Abstract
Chloroquine (CQ) and fansidar (sulphadoxine-pyrimethamine, SP) were widely used for treatment of Plasmodium falciparum for several decades in Malaysia prior to the introduction of Artemisinin-based Combination Therapy (ACT) in 2008. Our previous study in Kalabakan, located in south-east coast of Sabah showed a high prevalence of resistance to CQ and SP, suggesting the use of the treatment may no longer be effective in the area. This study aimed to provide a baseline data of antimalarial drug resistant markers on P. falciparum isolates in Kota Marudu located in the north-east coast of Sabah. Mutations on genes associated with CQ (pfcrt and pfmdr1) and SP (pfdhps and pfdhfr) were assessed by PCR amplification and restriction fragment length polymorphism. Mutations on the kelch13 marker (K13) associated with artemisinin resistance were determined by DNA sequencing technique. The assessment of pfmdr1 copy number variation associated with mefloquine resistant was done by real-time PCR technique. A low prevalence (6.9%) was indicated for both pfcrt K76T and pfmdr1 N86Y mutations. All P. falciparum isolates harboured the pfdhps A437G mutation. Prevalence of pfdhfr gene mutations, S108N and I164L, were 100% and 10.3%, respectively. Combining the different resistant markers, only two isolates were conferred to have CQ and SP treatment failure markers as they contained mutant alleles of pfcrt and pfmdr1 together with quintuple pfdhps/pfdhfr mutation (combination of pfdhps A437G+A581G and pfdhfr C59R+S108N+I164L). All P. falciparum isolates carried single copy number of pfmdr1 and wild type K13 marker. This study has demonstrated a low prevalence of CQ and SP resistance alleles in the study area. Continuous monitoring of antimalarial drug efficacy is warranted and the findings provide information for policy makers in ensuring a proper malaria control.
Introduction
Malaria remains one of the major health concerns where approximately half of the world's population is at risk. World Health Organization (WHO) reports show more than 214 million malaria cases globally and about 438,000 lives lost in Africa, South East Asia and Eastern Mediterranean region in 2014 [1]. Malaysia has executed better strategies for improvement in prevention and control of malaria since the introduction of the Malaria Eradication Programme in 1960. In 2011, the Malaria Control Programme was restructured from control to elimination, and the Ministry of Health, Malaysia has begun to implement the National Strategic Plan for Malaria Elimination in 2011–2020 [2].
Malaysia is in the pre-elimination phase and continues to progress towards elimination, reporting 3923 confirmed malaria cases in 2014 [1]. The incidence rate of malaria has declined from 16.1 per 100,000 populations in 2012 to 13.0 per 100,000 populations in 2014. Even though malaria control activities have significantly reduced malaria incidence in Malaysia, the disease still remain as main public health problem in the less developed areas of the country especially in Sabah. About forty-two percent of the malaria cases in Malaysia were reported from Sabah in the year 2013 [3].
In Malaysia, development of parasite resistance toward antimalarial drugs has led to increasing difficulties for sufficient malaria disease management and elimination. The widespread of resistance towards chloroquine (CQ) and sulphadoxine-pyrimethamine (SP) [4–9] has led Malaysia to change their antimalarial treatment policies to Riamet, a combination drug of artemether and lumefantrine (AL) for non-complicated P. falciparum malaria; while doxycycline are be given orally together with intravenous artesunate for complicated falciparum malaria treatment [2, 10].
Single nucleotide polymorphisms (SNPs) in P. falciparum CQ transporter (pfcrt) gene (K76T) and P. falciparum multidrug-resistance 1 (pfmdr1) gene (N86Y) have been associated with CQ resistance [11]. Additionally, several in vitro studies have demonstrated the association of high pfmdr1 copy number with lower parasites susceptibility to mefloquine and halofantrine [12, 13].
Resistance to SP drug combination has been shown to occur due to the alteration in the amino acid sequences of the P. falciparum dihydrofolate reductase (pfdhfr) [14] and P. falciparum dihydropteroate synthase (pfdhps) genes [15]. Specific changes of amino acid serine to asparagine at codon 108 (S108N) or isoleucine to leucine at codon 164 (I164L) on pfdhfr gene have been identified as the key determinants in the evolution of pyrimethamine (PYR) resistant in vitro [11]. The severity of pyrimethamine resistance often enhances by additional 51I and/or 59R mutation. Meanwhile, A437G and A581G point mutations on pfdhps gene confer resistance to sulphadoxine (SDX) in vitro enhanced by the presence of S436A, K540E and A613S [11]. Multiple mutation combinations of both pfdhps and pfdhfr were responsible in varying the degrees of SP resistance [11, 16, 17].
Resistance to artemisinin based combination therapies (ACT) has been observed in western Cambodia, Thailand, Vietnam, and Myanmar [18–21]. A previous study has identified mutations in the propeller domain of a kelch gene on chromosome 13 (PF3D7_1343700, K13 gene) as candidate molecular markers of ART resistance [22]. The prevalence of K-13 propeller region mutant alleles have been associated with parasite delayed clearance [22] played by Y493H, C580Y, M476I, R539T and I543T mutations [23].
