Abstract
Anti-cancer immunotherapies commonly rely on the use of interleukin-2 (IL-2) to promote the expansion of T lymphocytes. IL-2- dependent proliferation is the culmination of a complex network of phosphorylation-driven signaling events that impact on gene transcription through mechanisms that are not clearly understood. To study the role of IL-2 in the regulation of nuclear protein function we have performed an unbiased mass spectrometry-based study of the nuclear phosphoproteome of resting and IL-2-treated CD4+ T lymphocytes. We detected 8521distinct phosphosites including many that are not yet reported in curated phosphorylation databases. Although most phosphorylation sites remained unaffected upon IL-2 treatment, 391 sites corresponding to 288 gene products showed robust IL-2-dependent regulation. Importantly, we show that ATP-citrate lyase (ACLY) is a key phosphoprotein effector of IL-2-mediated T-cell responses. ACLY becomes phosphorylated on serine 455 in T lymphocytes upon IL-2-driven activation of AKT, and depletion or inactivation of ACLY compromises IL-2-promoted T-cell growth. Mechanistically, we demonstrate that ACLY is required for enhancing histone acetylation levels and inducing the expression of cell cycle regulating genes in response to IL-2. Thus, the metabolic enzyme ACLY emerges as a bridge between cytokine signaling and proliferation of T lymphocytes, and may be an attractive candidate target for the development of more efficient anti-cancer immunotherapies.
The underlying principle of cancer immunotherapy is to eliminate malignant cells by tuning the immune system (1–2). This revolutionary way of fighting tumors was originated three decades ago when a patient suffering from metastatic melanoma was treated with the T-cell growth promoting factor interleukin-2 (IL-2)1 (3). The success of IL-2 administration in fighting metastatic melanoma demonstrated for the first time that solely potentiating the activation of T lymphocytes could abrogate certain human cancers (4). Current immunotherapy approaches include the use of autologous gene-engineered T cells that, once expanded ex vivo with IL-2, are re-infused back into patients by the so-called adoptive cell transfer therapy (ACT) (5–7). Despite the promising results of this approach, a safe and long-lasting expansion of transferred T cells remains a major challenge because of the undesirable side effects derived from the use of IL-2. Continued exposure to high doses of IL-2 results in increased susceptibility of T cells to apoptosis (8). Moreover, IL-2 is also a critical component for regulatory T-cell (Treg) development and function (9–10), and as such it functions as a negative regulator of the immune response (11). Consequently, although IL-2 constitutes a key component of current immunotherapies, a great deal of effort is being devoted to the development of novel strategies that would boost the T-cell immune response more safely. In this regard, it has been shown that IL-2-related toxicity can be partially minimized by the use of gene-engineered T cells expressing IL-2 receptor chimeras capable of transducing signals in the absence of the cytokine (12–13). A synthetic version of IL-2 with increased affinity for IL-2 receptor beta chain, named superkine or super-2, has also been proven to successfully induce the proliferation of cytotoxic T cells while eliciting a reduced expansion of Treg populations (14). However, although great improvements have been achieved since IL-2 was first administered to boost the immune system of a cancer patient, IL-2-related toxicity issues still persist in current anti-cancer immunotherapies. Therefore, a deeper understanding of IL-2-mediated regulatory mechanisms could help to design safer therapies.
Signaling cascades activated in response to IL-2 have been extensively studied since the late ′70s when the cytokine was discovered (15–17). Antigen binding to the T-cell receptor promotes IL-2 secretion as well as expression of IL-2 receptor (IL-2R), which comprises a hetero-oligomeric complex consisting of three polypeptide subunits named α, β and γ. Engagement of IL-2/IL-2R induces receptor oligomerization which results in the activation of members of the JAK tyrosine kinase family (18–19). JAK kinases associate with discrete regions of the IL-2R β and γ subunits, thereby phosphorylating the receptor and initiating a complex network of downstream signaling cascades (20–21). JAK/STAT, RAS/MAPK and PI3K/AKT pathways are the three main signaling branches activated upon IL-2 stimulation of T cells that culminate in immune response modulation (22–24). Traditionally, effectors involved in signal transduction have been analyzed using classical biochemical methods. However, this approach is hampered by the limited capacity to analyze biological systems in their complexity. Signaling pathways are interconnected protein networks, therefore they need to be analyzed as a whole. In this regard, mass spectrometry (MS)-based quantitative proteomics/phosphoproteomics has emerged as a powerful tool to investigate signal transduction cascades (25–27). In fact, using quantitative mass spectrometry techniques we have elucidated the global tyrosine phosphoproteome of IL-2 and IL-15 signaling pathways in T lymphocytes (28–29).
It has been shown that besides playing a key role in signal transduction, protein phosphorylation is an essential regulatory mechanism altering gene expression (30–31). IL-2-triggered signals are known to be transmitted to the nucleus leading to changes in gene expression regulation that generate the adequate cellular response and immune regulation. For instance, IL-2 treatment results in phosphorylation of STAT5A, which is a pre-requisite for this transcription factor to translocate into the nucleus and regulate gene expression (32–33). Despite its relevance, the precise influence of IL-2 in the modulation of nuclear phosphorylation and its impact on cellular proliferation remains obscure. In the present study we have examined the nuclear phosphoproteome of resting and IL-2-treated T lymphocytes. We report the identification and quantification of thousands of site-specific phosphosites, which represents the largest dataset of nuclear phosphorylations reported on human CD4+ T lymphocytes. Importantly, over 300 phosphosites were found to be specifically regulated upon IL-2 stimulation, including Ser455 on ATP-citrate lyase (ACLY), a metabolic enzyme responsible for generating acetyl-CoA. By inhibiting or depleting ACLY in T cells we demonstrate the relevance of this enzyme in IL-2-dependent modulation of histone acetylation levels and T-cell proliferation. Moreover, our study provides a collection of candidates for future hypothesis-driven experiments that could clarify the regulatory networks that lead to IL-2-triggered expansion of T cells and may serve as targets for cancer immunotherapy.
MATERIALS AND METHODS
Reagents, Antibodies, Peptides, and Primers
Human recombinant IL-2 was kindly provided by “AIDS Research and Reference Reagent Program”, Division of AIDS (NIH, National Institute of Health, Bethesda, MD). The following antibodies from (Cell Signaling, Danver, MA) were used for Western blotting assays: AKT (#-9272), phospho-AKT (Ser473) (#-4060), ACLY (#-13390), phospho-ACLY (Ser455) (#-4331), ERK1/2 (#-9102), phospho-ERK1/2 pThr202/Tyr204 (#-9101), Histone H3 (#-9715), and phospho-STAT5 (Tyr695) (#-9351). α-tubulin (T6199) was purchased from (Sigma-Aldrich, Copenhaguen, Denmark). Anti-acetyl histone H3 (06–599) and anti-acetyl histone H4 (06–598) used for Western blotting and chromatin immunoprecipitation were purchased from (Millipore, Darmstadt, Munich, Germany). The HPR-conjugated secondary antibodies anti-mouse (NA931) and anti-rabbit (NA934) were purchased from GE Healthcare. AKT inhibitor MK-2206 and HDAC inhibitor SAHA were purchased from (Selleckchem, Munich, Germany) MEK inhibitor U0126 from (Promega, Madison, WI) and ACLY inhibitor SB-204990 from (Tocris, Bristol, UK). Information concerning stable isotope labeled (SIL) peptides purchased for PRM assay and sequences of the qRT-PCR primers as well as primers used for site-directed mutagenesis are provided in supplemental Table S1.
Cell culture and stimulation
Human leukemic Kit225 T lymphocytes were maintained in RPMI 1640 media (Gibco, Hvidovre, Denmark) supplemented with 10% FBS, 1% glutamine, 1% sodium pyruvate, 1% penicillin/streptomycin and 16 U/ml of IL-2 at a density of 1.106 cell/ml at 37 °C and 5% CO2. Cell number was estimated using NucleoCounter (ChemoMetec, Allerod, Denmark). For the large-scale SILAC experiments, Kit225 T cells were grown in media containing either the light (Arg0/Lys0) or heavy (Arg6/Lys4) isotopes of lysine and arginine. Prior to cytokine treatment, cells were IL-2-starved for 48 h to synchronize them at G1 phase of the cell cycle. Stimulation was performed by incubating cells with 200 U/ml of IL-2 for 5 min at 37 °C and rapidly quenched by placing cells on ice and washing with cold PBS. In the SILAC experiments, T cells grown in light media were kept unstimulated and served as control whereas heavily labeled Kit225 T cells were treated with the cytokine.
Human U2OS osteosarcoma cell line was maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS).
Plasmids, Cell Transfection and Generation of Stable T-cell Line
The lentiviral vector pSicoR was obtained from (Addgene, Cambridge, MA) (Addgene Plasmid 11579) in accordance with Material Transfer Agreements (MTAs) and the lentiviral vector pCDF1-MCS2-EF1-Puro was purchased from (System Biosciences Co, Palo Alto, CA). In order to create a DNA construct for ACLY expression, the pSicoR plasmid was modified so that the U6-ShRNA cloning cassette was exchanged with EF1alpha-eYFP construct by PCR methods and conventional cloning. Simiarly, the CMV-eGFP cassette on the pSicoR plasmid was exchanged by EF1-Puro cassette from vector pCDF1-MCS2-EF1-Puro and puromycin resistance gene was substituted by ORF of ACLY cDNA. ACLY cDNA containing each single point mutation of interest was generated by standard site-directed DNA mutagenesis. The sequences of several clones were verified by DNA sequencing and error free clones were chosen for transfections. U2OS cells were transfected using XtremeGENE HD (Roche, Hvidovre, Denmark) following manufacturer′s recommendations.
