Abstract
SNPs associated with disease susceptibility often reside in clusters of gene enhancers, or super enhancers. Constituents of these enhancer clusters cooperate to regulate target genes, and often extend beyond the linkage disequilibrium blocks containing GWAS risk SNPs. We identified “outside variants”, defined as SNPs in weak LD with GWAS risk SNPs that physically interact with risk SNPs as part of the target gene’s regulatory circuitry. These outside variants explain additional target gene expression variation beyond that of GWAS associated SNPs. Additionally, the clinical risk associated with the GWAS SNPs is considerably modified by the genotype of the outside variant. Collectively, these findings suggest a potential model whereby outside variants and GWAS SNPs that physically interact in 3D chromatin collude to influence target transcript levels as well as clinical risk. This model offers an additional hypothesis for the source of missing heritability of complex traits.
Introduction
Transcriptional regulatory elements are hotspots for genetic predisposition to disease. Single nucleotide polymorphisms (SNPs) associated with disease susceptibility by genome-wide association studies (GWAS) are heavily enriched in putative cell type-specific regulatory elements, mostly enhancers, demarcated through ChIP-seq studies of signature histone marks and associated transcription factors1–6. Of the heritability estimates for common disease made by GWAS studies, variants in regulatory elements are estimated to account for 79%7. The enrichment is particularly pronounced in regions of enhancer clusters, which have been described as super enhancers8,9, stretch enhancers10 and multiple enhancer variant (MEV) loci5. Enhancer clusters involve multiple, robust, cell type-specific enhancers arranged in cis and are often located near genes that function to establish and/or maintain cellular identity8–11. At enhancer clusters associated with disease-risk, it has been proposed that multiple SNPs distributed across the individual enhancer constituents cooperatively influence enhancer activity and effect expression of the target gene5,12–18.
Regulatory variants associated with disease susceptibility often impact target transcript levels, and expression quantitative trait loci (eQTL) studies have had great success in identifying functional variants. GWAS variants are enriched for eQTLs19–21 and this enrichment is particularly pronounced amongst eQTLs in tissues relevant to the pathogenesis of a given disorder22. However, to date eQTLs have not been identified for the majority of GWAS loci19–21,23,24. There are a variety of possible explanations: eQTLs may only be apparent in very specific cell types or conditions, or the effect sizes are too weak and large samples sizes are therefore required for their detection. An alternative explanation is that physical interactions among enhancer SNPs, dictated by higher-order chromatin folding at enhancer clusters, impact target transcript levels. Indeed, analysis of three-dimensional genomic architecture has demonstrated that multiple enhancers that are all part of a gene’s regulatory circuitry physically interact with one another and collectively engage a target promoter to facilitate transcription25,26. The SNPs within a gene’s regulatory circuit could cooperate in various ways to impact target gene expression, including additively27,28, synergistically29, conditionally29–33, epistatically or through currently unknown modalities that are locus and cell-context dependent. Regardless of the modality, SNPs within physically interacting enhancers could exert effects on target gene expression that may be missed through traditional eQTL analyses. Furthermore, given that a gene’s regulatory circuitry is independent of haplotype block structure, it is possible that SNPs in weak LD with GWAS risk SNPs, but within the same regulatory circuit, participate in the regulation of target gene expression and influence the overall clinical risk to disease.
Results
Regulatory circuitry at GWAS loci extends beyond LD blocks
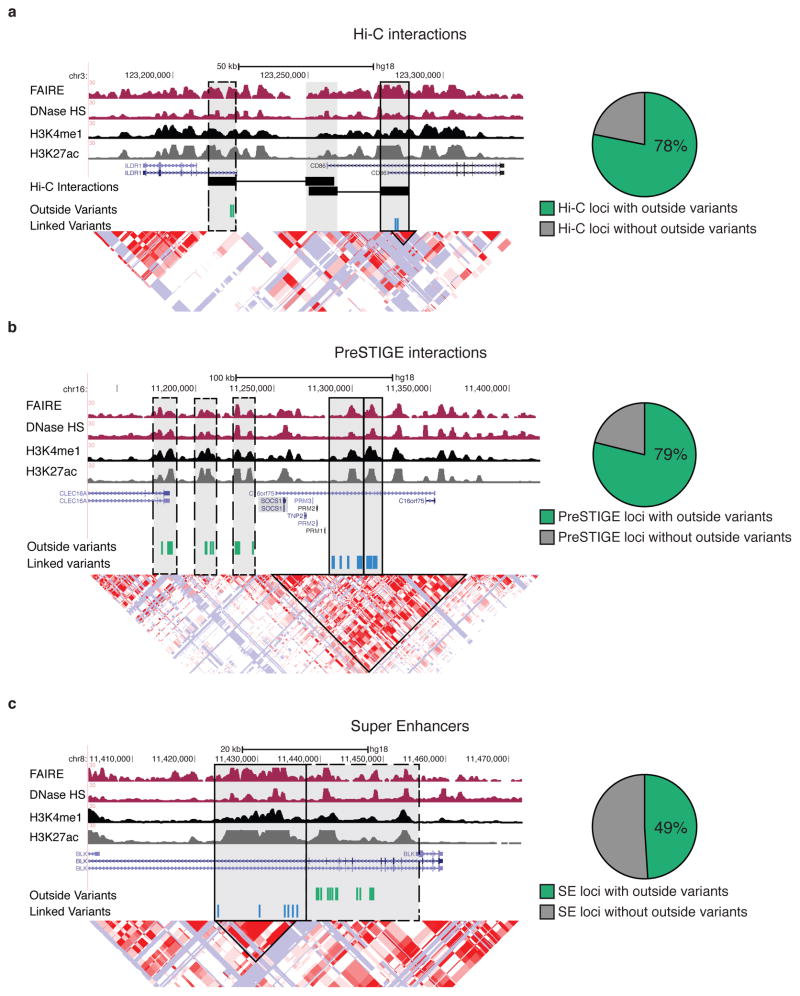
Compared to randomly sampled SNPs, SNPs associated with risk to six autoimmune diseases, rheumatoid arthritis, systemic lupus, Crohn’s disease, multiple sclerosis, ulcerative colitis and celiac disease are highly enriched in active gene enhancer elements in B-lymphoblasts, as well as B cells and T cells (which share a common regulatory landscape at risk loci)1,5. We identified high confidence interactions from B lymphoblast high resolution Hi-C data that associated putative regulatory elements (demarcated by H3K4me1) with promoters for 170 GWAS loci. For 78% of these loci, promoters associated with putative regulatory elements containing GWAS SNPs were also associated with regulatory elements that contained “outside variants”, i.e, SNPs in weak linkage disequilibrium with the GWAS linked SNPs (Supplementary Fig. 1a). An example is shown in Figure 1a, where Hi-C interactions associate multiple sclerosis risk SNP rs9282641 with the CD86 promoter. The CD86 promoter is also physically associated with an additional putative regulatory element (dotted box) that contains variants that are in weak LD with the GWAS SNP (D’<0.5 and r2<0.1). Thus the regulatory circuitry of CD86 extends beyond the haplotype block from which the GWAS association arose.
