Abstract
Cyclical expression of cell-autonomous circadian clock components and key metabolic regulators coordinate often discordant and distant cellular processes for efficient metabolism. Perturbation of these cycles, either by genetic manipulation, disruption of light/dark cycles, or, most relevant to the human population, via eating patterns, contributes to obesity and dysmetabolism. Time-restricted feeding (TRF), during which time of access to food is restricted to a few hours, without caloric restriction, supports robust metabolic cycles and protects against nutritional challenges that predispose to obesity and dysmetabolism. The mechanism by which TRF imparts its benefits is not fully understood but likely involves entrainment of metabolically active organs through gut signaling. Understanding the relationship of feeding pattern and metabolism could yield novel therapies for the obesity pandemic.
Circadian Rhythms and Metabolism
More than one-third of adults in the USA are obese [1], and this exerts a significant burden on national healthcare spending [2]. In the past, an underappreciation of the complexity of bodyweight regulation led to ineffective or scientifically unsound recommendations [3]. In recent years, tremendous resources have been committed to curtail obesity, and these efforts have had some success [4]. From 2003 to 2012 the prevalence of obesity in the USA has plateaued [1], after increasing consistently in the previous four decades. Recent findings have dramatically improved our understanding of the components of metabolic homeostasis and how their perturbation can lead to obesity. Whereas obesity has long been viewed as a simple behavioral problem of overnutrition, now it is appreciated as a phenomenon involving multiple genetic, epigenetic, and environmental factors, as well as multiple organs and commensal organisms. In this review we discuss one facet of the mammalian metabolic homeostasis – the impact of feeding time on metabolism as a whole.
Daily Rhythms: The Central Circadian Pacemaker and the Sleep/Wake Cycle
From cyanobacteria to humans, the majority of organisms have physiological rhythms that cycle with a period of approximately 24 h. The pervasiveness of these rhythms across organisms spanning kingdoms indicates that there is an evolutionary advantage to coordinate cellular machinery to anticipated external stimuli [5,6]. In mammals, virtually every tissue or physiological function exhibits diurnal oscillations. These patterns on a systemic level appear to be temporally coordinated by neuronal and chemical networks which are in turn maintained by various timekeeping mechanisms in the central nervous system (Figure 1) [7]. Central to these rhythms is the endogenous circadian rhythmicity of the hypothalamic suprachiasmatic nucleus (SCN), which is entrained by light [8]. The other dominant daily rhythm is the sleep/wake cycle driven by both circadian system and sleep homeostasis [9,10].
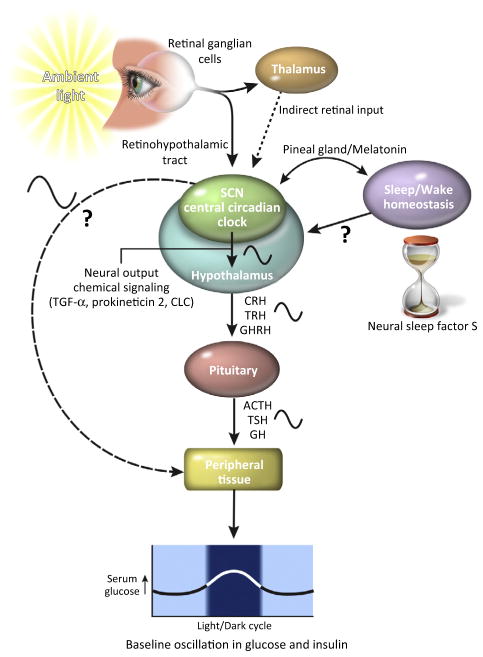
Figure 1.
Central Circadian Control of Endocrine Signals. Specialized retinal ganglion cells in the retina detect ambient light and send this information to the suprachiasmatic nucleus (SCN) through the retinohypothalamic tract. The SCN contains the central circadian clock. This area also receives information about light input indirectly thorough the thalamus. Sleep/wake homeostasis constitutes another oscillatory network. This system has an ‘hourglass’ oscillation that is dependent on the amount of neural sleep factor S (together with other sleep factors) that build up over time. The central circadian clock and the sleep/wake homeostasis systems are tightly regulated through unknown mechanisms. Melatonin from the pineal gland is one compound that can help to synchronize these two systems. Neural and chemical signaling from the SCN to the hypothalamus induces circadian release of multiple releasing hormones [e.g., corticotropin-releasing hormone (CRH), thyrotropin-releasing hormone (TRH), growth hormone releasing hormone (GHRH)]. These in turn cause the pituitary to release several hormones in a circadian manner [adrenocorticotropic hormone (ACTH), thyroid-stimulating hormone (TSH), growth hormone (GH)]. The circadian release of these hormones has an effect on peripheral tissues, but they respond differently to nutrients or stressors depending on the time of the day. For examples, blood glucose oscillates with time, rising above average in the late evening, early night, peaking in the middle of the night, and then at a nadir in the morning. The central circadian clock in the SCN also can modulate circadian cycles in the peripheral organs, although the process is not well understood. Abbreviations: CLC, cardiotrophin-like cytokine; TGF-α, transforming growth factor α.
The SCN is the dominant circadian pacemaker [11]. Ambient light is detected by specialized retinal ganglion cells that project to SCN neurons via the retinohypothalamic tract, as well as through multi-synaptic, indirect pathways via the thalamus [12]. In turn, SCN output is mediated by circadian variation of its neuronal firing, or release of a number of peptides, including arginine vasopressin (AVP), vasoactive intestinal polypeptide (VIP), gastrin-releasing peptide (GRP), transforming growth factor-α (TGFα), prokineticin 2, and cardiotrophin-like cytokine. These diffusible and synaptic outputs synchronize and modulate the activity of the autonomic nervous system and the release of hormones from the hypothalamus, which in turn help to maintain vigorous oscillation of circadian proteins in the peripheral tissue, although the mechanisms are not yet well understood.
Sleep/wake homeostasis has been far more difficult to study. However, sleep is essential for maintaining metabolic health [13–15], likely by facilitating the removal of waste byproducts and restoration of necessary metabolites [16]. Although the sleep/wake cycle and the signals from SCN are highly linked, sleep is heavily influenced by a sleep homeostat akin to an hourglass oscillation. For example, a neural sleep factor (‘S’) rises during waking, and decays during sleep, hence regulating the timing, amount, and intensity of sleep [17]. Melatonin, a pineal gland-derived hormone, plays an important role in synchronizing the sleep/wake cycle and the circadian clock [18,19].
The pathways by which the central circadian system and the sleep/wake cycles control hormonal release, peripheral clock oscillations, and metabolism in other tissues are still being actively investigated. Neurohumoral and neuronal signaling from SCN and brain regions involved in sleep regulation, however, can affect the activity of the hypothalamus and influence the pulsatile release of various hormones, which in turn affect the intermittent release of pituitary hormones [20]. Hence, many hormones that are released by the pituitary are circadian, with their levels oscillating throughout the day. These include plasma levels of cortisol (high in the morning), thyroid stimulating hormone (TSH, high at night), prolactin (high at night), and growth hormone (high at night), which all normally have ~24 h oscillation. In non-diabetic participants, the response to a glucose challenge is very different with the time of day, independently of the route of administration, resulting in higher plasma glucose levels in the evening than in the morning [21]. Post-meal insulin levels also oscillate during a 24 h period, increasing toward the evening and decreasing in the morning. Even sensitivity to insulin is circadian, although not mediated by the hypothalamus [22].
Together, the central circadian pacemaker and sleep/wake homeostasis have tremendous influence over the endocrine system. They can coordinate the physiological activity of various organs with the timing of the release of hormones. As a result, glucose homeostasis, as an example, is highly dynamic; baseline serum glucose and insulin levels, response to glucose or identical meal boluses, and sensitivity to insulin itself, all display circadian fluctuations [21].