The RNAi sequence potentially targeting human ACLY transcript was generated using available web resources (http://www.dharmacon.com) according to published recommendations for siRNA/shRNA design (34–35). The shRNA sequences were synthesized (DNA technology, Denmark) as two complementary DNA oligonucleotides: T(N19)TTCAAGAGA(rN19)TTTTTTC and TCGAGAAAAAA(N19)TCTCTTGAA(rN19)A where N19 is the 19-nt sense strand of the target sequence and rN19 is the complementary antisense strand. The targeting sequence (N19) we used for silencing ACLY was 5′-GGAGGAAGGGAATGAAACA-3′ whereas the random sequence 5′-GCAATATGACGAGTTAGTA-3′ was used as control. Annealed oligonucleotides were directly cloned into the lentiviral pSicoR-puro vector that allows puromycin resistance-based selection of shRNA-expressing cells. After DNA constructs were verified, lentiviral particles were generated as previously described (36). Briefly, HEK-293T cells were co-transfected with the lentiviral vector and the virus packaging plasmids using Metafectene and supernatants were collected 48 h and 72 h postinfection. Then viral particles were concentrated by ultracentrifugation and used for infection of Kit225 T cells to generate stable cell lines expressing the described RNAi constructs.
Nuclear and Phosphopeptide Enrichment
Unstimulated (Arg0/Lys0) and IL-2-treated (Arg6/Lys4) Kit225 T cells were subjected to cytosol and nuclear enrichment. Briefly, cells were resuspended in a lysis buffer (25 mm TrisHCl pH 7.5, 100 mm NaCl, 1% Nonidet P-40, complete protease inhibitor mixture tablets (Roche) and phosphoSTOP tablets (Sigma-Aldrich), and centrifuged for 15 min at 13,000 rpm. Cytosolic fraction was recovered and nuclei-containing pellet was disrupted by resuspending in urea buffer (8 m urea, 10 mm TrisHCl pH 7.5) and sonication. After estimating protein abundance by BCA method, light and heavy cell lysates were combined in 1:1 ratio according to their protein concentration and subjected to in-solution digestion using LysC and trypsin as previously described (37). Proteolytic digestion products were desalted on a Sep-Pak C18 cartridge (Waters, Milford), lyophilized for 2 days and enriched in phosphopeptides by consecutive incubations with TiO2 beads (38–39). Half sample of each fraction was directly analyzed by LC-MS/MS whereas the remaining halves were pooled together and further fractionated using high pH fractionation prior MS analysis (40).
Mass Spectrometry Analysis
Shotgun proteomic analyses were carried out using a reverse phase liquid chromatography system (EASY-nLC 1000 ultrahigh pressure, Thermo Fisher Scientific) interfaced with a Q Exactive HF mass spectrometer (Thermo Fisher Scientific, Waltham, MA) via a nanoelectrospray source (Thermo Fisher Scientific). Acidified peptides were loaded on an analytical in-house packed column (20 cm × 75 μm, ReproSil-Pur C18-AQ 3 μm resin (Dr. Maisch GmbH)) in solvent A (0.5% acetic acid) and eluted by a nonlinear 120 min solvent B gradient (0.5% acetic acid, 80% acetonitrile) at a flow rate of 250 nl/min. Q Exactive was operated in a top 10 data dependent mode. Survey scans were acquired at a resolution of 70,000 (m/z 400) and fragmentation spectra at 35,000 (m/z 400). Precursors were fragmented by higher energy C-trap dissociation (HCD) with normalized collision energy of 25 eV. The maximum injection time was 120 ms for survey and 124 ms for MS/MS scan whereas the AGC target values of 1e6 and 1e4 were used for survey scans and for MS/MS scans, respectively. Repeated sequencing of peptide was minimized by excluding the selected peptide candidates for 45 s.
All raw data files acquired were searched against the UniProt human database version 2014.01 (with 88,479 sequence entries) with MaxQuant proteomics computational platform version 1.3.0.5 and using Andromeda search engine (41). SILAC doublets were selected for the analysis where light and heavy labels were set as Arg0/Lys0 and Arg6/Lys4. Precursor and fragment mass tolerances were set to 7 and 20 ppm, respectively. Enzyme specificity was set to trypsin, allowing for cleavage N-terminal to proline and between aspartic acid and proline (with a maximum of 2 missed cleavages). Carbamidomethylation of C was set as fixed modification whereas oxidation of M, protein N-terminal acetylation, NQ deamidation and STY phosphorylation were selected as variable modifications for database searching. For the analysis of phosphopeptides, 1% FDR, a minimum localization probability of 0.75 and a score difference of at least 5 was used (42). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via PRIDE partner repository (43–44) with the data set identifier PXD002839.
PRM analyses were performed using a Q Exactive mass spectrometer (ThermoFisher Scientific) interfaced with an Easy-nLC 1000 nanoUPLC System (ThermoFisher Scientific). Phosphopeptides were air dried in a Speedvac and resuspended with the SIL phosphopeptide mixture. Samples were loaded onto an Acclaim PepMap100 precolumn (75 μm × 2 cm, ThermoFisher Scientific) connected to an Acclaim PepMap RSLC (50 μm × 15 cm, Thermo Scientific) analytical column. Peptides were eluted with a 90 min linear gradient from 3% to 30% of acetonitrile in 0.1% of formic acid at a flow rate of 300 nl/min directly onto the nanoES Emitter (ThermoFisher Scientific). The Q Exactive was operated in Targeted-MS2 mode and method optimization was achieved by analysis of SIL phosphopeptide by Full MS, Full MS/dd-MS2 (Top10) and Targeted-MS2. Then, selected m/z values were incorporated in an inclusion list and specific retention time windows were applied based on method optimization results. Spectra were acquired at a resolution of 17,500 (m/z 200). Peptide selection was done with an isolation window of 2.0 Th and normalized collision energy of 28 was applied for peptide fragmentation. The maximum injection time was 500 ms and an AGC target value of 5e5 was used. For peptide relative quantification Skyline v2.6.0.6851 software was used (45). All integrated peaks were manually inspected to ensure correct peak detection and total peak area values were exported into Microsoft Office Excel (Microsoft) for further data analysis.
Flow Cytometry
For cell proliferation assays, Kit225 T cells were incubated with 2.5 μm carboxyfluorescein diacetate succinimidyl ester (CFSE) (ThermoFisher Scientific) and U2OS cells with 0.5 μm CellTrace Far Red™ (CTFR) (ThermoFisher Scientific) for 15 min at 37 °C. Cells were washed with complete medium for 20 min, treated and cultured as indicated and finally fixed for 10 min in a solution of buffered formaldehyde (3.7%). Only YFP-expressing U2OS cells were selected for the dye dilution analysis and hence, estimation of the proliferation index. To assess cell cycle distribution, Kit225 T cells were fixed with chilled 70% ethanol and stained with 50 μg/ml propidium iodide. In both type of experiments fluorescence was detected using a FACSCalibur (BD Biosciences, San Jose, CA) flow cytometer. Proliferation Wizard software was used to identify cells in different generations and calculate the Proliferation Index whereas Summit 4.3 software was used to perform the cell cycle distribution analysis.
Crystal Violet Staining
U2OS cells were fixed for 10 min (buffered formaldehyde 3.7%), washed with PBS and subsequently stained with a 0.25% crystal violet solution. After removing excess stain, crystal violet was dissolved in 2% SDS solution for 30 min. The optical density of the extract was read with a spectrophotometer at 546 nm.
Acid Extraction of Histones
Cell pellets were resuspended in complete hypotonic buffer (10 mm Hepes, 10 mm KCl, 1.5 mm MgCl2, 0.5 mm DTT and phosphatase inhibitors and protease inhibitor mixture) supplemented with H2SO4 to a final concentration of 1% and incubated for 30 min at 4 °C with rotation. Samples were centrifuged for 10 min at 11,000 rpm at 4 °C and supernatant was collected to further precipitate protein by adding trichloroacetic acid (TCA) to 33% final concentration and incubating for 10 min at 4 °C. Following 5 min centrifugation at 14,000 rpm at 4 °C, pellets were washed twice with acetone and after allowing drying they were resuspended in MilliQ water.
Chromatin Immunoprecipitation (ChIP) Analyses
ChIP analyses were performed following previously published protocols with minor modifications (46–47). Briefly, differentially treated Kit225 T cells were crosslinked using formaldehyde and lysed. Nuclei were collected and subjected to sonication to fragment chromatin. DNA was immunoprecipitated using anti-H3 and anti-H4 antibodies and enriched chromatin was quantified by real-time PCR in the Applied Biosystems 7000 Real-Time PCR System using Power SYBR Green PCR Mastermix (Applied Biosystems).
Immunobloting
Equal amounts of protein lysates were loaded onto a precast gradient NuPAGE 4–12% Bis-Tris Gel (Invitrogen, Brondby, Denmark) and transferred into a nitrocellulose membrane using a semi-dry system. Membranes were blocked with 5% BSA in TTBS, incubated with the corresponding antibody overnight at 4 °C, washed prior incubation with the secondary antibody. Finally, ECL Western blotting detection reagents (PRN2106, GE Healthcare, Brondby, Denmark) and autoradiography films were used together with a Kodak Image Station 1000 to visualize the protein of interest. The intensity of the bands was quantified using ImageJ software (National Institutes of health, Bethesda, MD).
