Abstract
To study nucleolar involvement in brain development, the nuclear and nucleolar proteomes from the rat cerebral cortex at postnatal day 7 were analyzed using LC-MS/iTRAQ methodology. Data of the analysis are available via ProteomeXchange with identifier PXD002188. Among 504 candidate nucleolar proteins, the overrepresented gene ontology terms included such cellular compartmentcategories as “nucleolus”, “ribosome” and “chromatin”. Consistent with such classification, the most overrepresented functional gene ontology terms were related to RNA metabolism/ribosomal biogenesis, translation, and chromatin organization. Sixteen putative nucleolar proteins were associated with neurodevelopmental phenotypes in humans. Microcephaly and/or cognitive impairment were the most common phenotypic manifestations. Although several such proteins have links to ribosomal biogenesis and/or genomic stability/chromatin structure (e.g. EMG1, RPL10, DKC1, EIF4A3, FLNA, SMC1, ATRX, MCM4, NSD1, LMNA, or CUL4B), others including ADAR, LARP7, GTF2I, or TCF4 have no such connections known. Although neither the Alazami syndrome-associated LARP7nor the Pitt-Hopkins syndrome-associated TCF4 were reported in nucleoli of non-neural cells, in neurons, their nucleolar localization was confirmed by immunostaining. In cultured rat hippocampal neurons, knockdown of LARP7 reduced both perikaryal ribosome content and general protein synthesis. Similar anti-ribosomal/anti-translation effects were observed after knockdown of the ribosomal biogenesis factor EMG1 whose deficiency underlies Bowen-Conradi syndrome. Finally, moderate reduction of ribosome content and general protein synthesis followed overexpression of two Pitt-Hopkins syndrome mutant variants of TCF4. Therefore, dysregulation of ribosomal biogenesis and/or other functions of the nucleolus may disrupt neurodevelopment resulting in such phenotypes as microcephaly and/or cognitive impairment.
The nucleolus is the largest subnuclear structure and is the center of ribosomal biogenesis (1, 2). The RNA-polymerase-1 (Pol1)1-mediated transcription of nucleolar rRNA genes is the first step of ribosome production that is also essential for establishment and maintenance of the nucleolus. In addition, pre-rRNA is processed in the nucleolus into mature rRNA species and assembled with tens of ribosomal proteins to form pre-ribosomal subunits (3, 4). This complex process requires hundreds of trans-acting factors including proteins and small RNAs. Because ribosome production requires many resources, it is highly regulated (2).
Ribosomes are relatively stable structures with an estimated half-life time of several days (5). Hence, it is not surprising that a highly active nucleolus is often observed in intensely dividing cells which require many new ribosomes (1, 2). Conversely, nondividing cells usually display small nucleoli. However, prominent nucleoli are present in various types of mature neurons where initial observations of that structure were reported over 150 years ago (6). Moreover, neuronal nucleoli appear to increase in size as the nervous system develops (7–9). Given the importance of protein synthesis for neuron biology, including neuritic morphogenesis, neurite maintenance, and synaptic plasticity, ribosomal biogenesis may be required to maintain and regulate the adequate translation apparatus of a neuron. In support of such a possibility, brain derived neurotrophic factor (BDNF), which promotes neuronal morphogenesis, survival, and synaptic plasticity, stimulates Pol1-driven transcription (10). Therefore, despite their postmitotic status, neurons may maintain large nucleoli to rapidly produce additional ribosomes in response to neurotrophic stimulation.
Alternatively, the neuronal nucleolus may be involved in processes that are unrelated to ribosome biogenesis. For instance, as proficient ribosome biogenesis is required to suppress the p53 pathway, the nucleolus serves as a cellular stress sensor whose dysfunction triggers the p53-mediated cell cycle arrest and/or apoptosis (11). The stress sensing ability of the nucleolus has been also demonstrated in neurons (12, 13). In addition, the nucleolus is a site of non-ribosomal ribonucleoprotein complex biogenesis, including the signal recognition particle or components of the spliceosome (14). Finally, export and degradation of nuclear proteins, regulated protein sequestration, as well as establishment and maintenance of chromatin structure add to the expanding list of non-ribosomal functions of the nucleolus (14–17).
Proteomic analysis of nucleoli from various types of proliferating cells supports a notion of the plurifunctional nucleolus. Initial reports revealed that ribosomal biogenesis factors represent about 40–50% of roughly 300 proteins that were found in isolated nucleoli (18–20). Improved depth of peptide identification expanded the nucleolar proteome to nearly 700 proteins with 25% of them being initially implicated in ribosomal biogenesis (20, 21). However, an RNAi screen whose design was based on such extended proteomic data sets uncovered tens of new ribosomal biogenesis factors expanding their nucleolar proteome contribution to more than 40% (22). Processing of tRNA, mRNA metabolism including splicing, DNA repair, DNA replication, chromatin organization, and cell cycle regulation are among other functional categories that are abundantly represented in the nucleolar proteome. Hence, analysis of the nucleolar proteome can be used to identify potentially novel functions of the nucleolus as well as novel regulators of its well established roles such as ribosomal biogenesis.
The qualitative proteomic analysis of several human cell lines that were grown under diverse conditions suggested that 4500 proteins may be located in human cell nucleoli in various cellular contexts (23). However, as nucleolar preparations may also be contaminated by other cellular compartments, at least some of these proteins may represent false positive hits (20). Instead, quantitative analysis of nucleolar protein enrichment as compared with other cellular compartments provides a more specific approach to identify proteins with a localization preference for the nucleolus (24).
The functional organization of brain cell nucleoli remains mostly unexplored. In the nervous system, neither components nor regulators of the ribosomal biogenesis pathway have been systematically investigated with the exception of the Pol1 co-factor TIF1A whose knockdown or knockout blocks rRNA transcription in various neuronal populations (12, 13, 25, 26). Likewise, there is little information on nonribosomal functions of brain cell nucleoli beyond their involvement in the p53 stress response pathway. Therefore, the current study was initiated to identify nucleolar proteins of the developing rat brain using quantitative iTRAQ proteomics.
EXPERIMENTAL PROCEDURES
Animals
Sprague-Dawley pregnant female rats were purchased from Harlan (Indianapolis, IN). For proteomic analysis, three separate litters of neonate rats were used at postnatal day 7 (P7). The cortical tissue was collected from littermates of both sexes. All animal experiments strictly followed the protocols that were approved by the University of Louisville Institutional Animal Care and Use and Institutional Biosafety Committees.
Materials
All reagents were obtained from Sigma (St. Louis, MO), VWR (Radnor, PA), or Life Technologies-Invitrogen (Grand Island, NY) unless stated otherwise.
Nucleolar Isolation
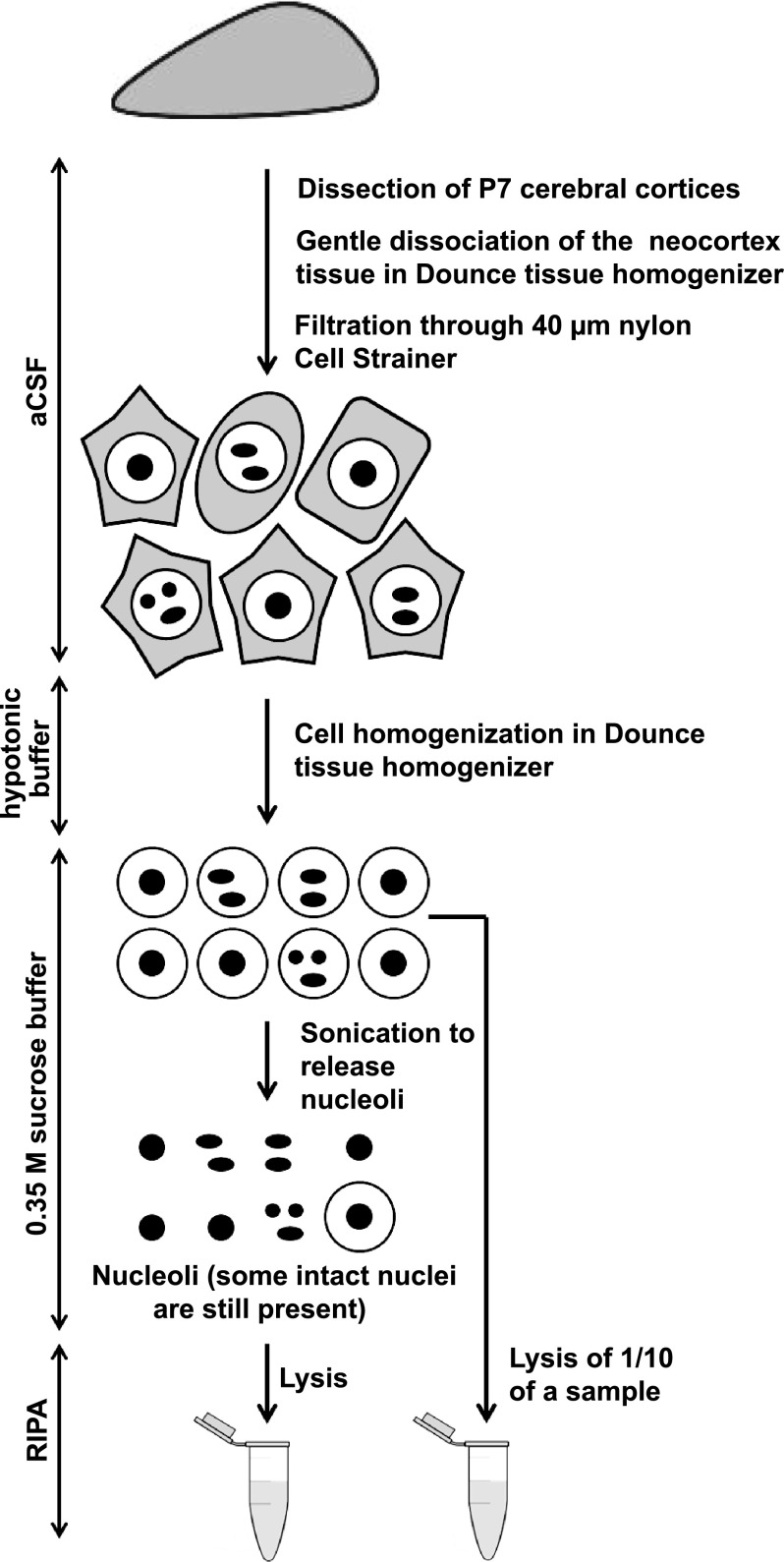
Rat neocortical nuclei and nucleoli were purified using a previously published procedure with modifications (18) (and http://www.lamondlab.com/pdf/noprotocol.pdf, Fig. 1A). For preparation of each set of samples 5–8 pups from a single litter were used. Pooling at least 5 animals/preparation was necessary to obtain sufficient amount of proteins in the nucleolar sample. Three independent preparations/sample sets from three different litters were used for the subsequent proteomic analysis. All steps were performed on ice or at 4 °C. The S2 solution as well as RIPA lysis buffer (Santa Cruz Biotechnology, Dallas, TX, #24948; 1xPBS (pH 7.4), 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 0.04% sodium azide) were supplemented with protease inhibitors. Dissected cerebral cortices were placed in the ice-cold artificial cerebrospinal fluid (aCSF, 126 mm NaCl, 25 mm NaHCO3, 25 mm Glucose, 1.25 mm NaH2PO4-H2O, 2.5 mm KCl, 2 mm CaCl2, 1 mm MgCl2, saturated with 5% CO2) and gently dissociated in a Dounce tissue homogenizer by turning the pestle to avoid generation of negative pressure. Cells were pelleted at 218 × g, resuspended in aCSF, and applied onto 40 μm nylon Cell Strainer (BD Falcon, Franklin Lakes, NJ) to separate tissue chunks and blood vessels. The filtrate was again centrifuged at 218 × g. The cell pellet was re-suspended in 5 ml of hypotonic Buffer A (10 mm Hepes, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.5 mm DTT) and incubated on ice for 5 min. The cell suspension was transferred directly to a pre-cooled Dounce tissue homogenizer and homogenized by 15 pestle strokes using a tight pestle until ∼90% of cells burst. After centrifugation at 218 × g for 5 min, whole nuclei were collected from the pellet in 5 ml of ice-cold S2 solution (0.35 m Sucrose, 0.5 mm MgCl2). One tenth of the sample was lysed in 1 ml RIPA buffer and used as a nuclear fraction for further analyses. The rest of the sample was sonicated at 40% power in 4 bursts, 10s each with 10s intervals to disrupt ∼90% of nuclei. The suspension was centrifuged at 1430 × g for 5 min. The resulting nucleolar pellet was re-suspended in 5 ml ice-cold S2 followed by centrifugation and lysis in 1 ml RIPA. The lysates were frozen on dry ice and stored at −80 °C.
Fig. 1.
Preparation of nucleolar proteins from the cerebral cortex of neonate rats. The schematic presents the nucleolar isolation procedure. First, whole nuclei were isolated from the neocortex of P7 rats. After sonication, the released nucleoli were purified by centrifugation; some intact nuclei were still present in such preparations (see Materials and Methods for more details).
Western Blotting
Western blotting was performed using standard procedures. The primary antibodies were as follows: anti-NeuN (1:1000, Millipore, Billerica, MA ABN78), anti-nucleophosmin/B23 (1:1000, Sigma #B0556), anti-fibrillarin (1:1000, Abcam, Cambridge, MA, AB4566), anti-UBF (1:1000, Santa Cruz Biotechnology, SC-13125), anti-actin (1:1000, Sigma, #A4700), anti-β-tubulin (1:1000, Sigma, T5293), anti-LARP7 (1:1000, Abcam, AB105683), anti-EMG1 (1:1000, Proteintech Group, 11965–1-AP), anti-GFP (MBL, 1:1000), anti-β-galactosidase (MP Biomedicals, 1:1000), anti-Tom20 (1:1500, a gift of Dr. Brian Wattenberg, University of Louisville, Louisville, KY), anti-calreticulin (1:1000, Enzo); secondary antibodies were horseradish peroxidase conjugated.
Dot Blot Analysis of Protein Concentration
Two μl of each sample were spotted onto chromatography paper filter (Whatman). After all spots dried (about an hour at RT), the membrane was incubated for at least 30 min at RT in fixing solution (40% methanol, 10% glacial acetic acid) on a platform shaker. Next, fixing solution was replaced by Coomassie G250 staining buffer (0,025% Coomassie, 10% glacial acetic acid) and incubated for at least 3 h. The membrane was then incubated with destaining buffer (25% methanol, 10% glacial acetic acid). Standard curve spots containing variable amount of BSA were used to estimate protein concentration of each sample.
