Significance
Large-scale initiatives toward personalized medicine are driving a massive expansion in the number of human genomes being sequenced. Therefore, there is an urgent need to define quality standards for clinical use. This includes deep coverage and sequencing accuracy of an individual’s genome. Our work represents the largest effort to date in sequencing human genomes at deep coverage with these new standards. This study identifies over 150 million human variants, a majority of them rare and unknown. Moreover, these data identify sites in the genome that are highly intolerant to variation—possibly essential for life or health. We conclude that high-coverage genome sequencing provides accurate detail on human variation for discovery and clinical applications.
Keywords: genomics, noncoding genome, human genetic diversity
Abstract
We report on the sequencing of 10,545 human genomes at 30×–40× coverage with an emphasis on quality metrics and novel variant and sequence discovery. We find that 84% of an individual human genome can be sequenced confidently. This high-confidence region includes 91.5% of exon sequence and 95.2% of known pathogenic variant positions. We present the distribution of over 150 million single-nucleotide variants in the coding and noncoding genome. Each newly sequenced genome contributes an average of 8,579 novel variants. In addition, each genome carries on average 0.7 Mb of sequence that is not found in the main build of the hg38 reference genome. The density of this catalog of variation allowed us to construct high-resolution profiles that define genomic sites that are highly intolerant of genetic variation. These results indicate that the data generated by deep genome sequencing is of the quality necessary for clinical use.
Recent technological advances have allowed for the large-scale sequencing of the whole human genome (1–7). Most studies generated population-based information on human diversity using low to intermediate coverage of the genome (4× to 20× sequencing depth). The highest coverage (30× or greater) was reported for the recent sequencing of 1,070 Japanese subjects (6), 129 trios from the 1000 Genomes Project (3), and 909 Icelandic subjects (4). High coverage, also described as deep coverage, may be needed for an adequate representation of the human genome. This shift in paradigm is only made stronger by the recent release of the Illumina HiSeq X Ten, which allows the sequencing of up to 160 genomes at 30× mean depth in 3-d cycles, at an average cost of $1,000–$2,000 per genome.
In an effort to evaluate the capabilities of whole human genome sequencing using short-read sequencing in full production mode, we first measured accuracy and generated quality standards by analysis of the reference material NA12878 from the CEPH Utah Reference Collection (8). We then assessed these quality standards across 10,545 human genomes sequenced to high depth and representative of the main human populations (SI Appendix, Fig. S1). This generated a reliable representation of human single-nucleotide variation (SNV) and the reporting of clinically relevant SNVs. We confronted, like other groups, the limitations of short-read sequencing for accurate calling of structural and copy-number variation; even with a variety of methods, resolving structural variation in a personal genome remains a challenge (9).
Results
Reproducibility of Sequencing on a Reference Genome.
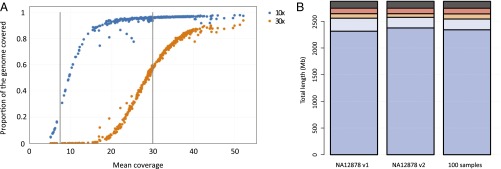
We assessed the extent of genome coverage using data from 325 technical replicates of NA12878 at different depths of read coverage. The canonical NA12878 Genome in a Bottle call set (GiaB v2.19; ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/NA12878_HG001/) defines a set of high-confidence regions that corresponds to ∼70% of the total genome. Regions of low complexity (e.g., centromeres, telomeres, and repetitive regions) and those challenging for sequencing, alignment, and variant-calling methods are excluded from the GiaB high-confidence region. At the target mean coverage of 30×, 95% of the high-confidence region of one NA12878 genome is covered at least at 10×. In contrast, at a target mean coverage of 7× used by several genome projects, only 23% of the high-confidence region of one NA12878 genome is sequenced at an effective 10× (Fig. 1A).
Fig. 1.
Effective genome coverage and sequence reproducibility. (A) Analysis of the relationship of mean coverage with effective genome coverage uses 100 NA12878 replicates with coverage <30×, 200 replicates with mean coverage 30× to 40×, and 25 replicates with coverage >40×. Vertical gray lines highlight mean target coverage of 7× and 30×. Each sequencing replicate is plotted at 10× (blue) and 30× (orange) effective minimal genome coverage. (B) Analysis of reproducibility uses NA12878 genomes at 30× to 40× mean coverage (two clustering chemistries, v1 and v2, each n = 100 replicas) to assess the consistency of base calling at each position in the whole genome. The analysis of reproducibility is then extended to 100 unrelated genomes (25 genomes per main ancestry group, African, European, and Asian, and for 25 admixed individuals). The color bars represent degree of consistency (blue, 100%; light blue, ≥90%; orange, ≥10 to <90%; red, <10%; black, failed).