Gene Expression Analyses
Total RNA was prepared as previously described (48). Briefly, RNA was purified from Kit225 T cells using TRIzol (Invitrogen) according to manufacturer′s instructions. cDNA was synthesized from 1 μg of total RNA by incubating with a reaction mixture containing 0.25 μg of random hexamers and 1 mm dNTP mix for 5 min at 65 °C followed by 1h incubation at 37 °C with Invitrogen 1st Strand Buffer, 10 mm dithiothreitol and 12 units of M-MLV reverse transcriptase (Promega). Prior PCR analysis samples were diluted with 200 μl of sterile MilliQ water. For mRNA expression analysis, cDNA was amplified in the Stratagene Mx3000P System using SYBR Green JumpStart Taq Ready Mix (Sigma-Aldrich) and quantified using Stratagene MxPro. Amplification of β-actin was used for normalization.
Bioinformatic Analyses
The Perseus software was employed for the calculation of the statistical significance B, a p value that depends on protein intensities and ratios. It was also used to perform the hierarchical clustering analysis. DAVID functional annotation tool (49) was used to detect the overrepresented gene ontology (GO) terms “cellular compartment” and “biological process” within the nuclear phosphoproteome of Kit225 T cells and IL-2-regulated phosphoproteins. Overrepresented phosphorylation motifs were searched by MotifX (50) using a sequence window of ±6 residues adjacent to the phosphorylated amino acid. Then, NetworKIN algorithm (51) version 3.0 was used to predict the kinases responsible for phosphorylating the defined motifs. For each motif, the frequency of putative kinases was calculated discarding the kinases predicted to phosphorylate less than 5% of the sites sharing a motif. Finally, the frequency of each kinase was normalized to the most frequently occurring kinase for each motif.
Experimental Design and Statistical Rationale
To define the nuclear phosphoproteome of resting and IL-2-treated T cells we analyzed three biological replicas that were separately SILAC-labeled, stimulated, processed and finally analyzed by LC-MS/MS. Acquired raw files were searched using the MaxQuant version 1.3.0.5 and specifying the raw files that corresponded to each replica. Data was filtered by FDR < 1% and only the phosphosites displaying a localization probability above 0.75 and a score difference of at least 5 were considered as confident phosphorylated sites (Class I sites). From those, the phosphosites that were quantified in two out of the three biological replicas were analyzed using the Perseus software to calculate their statistical significance B value, a p value that depends on phosphopeptide intensities and ratios. We considered as IL-2-dependent phosphosites the ones that were consistently regulated (IL-2/Ctr > 2 or IL-2/Ctr < 0.5 and Significance B p value 0.05) in at least two out of the three replicas performed. Proliferation experiments were performed at least three times and statistical analysis was performed using the Student′s t test.
RESULTS
Defining the Nuclear Phosphoproteome of CD4+ T lymphocytes
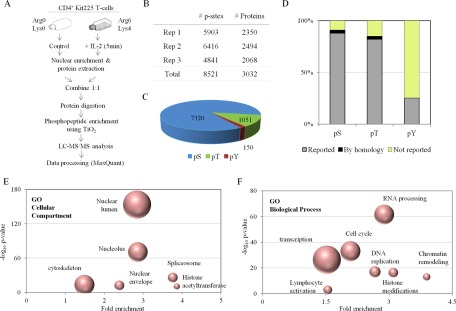
IL-2 signaling pathways converge into the nucleus to regulate gene expression and generate an adequate cellular response. Thus, a precise knowledge of the molecular mechanisms orchestrated in the nucleus of IL-2-treated T cells is essential for the better understanding of the molecular actions of the cytokine in CD4+ T lymphocytes. For that reason, we characterized on a global scale the IL-2-mediated regulation of nuclear function. We conducted an unbiased phosphoproteomic study based on the identification of site-specific phosphorylation events occurring primarily in the nucleus of resting and IL-2-treated Kit225 human T lymphocytes (Fig. 1A). Proteins were extracted from the nuclear-enriched fraction of SILAC-labeled T cells that were kept unstimulated (Arg0/Lys0) or were stimulated with IL-2 (Arg6/Lys4) for 5 min. Protein extracts derived from both conditions were combined at a 1:1 ratio according to protein concentration, digested with trypsin, and the resulting peptides were enriched in phosphopeptides using TiO2 beads prior to mass spectrometry (MS) analysis. We performed three biological replicates resulting in the identification of a total of 12,250 phosphorylated sites. Of these, 8521 could be confidently assigned to a specific position within a protein (median localization probability and score difference 0.999 and 35, respectively) (Fig. 1B). The 8,521 class I sites are comprised mainly by serine phosphorylations (pSer) although we also detected numerous phosphothreonine (pThr) and phosphotyrosine (pTyr) residues (Fig. 1C and supplemental Table S2). The majority of phosphosites that we identified have already been described and are reported in PhosphoSitePlus database. However, we now present evidence that a large number of phosphosites reported only in mice or rats in the database are also present in humans (termed as “by homology” in Fig. 1D). Furthermore, our study has identified many previously unreported phosphorylation sites (Fig. 1D), which represent a valuable source of novel information to understand the role of phosphorylation events in the nucleus of T lymphocytes.
Fig. 1.
Nuclear phosphoproteomic analysis of CD4+ T lymphocytes. A, Flow diagram of the MS-based quantitative phosphoproteomic strategy followed. B, Number of unique Class I phosphosites (p-sites) and corresponding proteins quantified in the three individual biological replicates. C, Distribution of all quantified phosphorylated serine, threonine and tyrosine residues. D, Proportion of phosphorylated sites reported or not in PhosphoSitePlus database. Gene Ontology analysis indicating the Cellular compartments (E) and biological processes (F) overrepresented within the phosphoproteome of CD4+ T lymphocytes. The fold enrichment and the statistical significance p value of the most indicative terms are indicated. The size of the dots correlate with the number of proteins grouped in the same term.
The 8521 class I sites detected in this study correspond to 3032 distinct gene products. These phosphoproteins were functionally classified by Gene Ontology (GO) analysis aiming to detect the most significantly enriched terms within the “Cellular Compartment” and “Biological Process” categories. Regarding the cellular compartment category, most of the phosphoproteins that we report belong to distinct nuclear localizations such as nuclear lumen, nucleolus or nuclear envelope, consistent with the aim of our study focused on depicting the nuclear phosphoproteome of T lymphocytes. Moreover, the spliceosome and histone acetyltransferase complexes were also highly enriched within our dataset of lymphocytic phosphoproteins (Fig. 1E). In line with these results, various biological processes related to key nuclear functions such as gene transcription and cell cycle regulation were significantly overrepresented among the phosphoproteins in our dataset (Fig. 1F).
IL-2-induced Changes in the Nuclear Phosphoproteome of T Cells
From the collection of unique class I phosphosites identified, a total of 8215 could be quantified and displayed a SILAC IL-2/control ratio. For subsequent analyses, we focused on 6145 phosphosites, which could be quantified in at least two out of the three biological replicates that we performed (Fig. 2A–2B, supplemental Table S3). These phosphosites belong to 2,382 proteins that were mainly monophosphorylated, although we also detected numerous nuclear proteins that were phosphorylated on multiple residues in Kit225 T lymphocytes (Fig. 2C).
Fig. 2.
Nuclear phosphoproteome in resting and IL-2-stimulated T lymphocytes. A, Overlap between the p-sites quantified in the three biological replicates performed. B, Number of unique p-sites and phosphoproteins overlapping between the different replicates. C, Distribution of number of p-sites identified and quantified for each phosphoprotein. D, Overall IL-2/Ctr SILAC ratio as a function of phosphopeptide intensity. In blue and red are indicated the p-sites down-regulated and up-regulated by IL-2, respectively. The MS spectra of ZNF292 pThr1017 and STAT5A pTyr694-bearing peptides that are down-regulated and up-regulated upon IL-2 stimulation respectively are shown. E, Phosphoproteins containing regulated p-sites may also have non-regulated sites. The size of the dots represents the amount of proteins following the same regulation. F, Biological processes over-represented among phosphoproteins with IL-2-regulated p-sites.
For deciphering IL-2-dependent nuclear phosphorylation events we only considered those sites that were consistently regulated (IL-2/Ctr ratio > 2 or IL-2/Ctr ratio < 0.5 and Significance B p value < 0.05) in at least 2 out of the three replicates. Following these criteria, we found that IL-2 triggers the phosphorylation of 331 sites and the de-phosphorylation of 59 sites (Fig. 2D, supplemental Table S4). Selected IL-2-dependent phosphorylations were validated by parallel reaction monitoring (PRM)-based targeted proteomics (supplemental Fig. S1). Several of the phosphorylation sites that are known to be induced upon IL-2 stimulation of T cells were also detected in our study, thus validating our approach (Table I). This is the case of STAT5A (Ser780/Tyr694), the two main representatives of the MAPK signaling cascades, MAPK1 (Thr185/Tyr187) and MAPK3 (Thr202/Tyr204), and AHNAK (Ser135) (28, 32–33, 52–53).