To achieve malaria elimination status, wide coverage of molecular data on antimalarial drug resistance in Malaysia is needed for proper implementation of antimalarial drug treatment policy. Therefore, the aim of this study is to assess the prevalence of point mutations in the genes associated with CQ and SP resistance such as pfcrt (codon 76), pfmdr1 (codon 86), pfdhfr (codons 16, 51, 59, 108 and 164) and pfdhps (codons 437, 540, 581) on P. falciparum isolated in Kota Marudu, Sabah. In addition, we have also assessed the status of K-13 propeller polymorphisms and high pfmdr1 copy number variation which have been associated with artemisinin and mefloquine resistance, respectively. The data from this study could also contribute to a baseline information on distribution of antimalarial drug resistance particularly in Sabah prior to malaria elimination.
Materials and Methods
Study Site
Kota Marudu is one of the districts in Kudat division of Sabah with approximately 19.17 square kilometres of land. The population in Kota Marudu as in 2009 is approximately 72,900 with the average population of five per household [24]. In the first quarter of 2011, malaria cases in Kota Marudu contributed 10% of total malaria cases in Sabah. The majority of malaria cases in this area was majorly caused by P. falciparum infection followed by P. vivax and P. malariae. The district also falls under the list of high malaria endemic area in Sabah with total malaria endemicity of 10,000–50,000 [25].
Ethics approval and consent to participate
The study protocol was reviewed and approved by the Research Review Committee (RRC) of the Institute for Medical Research (IMR) and the Medical Research Ethics Committee (MREC), Ministry of Health Malaysia. All individuals were given a detailed explanation of the study procedures. Written informed consent was obtained from adult individuals or from parents or guardians of children under the age of 18 years.
Sample Collection
Cross-sectional community malaria screening surveys were conducted in malaria endemic areas of Kota Marudu as suggested by the Sabah State Health Department and Kota Marudu District Health Office in 2011 and 2014. Each screening survey was conducted in different areas. All blood samples from 4049 individuals (symptomatic and asymptomatic) were randomly collected by active case detection in more than 50 sites in deep forested areas or villages in Kota Marudu such as Sonsogun Mogis, Sonsogun Magandai, Pintasan Darat, Mampakad, Pinatau, Lotong, Launa, Lembiding, Linkabungan, Sunsui and Gana. The individuals were recruited at meeting points in each villages. House to house screening survey was also conducted. In addition, 21 P. falciparum infected samples from Kalabakan, Sabah were included in this study but limited to K13 propeller domain mutations and pfmdr1 copy number variation assessment. Blood film for malaria parasite technique (BFMP) was also prepared to confirm the infection. Malaria infected individuals were advised and transported to the nearest public hospitals for treatment.
Blood sample was obtained by finger prick and malaria infection was diagnosed using rapid diagnostic test kit (Paramax-3™, Zephyr Biomedicals, India). Approximately 100 μl of malaria infected bloods were spotted onto 3MM® Whatman (Brentford, United Kingdom) filter paper. The dried filter papers were labelled and transferred into individual plastic bags before being transported to the IMR in Kuala Lumpur, Malaysia. The blood-spotted filter papers were stored at room temperature in a dessicator containing silica gel until further processing.
DNA Extraction and Species Identification
Malaria parasite genomic DNA was extracted from filter papers using QIAmp DNA Mini Kit (QIAGEN, Germany) according to the manufacturer's instructions. A similar protocol was used to extract genomic DNA from the laboratory clone strains of P. falciparum (3D7, K1, T9.96 and W2) for PCR controls. Plasmodium species identification was also performed on malaria-infected samples by PCR as previously described [11, 26].
Molecular Analysis of Pfcrt, Pfmdr1, Pfdhps, Pfdhfr and K13 Propeller Domain
All PCRs were performed by using an Eppendorf Mastercycler Gradient (Eppendorf, Germany). The DNA from established laboratory strains of P. falciparum served as controls for PCR and enzyme digestions. Water was used to replace the DNA template in the PCR reaction for negative control. All restriction enzymes were brought from New England Biolabs (Beverly, Massachusetts, USA). The PCR products were analyzed by using the Agilent 2100 Bioanalyzer and the Agilent DNA 1000 Kit (Agilent Technologies, Molecular Probes Inc, USA). The PCR products were cleaned using QIAquick PCR Purification Kit (QIAGEN) before sending for sequencing. The DNA sequences were analyzed by using DNASTAR (Lasergene) and by using Molecular Evolutionary Genetics Analysis (MEGA) version 6.0 software.
The pfcrt K76T mutation analysis was performed as described by Djimde et al. [27] with minor adjustments to the concentrations of the reagents used for the PCR reaction [6]. The N86Y mutation in pfmdr1 gene was performed as described elsewhere [28].
Detection of pfdhps mutations at residues 437, 540, and 581 and pfdhfr mutations at residues 6, 51, 59, 108 and 164 were done as previously described [29] with some modification to DNA and primer concentrations [30]. The secondary PCR products containing the target region were subjected to RLFP for the detection of mutations at the various sites. The enzyme digestions were carried out according to published methods by Duraisingh et al. [29] and Jelinek et al. [31] for both pfdhps and pfdhfr genes, respectively.
The K13-propeller domain was amplified by using nested PCR as described by Ariey et al. [22]. The nested PCR products were evaluated by using Bioanalyzer and sent for sequencing. Sequences were assembled and manually edited by using DNASTAR (WI, USA). DNA sequences alignments were performed with the K13 sequence of the 3D7 clone (PF3D7_1343700) retrieved from PlasmoDB as reference sequence by using Molecular Evolutionary Genetics Analysis (MEGA) version 6.0 software.
