Abstract
Most bacteria in nature exist as biofilms, which support intercellular signaling processes such as quorum sensing (QS), a cell-to-cell communication mechanism that allows bacteria to monitor and respond to cell density and changes in the environment. Because QS and biofilms are involved in the ability of bacteria to cause disease, there is a need for the development of methods for the non-invasive analysis of QS in natural bacterial populations. Here, by using surface-enhanced resonance Raman scattering spectroscopy, we report rationally designed nanostructured plasmonic substrates for the in-situ, label-free detection of a QS signaling metabolite in growing Pseudomonas aeruginosa biofilms and microcolonies. The in situ, non-invasive plasmonic imaging of QS in biofilms provides a powerful analytical approach for studying intercellular communication on the basis of secreted molecules as signals.
Quorum Sensing (QS) is a cell-to-cell communication phenomenon used by bacteria to monitor their local environment and population density by producing, secreting and sensing small diffusible signals called autoinducers, resulting in the synchronous regulation of gene expression on a population-wide scale.1, 2 In numerous bacterial genera, QS-regulated genes have been associated to a wide range of physiological processes such as cell differentiation, antibiotic production, stress tolerance, virulence or biofilm formation, all of which are crucial for survival and pathogenicity.3 Thus, understanding and controlling this chemical communication system could lead to medical and industrial applications.4 The elucidation of QS systems is based on planktonic (free-swimming) bacteria grown in shaken liquid cultures, given that bacteria are physiologically similar under these growth conditions. However, in natural environments most bacterial populations exist as densely packed multicellular aggregates embedded in self-produced extracellular polymeric substances (EPS) often referred to as biofilms. Bacterial biofilms represent a protected mode of growth that has been shown to profoundly affect human health and industrial productivity.5 In this environment, biofilm bacteria are distinct from their planktonic counterparts in terms of genetic program and physiology.6 Remarkably, although the high cell density within biofilms provides an optimal environment for intercellular signaling processes including QS, chemical communication in this habitat is also strongly influenced by numerous biological, chemical and physical parameters. 3, 7, 8
Pseudomonas aeruginosa (P. aeruginosa) is an opportunistic human pathogen responsible for acute and chronic infections, particularly in individuals with compromised immune systems and host defenses. Several lines of evidence indicate that the persistence and antibiotic resistance of this bacterium are due to its inherent capacity to form biofilms, which are often found in the lungs of CF patients, chronic wounds or in medical foreign bodies or indwelling devices.9 The QS network of P. aeruginosa is known to regulate the expression of key virulent factors that can contribute to the bacterial pathological process.10 Nevertheless, QS-deficient clinical isolates occur and are still capable of causing infections in humans.11 Due to the links between QS, biofilms and virulence, together with its clinical relevance, P. aeruginosa has become a model system to study these two important aspects of bacterial social behavior. However, despite much research, fundamental questions still remain unanswered regarding the role and mechanisms of QS signaling processes in the context of biofilms,8, 12 as well as their relevance in human infections.11
Traditionally, optical monitoring of QS dynamics employ bacterial biosensors bearing heterologous reporter genes that express bioluminescent or fluorescent proteins in response to QS signaling molecules.13, 14 Although this approach has greatly expanded our understanding of QS, it involves the use of genetically modified bacteria. Thus, the development of alternative methods to detect and image QS-regulated processes with no need for genetic manipulation or labeling is highly desirable toward understanding this form of bacterial communication in natural populations.
Surface-enhanced Raman scattering (SERS) spectroscopy is an ultrasensitive analytical technique15, 16, 17 that can be applied non-invasively for label-free detection and imaging of a wide range of molecules.18, 19, 20 SERS allows identification of the specific spectral fingerprint of a probe molecule in contact with a plasmonic nanostructure and its sensitivity can go as far as the single-molecule level,21 in particular when the frequency of the excitation laser is in resonance with an electronic transition of the molecule, which is known as surface-enhanced resonance Raman scattering (SERRS). Importantly, SERS/SERRS offers unambiguous identification of analytes, multiplexing capability, requires little or no sample preparation and provides high spatial resolution. The implementation of plasmonic nanoparticles as SERS sensors has set the basis for numerous high-performance in vitro analytical bioassays.22, 23 However, due to inherent limitations of SERS,19 the direct detection of target analytes in complex biological environments using plasmonics is still in its infancy. One of such limitations is related to the required access to the metal surface in the plasmonic substrate of the relevant analyte, while avoiding signal contamination by other biomolecules, such as proteins, lipids, etc.
We focus this work on pyocyanin, a heterocyclic nitrogen-containing compound of the phenazine family produced by P. aeruginosa, which is excreted into the environment where it displays a wide variety of biological activities.24, 25 This phenazine acts as an antibiotic and as a virulence factor in infected hosts, which is in general due to its capacity to generate reactive oxygen species. Remarkably, pyocyanin functions as an intercellular signaling molecule in the QS network of P. aeruginosa26 and plays important roles in biofilm morphogenesis in this organism.10, 27, 28, 29 The biosynthesis of pyocyanin is regulated by the interdependent QS circuits of P. aeruginosa in response to environmental factors including cell density.10, 30, 31 Since pyocyanin expression is controlled by QS and the possibility of SERS detection has been reported,32 we implemented a plasmonic approach toward in situ SERRS detection and imaging of pyocyanin, as a proxy of QS, in biofilms and microcolonies of P. aeruginosa. Our strategy involves the use of hybrid materials comprising a plasmonic component within a porous matrix that allows diffusion of small molecules only.33 With this concept in mind and aiming at providing different analytical tools to investigate this form of bacterial communication in live bacterial sessile communities, we fabricated three types of cell-compatible plasmonic platforms (Fig. 1). Macroporous poly-N-isopropylacrylamide (pNIPAM) hydrogels loaded with Au nanorods (Au@pNIPAM), devised as a highly porous platform with enhanced diffusivity, lead to plasmonic detection of pyocyanin homogenously in both colonized and non-colonized regions of the substrate. On the other hand, mesostructured Au@TiO2 substrates bearing a mesoporous TiO2 thin film over a sub-monolayer of Au nanospheres restricted pyocyanin detection to biofilm-colonized surfaces with a spatial resolution of ca. 20 μm. This platform allowed us to generate SERRS maps to reveal variation of QS plasmonic signal in bacterial biofilms up to millimeter-scale areas. Finally, mesoporous silica-coated micropatterned supercrystal arrays of Au nanorods (Au@SiO2), with extremely high electromagnetic enhancement factor, enabled plasmonic detection of QS-behavior (e.g. pyocyanin expression) at early stages of biofilm formation and allowed imaging of the phenazine produced by small clusters of bacteria colonizing micron-sized plasmonic features (25 μm2 on average). Details on substrate fabrication, characterization and analysis of the SERS performance with model analytes are provided in the Methods and Sections S1 and S2 in Supplementary Information (SI) section. This approach, combining purpose-designed nanostructured hybrid materials and SERS, provides an efficient and versatile tool for label-free molecular detection of QS communication in live microbial communities and may thus contribute to understanding biofilm sociomicrobiology and other cellular processes mediated by small, SERS active biomolecules.
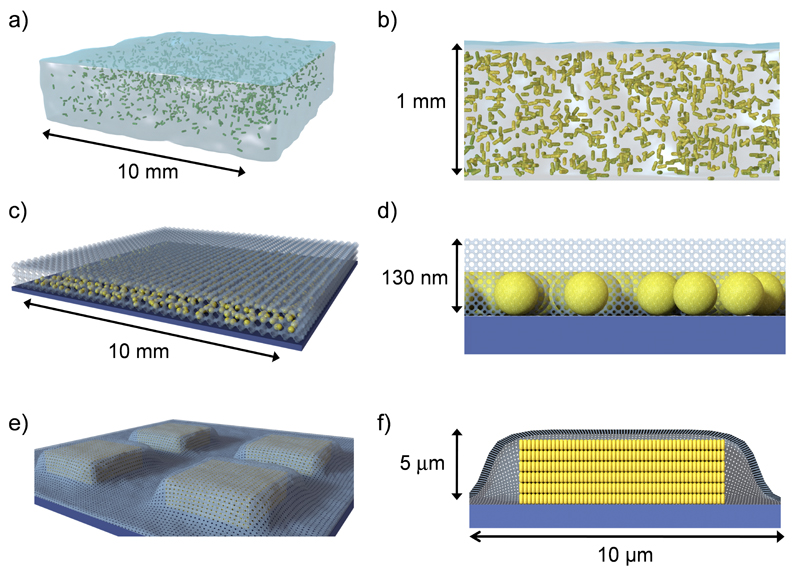
Figure 1. Schematic representation of nanostructured porous substrates for in-situ plasmonic detection and imaging of Quorum Sensing in Pseudomonas aeruginosa biofilms.
a,b) Au@pNIPAM hydrogel with embedded Au nanorods (drawn in green). c,d) Mesostructured Au@TiO2 substrate with a mesoporous TiO2 thin film over a submonolayer of Au nanospheres. e,f) Micropatterned Au@SiO2 supercrystal array comprising Au nanorods organized in micron-sized pedestal-like structures coated with mesoporous silica. b, d, f represent cross-sectional views.
