Abstract
Human galectin‐1 is a member of the galectin family, proteins with conserved carbohydrate‐recognition domains that bind galactoside. Galectin‐1 is highly expressed in various tumors and participates in various oncogenic processes. However, detailed descriptions of the function of galectin‐1 in urinary bladder urothelial carcinoma have not been reported. Our previous cohort investigation showed that galectin‐1 is associated with tumor invasiveness and is a possible independent prognostic marker of urinary bladder urothelial carcinoma. The present study aimed to clarify the relevance of galectin‐1 expression level to tumor progression and invasion. In order to decipher a mechanism for the contribution of galectin‐1 to the malignant behavior of urinary bladder urothelial carcinoma, two bladder cancer cell lines (T24 and J82) were established with knockdown of galectin‐1 expression by shRNA. Bladder cancer cells with LGALS1 gene silencing showed reduced cell proliferation, lower invasive capability, and lower clonogenicity. Extensive signaling pathway studies indicated that galectin‐1 participated in bladder cancer cell invasion by mediating the activity of MMP9 through the Ras–Rac1–MEKK4–JNK–AP1 signaling pathway. Our functional analyses of galectin‐1 in urinary bladder urothelial carcinoma provided novel insights into the critical role of galectin‐1 in tumor progression and invasion. These results revealed that silencing the galectin‐1‐mediated MAPK signaling pathway presented a novel strategy for bladder cancer therapy.
Keywords: Galectin‐1, invasion, JNK, shRNA, urothelial bladder urinary carcinoma
Urothelial carcinoma is the most common lesion of various heterogeneous tumor types that arise from the urothelium of the urinary bladder and ureter. Urinary bladder urothelial carcinoma (UBUC) is usually diagnosed as papillary/non‐invasive or superficially invasive lesions. Some UBUCs can develop relentless local recurrence followed by lethal distal spreading.1
Cystoscopy/urine cytology provides high specificity as well as acceptable sensitivity for the detection of high‐grade UBUCs but lacks sensitivity in low‐grade lesions.2 Furthermore, a subset of patients with tumor metastasis might not demonstrate detectable urinary bladder cancer recurrence, especially those who received radical cystectomy. Therefore, identification of prognostic biomarkers by uncovering the molecular carcinogenesis of UBUC may offer useful insights that aid in the exploration of better diagnosis/prognosis and novel targeted therapeutic strategies.
Human galectin‐1 (Gal‐1) is a member of the galectin family, proteins with conserved carbohydrate‐recognition domains that bind galactoside.3 Galectin‐1 is highly expressed in various tumors, including colon,4 breast,5 lung,6 head and neck,7 ovarian,8 and prostate carcinoma9 as well as in glioma,10 Kaposi's sarcoma,11 and Hodgkin's lymphoma.12 Intriguingly, in most lesions, Gal‐1 expression is associated with aggressiveness and metastasis of tumor. It participates in various oncogenic processes including cell transformation,13 metastasis,13, 14 cell proliferation,14, 15, 16, 17, 18 and cell migration.4 Galectin‐1 has also been shown to play an essential role in tumor angiogenesis.19 Immunohistochemical examination of Gal‐1 expression in human U87 and U373 glioblastoma xenografts from the brains of immunodeficient mice has shown a higher galectin‐1 expression level in invasive areas of the xenografts compared with non‐invasive areas.20 Paz et al.. reported that Gal‐1 can combine with H‐Ras and give rise to the membrane anchorage of H‐Ras. Galectin‐1 overexpression in tumor cell augments the membrane linkage of H‐Ras and cell transformation.21 Rubinstein et al.22 reported that melanoma cells can secret Gal‐1 protein to escape from T cell‐dependent immunity by inducing the apoptosis of activated T cells, thus conferring the immune privilege to tumor cells.
Our previous documented cohort study showed that the LGALS1 gene is amplified in UBUC specimens and Gal‐1 overexpression is significantly linked to UBUC invasion. Galectin‐1 overexpression in tumor significantly predicted disease‐specific survival at the univariate and multivariate levels.23 However, the molecular pathway underlying UBUC carcinogenesis in which Gal‐1 participates has not been uncovered. In the present study, we established a stable UBUC cell line with Gal‐1 expression knocked down by shRNA to decipher the role of Gal‐1 in UBUC invasion. We found that reduced Gal‐1 expression could interfere with the invasive ability of UBUC cells by Gal‐1‐evoked JNK signaling pathway.
Materials and Methods
Cell lines
Human urinary bladder urothelial carcinoma T24 cell, which is recognized as high grade and invasive, was purchased in 2010 from the Bioresource Collection and Research Center (Hsinchu, Taiwan) and cultured at 37°C in McCoy's 5A (Gibco, Life Technologies, Grand Island, NY, USA), supplemented with 10% FBS (Gibco). The J82 cell line was provided by Dr. Chien‐Feng Li (Department of Pathology, Chi‐Mei Medical Center (Tainan, Taiwan) and maintained at 37°C in DMEM (Gibco) with 10% (v/v) FBS. The TSGH8301 cell line (low grade) was derived from patients with superficial bladder cancer in Taiwan24 and also provided by Dr. Chien‐Feng Li. TSGH8301 cells were cultivated at 37°C in RPMI‐1640 (Gibco)/10% (v/v) FBS.
Establishment of Gal‐1 knockdown T24 and J82 cells with shRNA
DNA precursor oligonucleotide of siRNA was designed using Oligoengine software (Oligoengine, Seattle, WA, USA), which would be transcribed into shRNA processed into 19‐mer siRNA. Oligonucleotide DNA sequences are listed in Appendix S1. Custom DNA oligonucleotides were annealed and the resulting DNA duplex was cloned between BglII and HindIII restriction enzyme sites of the expression vector pSUPER‐NEO (Oligoengine) containing the neomycin resistance gene as the selection marker, as suggested by the manufacturer. T24 cells were transfected with the siRNA expression plasmid using PolyJet transfection reagent (SignaGen Laboratories, Rockville, MD, USA) according to the manufacturer's protocol. Stably transfected resistant cells were selected in medium containing 150 μg/mL G418 antibiotic. T24 cell clones with best targeting efficiency were chosen using Western immunoblotting and real‐time RT‐PCR from 20 cell clones with knockdown at +120 nt and 20 cell clones at +380 nt and named sh‐Gal‐1(+120) and sh‐Gal‐1(+380), respectively. Control cells were established by stable transfection of T24 cells with scrambled siRNA DNA precursor. T24 cell clones with the least effect on Gal‐1 expression were picked up from 20 cell clones scrambled at +120 nt and 20 clones at +380 nt and recognized as sh‐Sc(+120) and sh‐Sc(+380), respectively. J82 cells with Gal‐1 knockdown by siRNA DNA precursor were constructed similarly as described above.
