Abstract
The ocular surface is continuously exposed to the environment and, therefore, it is surprising that it harbors only few commensals with low degree of diversity. This unique aspect of the ocular surface physiology prompts the question whether there are core ocular commensal communities and how they affect ocular immunity. The purpose of this review is to provide an overview of what is known about the ocular surface commensals in health and disease and what we would like to learn in the near future. In addition, we discuss how microbiota at sites other than the eye may influence ocular immune responses. The information discussed in the review has been gathered using PubMed searches for literature published from January 1982 to December 2015.
Keywords: contact lenses, IgA, microbiota, ocular surface
I. Is there a core ocular commensal microbiota?
One of the most exciting discoveries made in the 21st century is undoubtedly the discovery of how microbiome affects human health.1 We now know that an average human body harbors as many microbial species as human cells.2 Many studies have linked microbiome to cancer, obesity, asthma, artherosclerosis, and diabetes, illustrating the significance of gut microbiota in health and disease.3–6 The initial studies of gut microbiota were followed by investigations describing core microbiota species at different sites such as skin, urinary tract, and oral mucosal surfaces.7,8 Therefore, the question that naturally arises is “What are the characteristics of the healthy ocular microbiota and how do they change during disease?”
Typically, microbiota is defined as microbial species that are present in the majority of the tested individuals at a particular location. Unlike any other body site, the healthy conjunctiva, lid margins, and tears have remarkably fewer microbial species than what has been reported for other mucosal sites, such as the oral mucosal surface. The most frequently identified species from the conjunctival surfaces in healthy humans are the Coagulase Negative Staphylococcus sp (CNS sp), which include Staphylococcus epidermidis. They are typically isolated from 20–80% of conjunctival swabs and from 30–100 % of swabs from the lid margin areas. Among the less frequently present microbial species are Propionibacterium sp (P. acnes), Corynebacterium sp, S. aureus, Streptococcus sp, Micrococcus sp, Baccilus sp, and Lactobacillus sp. Unlike the above-mentioned gram-positives, the gram-negatives are less frequently detected on the healthy ocular surface. These include P. aeruginosa, Enterobacter sp, E. coli, Proteus sp, and Acinetobacter sp.9–14 The data are mostly based on experiments where moistened ocular cotton swabs were used to sample the ocular tissues and aliquots were allowed to grow on selective agar-based media. What is striking across these studies is the huge variability in the number of samples showing positive bacterial growth, ranging from 16% to 89% (for CNS sp), for it also reveals that a significant number of the ocular swabs contain non-expanding in vitro microbial growth. Only a few studies reported the actual numbers of colony forming units (cfu) measured per individual swab seeding. For example, Ermis et al reported that 80% of the seeded samples showed microbial growth and only 17% of them had more than one microbial species, thus demonstrating that the sustainable in vitro ocular microbiota is not significantly diverse.15 This is in agreement with another study that measured 17–64 cfu per conjunctival swab in 10% of the swabs, 5–16 cfu/swab in 15 % of the swabs, and as few as 0–4 cfu/swab in 75% of the swabs, illustrating the measurable but infrequent bacterial presence in the conjunctival tissues.16 In contrast, lid swabs yielded a high number of cfu: 101–1000 cfu/swab in 3% of the cases, 11–100 cfu/swab in 38% of the swabs, and finally 0–10 cfu/swab in 59% of the swabs.17 Cumulatively, these experiments demonstrated that there is a limited abundance of the in vitro sustainable microbial species at the ocular surfaces, including the lid margin areas, which is remarkably different from what is present in the oral mucosa or saliva.18–21 Consistently, 100% of the swabs taken from the oral mucosa and saliva yielded bacterial presence and contained 107–108 cfu/ml of sustainable bacterial sp.22
The advent of the deep sequencing technique allowed an improved and significantly higher-resolution method for detection of microbial species. In particular, ocular microbiota revealed 12 genera that could be viewed as constituting the core of the conjunctival microbiome. These included Pseudomonas sp (20% of the detected genera), Propionibacterium (20%), Bradyrhizobium (16%), Corynebacteria (15%), Acinetobacter (12%), Brevundomonas (5%), Staphylococcus (4%), Aquabacterium (2%), Sphingomonas (1%), and Streptococcus (1%).23 In these experiments, it was surprising to see high numbers of Pseudomonas sp, because it did not correlate with the data originating from the probing of microbiota using culturing techniques. The elevated presence of P. aeruginosa may have been skewed by the increased abundance of these microorganisms in one of the four tested individuals. This differs from the findings reported by Graham et al.11 In the latter study, 16S ribosomal based sequencing of 57 samples from healthy subject's conjunctiva demonstrated presence of CNS sp, Baccilus sp, Rhodococcus sp, Corynebacterium sp, Propionebacterium sp, Klebsiella sp, and Ervinia sp. The differences between the two studies may also partly be a consequence of using cloned 16S fragments for sequencing leading to overrepresentation of detected species. Consistently, six genera, including Corynebacterium, Streptococcus, Propionibacterium, Staphylococcus, Bacillus, and Ralstonia, were reported present in more than 80% of the surveyed normal healthy conjunctiva in the study cohort from Gambia.24 This cohort did not reveal high relative abundance of Pseudomonas, Bradyrhizobium, and Acinetobacter as previously reported.23,24
In contrast to the low bacterial abundance and diversity detected in the prior studies, Shin et al showed that the conjunctival alpha diversity was significantly higher than that of the skin under the eye,25 suggestive of a more complex commensal repertoire. There was higher abundance of Haemophilus, Streptococcus, Staphylococcus, and Corynebacterium sp in the conjunctiva when compared to the skin of the eye, supportive of the concept of the ocular commensal signature.