Table I. Known IL-2-induced phosphorylation sites. Positions of phosphosites are presented according to Uniprot number and the phosphorylated residue is highlighted in bold within the sequence window.
| Protein | Uniprot ID | Modified residue | Normalized IL-2/Ctr ratio |
Sequence window (±6aa) | ||
|---|---|---|---|---|---|---|
| Rep1 | Rep2 | Rep3 | ||||
| MAPK1 | P28482 | Tyr187 | 17.64 | 15.68 | 7.23 | TGFLTEYVATRWY |
| Thr185 | 8.95 | 9.21 | 9.74 | DHTGFLTEYVATR | ||
| MAPK3 | P27361 | Thr202 | 10.45 | 10.40 | 4.61 | DHTGFLTEYVATR |
| Tyr204 | 6.48 | 9.18 | 4.61 | TGFLTEYVATRWY | ||
| STAT5A | P42229 | Tyr694 | 7.59 | 10.12 | 6.29 | AKAVDGYVKPQIK |
| Ser780 | 2.19 | 2.11 | – | SLDSRLSPPAGLF | ||
| AHNAK | Q09666 | Ser135 | 4.38 | 4.22 | 3.89 | IKPRLKSEDGVEG |
However, the majority of IL-2-regulated phosphosites identified in this study have not been described previously. In addition, we identified a large number of multiply phosphorylated proteins highlighting the complex regulation exerted by IL-2 on individual nuclear proteins in T lymphocytes (Fig. 2E).
Gene Ontology analysis of IL-2-regulated phosphoproteins in Kit225 T cells revealed that biological processes related to gene expression and cell cycle regulation are the most highly enriched terms, in line with the documented pro-proliferative function of IL-2 (Fig. 2F). Our dataset of IL-2-modulated phosphoproteins includes several transcription factors (supplemental Table S5) as well as numerous enzymes involved in chromatin remodeling. We found that the phosphorylation status of several residues corresponding to histone methyltransferases and demethylases was increased upon IL-2 stimulation of T cells (Table II). Additionally, we detected differentially regulated phosphosites corresponding to distinct histone acetyltransferases (HAT) whereas the overall phosphorylation status of histone deacetylases (HDAC) seemed to be unaltered upon IL-2 stimulation in Kit225 T cells (Table II and supplemental Table S6). Altogether, our data indicate that nuclear events triggering IL-2-mediated T-cell proliferation are readily induced following 5 min of stimulation with the cytokine.
Table II. IL-2-modulated enzymes involved in chromatin remodelling. Positions of phosphosites are presented according to Uniprot number and the phosphorylated residue is highlighted in bold within the sequence window.
| Protein | Uniprot ID | Modified residue | Normalized IL-2/Ctr ratio |
Sequence window (±6aa) | |||
|---|---|---|---|---|---|---|---|
| Rep1 | Rep2 | Rep3 | |||||
| Histone lysine N-methyltransferases | MLL | Q03164-3 | Ser261 | 11.33 | 5.51 | – | KIKRTPSATFQQA |
| Ser2152 | 3.45 | 3.83 | – | PRIRTPSYSPTQR | |||
| Ser2199 | 3.14 | 3.03 | – | IGSRRHSTSSLSP | |||
| Ser3039 | 2.57 | 2.35 | 3.17 | GLQVPVSPTVPIQ | |||
| MLL2 | O14686-3 | Ser2277 | 3.03 | 2.74 | – | ASEPLLSPPPFGE | |
| WHSC1L1 | Q9BZ95 | Thr456 | 3.55 | 3.74 | – | HSQRRHTSAEEEE | |
| Lysine-specific demethylases | PHF2 | O75151 | Ser655 | 2.87 | 2.85 | 2.05 | ALRPPTSPGVFGA |
| Ser705 | 11.81 | 9.72 | 8.48 | APKRDLSFLLDKK | |||
| Ser899 | 3.27 | 2.80 | – | SKKRKGSDDAPYS | |||
| Ser929 | 3.54 | 2.28 | – | EGTRVASIETGLA | |||
| KDM3B | Q7LBC6 | Ser798 | 5.20 | 6.08 | 4.29 | EAVKRFSLDERSL | |
| Histone acetyltransferases | KAT6A | Q92794 | Ser941 | 4.63 | 3.82 | – | LPKRRLSEGVEPW |
| KAT7 | O95251 | Ser50 | 2.72 | 2.59 | – | RSSARLSQSSQDS | |
| Ser57 | 0.15 | 1.04 | 0.15 | QSSQDSSPVRNLQ | |||
To have a better understanding of the mechanisms leading to changes in the nuclear phosphoproteome of IL-2-treated T cells, we searched for the presence of overrepresented linear kinase motifs within IL-2-modulated phosphosites. We processed all phosphopeptide sequences bearing the IL-2-modulated phosphosite (pThr/pSer ± 6 aa) using MotifX software and detected five significantly enriched pSer motifs and a single pThr motif (Fig. 3A). The sequence logos of the two most enriched motifs (.R.R…… and …RR.S…) and of the most abundant motif (……SP….) among IL-2-dependent phosphosites are shown in Fig. 3B. To identify the putative kinases responsible for the phosphorylation of those motifs, all phosphopeptide sequences sharing the same linear kinase motif were examined using the NetworKIN software. The analysis revealed that the phosphoserine motifs in which the modified residue is preceded by a single basic amino acid are likely to be phosphorylated by protein kinase C beta (PKCβ) and CDC-like kinase 1 (CDK1) (Fig. 3A). The most enriched basic motifs (.R.R..S…… and …RR.S……) were predicted to be targeted among others, by the AKT, mTOR, p70S6 kinase and the ribosomal protein S6 kinase alpha-1 (RSK1) which are downstream effectors of the PI3K and MAPK signaling pathways (54–55). Additionally, the proline-directed kinases MAPK1/3 and CDK1/2 were predicted to phosphorylate the majority of the IL-2-regulated phosphosites (31%). Given that the two key residues modulating the activity of MAPK1 and MAPK3 are phosphorylated in our nuclear samples of IL-2 stimulated T cells (Table I), most phosphorylation events triggered by IL-2 in the nucleus of CD4+ T lymphocytes appear to be conducted by members of the MAPK family. Furthermore, our data set includes hundreds of other kinases and phosphatases that may also contribute to IL-2-driven nuclear phosphorylation events (Table III and supplemental Table S3).
Fig. 3.
Linear phosphorylation motif analyses of IL-2-regulated nuclear phosphoproteome. A, Significantly overrepresented linear motifs (middle) were identified by MotifX and then, motifs were matched to putative kinases using NetworKIN (left). Fold enrichment and number of p-sites showing the same linear motif are also indicated (right). B, The two most enriched (.R.R..S…… and …RR.S……) and the most abundant (…..SP….) phosphorylation motifs among regulated p-sites.
Table III. IL-2-regulated serine/threonine kinases and phosphatases. Positions of phosphosites are presented according to Uniprot number and the phosphorylated residue is highlighted in bold within the sequence window.
| Protein | Uniprot ID | Modified residue | Normalized IL-2/Ctr ratio |
Sequence window (±6aa) | |||
|---|---|---|---|---|---|---|---|
| Rep1 | Rep2 | Rep3 | |||||
| Ser/Thr kinases | MAPK1 | P28482 | Tyr187 | 17.64 | 15.68 | 7.23 | TGFLTEYVATRWY |
| Thr185 | 8.95 | 9.21 | 9.74 | DHTGFLTEYVATR | |||
| MAPK3 | P27361 | Thr202 | 10.45 | 10.40 | 4.61 | DHTGFLTEYVATR | |
| Tyr204 | 6.48 | 9.18 | 4.61 | TGFLTEYVATRWY | |||
| CDKL5 | O76039 | Ser407 | 2.46 | 2.39 | 2.64 | NIPHLLSPKEAKS | |
| SRPK2 | P78362–2 | Ser391 | 2.58 | 3.45 | – | DPTWIESPKTNGH | |
| PRKG1 | Q13976–2 | Thr532 | – | 0.25 | 0.45 | FGKKTWTFCGTPE | |
| PRKAR2B | P31323 | Ser114 | 0.27 | 0.20 | – | RFTRRASVCAEAY | |
| CDK13 | Q14004 | Ser1054 | 0.38 | 0.55 | 0.30 | SLGLDDSRTNTPQ | |
| Ser/Thr phosphatases | PPP1R10 | Q96QC0 | Ser313 | 2.19 | 2.13 | 2.66 | KKKKVLSPTAAKP |
| CTDSPL2 | Q05D32 | Ser28 | 7.14 | 7.23 | – | RAKRKYSEVDDSL | |
| PPP1R12A | O14974 | Ser507 | 5.42 | 4.39 | – | IPRRLASTSDIEE | |
| SSH1 | Q8WYL5 | Ser897 | 5.73 | 4.19 | – | EGGSLKSPPPFFY | |
ACLY, a Novel Target of IL-2-driven Phosphorylation
Most of the IL-2-regulated phosphosites reported in this study have never been shown to be modulated by the cytokine. Hence, corresponding phosphoproteins are putative candidates to participate in IL-2-dependent T-cell responses. We focused our attention on ACLY (ATP-citrate lyase) whose phosphorylation levels appeared to be regulated by IL-2 (Fig. 4A). ACLY is an acetyl-CoA generating enzyme that is thought to play a key role in the proliferation of tumor cells (56–57). However, the contribution of this enzyme to IL-2-triggered T-cell growth is unknown. We identified three phosphorylation sites on the central region of ACLY: pSer455 and pSer481, which precede the CoA binding domain, and pSer646, located between the CoA binding and ligase CoA domains (Fig. 4A). Importantly, this is the first study demonstrating that ACLY Ser646 can be phosphorylated in CD4+ T cells. The precise assignment of the newly identified phosphosite was verified by manual annotation of the fragmentation spectra (supplemental Fig. S2). Our analysis revealed that, whereas pSer481 and pSer646 remain unaffected by cytokine treatment, phosphorylation on ACLY Ser455 is induced in IL-2-treated T lymphocytes (Fig. 4B). IL-2-dependent up-regulation of ACLY pSer455 was confirmed by PRM-based targeted proteomics (Fig. 4C, supplemental Fig. S3). Because phosphorylation of Ser455 has been shown to stimulate the enzymatic activity of ACLY (58), our results suggest that treatment of T lymphocytes with IL-2 triggers functional activation of ACLY.