We next assessed reproducibility on variant calling for the whole genome by restricting the analysis to a set of 200 samples of NA12878 that were sequenced at a mean coverage of 30× to 40×. Due to the manufacturer’s changes in clustering reagents, we analyzed 100 samples prepared with v1 and 100 with v2 (SI Appendix). After applying quality filters, passing genotype calls were compared for consistency (Fig. 1B). For v2 chemistry, 2.51 billion positions passed, and were called with 100% reproducibility in all replicates. An additional 210 Mb of genome positions yielded passing reproducible genotypes in more than 90%. Only 184 Mb of genome positions was sequenced with lower reproducibility (<90%). Similar results were obtained for v1 chemistry. The analysis of 100 unrelated genomes (25 individuals for each of the three main populations, African, Asian, and European, and 25 admixed individuals) confirmed the consistency of SNV calls across genomes (Fig. 1B). Overall, a total of 2,157 Mb (97.3%) of the GiaB high-confidence region could be sequenced with high reproducibility (SI Appendix, Table S1) with a low false discovery rate of 0.0008, precision of 0.999, and recall of 0.994. Details on the 2.7% of the GiaB high-confidence region that is not reliably sequenced are presented in SI Appendix. Overall, these analyses indicate that the current technology and sequencing conditions generate highly accurate sequence data and SNV calls over a large proportion of the genome.
The full extent of sequence generated for a single genome is greater than what is defined by the boundaries of GiaB. It should be noted that the various genome-sequencing initiatives use different reporting of what is sequenced (“accessible genome”), what is sequenced confidently, and whether these estimates are reported for an individual genome or for the collective analysis of multiple genomes. Our work specifically presents the genome calls for a single individual benchmarked against the complete sequence [total chromosomal length of autosomes and chromosome (Chr)X, 3,031 Mb] and against the community standard (GiaB; on autosomes + ChrX, 2,215 Mb) (SI Appendix, Table S2). For a single individual, we map the sequence on 90–95% of the genome—and 84% of a single genome is reported at high confidence (see below). In contrast, several published sequencing projects (2–5) describe genome coverage computed from the combination of all genomes—not for an individual genome. Using similar metrics as those in the current work for one 7× mean coverage 1000 Genomes Project sample (HG02541), we find that the loss of coverage genome-wide translates into severe loss of coverage of genes and variants (SI Appendix, Fig. S2). For example, the American College of Medical Genetics and Genomics recommends that laboratories performing clinical sequencing seek and report mutations of 56 genes (10). At 7× mean coverage, none of the exonic bases for those genes in HG02541 would be covered at 30×, 30% would be covered at 10×, and 84% would be covered at 5×. Therefore, low-coverage genomes are not suitable for clinical use because they can only generate confidence sequence for a fraction of the genome.
We also undertook the analysis of structural and copy-number variation using the set of 200 NA12878 replicas (SI Appendix). For short indels, the average precision and recall rates were 97.80% and 86.32%, respectively, but with unsatisfactory reproducibility (SI Appendix, Table S3). For structural variation larger than 50 bp and for copy-number variation, precision estimates were below 77%, recall was below 36%, and less than 53% of the calls could be highly reproduced (SI Appendix, Table S1). Overall, these results indicate that the identification of structural and copy-number variation using this short-read technology is unsatisfactory for clinical use if not supported by orthogonal technologies.
The Metrics of 10,000 Genomes.
The confidence regions established from sequencing of NA12878 and for 100 unrelated genomes served to guide the analysis of 10,545 human genomes. These samples cover various human populations, admixture, and ancestries (SI Appendix, Fig. S1). We first defined an extended confidence region (ECR) that includes the high-confidence GiaB regions and the highly reproducible regions extending beyond the boundaries of GiaB (SI Appendix, Fig. S3). The ECR encompasses 84% of the human genome, and includes 91.5% of the human exome sequence (GENCODE; 96 Mb), which is consistent with recent reports on coverage of the human exome in whole-genome analyses (11). We also examined the relevance for clinical variant calls: 28,831 of 30,288 (95.2%) unique ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and HGMD (www.hgmd.cf.ac.uk/ac/index.php) pathogenic variant positions are found in the ECR. We have now confirmed that 373 Mb (86%) of the additional 435 Mb of confident sequence in the ECR is also defined as high-confidence in the recently released GiaB v3.2.
