Abstract
Tris(1,3-dichloro-2-propyl)phosphate (TDCIPP) is a high-production volume organophosphate-based plasticizer and flame retardant widely used within the United States. Using zebrafish as a model, the objectives of this study were to determine whether (1) TDCIPP inhibits DNA methyltransferase (DNMT) within embryonic nuclear extracts; (2) uptake of TDCIPP from 0.75 h postfertilization (hpf, 2-cell) to 2 hpf (64-cell) or 6 hpf (shield stage) leads to impacts on the early embryonic DNA methylome; and (3) TDCIPP-induced impacts on cytosine methylation are localized to CpG islands within intergenic regions. Within this study, 5-azacytidine (5-azaC, a DNMT inhibitor) was used as a positive control. Although 5-azaC significantly inhibited zebrafish DNMT, TDCIPP did not affect DNMT activity in vitro at concentrations as high as 500 μM. However, rapid embryonic uptake of 5-azaC and TDCIPP from 0.75 to 2 hpf resulted in chemical- and chromosome-specific alterations in cytosine methylation at 2 hpf. Moreover, TDCIPP exposure predominantly resulted in hypomethylation of positions outside of CpG islands and within intragenic (exon) regions of the zebrafish genome. Overall, these findings provide the foundation for monitoring DNA methylation dynamics within zebrafish as well as identifying potential associations among TDCIPP exposure, adverse health outcomes, and DNA methylation status within human populations.
Introduction
Early embryonic development is a dynamic, complex process that is dependent on the timing and extent of epigenetic reprogramming of the zygotic genome. Immediately following fertilization, early embryogenesis progresses through two phases of the maternal-to-zygotic transition (MZT): (1) rapid degradation of maternally loaded transcripts and (2) minor and major waves of zygotic genome activation. The MZT is highly conserved across a wide range of invertebrate and vertebrate species, including sea urchins, nematodes, fruit flies, zebrafish, frogs, mice, and humans.1,2 In humans, the MZT occurs in utero at the 8-cell stage (3 d postfertilization) prior to implantation within the uterine wall2 whereas, in zebrafish, the MZT commences ex utero near the end of cleavage (2 h postfertilization, hpf) and terminates at approximately midblastula (2.75–3 hpf).3 After rapid erasure of maternal and paternal methylation marks following fertilization, zygotic genome activation, tissue-specific gene expression, and normal somatic development of mammalian and zebrafish embryos are dependent on steady, de novo genome-wide cytosine methylation by DNA methyltransferases (DNMT) during the MZT.4,5 Therefore, alterations in de novo DNA methylation marks, via impacts on DMNT activity or methyl donor concentrations, during early embryonic development have the potential to change the trajectory of normal development and, if inherited, persist across multiple generations.6
Over the last 10–15 years, the role of environmental factors, including nutrition, chemical exposures, and physical variables such as temperature, in mediating epigenetic modifications during early development has been intensely investigated, albeit the number of environment-related epigenetic studies pales in comparison to studies considering nonenvironmental factors.7 To date, the most well-studied environmental chemical known to alter DNA methylation and, as a result, offspring phenotypes is bisphenol A (BPA),8−13 a high-production volume chemical used in the production of polycarbonate plastics and epoxy resins. In a seminal study published by Dolinoy et al. in 2007,13 maternal exposure of yellow agouti (Avy) mice to BPA resulted in CpG hypomethylation at the Avy locus and a corresponding shift in coat color phenotypes within offspring, an effect that was rescued by maternal supplementation with methyl donors (folic acid or genistein). Importantly, this was one of the first animal studies to demonstrate that maternal exposure to an environmental chemical during in utero development has the potential to reprogram the offspring epigenome, leading to immediate phenotypic consequences (e.g., change in coat color) and potential long-term effects on adult disease susceptibility. These findings have since provided the basis for investigating the potential association of BPA exposure and epigenetic changes within human populations,14−16 raising the possibility that epigenetic marks within certain loci may be used as diagnostic indicators of prior environmental exposures.
Tris(1,3-dichloro-2-propyl)phosphate (TDCIPP) is a chlorinated phosphate ester used as a high-production volume plasticizer and flame retardant within polyurethane (rigid and flexible), plastics, resins, and acrylic latexes.17 As TDCIPP persists within indoor and outdoor environmental media,18 environmental exposure to TDCIPP may pose a health risk to humans and ecological species, particularly during sensitive windows of early development (e.g., during epigenetic reprogramming) that have the potential to coincide with higher internal doses of TDCIPP.19 Using zebrafish as a model, we previously demonstrated that the cleavage period (0.75–2.25 h postfertilization, hpf) during embryogenesis is susceptible to TDCIPP-induced delays in methylation of the zygotic genome.20 Within various life-stages of zebrafish, TDCIPP exposure also results in a wide range of adverse effects, including disruption of thyroid hormone regulation,21−23 gene expression,24,25 behavior,26−29 and reproduction.30−32 In addition, exposure of early zebrafish embryos to 3 μM TDCIPP, an identical nominal aqueous concentration used within our 2012 study, arrests epiboly and induces severe malformations later in development.24 However, to our knowledge, no studies have investigated the potential impacts of TDCIPP on DNMT activity and cytosine methylation within zebrafish embryos (or any other animal model). Therefore, the objectives of this study were to determine whether (1) TDCIPP inhibits zebrafish DNMT within embryonic nuclear extracts; (2) uptake of TDCIPP from 0.75 hpf (2-cell) to 2 hpf (64-cell) or 6 hpf (shield stage) leads to impacts on the DNA methylome during cleavage; and (3) TDCIPP-induced impacts on cytosine methylation are localized to CpG islands within intergenic regions. For DNMT activity assays and all embryonic exposures, 5-azacytidine (5-azaC), an inhibitor of DNMT activity, was used as a positive control.4
Materials and Methods
Animals
Adult wild-type (strain 5D) zebrafish were maintained and bred on a 14 h:10 h light:dark cycle within a five-shelf stand-alone system (Aquatic Habitats, Inc., Apopka, FL, USA) containing photoperiod light-cycle enclosures and recirculating conditioned reverse osmosis (RO) water as previously described.33 For all experiments described below, newly fertilized eggs were staged according to previously described methods.34 All fish were handled and treated in accordance with approved Institutional Animal Care and Use Committee protocols at the University of South Carolina, Columbia.
Chemicals
TDCIPP (99% purity) and 5-azaC (>98% purity) were purchased from ChemService (West Chester, PA) and Tocris (Bristol, United Kingdom), respectively. Stock and working solutions of each chemical were prepared and stored as previously described.33
DNMT Activity Assays
Newly fertilized eggs were collected immediately after spawning and placed in groups of approximately 100 per Petri dish within a light- and temperature-controlled incubator. Embryos (100 per time point) were collected at 2, 10, and 24 hpf and stored at −20 °C. Nuclear proteins were extracted from whole embryo pools using an EpiQuik Nuclear Extraction Kit (Epigentek Group, Farmingdale, NY). Nuclear extract was kept on ice and immediately quantified using a BCA Protein Assay (Pierce Biotechnology, Rockford, IL) following the manufacturer’s instructions. Optical density of the colorimetric reaction was quantified using a VICTOR X3Multilabel Plate Reader (PerkinElmer, Waltham, MA), and total protein was quantified using a standard curve generated from bovine serum albumin.