Raman and SERRS characterization of pyocyanin
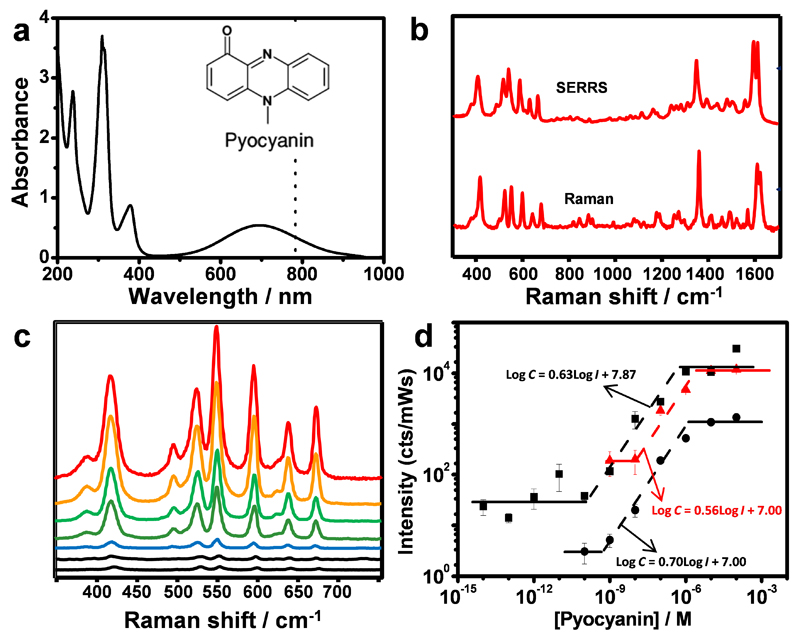
Pyocyanin exhibits a broad absorption band in the visible (550 to 900 nm, Fig. 2a), which has been ascribed to a HOMO-LUMO transition (Section S3 in SI). Illumination of an aqueous solution of commercial pyocyanin with either 633 nm or 785 nm laser lines leads to resonance Raman (RR) (Fig. 2b; see a detailed description of the optical properties and vibrational assignment in Section S3, SI). This band is not present in any of the other phenazines that may be produced in the P. aeruginosa culture (see Section S3, SI). Therefore, SERRS rather than SERS, spectra of commercial pyocyanin were recorded in the wet state upon immersion of the plasmonic composites in aqueous pyocyanin solutions of known concentration (between 10 fM and 10 μM, with 10-fold increments) and processed using the SERS spectrum of the plasmonic composite material as a blank. Upon equilibration, the SERRS spectrum of pyocyanin is clearly visible (Fig. 2b). It was noted that for the three selected plasmonic platforms the SERRS signal was found to have low variability from substrate to substrate. Quantitative SERRS detection can be achieved when changes in SERRS signal correspond to changes in analyte concentration in a predictable manner. Therefore, the quantitative detection range was evaluated for all three plasmonic substrates by measuring the SERRS spectra of pyocyanin solutions from 0.1 nM and 0.01 mM with 10-fold increase steps. Fig. 2c shows the SERRS spectra of pyocyanin at different concentrations measured from the Au@pNIPAM substrate with the 785 nm laser line (similar spectra were obtained for the other substrates). The SERRS measurements showed that quantitative detection of pyocyanin can be achieved in a concentration range that depends on the plasmonic substrate: from 0.1 µM down to 10 nM for mesostructured Au@TiO2 and Au@pNIPAM hydrogel; from 0.1 µM down to 1 nM for the micropatterned mesoporous Au@SiO2 substrate (Fig. 2d). Within these concentration ranges, pyocyanin concentration can be expressed quantitatively by empirical formulas: Log C = 0.70·Log I + 7.00 (R2=0.97) for Au@pNIPAM hydrogel, Log C = 0.56·Log I + 7.00 (R2=0.91) for Au@TiO2 substrate and, Log C = 0.63·Log I + 8.68 (R2=0.97) for Au@SiO2 substrate, where C is the pyocyanin concentration (in molar concentration) and I the SERRS intensity (in Kcounts/mW s).34 Above each concentration range the SERRS signal intensity will be saturated. Interestingly, for the micropatterned Au@SiO2 supercrystal array, when the concentration is lower than 0.1 nM the signal is not homogeneous across all pillars and detection of molecular binding events required SERRS mapping through several pillars. This led to ultrasensitive detection of pyocyanin down to a limit of detection (LOD) of 10−14 M (Fig. 2d), in agreement with the reported plasmonic properties for this type of nanostructure.35
Figure 2. Raman and SERRS spectra of pyocyanin.
a) UV-vis-NIR spectrum of aqueous pyocyanin solution (10−4 M) and molecular structure of pyocyanin (inset). The dotted line indicates 785 nm, corresponding to the excitation wavelength used for Raman and SERRS spectra. b) RR and SERRS spectra of pyocyanin measured in solid state and in aqueous solution (1µM, Au@pNIPAM hydrogel), respectively. Raman measurement was carried out with a 50× objective, a maximum power of 54.22 kW/cm2 and acquisition time of 10 s. SERS measurement was carried out with a 20× objective, a maximum power of 4.24 kW/cm2 and acquisition time of 10 s. c) Pyocyanin SERRS spectra at different concentrations (between 0.1 nM and 10 µM with 10-fold increase steps), recorded on Au@pNIPAM hydrogels. SERRS measurements were carried out with a 20× objective, a maximum power between 0.26-4.24 kW/cm2 and an acquisition time of 10 s. d) SERRS intensity (at 418 cm−1 for Au@pNIPAM -circles- and Au@TiO2 -triangles; at 1600 cm−1 for micropatterned Au@SiO2 -squares) as a function of pyocyanin concentration. The dashed lines are linear fits in the quantitative detection regions. Each data point corresponds to the average signal from five spectra collected from three different substrates. An excitation laser line at 785 nm was used for all measurements. A 20× objective, a maximum power of 0.98 kW/cm2 and an acquisition time of 0.1s was used for micropatterned Au@SiO2 substrate.
SERRS analysis of bacterial phenazines in planktonic cultures
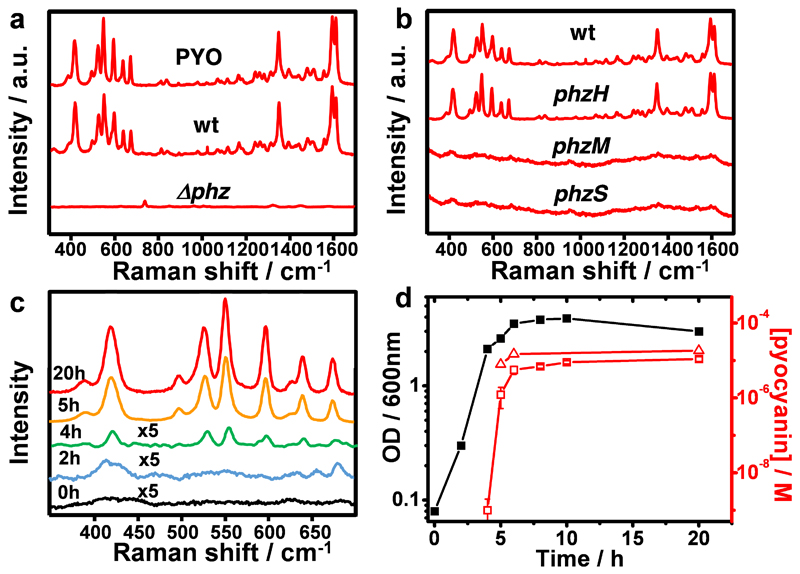
The high performance of our plasmonic substrates allowed us to focus on the detection of pyocyanin produced in vitro by P. aeruginosa PA14 bacteria. We first investigated pyocyanin expression in planktonic cultures grown under constant agitation, using Au@pNIPAM hydrogels as plasmonic substrates. To this end, bacteria-free supernatants of P. aeruginosa PA14 obtained from late stationary phase cultures grown for 20 hours were incubated for several hours with the plasmonic substrates prior to SERRS measurements. As shown in Fig. 3a, the sample obtained from wild type P. aeruginosa PA14 bacteria (wt) displayed a SERRS fingerprint almost identical to that of commercial pyocyanin, whereas no pyocyanin signal was detected in a sample from a phenazine-null mutant strain (Δphz1/2). P. aeruginosa PA14 produces at least five types of phenazines from phenazine-1-carboxylic acid (PCA), i.e. PCA can be converted into 1-hydroxyphenazine (1-HO-PHZ), phenazine-1-carboxamide (PCN), 5-methylphenazine-1-carboxylic acid betaine (5-MCA) and pyocyanin, by action of the enzymes PhzS, PhzH, and PhzM (see Fig. S4.1 in SI for details).10 To confirm that the SERRS signals specifically correspond to pyocyanin and not to any of the other four phenazines, cell-free samples obtained from planktonic cultures of different P. aeruginosa PA14 mutants were analyzed. The results (Fig. 3b) confirm that only the phzH mutant yielded the characteristic SERRS fingerprints of pyocyanin, meaning that the spectrum is indeed pyocyanin-specific, which is likely related to the additional enhancement under resonant Raman conditions.