Clearly, these findings suggest that the conjunctival commensal repertoire includes Haemophilus, Streptococcus, Staphylococcus, Propionibacterium, and Corynebacterium,11,23,25,26 justifying the need to resolve the lack of correlation between the significant diversity of the commensal community at the conjunctival surfaces detected by deep sequencing and the very limited diversity of bacterial species grown in culture. While there may be recognizable limitations in the culturing protocols, similar experiments using skin, lid margin, or oral mucosal swabs yielded a remarkably higher number of detectable commensal organisms, suggesting that the ocular microbiota is not relatively more abundant. It is also important to address whether bacteria actively colonize the ocular surfaces and replicate there or are only transiently introduced. A potential approach would be to employ transcriptome-based analysis of commensal communities in reconstitution experiments with germ-free mice exposed to different ocular commensals.
One of the limitations of the 16S sequencing approach is the inability to identify down to bacterial species level. Therefore, it is important to utilize alternative approaches, such as transcriptome-based analysis or culturing methods, to characterize the commensals. While a comprehensive, longitudinal quantification of the ocular microbiota is yet to be recognized, especially in the context of disease states, future studies of the ocular microbial communities should not be limited solely to the identification of bacterial species, and the potential presence of viriome should be considered.
One important consideration often overlooked in experiments like those described above is how the microbial presence changes with sex and aging. The majority of studies conducted did not evaluate sex or age-specific differences (cited above) with the exception of those reported in references 24 and 25. Expectedly, age-related changes in the composition of the ocular microbiota were observed among the individuals in the Gambian cohort24 with the diversity of the detected species changing from children (<10 years old) to adults. These observations are consistent with the age-dependent maturation of the immune system, which may define to some extent the composition of the micriobiota. As the strength of the immune system gradually declines with aging, further changes in the microbial communities of the eye are expected in the elderly. Evidence to support this inference comes from experiments with young and aged mice, where a significant trend of increase with age in the number of sustainable in vitro conjunctival species was observed.27
A number of unresolved questions remain. Are the detected sequences representing live bacterial species versus dead bacterial debri? Is the ocular surface colonized long-term, like other mucosal surfaces, by commensals, or is it undergoing continuous clearance via tearing and blinking, resulting in transient exposure? Can the small number of detected (in vitro) viable organisms have significant impact on maintaining ocular immunity? Are there alterations of the core microbial composition of the conjunctiva- and lid-associated microbiota in different disease states? These questions are challenging and also critical, as the number of ocular surface disorders with unknown etiologies is large and growing (e.g., dry eye, episcleritis, chronic follicular conjunctivitis, etc.).
II. What is the impact of contact lens wear on ocular commensals?
Many studies have been conducted to characterize how contact lens use affects ocular microbiota. For detailed information, see the excellent review by Willcox.12 Here we are listing only some of the classic examples, along with the newest observations. Sankaridurg et al employed longitudinal monitoring of microbial presence at the conjunctiva-lid margin area and reported that in contact lens-wearing children age 8–14, microbial growth was detected in 36% of the conjunctival swabs and in 54% of the lid swabs.28 The microbial types detected were CNS, Propionibacterium sp, Bacillus sp, Streptococcus sp, Micrococcus sp, S. aureus, and Corynebacterium sp (in the order of frequency). There were no differences in the microbial types recovered from non-contact lens wearers and wearers of HEMA-based soft lenses for a period of 2 years.29 In contrast, in adults, daily wear of soft contact lenses over a period of one year promoted alterations in the conjunctival microbiota by increasing the number of isolated commensal organisms.30 Consistently, an increase in the viable in vitro bacteria, including Corynebacterium sp and Propionibacterium acnes, were observed in the eyes of former contact lens wearers when compared to the control group.31 The extended wear of HEMA-based hydrogel contact lenses further expanded the conjunctival and lid margin microbiota. Individuals carrying gram-positive bacteria on lenses such as CNS and Corynebacterium sp were more likely to develop contact lens-induced peripheral ulcers, whereas carriers of gram-negative bacteria on lenses were more likely to develop contact-lens-induced acute red eye.29 Similarly, extended wear of contact lenses was associated with an increased number of pathogenic organisms in the conjunctival tissues.32
Utilizing 16S rRNA metagenomic sequencing to analyze samples from contact lens wearers versus non-lens wearers, Shin et al observed a shift in the microbiota of the conjunctiva of the wearers towards relatively higher abundance of Methylobacterium, Lactobacillus, Acinetobacter, and Pseudomonas, and lower relative abundance of Corynebacterium, Staphylococcus, Streptococcus, and Haemophilus in contact lens wearers compared to the controls, suggesting that contact lens wear altered ocular microbiota towards skin-like microbiota.25
In summary, the influence of contact lens wear on the microbial commensal communities of the eye depends on the type of contact lenses, the duration of their wear (e.g., daily wear versus extended wear), and the age group. These studies prompt several questions: Does the relative loss of commensal diversity consequent to the contact lens wear alter the ocular immunity to sensitize to infection? Does contact lens wear change epithelial responses to facilitate bacterial adhesion, as suggested in reference 33? Does contaminating the contact lenses with bacterial species from the skin promote the development of infection?25,33