Figure 1. Regulatory circuitry of GWAS loci frequently extends beyond the boundaries of haplotype blocks.
(a) Example of multiple sclerosis risk locus where Hi-C identifies physical interactions of the CD86 promoter with linked variants (black box), those in LD with the GWAS SNP rs9282641, and outside variants (dashed box), and those inherited independently from the GWAS SNP (left). Proportion of autoimmune-GWAS loci containing outside variants (D’<0.5 and r2<0.1 with GWAS SNP) for Hi-C identified enhancer-gene interactions (right, n= 412 total GWAS loci). (b) Example of a multiple sclerosis risk locus where the gene target SOCS1 is predicted to be regulated by enhancers (highlighted in grey) that contain variants linked to GWAS SNP rs7191700 and outside variants (left). Proportion of GWAS loci containing outside variants for PreSTIGE defined enhancer-gene interactions (right, n=156 total GWAS loci). (c) Example of a super enhancer lupus risk locus that contains both variants linked to GWAS SNP rs13277113 and outside variants (left). Proportion of GWAS loci containing outside variants for super enhancer loci (right, n= 159 total GWAS loci).
Given the limitations in resolution of Hi-C, we also employed a computational method, PreSTIGE5, to identify potential gene targets of putative regulatory elements containing risk SNPs at 112 autoimmune disease-associated loci in B lymphoblasts. Consistent with our findings using Hi-C-defined interactions, for 79% of loci we evaluated, putative regulatory elements containing risk SNPs were predicted to regulate target genes in cooperation with regulatory elements containing outside variants. An example is shown in Figure 1b, where the multiple sclerosis risk SNP rs7191700 is predicted to target SOCS1. SOCS1 is also predicted to be controlled by additional regulatory elements containing outside variants (dotted boxes).
Finally, we compared haplotype structure at risk loci to super enhancers; clusters of active gene regulatory elements that are proposed to act cooperatively on target gene expression8,9. We detected outside variants at 49% of risk loci containing super enhancers. An example is shown in Figure 1c, where a super enhancer containing a lupus associated SNP rs13277113 extends 17-kb beyond the associated block and contains outside variants. Thus, regardless of how regulatory circuitry was defined, either by using Hi-C interactions, computationally predicted enhancer-gene interactions, or super enhancers, DNA variants that are part of a common regulatory circuitry but are in weak LD of the risk locus were frequently observed (Supplementary Fig. 1b,c).
Physical interactions between SNPs impact gene expression
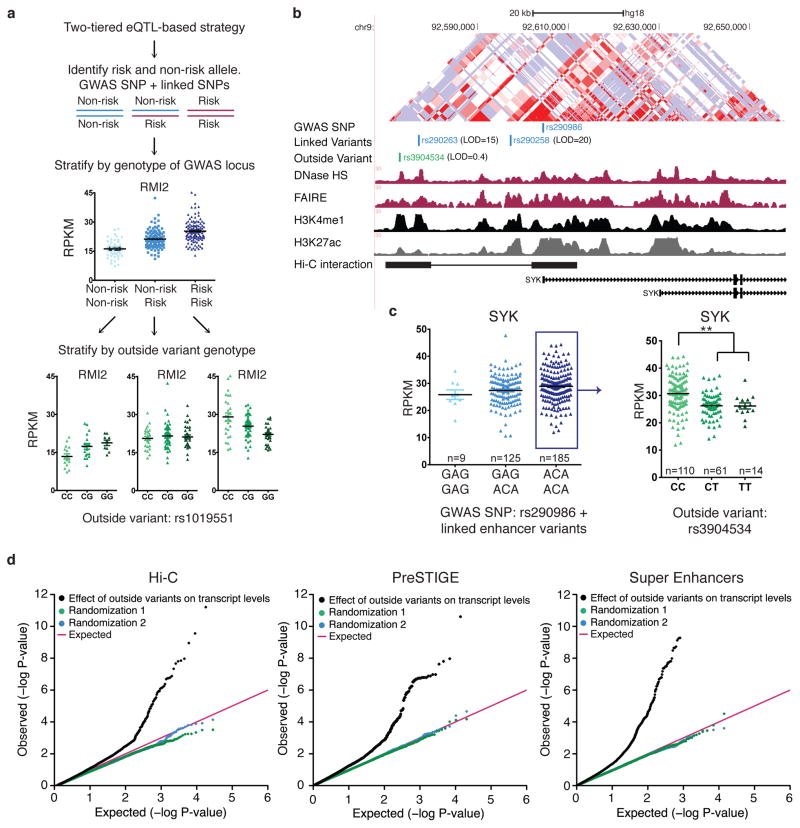
We next sought to determine if outside variants affect the levels of target gene expression. We utilized B lymphoblast transcriptome data along with corresponding SNP genotype data from 373 Europeans34. Given that there are diverse modalities by which multiple enhancers function, and these modalities are locus-dependent, we developed a two-tiered eQTL-based approach, whereby individuals were first stratified based on the genotype of GWAS linked variants, (risk versus non-risk), and then further subdivided based on the genotype of the outside variants (Fig. 2a). The two-tiered stratification approach is designed to be agnostic to the interaction modality of enhancers within a cluster, be it additive, epistatic, synergistic or a novel uncharacterized mechanism. The approach evaluates the impact of the outside variant on each GWAS genotype (e.g. non-risk/non-risk, non-risk/risk and risk/risk) separately and without regard to its effect in the context of the other GWAS genotypes. The approach is designed to capture variants that account for additional variation in gene expression beyond the effect of variants in tight LD (LOD >2 and D’ >0.6) with the GWAS allele. We started with the multiple sclerosis-associated SYK (spleen tyrosine kinase) locus on chromosome 9. SYK plays an important role in ITAM-mediated signaling transduction from B-cell receptors to downstream cellular functions35. We identified an outside variant (rs3904534) that lies in a putative regulatory element that is both computationally predicted to regulate SYK and determined through Hi-C analyses to be physically associated with the SYK promoter (Fig. 2b). There was no significant difference in SYK expression based on the genotype of the risk locus alone (Fig. 2c, left). When individuals homozygous for the risk allele were further stratified by the genotype of the outside variant, a significant difference in SYK transcript levels was observed (Fig. 2c, right).
