUkG1 protein was higher than that of tetrameric yZsGreen protein. Both ymUkG1 and yZsGreen were prepared as fusion reporters tagged to the Fig1 protein for use in monitoring signal transduction, and these fluorescent reporters exhibited significantly greater utility than the previously used, non-codon-optimized EGFPs (which were optimized for mammalian systems). Finally, the monomeric ymUkG1 was successfully used to improve the PPI detection system using the Fig1 reporter and increased the percentages of the detectable target cells using flow cytometry.
The most commonly used GFP in S. cerevisiae is probably the commercially available EGFP, whose codons are optimized for mammalian cells, not S. cerevisiae. Because gene and fragment synthesis services are readily available, it is easy to prepare a DNA fragment containing codon-optimized nucleic acid sequences. When strong protein expression is required, it might be best to measure the CAI and GC content and to test the codon-optimization of the protein of interest. We propose that the codon-optimized monomeric protein ymUkG1 (Supplementary Table S1) is a good choice for GFP experiments involving S. cerevisiae.
Methods
Strains and media
Details of the yeast S. cerevisiae strains BY474151, BY474251, MC-F144 and other recombinant strains used in this study and their genotypes are outlined in Table 3. The yeast strains were grown in YPD medium containing 1% (w/v) yeast extract, 2% peptone and 2% glucose or in SD media containing 0.67% yeast nitrogen base without amino acids (BD-Diagnostic Systems, Sparks, MD, USA) and containing 2% glucose. Amino acids and nucleotides (20 mg/L histidine, 60 mg/L leucine, 20 mg/L methionine, or 20 mg/L uracil) were supplemented into SD media lacking the relevant auxotrophic components. Agar (2%; w⁄v) was added to the medium described above to produce YPD and SD solid media.
Codon optimization and calculation of the codon adaptation index (CAI) and GC content
The nucleic acid sequences for codon-optimized GFPs for S. cerevisiae were designed using GeneArt® GeneOptimizer® software (Life Technologies/Thermo Fisher Scientific, San Jose, CA, USA). The DNA fragments for codon-optimized GFPs were prepared by using the GeneArt® Strings™ DNA fragments service. Codon adaptation indexes (CAIs) and GC contents before and after optimization were determined using GenScript Rare Codon Analysis Tool software (GenScript, Piscataway, NJ, USA).
Plasmid construction
All plasmids and primers used in this study are listed in Table 2 and Supplementary Table S8. The plasmids used for the expression of the tested GFPs were constructed as follows: DNA fragments encoding the GFPs AcGFP1, TagGFP2 and mUkG1 were PCR-amplified from pAcGFP1 (Clontech Laboratories/Takara Bio, Shiga, Japan), pTagGFP2-tubulin (Evrogen, Moscow, Russia) and pmUkG1-S1 (Medical & Biological Laboratories, Nagoya, Japan) using primer pairs 1 and 2, 3 and 4, and 5 and 6; the fragments were then digested with SalI + BamHI and inserted into the same sites between the PGK1 promoter (PPGK1) and the PGK1 terminator (TPGK1) on pGK41633, yielding the plasmids pGK416-AcGFP1, pGK416-TagGFP2 and pGK416-mUkG1. For EGFP and ZsGreen, the previously constructed plasmids pGK416-EGFP33 and pGK416-ZsGreen34 were used. The expression plasmids used for codon-optimized GFPs (yEGFP, yAcGFP1, yTagGFP2, ymUkG1, yZsGreen, ymWasabi and ymNeonGreen) were constructed as follows: DNA fragments encoding the codon-optimized GFPs were PCR-amplified from the GeneArt® Strings™ DNA fragments using primer pairs 7 and 8, 9 and 10, 11 and 12, 13 and 14, 15 and 16, 17 and 18, and 19 and 20; the fragments were then digested with SalI + BamHI and inserted into the same sites between the PPGK1 and the TPGK1 on pGK41633, yielding the plasmids pGK416-yEGFP, pGK416-yAcGFP1, pGK416-yTagGFP2, pGK416-ymUkG1, pGK416-yZsGreen, pGK416-ymWasabi and pGK416-ymNeonGreen.