DNMT inhibition was quantified using an EpiQuik DNMT Activity/Inhibition Assay Ultra Fluorometric Kit (Epigentek Group, Farmingdale, NY). DNMT inhibition within nuclear extracts (6.5 μg of protein per reaction) derived from 2-, 10-, and 24-hpf embryos was quantified in the presence of vehicle (0.1% dimethyl sulfoxide, DMSO), 250 μM 5-azaC, or TDCIPP (63, 125, 250, or 500 μM). All reactions were conducted in triplicate. Fluorescence was measured by a VICTOR X3Multilabel Plate Reader (PerkinElmer, Waltham, MA) and data were corrected for background and reported as relative fluorescent units.
Embryonic Exposures for Genomic DNA Extractions
Newly fertilized eggs were collected immediately after spawning and placed in groups of approximately 100 per Petri dish within a light- and temperature-controlled incubator until 0.75 hpf (2-cell stage). Prior to each experiment, 50 mL glass beakers were thoroughly rinsed with DMSO and RO water. Viable 5D embryos were exposed to 10 mL of vehicle (0.1% DMSO), 250 μM 5-azaC, or 2 μM TDCIPP in triplicate glass beakers (70 embryos per replicate beaker) under a 14 h:10 h light:dark cycle and static conditions at 28 °C from 0.75 hpf to 2 or 6 hpf. To minimize the potential for false negative findings and confounding effects of systemic toxicity at 2 and 6 hpf, 250 μM 5-azaC and 2 μM TDCIPP were selected based on the (1) absence of effects on cell cycle, overall embryo size, or cell morphology from 0.75 to 2 hpf; (2) absence of developmental delays from 0.75 to 6 hpf; but (3) presence of severe malformations (trunk curvature, tail malformations, craniofacial malformations, decreased body length, pericardial edema, and yolk sac edema) at 24 hpf (data not shown). Embryos (25/replicate) were collected at either 2 or 6 hpf, transferred from beakers to 2 mL cryovials, snap-frozen in liquid nitrogen, and stored at −80 °C. These experiments resulted in three independent replicate samples for each time point and treatment group.
Quantification of Internal Embryonic Doses of TDCIPP and BDCIPP
Embryos (50 per replicate) were treated with vehicle (0.1% DMSO) or 2 μM TDCIPP within six replicate beakers per treatment starting at 0.75 hpf. At 2 and 6 hpf, embryos were transferred from three replicate beakers to three 2 mL cryovials, snap-frozen in liquid nitrogen, and stored at −80 °C until analysis. Frozen embryos were then processed and analyzed for TDCIPP and BDCIPP concentrations using previously described protocols.20 Method detection limits (MDLs) were defined as three times the standard deviation of lab blanks (if present) or three times the noise. MDLs for TDCIPP and BDCIPP were 0.1 and 1.0 ng, respectively. Recoveries averaged 103 ± 8% and 118 ± 17% for the internal standards, D15-TDCIPP and D10-BDCIPP, respectively.
Quantification of 5-Methyl-2′-deoxycytidine (5-mdC) and 2′-Deoxyguanosine (dG)
Genomic DNA (1 μg per replicate sample) was digested and processed similar to previously described methods.35 Enzymes were removed by extraction with chloroform, and the resulting aqueous layer was subjected directly to LC-MS/MS and LC-MS/MS/MS analysis for quantification of 5-mdC and dG, respectively, as previously described.35 The amounts of 5-mdC and dG (in moles) in nucleoside mixtures were calculated from area ratios of peaks found in selected-ion chromatograms for the analytes over their corresponding isotope-labeled standards (SI Figures S1 and S2), amounts of the labeled standards added (in moles), and calibration curves (SI Figure S3). 5-mdC levels (% of dG) were calculated by comparing the moles of 5-mdC relative to the moles of dG.
DNA Methylome Profiling Using Whole-Genome Bisulfite Sequencing (WGBS)
Genomic DNA was extracted from pooled 2-hpf embryos (25 per replicate) using a Wizard Genomic DNA Purification Kit (Promega, Madison, WI). After elution of DNA in 25 μL of nuclease-free 1× Tris-EDTA buffer, the concentration and quality of all DNA samples were quantified using a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), respectively; as the DNA concentration within one of the vehicle (0.1% DMSO) samples was <0.5 ng/uL, only two vehicle control samples (vs three samples for 250 μM 5-azaC and 2 μM TDCIPP) were prepared for sequencing. DNA samples (8 total) were then processed using an EZ DNA Methylation-Lightning Kit (Zymo Research Corp., Irvine, CA) for bisulfite conversion, and nonbarcoded sequencing libraries were then prepared using a TruSeq DNA Methylation Kit (Illumina, San Diego, CA). The concentration and quality of sequencing libraries were quantified using a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), respectively, and eight libraries were then paired-end (2×50) sequenced on eight lanes (one library per lane) of a HiSeq 2500 Sequencing System (Illumina, San Diego, CA). This sequencing strategy generated 160–185 M short reads per replicate, providing sufficient coverage (∼6–7× per sample) for differential methylation analysis by WGBS.36
All bisulfite-converted, 2×50 sequence reads (Reads 1 and 2) were first checked for quality control using FastQC. Approximately 97% of Read 1 sequences resulted in a quality score ≥Q30, whereas only 76% of Read 2 sequences resulted in a quality score ≥Q30. Therefore, Read 1 sequences were aligned against the current zebrafish genome assembly (GRCz10) using Bismark v0.14.337 and Bowtie 2 v2.2.538 to determine the methylation status of all cytosines within each sample. Bismark’s output included BAM files (.bam) containing alignments, methylation call strings, and a report summarizing alignment statistics and percent methylated cytosines within a CpG, CHG, and CHH context, where H = A, T, or C. Bismark-generated BAM files were then sorted and converted into SAM files (.sam) using Samtools.39 SAM files were then imported into methylKit40 using the read.bismark function; the read.context option was set to “CpG”, whereas all other options were set to default values for the read.bismark function. This approach allowed us to determine percent cytosine methylation at base-pair resolution within a CpG context on each of 25 chromosomes. Using percent cytosine methylation data, signal-to-noise ratios (SNR = (μ1 – μ0)/(σ1 + σ0), where μ1 = treatment mean; μ0 = vehicle control mean; σ1 = treatment standard deviation; σ0 = vehicle control standard deviation) were then calculated at all available positions on each of 25 chromosomes to account for the magnitude and variation of cytosine methylation relative to vehicle controls. SNRs > 0 and SNRs < 0 represent hypermethylation and hypomethylation, respectively, relative to vehicle controls.