Figure 3. SERS analysis of phenazine production by P. aeruginosa PA14 strains grown in planktonic cultures, recorded with Au@pNIPAM hydrogels.
a) SERRS spectra of commercial pyocyanin (PYO) and of pyocyanin produced by the wild type (wt) and the phenazine-null Δphz1/2 (Δphz) strains. b) SERRS spectra of pyocyanin produced by the wt and the indicated phz mutant strains. c) SERRS analysis of pyocyanin release by P. aeruginosa PA14 strain at different growth times. d) Growth curve of P. aeruginosa PA14 by measuring optical density of the liquid culture at 600 nm (black squares) and pyocyanin concentration determined by SERRS (red squares) and UV-vis spectroscopy (red triangles). All SERS measurements were performed with a 785 nm laser line employing a 20× objective, maximum power between 1.72 kW/cm2 and an acquisition time of 10 s (intensity at 418 cm−1).
Next, the release of pyocyanin into the extracellular medium throughout the growth of planktonic wild type P. aeruginosa PA14 was investigated. Aliquots from the culture were collected at various times and used for measurement of the optical cell density at 600 nm (OD600), enumeration of viable bacteria (CFU/mL) and determination of pyocyanin concentration by SERRS and UV-Vis spectroscopy at 691 nm (λmax of pyocyanin)36 upon chloroform extraction (Section S4, SI). Extraction with chloroform is a common strategy for pyocyanin purification, which allowed us to interpolate SERRS intensities to the calibration curve and determine the corresponding concentrations. Representative sequential SERRS spectra shown in Fig. 3c demonstrate the presence of pyocyanin already at four hours after the start of a culture bearing an OD600 of 2.1 (Fig. 3d) and 2.9×109 CFU/mL. Interpolation of the SERRS signal to the calibration curve yielded a concentration of pyocyanin of approximately 1 nM (Fig. 3d). Interestingly, just one hour later, upon reaching an optical density of 2.6 and 3.2×109 CFU/mL, the SERRS signal increased significantly (Fig. 3c), indicating a concentration of 1 µM (Fig. 3d). This abrupt increase in pyocyanin concentration parallels the well-known exponential growth of bacterial cultures. At later times, the SERRS intensities and the amount of pyocyanin did not increase significantly and remained constant within the micromolar concentration range (Fig. 3c,d) as previously reported by UV-Vis spectroscopy.26 Indeed, the amount of pyocyanin measured by SERRS correlated to parallel measurements by UV-Vis (Fig. 3d). As expected, better sensitivity was achieved by SERRS, enabling the detection of pyocyanin in bacterial cultures at sub-nanomolar concentration, whereas UV-Vis spectroscopy was not reliable below micromolar concentration levels (Fig. 3d).26 These results are also in agreement with those of Wu and co-workers, who recently reported the use of a silver nanorod substrate for SERS quantification of pyocyanin (LOD of 2.38×10−8 M) extracted from planktonic cultures of a P. aeruginosa PAO1 strain.32 Mesostructured Au@TiO2 and micropattered Au@SiO2 substrates also allowed us to detect pyocyanin purified from planktonic cultures. Notably, whereas Au@TiO2 showed a similar sensitivity to that of Au@pNIPAM, Au@SiO2 supercrystals yielded pyocyanin detection from a planktonic culture having 3.4×107 CFU/mL, two orders of magnitude less than that required when the Au@pNIPAM hydrogel was used.
In situ detection of quorum sensing in biofilm communities
Once we confirmed the suitability of the proposed substrates toward plasmonic detection of pyocyanin, we explored their application as platforms for sustaining bacterial growth and in situ SERRS detection of the phenazine in developing biofilms. At the initial phase of biofilm formation, planktonic bacteria attach to biotic or abiotic surfaces within minutes, followed by aggregation into microcolonies, a step that is sometimes described as the first social trait in the development of a biofilm. Such bacterial aggregates support the production and accumulation of EPS that serve as scaffolds for the bacterial community and aid in the formation of a mature biofilm.5 Numerous approaches have been developed for the study of these sessile microbial communities.37, 38 The so-called “colony-biofilm assay” refers to a model for in vitro study of bacterial biofilms grown over an agar-based medium under static growth (i.e. no agitation). This model has been previously used for studying the role of phenazines in biofilm morphogenesis,27 as well as for the electrochemical characterization of QS in P. aeruginosa biofilms.39, 40
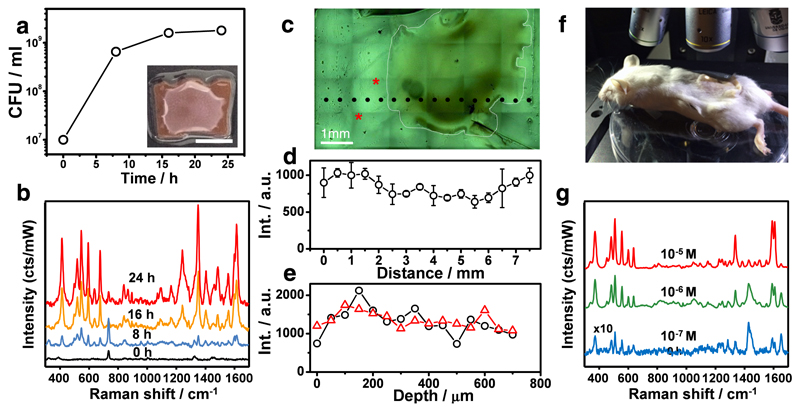
We adapted the colony-biofilm assay to assess the suitability of Au@pNIPAM hydrogels as platforms for sustaining bacterial growth and in situ SERRS detection of pyocyanin. Dry plasmonic hydrogels were infused with liquid nutrients by swelling in LB medium and then inoculated with a bacterial suspension of P. aeruginosa PA14 previously depleted of extracellular pyocyanin (Section S4, SI). As shown in Figure 4, the substrate indeed allowed bacterial proliferation as a colony on its surface, as well as plasmonic detection of pyocyanin secretion into the underlying sensor. SERRS measurements recorded from different spots over the substrate surface, both inside and outside the region colonized by the colony-biofilm (Fig. 4c), revealed horizontal pyocyanin diffusion toward regions located as far as 2 mm away from the biofilm margin (Fig. 4d). Interestingly, the intensity of the SERRS signals was slightly higher in non-colonized regions, most likely due to interferences caused by the biofilm matrix. Vertical diffusion was also evidenced by SERRS along the z-axis, showing that the SERRS signal intensity of the molecule remained constant from the surface through 700 μm in depth (Fig. 4e). A time-dependent (0, 8 and 24 hours) SERRS analysis of the diffusion of pyocyanin secreted by a growing biofilm shows homogeneous distribution of the molecule over the hydrogel surface. However, the diffusion is slightly compromised along the z-axis at 24 hours of growth, most likely due to interferences caused by biofilm matrix components (Fig. S4.2 in SI).
Figure 4. In situ detection of pyocyanin secreted by P. aeruginosa PA14 colony-biofilms grown on Au@pNIPAM hydrogels.
a) Graphical representation of viable bacteria (CFU/mL) quantified over time. The inset shows an image of the colony-biofilm (scale bar: 0.5 cm). b) SERRS spectra recorded at the indicated times. c) Optical microscopy image of a bacterial colony-biofilm, which is surrounded with a white striped line for clarity. d) SERRS intensities (418 cm−1) measured in Figure 4c at the places indicated with black dots and plotted as a function of distance. The error bars show the standard deviation from three different measurements. e) SERRS intensities (418 cm−1) measured in Figure 4c at the places indicated with asterisks and plotted as a function of the depth from the surface. SERRS measurements of colony-biofilms were done using 785 nm laser line for 10 s and using a maximum power of 0.91 kW/cm2 employing a 20x objective. f) Photograph showing the Raman experimental set-up for detection of pyocyanin in subcutaneous implants in mice. g) Under-skin SERRS spectra of pyocyanin spiked at the indicated concentrations on Au@pNIPAM hydrogel. SERRS measurements of pyocyanin-spiked hydrogels were performed using a 785 nm laser line for 10 s using a maximum power of 24.45 kW/cm2 employing a 10x objective.
