ABSTRACT
We report the isolation and characterization of three new cytochrome P450 monooxygenases: CYP101J2, CYP101J3, and CYP101J4. These P450s were derived from Sphingobium yanoikuyae B2, a strain that was isolated from activated sludge based on its ability to fully mineralize 1,8-cineole. Genome sequencing of this strain in combination with purification of native 1,8-cineole-binding proteins enabled identification of 1,8-cineole-binding P450s. The P450 enzymes were cloned, heterologously expressed (N-terminally His6 tagged) in Escherichia coli BL21(DE3), purified, and spectroscopically characterized. Recombinant whole-cell biotransformation in E. coli demonstrated that all three P450s hydroxylate 1,8-cineole using electron transport partners from E. coli to yield a product putatively identified as (1S)-2α-hydroxy-1,8-cineole or (1R)-6α-hydroxy-1,8-cineole. The new P450s belong to the CYP101 family and share 47% and 44% identity with other 1,8-cineole-hydroxylating members found in Novosphingobium aromaticivorans and Pseudomonas putida. Compared to P450cin (CYP176A1), a 1,8-cineole-hydroxylating P450 from Citrobacter braakii, these enzymes share less than 30% amino acid sequence identity and hydroxylate 1,8-cineole in a different orientation. Expansion of the enzyme toolbox for modification of 1,8-cineole creates a starting point for use of hydroxylated derivatives in a range of industrial applications.
IMPORTANCE CYP101J2, CYP101J3, and CYP101J4 are cytochrome P450 monooxygenases from S. yanoikuyae B2 that hydroxylate the monoterpenoid 1,8-cineole. These enzymes not only play an important role in microbial degradation of this plant-based chemical but also provide an interesting route to synthesize oxygenated 1,8-cineole derivatives for applications as natural flavor and fragrance precursors or incorporation into polymers. The P450 cytochromes also provide an interesting basis from which to compare other enzymes with a similar function and expand the CYP101 family. This could eventually provide enough bacterial parental enzymes with similar amino acid sequences to enable in vitro evolution via DNA shuffling.
INTRODUCTION
The bicyclic monoterpenoid 1,8-cineole (1,3,3-trimethyl-2-oxabicyclo[2,2,2]octane) is a chemically stable saturated ditertiary ether (1) that is an abundant natural resource. It is a major component of many essential oils, in particular Eucalyptus oil (2). While 1,8-cineole has its main applications in pharmaceutical, fragrance, food, and cleaning industries, its oxidation provides a potential route to value-added chemicals. Hydroxylated 1,8-cineole derivatives are chiral compounds that can serve as building blocks and precursors with uses in organic chemistry (3). Hydroxylation of 1,8-cineole can be achieved chemically; however, these reactions often use a range of environmentally hazardous reagents and/or yield product mixtures (4–6). On the other hand, biooxygenation of 1,8-cineole using enzymes or whole cells can be performed under mild reaction conditions with the potential to yield products with high enantiopurity (6, 7).
A potential route to bio-derived 1,8-cineole derivatives is the utilization of enzymes involved in bacterial 1,8-cineole metabolism. Bacterial 1,8-cineole metabolism was first examined in the late 1970s, when the isolation of a Pseudomonas flava strain capable of growing on 1,8-cineole as a sole source of carbon was reported (8). Subsequently, 1,8-cineole biohydroxylation has also been observed in several other bacterial species, including Rhodococcus sp. strain C1 (9), Bacillus cereus (10), Citrobacter braakii (11), Novosphingobium subterranea (12), and a Sphingomonas species (13). The first bacterial 1,8-cineole-hydroxylating enzyme which was isolated and characterized was isolated from C. braakii (11). The initial oxidation of 1,8-cineole by this Gram-negative bacterium is catalyzed by a P450 designated P450cin (CYP176A1), and with the aid of suitable electron transport partners it yielded (1R)-6β-hydroxy-1,8-cineole (7, 11). More recently, other P450s, including CYP101A1, CYP101B1, and the N252A P450cin mutant, were shown to hydroxylate 1,8-cineole and yield a range of hydroxy-1,8-cineole derivatives (14, 15).
P450 cytochromes are diverse oxidative hemoproteins that catalyze an enormous range of reactions, including hydroxylations of unactivated carbon-hydrogen bonds (16). These enzymes are widespread in nature, originating from all kingdoms of life, including more than 2,150 bacterial P450s (http://drnelson.uthsc.edu/CytochromeP450.html) (17). Several bacterial P450s are involved in monoterpenoid oxidation, including P450cam from Pseudomonas putida, which was enriched by selection for growth on (1R)-(+)-camphor (18). P450cam or CYP101A1 was the first P450 crystallized (19) and has served as a model system for mechanistic studies and structure-function relationships. Since then, many more monoterpene-oxidizing P450s have been isolated, characterized, crystallized, and used for laboratory-scale biooxidation reactions; the latest research in biooxidation of monoterpenes using bacterial monooxygenases has been reviewed recently (20).
Bacterial P450s (and their electron transport partners) are usually soluble proteins (21). For this reason, these proteins are easier to overexpress and purify than membrane-bound P450s of eukaryotic origin (20). They are particularly promising candidates for monoterpene biohydroxylations because of their relatively high activity and substrate specificity toward a number of monoterpenes (11, 20). With the recent availability of extensive metagenomic data, the number of gene sequences potentially encoding P450s has increased substantially (22); however, many of the identified genes remain uncharacterized. The isolation and detailed characterization of enzymes is a prerequisite for optimization and use in biocatalytic reactions, paving the way for applications in synthetic biology.
To identify and characterize new bacterial 1,8-cineole-hydroxylating P450s, we sequenced the genome of Sphingobium yanoikuyae strain B2, isolated from activated sludge (23), that was capable of using 1,8-cineole as the sole source of carbon and energy. Purification of native 1,8-cineole-binding proteins from S. yanoikuyae strain B2 followed by recombinant expression and enzyme characterization aided the identification of three new P450s capable of hydroxylating 1,8-cineole. This work has expanded the range of bacterial 1,8-cineole-oxidizing enzymes, creating an opportunity for their exploitation in industrial biocatalysis and further understanding of enzyme mechanisms.
MATERIALS AND METHODS
Reagents.
1,8-Cineole (99.5% purity) was obtained from Felton Grimwade & Bosisto's Pty Ltd. (Oakleigh South, Australia). Isopropyl-β-d-thiogalactopyranoside (IPTG), 2-adamantanone, β-ionone, α-terpineol, (1R)-(+)-camphor, (1S)-(−)-camphor, and sodium hydrosulfite (85% purity) were from Sigma-Aldrich. (1R)-6β-Hydroxy-1,8-cineole was produced using E. coli strain DH5α harboring the bicistronic plasmid pCW-P450cin/Cdx (7), which was a gift from James De Voss (University of Queensland). A mixture of chemically synthesized (1S)-2α-hydroxy-1,8-cineole and (1S)-2β-hydroxy-1,8-cineole was kindly provided by Stella Kyi (CSIRO). Restriction enzymes for molecular biology and Phusion high-fidelity DNA polymerase for PCRs were from New England BioLabs. General reagents for medium and buffer preparation were obtained from various commercial sources and were of reagent grade or better.
Strains and plasmid vector.
Microorganisms capable of metabolizing 1,8-cineole were isolated from activated sludge (obtained from Lilydale Sewage Treatment Plant, Victoria, Australia) using a modified continuous culture method (24) in combination with selection based on growth with 1,8-cineole as the sole source of carbon and energy (12). One isolate, S. yanoikuyae strain B2, was chosen for this study because of its ability to grow in relatively high concentrations of 1,8-cineole of up to 2 g liter−1 (23). The S. yanoikuyae type strain DSM 7462 (ATCC 51230) was obtained from the Leibniz Institute DSMZ–German Collection of Microorganisms and Cell Cultures GmbH. Unless stated otherwise, E. coli strain BL21(DE3) was used for protein expression and E. coli strain DH5α was used for cloning. The pET28a(+) vector was from Novagen.
Media.
Liquid media used in this study were defined medium [DM; 10.64 g liter−1 KH2PO4, 4 g liter−1 (NH4)2HPO4, 1.7 g liter−1 citric acid monohydrate, 0.62 g liter−1 MgSO4·7H2O, and 4.4 mg liter−1 thiamine hydrochloride supplemented with 5 ml liter−1 trace salts solution containing 2.0 g liter−1 Cu2SO4·5H2O, 0.08 g liter−1 NaI, 3.0 g liter−1 Mn2SO4·H2O, 0.2 g liter−1 Na2MoO4·2H2O, 0.02 g liter−1 boric acid, 0.5 g liter−1 CoCl2·6H2O, 7.0 g liter−1 ZnCl2, 22.0 g liter−1 FeSO4·7H2O, 0.5 g liter−1 CaSO4·2H2O, and 1 ml liter−1 H2SO4; DM was adjusted to pH 7.0 as required], DM 2 (461S) (12), terrific broth (TB; 12 g liter−1 tryptone, 24 g liter−1 yeast extract, 4 ml liter−1 glycerol, 9.4 g liter−1 K2HPO4, and 2.2 g liter−1 KH2PO4), and Miller's lysogeny broth (LB) (25). Carbon sources, antibiotics, and other additives were added as specified in the description of each experiment.