Quantitation of Pfmdr1 Copy Number using Real-time Quantitative PCR
Real time PCR was performed with Rotor-Gene® Q (QIAGEN). Amplification reactions were done in triplicate by multiplex PCR combining both pfmdr1 and β-tubulin primers with probes as previously described [32]. The reagents used for each sample were 1X QuantiTect Multiplex (2X, NoROX), 400 nM of each forward and reverse pfmdr1 primer, 200 nM of pfmdr1 probe, 400 nM of each forward and reverse β-tubulin primer, 200 nM of β-tubulin probe, 2.5 μl of template DNA and sterile water in total volume of 10 μl. β-tubulin served as an internal standard for the amout of sample added to the reactions. Pfmdr1 copy number was calculated by the following formula: Copy number = 2-ΔΔCt with ΔΔCt denoting the difference between ΔCt of the unknown sample and ΔCt of the reference sample. The Efficiency (E) of the β-tubulin was assumed to be 2. The 3D7 (1 copy number) and IC (2 copies number) laboratory clone was used as the reference DNA sample respectively.
Results
Total samples
A total number of 4049 individuals were screened for malaria infection. Eighty-seven (2.15%, 87/4049) were positive for malaria infection and twenty-nine (33.33%, 29/87 malaria infected individuals) were infected with P. falciparum. All isolates were successfully genotyped by PCR-RFLP for drug resistant genes pfcrt, pfmdr1, pfdhps and pfdhfr. We evaluated mutant alleles in various locus: pfcrt (K76T), pfmdr1 (N86Y), pfdhps (A437G, K540E and A581G) and pfdhfr (A16V, N51I, C59R, S108T/N and I164L). In addition, pfmdr1 copy number variation and K13 propeller region point mutations were also determined in this study. Besides 29 Kota Marudu samples, an additional of 21 Kalabakan P. falciparum infected samples collected between 2008 to 2009 were included for pfmdr1 copy number variation and K13 propeller region mutations analysis. The P. falciparum isolates from Kalabakan have been previously analyzed for CQ and SP resistances [5, 6].
Pfcrt and pfmdr1
As shown in Table 1, the frequency of the pure mutant allele for both pfcrt K76T and pfmdr1 N86Y were low with 6.9% (2/29) prevalence, respectively. Only one mutant genotype for pfcrt and pfmdr1 gene was identified among the isolates in which, two samples harboured both mutant alleles for pfcrt and pfmdr1 gene (Table 1).
Table 1. Frequency of wild type and mutant alleles of pfcrt, pfmdr1, pfdhfr and pfdhps in P. falciparum isolates from Kota Marudu, Sabah.
| Drug resistant | Chloroquine | Sulfadoxine | Pyrimethamine | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genes | Pfcrt | Pfmdr1 | Pfdhps | Pfdhfr | ||||||
| Mutant Codon | K76T | N86Y | A437G | K540E | A581G | A16V | N51I | C59R | S108N | I164L |
| Frequency (%) | 6.9 | 6.9 | 100 | 0 | 6.9 | 0 | 0 | 100 | 100 | 10.3 |
| No. of mutant samples | 2/29 | 2/29 | 29/29 | 0/29 | 2/29 | 0/29 | 0/29 | 29/29 | 29/29 | 3/29 |
Pfcrt, P. falciparum chloroquine transporter gene; pfmdr1, P. falciparum multidrug resistant gene; pfdhfr, P. falciparum dihydropteroate synthase, pfdhps, P. falciparum dihydrofolate reductase. Amino acids: A, alanine, C, cysteine, E, glutamic acid, G, glycine, I, isoleucine, K, lysine, L, leucine, N, asparagine, R, arginine, S, serine, T, threonine, V, valine.
Pfdhps and pfdhfr
For the pfdhps, all isolates (N = 29) carried the essential A437G mutant allele. None of the isolates carrying K540E mutant allele. Two isolates (6.9%, 2/29) carried mutant alleles for A581G (Table 1). Out of 29 samples, only 2 isolates were found to have double mutations in pfdhps specifically at A437G + A581G (6.9%, 2/29).
For the pfdhfr, 3 isolates (10.3%, 3/29) harboured a mutation at codon I164L. Double mutations at codon C59R and S108N was found in all samples while mutations at codon A16V and N51I were absent (Table 1). Three isolates harboured triple mutations at C59R + S108N + I164L (10.3%, 3/29).
Combining the pfdhps and pfdhfr mutations, 3 genotypes were identified. The most common combinations were triple mutation of A437G (Pfdhps) + C59R + S108N (Pfdhfr) (89.7%, 26/29) (Table 2). There were two isolates (6.9%, 2/29 isolates) with quintuple mutation of A437G + A581G (Pfdhps) + C59R + S108N + I164L (Pfdhfr) followed by 1 isolate (3.4%, 1/29) with quadruple mutation of A437G (Pfdhps) + C59R + S108N + I164L (Pfdhfr) (Table 2).