Current knowledge of the influence of QS in bacterial pathogenesis and biofilm development is largely derived from in vitro systems. However, as infection is a continuous interplay between the host and the microbes, experimental animal models are widely used in preclinical studies. Investigations of biofilm-related infections in animals are often based on bacteria previously adhered to organic and inorganic materials, which are implanted in the animal.37 For example, studies of chronic P. aeruginosa lung infection used embedded bacteria in natural hydrogels such as agar or alginate.37, 41 In vivo optical imaging combined with genetically-modified bacteria expressing bioluminescent or fluorescent reporters is an effective method for performing longitudinal studies of bacterial infection in animal models.42 Importantly, this imaging technique contributes towards the reduction in the number of experimental animals as it enables each animal to be analyzed over time with no need for sequential sacrifices performed for ex-vivo quantification of bacteria. However, optical imaging employing fluorescence or bioluminescence is often limited by autofluorescence and signal attenuation due to scattering and absorption of light by mammalian tissues.42
Since pyocyanin is detected by SERRS upon irradiation with a NIR (785 nm) laser (within the biological transparency window), it can be used as a SERS beacon for reporting QS in infection studies employing plasmonic hydrogels pre-colonized with natural (e.g. non-genetically modified) strains of P. aeruginosa. To test this idea, an Au@pNIPAM hydrogel was spiked with commercial pyocyanin at biologically relevant concentrations43 and the substrate was implanted in Swiss mice (Fig. 4f). As shown in the SERRS analysis of Figure 4g, pyocyanin was detected subcutaneously at concentrations as low as 0.1 µM, suggesting that it could be used as a reporter for label-free, non-invasive monitoring of QS and screening potential antimicrobial drugs in animal models for infections, using SERS. P. aeruginosa is one of the most common bacteria isolated from soft tissue infections, such as those in acute burns, bed sores, and diabetic ulcers.9 P. aeruginosa infections are often characterized by increasing occurrence of multi-resistance strains, leading to numerous deaths annually worldwide and thus cause a significant burden to healthcare systems. Hence, the development of new innovations toward their prevention and treatment has become imperative. Disruption of QS circuits with synthetic chemical compounds and natural products, is a strategy with potential applications to prevent or attenuate bacterial pathogenesis.44 Remarkably, the inhibition or decrease of pyocyanin expression is often used as an indicator of the treatment efficacy.44, 45, 46 Thus, plasmonically-active hydrogels could be furnished in standard animal models of wound infections47 as advanced in vivo optical sensors for providing new insights on the role of pyocyanin and QS in the pathogenesis of clinical strains of P. aeruginosa and to evaluate the efficacy of prophylactic and therapeutic treatments in longitudinal studies.
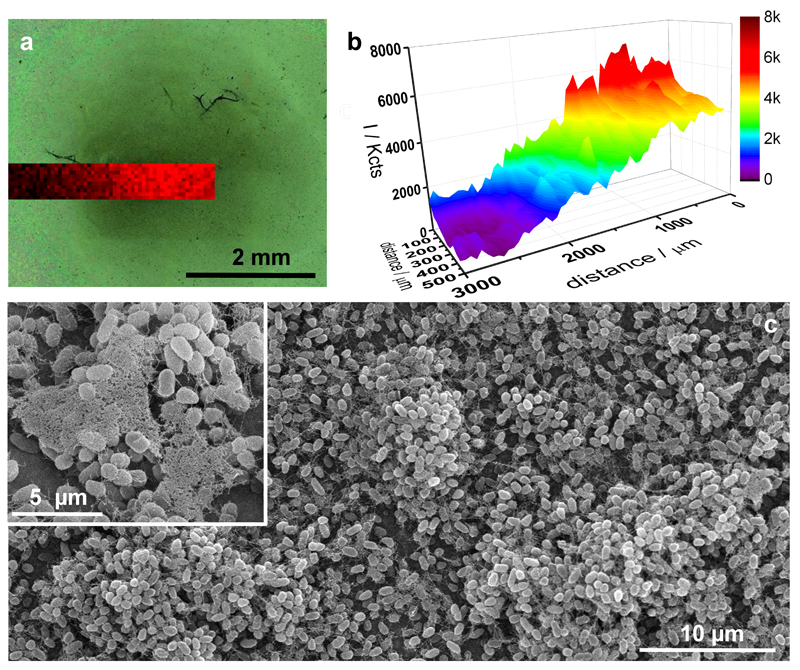
Microbial communities in biofilms are structurally organized entities, and the physical localization of signaling molecules within the cell population has profound impact on bacterial behavior and phenotypic heterogeneity.5, 48 Therefore, the capacity to image the chemical environment surrounding biofilms with spatial resolution may provide new insights to understand their physiology and behavior. We have demonstrated that Au@pNIPAM hydrogels allow non-invasive, in-situ SERRS detection of pyocyanin expressed by growing colony-biofilms. However, the enhanced diffusion of this platform impairs the analysis of pyocyanin distribution over large sessile bacterial populations with meaningful spatial resolution. Mesostructured Au@TiO2 thin films were evaluated as substrates for spatially resolved plasmonic imaging of pyocyanin in biofilm communities of P. aeruginosa. The highly organized mesoporous nature of this platform (interpore distance 12 nm; film thickness ca. 130 nm) (Fig. S1.2, SI) was hypothesized to restrict horizontal diffusion of the analyte, thereby enabling localized detection of the phenazine to biofilm-colonized regions only. In addition, the large area, averaging 1.0 cm2, allows plasmonic interrogation of pyocyanin expression over large (millimeter-scale) regions. A droplet-based assay was performed, in which a bacterial suspension (50 μL) of P. aeruginosa PA14 (1of 109 CFU/mL) depleted of extracellular pyocyanin was grown statically on Au@TiO2 at 30 ºC for 48 hours inside a humidified chamber. The substrate allowed bacterial growth, as evidenced by the increased number of viable bacteria (CFU/mL) over time, as well as SERRS detection of pyocyanin exclusively in those regions of the mesoporous substrate in contact with the bacterial culture (Fig. S4.3, SI). Interestingly, the intensity of the SERRS signals was observed to increase toward the innermost regions of the drop, as evidenced by plotting the SERRS intensity (418 cm−1) as a function of distance (Fig. S4.3, SI). In order to correlate the presence of pyocyanin with biofilm bacteria, the substrate was gently immersed in water so as to reveal substrate-adhered cells. SERRS mapping spanning the region shown in Figure 5a scanned with a 50 μm step-size, demonstrated high intensity pyocyanin SERRS signals at the central region, which was heavily colonized by bacteria as observed in the optical image acquired in situ. Significantly, the SERRS signal decreased towards less colonized regions and faded completely at non-colonized surfaces, indicating pyocyanin detection with a spatial resolution of ca. 50 μm. Analysis of the substrate by scanning electron microscopy (SEM) revealed the presence of bacterial microcolonies and self-produced extracellular polymeric substances (Fig. 5c and Fig. S4.4, SI), which are key features of biofilms. To further confirm the spatially-resolved detection of pyocyanin, rather than growing the biofilm in a liquid environment, a small colony of P. aeruginosa PA14 strain was grafted on the Au@TiO2 substrate, layered with a pad of 0.75% agar in LB medium and incubated for 24 hours. SERRS imaging performed in situ, without additional manipulation, revealed that pyocyanin signal was indeed restricted to the region colonized by bacteria (Fig. S4.5, SI).
Figure 5. In situ detection and imaging of pyocyanin produced by P. aeruginosa PA14 biofilms grown on mesostructured Au@TiO2 thin films.
a) Optical image of the bacterial biofilm (dark central region) captured with the Raman microscope and superimposed pyocyanin SERRS mapping (418 cm−1) acquired with excitation laser wavelength of 785 nm, 5× objective and a laser power of 0.94 mW for 10 s. b) Graphical representation of the SERRS intensity mapping shown in a. c) Representative SEM images of bacterial biofilm formed on Au@TiO2.
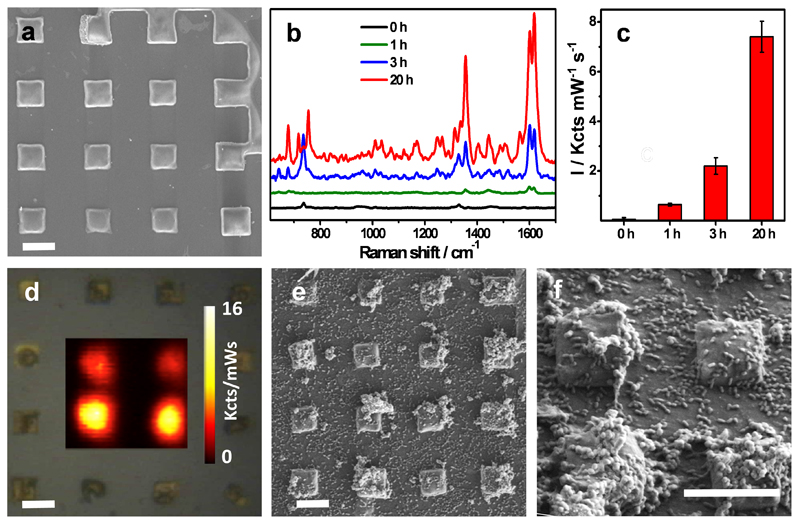
In an effort to increase the sensitivity of this approach, micropatterned Au@SiO2 supercrystal arrays (Fig. 6a), which showed the highest enhancement factor towards pyocyanin detection (LOD 10−14 M) (Fig. 2d), were evaluated. Again, 10 μL of a bacterial suspension (1×109 CFU/mL) devoid of pyocyanin was grown statically for 1, 3, or 20 hours on the patterned substrate and then analyzed. Strikingly, the SERRS fingerprint of pyocyanin was identified as early as one hour after initiating the culture, and the intensity of the signal increased over time, as shown by the SERRS spectra recorded at 3 and 20 hours (Fig. 6b,c). This indicates that starting with a cell density of approximately 1x107 cell/mL, P. aeruginosa can soon engage in QS behavior. When using this substrate we could also detect chloroform-extracted pyocyanin from a planktonic culture of 3.4×107 CFU/mL, which was not possible with either of the other two plasmonic substrates. This is reasonable, considering that the LOD of the Au@SiO2 substrate was shown to be 4 or 5 orders of magnitude lower than those of Au@pNIPAM and Au@TiO2 substrates, respectively (Fig. 2d). Analysis of the substrate by SEM performed at three hours of growth provides evidence of the low cell density present in the bacterial culture (Fig. S4.6a, SI). SERRS mappings recorded after 20 hours of growth demonstrated pyocyanin detection exclusively at the plasmonic features, 25 μm2 on average (Fig. 6d). Inspection of the substrates by SEM revealed the presence of microcolonies and small groups of cells on top of the pedestals (Fig. 6e,f and Fig. S4.6b,c, SI), which are likely responsible for the localized secretion of the phenazine. However, it cannot be ruled out that the SERRS signal may also arise from pyocyanin produced by nearby bacteria. This is in agreement with the report by Connell and collaborators, who employed scanning electrochemical microscopy and a lithographic technique to confine small populations of P. aeruginosa bacteria in low volumes and reported that pyocyanin can be produced by clusters containing only 500 bacterial cells.49
Figure 6. In situ detection and imaging of pyocyanin produced by P. aeruginosa PA14 grown on micropatterned Au@SiO2 substrates.
