Abstract
Organic cation transporter (OCT) function is critical for cellular homeostasis. C. elegans lacking OCT-1 displays a shortened lifespan and increased susceptibility to oxidative stress. We show that these phenotypes can be rescued by downregulating the OCT-1 paralogue, OCT-2. Herein, we delineate a biochemical pathway in C. elegans where uptake of genotoxic chemotherapeutics such as doxorubicin and cisplatin, and subsequent DNA damage-induced apoptosis of germ cells, are dependent exclusively upon OCT-2. We characterized OCT-2 as the main uptake transporter for doxorubicin, as well as a number of other therapeutic agents and chemical compounds, some identified through ligand-protein docking analyses. We provide insights into the conserved features of the structure and function and gene regulation of oct-1 and oct-2 in distinct tissues of C. elegans. Importantly, our innovative approach of exploiting C. elegans uptake transporters in combination with defective DNA repair pathways will have broad applications in medicinal chemistry.
The nematode Caenorhabditis elegans has a plethora of advantages as a useful in vivo model system1. Indeed, this organism exhibits a broad array of phenotypes that can be easily monitored for changes in various genetic/physiological pathways. For example, it can be utilized to understand the roles of Organic Cation Transporters (OCTs) in the uptake of therapeutic substrates2. In fact, evidence from mammalian systems dictates that OCTs mediate the uptake of chemotherapeutic drugs such as oxaliplatin and daunorubicin3,4. Hence, elucidating the molecular underpinnings of OCTs and consequent development of tools to modulate their transport activity in vivo are expected to improve chemotherapeutic outcome.
In C. elegans, little is known about the roles of OCTs and their affinity towards distinct substrates. OCT-1 was the first uptake transporter characterized from C. elegans and when expressed in mammalian cells was shown to mediate the transport of the organic cation tetraethylammonium, a prototypical substrate used for classifying OCTs5. C. elegans deleted for oct-1 exhibits a shortened lifespan and increased susceptibility to oxidative stress, which led to the proposition that OCT-1 facilitates the import of antioxidants required to protect oct-1 mutant animals from oxidative stress2. However, uptake of ergothioneine, the purported antioxidant substrate of OCT-1, was not reduced in oct-1 mutant animals as compared to the parent2. Therefore, it seems plausible that an alternative explanation could account for the oct-1 mutant animal phenotypes.
Recently, we documented that the expression of C. elegans OCT-1 can restore uptake of the chemotherapeutic drug doxorubicin into Saccharomyces cerevisiae cells lacking the regulator Agp2, an amino acid transporter that when deleted blocked the expression of several target genes including the polyamine transporters Dur3 and Sam36. No further studies were done to determine whether OCT-1 substituted for the regulatory function of Agp2 or directly for the roles of Dur3 and Sam3, as both of these transporters also mediate the transport of doxorubicin6. Furthermore, it remained unknown whether OCT-1 might mediate the transport of doxorubicin into C. elegans. Besides OCT-1, C. elegans possesses another related member of the SLC22 organic cation transporter family, i.e., OCT-2. OCT-2 shares 22.56% identity with OCT-1, but differs in having an extended N-terminal of 172 amino acid residues that is unrelated to OCT-1. To date, no studies have assigned a structural and functional role to the putative C. elegans OCT-2 transporter.
During the last decade, C. elegans has become instrumental in several drug discovery programs1,7. However, in many high-throughput screens performed so far to identify novel small molecules, e.g., those that act as antimicrobials, extend lifespan, inhibit oxidative stress or prevent multidrug resistance, the yield of bioactive compounds is typically in the range of 0.03% to less than 1%1,8. It is possible that the recovery rate could be higher if there is greater influx of the molecules. High-throughput screens at higher initial concentrations of the small molecules may alleviate this issue, but could be cost prohibitive. As such, we propose that characterization of the function and substrate specificities of uptake transporters in C. elegans will be advantageous towards improving the strategies employed to identify novel bioactive molecules.
In this study, we report a number of novel findings regarding the OCT-1 and OCT-2 transporters of C. elegans. We show for the first time that (i) unlike the downregulation of oct-1, depletion of oct-2 did not affect the lifespan of the animals and, instead, rescues the shortened lifespan of oct-1 deletion animals, (ii) oct-1 downregulation leads to oct-2 upregulation, which in turn mediates uptake of toxic environmental compounds and chemotherapeutic drugs, (iii) upregulation of OCT-2 increases uptake of prooxidants, as judged by the activation of the oxidative stress response reporter GST-4::GFP, leading to germ cell death, as well as to damages to other tissues that could account for the shortened lifespan of oct-1 deletion animals, (iv) oct-2 upregulation mediates the accumulation of clinically relevant genotoxic anticancer drugs that sensitizes DNA repair deficient animals to germ cell death and diminishes their survival, and (v) ligand-protein docking analysis can be exploited to define substrates such as DNA damaging agents that tightly bind to OCT-2 and which can be validated by suitable readouts. Our findings represent a robust OCT-based strategy to screen a plethora of new therapeutic drugs useful for treating human illnesses, and provide crucial information for rapid recognition of their pharmaceutical benefits and adverse effects.
Results
oct-2 deficiency rescues the shortened lifespan of oct-1 deletion mutants
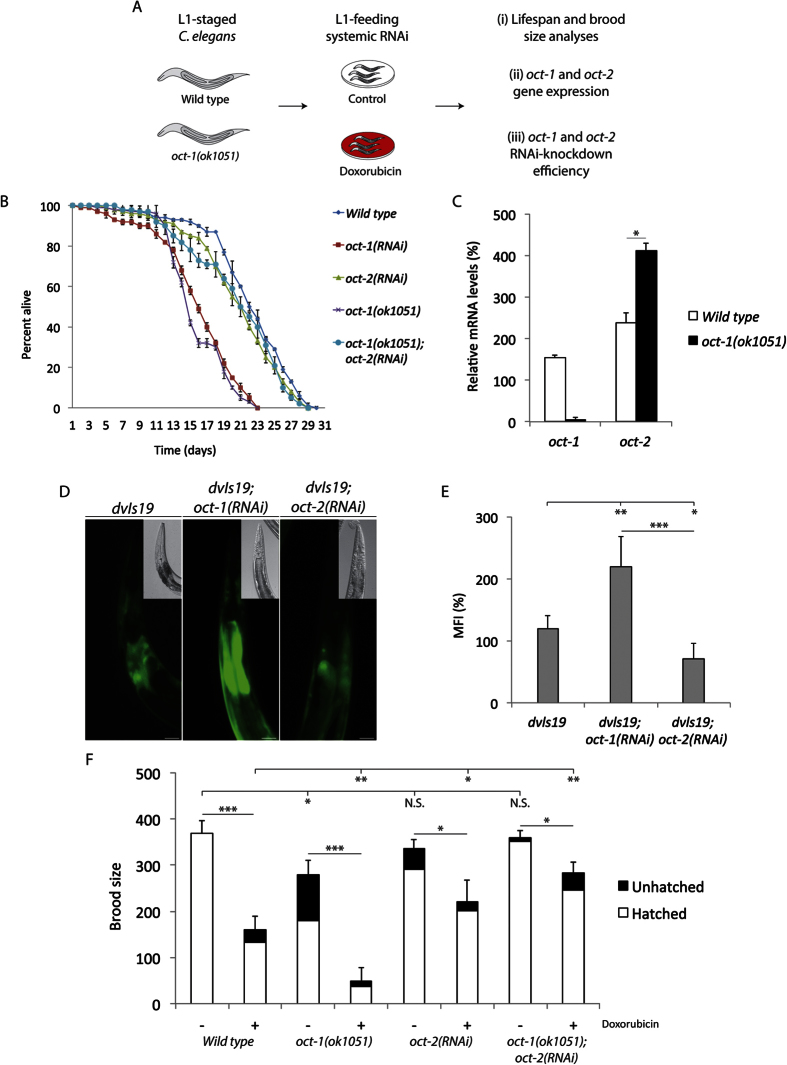
It has been postulated that C. elegans mutants deleted for oct-1 are defective in uptake of antioxidants and, as a consequence, exhibit shortened lifespan and increased susceptibility to oxidative stress2. However, the fact that oct-1 mutants show no defect in the uptake of the key antioxidant ergothioneine is inconsistent with this hypothesis. As such, we postulate that oct-1 gene deletion could instead lead to increased uptake of prooxidants from the environment if the loss of OCT-1 activates expression of a related transporter. This notion is based on the fact that OCT1 knockout mice manifest upregulation of two related transporter genes, OCT2 and OCT39. We therefore performed a homology search using C. elegans OCT-1 as a query for protein sequences in the C. elegans Wormbase. This analysis revealed a second C. elegans member of the organic cation transporter family SLC22, i.e., OCT-2, which shares 22.56% identity with OCT-1 (Figure S1). The predicted protein sequence of OCT-2 indicated that it possesses an extended N-terminal of 172 amino acid residues (Figure S1), suggesting that it is structurally distinct from OCT-1. To date, there is no previous report assigning a functional role to this putative transporter OCT-2 in C. elegans. We set out to characterize the molecular function of OCT-2 by first evaluating whether its RNA-interference (RNAi)-driven depletion might influence lifespan of C. elegans. In this experiment, L1-staged wild type animals were systematically fed bacteria harbouring the HT115 RNAi vector targeting oct-2 to measure adult lifespan, using oct-1 downregulation or the oct-1 gene deletion mutant oct-1(ok1051) for comparison (Fig. 1A)10. As expected, oct-1(RNAi) or oct-1 deletion mutants exhibited a shortened lifespan compared to wild type (Fig. 1B)2. In contrast, oct-2(RNAi) animals displayed a nearly normal lifespan (Fig. 1B). Quantitative real-time PCR (qRT-PCR) was used to ensure that oct-1 and oct-2 expression were indeed downregulated by the RNAi-driven approach (Figure S2A), and that this did not interfere with the expression of another transporter gene, namely pes-23 (Figure S2B), indicating that RNAi did not have off-target effects. Surprisingly, when oct-2 was downregulated in the oct-1 deletion mutant oct-1(ok1051), the resulting oct-1(ok1051); oct-2(RNAi) animals exhibited prolonged lifespan as compared to the oct-1(ok1051) mutants alone and approaching that of wild type worms (Fig. 1B). Thus, it would appear that the oct-1 mutant phenotype is dependent on OCT-2 function. A simple interpretation is that depletion of OCT-1 may cause the upregulation of oct-2 expression, such that OCT-2 mediates the uptake of toxic compounds that affect survival.
Figure 1. Upregulation of oct-2 gene expression shortens the lifespan and increases the sensitivity of C. elegans towards doxorubicin.