Secondary Analysis of TDCIPP-Specific WGBS Data
TDCIPP-specific data were sorted by SNR, and SNRs between −5 and +5 were removed from the data set. SNR thresholds of >5 or <5 were selected in order to (1) filter out highly variable vs consistent, TDCIPP-related impacts on cytosine methylation across independent treatment replicates and (2) prioritize regions of interest that were reliably impacted by TDCIPP exposure. Across all 25 chromosomes, 189 out of 25 270 positions were identified and mapped to the current zebrafish genome assembly (GRCz10) hosted by the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/genome?term=danio%20rerio). After mapping each position to the zebrafish genome, we then (1) determined whether the position mapped to an intragenic (exon or intron) or intergenic region; (2) identified the NCBI Gene ID if mapped to an intragenic region; (3) identified the NCBI Gene ID corresponding to genes immediately upstream and downstream of the mapped position; and 4) determined whether the position mapped to a NCBI-predicted CpG island. Genomic sequences for all identified CpG islands were also compared against NCBI’s nucleotide collection (nr/nt) database using Basic Local Alignment Search Tool (BLASTn) to identify potential human, mouse, or rat orthologs and, if mapped to an intragenic region, corresponding NCBI Gene IDs.
Statistical Analyses
Statistical procedures for DNMT data and internal embryonic doses were performed using SPSS Statistics 22.0 (Chicago, IL). A general linear model (GLM) analysis of variance (ANOVA) (α = 0.05) was used for all data, as these data did not meet the equal variance assumption for non-GLM ANOVAs. Pair-wise Tukey-based multiple comparisons of least-squares means were performed to identify significant differences among treatment groups.
Accession Number
Raw Illumina (fastq.gz) sequencing files (eight files totaling 70.98 GB) are available via NCBI’s BioProject database under BioProject ID PRJNA330715 (http://www.ncbi.nlm.nih.gov/bioproject/330715).
Results and Discussion
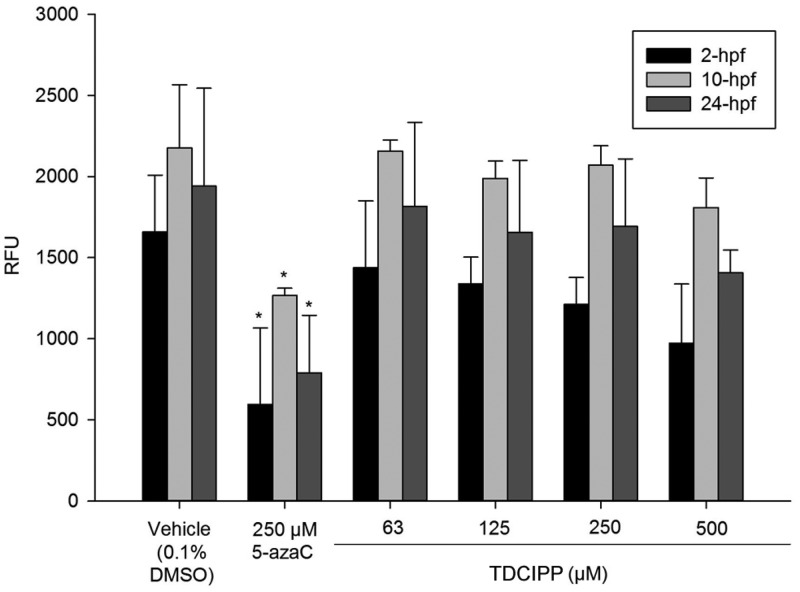
Since our previous study suggested that normal DNA methylation at the end of cleavage (2 hpf) was decreased in TDCIPP-treated embryos,20 we tested the hypothesis that TDCIPP inhibits zebrafish DNMT activity in vitro. We relied on an assay containing universal DNA substrate in order to identify potential impacts of TDCIPP on multiple zebrafish DNMTs present during early embryonic development.41−44 Although 5-azaC significantly inhibited zebrafish DNMT within 2-, 10-, and 24-hpf nuclear extracts, TDCIPP did not affect DNMT activity up to 500 μM (Figure 1), suggesting that TDCIPP-induced impacts on DNA methylation during cleavage are likely not due to direct DNMT inhibition. Rather, these findings point to the possibility that TDCIPP may be impacting the concentration of available methyl donors (e.g., S-adenosyl-l-methionine) during early embryonic development.
Figure 1.
TDCIPP does not inhibit zebrafish DNMT in vitro. Mean relative fluorescence units (RFU) ± standard deviation. N = three replicates/group.
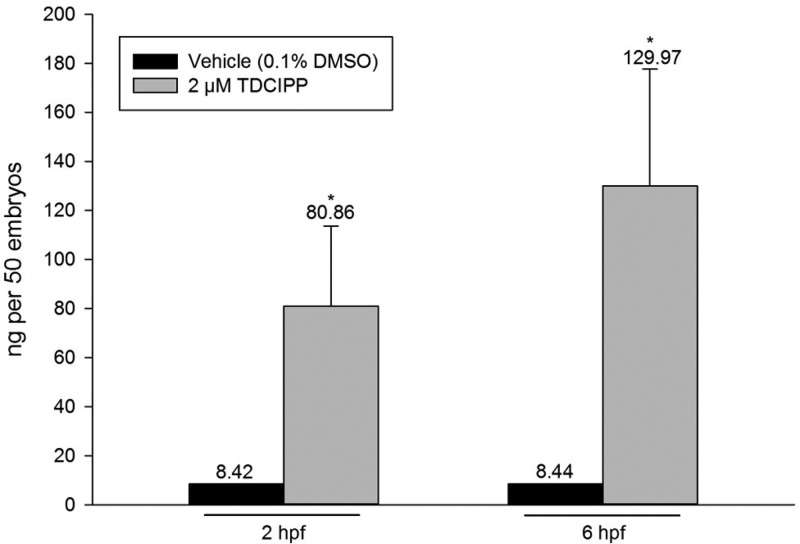
To help guide interpretation of our findings, we quantified mean internal doses of TDCIPP and BDCIPP within 2- and 6-hpf embryos following initiation of exposure to vehicle (0.1% DMSO) or 2 μM TDCIPP at 0.75 hpf. Although mean internal doses (per 50 embryos) of TDCIPP were <8.5 ng within vehicle controls, which is consistent with typical background concentrations, and BDCIPP was not detected across all treatment groups, mean internal doses (per 50 embryos) of TDCIPP at 2 and 6 hpf were 80.86 and 129.97 ng, respectively (Figure 2). Therefore, these data demonstrate that TDCIPP uptake is rapid, TDCIPP is not metabolized to BDCIPP during the first 6 h of embryonic development, and TDCIPP (but not BDCIPP) is likely responsible for any impacts on DNA methylation.