Table 2. Frequency of combined pfdhfr /pfdhps mutants in isolates from Kota Marudu.
| Pfdhps codons | Pfdhfr codons | Frequency of mutant genotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 437 | 540 | 581 | 16 | 51 | 59 | 108 | 164 | pfdhfr/ pfdhps combination | % of mutant genotype | No. of mutants samples |
| G | L | A | A | N | R | N | I | 1 pfdhps/ 2 pfdhfr | 89.7 | 26/29 |
| G | L | A | A | N | R | N | L | 1 pfdhps/ 3 pfdhfr | 3.4 | 1/29 |
| G | L | G | A | N | R | N | L | 2 pfdhps/ 3 pfdhfr | 6.9 | 2/29 |
Pfcrt, P. falciparum chloroquine transporter gene; pfmdr1, P. falciparum multidrug resistant gene; pfdhfr, P. falciparum dihydropteroate synthase, pfdhps, P. falciparum dihydrofolate reductase. Amino acids: A, alanine, C, cysteine, E, glutamic acid, G, glycine, I, isoleucine, K, lysine, L, leucine, N, asparagine, R, arginine, S, serine, T, threonine, V, valine. Mutation is represented by amino acid in italic bold.
K13 propeller region and pfmdr1 copy number
The propeller region of K13 gene was successfully sequenced for P. falciparum isolates from Kota Marudu (N = 29) and Kalabakan (N = 21). The sequencing results of these isolates revealed no polymorphisms were detected in all 17 locations in K13 propeller region which confer resistance to ACT (S1 Fig).
For pfmdr1 copy number variation analysis, only 22 Kota Marudu and 16 Kalabakan isolates were successfully amplified by RT-PCR. Both P. falciparum isolates from Kota Marudu (N = 22) and Kalabakan (N = 16) were found to carry one copy number of pfmdr1 gene (S1 Table).
Discussion
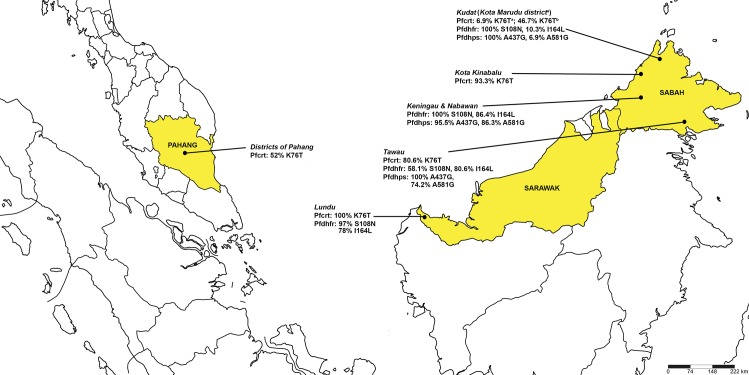
Our results demonstrated a low prevalence (6.9%) of pfcrt K76T mutation associated with resistance to CQ in P. falciparum isolated from Kota Marudu, Kudat Division of Sabah (Fig 1 and Table 3). Moreover, the same low percentage of mutation was detected for pfmdr1 N86Y, which is known to contribute to CQ resistance [33, 34]. Another study conducted in 2012 has detected 46.7% (7/15) of K76T mutation on clinical P. falciparum isolates in Kudat, Sabah (Fig 1 and Table 3). In contrast, a high prevalence rate of pfcrt K76T mutation was previously observed in Kalabakan and Kota Kinabalu, located in Tawau division and West Coast Division of Sabah, respectively (Fig 1 and Table 3) [6, 35]. A hundred percent prevalence of K76T mutation was reported in Lundu, Sarawak in a study conducted in 1999 and 2000 (Fig 1 and Table 3) [8]. A previous study by Atroosh et al.[9] have reported high prevalence rate of pfcrt K76T mutation in P. falciparum isolates in the Peninsular Malaysia (West Malaysia) area such as Pahang (Fig 1 and Table 3). The neighbouring countries including Thailand, Indonesia and Philippine [36–38] have also shown a high prevalence of the same mutation.
Fig 1. Distribution of essential mutations for CQ, PYR and SDX resistance in Malaysia.
The map shows the distribution of common SNPs for P. falciparum CQ, PYR and SDX resistant previously reported in Pahang [9], Lundu, Sarawak [8] and Sabah: Kalabakan (Tawau Division) [6, 30], Keningau & Nabawan (Interior Division) [39], Kota Kinabalu (West Coast Division) [35], aKota Marudu—present study, bKudat [35] (Kudat Division). This map was generated by using SimpleMappr online software [40]. Pfcrt, Plasmodium falciparum chloroquine resistant gene; Pfdhfr, Plasmodium falciparum dihydrofolate reductase gene; Pfdhps, Plasmodium falciparum dihydropteroate synthase gene; CQ, chloroquine; SDX, sulphadoxine; PYR, pyrimethamine. Amino acids: A, alanine; G, glycine; I, isoleucine; K, lysine; L, leucine N, asparagine; S, serine; T, threonine.
Table 3. Prevalence of CQ, PYR and SDX resistant marker genotypes in Malaysia reported from year 2003 to 2014.