Figure 2. Physical interactions between outside variants and GWAS alleles impact target gene expression.
(a) Two-tiered eQTL-based strategy to evaluate impact of outside variants on target transcript levels. Significant difference in RMI2 transcript levels is observed amongst individuals based on the genotype of the GWAS SNP rs4783055 (blue). The genotype of outside variant, rs1019551 explains additional variation in transcript levels (green) (b) The SNP rs290986 is associated with multiple sclerosis and is located in a putative enhancer element regulating SYK. rs3904534 is an “outside variant” that lies in an enhancer that regulates SYK. This interaction is both predicted by PreSTIGE and identified by Hi-C. (c) Individuals were stratified by the genotype of the GWAS locus, and the levels of SYK in B lymphoblasts were plotted (mean ± SEM). Individuals homozygous for the risk allele (blue box) were further stratified by the genotype of the outside variant. Outside variant, rs3904534, significantly alters the effect of the GWAS allele on SYK levels (Wilcox-test, **P<1.2E–6). (d) QQ plot showing distribution of P values for all tested interactions between outside variants and GWAS-linked loci on target transcript levels for Hi-C interactions (left), PreSTIGE predicted interactions (center), and super enhancers (right).
We expanded our two-tiered eQTL-based strategy to evaluate the impact of outside variants defined by our three methods of determining the chromatin regulatory circuitry for a total of 186 GWAS loci (see methods, Supplementary Fig. 2). We then compared P-values calculated from quantifying the effects of outside variants on target gene levels to random permutations and found that the outside variant genotype frequently alters transcript levels (Fig. 2d). 24–34% of all evaluated GWAS loci involved at least one outside variant significantly associated with gene expression (Supplementary Fig. 2g Table S1, methods). Hereafter we refer to these as “functional outside variants”. These estimates were based on two different methods for multi-test correcting: false discovery rate (FDR) and generation of null p-value distributions from permutations for each locus (methods)36,37. Outside variants were identified regardless of the approach used to define regulatory circuitry and the rates were comparable between all three methods. 61% of the functional outside variants were not previously identified as independent eQTLs (Supplementary Fig. 3).
Functional outside variants share key features of enhancers
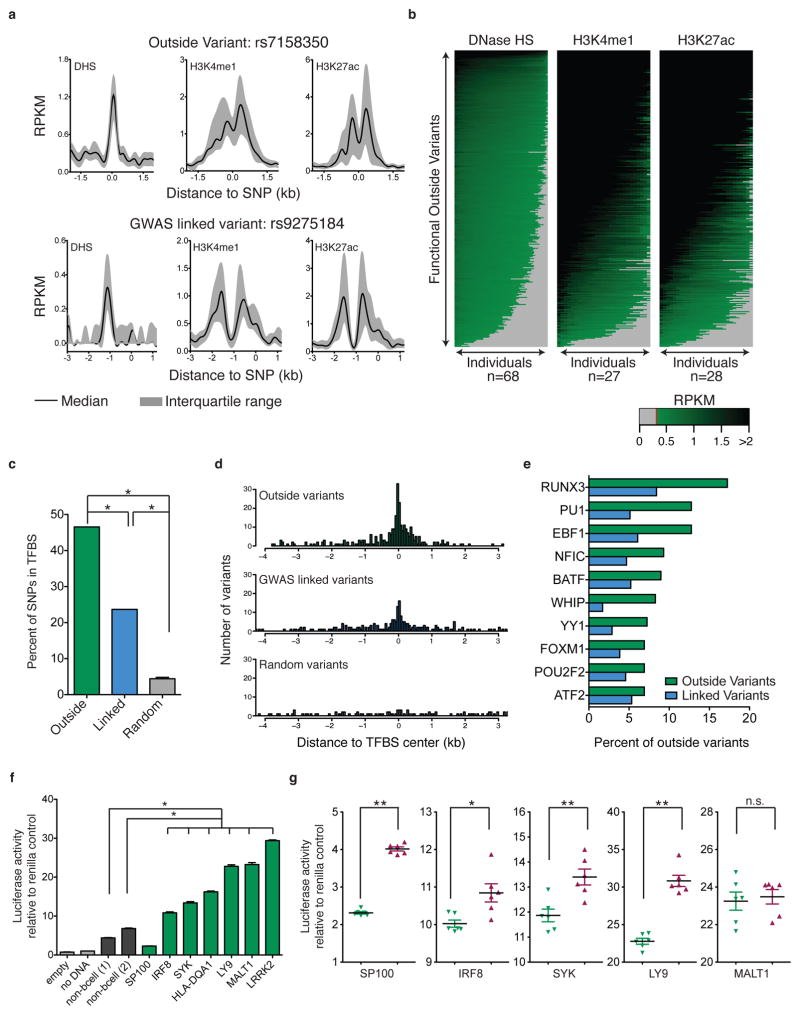
We hypothesized that outside variants altered the effect of GWAS alleles on target transcript levels by altering enhancer function within shared regulatory circuits. Using publicly available datasets from 27–68 B lymphoblast cell lines, we compared chromatin features associated with functional outside variants to those of disease-associated variants38–40. Example loci are shown in Figure 3a and while some inter-individual variability was evident, we detected enrichment for DHS, H3K4me1 and H3K27ac across individuals for outside variant rs7158350 and GWAS linked variant rs9275184. Similar enrichment was observed across all loci with functional outside variants (Fig. 3b). 85% of functional outside variants were enriched for H3K27ac, H3K4me1 and DNase hypersensitivity in more than two-thirds of all cell lines analyzed.
Figure 3. Functional outside variants share signature features of enhancer elements.
(a) B lymphoblast DNase-seq and ChIP-seq signal (RPKMs, reads per killobase per million mapped reads) surrounding functional outside variant rs7158350 (top) and GWAS linked variant rs9275184 (bottom) across a panel of individuals. Plotted are median and interquartile range of RPKMs for DHS (n=68), H3K4me1 (n=27) and H3K27ac (n=28). (b) RPKMs are shown for each functional outside variant locus (rows) for B lymphoblast cell lines (columns) profiled for DNase HS (left), H3K4me1 (center) and H3K27ac (right). Columns are sorted independently for each outside variant. Grey denotes below threshold of enrichment. (c) Proportion of outside variants (green) linked variants (blue), and randomly selected variants (grey) that lie in transcription factor binding sites (TFBS) identified through B lymphoblast TF ChIP-seq (Fisher’s exact test, *P<1E–4). (d) Distance of outside variants (green) linked variants (blue) and random control variants (grey) relative to the center of the nearest ChIP-seq identified TFBS. (e) Transcription factors that are most frequently bound at outside variant loci (green) and linked variant loci (blue). (f) Luciferase reporter activity for outside variant loci (green) and non-B-cell control enhancers and empty vector control enhancers (grey) (one-way ANOVA, *P<1E–4, mean ± SEM shown for 6 replicates) (g) Luciferase reporter activity (relative to co-transfection renilla control) for both alleles (red and green) at outside variant loci (t-test *P<0.01 **P<0.003, mean ± SEM).
