Significance
The T-cell antigen receptor (TCR) is an eight-subunit modular biosensor that detects the presence of pathogen-derived molecular fragments in infected cells and tissues. No atomic-resolution structure has been determined for the intact receptor complex, and the mechanism by which it transmits signals across the cell membrane remains poorly defined. Using a combination of biochemical, biophysical, and computational approaches to study the assembled complex in cellular membranes, we identified an evolutionarily conserved core transmembrane (TM) structure that organizes the complex into an intimately packed eight-helix bundle within the membrane and is critical for the structural integrity of the intact receptor. Our model provides insights into possible structural pathways involved in TM TCR signaling.
Keywords: T-cell receptor, transmembrane structure, NMR, MD simulation, cysteine cross-linking
Abstract
The T-cell antigen receptor (TCR) is an assembly of eight type I single-pass membrane proteins that occupies a central position in adaptive immunity. Many TCR-triggering models invoke an alteration in receptor complex structure as the initiating event, but both the precise subunit organization and the pathway by which ligand-induced alterations are transferred to the cytoplasmic signaling domains are unknown. Here, we show that the receptor complex transmembrane (TM) domains form an intimately associated eight-helix bundle organized by a specific interhelical TCR TM interface. The salient features of this core structure are absolutely conserved between αβ and γδ TCR sequences and throughout vertebrate evolution, and mutations at key interface residues caused defects in the formation of stable TCRαβ:CD3δε:CD3γε:ζζ complexes. These findings demonstrate that the eight TCR–CD3 subunits form a compact and precisely organized structure within the membrane and provide a structural basis for further investigation of conformationally regulated models of transbilayer TCR signaling.
The antigen-specific T-cell response depends critically upon the ability of the T-cell receptor (TCR) to signal the recognition of activating ligands. The variable TCR proteins (α, β, δ, and γ) that bind to these ligands have no intrinsic intracellular signaling capability and transmit information through the noncovalently associated CD3δε, CD3γε, and ζζ modules containing cytoplasmic immunoreceptor tyrosine-based activation motifs (ITAMs) that are phosphorylated by the Src-family kinase Lck. The mechanism by which antigen sensing is translated from ligand binding at the distal end of the TCR to phosphorylation events at the cytoplasmic tails of the associated signaling modules remains an important open question in T-cell biology. A mounting pool of evidence implicates ligand-induced alterations in receptor structure (1) and/or TCR–CD3 configuration (2–6) as the triggering event. Changes in the structured extracellular (EC) domains are proposed to translate, through an unknown mechanism, into alterations in the CD3 and ζζ cytoplasmic tails, converting them to a conformation that is receptive to phosphorylation by Lck and binding of other proximal signaling components (7–12). This type of signaling model implies a pathway between EC and cytoplasmic domains that may involve alterations in the subunit transmembrane (TM) domains with respect to each other and/or the lipid bilayer (13–16). Consistent with this view, Kuhns and colleagues (17) recently reported a ligand-induced change in intersubunit proximity at the TM–juxtamembrane juncture in the ζζ dimer and proposed that this “mechanical switch” is coupled to signal initiation at the ζζ cytoplasmic tails. However, the nature of the upstream structural changes that trigger this switch, and whether it is required for signal initiation, remain to be determined. Tracing possible transbilayer conformational pathways involved in TCR triggering will require a detailed view of subunit organization, particularly within and near the membrane.
The complexity of the octameric TCRαβ:CD3δε:CD3γε:ζζ assembly has made it an exceedingly challenging subject for structural studies, and our understanding of the specific molecular interfaces mediating communication between ligand-binding and signal-transducing modules is consequently incomplete. Data from a large collection of mutagenesis studies indicate that a continuum of interactions among EC, connecting peptide (CP)/stalk, and TM domains contribute to the assembly, stability, and function of the surface-expressed complex (reviewed in refs. 15, 16, and 18). The TM domains are particularly important sites of interactions that govern the assembly of the three dimeric signaling modules with TCR α- and β-chains through focused networks of polar interactions (Fig. 1A) (19). The solution NMR structure of the micelle-embedded ζζ module (20) revealed a highly specific TM helix dimer interface that is required to properly position the key polar residues (a pair of closely apposed aspartic acids) for interaction with an arginine in the TCRα TM domain. However, aside from the very similar polar interactions governing CD3δε association with TCRα and CD3γε association with TCRβ (19), little is known about the extent of structural organization that exists within the membrane-embedded region of this complex receptor. In particular, nothing at all is known about whether the proximity and orientation of TCRα and TCRβ TM domains are determined by a structured interface, yet this question is central to the problem of intramembrane TCR organization and transbilayer signaling because these proteins form the “hub” of the complex within the membrane.
Fig. 1.
Strategy for a TCRαβ TM disulfide scan. (A) Composition of the αβTCR–CD3 complex. The shaded disk represents the lipid bilayer. Arrows indicate the recognized intramembrane assembly steps orchestrated by basic (blue circles), acidic (red circles), and hydroxyl-bearing serine/threonine (yellow circles) residues. Yellow boxes represent immunoreceptor tyrosine-based activation motifs (ITAMs) in CD3 and ζ cytoplasmic tails. Ribbon diagrams of extracellular domains were prepared using the crystal structures of a human αβTCR [Protein Data Bank (PDB) ID code 1KGC] (78) and the crystal structures of human CD3δε (1XIW) (79) and human CD3γε (1SY6) (80). (B) Connecting peptide (CP), predicted transmembrane (TM), and cytosolic sequences of human TCRα and TCRβ proteins. Asterisks (*) indicate positions to be substituted with cysteine in a TM disulfide scan.
In this study, we combine intramembrane disulfide scanning, solution NMR, and computational modeling methods to interrogate the relationship between TCRα and TCRβ TM domains in the context of the assembled, membrane-embedded receptor complex. Our results identify a specific helical TCRαβ TM interface within the core of the assembled, untriggered receptor. We further demonstrate that formation of this TM interface is essential to the integrity of the assembled complex, and that its salient features are conserved between αβ and γδ TCRs and across species that span the entire range of vertebrates with recognizable TCR genes.
Results
Disulfide Scanning Identifies TCRα–TCRβ TM Contacts Within the Receptor Complex.