| Sampling site | Year of sampling | Sampling method | Resistance | Genes | Codon mutations | Frequency (%) | Total sample | References |
|---|---|---|---|---|---|---|---|---|
| Lundu, Sarawak | 1999 and 2000 | PCD | CQ | pfcrt | K76T | 100.0 | 47/47 | Cox-Singh et al. 2003 [8] |
| PYR | pfdhfr | C59R/S108N | 19.0 | 6/47 | ||||
| PYR | pfdhfr | C59R/S108N/I164L | 78.0 | 25/47 | ||||
| Pahang districts | 2010–2011 | ACD/PCD | CQ | pfcrt | K76T | 52.0 | 39/75 | Atroosh et al. 2012 [9] |
| Kalabakan (Tawau), Sabah | 2008–2009 | ACD | CQ | pfcrt | K76T | 80.6 | 25/31 | Norahmad et al. 2011 [6] |
| PYR | pfdhfr | C59R/S108N | 3.2 | 1/31 | Abdullah et al. 2013 [5] | |||
| C59R/I164L | 22.6 | 7/31 | ||||||
| C59R/S108N/I164L | 51.6 | 16/31 | ||||||
| A16V/C59R/I164L | 3.2 | 1/31 | ||||||
| A16V/C59R/S108N/I164L | 3.2 | 1/31 | ||||||
| SDX | pfdhps | A437G/A581G | 74.2 | 23/31 | ||||
| Keningau/Nabawan, Sabah | 2010 | PCD | PYR | pfdhfr | C59R/S108N/I164L | 86.4 | 19/22 | Lau et al. 2013 [39] |
| N51I/C59R/S108N | 4.5 | 1/22 | ||||||
| C59R/S108N | 9.1 | 2/22 | ||||||
| SDX | pfdhps | A437G/K540T/A581G | 72.7 | 16/22 | ||||
| A437G | 9.1 | 2/22 | ||||||
| A437G/A581G | 13.6 | 3/22 | ||||||
| Kota Kinabalu, Sabah | 2012 | PCD | CQ | pfcrt | K76T | 93.3 | 14/15 | Tan et al. 2014 [35] |
| Keningau, Sabah | 2012 | PCD | CQ | pfcrt | K76T | 100 | 1/1 | |
| Kudat, Sabah | 2012 | PCD | CQ | pfcrt | K76T | 46.7 | 7/15 | |
| Kota Marudu (Kudat), Sabah | 2011 and 2014 | ACD | CQ | pfcrt | K76T | 6.9 | 2/29 | This study |
| PYR | pfdhfr | C59R/S108N | 89.7 | 26/29 | ||||
| C59R/S108N/I164L | 10.3 | 3/29 | ||||||
| SDX | pfdhps | A437G | 93.1 | 27/29 | ||||
| A437G/A581G | 6.9 | 2/29 |
ACD, active case detection; PCD, passive case detection; Pfcrt, Plasmodium falciparum chloroquine resistant gene; Pfdhfr, Plasmodium falciparum dihydrofolate reductase gene; Pfdhps, Plasmodium falciparum dihydropteroate synthase gene; CQ, chloroquine; SDX, sulphadoxine; PYR, pyrimethamine. Amino acids: A, alanine; C, cysteine; G, glycine; I, isoleucine; K, lysine; L, leucine; N, asparagine; R, arginine; S, serine; T, threonine; V, valine. Bold fonts indicates strong determinant of resistance for each antimalarial drug.
Mutations on pfdhps and pfdhfr genes associated with SP resistance have been reported in most part of malaria endemic areas in Sabah and Sarawak (Fig 1 and Table 3) [5, 8, 41] and the neighbouring South Kalimantan, Indonesia [42]. Combination of triple pfdhfr mutation and double pfdhps mutation (quintuple mutant) have been associated with SP treatment failure [16]. Based on the previous study carried out by Cox-Singh et al. [8] in Lundu, Sarawak, triple pfdhfr mutation (C59R/S108N/I164L) could also lead to SP treatment failure. However, the status of pfdhps mutation was not reported in the latter study. Our study showed that all P. falciparum isolates were harbouring essential mutant alleles in pfdhps (A437G and A581G) and pfdhfr genes (S108N and I164L) which confer to SDX and PYR resistances, respectively [11]. Triple mutation involving a combination of 1 mutant allele of pfdhps and 2 mutant alleles of pfdhfr (A437G/C59R/S108N) were predominant followed by low prevalence of quadruple (A437G/C59R/S108N/I164L) and quintuple (A437G/A581G/C59R/S108N/I164L) mutations combinations (Table 2). These combinations have also been observed in Keningau and Nabawan district in the interior of Sabah [41] and Kalabakan in the south-eastern coast of Sabah [30]. Although the prevalence of quadruple and quintuple combining mutations of pfdhps and pfdhfr were low, double mutations on pfdhfr alone have been associated with longer parasite clearance time and higher gametocytemia, the presence of gametocytes responsible for transmission [17].
An increased copy number of pfmdr1 gene was associated with in vitro and in vivo resistance towards mefloquine (MQ) and AL antimalarial drugs [32, 43]. It has been reported that due to frequent usage of MQ monotherapy in some parts of Thailand and Cambodia, the prevalence of P. falciparum isolates with a pfmdr1 copy number greater than 1 was found to be high [32]. Currently, the ASMQ fixed-dose combination is recommended as alternative treatment to AL in treating uncomplicated P. falciparum malaria in Malaysia [2]. The absence of increased pfmdr1 copy number suggests the efficacy of MQ in the study area.
P. falciparum resistance towards artemisinin is a major setback for malaria control in Southeast Asia. The neighbouring countries such as Thailand, Myanmar and Cambodia have actively reported the prevalence of C580Y which was the marker for slow-clearing P. falciparum in malaria patients treated with artemisinin and ACT [19, 20, 22]. Although the delayed parasite clearance after treatment with ACT has not yet been reported in Malaysia, characterizing the diversity of this gene is important to assess the potential for ACT drug resistance and to provide a baseline for future surveillance. Therefore, we sequenced the K13 propeller domain in P. falciparum isolates from Kota Marudu and Kalabakan. As this treatment has just been introduced in Malaysia, the absence of K13 propeller mutations in P. falciparum isolates were expected in the study area.