For 10,545 genomes, the ECR included over 150 million SNVs at 146 million unique chromosomal positions. The mean SNV density in the ECR is 56.59 per 1 kb of sequence. However, there are differences across chromosomes: Chr1 is the least variable (55.12 SNVs per kb) and Chr16 the most variable (61.26 SNVs per kb) of the autosomal chromosomes. SNV density on ChrX is 35.60 SNVs per kb, but this estimate only considers female genomes (n = 6,320). A lower mutation rate of variation on the X chromosome than on autosomes is thought to reflect purifying selection of deleterious recessive mutations on hemizygous chromosomes (12). Diversity is further reduced by the effective population size of the X chromosome, because males only carry one copy (13). The SNV density on ChrY is 12.70 SNVs per kb, also consistent with previous research (14); however, only male genomes (n = 4,225) are considered here, and only 15% of the single Y chromosome is included in the ECR (SI Appendix, Fig. S4). The definition of ECR allowed for more high-confidence calls than those identified in GiaB (SI Appendix, Table S4). This is illustrated by the confident identification of 3,390 ClinVar and HGMD pathogenic variant sites identified in the 10,545 genomes: 2,628 (77.5%) were called in the GiaB region, whereas 3,191 (94.1%) could be called in the ECR (SI Appendix, Table S4).
Patterns of Genetic Variation in the Coding and Noncoding Genome.
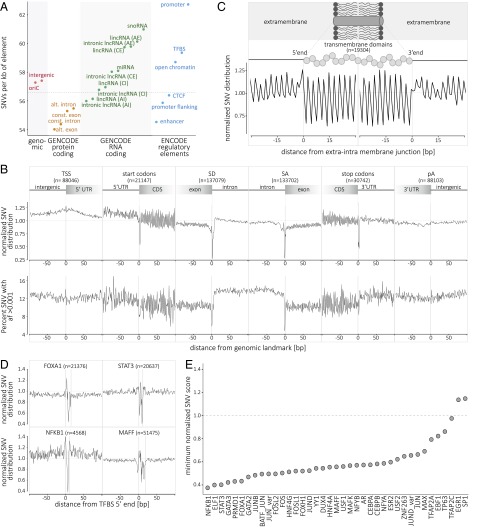
The volume of data presented here provides careful detail on the pattern of sequence conservation and SNVs across the human genome. We compared the rates of diversity in protein-coding, RNA-coding, and regulatory elements (Fig. 2A and SI Appendix, Fig. S5 and Table S5). On average, protein-coding elements are more conserved than intergenic regions and, as previously reported, alternative exons are the least variable (15). Alternative introns of long noncoding RNA (lncRNAs) are the most conserved, and small nucleolar RNA (snoRNA) is the most variable of RNA-coding elements. Among the analyzed DNA-regulatory elements, repressed chromatin is the most conserved and promoters are the least conserved (Fig. 2A). There is an extensive literature on the uneven distribution of SNV density across the genome. Positive selection, nucleotide composition, recombination hot spots, and replication timing are considered to be contributing factors (16–18). More recently, the sequence context has been shown to explain >81% of variability in substitution probabilities (19). These considerations notwithstanding, the pattern of SNV density is relatively stable across chromosomes (SI Appendix, Fig. S6). However, we identified three unique hypervariable megabase-long regions on autosomes (SI Appendix, Fig. S7). We observe the depletion of enhancer-associated histone marks (H3K4me1, H3K4me2, H3K4me3, H3K27me3, and H3K27ac) in these regions. The hypervariable regions are also gene-poor and depleted in chromatin loops, leading us to infer that these are domains that are not involved in long-distance interactions between regulatory elements and target genes. The enrichment of variation suggests there is limited purifying selection compared with other regions in the genome.
Fig. 2.