III. Is there an association between ocular microbiota and ocular surface disease?
Proteomic analysis of tear fluid from patients with dry eye disease showed specific alterations in the protein signature.34 Interestingly, several of the downregulated proteins had bactericidal activities, for example, lactotransferrin, lysozyme, polymeric immunoglobulin receptor, and lacritin.34 These data justify questioning whether there is a connection between the ocular commensal microbiota and the state of the ocular surface barrier. To date, only few studies have attempted to address this issue. Albeitz and Lenton reported more extensive bacterial loads in patients with Sjogren Syndrome than in healthy controls.35 Similarly, Graham et al reported increased bacterial presence of CNS species in addition to other common commensals such as Corynebacterium and Propionibacterium in non-autoimmune dry eye disease.11 Promisingly, the expanding applicability of deep sequencing approaches will provide us with insights to whether alterations of the ocular surface microbiota correlate with the development of dry eye disease.36
IV. What is the impact of the microbiome on ocular immunity?
One of the mechanisms of commensal-mediated protection of mucosal sites such as the gut is competing the pathobiont.37 Given that the ocular surface is paucibacterial, it is unlikely that the bacterial presence will interfere with the colonization of the mucosal surfaces. Thus, the impact of local ocular microbiota on ocular health may be very different from what has been reported for other mucosal surfaces. A possible scenario would present microbial stimulation of local immune responses. Currently, it is unclear whether local microbial communities have an effect on the immune responses of the eye. Because of the low abundancy and diversity of ocular microbiota, it is tempting to speculate that in health there is little impact of the ocular microbiota on ocular immunity. However, this should not negate the impact of microbiota resident at other sites on the immune responses occurring in the eye.
A. Impact of microbiome on IgA production
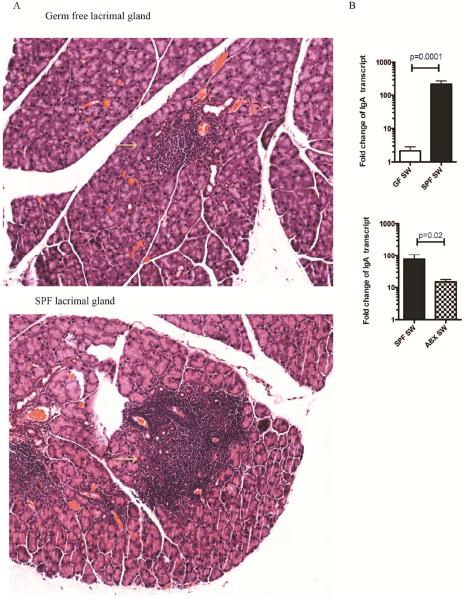
The predominant antibody at the ocular mucosal surface is secretory IgA.38,39 It is critical for maintaining mucosal homeostasis, as it neutralizes toxins, viruses, and bacteria; it has anti-inflammatory activities, promotes production of interleukin (IL)-10, and affects maturation of dendritic cells.40–43 Through these pleiotropic effects, secretory IgA induces tolerance at the mucosal sites. The generation of secretory IgA occurs through class-switch recombination of the Ig heavy chains. After the migration of the naïve B cells that express surface IgD and IgM from the bone marrow, further development of B cells occurs in the germinal centers of secondary lymphoid tissues through somatic hypermutation and class switching.44 The class-switching events replace the Ig heavy chains from the IgD and IgM to either IgA, IgG, or IgE. The IgA class switching is determined through T-cell dependent (TD) and T-cell–independent mechanisms.45 The TD mechanisms require cognate interactions between antigen-specific B cells and activated antigen-specific CD4+ T cells.46 These events can occur in the eye associated lymphoid tissues (EALT), such as those present in the conjunctiva or lacrimal glands.47 In pioneering experiments, Allansmith et al showed that the exposure of germ-free rats to the conventional environment resulted in a significant increase in the number of IgA- and IgM-producing B cells in the lacrimal glands. The conventionalization of the germ-free rats took four weeks at which point the tear levels of secretory IgA in the ex-germ-free rats reached levels comparable to those of the regular rats.48 While these studies did not specifically examine the impact of ocular microbiota on the generation of secretory IgA and IgM, they clearly showed a pronounced influence of the environment on B cell activation in the lacrimal glands, suggesting that the commensal communities may be important in promoting this process. We made similar observations in the germ-free mice (Figure1), where lacrimal glands had significantly diminished lymphoid infiltrates, lower levels of secretory IgA heavy chain transcripts, and lower levels of secretory IgA protein at the ocular surface (Table 1). Furthermore, the ocular surface levels of secretory IgA were decreased upon oral antibiotic treatment of conventional Swiss-Webster (SW) mice (Figure 1),49 confirming that commensal microbiome stimulates secretory IgA presence in tear film. These experiments prompted an important question, namely, whether ocular surface microbiota regulates B cell activation in the eye EALT. To this end, Sullivan et al showed that unilateral tarsorrhaphy of newborn rats before the eyelid opening did not influence the frequency of the lacrimal gland B cells, concluding that environmental antigen exposure at the ocular surface did not regulate B cell activation. These data also suggest that the commensal trigger may occur at a site different than the ocular surface. One potential approach to clarify this issue is through application of topical antibiotics versus oral antibiotics in conventional mice at the time of lid opening and evaluating the number and maturation state of B cells in the lacrimal glands.