The plasmids used for the western blotting were constructed as follows: DNA fragments encoding the GFPs fused the FLAG tag sequence were PCR-amplified from pGK416-EGFP, pGK416-AcGFP1, pGK416-TagGFP2, pGK416-mUkG1, pGK416-ZsGreen, pGK416-yEGFP, pGK416-yAcGFP1, pGK416-yTagGFP2, pGK416-ymUkG1 and pGK416-yZsGreen using primer pairs 21 and 22, 1 and 23, 3 and 24, 5 and 25, 26 and 27, 7 and 28, 9 and 29, 11 and 30, 13 and 31, and 15 and 32, 17 and 33, and 19 and 34 respectively. Primers 22, 23, 24, 25, 27, 28, 29, 30, 31 and 32 contain FLAG tag (24 bp). Then, the fragments were then digested with SalI + BamHI and inserted into the same sites between the PPGK1 and the TPGK1 on pGK41633, yielding the plasmids pGK416-EGFP-F, pGK416-AcGFP1-F, pGK416-TagGFP2-F, pGK416-mUkG1-F pGK416-ZsGreen, pGK416-yEGFP-F, pGK416-yAcGFP1-F, pGK416-yTagGFP2-F, pGK416-ymUkG1-F, pGK416-yZsGreen-F, pGK416-ymWasabi-F and pGK416-ymNeonGreen-F.
The plasmids used to integrate the GFP reporter genes at the FIG1 locus on the yeast chromosome were constructed as follows: A DNA fragment containing the homologous sequence of the FIG1 terminator (downstream of FIG1 gene; 200 bp) was PCR-amplified from pBlue-FIG1pt-ZsGreen34 using the primer pair 35 and 36. The amplified fragments were digested with XhoI + KpnI and inserted into the pBlueScript II KS(+) vector, yielding the plasmid pBlue-FIG1t. A DNA fragment containing the URA3 selectable marker was PCR-amplified from pBlue-FIG1pt-ZsGreen34 using the primer pair 37 and 38. DNA fragments containing the GFPs were PCR-amplified from pGK416-AcGFP1, pGK416-TagGFP2, pGK416-mUkG1, pGK416-ZsGreen, pGK416-yEGFP, pGK416-yAcGFP1, pGK416-yTagGFP2, pGK416-ymUkG1, and pGK416-yZsGreen34 using primer pairs 39 and 40, 41 and 42, 43 and 44, 45 and 46, 47 and 48, 49 and 50, 51 and 52, 53 and 54, and 55 and 56, respectively. Primers 39, 41, 43, 45, 47, 49, 40, 53 and 55 contain regions that are homologous to the C-terminus of FIG1 (50 bp). Then, the URA3 fragment and the GFP fragment were then linked by overlap PCR using primer pairs 39 and 38, 41 and 38, 43 and 38, 45 and 38, 47 and 38, 49 and 38, 51 and 38, 53 and 38, and 55 and 38, and the overlap fragments were digested with SacII + XhoI and inserted into pBlue-FIG1t, yielding the plasmids pBlue-UFt-AcGFP1, pBlue-UFt-TagGFP2, pBlue-UFt-mUkG1, pBlue-UFt-ZsGreen, pBlue-UFt-yEGFP, pBlue-UFt-yAcGFP1, pBlue-UFt-yTagGFP2, pBlue-UFt-ymUkG1 and pBlue-UFt-yZsGreen.
Yeast strain construction
The strains used in this study are listed in Table 3. DNA cassettes were integrated to express GFP-fused FIG1 as follow: DNA fragments containing FIG1(50 bp)-GFP-URA3-TFIG1 (TFIG1: FIG1 terminator) were amplified from pBlue-UFt-AcGFP1, pBlue-UFt-TagGFP2, pBlue-UFt-mUkG1, pBlue-UFt-ZsGreen, pBlue-UFt-yEGFP, pBlue-UFt-yAcGFP1, pBlue-UFt-yTagGFP2, pBlue-UFt-ymUkG1 and pBlue-UFt-yZsGreen using the primer pair 57 and 58. BY4741 was transformed with the amplified DNA fragments using the lithium acetate method52. The transformants were selected on SD-Ura plates (SD solid medium without uracil, but containing leucine, histidine and methionine). After confirming the correct integration, the URA3 marker was “popped-out” by homologous recombination using counter-selection with 5-fluoroorotic acid (5-FOA, Fluorochem, Derbyshire, UK), to yield BYFAG1, BYFTG1, BYFUG1, BYFZG1, BYFEG2, BYFAG2, BYFTG2, BYFUG2 and BYFZG2.