Figure 2.
TDCIPP uptake occurs by 2 hpf following initiation of exposure at 0.75 hpf. Mean internal dose (per 50 embryos) ± standard deviation. BDCIPP was less than the method detection limit (MDL) across all groups. N = three replicate pools/group. Asterisk (*) denotes significant treatment effect (p < 0.05) relative to vehicle control (0.1% DMSO).
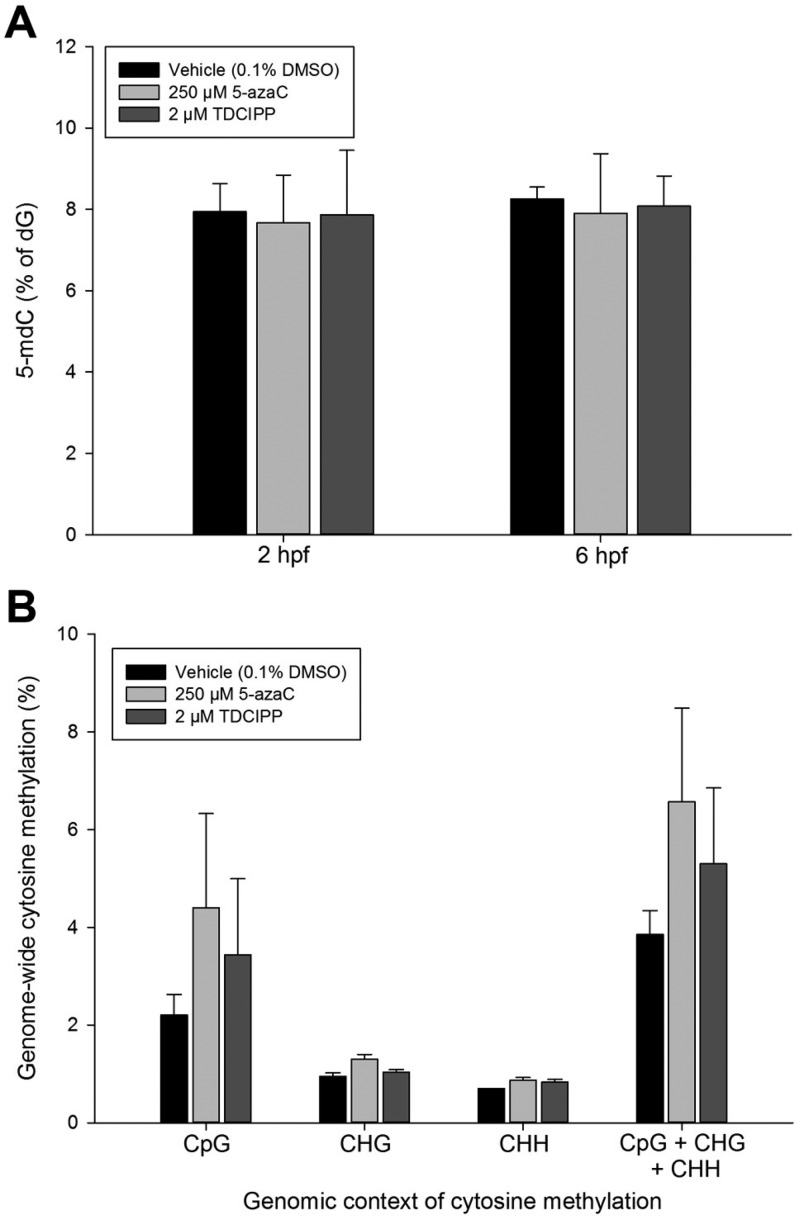
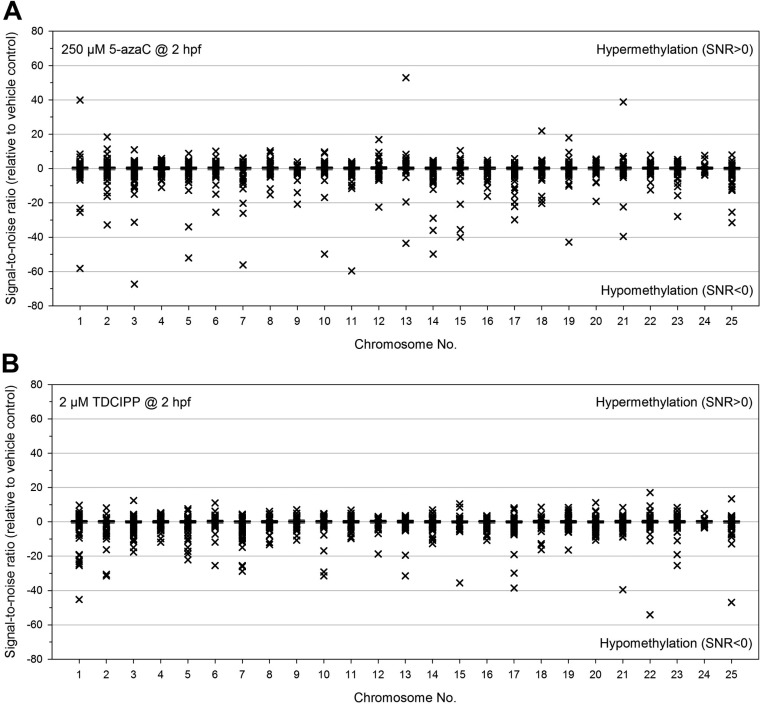
To address the potential for 5-azaC- and TDCIPP-induced impacts on DNA methylation, we relied on (1) LC-MS/MS and LC-MS/MS/MS to quantify 5-mdC and dG, respectively, within genomic DNA extracted from vehicle-, 5-azaC-, and TDCIPP-exposed 2- and 6-hpf embryos and (2) WGBS on Illumina’s HiSeq 2500 platform to identify the magnitude and extent, if any, of 5-azaC- and TDCIPP-induced effects on the entire DNA methylome at 2 hpf (the end of cleavage) at single base-pair resolution. Although 5-azaC and TDCIPP did not significantly impact 5-mdC levels at 2 or 6 hpf (Figure 3A) nor global cytosine methylation at 2 hpf (Figure 3B), 5-azaC and TDCIPP exposure resulted in chemical- and chromosome-specific alterations in cytosine methylation within a CpG context at 2 hpf (Figure 4), suggesting that aggregated analysis of global cytosine methylation lacks sufficient sensitivity to detect spatially resolved, position-specific effects. Interestingly, exposure to 5-azaC from 0.75 to 2 hpf resulted in a stronger impact than TDCIPP on cytosine methylation at 2 hpf, leading to strong hypermethylation (SNR > 20) within four chromosomes and strong hypomethylation (SNR < 20) within 19 chromosomes (Figure 4A). In contrast, although the overall magnitude of TDCIPP-induced impacts on cytosine methylation was less than 5-azaC, TDCIPP only resulted in strong hypomethylation (SNR < 20) across 13 chromosomes (in particular, chr1, chr22, and chr25) (Figure 4B).