We also analyzed B lymphoblast ChIP-seq datasets from >75 unique transcription factors41. Both functional outside variants and GWAS linked variants were bound by transcription factors significantly more often than expected (Fig. 3c). 77% of functional outside variants were located within 1-kb of TF binding sites mapped through ChIP-seq studies (Fig. 3d). By comparison, 12% of randomly selected SNPs were located within 1-kb of TF binding sites. Factors most frequently bound at outside variants were those with known roles in mounting immune responses and hematopoiesis, including RUNX3, PU.1, and EBF142–44 (Fig. 3e). To directly evaluate enhancer activity at outside variants, we cloned eight functional outside variant loci, and two control regions into luciferase reporter constructs and evaluated enhancer function of these regions in B lymphoblasts (Supplementary Fig. 4, Table S2). Seven of eight functional outside variant enhancer loci significantly enhanced luciferase activity compared to the two controls. We evaluated five of these eight loci for differential enhancer activity based on the outside variant genotype. Four showed a significant difference in luciferase activity (Fig. 3f,g). Altogether, the results suggest that outside variants functionally modify target transcript levels by altering enhancer activity in the B lymphoblast lineage.
Outside variants alter clinical risk to disease
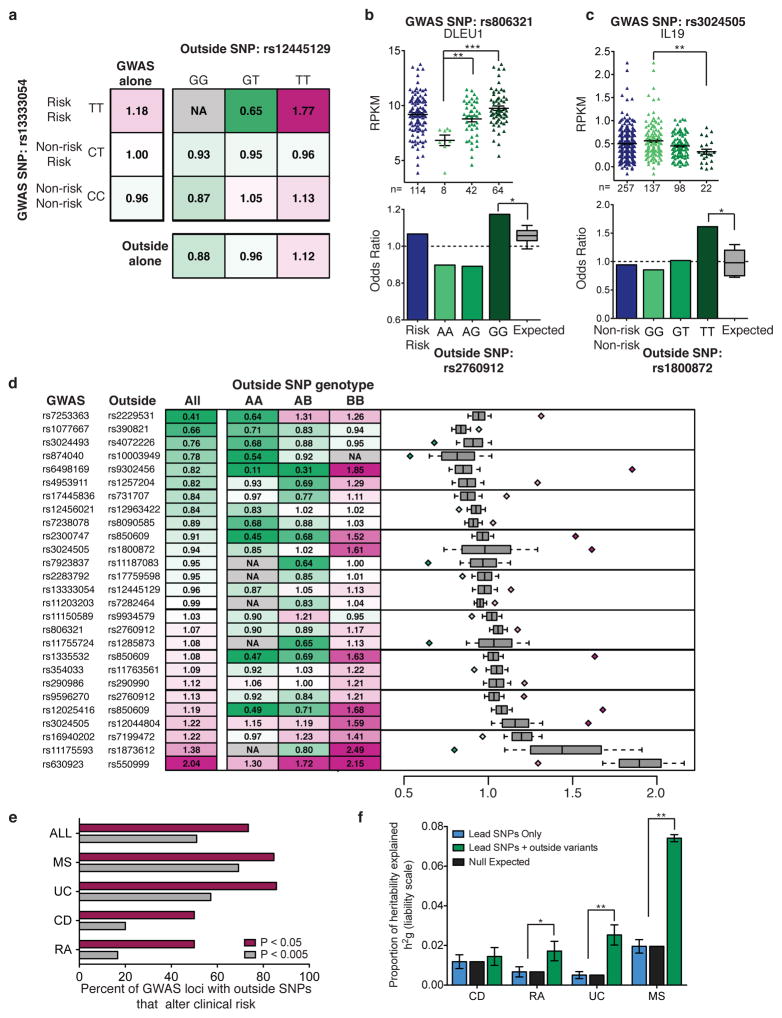
We set out to test if functional outside variants modify clinical disease risk. We utilized data generated by the Wellcome Trust Case Control Consortium45–48 to evaluate the impact of functional outside variants on clinical risk to multiple sclerosis, Crohn’s disease, ulcerative colitis and rheumatoid arthritis. We compared the clinical risk for each GWAS SNP and functional outside variant independently, to the clinical risk associated with each genotype combination. An example is shown in Figure 4a. We stratified the multiple sclerosis and control populations based on the genotype of the GWAS SNP, rs13333054, and found individuals homozygous for the risk SNP to have an odds ratio of 1.18 (leftmost column). We also stratified individuals solely by the genotype of the outside variant, rs12445129 and found an odds ratio of 1.12 for the TT genotype (bottom row). When we determined the odds ratio based on the genotype of both variants, we found an increase in clinical risk to 1.77 for individuals homozygous for the GWAS risk SNP and homozygous for the outside variant T-allele. Thus, the genotype of the outside variant alters the clinical risk associated with the locus.
Figure 4. Outside variants alter clinical risk.
(a) Example of odds ratio calculations for multiple sclerosis risk locus rs13333054. Odds ratios calculated considering only the lead GWAS SNP (leftmost column), only the outside variant (bottom row), and utilizing the genotype of both variants (square). (b) (Top) Impact of outside variant rs2760912 on DLEU1 transcript levels (wilcox-test, **P<0.002, ***<1E–5). (Bottom) Odds ratio for all individuals homozygous for the multiple sclerosis GWAS SNP rs806321 (blue) and odds ratios determined when homozygous individuals are stratified based on the outside variant genotype (green) compared to the expected distribution of odds ratios (median and quartiles (boxplot bars), 10–90th percentile (whiskers), *P<0.007). (c) Same as in (b) for ulcerative colitis GWAS locus rs3024505. (d) Odds ratios for all individuals with the same GWAS genotype compared to the odds ratios when individuals are stratified by the genotype of the outside SNP. (Right) Expected distribution of odds ratios (median and quartiles (boxplot bars), 10–90th percentile (whiskers)) compared to the most significant odds ratio from each row (diamonds, P<0.01). (e) Proportion of GWAS loci (n=49) for which an outside variant significantly alters clinical risk. MS = multiple sclerosis, UC = ulcerative colitis, CD = Crohn’s disease and RA = rheumatoid arthritis. (f) Narrow sense heritability (h2g ± standard error) explained by GWAS lead SNPs associated with functional outside variants (blue), the null expectation based on genomic coverage of outside variants (black) and h2g explained when lead SNPs are jointly modeled with functional outside variants (green) (two-sample z-test, *P<0.003, ** P<1E–30).