Circadian Regulators and Metabolism
Our understanding of the influence of the circadian clock on metabolism has significantly advanced with the discovery of clock proteins that serve as biological clocks. These proteins, expressed in nearly every eukaryotic cell as well as in some prokaryotic organisms, allow each individual cell to have an autonomous, self-sustained mechanism to keep track of a 24 h day [6]. These circadian oscillators are characterized by (i) sustained and persistent period length under stable conditions, (ii) the ability to entrain to external stimuli, and (iii) not being affected by wide variations in temperature. Circadian oscillations are driven by interlocked negative feedback circuits of activators such as CLOCK, brain and muscle ARNT-like 1 (BMAL1) and retinoic acid receptor-related orphan receptor (ROR), as well as by inhibitors such as cryptochrome (CRY), period (PER), and REV-ERB (nuclear receptor 1D1/2, NR1D1/2) that act as master regulators. Together these proteins produce a cell autonomous, self-sustained transcriptional rhythm of their own protein levels, and control the circadian expression of many target genes [23]. Importantly, they are able to synchronize to temporally relevant external events such as light, temperature, and feeding.
Transcriptomic studies show that up to 45% of all transcripts in mice display 24 h oscillations [24]. Many of these genes are key regulators of glucose and lipid metabolism, and of oxidative phosphorylation [25,26]. Metabolism in animals is highly circadian, coordinated not only by the cell-autonomous circadian rhythms but also by the feeding/fasting cycles which together drive genomic programs [27–29]. Regulators of nutrient homeostasis such as AMP-activated protein kinase (AMPK), cAMP response element-binding protein (CREB), and v-Akt murine thymoma viral oncogene homolog 1 (AKT/PKB), which are driven by daily rhythms of feeding and fasting, are coupled with circadian proteins. For example, fasting-sensitive protein kinase AMPK can phosphorylate CRY and target it for degradation, thereby constituting a mechanism by which fasting influences the circadian clock [30]. Feeding has a wide-ranging impact on rhythmic hepatic gene expression [28]. For example, cAMP-induced CREB activation upregulates Per transcription, thereby connecting the transcriptional circadian oscillation to G protein-coupled receptors (GPCRs) that use cAMP as a second messenger [31]. Several hormones, including glucagon, signal through a cAMP pathway. Moreover, post-prandial thermogenesis can also activate heat-shock factors, which subsequently modulate transcription of clock components. Consequently, feeding and fasting rhythms have a profound impact on rhythmic hepatic gene expression [28].
The contribution of cell-autonomous clock and feeding/fasting rhythms on the oscillation of the hepatic transcriptome has been systematically examined. Under ad libitum access to a standard diet, mice show a daily rhythm in food intake with the majority of food being consumed at night. In these mice, up to 3000 transcripts oscillate daily. However, in the absence of any food intake only ~350 hepatic transcripts are rhythmic in a 24 h period [28]. A large number of hepatic rhythmic transcripts under ad libitum feeding conditions are driven by the daily rhythm in food intake. The contribution of feeding- and fasting-induced genes to hepatic rhythms is more apparent in mice fed an isocaloric diet exclusively during the day. These mice with distinct periods of feeding and fasting show daily rhythms in up to 5000 hepatic transcripts. The phase of oscillation in the clock components and of other rhythmic transcripts is also changed in day-fed mice [28]. Hence, timing of food intake determines the phase of the hepatic clock. However, a distinct feeding/fasting rhythm alone cannot drive a vast majority of rhythmic transcription. While mice lacking CRY proteins show no discernible rhythm in daily feeding and have no significant hepatic transcript oscillation, under a isocaloric feeding/fasting cycle daily oscillations are restored in only 700 hepatic transcripts [28]. Similar effects are seen in liver metabolites. Hepatic lipidomic analysis of wild-type and Per1/2 null mice showed that a similar fraction of lipids (16–17%), including triglycerides, oscillated in both mouse strains [32]. However, the phases of these oscillations are different in the two mouse strains. Nighttime-restricted feeding increased the number of cycling lipids (~50%) in both conditions, and made their oscillation similar in both strains of mice. This implies that feeding/fasting rhythms can shape the phase of hepatic lipid accumulations [32].
These experiments conclude that a functional circadian clock, food availability, and the temporal pattern of feeding are important in determining the features of hepatic circadian transcriptome.
The Consequences of Dyssynchrony
Lessons from Animal Models
The importance of the interconnectedness between the circadian clock and dysmetabolism, such as obesity and diabetes, became clear in genetic mouse models. Mice with whole-body or tissue-specific loss of function or hypomorphic alleles of circadian genes often develop perturbation in glucose and lipid homeostasis, insulin resistance, and other hallmarks of metabolic disease (Table 1) [29,33,34]. Early observations of severe metabolic dysregulation in Bmal1−/−mice led to a systematic investigation of metabolic homeostasis in almost every circadian mutant mouse model [35]. Dysregulation of glucose homeostasis, with decreased gluconeogenesis, and increased insulin sensitivity occurs in Bmal1 knockout mice [36]. Double knockout of Rev-erbα and Rev-erbβ causes deregulated lipid metabolism [37]. Some of these genetic mouse models display aberrant feeding patterns, suggesting that a suboptimal feeding time may be contributing to metabolic disease. For example, Per1S714G mutant mice, where a serine to glycine mutation in PER1 causes acceleration of the molecular feedback loop, had a food intake time that differed with energy expenditure time by several hours [38].
Table 1.
Circadian Gene Knockout Mice and Their Metabolic Phenotypesa
| Mouse Strain, Background, and Age | Diet | Glucose and GTT | Insulin and ITT | Serum TG | Body Weight and Adiposity | Liver Phenotype | Refs |
|---|---|---|---|---|---|---|---|
| Rev-erba KO + sh Rev-erbb (Vennstrom; backcrossed to B6) | NC | Mild hypoglycemia | Hepatic steatosis | [103] | |||
| Rev-erbα/β tamoxifen-inducible double KO (Cho) | NC | Fasting hyperglycemia | [37] | ||||
| Rev-erbα KO (Schibler; backcrossed to B6) | NC | Hyperglycemia Normal GTT |
Mild hyperinsulinemia Normal ITT |
Normal Increased adiposity |
Higher hepatic TG | [104] | |
| HFD (53%) | Hyperglycemia | Hyperinsulinemia | Increased DIO | Higher hepatic TG | |||
| Per1/2 (129) | NC | Impaired GTT | Enhanced insulin sensitivity | Similar to control | [105] | ||
| Per1/2 | NC | Lower hepatic TG | [32] | ||||
| Per2ldc | NC | Elevated | Reduced (>4 months) Reduced adiposity |
[106] | |||
| Per2Brdm1 | NC | Hypoglycemia | [107] | ||||
| Per2Brdm1 (B6) | NC | Higher glucose clearance | Elevated Enhanced insulin sensitivity |
[108] | |||
| Per2Brdm1 (29S5/B6-Tyrc-Brd) | NC | Fasting hypoglycemia Normal GTT |
Normal | Normal Reduced adiposity |
[109] | ||
| Cry1/2 double KO (Sancar) | NC | Normal fasting glucose Impaired GTT |
Fasting hypoinsulinemia | Leaner | [110] | ||
| Cry1/2 double KO (Sancar) | NC | Normal fasting glucose Impaired GTT |
Normal ITT | [111] | |||