We designed a disulfide scanning approach that would allow analysis of TM interfaces within a receptor complex assembled from full-length subunits in a natural cellular membrane. Single cysteine substitutions were made at every position in the regions corresponding to the predicted TM domains of human TCRα and TCRβ proteins, including two to three additional amino acids on either side, but excluding the basic TM residues that are required for assembly with signaling modules (Fig. 1B) (19). To facilitate SDS/PAGE detection of combinations resulting in TCRαβ heterodimers that were disulfide bonded through their TM domains, all mutants were constructed on a background in which the cysteines responsible for a native intermolecular TCRαβ disulfide bond in the CP regions were mutated to serine (“cysteineless” TCR). This disulfide bond is not required for TCRαβ heterodimer formation or for surface expression of TCR–CD3 complexes, but its elimination is reported to result in “loose” association of ζζ such that it cannot be detected by coimmunoprecipitation (IP) from detergent extracts (21). As association of the ζζ dimer is the last step of TCR assembly in the endoplasmic reticulum (ER) (19, 22, 23) and is required for transport of the octameric TCRαβ:CD3δε:CD3γε:ζζ complex to the cell surface (23, 24), surface expression of cysteineless TCR complexes is an indication that this defect is not in ER assembly but rather in detergent stability of ζζ binding. We tested this in T-cell lines with and without ζ expression and observed that both disulfide-linked and non–disulfide-linked TCRs were absolutely dependent on the ζ-chain for surface expression (Fig. S1), confirming that ζζ associates with the cysteineless TCR. Based on these results, we concluded that the cysteineless TCR is capable of assembly with all signaling subunits and is therefore an appropriate background for our disulfide scan.
Fig. S1.
A “cysteineless” TCR requires ζζ for cell surface expression. The mouse OT-I TCR was encoded in a TCRβ-2A-TCRα retroviral construct using a pHAGE-IRES-ZsGreen vector. WT and CP-region cysteine-to-serine (Cys-less) versions of this TCR were transduced into a TCRαβ/CD3δ/ζ-deficient BW5147 murine thymoma line stably reconstituted with either CD3δ only (−ζ) or CD3δ and ζ (+ζ). ZsGreen+ cells were assessed for surface TCR expression by flow cytometry using PE-Cy7–conjugated anti-mouse TCRβ (clone H57-597).
Using a previously reported in vitro translation-based system to assemble 35S-labeled TCR–CD3 complexes in ER-derived membrane vesicles (19, 22), the TM cysteine mutants were tested in 104 different combinations to identify positions that were close enough to become disulfide bonded within the assembled complex (the entire primary screen is shown in Fig. S2). This system has been shown to faithfully recapitulate the cotranslational folding and assembly of multisubunit receptors (19, 22, 25–29), and the use of conformation-specific antibodies and specialized affinity tags allows isolation of receptor complexes of well-defined composition and stoichiometry with very high sensitivity and specificity (19, 22). For this screen, in vitro-transcribed mRNAs encoding each TCRαβ combination were cotranslated with a master mix of CD3 and ζ mRNAs and allowed to fold and assemble before treatment with copper(II)-o-phenanthroline (CuPhe) to catalyze intramembrane disulfide bond formation. Radiolabeled assembly products were then extracted from washed ER vesicles in 1% digitonin and analyzed by IP with the conformation-specific anti-CD3 antibody OKT3 to identify disulfide-trapped TCRαβ combinations that had assembled with properly folded CD3 heterodimers. The cross-link–positive TCRαβ pairs identified from this primary screen were collected and reanalyzed together in a separate experiment (Fig. 2 A and B). Because the OKT3-captured products could contain complexes of TCRαβ that coprecipitated with CD3δε or CD3γε but did not necessarily contain both CD3 modules (19, 22), we also performed a two-step sequential nondenaturing IP (snIP) (19) to select only those complexes containing both CD3δ and CD3γ, which represent minimally hexameric CD3δε:TCRαβ:CD3γε complexes (Fig. 2C). From this analysis, 17 TCRαβ combinations were identified that resulted in recovery of assembled complexes containing cross-linked TCR heterodimers [marked with asterisks (*) in Fig. 2C and connecting lines in Fig. 2A], demonstrating a very high degree of positional specificity. These varied significantly with regard to total complex recovery and the ratio of cross-linked to non–cross-linked TCR contained in the complex, which could be due to several different parameters including the distance and orientation between cysteines, depth in the membrane (affecting accessibility to oxidizing agent), and the ability of individual cysteine mutant TCR proteins to fold and assemble normally in the ER. Because these compounding factors are not easily deconvoluted, we have treated all of these as cross-link positive and have not attempted to distinguish among them on a quantitative basis in later analyses (Discussion). Cross-links were most abundant in the N- and C-terminal TM-flanking sequences, which may be unstructured, but those falling within the predicted TM domains display an approximately helical periodicity, spaced three to four residues apart. These results indicate that discrete helical faces of the TCRα and TCRβ TM domains are accessible to one another and in close proximity within the ER-assembled TCR–CD3 complex.
Fig. S2.
Primary disulfide scan of TCRα and TCRβ TM domains. All combinations tested for TCRαβ TM cross-links are shown in A–E. Analysis carried out as described in Fig. 2B with conformation-specific anti-CD3ε (OKT3) IP. The regions being interrogated in each panel are highlighted in orange. Cross-link–positive cysteine mutant combinations are indicated by lines between TCRα and TCRβ TM sequences and with * on the gels and quantitation.
Fig. 2.
Identification of specific TCRαβ TM contacts within the receptor complex. (A) Summary of observed TCRαβ TM cross-links indicated by solid lines between cysteine-substituted positions. The F×E cross-link highlighted in the text is marked in red. Residue numbering throughout the paper starts at the CP cysteines responsible for the native intermolecular TCRαβ disulfide bond. (B) Panel of cross-link–positive combinations from the primary disulfide scan shown in Fig. S2. The indicated control (WT, cysteineless) or cysteine mutant human A6 TCRα (HA-tagged) and TCRβ [streptavidin-binding peptide (SBP)-tagged] mRNAs were cotranslated with a master mix of human CD3 and ζ mRNAs in in vitro assembly reactions and treated with 1 mM copper(II):phenanthroline (CuPhe) in TBS to induce TM disulfide bond formation. Digitonin-extracted products were immunoprecipitated using mAb OKT3 (anti-CD3δε/CD3γε) and separated by nonreducing SDS/PAGE. IP control panels demonstrate the specificity of product capture using WT and two of the strongest cross-link combinations; each indicated assembly reaction was split and subjected to an IP with specific (OKT3) or isotype-matched irrelevant control (Ctrl) antibodies. (C) Aliquots of the same reactions shown in B were subjected to CD3δ (PC-tagged) → CD3γ (FLAG-tagged) sequential nondenaturing IP (snIP) (19) to isolate minimally hexameric (CD3δε:TCRαβ:CD3γε) complexes and analyzed as above. The IP controls contained isotype-matched irrelevant (Ctrl) antibodies in both steps of the snIP procedure. Asterisks (*) mark the combinations counted as cross-link positive. (D) CuPhe dependence of TCRα-F26C × TCRβ-E20C (F×E) cross-linking and ζζ association. The indicated TCR combinations were processed as in B either with (+) or without (−) CuPhe addition after assembly and analyzed by snIP to select minimally hexameric complexes (Left) or only fully assembled ζζ-containing complexes (Right). Control (Ctrl) snIPs were performed as above on a duplicate CuPhe-treated WT assembly reaction, but used biotin-blocked streptavidin (SA) in lieu of an isotype control in the first step of TCRβ → ζ snIP. (E) Comparison of TM cross-linking in the presence and absence of the native CP-region disulfide bond. Assembled and CuPhe-treated complexes containing WT, F×E on the cysteineless background (F×ECS) or F×E on an otherwise WT background with CP disulfide bond intact (F×EWT) were captured from digitonin lysates using SA beads to bind the SBP-tagged TCRβ chain. The final wash step was performed with (+) or without (−) 10 mM TCEP in digitonin solution to selectively reduce EC but not TM disulfide bonds.