Outside variant rs2760912 was found to significantly alter the impact of multiple sclerosis GWAS SNP rs806321 on expression of lymphocytic leukemia associated RNA-gene DLEU1. Inheriting the outside variant G-allele was associated with increased transcript levels (Fig. 4b, top). Likewise, individuals homozygous for the risk SNP and outside variant G-allele had a notable increase in risk to disease (Fig. 4b, bottom). Outside variant rs1800872 was found to significantly alter the impact of ulcerative colitis GWAS SNP rs3024505 on IL19 target transcript levels. In this instance, decrease in expression of IL19 was correlated with an increase in clinical risk (Fig. 4c). To evaluate the significance of the impact of the outside variant on clinical risk, we performed permutation analysis whereby individuals of each GWAS genotype (risk/risk, risk/non-risk and non-risk/non-risk) were randomly assigned an outside variant genotype while maintaining outside variant allele frequency. Thus the contribution of the GWAS SNP to risk was preserved in order to evaluate the ability of the outside variant to alter clinical risk (methods, grey boxplots Fig. 4b–d). Utilizing this metric, outside variants rs12445129, rs2760912, and rs1800872 (Fig. 4a–c) were found to significantly alter clinical risk (P<0.01). We expanded these analyses to include all functional outside variants detected across all four traits. The impact of functional outside variants on clinical risk for each significant GWAS locus (P<0.01) is shown in Figure 4d. In total, 73.5% of the GWAS loci evaluated were associated with a functional outside variant that significantly altered the clinical risk associated with the locus (P<0.05, 55% at P<0.01) (Fig. 4e).
While the majority of functional outside variants were observed to alter clinical risk, outside variants were not previously associated with these disorders by conventional GWAS. To evaluate why, we determined their impact on risk independent of the GWAS genotype. For less than one-quarter of the GWAS loci associated with one or more functional outside variants, at least one outside variant reached genome-wide significant association with risk when evaluated independently. The majority of these variants were the result of imputation analysis (methods) and were therefore not evaluated by the previous GWA studies. Another possibility is that outside variants are just below genome-wide significance and would be associated with clinical risk in larger cohorts. 21.4% of GWAS loci were associated with a functional outside variant that reached intermediate association with risk independently (1E–3 ≥ P > 1E–8). Thus for many of these GWAS loci (57%), the impact of the functional outside variant on risk appears to be contingent on the genotype of the GWAS locus.
Outside variants may explain additional heritability
We sought to evaluate the overall impact of functional outside variants on disease heritability. The co-localization of disease susceptibility loci amongst autoimmune diseases49,50 suggests these disorders may involve disruption of common pathways. Thus, we utilized functional outside variant loci associated with risk to all six autoimmune disorders to estimate genetic relationship matrices and narrow-sense heritability (h2g) for each trait (methods). We compared the heritability explained by the GWAS lead SNPs of all functional outside variant loci to the heritability explained when GWAS lead SNPs and functional outside variants are jointly modeled. Functional outside variants increased the total heritability explained by 2.6-fold for rheumatoid arthritis (P<0.03), 5-fold for ulcerative colitis (P<1E–4) and 3.8-fold for multiple sclerosis (P<1E–30) (Fig. 4f). Functional outside variants also increase h2g (the fraction of phenotypic variance explained by the SNPs) significantly more than is expected based on the genomic coverage of functional outside variants. Gusev et al. previously demonstrated that inclusion of local variants increases the total heritability attributed to GWAS loci51. Functional outside variants explain significantly more heritability than local controls for both ulcerative colitis and multiple sclerosis (Supplementary Fig. 5, UC P<0.03, MS P<1E–30). Thus functional outside variants are a distinct set of local variants that can account for a substantial increase in total heritability explained.
Evaluation of “third variants” at outside variant loci
Our results suggest that multiple SNPs within the same regulatory circuit may cooperate to influence expression and clinical risk. Alternatively, a single variant that is partially linked to both the GWAS and outside variant may be responsible for the observed effects. For example, SNPs recently identified as interacting and in statistical epistasis with one another were subsequently shown to also be in low LD with a single, “third SNP”. The presence of the third SNP calls into question whether the two interacting SNPs actually drive the effect on expression, or if the effect is driven solely by the single “third” SNP that is in LD with each of the interacting SNPs. We systematically looked for evidence of third SNPs at all loci containing functional outside variants. We first curated a list of “candidate third SNPs” by selecting all known common SNPs within 500-kb of gene targets with functional outside variants. A total of 158,083 SNPs were identified, averaging 4,863 SNPs per gene (Supplementary Table 3). At every locus, we identified a third variant that at least nominally correlated with expression. However, the third SNP was often insufficient to fully account for the effect of the outside variant. For example, after segregating individuals with the same third variant genotype, the outside variant often accounted for additional variation in gene expression (Supplementary Fig 6–7). We further tested whether any of the third variants could account for the effects on both gene expression and clinical risk. We found that the third SNP accounted for effects on both gene expression and clinical risk for ~13% (7/53) of genes evaluated. These loci were associated with risk to disease at a genome-wide P value threshold of <10−8. Because imputation can sometimes result in underestimation of effect sizes52,53, we also performed the analysis at less stringent thresholds. At an uncorrected P value of < 0.001, for 57% of outside variant gene targets, we did not identify a common SNP that could account for the effects of outside variants on both expression and risk. We also determined that the majority of third SNPs were not contained within open chromatin, nor did they overlap with either of the two canonical enhancer histone marks, H3K4me1 and H3K27ac (Supplementary Fig. 8). Based on this analysis, outside variants appear to account for both clinical risk and gene expression more often than any single third variant alone. We note however, that our analysis does not consider potential third variants that could be located >500-kb from the gene target, are poorly represented by the GWAS panel, or may have low minor allele frequencies.
Discussion
Some GWAS loci harbor a single causal variant that lies in an enhancer and influences spatiotemporal expression of the target gene54–57. However other GWAS loci, particularly those with enhancer clusters or “super enhancers”, contain multiple functional enhancer variants in LD that collude to impact target gene expression5,12–16,18,58,59. Here we demonstrate that the individual constituents of enhancer clusters physically interact and are rarely in LD, prompting the hypothesis that LD is perhaps not the best way to identify variants that explain disease heritability. We tested this hypothesis by integrating autoimmune-associated GWAS SNPs with epigenomic maps of regulatory elements, Hi-C chromatin interaction maps, and transcriptomic datasets. We identified numerous functional “outside variants” in weak LD with GWAS loci but lie within constituent enhancers of shared target genes and influence both target gene expression and the clinical risk to disease. The outside variants may as much as triple the total heritability explained, although one limitation is that these estimates are based on the widely used additive model of heritability and therefore may not account for the contribution of epistatic effects. Our findings emphasize the importance of chromatin state and a gene’s regulatory circuitry as a key determinant of heritable disease risk. Based on these findings, it is tempting to speculate that outside variants explain missing heritability for other GWAS traits besides the autoimmune disorders studied here, and that new disease-risk associations can be revealed by studying SNPs that interact in 3D chromatin to regulate gene expression.