The STE18 gene was substituted by kanMX4 in the yeast chromosome by amplifying the DNA fragment containing PSTE18–kanMX4–TSTE18 (PSTE18: STE18 promoter and TSTE18: STE18 terminator) from pGK426-GPTK42 using the primer pair 45 and 46. BYFUG2 was then transformed with the amplified DNA fragment, and transformants were selected on YPD solid medium containing G418 (500 ng/mL) (Nacalai Tesque, Kyoto, Japan) to yield the UGW2 strain.
DNA cassettes for the Gγcyto–Fc protein in the cytosol were integrated as follows. DNA fragments containing URA3-PPGK1-Gγcyto-Fc-TPGK1-THIS3 (THIS3: HIS3 terminator) were amplified from pUMGP-GγMFcH42 using the primer pair 61 (containing the homologous regions of the HIS3 promoter) and 62. UGW2 was transformed with the amplified DNA fragments using the lithium acetate method. The transformants were selected on SD-Ura plates (containing leucine, histidine and methionine) to yield UGFG2.
DNA cassettes used to express the membrane-associated Fc protein were integrated as follows: DNA fragments containing PSTE18-PPGK1-Fc-Ste18C-TPGK1-kanMX4-TSTE18 and PSTE18-PPGK1-Gpa1N-Fc-TPGK1-kanMX4-TSTE18 were amplified from pUMGPTK-Fc-Ste18C45 and pUMGPTK-Gpa1N-Fc45, respectively, using the primer pair 59 and 60. BYFUG2 was transformed with the amplified DNA fragments using the lithium acetate method. The transformants were selected on YPD solid medium containing G418 (500 ng/mL) to yield BYFUG2-FC and BYFUG2-FN. DNA cassettes for the Gγcyto–Z domain variants (ZWT, ZK35A, ZI31A and Z955) in the cytosol were integrated as follows: DNA fragments containing URA3-PPGK1-Gγcyto-ZWT(-ZK35A, -ZI31A and -Z955)-TPGK1-THIS3 and URA3-PPGK1-Gγcyto-TPGK1-THIS3 were amplified from pUSTE18p-Gγcyto-ZWT(-ZK35A, -ZI31A and -Z955)-HIS3t45 and pUSTE18p-Gγcyto-HIS3t45 using the primers 61 (containing the homologous regions of the HIS3 promoter) and 62. BYFUG2-FC and BYFUG2-FN were transformed with the amplified DNA fragments using the lithium acetate method52. The transformants were selected on SD-Ura plates containing leucine, histidine and methionine to yield UG2-FCGW, UG2-FCGK, UG2-FCGI, UG2-FCG9, UG2-FCG0, UG2-FNGW, UG2-FCGK, UG2-FCGI, UG2-FCG9 and UG2-FCG0.
The constructed strains were used for the assays or plasmids were introduced using the lithium acetate method. All strains and transformants used for the assays are listed in Supplementary Tables S2–S5.
Culture conditions for GFP expression in yeast cells
To express the tested GFPs (Fig. 1A), yeast transformants (Supplementary Table S2) were grown in 5 mL of SD-Ura medium at 30 °C overnight. The cultured cells were then inoculated into 2 mL of fresh SD-Ura medium to obtain an initial optical density at 600 nm (OD600) of 0.1. The cells were thereafter cultivated at 30 °C for 3 hours and then harvested.
Signal transduction assays using FIG1-GFP reporter genes (Fig. 2A) were conducted according to previously described methods40 with some modifications. The engineered yeast a-cells (Supplementary Table S3) were grown in 5 mL of YPD medium 30 °C overnight. The cultured cells were then inoculated into 2 mL of fresh YPD medium containing 5 μM α-factor (Zymo Research, Orange, CA, USA) to obtain an initial OD600 of 0.1. The cells were thereafter cultivated at 30 °C for 6 hours and then harvested.