(A) Scheme of the experimental design and the readouts analyses under different conditions. (B) Lifespan analysis of the indicated genotypes. L1-staged animals (n = 100) were fed the control and the indicated RNAi under normal growth conditions and lifespan was blindly assessed starting from young adult animals. The mean lifespan of two independent experiments is depicted. (C) The relative gene expression of the oct-1 and oct-2 transcripts in the wild type and the oct-1(ok1051) deletion mutant animals was measured and corrected on actin as an internal control. Synchronized young adult animals were collected and mRNA levels were assessed by qRT-PCR. Data shown represent the average ± s.d. and student T-test *P < 0.05 from a 60 × 15 mm petri dish of animals (n ~ 1000) pooled from two independent experiments. (D) Representative images of the oxidative stress-inducible GST-4::GFP reporter and DIC (higher right) of the indicated genotypes. All images were maximum projections of the whole dvsI19 strain, but cropped from the pharynx to the primary section of the intestine for representation. (E) Increased GFP intensity depicting uptake of prooxidants. The GST-4::GFP activation was measured with Fluoroskan Ascent Microplate reader set at λex544 nm–λem590 nm. The mean fluorescence intensity (MFI) is represented as percentage corrected on wild type. (F) Brood size analysis of the indicated animals under standard conditions (no treatment) and upon exposure to 100 μM doxorubicin. The data are mean ± S.D. No treatment: Wild type = 369.5 ± 27 (n = 17), oct-1(ok1051) = 280 ± 29 (n = 23), oct-2(RNAi) = 335.5 ± 20.5 (n = 20), oct-1(ok1051); oct-2(RNAi) = 360.5 ± 14.8 (n = 21). Doxorubicin treatment (100 μM): Wild type = 170 ± 29.1 (n = 24), oct-1(ok1051) = 58 ± 29.2 (n = 19), oct-2(RNAi) = 220.5 ± 47 (n = 21), oct-1(ok1051); oct-2(RNAi) = 283 ± 23.1 (n = 27). Error bars represent the S.D. Unpaired two-tail t-test *P < 0.03; **P < 0.01; ***P < 0.0005 were considered to be statistically significant. N.S. = Non Significant.
A gene expression analysis dataset available from the Wormbase for both oct-1 and oct-211, revealed that oct-2 expression is normally higher than oct-1 across C. elegans developmental stages (i.e. from the first larval stage (L1) to the fourth larval stage (L4)), as well as in the hermaphrodite gonads (Figure S3). We examined whether downregulation of oct-1 would alter oct-2 expression levels by comparing the oct-1 and oct-2 gene expression in the wild type to that of the oct-1(ok1051) mutant. We found in the latter that oct-2 gene expression was significantly augmented, while the act-1 mRNA levels, used as a control, were unchanged (Fig. 1C). This finding further supports the notion that the effects of oct-1 depletion on the lifespan of the worms are attributable to oct-2 upregulation.
The SKN-1 target GST-4::GFP is upregulated by oct-1(RNAi), and blocked by oct-2(RNAi)
We examined whether the elevated levels of oxidative damage reported for the oct-1 deletion mutant is dependent on OCT-2 function2. To test this, we used a reporter strain dvls19 (GST-4::GFP) in which the promoter of the gst-4 gene (encoding glutathione S-transferase 4) is fused to GFP. GST-4 is a target for the conserved SKN-1/Nrf2 transcriptional activator that plays a role in the defense against oxidative stress12,13. The dvls19 strain showed a basal level of GST-4::GFP expression in the intestine of a representative animal (Fig. 1D), which was upregulated following RNAi downregulation of oct-1 (Fig. 1D). In contrast, this dvls19 strain showed lower than basal levels of GST-4::GFP expression when oct-2 was downregulated by RNAi (Fig. 1D) and quantified by plate assay (Fig. 1E). These findings are consistent with a model whereby oct-2 upregulation, via oct-1 deletion, allows entry of toxic compounds such as prooxidants into C. elegans causing oxidative stress and leading to a shortened lifespan.
OCT-2, and not OCT-1, mediates the genotoxic effects of the anticancer drug doxorubicin
To determine whether OCT-1 regulation of OCT-2 would be involved in the differential uptake of toxic compounds, we treated worms with the chemotherapeutic drug doxorubicin at concentrations that did not lead to developmental arrest14, and monitored the survival of the animals by scoring brood size. Doxorubicin uptake depends upon cationic transporters in Saccharomyces cerevisiae6 and mammalian cells4, and has been shown to trigger germ cell apoptosis in C. elegans14. L1stage wild type animals treated with doxorubicin showed ~55% decrease in brood size as compared to the untreated animals (Fig. 1F). In contrast, the oct-1(ok1051) mutants displayed a significant level of unhatched or dead embryos, and when treated with doxorubicin showed a sharp decrease (nearly 80%) in brood size (Fig. 1F). While these observations were unexpected as the loss of the uptake transporter should cause drug resistance, we reasoned that the enhanced doxorubicin sensitivity of the oct-1(ok1051) mutant animals can be explained by an increase uptake of the drug due to the upregulation of oct-2. We therefore examined the sensitivity of worms depleted for oct-2 following exposure to doxorubicin. oct-2(RNAi) caused wild type animals to become less sensitive to doxorubicin with only 27% decrease in brood size, as compared to 55% for the control RNAi (Fig. 1F). Importantly, RNAi-driven depletion of oct-2 in the oct-1(ok1051) mutant partially suppressed embryonic arrest and abolished the hypersensitivity of these animals towards doxorubicin showing less than a 20% reduction in brood size compared to 80% in oct-1(ok1051) mutant RNAi control (Fig. 1F). oct-2(RNAi) did not completely block doxorubicin toxicity on brood size of the oct-1(ok1051) mutant, perhaps reflecting residual oct-2 expression. Also since this mutant was deleted for the oct-1 gene, a direct role for OCT-1 in doxorubicin uptake is excluded (Fig. 1F). Collectively, these observations strongly support the notion that OCT-2 has a predominant role over OCT-1 in uptake of doxorubicin.
OCT-2 allows the accumulation of doxorubicin in C. elegans tissues
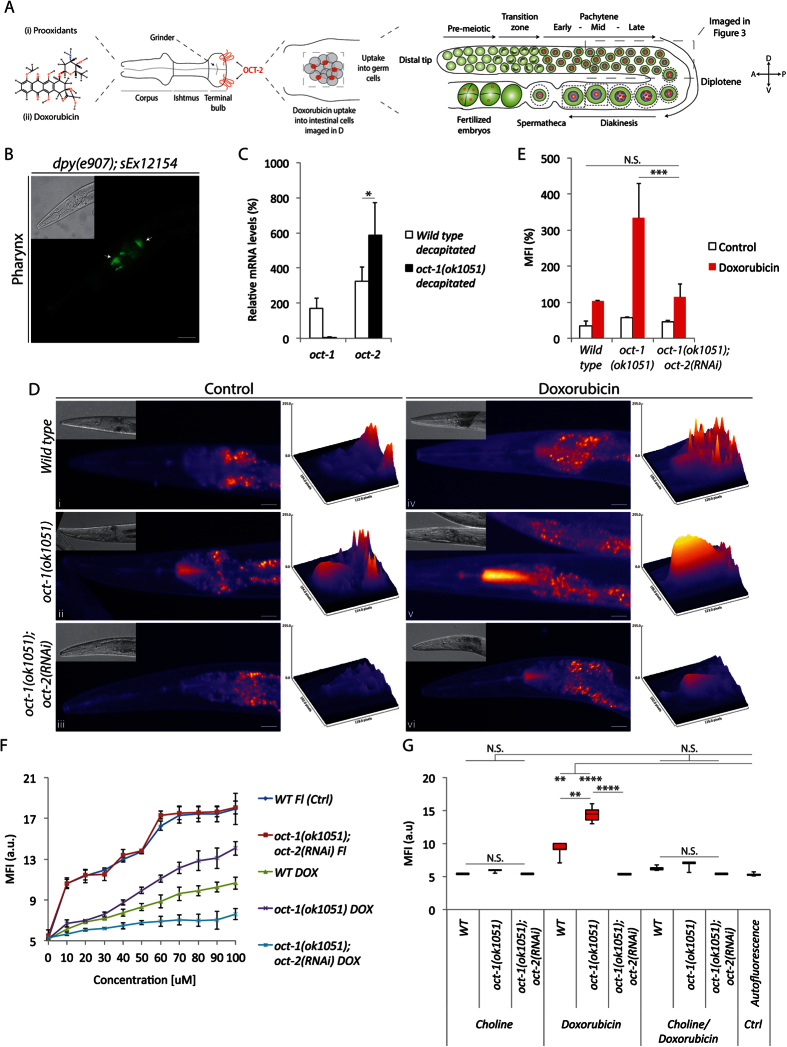
Based on the above findings, we examined whether OCT-2 might allow the accumulation of doxorubicin in C. elegans tissues. As target we chose the pharynx, a relatively large muscular organ that allow food consumption (Fig. 2A). We found that OCT-1 was localized at the anterior and posterior sides of the terminal bulb of the pharynx by imaging the dpy-5(e907); sEx12154 strain featuring an oct-1::GFP fusion (Fig. 2B), thereby validating this organ for uptake studies. Since no similar oct-2::GFP fusion has yet been constructed, we assessed the pharynx for oct-2 expression by measuring RNA levels in the heads, severed just posterior to the pharynx, of 300 each of wild type and oct-1(ok1051) animals. The results reveal that oct-2 is expressed at higher levels than oct-1 in the pharynx of the wild type animals and that its expression was stimulated nearly 2-fold in oct-1(ok1051) animals as compared to the wild type (Fig. 2C), suggesting that oct-2 is inducible in the pharynx.
Figure 2. Doxorubicin transport into the pharynx is stimulated by oct-2 upregulation and efficiently competed by choline.
(A) Model suggesting OCT-2 localization at the terminal bulb of the pharynx, the fluorescence (red dots) where the drug is detected, i.e., the posterior side of the pharynx and the initial part of the intestine (as imaged in F), and the region of germ cells analysed for apoptotic corpses in the gonad (see below). (B) DIC (upper left) and fluorescence image of dpy-5(e907); sEx12154 [OCT-1::GFP] genotype depicting OCT-1 localization in the pharynx (arrows in B). Enlargement of the pharynx is represented by a scale bar = 7 μm. (C) Relative gene expression of oct-1 and oct-2 transcripts measured in severed heads (n = 300) collected from wild type and oct-1(ok1051) animals and corrected to actin and pmp-3. (D) Representative ‘fire’ look-up images of the pharynx from untreated (control) and doxorubicin treated animals. The respective DIC images are shown in the upper left corner of each panel. Images to the right of each pharynx depict a 3D representation of the doxorubicin (100 μM) treatment signal intensity for the indicated genotypes. Data is representative of experiments performed in duplicates (n = 20). Enlargement of the pharynx is represented by a scale bar = 10 μm. Fluorescence posterior to the pharynx is autofluorescence detected from the intestine. (E) Quantification of doxorubicin uptake using a Fluoroskan plate reader. The mean fluorescence intensity (MFI) is represented as percentage corrected to wild type. White bars denote untreated animals where similar autofluorescence was detected in all genotypes. (F) oct-2(RNAi)diminishes the concentration-dependent uptake of doxorubicin (DOX) and not fluorescein (Fl) into the pharynx of oct-1(ok1051) mutant animals. (G) Comparison of doxorubicin uptake in the absence and presence of equimolar amounts of choline. Box and whisker plots represent duplicates (n = 10) of signal intensity measured from both compounds in the pharynx. Data is represented as ± S.D Student T-test ± S.D Student T-test *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 and N.S. = Non Significant.