TCRαβ TM Association Is Required for Stable Assembly with ζζ.
Because disulfide bond formation is irreversible under oxidizing conditions, transient encounters could result in accumulation of cross-linked products, and thus their observation does not necessarily indicate a specific and stable structure. Furthermore, the inability to coprecipitate ζζ in the absence of the native CP region disulfide bond (21) (also observed in our screen; Fig. 2 B and C) leaves open the possibility that the detection of TM interactions was the result of an altered TCRαβ structure. However, one of the 17 observed cross-links restored detergent-stable ζζ association (Fig. 2C, TCRα-F26C × TCRβ-E20C; termed “F×E” hereafter), providing strong evidence that this cross-linked product effectively reproduced the native TCRαβ TM structure. Co-IP of ζζ with the F×E heterodimer was CuPhe dependent (Fig. 2D, compare lanes 3 and 4 in both panels), showing that the disulfide cross-link, and not merely the presence of the cysteine substitutions, was what had restored the native-like ζζ association. To test whether this cross-link was compatible with the unaltered CP region, we further analyzed the F×E combination on the background of TCR proteins with the native CP cysteines intact (Fig. 2E). We were able to distinguish between the CP and TM disulfide bond locations by using the strong polar reducing agent tris(2-carboxyethyl)phosphine (TCEP) to selectively reduce extracellular disulfide bonds in the detergent-extracted complexes (Fig. 2E, compare lanes 1 and 4) while sparing the intramembrane cross-link (compare lanes 2 and 5). Introduction of the TM cysteines on the WT CP background did not alter complex assembly or formation of disulfide-bonded TCR (compare lanes 1 and 3), and the F×E cross-link was detected with similar efficiency whether the native disulfide bond was present or absent (compare lanes 5 and 6). Together, these data show that the F×E TM cross-link is equally compatible with WT and cysteineless backgrounds, and that detergent-resistant ζζ binding requires both close association of TCRα and TCRβ TM domains, presumably stabilized by the extracellular disulfide bond in the WT TCR, and a specific TM conformation that is uniquely replicated by the F×E intramembrane disulfide cross-link.
Structure of the F×E Disulfide-Bonded TCRαβ TM Heterodimer.
Based on the finding that the TCRαβ TM domains are closely associated within the assembled complex, and taking into account the unique characteristics of the F×E TM cross-link, we wanted to explore the possible conformations available to the F×E–cross-linked product within a lipid bilayer using replica exchange molecular dynamics (REMD) simulations (30). Accurate modeling of the individual TM domains requires knowledge of the regions of α-helical structure, which are commonly approximated from hydrophobicity-based TM predictions. To obtain a more precise definition of the α-helical limits for our simulations, we experimentally determined the regions of intrinsic helical structure in TCRα- and TCRβ-derived CP-TM peptides in lipid micelles using solution NMR (Fig. 3). Analysis of secondary backbone chemical shifts using TALOS+ (31) identified 25-residue (TCRα) and 23-residue (TCRβ) segments of continuous α-helical structure spanning the predicted TM domains, with an additional helical turn identified N-terminal to the probable extracellular TM limit of TCRα. We note that interactions between the monomeric TCRα and TCRβ peptides could not be detected in mixed samples under these micellar conditions, and generation of disulfide-stabilized heterodimeric peptide complexes in sufficient yield and purity for NMR studies has not been successful. However, with strong biochemical evidence that a specific interface exists within the assembled complex in a cellular membrane, we judged that a computational approach incorporating both biochemical and biophysical restraints would provide the most stringent analysis of in situ TCRαβ TM structure.
Fig. 3.
Determination of TCRα and TCRβ TM helix limits by solution NMR. (A) 15N-, 13C-, and 2H (70%)-labeled peptides corresponding to connecting peptide (CP) and predicted TM regions of TCRα and TCRβ were reconstituted to 0.5 mM in 250 mM LMPG and 20 mM phosphate buffer (pH 6.8) and 1H-15N TROSY–heteronuclear single-quantum coherence spectra were recorded at 600-MHz 1H frequency and 35 °C. Full backbone resonance assignments were obtained for residues 3–47 (TCRα) and 3–40 (TCRβ) using standard triple-resonance experiments (Experimental Procedures) and backbone secondary chemical shift analysis was carried out using the TALOS+ software package (31). (B) Identification of helical regions from TALOS+ analysis. Asterisks (*) indicate positions where substitutions were made in NMR peptide constructs to facilitate production and analysis: C → S to block covalent homodimerization during production and purification; K → V to stabilize unassembled TM peptides in lipid micelles; M → V to avoid internal cleavage by cyanogen bromide.