Figure 1.
Microbiota drives ocular surface IgA levels. A. Haematoxylin-eosin staining of lacrimal gland sections derived from conventional and germ--free Swiss-Webster (SW) mice. The individual lymphoid follicles are circled. The lacrimal glands from germ-free mice contain decreased presence of lymphocytes in the lymphoid follicles, signifying the role of microbiota in the formation of EALT. B. Germ-free mice have significantly lower levels of IgA transcripts in the lacrimal glands compared to conventional mice. Quantitative RT-PCR assay was used to compare the levels of IgA heavy chain transcripts between the individual cohorts of mice. The data are representative of two experiments including at least 5-7 mice per group. Mann Whitney test, P=.0001. C. Oral administration of antibiotic cocktail decreases significantly lacrimal gland IgA transcripts, signifying the role of bacterial driven-IgA synthesis. The data are representative of two experiments including at least 5-7 mice per group. Mann Whitney test, P=.02.
Table 1.
Quantitative LC-MS/MS analysis of the immunoglobulin peptides identified in the ocular surface washes of GF and SPF mice.
| Peptide identified | Fold change SPF/GF | p-value |
|---|---|---|
| Ig kappa V chain | 2.9 | 8.50783E-05 |
| Ig alpha chain C region | 2.4 | 0.004827752 |
| Ig kappa chain C region | 2.8 | 0.003947885 |
In addition, it is important to determine how ocular secretory IgA repertoire is dependent on commensal ocular, nasal, or gut microbiota. The low number of mutations detected in the class-switched transcripts in B cells in the lacrimal glands suggest that the majority of B cells in the lacrimal glands are effector cells, not undergoing activation and proliferation,50 providing evidence for the hypothesis that in health the microbiota-driven secretory IgA- committed B cells can be recruited from the nasal- or gut-associated mucosal tissues.
Unlike the lack of mechanistic understanding of how microbiota governs the production of secretory IgA-synthesizing B cells in the lacrimal glands, similar events in the lungs and the gut are known to depend on Toll-like receptor (TLR) signaling as the MyD88 and TRIF double KO mice show diminished IgA generation.42,51,52 Upon TLR activation, epithelial and dendritic cells release B-cell activating factor (BAFF) and proliferation-inducing ligand (APRIL), thereby promoting T-cell independent IgA class switching.53 These findings gave rise to the concept that T-independent secretory IgA production is important for protection against commensal intestinal microbes, while T-dependent secretory IgA are important for protection against pathogens. Similar mechanisms operate in the lungs, where the CD103+ CD24+CD11b+ dendritic cells induced IgA class-switch recombination to activate B cells through TD and T-cell–independent pathways51 that included retinoic acid and TGF-β stimulation of antigen-specific IgA class switching.51–54 It is highly likely that in the EALT alterations in the microbiota can regulate secretory IgA repertoire.
In summary, it is clear that the ocular secretory IgA levels depend on the commensal microbiome, reinforcing the tolerance to ocular commensals and providing much needed neutralization of the ocular surface antigens; however we still need information about the mechanisms of these processes in the eye.
B. Impact of microbiome on innate immunity functions
The microbiome-mediated maturation of immune cell functions is required for protection against ocular pathogens. Analysis of the mechanisms of protection mediated by Staphylococcus aureus-specific monoclonal antibody during keratitis revealed that the germ-free mice treated with the therapeutic antibody failed to mount a protective immunity against this pathogen, due to failure to recruit inflammatory cells to the site of infection.55 The lack of protective efficacy of the monoclonal antibody in infected RAG mice or CD4+ cell–depleted mice suggested stipulation of T cell functions. Consistently, there was a dependence on IL-17 and IL-22 in the antibody-induced protection.55 This paper prompted many important questions. Namely, what is the impact of microbiota on neutrophil maturation and activation and where does neutrophil priming occur? What is the interplay between neutrophils and commensal-primed CD4+ cells? How do IL-17 and IL-22 alter neutrophil recruitment to the cornea during infection, and how does microbiota-mediated priming of neutrophil activity alter the host responses to other pathogens at the ocular tissues? The answers are pending, but some insights may be gleaned from mechanisms and pathways characterizing the impact of microbiota at sites different than the eye.