Metabolite characterization.
The metabolites produced by S. yanoikuyae strain B2 were purified and characterized using techniques described previously (12). Briefly, compounds produced during growth on 1,8-cineole were initially detected using gas chromatography (GC) of filtered culture supernatants. The metabolites were then identified in dichloromethane extracts from 500-ml cultures. Cells were grown in DM2 with 1,8-cineole as the sole carbon source in 2-liter baffled flasks shaken at 200 rpm and 30°C. After purification, the compounds in the solvent extracts were identified using GC-mass spectrometry (GC-MS) and nuclear magnetic resonance (NMR).
Genome sequencing and strain identification.
Genomic DNA was extracted from the pellet of a 10-ml culture grown in DM2 containing 0.5 ml liter−1 1,8-cineole using the DNeasy blood and tissue kit (Qiagen) according to the manufacturer's instructions. Genomic DNA was prepared for Illumina MiSeq sequencing (University of New South Wales, Ramaciotti Centre) using the Nextera library preparation kit (Illumina). Quality filtering was performed using NESONI (version 0.128; https://github.com/Victorian-Bioinformatics-Consortium/nesoni) to remove low-quality base calls and residual adaptor sequences from the read sets. A draft genome sequence was constructed using the VELVET de novo genome assembler, version 1.2.07 (26); optimized assembly conditions were determined manually by evaluating assembly metrics from assemblies performed over a range of k-mer values. The draft genome sequence was annotated using Prokka version 1.7.1 (27); Prokka predicts both protein-coding genes and RNA-coding genes, with the function of protein-coding regions being inferred by protein similarity to characterized proteins. Isolate identity was determined using the RDP classifier (28) using the partial 16S rRNA gene sequence extracted from the draft strain B2 genome.
Isolation of 1,8-cineole-binding P450s from S. yanoikuyae strain B2.
S. yanoikuyae strain B2 cells were produced in a 2-liter stirred tank bioreactor (Biostat B, Sartorius Stedim) in fed-batch mode using glucose and 1,8-cineole as carbon sources. Using a 3-step purification procedure, 1,8-cineole-binding P450s were purified from crude cell extracts and identified using N-terminal sequencing and/or matrix-assisted laser desorption ionization (MALDI)-MS fingerprint mapping. Additional experimental details can be found in the supplemental material.
Molecular cloning.
Genes encoding proteins CYP101J2, CYP101J3, and CYP101J4 (systematic P450 names were assigned by David R. Nelson) were amplified by PCR using genomic DNA from S. yanoikuyae strain B2 as the template. The following primer pairs were used: fwCYP101J2 (5′-GGAATTCCATATGGAAGCAAGCGTGAAGGGC-3′) and revCYP101J2 (5′-CCGCTCGAGTTAGACTGGCCATTCGAGCAC-3′), fwCYP101J3 (5′-GGAATTCCATATGGCTGAACAGGCTGTTCTCG-3′) and revCYP101J3 5′-CCGCTCGAGTTACTGCCGCGGCCAGC-3′, and fwCYP101J4 (5′-GGAATTCCATATGGAAATCGGAACGATTGAACC-3′) and revCYP101J4 (5′-CCGCTCGAGTTAGACATCCCATTCGAGCATG-3′). Restriction sites NdeI and XhoI, introduced for cloning into the multiple cloning site of pET28a(+), are underlined. The thermocycler was preheated to 98°C. After initial denaturation at 98°C for 1 min, the genes were amplified by 30 cycles of denaturation at 98°C for 10 s, followed by annealing and extension at 72°C for 40 s and a final extension step at 72°C for 10 min. Original stop codons (TGA) were swapped to TAA. After ligation into the vector, DNA sequencing was used to confirm that the gene was inserted correctly.
Expression and purification of recombinant P450s.
Chemically competent E. coli BL21(DE3) cells were freshly transformed with pET28a(+) harboring genes encoding CYP101J2, CYP101J3, or CYP101J4, respectively, and plated on LB agar containing 50 μg ml−1 kanamycin sulfate (Kan). A single transformant was used to inoculate 50 ml of TB (in a 250-ml nonbaffled shake flask) containing 50 μg ml−1 Kan. Seed cultures were incubated with shaking at 180 rpm for 18 h at 30°C, and then this culture was used to inoculate 500 ml of TB supplemented with 50 μg ml−1 Kan in 2-liter baffled shake flasks to an optical density at 600 nm (OD600) of approximately 0.1. When the OD600 reached 0.6 to 0.8, cultures were induced with 1 mM IPTG, grown for a further 18 h, and then harvested by centrifugation at 6,000 × g for 20 min at 4°C. Cell pellets were washed in 50 mM Tris, 150 mM NaCl buffer, pH 7.4, and then stored at −80°C. Cells were resuspended in the same buffer containing 5 mM imidazole and then disrupted by two passages through an Avestin EmulsiFlex-C5 cell disruptor. After centrifugation, the clarified cell lysate was loaded onto a column with a bed volume of 2 to 3 ml of cobalt-charged resin (Talon Superflow metal affinity resin; Clontech) equilibrated with 50 mM Tris, 150 mM NaCl, and 5 mM imidazole, pH 7.4. After a washing step, the His6-tagged protein was eluted by a stepwise increase of imidazole. Fractions with similar purities as judged by SDS-PAGE were pooled and buffer exchanged into 50 mM Tris, pH 7.4, by repeated cycles of concentration and dilution using Amicon Ultra-15 centrifugal filter units with a molecular mass cutoff of 10 kDa. Concentrated protein solutions were then snap-frozen using a dry ice-ethanol (EtOH) bath and stored at −80°C.
P450 functionality.
Whole-cell biohydroxylation was achieved by expressing the P450 cytochromes in E. coli BL21(DE3) as described above, except for the following changes. Just after induction with 1 mM IPTG, 0.5 ml liter−1 1,8-cineole was added to the 500-ml cultures and the culture was incubated for a further 45 h. A 1-ml aliquot of the culture was then extracted with ethyl acetate, dried over Na2SO4, and analyzed using GC-MS. GC-MS was performed using a Thermo Scientific TSQ8000 GC mass spectrometer using electron impact ionization in the positive ion mode with an ionization energy of 70 eV. The gas chromatography was performed with a Thermo Scientific TG-5MT column (inner diameter of 15 m by 0.25 mm, 0.10-μm film thickness), with a temperature program of 50°C for 2 min and then heating at 25°C per min to 300°C, where the temperature was held for 3 min using an injection with a split of 10 and an injector temperature of 300°C, and the transfer line was set to 300°C. High-purity helium was used as the carrier gas at a flow rate of 1.0 ml min−1.
Spectroscopic characterization of P450s.
Spectra of oxidized protein solutions were prepared in 50 mM Tris, pH 7.4. 1,8-Cineole-induced absorbance shifts were demonstrated by adding 1 μl of 1,8-cineole to 1 ml of protein solution. The P450s were then reduced by the addition of a small amount of solid sodium dithionite and incubation for 1 min. Subsequently, carbon monoxide (CO) was bubbled through the cuvettes for 30 s. Spectra were recorded from 350 to 700 nm. The concentration of functional P450 was estimated using CO difference spectroscopy using an extinction coefficient (ε) of 91 mM−1 cm−1 (29, 30).
Determination of 1,8-cineole Kd.
The determination of dissociation constants (Kd) was performed using a procedure similar to that described by Bell et al. (31). Enzymes were diluted to ca. 2 μM in 50 mM Tris, pH 7.4, or 50 mM Tris, 200 mM KCl, pH 7.4, respectively. Stock solutions of 1,8-cineole in EtOH with a concentration range of 1 to 600 mM were prepared by serial dilution. To 1 ml of protein solution, 1-μl aliquots of 1,8-cineole stock solutions of various concentrations were added sequentially. After mixing, an absorbance spectrum between 350 and 450 nm was measured. Reference spectra of protein solutions containing EtOH instead of substrate solutions were subtracted from the sample absorbance spectra to obtain 1,8-cineole-induced difference spectra. Further, 1-μl aliquots of 1,8-cineole stock solutions were added until the absorbance difference between peak and trough (ΔA) reached its maximum; the maximum total amount of added substrate solution was 1% (vol/vol) of the sample. Kd values were calculated by fitting the ΔA against the 1,8-cineole concentration ([S]) to the hyberbolic function ΔA = (ΔAmax × [S])/(Kd + [S]). Alternative substrates were screened for the induction of absorbance shifts by adding solutions of 2-adamantanone, β-ionone, (1R)-(+)-camphor, (1S)-(−)-camphor, α-terpineol, and toluene in EtOH to the protein solutions to a final concentration of 1.2 mM (irrespective of substrate solubility) when the ΔA did not show any significant further increase. The spin-state shifts corresponding to the ΔA were measured in the absence and presence of 0.2 M KCl for each substrate tested. The spin-state shift with 1,8-cineole and 0.2 M KCl was set at 100% for each P450, and all other type I shifts were compared with this value.
