Abstract
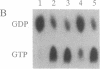
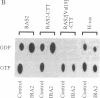
The ras GTPase-activating protein (GAP), identified and characterized in mammalian cells, stimulates the intrinsic GTPase activity of ras proteins. We have previously proposed that the IRA genes, negative regulators of RAS genes in Saccharomyces cerevisiae, encode yeast homologs of the mammalian GAP. In this paper, we present the following evidence that a product of the IRA2 gene exhibits GAP activity similar to that of the mammalian GAP protein. (i) Extracts of yeast cells overexpressing IRA2 stimulated the GTPase activity of the yeast RAS2 protein. (ii) An epitope for a monoclonal antibody (12CA5) was added to the N terminus of the IRA2 protein. The GAP activity of extracts prepared from cells expressing this fusion protein was shown to be immunoprecipitable by 12CA5. (iii) An IRA2 protein fused to glutathione S-transferase (GST) was produced and partially purified from Escherichia coli cells. GAP activity was detected with this purified GST-IRA2 fusion protein. (iv) The GAP activity of IRA2 proteins described above did not stimulate the GTPase activity of the RAS2Val19 protein, a protein having an amino acid alteration analogous to that found in mammalian oncogenic ras proteins. This result parallels studies showing that mammalian GAP is incapable of stimulating the GTPase activity of mammalian oncogenic proteins. The remarkable conservation between the GAP activity in mammalian and yeast cells supports the idea that the function of GAP is to negatively regulate ras proteins in mammalian cells.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballester R., Michaeli T., Ferguson K., Xu H. P., McCormick F., Wigler M. Genetic analysis of mammalian GAP expressed in yeast. Cell. 1989 Nov 17;59(4):681–686. doi: 10.1016/0092-8674(89)90014-7. [DOI] [PubMed] [Google Scholar]
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Field J., Nikawa J., Broek D., MacDonald B., Rodgers L., Wilson I. A., Lerner R. A., Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988 May;8(5):2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegold A. A., Schafer W. R., Rine J., Whiteway M., Tamanoi F. Common modifications of trimeric G proteins and ras protein: involvement of polyisoprenylation. Science. 1990 Jul 13;249(4965):165–169. doi: 10.1126/science.1695391. [DOI] [PubMed] [Google Scholar]
- Gibbs J. B., Schaber M. D., Allard W. J., Sigal I. S., Scolnick E. M. Purification of ras GTPase activating protein from bovine brain. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5026–5030. doi: 10.1073/pnas.85.14.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. ras and GAP--who's controlling whom? Cell. 1990 Jun 15;61(6):921–923. doi: 10.1016/0092-8674(90)90054-i. [DOI] [PubMed] [Google Scholar]
- Hata Y., Kikuchi A., Sasaki T., Schaber M. D., Gibbs J. B., Takai Y. Inhibition of the ras p21 GTPase-activating protein-stimulated GTPase activity of c-Ha-ras p21 by smg p21 having the same putative effector domain as ras p21s. J Biol Chem. 1990 May 5;265(13):7104–7107. [PubMed] [Google Scholar]
- Kataoka T., Powers S., McGill C., Fasano O., Strathern J., Broach J., Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984 Jun;37(2):437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- Marshall M. S., Gibbs J. B., Scolnick E. M., Sigal I. S. An adenylate cyclase from Saccharomyces cerevisiae that is stimulated by RAS proteins with effector mutations. Mol Cell Biol. 1988 Jan;8(1):52–61. doi: 10.1128/mcb.8.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall M. S., Hill W. S., Ng A. S., Vogel U. S., Schaber M. D., Scolnick E. M., Dixon R. A., Sigal I. S., Gibbs J. B. A C-terminal domain of GAP is sufficient to stimulate ras p21 GTPase activity. EMBO J. 1989 Apr;8(4):1105–1110. doi: 10.1002/j.1460-2075.1989.tb03480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick F. ras GTPase activating protein: signal transmitter and signal terminator. Cell. 1989 Jan 13;56(1):5–8. doi: 10.1016/0092-8674(89)90976-8. [DOI] [PubMed] [Google Scholar]
- Schaber M. D., Garsky V. M., Boylan D., Hill W. S., Scolnick E. M., Marshall M. S., Sigal I. S., Gibbs J. B. Ras interaction with the GTPase-activating protein (GAP). Proteins. 1989;6(3):306–315. doi: 10.1002/prot.340060313. [DOI] [PubMed] [Google Scholar]
- Shirakawa M., Tsurimoto T., Matsubara K. Plasmid vectors designed for high-efficiency expression controlled by the portable recA promoter-operator of Escherichia coli. Gene. 1984 Apr;28(1):127–132. doi: 10.1016/0378-1119(84)90096-9. [DOI] [PubMed] [Google Scholar]
- Sigal I. S., Gibbs J. B., D'Alonzo J. S., Scolnick E. M. Identification of effector residues and a neutralizing epitope of Ha-ras-encoded p21. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4725–4729. doi: 10.1073/pnas.83.13.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Matsumoto K., Toh-E A. IRA1, an inhibitory regulator of the RAS-cyclic AMP pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):757–768. doi: 10.1128/mcb.9.2.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Satoh T., Marshall M. S., Gibbs J. B., Matsumoto K., Kaziro Y., Toh-e A. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 1990 Mar 9;60(5):803–807. doi: 10.1016/0092-8674(90)90094-u. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Tamanoi F., Kaziro Y., Matsumoto K., Toh-e A. IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol Cell Biol. 1990 Aug;10(8):4303–4313. doi: 10.1128/mcb.10.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trahey M., McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987 Oct 23;238(4826):542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- Trahey M., Wong G., Halenbeck R., Rubinfeld B., Martin G. A., Ladner M., Long C. M., Crosier W. J., Watt K., Koths K. Molecular cloning of two types of GAP complementary DNA from human placenta. Science. 1988 Dec 23;242(4886):1697–1700. doi: 10.1126/science.3201259. [DOI] [PubMed] [Google Scholar]
- Vogel U. S., Dixon R. A., Schaber M. D., Diehl R. E., Marshall M. S., Scolnick E. M., Sigal I. S., Gibbs J. B. Cloning of bovine GAP and its interaction with oncogenic ras p21. Nature. 1988 Sep 1;335(6185):90–93. doi: 10.1038/335090a0. [DOI] [PubMed] [Google Scholar]
- Xu G. F., Lin B., Tanaka K., Dunn D., Wood D., Gesteland R., White R., Weiss R., Tamanoi F. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell. 1990 Nov 16;63(4):835–841. doi: 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]
- Xu G. F., O'Connell P., Viskochil D., Cawthon R., Robertson M., Culver M., Dunn D., Stevens J., Gesteland R., White R. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990 Aug 10;62(3):599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- de Vos A. M., Tong L., Milburn M. V., Matias P. M., Jancarik J., Noguchi S., Nishimura S., Miura K., Ohtsuka E., Kim S. H. Three-dimensional structure of an oncogene protein: catalytic domain of human c-H-ras p21. Science. 1988 Feb 19;239(4842):888–893. doi: 10.1126/science.2448879. [DOI] [PubMed] [Google Scholar]