Figure 3.
5-azaC and TDCIPP do not significantly impact total 5-mdC levels at 2 or 6 hpf (A) nor global cytosine methylation at 2 hpf (B) following initiation of exposure at 0.75 hpf. Mean 5-mdC (A) or cytosine methylation (B) ± standard deviation. N = three replicates per group within Panel A. N = two replicates per control group and three replicates per treatment group within Panel B.
Figure 4.
5-azaC (A) and TDCIPP (B) exposure results in chromosome-specific alterations in cytosine methylation at 2 hpf following initiation of exposure at 0.75 hpf. All signal-to-noise ratios (SNRs) are relative to vehicle controls and reflect variation among two replicates per control group and three replicates per treatment group. SNRs > 0 and SNRs < 0 represent hypermethylation and hypomethylation, respectively, relative to vehicle controls.
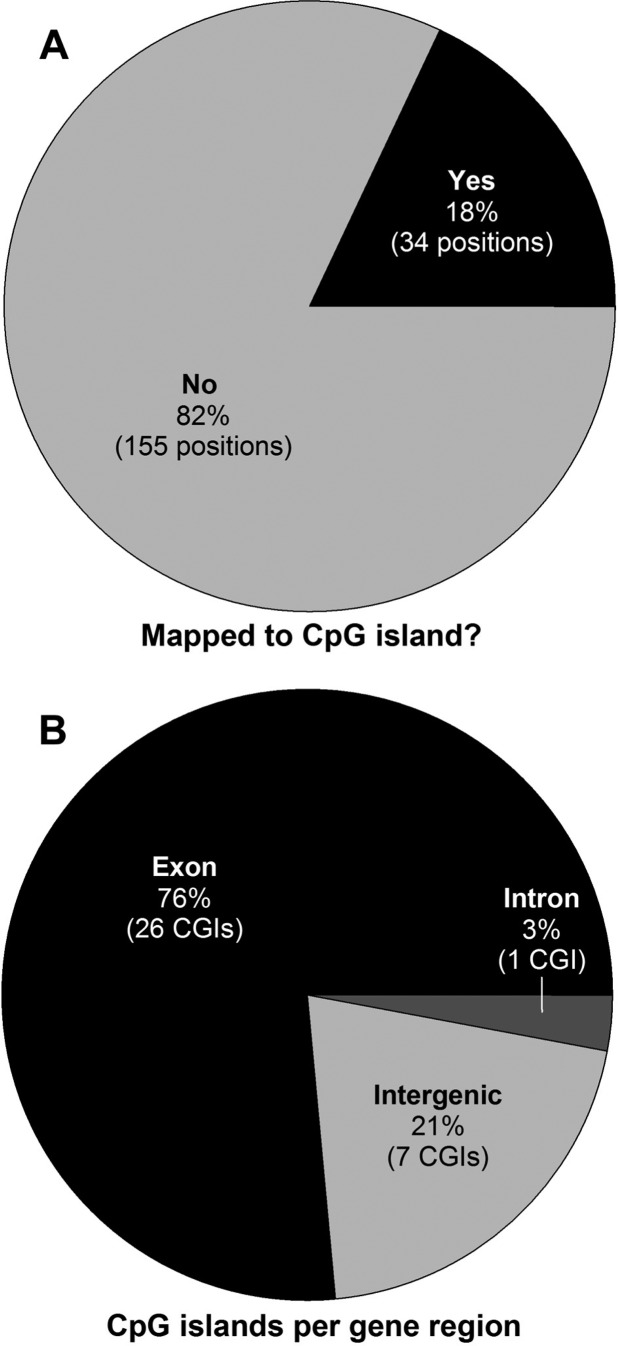
On the basis of secondary analysis of our TDCIPP-specific WGBS data, we identified 39 out of 189 positions (20%) with a SNR > 5 and 150 out of 189 positions (80%) with a SNR < 5 (Supplemental File 3), indicating that, based on a SNR threshold that filters out positions with highly variable responses, hypomethylation (relative to vehicle controls) represented the large majority of reproducible, TDCIPP-induced impacts on the zebrafish DNA methylome at 2 hpf. Moreover, with the exception of chr24, TDCIPP-induced alterations in cytosine methylation occurred on all other 24 chromosomes (in particular, chr1, chr 2, chr 3, chr 5, chr 7, and chr 20) (Supplemental File 3), suggesting that TDCIPP-induced epigenetic modifications are genome-wide and not localized to certain loci. Interestingly, contrary to our working hypothesis, 154 out of 189 positions (81.5%) mapped to intragenic regions of the zebrafish genome (151 positions mapped to exons, whereas only 3 positions mapped to introns), whereas only 35 out of 189 positions (18.5%) mapped to intergenic regions (which includes promoter regions) (Figure 5).
Figure 5.
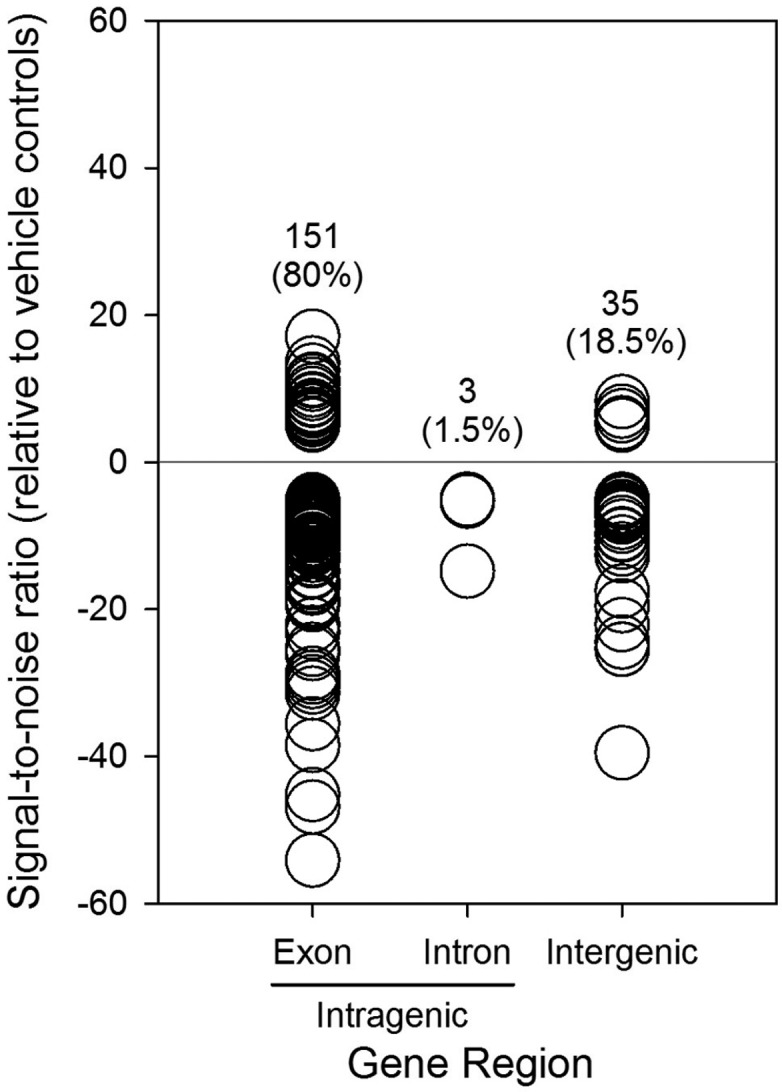
TDCIPP exposure from 0.75 to 2 hpf results in genome-wide hypomethylation within intragenic regions of the zebrafish genome. Intragenic regions include exons and introns, whereas intergenic regions represent sequences between genes.