Using the NMR-derived helix limits, we modeled the F×E disulfide-bonded TCRαβ TM dimer in an implicit lipid bilayer system (32) and ran a 10-ns REMD simulation (32 replicas) to evaluate the structural features of the interface. This simulation yielded a single dominant cluster encompassing 469 (94%) of 500 randomly selected structures that varied by only 0.5-Å backbone root-mean-square deviation (rmsd), indicating that a single unique TM packing orientation was consistent with the cross-linked product. The centroid structure representing this cluster (Fig. 4 A and B) reveals a left-handed coiled-coil with contacts along the entire length of the two helices. Several features of this structure indicate that it faithfully represents the interface detected in our disulfide scan: first, although only a single disulfide restraint was used in the simulation, the TCRαβ interface contains all of the positions where cysteine substitutions resulted in interhelical cross-links (Fig. 4A, shown in magenta), with the exception of the TCRα-N20 to TCRβ-S14 cross-link involving residues that face away from the interface. This is where the TM domains emerge from the membrane into the extracellular space and may represent a region of more dynamic structure. Second, this arrangement places the three basic residues in outward-facing positions (Fig. 4B) that are consistent with their known roles in assembly with the dimeric signaling modules (19). This is in line with our biochemical data showing that this cross-link formed in the context of a fully assembled TCRαβ:CD3δε:CD3γε:ζζ complex. Finally, the C-terminal portion of the interface contains three polar amino acids that form two potential interhelical hydrogen bonds: one between the TCRαT41 and TCRβY29 hydroxyl groups, and a second between the TCRαN37 side chain and a backbone carbonyl oxygen at TCRβA26 (Fig. 4C). This region was devoid of cross-links in our disulfide scan (Fig. 2A), indicating that cysteine substitutions in this part of the interface may have been disruptive to TCRαβ TM packing and therefore incompatible with detection of close helix–helix contacts. However, the detection of cross-links at interface positions before and after this region confirms their close proximity.
Fig. 4.
Structure of the TCRαβ TM interface within the receptor complex. (A) The centroid structure from the major cluster in a 10-ns REMD simulation of the F×E disulfide-bonded TCRαβ TM heterodimer in an implicit lipid bilayer model. Residues involved in the cross-links identified in Fig. 2 are highlighted in magenta. The basic residues required for assembly with dimeric signaling modules (blue) were replaced with leucine for stability in the bilayer during simulation, but are shown as the native arginine and lysine residues in the models to highlight their positions. (B) Top view (down the long axis of the helix dimer, from the extracellular side) of the structure shown in A. The experimental F×E disulfide bond is shown in magenta and yellow. The assembly points for dimeric signaling modules are labeled in blue. (C) Close-up view of the polar network formed by TCRα-N37, TCRα-T41, and TCRβ-Y29. (D) An independent REMD simulation (10 ns) was performed using helix restraints from the NMR data (Fig. 3) and eight distance restraints (Cβ–Cβ distance: 3.7- to 6.0-Å range) from disulfide scan data. Each plot shows the real Cβ–Cβ distance as a function of time during the simulation and is color-coded to the lines connecting cross-linked positions in the sequences above. (E) Side view of the aligned centroid structures from the F×E disulfide bond simulation (green and yellow with blue basic residues) and the distance-restrained simulation (light gray with blue basic residues). The backbone rmsd between these two structures is 0.5 Å.
A Distance-Restrained Simulation Using All Interhelical Cross-Links Yields the Same Structure.
In a separate and independent analysis, we tested whether the larger panel of experimentally derived interhelical restraints could be accommodated by a single TCRαβ TM structure. We selected all cross-links where both cysteine substitutions were located within the α-helical regions identified by NMR (Fig. 4D) and applied these as distance restraints rather than modeling them as disulfide bonds, following a strategy similar to NMR structure determination using nuclear Overhauser enhancement (NOE)-derived distances. Eight equally weighted distance restraints of 3.7 Å (lower bound) to 6.0 Å (upper bound) were applied between Cβ atoms of the native amino acids at positions where cross-links were observed. This range was chosen to approximate the Cβ–Cβ distance in real disulfide bonds (≤4.5 Å) with a flexible upper limit to accommodate the simultaneous enforcement of restraints that were observed separately and had variable cross-link efficiency. The results converged within the first 3 ns of a 10-ns REMD simulation (Fig. 4D), with the major cluster containing 475 (95%) of 500 randomly selected structures (backbone rmsd of 0.6 Å within the cluster). The centroid structure from this cluster was nearly identical to the F×E disulfide-bonded centroid structure, aligning with a backbone rmsd of 0.5 Å (Fig. 4E). The agreement between these two approaches provides very strong evidence that our model represents the TCRαβ TM structure that forms within the assembled receptor complex and suggests that the structural differences between the F×E cross-link that supports normal ζζ association and those that do not may be subtle.
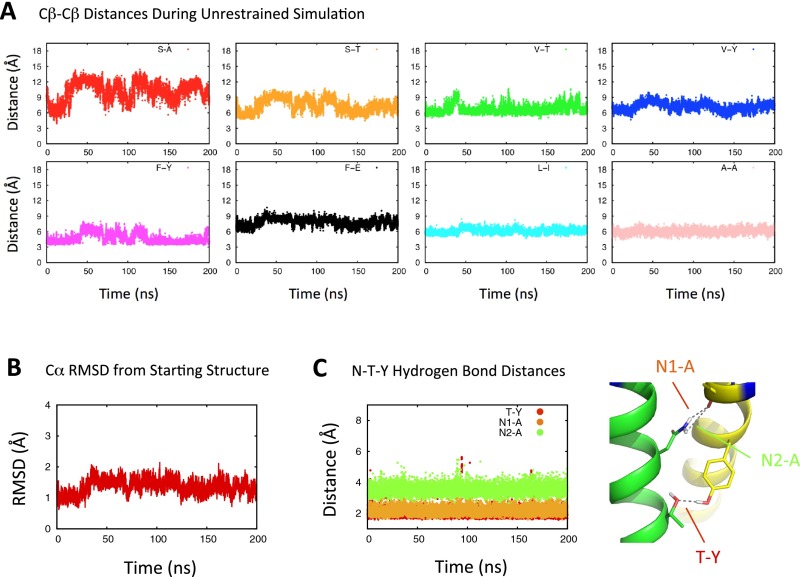
To test the favorability of the interface without restraints, the non–disulfide-bonded structure was inserted into an explicitly modeled 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer and subjected to a longer simulation (200 ns) in which all interhelical distance restraints were removed (Fig. S3). Variations from the starting structure were largely confined to the N-terminal half of the interface, whereas the C-terminal half was very stable and the polar interface contacts maintained consistent hydrogen-bonding distances throughout the simulation. Sequestration of polar residues at protein interfaces is particularly favorable in the hydrophobic environment of the lipid bilayer interior (33, 34), and polar interactions are already known to play important roles at other sites guiding TM associations within the TCR complex (18) (Fig. 1A) and other activating immune receptors (35). This prompted us to ask whether the polar residues identified in the TCRαβ TM interface are a common feature in TCR sequences from an evolutionary perspective.
Fig. S3.