We took advantage of the physical property of doxorubicin, which emits fluorescence at wavelengths of λex470 nm–λem585 nm, as a convenient means to monitor its uptake through OCT-1 and OCT-2 in situ by imaging the pharynx. Of note, C. elegans tissues have low levels of autofluorescence particularly at the posterior terminal bulb of the pharynx (Fig. 2D[i]) where most substances accumulate prior to consumption15. However, following exposure of wild type animals to doxorubicin, the emitted fluorescence was greater than background autofluorescence confirming consumption of the drug by the animals (Fig. 2D[iv]). Strikingly, the oct-1(ok1051) mutant did not reduce the fluorescence intensity of doxorubicin in the pharynx, instead depicting a markedly stimulated intensity (Fig. 2D[v]). To test whether the stimulated uptake of doxorubicin in the oct-1(ok1051) mutant could result from upregulation of oct-2, we downregulated oct-2 expression in this mutant. Under this condition, extremely low levels of doxorubicin accumulated in the oct-1(ok1051); oct-2(RNAi) animals (Fig. 2D[vi]). It is noteworthy that the deletion mutant eat-2(ad453), i.e., with a slow pumping pharynx16, also showed OCT-2-dependent uptake of doxorubicin, excluding the possibility that this transporter functions only when there is a surplus of nutrients (Figure S4).
Doxorubicin uptake in the pharynx of the oct-1(ok1051) strains was independently confirmed using another assay employing the Fluoroskan instrument, which measures the fluorescence intensity of the drug (Fig. 2E). Using this approach, oct-1(ok1051) animals showed a concentration-dependent uptake over a range (10–100 μM) of doxorubicin into the pharynx, which was blocked by RNAi-driven depletion of oct-2 (Fig. 2F). We tested whether uptake of another fluorescence compound, fluorescein which is anionic in nature, would be similarly dependent upon OCT-2. While fluorescein showed a concentration dependent uptake, ranging from 1 to 100 μM, into the pharynx of the oct-1(ok1051) mutant, it was not affected by oct-2 downregulation in oct-1(ok1051); oct-2(RNAi) animals (Fig. 2F). These findings suggest that OCT-2 may recognize differences in charges and display specificity in the uptake of substrates into C. elegans tissues.
OCT-2-mediated uptake of doxorubicin is blocked by choline in the pharynx
Human OCT1 and OCT2 were shown to transport other cationic molecules such as choline17. Thus if choline is also a substrate of C. elegans OCT-2 we would expect it to compete for doxorubicin uptake into the pharynx. Choline has the physical property of emitting fluorescence, but at lower wavelengths of λex290 nm–λem345 nm and is undetectable at the wavelength index used for monitoring doxorubicin uptake. Treatment of the oct-1(ok1051) animal with doxorubicin in the presence of equimolar amounts of choline (50 and 100 μM), impeded uptake of doxorubicin into the pharynx (Fig. 2G), clearly indicating that OCT-2 has the ability to recognize and compete for other cationic compounds. Therefore, we predict that the competition for doxorubicin uptake can be readily exploited as an assay to determine whether a putative ligand can serve as a substrate for OCT-2.
The oct-1(ok1051) mutant animals display increased spontaneous and drug-induced germ cell death that is suppressed by oct-2 downregulation
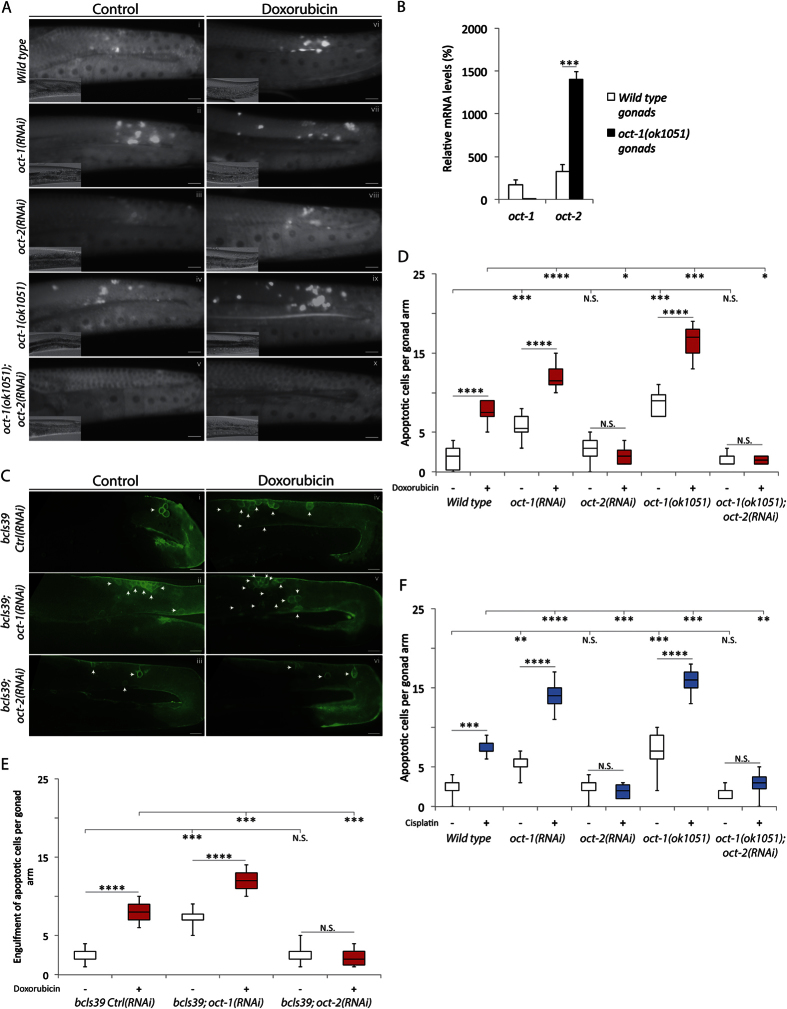
Analogous to many stem cell systems, C. elegans has a self-renewing germ cell population derived from a cellular niche located at the distal tip (see Fig. 2A)18. These germ cells progress through distinct stages of differentiation and must faithfully maintain the genome. They are very sensitive to genotoxic compounds and respond by using conserved DNA repair mechanisms to maintain genomic stability19. Germ cells with excessive DNA damage undergo apoptosis and are unable to form viable embryos18,20. We chose to monitor germ cell apoptosis as an experimental system to determine whether OCT-2 would be involved in the uptake of genotoxic anticancer drugs. This approach has the advantage of allowing assessment of the uptake of genotoxic anticancer drugs that do not emit fluorescence or are unavailable in radioactively labeled form. To monitor apoptotic germ cells, we first quantified in vivo germ cell corpses in the proximal zone of the gonad arm (see Fig. 2A) by utilizing differential interference contrast (DIC) microscopy and staining with the DNA dye acridine orange21. In wild type animals, one to four apoptotic cells were detected (Fig. 3A[i]), as previously reported22. In contrast, the oct-1(RNAi) or the oct-1(ok1051) mutants depicted an average of five to eight apoptotic cells (Fig. 3A[ii,iv]). Analysis of gene expression in the gonads from 100 each of dissected wild type and oct-1(ok1051) animals revealed that oct-1 gene deletion greatly stimulated the expression of oct-2 mRNA (Fig. 3B). Thus, the two-fold increase of apoptotic cells in the gonads of the oct-1(ok1051) animals may result from import of prooxidants, as assessed by activation of GST-4::GFP, that can damage the genome of the germ cells leading to embryos with hatching defects (Fig. 1F). Unlike the oct-1(RNAi), the oct-2(RNAi) animals displayed nearly the same average number of apoptotic cells as the wild type (Fig. 3A[iii vs. i]). Interestingly, RNAi-driven depletion of oct-2 in the oct-1(ok1051) mutant sharply reduced germ cell death, which was undetectable in some animals (Fig. 3A[v]).
Figure 3. OCT-2-mediated transport of genotoxic compounds induce apoptotic cell death of meiotic germ cells in C. elegans.
(A) Representative images of acridine orange-stained and DIC (lower left) of control and doxorubicin-induced apoptotic cell corpses from the indicated genotypes. Apoptotic cell corpses were identified as bright spots correlating with raised-bottom-like refractive corpses shown on DIC images. Posterior is right and dorsal is top. Scale bar = 15 μm. (B) Relative gene expression of oct-1 and oct-2 transcripts measured in dissected gonads (n = 100) collected from wild type and oct-2(ok1051) animals and corrected to actin and pmp-3 (see experimental procedures). (C) Representative images of control and doxorubicin-induced bcls39 [CED-1::GFP] clusters around apoptotic cell corpses indicated by white arrows. Data showing that OCT-2 mediated transport of compounds signal the apoptotic pathway in germ cells. Scale bar = 10 μm. (D) Box and whisker plots depicting quantification of apoptotic cell corpses from untreated and doxorubicin treated worms and showing maximum, minimum, upper & lower quartiles, and sample median. Statistical significance bars represent results of Mann-Whitney U-test of mean difference (*P < 0.05; **P < 0.01; ***P < 0.001 and ****P < 0.0001) computed from three independent experiments (n = 30). L1-stage animals were treated with doxorubicin (100 μM), and apoptotic cell corpses were analysed in young adult staged animals. Control is depicted as white and doxorubicin as red boxes. (E) Quantification of bcls39 engulfment of apoptotic cell corpses in the indicated genotypes without and with doxorubicin treatment. The results are presented as shown in (D). (F) Box and whisker plots depicting quantification of apoptotic cell corpses from untreated and cisplatin treated worms and quantified as in (D).
To confirm that the acridine orange-stained germ cells are undergoing apoptosis, as well as to avoid uptake differences of this dye, we evaluated a downstream step of the apoptotic pathway, i.e., engulfment of apoptotic cells by the CED-1 protein to signal phagocytic degradation23,24. We employed an imaging strategy that utilizes the bcls39 strain which carries CED-1::GFP as a reporter of engulfed apoptotic cells25. The bcls39 strain with control RNAi showed engulfment of 1 to 3 physiological apoptotic cells (Fig. 3C[i]), whereas downregulation of oct-1 engendered increased engulfment recapitulating the enhanced germ cell apoptosis observed in the oct-1(ok1051) mutant (Fig. 3A[iv] vs. C[ii]). As expected, downregulation of oct-2 in the bcls39; oct-2(RNAi) background displayed control levels of engulfment (Fig. 3C[iii]). Taken together, we propose that OCT-2 possesses the ability to transport toxic compounds such as prooxidants that cause germ cell death. These prooxidants are likely to cause damages to various tissues and may therefore account for reduced lifespan observed in the oct-1(RNAi) or oct-1(ok1051) animals (Fig. 1B).