| Cry1/2 double KO (van der Horst) | NC | Fasting hyperglycemia | Normal insulin | Leaner | [112] | ||
| HFD (45%) | Impaired GTT | Hyperinsulinemia Impaired ITT |
Increased DIO | Hepatic steatosis | |||
| Bmal1 KO (mixed B6×129) | NC | Normoglycemia loss of diurnal rhythm | Enhanced insulin sensitivity | Elevated | [35] | ||
| Bmal1 KO (mixed B6×129; Bradfield; 8–12 weeks) | NC | Normal fasting glucose Impaired GTT |
Hypoinsulinemia Insulin hypersensitivity |
Similar to control Increased adiposity |
[105] | ||
| Liver–Bmal1 KO (mixed B6×129; Weitz; albumin–Cre) | NC | Fasting hypoglycemia Higher glucose clearance |
Normal | Normal | |||
| Pancreas–Bmal1 KO (mixed B6×129×ICR; Bradfield; PDX1–Cre) | NC | Hyperglycemia Impaired GTT |
Hypoinsulinemia | Normal | [36] | ||
| Bmal1 KO(B6; Saito) | NC | Elevated | Leaner | [113] | |||
| HFD (60%) | Elevated | Protected from DIO Ectopic fat accumulation |
Hepatic steatosis | [113] | |||
| Adipose–Bmal1 KO | NC | Normal | Elevated in AP2–Cre model | [114] | |||
| (B6; Bradfield; AP2–Cre and adiponectin–Cre) | HFD | Increased DIO in both models | |||||
| Bmal1 KO (backcrossed to B6; Bradfield, 4–12 weeks) | NC | Fasting hyperglycemia | Loss of diurnal insulin sensitivity | [115] | |||
| HFD (60%) | Similar to control Increased adiposity |
||||||
| Bmal1 KO (mixed B6×129; Bradfield) | NC | Normal fasting glucose Normal GTT |
Hypoinsulinemia Normal ITT |
Elevated | Slightly lower Increased adiposity |
[116] | |
| HFD (22%) | Normal | Elevated | Similar to control Increased adiposity |
||||
| Bmal1 | NC | Fasting hyperglycemia | [79] | ||||
| ClockΔ19 (B6) | NC | Enhanced insulin sensitivity | [35] | ||||
| HFD (45%) | Protected from insulin intolerance | Similar to control | |||||
| ClockΔ19 (B6) | NC | Hyperglycemia | Normal | Elevated | Higher Increased adiposity |
Hepatic steatosis | [117] |
| HFD (45%) | Higher Increased adiposity |
||||||
| ClockΔ19 (ICR) | NC | Normoglycemia | Normal | Reduced | Similar to control | [118] | |
| HFD (32%) | Hypoinsulinemia | Protected from DIO | |||||
| ClockΔ19 | NC | Normal | Normal | [119] | |||
| (ICR) | HFD | Similar to control | Lower hepatic TG | ||||
| ClockΔ19+MEL (CBA; 6 months) | NC | Normoglycemia Impaired GTT |
Hypoinsulinemia Insulin hypersensitivity |
Reduced | Normal | [120] | |
| ClockΔ19 (B6; 8 months) | NC | Fasting hyperglycemia Impaired GTT |
Hypoinsulinemia Normal ITT and islet dysfunction |
[36] | |||
| ClockΔ19 (B6) | NC | Higher Increased adiposity |
[121] |
Abbreviations: AP2, adipocyte fatty acid binding protein (FABP4); GTT, glucose tolerance test; HFD, high-fat diet (with % fat); ITT, insulin tolerance test; KO, knockout; NC, normal chow; TG, triglyceride.
Disruption of feeding/fasting rhythms, circadian gene expression, or, in most cases, both, is commonly found in animal model of obesity and dysmetabolism. For example, in diet-induced obesity (DIO), animals consume a significant amount of food outside of their normal nocturnal window that dampens the normal feeding/fasting cycles [39,40]. This is accompanied by disruption of the oscillation of circadian clock genes and their transcription targets involved in metabolism. In addition, obese (ob/ob) mice, that lack leptin and thus exhibit hyperphagia, have dampened oscillation of their circadian clock genes before becoming obese [41,42]. Hence, animal models of obesity and dysmetabolism often have dyssynchrony between circadian clock genes and feeding/fasting driven metabolic regulators.
Light/Dark Cycles and Feeding Time
The role of maintaining a diurnal feeding rhythms in metabolism has also been demonstrated through disrupting normal circadian activity by altering external temporal cues such as light and feeding time [33]. For example, a classic ‘shift work’ experiment, in which nocturnal rats are required to be active during daytime, leads to discombobulation of circadian gene rhythmicity and with it obesity and other metabolic disturbances [43]. Adding a dim light to the dark phase of the light/dark cycle increased nighttime food consumption and led to dysmetabolism in wild-type mice [44]. In addition, altering feeding time by restricting access to food to a smaller time-window can separate the effects of temporal cues such as light on the SCN from the feeding/ fasting rhythms in peripheral tissue [45]. Restricting feeding of mice, which are nocturnal animals, to daytime leads to weight gain [46]. All these studies demonstrate that, even without genetic manipulation of the mice, obesity and dysmetabolism can be induced in wild-type animals when circadian clock and feeding/fasting rhythms are dyssynchronized.
There is strong evidence that this phenomenon also exists in humans. Individuals with specific single-nucleotide polymorphisms (SNPs) within the CLOCK gene (e.g., CLOCK 3111T/C, rs1554483G, rs4864548A) are more susceptible to metabolic syndrome and obesity, while other SNPs (e.g., rs1801260/CGC) are protective for the development of obesity [47–49]. The functional role of these SNPs is not yet fully understood.
Circadian and metabolic dyssynchrony has also been studied at a population level in shift workers, as well in an inpatient laboratory setting. Shift workers, or laborers that work outside typical daytime hours, comprise about 15–20% of the workforce population in the USA [50]. There is strong evidence that they are particularly predisposed to obesity and metabolic syndrome [51–54]. This is perhaps due to the uncoupling of clock systems and the hypothalamus–pituitary–adrenal (HPA) axis [8,55,56]. Other factors that affect the normal cyclical oscillation along this axis (e.g., chronic stress or treatment with high doses of corticosteroids) also lead to metabolic syndrome [57], although it is unclear whether this can specifically be related to circadian dyssynchrony.
Laboratory studies on humans have also shown that dyssynchrony between feeding/fasting time and the circadian clock can lead to obesity and dysmetabolism. Simulated night shift work induced reduced daily energy expenditure by approximately 12–16% [58]. There were also decreased levels of leptin and peptide YY which signal satiety. Individuals who have been subjected to circadian misalignment for 10 days had elevated post-prandial glucose, elevated insulin (that is, more insulin resistance), and increased mean arterial pressure [59].
In conclusion, there is mounting evidence in both animal models and humans that aberrantly timed feeding, out of synchrony with the light-entrained central circadian clock, leads to obesity and dysmetabolism. Synchronized oscillation of these two systems likely coordinates distant and often discordant cellular and physiological processes for more-optimal energy metabolism. At a cultural level, with the advent of electricity, modern humans are exposed to far more prolonged illumination [19]. There is also increasing evidence that aberrant feeding patterns are prevalent in our society today, wherein variation in eating patterns can be observed with people eating more frequently throughout the day and with a bias toward late-night eating [60]. Together these two phenomena are further environmental contributors to the rise in the prevalence of obesity over the past several decades.
TRF Induces Synchrony
Until recently it was not clear whether aiming to synchronize feeding/fasting times with the central circadian clock would prevent or even reverse obesity and its associated metabolic diseases. However, recent evidence has shown that synchrony between feeding rhythms and circadian oscillations can be restored.