Accession number(s).
The assembly described in the manuscript is available for download as “Sphingobium_yanoikuyae_strain_B2_scaffolds.zip” from the CSIRO Data Access Portal (https://data.csiro.au/dap/landingpage?pid=csiro:17150). Annotations for this assembly, along with contiguous sequences, sequences for putative and confirmed P450 cytochromes, and raw reads are also available for download at the CSIRO Data Access Portal. In addition, contiguous sequences over 1,000 bp were also uploaded to the NCBI database and are available under BioProject PRJNA316001. This whole-genome shotgun project has been deposited at DDBJ/ENA/NCBI under the accession number LVJD00000000. The version described in this paper is version LVJD01000000. Sequences for putative and confirmed cytochrome P450 monooxygenases are also available at GenBank under the accession numbers KX496991 (Sya_B2_00569), KX496992 (Sya_B2_01856), KX496993 (Sya_B2_02741), KX496994 (Sya_B2_02767; CYP101J4), KX496995 (Sya_B2_03538; CYP101J3), KX496996 (Sya_B2_03558; CYP101J2), KX496997 (Sya_B2_04893), KX496998 (Sya_B2_04918), and KX496999 (Sya_B2_05084). The partial 16S rRNA gene sequence used to classify strain B2 is available at GenBank under the accession number KX507119.
RESULTS
Strain identification and genomic analysis.
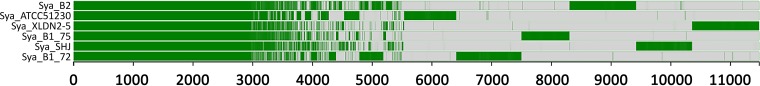
Previously, a Gram-negative, rod-shaped bacterial isolate, designated strain B2, which was capable of using 1,8-cineole as a sole carbon and energy source, was isolated from activated sludge (23). To characterize this strain further, genome sequence analysis was performed on this isolate. Genome sequencing and assembly of the filtered reads using Velvet yielded a draft genome sequence of approximately 5.9 Mb containing 263 scaffolds. These scaffolds varied in length and were distributed with mean, median, and N50 lengths of 22,468 bp, 7,012 bp, and 60,180 bp, respectively. The longest scaffold was 178,243 bp in length, while the average GC content of the genome was 62.3%. A 1,484-bp segment of the 16S rRNA gene sequence was extracted from the draft strain B2 genome and used to classify the strain as Sphingobium using the RDP classifier (28). All characteristic 16S rRNA signatures that distinguish this genus (32) were present in the sequence of strain B2, and it matched closely (≥99%) with members of the species S. yanoikuyae, including the type strain (ATCC 51230 [DSM 7462]). Hence, it was designated S. yanoikuyae strain B2. A comparison of 16S rRNA gene sequences from a selection of strains belonging to the genus Sphingobium is shown in Fig. S2 in the supplemental material. The S. yanoikuyae isolates appear as a single clade which contains no taxa from other species. The percent identity of the S. yanoikuyae strains for which a draft genome sequence is available to an outgroup taxon (Sphingobium indicum B90A) was calculated by counting the number of k-mers (25-mers) shared between each pair of organisms, effectively the D2 metric (33, 34). The code used to calculate this relatedness metric differs from conventional D2 tools in that it compensates for the effects of single mismatched bases, giving counts that more closely approximate those obtained from multiple-alignment tools. The comparison shows that the S. yanoikuyae strains share 68 to 80% genome identity (see Table S4). A comparison of the distribution of orthologous proteins in each of the six S. yanoikuyae strains (annotated using Prokka) was derived using Roary version 3.6.0 (35), and the data were displayed using FriPan (http://drpowell.github.io/FriPan/). Approximately 1,000 proteins are unique to strain B2 (Fig. 1).
FIG 1.
Overview of the distribution of orthologous proteins in each of the six sequenced S. yanoikuyae strains (Sya_B2, BioProject ID PRJNA316001; Sya_ATCC51230, BioProject ID PRJNA52201; Sya_XLDN2-5, BioProject ID PRJNA71691; Sya_B1_75, BioProject ID PRJNA255061; Sya_SHJ, BioProject ID PRJNA239177; and Sya_B1_72, BioProject ID PRJNA241283) displayed using FriPan (http://drpowell.github.io/FriPan/). The region of continuous green across the six lines (each representing an S. yanoikuyae strain) spanning from 0 to ca. 3,000 indicates the proteins that are present in each of the strains. The region from ca. 3,000 through ca. 5,500 shows the set of proteins that are present in two to five strains. The region from ca. 5,500 through ca. 11,400 is an overview of the proteins that are unique to particular strains.
Metabolite characterization.
To determine the metabolic intermediates produced by S. yanoikuyae strain B2 from 1,8-cineole, GC analysis of aqueous samples taken from shake flask culture supernatants was performed. In exponential phase, residual 1,8-cineole was detected, as well as three other compounds with different retention times. No 1,8-cineole or compounds found in exponential cultures were detected in stationary-phase cultures; however, a further two compounds with longer retention times were observed (data not shown). Solvent extraction of 500-ml cultures provided a sufficient quantity of each compound to enable identification. Using previously described methods (NMR and GC-MS) (12), the compounds found in the dichloromethane extracts from exponential-phase cultures were identified as the α and β forms of hydroxylated 1,8-cineole and oxo-1,8-cineole (oxidized at position 2 or 6); only 5,5-dimethyl-4-(3′-oxobutyl)-4,5-dihydrofuran-2(3H)-one) was identified in solvent extracts from stationary-phase cultures. These compounds have been described previously and are consistent with 1,8-cineole-derived products extracted from cultures of N. subterranea (12).
Identification of P450s. (i) Genes encoding P450s in genome sequence.
After annotation of the draft genome, nine P450-like proteins were identified. The nine proteins included six putative camphor-hydroxylating P450s, one putative α-terpineol-oxidizing P450, one putative cytochrome P450 PikC, and one putative pentalenene oxygenase. A BLAST search of the UniProt database (http://www.uniprot.org/blast) showed that these putative P450s are similar to a number of proteins, including putative P450s from other S. yanoikuyae strains, but four of the P450s were absent from the type strain genome (see Table S1 in the supplemental material). Two of these genes, CYP101J2 and CYP101J3, appear to be located on a single contiguous sequence with complementary, overlapping ends that appears to be a plasmid. This plasmid was 73,051 bp in length and had a GC content of 61.2%. The sequence for the putative plasmid is available at the CSIRO Data Access Portal (https://data.csiro.au/dap/landingpage?pid=csiro:17150). BLASTn analyses of the putative plasmid indicate its closest matches are to plasmids from Sphingobium chlorophenolicum (pSPHCH01), Sphingobium sp. strain YBL2 (2pYBL2-2), and Sphingomonas wittichii RW1 (pSWIT02), although it shares only low identity (73%; over 10 to 20% query coverage) with these plasmids.
(ii) Growth of S. yanoikuyae strains B2 and DSM 7462 on selected monoterpenoids.
To determine whether the ability to grow on 1,8-cineole is ubiquitous in S. yanoikuyae strains, strain B2 and the type strain S. yanoikuyae DSM 7462 (ATCC 51230) were tested for growth in DM2 supplemented with 0.25 to 1 ml liter−1 1,8-cineole, 0.25 to 1 ml liter−1 α-terpineol, or 0.25 to 1 g liter−1 (1R)-(+)-camphor or (1S)-(−)-camphor as sole sources of carbon and energy. Growth in the supplemented media at 30°C and 200 rpm was monitored over a 7-day time period. Following 7 days of incubation, visual examination showed that S. yanoikuyae DSM 7462 did not grow on any of the substrates tested, while S. yanoikuyae strain B2 grew on both 1,8-cineole and α-terpineol but showed no growth on either of the camphor enantiomers.
(iii) Purification of 1,8-cineole-binding P450s from S. yanoikuyae strain B2.