a) SEM images of silica-coated Au nanorod supercrystals. Scale bar: 5 μm. b) Representative SERRS spectra measured at 0, 1, 3 and 20 hours. c) Relative SERRS intensities (1600 cm−1) recorded at 0, 1, 3 and 20 hours. d) Optical image of the substrate and SERRS mapping of pyocyanin (1600 cm−1) recorded at 20 hours of growth. Scale bar: 5 μm. e,f) SEM images of silica-coated Au nanorod supercrystals colonized by P. aeruginosa (20 h) at different magnifications. Scale bar: 5 μm. All SERS measurements were carried out with a 785 nm laser line, 50× objective and a maximum power of 0.98 kW/cm2. Acquisition time was 0.1s.
Outlook
The engagement of bacteria in social behavior, including the coordination of group activities via QS, such as biofilm formation, has become one of the most intensively studied topics in microbiology during the past few decades. In addition, the huge impact of bacterial biofilms in industrial fouling, contamination and infection have made this form of microbial growth a priority research topic. The strict QS control of pyocyanin expression, together with its high Raman cross-section, allowed us to use it as a beacon to demonstrate label-free plasmonic detection and imaging of QS communication in live biofilm communities of P. aeruginosa grown on three types of rationally designed SERS-active substrates. Au@pNIPAM hydrogels allowed proliferation of biofilms and homogeneous SERS sensing of the secreted phenazine across the substrate. The high water content and hydrophilic nature of hydrogels are similar to the void-filling component of microbial biofilms or the extracellular matrix of mammalian cells, which renders them intrinsically biocompatible. Thus, hydrogels are widely used in diverse biomedical applications as cellular scaffolds and bioactive materials.50 For instance, hydrogels of controlled diffusivity and mass transfer properties have been fabricated by soft lithography for engineering cellular microenvironments in which cell populations can maintain chemical exchange and communication.48, 50, 51 Our results suggest that, besides providing scaffolding and cellular support, SERS-active nanostructured hydrogels could be implemented as plasmonic sensors for label-free characterization and monitoring of secreted diffusible biomolecules. Additionally, the results show that plasmonically-active hydrogels can be used as implantable materials in experimental animal models, to investigate QS triggered by natural populations of P. aeruginosa and to evaluate anti-virulence therapies by pharmacologic intervention.44
Imaging QS in biofilm communities with spatial resolution is important toward gaining new understanding of this form of bacterial communication. It was demonstrated with mesostructured Au@TiO2 and micropatterned supercrystal Au@SiO2 platforms that P. aeruginosa not only engage QS in densely-populated biofilms, but also at early stages of biofilm development. The mesostructured Au@TiO2 platform enabled us to interrogate up to millimeter-scale regions of the colonized substrate with high spatial resolution (20-50 μm). The micropatterned supercrystal Au@SiO2 enabled ultrasensitive SERS detection of QS events in low-density bacterial cultures at the initial hours of growth and imaging of bacterial communication, owing to its high enhacement factor. The Au@SiO2 substrates exhibited pyocyanin detection down to 10−14 M, possibly triggered by small clusters of cells colonizing the micron-sized plasmonic features. The high performance of this substrate is most likely due to a high density of efficient hot spots and collective plasmon modes in the supercrystal.35 Since it is well known that the electric field enhancement decays very rapidly from the plasmonic nanoparticles surface,52 it is essential to consider that the mesoporous silica coating infiltrates within the highly ordered nanorod structure,53 thereby increasing the “plasmonically-active space” and improving the sensitivity of the Au@SiO2 optical enhancer. Microfabrication of ordered plasmonic nanoparticle supercrystals provides unique optical and photonic coupling effects but also patterning is an interesting strategy for fabricating complex nanostructured sensing materials. For example, micropatterning mediated by nano- and micro-fabrication techniques has been applied to the analysis of cell behavior on substrates with ordered topographical features.54, 55, 56 Thus, the ability to selectively immobilize cells and/or modulate cellular responses at plasmonically-active micropatterned features, combined with ultrasensitive SERS detection, will provide powerful analytical platforms for providing more insight into cell-to-cell communication in microenvironments based on diffusible SERS-active signals.
The high resolution displayed by the Au@TiO2 and Au@SiO2 plasmonic substrates (20-50 μM) is in the range of that obtained with most imaging mass spectrometry (IMS) techniques (1-100 μm), which have been successfully applied to probe single cells and colonies in microbial research.57, 58 A key advantage of IMS is that it enables the label-free spatial imaging of the distribution of biomolecules in biological samples. However, IMS techniques often require various forms of sample preparation that may disturb the specimen under investigation (e.g. layering with an inorganic compound in matrix-assisted laser desorption/ionization, or coating with an organic matrix in MALDI-TOF), as well as the limited mass resolving power of IMS.57 As we have shown, our SERS-based approach, focusing on the detection of pyocyanin released from bacterial biofilms and small clusters of cells, demonstrates the potential of plasmonics as an alternative for imaging biomolecules from undisturbed biological samples. The porous nature of the substrates plays an important role in restricting the contact of the plasmonic component with (bio)molecules that could likely contaminate the SERS spectrum and thereby hinder the selectivity and sensitivity of the assay. Large molecules may also contaminate the plasmonic surface and passivate it from interaction with the analyte. The SERS-based sensing approaches described in this work not only provide the necessary tools to probe important questions in QS, but can also serve as general detection systems to be applied for the investigation of other cellular communication processes based on SERS-active diffusible molecules.
Methods
Materials
Tetrachloroauric(III) acid trihydrate (HAuCl4· 3H2O), L- ascorbic acid (ACS reagent 99%), N-isopropylacrylamide (NIPAM, 97%), N,N’-methylenebis(acrylamide) (BIS, 99%), N,N,N’,N’-tetramethylethylenediamine (TEMED), 5-bromosalicylic acid (technical grade 90%), silver nitrate (AgNO3, ACS reagent 99.0%), sodium borohydride (NaBH4, ReagentPlus 99%), O-[2-(3-mercaptopropionylamino)ethyl]-O’-methyl-poly(ethylene glycol) (mPEG-SH, Mw 5000), trisodium citrate dihydrate, titanium tetrachloride (TiCl4), (3-aminopropyl)trimethoxysilane (APS), Pluronic F127 (HO(CH2CH2O)106(CH2CH(CH3)-O)70(CH2CH2O)106OH), hydrogen peroxide (H2O2, 28%), sulfuric acid (H2SO4, 98%), chloroform (99.8%) and pyocyanin from Pseudomonas aeruginosa were supplied by Sigma Aldrich. Hydrochloric acid (HCl, 37 wt % in water) was supplied by Panreac. Cetyltrimethylammonium bromide (CTAB, 96%) was supplied by Fluka. All chemicals were used as received. Pure grade ethanol and Milli-Q water were used as solvents.
Characterization
Optical characterization was carried out by UV-vis-NIR spectroscopy with a Cary 5000 spectrophotometer and Agilent 8453 spectrophotometer. Transmission electron microscopy (TEM) analysis was performed by using a JEOL JEM 1010 microscope operating at an acceleration voltage of 100 kV. Samples for TEM in the case of Au@TiO2 substrates were obtained by scratching the films from the substrate and depositing them on carbon- and FORMVAR-coated copper grids. The surface morphology of the dried gels was investigated by using a scanning electron microscope (JEOL JSM-6700F FEG).