We examined whether the OCT-2 dependent accumulation of doxorubicin would lead to the stimulation of germ cell death. Treatment of wild type worms with doxorubicin elevated the levels of apoptotic cell corpses as visualized by both acridine orange staining (Fig. 3A[vi]) and engulfment of the cells (Fig. 3C[iv]) (and quantified as in Fig. 3D,E, respectively). oct-1(RNAi) or the oct-1 deletion mutants treated with doxorubicin showed substantially higher levels of acridine orange-stained apoptotic cells (Fig. 3A [vii,ix] and quantified as in Fig. 3D), which paralleled increased CED-1::GFP engulfment of the cells as compared to the wild type (Fig. 3C[v vs. iv] and quantified as in Fig. 3E). In contrast, far fewer engulfed apoptotic corpses appeared in doxorubicin exposed animals downregulated for oct-2 (Fig. 3C[vi]). As expected based on acridine orange staining and engulfment of apoptotic cells, mutants deleted for cep-1, egl-1, ced-9, ced-4 or ced-3, which manifest defects in the apoptotic pathway did not show enhanced apoptotic corpses (Figure S5[ii–vi], respectively), i.e., unlike wild type animals upregulated for OCT-2 and treated with doxorubicin (Figure S5[i]). Several conclusions can be derived from these observations: (i) OCT-2 has a predominant role over OCT-1 in the uptake of doxorubicin, (ii) doxorubicin uptake leads to induced germ cell death that correlates with decrease survival, and (iii) both the drug uptake and the induced-germ cell death are OCT-2 dependent.
Cisplatin-induced germ cell death requires OCT-2 function
We assessed whether germ cell death in our experimental model can be used to monitor the uptake of genotoxic drugs that are not readily available as either fluorescently- or radioactively-labeled form. Since human OCT1 has been shown to transport members of the platinum family of anticancer drugs that act by creating intra- and inter-strand DNA cross-links26, we tested the role of C. elegans OCT-1 and OCT-2 in the uptake of cisplatin using germ cell death as a reporter. We monitored germ cell apoptosis in the absence and presence of cisplatin in the following four genotypes: wild type, oct-1(ok1051), oct-2(RNAi) and oct-1(ok1051); oct-2(RNAi). Cisplatin induced an increased level of germ cell death in the wild type, which was further greatly stimulated in the oct-1(ok1051) mutants (Fig. 3F). Downregulation of oct-2 in the oct-1(ok1051) mutants prevented cisplatin-induced germ cell death (Fig. 3F). These observations strongly indicate that germ cell death induced by cisplatin primarily depends upon its uptake via OCT-2. We noted that the DNA damaging agent methyl methanesulfonate (MMS), which alkylates DNA bases resulting in both DNA-single and -double strand breaks, induced germ cell death, but independently of OCT-1 and OCT-2 function (Figure S6A). Likewise, γ-rays that create multiple DNA lesions also induced germ cell death (Figure S6B) independently of these transporters (Figure S6C). Thus, the transporter function of OCT-2 is not directly involved in the process of germ cell death induced by the DNA damaging agents.
OCT-2-dependent transport of doxorubicin or cisplatin stimulates germ cell death in C. elegans mutants defective in DNA repair
We next systematically examined whether mutants defective in major DNA repair pathways would show OCT-2 dependent sensitization of DNA damage-induced germ cell death. As shown in Figure S7B, the rad-51 deletion mutant, rad-51(ok2218), lacking the RAD-51 protein needed for DNA strand invasion during homologous recombination (HR)-dependent double strand break repair, exhibited higher endogenous levels of apoptotic cell death due to spontaneous unrepaired meiotic breaks27 as compared to wild type (Figure S7A). Treatment of the rad-51(ok2218) mutant with doxorubicin greatly stimulated apoptosis, which was further induced upon downregulation of oct-1 in the rad-51(ok2218); oct-1(RNAi) genotype (Figure S7C). Consistent with this data, oct-2 gene expression was indeed upregulated in the rad-51(ok2218); oct-1(RNAi) genetic background (Figure S7J). In contrast, depletion of oct-2 by RNAi in rad-51(ok2218) animals suppressed the high level of apoptotic cells observed in this mutant upon exposure to doxorubicin (Figure S7C). These data indicate that upregulation of oct-2 burdens the rad-51(ok2218) animals with doxorubicin-induced DNA lesions leading to enhanced germ cell death.
We also found that apn-1(tm6691) animals lacking the key enzyme APN-1, required for removing a variety of DNA lesions including oxidized bases via the base-excision DNA repair (BER) pathway28, showed enhance germ cell death by doxorubicin when oct-2 expression was stimulated (Figure S7E,K). This induced apoptosis was strongly attenuated following depletion of oct-2 by RNAi (Figure S7E). These data suggest that BER in C. elegans is also involved in processing doxorubicin-induced oxidative DNA lesions29 but, more importantly, OCT-2 controls the toxicity of the drug in apn-1(tm6691) mutant animals.
Unlike doxorubicin, cisplatin damages the DNA by creating DNA cross-links most of which are processed by the nucleotide excision repair (NER) and DNA mismatch repair (MMR) pathways30. We set out to investigate whether oct-2 expression levels modulate cisplatin-induced germ cell death in mutants defective in either NER or MMR. Remarkably, cisplatin induced substantial levels of apoptotic cells in the xpa-1(ok698); oct-1(RNAi) (Figure S7G) and msh-2(ok2410); oct-1(RNAi) (Figure S7I) mutants, when oct-2 was upregulated (Figure S7L,M, respectively). These two DNA repair defective mutants were effectively protected from the onslaught of cisplatin-induced DNA lesions upon RNAi-driven depletion of oct-2 (Figure S7G,I, respectively), consistent with the involvement of OCT-2 in the transport of cisplatin.
It is noteworthy that amongst the DNA repair deficient animals, only in the case of the apn-1(tm6691); oct-1(RNAi) genotype was there a significant increase in spontaneous germ cell death (Figure S7D). One possible explanation for this observation is that OCT-2 mediated uptake of prooxidants may lead to oxidative lesions that must be repaired by BER28. Collectively, the above data suggest that by combining defects in DNA repair pathways with functional organic cation transporters such as OCT-2, it is possible to determine whether an unknown compound has genotoxic effects and the type of lesions it may create.
Ligand-protein docking analysis predicts the substrate specificity of OCT-2
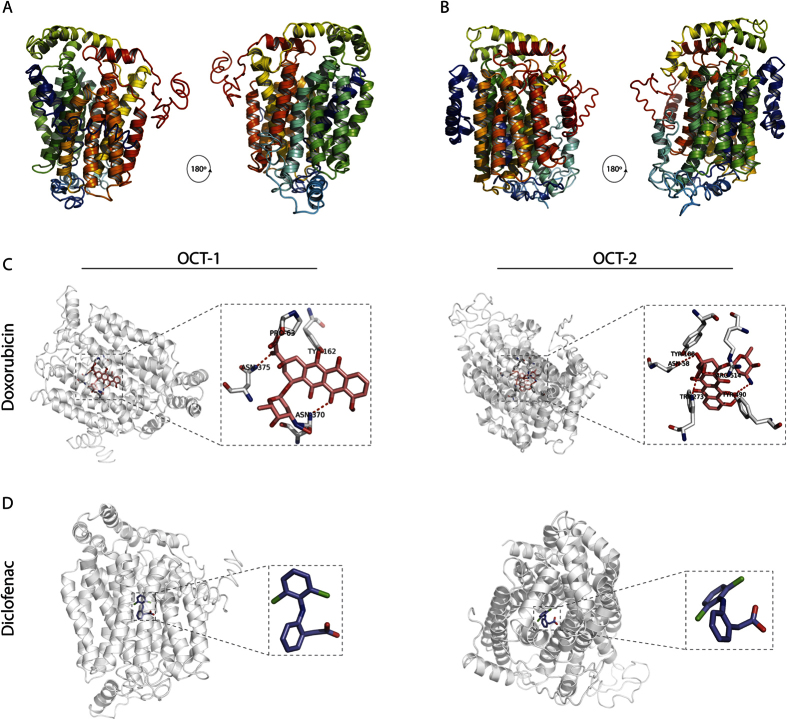
To gather insights into the substrates that can be recognized by OCT-2, we first made predictions of its protein structure relative to OCT-1 by inputting the respective primary protein sequences into the I-TASSER protein structure prediction server for in silico analyses31. The I-TASSER server employed known Protein Data Bank (PDB) structures as threading templates to predict the OCT-1 and OCT-2 structures (Table S1A,B). OCT-1 and OCT-2 were both modeled based on the X-ray diffraction structure of the glucose transporter GLUT3/SLC2A3 from Homo sapiens (PDB ID: 5c65) and validated with the glucose transporters GLUT1–4 structures (PDB ID: 4gc0) (Table S2A,B)32. The analysis revealed that OCT-1 and OCT-2 have predicted structures similar to each other (Fig. 4A,B). The comparative models of OCT-1 and OCT-2 were computed by utilizing several criteria as described in the materials and methods (Table S3). The final predicted 3D structure of OCT-1 and OCT-2 featured the entire 12 transmembrane domain helices (Figure S8A,B). Overall, the analysis predicted structures for both OCT-1 and OCT-2 that belong to solute carrier transporter family (Fig. 4A,B).
Figure 4. Structural modelling and protein-ligand docking of C. elegans OCT-1 and OCT-2.
(A,B) Structural models of OCT-1 and OCT-2 were generated on the basis of GLUT3 (PDB ID: 5c65) structure and sequence conservation. N-terminal and C-terminal are coloured blue and red, respectively. (C,D) Predicted binding models of OCT-1 and OCT-2 with cationic and anionic ligands doxorubicin (pink) and diclofenac (purple), respectively. Residues making polar contacts with the ligands are depicted with sticks and represented with dotted red lines; oxygen atoms are coloured in red, nitrogen atoms in blue and carbon atoms in white. All representative figures were rendered with PyMol.
We next determined whether the anticancer drugs, doxorubicin and cisplatin, would dock onto the predicted structures of OCT-1 and OCT-2. We utilized the BSP-SLIM and COACH algorithms33,34 to predict the amino acid residues of the transporters constituting the ligand–protein docking sites (Fig. 4C and Table S4). Since the BSP-SLIM server did not recognize the cisplatin chemical structure, a related member of the platinum drug family, carboplatin, was used. The OCT-1 and OCT-2 models interacting with doxorubicin computed a C-score of 0.38 and 0.75, respectively (Fig. 4C) (see materials and methods, Table S4). The predicted key residues mediating interaction between the drugs and the transporters suggest that the binding pockets of OCT-1 and OCT-2 are structurally different (Fig. 4C), making the ligand-protein binding highly selective depending upon the chemical structure of the ligand.