Fig. 4.
Characterization of IL-2-induced ACLY phosphorylation. A, Graphical representation of ACLY. The localization and sequence of the three phosphopeptides quantified in this study are indicated. B, IL-2/Ctr SILAC ratios obtained for each ACLY phosphopeptide in the three biological experiments. C, PRM-based validation of IL-2-induced phosphorylation of ACLY Ser455. D, Time-dependent phosphorylation of nuclear and cytoplasmic ACLY in response to IL-2 stimulation. E, IL-2 dose-dependent response of ACLY pSer455. F, IL-2-induced pSer455 on ACLY is mediated by AKT.
ACLY has been traditionally considered a cytosolic enzyme, although it was recently found in the nucleus of several types of mammalian cells as well (59). Our large-scale phosphoproteomic analysis using a nuclear-enriched fraction of T cells also suggested that ACLY could be present in the nucleus of CD4+ T lymphocytes. To confirm this, we followed an exhaustive nuclear enrichment protocol and investigated the presence of ACLY by Western blotting both in the nuclear and cytosolic fractions obtained from resting or IL-2-treated T cells. As shown in Fig. 4D, ACLY could be clearly detected in the nucleus and cytoplasm of Kit225 T cells independently of the stimulus. Importantly, ACLY phosphorylation on Ser455 was nearly undetectable in cytokine-starved T cells but was induced upon 5 min of stimulation with IL-2. In addition, we monitored the dynamics of ACLY pSer455 and showed that IL-2-induced phosphorylation of this site reached the maximum level after 20 min of stimulation both in the nucleus and cytoplasm and decreased subsequently (Fig. 4D).
To characterize further the regulation mediated by IL-2 on ACLY we performed a dose-dependent assay. We found that ACLY pSer455 is significantly induced in T cells treated with a saturating IL-2 concentration (6.25 nM) coinciding with the activation of the MAPK (or ERK) and PI3K signaling networks, as shown by the increased phosphorylation of their representative effectors MAPK1/3 and AKT, respectively (Fig. 4E). To disclose if any of these two signaling pathways is responsible for mediating IL-2-induced phosphorylation of ACLY, T cells were pre-treated with several concentrations of U0126 and MK-2206, specific inhibitors of MEK1/2 and AKT, respectively, and subsequently stimulated with IL-2. As shown in Fig. 4F, U0126 dramatically hampered IL-2-induced phosphorylation of MAPK1/3 or ERK1/2 without altering the phosphorylation status of ACLY. By contrast, pre-treatment with the AKT inhibitor resulted in a reduction of AKT phosphorylation and a notable decrease in IL-2-triggered ACLY pSer455. Altogether, our data indicate that phosphorylation of ACLY Ser455 is mediated by IL-2-induced activation of AKT.
ACLY Plays a Key Role in IL-2-dependent Proliferation of Kit225 T Cells
We and others have previously demonstrated that IL-2, or its closely related family member IL-15, is required for promoting the proliferation of Kit225 T cells (29, 53). In the present study we aimed to investigate the role of ACLY in IL-2-mediated T-cell growth. We generated a stable Kit225 T-cell line with efficient shRNA-mediated suppression of ACLY and a control cell line (Fig. 5A). Both were deprived of IL-2 for 2 days and then maintained in the presence of the cytokine for an additional 48 h. After this period, cell number was estimated. As shown in Fig. 5B, ACLY-depleted cells grew significantly less than control T lymphocytes. A similar effect was observed by estimating T-cell proliferation index by flow cytometry using the intracellular fluorescent dye CFSE, which is diluted twofold with each cell division. Proliferation of shACLY-expressing Kit225 T cells was clearly hampered in comparison to control cells 48 h after re-introduction of the cytokine, an effect that was even more dramatic 72 h after cytokine addition (Fig. 5C). To strengthen these data pointing to ACLY as a relevant mediator of IL-2-induced T-cell proliferation, we carried out additional experiments using a commercially available ACLY inhibitor (SB-204990). In agreement with the data gathered with shACLY, cell counting assay showed that T cells cultured with IL-2 in the presence of the ACLY inhibitor lack the capacity to proliferate (Fig. 5D). Moreover, CFSE-based flow cytometry analysis showed that the inhibitor dramatically compromised the capacity of the T lymphocytes to respond to IL-2 in a dose-dependent manner (Fig. 5E). Altogether, these results indicate that functional ACLY is required for Kit225 T cells to proliferate in the presence of IL-2.
Fig. 5.
ACLY activity is required for IL-2-triggered T-cell proliferation. A, Western blot showing total levels of ACLY in shCtr- and shACLY-expressing cells. B, Cell number was estimated in control and ACLY silencing Kit225 cells deprived with IL-2 and 48 h after re-introduction of the cytokine. C, Resting shCtr- and shACLY-expressing cells were stained with CFSE and cultured with IL-2 up to 48 h and 72 h. Then fluorescence was measured by flow cytometry and Proliferation Wizard software was used to calculate the proliferation index. D, Kit225 T-cell number was estimated in IL-2-deprived and IL-2-treated T cells that were grown in the absence or presence of the ACLY inhibitor. E, Proliferation index of Kit225 T cells cultured with IL-2 in the absence or presence of increasing concentrations of ACLY inhibitor for 2 days was estimated using CFSE dye dilution method. F, Proliferation index of YFP-ACLY phosphomimetic Ser455Asp (S455D) and Ser455Glu (S455E) and phosphomutant expressing U2OS cells in the presence of 5 μm AKT inhibitor was estimated by flow cytometry; mean ± S.D. of triplicates (*, p < 0.05). G, Images of crystal violet stained U2OS cells transfected with the indicated YFP-ACLY mutant and cultured in the presence of 5 μm AKT inhibitor. Relative quantification of the optical density is also shown; mean ± S.D. of triplicates (**, p < 0.0005; ***, p < 0.0001).
We next tested the impact of ACLY phosphorylation in cellular proliferation. We expressed YFP-ACLY Ser455 phosphomimetic Ser455Asp (S455D) or Ser455Glu (S455E) and phosphomutant Ser455Ala (S455A) proteins in U2OS cells. Transfected cells were grown in the presence of the AKT inhibitor MK-2206 for 24 h and subsequently the proliferation index of YFP+ U2OS cells was estimated by dye dilution assay using flow cytometry. As shown in Fig. 5F, ACLY S455D- and S455E-expressing cells proliferated significantly more than ACLY S455A-expressing cells. Accordingly, crystal violet staining revealed that ectopic expression of phosphorylation-mimicking mutants (ACLY S455D and S455E) promote higher cellular proliferation rates relative to the expression of ACLY S455A (Fig. 5G). These results suggest that ACLY is a relevant downstream effector of AKT involved in promoting the proliferation of cells.
To examine further the effect of ACLY depletion or inhibition in cellular proliferation, we monitored the distribution of cell cycle phases in Kit225 T cells. As expected, cytokine deprived T cells were arrested in the G1 phase of the cell cycle (G1 82%; S 9%; G2/M 7%) and entered the S phase 24 h after addition of IL-2 (G1 46%; S 41%; G2/M 12.5%) (Fig. 6A). By contrast, in the presence of ACLY inhibitor fewer cells could enter S phase, an effect that was more dramatic as the concentration of the inhibitor increased. In agreement with these results, we observed that the expression of several key regulators of the G1/S transition such as CDC25, CDK1, E2F2 and cyclin A (CYCA) was induced in IL-2-treated T cells but dramatically reduced in cells treated with the ACLY inhibitor (Fig. 6B). Collectively, our data show that inhibition of ACLY compromises IL-2-induced T-cell proliferation by reducing the expression of key genes required for G1/S transition of the cell cycle.
Fig. 6.
Inhibition of ACLY results in G1/S cell cycle arrest. A, Cell cycle distribution analysis of Kit225 T cells supplemented with IL-2 in the absence or presence of indicated concentration of ACLY inhibitor. B, mRNA expression levels of the genes (CDC25, CDK1, E2F1 and CYCA) was measured by qRT-PCR in control and IL-2-treated T cells that were or were not simultaneously treated with the ACLY inhibitor. Results were normalized against the expression levels of actin.
ACLY Influences Histone Acetylation Levels on CD4+ T lymphocytes
Acetyl-CoA is impermeable to membranes and consequently it is assumed that it is generated in the same subcellular compartment where it is required. Given the nuclear localization of ACLY in Kit225 T lymphocytes, we postulated that this enzyme could be a source of acetyl-CoA in the nucleus of IL-2-treated T cells. We observed that IL-2 stimulation leads to an increase in global acetylation of both histone 3 and 4 (H3 and H4) in human T lymphocytes (Fig. 7A). The increase in H4 acetylation observed is in line with the data presented by Taplick and colleagues showing that IL-2 promotes changes in the acetylation of H4 in murine T cells deprived of IL-2 (60). More interestingly, we found that IL-2-induced H3 and H4 acetylation was dramatically reduced upon inhibition of ACLY (Fig. 7A), suggesting that ACLY is a key player modulating histone acetylation in IL-2-treated T lymphocytes.
Fig. 7.
ACLY influences the histone acetylation level of IL-2-treated T cells. A, Acetylation of acid-extracted histones from starved T cells and T cells grown with IL-2 in the presence or absence of ACLY inhibitor. SAHA deacetylase inhibitor was used as control. B, Chromatin immunoprecipitation was performed with antibodies against acetylated H3 (AcH3) and AcH4 using starved T cells and T cells grown IL-2 in the presence or absence of ACLY inhibitor. Purified DNA was analyzed by Q-PCR using primers close to transcription start site of CDC25, CDK1, E2F2 and CYCA genes. Data are presented as percentage of input chromatin.