Raman Spectroscopy experiments were conducted with a Renishaw InVia Reflex system. The spectrograph used a high-resolution grating (1200 or 1800 grooves mm−1) with additional band-pass filter optics, a confocal microscope, and a 2D-CCD camera. Several laser excitation energies were employed, including laser lines at 633 (HeNe) and 785 and 830 nm (diode).
Scanning electron microscopy analysis was performed with a JEOL JSM-6700F or with a Helios NanoLab 450S Scanning Electron Microscopes.
Fabrication of the plasmonic substrates
Au@pNIPAM hydrogels (1 mm thickness) were prepared by free-radical crosslinking copolymerization of N-isopropylacrylamide and bisacrylamide (as crosslinker) in an aqueous solution containing PEGylated Au nanorods.59 A solution of 1mL of the monomer NIPAM (0.85M), crosslinker BIS (4.24 mM) and APS (0.085M) as the initiator was purged with nitrogen to remove the dissolved oxygen in the system. Subsequently, a 0.5 mL dispersion of gold nanorods (average dimensions: 91.3 ± 8.9 nm long; 27.0 ± 2.3 nm thick)60 functionalized with mPEG-SH (5 molecules/nm2) and accelerator TEMED (1.5 µL) were mixed with the solution, and were quickly transferred to a mold consisting of two glass plates separated by 1.0 mm-thick spacers. Polymerization was carried out at room temperature for 1h. The resulting hydrogel was removed from the mold, and gels were cut in pieces of 1 cm2. The gels pieces were washed extensively in an excess of deionized water. The water was replaced every day to remove the residual unreacted components and the washing was continued for at least 3 days.
The optical characterization of the Au@pNIPAM substrate reveals that the hydrogel retains the main optical features of the Au nanorods indicating uniform nanoparticle distribution (Fig. S1.1.a, SI), which was also confirmed by scanning electron microscopy (SEM) analysis (Fig. S1.1.b, SI).
Mesostructured Au@TiO2 substrates were synthesized depositing a mesoporous thin film on a submonolayer of 80 nm Au nanospheres by spin coating using block copolymer Pluronic F127 micelles as templates.33 This procedure leads to highly ordered body-centered cubic (Im3m) pore system, which is commonly obtained when templating with F127.61 The Vis-NIR spectrum of the Au@TiO2 substrate (Fig. S1.1.c, SI) shows the presence of two bands, one at 580 nm from the single particles and a broad band at around 830 nm due to plasmon coupling. As can be observed in Figure S1.1.d (SI), the transmission electron microscopy (TEM) image of the composite substrate shows ordered mesoporous structures over the metal nanoparticles. The pore diameter and wall thickness (i.e. interpore distance) measured from TEM images yielded average values of 12 nm, and the film thickness measured from SEM cross section images led to average values of 130 ± 6 nm (Fig. S1.2, SI).
Micropatterned Au@SiO2 supercrystal arrays
Mold fabrication
The microtextured mold for the supercrystals was manufactured in PDMS (Sylgard 184, Dow Corning), following standard soft lithography techniques.62, 63 The master was composed of pillars having different shapes and sizes on a 4 inch silicon wafer covered with SU-8 resin. PDMS (10:1 elastomer to curing agent) was cured at 60 °C for 2h. In this work, assembly experiments were performed on <100> silicon wafers.
GNR supercrystal assembly
Patterned substrates were fabricated by drying a GNR dispersion within an array of micron-sized cavities with a fixed height of 4.8 μm and varying lateral dimensions and shapes, as previously reported.35 By adjusting the cavity morphology of the template together with the particle concentration, the morphology and height of the supercrystals can be tuned. In all cases, a 2 μL drop of MUDOL-GNR dispersion with the selected concentration was deposited on the micro-textured PDMS template and then covered by the substrate. The solvent was subsequently allowed to evaporate to dryness (within 12h) and the template removed resulting in the formation of arrays of GNR supercrystals spread over millimeter-sized areas.
Description of the silica coating procedure
Prior to silica growth, the substrates were cleaned with a combination of oxygen plasma (2 min, 0.4 mbar, 200 W) and UV/Ozone (1h). Oxygen plasma surface treatment was performed in a low pressure plasma system (PICO, Diener Electronic). UV/Ozone surface treatment was performed in a UV/Ozone Cleaner (ProCleaner).
For the mesoporous silica film growth, 17.5 mL of a 6 mM CTAB solution in water was mixed with 7.5 mL of ethanol and stirred at 35 °C for ten minutes. 5 µL of NH3 (25 vol%) was added to achieve a pH value between 9 and 10. Finally, 20 µL of TEOS was added drop-wise while stirring vigorously. 1 mL aliquots of growth solution were immediately poured in Eppendorf centrifuge tubes, where a clean gold substrate had previously been introduced in each of the tubes. The tubes were subsequently closed and sealed with Parafilm to avoid evaporation of the synthesis solution and placed in an oven to incubate at 60 °C for three days. The GNR silica substrates were then rinsed with ethanol and immersed in a 0.1 M HCl solution (in ethanol) for 10 min to get rid of excess CTAB. A further rinsing step in ethanol was performed to remove any trace of HCl. The films were then treated at 130 °C for 2 hours to improve silica stability in water without significant reshaping of the nanorods. Scanning electron microscopy (SEM) images of the silica-coated composite substrate (Figure S1.3, SI) show the internal nanorod organization and the presence of mesoporous silica infiltrating nanorod monolayers.
Analysis of the SERS performance of the plasmonic substrates
Au@pNIPAM hydrogels
An Au@pNIPAM hydrogel was immersed overnight in an aqueous solution of 1-naphtalenethiol (1-NAT) and SERS spectra were recorded under excitation with three different laser lines (633, 785 and 803 nm), obtaining in all cases well-defined bands characteristic of 1-NAT vibrations. SERS mapping additionally demonstrated that the SERS enhancing ability of hydrogel is uniform over extended areas (Fig. S2.1). We therefore conclude that Au@pNIPAM hydrogels are efficient SERS substrates offering a wide excitation wavelength range.
Mesostructured Au@TiO2 substrates
The efficiency of Au@TiO2 mesoporous thin films was confirmed using 4-nitrobenzenethiol (4-NBT) as analyte (data not shown), as well as its ability to exclude large biomolecules from the interior of the mesoporous films when analyzing complex matrixes (i.e. molecular sieve effect), as previously reported.33
Micropatterned Au@SiO2 supercrystal arrays
The efficiency of micropatterned Au@SiO2 supercrystal arrays was tested using commercial pyocyanin as analyte. The SERS spectrum was recorded under 785 nm laser line obtaining a well-defined bands characteristic of pyocyanin vibrations. SERS mapping additionally demonstrated that the SERS enhancing ability of the supercrystals is uniform and confined to the supercrystal (Fig. S2.2, SI).
Raman Enhancement Factors calculations
We estimated the enhancement factors (EF) of the different plasmonic substrates by applying the following equation:64
| (1) |
Where VA and VB represent the probed volumes, and IA and IB the intensities in SERS and Raman, respectively; f, is a correction factor that considers the concentration ratio of the probed molecule in both experiments under the same conditions. Since pyocianin is a solid Raman spectra was collected from a 10−4 M solution. Provided that VA and VB are similar, Equation (1) can be reduced to EF = (IA / IB) x f, where f is 5, 4 and 6 for the Au@pNIPAM hydrogels, the Au@TiO2 mesostructured substrate and the micropatterned Au@SiO2 supercrystal array, respectively. For a reliable estimation of the EF we have considered the SERS intensity of the lowest concentration of pyocianin within the quantitative detection region.
The estimated enhancement factors (EF) are 3.99 x 105, 1.55 x 106 and 1.67 x 106 for the Au@pNIPAM hydrogels, the Au@TiO2 mesostructured substrate and the micropatterned Au@SiO2 supercrystal array, respectively. The calculated EF can be considered as high since pyocianin does not contain any thiol or amine functional group, so that its affinity for gold should be low.
Plasmonic detection of pyocyanin in planktonic and biofilm cultures
Bacterial strains and growth conditions of planktonic cultures
PA14 Pseudomas aeruginosa bacterial strains used are indicated in Table S4.1, SI. Bacteria were streaked onto Luria-Bertani (LB) agar (1.5% w/v) plates and incubated overnight at 37°C. A single colony was used to inoculate a 10 ml culture in LB medium and grown at 37°C with agitation (210 rpm). The optical density (OD) of the liquid culture was measured at OD600 nm for monitoring bacterial growth.