Cell proliferation and cell counting assays
A 96‐well plate was seeded with 5000 sh‐Sc or sh‐Gal‐1 T24 cells per well. After cultivation for the indicated time period, 20 μL MTT solution (Merck, Darmstadt, Germany) (5 mg/mL PBS) was added to each well and the plate was incubated at 37°C for 4 h. After medium removal, 200 μL DMSO was applied to each well and the plate was gently shaken for 5 min. The absorbance was measured at 540 nm. Quadruplicate wells were used for a specific time period. Six thousand J82 cells were seeded per well for the MTT assay.
A 6‐well plate was seeded with 8 × 104 sh‐Sc or shGal‐1 T24 cells per well. After cultivation for the indicated time period, cells were counted with a Beckman Coulter Z1 Counter (Beckman Coulter, Indianapolis, IN, USA). Three wells were assessed for a specific time period.
Soft agar assay
Human sh‐Sc or shGal‐1 T24 cells (1 × 105) were resuspended in 1.0 mL DMEM with 20% (v/v) FBS and 0.33% (w/v) agar and plated over a layer of solidified DMEM medium/20% (v/v) FBS/0.66% (w/v) agar (2.0 mL per well) in a 6‐well plate. Plates are incubated at 37°C until clone formation, and the resultant clones were stained with 0.005% (w/v) crystal violet and scored in a blinded fashion.
Wound scratch assay
Human sh‐Sc or sh‐Gal‐1 T24 cells (2.1 × 104) were plated onto each well of a 6‐well plate with a culture insert (Ibidi, Planegg / MartinsriedGermany) and cultivated overnight in McCoy's 5A medium containing 1% FBS. The culture insert was removed by sterile tweezers and 2 mL fresh medium was added to each well. We measured the distances of the initial gaps at 0 h and the final gaps at 10 h under a phase contrast microscope (Nikon, Tokyo, Japan). The relative cell migration capability was transformed to absolute migration capability. Absolute migration capability (μm/h) = (G 0 − G t) / 2t, where G 0 is the initial gap (μm) at 0 h and G t is the final gap for a time period of 10 h. Experiments were carried out in triplicate. For basic fibroblast growth factor (bFGF) effects, sh‐Sc or sh‐Gal‐1 T24 cells were incubated with 1 μg/mL bFGF dissolved in 20 mM Tris (pH 7.0) for 12 h. Then treated cells were evaluated for cell migration. Cells incubated with 20 mM Tris (pH 7.0) were recognized as 0 μg/mL. For Ras‐related C3 botulinum toxin substrate 1 (Rac1) activator effects, sh‐Sc or sh‐Gal‐1 T24 cells were incubated with 1 unit/mL Rac1 activator dissolved in H2O for 24 h. H2O‐treated cells were recognized as 0 unit/mL.
Cell invasion assays
Human sh‐Sc, sh‐Gal‐1 T24, or sh‐Gal‐1 J82 cells (1 × 105) were seeded onto a matrix gel made with serum‐free McCoy's 5A media in a Transwell migration chamber (Falcon/Thermo Fisher Scientific, sugar land, Tx, USA) with 8‐μm pore size membrane on the bottom. The chamber was inserted in a well of a 24‐well plate containing the corresponding media with 10% (v/v) FBS. The cells were incubated at 37°C for 24 h. After incubation, the matrix gel in the chamber was removed by carefully wiping with a cotton swab. The cells attached on the other side of membrane were incubated with MTT solution (5 mg/mL PBS) in the well for 4 h and then lysed with DMSO. The absorbance was determined at 540 nm.
Gelatin zymography assay
The activity of MMP9 was assayed by gelatin zymography. In brief, sh‐Gal‐1 or sh‐Sc T24 cells were incubated with serum‐free McCoy's 5A medium supplemented with 2 μg/mL PMA for 24 h at 37°C. The conditioned medium was collected and mixed with β‐mercaptoethanol‐free sample loading buffer without boiling, and then electrophoresed on 7% (w/v) SDS‐PAGE containing 0.1% (w/v) gelatin (Sigma, St. Louis, MO, USA). After electrophoresis, the gels were washed twice for 15 min with 2.5% (v/v) Triton X‐100 (Merck) to remove SDS and then incubated overnight at 37°C in zymography reaction buffer (50 mM Tris [pH 8.8], 5 mM CaCl2). Bands corresponding to activity were visualized by negative staining with 0.1% (w/v) Coomassie brilliant blue R‐250 (Merck), 40% (v/v) methanol, and 10% (v/v) acetic acid. The activity of MMP9 was quantified using Quantity One software (Bio‐Rad Laboratories, Hercules, CA, USA) and actin detected using Western immunoblotting as an internal control.
Western immunoblotting
Appropriate amounts of sh‐Sc cells, shGal‐1 T24 cells, or shGal‐1 J82 cells were harvested and lysed in lysis buffer (10 mM Tris [pH 8.0], 0.32 M sucrose, 1% [v/v] Triton X‐100, 5 mM EDTA, 2 mM DTT, and 1 mM PMSF). After determining its protein concentration using a Bio‐Rad DC protein assay kit, equal volume of 2× sample buffer (0.1 M Tris [pH 6.8], 2% SDS, 0.2% [v/v] β‐mercaptoethanol, 10% [v/v] glycerol, and 0.0016% [w/v] bromophenol blue) was combined with the cytoplasmic extract. Equal amounts of total proteins were separated by electrophoresis at 100 V with 10% (w/v) SDS–polyacrylamide gels, and further transferred onto a PVDF membrane (Strategene, La Jolla, CA, USA). After blocking for 1 h in 3% (w/v) BSA at room temperature, membranes were hybridized overnight at 4°C with primary antibodies. The following primary antibodies were used: Gal‐1 (1:2500; Epitomics, Burlingame, CA, USA), Erk (1:2000; AbFrontier, Seoul, Korea), p‐Erk (1:5000; AbFrontier), JNK (1:1000; BD Biosciences, San Jose, CA, USA), p‐JNK (1:1000; Cell Signaling Technology, Danvers, MA, USA), MMP9 (1:1500; Gene Tex, Irvine, CA, USA), c‐Jun (1:5000; Millipore, Billerica, MA, USA), Rac1 (1:1000; Millipore), MEKK4 (1:1000; Millipore), H‐Ras (1:300; Millipore), flotillin‐2 (1:5000, BD Biosciences, Bedford, MA, USA), and nucleolin (Millipore). The membranes were washed and probed with suitable secondary antibodies for 1 h at room temperature. Secondary antibodies binding on the membrane were detected by an ECL detection system (GE Healthcare, Little Chalfont, UK) using a Fujifilm LAS‐3000 Luminescent Image Analyzer (Fujifilm, Tokyo, Japan). The intensity of each protein band was quantified by PDQUEST Quantity One software (Bio‐Rad Laboratories) and normalized with actin protein expression level. The membranes were also stained with anti‐actin (1:5000; Millipore) or GAPDH antibody (1:5000; Millipore) as a loading control. The Rac1 protein was detected by a Rac1/Cdc42 Activation Assay Kit (Millipore) according to manufacturer's protocol.