TRF in Mice
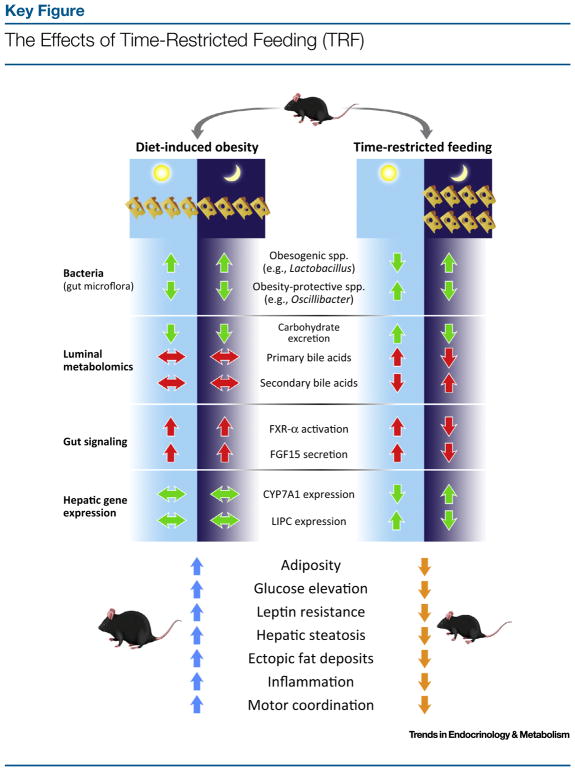
Using TRF to allow mice access to food only in the nocturnal phase can promote natural feeding rhythms and restore synchrony with circadian oscillations [61]. TRF restores cycling of metabolic regulators such as CREB, mTOR (mechanistic target of rapamycin), and AMPK, and downstream signaling pathways, as well as oscillations of the circadian clock and expression of their target genes, which are dampened or obliterated by DIO. The metabolic benefits of restoring these rhythms are tremendous (Figure 2, Key Figure). Although the mice consume similar amount of calories and have similar activity levels, they do not become obese on a high-fat diet (HFD). TRF mice display significantly less adiposity and improved glucose and lipid homeostasis. This is accompanied with decreased leptin resistance, decreased hepatic inflammation and steatosis, decreased ectopic lipid deposition, and even improved motor coordination [62–64].
Figure 2.
Green arrows indicate findings published in the literature. Red arrows signify potential pathways. Mice on a diet-induced obesity (DIO) protocol have ad libitum access to a high-fat diet (HFD). These mice have changes in the gut microbiome, stool metabolomics, and hepatic gene expression. There are presumed changes in gut signaling, particularly in the bile acid signaling pathway. As a result, these mice are particularly predisposed to obesity and dysmetabolism. With TRF, cyclical fluctuation is restored in the gut microflora, luminal metabolites, and hepatic gene expression. Presumably TRF also restores oscillations in gut signaling. As a result, TRF protects against obesity and dysmetabolism. Abbreviations: CYP7A1, cytochrome P450 7A1; FGF15, fibroblast growth factor 15; FXR, farnesoid X receptor; LIPC, hepatic lipase.
Although TRF affords protection against the development of obesity from the nutritional challenge of HFD (61% dietary fat), it was unclear if it was effective against dysmetabolism caused by other types of nutritional challenges. However, in a later study Chaix et al. showed that the TRF protocol attenuated the metabolic consequences from a variety of obesogenic diets, not only high-fat, but also high-fructose and high-sucrose diets [65]. Furthermore, TRF reversed the progression of dysmetabolism in mice with pre-existing obesity and insulin resistance. The benefits of TRF were directly proportional to the length of time of food restriction. That is, mice with restricted access of 9 h fared better than those who had access for 15 h.
These initial studies showed that TRF is particularly effective against DIO and other nutritional challenges that cause obesity. TRF also proved to be effective against at least one genetic cause of obesity. In Per1S714G mutated mice, where feeding rhythms are perturbed and the animals are particularly susceptible to DIO [38], TRF prevented the weight gain displayed by PER1 mutated mice on a HFD. Furthermore, PER1 mutated mice with ad libitum access to HFD had significant misalignment of various behavior measurements such as feeding phase and oxygen consumption, although their overall locomotor activity, food intake, and oxygen consumption were not significantly different from controls. However, TRF improved alignment between feeding phase and oxygen consumption [38].
In murine models, TRF also leads to improvement of fatty acid turnover in the liver, brown and white adipose tissue, and cardiac muscle [61,64,65]. Furthermore, nighttime-restricted feeding considerably reduced hepatic lipid levels (including triglyceride) in wild-type mice fed a normal chow diet. This suggests that fasting/feeding rhythms also affect the amount of hepatic lipid accumulation [32].
The function of TRF in aging and heart health was recently assessed in a Drosophila model [66]. As in murine models, 12 h TRF in Drosophila, where caloric intake and activity were unchanged, led to prevention of weight gain and deceleration of age- and diet-induced decline in cardiac function including arrhythmia. These benefits appeared to be mediated not only by the oscillation of circadian clock gene but also by the mitochondrial electron transport chain, and by the TCP-1 ring complex chaperonin that assists the folding of proteins upon ATP hydrolysis.
TRF in Humans
Although no studies in humans have yet specifically tested TRF, some small studies about timed eating suggest that restoring feeding/fasting cycles with light/dark circadian rhythms can have weight-loss benefits [67]. For example, in a cohort of patients undergoing a behavioral weight-loss treatment, those who consumed their calories earlier in the day were more likely to lose weight, compared to those who ate later [68]. Another study, where two groups of obese women were randomized to consuming an isocaloric meal during breakfast or dinner, showed that high-calorie breakfast consumers had much better fasting glucose, insulin sensitivity, and improved lipid profile compared to the high-calorie dinner consumers [69]. As discussed above, recent studies have demonstrated that the timing of feeding in the general population is diverse [60], suggesting that more precision medicine is needed to identify those who have poor feeding patterns, and selectively recommend behavioral changes in these individuals to improve their metabolism. Ongoing effort to monitor the daily pattern of food intake, sleep, and activity using smartphones (http://www.mycircadianclock.org) in a large cohort will quantify the epidemic of circadian disruption and identify at-risk individuals.
Multiple studies show that TRF, where feeding has been restricted to the active time phase, has significant metabolic benefits. TRF prevents obesity and improves glucose and lipid homeosta-sis, and has beneficial effects on other metabolic organs such as the liver, heart, and brown adipose tissue. These affects are accompanied with synchrony and more robust oscillation between circadian effectors and metabolic regulators that are entrained to feeding/fasting cycles. However, although restoration of circadian and metabolic synchrony is correlated with improved metabolism, the mechanism by which this improvement occurs is not yet fully understood. There are still many gaps in understanding the relationship between light-entrained central circadian oscillations with those in the periphery that may be entrained by other external stimuli such as feeding. In addition, although it is clear that light-entrainment occurs with specialized retinal ganglion cell signaling to the SCN, feeding entrainment from the gut to metabolically active tissues such as the liver, muscle, and adipose tissue is not entirely understood.
Gut Signaling and Feeding Entrainment
The impact of TRF on obesity has important implications in our understanding of metabolism and circadian rhythms, adding a further level of complexity. The timing of food intake has as much of an impact, if not more, on energy homeostasis as the nutritional content of the diet and gut signaling. Recent advances in our understanding of the effect of bariatric surgery on gut hormones such as incretins, or the impact of obesity on the composition and function of the gut microbiome, has led to tremendous interest in the role of gut signaling in metabolism. Orexigenic or anorexigenic peptides from the gut have the ability to affect energy utilization in other organs in a significant manner. Our understanding of how peripheral circadian genes are entrained by appetite hormones is currently limited, and future investigations in this area could lead to novel therapies for obesity and its associated metabolic disorders [70].
The Gut–Brain Axis
The SCN itself can likely influence appetite and feeding behavior through direct connections with hypothalamic nuclei that control appetite [71]. However, if light entrains the circadian cycling of SCN neurons, what agent conveys the feeding cycles from the gut? The gut is a mediator between ingested food and all other organs. It has an elaborate mechanism in place that transmits crucial information about the size and composition of the meal through endocrine, inflammatory, and neuronal signals. It is a highly circadian organ [72]. Furthermore, the gastrointestinal (GI) track houses a reservoir of trillions of commensal bacteria that are involved in sophisticated, direct and indirect (by secondary metabolites) interactions with the host. Candidate entraining agents can include any substance that oscillates with feeding within the gut lumen, or in the gut tissue, and has an impact on host metabolism. Potential agents identified that fit this profile include luminal nutrients, incretins, bile acids (BA), and the gut microbiome. Some of these candidates will be discussed in further detail.