Stability of the TCRαβ TM interface in a lipid bilayer. The TCRαβ TM centroid structure from distance-restrained REMD simulations was placed in an explicitly modeled POPC bilayer and subjected to a 200-ns molecular dynamics simulation in which distance restraints were removed. Distances between the originally restrained atom pairs are traced in A over the simulation time frame, and total rmsd with respect to the starting structure is traced in B. C shows the distance between potential hydrogen bond donor–acceptor pairs as a function of simulation time, and the distances being monitored are indicated on the structure graphic for reference.
The Polar Residues in the TCRαβ TM Interface Are a Conserved Feature of All TCRs.
The predicted TM domains of TCR proteins have previously been noted for their high degree of sequence conservation (36). The variation in TM sequences among placental mammals is indeed very low, particularly for TCRβ. Considerably greater variation is observed when sequences from marsupials, birds, reptiles, amphibians, and fish are included (Fig. S4), yet the polar N, T, and Y residues in the TCRαβ TM interface identified here are conserved. Moreover, these features are also present in vertebrate TCRδ and TCRγ sequences, which otherwise diverge significantly from their TCRαβ counterparts. Fig. 5 illustrates the variation among TCRα/δ sequences (A) and TCRβ/γ sequences (B) representing vertebrate species from humans to sharks. The most conserved features are the basic residues making key contacts to signaling modules (R and K) and the polar residues found in the TCRαβ interface (N, T[S], Y). The pre-Tα chain, which forms receptor complexes containing newly synthesized TCRβ chains at an early stage of T-cell development in the thymus, contains a similar motif to TCRα/δ in which the asparagine is replaced with aspartic acid (Fig. S4E). This level of conservation suggests that the C-terminal portion of the TM interface plays a particularly important role in TCR structure and/or function. We also noted that a second (N-terminal) conserved tyrosine in TCRβ/γ is positioned in our model such that it is directly adjacent to the TCRα arginine that contacts ζζ (Fig. 4). This is consistent with reports that mutation of this residue caused loss of ζζ co-IP and signaling defects in cell lines (37, 38) and indicates that it is likely to make a direct contribution to ζζ stabilization within the complex.
Fig. S4.
Individual TCR chain transmembrane sequence logos and all sequences used in alignments. Sequence logos as in Fig. 5 are shown for individual TCRα (A), TCRδ (B), TCRβ (C), TCRγ (D), and pre-Tα (E) predicted TM domains with the human sequences below the graphic for reference. The aligned sequences are shown with conserved residues highlighted in yellow.
Fig. 5.
Evolutionary conservation of key TCR TM interface features. Sequence logos illustrate the degree of amino acid conservation within TM sequences based on vertebrate TCRα/TCRδ (A; 65 sequences total) and TCRβ/TCRγ (B; 66 sequences total) alignments. The sequences of human TCRα and TCRβ TM domains are shown below sequence logos for reference. The height of each letter stack indicates relative conservation at that position, whereas the height of each individual letter indicates its relative prevalence at that position. Colors represent basic (blue), acidic (red), hydroxyl/thiol-containing (yellow), carboxamide (purple), small (green), and aromatic/hydrophobic (black) side-chain categories. Labels below the reference sequences indicate the role of each conserved polar residue. Logo graphics were generated using the WEBLOGO internet application (weblogo.berkeley.edu). Additional sequence logos for individual TCR proteins and all sequences included in the analysis are provided in Fig. S4.
Mutations in the Conserved TCRαβ TM Interface Result in Altered Receptor Complex Assembly.
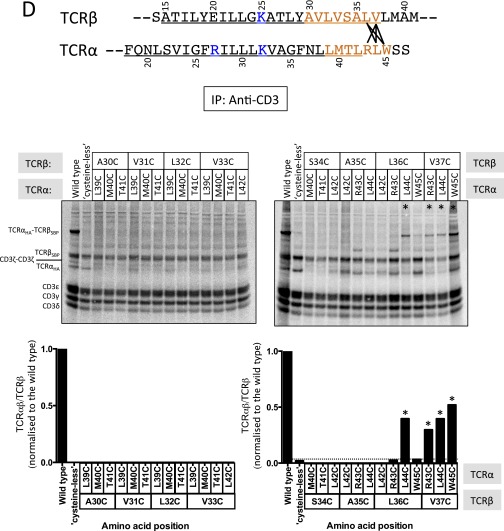
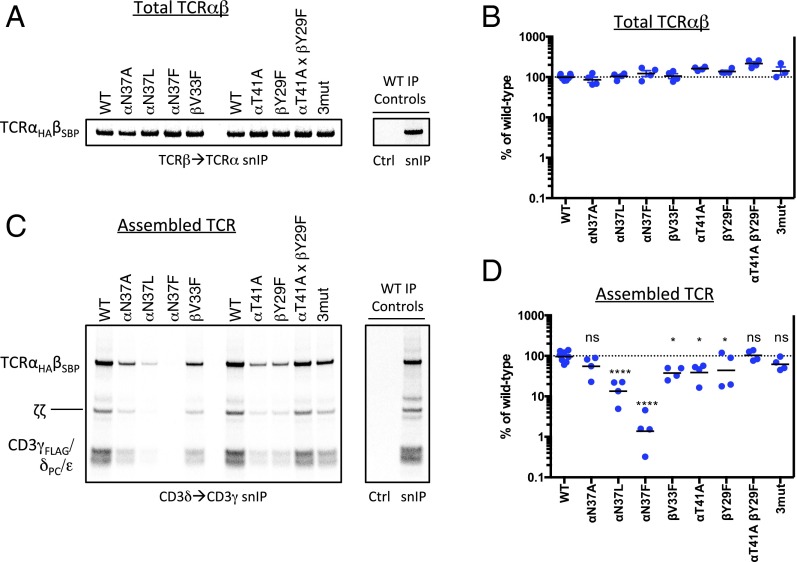
Finally, we examined the effects of mutations in the conserved C-terminal region of the TCRαβ TM interface on receptor complex assembly. We generated a panel of mutants predicted to alter the structure or stability of the interface and examined their ability to form TCRαβ heterodimers and assembled receptor complexes (Fig. 6). None of the mutants exhibited significant defects in formation of disulfide-bonded TCRαβ heterodimers (Fig. 6 A and B), which is driven by folding of the Ig EC domains, but large variations were observed in the recovery of stably assembled TCR–CD3 complexes (Fig. 6 C and D). We probed aliquots of the same reactions shown in A for assembled complexes using a two-step hexamer selection (as in Fig. 2C) and found that substitution of TCRα-N37 with the large hydrophobic residues leucine and phenylalanine resulted in severe reductions in complex recovery (Fig. 6D, quantitation from four independent experiments: mean 13% and 1.4% of WT for leucine and phenylalanine substitutions, respectively). A defect was also observed with the smaller alanine substitution at this position, although this difference did not reach statistical significance in our quantitative analysis. As this result indicated a steric effect related to introduction of bulky hydrophobic residues between helices, we tested a phenylalanine substitution at TCRβ-V33, another position that is buried in the closely packed C-terminal interface. A significant defect was also observed with this mutation (mean 38% of WT), and we noted that phenylalanine never occurs at this position in our survey of 66 vertebrate TCRβ/γ sequences (Fig. 5B and Fig. S4 C and D). This series of mutants provides strong support for the location of these residues at a helix–helix interface and demonstrates that introduction of a steric barrier to close association was sufficient to cause a near-complete block in stable complex formation. Thus, the ability of the TCRαβ TM domains to adopt a closely packed structure also impacts on their ability to productively assemble with the two CD3 heterodimers.