Microbiota is a source for peptidoglycans that systemically prime the neutrophils for opsonophagocytic-mediated bactericidal activity against Staphylococcus aureus and Streptococcus pneumoniae.56 This phenomenon was dependent on nucleotide-binding oligomerization domain containing protein-1 (NOD1), but not on Toll-like receptor 4 (TLR4) and nucleotide-binding oligomerization domain containing protein-2 (NOD2). Consistently, in vitro priming of human neutrophils with meso-diaminopimelic acid–containing peptidoglycan, a ligand for NOD1, improved the opsonophagocytic bactericidal activity of human neutrophils against Staphylococcus aureus and Streptococcus pneumoniae. These findings highlight a direct effect of microbiota-derived products on neutrophil activity.
Furthermore, Deshmukh et al reported an alternative mechanism for microbiota-mediated regulation of neutrophil functions. In these studies, commensal microbiota protected neonatal mice against E. coli K1–induced sepsis by promoting granulopoiesis via IL-17 release from group 3 innate lymphoid cells (ILC-3). Specific depletion of the innate lymphoid cells abrogated IL-17 and emanating granulocyte colony-stimulating factor (G-CSF)–dependent granulopoiesis.57 The ILC-3 produced IL-17A in response to gut-derived LPS. However, unlike in the case of peptidoglycan treatment, PMNs derived from the antibiotic-treated mice had bactericidal activities against E. coli not different from the PMNs derived from the commensal-exposed animals, suggesting that the mechanism of commensal-induced protection was different. Therefore, it appears that the effects of microbiota on regulating neutrophil generation and activation status may be redundant and context-dependent. While further work is needed to characterize the commensal site-dependent impact on local immune responses, one possible scenario that is worth investigating is the development of more efficient neutrophil priming as a result of the effect of local microbiota on the activities of ILCs, Th17, and γδT cells through, and the production of IL-17 and/or IL-22.
V. What is the influence of microbiome on ocular autoimmunity?
As discussed above, whether or not the commensal presence at the ocular surface changes during chronic inflammation remains an open question. To address this issue, we compared the conjunctival microbial presence in healthy C57BL6 mice and age- and sex-matched thrombospondin-1 (TSP-1) knockout (TSP KO) mice, which are known to have chronic conjunctival inflammation consistent with Sjogren syndrome–like disease.27 We observed that the commensal frequency increased with the aging of the TSP-1 KO mice, which correlated with spontaneous disease development, suggesting that the elevated presence of commensals might serve as exacerbating factors for disease progression.
Using microbiological typing technique, we identified that CNS xylosus, a commensal opportunistic pathobiont, had elevated prevalence in the conjunctiva of the TSP-1 KO. The measurements of elevated anti-CNS xylosus specific antibody IgG titers in the blood circulation of the TSP-1 KO in comparison to the wild type controls verified that the loss of ocular barrier and tolerance to this commensal occurred during chronic inflammation. The increased abundance of the CNS xylosus may be attributed to the increased fitness of these microorganisms to the altered tear film conditions characterized with elevated concentration of salt. Interestingly, the increased presence of CNS sp in the TSP-1 KO mice resembled what was reported by Lenton et al, who showed elevated numbers of CNS and Staphylococcus aureus species isolated from the lid and conjunctiva in primary Sjogren syndrome patients.35 The enriched commensal presence in the TSP-1 KO mice followed the secretory dysfunction of lacrimal glands and the decrease of the conjunctival goblet cell population, suggesting that it was co-occurring with the alterations in the ocular surface barrier composition in these animals. These data justify the need to evaluate how ocular commensal presence is changing in other forms of autoimmune and non-autoimmune dry eye disease and define how commensals contribute to the development of the ocular surface inflammatory responses.
Appreciation of the essential connection between gut microbiota and ocular autoimmunity comes from the elegant studies done by Caspi and colleagues.58 Autoreactive retinal antigen-specific T cells drive autoimmune uveitis, but it is unclear how retina-specific T cells become activated. The authors used the R161H mice expressing a transgenic T cell receptor (TCR) specific for the interphotoreceptor retinoid-binding protein (IRBP), an autoantigen associated with uveitis, and showed that the autoreactive T-cells become primed by microbiota in the gut. In keeping with this, treatment with broad-range antibiotics reduced the inflammation of the eye. Compared with wild type, the R161H mice showed increased frequency of IL-17-producing IRBP-specific T-cells in the gut, which were decreased upon antibiotic treatment. It is gratifying to see this data, given that Th17 cells are pathogenic in experimental autoimmune uveitis models, and the study clearly demonstrates a dependence of these pathogenic T-cells on microbiota. While the intestinal antigen involved in the activation of the IRBP-specific T-cells is yet to be identified, the authors suggested that the stimulant is likely a protein component of microbiota. Consistent with this, R161H T cells cultured with protein preparation from the intestinal microbiota derived from the diseased mice caused uveitis when transferred to the wild type recipient mice, whereas control R161H T cells cultured without this stimulation did not promote the disease. These experiments clearly demonstrate a connection between gut microbiota, its impact on CD4+ T cell activation, and development of ocular autoimmunity. We are eagerly awaiting follow- up studies that will shed light on the identity of the commensals that trigger this pathology.