Mass Spectrometry
All the samples were brought to the following reagent concentrations: 2% of SDS and 100 mm DTT. Protein concentration was estimated with dot-blot. and 100 μg of protein from each sample was taken for further preparation. Proteins were denatured and reduced at 95 °C for 5 min, alkylated with iodoacetamide and subsequently digested with trypsin (Promega) in filter devices (10 MWCO, Sartorius Stedim) using the Filter-Aided Sample Preparation (FASP) method (27). The digestion was performed for 16 h at enzyme/protein ratio of 1/100. For better compatibility with iTRAQ, we used triethylammonium bicarbonate buffer (TEAB, Sigma) instead of ammonium bicarbonate. Further on, all samples were lyophillized and incubated with iTRAQ mass tags as recommended in the manufacturer's protocol (Agilent, Santa Clara, CA). iTRAQ 8-plex was used and the labeling pattern was as follows: nuclear fractions in channels 113, 115, and 117, nucleolar fractions in channels 114, 116, and 118. After 1 h the labeling reaction was stopped by diluting each sample with an appropriate amount of sample buffer (8 m urea, 0.2% IPG buffer pH 3–11 NL (GE Healthcare), 0.002% bromphenol blue in 50 mm Tris-HCl, pH 8.0). Labeled samples were then pooled, concentrated in a Speedvac and fractionated on the IEF strip. The strip was then cut into 18 fractions and from each fraction peptides were extracted. Peptide mixtures were analyzed with an LC-MS system, composed of a UPLC chromatograph (nanoAcquity, Waters) directly coupled to a Q Exactive mass spectrometer (Thermo), working in the data-dependent acquisition mode.
Proteomic Data Analysis
The acquired MS/MS data were preprocessed with Mascot Distiller software (v.2.4.3, MatrixScience, Boston, MA) and a search was performed with the Mascot Search Engine (MatrixScience, Mascot Server 2.4.1) against the UniProt mouse protein database (UniProt 2013_11; 43,236 sequences). To reduce mass errors, the peptide and fragment mass tolerance settings were established separately for individual LC-MS/MS runs after a measured mass recalibration, as described previously (28). Subsequently, data from individual MS runs were merged into a single Mascot search. The parameters were as follows: enzyme, Trypsin; missed cleavages, 1; fixed modifications, Carbamidomethyl(C); variable modifications, Oxidation (M); quantitation, iTRAQ 8plex; taxonomy, unrestricted; instrument, HCD; parent MMD, 5 ppm; fragment MMD, 0.08 Da; Decoy option, active. A statistical assessment of the confidence of peptide assignments was based on the target/decoy database search strategy (29). This procedure provided q-value estimates for each peptide spectrum match in the data set. All queries with q-values >0.01 were removed from further analysis. A protein was regarded as identified confidently when at least two peptides of this protein were found. Proteins identified by a subset of peptides from another protein were excluded from the analysis. Proteins that exactly matched the same set of peptides were combined into a single group (family). The mass calibration and data filtering described above were carried out with MScan software, developed in-house (http://proteom.ibb.waw.pl/mscan/). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (http://www.ebi.ac.uk/pride/archive/) with the data set identifier PXD002188. In addition, detailed peptide/protein identification data are presented as a supplemental material (supplemental Table S5).
Nucleolar Classifier
iTRAQ results were also incorporated into features used to create a classifier for prediction of nucleolar localization of identified proteins. The Multilayer Perceptron neural network-based classifier was constructed using WEKA suite (30). It included the following features: iTRAQ protein ratio (each sample pair separately), number of peptides, nucleolar localization sequences predicted by the Nod software (31), number of various types of amino acids present in protein sequence, length of amino acid sequence, gene ontology and interaction data (from the IntAct database) and presence of various protein domains. Out of them, the following were selected by the software to perform the classification: iTRAQ protein ratio, interaction data, presence of lysine-rich domain (PS50318). The training set—which included proteins present in nucleus but not nucleolus and known nucleolar proteins—was constructed using published data (32). The classifier achieved 93% of correctly assigned instances and precision of 0.88 in cross-validation. Two hundred two proteins identified in all preparations were assigned nucleolar localization by the classifier, 88 of which acquired q-value below 0.05 in the statistical analysis. The remaining 114 proteins had on average less peptides identified (2–5, compared with median of 9 peptides for significantly enriched proteins) but higher No/Nu enrichment ratios.
Gene Ontology Analysis
Gene ontology analysis was performed with the NCBI's DAVID analysis tool. GO_CC_FAT, GO_BP_FAT, and GO_MF_FAT algorithms were used (33)(http://david.abcc.ncifcrf.gov/). The default parameters of the GO enrichment searches were used with a count threshold set at 2 and the EASE (p value) threshold set at 0.1; fold enrichment as well as FDR values were displayed in search results. Human phenotype analysis was performed with the ToppGene using the ToppFun algorithm (34) (https://toppgene.cchmc.org/).
Plasmid DNA
The following plasmids were previously described: pCDNA3.1-based expression vectors for human wild type TCF4 B- isoform and its mutants (Arg580Trp or Arg582Pro) (35); chicken β-actin promoter-driven-RFP or GFP (36), CRE-Luc (cAMP Response Element-driven luciferase reporter plasmid) and EF1α-LacZ (EF1α promoter-driven β-galactosidase) (37), p3XFLAG-CMV10-based expression vector for human LARP7 (38). To prepare EMG1-EGFP expression vector a 720 bp-long cDNA encoding rat EMG1 was cloned into pEGFP-N1 plasmid that was cut with XhoI/BamHI. The shEMG1–1-insensitive mutant variant of rat EMG1 was prepared by introducing two silent mutations (A319T,C322G) using QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Cat. 200523–5).
Cell Culture and Transfections
Cortical and hippocampal neurons were prepared from newborn Sprague-Dawley rats at postnatal day 0 as described (39). Cells were plated onto poly-d-lysine- and laminin-coated 12 mm diameter plastic coverslips that were produced in the lab from electron microscopy-grade mylar masks (Electron Microscopy Sciences, Hatfield, PA, #50425). Cells were grown in Neurobasal medium (supplemented with the B-27 supplement (Invitrogen), 1 mm l-glutamine, 100 units/ml penicillin, and 0.1 mg/ml streptomycin) half of which was changed every other day. Neurons were transfected on day in vitro 3 (DIV3) or DIV6 using Lipofectamine 2000 (Invitrogen) as previously described (40). Cerebrocortical astrocyte-enriched glial cultures were prepared from neonate rats as reported elsewhere (41).
Immunostaining
All stainings of cultured neurons were performed following 20 min fixation in 4% formaldehyde at room temperature followed by permeabilization by 0.5% Nonidet P-40 in PBS for 15 min. The following primary antibodies were used: rabbit anti-NeuN (1:1000, Millipore, ABN78), mouse anti-nucleophosmin/B23 (1:750, Sigma #B0556), mouse anti-fibrillarin (1:750, Abcam AB4566), mouse anti-UBF (1:750, St. Cruz, sc-13125), rabbit anti-LARP7 (1:100, Abcam, AB105682), rabbit anti-TCF4 (1:100, Abcam AB185736), rabbit anti-GFP (1:1000, MBL #598) and mouse anti-FLAG (1:1000, Sigma, F3165). The following secondary antibodies were used Alexa Fluor 488 goat anti-rabbit IgG (1:300, Invitrogen, A11034), Alexa Fluor 488 goat anti-mouse IgG (1:300, Invitrogen, A11029), Alexa Fluor 594 anti-mouse IgG (1:300, Invitrogen, A11005) and Alexa Fluor 555 anti-rabbit IgG (1:300, Invitrogen, A21429). Brain tissue was obtained from P7 rats; immersion-fixed in 4% paraformaldehyde for 24h and 20 μm sections were prepared using as previously described (42). Antigen recovery was performed by incubating brain sections with 10 mm sodium citrate, 0.05% Tween 20, pH 6.0 at 90–95 °C for 20 min followed by standard immunostaining protocol.
Generation of shRNA Expression Constructs
To generate shRNAs against rat Larp7 and Emg1 each mRNA sequence was analyzed using shRNA design software (rnaidesigner.lifetechnologies.com) followed by off-target exclusion using Blastn (NCBI). Oligonucleotides were designed:
Larp7–1: 5′gatccccgccagtcagcacattcgatttcaagagaatcgaatgtgctgactggcttttta3′,
Larp7–2: 5′gatccccgctaatcaccaaagctgagttcaagagactcagctttggtgattagcttttta3′,
Larp7–3: 5′gatccccggccaaagctaaagaagagttcaagagactcttctttagctttggccttttta3′,
Emg1–1: 5′gatcccccttacgagctactcaactgttcaagagacagttgagtagctcgtaagttttta3′,
Emg1–2: 5′gatccccgaatgtgctcattgaagtgttcaagagacacttcaatgagcacattctttta3′ together with their complementary counterparts, annealed and subcloned into the pSuper vector (Oligoengine) digested with BglII and HindIII.
Quantitative Reverse Transcriptase PCR (qRT-PCR)
Tri Reagent-based (Sigma) RNA isolation, cDNA synthesis and quantitative PCR were performed as described previously (12). The following oligonucleotide primers were used: bacterial β-galactosidase forward 5′tactgacgaaacgcctgccagtat3′ and reverse 5′gctgtgccgaaatggtccatcaaa3′; human TCF4-specific forward: 5′acttgcttcgatctattctccag3′ and reverse 5′cataattaggagacgatgaggcc3′. The RNA levels were analyzed using ΔΔct method. Expression values obtained from triplicate runs of each cDNA sample were calculated relative to the triplicate value for the bacterial β-galactosidase from the same cDNA preparation.
Quantification of Ribosomes
Cultured hippocampal neurons were fixed with 4% formaldehyde for 20 min. Ribosomes were visualized with the NeuroTrace 500/525 Green Fluorescent Nissl Stain (Invitrogen, N21480, 1:500) or the NeuroTrace 530/615 Red Fluorescent Nissl Stain (Invitrogen, N21482, 1:500); DNA was counterstained with 2.5 μg/ml Hoechst 33258. To control for the specificity of NeuroTrace staining, some coverslips with attached cells were pre-treated with RNase A (Qiagen, 20 u in PBS, 1 h at 37 °C). Cells were imaged using a 40x objective lens and a Zeiss Axioobserver inverted microscope system. Digitalized pictures were captured using the AxioVision software (Zeiss) followed by conversion to gray scale TIFF files. Identical exposure times were used for all pictures that were used for comparative analyses. Quantification of the fluorescence intensity was prepared by using the ,,Integrated Density” parameter in the ImageJ software with background correction.
Protein Synthesis Assay
To visualize protein synthesis in individual neurons O-propargyl-puromycin (OPP) labeling method was used followed by Click-iT chemistry detection of nascent peptides (43). A commercial kit was employed according to manufacturer's recommendations (Click-iT Plus OPP Alexa Fluor 594 Protein Synthesis Assay Kit, Molecular Probes, C10457). To control for the specificity of OPP staining, some coverslips with attached cells were pre-treated with cycloheximide (CHX, Sigma, 50 μm, 1h at 37 °C). Quantification of newly synthesized, OPP-labeled peptides was performed as described for ribosome content analysis.
Transcription Luciferase Assay
Activities of luciferase and β-gal were determined using a standard procedure (37). Luciferase activity was normalized against β-gal.
Statistical Analysis
Statistical analysis of quantitative proteomic results was performed with in-house software (Diffprot) using the following parameters: data normalization - LOWESS, clustering of peptide sets with over 80% similarity, statistics - Wilcoxon test (28). To ensure that individual variability between biological replicates is not a major contributor to quantitative differences between nucleolar- and nuclear data sets, boxplot analysis of interquartile range and principal component analysis of quantitative data from individual samples were performed (supplemental Fig. S1). In addition, the Diffprot algorithm penalizes quantitative inconsistencies in control samples, which further reduces the risk of obtaining artifacts. Statistical analysis of ribosome content and protein synthesis activity was performed using one-way ANOVA that was followed by Fisher's LSD posthoc tests.
RESULTS
Isolation of Brain Nucleolar Proteins
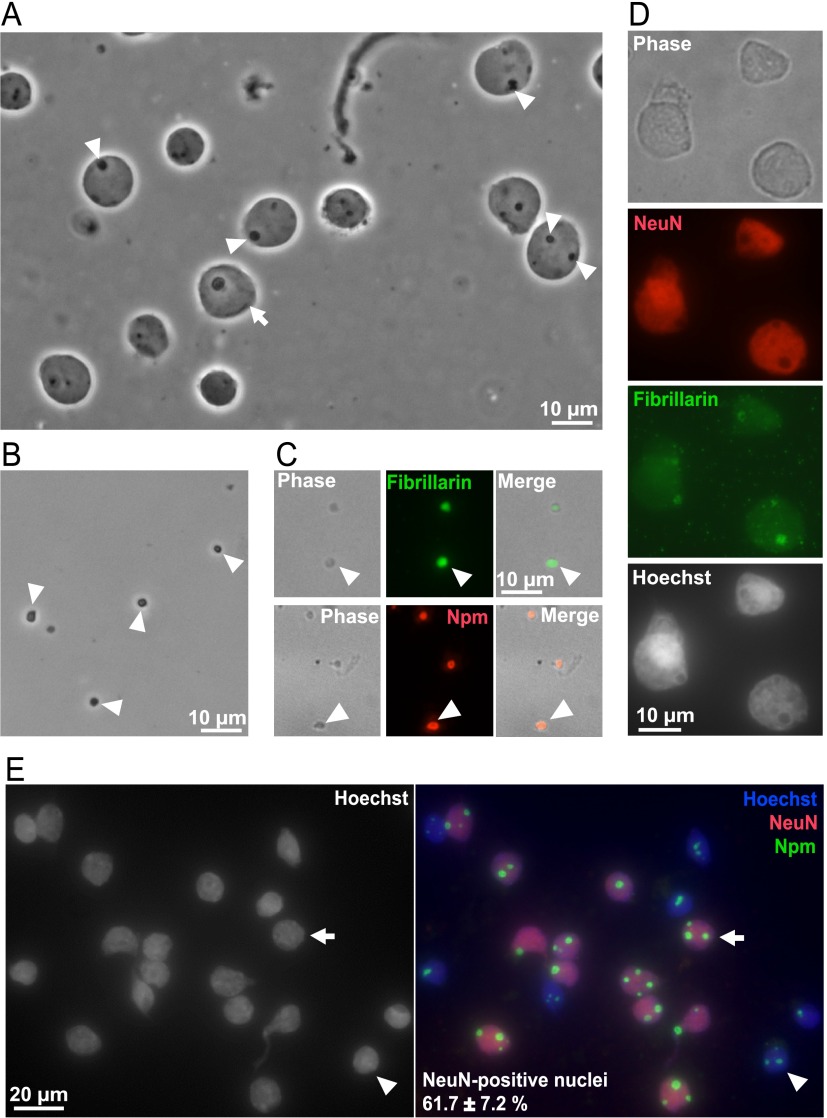
To obtain the nucleolus-enriched protein fraction, whole nuclei were first isolated from the neocortex of P7 rats (Fig. 1). Then, nucleoli were obtained from the whole nuclei by sonication and centrifugation in sucrose solution (Fig. 1). Relative purity of the isolated whole nuclei- and nucleoli was verified by phase contrast microscopy (Fig. 2A, 2B). Immunofluorescence for the nucleolar markers fibrillarin and nucleophosmin/B23 (Npm) confirmed successful isolation of the nucleoli (Fig. 2C). Sixty-one percent of the nuclei from which nucleoli were isolated were of neuronal origin as determined by the presence of the neuronal nuclear marker NeuN (Fig. 2D, 2E). Importantly, as in P7 cerebral cortex nucleolar presence of Npm is disrupted by stressors that interfere with rRNA transcription (42), maintenance of the nucleolar Npm staining throughout the nucleolar isolation procedure suggest well preserved integrity of the isolated nucleoli (Fig. 2C, 2E).
Fig. 2.