Fig. 6.
Alteration of TCR complex stability by mutations in the C-terminal conserved TCRαβ TM interface. Assembly reactions containing the indicated TCRαβ mutants (on the WT CP region background) were performed as in Fig. 2 but were not subjected to a CuPhe treatment step. All reactions received the same master mix of CD3/ζ mRNAs. Each completed assembly reaction was split for analysis by snIP targeting TCRβ (SBP-tagged) followed by TCRα (HA-tagged) to quantitate total disulfide-linked TCRαβ heterodimer (A), or targeting CD3δ (PC-tagged) followed by CD3γ (FLAG-tagged) to isolate minimally hexameric CD3δε:TCRαβ:CD3γε complexes (C). Control (Ctrl) IPs used isotype-matched irrelevant antibodies or biotin-blocked SA in both steps of the snIP procedure as in Fig. 2. A and C show a single representative experiment. B and D show quantitative analysis of four independent experiments from densitometry data. Each plotted value represents the raw intensity of the TCRαβ band for the mutant, expressed as a percentage of WT in that experiment (an average of two independent WT controls in each experiment). Significance in D was determined in an ordinary one-way ANOVA uncorrected Fisher’s least significant difference test with single pooled variance. Means and P values for each mutant are as follows: (TCRα-N37A) 55.1%, P = 0.1512 (ns); (TCRα-N37L) 13.4%, P < 0.0001; (TCRα-N37F) 1.4%, P < 0.0001; (TCRβ-V33F) 37.6%, P = 0.0188; (TCRα-T41A) 38.9%, P = 0.0231; (TCRβ-Y29F) 43.8%, P = 0.0465; (TCRα-T41A, TCRβ-Y29F) 102.4%, P = 0.8681 (ns); (TCRα-N37A,T41A; TCRβ-Y29F 3mut) 62.0%, P = 0.2562 (ns).
Substitutions that eliminated the hydrogen-bonding hydroxyl groups at TCRα-T41 (mutated to alanine) or TCRβ-Y29 (mutated to phenylalanine) also caused defects (mean 39% and 44% of WT, respectively). However, because the combination of both mutations restored the assembled complex recovery to WT levels, these defects likely derive from leaving an unpaired polar group rather than to elimination of the intermolecular hydrogen bond per se. This is also supported by the observation that the combination of these two mutations with the TCRα-N37A substitution (“3mut”) was indistinguishable from TCRα-N37A alone, showing that elimination of the tyrosine–threonine interaction caused no additional defect. Because our biochemical analysis relies on recovery of detergent-extracted noncovalent protein complexes, we cannot distinguish between failure to assemble within the ER and formation of an altered, detergent-sensitive structure. Nonetheless, these results demonstrate that TCRαβ heterodimers with altered TM interfaces are less able to form “normal” receptor complex structures. Taken together, our data show that the TCRαβ TM domains are closely associated through a specific and conserved interface, and that formation of this core structure is an integral feature of the assembled TCR complex.
Discussion
In this study, we have applied a combination of biochemical, biophysical, and computational techniques to analyze the relationship between TCRα and TCRβ TM domains within the membrane-embedded receptor complex and evaluate its relevance in establishing the overall structure and stability of the intact, untriggered receptor. The interaction of two α-helices oriented in a lipid bilayer is a problem that is highly amenable to computational analysis, and inclusion of high-quality experimental restraints such as our NMR-derived secondary structure analysis and intermolecular cysteine cross-linking data results in a significant contraction of the relevant conformational space to be searched. Cysteine cross-linking has previously been applied to study TM interactions within homodimeric and heterodimeric type I membrane protein complexes, and these studies have established a strong correlation between cross-link–derived models and independent biochemical, biophysical, computational, and functional data (39–41). Our stringent analysis of assembly products identified one TCRαβ cysteine combination (F×E) that we could confidently model as an explicitly disulfide-bonded, membrane-embedded TM heterodimer in REMD simulations, because we showed that it supports a stable octameric receptor complex assembly and is compatible with the native, disulfide-bonded CP regions. The excellent agreement of this model with the results of an independent simulation using multiple interhelical cross-links as distance restraints confirmed that our biochemical data are internally consistent and supports the conclusion that our model represents the structure that is formed within the assembled complex in a native lipid bilayer. Disulfide cross-links as structural restraints have been given a quantitative treatment by others, relating cross-link efficiency to distance over a continuous scale (41). However, the interpretation of disulfide bond efficiency within a lipid bilayer is subject to many compounding factors, including distance and orientation between cysteine thiol groups, depth in the membrane, and, in the present case, the ability of the cross-linked TCR product to maintain relevant higher-order structures. For this reason, we took a conservative approach by treating all cross-links equally and applied a generous upper bound in our distance restraints.
Our results identified an interface between TCRα and TCRβ TM domains that forms a structured hub for the octameric receptor complex within the lipid bilayer. Based on the locations of lysine and arginine residues that organize the assembly with dimeric signaling modules (19), this structure fixes the positions of the two CD3 modules on opposite sides of the TCR within an intimately associated eight-TM–helix bundle (Fig. 7). The ζζ homodimer bridges and possibly contacts both CD3 modules as well as TCRαβ. This close association with multiple subunit TM domains provides a possible explanation for the observation that ζζ is the last module to join the complex (23), suggesting that its stable assembly is contingent on recognition of a composite surface with contributions from as many as four other TM helices that only become available in the preassembled hexameric intermediate. Our model also shows that there is only one significant TM surface in the TCRαβ heterodimer (on TCRβ) that is not likely to be buried at an interface with other subunit TM domains. This surface features a glutamic acid residue (Fig. 7, E20) that is present in all mammalian TCRβ sequences and is unlikely to be found unshielded in the hydrophobic lipid bilayer interior (42). The function of this acidic TM residue is unknown (discussed in ref. 16), but its location identifies the “exposed” TCRβ helix face as a region of interest in potential oligomeric TCR interactions (43–45) and associations with particular lipid or sterol moieties that have been shown to regulate receptor activation (46, 47).
