Significance
Alzheimer’s disease (AD) affects 10% of the elderly population. The disease remains poorly understood with no cure. The main symptom is memory loss, but other symptoms might include impaired executive function (ability to plan and accomplish goals; e.g., grocery shopping). The severity of behavioral symptoms and brain atrophy (gray matter loss) can vary widely across patients. This variability complicates diagnosis, treatment, and prevention. A mathematical model reveals distinct brain atrophy patterns, explaining variation in gray matter loss among AD dementia patients. The atrophy patterns can also explain variation in memory and executive function decline among dementia patients and at-risk nondemented participants. This model can potentially be applied to understand brain disorders with varying symptoms, including autism and schizophrenia.
Keywords: mental disorder subtypes, Alzheimer’s disease subtypes, Alzheimer’s disease heterogeneity, voxel-based morphometry, unsupervised machine learning
Abstract
We used a data-driven Bayesian model to automatically identify distinct latent factors of overlapping atrophy patterns from voxelwise structural MRIs of late-onset Alzheimer’s disease (AD) dementia patients. Our approach estimated the extent to which multiple distinct atrophy patterns were expressed within each participant rather than assuming that each participant expressed a single atrophy factor. The model revealed a temporal atrophy factor (medial temporal cortex, hippocampus, and amygdala), a subcortical atrophy factor (striatum, thalamus, and cerebellum), and a cortical atrophy factor (frontal, parietal, lateral temporal, and lateral occipital cortices). To explore the influence of each factor in early AD, atrophy factor compositions were inferred in beta-amyloid–positive (Aβ+) mild cognitively impaired (MCI) and cognitively normal (CN) participants. All three factors were associated with memory decline across the entire clinical spectrum, whereas the cortical factor was associated with executive function decline in Aβ+ MCI participants and AD dementia patients. Direct comparison between factors revealed that the temporal factor showed the strongest association with memory, whereas the cortical factor showed the strongest association with executive function. The subcortical factor was associated with the slowest decline for both memory and executive function compared with temporal and cortical factors. These results suggest that distinct patterns of atrophy influence decline across different cognitive domains. Quantification of this heterogeneity may enable the computation of individual-level predictions relevant for disease monitoring and customized therapies. Factor compositions of participants and code used in this article are publicly available for future research.
Alzheimer’s disease (AD) dementia is a devastating neurodegenerative disease that affects 11% of individuals over age 65 with no disease-modifying treatment available. Accurate in vivo biomarkers are urgently needed to assist in early detection of at-risk individuals, improve diagnosis, monitor disease progression, and serve as outcome measures in clinical trials.
Although AD is typically associated with an amnestic clinical presentation and disruption of the medial temporal lobe (1), it has become increasingly clear that heterogeneity exists within this disease. Specifically, heterogeneity has been observed in the clinical presentation of AD (2) and the spatial distribution of neurofibrillary tangles (NFTs) (3, 4) as well as the presence of comorbid pathologies, such as vascular disease, Lewy bodies, and transactive response DNA binding protein 43 kDa (TDP-43) (5, 6). Interestingly, the spatial distribution of atrophy varies across AD subtypes defined on the basis of NFT distribution (7), suggesting that analyses of gray matter (GM) patterns are useful to characterize heterogeneity in AD. Furthermore, although distinct atrophy patterns have been observed in patients who clearly show atypical clinical presentations (8), heterogeneity in GM atrophy has also been reported among late-onset AD cases (9). It is, therefore, likely that the ability to quantify varying patterns of atrophy among AD patients will help inform our understanding of fundamental disease processes.
In this study, we sought to explore the heterogeneity of atrophy patterns in late-onset AD using a data-driven Bayesian framework that accounted for and estimated latent AD atrophy factors derived from structural MRI data. The mathematical framework that we used, latent Dirichlet allocation (LDA) (10), has been successfully used to extract overlapping brain networks from functional MRI (11) and metaanalytic data (12, 13). Importantly, this approach does not require the atrophy pattern of an individual to be determined by a single atrophy factor. Instead, the model allows the possibility that multiple latent factors are expressed to varying degrees within an individual. For example, the atrophy pattern of a patient might be 90% owing to factor 1 and 10% owing to factor 2, whereas the atrophy pattern of another patient might be 60% owing to factor 1 and 40% owing to factor 2. Given that multiple contributors that are not mutually exclusive may influence heterogeneity in AD, such as the spatial location of NFT pathology (3, 4), coexisting non-AD pathologies (5, 14), and genetics (9), we believe that it is more biologically plausible that individuals express varying degrees of distinct atrophy factors rather than one single factor. Thus, the LDA approach is particularly well-suited for these analyses and will provide insight into whether expressing multiple atrophy factors is common among late-onset AD patients.
Most studies investigating the heterogeneity of AD have examined patients soon after AD onset or at advanced AD stages (3, 7, 15–17). However, the pathophysiological processes of AD begin at least a decade before clinical diagnosis (18), suggesting that the emergence of this heterogeneity may occur before the onset of clinical dementia. In this study, we, therefore, examined how distinct atrophy factors identified in AD dementia patients were associated with longitudinal cognitive decline early in nondemented participants who were at risk for AD dementia based on elevated beta-amyloid (Aβ) (19–21).
Our study makes three significant contributions. First, we introduced an innovative modeling strategy where expressions of multiple atrophy patterns are estimated rather than assigning each participant to a single subtype. Second, our approach harnesses the rich multidimensional information across all GM voxels, avoiding the need for a priori selection of regions and enabling an in-depth exploration of atrophy patterns. Third, application of this approach to participants spanning the clinical spectrum revealed that latent atrophy factors are associated with distinct memory and executive function trajectories, providing insights into the impact of disease heterogeneity throughout the prolonged course of AD.
Results
Overall Approach.
Our approach involved three main steps. In step I, we performed LDA (a Bayesian model) (10) to estimate latent atrophy factors in 188 AD dementia patients and used this model to extract factor compositions in two independent samples of nondemented participants: 147 Aβ+ mild cognitively impaired (MCI) and 43 Aβ+ cognitively normal (CN) participants. In step II, we examined robustness across different analytic approaches and investigated characteristics of the factor compositions across participants. Third, in step III, we examined the associations between atrophy factors and different cognitive domains (memory and executive function). The results of each step are described in detail below.
Step I. Discovering Latent Atrophy Factors in AD Dementia Patients.
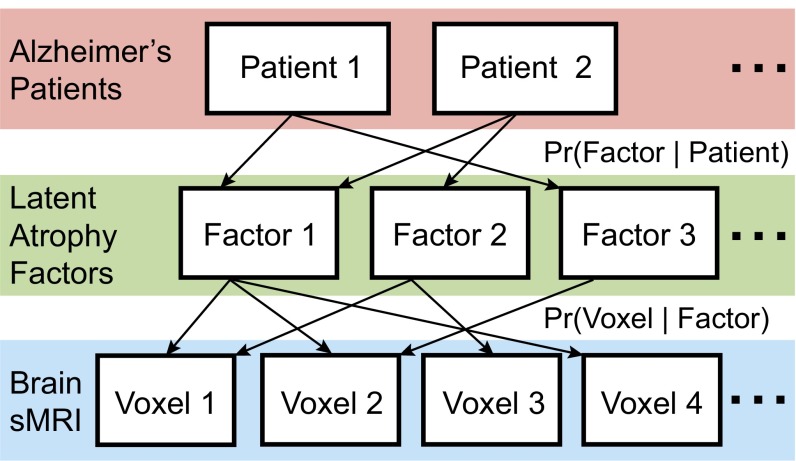
We used the Bayesian LDA model (10) to encode our assumption that a patient expresses one or more latent atrophy factors (Fig. 1). The LDA model was applied to the structural MRI of 188 AD dementia patients. Given the voxelwise GM density values derived from structural MRI (FSL-VBM) (22) and a predefined number of factors K, the model is able to estimate the probability that a particular factor is associated with atrophy at a specific spatial location [i.e., Pr(Voxel | Factor) or probabilistic atrophy map of the factor] and the probability that an individual expresses each atrophy factor [i.e., Pr(Factor | Patient) or atrophy factor composition of the individual]. Importantly, resulting atrophy factors were not predetermined but estimated from data (Materials and Methods).
Fig. 1.
A Bayesian model of AD dementia patients, latent atrophy factors, and brain structural MRI (sMRI). Underpinning our approach is the premise that each participant expresses one or more latent factors. Each factor is associated with distinct but possibly overlapping patterns of brain atrophy. The framework can be instantiated with a mathematical model (LDA) (10), with parameters that can be estimated from the structural MRI data of AD dementia patients. The estimated parameters are the probability that a patient expresses a particular factor [i.e., Pr(Factor | Patient)] and the probability that a factor is associated with atrophy at an MRI voxel [i.e., Pr(Voxel | Factor)].