Protein–protein interaction assays using FIG1-GFP reporter genes were conducted as described previously40. In brief, for the Gγ recruitment system involving soluble cytosolic target protein (Fig. 3A), the engineered yeast a-cells (Supplementary Table S4) were grown in 5 mL of SD-His medium at 30 °C overnight. The cultured cells were then inoculated into 2 mL of fresh SD-His medium containing 5 μM α-factor to obtain an initial OD600 of 0.1. For the Gγ recruitment system involving membrane target proteins (Fig. 3B and Supplementary Fig. S4), the engineered yeast a-cells (Supplementary Table S5) were grown in 5 mL of YPD medium at 30 °C overnight. The cultured cells were then inoculated in 2 mL of fresh YPD medium containing 5 μM α-factor to obtain an initial OD600 of 0.1. In both cases, the cells were thereafter cultivated at 30 °C for 6 hours and then harvested.
Fluorescence microscopy
Fluorescence microscopy observation was performed as described previously53. The harvested cells were washed and resuspended in distilled water; the cell suspensions were then observed using a BIOREVO BZ-9000 fluorescence microscope equipped with a 60× objective lens (Keyence, Osaka, Japan). Green fluorescence images were acquired with a 470/40 band-pass filter for excitation and a 535/50 band-pass filter for emission.
Flow cytometry
Flow cytometry analysis was conducted as described previously54. The harvested cells were diluted into test tubes containing the sheath solution, and GFP fluorescence was measured by using a BD FACSCanto II flow cytometer (BD Biosciences, San Jose, CA). The green fluorescence signal from 10,000 cells was excited with a 488-nm blue laser and collected through a 530/30-nm band-pass (GFP) filter. The data were analyzed by using BD FACSDiva software (version 5.0, BD Biosciences) and FlowJo software version 7.2.2 (Treestar, Inc., San Carlos, CA).
FACS analysis of target cells in cell mixtures
To evaluate the Fig1-EGFP and Fig1-ymUkG1 reporters (Supplementary Fig. S5), yeast transformants (Supplementary Table S4) were grown in 5 mL of SD-His medium at 30 °C overnight. Cell mixtures were prepared by mixing target yeast cells (BFG2118 + pGK413-ZWTmem, UGFG2 + pGK413-ZWTmem, BFG2118 + pGK413-ZI31Amem or UGFG2 + pGK413-ZI31Amem) and negative yeast cells (BFG2118 + pGK413-Z955mem or UGFG2 + pGK413-Z955mem) with 0.1% and 0.01% of the initial target cell ratios. The cell mixtures were then inoculated into 2 mL of fresh SD-His medium containing 5 μM α-factor to obtain an initial OD600 of 0.1. The cells were thereafter cultivated at 30 °C for 6 hours, and then 106 cells were acquired by using a BD FACSCanto II flow cytometer and analyzed by using BD FACSDiva software.
Western blot analysis
After yeast transformants (Supplementary Table S2) were grown in 5 mL of SD-Ura medium at 30 °C for 16 hours, samples were prepared using an alkaline lysis method55. Protein extracts (3 μl) were resolved in a 15% e-PAGEL (Atto Co., Tokyo, Japan). Western blot analysis was performed using the monoclonal anti-FLAG M2 antibody (Sigma–Aldrich, Irvine, UK) for the GFPs fused FLAG tag. Alkaline phosphatase-conjugated anti-mouse IgG (Promega, Madison, WI, USA) was used as the secondary antibody, and colorimetric detection of alkaline phosphatase activity was performed using CDP-Star detection reagent (GE Healthcare Life Sciences, UK).
Additional Information
How to cite this article: Kaishima, M. et al. Expression of varied GFPs in Saccharomyces cerevisiae: codon optimization yields stronger than expected expression and fluorescence intensity. Sci. Rep. 6, 35932; doi: 10.1038/srep35932 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported in part by a Research Fellowship for Young Scientists (253862), a Grant-in-Aid for Young Scientists (B) from the Japan Society for the Promotion of Science (JSPS), a Special Coordination Funds for Promoting Science and Technology, Creation of Innovation Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe; iBioK) from the Ministry of Education, Culture, Sports and Technology (MEXT), and the commission for Development of Artificial Gene Synthesis Technology for Creating Innovative Biomaterial from the Ministry of Economy, Trade and Industry (METI), Japan.