Luminal Nutrients and Their Sensors
Luminal nutrients, such as particular fatty acids and starches, can elicit a complex signaling cascade in the gut–brain axis that provides feedback to satiety centers in the brain by suppressing feeding and appetite, and by altering the endogenous production of specific nutrients in other organs [73]. This signaling is either conveyed by the nutrients alone or in conjunction with the release of GI peptides from enteroendocrine cells (EECs). EECs contain multiple nutrient-dependent receptors, many of which are GPCRs, that can trigger depolarization and fluctuation in intracellular calcium and, hence, the secretion of gut peptides such as incretins, peptide YY, cholecystokinin, gastrin, serotonin, neurotensin, or secretin. Many of these peptides can then activate afferent enteric neurons, as well as work in an endocrine fashion. Such receptors include GPR41/FFAR3 and GPR43/FFAR2, which can detect short-chain fatty acids (SCFAs) [74]. Upon activation by SCFAs derived from the gut during feeding, GPR43, in particular, is able to modulate adipocyte sensitivity to insulin and hence affect lipid homeostasis [75]. In addition, microbiota-produced SCFAs that activate GPR41 can affect host energy homeostasis by stimulating adipocytes into making leptin [76], and inducing glucagon-like peptide 1 (GLP-1) secretion by intestinal EECs [74].
Novel Supplements
Recent studies now show that orally ingested substances can affect circadian clock function and energy homeostasis, and this has broadened our understanding of how luminal substances can affect host metabolism. Circadian proteins control the biosynthesis of NAD+ which feeds back to modulate the activity of core clock components through sirtuin 1 (SIRT1) and regulates rhythmic mitochondrial respiration through SIRT3 [77–79]. Bmal1 knockout mice have significantly decreased SIRT3 activity, which affects mitochondrial oxidative function [79]. However, supplementation with nicotinamide mononucleotide (NMN), a NAD+ precursor, restored the function of SIRT3 as well as its effects on mitochondrial function [79].
In addition, circadian clock and feeding/fasting patterns regulate oscillations in polyamines, which are essential polycations present in all living cells that regulate essential cellular processes including gene transcription and translation, and cell growth [80]. Polyamines also regulate the activity of core clock proteins by modulating the interaction of PER2 and CRY1 [81]. Nevertheless, their biological levels decrease with age, leading to longer circadian periods [81]. Supplementation with dietary polyamine results in reversal of age-related changes to the circadian period [81]. These new studies not only suggest that circadian gene expression and, with it, host metabolism can be affected with ingestible supplements, but that other, undiscovered compounds in the diet can potentially have a tremendous influence in the rhythmicity of the circadian clock program.
GI-Derived Peptides
Incretins such as GLP-1 can also serve as the agent that entrains feeding behavior. GLP-1 is a gut hormone that boosts glucose-dependent insulin secretion and is released during feeding, specifically when food enters the small intestine. In mice, GLP-1 excretion is circadian, with larger amounts being released in the evening, and is highly entrained by feeding behavior [82]. GLP-1 affects satiety in the central nervous system [83] and can influence metabolic homeostasis by affecting hepatic gene expression. GLP-1 receptor activation in hepatocytes increases cAMP production as well as peroxisome proliferator-activated receptor α (PPARα) and acetyl-CoA oxidase expression, both of which are involved in fatty acid oxidation. In addition, there is decreased expression of stearoyl-coA desaturase 1, sterol regulatory element-binding protein 1 (SREBP-1), acetyl-CoA carboxylase (ACC), and fatty acid synthase (FAS), collectively suggesting the GLP-1 impairs lipogenesis and improves β-oxidation of fatty acids [84,85]. Hence, GLP-1 or other EEC-released hormones could serve as feeding entraining agents to both the central nervous system and metabolically active tissues.
Bile Acids
BAs are secreted into the GI lumen to facilitate lipid digestion. They are the endogenous ligands for the orphan nuclear receptor farnesoid X receptor (FXR) [86]. BA activation of FXR is the main regulatory step in the control of cholesterol and BA metabolism. Furthermore, in mice, FXR activation in hepatocytes, or FXR-mediated release of fibroblast growth factor 15 (FGF15) from the terminal ileum, play a modulatory role in lipid and glucose homeostasis [87]. TGR5 (G protein-coupled bile acid receptor 1, GPBAR1), another BA receptor, also plays an important regulatory role in host metabolism [88]. For example, BA activation of TGR5 can lead to weight loss. This is mediated by increased activation of thyroid hormone receptor in brown adipose tissue and, hence, increased energy expenditure in the setting of unchanged caloric intake [89]. Finally, BAs can affect host metabolism indirectly by inducing GLP-1 secretion through TGR5 activation [90]. Importantly, both serum BA levels and the FXR pathway, including its gene targets, exhibit circadian behavior and are induced by feeding [91].
The Gut Microbiota and Circadian Rhythms
The most recent addition to the list of potential feeding entrainment agents is the gut micro-biome. Recent studies show that the gut microbiome is far more dynamic than previously imagined. It is also now appreciated that the gut microbiota is a major environmental contributor to obesity, although the full mechanisms by which it affects host metabolism are unknown [92]. In mice, the composition of the gut microbiome fluctuates cyclically. However, in DIO mice this cyclical fluctuation is dampened dramatically [63,93]. In addition to diet, the dynamic structure of the gut microbiome responds to a myriad of separate host controls, including gender, feeding time, clock gene mutation, diet, and jet-lag [63,93–95]. The gut microbiota is essential for normal circadian gene expression in the gut [94,96], and clock mutant mice also have a far less dynamic structure to their gut microbiome compared to control mice [93,95]. Behavioral disruption of the circadian clock, such as reversals of the light/dark cycle, leads to changes in the gut microbiome composition in mice [97]. TRF restores oscillating fluctuations in key bacterial families that are thought to affect host metabolism [63]. Dysfunctional gut microbiota can adversely affect host metabolism either via direct interactions with the host through activation of inflammatory pathways [98], by influencing the activation of GLP-1 [99], changing the luminal BA profile [63,100,101], or by affecting gut corticosteroid release [96].
We now begin to understand that the idea of a single agent that entrains feeding rhythms is false. Instead, it is entirely possible that a cascade of events is necessary for feeding entrainment. For example, dynamic fluctuation of the gut microbiome could induce fluctuations in the levels of SCFAs. This in turn could cause cyclical fluctuation of GPR43 activation, which would then suppress insulin signaling in adipocytes, promoting the utilization of lipids in other ways, hence controlling metabolic homeostasis [75]. In addition, the dynamic fluctuations in the gut micro-biome can lead to cyclical fluctuation of primary and secondary BAs which can modulate FXR and TGR5 activation and downstream signaling, affecting energy expenditure and influencing cholesterol, lipid, and glucose homeostasis. One can imagine that when the normal feeding rhythms are perturbed with DIO [63] this leads to a cascade of disrupted cyclical fluctuations of the gut microbiome that, in turn, alter the luminal SCFA profile [102] or BA profile, leading to constant suppression of GPR43 signaling and a continuous increase in insulin-mediated lipid uptake in adipocytes [75]. Few studies have investigated how each single agent discussed can affect circadian biology. It will be exciting to determine, by investigating each of these complex and intertwined pathways, the mechanism by which meal timing can promote optimal metabolic health
Concluding Remarks and Future Perspectives
Recent advances in circadian biology have had a tremendous impact on our understanding of the temporal regulation of metabolism. Metabolism is a highly dynamic process, controlled by three oscillating systemic networks: light-entrained central circadian neural oscillations, sleep/ wake homeostasis, and feeding/fasting rhythms. These networks affect cellular processes through circadian proteins, which are exquisitely coordinated with key cellular metabolic regulators. The synchrony of these oscillating networks with endocrine and enteroendocrine hormones, and neuronal input at the system level, and with the cellular machinery at the single-cell level, likely synchronizes distant organs and discordant cellular processes.
Dyssynchrony from altered genetics, disruption of normal cycling, or the introduction of obesogenic diets leads to dysmetabolism. Synchronizing feeding/fasting times with the light-entrained circadian clock by TRF can, in some cases, prevent and treat obesity and dysmetabolism. The gut signals that impart the benefits of TRF are not yet well understood (see Outstanding Questions). However, deciphering how the gut can entrain the feeding/fasting rhythms could lead to the development of much needed novel therapies for obesity and its associated metabolic diseases. The circadian rhythm of metabolism has highlighted the necessity for temporal separation of catabolic and anabolic metabolism to distinct times of the day. Such separation is now increasingly appreciated as a dominant component of metabolic homeostasis.