In conclusion, exciting new data is accumulating to signify the impact of microbiota in promoting immune system activation and ensuring the maintenance of homeostasis. Understanding how commensals govern innate and adaptive immune activation will allow us to produce improved vaccines to fight infectious diseases and find alternative strategies to combat autoimmune states.
VI. Key Questions for Future Investigations
Is the ocular surface long-term colonized, like other mucosal surfaces, by commensals or is it undergoing continuous clearance via tearing and blinking resulting in transient exposure?
Can the small number of detected in vitro viable organisms have significant impact on maintaining ocular immunity?
Are there alterations of the core microbial composition of the conjunctiva- and lid margin-associated microbiota in different disease states?
Does the relative loss of commensal diversity consequent to the contact lens wear alter the ocular immunity to sensitize to infection?
What is the impact of microbiota on immune responses during ocular infections? Is there a role for commensal species to regulate neutrophil priming, where does it occur, how does it affect susceptibility to the different types of pathogens?
How does ocular commensal presence change in autoimmune and non-autoimmune dry eye disease and what is its impact on ocular inflammation?
Outline.
Is there a core ocular commensal microbiota?
What is the impact of contact lens wear on ocular commensals?
Is there an association between ocular microbiota and ocular surface disease?
-
What is the impact of the microbiome on ocular immunity?
Impact of microbiome on IgA production
Impact of microbiome on innate immunity functions
What is the influence of microbiome on ocular autoimmunity?
Key questions for future investigations
Acknowledgments
Sources of support: The authors are supported by Public Health Service grant EY022054 (to MG) from the National Eye Institute and American Association of Immunology Fellowship grant (to MG and AK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no commercial or proprietary interest in any concept or product discussed in this article.
INSTRUCTION TO COMPOSITOR: Because most of the section heads are formatted as complete sentences (questions), please do not follow usual style of capitalizing all words in heading. Capitalize first word only, as shown.
References
- 1.Peterson CT, Sharma V, Elmen L, Peterson SN. Immune homeostasis, dysbiosis and therapeutic modulation of the gut microbiota. Clin Exp Immunol. 2015;179:363–77. doi: 10.1111/cei.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164:337–40. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 3.Viaud S, Daillere R, Boneca IG, et al. Gut microbiome and anticancer immune response: really hot Sh*t! Cell Death Differ. 2015;22:199–214. doi: 10.1038/cdd.2014.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohtani N. Microbiome and cancer. Sem Immunopathol. 2015;37:65–72. doi: 10.1007/s00281-014-0457-1. [DOI] [PubMed] [Google Scholar]
- 5.Sanz Y, Olivares M, Moya-Perez A, Agostoni C. Understanding the role of gut microbiome in metabolic disease risk. Pediatr Res. 2015;77:236–44. doi: 10.1038/pr.2014.170. [DOI] [PubMed] [Google Scholar]
- 6.SanMiguel A, Grice EA. Interactions between host factors and the skin microbiome. Cell Mol Life Sci. 2015;72:1499–515. doi: 10.1007/s00018-014-1812-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whiteside SA, Razvi H, Dave S, et al. The microbiome of the urinary tract--a role beyond infection. Nat Rev Urol. 2015;12:81–90. doi: 10.1038/nrurol.2014.361. [DOI] [PubMed] [Google Scholar]
- 8.He J, Li Y, Cao Y, et al. The oral microbiome diversity and its relation to human diseases. Folia Microbiol (Praha) 2015;60:69–80. doi: 10.1007/s12223-014-0342-2. [DOI] [PubMed] [Google Scholar]
- 9.Campos MS, Campos e Silva Lde Q, Rehder JR, et al. Anaerobic flora of the conjunctival sac in patients with AIDS and with anophthalmia compared with normal eyes. Acta Ophthalmol. 1994;72:241–5. doi: 10.1111/j.1755-3768.1994.tb05023.x. [DOI] [PubMed] [Google Scholar]
- 10.Elander TR, Goldberg MA, Salinger CL, et al. Microbial changes in the ocular environment with contact lens wear. CLAO J. 1992;18:53–5. [PubMed] [Google Scholar]
- 11.Graham JE, Moore JE, Jiru X, et al. Ocular pathogen or commensal: a PCR-based study of surface bacterial flora in normal and dry eyes. Invest Ophthalmol Vis Sci. 2007;48:5616–23. doi: 10.1167/iovs.07-0588. [DOI] [PubMed] [Google Scholar]