Morphology of isolated nuclei and nucleoli from the cerebral cortex of neonate rats. A, B, Phase contrast micrographs of unfixed nuclei (arrows) and nucleoli (arrowheads). C, D, Isolated nucleoli (C) or nuclei (D, E) were fixed and immunostained as indicated; DNA was visualized by counterstaining with Hoechst-33258. In isolated nucleoli (C), or, in intact nuclei (D, E), nucleolar markers fibrillarin and nucleophosmin (Npm) remained nucleolar suggesting good preservation of the nucleolus throughout the fractionation procedure. In D, the isolated whole nuclei were co-immunostained for the neuronal nuclear marker NeuN and the Npm. The arrow points a NeuN-reactive (i.e. neuronal) nucleus; the arrowhead marks a non-neuronal nucleus. Note that in neuronal nuclei, nucleoli were usually larger than in non-neuronal nuclei. The results represent two independent nucleolar isolations that were not included in the LC/MS proteomic analysis. However, similar morphological characteristics of the nucleolar- and the whole nuclear fraction were verified by phase contrast microscopy for each sample set that was used for proteomic experiments.
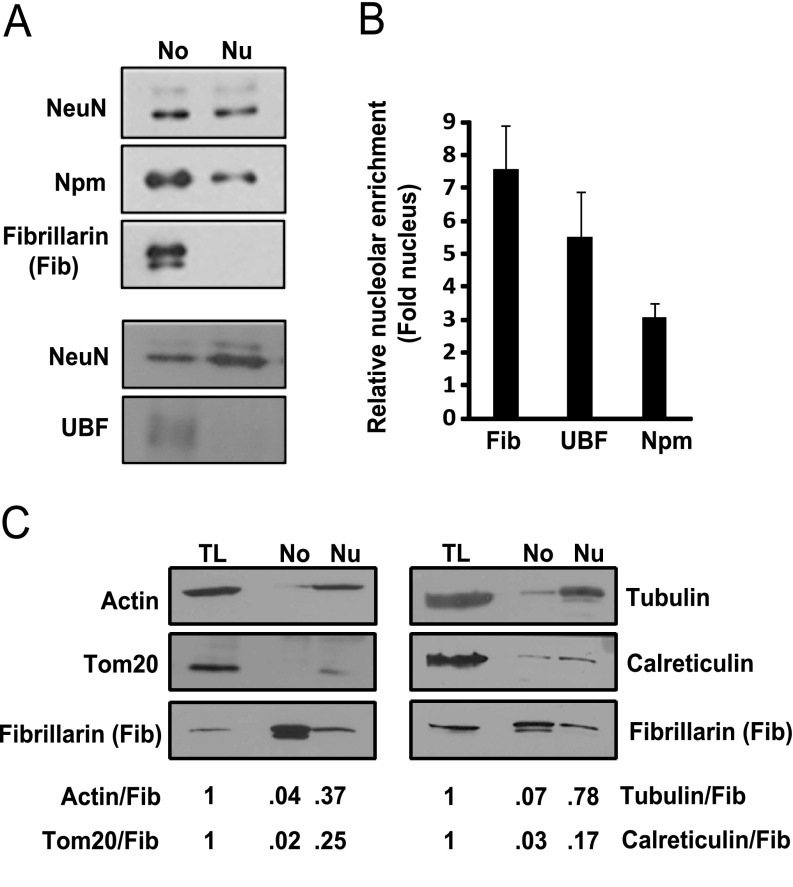
Next, protein lysates were prepared and relative nucleolar enrichment was evaluated by Western blotting for several nucleolar marker proteins including Npm, fibrillarin, and upstream binding factor (UBF) (Fig. 3A, 3B). As compared with the nuclear lysates, all the nucleolar markers were enriched in the nucleolar fractions. In addition, relative purity of the nucleolar fraction was further verified by Western blotting for markers of the cytoskeleton (actin, tubulin), the mitochondrion (Tom20), and the endoplasmic reticulum (calreticulin). In nucleolar fractions, their fibrillarin-normalized signals were 14–50 times lower than in whole cortical lysates (Fig. 3C). Thus, enriched nucleolar protein samples can be obtained from the cerebral cortex of neonate rats.
Fig. 3.
Enrichment of nucleolar marker proteins in isolated nucleoli from the cerebral cortex of neonate rats. Enrichment of nucleolar proteins in nucleolar preparations (No) was verified by Western blotting. A, Note that nucleolar proteins including fibrillarin, Npm and UBF were more abundant in No lysates than in total nuclear lysate (Nu) as compared with a nuclear marker NeuN. The membrane that was used to detect UBF contained twice as much nucleolar material as that for detection of Npm/Fib; each membrane represents an independent preparation of nucleolar/nuclear proteins; the Npm/Fib membrane was loaded with 1/4- or 1/40- of the nucleolar- or the whole nuclear protein lysate from cortices of one neonate rat, respectively; the UBF membrane contained 1/2- or 1/20- of the nucleolar- or the whole nuclear protein lysate from cortices of one neonate rat, respectively. Such a loading ratio of whole nuclear to total nucleolar lysate was established empirically to ensure linear signal of the detected nucleolar proteins in both fractions; hence, significant amounts of proteins were lost during nucleolar fractionation. B, Averages ±S.E. of NeuN-normalized nucleolar enrichments from 9 (Npm), 5 (Fib), or 5 (UBF) independent preparations are presented on the graph. All sample sets that were used for the LC/MS proteomic analysis were verified by Western blotting for the nucleolar markers and are included in this data. C, Nucleolar enrichment was further evaluated by Western blotting for markers of other cellular components including microfilaments (actin), microtubules (tubulin), mitochondria (Tom20) and endoplasmic reticulum (calreticulin). Total cortical lysate (TL), nucleolar lysate (No) and whole nuclear lysate (Nu) from 1/30, 1/3 and 1/30 of cortices of one neonate rat, respectively were loaded on the gel as indicated. When compared with total cortical-, or, whole nuclear lysates, the fibrillarin-normalized content of the potential contaminants was lowest in nucleolar lysates (No). Similar results were observed with lysates from two independent experiments/isolations; these isolations were not used for the LC/MS proteomic analysis. Band intensities were quantified by densitometry and normalized against intensity of NeuN-(B) or fibrillarin (C) signals.
Proteomic Analysis of Nucleoli from the Developing Cerebral Cortex
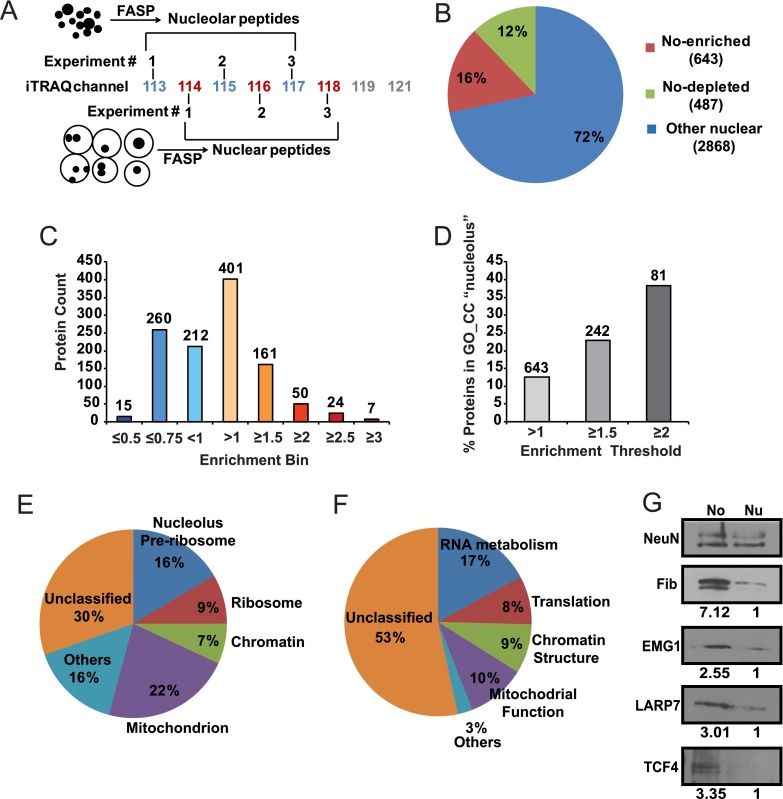
Quantitative proteomics using the iTRAQ labeling method was applied to compare samples of intact nuclei and isolated nucleoli (Fig. 4A). Three sets of such samples, each prepared independently from different litters of neonate rats, were evaluated. Altogether, 3997 proteins were identified (supplemental Tables S1 and S5). Six hundred forty-three proteins were significantly enriched in nucleoli and 487 proteins were significantly depleted from this compartment (supplemental Table S2, Fig. 4B, 4C). The gene ontology (GO) term “nucleolus” was associated with 12.6% of all nucleolus-enriched proteins (Fig. 4D). That fraction increased to 22.9 and 38.3% when the analysis was limited to nucleolar proteins with at least 1.5- and twofold enrichment, respectively (Fig. 4D).
Fig. 4.
Analysis of the nucleolar proteome from the developing rat cerebral cortex. A, Three independent sets of total nuclear/nucleolar protein samples from the cerebral cortex of P7 rats were analyzed using quantitative iTRAQ proteomics. Filter Aided Sample Preparation (FASP) was applied to obtain peptides that were tagged using an 8-plex iTRAQ system for quantitative LC/MS analysis. B, The pie chart illustrates the numbers of proteins with differential abundance in the nucleolus (No). C, Distribution of relative No enrichments among 1130 differentially localized proteins. Numbers over the bars correspond to numbers of proteins in each enrichment bin. D, Increased No enrichment correlates with greater fraction of proteins that are associated with the gene ontology (GO) term “nucleolus”. Numbers over the bars correspond to numbers of proteins that are passing each enrichment threshold. E–F, Gene ontology (GO) analysis of 504 mapped proteins whose nucleolar enrichment was at least 1.3 fold. The significance threshold for GO enrichment was FDR<0.05. The pie charts present fractions of No proteins that were assigned to enriched GO_CC- (cellular compartment, E) or GO_BP (biological process, F) categories. Related terms were grouped together; overlapping GO assignments were avoided to accurately determine the fraction of unclassified proteins. More details about GO enrichment are in Tables I–III. G, Nucleolar enrichment of the candidate nucleolar proteins EMG1, LARP7, and TCF4 was verified by Western blotting. The nucleolar marker fibrillarin was used as a positive control. Numbers under the blots indicate relative nucleolar enrichment as determined by NeuN-normalized band intensity; for each analyzed protein, its NeuN-normalized content in the total nuclear lysate was defined as 1. Similar trends were observed in two independent experiments/isolations. The gel loading was as for Fig. 3C.
Next an artificial neural network-based classifier algorithm was built to investigate whether apparent but non-significant nucleolar enrichment/depletion of some proteins may represent real differences in their subnuclear distribution (see Experimental Procedures for more details). Using such an approach, an additional 114 proteins were identified as likely candidates for differential nucleolar distribution (supplemental Table S3). This set included 102 proteins with apparent nucleolus-enrichment; 75 of them were enriched at least 1.3-fold.
Gene Ontology Analysis of Nucleolar Proteome from the Developing Cerebral Cortex
For Gene Ontology (GO) analysis, a list of candidate nucleolar proteins with a minimum 1.3 fold nucleolar enrichment was prepared by combining significantly enriched proteins (437 of 643) with those whose nucleolar enrichment trend was supported by the classifier (75 of 102). For simplicity, proteins from this list will be further referred to as nucleolar.
GO terms associated with the cellular compartment (GO_CC) were assigned to 433 nucleolar proteins with several GO_CCs being significantly enriched (FDR p < 0.05, Table I, Fig. 4E). The GO_CC “nucleolus” was assigned to 17% proteins whereas 9% was associated with “ribosome”/“ribosomal subunit.” In addition, 7% proteins were assigned to chromatin-related GO_CCs and 3% were spliceosomal. Of note, with more than 22% coverage, the GO_CC “mitochondrion” had the largest presence among nucleolar proteins of all enriched GO_CCs that were organelle/structure specific. Large representation of mitochondrial proteins can be also found in other nucleolar proteome data sets that were obtained using similar fractionation methodology (21). Indeed, this mitochondrial enrichment appears to be a consequence of contamination whose origins are in the whole nuclei fraction (44). In support of that notion, similar fold enrichment of mitochondrial proteins was present in nucleolar- and nuclear samples from the brain (2.47-, and 1.89-fold, respectively, Table I). As nucleolar proteins represent a subset of whole nuclear proteins, similar enrichment indicates that only a relatively small fraction of mitochondrial proteins was enriched in the nucleolus (80 of 574 or 14%). The mitochondrial marker Tom20 that was used to evaluate purity of nucleolar/whole nuclear lysates using Western blotting was not identified by the LC/MS analysis (supplemental Table S1). However, its relative depletion from the nucleolar material as compared the whole nuclear lysate indicates that it belongs to the majority of mitochondrial proteins that do not contaminate nucleolar fractions (Fig. 3C). Contamination may also account for a smaller, yet significantly enriched group of extracellular matrix/basal membrane components (3%, Table I). Other enriched GO_CC terms were unrelated to a specific subcellular organelle/structure and largely overlapped with the aforementioned specific GO_CCs (e.g. “ribonucleoprotein complex,” “nonmembrane-bounded organelle,” data not shown). Of note, 30% nucleolar proteins did not associate with any of the enriched GO_CC terms.
Table I. The FDR-verified enrichment of GO_CC categories. NS, p > 0.05. NE, not enriched.
| GOTERM_CC_FATa | NUCLEOLUSb |
NUCLEUS (all identified)c |
||||
|---|---|---|---|---|---|---|
| Fold Enriched | % List | FDR | Fold Enriched | % List | FDR | |
| GO:0030684∼preribosome | 17.77 | 1.59 | 1.10E-04 | 3.36 | 0.26 | NS |
| GO:0005730∼nucleolus | 7.73 | 16.47 | 2.94E-47 | 1.93 | 3.58 | 1.67E-13 |
| GO:0000786∼nucleosome | 7.45 | 3.17 | 2.70E-06 | 2.32 | 0.86 | 0.001155 |
| GO:0005840∼ribosome | 7.22 | 9.52 | 3.96E-24 | 2.52 | 2.90 | 5.58E-22 |
| GO:0033279∼ribosomal subunit | 6.56 | 2.98 | 5.76E-05 | 2.51 | 0.99 | 8.55E-06 |
| GO:0000792∼heterochromatin | 6.23 | 2.18 | 0.010759 | 2.22 | 0.68 | NS |
| GO:0005681∼spliceosome | 4.66 | 3.97 | 6.36E-05 | 2.92 | 2.17 | 1.04E-21 |
| GO:0000785∼chromatin | 4.38 | 4.96 | 3.13E-06 | 1.85 | 1.83 | 6.27E-05 |
| GO:0005694∼chromosome | 3.13 | 8.13 | 3.42E-07 | 1.49 | 3.37 | 8.18E-04 |
| GO:0044427∼chromosomal part | 2.72 | 5.95 | 0.002514 | 1.47 | 2.79 | 0.01671 |
| GO:0005604∼basement membrane | 6.72 | 3.37 | 3.71E-06 | 1.43 | 0.63 | NS |
| GO:0044420∼extracellular matrix part | 5.34 | 3.37 | 1.21E-04 | NE | NE | NE |
| GO:0005739∼mitochondrion | 2.47 | 22.42 | 3.12E-17 | 1.89 | 14.99 | 6.36E-66 |
a only organelle/structure/complex-specific GO_CC categories are not shown.
b the analyzed list included 504 mapped IDs of proteins with at least 1.3 fold No enrichment; of them 433 had assigned GO_CC terms.
c the analyzed list included 3830 mapped IDs, of them 2865 had assigned GO_CC terms.