Online Methods
Definition of chromatin regulatory circuits
We utilized high-depth GM12878 Hi-C datasets (1.2 billion paired-end reads60) to define Hi-C chromatin interactions. Both sequences from paired-end reads were aligned to hg18 independently using bowtie261. Hi-C analysis package available through Homer62 was utilized to define significant interactions between genomic loci at 10-kb resolution (P < 2.5E–5). H3K4me1-enriched loci were called using BWA63 and MACS64. We identified Hi-C significant interactions for which there was a transcription start site in one locus that was paired to an H3K4me1 ChIP peak in the other. These pairs were utilized to define the list of H3K4me1 putative enhancer sites that were associated with the same gene target, i.e. define the chromatin regulatory circuitry of each locus. Computational prediction of enhancer-gene interactions was also utilized to define chromatin regulatory circuitry. The PreSTIGE (Predicting Specific Tissue Interactions of Genes and Enhancers) algorithm was used to predict enhancer-gene interactions from GM12878 H3K4me1 ChIP-seq and RNA-seq data as previously described5. Briefly, PreSTIGE utilizes a comparative analysis across multiple tissues types to identify enhancers and gene with concordant cell type-specificity within a defined linear domain. Super enhancers previously defined for GM128788,9 were also utilized to define chromatin regulatory circuits. All genes within 100-kb of super enhancers were evaluated in transcriptional analysis.
Definition of outside variants for transcriptional analysis
GWAS variants associated with multiple sclerosis, celiac disease, Crohn’s disease, ulcerative colitis, systemic lupus and rheumatoid arthritis in European populations were downloaded from NHGRI’s catalog of GWAS variants65. SNPs in tight LD (LOD > 2 and D’ > 0.6) with GWAS SNPs were defined as linked variants. All GWAS SNPs in tight LD with variants found in protein-coding regions were excluded from all subsequent analyses. Noncoding linked variants were compared to chromatin regulatory circuits (defined above) to identify potential gene targets. All common variants within putative enhancers associated with these gene targets were identified. Variants in tight LD with GWAS SNPs were removed (LOD > 2 and D’ > 0.6) to create a list of candidate outside variants. This list was further pruned by removing all candidate variants that were in tight LD (LOD > 2 and D’ > 0.6) with a third variant that was in tight LD (LOD > 2 and D’ > 0.6) with a GWAS SNP. Thus there was no overlap between the variants in tight LD with the GWAS risk SNPs and those in tight LD with the putative outside variants (diagram in Supplementary Figure 2a). The resultant r2, D’ and LOD scores for GWAS and outside variant pairs are described in Supplementary Fig. 2c–f. Finally, only alleles (GWAS SNP + linked variants + outside variant genotypes) present in >1% of the gene expression panel (373 individuals) were utilized (Supplementary Fig. 2) in order to ensure sufficient power.
Impact of outside variants on target transcript levels
We obtained publicly available genotypes and RNA-seq from B lymphoblasts of 373 European individuals34. The reported PEER normalized expression was utilized to control for technical variance34. We first stratified this panel by the genotype of the GWAS haplotype (lead SNPs + all linked variants) and then divided each GWAS genotype subgroup by the genotype of the outside variant. Transcript levels of gene targets defined by regulatory circuitry analysis were compared (Wilcox-test) to determine the impact of the outside variant genotype on expression for each GWAS genotype (risk/risk, risk/non-risk, non-risk/non-risk). The P-values generated were pruned so that outside variants that stratified individuals into the same groups were only represented once in the construction of QQ-plots.
Definition of functional outside variants
Permutation analysis was utilized in order to define outside variants that significantly impact the effect of GWAS alleles on target transcript levels. Permutations randomly associated an individual’s genotypes to a different individual’s RNA-seq profile. Thus, the linkage disequilibrium and allele frequencies were maintained, but association with gene expression was randomized (5,000 permutations). Multiple test correction was performed with two methodologies. The number of significant tests for each permutation was compared to the number of significant tests in the non-randomized data in order to define the false discovery rate (FDR) for each p-value threshold. Alternatively, the lowest p-value generated for each GWAS haplotype (risk/risk, non-risk/risk and non-risk/non-risk) was identified for all GWAS loci. The lowest p-values from each of the 5,000 permutations were utilized to generate an expected distribution for each locus and haplotype. P-values below the 1st percentile of the expected distribution for the locus were defined as significant (referred to as permutation P<0.01). ‘Functional outside variants’ include variants that were determined as significant by the FDR methodology (q < 0.10) or permutation methodology (P<0.01).
Chromatin state of outside and linked variants
H3K4me1 and H3K27ac B lymphoblast ChIP-seq and B lymphoblast DNase Hypersensitive data were aligned to hg18 using BWA63. RPKMs (reads per kilobase per million mapped read) were calculated for the 1-kb region surrounding functional outside variants and GWAS linked variants and results were quantile-normalized across individuals for each mark.
Luciferase reporter assay
GM11993 and GM12005 B lymphoblast cell lines, mycoplasma-negative, were obtained from Coriell Institute Biorepository. Eight functional outside variant loci (~1–2 kb, Supplementary Table 2) were cloned from B lymphoblast cell lines that were heterozygous for the outside variant allele of interest into a luciferase reporter construct (pGL4 from Promega) for which the luciferase gene was driven by the ubiquitous mSox9 promoter. Sanger sequencing was utilized to identify the genotype of the outside variant allele in each clone (Supplementary Fig. 4). Two control loci, ~1–2 kb regions with no expected enhancer activity in B-lymphoblasts, were also cloned into the same construct to generate size-matched constructs to control for basal promoter activity. Reporter constructs were transfected into B lymphoblast cell line GM12005 using the transfection reagent DMRIE-C (Life Technologies). As an internal control, a renilla luciferase plasmid (pRL-SV40 from Promega) was co-transfected. After five hours, transfection reagents were replaced with fresh media. 24-hours post transfection, cells were harvested and luciferase reporter levels were compared to renilla reporter activity using Dual-Luciferase Reporter Assay System (Promega).