The OCT-1 and OCT-2 models were also evaluated for their ability to discriminate between the interacting ligands and probable non-binder compounds. As non-binder compounds, we targeted the nonsteroidal anti-inflammatory drug diclofenac, which is classified as an organic anion. The predicted models showed that no residues of OCT-1 or OCT-2 interact with diclofenac (Fig. 4D and Table S5). Thus, it is possible to perform protein-ligand modeling studies in order to predict and uncover novel substrates for uptake by these transporters, which could be further corroborated by the above in vivo assays.
Predicted ligands of OCT-1 and OCT-2 and experimental validation
So far, the roles of transporters in the uptake of a vast majority of genotoxic cationic drugs have not been tested in C. elegans. We sought to identify which of the known cationic drugs would be taken up by OCT-1 and/or OCT-2. A number of drugs were selected based on two criteria (i) mechanism of action and (ii) biological response. We assessed the docking ability of each compound with OCT-1 and OCT-2. From 19 tested ligands, four resulted with a docking score of zero, whereas the remaining 15 revealed a docking score > 3.5 favouring avid binding with OCT-2 (Table 1 dataset, docking score columns). Amongst these 15 ligands, some possess distinct pharmacological attributes such as creating different types of DNA lesions (Table S6). We next experimentally validated the in silico analyses using our drug-induced apoptosis assay as readout. We found that a number of the compounds that docked onto OCT-2 were capable of triggering high level of apoptotic cell death when oct-2 was upregulated by oct-1(RNAi), as compared to the control RNAi (Table 1 dataset; numbers in the bracket are from control RNAi). There were also ligands that act by damaging the DNA such as melphalan and methoxyamine, which did not dock onto OCT-2, but induced apoptotic corpses (Table 1 dataset). We postulate that these non-binders might use alternative transporters to enter the animal.
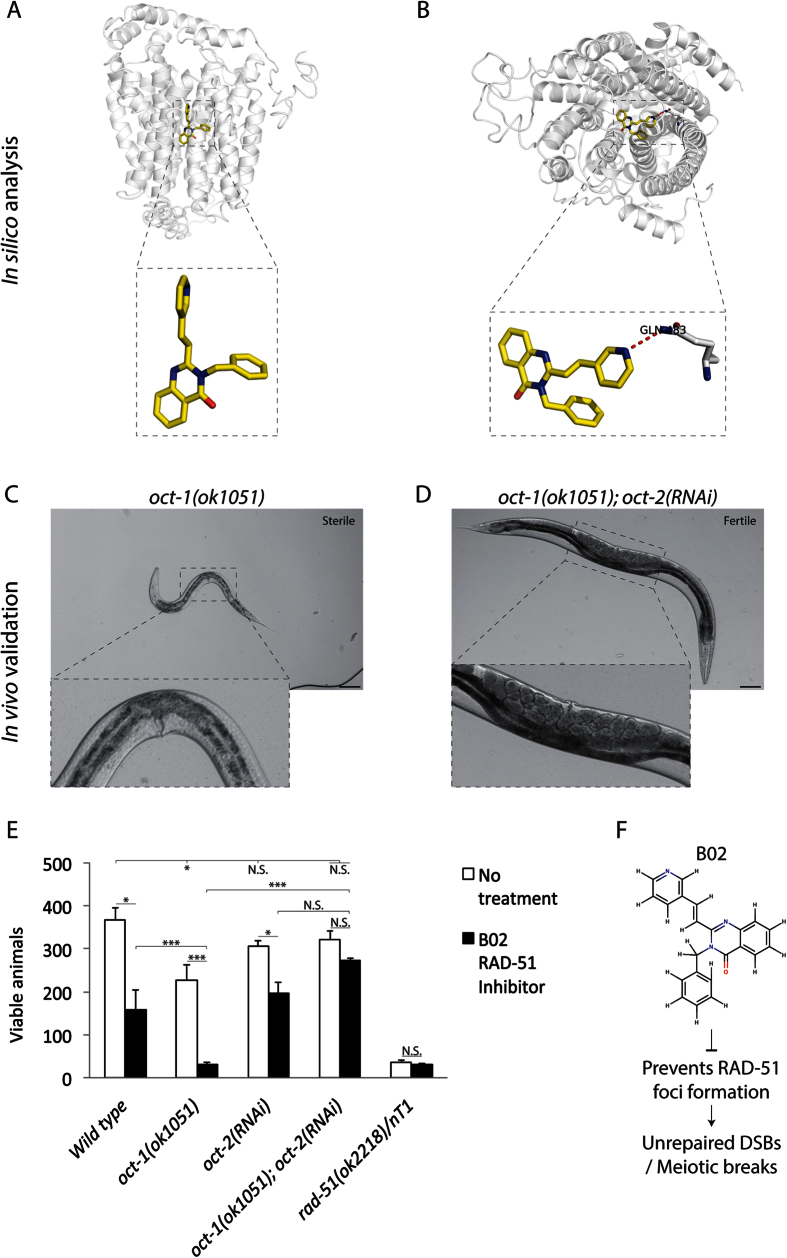
As a final validation, we focused on the ligand B02, which robustly docked only onto OCT-2 with a docking score of 5.4 (Table 1 dataset, Fig. 5A vs. B). B02 was shown to interfere with human RAD51 in DNA strand exchange and nuclear focus formation in response to DNA damage35,36,37. However, to our knowledge the pharmacological effect of B02 has not been tested in C. elegans. To confirm that B02 enters via the OCT-2 transporter, we tested the effect of the compound on wild type, oct-1(ok1051) and the oct-1(ok1051); oct-2(RNAi) backgrounds at concentrations ranging from 1 to 75 μM. Notably, extreme toxicity was observed with 75 μM B02 for all the genotypes. Therefore, we reduced the concentrations to 5 μM and observed that oct-2 upregulation caused sterility (Fig. 5C) resulting in a decrease in viable animals (Fig. 5E) and phenocopying the rad-51 homozygotes as previously reported38. As predicted from our model, the B02 ligand did not cause reduction in viable animals in the oct-1(ok1051); oct-2(RNAi) genetic background (Fig. 5D) and restored the number of broods to nearly untreated levels (Fig. 5E). These compelling data revealed for the first time that the B02 inhibitor of RAD51 is functionally active in C. elegans and requires exclusive uptake by OCT-2. We conclude that the docking scores of ligands and quantifiable endpoints provide valuable tools to monitor transporter-mediated drug uptake into C. elegans. This approach is particularly suitable for newly developed drugs that cannot be readily labeled or lack fluorescent properties for uptake studies.
Figure 5. The B02 inhibitor of RAD-51 mimics the rad-51/rad-51 homozygotes phenotypes when oct-2 is upregulated.
(A,B) Depiction of the ligand-protein docking of OCT-1 and OCT-2 modelled with B02 and showing exclusive binding of the drug to OCT-2. (C) DIC image showing that B02 causes embryonic lethality (diminishing number of germ cells) in the oct-1(ok1051) mutant. (D) DIC image showing that oct-2 downregulation in the oct-1(ok1051) mutant animals restores fertility. In (C,D) Scale bar = 35 μm. (E) B02 diminishes the number of viable animals when oct-2 is expressed. Brood size analysis of wild type, oct-1(ok1051) and oct-1(ok1051); oct-2(RNAi) and rad-51(ok2218)/nT1 animals under standard conditions (no treatment) and upon exposure to 5 μM B02. Data are mean ± S.D. No treatment: Wild type = 366 ± 26.7 (n = 20), oct-1(ok1051) = 296 ± 7.1 (n = 29), oct-2(RNAi) = 307 ± 11.3 (n = 23), oct-1(ok1051); oct-2(RNAi) = 321 ± 19.7 (n = 25), rad-51(ok2218)/nT1 = 35 ± 5.6 (n = 9). B02 treatment (5 μM): Wild type = 158 ± 29.7 (n = 21), oct-1(ok1051) = 32 ± 7.0 (n = 17), oct-2(RNAi) = 196 ± 26.8 (n = 25), oct-1(ok1051); oct-2(RNAi) = 272 ± 7.1 (n = 25), rad-51(ok2218)/nT1 = 30 ± 4.2 (n = 10). Error bars represent the S.D. Unpaired two-tail t-test *P < 0.03; **P < 0.01; ***P < 0.0005 were considered to be statistically significant. N.S. = Non Significant. (F) The RAD-51 inhibitor, B02, mode of action in C. elegans germline.
Discussion
In this study, we established for the first time the function of OCT-2 in C. elegans, as well as the specific roles for OCT-1 and OCT-2 in mediating tissue transport of distinct compounds, such as the chemotherapeutic drugs anthracyclines and cisplatin. We show that OCT-1 has no direct role in the transport of these compounds, however, it exerts control on oct-2 expression, and it is OCT-2 that is primarily involved in the uptake of these agents. This conclusion is derived from three key findings. First, the oct-1 deletion mutant oct-1(ok1051) or RNAi-driven knockdown of oct-1 triggered the upregulation of oct-2 in the whole body, the head and gonads of the animal, causing hypersensitivity to chemotherapeutic drugs. This phenotype is observed only when OCT-2 is present. Second, RNAi-driven knockdown of oct-2 blocked uptake of chemotherapeutic drugs thus preventing their genotoxic effects. Since no additional drug resistance was observed in oct-1(ok1051); oct-2(RNAi) double mutant animals as compared to ones depleted for oct-2 alone, a role for OCT-1 in the uptake of these chemotherapeutic drugs can be excluded; nonetheless OCT-1 could act as a transporter for selective ligands. And third, our in silico modeling-based screening of OCT-1 and OCT-2 selectively discriminated amongst DNA damaging agents those that are potential ligands for OCT-2. By validating this approach in vivo, we show that OCT2-dependent transport of the DNA damaging agents can sensitize mutant animals that are defective in DNA repair pathways. Collectively, these results underscore the importance of uptake transporters in regulating the entry of chemotherapeutic drugs into cells and raise the possibility that the drug-resistance and drug-sensitive responses observed by cancer patients could be governed at the level of drug uptake.