Gene ontology terms for Biological Process (GO_BP) were assigned to 411 nucleolar proteins. The greatest enrichment was observed for categories related to RNA metabolism including “ribosome biogenesis,” “ribonucleoprotein complex biogenesis,” “RNA processing” and several other similar terms which all together spanned 17% of the entire nucleolar protein list (Table II, Fig. 4F). “Translation” was another highly enriched GO_BP term followed by several GO_BPs that were associated with chromatin organization (e.g.“chromosome organization,” “chromatin assembly or disassembly,” “chromatin organization,” “DNA packaging,” “nucleosome assembly,” “nucleosome organization”). Their contribution to the nucleolar proteome was 8 and 9%, respectively. Ten % of the identified proteins were associated with mitochondrial GO_BPs (e.g.“cellular respiration”, “generation of precursor metabolites and energy,” “electron transport chain,” “oxidation reduction,” “acetyl-CoA metabolic process,” “tricarboxylic acid cycle,” and other related terms, Fig. 4F and data not shown). Finally, 53% of nucleolar proteins were not associated with any significantly enriched GO_BP.
Table II. The FDR-verified enrichment of GO_BP categories. NS, p > 0.05.
| GO_BP_FAT Terma | Nucleolusb |
Nucleus (all identified)c |
||||
|---|---|---|---|---|---|---|
| Fold enriched | % list | FDR | Fold enriched | % list | FDR | |
| GO:0042254∼ribosome biogenesis | 13.85 | 8.73 | 1.90E-23 | 2.69 | 1.70 | 4.1078E-13 |
| GO:0006396∼RNA processing | 12.99 | 13.89 | 2.18E-33 | 2.36 | 5.80 | 5.7144E-39 |
| GO:0006364∼rRNA processing | 11.50 | 6.15 | 0.005802 | 2.76 | 1.15 | 1.5491E-08 |
| GO:0016072∼rRNA metabolic process | 11.02 | 6.15 | 0.007979 | 2.72 | 1.15 | 2.8768E-08 |
| GO:0006412∼translation | 10.86 | 9.92 | 2.61E-30 | 2.60 | 4.67 | 2.3355E-38 |
| GO:0034470∼ncRNA processing | 9.80 | 6.75 | 0.018929 | 1.67 | 1.49 | NS |
| GO:0034660∼ncRNA metabolic process | 9.60 | 6.94 | 0.004574 | 1.88 | 2.14 | 2.82E-06 |
| GO:0051276∼chromosome organization | 9.45 | 8.53 | 0.024588 | 1.53 | 3.47 | 0.0001808 |
| GO:0006397∼mRNA processing | 9.12 | 6.35 | 0.031575 | 2.76 | 4.07 | 2.5363E-37 |
| GO:0006333∼chromatin assembly or disassembly | 8.97 | 4.17 | 2.39E-07 | 2.38 | 1.46 | 4.48E-08 |
| GO:0006325∼chromatin organization | 7.25 | 6.94 | 5.93E-06 | 1.62 | 2.87 | 9.0115E-05 |
| GO:0006323∼DNA packaging | 7.05 | 3.97 | 8.79E-06 | 2.25 | 1.28 | 1.062E-05 |
| GO:0016071∼mRNA metabolic process | 6.96 | 6.55 | 1.07E-05 | 2.55 | 4.33 | 8.101E-34 |
| GO:0008380∼RNA splicing | 6.96 | 5.16 | 1.07E-05 | 2.98 | 3.37 | 3.5164E-35 |
| GO:0006334∼nucleosome assembly | 6.55 | 3.17 | 2.39E-07 | 2.48 | 1.02 | 1.759E-05 |
| GO:0031497∼chromatin assembly | 6.37 | 3.17 | 1.25E-07 | 2.47 | 1.04 | 1.1704E-05 |
| GO:0034728∼nucleosome organization | 5.40 | 3.17 | 3.74E-04 | 2.38 | 1.02 | 7.316E-05 |
a all mitochdria related GO_BP categories were excluded as a potential contamination.
b the analyzed list included 504 mapped IDs of proteins with at least 1.3 fold No enrichment; of them 411 had assigned GO_BP terms.
c the analyzed list included 3830 mapped IDs, of them 2930 had assigned GO_BP terms.
As expected for the center of ribosomal biogenesis, the most nucleolus-enriched gene ontology terms of molecular function (GO_MF) were associated with rRNA binding and ribosome structure (Table III). In addition other enriched GO_MFs included helicases, RNA binding proteins, ATPases and nucleotide binding proteins. Taken together, GO analysis supports functional specialization of brain cell nucleoli confirming their involvement in ribosome synthesis.
Table III. The FDR-verified enrichment of GO_MF categories.
| GOTERM_MF_FATa | Nucleolusb |
Nucleus (all identified)c |
||||
|---|---|---|---|---|---|---|
| Fold enriched | % List | FDR | Fold enriched | % List | FDR | |
| GO:0019843∼rRNA binding | 16.69 | 2.38 | 3.8E-08 | 3.07 | 0.42 | 0.030584 |
| GO:0003735∼structural constituent of ribosome | 9.95 | 8.93 | 1.6E-28 | 2.90 | 2.48 | 4.82E-24 |
| GO:0070035∼purine NTP-dependent helicase activity | 6.60 | 3.37 | 6.2E-06 | 2.31 | 1.12 | 3.67E-05 |
| GO:0008026∼ATP-dependent helicase activity | 6.60 | 3.37 | 6.2E-06 | 2.31 | 1.12 | 3.67E-05 |
| GO:0004386∼helicase activity | 5.69 | 4.37 | 3.1E-07 | 2.18 | 1.59 | 4.60E-07 |
| GO:0005198∼structural molecule activity | 4.53 | 12.10 | 5.6E-20 | 1.93 | 4.91 | 3.91E-19 |
| GO:0003723∼RNA binding | 3.83 | 15.28 | 1.7E-21 | 2.13 | 8.12 | 1.27E-44 |
| GO:0042623∼ATPase activity, coupled | 3.14 | 4.17 | 1.8E-02 | 2.03 | 2.56 | 2.73E-10 |
| GO:0000166∼nucleotide binding | 1.54 | 20.04 | 5.8E-03 | 1.87 | 23.11 | 1.85E-104 |
a mitochondrial protein-enriched categories are not shown.
b the analyzed list included 504 mapped IDs of proteins with at least 1.3 fold No enrichment; of them 398 had assigned GO_MF terms.
c the analyzed list included 3830 mapped IDs, of them 2881 had assigned GO_MF terms.
Ribosomal Proteins of the Developing Cerebral Cortex
Ribosome diversity has been previously proposed as a possible mechanism of translational regulation that may be active in the nervous system (45). Such a ribosomal diversity may originate from differential use of ribosomal proteins (RPs), as demonstrated for RPL38 (46). In addition, mutations of some ribosomal proteins including S3, S4, S15A, and L10 have relatively selective impact on forebrain development (47, 48). In the current data set, 75 nuclear ribosomal proteins were detected (supplemental Fig. S2). Of them, 51 were nucleolus-enriched (supplemental Fig. S2). The abundant nucleolar presence of S3, S4, S15A, and L10 suggests that these RPs are included into newly made brain ribosomes. Interestingly, two pairs of RP paralogs were also detected (L7/L7l1 and L22/L22l1). Thus, differential use of these paralogs as well as inclusion of S3, S4, S15, and L10 may support ribosomal specialization in the developing cerebral cortex.
Human Neurological Phenotypes Associated with Nucleolar Proteins of the Cerebral Cortex
ToppFun analysis of proteins with a minimum 1.3-fold nucleolus-enrichment revealed significant over-representation of such overlapping human phenotype (HP) terms as “Abnormality of the cerebellum,” “abnormality of hindbrain morphology,” “abnormality of brain morphology” (FDR p < 0.05). Of 69 proteins that were in these categories, 48 were associated with GO_CCs “mitochondrion” and “ECM part” suggesting that they represent contaminations rather than bona fide nucleolar proteins. Three additional potential contaminants were two lysosomal hydrolases (NAGA, GLB1) and an ER glycosylase (STT3A). The remaining proteins included 14 whose mutations result in neurodevelopmental syndromes (Table IV). Two additional nucleolar proteins that are linked to neurodevelopmental disorders were identified by analyzing literature (RPL10 and EIF4A3, Table IV). Adult onset neurodegeneration was associated with mutations of three candidate nucleolar proteins (Table V).
Table IV. Human counterparts of No-enriched rat brain proteins whose mutations cause neurodevelopmental diseases.
| Protein | No/Nu | Function | Disease/neurological phenotype | OMIM # |
|---|---|---|---|---|
| EMG1 | 1.66 | Ribosomal biogenesis | Bowen-Conradi Syndrome/microcephaly, developmental delay | 211180 |
| RPL10 | 1.43 | Structural protein of the ribosome | X-linked microcephaly, growth retardation, and seizures | 300847 |
| DKC1 | 2.07 | Ribosomal biogenesis/telomere maintenance | Dyskeratosis Congenita, X-Linked/learning difficulties, microcephaly, cerebellar hypoplasia | 305000 |
| EIF4A3 | 1.36 | Ribosomal biogenesis, mRNA splicing and decay | Richieri-Costa-Pereira Syndrome/developmental delay, learning disability, language delay/disorder | 268305 |
| FLNA | 1.63 | Actin regulation, negative regulation of PI3K signaling, negative regulation of Pol1, pre-rRNA processing | Heterotopia, Periventricular, X-Linked Dominant/ seizures, cortical dysplasia | 300049 |
| SMC1A | 1.42 | Chromatid cohesion, general transcription regulation | Cornelia De Lange Syndrome 2/microcephaly, mental retardation, dilated ventricles, seizures | 300590 |
| MCM4 | 1.93a | Prevents DNA re-replication, maintains genomic stability | Natural Killer Cell And Glucocorticoid Deficiency With Dna Repair Defect/microcephaly | 609981 |
| NSD1 | 1.3 | Transcription factor and chromatin modifier | Sotos Syndrome 1/ macrocephaly, ventriculomegaly, seizures, developmental delay, mental retardation, poor coordination | 117550 |
| CUL4B | 1.31 | DNA damage response | Mental Retardation, X-Linked, With Short Stature | 300354 |
| PRKDC | 2.2a | DNA damage response | Immunodeficiency 26 With Or Without Neurologic Abnormalities/ Microcephaly, mental retardation) | 615966 |
| ATRX | 1.43 | Chromatin remodeling | Alpha-Thalassemia and/or Mental Retardation Syndromes, X-Linked; Linked/mental retardation, microcephaly | 301040; 309580 |
| LMNA | 1.54 | Nuclear envelop protein | Charcot-Marie-Tooth Disease, Axonal, Type 2b1 (LMNA loss of function)/axonal atrophy in peripheral nerves, distal limb muscle weakness/atrophy due to peripheral neuropathy; Malouf Syndrome (heterozygous point mutation in LMNA, possible gain of function)/mental retardation | 605588; 212112 |
| LARP7 | 2.05 | Stabilizes 7SK ncRNA that inhibits the p-TEFb transcription factor | Alazami Syndrome/ intelectual disability and microcephaly | 615071 |
| TCF4 | 2.23a | Transcription factor | Pitt-Hopkins Syndrome/postnatal microcephaly, cerebral atrophy, mental retardation, poor or absent speech development, seizures, hypotonia | 610954 |
| GTF2I | 1.32 | General transcription factor | Williams-Beuren Syndrome/mental retardation, poor visual-motor integration, hypotonia, poor balance and coordination | 194050 |
| ADAR | 1.47 | RNA editing | Aicardi-Goutieres Syndrome 6/atrophy of striatum, leukodystrophy, dystonia, tremor, loss of motor skils | 615010 |
a Nonsignificant enrichment that was supported by the classifier.
Table V. Human counterparts of No-enriched rat brain proteins whose mutations cause neurodegenerative diseases.
| Protein | No/Nu | Function | Disease/neurological phenotype | OMIM#/(Reference) |
|---|---|---|---|---|
| EWSR1 | 1.9 | RNA binding protein, RNA splicing, ribosomal biogenesis | Amyotrophic Lateral Sclerosis | (Couthouis et al. 2012)a |
| TARDBP | 1.45 | DNA/RNA binding protein, RNA splicing | Amyotrophic Lateral Sclerosis 10 With Or Without Frontotemporal Dementia; Als10 | 612069 |
| LMNB1 | 1.47 | nuclear envelop protein | Leukodystrophy, Demyelinating, Adult-Onset, Autosomal Dominant (LMNB1 duplication); ADLD; cerebellar ataxia, cognitive decline, personality changes due to degeneration of the CNS white matter | 169500 |
a Couthouis, J. et al. (2012) Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 21, 2899–911.
Microcephaly and/or cognitive impairment were the most common elements of the neurodevelopmental phenotypes (14 of 16). The target proteins for these phenotypes included a ribosomal protein (RPL10), four established ribosomal biogenesis factors (EMG1, DKC1, EIF4A3, FLNA), and seven chromatin organization/genome maintenance regulators (SMC1A, MCM4, CUL4B, PRKDC, ATRX, NSD1, LMNA). Moreover, neurodevelopmental phenotypes were associated with several nucleolus-enriched proteins that did not have any demonstrated connections to ribosomal biogenesis and/or chromatin structure. That group included transcription regulators (LARP7, TCF4, and GTF2I, Table IV) and an RNA editing enzyme (ADAR, Table IV).
Verification of Nucleolar Enrichment of LARP7 and TCF4
Previously published proteomic studies of human cell lines of various origins did not report LARP7 or TCF4 to be nucleolar (24, 49, 50). Moreover, immunofluorescence analysis of several other human cell lines did not support their nucleolar localization either (http://www.proteinatlas.org/). Therefore, in the developing rat brain, the apparent nucleolar enrichment of LARP7 and TCF4 were further examined using specific antibodies. First, Western blot analysis of the nucleolar- and the whole nuclear lysates of the P7 cerebral cortex confirmed nucleolar enrichment of either protein (Fig. 4G). In addition, nucleolar enrichment of the established ribosomal biogenesis factor EMG1 was also confirmed by Western blotting (Fig. 4G).
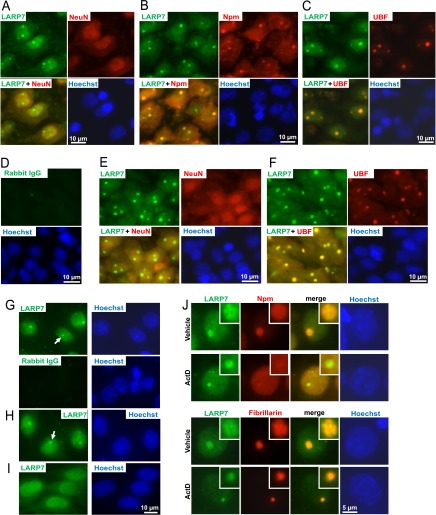
Second, immunofluorescence analysis of P7 rat brain sections revealed a strong nucleolar-like signal that was obtained with the anti-LARP7 antibody (Fig. 5A–5F). In the neocortex and the hippocampus, nucleolar presence of LARP7 was observed in neurons but not in non-neuronal cells as determined by co-staining for the neuronal marker NeuN (Fig. 5A, 5E, data not shown). Its nucleolar character was also confirmed by co-stainings for the nucleolar markers Npm and UBF (Fig. 5B, 5C, 5F). In addition, a diffused LARP7 signal was observed in the nucleoplasm (Fig. 5A-5F). A similar pattern of LARP7 immunofluorescence was found in other analyzed structures including the thalamus (data not shown).