Extraction of phenazines and spectrophotometric quantification
Cells from a bacterial culture grown in LB were pelleted by centrifugation (8,000 g / 3min) and the supernatant was filtered (0.2 µm filter pore size). The filtrate (2 mL) was mixed thoroughly with the 4 mL of chloroform and incubated for 20 min, after which the aqueous phase was removed. Phenazines were recovered by air or nitrogen-drying the solvent and re-dispersion in 2 mL of water. The concentration of pyocyanin was determined spectrophotometrically based on its absorbance at 691 nm (A691), according to the Beer-Lambert law, knowing the εPYO= 4310 M−1 cm−1.65
SERS detection of pyocyanin in planktonic cultures
Bacteria-free supernatants of P. aeruginosa PA14 obtained from a late stationary phase culture grown for 20 hours were incubated for at least five hours with the different plasmonic substrates. In the case of Au@pNIPAM hydrogels 1:100 dilution in water of the filtered supernatant was performed to reduce interference (SERS background), whereas this was not necessary in the mesostructured Au@TiO2 and micropatterned Au@SiO2 substrates, possibly due to the molecular sieve effect of their mesoporous layer.33
SERS detection of pyocyanin in biofilms grown on plasmonic substrates
The Au@pNIPAM hydrogel was sterilized under an ultraviolet light lamp for 16 hours. The mesostructured Au@TiO2 and micropatterned Au@SiO2 substrates were sterilized by immersion in 70% ethanol for 3 hours. PA14 Pseudomas aeruginosa bacteria were streaked onto LB agar plates and incubated overnight at 37°C. Single colonies from the agar plates were used to inoculate 10 mL culture in LB medium at 37°C, 210 rpm and grown for 20 hours. The bacterial culture (1 mL) was washed three times by centrifugation (7000 rpm/3 min) with 10 mL of LB for removing extracellular pyocyanin, and the final cell pellet was re-suspended in LB to an OD600 nm of 3.0. The procedure for rinsing off any pyocyanin carry-over from the bacterial suspension was assessed by SERRS for each substrate. The bacterial suspension depleted of pyocyanin was used to start the cultures on the plasmonic platforms with the indicated cell densities at the following final volumes: 5 µL on Au@pNIPAM, 50 µL on Au@TiO2 and 10 µL on Au@SiO2. The bacterial cultures were grown in a humidified chamber at 28°C (Au@pNIPAM), 30°C (Au@TiO2), and 37°C (Au@SiO2). For growing P. aeruginosa on the Au@TiO2 substrate in a semi-solid environment embedded on agar, a small colony of bacteria was grafted with a sterile tip onto the surface of the plasmonic platform. Subsequently, 50 μL of molten LB agar (0.75% w/v) was solidified on top and the bacterial colony was incubated in a humidified chamber for 24 hours at 30°C.
Viable bacteria were quantified by counting colony-forming units (CFUs) as follows: Samples were removed from planktonic cultures, serially diluted in LB, spread on LB-Agar plates and incubated at 37°C overnight to determine CFUs. For determination of viable bacteria growing as biofilms on plasmonic platforms, bacteria were scraped off the substrate in 100 µL of sterile PBS using an inoculating loop. The samples were bath-sonicated to disperse bacterial aggregates. Next, the bacterial suspension was serially diluted in LB, spread onto LB-Agar plates and incubated at 37°C overnight to determine CFUs. Bacterial titers were expressed as CFUs/mL.
Detection of spiked pyocyanin in plasmonic substrates implanted in mice
The Au@pNIPAM substrates were doped with 10−5, 10−6 or 10−7 M concentrations of commercial pyocyanin. The substrates were implanted subcutaneously in cadavers of Swiss mice, kindly provided by Dr. Jose Antonio Lamas (University of Vigo). SERS measurements were performed with a 20x objective for 10 s and using a maximum power at the sample of 108.6 mW. Animal handling procedures were approved by the Spanish Research Council and the University of Vigo Committee for Animal Experimentation, and they observed the Spanish and European directives for the protection of experimental animals (RD1201/2005; 86/609/EEC).
Scanning electron microscopy analysis of bacterial biofilms
Sessile bacteria on Au@TiO2 substrate were fixed in 2% glutaraldehyde with 0.1 M cacodylate buffer (pH 7.4) for 2 h at 4 ºC. The fixing solution was rinsed with cacodylate buffer (3 x 30 min), and samples were dehydrated with a series of increasing concentrations of acetone: 30%, 50%, 70%, 80% and 95% for 30 min, and absolute acetone for 1 h. Next, the sample was subjected to critical point drying with CO2 (73 atm; 31.3 ºC) and coated with gold (10-20 nm). The sample was observed with a JEOL JSM-6700F Scanning Electron Microscope. Sessile bacteria on Au@SiO2 substrate were treated with 4% formaldehyde at 4 ºC for 12 hours. The fixative was washed out with milli-Q water and the sample was observed with a Helios NanoLab 450S Scanning Electron Microscope.
Supplementary Material
Acknowledgements
This work has been funded by the European Research Council (ERC Advanced Grant #267867 Plasmaquo). We thank Professor Dianne K. Newman, Department of Biology, California Institute of Technology and Howard Hughes Medical Institute for providing us with P. aeruginosa PA14 and PA14 Δphz1/2 strains. We thank Professor Frederick M. Ausubel, Department of Genetics, Harvard Medical School for kindly providing P. aeruginosa PA14 phzH, phzM and phzS strains. Evgeny Modin and Prof. Andrey Chuvilin are thanked for their work on FIB/SEM Of supercrystals. EHH gratefully acknowledges the Spanish Ministry of Economy and Competitiveness for funding a Juan de la Cierva Fellowship. VMG acknowledges an FPU scholarship from the Spanish MINECO.
Footnotes
Author contributions
G. Bodelón designed and executed bacterial growth experiments, V. Montes-García performed SERS experiments with Au@pNIPAM substrates, V. López-Puente fabricated Au@TiO2 substrates and measured SERS, C. Costas executed bacterial growth experiments, S. Rodal-Cedeira and S. Celiksoy participated in the fabrication of Au@pNIPAM substrates, I. Pérez-Juste carried out DFT calculations, E.H. Hill executed bacterial growth experiments and measured SERS on Au@SiO2 substrates, C. Hamon, M.N. Sanz-Ortiz and L. Scarabelli participated in the fabrication of Au@SiO2 substrates, A. La Porta carried out SEM imaging, I. Pastoriza-Santos and J. Pérez-Juste contributed to substrate design and discussion, L.M. Liz-Marzán designed and supervised the project and the discussions. All authors participated in writing of the manuscript.
References
- 1.Waters CM, Bassler BL. Quorum sensing: Cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 2.Lazdunski AM, Ventre I, Sturgis JN. Regulatory circuits and communication in gram-negative bacteria. Nat Rev Microbiol. 2004;2:581–592. doi: 10.1038/nrmicro924. [DOI] [PubMed] [Google Scholar]
- 3.Rutherford ST, Bassler BL. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. CSH Perspect Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LaSarre B, Federle MJ. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol Mol Biol Rev. 2013;77:73–111. doi: 10.1128/MMBR.00046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 6.Davey ME, O'toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li YH, Tian XL. Quorum Sensing and Bacterial Social Interactions in Biofilms. Sensors. 2012;12:2519–2538. doi: 10.3390/s120302519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Rybtke M, Hultqvist LD, Givskov M, Tolker-Nielsen T. Pseudomonas aeruginosa Biofilm Infections: Community Structure, Antimicrobial Tolerance and Immune Response. J Mol Biol. 2015;427:3628–3645. doi: 10.1016/j.jmb.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 10.Jimenez PN, et al. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 2012;76:46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castillo-Juarez I, et al. Role of quorum sensing in bacterial infections. World J Clin Cases. 2015;3:575–598. doi: 10.12998/wjcc.v3.i7.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Decho AW, Norman RS, Visscher PT. Quorum sensing in natural environments: emerging views from microbial mats. Trends Microbiol. 2010;18:73–80. doi: 10.1016/j.tim.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Steindler L, Venturi V. Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol Lett. 2007;266:1–9. doi: 10.1111/j.1574-6968.2006.00501.x. [DOI] [PubMed] [Google Scholar]
- 14.Jansson JK. Marker and reporter genes: illuminating tools for environmental microbiologists. Curr Opin Microbiol. 2003;6:310–316. doi: 10.1016/s1369-5274(03)00057-2. [DOI] [PubMed] [Google Scholar]
- 15.Moskovits M. Surface-enhanced Raman spectroscopy: a brief retrospective. J Raman Spectrosc. 2005;36:485–496. [Google Scholar]
- 16.Stiles PL, Dieringer JA, Shah NC, Van Duyne RR. Surface-Enhanced Raman Spectroscopy. Annu Rev Anal Chem. 2008;1:601–626. doi: 10.1146/annurev.anchem.1.031207.112814. [DOI] [PubMed] [Google Scholar]
- 17.Schlucker S. Surface-Enhanced Raman Spectroscopy: Concepts and Chemical Applications. Angew Chem Int Ed. 2014;53:4756–4795. doi: 10.1002/anie.201205748. [DOI] [PubMed] [Google Scholar]