Chromatin immunoprecipitation assay
Either sh‐Sc or shGal‐1 T24 cells (1 × 106) were seeded in a 10‐cm dish and treated with 2 μg/mL PMA at 37°C for 24 h. Chromatin immunoprecipitation analysis was carried out according to the CHIP‐ITTM Express Enzymatic kit (Active Motif, Carlsbad, CA, USA). DNA immunoprecipitation was undertaken with c‐Jun antibody (1:50; Millipore) and negative control mouse IgG (1:40; Active Motif). Immunoprecipitated DNA was amplified by real‐time RT‐PCR (7500 Real‐Time PCR System; Applied Biosystems, Carlsbad, CA USA) using the following primers: activator protein 1 (AP1) binding site of MMP9 promoter (forward, 5′‐CAGCCTGGTCAACGTAGTGA‐3′; reverse, 5′‐GGGTTCAAGCAATTCTCCTG‐3′) and GAPDH primer mix (Active Motif) was used as an internal control. The PCR reaction was carried out using SYBR GreenER qPCR SuperMix Universal kits (Invitrogen, Carlsbad, CA, USA) with 10 pmol each primer. The PCR amplification conditions were one cycle at 50°C for 2 min, one cycle at 95°C for 10 min, 50 cycles at 95°C for 15 s, 55°C or 60°C for 60 s. Relative quantitative fold change compared with control was calculated using the comparative C t method; C t is the cycle number at which fluorescence intensity first exceeds the threshold level. = C t (MMP9) − C t (GAPDH). is (sh‐Gal‐1) − (sh‐Sc). is expressed as a fold difference of MMP9 of sh‐Gal‐1 compared to that of sh‐Sc cells.
Statistical analysis
Statistical analysis was carried out using Student's t‐test (for two‐group comparisons) (statistica; StatSoft, Tulsa, OK, USA). Differences between groups with P‐values <0.05 were considered statistically significant.
Results
Association of Gal‐1 with the malignant behavior of UBUC cells
In order to clarify the role of Gal‐1 in UBUC invasiveness, a Gal‐1 knockdown cell line was established as described above. As Gal‐1 protein was overexpressed in T24 and J82 cells,23 they were chosen to construct Gal‐1 knockdown cell lines sh‐Gal‐1(+120) and sh‐Gal‐1(+380). As indicated in Figure S1, the amounts of Gal‐1 protein and mRNA were reduced in sh‐Gal‐1(+120) and sh‐Gal‐1(+380) T24 cells respectively as compared to sh‐Sc(+120) and sh‐Sc(+380). However, the knockdown efficiency in sh‐Gal‐1(+120) T24 cells was much better than that in sh‐Gal‐1(+380) T24 cells. Expression of Gal‐1 was barely affected in sh‐Sc(+120) and sh‐Sc(+380) cells. Galectin‐1 expression was also knocked down in sh‐Gal‐1(+120) J82 cells (Fig. S1).
Characterization of tumor cell behavior indicated that sh‐Gal‐1(+120) and sh‐Gal‐1(+380) T24 cells proliferated more slowly than sh‐Sc cells (Fig. 1a,b). But sh‐Gal‐1(+380) cells proliferated faster than sh‐Gal‐1(+120) cells, implying that T24 cell proliferation showed a dose‐dependent response to Gal‐1 knockdown. In addition, Gal‐1 dampening could impede clone formation capability of sh‐Gal‐1(+120) and sh‐Gal‐1(+380) T24 cells compared to those of sh‐Sc(+120) and sh‐Sc(+380) cells (Fig. 1c). T24 cells also showed a dose‐dependent response to Gal‐1 knockdown with regard to clone formation capability. The above phenomena showed that Gal‐1 was significantly tied to UBUC cell proliferation. Moreover, lower Gal‐1 levels could interfere with T24 cell migration and invasion. Dose‐dependent cell behavior was also observed in T24 cell migration and invasion examinations (Fig. 1d,e). Galectin‐1 knockdown also reduced the migration and invasion capabilities of sh‐Gal‐1(+120) J82 cells (Fig. 1f,g). The above results implied that Gal‐1 was intimately link to UBUC invasiveness.
Figure 1.
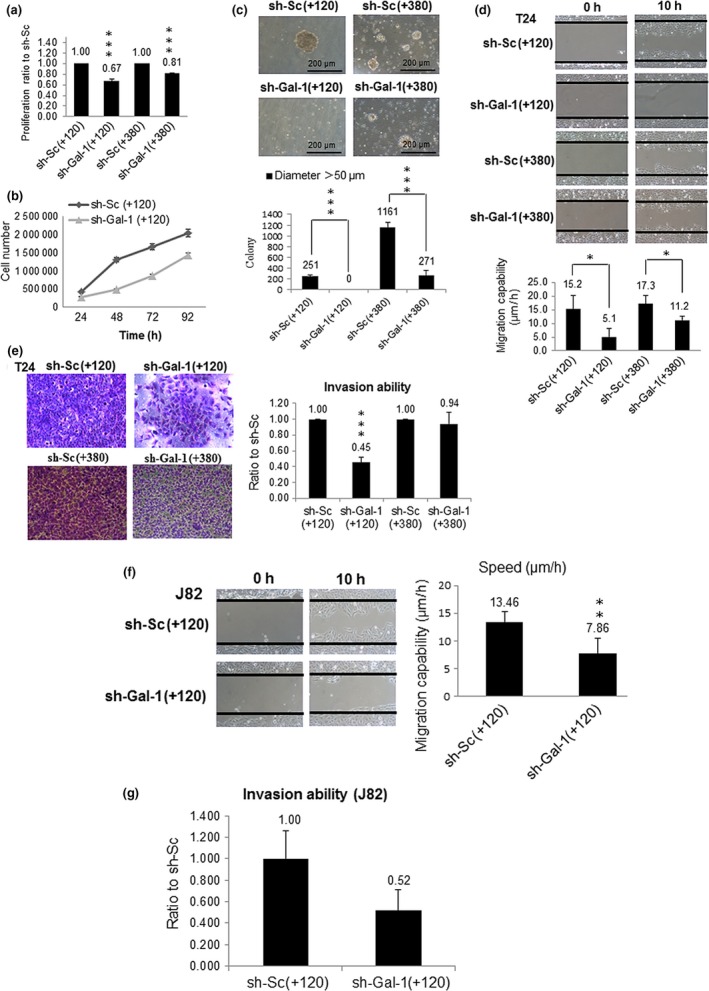
Effects of galectin‐1 (Gal‐1) knockdown on the malignant characteristics of urinary bladder urothelial carcinoma T24 and J82 cells. (a) Cell survival assays of T24 cell clones with knockdown at +120 nt (sh‐Gal‐1(+120)) and +380 nt (sh‐Gal‐1(+380)). (b) Cell number counting assays of sh‐Gal‐1(+120) T24 cells. (c–e) Soft agar (c), cell migration (d), and cell invasion (e) assays of sh‐Gal‐1(+120) and sh‐Gal‐1(+380) T24 cells. (f,g) Cell migration (f) and cell invasion (g) assays of sh‐Gal‐1(+120) J82 cells. The results of MTT, cell number counting, soft agar, migration, and invasion assays show representative data of three independent experiments. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Participation of Gal‐1 in signaling pathways linked to invasiveness of UBUC cells
Our previous cohort study reported that Gal‐1 overexpression is significantly linked to UBUC invasion.23 In addition, Gal‐1 silencing inhibited the invasive ability of T24 cells and MMP9 is closely associated with tumor metastasis.25 Thus, we hypothesized that the reduced invasive ability of sh‐Gal‐1(+120) T24 and sh‐Gal‐1(+120) J82 cells might be attributed to decreased MMP9 expression. In accordance with our expectation, the results of the MMP9 assay revealed that MMP9 mRNA as well as protein syntheses were suppressed and MMP9 activity was also inhibited in sh‐Gal‐1(+120) T24 and J82 cells (Fig. 2a,b).