Fig. 5.
Nucleolar presence of the RNA-binding protein LARP7 in forebrain neurons. A–F, LARP7 immunofluorescence in P7 rat brain sections that were cut through the dorsal hippocampus; co-stainings with neuronal- or nucleolar markers are shown as indicated; DNA was counterstained with Hoechst-33258; in D, LARP7 antibody was replaced by control rabbit IgG. Representative micrographs of cells from the neocortical layer V (A–D) and the hippocampal field CA1 (E–F, the pyramidal layer) are shown. Both nuclear and nucleolar signals of LARP7 are visible in neurons from each region. G–J, LARP7 Immunostaining in cultured primary rat brain cells including DIV6 cortical neurons (G, J), DIV7 hippocampal neurons (H) and DIV10 astrocytes (I). Note nucleolus-like staining in neurons but not astrocytes; both cell types display nuclear presence of LARP7. In J, representative nuclear profiles are shown following 6 h treatment with 0.1% DMSO (vehicle) or 1 μm ActinomycinD (ActD); magnified images of the nucleoli are shown in the insets. In vehicle-treated cells, overlapping stainings of LARP7 and fibrillarin suggest LARP7 localization to the dense fibrillar component (DFC) of the nucleolus. After transcriptional inhibition, Npm but not LARP7 or fibrillarin were released into nucleoplasm; moreover, the persistent overlap of LARP7 and fibrillarin indicates co-localization to light nucleolar caps which sequester early rRNA processing factors including snoRNAs (see text for details). The colocalization and/or separation of LARP7 and nucleolar markers was confirmed by quantitative analysis of images from panel J (supplemental Fig. S3).
Also, cultured rat neurons displayed nucleolar enrichment of LARP7 (Fig. 5G, 5H). However, as compared with cells from brain sections, stronger nuclear signal was observed in cultured cells. Conversely, in cultured rat primary astrocytes, no nucleolar enrichment was observed despite a strong nuclear signal of LARP7 (Fig. 5I). Therefore, nucleolar accumulation of LARP7 appears to be neuron-specific and may be sensitive to culture conditions.
When cortical neurons were imaged at high magnification, diffused- and fine granular staining of LARP7 was visible throughout the nucleus (Fig. 5J). Such a distribution is consistent with a previously proposed role of LARP7 in regulation of RNA-Polymerase-2 (51–53). In the nucleolus, LARP7 signal only partially overlapped with that of Npm (Fig. 5J and supplemental Fig. S3) (1). Conversely, LARP7 appeared to co-localize with fibrillarin which marks the dense fibrillar component (DFC) of the nucleolus where rRNA genes are transcribed and where their primary transcripts undergo initial processing (Fig. 5J and supplemental Fig. S3) (1).
The transcriptional inhibitor Actinomycin D (ActD) led to nucleoplasmic release of Npm but not LARP7 suggesting that rRNA-protein interactions are dispensable for its nucleolar localization (Fig. 5J and supplemental Fig. S3). In ActD-treated neurons, LARP and fibrillarin separated into apparently overlapping structures that were highly similar to nucleolar caps (Fig. 5J and supplemental Fig. S3) (54). Hence, the LARP7-containing caps resemble the previously reported light nucleolar caps (LNC) which are also known to sequester several snoRNAs (54).
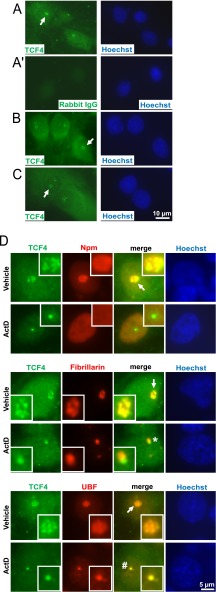
Unfortunately, in brain section, immunofluorescence analysis with the anti-TCF4 antibody was not conclusive as no specific signal was obtained from cell nuclei or perikarya. However, in cultured primary rat brain cells, a specific signal was observed in these compartments (Fig. 6). In cortical- and hippocampal neurons as well as astrocytes, the nuclear signal was concentrated in the nucleolus (Fig. 6A–6C). Besides nucleolus, both a diffuse- and a fine granular signal appeared throughout the nucleoplasm and in the perikarya (Fig. 6A–6D). Hence, only a relatively minor fraction of cellular TCF4 may reside in the nucleolus.
Fig. 6.
Nucleolar presence of the transcription factor TCF4 in primary cultures of forebrain neurons and astrocytes. Cultured rat brain cells were immunostained for TCF4 and the nucleolar markers as indicated. A, Representative images depicting TCF4 immunofluorescence in DIV6 cortical neurons. Both diffused and fine granular signals were observed in the nucleus and the perikaryon. In addition, nucleolus-like structures were detected (arrows). A', No signal was observed with a control IgG. Similar TCF4 staining pattern including nucleolar-like structures was found in DIV7 hippocampal neurons (B) and in DIV10 primary astrocytes (C). D, Representative nuclear profiles of DIV6 cortical neurons following 6h treatment with 0.1% DMSO (vehicle) or 1 μm ActinomycinD (ActD); magnified images of the nucleoli are shown in the insets. Note that the TCF4 signal shows the most extensive overlap with fibrillarin and only a partial overlap with Npm or UBF suggesting its localization to the transcriptionally active DFC. After transcriptional inhibition, TCF4, fibrillarin and UBF remained associated with the nucleolus. Note that fibrillarin and TCF4 segregated into distinct structures (indicated by a star) whereas UBF and TCF4 overlapped (indicated by a hash sign). Such a segregation pattern resembles fibrillar caps that sequester core components of the nucleolar transcription machinery (see text for details). The co-localization and/or separation of TCF4 and nucleolar markers was confirmed by quantitative analysis of images from panel D (supplemental Fig. S4).
In nucleoli of cortical neurons, TCF4 overlapped nearly completely with fibrillarin but only partially with Npm or UBF (Fig. 6D and supplemental Fig. S4). As in the case of LARP7, ActD did not trigger nucleoplasmic release of TCF4 which, instead, appeared sequestered in nucleolar cap-like structures (Fig. 6D and supplemental Fig. S4). However, the TCF4-containing caps were fibrillarin-negative but UBF-positive suggesting they were the fibrillar caps that sequester various component of the Pol1 machinery including UBF (54). Thus, at least in rat cortical neurons, LARP7 and TCF4 appear to be concentrated in the DFC compartment of the nucleolus and undergo nucleolar cap sequestration after inhibition of Pol1. However, they segregate into distinct types of caps which may suggest different roles at the early stages of ribosomal biogenesis.
Requirement of LARP7 for Ribosomal Biogenesis
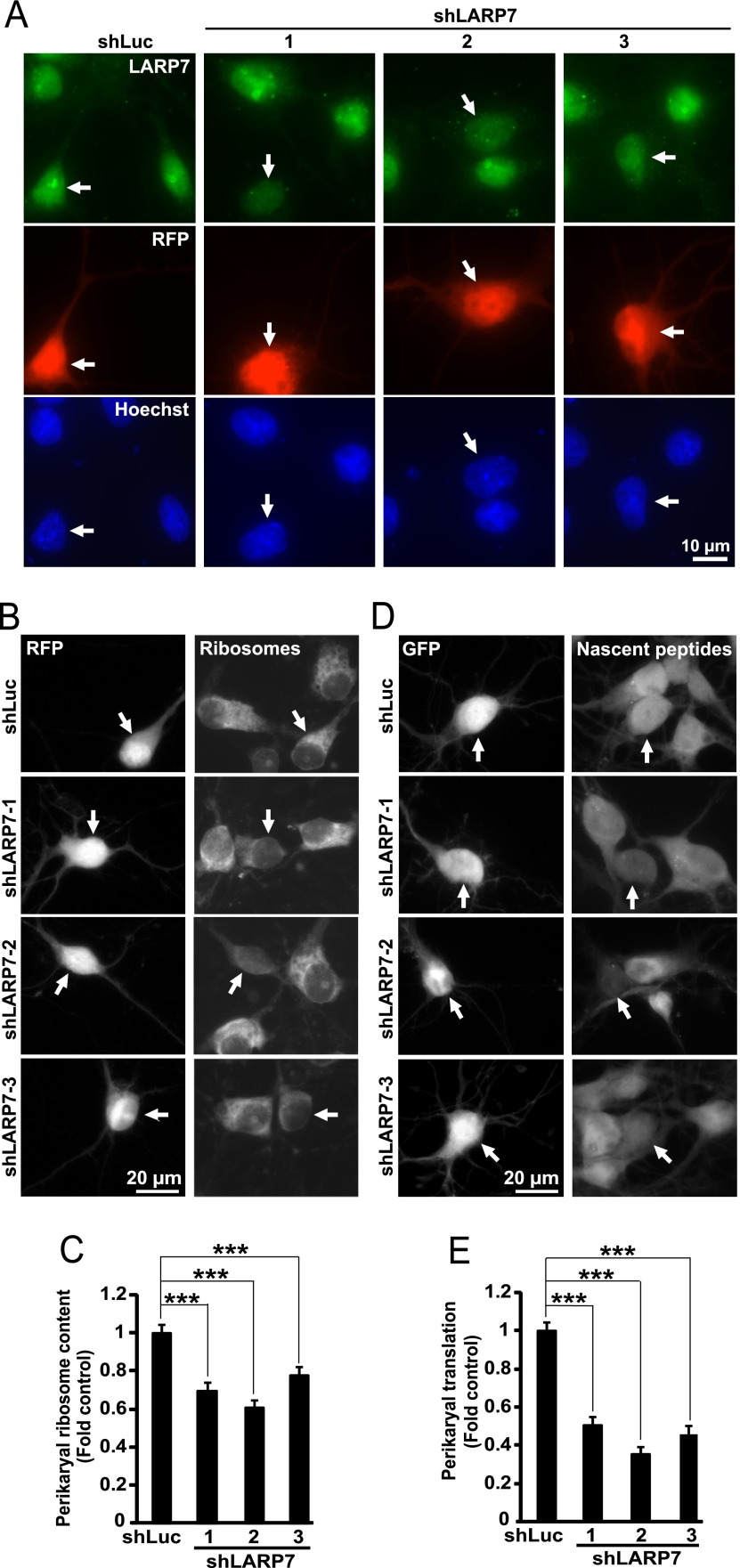
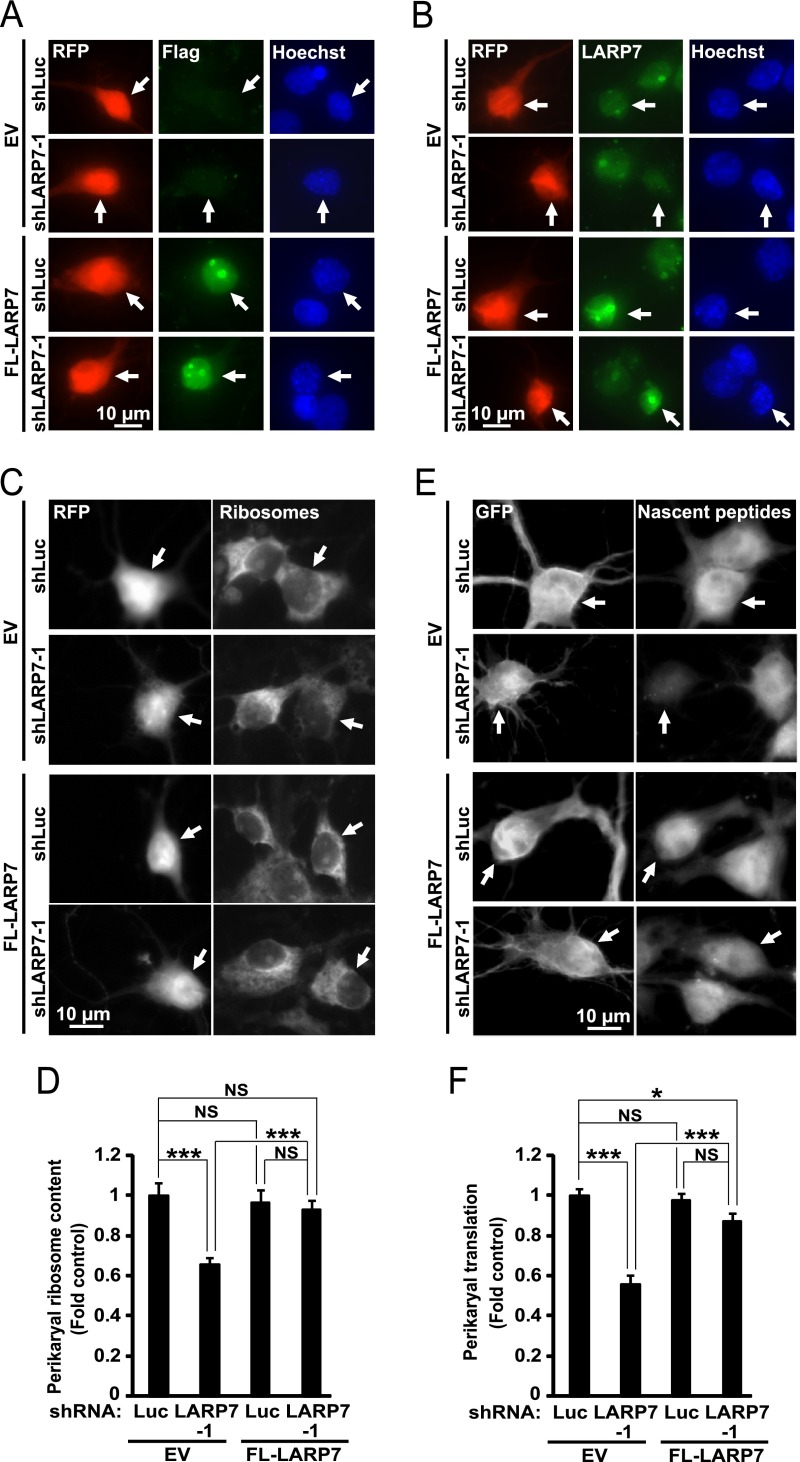
To test the role of LARP7 in ribosomal biogenesis, three different shRNAs against rat LARP7 were designed. When overexpressed in hippocampal neurons, these constructs reduced expression of endogenous LARP7 (Fig. 7A). Importantly, in transfected neurons, shRNAs against LARP7 prevented nucleolar staining with the LARP7 antibody further confirming specificity of the nucleolar signal (Fig 7A). Moreover, each of these constructs reduced perikaryal ribosome content by 22–40% as compared with a control shRNA (Fig. 7B, 7C). As reduction in ribosome content is expected to compromise protein synthesis activity, perikaryal accumulation of nascent peptides was monitored in LARP7-deficient neurons using co-translational labeling with puromycin (Fig. 7D, 7E). Indeed, shLARP7 reduced general protein synthesis. However, the observed declines by 50–65% were greater than those in ribosome content (Fig. 7D, 7E). When the rat-specific shLARP7–1 was combined with the expression vector for FLAG-tagged human LARP7 (FL-LARP7), LARP7 remained expressed including its enrichment in nucleolus-like structures (Fig. 8A, 8B). Moreover, the anti-ribosomal and anti-translational effects of shLARP7–1 were attenuated confirming that they are because of LARP7 deficiency (Fig. 8C–8F).