Single-nucleotide variant distribution and metaprofiles in the coding and noncoding genome. (A) Distribution of SNVs in selected genomic elements (genomic, protein-coding, RNA-coding, and regulatory elements; see SI Appendix for details). The genome average of 56.59 SNVs per kb is indicated by the horizontal dashed line. (B) The metaprofiles of protein-coding genes are created by aligning all elements of six different genomic landmarks (TSS, start codon, SD, SA, stop codon, and pA) for all 10,545 genomes. The y axis (Upper) describes the enrichment/depletion of SNV occurrence per position (count score; SI Appendix, Fig. S7), normalized to the mean of the protein-coding score (indicated by the horizontal dashed line); the y axis (Lower) describes the percent of SNVs at each position with an allelic frequency higher than 1 in 1,000 (frequency score; SI Appendix, Fig. S8). The x axis represents the distance from the genomic landmark. The vertical lines indicate the genomic landmark position. The SD and SA metaprofiles highlight the strong conservation of the splice sites (Upper) and the difference in SNV allele frequency between exons and introns (Lower). (C) The metaprofile of transmembrane domains is created by aligning all single domains at their 5′ and 3′ ends. The figure highlights that every amino acid in the transmembrane domain is conserved compared with the surrounding structure of the protein. (D) The metaprofiles of TFBSs are created by aligning all of the binding sites of four transcription factors (FOXA1, STAT3, NFKB1, and MAFF) for all 10,545 genomes. The x axis represents the distance from the 5′ end of the TFBS. The vertical lines indicate the 5′ and 3′ ends of the TFBS. (E) Ranking of 39 TFBSs by conservation (minimum score for the motif; i.e., the nucleotide with the lowest tolerance to variation). For C–E, the y axis describes the normalized enrichment/depletion of SNV occurrence per position, normalized to the mean of the protein-coding score (indicated by the horizontal dashed line). AE, alternative exon; AI, alternative intron; CE, constitutive exon; CI, constitutive intron; oriC, origin of replication; pA, polyadenylation site; SA, splice acceptor site; SD, splice donor site; TSS, transcription start site.
To explore the pattern of variation in the human genome in depth, we built “SNV metaprofiles” by collapsing all members of a family of genomic elements into a single alignment (SI Appendix, Fig. S8). Metaprofiles of protein-coding genes used GENCODE-annotated TSS, transcription start sites (TSSs) (n = 88,046), start codons (n = 21,147), splice donor and acceptor sites (n = 137,079 and 133,702, respectively), stop codons (n = 30,742), and polyadenylation sites (n = 88,103) (see SI Appendix for details). For each nucleotide aligned against these landmark positions, all of the genomes in this dataset (n = 10,545) were used to generate a precise representation of the pattern of conservation and allele spectra (Fig. 2B). A pattern is built by incorporating up to 1.4 billion data points (number of aligned elements × 10,545 samples) per genomic position. For example, the analysis captures the decrease in variant-allele frequency in exons, with the maximum drop occurring at the splice donor site (Fig. 2B). Positions that do not tolerate human variation can be interpreted as essential and possibly linked to embryonic lethality. In addition, the metaprofiles reveal discreet patterns, including with great precision the periodicity of conservation in coding regions due to the degeneracy of the third nucleotide in the codon in every exon window. The precision of the approach is also illustrated by the metaprofile of 19,304 transmembrane domains from 4,719 proteins. The constraint of maintaining alpha-helices (or other structures) and the hydrophobic (or polar) nature of the transmembrane domain result in all amino acids being distinctively conserved (Fig. 2C).
Many differences across individuals and species occur at the level of transcription-factor binding (20). We use the binding-site core motifs for metaprofile landmarking to identify signatures that include both variation-intolerant and hypertolerant positions at the binding site (Fig. 2D). Ranking of 39 transcription-factor binding sites (TFBSs) by the minimum score of the metaprofile (i.e., the nucleotide with the lowest tolerance to variation) emphasizes profound differences in the requirements for conservation across transcription factors (Fig. 2E). Although the identification of conserved, intolerant sites is expected, the biology behind unique hypertolerant positions at transcription binding sites remains to be investigated.