Outstanding Questions.
How do circadian rhythms and feeding/fasting cycles ‘synergize’ to drive oscillations at a genomic level? Circadian clock or feeding/fasting cycles alone drive oscillations in a small set of transcripts, but the mechanism by which they exert their synergistic effect to regulate daily oscillations in a large number of transcripts or proteins is largely unknown.
What effect does the gut microbiome have on local and systemic rhythms in metabolism and physiology? Are there gut microbiome-derived metabolites that affect the circadian oscillators in distant organs or hunger/satiety circuits in the brain? These effects can affect daily rhythms of nutrient metabolism in the whole organism.
Why do particular foods, such as HFD, dampen the feeding/fasting rhythms?
How does dyssynchrony between the light/dark cycles and feeding/fasting rhythms result in dysmetabolism?
Can obesity and dysmetabolism develop without dyssynchrony of the multiple oscillatory networks? Although dyssynchrony of oscillators predisposes to metabolic diseases, it is not known whether this is a hallmark feature of metabolic disease.
Does TRF protect against obesity and dysmetabolism in humans?
How do the circadian oscillations affect the homeostatic control of satiety to generate daily rhythms in feeding and fasting?
Could the relationship between obesity and specific cancers be linked to metabolic dysregulation caused by the dyssynchrony of circadian oscillatory networks?
Trends.
Central circadian pacemakers, sleep/ wake homeostasis, and feeding/fasting rhythms coordinate distant metabolic processes for efficient metabolism through endocrine and neuronal signals.
Each cell has autonomous circadian clock components that interact with key metabolic regulators and effect cellular metabolic efficiency.
Obesity and dysmetabolism can be induced by perturbing these physiological or cellular cycles either via genetic manipulation, disruption of light/dark cycles, and by feeding patterns.
TRF promotes synchrony of feeding/ fasting rhythms with the central circadian pacemaker, resulting in more-robust circadian and metabolic cycles, and prevents and treats obesity and its metabolic consequences.
Entrainment of feeding/fasting rhythms is entrained by gut signaling which can by mediated by multiple potential candidates, including the gut microbiome, bile acids, incretins, nutrients, and secondary luminal metabolites.
Acknowledgments
A.Z. received support from National Institutes of Health (NIH) grant K08 DK102902, an American Association for the Study of Liver Diseases (AASLD) Liver Scholar Award, and an American Gastroenterological Association Research Foundation Microbiome Junior Investigator Research Award. A.C. received salary support from an American Diabetes Association Mentor-Based Postdoctoral Fellowship (7-12-MN-64). S.P. received support from NIH DK091618.
Glossary
- Ad libitum
literally meaning ‘at liberty’, where mice have access to food at all times. Mice with ad libitum access to normal chow have a diurnal feeding pattern, consuming nearly 75% of their daily calories during the dark/active period. Mice with ad libitum access to HFD spread their caloric intake more widely, consuming approximately 50% of their daily calories during the dark/ active period.
- AMP-activated protein kinase (AMPK)
a cellular metabolic regulator important for energy homeostasis. AMPK promotes ATP production and increases the catabolic drive of cells during periods of fasting.
- Brain and muscle ARNT-like 1 (BMAL1)
also known as ARNTL (aryl hydrocarbon receptor nuclear translocator-like protein 1), is a transcription factor that acts as a positive element in the mammalian cell-autonomous circadian feedback loop. BMAL1 dimerizes with CLOCK to activate Per and Cry transcription.
- CLOCK (circadian locomotor output cycles kaput)
a positive transcription factor in the mammalian cell autonomous circadian feedback loop. CLOCK dimerizes with BMAL1 to activate transcription of Per and Cry.
- cAMP response element-binding protein (CREB)
a transcription factor that plays a key role in energy homeostasis. It increases the anabolic drive of cells during feeding.
- Cryptochrome (CRY)
a transcription factor that acts as an inhibitory element in the mammalian cell-autonomous circadian feedback loop. CRY dimerizes with PER and inhibits the transcription of the Clock/ Bmal1 components.
- Diet-induced obesity (DIO)
wild-type mice fed a HFD ad libitum constitute a model used to study dysmetabolism. The mice develop increased adiposity, insulin resistance, leptin resistance, and hepatic steatosis, among other metabolic problems.
- Isocaloric diets
two diets that share the same amount of calories, but may differ in their nutrient composition or the timing of their intake.
- Period (PER)
an inhibitory transcription factor in the mammalian cell-autonomous circadian feedback loop. PER dimerizes with CRY and inhibits the transcription of Clock/ Bmal1 components.
- Retinoic acid related-related orphan receptor (ROR)
also known as NR1F (nuclear receptor subfamily 1, group F), ROR is a member of the nuclear receptor family of transcription factors that serves as an activator of Bmal1.
- REV-ERB
also known as NR1D1/2 (nuclear receptor subfamily 1, group D, members 1/2), a member of the nuclear receptor family of transcription factors that serves as an inhibitor of Bmal1. It also plays a major role in lipid homeostasis.
- Suprachiasmatic nucleus (SCN)
a group of specialized neurons in the hypothalamus that receive light information from specialized cells in the retina and control circadian cycles. They can influence many physiological and behavioral rhythms that occur over a 24 h period and coordinate them with the change in light they detect.
- v-Akt murine thymoma viral oncogene homolog 1 (AKT)
also known as protein kinase B (PKB), AKT is a serine/threonine kinase and central regulator of cellular metabolism, cell survival, and health. It plays a prominent role in glucose metabolism, promoting the catabolic drive of cells during fasting.
Footnotes
Disclaimer Statement
The authors declare no competing financial interests.