Figure 2.
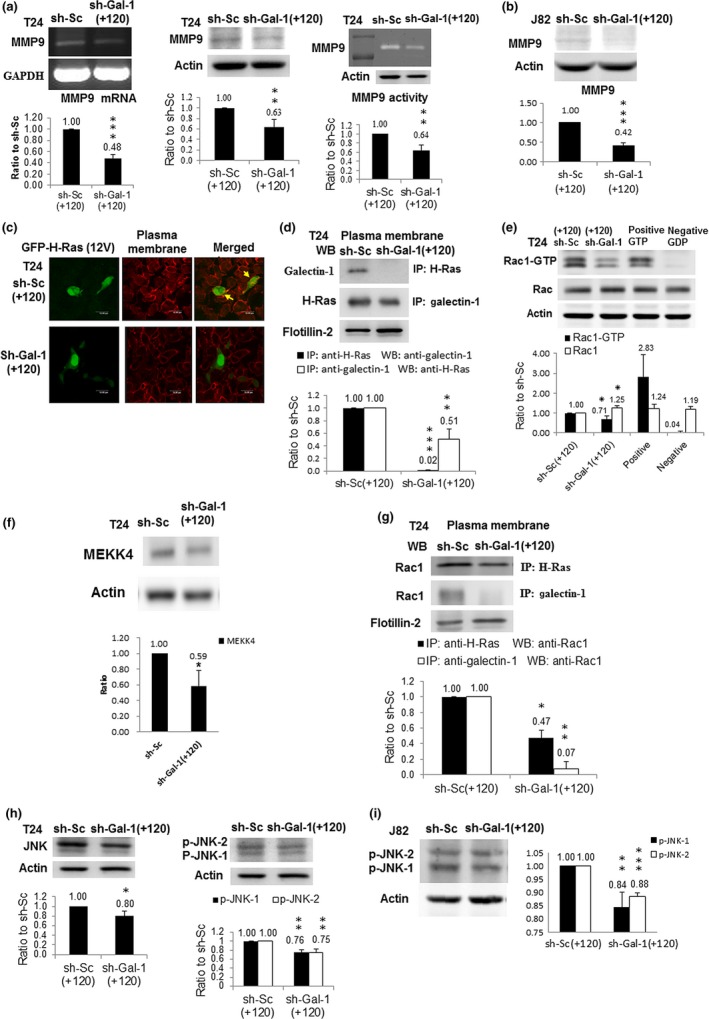
Impacts of galectin‐1 (Gal‐1) silencing on the Gal‐1‐mediated signaling pathway. (a) MMP9 protein (left panel) and mRNA (middle panel) amounts in T24 cell clones with knockdown at +120 nt (shGal‐1(+120)) measured by Western immunoblotting and RT‐PCR, respectively. MMP9 activities (right panel) in shGal‐1(+120) T24 cells were evaluated by zymography. The blot in each figure was the typical result of at least three independent studies. The diagram (ratio [mean ± SD]) under each blot shows the differential protein expression that was expressed as the ratio of normalized intensity (observed protein/actin) of sh‐Gal‐1(+120) cells divided by that of scrambled sh‐Sc(+120) cells. The number above each bar is the average ratio. (b) MMP9 protein level in shGal‐1(+120) J82 cells. (c) Confocal microscopic image of shGal‐1(+120) T24 cells, typical of three independent experiments. shGal‐1(+120) and sh‐Sc(+120) T24 cells were transfected with H‐Ras expression vector (pMSCV‐H‐Ras(12V)‐GFP). H‐Ras protein is shown in green and the cell membrane is stained in red in T24 sh‐Sc(+120) cells. Yellow (arrow), shown on the cell membrane in the merged image, indicates that H‐Ras was located on cell membrane of T24 sh‐Sc(+120) cells. However, the cell membrane remained red in the merged image of T24 sh‐Gal‐1(+120) cells, indicating that H‐Ras did not appear on the cell membrane because of Gal‐1 knockdown. (d) Interaction of Gal‐1 with H‐Ras in plasma membranes of shGal‐1(+120) cells. Protein lysate from purified plasma membrane immunoprecipitated (IP) with antibodies against H‐Ras and Gal‐1 proteins present in the immunocomplex was detected by western immunoblotting and vice versa. (e) Rac1 protein level in shGal‐1(+120) T24 cells. Protein lysates were collected for Western immunoblotting (WB). GTP or GDP‐incorporated protein lysates were used as positive or negative controls. (f) MEKK4 protein amount in shGal‐1(+120) T24 cells. (g) Interaction of Rac1 with H‐Ras or Gal‐1 in plasma membranes of shGal‐1(+120) cells. Protein lysate from purified plasma membrane was immunoprecipitated with antibody against H‐Ras or Gal‐1 protein and Rac1 protein present in the immunocomplex was detected by Western immunoblotting. Flotillin‐2 was the loading control. (h) JNK (left panel) and phosphorylated (p‐)JNK) (right panel) protein levels in shGal‐1(+120) T24 cells. (i) p‐JNK protein amount in shGal‐1(+120) J82 cells. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Galectin‐1 has been shown to contribute to H‐Ras membrane anchorage,26 and thus in turn activates the Rac1–MEKK4–JNK pathway.27 Results of in situ hybridization indicated that Gal‐1 silencing resulted in GFP–H‐Ras (12V) mislocalization (Fig. 2c). Immunoprecipitation results further showed that Gal‐1 knockdown reduced the interaction between Gal‐1 and H‐Ras (Fig. 2d), suggesting that Gal‐1 was involved in H‐Ras membrane anchorage in T24 cells. Further signaling pathway investigations showed that, in sh‐Gal‐1(+120) T24 cells, Rac1 and MEKK4 activities were reduced (Fig. 2e,f) and the interaction between Rac1 and Ras or Gal‐1 was also diminished (Fig. 2g). Furthermore, the expression levels of JNK and phosphorylated‐JNK protein were downregulated in sh‐Gal‐1(+120) T24 and J82 cells (Fig. 2h,i). The above results implied that Gal‐1 participated in the Ras–Rac1–MEKK4–JNK pathway in T24 cells.