Odds ratio analysis
Primary GWAS data was obtained from Wellcome trust case control consortium for multiple sclerosis (9,772 cases, 2,679 controls), Crohn’s disease (1,753 cases, 1,461 controls), ulcerative colitis (2,366 cases, 2,679 controls) and rheumatoid arthritis (1,865 cases, 1,461 controls). Quality control and filtering of SNPs and individuals was performed as previously described45–48. Imputation analysis was performed for all functional outside variant loci associated with these disorders using IMPUTE266 and an integrated reference panel from 1000 Genomes (Phase 1)67. Imputation output was filtered to include only genotypes with a probability greater than 0.90, while the remaining two genotypes had probabilities less than 0.3. Odds ratios were calculated for individuals who were stratified by the lead SNP genotype, outside variant genotype or by the genotype of both variants. To determine which outside variants significantly altered clinical risk, permutation analysis was utilized. Permutations were performed such that individuals (cases and controls) of each GWAS genotype (risk/risk, risk/non-risk and non-risk/non-risk) were randomly assigned an outside variant genotype while maintaining the allele frequency of the outside variant. The distributions of the resulting odds ratios were then utilized to define a p-value for each odds ratio.
Narrow-sense heritability
GWAS lead SNPs associated with functional outside variants for all six autoimmune traits were utilized to determine genetic relationship matrices (GRM) utilizing GCTA. GCTA restricted maximum likelihood analysis68,69 was then utilized to determine the proportion of phenotypic variance explained by each SNP subset. h2g estimates are reported on a liability-scale that estimates European disease prevalence of 0.25% for Crohn’s Disease, 0.5% for rheumatoid arthritis, 0.28% for ulcerative colitis and 0.13% for multiple sclerosis. We performed two sample z-tests to compare h2g estimates from jointly modeling GWAS lead SNPs only to h2g estimates from jointly modeling GWAS lead SNPs and functional outside variants. We also calculated the null expected h2g based on the fraction of the genome represented by the inclusion of outside variants in the heritability estimates. As previously described51, h2null = h2lead SNPs + x*(total h2g − h2lead SNPs), where x is the fraction of the genome covered by the outside variants. Z-tests were also performed to compare h2g estimates from jointly modeling GWAS lead SNPs and functional outside variants to the null expected heritability estimates.
To investigate whether the increase in heritability was specific to outside variants, we compared outside variants to “local controls.” Local control variants were defined for each GWAS locus by three requirements. Control variants (1) were within 200-kb of GWAS lead SNP (2) did not lie within the regulatory circuitry defined using any of the three methods (see above) and (3) had r2 < 0.3 with all variants in tight LD (LOD>2, D’>0.6) with GWAS SNPs or outside variants. Given the proximity of these controls, many are in tight LD with one another. To compare these controls to outside variants we employed LD pruning. From the list of potential control SNPs, we removed the SNP with the most LD partners (r2>0.3) one at a time, until no SNP pairs with r2 > 0.3 remained. We pruned the outside variant list by the same method. We selected 1,000 random subsets of controls, such that the number of controls per locus was proportionate to the number of functional outside variants for that locus. We compared the heritability distribution generated from 1,000 randoms and found the both UC and MS outside variants explain significantly more heritability than the local controls (UC: P=0.004 and MS: P<0.001).
Analysis of “third variant” hypothesis
We identified all known common SNPs within 500-kb of gene targets with functional outside variants. Individuals were stratified based on the genotype of “third” SNPs and expression levels were compared by Mann Whitney Wilcoxon test. All genotypes present in >1% of the 373-individual panel were assessed. A total of 158,083 SNPs were evaluated, averaging 4,863 SNPs per gene (Supplementary Table 3). In Supplementary Figures 6 and 7, we present three different p-value thresholds for evaluating the impact of the third variant on expression. These threshold include (1) multi-test correction (MTC) for the total number of Mann Whitney Wilcoxon tests performed for the analysis (P-value <7.5E–8) (2) multi-test correction for the number of tests performed for the given gene (P-value threshold varies per gene, corrected P<0.05) (3) multi-test correction for 10 tests (P-value < 5E–3) utilized to demonstrate loci with third variants that have modest effects.
We next asked whether the third variant is sufficient to explain the observed effect of the outside variants and GWAS allele. In order to evaluate this, we applied our two-tiered approach and stratified first by the genotype of each third variant. We then asked, given the effect of the third variant, can the outside variant or GWAS allele explain additional variation in gene expression? (diagram Supplementary Fig. 6). If the third SNP is sufficient to explain the observed effect, further stratification of individuals with the same third SNP genotype would not distinguish cohorts with significantly different transcript levels. We applied this approach to all third SNPs that achieved each of the three thresholds of significance. The number of genes for which the outside variant or GWAS allele could explain more variance for every significant third SNP were counted (i.e. if the outside variant or GWAS allele could not explain additional variance for all third SNPs, then that locus was considered to be potentially explained by the third SNP and this SNP carried through to the evaluation on clinical risk).
We next assessed whether the remaining third SNPs were associated with clinical risk. To test this, we took all third SNPs that correlated with expression (at the three significance thresholds) where the outside variant or GWAS allele could not account for additional variation and quantified their effect on clinical risk. For this analysis we evaluated loci associated with risk to four traits, multiple sclerosis, ulcerative colitis, rheumatoid arthritis and Crohn’s disease. Approximately, two-thirds of the potential third SNPs were imputed (methods) in the respective study. For the two most stringent p-value thresholds, at least one third SNP was represented on the appropriate GWAS panel for each gene. For one outside variant locus associated with rheumatoid arthritis (gene target FLVCR2), three third SNPs that had nominal association with an effect on expression (P<5E–3, uncorrected) were detected. None of these third SNPs were successfully imputed in the RA GWAS panel. This gene was excluded from the analysis (Supplementary Figure 6 and 7, bottom row).
We also compared third variants to DNase Hypersensitivity, H3K4me1 and H3K27ac profiles for B-lymphoblasts (GM12878). We evaluated all third variants that have the potential to explain the effect of outside variants on expression (see Supplementary Fig 6 and 7, third arrow) and quantified the proportion that overlapped with regions significantly enriched for each marker (called peaks) of active chromatin (Supplementary Fig. 8).
Supplementary Material
Acknowledgments
We thank Paul Tesar, Tom LaFramboise, Angela Lager and James Morrow for helpful comments and discussion. This research was supported by NIH grants R01CA160356, R01CA143237, 1R01CA193677, and P50CA150964 (P.C.S.) and T32GM008056 (O.C.).
Footnotes
Author Contributions
O.C. and P.C.S. designed the study, performed the analyses and interpreted the results. A.J.C. and F.R.S. aided statistical analyses and discussion. O.C., J.M.L., and I.M.B. performed cell culture and luciferase experiments.