It is assumed that histone acetylation renders chromatin more accessible to nuclear factors, leading to enhanced gene expression (61–62). We aimed to determine if the impaired gene expression detected upon ACLY inhibition (Fig. 6B) could be explained by alterations in promoter histone acetylation levels. Using chromatin immunoprecipitation assays, we observed that H3 and H4 acetylation was increased on the promoters of CDC25, CDK1, E2F2, and CYCA upon IL-2 stimulation. By contrast, the acetylation levels of H3 and H4 were significantly reduced in these four promoters upon ACLY inhibition (Fig. 7B). These results correlate well with gene expression data presented above. Altogether, we provide evidence indicating that ACLY is involved in modulating histone acetylation levels that result in induction of gene expression to promote IL-2-dependent T-cell growth.
DISCUSSION
IL-2-induced proliferation of T lymphocytes is vital in the generation of the immune response and a key process in current anti-cancer immunotherapy (63–64). Nevertheless, IL-2 administration is usually accompanied by severe side effects and it is considered that a more detailed knowledge of IL-2-dependent signaling events should help improve the existing anti-cancer therapies. We have evaluated the nuclear phosphoproteome of resting and IL-2-treated T lymphocytes to gain an insight into the phosphorylation events occurring in the nucleus of T cells upon IL-2 stimulation. We report the identification of a large number of IL-2-regulated phosphoproteins, including the metabolic enzyme ACLY. Importantly, we present mechanistic evidence suggesting that ACLY is a crucial player in IL-2-induced T-cell proliferation.
Our nuclear phosphoproteomic screen has uncovered over 8000 phosphosites, including many previously not reported ones and thus, represents the most extensive phosphoproteomic screen performed to date in CD4+ T lymphocytes. In addition, applying a SILAC-based quantification strategy allowed us to discern between IL-2-dependent and -independent phosphorylation events. In agreement with most differential phosphoproteomic studies performed to date the vast majority of phosphosites detected were not affected by cytokine stimulation, but they could nevertheless play a role in T-cell biology. This could be the case of constitutive phosphorylations on histone deacetylases (HDACs). Indeed, Navarro and colleagues demonstrated that basal serine-threonine phosphorylations on HDACs play a central role in dictating the capacity of cytotoxic T lymphocytes (CTL) to respond to T-cell receptor (TCR) signaling (65). Interestingly, phosphorylation status of HDACs also remains unaffected by IL-2 stimulation (supplemental Table S7), although their role in T-cell fate has not been examined in the present work.
Without underestimating the relevance of basal phosphorylations, we focused our efforts on characterizing the fraction of phosphosites that was modulated by IL-2. We detected five times more phosphosites that were up-regulated by IL-2 relative to those down-regulated by the cytokine. A similar distribution was observed upon TCR engagement on CTLs (65). Kinase prediction tools anticipated that the majority of the IL-2 induced phosphosites detected were likely to be phosphorylated by members of the MAPK family, which we found phosphorylated and therefore activated. Although MAP kinases also become active upon TCR engagement, Navarro and co-workers showed that TCR-induced phosphorylations do not depend so much on MAPK activity, unveiling a relevant difference between TCR and IL-2 mediated phosphorylations (65). In addition, the phosphorylation status of certain chromatin re-modeling factors is also differentially affected upon stimulation of TCR or IL-2R. For instance, CBX3 Ser93 becomes phosphorylated in IL-2-treated CD4+ T cells but not in CTLs (66). By contrast, SMARCC2 pSer283 and CBX3 pSer95 remain unaffected in TCR- and IL-2-treated cells (supplemental Table S7) whereas TRIM28 pSer473 and ACLY pSer455 are induced in both cases.
Metabolic enzymes have emerged as pivotal linkers between signaling networks and proliferative responses (67–68). ACLY is a metabolic enzyme that mediates the conversion of mitochondrial-derived citrate into acetyl-CoA, an essential metabolite at the intersection of anabolism and catabolism. Its enzymatic activity is modulated by phosphorylation, and in addition to the catalytic autophosphorylation site (His760), at least three site-specific phosphorylations (Thr447, Ser451 and Ser455) are known to influence the activity of ACLY (58, 69). ACLY Ser451-containing peptide was repeatedly detected in our study but never identified as phosphorylated on that specific residue, suggesting that Ser451 is unlikely to be phosphorylated in resting or cytokine-treated Kit225 T cells. Conversely, we demonstrated that ACLY Ser455 becomes phosphorylated in IL-2-treated T cells, a modification that is known to increase up to sixfold the catalytic activity of the enzyme. ACLY Ser455 phosphorylation has also been demonstrated in TCR-stimulated cytotoxic T lymphocytes (65) and also in IL-3-treated hematopoietic cells (56). Therefore, distinct mitogenic signals affecting immune cells may converge on the phosphorylation on this specific residue of ACLY to elicit a related cellular response. Notably, among the signaling pathways that are activated in response to IL-2, our results indicate that PI3K/AKT pathway is responsible for the phosphorylation of ACLY Ser455 in T cells (Fig. 8).
Fig. 8.
IL-2 Signaling to histone acetylation via ACLY. Our working model suggests that ACLY pSer455 is induced upon IL-2-triggered AKT activation. Consequently, the enzyme becomes active and generates acetyl-CoA which serves as substrate for HATs that induce acetylation of histones in the promoters of cell cycle-related genes. Accordingly, the transcription of these genes is induced and T cells proliferate.
ACLY has recently emerged as a relevant therapeutic target in anti-cancer treatments because of the capacity of its product acetyl-CoA to promote proliferation in yeast and mammalian cells (70–72). The acetyl-CoA generated through ACLY activity is the building block for the biosynthesis of fatty acids and cholesterol (73) and also serves as substrate for acetylation reactions that affect a plethora of cellular functions both in the cytoplasm and nucleus (74–76). In line with this, we demonstrate here that ACLY inhibition is accompanied by a reduction of overall histone acetylation levels that could lead to suppression of IL-2-induced T-cell growth. A similar effect was observed upon down-regulation of AKT, the kinase involved in the phosphorylation of ACLY Ser455 in Kit225 T-cells. Interestingly, Lee and colleagues have recently shown that AKT-dependent phosphorylation of ACLY is sufficient to sustain high histone acetylation levels in LN229 glioblastoma cells even when nutrients are limited (71). In line with this, we observed that inhibition of ACLY abolished IL-2-induced histone acetylation on the promoter of key proliferation-related genes such as E2F2 and CDK1, which correlated with a reduction of gene expression and subsequent arrest at the G1/S transition of the cell cycle (Fig. 7). Nevertheless, it is unlikely that the function of nuclear ACLY is restricted to modulating the acetylation of histones; acetylation of additional proteins such as transcription factors may also be affected by ACLY activity. Furthermore, ACLY is also present in the cytoplasm of Kit225 T cells where it follows the same IL-2-dependent regulation of phosphorylation as the nuclear population. Thus, in addition to its nuclear function, ACLY probably performs a cytoplasmic function by triggering the lipogenic pathway in IL-2-treated Kit225 T cells. The anti-proliferative effects observed upon ACLY inhibition in T cells may partially be the result of a disrupted lipid metabolism. Indeed, several studies have shown that inhibition or depletion of the enzyme results in impaired lipid synthesis and subsequent tumor growth suppression (56–57, 73). Further investigation will clarify the role of cytoplasmic ACLY in mediating IL-2 initiated proproliferative pathways.
Recently, two additional sources of nuclear acetyl-CoA have been described and both were detected in our large-scale phosphoproteomics study. Mitochondrial pyruvate dehydrogenase complex (PDC) has been shown to translocate into the nucleus of human lung carcinoma cells, and to generate acetyl-CoA from pyruvate, which can be used for epigenetic regulation (77). According to our data the three inhibitory phosphorylations on the E1 alpha subunit of PDC (Ser232, Ser293, and Ser300) remain unaffected by IL-2 treatment, indicating that although inactive, PDC localizes to the nucleus of human leukemic Kit225 T lymphocytes. Similarly, we detected two non-regulated phosphosites of unknown function corresponding to acetyl-CoA synthetase (ACSS2), which generates acetyl-CoA using acetate as substrate. Although we cannot come to any conclusion regarding the activity of ACSS2, our data suggests that this metabolic enzyme is present in the nucleus of CD4+ T cell, consistent with other reports (59). Despite the coexistence of several acetyl-CoA-generating enzymes in the nucleus of Kit225 T cells, considering the impact of ACLY inhibition in IL-2-triggered histone acetylation, it is likely that ACLY is the principal source of nuclear acetyl-CoA in the nucleus of Kit225 T cells.
In summary, we have conducted a quantitative phosphoproteomic screen that has uncovered the complexity of phosphorylation-dependent nuclear IL-2 regulatory networks. A large set of putative candidates have been identified, which could be used in future studies. Characterization of the role of additional players in IL-2-induced T-cell growth should give a better understanding of the molecular events initiated by the cytokine that may help design more efficient and less harmful therapies.
Supplementary Material
Footnotes
Author contributions: N.O., B.B., and I.K. designed research; N.O., J.M., and K.A. performed research; N.O., V.A., and B.B. contributed new reagents or analytic tools; N.O., J.M., and K.A. analyzed data; N.O., V.S., and I.K. wrote the paper; J.M.A., A.M.Z., B.B., and I.K. evaluate and discuss research.