Metaprofile Tolerance Score and Variant Pathogenicity.
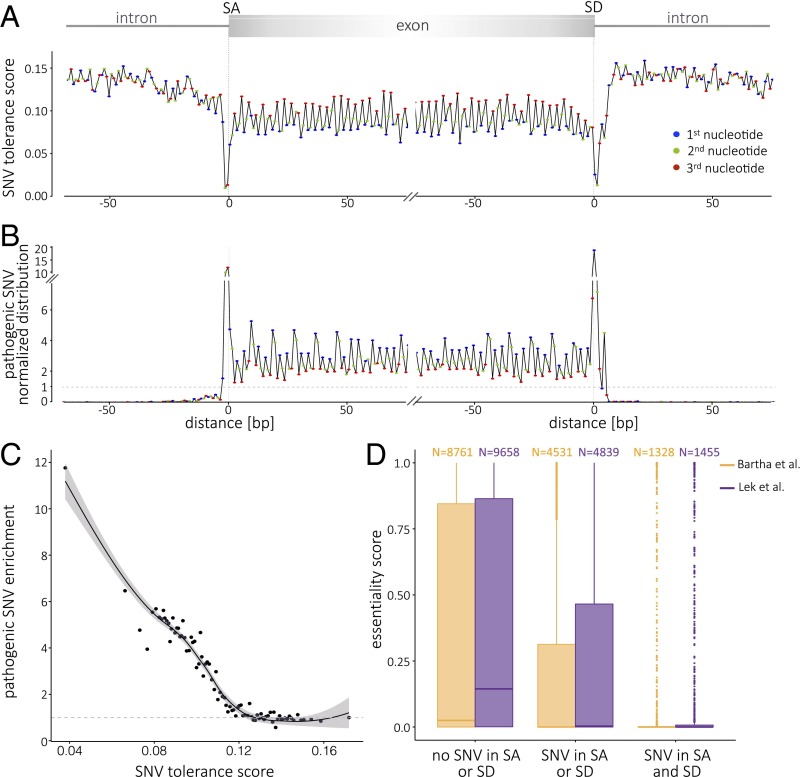
Rare human variants at intolerant sites may carry a greater fitness cost and associate with greater phenotypic consequences, and thus can be prioritized for clinical assessment. To apply metaprofiles for scoring of functional severity of variants, we established a tolerance score (Fig. 3A) that summarizes the rates and frequency of variation at a given position and for a given landmark. Using this approach, Fig. 3B illustrates the accumulation of pathogenic variant calls at sites with the lowest metaprofile tolerance scores. To formalize this analysis, we calculated the tolerance score at positions aligned to the main coding-region landmarks:10 positions upstream and downstream of the TSS, start codon, splice donor and acceptor, stop codon, and polyadenylation site. At the lowest tolerance score, we observe up to 12-fold enrichment for pathogenic variants (Fig. 3C). To understand the characteristics of genes that tolerate variants at privileged sites, we used an orthogonal assessment of gene essentiality (21, 22). The set of essential genes includes highly conserved genes that have fewer paralogs and are part of larger protein complexes. Essential genes also display a higher probability of CRISPR-Cas9 editing compromising cell viability (23), and knockouts in the mouse model are associated with increased mortality (24). Essential genes are endowed with a distinct coding metaprofile (SI Appendix, Fig. S9). Fig. 3D supports the concept that the less essential genes can tolerate variation at sites with low metaprofile tolerance scores.
Fig. 3.
Relationship of a metaprofile tolerance score with variant pathogenicity and gene essentiality. (A) Metaprofile of the transition between introns and exons expressed as the tolerance score (TS). The TS is the product of the normalized SNV distribution value by the proportion of SNVs with allele frequency ≥0.001 (Fig. 2B). The exon sequence highlights the conservation and tolerance to variation of the third position in codons (red). The pattern of higher tolerance to variation every third nucleotide is lost in introns. The TS is lowest at the splice donor and acceptor sites and highest in introns. (B) The distribution of ClinVar and HGMD pathogenic SNVs (n = 29,808 in SD; n = 30,369 in SA metaprofiles) reflects a significant enrichment of pathogenic variants at the sites of lowest TS. Consistently, the exon sequence highlights the enrichment for variation at the first position in codons (blue), as it results in amino acid change or truncation. (C) Relationship of tolerance score and enrichment for pathogenic variants. Represented on the x axis are the mean TS values for the coding region (±10 bp of intergenic or intronic boundaries); each dot represents the mean of 10 positions. The y axis represents the fold enrichment in pathogenic variants. local regression (LOESS) curve fitting is represented by the solid line; the shaded area indicates the 95% confidence interval. (D) Less essential genes tolerate variation at sites with lowest TS values. The x axis represents three different classes of genes according to their having evidence for splice acceptor/donor variation. The y axis represents essentiality scores of Bartha et al. (21) (yellow) and Exome Aggregation Consortium (ExAC) pLI (probability that a gene is intolerant to a loss of function mutation) (22) (purple). The large majority of genes that tolerate splice-site variants are not essential; in contrast, there is a marked shift to higher essentiality values for genes that are not observed to be variant at the splice sites.
An important feature of metaprofiling is that it predicts functional consequences of variation solely on the basis of human diversity. In contrast, the Combined Annotation-Dependent Depletion (CADD) score (25) uses evolutionary information, annotation from the Ensembl Variant Effect Predictor, and extensive information from University of California Santa Cruz (UCSC) Genome Browser tracks. Despite these profoundly different approaches, the tolerant scores obtained from metaprofiles in protein-coding regions perform similarly to CADD for the identification of functional variants (SI Appendix, Fig. S10). This observation underscores the potential of metaprofiling to analyze the genome with minimal preexisting knowledge—in particular in the noncoding genome, as metaprofile tolerance scores only rely on human variation.
Variant Discovery Rates per Individual.