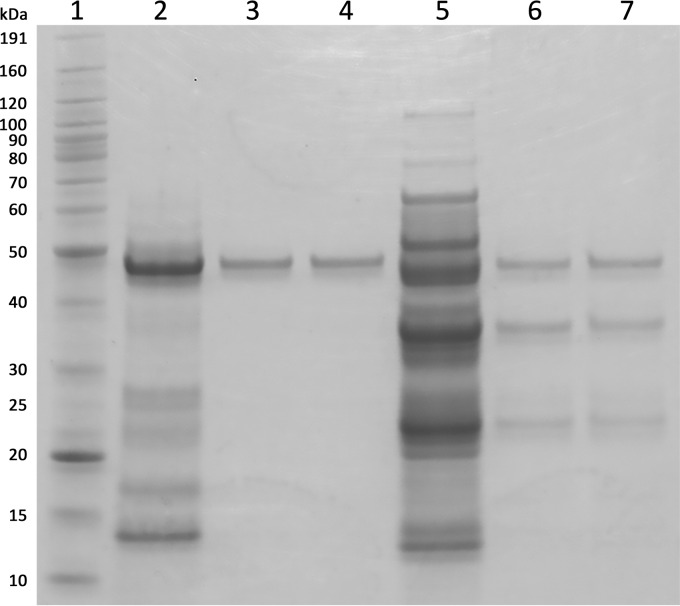
Confirmation of the genes encoding 1,8-cineole-hydroxylating P450s required purification of 1,8-cineole-binding P450s from S. yanoikuyae strain B2. Proteins were purified from extracts of S. yanoikuyae strain B2 cells grown in fed-batch mode using glucose and/or 1,8-cineole as carbon sources. Binding of 1,8-cineole was tested in partially purified protein fractions. Transition from the low-spin to the high-spin state (36) upon 1,8-cineole binding was detected in multiple peaks after ion-exchange chromatography (IEX) of ammonium sulfate (AS)-precipitated fractions. Two distinct peak fractions (PF), PF 1 and PF 2 from the 40 to 60% AS cut, were further purified using gel filtration (GF). After the 3-step purification procedure, PF 1 was purified to apparent homogeneity by SDS-PAGE analysis with an estimated mass of ca. 46 kDa, but PF 2 appeared to still contain impurities (Fig. 2). The purity was also reflected by the Reinheitszahl (RZ), which is defined as the ratio between P450 absorbance at its typical Soret absorbance for the LS state (in this case 417 nm) and the total protein absorbance at 280 nm (A417/A280). A higher RZ is indicative of higher protein purity (11). PF 1 and PF 2 had an RZ of 1.4 and 0.6, respectively, which correlated with the purity observed by SDS-PAGE analysis (Fig. 2).
FIG 2.
Coomassie-stained SDS-PAGE (4 to 12% Bis-Tris, morpholineethanesulfonic acid running buffer) of PF 1 and 2 after IEX and GF. Lane 1, molecular mass marker (BenchMark; Invitrogen); lane 2, PF 1 after IEX; lanes 3 and 4, PF 1 after GF; lane 5, PF 2 after IEX; lanes 6 and 7, PF 2 after GF.
After concentration, reduction, and alkylation, PF1 was further purified using reversed-phase high-performance liquid chromatography. N-terminal sequencing of the fraction with a molecular mass of approximately 46 to 47 kDa, the typical size for P450s, produced a readable sequence of 22 amino acids. Protein identity was confirmed by a tryptic digest of the reduced and alkylated protein followed by MALDI-MS fingerprint mapping. Together with the genome sequence, the MS data were used to identify a gene encoding a P450 (Sya_B2_03558; CYP101J2) with a length of 410 amino acids.
Protein identity for PF2 was determined by an in-gel tryptic digest of the protein with a mass of ca. 46 kDa, followed by MALDI-MS fingerprint mapping. The peptide sequences gave more than 30% coverage of a P450 (Sya_B2_03538; CYP101J3) with a length of 406 amino acids. A third protein (Sya_B2_02767; CYP101J4) was identified as a potential 1,8-cineole-hydroxylating P450 based on high amino acid sequence identity (74%) to CYP101J2.
Enzyme characterization. (i) Recombinant expression and purification of P450s.
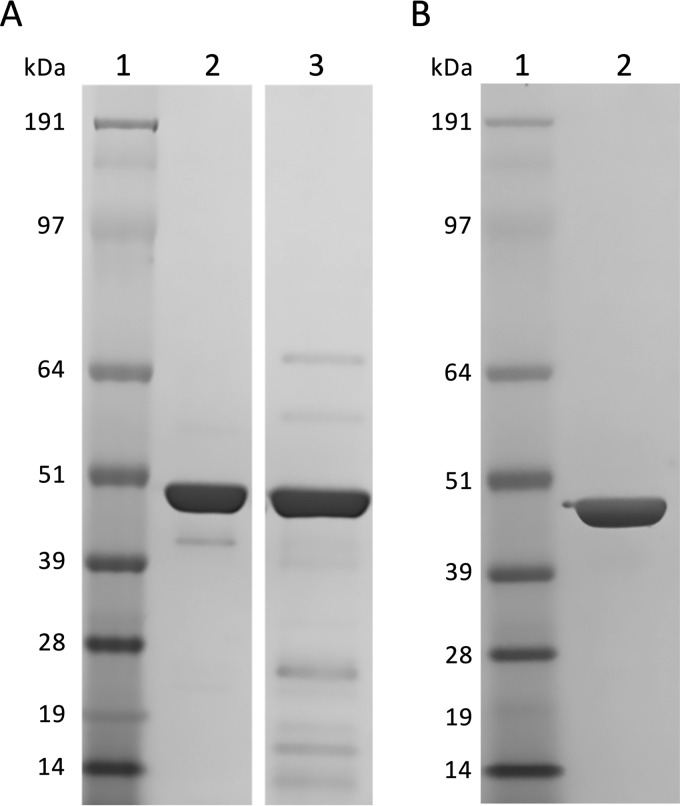
To further characterize the identified P450s, the genes encoding the three identified 1,8-cineole-binding P450s were cloned into the pET28a(+) expression vector and expressed in E. coli BL21(DE3). This yielded N-terminally His6-tagged proteins, which were purified using immobilized metal affinity chromatography (IMAC). The molecular masses estimated from the protein sequences, including tags (CYP101J2, 48.7 kDa; CYP101J3, 47.8 kDa; CYP101J4, 48.4 kDa), matched the main expression products (Fig. 3). The yield of active protein was estimated in the purified pools by performing CO-difference spectroscopy in the presence of 1,8-cineole. CYP101J2 and CYP101J3 were both recovered in good yields (>30 mg purified protein per liter of original culture). CYP101J4 was expressed in lower quantities as judged by SDS-PAGE of IMAC fractions (data not shown), and this was reflected by the amount of functional protein recovered (ca. 1.5 mg purified protein per liter of original culture).
FIG 3.
Coomassie-stained SDS-PAGE (4 to 12% Bis-Tris, morpholinepropanesulfonic acid running buffer) of purified His6-tagged P450s (ca. 2 μg) originating from S. yanoikuyae strain B2 recombinantly expressed in E. coli BL21(DE3). (A) Lane 1, molecular mass marker (SeeBlue plus 2 prestained; Invitrogen); lane 2, purified CYP101J2; lane 3, purified CYP101J4. (B) Lane 1, molecular mass marker (SeeBlue plus 2 prestained; Invitrogen); lane 2, purified CYP101J3.
(ii) Spectroscopic characterization.
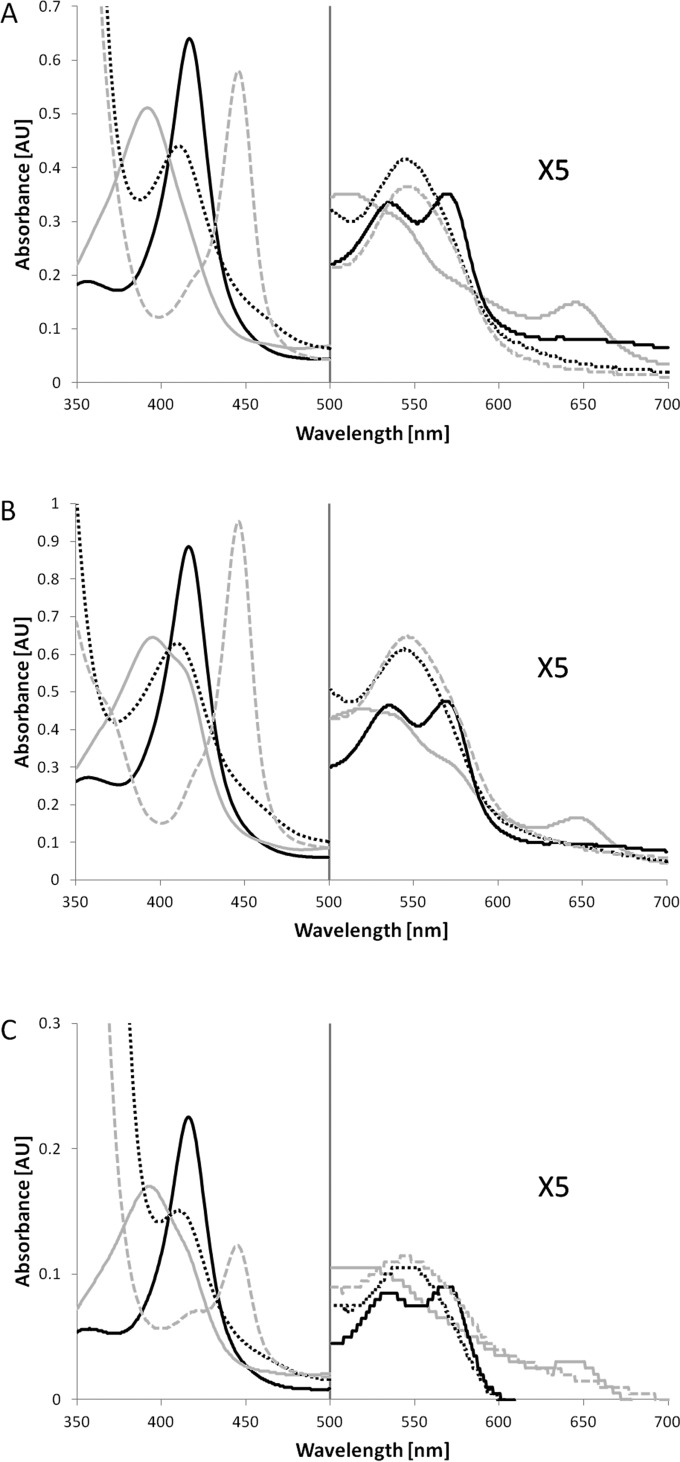
All three isolated and purified P450s showed the characteristic spectroscopic properties expected for P450s (Fig. 4). Most importantly, the typical Soret absorbance maximum (416 to 417 nm) largely shifted to lower wavelengths (392 to 396 nm) upon addition of 1,8-cineole. After reduction and CO binding, the P450s exhibited the typical absorbance maximum at ca. 450 nm (445 to 446 nm).