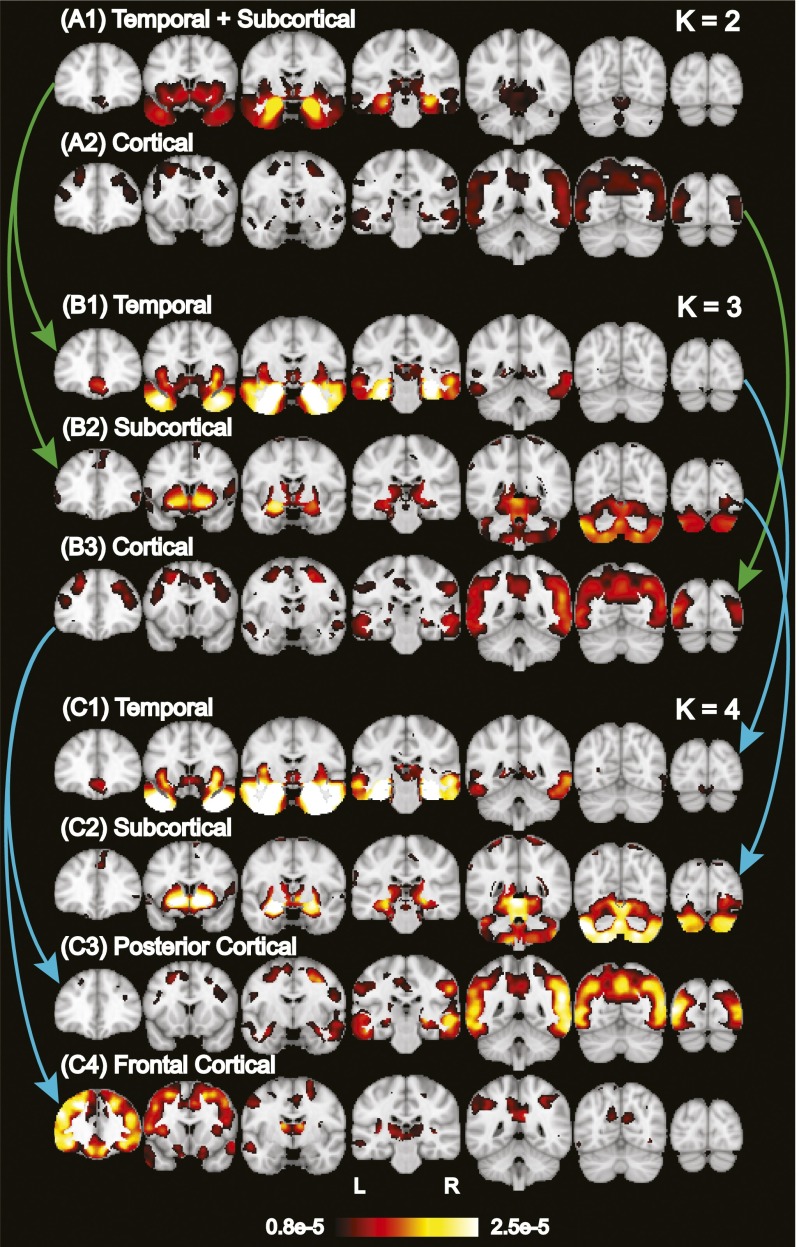
An important model parameter is the number of latent atrophy factors K. Therefore, we first determined how factor estimation changed from K = 2 to 10. Visual inspection of the spatial distribution of each atrophy factor suggested that factor estimates from K = 2 to 10 were organized in a hierarchical fashion (Fig. 2 and SI Appendix, Fig. S1). For instance, the two-factor model revealed one factor associated with atrophy in temporal and subcortical regions (“temporal+subcortical”) (Fig. 2A1) and another factor associated with atrophy throughout cortex (“cortical”) (Fig. 2A2). The three-factor model resulted in a similar cortical factor (Fig. 2B3 and SI Appendix, Table S1C), whereas the temporal+subcortical factor split into a “temporal” factor associated with extensive atrophy in the medial temporal lobe (Fig. 2B1 and SI Appendix, Table S1A) and a “subcortical” factor associated with atrophy in the cerebellum, striatum, and thalamus (Fig. 2B2 and SI Appendix, Table S1B). Likewise, the four-factor model resulted in the cortical factor splitting into “frontal cortical” and “posterior cortical” factors, whereas the temporal and subcortical factors remained the same (Fig. 2C). Sagittal and axial slices of these probabilistic atrophy maps are available in SI Appendix, Fig. S1.
Fig. 2.
Hierarchy of latent atrophy factors with distinct atrophy patterns in AD. Bright color indicates higher probability of atrophy at that spatial location for a particular atrophy factor [i.e., Pr(Voxel | Factor)]. Each of the (A) two, (B) three, and (C) four factors was associated with a distinct pattern of brain atrophy and named accordingly. A nested hierarchy of atrophy factors was observed, although the model did not mandate such a hierarchy. For example, when going from two to three factors, the temporal+subcortical factor (A1) split into temporal (B1) and subcortical (B2) factors, whereas the cortical factor remained the same (A2 and B3). From three to four factors, the temporal and subcortical factors remained the same (B1 and C1; B2 and C2), whereas the cortical factor (B3) split into posterior cortical (C3) and frontal cortical (C4) factors. This hierarchical phenomenon was quantified for 2–10 factors (SI Appendix, Fig. S2).
To quantify the hierarchical phenomenon, we used an exhaustive search to assess the possibility that two unknown factors in the (K + 1)-factor model were subdivisions of an unknown factor in the K-factor model (whereas the other factors remained the same). The exhaustive search yielded a hypothesized factor hierarchy with associated correlation values quantifying the subdivision quality (SI Appendix, SI Methods). The high correlation values (SI Appendix, Fig. S2) confirmed that additional factors emerged as subdivisions of lower-order factors, corresponding to a nested hierarchy of atrophy factors.
This nested hierarchy suggested that specification of different numbers of estimated factors might yield distinct insights into AD. In the remainder of this paper, we highlighted the results of three-factor model (Fig. 2B), because the emergence of the temporal and cortical factors were consistent with the “limbic-predominant” and “hippocampal-sparing” pathologically defined AD subtypes previously reported (3, 7). We additionally repeated analyses for two- and four-factor models, which yielded behavioral insights consistent with the three-factor model. These additional results are reported in SI Appendix, Figs. S6 and S8.
To explore the influence of atrophy factors in early AD, probabilistic atrophy maps Pr(Voxel | Factor) estimated from the AD dementia patients were used to infer factor compositions Pr(Factor | Participant) of 190 Aβ+ nondemented participants using the standard variational expectation–maximization (VEM) algorithm (10).
Step II. Examining Factor Robustness and Characteristics of Factor Compositions.
Among the 188 AD dementia patients, 100 had their cerebrospinal fluid (CSF) amyloid data available; 91 of 100 patients were Aβ+ (CSF amyloid concentration <192 pg/mL) (23). We performed LDA on the subset of Aβ+ AD dementia patients (and Aβ+ MCI participants) (SI Appendix, SI Results) and compared atrophy patterns of the resulting factors with those derived using the larger sample (SI Appendix, Fig. S3). Atrophy factors were similar across these methods, with an average correlation across all pairwise comparisons of r = 0.89. Given this similarity and to improve our estimates of the atrophy factors, we elected to use the atrophy factors derived from the larger sample of 188 AD dementia patients for subsequent analyses. Furthermore, resulting atrophy patterns were consistent between FreeSurfer (24) and FSL-VBM, suggesting that the atrophy factors were robust to variations in image preprocessing software (SI Appendix, SI Results and SI Methods).
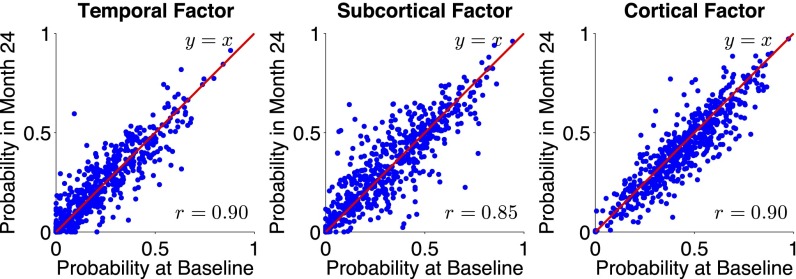
To determine whether expression of atrophy factors remained stable over time, we examined the subset of the Alzheimer's Disease Neuroimaging Initiative 1 (ADNI 1) participants who had a two-year follow-up scan available (n = 560 of 810). We were specifically interested in whether atrophy factors reflected different disease stages rather than different atrophy subtypes (for instance, high expression of the temporal factor may lessen over time with greater expression of the cortical factor). Therefore, we compared factor compositions after two years with baseline compositions. The factor probabilities were positivity correlated and highly consistent (r > 0.85 across all three factors; Fig. 3) (SI Appendix, Fig. S4 shows results by diagnostic group with additional amyloid information), suggesting that these factors do not merely reflect a sequence of atrophy patterns.
Fig. 3.
Stability of factor compositions over two years. Each blue dot represents a participant. For each plot, x and y axes represent the probabilities of factor at baseline and two years after baseline, respectively. In the ideal case, where factor probability estimates remain exactly the same after two years, one would expect a y = x linear fit as well as an r = 1 correlation. In our case, the linear fits were close to y = x with r > 0.85 for all three atrophy factors, suggesting that the factor compositions were stable, despite disease progression.
Examination of atrophy factor compositions among AD dementia patients revealed that the majority expressed multiple latent atrophy factors rather than predominantly expressing a single atrophy factor (Fig. 4). Examination of factor compositions of 190 Aβ+ nondemented participants revealed a similar pattern, such that the majority of participants expressed multiple atrophy factors (SI Appendix, Fig. S5A). Factor compositions for the two- and four-factor models also suggest that most participants expressed multiple atrophy factors (SI Appendix, Fig. S5 B and C).
Fig. 4.
Factor compositions of 188 AD dementia patients. Each patient corresponds to a dot, with location (in barycentric coordinates) that represents the factor composition. Color indicates amyloid status: red for Aβ+, green for Aβ−, and blue for unknown. Corners of the triangle represent pure factors; closer distance to the respective corner indicates higher probability for the respective factor. Most dots are far from the corners, suggesting that most patients expressed multiple factors.
To understand the association between atrophy factors and demographic variables, general linear model (GLM; for continuous variables) and logistic regression (for binary variables) were conducted in 188 AD dementia patients (SI Appendix, Table S2). Briefly, the response variable was the variable of interest (e.g., age at AD onset), and the explanatory variables consisted of two columns encoding participants’ loading on the cortical and subcortical factors. The temporal factor was implicitly modeled, because the factor probabilities summed to one (Materials and Methods).
There were no significant differences in years from AD onset, education, sex, or apolipoprotein E (APOE) ε4 loadings across the three factors. Importantly, amyloid level was not significantly different across factors. The cortical factor was associated with significantly younger baseline age than the temporal factor (P = 1e−5) and subcortical factor (P = 2e−6) as well as younger age at AD onset than the temporal factor (P = 3e−4) and subcortical factor (P = 7e−6). In addition, the subcortical factor was associated with higher APOE ε2 loading than the temporal factor (P = 0.01) and cortical factor (P = 0.04), but these associations were not significant when corrected for multiple comparisons.
Similar analyses were conducted for the Aβ+ MCI and CN groups. The only significant association was that, among Aβ+ MCI participants, the cortical factor was associated with younger age at baseline compared with the temporal factor (P = 0.05) and subcortical factor (P = 0.02). However, this association did not survive after correcting for multiple comparisons.