References
- 1.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–9. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trynka G, et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet. 2013;45:124–30. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akhtar-Zaidi B, et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science. 2012;336:736–9. doi: 10.1126/science.1217277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corradin O, et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farh KK, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2014 doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gusev A, et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet. 2014;95:535–52. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parker SC, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110:17921–6. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasquali L, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet. 2014;46:136–43. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biancolella M, et al. Identification and characterization of functional risk variants for colorectal cancer mapping to chromosome 11q23.1. Hum Mol Genet. 2014;23:2198–209. doi: 10.1093/hmg/ddt584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fortini BK, et al. Multiple functional risk variants in a SMAD7 enhancer implicate a colorectal cancer risk haplotype. PLoS One. 2014;9:e111914. doi: 10.1371/journal.pone.0111914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glubb DM, et al. Fine-Scale Mapping of the 5q11.2 Breast Cancer Locus Reveals at Least Three Independent Risk Variants Regulating MAP3K1. Am J Hum Genet. 2015;96:5–20. doi: 10.1016/j.ajhg.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo C, et al. Coordinated regulatory variation associated with gestational hyperglycaemia regulates expression of the novel hexokinase HKDC1. Nat Commun. 2015;6:6069. doi: 10.1038/ncomms7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He H, et al. Multiple functional variants in long-range enhancer elements contribute to the risk of SNP rs965513 in thyroid cancer. PNAS. 2015;112:6128–6133. doi: 10.1073/pnas.1506255112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roman TS, et al. Multiple Hepatic Regulatory Variants at the GALNT2 GWAS Locus Associated with High-Density Lipoprotein Cholesterol. Am J Hum Genet. 2015;97:801–15. doi: 10.1016/j.ajhg.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaw AD, et al. Characterisation of genetic variation in ST8SIA2 and its interaction region in NCAM1 in patients with bipolar disorder. PLoS One. 2014;9:e92556. doi: 10.1371/journal.pone.0092556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 20.Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet. 2013;93:779–97. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dimas AS, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–50. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright FA, et al. Heritability and genomics of gene expression in peripheral blood. Nat Genet. 2014;46:430–7. doi: 10.1038/ng.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bryois J, et al. Cis and trans effects of human genomic variants on gene expression. PLoS Genet. 2014;10:e1004461. doi: 10.1371/journal.pgen.1004461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ing-Simmons E, et al. Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Res. 2015;25:504–13. doi: 10.1101/gr.184986.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dowen JM, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–87. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam DD, et al. Partially redundant enhancers cooperatively maintain Mammalian pomc expression above a critical functional threshold. PLoS Genet. 2015;11:e1004935. doi: 10.1371/journal.pgen.1004935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bothma JP, et al. Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. Elife. 2015;4 doi: 10.7554/eLife.07956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry MW, Boettiger AN, Levine M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc Natl Acad Sci U S A. 2011;108:13570–5. doi: 10.1073/pnas.1109873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiersma EJ, Ronai D, Berru M, Tsui FW, Shulman MJ. Role of the intronic elements in the endogenous immunoglobulin heavy chain locus. Either the matrix attachment regions or the core enhancer is sufficient to maintain expression. J Biol Chem. 1999;274:4858–62. doi: 10.1074/jbc.274.8.4858. [DOI] [PubMed] [Google Scholar]
- 31.Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development. 2006;133:761–72. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- 32.Montavon T, et al. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–45. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 33.Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20:1562–7. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–11. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mocsai A, Ruland J, Tybulewicz VL. The SYK tyrosine kinase: a crucial player in diverse biological functions. Nat Rev Immunol. 2010;10:387–402. doi: 10.1038/nri2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stranger BE, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kasowski M, et al. Extensive variation in chromatin states across humans. Science. 2013;342:750–2. doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kilpinen H, et al. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science. 2013;342:744–7. doi: 10.1126/science.1242463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–4. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puig-Kroger A, Corbi A. RUNX3: a new player in myeloid gene expression and immune response. J Cell Biochem. 2006;98:744–56. doi: 10.1002/jcb.20813. [DOI] [PubMed] [Google Scholar]
- 43.Busslinger M. Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- 44.Carotta S, Wu L, Nutt S. Surprising new roles for PU.1 in the adaptive immune response. Immunological Reviews. 2010;238:63–75. doi: 10.1111/j.1600-065X.2010.00955.x. [DOI] [PubMed] [Google Scholar]
- 45.Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barrett JC, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–4. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The Wellcome Trust Case Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plenge RM, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39:1477–82. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cotsapas C, et al. Pervasive Sharing of Genetic Effects in Autoimmune Disease. PloS Genetics. 7 doi: 10.1371/journal.pgen.1002254. (20011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fortune MD, et al. Statistical colocalization of genetic risk variants for related autoimmune diseases in the context of common controls. Nat Genet. 2015;47:839–46. doi: 10.1038/ng.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gusev A, et al. Quantifying missing heritability at known GWAS loci. PLoS Genet. 2013;9:e1003993. doi: 10.1371/journal.pgen.1003993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zaitlen N, Eskin E. Imputation aware meta-analysis of genome-wide association studies. Genet Epidemiol. 2010;34:537–42. doi: 10.1002/gepi.20507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 54.Guenther CA, Tasic B, Luo L, Bedell MA, Kingsley DM. A molecular basis for classic blond hair color in Europeans. Nat Genet. 2014;46:748–52. doi: 10.1038/ng.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cowper-Sal·lari R, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet. 2012;44:1191–8. doi: 10.1038/ng.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alcina A, et al. Identification of a functional variant in the KIF5A-CYP27B1-METTL1-FAM119B locus associated with multiple sclerosis. Journal of Medical Genetics. 2012;50:25–33. doi: 10.1136/jmedgenet-2012-101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaulton KJ, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–9. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spieler D, et al. Restless legs syndrome-associated intronic common variant in Meis1 alters enhancer function in the developing telencephalon. Genome Res. 2014;24:592–603. doi: 10.1101/gr.166751.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stadhouders R, et al. HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J Clin Invest. 2014;124:1699–710. doi: 10.1172/JCI71520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Selvaraj S, JRD, Bansal V, Ren B. Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nat Biotechnol. 2013;31:1111–8. doi: 10.1038/nbt.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hindorff LA, MJ, Morales J, Junkins HA, Hall PN, Klemm AK, Manolio TA. A Catalog of Published Genome-Wide Association Studies. [Accessed: Jan. 10th, 2014]. [Google Scholar]
- 66.Howie BN, Donnelly P, Marchini J. A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. PloS Genetics. 2009;5 doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Howie BN, Marchini J, Stephens J. Genotype Imputation with Thousands of Genomes. G3. 2011;1:457–469. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–9. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.