Fig. 7.
Reduced neuronal ribosome content and inhibition of protein synthesis after knockdown of LARP7 in hippocampal neurons. DIV4 (A) or DIV6 (B-E) rat hippocampal neurons were cotransfected with expression vectors for RFP and shRNAs against LARP7 or a control shRNA (shLuc, 0.2 + 0.6 μg of plasmid DNAs/2*105 neurons, respectively); in d-E, 0.1 μg EGFP expression vector was used instead of RFP. A, Representative images of transfected neurons (i.e. RFP-positive) that were fixed and immunostained for endogenous LARP7 at 72h post-transfection (arrows). Note that shLARP7 constructs reduced expression of endogenous LARP7 including disruption of the LARP7 signal in the nucleoli. B–C, After 3 days, perikaryal ribosome content was evaluated in RFP-positive cells by measuring intensity of ribosomal staining with NeuroTrace Green. For quantification, the ribosomal signal was normalized against DNA content in the nucleus as determined by counterstaining with Hoechst-33258. B, Representative images of transfected (i.e. RFP-positive) neurons whose ribosomes were stained with NeuroTrace Green (arrows). C, Quantification of ribosomal content in transfected neurons. shRNAs reduced mean ribosomal content. Data represent means ±S.E. of at least 72 cells from three independent experiments; ***, p < 0.001. D–E, Three days after transfection, O-propargyl-puromycin (OPP) was added to the culture medium to label nascent peptides which were visualized after fixation by fluorescent Click iT chemistry that was followed by GFP immunostaining. D, Representative images of transfected (i.e. GFP-positive) neurons with nascent peptide accumulation (arrows). E, Nascent peptide accumulation in the perikarya was reduced by shLARP7 constructs suggesting global translation deficiency. Data represent means ±S.E. of at least 54 individual cells from two independent experiments; ***, p < 0.001. Signal specificity controls for NeuroTrace staining of ribosomes (loss of staining after RNase A treatment) and OPP labeling of nascent peptides (loss of signal in cycloheximide-treated neurons) are presented in supplemental Fig. S5.
Fig. 8.
Overexpression of human LARP7 rescues anti-ribosomal and anti-tranlsational effects of an shRNA against rat LARP7. DIV4 (A–B) or DIV6 (C–D) rat hippocampal neurons were cotransfected with expression vectors for RFP, human FLAG-tagged LARP7 (FL-LARP7) and rat-specific shLARP7–1 or a control shRNA (shLuc, 0.1 + 0.2 + 0.6 μg of plasmid DNAs/2*105 neurons, respectively); pcDNA3.1 was used as an empty vector control (EV) for the FL-LARP7 construct; in E-F, the EGFP expression vector was used instead of RFP. A–B, Representative images of transfected (i.e. RFP-positive) neurons that were fixed and immunostained to detect the FLAG epitope tag (A, arrows) or human/rat LARP7 (B, arrows) at 48 h post-transfection. Distribution of FL-LARP7 resembled that of endogenous LARP7 including enrichment in nucleolus-like structures (A, B). Negative effects of shLARP7–1 on LARP7 expression were rescued by FL-LARP7 (B). C–D, After 3 days, perikaryal ribosome content was evaluated in RFP-positive cells as in Fig. 7B–7C. C, Representative images of transfected neurons whose ribosomes were stained with NeuroTrace Green (arrows). D, Quantification of ribosomal content in transfected neurons. Over expression of FL-LARP7 rescued anti-ribosomal effects of shLARP7–1. E–F, Three days after transfection, nascent peptides were metabolically labeled using OPP followed by GFP immunostaining as described for Fig. 7D–7E. E, Representative images depicting accumulation of nascent peptides in transfected (i.e. GFP-positive) neurons (arrows). F, Quantification of the nascent protein signal revealed that overexpression of FL-LARP7 attenuated anti-translational effects of shLARP7–1. Data represent means ±S.E. of at least 76 individual cells/condition from three independent experiments; ***, p < 0.001; *, p < 0.05; NS, p > 0.05.
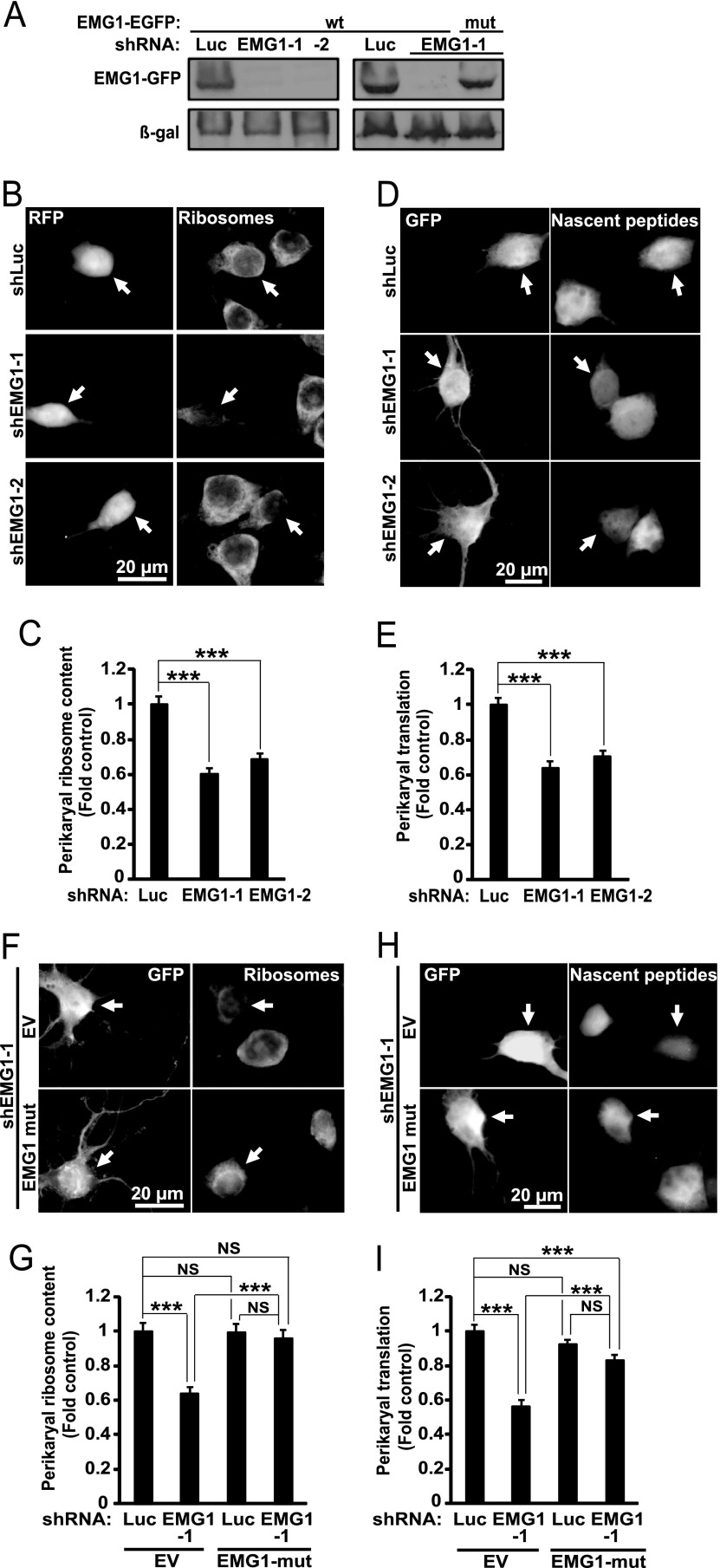
The effects of shLARP7 were compared with those of knocking down EMG1. EMG1 deficiency is a cause of Bowen-Conradi syndrome that combines neurodevelopmental delay and microcephaly. Two different shRNAs against EMG1 reduced ribosome content and protein synthesis by 32–40% and 30–36%, respectively (Fig. 9A–9E). The expression vector for rat EMG1 that was mutated to make it insensitive to shEMG1–1 attenuated the anti-ribosomal and anti-translational effects of the latter shRNA (Fig. 9A, 9F–9I). Thus, knockdowns of LARP7 and EMG1 produced similar anti-ribosomal effects. However, translational inhibition was stronger with shRNAs against LARP7. Therefore, at least in hippocampal neurons LARP7 is required for ribosomal biogenesis. It may also have additional functions in regulation of general protein synthesis.
Fig. 9.
Reduced neuronal ribosome content and inhibition of protein synthesis after knockdown of EMG1 in hippocampal neurons. A, COS7 cells were cotransfected with expression vectors for rat wild type (wt) EMG1-EGFP, β-galactosidase (β-gal) and shRNAs against EMG1 or a control shRNA (shLuc, 1 + 1 + 2 μg of plasmid DNA/60 mm plate, respectively). A mutant variant of rat EMG1-EGFP (mut) that was modified to become resistant to shEMG1–1 was also tested. Western blot for GFP revealed efficient knockdown of rat EMG1wt but not rat EMG1mut 48 h post transfection; the membrane was also probed for β-gal to ensure similar transfection efficiency. B–E, DIV6 rat hippocampal neurons were transfected and stained as described for experiments in Fig. 7B–7E except use of different shRNAs as indicated. B–C, Knockdown of EMG1 reduced mean ribosomal content in the perikarya. D–E, Knockdown of EMG1 reduced general protein synthesis. F–I, Transfections and stainings were as in Fig. 8 except use of different constructs as indicated; pcDNA3.1 was used as an empty vector control (EV) for EMG1-EGFPmut. Note that in F–G, the EGFP expression vector was used instead of RFP (Fig 8C–8D) and ribosomes were stained with NeuroTrace 530/615 Red Nissl Stain to avoid GFP interference. Anti-ribosomal (G) or anti-translational (I) effects of shEMG1–1 are blocked or attenuated by EMG1-EGFPmut. Data represent means ±S.E. of at least 58 cells from three independent experiments (C), 48 cell from two independent experiments (E), 52 cells from two independent experiments (G), or 91 cells from three independent experiments (I); ***, p < 0.001; NS, p > 0.05.
Previous work in non-neuronal cells has documented that by stabilizing 7SK RNA LARP7 negatively regulates RNA-Polymerase-2 (Pol2) driven transcription (51, 52, 55). However, in neurons, knockdown of LARP7 did not affect BDNF-mediated stimulation of Pol2-driven transcription of a cAMP Response Element (CRE)-regulated luciferase reporter (5.1-, 4.3-, or fourfold unstimulated controls with control shRNA, or shLARP7–1 or shLARP7–2, respectively, p > 0.05, supplemental Fig. S6). Therefore, although neuronal LARP7 affects production of ribosomes and protein synthesis it does not appear to regulate Pol2 activity at a CRE-regulated reporter gene.
Potential Role of TCF4 in Ribosomal Biogenesis
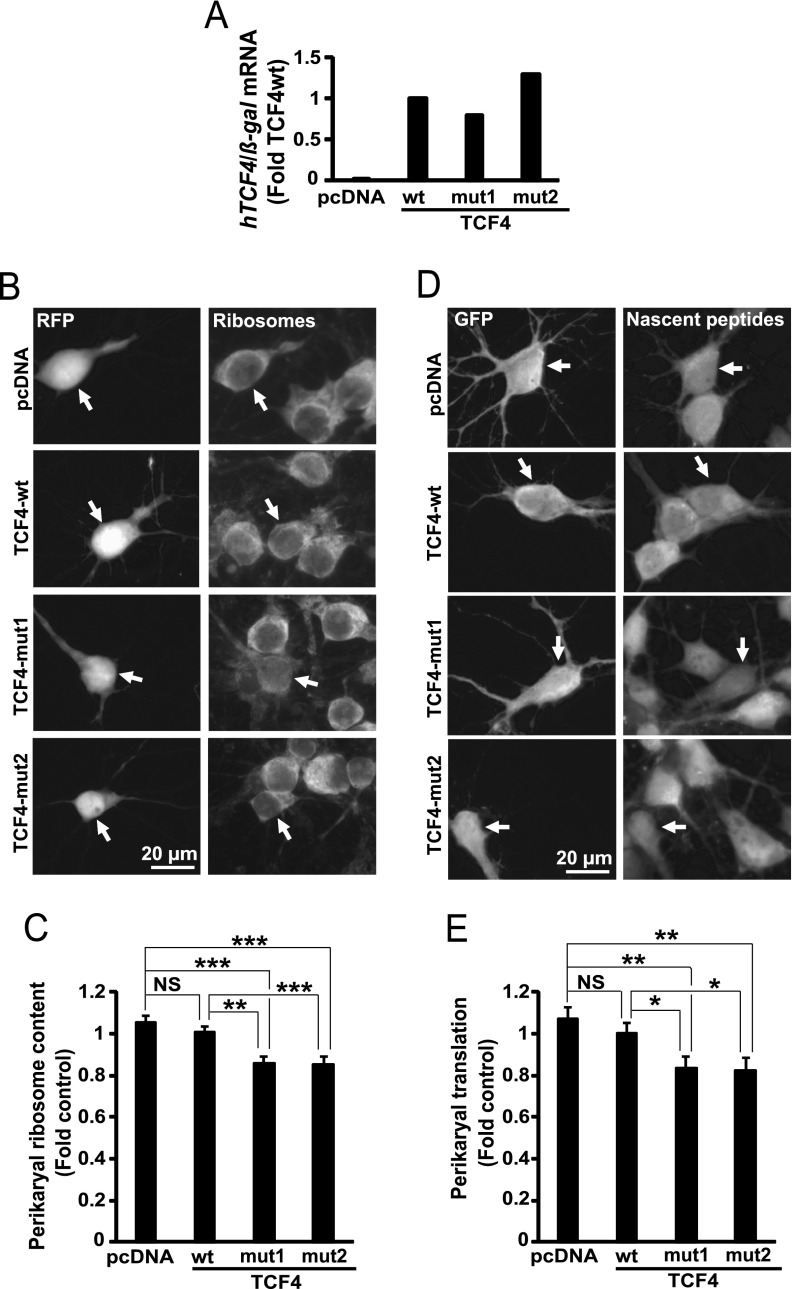
To achieve loss-of-function of TCF4, two mutant forms of human TCF4 were overexpressed in cultured cortical neurons (Fig. 10A). These mutants (Arg580Trp and Arg582Pro) disrupt the bHLH domain producing dominant-negative effects against wtTCF4 and the related members of the E-box family (35). Moreover, these mutants cause a neurodevelopmental disease, PTHS which may also result from TCF4 haploinsufficiency. After overexpressing the mutants for 3 days, perikaryal ribosome content was reduced by 20% or 15% as compared with empty vector (EV) or wtTCF4, respectively (Fig. 10B-10C). Similar reductions of general protein synthesis were also observed (Fig. 10D-10E). These findings suggest that in hippocampal neurons TCF4 and/or its relatives from the E-box family of transcription factors play a role in ribosomal biogenesis.
Fig. 10.