FIG 4.
Absorbance spectra of CYP101J2 (A), CYP101J3 (B), and CYP101J4 (C) in 50 mM Tris, pH 7.4, of the different forms: oxidized (full black line), oxidized in the presence of 1,8-cineole (full gray line), reduced by the addition of sodium dithionite in the presence of 1,8-cineole (broken black line), and reduced and in complex with CO in the presence of 1,8-cineole (broken gray line). AU, arbitrary units.
(iii) Substrate binding.
The purified P450s were tested for their ability to bind 1,8-cineole. The dissociation constants (Kd) determined with 1,8-cineole as the substrate are summarized in Table 1. Kd values are in the same order of magnitude for all three P450s. CYP101J4 shows slightly tighter substrate binding (by a factor of ca. 2). Binding of 1,8-cineole was increased by a factor of approximately 2 when 0.2 M KCl was present. Under the tested conditions, ΔAmax was largest using 1,8-cineole in the presence of KCl for CYP101J2, CYP101J3, or CYP101J4. Compared to the shift of each individual P450 with 1,8-cineole and 0.2 M KCl, addition of 1,8-cineole resulted in a 95% (CYP101J2), 85% (CYP101J3), and 90% (CYP101J4) shift in the absence of potassium chloride.
TABLE 1.
Kd values for 1,8-cineole binding in the absence and presence of KCl
| P450 |
Kda (μM) |
|
|---|---|---|
| Without KCl | With KCl | |
| CYP101J2 | 20.4 ± 2.6 | 10.1 ± 1.0 |
| CYP101J3 | 22.1 ± 0.8 | 9.5 ± 0.5 |
| CYP101J4 | 8.4 ± 0.6 | 5.1 ± 0.1 |
Kd values were determined for 1,8-cineole binding in the absence and presence of 0.2 M potassium chloride in 50 mM Tris, pH 7.4. Kd values are reported as means ± standard deviations (n ≥ 3).
CYP101J2 and CYP101J3 were also screened for binding of alternative substrates [2-adamantanone, toluene, β-ionone, (1S)-(−)-camphor, and (1R)-(+)-camphor], and percent spin-state shifts are summarized in Table 2. The percentages of type I spin-state shifts induced by these substrates were estimated to within approximately ±5% by comparison to the shift of each individual P450 with 1,8-cineole in the presence of 0.2 M KCl. While β-ionone triggered a 40% (CYP101J2) or 35% (CYP101J3) shift, binding of 2-adamantanone, (1S)-(−)-camphor, and (1R)-(+)-camphor resulted in small shifts between 5 and 20%. Toluene addition resulted in no detectable spin-state shift. The enzymes did not show a type I shift with α-terpineol. However, there was a shift from ca. 410 nm to ca. 426 nm, indicating a type II shift. Due to the low expression levels of CYP101J4, alternative substrates were not tested with this enzyme.
TABLE 2.
Percent absorbance difference induced by alternative substrates compared to ΔAmax induced by 1,8-cineole
| Substrate | % spin-state shifta (type I) |
|
|---|---|---|
| CYP101J2 | CYP101J3 | |
| β-Ionone | 40 (40) | 35 (35) |
| (1R)-(+)-Camphor | 10 (15) | 15 (15) |
| 2-Adamantanone | 10 (10) | 15 (15) |
| (1S)-(−)-Camphor | 5 (10) | 5 (5) |
Values shown are percent absorbance differences induced by alternative substrates in 50 mM Tris, pH 7.4, compared to ΔAmax induced by 1,8-cineole in the presence of 0.2 M KCl (estimated to within ca. ±5%). Data obtained in the same buffer in the presence of 0.2 M KCl are in parentheses.
(iv) Functionality of recombinant P450s in E. coli.
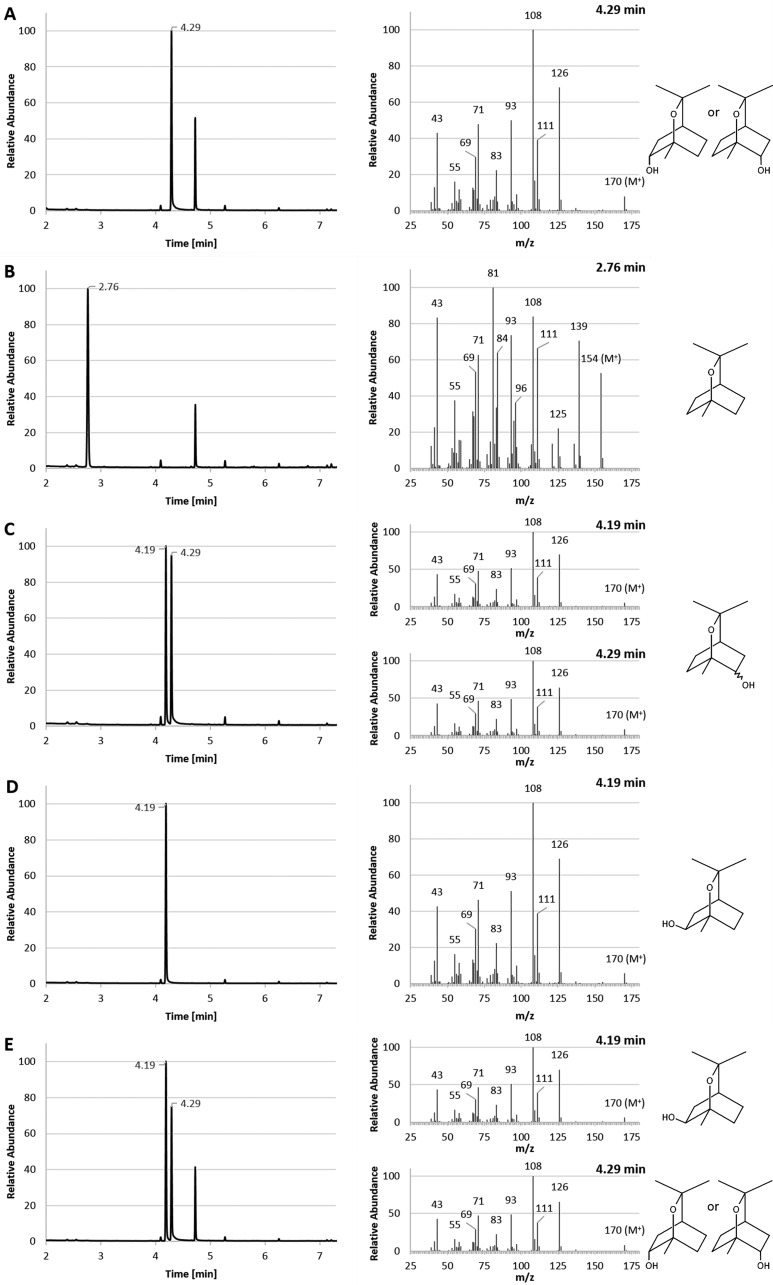
E. coli cultures expressing recombinant CYP101J2 in the presence of 1,8-cineole were solvent extracted and analyzed using GC-MS. Forty-five h after addition of 1,8-cineole, a peak with a retention time (RT) of 4.29 min was detected (Fig. 5A). This peak had a mass increase of 16 U with a molecular peak at m/z = 170 and the typical fragmentation pattern of hydroxylated 1,8-cineole (6). It was identified with a 91 to 94% probability match to 1,3,3-trimethyl-2-oxabicyclo[2.2.2]octan-6-ol in the National Institute of Standards and Technology (NIST) 14 (MS Database and MS Search Program v.2.2) library, containing spectra for 242,466 chemical compounds. This peak was not detected in the negative control [E. coli BL21(DE3) harboring the pET28a(+) vector alone] (Fig. 5B). Analysis of a mixture of chemically synthesized α and β forms of (1S)-2-hydroxy-1,8-cineole showed that these two stereoisomers can be separated using nonchiral GC-MS (RT of 4.19 and 4.29 min) (Fig. 5C). Analysis of (1R)-6β-hydroxy-1,8-cineole (produced using P450cin isolated from C. braakii) using the same technique yielded a peak with an RT of 4.19 min (Fig. 5D). When (1R)-6β-hydroxy-1,8-cineole was mixed with a solvent extract from a culture expressing CYP101J2, two distinct GC peaks were detected (RT of 4.19 min and 4.29 min) with highly similar MS data (Fig. 5E). Since monohydroxylated 1,8-cineole from this study was separated from (1R)-6β-hydroxy-1,8-cineole) using nonchiral GC and had the same RT as one of the α and β forms of (1S)-2-hydroxy-1,8-cineole, we putatively identified the product of CYP101J2 as (1S)-2α-hydroxy-1,8-cineole or (1R)-6α-hydroxy-1,8-cineole. The same data were obtained when testing CYP101J3 and CYP101J4 under identical conditions (data not shown).