Step III. Examining Associations Between Atrophy Factors and Cognition.
We first examined diagnostic group differences in memory (ADNI-Mem) (25) and executive function (ADNI-EF) (26) without considering factor compositions. As expected, cross-sectional memory was worse for AD dementia patients (mean = −0.84) compared with Aβ+ MCI participants (mean = −0.21; t test P = 5e−23). Aβ+ MCI participants had worse memory than Aβ+ CN participants (mean = 0.93; t test P = 2e−26). Likewise, cross-sectional executive function was worse for AD dementia patients (mean = −0.92) compared with Aβ+ MCI participants (mean = −0.17; t test P = 3e−16). Aβ+ MCI participants had worse executive function than Aβ+ CN participants (mean = 0.50; t test P = 1e−7).
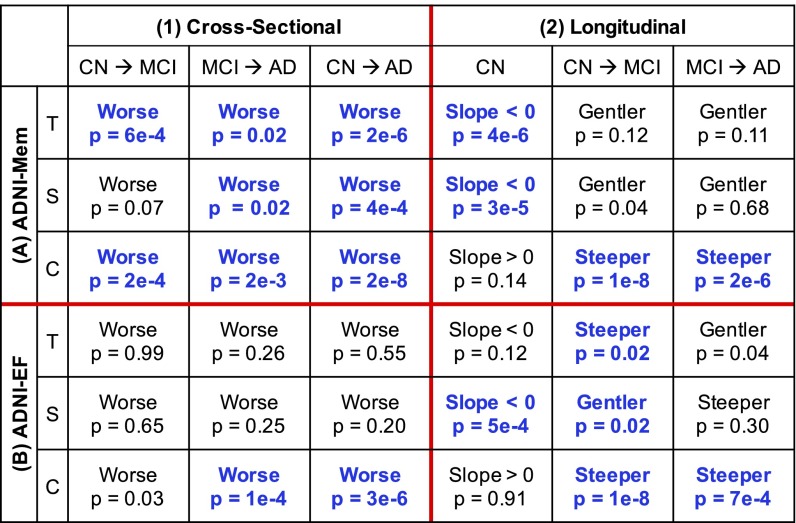
We then examined a GLM predicting cross-sectional memory and executive function, which included both diagnosis and factor compositions as well as their interactions as predictors (Materials and Methods shows model details; Fig. 5 A1 and B1). This analysis revealed that all factors were associated with baseline memory, and these associations continued to worsen across the disease spectrum (Fig. 5A1). For cross-sectional executive function, there was only an association with the cortical factor, and this association also worsened across the disease spectrum (Fig. 5B1).
Fig. 5.
Differences by diagnosis and atrophy factor in (1) cross-sectional baseline and (2) longitudinal decline rates of (A) memory and (B) executive function. Comparisons remaining significant after FDR (q = 0.05) control are highlighted in blue. T, S, and C indicate temporal, subcortical, and cortical factors, respectively. For example, the top left cell of A, 1 suggests that Aβ+ MCI participants with high loading on the temporal factor had worse baseline memory than Aβ+ CN participants with high loading on the same factor (P = 6e−4). However, the bottom left cell of B, 2 suggests that Aβ+ CN participants expressing the cortical factor did not exhibit executive function decline (P = 0.91), whereas the bottom right cell of B, 2 suggests that AD dementia patients expressing the cortical factor showed faster executive function decline than Aβ+ MCI participants expressing the same factor (P = 7e−4).
Next, we examined a linear mixed effects (LME) model predicting longitudinal change in memory and executive function (Fig. 5 A2 and B2). The LME model provides significantly improved exploitation of longitudinal measurements (27) by accounting for both intraindividual measurement correlations and interindividual variability. The model setup was the same as the GLM above, except that time and its interactions with diagnosis and factor compositions were included as predictors (SI Appendix, SI Methods).
This analysis revealed that the temporal and subcortical factors exhibited memory decline that began in CN and maintained similar memory decline rates in MCI and AD (Fig. 5A2). In contrast, the cortical factor was not associated with memory decline in CN but showed faster decline in MCI compared with CN and AD compared with MCI (Fig. 5A2). The cortical factor was not associated with executive function decline in CN but showed faster longitudinal executive function decline in MCI compared with CN and AD compared with MCI (Fig. 5B2).
In our final set of analyses examining cognition, we directly compared the three factors. The GLM and LME model were exactly the same as the previous sections, but we instead focused on the contrasts between factors.
For cross-sectional memory, the temporal factor was associated with worse performance than the subcortical (P = 3e−6) and cortical (P = 7e−3) factors among AD dementia patients (Fig. 6A). Similar results were found for Aβ+ MCI participants (SI Appendix, Fig. S7A1). Among Aβ+ CN participants, there was no memory difference across the atrophy factors (SI Appendix, Fig. S7A1). For cross-sectional executive function, the cortical factor was associated with worse performance than the temporal (P = 0.01) and subcortical (P = 1e−5) factors among AD dementia patients (Fig. 6B). There was no executive function difference across the factors among Aβ+ CN and MCI participants (SI Appendix, Fig. S7B1).
Fig. 6.
Comparisons of baseline (A) memory and (B) executive function in AD dementia patients across factors. Comparisons remaining significant after FDR (q = 0.05) control are highlighted in blue. T, S, and C indicate temporal, subcortical, and cortical factors, respectively. Blue dots are estimated differences between “pure atrophy factors,” and red bars show the SEs (Materials and Methods). For example, the top row in A suggests that AD dementia patients expressing the temporal factor had worse baseline memory than AD dementia patients expressing the subcortical factor (P = 3e−6).
For longitudinal change in memory (Fig. 7A), the cortical factor was associated with faster longitudinal memory decline than the temporal (P = 1e−4) and subcortical (P = 4e−6) factors among AD dementia patients. Among Aβ+ MCI participants, the subcortical factor was associated with slower decline rate than the cortical (P = 8e−4) and temporal (P = 4e−3) factors. Finally, among Aβ+ CN participants, the cortical factor showed slower memory decline than the temporal (P = 1e−4) and subcortical (P = 3e−3) factors.
Fig. 7.
Comparisons of (A) memory and (B) executive function decline rates across factors by clinical group. Comparisons remaining significant after FDR (q = 0.05) control are highlighted in blue. T, S, and C indicate temporal, subcortical, and cortical factors, respectively. Blue dots are estimated differences between pure atrophy factors, and red bars show the SEs (SI Appendix, SI Methods). For example, the second row in A suggests that AD dementia patients expressing the cortical factor showed faster memory decline than patients expressing the temporal factor (P = 1e−4).
For longitudinal change in executive function (Fig. 7B), the cortical factor was associated with faster executive function decline than temporal (P = 2e−3) and subcortical (P = 2e−4) factors among AD dementia patients. Among Aβ+ MCI participants, the subcortical factor had slower decline than the cortical (P = 8e−9) and temporal (P = 1e−8) factors. There was no executive function decline difference across the factors among Aβ+ CN participants.
All cognitive analyses were repeated using the two- and four-factor LDA atrophy factors (SI Appendix, Figs. S6 and S8). The results were consistent with the three-factor model (SI Appendix, SI Results). In addition, associations between minimental state examination (MMSE) and the three atrophy factors are reported in SI Appendix, Fig. S7C.
Discussion
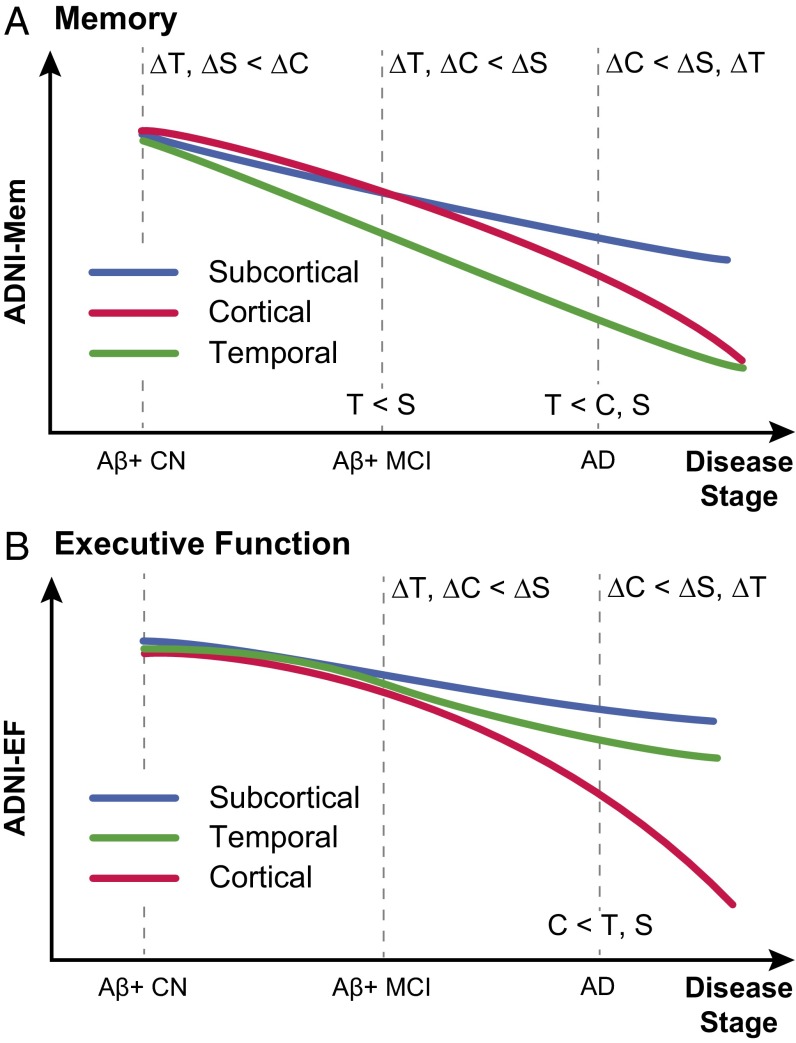
In this study, we identified distinct atrophy factors within AD dementia patients using Bayesian LDA modeling of MRI GM density maps. This approach estimated the factor composition of multiple atrophy factors for each participant rather than assuming membership to a single atrophy subtype (Fig. 1). Our analysis yielded a nested hierarchy of atrophy factors (Fig. 2), which corresponded to distinct trajectories of memory and executive function decline across the disease spectrum (Fig. 8). Overall, these results provide evidence that heterogeneity in patterns of atrophy exists in late-onset AD and that these atrophy patterns are associated with distinct cognitive trajectories.