* This project has been funded by a grant from the Novo Nordisk Foundation and the Lundbeck Foundation to IK, and grants from the Basque Government (IT634–13) and the University of the Basque Country UPV/EHU (UFI11/20) to AMZ. N.O. is supported by the Lundbeck Foundation. I.K. is supported by grants from the Danish Natural Science Research Council and the Danish Medical Research Council. JMA's lab is a member of Proteored, PRB2-ISCIII and is supported by grant PT13/0001 funded by ISCIII and FEDER (European Regional Development Fund). Targeted proteomic analysis was performed in the Proteomics Core Facility-SGIker at the University of the Basque Country.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- IL
- Interleukin
- ACLY
- ATP-citrate lyase
- CTL
- cytotoxic T lymphocytes
- CYCA
- Cyclin A
- HDAC
- histone deacetylase
- IP
- immunoprecipitation
- NK-cell
- natural killer cell
- pSer
- phosphoserine
- pThr
- phosphothreonine
- pTyr
- phosphotyrosine
- PRM
- Parallel Reaction Monitoring
- Q-MS
- quantitative mass spectrometry
- SILAC
- stable isotope labeling by amino acids in cell culture
- TCR
- T-cell receptor.
REFERENCES
- 1. Restifo N. P., Dudley M. E., and Rosenberg S. A. (2012) Adoptive immunotherapy for cancer: harnessing the T cell response. Nat. Rev. Immunol. 12, 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vanneman M., and Dranoff G. (2012) Combining immunotherapy and targeted therapies in cancer treatment. Nat. Rev. Cancer 12, 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rosenberg S. A. (2014) IL-2: the first effective immunotherapy for human cancer. J. Immunol. 192, 5451–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fyfe G., Fisher R. I., Rosenberg S. A., Sznol M., Parkinson D. R., and Louie A. C. (1995) Results of treatment of 255 patients with metastatic renal cell carcinoma who received high-dose recombinant interleukin-2 therapy. J. Clin. Oncol 13, 688–696 [DOI] [PubMed] [Google Scholar]
- 5. Charo J., Finkelstein S. E., Grewal N., Restifo N. P., Robbins P. F., and Rosenberg S. A. (2005) Bcl-2 overexpression enhances tumor-specific T-cell survival. Cancer Res. 65, 2001–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Di Stasi A., De Angelis B., Rooney C. M., Zhang L., Mahendravada A., Foster A. E., Heslop H. E., Brenner M. K., Dotti G., and Savoldo B. (2009) T lymphocytes coexpressing CCR4 and a chimeric antigen receptor targeting CD30 have improved homing and antitumor activity in a Hodgkin tumor model. Blood 113, 6392–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kerkar S. P., Goldszmid R. S., Muranski P., Chinnasamy D., Yu Z., Reger R. N., Leonardi A. J., Morgan R. A., Wang E., Marincola F. M., Trinchieri G., Rosenberg S. A., and Restifo N. P. (2011) IL-12 triggers a programmatic change in dysfunctional myeloid-derived cells within mouse tumors. J. Clin. Invest. 121, 4746–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lenardo M. J. (1991) Interleukin-2 programs mouse alpha beta T lymphocytes for apoptosis. Nature 353, 858–861 [DOI] [PubMed] [Google Scholar]
- 9. Malek T. R. (2003) The main function of IL-2 is to promote the development of T regulatory cells. J. Leukoc Biol. 74, 961–965 [DOI] [PubMed] [Google Scholar]
- 10. Sakaguchi S. (2004) Naturally arising CD4+ regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu. Rev Immunol. 22, 531–562 [DOI] [PubMed] [Google Scholar]
- 11. Fehervari Z., Yamaguchi T., and Sakaguchi S. (2006) The dichotomous role of IL-2: tolerance versus immunity. Trends Immunol 27, 109–111 [DOI] [PubMed] [Google Scholar]
- 12. Hunter M. R., Prosser M. E., Mahadev V., Wang X., Aguilar B., Brown C. E., Forman S. J., and Jensen M. C. (2013) Chimeric gammac cytokine receptors confer cytokine independent engraftment of human T lymphocytes. Mol. Immunol. 56, 1–11 [DOI] [PubMed] [Google Scholar]
- 13. Sogo T., Kawahara M., Ueda H., Otsu M., Onodera M., Nakauchi H., and Nagamune T. (2009) T cell growth control using hapten-specific antibody/interleukin-2 receptor chimera. Cytokine 46, 127–136 [DOI] [PubMed] [Google Scholar]
- 14. Levin A. M., Bates D. L., Ring A. M., Krieg C., Lin J. T., Su L., Moraga I., Raeber M. E., Bowman G. R., Novick P., Pande V. S., Fathman C. G., Boyman O., and Garcia K. C. (2012) Exploiting a natural conformational switch to engineer an interleukin-2 ‘superkine’. Nature 484, 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arima N., Kamio M., Imada K., Hori T., Hattori T., Tsudo M., Okuma M., and Uchiyama T. (1992) Pseudo-high affinity interleukin 2 (IL-2) receptor lacks the third component that is essential for functional IL-2 binding and signaling. J. Exp. Med. 176, 1265–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Evans G. A., Goldsmith M. A., Johnston J. A., Xu W., Weiler S. R., Erwin R., Howard O. M., Abraham R. T., O'Shea J. J., Greene W. C., and et al. (1995) Analysis of interleukin-2-dependent signal transduction through the Shc/Grb2 adapter pathway. Interleukin-2-dependent mitogenesis does not require Shc phosphorylation or receptor association. J. Biol. Chem. 270, 28858–28863 [DOI] [PubMed] [Google Scholar]
- 17. Weissman A. M., Harford J. B., Svetlik P. B., Leonard W. L., Depper J. M., Waldmann T. A., Greene W. C., and Klausner R. D. (1986) Only high-affinity receptors for interleukin 2 mediate internalization of ligand. Proc. Natl. Acad. Sci. U.S.A. 83, 1463–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Friedmann M. C., Migone T. S., Russell S. M., and Leonard W. J. (1996) Different interleukin 2 receptor beta-chain tyrosines couple to at least two signaling pathways and synergistically mediate interleukin 2-induced proliferation. Proc. Natl. Acad. Sci. U.S.A. 93, 2077–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnston J. A., Bacon C. M., Finbloom D. S., Rees R. C., Kaplan D., Shibuya K., Ortaldo J. R., Gupta S., Chen Y. Q., Giri J. D., and et al. (1995) Tyrosine phosphorylation and activation of STAT5, STAT3, and Janus kinases by interleukins 2 and 15. Proc. Natl. Acad. Sci. U.S.A. 92, 8705–8709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miyazaki T., Kawahara A., Fujii H., Nakagawa Y., Minami Y., Liu Z. J., Oishi I., Silvennoinen O., Witthuhn B. A., Ihle J. N., and et al. (1994) Functional activation of Jak1 and Jak3 by selective association with IL-2 receptor subunits. Science 266, 1045–1047 [DOI] [PubMed] [Google Scholar]
- 21. Nelson B. H., Lord J. D., and Greenberg P. D. (1996) A membrane-proximal region of the interleukin-2 receptor gamma c chain sufficient for Jak kinase activation and induction of proliferation in T cells. Mol. Cell. Biol. 16, 309–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dybkaer K., Iqbal J., Zhou G., Geng H., Xiao L., Schmitz A., d'Amore F., and Chan W. C. (2007) Genome wide transcriptional analysis of resting and IL2 activated human natural killer cells: gene expression signatures indicative of novel molecular signaling pathways. BMC Genomics 8, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Merida I., Diez E., and Gaulton G. N. (1991) IL-2 binding activates a tyrosine-phosphorylated phosphatidylinositol-3-kinase. J. Immunol. 147, 2202–2207 [PubMed] [Google Scholar]
- 24. Remillard B., Petrillo R., Maslinski W., Tsudo M., Strom T. B., Cantley L., and Varticovski L. (1991) Interleukin-2 receptor regulates activation of phosphatidylinositol 3-kinase. J. Biol. Chem. 266, 14167–14170 [PubMed] [Google Scholar]
- 25. Blagoev B., Kratchmarova I., Ong S. E., Nielsen M., Foster L. J., and Mann M. (2003) A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat. Biotechnol. 21, 315–318 [DOI] [PubMed] [Google Scholar]
- 26. Kratchmarova I., Blagoev B., Haack-Sorensen M., Kassem M., and Mann M. (2005) Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science 308, 1472–1477 [DOI] [PubMed] [Google Scholar]
- 27. Kruger M., Kratchmarova I., Blagoev B., Tseng Y. H., Kahn C. R., and Mann M. (2008) Dissection of the insulin signaling pathway via quantitative phosphoproteomics. Proc. Natl. Acad. Sci. U.S.A. 105, 2451–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Osinalde N., Moss H., Arrizabalaga O., Omaetxebarria M. J., Blagoev B., Zubiaga A. M., Fullaondo A., Arizmendi J. M., and Kratchmarova I. (2011) Interleukin-2 signaling pathway analysis by quantitative phosphoproteomics. J. Proteomics 75, 177–191 [DOI] [PubMed] [Google Scholar]
- 29. Osinalde N., Sanchez-Quiles V., Akimov V., Guerra B., Blagoev B., and Kratchmarova I. (2015) Simultaneous dissection and comparison of IL-2 and IL-15 signaling pathways by global quantitative phosphoproteomics. Proteomics 15, 520–531 [DOI] [PubMed] [Google Scholar]
- 30. Baek S. H. (2011) When signaling kinases meet histones and histone modifiers in the nucleus. Mol. Cell 42, 274–284 [DOI] [PubMed] [Google Scholar]
- 31. Suganuma T., and Workman J. L. (2013) Chromatin and signaling. Curr Opin Cell Biol. 25, 322–326 [DOI] [PubMed] [Google Scholar]