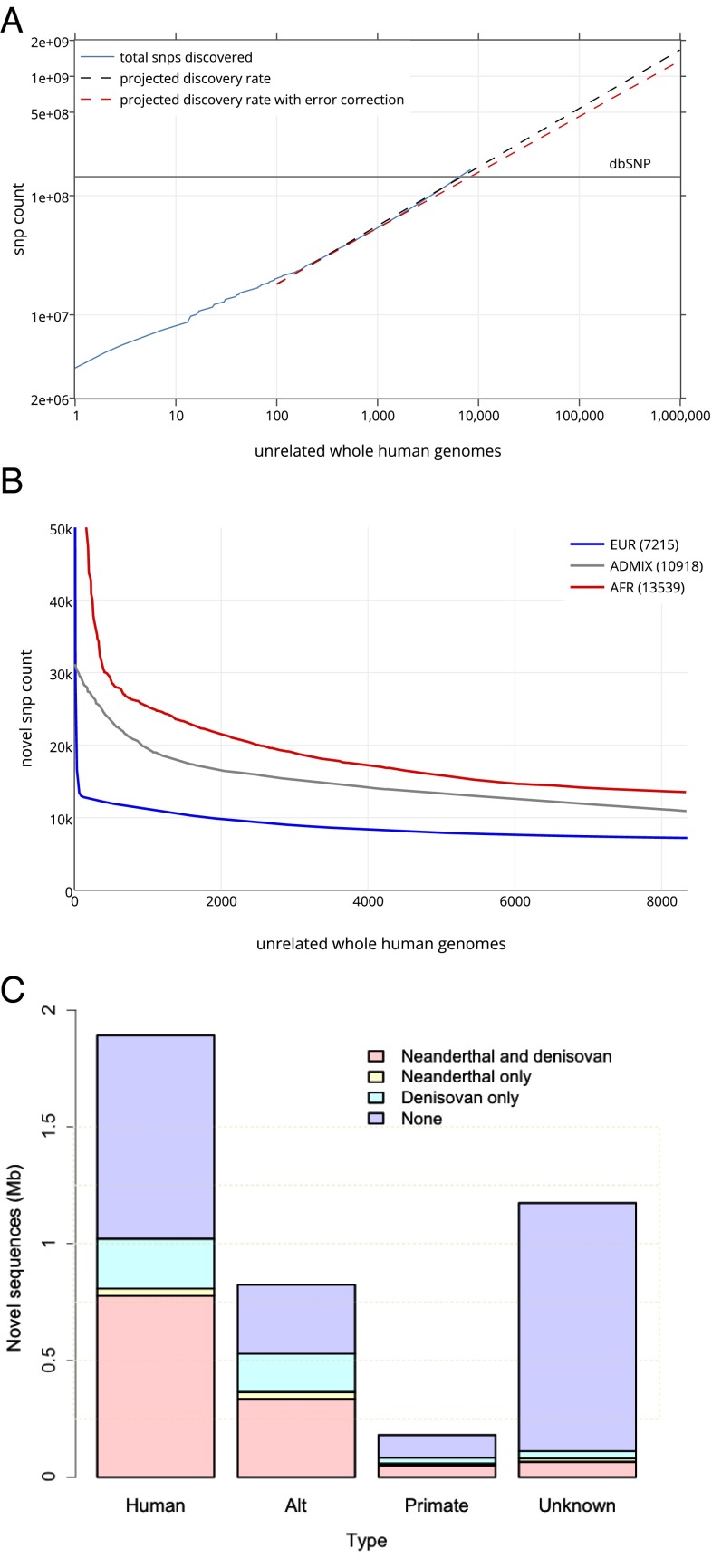
The large number of genomes, and the coverage of various human populations, served to describe the rate of newly observed, unshared SNVs for each additional sequenced genome. We restricted the analysis to the 8,096 unrelated individuals among the 10,545 genomes (SI Appendix). There is an expectation of 500 million variants identified after sequencing the genomes of 100,000 individuals (Fig. 4A). This analysis establishes at the whole-genome level prior estimates from the study of a limited set of genes or using exome analysis (22, 26).
Fig. 4.
Novel variants and genome sequences. (A) SNV discovery rate for 8,096 unrelated individual genomes contributing over 150 million SNVs (blue line). The projection for discovery rates as more genomes are sequenced is represented without (dashed black line) and with correction for the empirical false discovery rate of 0.0025 (dashed orange line). The number of SNVs in dbSNP is represented by the horizontal gray line. (B) The number of newly observed variants as more individuals are sequenced is determined by the ancestry background and number of participants in the study. Shown are the rates of identification of novel variants for each additional African genome (13,539 SNVs) and for each additional genome of admixed individuals (10,918 SNVs). The most numerous population in the study, Europeans, contributes the lowest number of novel variants (7,215 SNVs). (C) Unmapped sequence from the analysis of 8,096 unrelated individual genomes contributing over 3.2 Mb of nonreference genome. The 4,876 unique nonreference contigs had matches in the NCBI nt database as human, or nonhuman primate, and with hominins. There are contigs with human-like features that do not have a known match in databases.