Reduced neuronal ribosome content and inhibited protein synthesis after overexpression of the PTHS-associated dominant-negative mutant form of TCF4. DIV3 (A) or DIV6 (B–D) rat hippocampal neurons were cotransfected with expression vectors for human TCF4 including its wt- and two PTHS-associated mutant variants (mut1 - Arg582Pro, mut2 - Arg580Trp) together with expression constructs for β-galactosidase (β-gal, A) or RFP (B, C) or EGFP (D, E) (0.4 + 0.2 μg of plasmid DNAs/2 × 105 neurons, respectively). A, After 24 h, human TCF4 mRNA levels were determined by qRT-PCR and normalized to β-gal mRNA. Human TCF4 was detected in neurons that received TCF4 plasmids but not the empty vector control. B–E, After 3 days, perikaryal ribosome content (B–C) and general protein synthesis (D–E) were evaluated as described for Fig. 7. The mutants but not the wt reduced mean ribosomal content and general protein synthesis. Data represent means ±S.E. of at least 80 cells from five independent experiments (C) or 68 cells from three independent experiments (E); *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, p > 0.05.
DISCUSSION
Quantitative proteomic analysis uncovered several hundred proteins that are enriched in nucleoli of the cerebral cortex of neonate rats. Because of limitations of the applied methodology, it is likely that in many cases the nucleolar enrichment was underestimated. Thus, protein and peptide ratios derived from the multiplexed quantification (iTRAQ or TMT) of highly complicated mixtures are known to be distorted because of the interference from the co-fragmenting ions. High fold changes tend to be substantially underestimated, and, in consequence, the distribution of peptide/protein ratios is artificially narrowed (56). This effect might be dealt with using several methods. They are, however, either moderately effective (computing isolation specificity, fractionation) or costly in terms of protein identification efficiency (12–22% reduction of non-redundant protein identifications when isolation window is narrowed or additional MS3 fragmentation is employed). Increasing the accuracy of protein ratio at the expense of proteome coverage might be in some cases a prudent solution. However, this study was more focused on the enrichment/depletion status of proteins than on actual protein ratio and was also aiming at possibly comprehensive protein sequencing. Therefore, the iTRAQ ratio compression was considered acceptable, and the assessment of accurate protein ratios was left for the follow-up experiments. However, it is possible that the currently used proteomic approach missed a significant number of proteins that are enriched in brain cell nucleoli.
Despite these limitations, the emerging nucleolar proteome landscape of the cerebral cortex of neonate rats suggests that ribosomal biogenesis remains active during postnatal stages of brain development. In addition, the importance of nucleolar contributions to neurodevelopment is suggested by human phenotypes that are associated with 16 proteins that are enriched in cerebro-cortical nucleoli including the ribosomal biogenesis factor EMG1, the RNA binding protein LARP7, and, the transcription factor TCF4.
Surprisingly, only 106 of 286 human ribosomal biogenesis factors were represented by rodent homologs among all Nucleolus-enriched proteins from rat brain (including 643 proteins with significant Nucleolus-enrichment and 102 proteins with Nucleolus-enrichment trends that were supported by the Nucleolus-classifier). Such observations suggest a unique organization of the ribosomal biogenesis pathway in brain cells, including neurons. That concept is consistent with a report that in mouse brain there are different species of rRNA processing intermediates than in mouse kidney (57). In addition, as brain ribosomal production is relatively rapidly down-regulated during the maturation phase of neurodevelopment (58–61), lesser number of ribosomal biogenesis factors may be required to satisfy reduced needs for new ribosomes. In contrast, in proliferating cells, many additional enzymes/regulators may be necessary to maximize rate of ribosome synthesis.
Overall, the overlap between nucleolar proteomes of the brain and proliferating cells was relatively limited. For instance, when the list of putative brain nucleolar proteins with at least 1.3-fold No/Nu enrichment was compared with 344 such proteins from human HCT116 colon carcinoma cells (24), only 114 proteins were found in both data sets. On that short list, the overrepresented GO_BPs were mostly related to ribosomal biogenesis and RNA metabolism.
In addition, the nucleolus-enriched brain proteome was compared with non-quantitative data sets of 726 or 873 nucleolar proteins from human cervical carcinoma HeLa- or human T-cell lymphoma Jurkat cells, respectively. The overlaps of 192 or 189 proteins were observed for each comparison (49, 50). The most highly enriched GO_BPs among the shared proteins were related to ribosomal biogenesis and RNA metabolism. Enrichment was also found for GO_BPs that are related to chromatin organization. When 263 brain-unique components of the nucleolar proteome (e.g. not present in either HeLa or Jurkat data sets) were analyzed for GO_BP enrichment, chromatin organization- but not ribosomal biogenesis remained overrepresented. The GO_BP term “chromosome organization” was associated with 21 proteins that were found in brain nucleoli but not in any other of the three nucleolar data sets from human cell lines (supplemental Table S4). Overall, it appears that nucleoli of mostly post-mitotic brain cells and proliferating cells share various proteins of the ribosomal biogenesis pathway and chromatin organization machinery.
Several putative nucleolar proteins from the developing brain are associated with human neurodevelopmental diseases including four established ribosomal biogenesis factors and a ribosomal protein (Table IV). A question that emerges from such an association is whether the nucleolus contributes to neurodevelopment? Rapidly proliferating cells are considered to be most sensitive to deficiencies in ribosome biogenesis (62, 63). Hence, neuroprogenitor proliferation could be impaired if ribosome production declines. In addition, the nucleolar/ribosomal stress response could initiate p53-dependent apoptosis eliminating neuroprogenitors with suboptimal ribosome production (13). Such a defective neurogenesis could account for microcephaly that is observed in 9 out of 16 syndromes that are associated with mutations of the putative nucleolar brain proteins (Table IV).
In addition, neuronal maturation is associated with intense neurite outgrowth that involves ribosomal biogenesis (10, 61). In fact, on postnatal day 7, when the nucleolar fractions of the cerebral cortex were obtained, there is neuritic growth and maturation but little neurogenesis in that region of the rodent brain (64). Therefore, it is tempting to speculate that ribosome depletion and consequent dysregulation of neuronal maturation including reduced neuritic morphogenesis contribute to at least some neurodevelopmental diseases that are associated with the nucleolar proteins.
The newly identified components of neuronal nucleolar proteome include the RNA binding protein LARP7 and the E-box transcription factor TCF4 whose deficiencies in humans are associated with cognitive impairment and microcephaly (65, 66). By binding and stabilizing the non-coding 7SK RNA, LARP7 negatively regulates the general Pol2 transcription elongation factor P-TEFb as well as the transcription factor HMGA1 (51–53, 55). However, it is unclear whether dysregulation of these transcription factors contributes to pathology of Alazami syndrome that besides sever cognitive impairment also includes dwarfism (65). In fact, such a pathogenic contribution appears counterintuitive as excessive activity of P-TEFb or HMGA1 may underlie increased proliferative growth following transformation (see discussion in (65)). Moreover, our results suggest that in neurons, knockdown of LARP7 does not increase Pol2-driven transcription of a CRE-regulated reporter construct. Likewise, no general effects on mRNA expression were observed in LARP7-depleted mouse ES cells (67). Conversely, our findings suggest that, at least, neurological symptoms of Alazami syndrome are caused by translational deficits and/or nucleolar stress.
There are several possible mechanisms by which LARP7 could support ribosome biogenesis and/or protein synthesis. First, as Pol2-driven non-coding transcription within rRNA genes interferes with Pol1-mediated expression of rRNA, LARP7 could support rRNA transcription by inhibiting Pol2 activity in the nucleolus (68). Second, it is possible that besides 7SK RNA, LARP7 stabilizes other RNAs including snoRNAs, many of which play a critical role in ribosomal biogenesis. A potential role of LARP7 in stabilization of C/D box snoRNAs is supported by its binding to the C/D box RNP core proteins including fibrillarin and NOP56 (51). Moreover, LARP7 and fibrillarin are colocalized in neuronal nucleoli.
Although both LARP7 and EMG1 support ribosome production in neurons, data from nonneuronal systems suggest that they are engaged in mostly nonoverlapping protein-protein interactions (thebiogrid.org). Therefore, LARP7 and EMG1 may regulate different aspects of the complex process of ribosomal biogenesis. However, interference with either activity could result in similar ribosomal deficits as shown in rat hippocampal neurons whose ribosomal biogenesis was perturbed at the levels of rRNA transcription or post-transcriptional assembly of the large- or small ribosomal subunit (61).
Lastly, knockdown of LARP7 had stronger anti-translational consequences than EMG1 despite similar depletion of ribosomes. This finding suggests that LARP7 may play a role in supporting expression and/or activity of other components of the translation machinery besides ribosomes. For instance, in mouse ES cells, LARP7-stimulates poly-adenylation of LIN28 mRNA (67) whose protein product enhances translation of various RPs (69). Thus, translational regulation adds to the list of potential links between ribosomal biogenesis and LARP7.
Similarly to Alazami syndrome, little is known about pathogenic mechanisms that underlie PTHS. The disease is caused by heterozygous deletions or point mutations disrupting the function of TCF4 by a loss of one active allele, generation of hypomorphic alleles or dominant negative effects (35). Our data indicate that the PTHS-associated dominant-negative mutants of TCF4 moderately reduce ribosomal biogenesis and general protein synthesis. Although future experiments are needed to clarify role of TCF4 in neuronal ribosome biogenesis, it is noteworthy that ribosomal proteins as well as many trans-acting factors of ribosome synthesis and nucleolus-associated proteins were shown to have their gene regulatory regions bound to TCF4 (70). In addition, the GO_BP term “ribosomal biogenesis” appeared to be overrepresented among genes that were down-regulated after TCF4 knockdown in human neuroblastoma cells (71). In proliferating cells, many of these genes, as well as rDNA, are regulated by E-boxes that interact with c-Myc (72, 73). Conversely, TCF4 and/or a TCF4-related transcription factors may regulate ribosomal biogenesis by engaging the same E-boxes in postmitotic and/or differentiating cells.
Putative proteins of the nucleolar brain proteome include 7 chromatin organization factors that are also associated with neurodevelopmental diseases. At least for one of such factors, the cohesin SMC1A, deficient rRNA transcription has been proposed as a mechanism that could explain cohesonipathy pathology in non-dividing cells including neurons (74, 75). Of note, two other cohesinopathy-associated proteins including NIPBL and SMC3 were also enriched in cerebro-cortical nucleoli, albeit less than 1.3-fold (OMIM #122470 and #610759, supplemental Table S2). Critical role of chromatin organization factors in rRNA transcription is not limited to cohesins but has been also reported for the chromatin remodeling complexes B-WICH and FACT whose various components are enriched in brain cell nucleoli (76, 77) (supplemental Table S2). Future studies are needed to investigate the role of nucleolus-enriched chromatin organizers in ribosome synthesis in the brain.
Taken together, the presented proteomic characterization of the nucleoli from the cerebral cortex of neonate rats supports the notion that the nucleolus is critical for neurodevelopment. Ribosomal biogenesis appears as top candidate function of the nucleolus through which it may affect formation and maturation of the nervous system. Disruption of ribosomal biogenesis by genetic and/or environmental factors may contribute to pathogenesis of various neurodevelopmental disorders. Finally, the current description of the nucleolar proteome offers a starting point to examine brain cell specific regulation of ribosomal synthesis, brain ribosome diversity, and nucleolar impact on neuronal chromatin organization and genomic maintenance.
Supplementary Material
Acknowledgments
We thank Dr. Marta Tkaczyk for advice on design of the initial proteomic experiments, and Dr. Scott R. Whittemore for critical reading of the manuscript. Drs. Tamas Kiss and Sylvian Egloff (Universite Paul Sabatier, Toulouse, France) donated the expression vector for human LARP7. The equipment used was sponsored in part by the Centre for Preclinical Research and Technology (CePT), a project co-sponsored by European Regional Development Fund and Innovative Economy, The National Cohesion Strategy of Poland. We declare no competing financial interests.
Footnotes
Author contributions: L.S., A.M., M.D., and M.H. designed research; L.S., A.M., A.P., and J.Z. performed research; M.S. and T.T. contributed new reagents or analytic tools; L.S., A.M., M.K., and M.H. analyzed data; L.S., A.M., and M.H. wrote the paper.
* This work was supported by NIH (R01 NS073584 and P30 GM103507 to MH), NSF (IOS1021860 to MH), the Commonwealth of Kentucky Challenge for Excellence Fund (MH), Estonian Research Council (IUT19–18 to TT) and Estonian Academy of Sciences (TT). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- Pol1
- RNA polymerase I
- aCSF
- artificial cerebrospinal fluid
- ActD
- Actinomycin D
- AS
- Alazami Syndrome
- bHLH
- basic helix-loop-helix motif
- BCS
- Bowen-Conradi Syndrome
- BP
- biological function
- CC
- cellular compartment
- CDLS
- Cornelia De Lange syndrome
- DFC
- dense fibrillar component
- DIV7
- day in vitro 7
- ECM
- extracellular matrix
- ER
- endoplasmic reticulum
- FASP
- Filter-Aided Sample Preparation
- GC
- granular component
- GFP
- green fluorescent protein
- GO
- Gene Ontology
- HP
- human phenotype
- iTRAQ
- isobaric Tags for Relative and Absolute Quantitation
- LARP7
- La-related protein 7
- MF
- molecular function
- NeuN
- neuronal nuclei
- No
- nucleolus
- Npm
- nucleophosmin/B23
- Nu
- nucleus
- OPP
- O-propargyl-puromycin
- P7
- postnatal day 7
- PTHS
- Pitt-Hopkins syndrome
- rDNA
- ribosomal DNA
- RFP
- red fluorescent protein
- RP
- ribosomal protein
- TCF4
- transcription factor 4
- TIF1A
- transcription initiation factor 1A
- UBF
- upstream binding factor
- UPLC
- Ultra Performance Liquid Chromatography
- wt
- wild type
- β-gal
- β-galactosidase.