Fig. 7.
Model of TM arrangement within the octameric TCR complex. Positions of signaling modules are inferred from the locations of key basic TM residues (19) around the TCRαβ TM coiled-coil structure. Polar side chains discussed in the text are shown in stick representation. The ζζ TM structure was generated from PDB entry 2HAC (20). CD3δε and CD3γε are shown as gray circles because no structural information is available and the subunit positions within each dimer are unknown. Red crescents represent acidic residues involved in assembly with TCR chains (19).
Several groups have proposed models of TCR–CD3 arrangements based on extracellular domain interactions. Antibody epitope mapping and computational docking studies (2, 48) suggested association of CD3 heterodimers on opposing faces of TCRαβ, whereas the locations of Cα and Cβ mutations that destabilize TCR–CD3 associations (3, 49) point to colocation of CD3 modules on one face of the TCR heterodimer. Two recent solution NMR-based studies proposed two-sided (50) or one-sided (51) models using chemical shift and resonance intensity perturbations measured among soluble dimeric ectodomain fragments. As it is unclear how well the low-affinity (hundreds of micromolar to millimolar), unoriented interactions observed in solution represent the contacts present within assembled, membrane-embedded complexes, models derived from these data should be considered with care. Kuhns et al. (52) have also used cytoplasmic erythropoietin receptor (EpoR) signaling domain fusions to show that the two CD3ε subunits are in close proximity to one another at the intracellular membrane surface. This result appears to favor a one-sided arrangement; however, we note that the close proximity among subunits within the eight-TM bundle in our model indicates that any two TM domains could exit the membrane separated by as little as one or two α-helix widths (∼15–30 Å), a distance range that is well within the ∼40-Å limit for transactivation of EpoR-associated JAK kinases (53). Although the short, rigid stalk regions of the CD3 proteins make it likely that their folded EC domains sit directly above their TM domains, the positions of TCRαβ EC domains cannot be extrapolated from the TM arrangement because of the long CP sequences in TCR subunits (∼22 and 16 residues in TCRα and TCRβ, respectively). The TCR heterodimer could therefore sit above the two closely positioned CD3 modules, consistent with the compact, elongated structure observed in low-resolution electron microscopy images of intact, membrane-bound complexes (43). Higher-resolution data on intact complexes will be required to definitively determine how the orientation of extracellular domains relates to the TM arrangement we report here.
In addition to these insights into subunit organization within the membrane, our study reveals a high degree of structural interdependence among TM interactions within the octameric receptor complex. Although the conserved basic residues in the TCR TM domains are well known to act as key determinants of the modular assembly (16, 18) (the “spokes” around the TCRαβ hub in our model), we find that alterations to the internal TCRαβ TM interface can have a profound impact on the assembly and/or stability of the full complex even when these key contacts are not mutated. This interdependence points to a highly structured unit within the membrane and suggests that the TM domains of the TCR complex may be broadly more similar to the closely packed TM helix bundle of a G-protein–coupled receptor than to a loosely associated group of helices pinned together at a few focused contact points; thus, it may represent a structure that is capable of transmitting signals across the membrane through concerted conformational changes. The relevance of structural transitions in TCR signal initiation is an area of active research, and the recently recognized role for force in generating TCR-mediated responses (2, 4, 54–56) requires a mechanistic explanation of how force transmission to the cytoplasmic signaling domains is achieved. The potential involvement of changes in TM orientation in this process has been proposed by several groups (13–15, 36), but the first direct experimental test of a TM change model comes from the recent work of Lee et al. (17), who applied three different proximity-based techniques to document a ligand-induced change in the distance between the membrane-proximal intracellular ends of the two chains within a ζζ dimer. The nature of upstream structural changes that may trigger this mechanical switch was not experimentally addressed in this study, but our data suggest that alterations in the TCRαβ TM structure could be directly communicated to some or all of the signaling modules at the level of TM helix interactions. This will be an important area for further investigation.
The conservation of the polar N-T-Y residues in the C-terminal TCR TM interface is striking: they are present in species ranging from sharks to humans, encompassing ∼450 My (57) of vertebrate evolution, in both αβ and γδ TCR sequences. Although our mutagenesis experiments (Fig. 6) showed that introducing a steric block to close packing in this region was highly disruptive, the milder structural effects of eliminating all three polar groups raises the question of whether their primary role is in complex assembly within the ER, providing a stable core around which the signaling modules are gathered, or in a later stage such as receptor triggering or regulation. Evidence that this site is functionally relevant comes from a series of studies showing that mutation of the tyrosine in this triad (to leucine or phenylalanine) caused signaling defects in mutant T-cell clones (37, 58–60). Teixeiro et al. (61) also produced OT-I transgenic mice bearing a TCR with the tyrosine-to-leucine mutation and reported defects in NF-κB activation, TCR polarization, and CD8+ memory T-cell generation. Although the mechanism underlying these effects has not been elucidated, our results predict that alteration of the core TCRαβ TM interface and the network of conserved interface residues are involved. Further studies will be required to establish whether the structure we identified here in the untriggered receptor changes upon antigen ligation and how this may be related to other extracellular and intracellular structural transitions that influence transbilayer signal transmission.
Experimental Procedures
Cysteine Scanning Assay.