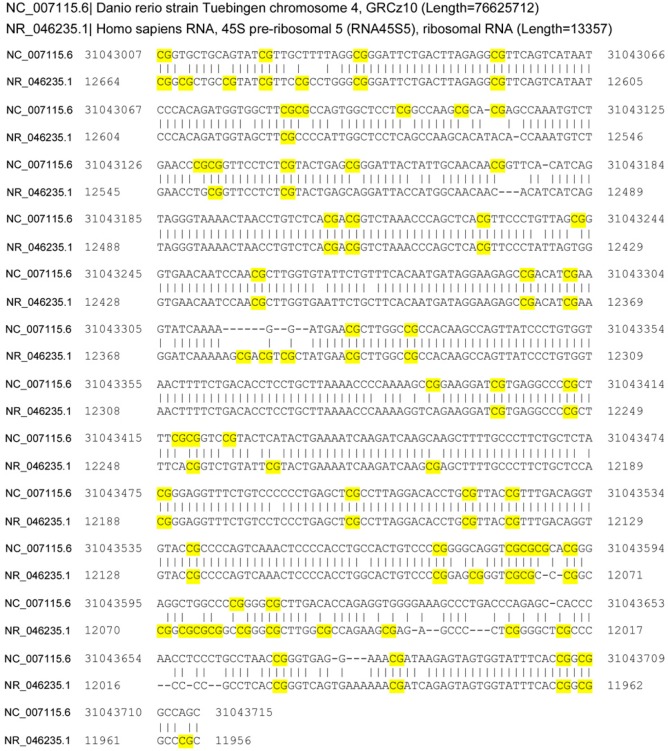
In addition to revealing the location of TDCIPP-induced impacts on the zebrafish genome, we also determined whether these effects occurred within or outside of CpG islands. Surprisingly, 155 out of 189 positions (82%) mapped outside of CpG islands (Figure 6A). For the remaining positions (34 out of 189) that mapped within CpG islands, 79% of CpG islands identified (27 out of 34) were localized to intragenic regions of the zebrafish genome (with the majority being mapped to exons) (Figure 6B). For all 34 CpG islands, we then used BLASTn to identify potential orthologs within the human, mouse, and rat genome. Although no orthologs were identified within the rat or mouse genome, one ortholog was identified within the human genome (Supplemental File 3), where a zebrafish-specific 726-bp fragment of a 1373-bp intergenic CpG island on chr4 was 87% identical to an exon-localized CpG island within human 45S preribosomal 5 (RNA45S5, NCBI GeneID: 100861532) on chr22 (Figure 7).
Figure 6.
TDCIPP exposure from 0.75 to 2 hpf predominantly impacts positions mapped outside of CpG islands (A) and, for the remaining positions mapped within CpG islands, TDCIPP-induced effects were localized to intragenic regions of the zebrafish genome (B).
Figure 7.
BLASTn identifies a CpG island localized to human 45S preribosomal 5 as an ortholog (87% identity) to a zebrafish-specific, TDCIPP-susceptible 726-bp fragment of a 1373-bp intergenic CpG island on chr4. CpG sites are highlighted yellow.
This study yielded two important yet unexpected conclusions: (1) TDCIPP exposure during cleavage predominantly results in non-CpG island hypomethylation localized to intragenic regions of the zebrafish genome; and (2) WGBS-based identification of contaminant-susceptible CpG islands within the zebrafish methylome has the potential to identify human orthologs for epigenetic-focused environmental epidemiology studies. To date, the majority of the scientific literature has focused on the role of CpG island methylation within promotor (intergenic) regions in regulating gene transcription.7 Moreover, most investigations have been limited to studying environmentally induced changes in DNA methylation within small, targeted regions of the genome rather than nontargeted, unbiased profiling of the entire methylome.45 By using high-resolution WGBS to scan the entire zebrafish DNA methylome, we revealed that, contrary to our working hypothesis, non-CpG island hypomethylation within exons accounted for the majority of TDCIPP-induced effects, suggesting that strategies for enrichment of CpG islands prior to sequencing, such as methylated DNA affinity capture coupled with sequencing (MethylCap-seq) or reduced representation bisulfite sequencing (RRBS), may, although more cost-effective, lead to a high proportion of false-negative results. Moreover, the use of methods to eliminate non-CpG islands prior to sequencing may limit the probability of identifying potential candidate loci for future investigations in zebrafish, other animal models, or human populations.
In conclusion, we have revealed that TDCIPP-induced impacts to the zebrafish DNA methylome at 2 hpf are complex and primarily localized to regions outside of classical CpG islands. As our data are based on a single TDCIPP concentration and developmental stage, future research should focus on monitoring DNA methylation dynamics within WGBS-identified, TDCIPP-susceptible loci across multiple TDCIPP concentrations and developmental stages using a cost-effective, targeted sequencing-based strategy such as bisulfite amplicon sequencing.46 In addition, as we have identified a CpG island within the human genome that may be susceptible to TDCIPP (or other flame retardants and plasticizers), future environmental epidemiological studies should consider measuring DNA methylation within this region to determine whether there are potential associations between chemical exposure, adverse health outcomes, and DNA methylation status.
Acknowledgments
Funding was provided by the National Institutes of Health (R21ES022797 and R21ES025392). We gratefully thank Dr. Robert Tanguay (Oregon State University) for providing founder fish to establish our 5D zebrafish colony, Dr. R. Sean Norman (University of South Carolina) for use of the VICTOR X3Multilabel Plate Reader, and John Weger and Clay Clark (Institute for Integrative Genome Biology, University of California, Riverside) for Illumina sequencing services.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.est.6b03656.
Spreadsheets containing processed and analyzed WGBS data (XLSX)
Supporting data for 5-mdC and dG analysis (Figures S1–S3), and position-specific signal-to-noise ratios for cytosine methylation on each of 25 chromosomes (Figures S4–S28) (PDF)
Spreadsheets containing secondary analysis of TDCIPP-specific cytosine methylation data (XLSX)
Author Present Address
# J. K. Leet. Columbia Environmental Research Center, U.S. Geological Survey, Columbia, Missouri 65201, United States.
The authors declare no competing financial interest.