REFERENCES
- 1. Sirri V., Urcuqui-Inchima S., Roussel P., and Hernandez-Verdun D. (2008) Nucleolus: the fascinating nuclear body. Histochem. Cell Biol. 129, 13–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Drygin D., Rice W. G., and Grummt I. (2010) The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 50, 131–156 [DOI] [PubMed] [Google Scholar]
- 3. Freed E. F., Bleichert F., Dutca L. M., and Baserga S. J. (2010) When ribosomes go bad: diseases of ribosome biogenesis. Mol. Biosyst. 6, 481–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kressler D., Hurt E., and Bassler J. (2010) Driving ribosome assembly. Biochim. Biophys. Acta 1803, 673–683 [DOI] [PubMed] [Google Scholar]
- 5. Stoykova A. S., Dudov K. P., Dabeva M. D., and Hadjiolov A. A. (1983) Different rates of synthesis and turnover of ribosomal RNA in rat brain and liver. J. Neurochem. 41, 942–949 [DOI] [PubMed] [Google Scholar]
- 6. Hetman M., and Pietrzak M. (2012) Emerging roles of the neuronal nucleolus. Trends Neurosci. 35, 305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lafarga M., Villegas J., and Crespo D. (1985) Changes in nucleolar morphology and volume of the supraoptic nucleus neurons during postnatal development of the rat. Brain Res. 354, 310–313 [DOI] [PubMed] [Google Scholar]
- 8. Clark P., Jones K. J., and LaVelle A. (1990) Ultrastructural and morphometric analysis of nucleolar and nuclear changes during the early growth period in hamster facial neurons. J. Comp. Neurol. 302, 749–760 [DOI] [PubMed] [Google Scholar]
- 9. Berciano M. T., Novell M., Villagra N. T., Casafont I., Bengoechea R., Val-Bernal J. F., and Lafarga M. (2007) Cajal body number and nucleolar size correlate with the cell body mass in human sensory ganglia neurons. J. Struct. Biol. 158, 410–420 [DOI] [PubMed] [Google Scholar]
- 10. Gomes C., Smith S. C., Youssef M. N., Zheng J. J., Hagg T., and Hetman M. (2011) RNA polymerase 1-driven transcription as a mediator of BDNF-induced neurite outgrowth. J. Biol. Chem. 286, 4357–4363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boulon S., Westman B. J., Hutten S., Boisvert F. M., and Lamond A. I. (2010) The nucleolus under stress. Mol. Cell 40, 216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kalita K., Makonchuk D., Gomes C., Zheng J. J., and Hetman M. (2008) Inhibition of nucleolar transcription as a trigger for neuronal apoptosis. J. Neurochem. 105, 2286–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parlato R., Kreiner G., Erdmann G., Rieker C., Stotz S., Savenkova E., Berger S., Grummt I., and Schutz G. (2008) Activation of an endogenous suicide response after perturbation of rRNA synthesis leads to neurodegeneration in mice. J. Neurosci. 28, 12759–12764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pederson T., and Tsai R. Y. (2009) In search of nonribosomal nucleolar protein function and regulation. J. Cell Biol. 184, 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boyd M. T., Vlatkovic N., and Rubbi C. P. (2011) The nucleolus directly regulates p53 export and degradation. J. Cell Biol. 194, 689–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Latonen L., Moore H. M., Bai B., Jaamaa S., and Laiho M. (2011) Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability. Oncogene 30, 790–805 [DOI] [PubMed] [Google Scholar]
- 17. Guetg C., and Santoro R. (2012) Formation of nuclear heterochromatin: the nucleolar point of view. Epigenetics 7, 811–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Andersen J. S., Lyon C. E., Fox A. H., Leung A. K., Lam Y. W., Steen H., Mann M., and Lamond A. I. (2002) Directed proteomic analysis of the human nucleolus. Curr. Biol. 12, 1–11 [DOI] [PubMed] [Google Scholar]
- 19. Scherl A., Coute Y., Deon C., Calle A., Kindbeiter K., Sanchez J. C., Greco A., Hochstrasser D., and Diaz J. J. (2002) Functional proteomic analysis of human nucleolus. Mol. Biol. Cell 13, 4100–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Coute Y., Burgess J. A., Diaz J. J., Chichester C., Lisacek F., Greco A., and Sanchez J. C. (2006) Deciphering the human nucleolar proteome. Mass Spectrom. Rev. 25, 215–234 [DOI] [PubMed] [Google Scholar]
- 21. Andersen J. S., Lam Y. W., Leung A. K., Ong S. E., Lyon C. E., Lamond A. I., and Mann M. (2005) Nucleolar proteome dynamics. Nature. 433, 77–83 [DOI] [PubMed] [Google Scholar]
- 22. Tafforeau L., Zorbas C., Langhendries J. L., Mullineux S. T., Stamatopoulou V., Mullier R., Wacheul L., and Lafontaine D. L. (2013) The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol. Cell 51, 539–551 [DOI] [PubMed] [Google Scholar]
- 23. Ahmad Y., Boisvert F. M., Gregor P., Cobley A., and Lamond A. I. (2009) NOPdb: Nucleolar Proteome Database–2008 update. Nucleic Acids Res. 37, D181–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boisvert F. M., Lam Y. W., Lamont D., and Lamond A. I. (2010) A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol. Cell. Proteomics 9, 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rieker C., Engblom D., Kreiner G., Domanskyi A., Schober A., Stotz S., Neumann M., Yuan X., Grummt I., Schutz G., and Parlato R. (2011) Nucleolar disruption in dopaminergic neurons leads to oxidative damage and parkinsonism through repression of mammalian target of rapamycin signaling. J. Neurosci. 31, 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kreiner G., Bierhoff H., Armentano M., Rodriguez-Parkitna J., Sowodniok K., Naranjo J. R., Bonfanti L., Liss B., Schutz G., Grummt I., and Parlato R. (2013) A neuroprotective phase precedes striatal degeneration upon nucleolar stress. Cell Death Differ. 20, 1455–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wisniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 28. Malinowska A., Kistowski M., Bakun M., Rubel T., Tkaczyk M., Mierzejewska J., and Dadlez M. (2012) Diffprot - software for non-parametric statistical analysis of differential proteomics data. J. Proteomics 75, 4062–4073 [DOI] [PubMed] [Google Scholar]
- 29. Elias J. E., Haas W., Faherty B. K., and Gygi S. P. (2005) Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods 2, 667–675 [DOI] [PubMed] [Google Scholar]
- 30. Hall M., Frank E., Holmes G., Pfaringer B., Reutermann P., and Witten I. H. (2009) The WEKA Data Mining Software: An Update. SIGKDD Explorations 11 [Google Scholar]
- 31. Scott M. S., Troshin P. V., and Barton G. J. (2011) NoD: a Nucleolar localization sequence detector for eukaryotic and viral proteins. BMC Bioinform. 12, 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sutherland H. G., Mumford G. K., Newton K., Ford L. V., Farrall R., Dellaire G., Caceres J. F., and Bickmore W. A. (2001) Large-scale identification of mammalian proteins localized to nuclear sub-compartments. Hum. Mol. Genet. 10, 1995–2011 [DOI] [PubMed] [Google Scholar]
- 33. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen J., Bardes E. E., Aronow B. J., and Jegga A. G. (2009) ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sepp M., Pruunsild P., and Timmusk T. (2012) Pitt-Hopkins syndrome-associated mutations in TCF4 lead to variable impairment of the transcription factor function ranging from hypomorphic to dominant-negative effects. Hum. Mol. Genet. 21, 2873–2888 [DOI] [PubMed] [Google Scholar]
- 36. Jaworski J., Spangler S., Seeburg D. P., Hoogenraad C. C., and Sheng M. (2005) Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin pathway. J. Neurosci. 25, 11300–11312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kalita K., Kharebava G., Zheng J. J., and Hetman M. (2006) Role of megakaryoblastic acute leukemia-1 in ERK1/2-dependent stimulation of serum response factor-driven transcription by BDNF or increased synaptic activity. J. Neurosci. 26, 10020–10032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Muniz L., Egloff S., and Kiss T. (2013) RNA elements directing in vivo assembly of the 7SK/MePCE/Larp7 transcriptional regulatory snRNP. Nucleic Acids Res. 41, 4686–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Habas A., Kharebava G., Szatmari E., and Hetman M. (2006) NMDA neuroprotection against a phosphatidylinositol-3 kinase inhibitor, LY294002 by NR2B-mediated suppression of glycogen synthase kinase-3beta-induced apoptosis. J. Neurochem. 96, 335–348 [DOI] [PubMed] [Google Scholar]
- 40. Hetman M., Hsuan S. L., Habas A., Higgins M. J., and Xia Z. (2002) ERK1/2 Antagonizes Glycogen Synthase Kinase-3beta -induced Apoptosis in Cortical Neurons. J. Biol. Chem. 277, 49577–49584 [DOI] [PubMed] [Google Scholar]
- 41. Yang P., Arnold S. A., Habas A., Hetman M., and Hagg T. (2008) Ciliary neurotrophic factor mediates dopamine D2 receptor-induced CNS neurogenesis in adult mice. J. Neurosci. 28, 2231–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pietrzak M., Smith S. C., Geralds J. T., Hagg T., Gomes C., and Hetman M. (2011) Nucleolar disruption and apoptosis are distinct neuronal responses to etoposide-induced DNA damage. J. Neurochem. 117, 1033–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu J., Xu Y., Stoleru D., and Salic A. (2012) Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc. Natl. Acad. Sci. U.S.A. 109, 413–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liang Y. M., Wang X., Ramalingam R., So K. Y., Lam Y. W., and Li Z. F. (2012) Novel nucleolar isolation method reveals rapid response of human nucleolar proteomes to serum stimulation. J. Proteomics 77, 521–530 [DOI] [PubMed] [Google Scholar]
- 45. Holt C. E., and Schuman E. M. (2013) The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 80, 648–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kondrashov N., Pusic A., Stumpf C. R., Shimizu K., Hsieh A. C., Xue S., Ishijima J., Shiroishi T., and Barna M. (2011) Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145, 383–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Uechi T., Nakajima Y., Nakao A., Torihara H., Chakraborty A., Inoue K., and Kenmochi N. (2006) Ribosomal protein gene knockdown causes developmental defects in zebrafish. PLoS ONE 1, e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brooks S. S., Wall A. L., Golzio C., Reid D. W., Kondyles A., Willer J. R., Botti C., Nicchitta C. V., Katsanis N., and Davis E. E. (2014) A novel ribosomopathy caused by dysfunction of RPL10 disrupts neurodevelopment and causes X-linked microcephaly in humans. Genetics 198, 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leung A. K., Trinkle-Mulcahy L., Lam Y. W., Andersen J. S., Mann M., and Lamond A. I. (2006) NOPdb: Nucleolar Proteome Database. Nucleic Acids Res. 34, D218–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jarboui M. A., Wynne K., Elia G., Hall W. W., and Gautier V. W. (2011) Proteomic profiling of the human T-cell nucleolus. Mol. Immunol. 49, 441–452 [DOI] [PubMed] [Google Scholar]
- 51. Krueger B. J., Jeronimo C., Roy B. B., Bouchard A., Barrandon C., Byers S. A., Searcey C. E., Cooper J. J., Bensaude O., Cohen E. A., Coulombe B., and Price D. H. (2008) LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 36, 2219–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. He N., Jahchan N. S., Hong E., Li Q., Bayfield M. A., Maraia R. J., Luo K., and Zhou Q. (2008) A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell 29, 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Eilebrecht S., Brysbaert G., Wegert T., Urlaub H., Benecke B. J., and Benecke A. (2011) 7SK small nuclear RNA directly affects HMGA1 function in transcription regulation. Nucleic Acids Res. 39, 2057–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shav-Tal Y., Blechman J., Darzacq X., Montagna C., Dye B. T., Patton J. G., Singer R. H., and Zipori D. (2005) Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell 16, 2395–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Markert A., Grimm M., Martinez J., Wiesner J., Meyerhans A., Meyuhas O., Sickmann A., and Fischer U. (2008) The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 9, 569–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ting L., Rad R., Gygi S. P., and Haas W. (2011) MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods 8, 937–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Akhmanova A., Verkerk T., Langeveld A., Grosveld F., and Galjart N. (2000) Characterisation of transcriptionally active and inactive chromatin domains in neurons. J. Cell Sci. 113, 4463–4474 [DOI] [PubMed] [Google Scholar]
- 58. Adams D. H. (1966) The relationship between cellular nucleic acids in the developing rat cerebral cortex. Biochem. J. 98, 636–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Johnson T. C. (1967) The effects of maturation on in vitro RNA synthesis by mouse brain cells. J. Neurochem. 14, 1075–1081 [DOI] [PubMed] [Google Scholar]
- 60. Berthold W., and Lim L. (1976) Nucleo-cytoplasmic relationships of high-molecular-weight ribonucleic acid, including polyadenylated species, in the developing rat brain. Biochem. J. 154, 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Slomnicki L. P., Pietrzak M., Vashishta A., Jones J., Lynch N., Elliot S., Poulos E., Malicote D., Morris B. E., Hallgren J., and Hetman M. (2016) Requirement of Neuronal Ribosome Synthesis for Growth and Maintenance of the Dendritic Tree. J. Biol. Chem. 291, 5721–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Trainor P. A., and Merrill A. E. (2014) Ribosome biogenesis in skeletal development and the pathogenesis of skeletal disorders. Biochim. Biophys. Acta 1842, 769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ellis S. R. (2014) Nucleolar stress in Diamond Blackfan anemia pathophysiology. Biochim. Biophys. Acta 1842, 765–768 [DOI] [PubMed] [Google Scholar]
- 64. Bayer S. A., and Altman J. (1987) Directions in neurogenetic gradients and patterns of anatomical connections in the telencephalon. Prog. Neurobiol. 29, 57–106 [DOI] [PubMed] [Google Scholar]
- 65. Alazami A. M., Al-Owain M., Alzahrani F., Shuaib T., Al-Shamrani H., Al-Falki Y. H., Al-Qahtani S. M., Alsheddi T., Colak D., and Alkuraya F. S. (2012) Loss of function mutation in LARP7, chaperone of 7SK ncRNA, causes a syndrome of facial dysmorphism, intellectual disability, and primordial dwarfism. Hum. Mutat. 33, 1429–1434 [DOI] [PubMed] [Google Scholar]
- 66. Sweatt J. D. (2013) Pitt-Hopkins Syndrome: intellectual disability due to loss of TCF4-regulated gene transcription. Exp. Mol. Med. 45, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dai Q., Luan G., Deng L., Lei T., Kang H., Song X., Zhang Y., Xiao Z. X., and Li Q. (2014) Primordial dwarfism gene maintains Lin28 expression to safeguard embryonic stem cells from premature differentiation. Cell Rep. 7, 735–746 [DOI] [PubMed] [Google Scholar]
- 68. Gagnon-Kugler T., Langlois F., Stefanovsky V., Lessard F., and Moss T. (2009) Loss of human ribosomal gene CpG methylation enhances cryptic RNA polymerase II transcription and disrupts ribosomal RNA processing. Mol. Cell 35, 414–425 [DOI] [PubMed] [Google Scholar]
- 69. Peng S., Chen L. L., Lei X. X., Yang L., Lin H., Carmichael G. G., and Huang Y. (2011) Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells 29, 496–504 [DOI] [PubMed] [Google Scholar]
- 70. Ghosh H. S., Cisse B., Bunin A., Lewis K. L., and Reizis B. (2010) Continuous expression of the transcription factor e2–2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity 33, 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Forrest M. P., Waite A. J., Martin-Rendon E., and Blake D. J. (2013) Knockdown of human TCF4 affects multiple signaling pathways involved in cell survival, epithelial to mesenchymal transition and neuronal differentiation. PLoS ONE 8, e73169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Grandori C., Gomez-Roman N., Felton-Edkins Z. A., Ngouenet C., Galloway D. A., Eisenman R. N., and White R. J. (2005) c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 7, 311–318 [DOI] [PubMed] [Google Scholar]
- 73. Poortinga G., Wall M., Sanij E., Siwicki K., Ellul J., Brown D., Holloway T. P., Hannan R. D., and McArthur G. A. (2011) c-MYC coordinately regulates ribosomal gene chromatin remodeling and Pol I availability during granulocyte differentiation. Nucleic Acids Res. 39, 3267–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Xu B., Lee K. K., Zhang L., and Gerton J. L. (2013) Stimulation of mTORC1 with L-leucine rescues defects associated with Roberts syndrome. PLoS Genet. 9, e1003857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xu B., Sowa N., Cardenas M. E., and Gerton J. L. (2014) l-leucine partially rescues translational and developmental defects associated with zebrafish models of Cornelia de Lange syndrome. Hum. Mol. Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tsai Y. C., Greco T. M., Boonmee A., Miteva Y., and Cristea I. M. (2012) Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol. Cell. Proteomics 11, 60–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tessarz P., Santos-Rosa H., Robson S. C., Sylvestersen K. B., Nelson C. J., Nielsen M. L., and Kouzarides T. (2014) Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature 505, 564–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.