Fig. 8.
Schematics of distinct (A) memory and (B) executive function trajectories for temporal, subcortical, and cortical atrophy factors. T, S, and C indicate temporal, subcortical, and cortical factors, respectively. Labels on dotted lines indicate cross-sectional differences. For example, T < C, S in A indicates that the temporal factor was associated with the worst baseline memory among AD dementia patients. Labels in the intervals indicate differences in longitudinal decline rates. For example, ΔT, ΔC < ΔS in B indicates that, among Aβ+ MCI participants, the temporal and cortical factors were associated with faster executive function decline than the subcortical factor. The schematics summarize the behavioral results of Figs. 5, 6, and 7 (more discussion is in SI Appendix, SI Results). Within each cognitive domain, the atrophy factors were associated with distinct trajectories across the stages. The trajectories of the cortical and subcortical factors transpose between the two cognitive domains. Divergence in memory trajectories existed even at the asymptomatic stage of the disease (i.e., among Aβ+ CN participants).
Atrophy Patterns in AD Dementia.
Our model revealed a hierarchy of atrophy patterns within AD dementia patients (Fig. 2). As the number of estimated atrophy factors was increased from K to K + 1, one atrophy pattern fractionated into two atrophy patterns, whereas the remaining patterns remained unchanged (SI Appendix, Fig. S2). It is noteworthy that the atrophy patterns extracted using K = 3 were similar to results from other groups investigating AD subtypes (7, 15, 16), although notable differences did emerge.
Specifically, our three-factor model revealed a temporal factor associated with atrophy in the temporal cortex, hippocampus, and amygdala; a cortical pattern associated with atrophy in the frontal, parietal, lateral temporal, and lateral occipital cerebral cortices; and a subcortical factor associated with atrophy in the cerebellum, striatum, and thalamus (Fig. 2B). Our temporal factor was similar to the previously described limbic-predominant subtype, whereas the cortical factor was similar to the hippocampal-sparing subtype (3, 7). More specifically, previous pathologically defined subtypes were identified based on the ratio of NFT burden in hippocampal subregions versus association cortex, resulting in a limbic-predominant subtype and a hippocampal-sparing subtype. Follow-up VBM analyses (7) suggested GM loss in the temporoparietal cortex, frontal cortex, insula, and precuneus in the hippocampal-sparing subtype, consistent with our cortical atrophy factor. However, Whitwell et al. (7) identified predominant atrophy in the medial temporal lobe of the limbic-predominant subtype, consistent with our temporal atrophy factor.
A benefit of our approach is that the nested hierarchy of atrophy patterns was not mandated by our model but completely data-driven. Thus, although not mandated, our results revealed a nested hierarchy in contrast with previous approaches where hierarchy was imposed (15). Specifically, Noh et al. (15) identified three subtypes: a “medial temporal” subtype, a “parietal frontal-dominant” subtype, and a “diffuse” subtype. Our temporal atrophy factor might correspond to their medial temporal subtype, whereas our cortical factor might correspond to their parietal frontal-dominant subtype, although direct comparison was difficult, because their analyses were restricted to the cerebral cortex.
Our model suggests that atrophy patterns in AD patients follow a nested hierarchy structure. Given the nested hierarchy of cognitive functions revealed by a recent large-scale metaanalysis of 10,000 brain imaging experiments (12) as well as brain network analyses (28–31), one might speculate that the nested hierarchy of atrophy factors arises from a natural hierarchy of brain functions and networks.
Atrophy Factors Reflect Subtypes Rather than Disease Stages.
A potential pitfall of AD subtype analyses (32) is that the observed heterogeneity might correspond to different disease stages (stage hypothesis) rather than heterogeneity in disease expression (subtype hypothesis). There are various reasons why the atrophy factors discussed in this manuscript likely correspond to subtypes rather than disease stages (33). First, there was not a single factor associated with the worst memory and executive function. Instead, decline trajectories of the temporal and cortical factors varied in their associations with the two cognitive domains (Fig. 8). Furthermore, analysis of follow-up MRI scans revealed that factor compositions were stable over time (Fig. 3), suggesting that individuals were not progressing from one factor to another [e.g., from temporal factor to cortical factor as predicted under the Braak staging scheme (34)].
Factor-Dependent Characteristics.
There were significant differences across the atrophy factors in baseline age (P = 8e−7) and age at AD onset (P = 1e−5). Baseline age is dependent on study design, and therefore, drawing meaningful comparisons with the literature is difficult. Nevertheless, the cortical factor was associated with younger age at AD onset, consistent with previous studies describing subtypes with predominant cortical atrophy (3, 8, 15). Importantly, years from AD onset to baseline did not differ across the three latent factors (P = 0.29) (SI Appendix, Table S2), providing additional evidence that these factors were not simply disease stages. The subcortical factor was associated with a higher prevalence of the APOE ε2 allele (P = 0.03; not significant when corrected for multiple comparisons). The protective effects of the ε2 allele (35) might potentially contribute to the observation that the subcortical factor was associated with the mildest decline in both memory and executive function across all stages (Fig. 8).
Importantly, a lack of association between each factor and amyloid status suggests that atrophy factors do not merely reflect patterns associated with non-AD dementia patients who may have been “misdiagnosed” as AD dementia within the ADNI dataset (36). However, although repeating our factor estimation with Aβ+ AD dementia patients revealed consistent atrophy patterns with the model using all AD patients, we are not able to determine whether atrophy patterns are a result of Aβ pathology or precede Aβ pathology. For instance, these atrophy patterns may emerge through processes not directly linked to Aβ pathology but instead, converge with AD pathology to influence disease progression. It is possible that factors, such as comorbid TDP-43 pathology and genetics, as well as development differences contribute to this heterogeneity. Along these lines, recent work suggests that different pathologies have distinct impacts on cognitive trajectories (37). Interestingly, TDP-43 was shown to have a very early impact on cognitive trajectories compared with other pathologies, such as hippocampal sclerosis and Lewy bodies. Given that TDP-43 is known to impact the medial temporal lobe (6), it is possible that the temporal atrophy factor is influenced by the involvement of this pathology (because the temporal factor shows an early impact on memory among Aβ+ CN in our study).
A fundamental question that remains is why the expression of these atrophy patterns varies across individuals, especially because the spatial distribution of Aβ tends to be very diffuse throughout cortex. A similar dissociation is observed among AD patients with atypical clinical presentations, such that, although the spatial pattern of Aβ is diffuse, the underlying pattern of NFTs and GM atrophy aligns with clinical symptoms (4). Future work should investigate the time course of these atrophy patterns using longitudinal MRI as well as longitudinal assessment of Aβ and also investigate the prevalence of atrophy patterns among Aβ− participants to understand whether these patterns are specific for AD or merely converge with AD processes to influence disease progression.
Distinct Memory and Executive Function Decline Trajectories.
The behavioral results (Figs. 5, 6, and 7) are summarized in Fig. 8. Overall, we found that the associations between atrophy factors and cognition varied by domain as well as time course in the disease. Specifically, the temporal factor showed the greatest association with memory, a relationship that emerged early among Aβ+ CN participants and remained consistent in later disease stages. Conversely, the cortical factor was associated with both memory and executive function but exerted greater impact later in the disease among Aβ+ MCI participants and AD patients.
Overall, the trajectories (Fig. 8) revealed several salient points. First, memory decline in the context of late-onset AD occurred earlier than decline in executive function, which is in line with previous studies (38). Second, divergence of memory trajectories among atrophy factors appeared as early as the asymptomatic (CN) stage of the disease, whereas divergence of executive function trajectories was not detectable until the MCI stage (Fig. 8). Specifically, the temporal and subcortical factors showed faster memory decline than the cortical factor among Aβ+ CN participants, and by MCI, the temporal factor was already associated with worse memory at baseline than the subcortical factor. In contrast, there was no difference in executive function decline rates among Aβ+ CN participants or cross-sectional difference among Aβ+ MCI participants. Interestingly, AD dementia patients expressing the cortical factor exhibited the fastest decline rates in both executive function and memory. Third, the subcortical factor (blue curves in Fig. 8) was the mildest factor in terms of both memory and executive function deterioration. In both Aβ+ MCI and AD dementia participants, the subcortical factor was associated with the best memory and executive function scores as well as the slowest decline rates.
Correspondence and Extensions of AD Heterogeneity Literature.
Our results were consistent with the preponderance of literature on heterogeneity among AD dementia patients. For example, our atrophy factors show overlap with the pathologically defined hippocampal-sparing and limbic-predominant subtypes (7) as well as the subtypes described by Noh et al. (15). Our analyses suggested that the cortical factor was associated with faster decline in both memory and executive function than the temporal factor at the dementia stage, which is consistent with the hippocampal-sparing subtype exhibiting faster MMSE decline than the limbic-predominant subtype among AD dementia patients (3). Similarly, our finding that the cortical factor was associated with the most rapid memory and executive function decline among AD dementia patients was also consistent with the work by Byun et al. (16). Among AD dementia patients, the cortical factor was associated with the worst baseline executive function, whereas the temporal factor was associated with the worst baseline memory. This result is consistent with previous work showing that thinning of frontoparietal cortical regions was associated with nonamnestic presentations and dysexecutive phenotypes (9) and that the “cortical atrophy-only” subtype had worse baseline executive function than the “hippocampal atrophy-only” subtype (16). Thus, our data-driven approach provides additional evidence that distinct atrophy patterns among AD patients impact different cognitive domains.