Supplementary Material
References
- Tadros W.; Lipshitz H. D. The maternal-to-zygotic transition: a play in two acts. Development 2009, 136 (18), 3033–42. 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Niakan K. K.; Han J.; Pedersen R. A.; Simon C.; Pera R. A. Human pre-implantation embryo development. Development 2012, 139 (5), 829–41. 10.1242/dev.060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhanni A. A.; McGowan R. A. Global changes in genomic methylation levels during early development of the zebrafish embryo. Dev. Genes Evol. 2004, 214 (8), 412–7. 10.1007/s00427-004-0418-0. [DOI] [PubMed] [Google Scholar]
- Martin C. C.; Laforest L.; Akimenko M. A.; Ekker M. A role for DNA methylation in gastrulation and somite patterning. Dev. Biol. 1999, 206 (2), 189–205. 10.1006/dbio.1998.9105. [DOI] [PubMed] [Google Scholar]
- Ding Y. B.; Long C. L.; Liu X. Q.; Chen X. M.; Guo L. R.; Xia Y. Y.; He J. L.; Wang Y. X. 5-aza-2′-deoxycytidine leads to reduced embryo implantation and reduced expression of DNA methyltransferases and essential endometrial genes. PLoS One 2012, 7 (9), e45364. 10.1371/journal.pone.0045364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L.; Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat. Rev. Genet. 2012, 13 (3), 153–62. 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- Burris H. H.; Baccarelli A. A. Environmental epigenetics: from novelty to scientific discipline. J. Appl. Toxicol. 2014, 34 (2), 113–6. 10.1002/jat.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M.; Gudsnuk K.; Franks B.; Madrid J.; Miller R. L.; Perera F. P.; Champagne F. A. Sex-specific epigenetic disruption and behavioral changes following low-dose in utero bisphenol A exposure. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (24), 9956–61. 10.1073/pnas.1214056110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson O. S.; Nahar M. S.; Faulk C.; Jones T. R.; Liao C.; Kannan K.; Weinhouse C.; Rozek L. S.; Dolinoy D. C. Epigenetic responses following maternal dietary exposure to physiologically relevant levels of bisphenol A. Environ. Mol. Mutagen 2012, 53 (5), 334–42. 10.1002/em.21692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R.; Chen F.; Chang F.; Bai Y.; Chen L. Persistent overexpression of DNA methyltransferase 1 attenuating GABAergic inhibition in basolateral amygdala accounts for anxiety in rat offspring exposed perinatally to low-dose bisphenol A. J. Psychiatr. Res. 2013, 47 (10), 1535–44. 10.1016/j.jpsychires.2013.05.013. [DOI] [PubMed] [Google Scholar]
- Strakovsky R. S.; Wang H.; Engeseth N. J.; Flaws J. A.; Helferich W. G.; Pan Y. X.; Lezmi S. Developmental bisphenol A (BPA) exposure leads to sex-specific modification of hepatic gene expression and epigenome at birth that may exacerbate high-fat diet-induced hepatic steatosis. Toxicol. Appl. Pharmacol. 2015, 284 (2), 101–12. 10.1016/j.taap.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z.; Xia W.; Chang H.; Huo W.; Li Y.; Xu S. Paternal BPA exposure in early life alters Igf2 epigenetic status in sperm and induces pancreatic impairment in rat offspring. Toxicol. Lett. 2015, 238 (3), 30–8. 10.1016/j.toxlet.2015.08.009. [DOI] [PubMed] [Google Scholar]
- Dolinoy D. C.; Huang D.; Jirtle R. L. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (32), 13056–61. 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar M. S.; Liao C.; Kannan K.; Harris C.; Dolinoy D. C. In utero bisphenol A concentration, metabolism, and global DNA methylation across matched placenta, kidney, and liver in the human fetus. Chemosphere 2015, 124, 54–60. 10.1016/j.chemosphere.2014.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar M. S.; Kim J. H.; Sartor M. A.; Dolinoy D. C. Bisphenol A-associated alterations in the expression and epigenetic regulation of genes encoding xenobiotic metabolizing enzymes in human fetal liver. Environ. Mol. Mutagen 2014, 55 (3), 184–95. 10.1002/em.21823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H.; Rozek L. S.; Soliman A. S.; Sartor M. A.; Hablas A.; Seifeldin I. A.; Colacino J. A.; Weinhouse C.; Nahar M. S.; Dolinoy D. C. Bisphenol A-associated epigenomic changes in prepubescent girls: a cross-sectional study in Gharbiah, Egypt. Environ. Health 2013, 12, 33. 10.1186/1476-069X-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, Environmental Health Criteria 209-Flame Retardants: Tris(chloropropyl)phosphate and tris(2-chloroethyl)phosphate. In Geneva, Switzerland, 1998; p 129pp.