As Malaysia has been listed as one of the pre-elimination phase countries, a fragmented population structure of the P. falciparum populations is expected [44]. For example, the frequency of CQ resistant parasite in Kota Marudu were extremely low as compared to other areas such as Kalabakan. This suggested that the spread of resistant P. falciparum genotype in the region was contained within the malaria focus area. In addition, the genetic differentiation analysis has assured the large variation of genetic pattern between the P. falciparum populations in the areas of Sabah [45, 46]. Another possible explanation to this is the different sampling period between present study and previous studies conducted in Kalabakan [6, 30] and Pahang [9] areas where samples were collected before the implementation of ACT in the country. The effect of antimalarial drug interventions such as the use of AL in 2008 might have caused the extinction of CQ resistant P. falciparum lineages. The selection of the strongest P. falciparum population after ACT interventions had been demonstrated in Ghana and South American [47, 48].
As a developing area, malaria cases in Kota Marudu have become more confined to the rural population living in less accessible, hilly, forested hinterland, and to areas with inadequate transportation and communication facilities. These situations have made the ACD malaria screening in the study areas more challenging. The limitation of this study is the small number of sample (N = 29) collected in year 2011 and 2014. However, the study has randomly screened more than 4000 individuals living in the foci areas by ACD and it reflects the declining of malaria cases reported in Malaysia during the period of the study. Continuous molecular surveillance of antimalarial drug resistant markers is recommended to track the emergence and spread of P. falciparum mutation towards CQ, SP and ART resistance. This effort is also crucial to ensure the efficacy of malaria treatment and control programs, particularly in East Malaysia; Sabah and Sarawak.
Conclusion
This was the first molecular study carried out in this geographical area focusing on mutations of pfcrt, pfmdr1, pfdhps and pfdhfr genes that were strongly associated to CQ and SP resistance. This study showed low prevalence of resistance markers to CQ and SP that dramatically contrasted with the pattern observed with our previous study in Kalabakan, where higher pfcrt mutant allele and quintuple pfdhfr/pfdhps mutation were observed. Additionally, absence of increased pfmdr1 copy number and K-13 propeller domain mutations are expected due to limited usage of MQ and ACT in the study area.
Supporting Information
(PDF)
(PDF)
Acknowledgments
The authors would like to thank the Director General of Health Malaysia for the permission to publish this paper. We also thank the Director for the Institute for Medical Research (IMR), Kuala Lumpur for her critical review and support in publishing this paper; all the volunteers participating in this study; the officers and staffs of Kota Marudu and Kalabakan Health District Department for their efforts in helping us reaching to the study sites and recruiting the volunteers; and Dr Mallika’s lab staffs for their consultation in genotyping and interpretation of data. This study was supported by the National Institute of Health, Ministry of Health Malaysia.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The source of funding is National Institute of Health, Ministry of Health Malaysia. The sponsor’s role is to provide funding for the research only. It is the requirement of the sponsor (funder) that findings from the study should be published.
References
- 1.WHO. World Malaria Report 2015. Switzerland: World Health Organization, 2015. [Google Scholar]
- 2.Division VBDSDC. Management Guidelines Of Malaria In Malaysia. Malaysia: Ministry of Health Malaysia; 2014. [Google Scholar]
- 3.Centre HI. Health Indicators 2014. Malaysia: Ministry of Health Malaysia, 2014. 1511–4589. MOH/S/RAN/74.14(TR). [Google Scholar]
- 4.Montgomery R, Eyles DE. Chloroquine resistant falciparum malaria in Malaya. Trans R Soc Trop Med Hyg. 1963;57:409–16. . [DOI] [PubMed] [Google Scholar]
- 5.Abdullah NR, Norahmad NA, Jelip J, Sulaiman LH, Mohd Sidek H, Ismail Z, et al. High prevalence of mutation in the Plasmodium falciparum dhfr and dhps genes in field isolates from Sabah, Northern Borneo. Malar J. 2013;12(1):198 10.1186/1475-2875-12-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Norahmad NA, Abdullah NR, Yaccob N, Jelip J, Dony JF, Ruslan KF, et al. High prevalence of pfcrt K76t mutants among Plasmodium falciparum isolates from Sabah, Malaysia. The Southeast Asian journal of tropical medicine and public health. 2011;42(6):1322–6. . [PubMed] [Google Scholar]
- 7.Lokman Hakim S, Sharifah Roohi SW, Zurkurnai Y, Noor Rain A, Mansor SM, Palmer K, et al. Plasmodium falciparum: increased proportion of severe resistance (RII and RIII) to chloroquine and high rate of resistance to sulfadoxine-pyrimethamine in Peninsular Malaysia after two decades. Trans R Soc Trop Med Hyg. 1996;90(3):294–7. . [DOI] [PubMed] [Google Scholar]
- 8.Cox-Singh J, Lu HY, Davis TM, Ilett KF, Hackett LP, Matusop A, et al. Application of a multi-faceted approach for the assessment of treatment response in falciparum malaria: a study from Malaysian Borneo. Int J Parasitol. 2003;33(13):1545–52. . [DOI] [PubMed] [Google Scholar]
- 9.Atroosh WM, Al-Mekhlafi HM, Mahdy MA, Surin J. The detection of pfcrt and pfmdr1 point mutations as molecular markers of chloroquine drug resistance, Pahang, Malaysia. Malar J. 2012;11:251 10.1186/1475-2875-11-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rundi C. Malaria Elimination in Malaysia. Third annual meeting of the Asia Pacific Malaria Elimination Network (APMEN); Kota Kinabalu, Sabah, Malaysia: Ministry of Health Malaysia; 2011.