- 18.Anker JN, et al. Biosensing with plasmonic nanosensors. Nat Mater. 2008;7:442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 19.Bantz KC, et al. Recent progress in SERS biosensing. Phys Chem Chem Phys. 2011;13:11551–11567. doi: 10.1039/c0cp01841d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abalde-Cela S, et al. Surface-enhanced Raman scattering biomedical applications of plasmonic colloidal particles. J R Soc Interface. 2010;7:S435–S450. doi: 10.1098/rsif.2010.0125.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alvarez-Puebla RA, Liz-Marzan LM. SERS-Based Diagnosis and Biodetection. Small. 2010;6:604–610. doi: 10.1002/smll.200901820. [DOI] [PubMed] [Google Scholar]
- 22.Howes PD, Chandrawati R, Stevens MM. Colloidal nanoparticles as advanced biological sensors. Science. 2014;346:1247390–1247390. doi: 10.1126/science.1247390. [DOI] [PubMed] [Google Scholar]
- 23.Zeng SW, Baillargeat D, Ho HP, Yong KT. Nanomaterials enhanced surface plasmon resonance for biological and chemical sensing applications. Chem Soc Rev. 2014;43:3426–3452. doi: 10.1039/c3cs60479a. [DOI] [PubMed] [Google Scholar]
- 24.Pierson LS, Pierson EA. Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl Microbiol Biotechnol. 2010;86:1659–1670. doi: 10.1007/s00253-010-2509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mavrodi DV, et al. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol. 2001;183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dietrich LEP, et al. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol Microbiol. 2006;61:1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- 27.Dietrich LEP, Teal TK, Price-Whelan A, Newman DK. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science. 2008;321:1203–1206. doi: 10.1126/science.1160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramos I, Dietrich LEP, Price-Whelan A, Newman DK. Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res Microbiol. 2010;161:187–191. doi: 10.1016/j.resmic.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dietrich LEP, et al. Bacterial Community Morphogenesis Is Intimately Linked to the Intracellular Redox State. J Bacteriol. 2013;195:1371–1380. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price-Whelan A, Dietrich LE, Newman DK. Rethinking 'secondary' metabolism: physiological roles for phenazine antibiotics. Nat Chem Biol. 2006;2:71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- 31.Lee J, Zhang LH. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein & Cell. 2015;6:26–41. doi: 10.1007/s13238-014-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu XM, et al. Culture-free diagnostics of Pseudomonas aeruginosa infection by silver nanorod array based SERS from clinical sputum samples. Nanomedicine. 2014;10:1863–1870. doi: 10.1016/j.nano.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Puente V, et al. Plasmonic Mesoporous Composites as Molecular Sieves for SERS Detection. J Phys Chem Lett. 2013;4:2715–2720. [Google Scholar]
- 34.Yang S, Dai X, Boschitsch Stogin B, Wong T. Ultrasensitive surface-enhanced Raman scattering detection in common fluids. Proc Natl Acad Sci USA. 2015;113:268–273. doi: 10.1073/pnas.1518980113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamon C, et al. Hierarchical Self-Assembly of Gold Nanoparticles into Patterned Plasmonic Nanostructures. ACS Nano. 2014;8:10694–10703. doi: 10.1021/nn504407z. [DOI] [PubMed] [Google Scholar]
- 36.Reszka KJ, et al. Oxidation of pyocyanin, a cytotoxic product from Pseudomonas aeruginosa, by microperoxidase 11 and hydrogen peroxide. Free Radical Biol Med. 2004;36:1448–1459. doi: 10.1016/j.freeradbiomed.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 37.Lebeaux D, Chauhan A, Rendueles O, Beloin C. From in vitro to in vivo Models of Bacterial Biofilm-Related Infections. Pathogens. 2013;2:288–356. doi: 10.3390/pathogens2020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coenye T, Nelis HJ. In vitro and in vivo model systems to study microbial biofilm formation. J Microbiol Methods. 2010;83:89–105. doi: 10.1016/j.mimet.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 39.Koley D, Ramsey MM, Bard AJ, Whiteley M. Discovery of a biofilm electrocline using real-time 3D metabolite analysis. Proc Natl Acad Sci U S A. 2011;108:19996–20001. doi: 10.1073/pnas.1117298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bellin DL, et al. Integrated circuit-based electrochemical sensor for spatially resolved detection of redox-active metabolites in biofilms. Nat Commun. 2014;5:3256. doi: 10.1038/ncomms4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedersen SS, Shand GH, Hansen BL, Hansen GN. Induction of Experimental Chronic Pseudomonas-Aeruginosa Lung Infection with Pseudomonas-Aeruginosa Entrapped in Alginate Microspheres. APMIS. 1990;98:203–211. [PubMed] [Google Scholar]
- 42.Leevy WM, Serazin N, Smith BD. Optical Imaging of Bacterial Infection Models. Drug Discov Today Dis Models. 2007;4:91–97. doi: 10.1016/j.ddmod.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunter RC, et al. Phenazine Content in the Cystic Fibrosis Respiratory Tract Negatively Correlates with Lung Function and Microbial Complexity. Am J Respir Cell Mol Biol. 2012;47:738–745. doi: 10.1165/rcmb.2012-0088OC. [DOI] [PubMed] [Google Scholar]
- 44.Reuter K, Steinbach A, Helms V. Interfering with Bacterial Quorum Sensing. Perspect Medicin Chem. 2016;8:1–15. doi: 10.4137/PMC.S13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Loughlin CT, et al. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci U S A. 2013;110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welsh MA, Eibergen NR, Moore JD, Blackwell HE. Small Molecule Disruption of Quorum Sensing Cross-Regulation in Pseudomonas aeruginosa Causes Major and Unexpected Alterations to Virulence Phenotypes. J Am Chem Soc. 2015;137:1510–1519. doi: 10.1021/ja5110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dai TH, et al. Animal models of external traumatic wound infections. Virulence. 2011;2:296–315. doi: 10.4161/viru.2.4.16840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wessel AK, Hmelo L, Parsek MR, Whiteley M. Going local: technologies for exploring bacterial microenvironments. Nat Rev Microbiol. 2013;11:337–348. doi: 10.1038/nrmicro3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Connell JL, et al. Real-time monitoring of quorum sensing in 3D-printed bacterial aggregates using scanning electrochemical microscopy. Proc Natl Acad Sci U S A. 2014;111:18255–18260. doi: 10.1073/pnas.1421211111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buenger D, Topuz F, Groll J. Hydrogels in sensing applications. Prog Polym Sci. 2012;37:1678–1719. [Google Scholar]
- 51.Weibel DB, DiLuzio WR, Whitesides GM. Microfabrication meets microbiology. Nat Rev Microbiol. 2007;5:209–218. doi: 10.1038/nrmicro1616. [DOI] [PubMed] [Google Scholar]
- 52.Tian L, et al. Plasmonic Biofoam: A Versatile Optically Active Material. Nano Lett. 2016;16:609–616. doi: 10.1021/acs.nanolett.5b04320. [DOI] [PubMed] [Google Scholar]
- 53.Hamon C, et al. Hierarchical organization and molecular diffusion in gold nanorod/silica supercrystal nanocomposites. Nanoscale. 2016;8:7914–7922. doi: 10.1039/c6nr00712k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Falconnet D, Csucs G, Grandin HM, Textor M. Surface engineering approaches to micropattern surfaces for cell-based assays. Biomaterials. 2006;27:3044–3063. doi: 10.1016/j.biomaterials.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 55.Shao Y, Fu JP. Integrated Micro/Nanoengineered Functional Biomaterials for Cell Mechanics and Mechanobiology: A Materials Perspective. Adv Mater. 2014;26:1494–1533. doi: 10.1002/adma.201304431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mitragotri S, Lahann J. Physical approaches to biomaterial design. Nat Mater. 2009;8:15–23. doi: 10.1038/nmat2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watrous JD, Dorrestein PC. Imaging mass spectrometry in microbiology. Nat Rev Microbiol. 2011;9:683–694. doi: 10.1038/nrmicro2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fang JS, Dorrestein PC. Emerging mass spectrometry techniques for the direct analysis of microbial colonies. Curr Opin Microbiol. 2014;19:120–129. doi: 10.1016/j.mib.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang C, Flynn NT, Langer R. Controlled Structure and Properties of Thermoresponsive Nanoparticle–Hydrogel Composites. Adv Mater. 2004;16:1074–1079. [Google Scholar]
- 60.Scarabelli L, Grzelczak M, Liz-Marzan LM. Tuning Gold Nanorod Synthesis through Prereduction with Salicylic Acid. Chem Mater. 2013;25:4232–4238. [Google Scholar]
- 61.Crepaldi EL, et al. Controlled formation of highly organized mesoporous titania thin films: From mesostructured hybrids to mesoporous nanoanatase TiO2. J Am Chem Soc. 2003;125:9770–9786. doi: 10.1021/ja030070g. [DOI] [PubMed] [Google Scholar]
- 62.Qin Dong, X Y, Whitesides George M. Soft lithography for micro- and nanoscale patterning. Nature potocols. 2010 doi: 10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- 63.McDonald JC, et al. Fabrication of microfluidic systems in poly(dimethylsiloxane) Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 64.Alvarez-Puebla RA, et al. Au@pNIPAM Colloids as Molecular Traps for Surface-Enhanced, Spectroscopic, Ultra-Sensitive Analysis. Angew Chem Int Edit. 2009;48:138–143. doi: 10.1002/anie.200804059. [DOI] [PubMed] [Google Scholar]
- 65.Kern SE, Newman DK. Measurement of phenazines in bacterial cultures. Methods Mol Biol. 2014;1149:303–310. doi: 10.1007/978-1-4939-0473-0_25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.