Much well‐documented evidence has established that the JNK signaling pathway activates c‐Jun by phosphorylation and in turn phosphorylated c‐Jun moves to the nucleus and combines with c‐fos to form AP1 transcription factor to activate the expression of many target genes related to cell proliferation and invasion,28, 29 for example, MMP9. In this study we showed that the c‐Jun protein level was reduced in the nucleus of sh‐Gal‐1(+120) T24 cells while that in the cytosol was slightly increased compared to sh‐Sc(+120) T24 cells (Fig. 3a), suggesting that Gal‐1 attenuation restricted the movement of activated c‐Jun. Using ChIP that measures whether a transcription factor binds to its cognate promoter in a viable cell, we found that Gal‐1 silencing could block the binding of AP1 to MMP9 promoter in sh‐Gal‐1(+120) T24 cells (Fig. 3b) and thus decreased MMP9 gene expression.
Figure 3.
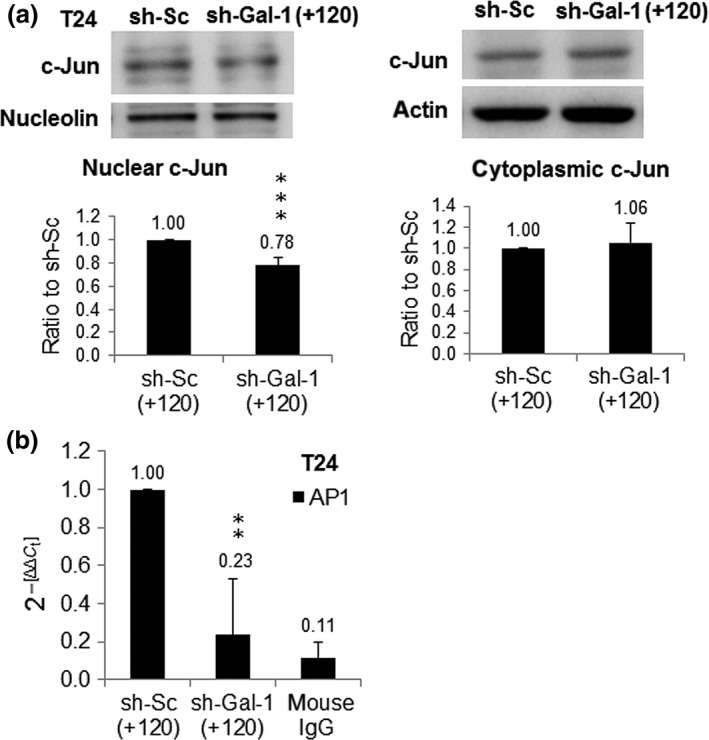
Influences of galectin‐1 (Gal‐1) knockdown on c‐Jun and activator protein 1 (AP1) activities in urinary bladder urothelial carcinoma cells. (a) c‐Jun protein levels in nucleus (left panel) and cytoplasm (right panel) of cell clones with knockdown at +120 nt (shGal‐1(+120)). shGal‐1(+120) or scrambled (sh‐Sc(+120)) cells were treated with 2 μg/mL PMA at 37°C for 24 h, then the nuclear and cytoplasmic extracts were extracted from cells. (b) AP1 activity was observed by ChIP assay. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
To further characterize the link of Gal‐1–JNK to UBUC cell invasiveness, we observed the impacts of bFGF (JNK pathway agonist)30 on invasive/migratory capabilities of sh‐Gal‐1(+120) and sh‐Sc(+120) T24 cells. The results in Figure 4 showed that bFGF treatment could boost the invasive/migratory abilities of sh‐Sc(+120) T24 cells, but those of sh‐Gal‐1(+120) T24 cells were hardly affected. Treatment with bFGF also increased the expressions of phosphorylated (p‐)JNK and MMP9 in sh‐Sc(+120) T24 cells, but not in sh‐Gal‐1(+120) T24 cells (Fig. 4c,d). In addition, Rac1 agonist treatment could evoke the invasive/migratory abilities of sh‐Gal‐1(+120) T24 cells (Fig. 4e,f). It also augmented the expressions of p‐JNK, p‐Erk, and MMP9, suggesting that Gal‐1 functioned upstream of Rac1 in the Gal‐1 signaling pathway (Fig. 4g–i). The effect of JNK inhibitor SP600125 on migratory capability was also examined. Results in Fig. 4(j) indicate that Gal‐1 silencing deteriorated the inhibition of sh‐Gal‐1(+120) T24 cell migration by SP600125 treatment.
Figure 4.