FIG 5.
GC-MS analysis and comparison to known chemical standards with solvent extracts from E. coli cultures expressing CYP101J2. Only relevant peaks are labeled with RT. Unlabeled peaks are impurities. (A) The solvent extract from a culture expressing CYP101J2 in the presence of 1,8-cineole showed a peak at 4.29 min. The mass spectrum of this peak was identical to the mass spectra of the hydroxy-1,8-cineole standards. (B) The negative control [solvent extract from E. coli BL21(DE3) culture harboring the pET28a(+) vector alone] showed a peak at an RT of 2.76 min, corresponding to 1,8-cineole; no hydroxy-1,8-cineole was detected. (C) The α and β forms of chemically synthesized (1S)-2-hydroxy-1,8-cineole separated using nonchiral GC-MS (RT of 4.19 and 4.29 min) and showed the characteristic mass spectra. (D) Purified (1R)-6β-hydroxy-1,8-cineole produced using P450cin showed a peak at an RT of 4.19 min and the characteristic mass spectrum. (E) A mixture of purified (1R)-6β-hydroxy-1,8-cineole and CYP101J2 solvent extract showed that the product of CYP101J2 separates from (1R)-6β-hydroxy-1,8-cineole.
DISCUSSION
The industrial application of biocatalytic processes requires enzymes with specific catalytic ability, and on many occasions these enzymes are discovered in microbial isolates with particular metabolic capabilities. To enable development of cost-effective production of 1,8-cineole derivatives, we previously isolated a number of microorganisms with the ability to utilize 1,8-cineole as a sole carbon and energy source (23). Unlike earlier studies, during which microorganisms were isolated from environments that were likely to contain 1,8-cineole (e.g., soil proximate to Eucalyptus trees and Eucalyptus leaves) (8, 9, 11, 13), our approach was to interrogate a potentially more diverse microbial consortium: activated sludge. This approach is not without precedent. Terpene oxidation in a microorganism from sewage sludge was reported as early as 1959, when an isolate that was identified as a pseudomonad was shown to utilize camphor as a carbon source (37).
In this study, one isolate, strain B2, demonstrating a relatively high tolerance to 1,8-cineole (23), was chosen for further study. A draft genome sequence was determined and the strain B2 was identified as S. yanoikuyae based on its partial 16S rRNA gene sequence extracted from the genome sequence. While there may be more than one copy of the 16S rRNA gene, this was not determined, and it is unlikely that potential sequence variation would be significant enough to change the taxonomic classification. Interestingly, compared to other S. yanoikuyae draft genomes, the sequence of strain B2 is relatively large. A comparison of the sequenced strains of S. yanoikuyae is shown in Table S4 in the supplemental material. There appears to be more genetic difference between Sphingobium yanoikuyae strains than is normal for a typically defined bacterial species. This likely accounts for the differences in genome size and indicates further taxonomic scrutiny is warranted.
The isolate belongs to the group of sphingomonads, bacteria that are known to degrade a diverse range of recalcitrant compounds, in particular environmental pollutants (38). This potential metabolic diversity may also be related to modification and degradation of terpenes. Previously, P450s isolated from N. aromaticivorans DSM12444 were shown to hydroxylate camphor (39), and a Sphingomonas isolate was capable of metabolizing 1,8-cineole (13). However, to our knowledge this is the first report of 1,8-cineole metabolism by a sphingomonad from the genus Sphingobium. A comparison of the distribution of orthologous proteins in each of six sequenced S. yanoikuyae strains also indicates metabolic flexibility among members of this species. A significant fraction of proteins was unique to each strain, with approximately 1,000 proteins being unique to strain B2 (Fig. 1).
Analysis of S. yanoikuyae strain B2 cultures grown with 1,8-cineole as a sole carbon and energy source enabled identification of a series of compounds in solvent extracts of culture supernatants. Three compounds were identified in exponential-phase cultures (the α and β forms of hydroxylated 1,8-cineole and oxo-1,8-cineole). The presence of two forms of hydroxy-1,8-cineole may be due to the reduction of oxo-1,8-cineole by the organisms rather than a direct product of 1,8-cineole hydroxylation (1). These compounds are typical metabolites derived from 1,8-cineole (8, 9, 12, 13). Based on the analytical techniques used, the enantioselectivity of the initial oxidation could not be established; therefore, the oxidation may occur at position 2 or 6. Although two different compounds were identified in aqueous samples from stationary-phase cultures, only one compound was recovered from solvent extracts of these cultures. The single compound was identified as 5,5-dimethyl-4-(3′-oxobutyl)-4,5-dihydrofuran-2(3H)-one). Williams et al. (9) also detected this compound in solvent extracts from crude extracts of Rhodococcus sp. strain C1 incubated with 6-oxo-1,8-cineole and concluded that this compound is not derived from metabolism of 1,8-cineole but is in fact an artifact of the acidification and solvent extraction procedure. A more likely product is 3-(1-hydroxy-1-methylethyl)-6-oxoheptanoic acid, which could be one of the compounds detected in aqueous supernatant samples; however, production of this compound from 1,8-cineole by S. yanoikuyae strain B2 was not confirmed.
The metabolites produced by S. yanoikuyae B2 included hydroxylated 1,8-cineole; hydroxylation is typically the first step in the metabolism of 1,8-cineole. To gain greater insight into this initial reaction, we sought to identify and characterize the enzyme(s) involved in catalysis. Through sequencing of the selected strain's genome, several genes potentially encoding P450s were identified based on similarity to known P450 sequences. These included six genes putatively encoding P450s that were similar to known camphor-hydroxylating P450s and one which was similar to an α-terpineol-oxidizing P450 (see Table S1 in the supplemental material). This was encouraging, as both camphor and α-terpineol are monoterpenoids like 1,8-cineole, and camphor in particular shares structural similarity to 1,8-cineole. However, sequences of P450s are diverse (40), and the hydroxylation of similar substrates is often catalyzed by a range of divergent P450s from different families (41). In all, nine P450s (putatively annotated) were found; consequently, further characterization of the native proteins was required to determine function.
As a first step toward elucidating the P450s involved in 1,8-cineole hydroxylation, the metabolic ability of S. yanoikuyae strain B2 was compared to that of the type strain (DSM 7462 [ATCC 51230]). Unlike S. yanoikuyae B2, strain DSM 7462 was not able to grow on 1,8-cineole or α-terpineol. Through comparison of the genomes from the two strains, this observation correlated with the absence of orthologous proteins of the four P450s in the type strain that were present in S. yanoikuyae B2 (see Table S1 in the supplemental material). Therefore, these genes were implicated in 1,8-cineole (and/or α-terpineol) hydroxylation. However, it is also possible that enzymes catalyzing subsequent steps in the degradation of the tested monoterpenoids are absent from the S. yanoikuyae strain DSM 7462; hence, further investigation was required to confirm the function of these genes.
Other monoterpenoid-oxidizing P450s, such as CYP176A1 (P450cin) and CYP108A1 (P450terp), were isolated and identified via purification from native cell extracts (11, 42). To facilitate a similar approach, a 3-step purification procedure was developed to isolate 1,8-cineole-binding P450s from S. yanoikuyae strain B2 cell extracts. Two proteins were purified, and partial amino acid sequencing and subsequent comparison with translated genes from the genome sequence resulted in the identification of two 1,8-cineole-binding proteins, CYP101J2 and CYP101J3. A third P450, CYP101J4, was identified based on sequence identity to CYP101J2. These protein sequences represent three of the four P450s which were absent from the strain ATCC 51230 draft genome. The three identified proteins belong to the cytochrome P450 family CYP101. Pairwise sequence alignments of all characterized P450s from the CYP101 family as well as P450cin (CYP176A1) showed that CYP101J2 shares the highest sequence identity with CYP101J4 (74%). CYP101J3 shares sequence identity with CYP101J2 (53%) and CYP101J4 (54%). Compared to previously characterized members of the CYP101 family, all three newly discovered P450s are most closely related to CYP101B1 and CYP101A1, sharing 45 to 47% and 44% sequence identity, respectively. CYP101B1 has been shown to oxidize a broad range of substrates, including norisoprenoids (e.g., β-ionone), various aromatic compounds, and camphor (39, 43). CYP101A1 was characterized based on its activity with camphor (18). More recently, both CYP101B1 and CYP101A1 also have been shown to hydroxylate 1,8-cineole, producing a range of hydroxy-1,8-cineole isomers (14). The P450 from C. braakii is unique, as no other bacterial enzymes have been found that belong to the CYP176 family or use similar electron transfer partners. However, despite the enzymes from S. yanoikuyae strain B2 and C. braakii sharing less than 30% sequence identity, all of them oxidize the same substrate, indicating a high degree of flexibility within this large group of enzymes.