In addition to characterizing heterogeneity among AD dementia patients, we extended our approach to participants who were presumably in very early stages of AD development (i.e., Aβ+ but without the clinical symptoms of dementia) (19, 20). By examining earlier stages, we found that the temporal factor showed the greatest association with memory decline among Aβ+ CN participants but that the cortical factor was a stronger predictor of memory decline among AD dementia patients (SI Appendix, Fig. S7A2). Likewise, although the cortical factor was not associated with either cognitive domain among Aβ+ CN participants, this factor was associated with executive function decline in Aβ+ MCI participants and AD patients (Figs. 7B and 8). The impact of these atrophy factors at different points along the clinical spectrum has important implications for measuring decline and understanding the progression of AD. Furthermore, consideration of this heterogeneity may improve the ability to identify individuals most at risk for cognitive decline compared with approaches that measure atrophy using the same regional metric across all participants.
Mixed Membership Modeling and Precision Medicine.
One key advantage of our modeling strategy is that individuals can express multiple latent atrophy factors (i.e., mixed membership) rather than being assigned to a single subtype. Therefore, patients classified by Murray et al. (3) as hippocampal-sparing (or limbic-predominant) might correspond to the few patients in our study who predominantly expressed the cortical (or temporal) atrophy factor. Murray et al. (3) also defined a third group of patients who were considered “typical” by virtue of being neither hippocampal-sparing nor limbic-predominant. These typical patients might correspond to the majority of AD dementia patients in our study who expressed multiple latent factors to similar degrees.
The use of mixed membership modeling has implications for estimation of factor-dependent atrophy maps and cognitive decline. For example, consider a hypothetical patient who expressed 50% subcortical, 40% temporal, and 10% cortical factors. In our analyses, 50%, 40%, and 10% of the patient’s atrophy map would contribute to the estimation of the probabilistic atrophy maps of the subcortical, temporal, and cortical factors, respectively. This method extends previous approaches (7, 15, 16) that classified each patient into one single subtype and then, performed group comparisons to obtain differential atrophy patterns, despite the fact that each patient might express multiple latent atrophy factors. Thus, more information about each participant is retained by treating factor compositions continuously rather than assigning participants to a single group.
Similarly, 50%, 40%, and 10% of the hypothetical patient’s cognitive decline rate would contribute to our estimation of the memory decline rates associated with the subcortical, temporal, and cortical factors, respectively. Indeed, when such a patient was simply assigned to a single factor based on the highest probability (i.e., assigned to a pure subtype), the estimated differences in cognitive decline rates across subtypes were found to be substantially weaker. The reason should be clear when considering the hypothetical patient. Because the patient expressed 50% subcortical, 40% temporal, and 10% cortical factors, one would expect the memory decline rate to be faster than a pure subcortical subtype (and slower than a pure temporal factor). By assigning this patient to be a pure subcortical subtype, one would overestimate the decline rate of the subcortical subtype.
Although we observe some participants with extreme probabilities of a single atrophy factor, these participants are infrequent. Instead, the majority of the participants expressed intermediate probabilities across multiple latent atrophy factors. We can potentially use the factor decomposition to predict the memory and executive function decline trajectories of individual participants. For example, we might predict the hypothetical patient who expressed 50% subcortical, 40% temporal, and 10% cortical factors to have decline trajectories corresponding to 50% times the blue curve plus 40% times the green curve plus 10% times the red curve from Fig. 8. Therefore, the factor composition can be thought of as an individualized subtype diagnosis of the participant, representing a small but crucial step toward precision medicine.
Limitations.
Our study has multiple limitations. First, direct comparisons with other subtype studies were difficult because of methodological differences, including the utilization of mixed membership modeling and participant selection. Another limitation is the arbitrary choice of the number of latent atrophy factors to estimate using LDA. Given consistency with previous studies and a limited sample size, we focused on K = 2–4 factors, but atrophy factors beyond K = 4 may be biologically relevant.
Conclusion
By using a Bayesian modeling framework, our study revealed three latent AD atrophy factors with distinct memory and executive function trajectories. Across the clinical spectrum, the cortical atrophy factor was associated with the worst executive function performance, whereas the temporal atrophy factor was associated with the worst memory performance. The subcortical atrophy factor has not been discussed in the literature and was associated with the slowest memory and executive function decline. Our approach allowed each individual to express multiple atrophy factors to various degrees rather than assigning the individual to a single subtype. Therefore, each participant exhibited his or her own unique factor composition, which can potentially be exploited to predict individual-specific cognitive decline trajectories, with potential implications for prevention and monitoring disease progression. Finally, our methodological framework is general and can be used to discover subtypes in other brain disorders. Factor compositions of ADNI participants and code used in this manuscript are publicly available (https://github.com/ThomasYeoLab/CBIG/tree/master/stable_projects/disorder_subtypes/Zhang2016_ADFactors).
Materials and Methods
Overview.
Voxelwise atrophy of 188 AD dementia patients was derived from their structural MRI data (22, 39). Subsequent analyses proceeded in three steps. In step I, a Bayesian model (Fig. 1) (10) was applied to estimate the probabilistic atrophy maps of latent factors Pr(Voxel | Factor) and the factor composition of each patient Pr(Factor | Patient). The probabilistic atrophy maps were then used to infer the factor compositions of 43 Aβ+ CN participants and 147 Aβ+ MCI participants. In step II, stability of the factor decomposition over a period of two years was analyzed. In addition, characteristics (demographics, age at AD onset, years from AD onset to baseline, amyloid burden, and APOE genotype) of all participants were compared across the factors. Finally, in step III, we analyzed the atrophy factors’ relationships with cross-sectional baseline and longitudinal decline of memory and executive function. Each step is described in detail below.
Data.
Data used in this study were obtained from the ADNI database (adni.loni.usc.edu), which was launched in 2003 as a public–private partnership and led by Principal Investigator Michael W. Weiner. The primary goal of the ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD (up to date information is at www.adni-info.org/). Institutional review boards approved study procedures across participating institutions (the complete list of the institutions is in SI Appendix). Written informed consent was obtained from all participants.
This study considered the structural MRI (T1-weighted, 1.5 T) of 810 participants enrolled in the ADNI 1, comprising 188 AD dementia (at baseline; same hereinafter) patients, 394 MCI participants, and 228 CN participants. Of the 188 AD dementia patients, 100 had their CSF amyloid data available, and 91 of 100 were Aβ+. AD onset was, on average, 3.6 years (SD = 2.5, minimum = 0, maximum = 13) before baseline. Of 394 MCI participants, 197 had their CSF amyloid data available, and 147 of 197 were Aβ+. Of 228 CN participants, 114 had their CSF amyloid data available, and 43 of 114 were Aβ+. The Aβ+ CN elderly participants and the Aβ+ MCI participants are referred to as the Aβ+ nondemented group (n = 190) in this study.
According to the ADNI protocol, AD dementia patients had their cognition examined at baseline and in months 6, 12, and 24. In addition, normal participants were examined in month 36 and annually afterward. MCI participants underwent another extra examination in month 18. Although this study only considered participants enrolled in the ADNI 1, to increase statistical power, their neuropsychological scores (ADNI-Mem, ADNI-EF, and MMSE) from ADNI Grand Opportunity (GO) and ADNI 2 were also included in the longitudinal analyses of cognitive decline.
Voxel-Based Morphometry.
Structural MRI data of all 810 participants were analyzed with FSL-VBM (fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSLVBM) (22), a VBM protocol (40) carried out with FSL tools (41). First, structural images were brain-extracted and GM-segmented before being registered to the Montreal Neurological Institute (MNI152) standard space using affine registration. Second, the affine-registered images were flipped about the x axis and averaged to create a left–right symmetric, study-specific affine GM template. Third, the GM images were nonlinearly registered to the affine GM template, and again, they were flipped and averaged to create a final left–right symmetric, study-specific nonlinear GM template in MNI152 space. Fourth, all native GM images were nonlinearly registered to this final template and modulated to account for local expansion (or contraction) because of the nonlinear component of the spatial transformation. The resulting GM density images were smoothed with a Gaussian kernel of 10-mm FWHM, consistent with standard VBM practices (42, 43). Finally, we applied log10 to the smoothed GM density images and regressed out possible effects of age, sex, and intracranial volume (ICV) with a GLM estimated from just 228 CN participants.
Quality Control for Voxel-Based Morphometry.
The outputs of each VBM step were visually checked by authors X.Z. and N.S. Details are found in SI Appendix, SI Methods.
Bayesian Model.
We sought a mathematical model that captured the premise that each AD patient expresses one or more latent atrophy factors, each of which is associated with distinct but possibly overlapping atrophy patterns (Fig. 1). Among many possible models, the LDA model (10) is probably the simplest and was applied to the ADNI data.
The LDA model was originally developed to automatically discover latent topics in a collection of text documents. The model assumes that each document is an unordered collection of words associated with a subset of K latent topics. Each topic is represented by a probability distribution over a dictionary of words. Given a collection of documents, there exist algorithms (10) to estimate the probability of a dictionary word given a topic Pr(Word | Topic) and the probability that a topic is associated with a particular document Pr(Topic | Document). The LDA model is useful, because it allows a document to be associated with multiple topics (which can be shared across documents) and each topic to be associated with multiple words (which can be shared across topics).
To map the LDA model to the ADNI data, one can think of AD patients as text documents, atrophy factors as topics, and MNI152 voxels as dictionary words. Correspondingly, each patient expresses one or more latent atrophy factors to different extents [Pr(Factor | Patient)], and each factor is associated with atrophy at multiple voxels to different extents [Pr(Voxel | Factor)].
LDA assumes that a document is summarized by the number of times that a dictionary word appears in the document. Because dictionary words correspond to MNI voxels, the continuous log-transformed GM density images (in the previous section) were discretized, so that greater atrophy corresponded to larger word counts. More specifically, for each voxel of the log-transformed GM density images, z transformation (with respect to 228 CN participants) was performed for each of 810 participants. Therefore, a z score of <0 at a given voxel of a particular individual would imply above-average atrophy at the voxel relative to the CN participants. z Scores above zero were set to zero, equivalent to regarding the voxels as atrophy-free. Finally, the z scores were multiplied by −10 and rounded to the nearest integer, so that larger positive values (greater word count) indicated more severe atrophy.
The LDA model assumes that the ordering of words within a document is exchangeable. In the context of our application, the corresponding assumption is that the ordering of atrophied voxels is exchangeable. Although word order in real documents is important, the ordering of atrophied regions (e.g., prefrontal vs. parietal) reported in an experiment is arbitrary and thus, consistent with the assumption. Consequently, the LDA model appears particularly well-suited for applications in this context.