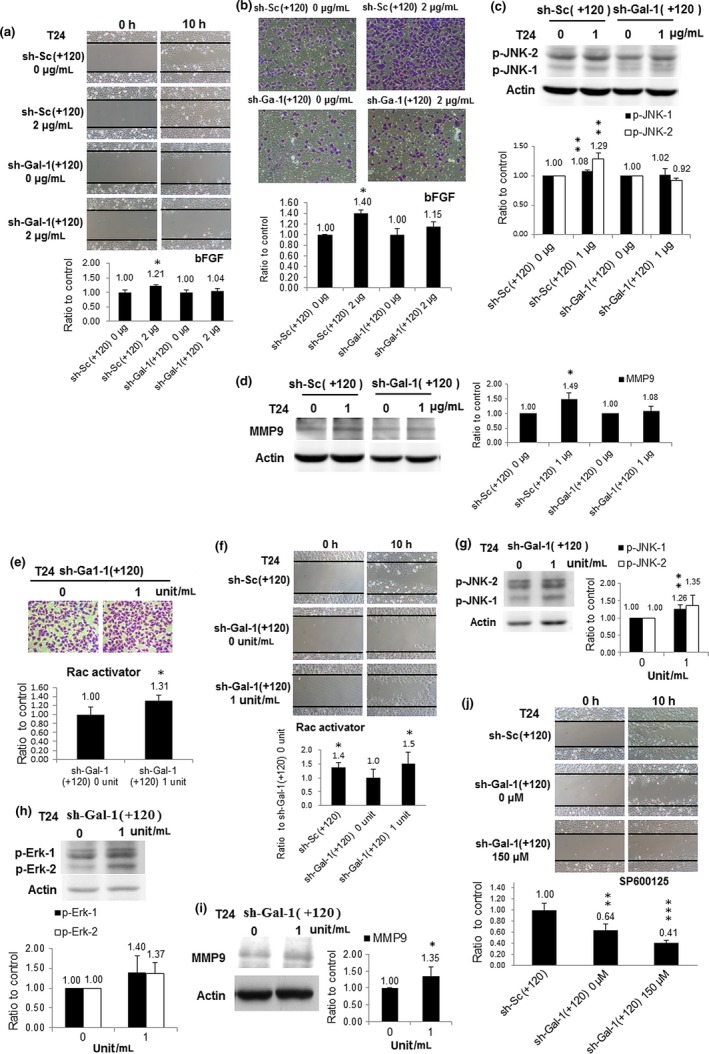
Effects of basic fibroblast growth factor (bFGF), Rac1 activator, and SP600125 on T24 cell clones with knockdown at +120 nt (sh‐Gal‐1(+120)). (a) Impacts of bFGF on sh‐Gal‐1(+120) T24 cell migration. Cells were treated with 1 μg/mL bFGF for 12 h then cells were evaluated for migration. (b) Influence of bFGF on sh‐Gal‐1(+120) T24 cell invasion. (c) Expression of phosphorylated (p‐)JNK in bFGF‐treated sh‐Gal‐1(+120) T24 cells. (d) MMP9 protein level in bFGF‐treated sh‐Gal‐1(+120) T24 cells. (e) Rac1 activator‐incubated sh‐Gal‐1(+120) T24 cell invasion. (f) Rac1 activator‐treated sh‐Gal‐1(+120) T24 cell migration. (g) p‐JNK protein level in Rac1 activator‐treated sh‐Gal‐1(+120) T24 cells. (h) MMP9 protein amount in Rac1 activator‐treated sh‐Gal‐1(+120) T24 cells. (i) p‐Erk protein level in Rac1 activator‐treated sh‐Gal‐1(+120) T24 cells. (j) SP600125‐incubated sh‐Gal‐1(+120) T24 cell migration. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Galectin‐1 overexpression reinforced the invasiveness of TSGH8301 cells
To further determine that Gal‐1 was closely related to UBUC cell invasion, TSHG8301 cells overexpressing Gal‐1 were established as described in Appendix S1 and shown in Figure S2. Cell invasion assay indicated that Gal‐1 overexpression could increase the migratory and invasive capabilities of TSGH8301 cells (Fig. 5a,b). Moreover, it could also increase the expression levels of p‐JNK and MMP9 (Fig. 5c). The above results implied that Gal‐1 mediated UBUC cell invasiveness by regulating MMP9 gene expression through the Gal‐1−Ras−Rac1−MEKK4−JNK−c‐Jun−AP1 signaling pathway.
Figure 5.
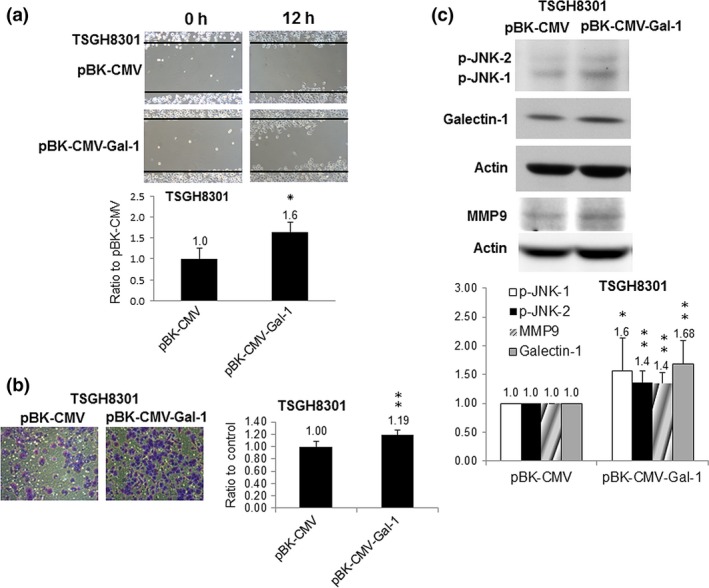
Impacts of galectin‐1 (Gal‐1) overexpression on TSGH8301 low‐grade urinary bladder urothelial carcinoma cells. (a) Migration of TSGH8301 cells overexpressing Gal‐1, constructed by transfection of PBK‐CMV‐Gal‐1 vector inserted with Gal‐1 cDNA. (b) Invasion of TSGH8301 cells overexpressing Gal‐1. (c) Phosphorylated (p‐)JNK and MMP9 protein levels in TSGH8301 cells overexpressing Gal‐1. Blot shows the typical result of three independent experiments. Right panel shows densitometer intensity data. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Discussion
Increased Gal‐1 expression has been reported in various tumors.3, 4, 5, 6, 7, 8, 9, 10, 11, 12 It is well documented that Gal‐1 expression is linked to tumor stage, tumor grade, and lymph node metastasis in gastric and cervical cancer.31, 32 In our previous cohort investigation, we found that there is a significant correlation between Gal‐1 overexpression and tumor stage, histological grade, vascular invasion, and lymph node metastasis. Increased Gal‐1 expression in UBUC significantly predicted disease‐specific survival at univariate and multivariate levels.23 The aforementioned study is the first to provide the clinical correlation between Gal‐1 expression, patient survival, and prognosis of UBUC.33 The correlation between Gal‐1 expression and survival is also observed in Kaposi's sarcoma, colon, breast, prostate, and gastric cancer.33
In the present study we intended to uncover the Gal‐1‐involved molecular mechanism underlying UBUC carcinogenesis. To decipher the clinical link of Gal‐1 to tumor progression and invasiveness, T24 cells with Gal‐1 knockdown by shRNA were evaluated for the influence of Gal‐1 intervention on tumor cell behavior. Characterization of T24 cells with Gal‐1 knockdown indicated that sh‐Gal‐1(+120) T24 cells with better knockdown efficiency had poorer cell proliferation and clone formation than sh‐Gal‐1(+380) cells. Silencing Gal‐1 in T24 cells also attenuated the migration capabilities and invasive abilities of T24 cells and showed the dose‐dependent impacts on T24 invasive capability (sh‐Gal‐1(+120) vs sh‐Gal‐1(+380)). The above findings suggested that Gal‐1 plays an important role in supporting UBUC cell behavior and reinforced the notion that Gal‐1 level was related to UBUC progression and invasiveness.