- 11.Wernsdorfer WH, Noedl H. Molecular markers for drug resistance in malaria: use in treatment, diagnosis and epidemiology. Current opinion in infectious diseases. 2003;16(6):553–8. 10.1097/01.qco.0000104295.87920.fd . [DOI] [PubMed] [Google Scholar]
- 12.Peel SA, Bright P, Yount B, Handy J, Baric RS. A strong association between mefloquine and halofantrine resistance and amplification, overexpression, and mutation in the P-glycoprotein gene homolog (pfmdr) of Plasmodium falciparum in vitro. The American journal of tropical medicine and hygiene. 1994;51(5):648–58. . [DOI] [PubMed] [Google Scholar]
- 13.Barnes DA, Foote SJ, Galatis D, Kemp DJ, Cowman AF. Selection for high-level chloroquine resistance results in deamplification of the pfmdr1 gene and increased sensitivity to mefloquine in Plasmodium falciparum. The EMBO journal. 1992;11(8):3067–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(23):9114–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Triglia T, Menting JG, Wilson C, Cowman AF. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(25):13944–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, et al. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. The Journal of infectious diseases. 2002;185(3):380–8. Epub 2002/01/25. 10.1086/338566 . [DOI] [PubMed] [Google Scholar]
- 17.Mendez F, Munoz A, Carrasquilla G, Jurado D, Arevalo-Herrera M, Cortese JF, et al. Determinants of treatment response to sulfadoxine-pyrimethamine and subsequent transmission potential in falciparum malaria. Am J Epidemiol. 2002;156(3):230–8. Epub 2002/07/27. . [DOI] [PubMed] [Google Scholar]
- 18.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM, et al. Evidence of artemisinin-resistant malaria in western Cambodia. The New England journal of medicine. 2008;359(24):2619–20. 10.1056/NEJMc0805011 . [DOI] [PubMed] [Google Scholar]
- 19.Imwong M, Jindakhad T, Kunasol C, Sutawong K, Vejakama P, Dondorp AM. An outbreak of artemisinin resistant falciparum malaria in Eastern Thailand. Sci Rep. 2015;5:17412 Epub 2015/12/01. 10.1038/srep17412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tun KM, Imwong M, Lwin KM, Win AA, Hlaing TM, Hlaing T, et al. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. The Lancet Infectious diseases. 2015;15(4):415–21. Epub 2015/02/24. 10.1016/S1473-3099(15)70032-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, et al. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet. 2012;379(9830):1960–6. Epub 2012/04/10. 10.1016/S0140-6736(12)60484-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505(7481):50–5. 10.1038/nature12876 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Straimer J, Gnadig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, et al. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347(6220):428–31. 10.1126/science.1260867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Department of Statistics M. Population Distribution and Basic Demographic Characteristics Kuala Lumpur: Population and Housing Census of Malaysia 2010. Department of Statistics, Malaysia, 2011. [Google Scholar]
- 25.Vector Borne Disease Control Sector M. Malaria and Filariasis Technical Meeting. Putrajaya: Ministry of Health, Malaysia, Vector Borne Disease Control Sector DCD; 2014 15 May 2014. Report No.
- 26.Padley D, Moody AH, Chiodini PL, Saldanha J. Use of a rapid, single-round, multiplex PCR to detect malarial parasites and identify the species present. Annals of tropical medicine and parasitology. 2003;97(2):131–7. 10.1179/000349803125002977 . [DOI] [PubMed] [Google Scholar]
- 27.Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, et al. A molecular marker for chloroquine-resistant falciparum malaria. The New England journal of medicine. 2001;344(4):257–63. 10.1056/NEJM200101253440403. . [DOI] [PubMed] [Google Scholar]
- 28.Tinto H, Ouedraogo JB, Erhart A, Van Overmeir C, Dujardin JC, Van Marck E, et al. Relationship between the Pfcrt T76 and the Pfmdr-1 Y86 mutations in Plasmodium falciparum and in vitro/in vivo chloroquine resistance in Burkina Faso, West Africa. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2003;3(4):287–92. . [DOI] [PubMed] [Google Scholar]
- 29.Duraisingh MT, Curtis J, Warhurst DC. Plasmodium falciparum: detection of polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes by PCR and restriction digestion. Experimental parasitology. 1998;89(1):1–8. 10.1006/expr.1998.4274 . [DOI] [PubMed] [Google Scholar]
- 30.Abdullah NR, Norahmad NA, Jelip J, Sulaiman LH, Mohd Sidek H, Ismail Z, et al. High prevalence of mutation in the Plasmodium falciparum dhfr and dhps genes in field isolates from Sabah, Northern Borneo. Malaria journal. 2013;12:198 10.1186/1475-2875-12-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jelinek T, Ronn AM, Lemnge MM, Curtis J, Mhina J, Duraisingh MT, et al. Polymorphisms in the dihydrofolate reductase (DHFR) and dihydropteroate synthetase (DHPS) genes of Plasmodium falciparum and in vivo resistance to sulphadoxine/pyrimethamine in isolates from Tanzania. Tropical medicine & international health: TM & IH. 1998;3(8):605–9. . [DOI] [PubMed] [Google Scholar]