- 12.Willcox MD. Characterization of the normal microbiota of the ocular surface. Exp Eye Res. 2013;117:99–105. doi: 10.1016/j.exer.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Hsu HY, Lind JT, Tseng L, Miller D. Ocular flora and their antibiotic resistance patterns in the midwest: a prospective study of patients undergoing cataract surgery. Am J Ophthalmol. 2013;155:36–44 e2. doi: 10.1016/j.ajo.2012.06.024. [DOI] [PubMed] [Google Scholar]
- 14.Ozkan J, Zhu H, Gabriel M, et al. Effect of prophylactic antibiotic drops on ocular microbiota and physiology during silicone hydrogel lens wear. OptomVis Sci. 2012;89:326–35. doi: 10.1097/OPX.0b013e318243280e. [DOI] [PubMed] [Google Scholar]
- 15.Ermis SS, Aktepe OC, Inan UU, et al. Effect of topical dexamethasone and ciprofloxacin on bacterial flora of healthy conjunctiva. Eye (Lond) 2004;18:249–52. doi: 10.1038/sj.eye.6700631. [DOI] [PubMed] [Google Scholar]
- 16.Hart DE, Hosmer M, Georgescu M, Farris RL. Bacterial assay of contact lens wearers. OptomVis Sci. 1996;73:204–7. doi: 10.1097/00006324-199603000-00014. [DOI] [PubMed] [Google Scholar]
- 17.Larkin DF, Leeming JP. Quantitative alterations of the commensal eye bacteria in contact lens wear. Eye (Lond) 1991;5(Pt 1):70–4. doi: 10.1038/eye.1991.14. [DOI] [PubMed] [Google Scholar]
- 18.Nasidze I, Li J, Quinque D, et al. Global diversity in the human salivary microbiome. Genome Res. 2009;19:636–43. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avila M, Ojcius DM, Yilmaz O. The oral microbiota: living with a permanent guest. DNA Cell Biol. 2009;28:405–11. doi: 10.1089/dna.2009.0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen T, Yu WH, Izard J, et al. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database. 2010;2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evaldson G, Heimdahl A, Kager L, Nord CE. The normal human anaerobic microflora. Scand J Infect Dis Suppl. 1982;35:9–15. [PubMed] [Google Scholar]
- 23.Dong Q, Brulc JM, Iovieno A, et al. Diversity of bacteria at healthy human conjunctiva. Invest Ophthalmol Vis Sci. 2011;52:5408–13. doi: 10.1167/iovs.10-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Holland MJ, Makalo P, et al. The conjunctival microbiome in health and trachomatous disease: a case control study. Genome Med. 2014;6:99. doi: 10.1186/s13073-014-0099-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin H, Price K, Albert L, et al. Changes in the eye microbiota associated with contact lens wearing. mBio. 2016;7 doi: 10.1128/mBio.00198-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SH, Oh DH, Jung JY, et al. Comparative ocular microbial communities in humans with and without blepharitis. Invest Ophthalmol VisSci. 2012;53:5585–93. doi: 10.1167/iovs.12-9922. [DOI] [PubMed] [Google Scholar]
- 27.Terzulli M, Ruiz LC, Kugadas A, et al. TSP-1 deficiency alters ocular microbiota: implications for Sjogren's syndrome pathogenesis. J Ocul Pharmacol Ther. 2015;31:413–8. doi: 10.1089/jop.2015.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sankaridurg PR, Markoulli M, de la Jara PL, Harmis N, Varghese T, Willcox MD, et al. Lid and conjunctival micro biota during contact lens wear in children. Optom Vis Sci. 2009;86:312–7. doi: 10.1097/opx.0b013e318199d20c. [DOI] [PubMed] [Google Scholar]
- 29.Zhong W, Zhou Z. Alterations of the gut microbiome and metabolome in alcoholic liver disease. World J Gastrointest Pathophysiol. 2014;5:514–22. doi: 10.4291/wjgp.v5.i4.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang YE, Wang Y, He Y, et al. Homogeneity of the vaginal microbiome at the cervix, posterior fornix, and vaginal canal in pregnant Chinese women. Microb Ecol. 2015;69:407–14. doi: 10.1007/s00248-014-0487-1. [DOI] [PubMed] [Google Scholar]
- 31.Callender MG, Tse LS, Charles AM, Lutzi D. Bacterial flora of the eye and contact lens. Cases during hydrogel lens wear. Am J Optom Physiol Opt. 1986;63:177–80. doi: 10.1097/00006324-198603000-00002. [DOI] [PubMed] [Google Scholar]
- 32.Fleiszig SM, Efron N. Microbial flora in eyes of current and former contact lens wearers. J Clin Microbiol. 1992;30:1156–61. doi: 10.1128/jcm.30.5.1156-1161.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fleiszig SM, Efron N, Pier GB. Extended contact lens wear enhances Pseudomonas aeruginosa adherence to human corneal epithelium. Invest Ophthalmol Vis Sci. 1992;33:2908–16. [PubMed] [Google Scholar]
- 34.Karnati R, Laurie DE, Laurie GW. Lacritin and the tear proteome as natural replacement therapy for dry eye. Exp Eye Res. 2013;117:39–52. doi: 10.1016/j.exer.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albietz JM, Lenton LM. Effect of antibacterial honey on the ocular flora in tear deficiency and meibomian gland disease. Cornea. 2006;25:1012–9. doi: 10.1097/01.ico.0000225716.85382.7b. [DOI] [PubMed] [Google Scholar]