Given the discretized voxelwise atrophy of 188 AD dementia patients and the number of latent atrophy factors K, the VEM algorithm (www.cs.princeton.edu/∼blei/lda-c/) (10) was applied to estimate Pr(Factor | Patient) and Pr(Voxel | Factor). For each K, the algorithm was rerun with 40 different random initializations, and the solution with the highest likelihood (bound) was selected. The random initializations led to highly similar solutions, suggesting that 40 random initializations were sufficient for robust factor estimations.
The probabilistic atrophy maps Pr(Voxel | Factor) estimated from the AD dementia patients were used to infer factor compositions Pr(Factor | Participant) of 190 Aβ+ nondemented participants using the standard VEM algorithm (10).
Interpreting Pr(Voxel | Factor) and Pr(Factor | Patient).
For a given latent factor, Pr(Voxel | Factor) is a probability distribution over all of the GM voxels, which can be visualized as a probabilistic atrophy map overlaid on the FSL MNI152 template (each row of Fig. 2).
Pr(Factor | Patient) is a probability distribution over latent atrophy factors, representing the factor composition of the patient, and can be visualized as a dot inside a “factor triangle” (for K = 3 factors) with barycentric coordinates that equal Pr(Factor | Patient) as shown in Fig. 4 and SI Appendix, Fig. S5A. For example, Pr(Factor | Patient) = [0.7, 0.2, 0.1] implies that the patient expresses a pattern of brain atrophy caused by 70% temporal, 20% subcortical, and 10% cortical factors, respectively, and that the dot representing this patient falls closer to the “temporal corner” of the factor triangle. This approach contrasts with work in the literature that assigns each individual to a single subtype (3, 15, 16).
Quantifying the Nested Hierarchy of Atrophy Factors.
An important model parameter is the number of latent factors K. Therefore, we determined how factor estimation changed from K = 2 to 10 factors. The detailed description is in SI Appendix, SI Methods.
Top Anatomical Structures Associated with Each Factor.
This manuscript focuses on three atrophy factors. To automatically identify the GM anatomical structures most associated with each atrophy factor, the MNI152 template was first processed using FreeSurfer 4.5.0 (24). The FreeSurfer software automatically segmented the MNI152 template into multiple cortical (44, 45) and subcortical (44, 46) structures, such as the inferior parietal cortex and hippocampus. For each anatomical structure, we averaged Pr(Voxel | Factor) over all of its voxels. The structure was assigned to the factor with the largest average probability. For each factor, we tabulated the assigned brain structures and ranked them in the descending order of average probability. The results are in SI Appendix, Table S1.
Cross-Pipeline Validation of Atrophy Patterns.
To ensure that the atrophy factors were robust to choice of VBM software (FSL), we performed post hoc analyses using FreeSurfer. Details are found in SI Appendix, SI Results and SI Methods.
Atrophy Factor Stability.
To examine the atrophy factor stability during disease progression, we considered all 810 participants who had their two-year follow-up scans available (n = 560). First, their baseline factor compositions Pr(Factor | Participant) were extracted using their baseline MRI data. Second, VBM was performed on the follow-up structural MRI data using the VBM template previously created with all 810 participants. Subsequent processing (e.g., z normalization) adopted parameters used in processing 810 baseline scans. Factor compositions were then inferred with the processed VBM results (same procedure as inferring factor compositions of Aβ+ CN and MCI participants). The factor stability was visualized with a scatter plot for each factor (Fig. 3 and SI Appendix, Fig. S4). Each participant is represented by a dot with an x coordinate that is the factor composition at baseline and a y coordinate that is the factor composition after two years. Therefore, if the factor estimation is stable over disease progression, one would expect a close-to-one correlation coefficient and a y = x linear fit.
Comparing Patient Characteristics by Atrophy Factor.
We explored how patient characteristics (baseline age, age at AD onset, years from onset to baseline, education, sex, amyloid, and APOE genotype) varied across the three latent factors (SI Appendix, Table S2) using GLM (and logistic regression for binary variables).
GLM was applied to baseline age, age at AD onset, years from onset to baseline, education, amyloid, and APOE: the characteristic of interest served as response y, and the subcortical factor probability s and cortical factor probability c were included as explanatory variables. Hence, the GLM was y = β0 + βs·s + βc·c + ε, where β indicates the regression coefficients, and ε is the residual. The temporal factor probability t was implicitly modeled, because t + s + c = 1. Intuitively, β0 reflected the response of the temporal factor, βs reflected the response difference between the subcortical and temporal factors, and βc reflected the difference between the cortical and temporal factors.
Statistical tests of whether the characteristic y varied across factors involved null hypotheses of the form Hβ = 0, where β = [β0, βs, βc]T, and H is the linear contrast (47). We first performed a statistical test of overall differences across all factors with H = [0, 1, 0; 0, 0, 1]. We then tested for differences between the factors. For example, H = [0, 1, −1] tested possible differences between the subcortical and cortical factors. H = [0, 1, 0] compared the subcortical and temporal factors. Similarly, H = [0, 0, 1] compared the cortical and temporal factors.
Because sex is a binary variable, logistic regression was applied. In this case, response y was sex (zero for male, and one for female), and explanatory variables consisted of the subcortical factor probability s and cortical factor probability c. Therefore, the regression model was log(µ/(1 − µ)) = β0 + βs·s + βc·c + ε, where µ is the probability of female, β indicates the regression coefficients, and ε is the residual. Intuitively, the linear combination β0 + βs·s + βc·c predicts the probability of female (y = 1); exp(β0) reflects the odds ratio for the temporal factor, exp(βs) reflects the ratio of odds ratio between the subcortical and temporal factors, and exp(βc) reflects the ratio of odds ratio between the cortical and temporal factors.
Likelihood ratio test was used to determine whether sex varied across the latent atrophy factors. In short, the test involved comparing the likelihood of an appropriately restricted model with the original model (47). We first performed a statistical test of overall differences across factors. In this case, the restricted model log(µ/(1 − µ)) = β0 + ε was fitted to the data, and the resulting likelihood was compared with the likelihood of the original model log(µ/(1 − µ)) = β0 + βs·s + βc·c + ε. We then tested for possible differences between atrophy factors. For example, to compare the subcortical and cortical factors, the restricted model was log(µ/(1 − µ)) = β0 + βs·(s + c) + ε, because βs = βc under the null hypothesis. To compare the subcortical and temporal factors, the restricted model became log(µ/(1 − µ)) = β0 + βc·c + ε, because βs = 0 under the null hypothesis. To compare the cortical and temporal factors, the restricted model was log(µ/(1 − µ)) = β0 + βs·s + ε, because βc = 0 under the null hypothesis.
General Linear Modeling of Cross-Sectional Cognition Among Aβ+ CN, Aβ+ MCI, and AD Dementia Participants.
A single GLM was used to examine cross-sectional differences in memory (ADNI-Mem) (25) across the atrophy factors in 43 Aβ+ CN, 147 Aβ+ MCI, and 188 AD dementia participants. The same model was estimated for K = 2, 3, and 4 factors as well as for executive function (ADNI-EF) (26) and MMSE.
For ease of explanation, we will focus on explaining the GLM for the case of three atrophy factors and ADNI-Mem. Response y of the GLM consisted of 378 (=43 CN + 147 MCI + 188 AD) participants’ baseline ADNI-Mem. Explanatory variables consisted of binary MCI group indicator m, binary AD dementia group indicator d, subcortical factor probability s, cortical factor probability c, and interactions between group indicators and factor probabilities (i.e., m·s, m·c, d·s, and d·c), whereas nuisance variables consisted of baseline age x1, sex x2, education x3, and total atrophy x4 (defined as ICV divided by total GM volume as estimated by FSL).
Therefore, the GLM was y = β0 + βm·m + βd·d + βs·s + βc·c + βms·m·s + βmc·m·c + βds·d·s + βdc·d·c + β1·x1 + β2·x2 + β3·x3 + β4·x4 + ε, where β indicates the regression coefficients, and ε is the residual. Temporal factor probability t was implicitly modeled, because t + s + c = 1. Similarly, the CN group indicator n was also implicitly modeled, because only one of n, m, and d is one, with the other two being zero. Intuitively, β0 reflected the temporal factor’s contribution to ADNI-Mem at the CN baseline (because m = d = s = c = 0), β0 + βm reflected the temporal factor’s contribution to ADNI-Mem at the MCI baseline (because m = 1 and d = s = c = 0), and β0 + βm + βs + βms reflected the subcortical factor’s contribution to ADNI-Mem at the MCI baseline (because m = s = 1 and d = c = 0). With this model setup, variations in age, sex, education, and total atrophy were controlled for across participants.
Statistical tests involved null hypotheses of the form Hβ = 0, where β = [β0, βm, βd, βs, βc, βms, βmc, βds, βdc, β1, β2, β3, β4]T, and H is the linear contrast (47). We tested whether ADNI-Mem deteriorated across disease stages (i.e., from CN to MCI to AD) for each factor. Specifically, for each factor, we tested possible differences in ADNI-Mem between the CN and MCI baselines, between the MCI and AD baselines, and between the CN and AD baselines. For example, to test whether ADNI-Mem deteriorated significantly from the CN to MCI baseline for the temporal factor, H was specified, such that Hβ = βm = 0. As another example, Hβ = βd + βdc – βm – βmc = 0 tested whether ADNI-Mem degraded greatly from the MCI to AD baseline for the cortical factor. The test results for both memory and executive function are tabulated in Fig. 5 A1 and B1.
To foreshadow the results, the hypothesis tests in the previous paragraph hinted at differences in cross-sectional ADNI-Mem across the factors. Therefore, statistical tests of whether cross-sectional ADNI-Mem y varied across factors at each disease stage were performed. For each stage baseline, we first performed a statistical test of overall differences across all factors and then tested for pairwise differences. Take the AD baseline as an example. To test whether baseline memory differed across all factors among AD dementia patients, H was specified, such that Hβ = 0 translated to βs + βds = βc + βdc = 0. For pairwise comparisons, βs + βds = 0 tested possible differences between the temporal and subcortical factors at the AD baseline, βc + βdc = 0 compared the temporal and cortical factors at the AD baseline, and βs + βds = βc + βdc tested possible differences between the subcortical and cortical factors at the AD baseline.