Footnotes
Author Contributions M.K., J.I., N.F. and A.K. designed the experiments; M.K. and T.M. performed the experiments and collected the data; M.K. and J.I. analyzed and interpreted the data; and M.K. and J.I. wrote the manuscript with contributions from N.F. and A.K.
References
- Shimomura O., Johnson F. H. & Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell. Comp. Physiol. 59, 223–239, doi: 10.1002/jcp.1030590302 (1962). [DOI] [PubMed] [Google Scholar]
- Tsien R. Y. The Green Fluorescent Protein. Annu. Rev. Biochem. 67, 509–544, doi: 10.1146/annurev.biochem.67.1.509 (1998). [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S. et al. Global analysis of protein expression in yeast. Nature 425, 737–741, doi: 10.1038/nature02046 (2003). [DOI] [PubMed] [Google Scholar]
- Chudakov D. M. & Lukyanov S. & Lukyanov, K. a. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 23, 605–613, doi: 10.1016/j.tibtech.2005.10.005 (2005). [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J. & Patterson G. H. Development and use of fluorescent protein markers in living cells. Science 300, 87–91, doi: 10.1126/science.1082520 (2003). [DOI] [PubMed] [Google Scholar]
- Chalfie M., Tu Y., Euskirchen G., Ward W. W. & Prasher D. C. Green fluorescent protein as a marker for gene expression. Science (80-.). 263, 802–805, doi: 10.1126/science.8303295 (1994). [DOI] [PubMed] [Google Scholar]
- Huh W.-K. et al. Global analysis of protein localization in budding yeast. Nature 425, 686–691, doi: 10.1038/nature02026 (2003). [DOI] [PubMed] [Google Scholar]
- Giepmans B. N. G., Adams S. R., Ellisman M. H. & Tsien R. Y. The fluorescent toolbox for assessing protein location and function. Science 312, 217–224, doi: 10.1126/science.1124618 (2006). [DOI] [PubMed] [Google Scholar]
- Zid B. M. & O’Shea E. K. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature 514, 117–121, doi: 10.1038/nature13578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa P. a., DeRisi J. L., Wilhelm J. E. & Vale R. D. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290, 341–344, doi: 10.1126/science.290.5490.341 (2000). [DOI] [PubMed] [Google Scholar]
- Felice M. R. et al. Post-transcriptional regulation of the yeast high affinity iron transport system. J. Biol. Chem. 280, 22181–22190, doi: 10.1074/jbc.M414663200 (2005). [DOI] [PubMed] [Google Scholar]
- Newman J. R. S. et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441, 840–846, doi: 10.1038/nature04785 (2006). [DOI] [PubMed] [Google Scholar]
- Enenkel C., Lehmann A. & Kloetzel P. M. Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J. 17, 6144–6154, doi: 10.1093/emboj/17.21.6144 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr M., Frommer W. B. & Lalonde S. Visualization of maltose uptake in living yeast cells by fluorescent nanosensors. Proc. Natl. Acad. Sci. USA 99, 9846–9851, doi: 10.1073/pnas.142089199 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. et al. A yEGFP-Based Reporter System for High-Throughput Yeast Two-Hybrid Assay by Flow Cytometry. Cytometry A 73, 312–320, doi: 10.1002/cyto.a.20525 (2008). [DOI] [PubMed] [Google Scholar]
- Cubitt A. B. et al. Understanding, improving and using green fluorescent proteins. Trends Biochem. Sci. 20, 448–455, doi: 10.1016/S0968-0004(00)89099-4 (1995). [DOI] [PubMed] [Google Scholar]
- Heim R., Prasher D. C. & Tsien R. Y. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. USA 91, 12501–12504, doi: 10.1073/pnas.91.26.12501 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R., Cubitt A. B. & Tsien R. Y. Improved green fluorescence. Nature 373, 663–664, doi: 10.1038/373663b0 (1995). [DOI] [PubMed] [Google Scholar]
- Cormack B. P., Valdivia R. H. & Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38, doi: 10.1016/0378-1119(95)00685-0 (1996). [DOI] [PubMed] [Google Scholar]