To characterize the cineole-binding proteins, CYP101J2, CYP101J3, and CYP101J4 were heterologously expressed in E. coli BL21(DE3) as N-terminally His6-tagged fusions and purified using IMAC. Recovery of functional proteins varied significantly. While CYP101J2 and CYP101J3 were obtained in good yield, expression of CYP101J4 under the selected conditions resulted in lower protein recovery. This may be due to improper folding (and/or the formation of insoluble aggregates), which could be resolved by modifying protein expression conditions. Spectroscopic characterization of the three purified recombinant proteins revealed the distinctive P450 properties. All 3 recombinant enzymes showed the typical P450 absorbance peaks and shifts, and most importantly, 1,8-cineole binding resulted in a large spin-state shift, indicating that 1,8-cineole is the preferred substrate. Kd values using 1,8-cineole as the substrate were in the same order of magnitude for all three P450s, and the presence of KCl appeared to result in tighter substrate binding (by a factor of ca. 2). Potassium ions have previously been shown to have a positive effect on substrate binding by other P450s (39, 44). Within the structure of CYP101A1, Glu84, Gly93, Glu94, and Tyr96 have been shown to be involved in binding of potassium ions (19, 45), with the latter three residues being conserved in the CYP101J enzymes from S. yanoikuyae B2. The presence of potassium ions has a small effect on 1,8-cineole binding by CYP101J2, CYP101J3, and CYP101J4. It is not clear whether the effect of potassium ions is specifically related to the interaction of potassium ions with the enzymes or a nonspecific effect due to altered ionic strength causing a change in substrate solubility. The affinity of CYP101J2, CYP101J3, and CYP101J4 for 1,8-cineole is less than that of P450cin (11) and is also lower than those for other monoterpenoid-hydroxylating P450s for their preferred substrates (18, 42).
Testing CYP101J2 and CYP101J3 with alternative substrates, including compounds which are converted by other P450s from the CYP101 family, did not lead to a type I spin-state shift greater than that induced by 1,8-cineole. For example, β-ionone and camphor triggered spin-state shifts of 35 to 40% and 10 to 15% with CYP101J2 and CYP101J3, respectively. These shifts are relatively small compared to the large shift observed when β-ionone binds to CYP101B1 (39) and camphor to CYP101A1 (46). S. yanoikuyae B2 was able to use both 1,8-cineole and α-terpineol as carbon sources for growth; however, α-terpineol did not induce a type I spin-state shift in either CYP101J2 or CYP101J3. This observation, combined with the minimal shifts triggered by the camphor enantiomers, indicates that 1,8-cineole is the physiological substrate for the newly discovered P450s. Furthermore, despite being shown to oxidize 1,8-cineole, CYP101A has a Kd value that is higher and a spin-state shift that is smaller (14) than those of the P450s from S. yanoikuyae. These functional variations due to sequence divergence and presumably structural differences warrant further investigations and comparison.
Testing of CYP101J2, CYP101J3, and CYP101J4 in a recombinant whole-cell biotransformation confirmed their proposed function. Presumably with the aid of electron transport partners from E. coli, all three enzymes catalyzed the oxidation of 1,8-cineole to a product tentatively identified as (1S)-2α-hydroxy-1,8-cineole or (1R)-6α-hydroxy-1,8-cineole. The product from the S. yanoikuyae enzymes is hydroxylated in the α-orientation compared to the product of the wild-type P450cin from C. braakii, which hydroxylates in the ß-orientation (7). Due to the symmetric nature of 1,8-cineole, NMR also would not enable differentiation between derivatives that are hydroxylated in either the 2 or 6 position. Interestingly, even though the 1,8-cineole-hydroxylating proteins from the two different microorganisms share limited sequence similarity and hydroxylate in different orientations, both are able to use electron transfer chains present in E. coli (11). Compared to the N242A P450cin mutant CYP101A1 and CYP101B1, which each produce a series of hydroxylated 1,8-cineoles, the CYP101J enzymes from S. yanoikuyae B2 appear to hydroxylate 1,8-cineole more specifically (14).
This research has shown that S. yanoikuyae strain B2 harbors at least three genes encoding enzymes with similar function; the presence of multiple genes that encode functionally similar proteins suggests that the action of these proteins is essential for growth or survival of the bacterium within specific environments. Functional redundancy is often the result of this essential need. Unlike other monoterpene-oxidizing P450s, including P450cin (11), P450terp (42), and P450cam (47), the genes encoding the P450s were not adjacent to potential electron transport partners in the draft genome sequence. This observation is typical, as genes encoding the enzymes involved in degradative pathways in sphingomonads are often scattered throughout a number of gene clusters within the genome (including plasmids) (31, 48, 49).
In this study, three new bacterial P450s capable of 1,8-cineole hydroxylation have been isolated from S. yanoikuyae strain B2. While these P450s provide a basis of comparison to other bacterial monoterpenoid-oxidizing P450s for mechanistic studies, these enzymes can also be used for process development and scaling up biochemical production of hydroxylated 1,8-cineole derivatives. Furthermore, as the number of enzymes in the CYP101 family grows, enzyme modification using DNA shuffling can be undertaken to engineer proteins with improved properties and altered substrate specificities. We are currently identifying and testing potential native electron transport partners from S. yanoikuyae strain B2 to increase product yields in recombinant E. coli. Scale-up of the biotransformation will also enable the isolation of the hydroxylated 1,8-cineole derivatives for a definitive structural characterization and determination of the hydroxylation position.
Supplementary Material
ACKNOWLEDGMENTS
We thank Carl Braybrook and Jo Cosgriff for GC-MS analyses and Lindsay Sparrow and Nick Bartone for amino acid sequencing. We also acknowledge Stella Kyi for synthesis of a monohydroxylated 1,8-cineole standard and James de Voss for providing the bicistronic plasmid containing the P450cin gene.
Funding Statement
Dena Lyras was funded by Australian Research Council (ARC) (FT120100779).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02067-16.