- van der Veen I.; de Boer J. Phosphorus flame retardants: properties, production, environmental occurrence, toxicity and analysis. Chemosphere 2012, 88 (10), 1119–53. 10.1016/j.chemosphere.2012.03.067. [DOI] [PubMed] [Google Scholar]
- Hoffman K.; Butt C. M.; Chen A.; Limkakeng A. T. Jr.; Stapleton H. M. High Exposure to Organophosphate Flame Retardants in Infants: Associations with Baby Products. Environ. Sci. Technol. 2015, 49 (24), 14554–9. 10.1021/acs.est.5b03577. [DOI] [PubMed] [Google Scholar]
- McGee S. P.; Cooper E. M.; Stapleton H. M.; Volz D. C. Early zebrafish embryogenesis is susceptible to developmental TDCPP exposure. Environ. Health Perspect 2012, 120 (11), 1585–91. 10.1289/ehp.1205316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T.; Wang Q.; Shi Q.; Fang Q.; Guo Y.; Zhou B. Bioconcentration, metabolism and alterations of thyroid hormones of Tris(1,3-dichloro-2-propyl) phosphate (TDCPP) in Zebrafish. Environ. Toxicol. Pharmacol. 2015, 40 (2), 581–6. 10.1016/j.etap.2015.08.020. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Lai N. L.; Wang X.; Guo Y.; Lam P. K.; Lam J. C.; Zhou B. Bioconcentration and transfer of the organophorous flame retardant 1,3-dichloro-2-propyl phosphate causes thyroid endocrine disruption and developmental neurotoxicity in zebrafish larvae. Environ. Sci. Technol. 2015, 49 (8), 5123–32. 10.1021/acs.est.5b00558. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Liang K.; Liu J.; Yang L.; Guo Y.; Liu C.; Zhou B. Exposure of zebrafish embryos/larvae to TDCPP alters concentrations of thyroid hormones and transcriptions of genes involved in the hypothalamic-pituitary-thyroid axis. Aquat. Toxicol. 2013, 126, 207–13. 10.1016/j.aquatox.2012.11.009. [DOI] [PubMed] [Google Scholar]
- Fu J.; Han J.; Zhou B.; Gong Z.; Santos E. M.; Huo X.; Zheng W.; Liu H.; Yu H.; Liu C. Toxicogenomic responses of zebrafish embryos/larvae to tris(1,3-dichloro-2-propyl) phosphate (TDCPP) reveal possible molecular mechanisms of developmental toxicity. Environ. Sci. Technol. 2013, 47 (18), 10574–10582. 10.1021/es401265q. [DOI] [PubMed] [Google Scholar]
- Liu C.; Wang Q.; Liang K.; Liu J.; Zhou B.; Zhang X.; Liu H.; Giesy J. P.; Yu H. Effects of tris(1,3-dichloro-2-propyl) phosphate and triphenyl phosphate on receptor-associated mRNA expression in zebrafish embryos/larvae. Aquat. Toxicol. 2013, 128–129, 147–57. 10.1016/j.aquatox.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Jarema K. A.; Hunter D. L.; Shaffer R. M.; Behl M.; Padilla S. Acute and developmental behavioral effects of flame retardants and related chemicals in zebrafish. Neurotoxicol. Teratol. 2015, 52 (Pt B), 194–209. 10.1016/j.ntt.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes P. D.; Haggard D. E.; Gonnerman G. D.; Tanguay R. L. Advanced morphological - behavioral test platform reveals neurodevelopmental defects in embryonic zebrafish exposed to comprehensive suite of halogenated and organophosphate flame retardants. Toxicol. Sci. 2015, 145 (1), 177–95. 10.1093/toxsci/kfv044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Lam J. C.; Man Y. C.; Lai N. L.; Kwok K. Y.; Guo Y.; Lam P. K.; Zhou B. Bioconcentration, metabolism and neurotoxicity of the organophorous flame retardant 1,3-dichloro 2-propyl phosphate (TDCPP) to zebrafish. Aquat. Toxicol. 2015, 158, 108–15. 10.1016/j.aquatox.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Dishaw L. V.; Hunter D. L.; Padnos B.; Padilla S.; Stapleton H. M. Developmental exposure to organophosphate flame retardants elicits overt toxicity and alters behavior in early life stage zebrafish (Danio rerio). Toxicol. Sci. 2014, 142 (2), 445–54. 10.1093/toxsci/kfu194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Lam J. C.; Han J.; Wang X.; Guo Y.; Lam P. K.; Zhou B. Developmental exposure to the organophosphorus flame retardant tris(1,3-dichloro-2-propyl) phosphate: estrogenic activity, endocrine disruption and reproductive effects on zebrafish. Aquat. Toxicol. 2015, 160, 163–71. 10.1016/j.aquatox.2015.01.014. [DOI] [PubMed] [Google Scholar]
- Liu X.; Ji K.; Jo A.; Moon H. B.; Choi K. Effects of TDCPP or TPP on gene transcriptions and hormones of HPG axis, and their consequences on reproduction in adult zebrafish (Danio rerio). Aquat. Toxicol. 2013, 134–135, 104–11. 10.1016/j.aquatox.2013.03.013. [DOI] [PubMed] [Google Scholar]
- Zhu Y.; Ma X.; Su G.; Yu L.; Letcher R. J.; Hou J.; Yu H.; Giesy J. P.; Liu C. Environmentally Relevant Concentrations of the Flame Retardant Tris(1,3-dichloro-2-propyl) Phosphate Inhibit Growth of Female Zebrafish and Decrease Fecundity. Environ. Sci. Technol. 2015, 49 (24), 14579–87. 10.1021/acs.est.5b03849. [DOI] [PubMed] [Google Scholar]
- Isales G. M.; Hipszer R. A.; Raftery T. D.; Chen A.; Stapleton H. M.; Volz D. C. Triphenyl phosphate-induced developmental toxicity in zebrafish: potential role of the retinoic acid receptor. Aquat. Toxicol. 2015, 161, 221–30. 10.1016/j.aquatox.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C. B.; Ballard W. W.; Kimmel S. R.; Ullmann B.; Schilling T. F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203 (3), 253–310. 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Yu Y.; Guerrero C. R.; Liu S.; Amato N. J.; Sharma Y.; Gupta S.; Wang Y. Comprehensive Assessment of Oxidatively Induced Modifications of DNA in a Rat Model of Human Wilson’s Disease. Mol. Cell. Proteomics 2016, 15 (3), 810–7. 10.1074/mcp.M115.052696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller M. J.; Hansen K. D.; Meissner A.; Aryee M. J. Coverage recommendations for methylation analysis by whole-genome bisulfite sequencing. Nat. Methods 2015, 12 (3), 230–232. 1 p following 232 10.1038/nmeth.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F.; Andrews S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27 (11), 1571–2. 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B.; Salzberg S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9 (4), 357–9. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Handsaker B.; Wysoker A.; Fennell T.; Ruan J.; Homer N.; Marth G.; Abecasis G.; Durbin R. Genome Project Data Processing, S., The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25 (16), 2078–9. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akalin A.; Kormaksson M.; Li S.; Garrett-Bakelman F. E.; Figueroa M. E.; Melnick A.; Mason C. E. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome biology 2012, 13 (10), R87. 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluru N.; Kuo E.; Helfrich L. W.; Karchner S. I.; Linney E. A.; Pais J. E.; Franks D. G. Developmental exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin alters DNA methyltransferase (dnmt) expression in zebrafish (Danio rerio). Toxicol. Appl. Pharmacol. 2015, 284 (2), 142–51. 10.1016/j.taap.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X.; Corrales J.; Thornton C.; Scheffler B. E.; Willett K. L. Global and gene specific DNA methylation changes during zebrafish development. Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol. 2013, 166 (1), 99–108. 10.1016/j.cbpb.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K.; Shimoda N.; Takanaga S.; Hozumi S.; Kikuchi Y. Expression patterns of dnmt3aa, dnmt3ab, and dnmt4 during development and fin regeneration in zebrafish. Gene Expression Patterns 2014, 14 (2), 105–10. 10.1016/j.gep.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Mhanni A. A.; McGowan R. A. Variations in DNA (cytosine-5)-methyltransferase-1 expression during oogenesis and early development of the zebrafish. Dev. Genes Evol. 2002, 212 (11), 530–3. 10.1007/s00427-002-0275-7. [DOI] [PubMed] [Google Scholar]
- Rozek L. S.; Dolinoy D. C.; Sartor M. A.; Omenn G. S. Epigenetics: relevance and implications for public health. Annu. Rev. Public Health 2014, 35, 105–22. 10.1146/annurev-publhealth-032013-182513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masser D. R.; Stanford D. R.; Freeman W. M.. Targeted DNA methylation analysis by next-generation sequencing. J. Visualized Exp. 2015, ((96)), , e52488. 10.3791/52488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.