- Patterson G. H., Knobel S. M., Sharif W. D., Kain S. R. & Piston D. W. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 73, 2782–2790, doi: 10.1016/S0006-3495(97)78307-3 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurskaya N. G. et al. A colourless green fluorescent protein homologue from the non-fluorescent hydromedusa Aequorea coerulescens and its fluorescent mutants. Biochem. J. 373, 403–408, doi: 10.1042/BJ20021966 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach O. M. et al. Conversion of Red Fluorescent Protein into a Bright Blue Probe. Chem. Biol. 15, 1116–1124, doi: 10.1016/j.chembiol.2008.08.006 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt M. E., Scherrer M. P., Ferrari F. D. & Matz M. V. Very bright green fluorescent proteins from the pontellid copepod Pontella mimocerami. PLoS One 5, 3–10, doi: 10.1371/journal.pone.0011517 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C. et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407–409, doi: 10.1038/nmeth.2413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deheyn D. D. et al. Endogenous green fluorescent protein (GFP) in amphioxus. Biol. Bull. 213, 95–100, doi: 213/2/95 [pii] (2007). [DOI] [PubMed] [Google Scholar]
- Tsutsui H., Karasawa S., Okamura Y. & Miyawaki A. Improving membrane voltage measurements using FRET with new fluorescent proteins. Nat. Methods 5, 683–685, doi: 10.1038/nmeth.1235 (2008). [DOI] [PubMed] [Google Scholar]
- Matz M. V. et al. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 17, 969–973, doi: 10.1038/13657 (1999). [DOI] [PubMed] [Google Scholar]
- Karasawa S., Araki T., Yamamoto-Hino M. & Miyawaki A. A Green-emitting Fluorescent Protein from Galaxeidae Coral and Its Monomeric Version for Use in Fluorescent Labeling. J. Biol. Chem. 278, 34167–34171, doi: 10.1074/jbc.M304063200 (2003). [DOI] [PubMed] [Google Scholar]
- Ai H., Olenych S. G., Wong P., Davidson M. W. & Campbell R. E. Hue-shifted monomeric variants of Clavularia cyan fluorescent protein: identification of the molecular determinants of color and applications in fluorescence imaging. BMC Biol. 6, 13, doi: 10.1186/1741-7007-6-13 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza A. M., Curran K. a., Rey L. G. & Alper H. S. A condition-specific codon optimization approach for improved heterologous gene expression in Saccharomyces cerevisiae. BMC Syst. Biol. 8, 33, doi: 10.1186/1752-0509-8-33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack B. P. et al. Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology 143, 303–311, doi: 10.1099/00221287-143-2-303 (1997). [DOI] [PubMed] [Google Scholar]
- Sheff M. a. & Thorn, K. S. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21, 661–670, doi: 10.1002/yea.1130 (2004). [DOI] [PubMed] [Google Scholar]
- Ishii J. et al. A simple and immediate method for simultaneously evaluating expression level and plasmid maintenance in yeast. J. Biochem. 145, 701–708, doi: 10.1093/jb/mvp028 (2009). [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Ishii J. & Kondo A. Bright fluorescence monitoring system utilizing zoanthus sp. green fluorescent protein (ZsGreen) for human g-protein-coupled receptor signaling in microbial yeast cells. PLoS One 8, 1–18, doi: 10.1371/journal.pone.0082237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabantous S., Terwilliger T. C. & Waldo G. S. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 23, 102–107, doi: 10.1038/nbt1044 (2005). [DOI] [PubMed] [Google Scholar]
- Lee S., Lim W. A. & Thorn K. S. Improved Blue, Green, and Red Fluorescent Protein Tagging Vectors for S. cerevisiae. 8, 4–11, doi: 10.1371/journal.pone.0067902 (2013). [DOI] [PMC free article] [PubMed]
- Erdman S., Lin L., Malczynski M. & Snyder M. Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 140, 461–483, doi: 10.1083/jcb.140.3.461 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii J., Fukuda N., Tanaka T., Ogino C. & Kondo A. Protein-protein interactions and selection: Yeast-based approaches that exploit guanine nucleotide-binding protein signaling. FEBS J. 277, 1982–1995, doi: 10.1111/j.1742-4658.2010.07625.x (2010). [DOI] [PubMed] [Google Scholar]