References
- 1.Ogden CL, et al. Prevalence of childhood and adult obesity in the United States, 2011–2012. J Am Med Assoc. 2014;311:806–814. doi: 10.1001/jama.2014.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finkelstein EA, et al. Annual medical spending attributable to obesity: payer-and service-specific estimates. Health Aff. 2009;28:w822–w831. doi: 10.1377/hlthaff.28.5.w822. [DOI] [PubMed] [Google Scholar]
- 3.Casazza K, et al. Myths, presumptions, and facts about obesity. N Engl J Med. 2013;368:446–454. doi: 10.1056/NEJMsa1208051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levi J, et al. Trust for America's Health. 2014. The State of Obesity: Better Policies for a Healthier America 2014. [Google Scholar]
- 5.Gerhart-Hines Z, Lazar MA. Circadian metabolism in the light of evolution. Endocr Rev. 2015;36:289–304. doi: 10.1210/er.2015-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 7.van Cauter E. Endocrine rhythms. In: Becker KL, editor. Principles and Practice of Endocrinology and Metabolism. 3. Lippincott Williams & Wilkins; 2001. pp. 57–68. [Google Scholar]
- 8.Nicolaides NC, et al. Circadian endocrine rhythms: the hypothalamic–pituitary–adrenal axis and its actions. Ann N Y Acad Sci. 2014;1318:71–80. doi: 10.1111/nyas.12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nohara K, et al. Manipulating the circadian and sleep cycles to protect against metabolic disease. Front Endocrinol. 2015;6:35. doi: 10.3389/fendo.2015.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franken P, Dijk DJ. Circadian clock genes and sleep homeostasis. Eur J Neurosci. 2009;29:1820–1829. doi: 10.1111/j.1460-9568.2009.06723.x. [DOI] [PubMed] [Google Scholar]
- 11.Welsh DK, et al. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mure LS, Panda S. Fear of the light or need for action: the IGL will judge. Neuron. 2012;75:546–548. doi: 10.1016/j.neuron.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 13.von Ruesten A, et al. Association of sleep duration with chronic diseases in the European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam study. PLoS ONE. 2012;7:e30972. doi: 10.1371/journal.pone.0030972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, et al. Sleep duration and chronic diseases among U. S. adults age 45 years and older: evidence from the 2010 Behavioral Risk Factor Surveillance System. Sleep. 2010;36:1421–1427. doi: 10.5665/sleep.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spiegel K, et al. Effects of poor and short sleep on glucose metabolism and obesity risk. Nat Rev Endocrinol. 2009;5:253–261. doi: 10.1038/nrendo.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siegel JM. Clues to the functions of mammalian sleep. Nature. 2005;437:1264–1271. doi: 10.1038/nature04285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris CJ, et al. Circadian system, sleep and endocrinology. Mol Cell Endocrinol. 2012;349:91–104. doi: 10.1016/j.mce.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reiter RJ. The pineal gland and melatonin in relation to aging: a summary of the theories and of the data. Exp Gerontol. 1995;30:199–212. doi: 10.1016/0531-5565(94)00045-5. [DOI] [PubMed] [Google Scholar]
- 19.Wright KP, Jr, et al. Entrainment of the human circadian clock to the natural light–dark cycle. Curr Biol. 2013;23:1554–1558. doi: 10.1016/j.cub.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albrecht U. Timing to perfection: the biology of central and peripheral circadian clocks. Neuron. 2012;74:246–260. doi: 10.1016/j.neuron.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Van Cauter E, et al. Roles of circadian rhythmicity and sleep in human glucose regulation. Endocr Rev. 1997;18:716–738. doi: 10.1210/edrv.18.5.0317. [DOI] [PubMed] [Google Scholar]
- 22.Zhou B, et al. CLOCK/BMAL1 regulates circadian change of mouse hepatic insulin sensitivity by SIRT1. Hepatology. 2014;59:2196–2206. doi: 10.1002/hep.26992. [DOI] [PubMed] [Google Scholar]
- 23.Reddy AB, O’Neill JS. Healthy clocks, healthy body, healthy mind. Trends Cell Biol. 2010;20:36–44. doi: 10.1016/j.tcb.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang R, et al. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci USA. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 26.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 27.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Vollmers C, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asher G, Sassone-Corsi P. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015;161:84–92. doi: 10.1016/j.cell.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 30.Lamia KA, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Neill JS, et al. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adamovich Y, et al. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 2014;19:319–330. doi: 10.1016/j.cmet.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arble DM, et al. Circadian disruption and metabolic disease: findings from animal models. Best Pract Res Clin Endocrinol Metab. 2010;24:785–800. doi: 10.1016/j.beem.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalsbeek A, et al. Circadian control of glucose metabolism. Mol Metab. 2014;3:372–383. doi: 10.1016/j.molmet.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudic RD, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marcheva B, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho H, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z, et al. PER1 phosphorylation specifies feeding rhythm in mice. Cell Rep. 2014;7:1509–1520. doi: 10.1016/j.celrep.2014.04.032. [DOI] [PubMed] [Google Scholar]
- 39.Kohsaka A, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 40.Mendoza J, et al. High-fat feeding alters the clock synchronization to light. J Physiol. 2008;586:5901–5910. doi: 10.1113/jphysiol.2008.159566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ando H, et al. Impairment of peripheral circadian clocks precedes metabolic abnormalities in ob/ob mice. Endocrinology. 2011;152:1347–1354. doi: 10.1210/en.2010-1068. [DOI] [PubMed] [Google Scholar]
- 42.Ho A, Chin A. Circadian feeding and drinking patterns of genetically obese mice fed solid chow diet. Physiol Behav. 1988;43:651–656. doi: 10.1016/0031-9384(88)90221-1. [DOI] [PubMed] [Google Scholar]
- 43.Salgado-Delgado R, et al. Internal desynchronization in a model of night-work by forced activity in rats. Neuroscience. 2008;154:922–931. doi: 10.1016/j.neuroscience.2008.03.066. [DOI] [PubMed] [Google Scholar]
- 44.Fonken LK, et al. Light at night increases body mass by shifting the time of food intake. Proc Natl Acad Sci USA. 2010;107:18664–18669. doi: 10.1073/pnas.1008734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arble DM, et al. Circadian timing of food intake contributes to weight gain. Obesity. 2009;17:2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez-Guimera G, et al. CLOCK 3111 T/C SNP interacts with emotional eating behavior for weight-loss in a Mediterranean population. PLoS ONE. 2014;9:e99152. doi: 10.1371/journal.pone.0099152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott EM, et al. Association between polymorphisms in the Clock gene, obesity and the metabolic syndrome in man. Int J Obes. 2008;32:658–662. doi: 10.1038/sj.ijo.0803778. [DOI] [PubMed] [Google Scholar]
- 49.Sookoian S, et al. Genetic variants of Clock transcription factor are associated with individual susceptibility to obesity. Am J Clin Nutr. 2008;87:1606–1615. doi: 10.1093/ajcn/87.6.1606. [DOI] [PubMed] [Google Scholar]
- 50.McMenamin TM. A time to work: recent trends in shift work and flexible schedules. Mon Labor Rev. 2007;130:3–15. [Google Scholar]
- 51.Canuto R, et al. Metabolic syndrome and shift work: a systematic review. Sleep Med Rev. 2013;17:425–431. doi: 10.1016/j.smrv.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 52.van Drongelen A, et al. The effects of shift work on body weight change – a systematic review of longitudinal studies. Scand J Work Environ Health. 2011;37:263–275. doi: 10.5271/sjweh.3143. [DOI] [PubMed] [Google Scholar]
- 53.Wang F, et al. Meta-analysis on night shift work and risk of metabolic syndrome. Obes Rev. 2014;15:709–720. doi: 10.1111/obr.12194. [DOI] [PubMed] [Google Scholar]
- 54.Brum MC, et al. Shift work and its association with metabolic disorders. Diabetol Metab Syndr. 2015;7:45. doi: 10.1186/s13098-015-0041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sookoian S, et al. Effects of rotating shift work on biomarkers of metabolic syndrome and inflammation. J Intern Med. 2007;261:285–292. doi: 10.1111/j.1365-2796.2007.01766.x. [DOI] [PubMed] [Google Scholar]
- 56.Fujino Y, et al. A prospective cohort study of shift work and risk of ischemic heart disease in Japanese male workers. Am J Epidemiol. 2006;164:128–135. doi: 10.1093/aje/kwj185. [DOI] [PubMed] [Google Scholar]
- 57.Bose M, et al. Stress and obesity: the role of the hypothalamic–pituitary–adrenal axis in metabolic disease. Curr Opin Endocrinol Diab Obes. 2009;16:340–346. doi: 10.1097/MED.0b013e32832fa137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McHill AW, et al. Impact of circadian misalignment on energy metabolism during simulated nightshift work. Proc Natl Acad Sci USA. 2014;111:17302–17307. doi: 10.1073/pnas.1412021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheer FA, et al. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA. 2009;106:4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gill S, Panda S. A smartphone app reveals erratic diurnal eating patterns in humans that can be modulated for health benefits. Cell Metab. 2015;22:789–798. doi: 10.1016/j.cmet.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hatori M, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sherman H, et al. Timed high-fat diet resets circadian metabolism and prevents obesity. FASEB J. 2012;26:3493–3502. doi: 10.1096/fj.12-208868. [DOI] [PubMed] [Google Scholar]