- 32. Beadling C., Guschin D., Witthuhn B. A., Ziemiecki A., Ihle J. N., Kerr I. M., and Cantrell D. A. (1994) Activation of JAK kinases and STAT proteins by interleukin-2 and interferon alpha, but not the T cell antigen receptor, in human T lymphocytes. EMBO J. 13, 5605–5615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Frank D. A., Robertson M. J., Bonni A., Ritz J., and Greenberg M. E. (1995) Interleukin 2 signaling involves the phosphorylation of Stat proteins. Proc. Natl. Acad. Sci. U.S.A. 92, 7779–7783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Birmingham A., Anderson E., Sullivan K., Reynolds A., Boese Q., Leake D., Karpilow J., and Khvorova A. (2007) A protocol for designing siRNAs with high functionality and specificity. Nat. Protoc. 2, 2068–2078 [DOI] [PubMed] [Google Scholar]
- 35. Taxman D. J., Livingstone L. R., Zhang J., Conti B. J., Iocca H. A., Williams K. L., Lich J. D., Ting J. P., and Reed W. (2006) Criteria for effective design, construction, and gene knockdown by shRNA vectors. BMC Biotechnol. 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chylek L. A., Akimov V., Dengjel J., Rigbolt K. T., Hu B., Hlavacek W. S., and Blagoev B. (2014) Phosphorylation site dynamics of early T-cell receptor signaling. PLoS ONE 9, e104240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rigbolt K. T., Prokhorova T. A., Akimov V., Henningsen J., Johansen P. T., Kratchmarova I., Kassem M., Mann M., Olsen J. V., and Blagoev B. (2011) System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 4, rs3. [DOI] [PubMed] [Google Scholar]
- 38. Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., and Jorgensen T. J. (2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873–886 [DOI] [PubMed] [Google Scholar]
- 39. Osinalde N., Sanchez-Quiles V., Akimov V., Blagoev B., and Kratchmarova I. (2015) SILAC-based quantification of changes in protein tyrosine phosphorylation induced by Interleukin-2 (IL-2) and IL-15 in T-lymphocytes. Data Brief 5, 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Batth T. S., Francavilla C., and Olsen J. V. (2014) Off-line high-pH reversed-phase fractionation for in-depth phosphoproteomics. J. Proteome Res. 13, 6176–6186 [DOI] [PubMed] [Google Scholar]
- 41. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 42. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., and Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 43. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., and Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., and MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Laresgoiti U., Apraiz A., Olea M., Mitxelena J., Osinalde N., Rodriguez J. A., Fullaondo A., and Zubiaga A. M. (2013) E2F2 and CREB cooperatively regulate transcriptional activity of cell cycle genes. Nucleic Acids Res. 41, 10185–10198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Osinalde N., Olea M., Mitxelena J., Aloria K., Rodriguez J. A., Fullaondo A., Arizmendi J. M., and Zubiaga A. M. (2013) The nuclear protein ALY binds to and modulates the activity of transcription factor E2F2. Mol. Cell. Proteomics 12, 1087–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hallenborg P., Petersen R. K., Feddersen S., Sundekilde U., Hansen J. B., Blagoev B., Madsen L., and Kristiansen K. (2014) PPARgamma ligand production is tightly linked to clonal expansion during initiation of adipocyte differentiation. J. Lipid Res. 55, 2491–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schwartz D., and Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 51. Linding R., Jensen L. J., Pasculescu A., Olhovsky M., Colwill K., Bork P., Yaffe M. B., and Pawson T. (2008) NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res. 36, D695–D699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Arneja A., Johnson H., Gabrovsek L., Lauffenburger D. A., and White F. M. (2014) Qualitatively different T cell phenotypic responses to IL-2 versus IL-15 are unified by identical dependences on receptor signal strength and duration. J. Immunol. 192, 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zambricki E., Shigeoka A., Kishimoto H., Sprent J., Burakoff S., Carpenter C., Milford E., and McKay D. (2005) Signaling T-cell survival and death by IL-2 and IL-15. Am. J. Transplant. 5, 2623–2631 [DOI] [PubMed] [Google Scholar]
- 54. Cargnello M., and Roux P. P. (2011) Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 75, 50–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hsu P. P., Kang S. A., Rameseder J., Zhang Y., Ottina K. A., Lim D., Peterson T. R., Choi Y., Gray N. S., Yaffe M. B., Marto J. A., and Sabatini D. M. (2011) The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bauer D. E., Hatzivassiliou G., Zhao F., Andreadis C., and Thompson C. B. (2005) ATP citrate lyase is an important component of cell growth and transformation. Oncogene 24, 6314–6322 [DOI] [PubMed] [Google Scholar]
- 57. Hatzivassiliou G., Zhao F., Bauer D. E., Andreadis C., Shaw A. N., Dhanak D., Hingorani S. R., Tuveson D. A., and Thompson C. B. (2005) ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell 8, 311–321 [DOI] [PubMed] [Google Scholar]
- 58. Potapova I. A., El-Maghrabi M. R., Doronin S. V., and Benjamin W. B. (2000) Phosphorylation of recombinant human ATP:citrate lyase by cAMP-dependent protein kinase abolishes homotropic allosteric regulation of the enzyme by citrate and increases the enzyme activity. Allosteric activation of ATP:citrate lyase by phosphorylated sugars. Biochemistry 39, 1169–1179 [DOI] [PubMed] [Google Scholar]
- 59. Wellen K. E., Hatzivassiliou G., Sachdeva U. M., Bui T. V., Cross J. R., and Thompson C. B. (2009) ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Taplick J., Kurtev V., Lagger G., and Seiser C. (1998) Histone H4 acetylation during interleukin-2 stimulation of mouse T cells. FEBS Lett. 436, 349–352 [DOI] [PubMed] [Google Scholar]
- 61. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 62. Lim P. S., Li J., Holloway A. F., and Rao S. (2013) Epigenetic regulation of inducible gene expression in the immune system. Immunology 139, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Malek T. R. (2008) The biology of interleukin-2. Annu. Rev. Immunol. 26, 453–479 [DOI] [PubMed] [Google Scholar]
- 64. Sim G. C., and Radvanyi L. (2014) The IL-2 cytokine family in cancer immunotherapy. Cytokine Growth Factor Rev. 25, 377–390 [DOI] [PubMed] [Google Scholar]
- 65. Navarro M. N., Goebel J., Feijoo-Carnero C., Morrice N., and Cantrell D. A. (2011) Phosphoproteomic analysis reveals an intrinsic pathway for the regulation of histone deacetylase 7 that controls the function of cytotoxic T lymphocytes. Nat. Immunol. 12, 352–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lomberk G., Bensi D., Fernandez-Zapico M. E., and Urrutia R. (2006) Evidence for the existence of an HP1-mediated subcode within the histone code. Nat. Cell Biol. 8, 407–415 [DOI] [PubMed] [Google Scholar]
- 67. Arrizabalaga O., Lacerda H. M., Zubiaga A. M., and Zugaza J. L. (2012) Rac1 protein regulates glycogen phosphorylase activation and controls interleukin (IL)-2-dependent T cell proliferation. J. Biol. Chem. 287, 11878–11890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhao Y., Butler E. B., and Tan M. (2013) Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 4, e532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ramakrishna S., D'Angelo G., and Benjamin W. B. (1990) Sequence of sites on ATP-citrate lyase and phosphatase inhibitor 2 phosphorylated by multifunctional protein kinase (a glycogen synthase kinase 3 like kinase). Biochemistry 29, 7617–7624 [DOI] [PubMed] [Google Scholar]
- 70. Cai L., Sutter B. M., Li B., and Tu B. P. (2011) Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 42, 426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee J. V., Carrer A., Shah S., Snyder N. W., Wei S., Venneti S., Worth A. J., Yuan Z. F., Lim H. W., Liu S., Jackson E., Aiello N. M., Haas N. B., Rebbeck T. R., Judkins A., Won K. J., Chodosh L. A., Garcia B. A., Stanger B. Z., Feldman M. D., Blair I. A., and Wellen K. E. (2014) Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 20, 306–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shi L., and Tu B. P. (2013) Acetyl-CoA induces transcription of the key G1 cyclin CLN3 to promote entry into the cell division cycle in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 110, 7318–7323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Migita T., Okabe S., Ikeda K., Igarashi S., Sugawara S., Tomida A., Soga T., Taguchi R., and Seimiya H. (2014) Inhibition of ATP citrate lyase induces triglyceride accumulation with altered fatty acid composition in cancer cells. Int. J. Cancer 135, 37–47 [DOI] [PubMed] [Google Scholar]
- 74. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., and Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 75. Fujita Y., Fujiwara K., Zenitani S., and Yamashita T. (2015) Acetylation of NDPK-D Regulates Its Subcellular Localization and Cell Survival. PLoS ONE 10, e0139616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hwang J. S., Choi H. S., Ham S. A., Yoo T., Lee W. J., Paek K. S., and Seo H. G. (2015) Deacetylation-mediated interaction of SIRT1-HMGB1 improves survival in a mouse model of endotoxemia. Sci. Rep. 5, 15971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sutendra G., Kinnaird A., Dromparis P., Paulin R., Stenson T. H., Haromy A., Hashimoto K., Zhang N., Flaim E., and Michelakis E. D. (2014) A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell 158, 84–97 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.