The results of the above statistical tests are shown in Figs. 5 A1 and B1 and 6 and SI Appendix, Figs. S6 A1 and B1; S7 A1, B1, and C1; and S8 A1 and B1, where (except in Figs. 5 A1 and B1) the blue dots correspond to the estimated difference in baseline scores between two “pure factors” after controlling for age, sex, education, and total atrophy. For example, when comparing subcortical and cortical factors at the MCI baseline, the estimated difference in baseline cognition is given by βs + βms – βc – βmc. The red bars correspond to the SE of this estimation given by SD(βs + βms – βc – βmc).
LME Modeling of Longitudinal Cognitive Decline Among Aβ+ CN, Aβ+ MCI, and AD Dementia Participants.
To analyze variations in cognitive decline rates across atrophy factors, we used the LME model, which had a setup that was similar to the GLM setup (in the previous section). Details are found in SI Appendix, SI Methods. Results of the LME statistical tests are illustrated in Figs. 5 A2 and B2 and 7 and SI Appendix, Figs. S6 A2 and B2; S7 A2, B2, and C2; and S8 A2 and B2.
False Discovery Rate Correction for Behavioral Tests.
Because of the many statistical tests performed in the behavioral analyses, multiple testing was corrected using false discovery rate (FDR) (48) at q = 0.05 for all behavioral comparisons. In detail, included tests are diagnostic group comparisons in memory and executive function regardless of factors as well as all comparisons of baseline and longitudinal decline rates of memory, executive function, and MMSE at all disease stages for K = 2, 3, and 4 factors. In total, we corrected for 240 statistical tests. P values that remained significant after FDR control were highlighted in blue in Figs. 5, 6, and 7 and SI Appendix, Figs. S6–S8.
Supplementary Material
Acknowledgments
We thank Maxwell Bertolero, Mark D'Esposito, Rik Ossenkoppele, Daniel Alexander, and Christopher Asplund for their constructive comments as well as Gia Ngo, Muhammad Anwar, Ryan Fong, and Xilin Jiang for assistance with public release of code and data. This work was supported by National University of Singapore (NUS) Tier 1; Singapore Ministry of Education Tier 2 Grant MOE2014-T2-2-016; NUS Strategic Research Grant DPRT/944/09/14; NUS School of Medicine Aspiration Fund R185000271720; Singapore National Medical Research Council Grant CBRG14nov007, NMRC/CG/013/2013; NUS Young Investigator Award; and NIH Grants 1K25EB013649-01, 1R21AG050122-01A1, P01AG036694, and F32AG044054. The research also used resources provided by Center for Functional Neuroimaging Technologies Grant P41EB015896 and instruments supported by Grants 1S10RR023401, 1S10RR019307, and 1S10RR023043 from the Athinoula A. Martinos Center for Biomedical Imaging at the Massachusetts General Hospital. Data collection and sharing for this project was funded by the ADNI (NIH Grant U01 AG024904) and the Department of Defense (DOD) ADNI (DOD Grant W81XWH-12-2-0012). The ADNI is funded by the National Institute on Aging and the National Institute of Biomedical Imaging and Bioengineering and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. The ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. Data used in preparation of this article were obtained from the ADNI database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report.
Footnotes
The authors declare no conflict of interest.
A complete list of the Alzheimer's Disease Neuroimaging Initiative can be found in SI Appendix.
This article is a PNAS Direct Submission.
Data deposition: The Alzheimer's Disease Neuroimaging Initiative data are publicly available at adni.loni.usc.edu/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611073113/-/DCSupplemental.
Contributor Information
Collaborators: Michael W. Weiner, Paul Aisen, Michael Weiner, Paul Aisen, Ronald Petersen, Clifford R. Jack, Jr., William Jagust, John Q. Trojanowki, Arthur W. Toga, Laurel Beckett, Robert C. Green, Andrew J. Saykin, John Morris, Leslie M. Shaw, Zaven Khachaturian, Greg Sorensen, Maria Carrillo, Lew Kuller, Marc Raichle, Steven Paul, Peter Davies, Howard Fillit, Franz Hefti, David Holtzman, M. Marcel Mesulam, William Potter, Peter Snyder, Adam Schwartz, Robert C. Green, Tom Montine, Ronald Petersen, Paul Aisen, Ronald G. Thomas, Michael Donohue, Sarah Walter, Devon Gessert, Tamie Sather, Gus Jiminez, Archana B. Balasubramanian, Jennifer Mason, Iris Sim, Laurel Beckett, Danielle Harvey, Michael Donohue, Clifford R. Jack, Jr., Matthew Bernstein, Nick Fox, Paul Thompson, Norbert Schuff, Charles DeCArli, Davis Bret Borowski, Jeff Gunter, Matt Senjem, Prashanthi Vemuri, David Jones, Kejal Kantarci, Chad Ward, William Jagust, Robert A. Koeppe, Norm Foster, Eric M. Reiman, Kewei Chen, Chet Mathis, Susan Landau, John C. Morris, Nigel J. Cairns, Erin Franklin, Lisa Taylor-Reinwald, Leslie M. Shaw, John Q. Trojanowki, Virginia Lee, Magdalena Korecka, Michal Figurski, Arthur W. Toga, Karen Crawford, Scott Neu, Andrew J. Saykin, Tatiana M. Foroud, Steven Potkin, Li Shen, Kelley Faber, Sungeun Kim, Kwangsik Nho, Michael W. Weiner, Lean Thal, Zaven Khachaturian, Leon Thal, Neil Buckholtz, Michael W. Weiner, Peter J. Snyder, William Potter, Steven Paul, Marilyn Albert, Richard Frank, Zaven Khachaturian, John Hsiao, Jeffrey Kaye, Joseph Quinn, Lisa Silbert, Betty Lind, Raina Carter, Sara Dolen, Lon S. Schneider, Sonia Pawluczyk, Mauricio Becerra, Liberty Teodoro, Bryan M. Spann, James Brewer, Helen Vanderswag, Adam Fleisher, Judith L. Heidebrink, Joanne L. Lord, Ronald Petersen, Sara S. Mason, Colleen S. Albers, David Knopman, Kris Johnson, Rachelle S. Doody, Javier Villanueva-Meyer, Valory Pavlik, Victoria Shibley, Munir Chowdhury, Susan Rountree, Mimi Dang, Yaakov Stern, Lawrence S. Honig, Karen L. Bell, Beau Ances, John C. Morris, Maria Carroll, Mary L. Creech, Erin Franklin, Mark A. Mintun, Stacy Schneider, Angela Oliver, Daniel Marson, David Geldmacher, Marissa Natelson Love, Randall Griffith, David Clark, John Brockington, Erik Roberson, Hillel Grossman, Effie Mitsis, Raj C. Shah, Leyla deToledo-Morrell, Ranjan Duara, Maria T. Greig-Custo, Warren Barker, Marilyn Albert, Chiadi Onyike, Daniel D’Agostino, II, Stephanie Kielb, Martin Sadowski, Mohammed O. Sheikh, Anaztasia Ulysse, Mrunalini Gaikwad, P. Murali Doraiswamy, Jeffrey R. Petrella, Salvador Borges-Neto, Terence Z. Wong, Edward Coleman, Steven E. Arnold, Jason H. Karlawish, David A. Wolk, Christopher M. Clark, Charles D. Smith, Greg Jicha, Peter Hardy, Partha Sinha, Elizabeth Oates, Gary Conrad, Oscar L. Lopez, MaryAnn Oakley, Donna M. Simpson, Anton P. Porsteinsson, Bonnie S. Goldstein, Kim Martin, Kelly M. Makino, M. Saleem Ismail, Connie Brand, Steven G. Potkin, Adrian Preda, Dana Nguyen, Kyle Womack, Dana Mathews, Mary Quiceno, Allan I. Levey, James J. Lah, Janet S. Cellar, Jeffrey M. Burns, Russell H. Swerdlow, William M. Brooks, Liana Apostolova, Kathleen Tingus, Ellen Woo, Daniel H.S. Silverman, Po H. Lu, George Bartzokis, Neill R Graff-Radford, Francine Parfitt, Kim Poki-Walker, Martin R. Farlow, Ann Marie Hake, Brandy R. Matthews, Jared R. Brosch, Scott Herring, Christopher H. van Dyck, Richard E. Carson, Martha G. MacAvoy, Pradeep Varma, Howard Chertkow, Howard Bergman, Chris Hosein, Sandra Black, Bojana Stefanovic, Curtis Caldwell, Ging-Yuek Robin Hsiung, Benita Mudge, Vesna Sossi, Howard Feldman, Michele Assaly, Elizabeth Finger, Stephen Pasternack, Irina Rachisky, Dick Trost, Andrew Kertesz, Charles Bernick, Donna Munic, Marek-Marsel Mesulam, Emily Rogalski, Kristine Lipowski, Sandra Weintraub, Borna Bonakdarpour, Diana Kerwin, Chuang-Kuo Wu, Nancy Johnson, Carl Sadowsky, Teresa Villena, Raymond Scott Turner, Kathleen Johnson, Brigid Reynolds, Reisa A. Sperling, Keith A. Johnson, Gad Marshall, Jerome Yesavage, Joy L. Taylor, Barton Lane, Allyson Rosen, Jared Tinklenberg, Marwan N. Sabbagh, Christine M. Belden, Sandra A. Jacobson, Sherye A. Sirrel, Neil Kowall, Ronald Killiany, Andrew E. Budson, Alexander Norbash, Patricia Lynn Johnson, Thomas O. Obisesan, Saba Wolday, Joanne Allard, Alan Lerner, Paula Ogrocki, Curtis Tatsuoka, Parianne Fatica, Evan Fletcher, Pauline Maillard, John Olichney, Charles DeCarli, Owen Carmichael, Smita Kittur, Michael Borrie, T-Y Lee, Dr Rob Bartha, Sterling Johnson, Sanjay Asthana, Cynthia M. Carlsson, Steven G. Potkin, Adrian Preda, Dana Nguyen, Pierre Tariot, Anna Burke, Ann Marie Milliken, Nadira Trncic, Adam Fleisher, Stephanie Reeder, Vernice Bates, Horacio Capote, Michelle Rainka, Douglas W. Scharre, Maria Kataki, Brendan Kelley, Earl A. Zimmerman, Dzintra Celmins, Alice D. Brown, Godfrey D. Pearlson, Karen Blank, Karen Anderson, Laura A. Flashman, Marc Seltzer, Mary L. Hynes, Robert B. Santulli, Kaycee M. Sink, Leslie Gordineer, Jeff D. Williamson, Pradeep Garg, Franklin Watkins, Brian R. Ott, Geoffrey Tremont, Lori A. Daiello, Stephen Salloway, Paul Malloy, Stephen Correia, Howard J. Rosen, Bruce L. Miller, David Perry, Jacobo Mintzer, Kenneth Spicer, David Bachman, Elizabeth Finger, Stephen Pasternak, Irina Rachinsky, John Rogers, Andrew Kertesz, Dick Drost, Nunzio Pomara, Raymundo Hernando, Antero Sarrael, Susan K. Schultz, Karen Ekstam Smith, Hristina Koleva, Ki Won Nam, Hyungsub Shim, Norman Relkin, Gloria Chiang, Michael Lin, Lisa Ravdin, Amanda Smith, Balebail Ashok Raj, and Kristin Fargher
References