Unrelated individuals were assigned to five superpopulations or to an admixed or “other” population group on the basis of genetic ancestry (SI Appendix, Fig. S1). Each subsequently sequenced genome contributes on average 8,579 novel variants, which varied from 7,215 in Europeans and 10,918 in admixed to 13,539 in individuals of African ancestry (Fig. 4B). This reflects the current understanding of Africa as the most genetically diverse region in the world (5). Of the 150 million SNVs observed in the ECR, 82 million (54.7%) have not been reported in dbSNP of the National Center for Biotechnology Information (NCBI) or in the most recent phase 3 of the 1000 Genomes Project (3). The proportion of novel variants increases with decreasing allele frequency—as expected, there is a negligible number of “novel” variants with allele frequencies greater than 1% (SI Appendix, Fig. S11).
Unmapped Human Genome Sequences.
In addition to new variants, we identified 4,876 unique human, or human-like, contigs (SI Appendix) assembled from 3.26 Mb of nonreference (hg38 build) sequences (“unmapped reads”). On average, we identified 0.71 Mb of nonreference sequences per genome. A total of 1.89 Mb of the nonredundant sequences could be mapped to known human sequences in GenBank (although not in the hg38 reference assembly). An additional 0.18 Mb mapped to primate sequences in the NCBI nucleotide (nt) database. There is 1.17 Mb that did not have a known match in the nt or nonredundant sequence (nr) databases. The GC content and dinucleotide bias of the unknown contigs reflect the patterns of human sequences. However, we also identified successfully mapped eukaryotic, prokaryotic, and viral contigs that had indistinguishable metrics from human contigs (SI Appendix, Fig. S12). Therefore, it remains difficult to solve bioinformatically the nature of unmapped human-like reads—they may simply result from contamination (27). Much of the nonreference sequence is shared with hominins. The unmapped contigs were compared with Neanderthal and Denisovan sequencing reads that did not map to hg38. There were 0.96 Mb covered by Neanderthal reads and 1.18 Mb covered by Denisovan reads. In addition, 0.82 Mb is not in the hg38 primary assembly but in the “alt” sequences or subsequent patches (Fig. 4C). The presence in some individuals of novel sequence content that is also found among unmapped reads from Denisovan and Neanderthal genomes and in nonhuman primates reinforces the notion that the human genome is larger and more distributed than what is currently represented by a single (hg38) reference genome.
Conclusions
The goal of clinical use of the genome requires standards for sequencing, analysis, and interpretation. Our work specifically addresses the first two steps: sequencing and sequence analysis. The performance of the platform, implemented in full production mode, improves on recent benchmarks for the accurate interpretation of next-generation DNA sequencing in the clinical setting (22, 28, 29). This is needed for laboratory standards, regulatory purposes, and clinical diagnostics and research. The third step—interpretation—remains a major issue given the many types of genetic evidence that laboratories consider. Initiatives such as ClinVar and policies and guidelines (10, 30) set standards for clinical interpretation.
This report also extends prior efforts at genome and exome sequencing by detailing the distribution of human variation in the noncoding genome. The amount of data supports the discovery of sites in the genome that are intolerant to variation. The 10,545 genomes provide estimates of the rate of discovery of new SNVs, and complements the human genome by more than 3 Mb through the identification of nonreference and putative human-like sequences. These data anticipate the relentless accumulation of rare variants and the scale of observable mutagenesis of the human genome.
Materials and Methods
Detailed information is provided in SI Appendix, Materials and Methods. All research involving human subjects was performed, and informed consent was obtained, under protocols approved by the Western Institutional Review Board. Participants were representative of major human populations and ancestries. The study population was not ascertained for a specific health status. Institutional review board-approved consent forms for participation in research and collection of biological specimens and other data used in this publication were confirmed to be appropriate for use. All samples were sequenced on the Illumina HiSeq X sequencer using a 150-base paired-end single-index read format. Reads were mapped to human reference hg38 using ISIS analysis software. Bam files were characterized using Picard and input to the ISIS Isaac Variant Caller to generate genomic variant call format (VCF) files. Admixture analysis used ADMIXTURE. Kinship analysis used KING. Sample contamination was assessed with verifyBamID. Structural variation analysis used MANTA; copy-number variation analysis used CANVAS. Annotation was based on the ClinVar and HGMD databases. SnpEff was used for genomic annotation and predicting effects of SNVs. Exomic regions for protein-coding genes were extracted from GENCODE. Identification and assembly of nonreference sequences used SOAPdenovo2, BLASTN, and DIAMOND. Web addresses and references for the software described above are found in SI Appendix. Neanderthal and Denisovan sequence data were downloaded from cdna.eva.mpg.de.