Compelling evidence supports the involvement of Gal‐1 in the biological processes linked to tumor cell invasiveness. Galectin‐1 silencing in primary breast or colon cancer tumors reduces the number of lung metastatic tumors.5, 34 Furthermore, Gal‐1 is also reported to promote lung tumor invasion and migration by increasing the integrin α6β4–Notch 1 signaling pathway.35 The findings by Wu et al. showed that reduced Gal‐1 expression by siRNA decreased cell invasion of highly invasive oral squamous carcinoma cells and Gal‐1 is overexpressed in the invasive front of primary tumors of patients with oral squamous cell carcinoma. Upregulated Gal‐1 expression stimulates tumor invasion by upregulating MMP9 and MMP2 and reorganizes the cytoskeleton by increasing cell division cycle 42 activity, a member of Rho GTPase family.36 Matrix metalloproteinases are a family of zinc‐dependent enzymes that degrade the ECM, including gelatin and collagen. Matrix metalloproteinase‐9 is closely tied to tumor invasion and metastasis.25 In accordance with the previously documented observations, this study showed that inhibition of LGALS1 gene expression dwindled MMP9 gene expression and its enzymatic activity in T24 cells. Our further results revealed that reduced MMP9 gene expression, mediated by Gal‐1 knockdown in T24 cells, might be attributed to the intervention of the JNK signaling pathway to activate c‐Jun, which in turn impaired the movement of activated c‐Jun to the nucleus to form AP1 transcription factor. Activator protein 1 can provoke the expression of many target genes involved in cell proliferation, differentiation, cytoskeleton organization, and invasion.37 In addition to upregulation of MMP9 gene expression, Gal‐1 can promote invasion through augmentation of cell–cell adhesion or cell–ECM interaction. Galectin‐1 silencing causes protein kinase Cε and vimentin to accumulate around the nucleus in glioblastoma cells,33 which interferes with β‐integrin trafficking and recycling to the cell membrane, and thus leads to a decreased amount of integrin receptors on the cell membrane. Galectin‐1 is also reported to stimulate lung tumor invasion and migration by provoking the integrin α6β4/NOTCH1 biochemical cascade.33
Based on the present data and previous studies,23 we propose that LGALS1 gene expression ramped up in UBUC specimens might provoke the Rac1−MEKK4−JNK signaling pathway to augment the JNK−c‐Jun−AP1−MMP9 pathway, which in turn stimulates tumor cell invasion. Our observations provide unprecedented insight into the molecular mechanism of UBUC invasiveness and metastasis.
Our functional analyses of Gal‐1 in this study provide the first insight into the critical role of Gal‐1 in the progression and invasion of urinary bladder urothelial carcinoma. These results reveal that silencing the Gal‐1‐mediated MAPK signaling pathway presents a novel strategy for bladder cancer therapy.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Appendix S1. Extended materials and methods.
Fig. S1. LGALS1 gene expression in galectin‐1 (Gal‐1) knockdown T24 and J82 cells.
Fig. S2. LGALS1 gene expression in galectin‐1 (Gal‐1) overexpressing TSGH8301 cells.
Acknowledgments
This work was supported by grants from the Taiwan Ministry of Science and Technology (Grant Nos. NSC 101‐2632‐B‐218‐001‐MY3, MOST 104‐2320‐B‐218‐001, and MOST 104‐2632‐E‐218‐002). The Authors also would like to thank the Center for Bioscience and Biotechnology of National Cheng Kung University for use of the laser scanning confocal microscope.
Cancer Sci 107 (2016) 1390–1398
Funding Information
Taiwan Ministry of Science and Technology.
References
- 1. Stein JP, Grossfeld GD, Ginsberg DA et al Prognostic markers in bladder cancer: a contemporary review of the literature. J Urol 1998; 160: 645–59. [DOI] [PubMed] [Google Scholar]
- 2. Dey P. Urinary markers of bladder carcinoma. Clin Chim Acta 2004; 340: 57–65. [DOI] [PubMed] [Google Scholar]
- 3. Ahmad N, Gabius HJ, Sabesan S, Oscarson S, Brewer CF. Thermodynamic binding studies of bivalent oligosaccharides to galectin‐1, galectin‐3 and the carbohydrate recognition domain of galectin‐3. Glycobiol 2004; 14: 817–25. [DOI] [PubMed] [Google Scholar]
- 4. Barrow H, Rhodes JM, Yu LG. The role of galectins in colorectal cancer progression. Int J Cancer 2011; 129: 1–8. [DOI] [PubMed] [Google Scholar]
- 5. Dalotto‐Moreno T, Croci DO, Cerliani JP et al Targeting galectin‐1 overcomes breast cancer associated immunosuppression and prevents metastatic disease. Cancer Res 2013; 73: 1107–17. [DOI] [PubMed] [Google Scholar]
- 6. Szoke T, Kayser K, Baumhakel JD et al Prognostic significance of endogenous adhesion/growth‐regulatory lectins in lung cancer. Oncology 2005; 69: 67–74. [DOI] [PubMed] [Google Scholar]
- 7. Saussez S, Camby I, Toubeau G, Kiss R. Galectins as modulators of tumor progression in head and neck squamous cell carcinomas. Head Neck 2007; 29: 874–84. [DOI] [PubMed] [Google Scholar]
- 8. Chow SN, Chen RJ, Chen CH et al Analysis of protein profiles in human epithelial ovarian cancer tissues by proteomic technology. Eur J Gynaecol Oncol 2010; 31: 55–62. [PubMed] [Google Scholar]
- 9. Laderach DJ, Gentilini LD, Giribaldi L et al A unique galectin signature in human prostate cancer progression suggests galectin‐1 as a key target for treatment of advanced disease. Cancer Res 2013; 73: 86–96. [DOI] [PubMed] [Google Scholar]
- 10. Rorive S, Belot N, Decaestecker C et al Galectin‐1 is highly expressed in human gliomas with relevance for modulation of invasion of tumor astrocytes into the brain parenchyma. Glia 2001; 33: 241–55. [DOI] [PubMed] [Google Scholar]