- 63.Zarrinpar A, et al. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014;20:1006–1017. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsai JY, et al. Influence of dark phase restricted high fat feeding on myocardial adaptation in mice. J Mol Cell Cardiol. 2013;55:147–155. doi: 10.1016/j.yjmcc.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chaix A, et al. Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 2014;20:991–1005. doi: 10.1016/j.cmet.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gill S, et al. Time-restricted feeding attenuates age-related cardiac decline in Drosophila. Science. 2015;347:1265–1269. doi: 10.1126/science.1256682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mattson MP, et al. Meal frequency and timing in health and disease. Proc Natl Acad Sci USA. 2014;111:16647–16653. doi: 10.1073/pnas.1413965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garaulet M, et al. Timing of food intake predicts weight loss effectiveness. Int J Obes. 2013;37:604–611. doi: 10.1038/ijo.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jakubowicz D, et al. High caloric intake at breakfast vs. dinner differentially influences weight loss of overweight and obese women. Obesity. 2013;21:2504–2512. doi: 10.1002/oby.20460. [DOI] [PubMed] [Google Scholar]
- 70.Zarrinpar A, Loomba R. Review article: the emerging interplay among the gastrointestinal tract, bile acids and incretins in the pathogenesis of diabetes and non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2012;36:909–921. doi: 10.1111/apt.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bechtold DA, Loudon AS. Hypothalamic clocks and rhythms in feeding behaviour. Trends Neurosci. 2013;36:74–82. doi: 10.1016/j.tins.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 72.Scheving LA. Biological clocks and the digestive system. Gastroenterology. 2000;119:536–549. doi: 10.1053/gast.2000.9305. [DOI] [PubMed] [Google Scholar]
- 73.Duca FA, et al. Glucoregulatory relevance of small intestinal nutrient sensing in physiology, bariatric surgery, and pharmacology. Cell Metab. 2015;22:367–380. doi: 10.1016/j.cmet.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 74.Nohr MK, et al. GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology. 2013;154:3552–3564. doi: 10.1210/en.2013-1142. [DOI] [PubMed] [Google Scholar]
- 75.Kimura I, et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 2013;4:1829. doi: 10.1038/ncomms2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Samuel BS, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci USA. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakahata Y, et al. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peek CB, et al. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science. 2013;342:1243417. doi: 10.1126/science.1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller-Fleming L, et al. Remaining mysteries of molecular biology: the role of polyamines in the cell. J Mol Biol. 2015;427:3389–3406. doi: 10.1016/j.jmb.2015.06.020. [DOI] [PubMed] [Google Scholar]
- 81.Zwighaft Z, et al. Circadian clock control by polyamine levels through a mechanism that declines with age. Cell Metab. 2015;22:874–885. doi: 10.1016/j.cmet.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 82.Gil-Lozano M, et al. Circadian secretion of the intestinal hormone GLP-1 by the rodent L cell. Diabetes. 2014;63:3674–3685. doi: 10.2337/db13-1501. [DOI] [PubMed] [Google Scholar]
- 83.Shah M, Vella A. Effects of GLP-1 on appetite and weight. Rev Endocr Metab Disord. 2014;15:181–187. doi: 10.1007/s11154-014-9289-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ding X, et al. Exendin-4, a glucagon-like protein-1 (GLP-1) receptor agonist, reverses hepatic steatosis in ob/ob mice. Hepatology. 2006;43:173–181. doi: 10.1002/hep.21006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu Y, et al. Potential roles of glucagon-like peptide-1-based therapies in treating non-alcoholic fatty liver disease. World J Gastroenterol. 2014;20:9090–9097. doi: 10.3748/wjg.v20.i27.9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matsubara T, et al. FXR signaling in the enterohepatic system. Mol Cell Endocrinol. 2013;368:17–29. doi: 10.1016/j.mce.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cicione C, et al. Emerging role of fibroblast growth factors 15/19 and 21 as metabolic integrators in the liver. Hepatology. 2012;56:2404–2411. doi: 10.1002/hep.25929. [DOI] [PubMed] [Google Scholar]
- 88.Pols TW, et al. The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J Hepatol. 2011;54:1263–1272. doi: 10.1016/j.jhep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Watanabe M, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 90.Katsuma S, et al. Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem Biophys Res Commun. 2005;329:386–390. doi: 10.1016/j.bbrc.2005.01.139. [DOI] [PubMed] [Google Scholar]
- 91.Zhang YK, et al. Diurnal variations of mouse plasma and hepatic bile acid concentrations as well as expression of biosynthetic enzymes and transporters. PLoS ONE. 2011;6:e16683. doi: 10.1371/journal.pone.0016683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Greiner T, Backhed F. Effects of the gut microbiota on obesity and glucose homeostasis. Trends Endocrinol Metab. 2011;22:117–123. doi: 10.1016/j.tem.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 93.Thaiss CA, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 94.Leone V, et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17:681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liang X, et al. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc Natl Acad Sci USA. 2015;112:10479–10484. doi: 10.1073/pnas.1501305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mukherji A, et al. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 97.Voigt RM, et al. Circadian disorganization alters intestinal microbiota. PLoS ONE. 2014;9:e97500. doi: 10.1371/journal.pone.0097500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Everard A, Cani PD. Gut microbiota and GLP-1. Rev Endocr Metab Disord. 2014;15:189–196. doi: 10.1007/s11154-014-9288-6. [DOI] [PubMed] [Google Scholar]
- 100.De Vadder F, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 101.Sayin SI, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 102.Schulz MD, et al. High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature. 2014;514:508–512. doi: 10.1038/nature13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bugge A, et al. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Delezie J, et al. The nuclear receptor REV-ERBalpha is required for the daily balance of carbohydrate and lipid metabolism. FASEB J. 2012;26:3321–3335. doi: 10.1096/fj.12-208751. [DOI] [PubMed] [Google Scholar]
- 105.Lamia KA, et al. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grimaldi B, et al. PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell Metab. 2010;12:509–520. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schmutz I, et al. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24:345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhao Y, et al. Loss of mPer2 increases plasma insulin levels by enhanced glucose-stimulated insulin secretion and impaired insulin clearance in mice. FEBS Lett. 2012;586:1306–1311. doi: 10.1016/j.febslet.2012.03.034. [DOI] [PubMed] [Google Scholar]
- 109.Zani F, et al. PER2 promotes glucose storage to liver glycogen during feeding and acute fasting by inducing Gys2 PTG and GL expression. Mol Metab. 2013;2:292–305. doi: 10.1016/j.molmet.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ikeda H, et al. Clock gene defect disrupts light-dependency of autonomic nerve activity. Biochem Biophys Res Commun. 2007;364:457–463. doi: 10.1016/j.bbrc.2007.10.058. [DOI] [PubMed] [Google Scholar]
- 111.Lamia KA, et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480:552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Barclay JL, et al. High-fat diet-induced hyperinsulinemia and tissue-specific insulin resistance in Cry-deficient mice. Am J Physiol Endocrinol Metab. 2013;304:E1053–E1063. doi: 10.1152/ajpendo.00512.2012. [DOI] [PubMed] [Google Scholar]
- 113.Shimba S, et al. Deficient of a clock gene, brain and muscle Arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS ONE. 2011;6:e25231. doi: 10.1371/journal.pone.0025231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Paschos GK, et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med. 2012;18:1768–1777. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shi SQ, et al. Circadian disruption leads to insulin resistance and obesity. Curr Biol. 2013;23:372–381. doi: 10.1016/j.cub.2013.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kennaway DJ, et al. Global loss of bmal1 expression alters adipose tissue hormones, gene expression and glucose metabolism. PLoS ONE. 2013;8:e65255. doi: 10.1371/journal.pone.0065255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Oishi K, et al. Disrupted fat absorption attenuates obesity induced by a high-fat diet in Clock mutant mice. FEBS Lett. 2006;580:127–130. doi: 10.1016/j.febslet.2005.11.063. [DOI] [PubMed] [Google Scholar]
- 119.Kudo T, et al. Attenuating effect of clock mutation on triglyceride contents in the ICR mouse liver under a high-fat diet. J Biol Rhythms. 2007;22:312–323. doi: 10.1177/0748730407302625. [DOI] [PubMed] [Google Scholar]
- 120.Kennaway DJ, et al. Metabolic homeostasis in mice with disrupted Clock gene expression in peripheral tissues. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1528–R1537. doi: 10.1152/ajpregu.00018.2007. [DOI] [PubMed] [Google Scholar]
- 121.Shostak A, et al. Circadian regulation of lipid mobilization in white adipose tissues. Diabetes. 2013;62:2195–2203. doi: 10.2337/db12-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]