Supplementary Material
Acknowledgments
We thank Drs. T. Caskey and M. Hicks for useful commentaries, and Drs. C. Maher and M. Schultz for contributions to earlier work.
Footnotes
The authors are employees of Human Longevity, Inc.
Data deposition: Data access is granted through the Human Longevity, Inc. gene browser (HLI-OpenSearch.com). In addition, 325 NA12878 reference genome sequences have been donated to PrecisionFDA (https://precision.fda.gov).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613365113/-/DCSupplemental.
References
- 1.Walter K, et al. UK10K Consortium The UK10K project identifies rare variants in health and disease. Nature. 2015;526(7571):82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genome of the Netherlands Consortium Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet. 2014;46(8):818–825. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 3.Auton A, et al. 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gudbjartsson DF, et al. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet. 2015;47(5):435–444. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- 5.Gurdasani D, et al. The African Genome Variation Project shapes medical genetics in Africa. Nature. 2015;517(7534):327–332. doi: 10.1038/nature13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagasaki M, et al. ToMMo Japanese Reference Panel Project Rare variant discovery by deep whole-genome sequencing of 1,070 Japanese individuals. Nat Commun. 2015;6:8018. doi: 10.1038/ncomms9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sidore C, et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat Genet. 2015;47(11):1272–1281. doi: 10.1038/ng.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zook JM, et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci Data. 2016;3:160025. doi: 10.1038/sdata.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.English AC, et al. Assessing structural variation in a personal genome—Towards a human reference diploid genome. BMC Genomics. 2015;16:286. doi: 10.1186/s12864-015-1479-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards S, et al. ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belkadi A, et al. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci USA. 2015;112(17):5473–5478. doi: 10.1073/pnas.1418631112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McVean GT, Hurst LD. Evidence for a selectively favourable reduction in the mutation rate of the X chromosome. Nature. 1997;386(6623):388–392. doi: 10.1038/386388a0. [DOI] [PubMed] [Google Scholar]
- 13.Schaffner SF. The X chromosome in population genetics. Nat Rev Genet. 2004;5(1):43–51. doi: 10.1038/nrg1247. [DOI] [PubMed] [Google Scholar]
- 14.Wilson Sayres MA, Lohmueller KE, Nielsen R. Natural selection reduced diversity on human Y chromosomes. PLoS Genet. 2014;10(1):e1004064. doi: 10.1371/journal.pgen.1004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: Diversification, exon definition and function. Nat Rev Genet. 2010;11(5):345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 16.Hellmann I, et al. Why do human diversity levels vary at a megabase scale? Genome Res. 2005;15(9):1222–1231. doi: 10.1101/gr.3461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaibley VM, et al. The influence of genomic context on mutation patterns in the human genome inferred from rare variants. Genome Res. 2013;23(12):1974–1984. doi: 10.1101/gr.154971.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francioli LC, et al. Genome of the Netherlands Consortium Genome-wide patterns and properties of de novo mutations in humans. Nat Genet. 2015;47(7):822–826. doi: 10.1038/ng.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aggarwala V, Voight BF. An expanded sequence context model broadly explains variability in polymorphism levels across the human genome. Nat Genet. 2016;48(4):349–355. doi: 10.1038/ng.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasowski M, et al. Variation in transcription factor binding among humans. Science. 2010;328(5975):232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartha I, et al. The characteristics of heterozygous protein truncating variants in the human genome. PLoS Comput Biol. 2015;11(12):e1004647. doi: 10.1371/journal.pcbi.1004647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. Mouse Genome Database Group The Mouse Genome Database (MGD): Facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 2015;43(Database issue):D726–D736. doi: 10.1093/nar/gku967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson MR, et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337(6090):100–104. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lusk RW. Diverse and widespread contamination evident in the unmapped depths of high throughput sequencing data. PLoS One. 2014;9(10):e110808. doi: 10.1371/journal.pone.0110808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linderman MD, et al. Analytical validation of whole exome and whole genome sequencing for clinical applications. BMC Med Genomics. 2014;7:20. doi: 10.1186/1755-8794-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldfeder RL, et al. Medical implications of technical accuracy in genome sequencing. Genome Med. 2016;8:24. doi: 10.1186/s13073-016-0269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amendola LM, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet. 2016;98(6):1067–1076. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.