- 11. Croci DO, Salatino M, Rubinstein N et al Disrupting galectin‐1 interactions with N‐glycans suppresses hypoxia‐driven angiogenesis and tumorigenesis in Kaposi's sarcoma. J Exp Med 2012; 209: 1985–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. D'Haene N, Maris C, Sandras F et al The differential expression of galectin‐1 and galectin‐3 in normal lymphoid tissue and non‐Hodgkins and Hodgkins lymphomas. Int J Immunopathol Pharmacol 2005; 18: 431–43. [DOI] [PubMed] [Google Scholar]
- 13. Rabinovich GA. Galectin‐1 as a potential cancer target. Br J Cancer 2005; 92: 1188–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Banh A, Zhang J, Cao H et al Tumor galectin‐1 mediates tumor growth and metastasis through regulation of T‐cell apoptosis. Cancer Res 2011; 71: 4423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scott K, Weinberg C. Galectin‐1: a bifunctional regulator of cellular proliferation. Glycoconj J 2004; 19: 467–77. [DOI] [PubMed] [Google Scholar]
- 16. He J, Baum LG. Galectin interactions with extracellular matrix and effects on cellular function. Methods Enzymol 2006; 417: 247–56. [DOI] [PubMed] [Google Scholar]
- 17. Hsu DK, Liu FT. Regulation of cellular homeostasis by galectins. Glycoconj J 2004; 19: 507–15. [DOI] [PubMed] [Google Scholar]
- 18. Yang RY, Liu FT. Galectins in cell growth and apoptosis. Cell Mol Life Sci 2003; 60: 267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thijssen VL, Postel R, Brandwijk RJ et al Galectin‐1 is essential in tumor angiogenesis and is a target for antiangiogenesis therapy. Proc Natl Acad Sci USA 2006; 103: 15975–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Camby I, Belot N, Lefranc F et al Galectin‐1 modulates human glioblastoma cell migration into the brain through modifications to the actin cytoskeleton and levels of expression of small GTPases. J Neuropathol Exp Neurol 2002; 61: 585–96. [DOI] [PubMed] [Google Scholar]
- 21. Paz A, Haklai R, Elad‐Sfadia G, Ballan E, Kloog Y. Galectin‐1 binds oncogenic H‐Ras to mediate Ras membrane anchorage and cell transformation. Oncogene 2001; 20: 7486–93. [DOI] [PubMed] [Google Scholar]
- 22. Rubinstein N, Alvarez M, Zwirner NW et al Targeted inhibition of galectin‐1 gene expression in tumor cells results in heightened T cell‐mediated rejection; a potential mechanism of tumorimmune privilege. Cancer Cell 2004; 5: 241–51. [DOI] [PubMed] [Google Scholar]
- 23. Wu TF, Li CF, Chien LH et al Galectin‐1 dysregulation independently predicts disease‐specific survival in urinary bladder urothelial carcinoma. J Urol 2015; 193: 1002–8. [DOI] [PubMed] [Google Scholar]
- 24. Yeh MY, Yü DS, Chen SC et al Establishment and characterization of a human urinary bladder carcinoma cell line (TSGH‐8301). J Surg Oncol 1988; 37: 177–84. [DOI] [PubMed] [Google Scholar]
- 25. Fridman R, Toth M, Peña D, Mobashery S. Activation of progelatinase B (MMP‐9) by gelatinase A (MMP‐2). Cancer Res 1995; 55: 2548–55. [PubMed] [Google Scholar]
- 26. Rotblat B, Niv H, André S, Kaltner H, Gabius HJ, Kloog Y. Galectin‐1(L11A) predicted from a computed galectin‐1 farnesyl‐binding pocket selectively inhibits Ras‐GTP. Cancer Res 2004; 64: 3112–8. [DOI] [PubMed] [Google Scholar]
- 27. Fanger GR, Johnson NL, Johnson GL. MEK kinases are regulated by EGF and selectively interact with Rac/Cdc42. EMBO J 1997; 16: 4961–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature 2001; 410: 37–40. [DOI] [PubMed] [Google Scholar]
- 29. Milde‐Langosch K, Röder H, Andritzky B et al The role of the AP‐1 transcription factors c‐Fos, FosB, Fra‐1 and Fra‐2 in the invasion process of mammary carcinomas. Breast Cancer Res Treat 2004; 86: 139–52. [DOI] [PubMed] [Google Scholar]
- 30. Kanazawa S, Fujiwara T, Matsuzaki S et al bFGF regulates PI3‐kinase‐Rac1‐JNK pathway and promotes fibroblast migration in wound healing. PLoS ONE 2010; 5: e12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim HJ, Do IG, Jeon HK et al Galectin 1 expression is associated with tumor invasion and metastasis in stage IB to IIA cervical cancer. Hum Pathol 2013; 44: 62–8. [DOI] [PubMed] [Google Scholar]
- 32. Chen J, Zhou SJ, Zhang Y et al Clinicopathological and prognostic significance of galectin‐1 and vascular endothelial growth factor expression in gastric cancer. World J Gastroenterol 2013; 19: 2073–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Astorgues‐Xerri L, Riveiro ME, Tijeras‐Raballand A et al Unraveling galectin‐1 as a novel therapeutic target for cancer. Cancer Treat Rev 2014; 40: 307–19. [DOI] [PubMed] [Google Scholar]
- 34. Ito K, Ralph SJ. Inhibiting galectin‐1 reduces murine lung metastasis with increased CD4(+) and CD8(+) T cells and reduced cancer cell adherence. Clin Exp Metastasis 2012; 29: 561–72. [DOI] [PubMed] [Google Scholar]
- 35. Hsu YL, Wu CY, Hung JY et al Galectin‐1 promotes lung cancer tumor metastasis by potentiating integrin α6β4 and Notch1/Jagged2 signaling pathway. Carcinogenesis 2013; 34: 1370–81. [DOI] [PubMed] [Google Scholar]
- 36. Wu MH, Hong TM, Cheng HW et al Galectin‐1‐mediated tumor invasion and metastasis, up‐regulated matrix metalloproteinase expression, and reorganized actin cytoskeletons. Mol Cancer Res 2009; 7: 311–8. [DOI] [PubMed] [Google Scholar]
- 37. Hartl M, Bader AG, Bister K et al Molecular targets of the oncogenic transcription factor jun. Curr Cancer Drug Targets 2003; 3: 41–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Extended materials and methods.
Fig. S1. LGALS1 gene expression in galectin‐1 (Gal‐1) knockdown T24 and J82 cells.
Fig. S2. LGALS1 gene expression in galectin‐1 (Gal‐1) overexpressing TSGH8301 cells.