The downregulation of oct-1 leading to upregulation of oct-2 was an unexpected finding, and provides a compelling argument that the animal has evolved tight regulation of OCT-2. So how might OCT-1 depletion lead to the activation of OCT-2? One possibility is that both transporters have the ability to transport common essential nutrients and thus deletion of oct-1 would stimulate OCT-2 to compensate for the deprivation of pivotal nutrients. However, we argue against this possibility, as depletion of both oct-1 and oct-2 expression did not result in animals with any dramatic phenotypes under normal growth conditions. A more likely possibility is that OCT-1 might belong to the recent characterized class of surface sensors that act as non-transporting transceptors by sensing the availability of nutrients and signal the regulation of downstream plasma membrane transporters39,40. In this model, when nutrients become scarce, OCT-1 might serve as a sensor to promote the upregulation of OCT-2 to scavenge limiting resources. Conversely, when nutrients are plentiful OCT-1 might function to sustain the basal expression of OCT-2. Precedence for this mode of regulation exists in S. cerevisiae, Drosophila melanogaster and Homo sapiens41. For instance, in S. cerevisiae the Ssy1 sensor, a plasma membrane protein belonging to the amino acid permease family, is endowed with no, or only limited, transport function42. Ssy1 senses amino acid availability by direct interaction with extracellular amino acids and transmitting the signal to zinc-finger transcription factors that trigger the expression of several downstream target genes encoding amino acid permeases42,43,44. Similar sensors exist in mammalian cells, e.g., the SGLT3 glucose sensor that binds, but does not transport, sugar molecules45. Thus, in view of the increasing number of sensors that are currently being identified, it is plausible that OCT-1 may indeed act either as a non-transporting or transporting sensor leading to regulation of OCT-2 expression. The exact role by which OCT-1 exerts control on OCT-2 will need further investigation, but it is noteworthy that a similar regulation appears to occur in mice where deletion of OCT1 causes significant upregulation of mRNA transcripts of its homologues OCT2 and OCT39.
Remarkably, oct-2 downregulation rescued shortened lifespan and sharply decreased spontaneous apoptosis observed in the oct-1(ok1051) deletion mutant (Figs 1B and 3B). The most direct interpretation is that OCT-2 indiscriminately transports toxic compounds such as prooxidants. Although the source of these compound(s) is unknown, i.e., whether they originate from the feeding bacteria or re-adsorption of metabolites secreted by C. elegans, they are capable of inducing DNA lesions that must be removed by BER. This is supported by the observation that only the BER defective apn-1 deletion mutant exhibited higher levels of spontaneous apoptosis when oct-2 expression is upregulated. Thus, the previous report showing that oct-1 deletion animal exhibit a decline in lifespan due to reduced uptake of the antioxidant ergothioneine can alternatively be explained if the sulphur atom on the imidazole ring of ergothioneine serves to detoxify the OCT-2-dependent uptake of prooxidants2. Nonetheless, our findings raise a very important concern regarding genetic variations leading to hyperactivation of uptake transporters as previously reported46. This hyperactivation likely to cause accumulation of abnormally high concentrations of genotoxic compounds and metabolites. Consequently, such toxic agents could induce substantial DNA damage over the lifetime of an individual causing genomic instability and eventually cancer.
Our study is the first to demonstrate that C. elegans OCT-2 plays a role in ligand uptake. We tested doxorubicin and cisplatin as the initial cationic ligands because they are first-line chemotherapeutics believed to be transported by OCTs in human cells4,47. Besides these anticancer drugs, we postulate that OCT-2 may recognize a vast array of other cationic compounds. Per se, we implemented the OCT-based ligand-protein docking approach and explore a short list of selected cationic compounds to deduce that OCT-2 was promiscuous compared to OCT-1 and that it interacts robustly with several cationic ligands. Importantly, the analysis produced a refined list of genotoxic compounds that display high protein-ligand docking scores all of which show an OCT-2 dependent in vivo effect of triggering germ cells apoptosis. We believe that exploiting OCT-2 in C. elegans could have far reaching applications and supersede other whole model systems in drug discovery programs with respect to cost and time. Thus, maintaining the OCT-2 transporter at optimal levels by deleting oct-1 should represent a useful step for incorporation into any high-throughput screens to more efficiently identify bioactive molecules from chemical libraries. A key aspect of this strategy is that overexpressed OCT-2 is expected to operate with significantly lower chemical concentrations as observed with cisplatin where a fixed lower concentration of the drug had no effect on the wild type, but significantly induced apoptosis in the oct-1(ok1051) mutant (Figure S9). Thus, the previous barriers posed by C. elegans to find bioactive molecules could be explained by the lack of an activated mechanism to efficiently take up the compounds at lower concentrations. In short, we now provide a comprehensive readout of the OCT-2 functional selectivity towards cationic molecules that have a deleterious effect on C. elegans, and therefore provide a foundation to understand the regulatory control of drug uptake to circumvent genotoxicities. In addition, we hypothesize that OCT-2 could be exploited either through the oct-1 gene deletion mutant or oct-2 over-expression transgenic animals to generate a hypersensitive ‘screening’ C. elegans to facilitate high-throughput drug screening.
Materials and Methods
Nematode strains and culture conditions
The Bristol N2 (wild type), RB1084 [oct-1(ok1051) I], VC1973 [rad-51(ok2218) IV/nT1 [qIs51] (IV;V)]*, RB864 [xpa-1(ok698) I], RB1864 [msh-2(ok2410) I], MD701 [bcIs39 [lim-7p::ced-1::GFP + lin-15(+)] and DA453 [eat-2(ad453) II], TJ1 [cep-1(gk138) I], MT1082 [egl-1(n487) V], MT4770 [ced-9(n1950) III], MT5287 [ced-4(n1894) III], MT3002 [ced-3(n1286) IV] and CL2166 [dvIs19 [(pAF15) gst-4p::GFP::NLS] III] Caenorhabditis elegans strains were obtained from the CGC Stock center (Caenorhabditis Genetics Centre, University of Minnesota, Minneapolis, USA). The [apn-1(tm6691) II] were obtained from Shohei Mitani (Tokyo Women’s Medical University School of Medicine, Japan and the National Bioresource Project for the nematode C. elegans). The alleles used in this study were all previously validated to be null. All C. elegans strains were maintained at 20 °C on nematode growth medium (NGM) agar (2.5 g/L peptone, 51.3 mM NaCl, 17 g/L agar, 1 mM CaCl2, 1 mM MgSO4, 25 mM KPO4, and 12.9 μM cholesterol) enriched with a lawn of streptomycin-resistant Escherichia coli OP50 bacterial strain as a source of food. For all in vivo experiments, developmental staged-synchronized nematodes were obtained by hypochlorite treatment of gravid adult hermaphrodites. Eggs were allowed to hatch on M9 buffer (6g Na2HPO4, 3g KH2PO4, 5g NaCl, 0.25g MgSO4 · 7H2O per liter filter sterilized). In all experiments, animals were monitored from day 1 post-L1 larvae stage and from L4 to avoid experimental bias. *Homozygous rad-51/rad-51 nematodes27 show almost complete inviability due to high embryonic lethality in their progeny, thus we analyzed heterozygote nematodes due to the ease of RNAi-feeding for further analyses. C. elegans strains were backcrossed at least three times.
Lifespan assay
Lifespan assays were performed at 20 °C in standard conditions and assessed blindly as previously described10.
Drug treatment
The anthracycline doxorubicin, water-soluble platinum complex cisplatin and alkylating agent methyl methanesulfonate (Sigma Cat. N° 129925) were added to the NGM agar medium before solidification (~55 °C) to obtain a final concentration of 100 μM for doxorubicin and cisplatin and 0.25 μM for methyl methanesulfonate, respectively. For all experiments, L1-staged from F1 synchronized nematodes were transferred to NGM control agar plates and containing doxorubicin, cisplatin and methyl methanesulfonate. Doxorubicin and cisplatin working concentrations were chosen based on previously reported assays14. All drug-containing plates were freshly made prior to each experiment. Our oncology pharmacy department (Maisonneuve-Rosemont Hospital (HMR) provided doxorubicin and cisplatin.
Microscopy and imaging
All microscopy was performed utilizing a DeltaVision Elite Image Restoration System (Applied Precision) with either 40x/0.65–1.35 or 63x/1.42 oil objective. The worms were anesthetized with levamisole (5 μM, Sigma Cat. N° L0380000) and mounted on 2% agarose pads for their respective imaging and quantification. Images were processed utilizing ImageJ imaging software48.
DNA damage response assay and germ cells imaging
The methods previously described were used21. Briefly, to quantify the number of apoptotic corpses in nematodes, L1-staged synchronized N2 wild type, oct-1(ok1051) and DNA repair deficient mutants were exposed to different doses of drugs followed by germ cells apoptosis assay. Between 18 to 24 hours past L4-staged nematodes, adult moult staged nematodes were assayed with differential interference contrast (DIC) microscopy (Nomarski) optics and the vital DNA dye acridine orange (Sigma Cat. N° A6024). Nematodes were incubated in the dark for 2 hours at 20 °C on NGM plates containing 1 ml of 50 μg/μl of acridine orange DNA dye dissolved in M9 buffer. Stained nematodes were transferred to fresh OP50-seeded NGM plates to incubate for 2 hours in order to clear off the stained bacteria. The acridine orange-stained and DIC-visible apoptotic corpses were counted with an exposure time of 1 second and 0.8 seconds, respectively. The engulfment of apoptotic corpses was scored utilizing the CED-1::GFP reporter and imaged similarly with an exposure time of 1 second utilizing the GFP channel. Images were collected as a series of 25/0.5 μm optical sections covering the complete thickness of the gonad arm.
Imaging the oxidative stress-inducible GFP reporter GST-4::GFP
The uptake of prooxidants was detected by imaging the oxidative stress-inducible GFP reporter GST-4::GFP. L4-staged synchronized nematodes were maintained in NGM plates at 20 °C overnight and imaged the next day at the adult moult stage. The image analysis was performed by measuring the GFP fluorescence intensity of the whole animal utilizing the GFP channel exposed for 0.8 seconds and DIC exposed for 0.05 seconds.
Imaging the pharynx to measure the uptake of doxorubicin
To measure the uptake of doxorubicin, an approach focusing on the pharynx of C. elegans was developed. L4-staged synchronized nematodes were treated with doxorubicin 100 μM in NGM plates, incubated at 20 °C overnight and imaged the next day at the adult moult stage. The image analysis was performed by measuring the doxorubicin fluorescence intensity (λex470 nm–λem585 nm) localized at the pharynx by utilizing the GFP channel and observed under a 40x/0.65–1.35 oil objective exposed for a period of time of 0.5 seconds. ImageJ48 imaging software was utilized to determine the level of fluorescence in the pharynx region. The data of measured fluorescence intensity for doxorubicin uptake was depicted by implementing the custom built-in interactive 3D Surface Plot featured in ImageJ, which display the intensities of pixels from a region of interest of a given image. The uptake was corroborated by two means: (i) The area, the integrated density and the mean gray value were considered to calculate the corrected total pharynx fluorescence [CTPF = Integrated Density − (Area of selected pharynx X Mean fluorescence of background readings)]49, and (ii) Fluoroskan analysis (see below). To measure doxorubicin uptake in a dose-dependent manner, the nematodes were treated with 1 ml of varying concentrations (10 to 100 μM) of the drug in M9 buffer. Uptake of Fluorescein (Sigma Cat. N° F2456), at concentrations ranging from 1 to 100 μM, was used as a control.