- Ishii J. et al. Cell wall trapping of autocrine peptides for human G-protein-coupled receptors on the yeast cell surface. PLoS One 7, e37136, doi: 10.1371/journal.pone.0037136 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaishima M., Fukuda N., Ishii J. & Kondo A. Desired Alteration of Protein Affinities: Competitive Selection of Protein Variants Using Yeast Signal Transduction Machinery. PLoS One 9, e108229, doi: 10.1371/journal.pone.0108229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda N., Ishii J., Kaishima , M. & Kondo , A. Amplification of agonist stimulation of human G-protein-coupled receptor signaling in yeast. Anal. Biochem. 417, 182–187, doi: 10.1016/j.ab.2011.06.006 (2011). [DOI] [PubMed] [Google Scholar]
- Fukuda N., Ishii J., Tanaka T., Fukuda H. & Kondo A. Construction of a novel detection system for protein-protein interactions using yeast G-protein signaling. FEBS J. 276, 2636–2644, doi: 10.1111/j.1742-4658.2009.06991.x (2009). [DOI] [PubMed] [Google Scholar]
- Fukuda N. & Honda S. Rapid Evaluation of Tyrosine Kinase Activity of Membrane-Integrated Human Epidermal Growth Factor Receptor Using the Yeast Gγ Recruitment System. ACS Synth. Biol. 4, 421–429, doi: 10.1021/sb500083t (2015). [DOI] [PubMed] [Google Scholar]
- Fukuda N., Ishii J. & Kondo A. Gγ recruitment system incorporating a novel signal amplification circuit to screen transient protein-protein interactions. FEBS J. 278, 3086–3094, doi: 10.1111/j.1742-4658.2011.08232.x (2011). [DOI] [PubMed] [Google Scholar]
- Kaishima M., Ishii J., Fukuda N. & Kondo A. Gγ recruitment systems specifically select PPI and affinity-enhanced candidate proteins that interact with membrane protein targets. Sci. Rep. 5, 16723, doi: 10.1038/srep16723 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manahan C. L., Patnana M., Blumer K. J. & Linder M. E. Dual lipid modification motifs in G(alpha) and G(gamma) subunits are required for full activity of the pheromone response pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 11, 957–968 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson B. et al. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. Des. Sel. 1, 107–113, doi: 10.1093/protein/1.2.107 (1987). [DOI] [PubMed] [Google Scholar]
- Cedergren L., Andersson R., Jansson B., Uhlén M. & Nilsson B. Mutational analysis of the interaction between staphylococcal protein A and human IgG1. Protein Eng. 6, 441–448 (1993). [DOI] [PubMed] [Google Scholar]
- Nordberg E. et al. Cellular studies of binding, internalization and retention of a radiolabeled EGFR-binding affibody molecule. Nucl. Med. Biol. 34, 609–618, doi: 10.1016/j.nucmedbio.2007.05.010 (2007). [DOI] [PubMed] [Google Scholar]
- Peter M., Gartner A., Horecka J., Ammerer G. & Herskowitz I. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell 73, 747–760, doi: 10.1016/0092-8674(93)90254-N (1993). [DOI] [PubMed] [Google Scholar]
- Brachmann C. B. et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132, doi: (1998). [DOI] [PubMed] [Google Scholar]
- Gietz D., St Jean A., Woods R. a. & Schiestl R. H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii J. et al. Three gene expression vector sets for concurrently expressing multiple genes in Saccharomyces cerevisiae. FEMS Yeast Res. 14, 399–411, doi: 10.1111/1567-1364.12138 (2014). [DOI] [PubMed] [Google Scholar]
- Ishii J. et al. Microbial fluorescence sensing for human neurotensin receptor type 1 using Gα-engineered yeast cells. Anal. Biochem. 446, 37–43, doi: 10.1016/j.ab.2013.10.016 (2014). [DOI] [PubMed] [Google Scholar]
- von der Haar T. Optimized protein extraction for quantitative proteomics of yeasts. PLoS One 2, e1078, doi: 10.1371/journal.pone.0001078 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.