REFERENCES
- 1.Carman RM, MacRae IC, Perkins MV. 1986. The oxidation of 1,8-cineole by Pseudomonas flava. Aust J Chem 39:1739–1746. doi: 10.1071/CH9861739. [DOI] [Google Scholar]
- 2.Simonsen JL, Owen LN. 1953. The terpenes, 2nd ed, vol 1 Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 3.Farlow AJ, Bernhardt PV, De Voss JJ. 2013. Synthesis of oxygenated cineole derivatives from cineole: utility of cytochrome P450cin as an enantioselective catalyst. Tetrahedron Asymmetry 24:324–333. doi: 10.1016/j.tetasy.2013.02.004. [DOI] [Google Scholar]
- 4.de Boggiatto MV, de Heluani CS, de Fenik IJS, Catalán CAN. 1987. Regiospecific functionalization of the monoterpene ether 1,3,3-trimethyl-2-oxabicyclo[2.2.2]octane (1,8-cineole). Synthesis of the useful bridged gamma γ-lactone 1,3-dimethyl-2-oxabicyclo[222]octan-3→5-olide. J Org Chem 52:1505–1511. [Google Scholar]
- 5.Asakawa Y, Matsuda R, Tori M. 1986. Hydroxylation of menthols and cineoles with m-chloroperbenzoic acid. Experientia 42:201–203. doi: 10.1007/BF01952469. [DOI] [Google Scholar]
- 6.Azerad R. 2014. 1,8-Cineole: chemical and biological oxidation reactions and products. Chempluschem 79:634–655. doi: 10.1002/cplu.201300422. [DOI] [Google Scholar]
- 7.Slessor KE, Hawkes DB, Farlow A, Pearson AG, Stok JE, De Voss JJ. 2012. An in vivo cytochrome P450cin (CYP176A1) catalytic system for metabolite production. J Mol Catal B Enzym 79:15–20. doi: 10.1016/j.molcatb.2012.03.007. [DOI] [Google Scholar]
- 8.MacRae IC, Alberts V, Carman RM, Shaw IM. 1979. Products of 1,8-cineole oxidation by a pseudomonad. Aust J Chem 32:917–922. doi: 10.1071/CH9790917. [DOI] [Google Scholar]
- 9.Williams RD, Trudgill PW, Taylor DG. 1989. Metabolism of 1,8-cineole by a Rhodococcus species: ring cleavage reactions. J Gen Microbiol 135:1957–1967. [Google Scholar]
- 10.Liu WG, Rosazza JPN. 1990. Stereospecific hydroxylation of 1,8-cineole using a microbial biocatalyst. Tetrahedron Lett 31:2833–2836. doi: 10.1016/0040-4039(90)80160-N. [DOI] [Google Scholar]
- 11.Hawkes DB, Adams GW, Burlingame AL, Ortiz de Montellano PR, De Voss JJ. 2002. Cytochrome P450cin (CYP176A): isolation, expression, and characterization. J Biol Chem 277:27725–27732. doi: 10.1074/jbc.M203382200. [DOI] [PubMed] [Google Scholar]
- 12.Rasmussen J-AM, Henderson KA, Straffon MJ, Dumsday GJ, Coulton J, Zachariou M. 2005. Two new biocatalysts for improved biological oxidation of 1,8-cineole. Aust J Chem 58:912–916. doi: 10.1071/CH05204. [DOI] [Google Scholar]
- 13.Knight AR. 2009. Ph.D. thesis. Murdoch University, Perth, Western Australia, Australia. [Google Scholar]
- 14.Stok JE, Hall EA, Stone ISJ, Noble MC, Wong SH, Bell SG, De Voss JJ. 2016. In vivo and in vitro hydroxylation of cineole and camphor by cytochromes P450CYP101A1, CYP101B1 and N242A CYP176A1. J Mol Catal B Enzym 128:52–64. doi: 10.1016/j.molcatb.2016.03.004. [DOI] [Google Scholar]
- 15.Meharenna YT, Slessor KE, Cavaignac SM, Poulos TL, De Voss JJ. 2008. The critical role of substrate-protein hydrogen bonding in the control of regioselective hydroxylation in P450cin. J Biol Chem 283:10804–10812. doi: 10.1074/jbc.M709722200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urlacher VB, Lutz-Wahl S, Schmid RD. 2004. Microbial P450 enzymes in biotechnology. Appl Microbiol Biotechnol 64:317–325. doi: 10.1007/s00253-003-1514-1. [DOI] [PubMed] [Google Scholar]
- 17.Nelson DR. 2009. The cytochrome P450 homepage. Hum Genomics 4:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katagiri M, Ganguli BN, Gunsalus IC. 1968. A soluble cytochrome P-450 functional in methylene hydroxylation. J Biol Chem 243:3543–3546. [PubMed] [Google Scholar]
- 19.Poulos TL, Finzel BC, Howard AJ. 1987. High-resolution crystal structure of cytochrome P450cam. J Mol Biol 195:687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 20.Schewe H, Mirata MA, Holtmann D, Schrader J. 2011. Biooxidation of monoterpenes with bacterial monooxygenases. Process Biochem 46:1885–1899. doi: 10.1016/j.procbio.2011.06.010. [DOI] [Google Scholar]
- 21.Hannemann F, Bichet A, Ewen KM, Bernhardt R. 2007. Cytochrome P450 systems- biological variations of electron transport chains. Biochim Biophys Acta 3:330–344. [DOI] [PubMed] [Google Scholar]
- 22.Yang G, Ding Y. 2014. Recent advances in biocatalyst discovery, development and applications. Bioorg Med Chem 22:5604–5612. doi: 10.1016/j.bmc.2014.06.033. [DOI] [PubMed] [Google Scholar]
- 23.Dumsday G, Pilkington N, Straffon M, Zachariou M. 2007. Using eucalypt oil as a carbon source for deriving other compounds—development of a microbial library. RIRDC publication no. 07/086. Rural Industries Research and Development Corporation, Kingston, Australia. [Google Scholar]
- 24.Dumsday GJ, Ocal G, Bridger JS, Zachariou M. 2009. The use of oxygen uptake rate to monitor discovery of microbial and enzymatic biocatalysts. Biotechnol Bioeng 102:673–683. doi: 10.1002/bit.22115. [DOI] [PubMed] [Google Scholar]
- 25.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 26.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 28.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Omura T, Sato R. 1964. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239:2370–2378. [PubMed] [Google Scholar]
- 30.Schenkman JB, Jansson I. 2006. Spectral analyses of cytochromes P450, p 11–18. In Phillips IR, Shephard EA (ed), Cytochrome P450 protocols, 2nd ed Humana Press, Totowa, NJ. [Google Scholar]
- 31.Bell SG, Dale A, Rees NH, Wong L-L. 2010. A cytochrome P450 class I electron transfer system from Novosphingobium aromaticivorans. Appl Microbiol Biotechnol 86:163–175. doi: 10.1007/s00253-009-2234-y. [DOI] [PubMed] [Google Scholar]
- 32.Takeuchi M, Hamana K, Hiraishi A. 2001. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int J Syst Evol Microbiol 51:1405–1417. doi: 10.1099/00207713-51-4-1405. [DOI] [PubMed] [Google Scholar]
- 33.Song K, Ren J, Reinert G, Deng M, Waterman MS, Sun F. 2014. New developments of alignment-free sequence comparison: measures, statistics and next-generation sequencing. Brief Bioinform 15:343–353. doi: 10.1093/bib/bbt067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan CX, Bernard G, Poirion O, Hogan JM, Ragan MA. 2014. Inferring phylogenies of evolving sequences without multiple sequence alignment. Sci Rep 4:6504. doi: 10.1038/srep06504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luthra A, Denisov IG, Sligar SG. 2011. Spectroscopic features of cytochrome P450 reaction intermediates. Arch Biochem Biophys 507:26–35. doi: 10.1016/j.abb.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bradshaw WH, Conrad HE, Corey EJ, Gunsalus IC, Lednicer D. 1959. Microbiological degradation of (+)-camphor. J Am Chem Soc 81:5507. [Google Scholar]
- 38.Aylward FO, McDonald BR, Adams SM, Valenzuela A, Schmidt RA, Goodwin LA, Woyke T, Currie CR, Suen G, Poulsen M. 2013. Comparison of 26 sphingomonad genomes reveals diverse environmental adaptations and biodegradative capabilities. Appl Environ Microbiol 79:3724–3733. doi: 10.1128/AEM.00518-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bell SG, Wong L-L. 2007. P450 enzymes from the bacterium Novosphingobium aromaticivorans. Biochem Biophys Res Commun 360:666–672. doi: 10.1016/j.bbrc.2007.06.119. [DOI] [PubMed] [Google Scholar]
- 40.Sezutsu H, Le Goff G, Feyereisen R. 2013. Origins of P450 diversity. Philos Trans R Soc Lond B Biol Sci 368:20120428. doi: 10.1098/rstb.2012.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuler MA, Sligar SG. 2007. Diversities and similarities in P450 systems: an introduction, p 1–26. In Sigel A, Sigel H, Sigel R (ed), The ubiquitous roles of cytochrome P450 proteins. John Wiley & Sons, Ltd, Chichester, England. [Google Scholar]
- 42.Peterson JA, Lu J-Y, Geisselsoder J, Graham-Lorence S, Carmona C, Witney F, Lorence MC. 1992. Cytochrome P-450terp. Isolation and purification of the protein and cloning and sequencing of its operon. J Biol Chem 267:14193–14203. [PubMed] [Google Scholar]
- 43.Hall EA, Bell SG. 2015. The efficient and selective biocatalytic oxidation of norisoprenoid and aromatic substrates by CYP101B1 from Novosphingobium aromaticivorans DSM12444. RSC Adv 5:5762–5773. doi: 10.1039/C4RA14010A. [DOI] [Google Scholar]
- 44.Deprez E, Gill E, Helms V, Wade RC, Hui Bon Hoa G. 2002. Specific and non-specific effects of potassium cations on substrate-protein interactions in cytochromes P450cam and P450lin. J Inorg Biochem 91:597–606. doi: 10.1016/S0162-0134(02)00467-1. [DOI] [PubMed] [Google Scholar]
- 45.Westlake ACG, Harford-Cross CF, Donovan J, Wong LL. 1999. Mutations of glutamate-84 at the putative potassium-binding site affect camphor binding and oxidation by cytochrome P450cam. Eur J Biochem 265:929–935. [DOI] [PubMed] [Google Scholar]
- 46.Sligar SG. 1976. Coupling of spin, substrate, and redox equilibria in cytochrome P450. Biochemistry 15:5399–5406. doi: 10.1021/bi00669a029. [DOI] [PubMed] [Google Scholar]
- 47.Koga H, Yamaguchi E, Matsunaga K, Aramaki H, Horiuchi T. 1989. Cloning and nucleotide sequences of NADH-putidaredoxin reductase gene (camA) and putidaredoxin gene (camB) involved in cytochrome P-450cam hydroxylase of Pseudomonas putida. J Biochem 106:831–836. [DOI] [PubMed] [Google Scholar]
- 48.Stolz A. 2009. Molecular characteristics of xenobiotic-degrading sphingomonads. Appl Microbiol Biotechnol 81:793–811. doi: 10.1007/s00253-008-1752-3. [DOI] [PubMed] [Google Scholar]
- 49.Basta T, Keck A, Klein J, Stolz A. 2004. Detection and characterization of conjugative degradative plasmids in xenobiotic-degrading Sphingomonas strains. J Bacteriol 186:3862–3872. doi: 10.1128/JB.186.12.3862-3872.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.