Relative RNA quantification to monitor gene expression
Total RNA (RNeasy mini kit Qiagen Cat. N° 74104) was prepared from ~1000 L4 synchronized nematodes and used for cDNA synthesis (Invitrogen Cat. N° 28025-013) followed by quantitative real-time PCR (qRT-PCR). qRT-PCR was performed with the Supergreen Mastermix (Wisent Bioproducts Cat. N° 800-431-UL) starting at 95 °C for 2 min, followed by 40 cycles at 95 °C for 5 sec, 63 °C for 30 sec and 72 °C for 30 sec. Transcript levels were normalized to the internal controls act-1 and pmp-3 encoding actin and the peroxisomal membrane protein, respectively. Because there is a putative third organic cationic transporter PES-23, which shares 21.31% and 17.18% identity with OCT-1 and OCT-2, respectively, we monitored pes-23 gene expression for off targets. The forward and reverse primer sequences utilized in this study were: oct-1: 5′-TTTGGAGCAGCTATGGCTTT-3′ and 5′-CTTAGCGTCAGCCCATTTTC-3′; oct-2: 5′-TTGGAGTCGTGCTCACGTTC-3′ and 5′-GAGTATGTGAGAAGAAAGCC-3′; act-1: 5′-TGCTGATCGTATGCAGAAGG-3′ and 5′-TAGATCCTCCGATCCAGACG-3′; pmp-3: 5′-GTTCCCGTGTTCATCACTCAT-3′ and 5′-ACACCGTCGAGAAGCTGTAGA-3′; pes-23: 5′-TTCTTGCCGGAGTACCTGCC-3′ and 5′-GCACACATGGAGATTCCGTT-3′.
Dissection of C. elegans heads and gonads for relative RNA quantification
Nematodes were first transferred to a dried 2% agarose pad in 10 μl of M9 buffer. Exactly 300 N2 wild type and 300 oct-1(ok1051) L4 ~ young adult moult staged nematodes were decapitated just posterior to the pharynx by utilizing a 261/2 gauge syringe. Severed heads were washed and collected with M9 buffer in a micro-centrifuge tube and rapidly stored at −80 °C in Trizol (Ambion Life Technologies Cat. N° 15596-018). The same procedure was followed to dissect 100 gonads from L4 ~ young adult staged nematodes. In order to preserve optimum tissue integrity, the collection was made every 15 severed heads and every 10 gonads respectively. RNA extraction was performed from the pool of all collected severed heads and dissected gonads and stored at −80 °C for further qRT-PCR analysis.
RNA interference analysis
Escherichia coli HT115DE3 strain harboring specific RNAi constructs against oct-1 and oct-2 was grown on lysogeny broth (LB) agar plates containing ampicillin and tetracycline. Overnight cultures were grown in LB media containing ampicillin. For oct-1 and oct-2 RNAi-driven knockdown experiments, nematodes were maintained until first generation (F1) on NGM agar plates containing 1 mM IPTG (isopropyl-β-D-1-thiogalactopyranoside) enriched with a lawn of E. coli HT115DE3 expressing RNAi constructs in the pL4440-feeding vector at standard temperature 20 °C. For oct-1 and oct-2 RNAi-driven knockdown efficiency, mRNA expression levels were measured in synchronized L4-staged collected from the F1 generation of nematodes fed with E. coli expressing RNAi targeted to the indicated genes. The RNAi clones were obtained from the Ahringer laboratory library50 and verified by sequencing. The depletion efficiency of oct-1 and oct-2 genes was validated by qRT-PCR. In all experiments synchronized L4 animals were fed RNAi expressing bacteria and the resulting F1 animals were analyzed for phenotypes.
Choline-based competition assay
L4-staged F1 nematodes were treated overnight with 50 μM and 100 μM concentrations of choline and doxorubicin as separate conditions and together in equimolar amounts in NGM agar plates. The differences in wavelength indexes between doxorubicin (λex470 nm–λem585 nm) and choline (λex290 nm–λem345 nm), allowed us to measure the competitive uptake in the pharynx utilizing the GFP wavelength index where only doxorubicin is detectable and not choline.
Statistical analyses
Lifespan analyses were performed utilizing the Kaplan-Meier estimator calculating the Log-rank test for statistical significance utilizing OASIS software (Online Application for the Survival Analysis of Lifespan Assays Performed in Aging Research)51. Germ cell death statistical significance was assessed with the Mann-Whitney U-test calculator Mean values ± s.e.m were calculated for each condition. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 were considered to be statistically significant. For the Brood size analysis, statistical differences were calculated by the unpaired two-tail t-test (*P < 0.03; **P < 0.01; ***P < 0.0005) and represented as ± S.D. The Fluoroskan data and the Mean Fluorescence Intensity measurements extracted from the competition assay, Student T-test was calculated and represented as ± S.D (***P < 0.001 significant). N.S. = Non Significant. Statistical differences were calculated by using the GraphPad Prism Statistical Software Mac Version 6.
Comparative structure and model construction
C. elegans OCT-1 (F52F12.1) and OCT-2 (ZK455.8) putative protein sequences were first obtained from the Wormbase. OCT-1 and OCT-2 were modeled using I-TASSER (Iterative Threading ASSEmbly Refinement)31 which utilized the best 10 threading template structures from distinct species from the PDB database (Table S1a,b). We relied on the top 5 threading templates ranked by their identity and Z-score, where a Z-score higher than 1 signified a correct alignment. The final models of OCT-1 and OCT-2 were assessed based on the X-ray diffraction structural analogs of the glucose transporter GLUT3 (PDB ID: 5c65) from Homo sapiens (Table S2a,b). The models took into account the following (i) the C-score criteria, a confidence score for estimating the quality of predicted models and TM-score criteria, a metric measurement of the structural similarity between two protein models from I-TASSER52, (ii) the TM-score from ModRefiner53 and (iii) the Z-DOPE from Modeller, an atomic distance-dependent statistical calculation from samples of native protein structures that does not depend on any adjustable criteria54. These final predicted structures were also assessed based on primary sequence alignment and evolutionary conservation profiles using the PROMALS3D multiple sequence and structure alignment server55. Finally, for the predictions of the transmembrane domains, the ResQ B-factor profile provided a consensus prediction where the secondary structure helices (SS) are depicted as red tubes56 (Figure S8A,B), and corroborated with the Orientation of Protein in Membranes (OPM) server57, the Dense Alignment Surface (DAS) method58 and the Open-source tool for visualization of proteoforms (PROTTER)59.
Ligand-protein docking
The ligand chemical structures for doxorubicin (ID: 31703) and diclofenac (ID: 3033) were obtained from the PubChem database60. Ligand-protein docking were performed through the BSP-SLIM (Binding Site Prediction with Shape-based Ligand Matching with binding pocket) and COACH algorithms33,34, featured in the I-TASSER unified platform, to predict the residues constituting the ligand–protein docking sites and conformations of OCT-1 and OCT-2 with doxorubicin and diclofenac.
Structure visualization
OCT-1 and OCT-2 three-dimensional structures and protein-ligand interacting structures were visualized using the OpenGL PyMOL Molecular Graphics System, Mac Version 1.7.4 Schrödinger, LLC.
In vivo validation of the predicted ligand-protein docking models
The ligand-protein docking was performed utilizing the BSP-SLIM server and validated following the same strategy as described in the drug treatment and DNA damage response assay and germ cells imaging sections. Synchronized L1-staged worms were exposed to the following compounds dissolved in DMSO; the RAD-51 inhibitor B02 (5 μM) (ID: 5738263) (EMD Millipore Cat. N° 553525), Camptothecin (75 μM) (ID: 24360) (Sigma Cat. N° C9911), Cycloheximide (50 μM) (ID: 6197) (Sigma Cat. N° C7698), Ketamine (ID: 3821) (predicted virtually), Melphalan (ID: 460612) (predicted virtually), and these additional compounds dissolved in water; Metformin (75 μM) (ID: 4091), Methotrexate (50 μM) (ID: 126941), Methoxyamine (10 μM) (ID: 4113) (Santa Cruz Biotechnology Cat. N° SC257710), Methyl methane sulfonate (0.25 μM) (ID: 5156) (Sigma Cat, N° 129925), Nicotinamide (100 μM) (ID: 936) (Sigma Cat. N° N3376), 4-Nitroquinoline N-oxide (75 μM) (ID: 5955) (ICN Biomedicals Cat. N° 15596), Olaparib (ID: 23725625) (predicted virtually), Paraquat (100 μM) (ID: 15939) (Sigma Cat. N° 856177), Phenformin (75 μM) (Sigma Cat. N° P7045), Puromycin (100 μM) (ID: 439530) (Sigma Cat. N° P9620), Zeocin (25 μM) (ID: 71668282) (Santa Cruz Biotechnology Cat. N° SC496345), and analyzed at the young adult stage. Methotrexate and Metformin were obtained from our oncology pharmacy department (Maisonneuve-Rosemont Hospital (HMR). A concentration of 0.2% DMSO was used for control plates. All drug-treated plates were made fresh and seeded with bacteria 12 hours prior to each experiment.
Brood size quantification to validate the effects of the RAD-51 inhibitor, B02
Single L1-staged worm from wild type and mutant genotypes were transferred to seeded NGM plates without and with B02 and maintained at 20 °C. Animals were transferred to fresh plates each day until they stopped laying eggs. The hatched larvae on each plate were counted and total number of viable larvae that developed to the L1 stage descended from a single hermaphrodite was counted. The average number of viable larvae from 10 to 25 animals of a strain was plotted as brood size where the progeny is allowed to reach adulthood and scored as being fertile or sterile. The brood size of doxorubicin-treated animals was performed similarly where unhatched and hatched progeny was plotted as the total brood size. Statistical differences were calculated by unpaired two-tail t-test (*P < 0.03; **P < 0.01; ***P < 0.0005; N.S. = Non Significant) and represented as ± S.D using GraphPad Prism Statistical Software.
Fluoroskan analysis
Synchronized L1-staged nematodes were exposed to doxorubicin (100 μM) as described above. L4 ~ young adult-staged nematodes were washed at least two times with M9 buffer prior to quantification. A total of 50 worms were placed into each well in duplicate in a 96 format black-well plate with optical bottom (Fisher Scientific). Doxorubicin uptake was measured with a microplate fluorometer (Fluoroskan Ascent, Thermo Scientific, USA) utilizing λex544 nm–λem590 nm filters. Fluorescein 1 μM was used as an internal control for all genotypes. Student T-test was calculated and represented as ± S.D ***P < 0.001 statistical significance and N.S. = Non Significant.