- 32.Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364(9432):438–47. 10.1016/S0140-6736(04)16767-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Babiker HA, Pringle SJ, Abdel-Muhsin A, Mackinnon M, Hunt P, Walliker D. High-level chloroquine resistance in Sudanese isolates of Plasmodium falciparum is associated with mutations in the chloroquine resistance transporter gene pfcrt and the multidrug resistance Gene pfmdr1. The Journal of infectious diseases. 2001;183(10):1535–8. 10.1086/320195 . [DOI] [PubMed] [Google Scholar]
- 34.Setthaudom C, Tan-ariya P, Sitthichot N, Khositnithikul R, Suwandittakul N, Leelayoova S, et al. Role of Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes on in vitro chloroquine resistance in isolates of Plasmodium falciparum from Thailand. The American journal of tropical medicine and hygiene. 2011;85(4):606–11. 10.4269/ajtmh.2011.11-0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan LL, Lau TY, Timothy W, Prabakaran D. Full-length sequence analysis of chloroquine resistance transporter gene in Plasmodium falciparum isolates from Sabah, Malaysia. TheScientificWorldJournal. 2014;2014:935846 10.1155/2014/935846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rungsihirunrat K, Chaijareonkul W, Seugorn A, Na-Bangchang K, Thaithong S. Association between chloroquine resistance phenotypes and point mutations in pfcrt and pfmdr1 in Plasmodium falciparum isolates from Thailand. Acta tropica. 2009;109(1):37–40. 10.1016/j.actatropica.2008.09.011 . [DOI] [PubMed] [Google Scholar]
- 37.Maguire JD, Susanti AI, Krisin, Sismadi P, Fryauff DJ, Baird JK. The T76 mutation in the pfcrt gene of Plasmodium falciparum and clinical chloroquine resistance phenotypes in Papua, Indonesia. Annals of tropical medicine and parasitology. 2001;95(6):559–72. 10.1080/00034980120092516 . [DOI] [PubMed] [Google Scholar]
- 38.Chen N, Kyle DE, Pasay C, Fowler EV, Baker J, Peters JM, et al. pfcrt Allelic types with two novel amino acid mutations in chloroquine-resistant Plasmodium falciparum isolates from the Philippines. Antimicrobial agents and chemotherapy. 2003;47(11):3500–5. 10.1128/AAC.47.11.3500-3505.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau TY, Sylvi M, William T. Mutational analysis of Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase genes in the interior division of Sabah, Malaysia. Malar J. 2013;12:445 Epub 2013/12/11. 10.1186/1475-2875-12-445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shorthouse DP. SimpleMappr, an online tool to produce publication-quality point maps. 2010;Available: http://www.simplemappr.net.
- 41.Lau TY, Sylvi M, William T. Mutational analysis of Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase genes in the interior division of Sabah, Malaysia. Malaria journal. 2013;12:445 10.1186/1475-2875-12-445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basuki S, Fitriah, Riyanto S, Budiono, Dachlan YP, Uemura H. Two novel mutations of pfdhps K540T and I588F, affecting sulphadoxine-pyrimethamine-resistant response in uncomplicated falciparum malaria at Banjar district, South Kalimantan Province, Indonesia. Malaria journal. 2014;13:135 10.1186/1475-2875-13-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price RN, Uhlemann AC, van Vugt M, Brockman A, Hutagalung R, Nair S, et al. Molecular and pharmacological determinants of the therapeutic response to artemether-lumefantrine in multidrug-resistant Plasmodium falciparum malaria. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2006;42(11):1570–7. 10.1086/503423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Escalante AA, Ferreira MU, Vinetz JM, Volkman SK, Cui L, Gamboa D, et al. Malaria Molecular Epidemiology: Lessons from the International Centers of Excellence for Malaria Research Network. Am J Trop Med Hyg. 2015;93(3 Suppl):79–86. Epub 2015/08/12. 10.4269/ajtmh.15-0005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anthony TG, Conway DJ, Cox-Singh J, Matusop A, Ratnam S, Shamsul S, et al. Fragmented population structure of Plasmodium falciparum in a region of declining endemicity. The Journal of infectious diseases. 2005;191(9):1558–64. 10.1086/429338 . [DOI] [PubMed] [Google Scholar]
- 46.Mohd Abd Razak MR, Sastu UR, Norahmad NA, Abdul-Karim A, Muhammad A, Muniandy PK, et al. Genetic Diversity of Plasmodium falciparum Populations in Malaria Declining Areas of Sabah, East Malaysia. PloS one. 2016;11(3):e0152415 Epub 2016/03/31. 10.1371/journal.pone.0152415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alam MT, de Souza DK, Vinayak S, Griffing SM, Poe AC, Duah NO, et al. Selective sweeps and genetic lineages of Plasmodium falciparum drug -resistant alleles in Ghana. The Journal of infectious diseases. 2011;203(2):220–7. Epub 2011/02/04. 10.1093/infdis/jiq038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Griffing SM, Mixson-Hayden T, Sridaran S, Alam MT, McCollum AM, Cabezas C, et al. South American Plasmodium falciparum after the malaria eradication era: clonal population expansion and survival of the fittest hybrids. PloS one. 2011;6(9):e23486 Epub 2011/09/29. 10.1371/journal.pone.0023486 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.