- 36.Narayanan S, Redfern RL, Miller WL, et al. Dry eye disease and microbial keratitis: is there a connection? Ocul Surf. 2013;11:75–92. doi: 10.1016/j.jtos.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivanov II, Honda K. Intestinal commensal microbes as immune modulators. Cell Host Microbe. 2012;12:496–508. doi: 10.1016/j.chom.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macpherson AJ, Koller Y, McCoy KD. The bilateral responsiveness between intestinal microbes and IgA. Trends Immunol. 2015;36:460–70. doi: 10.1016/j.it.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Wells PA, Hazlett LD. Immunocytochemical localization of immunoglobulins at the corneal surface of the mouse. Exp Eye Res. 1985;40:779–96. doi: 10.1016/0014-4835(85)90123-x. [DOI] [PubMed] [Google Scholar]
- 40.Cerutti A. The regulation of IgA class switching. Nat Rev Immunol. 2008;8:421–34. doi: 10.1038/nri2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazanec MB, Nedrud JG, Kaetzel CS, Lamm ME. A three-tiered view of the role of IgA in mucosal defense. Immunol Today. 1993;14:430–5. doi: 10.1016/0167-5699(93)90245-G. [DOI] [PubMed] [Google Scholar]
- 42.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–39. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 43.Pilette C, Detry B, Guisset A, et al. Induction of interleukin-10 expression through Fcalpha receptor in human monocytes and monocyte-derived dendritic cells: role of p38 MAPKinase. Immunol Cell Biol. 2010;88:486–93. doi: 10.1038/icb.2009.120. [DOI] [PubMed] [Google Scholar]
- 44.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–52. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 45.Casola S, Otipoby KL, Alimzhanov M, et al. B cell receptor signal strength determines B cell fate. Nat Immunol. 2004;5:317–27. doi: 10.1038/ni1036. [DOI] [PubMed] [Google Scholar]
- 46.Dullaers M, Li D, Xue Y, et al. A T cell-dependent mechanism for the induction of human mucosal homing immunoglobulin A-secreting plasmablasts. Immunity. 2009;30:120–9. doi: 10.1016/j.immuni.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gill RF, Pirockinaite G, O'Sullivan NL, Montgomery PC. Nasal-associated lymphoid tissue is not an absolute requirement for the induction of rat tear IgA antibody responses. Curr Eye Res. 2010;35:1–8. doi: 10.3109/02713680903395281. [DOI] [PubMed] [Google Scholar]
- 48.Allansmith MR, Gudmundsson OG, Hann LE, et al. The immune response of the lacrimal gland to antigenic exposure. Curr Eye Res. 1987;6:921–7. doi: 10.3109/02713688709034860. [DOI] [PubMed] [Google Scholar]
- 49.Ng KM, Ferreyra JA, Higginbottom SK, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–9. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin M, Du L, Brandtzaeg P, Pan-Hammarstrom Q. IgA subclass switch recombination in human mucosal and systemic immune compartments. Mucos Immunol. 2014;7:511–20. doi: 10.1038/mi.2013.68. [DOI] [PubMed] [Google Scholar]
- 51.Ruane D, Chorny A, Lee H, et al. Microbiota regulate the ability of lung dendritic cells to induce IgA class-switch recombination and generate protective gastrointestinal immune responses. J Exp Med. 2016;213:53–73. doi: 10.1084/jem.20150567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Neill LA, Golenbock D, Bowie AG. The history of Toll-like receptors - redefining innate immunity. Nat Rev Immunol. 2013;13:453–60. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- 53.He B, Santamaria R, Xu W, et al. The transmembrane activator TACI triggers immunoglobulin class switching by activating B cells through the adaptor MyD88. Nat Immunol. 2010;11:836–45. doi: 10.1038/ni.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tezuka H, Abe Y, Iwata M, et al. Regulation of IgA production by naturally occurring TNF/iNOS-producing dendritic cells. Nature. 2007;448:929–33. doi: 10.1038/nature06033. [DOI] [PubMed] [Google Scholar]
- 55.Zaidi T, Zaidi T, Cywes-Bentley C, Lu R, Priebe GP, Pier GB. Microbiota-driven immune cellular maturation is essential for antibody-mediated adaptive immunity to Staphylococcus aureus infection in the eye. Infect Immunol. 2014;82:3483–91. doi: 10.1128/IAI.01951-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–31. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deshmukh HS, Liu Y, Menkiti OR, et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med. 2014;20:524–30. doi: 10.1038/nm.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horai R, Zarate-Blades CR, Dillenburg-Pilla P, et al. Microbiota-dependent activation of an autoreactive T cell receptor provokes autoimmunity in an immunologically privileged site. Immunity. 2015;43:343–53. doi: 10.1016/j.immuni.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]