Additional Information
How to cite this article: Papaluca, A. and Ramotar, D. A novel approach using C. elegans DNA damage-induced apoptosis to characterize the dynamics of uptake transporters for therapeutic drug discoveries. Sci. Rep. 6, 36026; doi: 10.1038/srep36026 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We acknowledge Dr. John Pascal (Département de Biochimie et Médecine Moléculaire, Université de Montréal) for guidance with the protein structure modeling analysis. We thank Drs Marie-Eve Lalonde and Florence Coteau (Centre de Recherche Hôpital Maisonneuve-Rosemont) for technical assistance with the gene expression and C. elegans genetic analyses, respectively. We thank Drs Ivan Topisirovic (Lady Davis Institute Jewish General Hospital, McGill University), Nigel O’Neil (Michael Smith Laboratories, Vancouver, BC) and Elliot Drobetsky (Université de Montréal) for critically reading the manuscript. We also thank the CGC and Shohei Mitani for strains. This work was funded by the research grant (RGPIN/202432–2012) to D.R. from the Natural Science and Engineering Research Council of Canada. A.P. was awarded with a Ph.D bourse d’excellence du Programme en Médecine Expérimentale, Université de Montréal.
Footnotes
The authors declare no competing financial interests.
Author Contributions A.P. and D.R. designed the experiments. A.P. performed 100% of the experiments. A.P. and D.R. analyzed the data. A.P. wrote the manuscript. A.P. and D.R. corrected the final version of the manuscript.
References
- O’Reilly L. P., Luke C. J., Perlmutter D. H., Silverman G. A. & Pak S. C. C. elegans in high-throughput drug discovery. Adv Drug Deliv Rev 69–70, 247–253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah I. K. et al. Knockout of a putative ergothioneine transporter in Caenorhabditis elegans decreases lifespan and increases susceptibility to oxidative damage. Free Radic Res 47, 1036–1045 (2013). [DOI] [PubMed] [Google Scholar]
- Koepsell H. The SLC22 family with transporters of organic cations, anions and zwitterions. Mol Aspects Med 34, 413–435 (2013). [DOI] [PubMed] [Google Scholar]
- Andreev E., Brosseau N., Carmona E., Mes-Masson A. M. & Ramotar D. The human organic cation transporter OCT1 mediates high affinity uptake of the anticancer drug daunorubicin. Sci Rep 6, 20508 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. et al. Identity of the F52F12.1 gene product in Caenorhabditis elegans as an organic cation transporter. Biochim Biophys Acta 1418, 239–244 (1999). [DOI] [PubMed] [Google Scholar]
- Brosseau N., Andreev E. & Ramotar D. Complementation of the Yeast Model System Reveals that Caenorhabditis elegans OCT-1 Is a Functional Transporter of Anthracyclines. PLoS One 10, e0133182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletta T. & Hengartner M. O. Finding function in novel targets: C. elegans as a model organism. Nat Rev Drug Discov 5, 387–398 (2006). [DOI] [PubMed] [Google Scholar]
- Burns A. R. et al. A predictive model for drug bioaccumulation and bioactivity in Caenorhabditis elegans. Nat Chem Biol 6, 549–557 (2010). [DOI] [PubMed] [Google Scholar]
- Chen L. et al. OCT1 is a high-capacity thiamine transporter that regulates hepatic steatosis and is a target of metformin. Proc Natl Acad Sci USA 111, 9983–9988 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionaki E. & Tavernarakis N. Assessing aging and senescent decline in Caenorhabditis elegans: cohort survival analysis. Methods Mol Biol 965, 473–484 (2013). [DOI] [PubMed] [Google Scholar]
- McKay S. J. et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb Symp Quant Biol 68, 159–169 (2003). [DOI] [PubMed] [Google Scholar]
- Fishel M. L. et al. Apurinic/apyrimidinic endonuclease/redox factor-1 (APE1/Ref-1) redox function negatively regulates NRF2. J Biol Chem 290, 3057–3068 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabreiro F. et al. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 153, 228–239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendoel A. et al. DEPDC1/LET-99 participates in an evolutionarily conserved pathway for anti-tubulin drug-induced apoptosis. Nat Cell Biol 16, 812–820 (2014). [DOI] [PubMed] [Google Scholar]
- Hall D. & Altun Z. C. elegans Atlas, (Cold Spring Harbor Laboratory Press, 2008). [Google Scholar]
- McKay J. P., Raizen D. M., Gottschalk A., Schafer W. R. & Avery L. eat-2 and eat-18 are required for nicotinic neurotransmission in the Caenorhabditis elegans pharynx. Genetics 166, 161–169 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann K., Grafe F., Wohlrab W., Neubert R. H. & Brandsch M. Functional characterization of a high-affinity choline transport system in human keratinocytes. J Invest Dermatol 119, 118–121 (2002). [DOI] [PubMed] [Google Scholar]
- Gartner A., Boag P. R. & Blackwell T. K. Germline survival and apoptosis. WormBook, 1–20 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lans H. & Vermeulen W. Tissue specific response to DNA damage: C. elegans as role model. DNA Repair (Amst) 32, 141–148 (2015). [DOI] [PubMed] [Google Scholar]
- Horvitz H. R. Genetic control of programmed cell death in the nematode Caenorhabditis elegans. Cancer Res 59, 1701s–1706s (1999). [PubMed] [Google Scholar]
- Craig A. L., Moser S. C., Bailly A. P. & Gartner A. Methods for studying the DNA damage response in the Caenorhabdatis elegans germ line. Methods Cell Biol 107, 321–352 (2012). [DOI] [PubMed] [Google Scholar]
- Gumienny T. L., Lambie E., Hartwieg E., Horvitz H. R. & Hengartner M. O. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126, 1011–1022 (1999). [DOI] [PubMed] [Google Scholar]
- Savill J. & Fadok V. Corpse clearance defines the meaning of cell death. Nature 407, 784–788 (2000). [DOI] [PubMed] [Google Scholar]
- Zhou Z., Hartwieg E. & Horvitz H. R. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104, 43–56 (2001). [DOI] [PubMed] [Google Scholar]
- Li Z., Lu N., He X. & Zhou Z. Monitoring the clearance of apoptotic and necrotic cells in the nematode Caenorhabditis elegans. Methods Mol Biol 1004, 183–202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Yee S. W., Kim R. B. & Giacomini K. M. SLC transporters as therapeutic targets: emerging opportunities. Nat Rev Drug Discov 14, 543–560 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo C., Bazzicalupo P., Ederle S., Hilliard M. & La Volpe A. Roles for Caenorhabditis elegans rad-51 in meiosis and in resistance to ionizing radiation during development. Genetics 160, 471–479 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. et al. Functional characterization of the Caenorhabditis elegans DNA repair enzyme APN-1. DNA Repair (Amst) 11, 811–822 (2012). [DOI] [PubMed] [Google Scholar]
- Minotti G., Menna P., Salvatorelli E., Cairo G. & Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev 56, 185–229 (2004). [DOI] [PubMed] [Google Scholar]
- Wang D. & Lippard S. J. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov 4, 307–320 (2005). [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A. & Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. et al. Crystal structure of a bacterial homologue of glucose transporters GLUT1-4. Nature 490, 361–366 (2012). [DOI] [PubMed] [Google Scholar]
- Lee H. S. & Zhang Y. BSP-SLIM: a blind low-resolution ligand-protein docking approach using predicted protein structures. Proteins 80, 93–110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Roy A. & Zhang Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 29, 2588–2595 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F. & Mazin A. V. A small molecule inhibitor of human RAD51 potentiates breast cancer cell killing by therapeutic agents in mouse xenografts. PLoS One 9, e100993 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F. & Mazin A. V. Targeting the homologous recombination pathway by small molecule modulators. Bioorg Med Chem Lett 24, 3006–3013 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagpulinsa D. A., Ayyadevara S. & Shmookler Reis R. J. A Small-Molecule Inhibitor of RAD51 Reduces Homologous Recombination and Sensitizes Multiple Myeloma Cells to Doxorubicin. Front Oncol 4, 289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpi A., Pasierbek P., Gartner A. & Loidl J. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112, 6–16 (2003). [DOI] [PubMed] [Google Scholar]
- Aouida M., Rubio-Texeira M., Thevelein J. M., Poulin R. & Ramotar D. Agp2, a member of the yeast amino acid permease family, positively regulates polyamine transport at the transcriptional level. PLoS One 8, e65717 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova Y., Thayumanavan P., Lonati E., Agrochao M. & Thevelein J. M. Transport and signaling through the phosphate-binding site of the yeast Pho84 phosphate transceptor. Proc Natl Acad Sci USA 107, 2890–2895 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schothorst J. et al. Yeast nutrient transceptors provide novel insight in the functionality of membrane transporters. Curr Genet 59, 197–206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaber R. F., Ottow K., Andersen H. A. & Kielland-Brandt M. C. Constitutive and hyperresponsive signaling by mutant forms of Saccharomyces cerevisiae amino acid sensor Ssy1. Eukaryot Cell 2, 922–929 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didion T., Regenberg B., Jorgensen M. U., Kielland-Brandt M. C. & Andersen H. A. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol Microbiol 27, 643–650 (1998). [DOI] [PubMed] [Google Scholar]
- Ljungdahl P. O. Amino-acid-induced signalling via the SPS-sensing pathway in yeast. Biochem Soc Trans 37, 242–247 (2009). [DOI] [PubMed] [Google Scholar]
- Bianchi L. & Diez-Sampedro A. A single amino acid change converts the sugar sensor SGLT3 into a sugar transporter. PLoS One 5, e10241 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y. et al. Evolutionary conservation predicts function of variants of the human organic cation transporter, OCT1. Proc Natl Acad Sci USA 100, 5902–5907 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. et al. Organic cation transporters are determinants of oxaliplatin cytotoxicity. Cancer Res 66, 8847–8857 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S. & Eliceiri K. W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloy R. A. et al. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle 13, 1400–1412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S. & Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313–321 (2003). [DOI] [PubMed] [Google Scholar]
- Yang J. S. et al. OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS One 6, e23525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. & Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins 57, 702–710 (2004). [DOI] [PubMed] [Google Scholar]
- Xu D. & Zhang Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J 101, 2525–2534 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colubri A. et al. Minimalist representations and the importance of nearest neighbor effects in protein folding simulations. J Mol Biol 363, 835–857 (2006). [DOI] [PubMed] [Google Scholar]
- Pei J. & Grishin N. V. PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods Mol Biol 1079, 263–271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Wang Y. & Zhang Y. ResQ: An Approach to Unified Estimation of B-Factor and Residue-Specific Error in Protein Structure Prediction. J Mol Biol (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomize M. A., Pogozheva I. D., Joo H., Mosberg H. I. & Lomize A. L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res 40, D370–D376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol 225, 487–494 (1992). [DOI] [PubMed] [Google Scholar]
- Omasits U., Ahrens C. H., Muller S. & Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886 (2014). [DOI] [PubMed] [Google Scholar]
- Kim S. et al. PubChem Substance and Compound databases. Nucleic Acids Res (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.