- 1.Salmon DP, Bondi MW. Neuropsychological assessment of dementia. Annu Rev Psychol. 2009;60:257–282. doi: 10.1146/annurev.psych.57.102904.190024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam B, Masellis M, Freedman M, Stuss DT, Black SE. Clinical, imaging, and pathological heterogeneity of the Alzheimer’s disease syndrome. Alzheimers Res Ther. 2013;5(1):1. doi: 10.1186/alzrt155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murray ME, et al. Neuropathologically defined subtypes of Alzheimer’s disease with distinct clinical characteristics: A retrospective study. Lancet Neurol. 2011;10(9):785–796. doi: 10.1016/S1474-4422(11)70156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ossenkoppele R, et al. Tau PET patterns mirror clinical and neuroanatomical variability in Alzheimer’s disease. Brain. 2016;139(Pt 5):1551–1567. doi: 10.1093/brain/aww027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider JA, Arvanitakis Z, Leurgans SE, Bennett DA. The neuropathology of probable Alzheimer disease and mild cognitive impairment. Ann Neurol. 2009;66(2):200–208. doi: 10.1002/ana.21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Josephs KA, et al. Staging TDP-43 pathology in Alzheimer’s disease. Acta Neuropathol. 2014;127(3):441–450. doi: 10.1007/s00401-013-1211-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitwell JL, et al. Neuroimaging correlates of pathologically defined subtypes of Alzheimer’s disease: A case-control study. Lancet Neurol. 2012;11(10):868–877. doi: 10.1016/S1474-4422(12)70200-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ossenkoppele R, et al. Atrophy patterns in early clinical stages across distinct phenotypes of Alzheimer’s disease. Hum Brain Mapp. 2015;36(11):4421–4437. doi: 10.1002/hbm.22927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickerson BC, Wolk DA. Alzheimer’s Disease Neuroimaging Initiative Dysexecutive versus amnesic phenotypes of very mild Alzheimer’s disease are associated with distinct clinical, genetic and cortical thinning characteristics. J Neurol Neurosurg Psychiatry. 2011;82(1):45–51. doi: 10.1136/jnnp.2009.199505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blei DM, Ng AY, Jordan MI. Latent Dirichlet allocation. J Mach Learn Res. 2003;3:993–1022. [Google Scholar]
- 11.Yeo BTT, Krienen FM, Chee MW, Buckner RL. Estimates of segregation and overlap of functional connectivity networks in the human cerebral cortex. Neuroimage. 2014;88:212–227. doi: 10.1016/j.neuroimage.2013.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeo BTT, et al. Functional specialization and flexibility in human association cortex. Cereb Cortex. 2015;25(10):3654–3672. doi: 10.1093/cercor/bhu217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertolero MA, Yeo BTT, D’Esposito M. The modular and integrative functional architecture of the human brain. Proc Natl Acad Sci USA. 2015;112(49):E6798–E6807. doi: 10.1073/pnas.1510619112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coutu JP, Goldblatt A, Rosas HD, Salat DH. Alzheimer’s Disease Neuroimaging Initiative (ADNI) White matter changes are associated with ventricular expansion in aging, mild cognitive impairment, and Alzheimer’s disease. J Alzheimers Dis. 2015;49(2):329–342. doi: 10.3233/JAD-150306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noh Y, et al. Anatomical heterogeneity of Alzheimer disease: Based on cortical thickness on MRIs. Neurology. 2014;83(21):1936–1944. doi: 10.1212/WNL.0000000000001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byun MS, et al. Alzheimer’s Disease Neuroimaging Initiative Heterogeneity of regional brain atrophy patterns associated with distinct progression rates in Alzheimer’s disease. PLoS One. 2015;10(11):e0142756. doi: 10.1371/journal.pone.0142756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheltens NM, et al. The identification of cognitive subtypes in Alzheimer’s disease dementia using latent class analysis. J Neurol Neurosurg Psychiatry. 2016;87(3):235–243. doi: 10.1136/jnnp-2014-309582. [DOI] [PubMed] [Google Scholar]
- 18.Villemagne VL, et al. Australian Imaging Biomarkers and Lifestyle (AIBL) Research Group Amyloid β deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer’s disease: A prospective cohort study. Lancet Neurol. 2013;12(4):357–367. doi: 10.1016/S1474-4422(13)70044-9. [DOI] [PubMed] [Google Scholar]
- 19.Sperling RA, et al. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albert MS, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rowe CC, et al. Predicting Alzheimer disease with β-amyloid imaging: Results from the Australian imaging, biomarkers, and lifestyle study of ageing. Ann Neurol. 2013;74(6):905–913. doi: 10.1002/ana.24040. [DOI] [PubMed] [Google Scholar]
- 22.Douaud G, et al. Anatomically related grey and white matter abnormalities in adolescent-onset schizophrenia. Brain. 2007;130(Pt 9):2375–2386. doi: 10.1093/brain/awm184. [DOI] [PubMed] [Google Scholar]
- 23.Shaw LM, et al. Alzheimer’s Disease Neuroimaging Initiative Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009;65(4):403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crane PK, et al. Alzheimer’s Disease Neuroimaging Initiative Development and assessment of a composite score for memory in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Brain Imaging Behav. 2012;6(4):502–516. doi: 10.1007/s11682-012-9186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibbons LE, et al. Alzheimer’s Disease Neuroimaging Initiative A composite score for executive functioning, validated in Alzheimer’s Disease Neuroimaging Initiative (ADNI) participants with baseline mild cognitive impairment. Brain Imaging Behav. 2012;6(4):517–527. doi: 10.1007/s11682-012-9176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernal-Rusiel JL, Greve DN, Reuter M, Fischl B, Sabuncu MR. Alzheimer’s Disease Neuroimaging Initiative Statistical analysis of longitudinal neuroimage data with Linear Mixed Effects models. Neuroimage. 2013;66:249–260. doi: 10.1016/j.neuroimage.2012.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou C, Zemanová L, Zamora G, Hilgetag CC, Kurths J. Hierarchical organization unveiled by functional connectivity in complex brain networks. Phys Rev Lett. 2006;97(23):238103. doi: 10.1103/PhysRevLett.97.238103. [DOI] [PubMed] [Google Scholar]
- 29.Bassett DS, et al. Hierarchical organization of human cortical networks in health and schizophrenia. J Neurosci. 2008;28(37):9239–9248. doi: 10.1523/JNEUROSCI.1929-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meunier D, Lambiotte R, Fornito A, Ersche KD, Bullmore ET. Hierarchical modularity in human brain functional networks. Front Neuroinform. 2009;3:37. doi: 10.3389/neuro.11.037.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeo BTT, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106(3):1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie K, Touchon J. Heterogeneity in senile dementia of the Alzheimer type: Individual differences, progressive deterioration or clinical sub-types? J Clin Epidemiol. 1992;45(12):1391–1398. doi: 10.1016/0895-4356(92)90201-w. [DOI] [PubMed] [Google Scholar]
- 33.Young AL, et al. Alzheimer’s Disease Neuroimaging Initiative A data-driven model of biomarker changes in sporadic Alzheimer’s disease. Brain. 2014;137(Pt 9):2564–2577. doi: 10.1093/brain/awu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 35.Corder EH, et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7(2):180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- 36.Lowe VJ, et al. Application of the National Institute on Aging-Alzheimer’s Association AD criteria to ADNI. Neurology. 2013;80(23):2130–2137. doi: 10.1212/WNL.0b013e318295d6cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson RS, Capuano AW, Bennett DA, Schneider JA, Boyle PA. Temporal course of neurodegenerative effects on cognition in old age. Neuropsychology. 2016;30(5):591–599. doi: 10.1037/neu0000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grober E, et al. Memory impairment, executive dysfunction, and intellectual decline in preclinical Alzheimer’s disease. J Int Neuropsychol Soc. 2008;14(2):266–278. doi: 10.1017/S1355617708080302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashburner J, Friston KJ. Voxel-based morphometry--the methods. Neuroimage. 2000;11(6 Pt 1):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 40.Good CD, et al. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001;14(1 Pt 1):21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- 41.Smith SM, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 42.Dole M, Meunier F, Hoen M. Gray and white matter distribution in dyslexia: A VBM study of superior temporal gyrus asymmetry. PLoS One. 2013;8(10):e76823. doi: 10.1371/journal.pone.0076823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pardoe H, Pell GS, Abbott DF, Berg AT, Jackson GD. Multi-site voxel-based morphometry: Methods and a feasibility demonstration with childhood absence epilepsy. Neuroimage. 2008;42(2):611–616. doi: 10.1016/j.neuroimage.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischl B, et al. Sequence-independent segmentation of magnetic resonance images. Neuroimage. 2004;23(Suppl 1):S69–S84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 45.Desikan RS, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 46.Fischl B, et al. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 47.Koch KR. Parameter Estimation and Hypothesis Testing in Linear Models. Springer; Berlin: 1999. [Google Scholar]
- 48.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.