Single cysteine substitutions were generated on the human A6 TCR (62) sequence by standard PCR-based mutagenesis. In vitro translation/assembly constructs and reaction conditions have been described in detail elsewhere (19). Briefly, in vitro-transcribed mRNAs encoding all full-length receptor subunits and mutants were pretested for matched translation and ER microsome import. Combinations of TCRα and TCRβ mRNAs were then cotranslated with a master mix of CD3δ, CD3γ, CD3ε, and ζ mRNAs in the presence of ER microsomes for 30 min at 30 °C before addition of oxidized glutathione (4 mM final concentration) to initiate oxidative folding and assembly. After a further 4-h incubation at 30 °C, the completed assembly reactions were stopped by dilution in 0.5 mL of ice-cold Tris-buffered saline (TBS) (pH 8) containing 1 mM copper(II)-o-phenanthroline, subjected to one freeze/thaw cycle, and incubated at 30 °C for 30 min. The membrane fraction was collected by centrifugation and washed with cold TBS before extraction with 1% digitonin in TBS containing 10 mM iodoacetamide to block disulfide bond formation after extraction and during handling. Cleared lysates were immunoprecipitated with antibody-coupled agarose beads (4 °C for 2 h or overnight). For two-step CD3δ → CD3γ IP analysis (hexamer selection), products were captured using the calcium-dependent anti-protein C antibody HPC4 (Roche) with 1 mM CaCl2, washed and eluted with 5 mM EDTA, and then recaptured using anti-FLAG M2 mAb (Sigma-Aldrich). Alternatively, streptavidin-binding peptide (SBP)-tagged products were initially captured with streptavidin, washed and eluted with 100 μM biotin, and then recaptured with anti-ζ mAb 6B10.2 (Santa Cruz) or anti-HA mAb HA-7 (Sigma-Aldrich). Final products were eluted in SDS, treated with Endonuclease H (New England Biolabs), separated on 12% NuPAGE gels (Life Technologies) under nonreducing conditions and transferred to PVDF membranes for phosphoimagery.
Peptide Production and NMR Analysis.
Human TCRα and TCRβ CP-TM peptides were produced in Escherichia coli as trpLE fusions as previously reported (63), purified on nickel-NTA affinity resin (Sigma), and released from the histidine-tagged trpLE sequence by cyanogen bromide digest [0.5 M CNBr in 80% (vol/vol) trifluoroacetic acid] overnight at room temperature. Digest products were separated by reversed-phase HPLC on an Agilent Zorbax Stable Bond C3 column. NMR samples were prepared by dissolving lyophilized, stable isotope-labeled peptides to 0.5 mM in 250 mM lysomyristoyl phosphatidylglycerol, 20 mM phosphate buffer (pH 6.8), and 5% (vol/vol) D2O. Data were collected on a Bruker Avance III 600-MHz spectrometer equipped with triple-resonance cryoprobe. Backbone assignments were obtained from transverse relaxation-optimized (TROSY) (64) versions of the HNCO, HN(CO)CA, and HNCACB experiments (65) using the CARA software package (66). Backbone chemical shift-based secondary structure analysis was done using the TALOS+ web server (31).
TCRαβ TM Modeling in Implicit Bilayer.
REMD simulations (30) were used to model TCRαβ TM structures. The TM sequences used in this study are LNFQNLSVIGFRILLLKVAGFNLLMTLRL (human TCRα) and LSATILYEILLGKA-TLYAVLVSALVLMA (human TCRβ), where charged arginine and lysine residues (bold) were mutated to leucine for helix stability in the hydrophobic membrane. TCRα and TCRβ TM helices were built based on backbone chemical shift analysis and were initially separated by 30 Å in simulations using distance restraints. For both disulfide-bonded and distance-restrained models, a 10-ns simulation with 32 replicas in a temperature range of 300–750 K was performed in a GBSW implicit membrane model (32) using CHARMM (67). The GBSW default options provided in Implicit Solvent Modeler in CHARMM-GUI (68) were used with an empirical surface tension coefficient (0.03 kcal⋅mol−1⋅Å−2) for the nonpolar solvation contribution. During the simulation without the disulfide bond, distance restraints were applied between Cβ atoms of eight residue pairs showing cross-links when mutated to cysteine. The CHARMM default options for NOE restraints with a soft asymptote were used with RMIN = 3.7 Å and RMAX = 6.0 Å to mimic disulfide bond formation while allowing some additional flexibility to satisfy multiple restraints. We used a time step of 2 fs and a collision frequency of γ = 5 ps−1 for the Langevin dynamics simulation with the SHAKE algorithm (69). Replica exchanges were attempted every 1 ps, controlled by the CHARMM REPDSTR module (70). The centroid structure obtained from the cluster analysis (see below) was used to model initial structures of TCRαβ in longer, unrestrained simulations.
Molecular Dynamics Simulations in Explicit Lipid Bilayer.
The TCRαβ centroid structure from distance-restrained REMD simulations was inserted into an explicit POPC bilayer using Membrane Builder (71, 72) at the CHARMM-GUI website (68). The system was composed of the TM model with 60 POPC lipids in each leaflet and bulk water with 150 mM KCl. This system was equilibrated using CHARMM by following Membrane Builder’s six-step protocol (71), where the restraints on the components were gradually relaxed during simulations. For better statistics, we repeated the above procedure to prepare five independent replicas. After equilibration, a 200-ns restraint-free production run was performed for each system using NAMD (73) under the constant temperature and pressure condition (NPT) at 303.15 K and 1 bar, where temperature was controlled by Langevin dynamics with a coupling coefficient of 1 ps−1 and the pressure was controlled by a Nosé–Hoover Langevin piston (74, 75) with a period of 50 fs and a decay of 25 fs. The van der Waals interactions were smoothly switched off over 10–12 Å by a force-switching function (76), and the electrostatic interactions were calculated by the particle-mesh Ewald method (77) with a mesh size of ∼1 Å for fast Fourier transformation and sixth-order B-spline interpolation. We used an integration time step of 2 fs using the SHAKE algorithm (69).
Cluster Analysis for Modeling.
To evaluate the sampled conformations, we performed hierarchical clustering, where initially all sample conformations were assigned into different clusters. Starting from these initial clusters, pairs were merged when the average distance metric between the cluster pair (e.g., rmsd between conformations) was less than a predefined cutoff value. The clustering was iterated until there was no cluster closer than the cutoff distance metric. Sample size was limited to 500 conformations in each cluster analysis to minimize computational cost. Cluster analysis was performed based on Cα rmsd with a cutoff value of 3 Å using 3- to 10-ns REMD simulation trajectory at 300 K.
Acknowledgments
This work was supported by National Health and Medical Research Council (NHMRC) Grant 1011352 (to M.E.C. and M.J.C.) and NHMRC Infrastructure Support (Independent Research Institute Infrastructure Support Scheme grant [to Walter and Eliza Hall Institute of Medical Research (WEHI)]), the Victorian Government [Veski Innovation Fellowship VIF12 (to M.E.C.); Operational Infrastructure Support (to WEHI)], Extreme Science and Engineering Discovery Environment Grant MCB070009 (to W.I.), National Science Foundation Grant MCB-1157677 (to W.I.), and National Institutes of Health Grant R01-GM092950 (to W.I.). L.K. was supported by Melbourne International Research and Fee Remission Scholarships (University of Melbourne) and an Excellent Student Fund Scholarship from Federal Land and Development Authority (Malaysia). M.E.C. was supported by Queen Elizabeth II Fellowship DP110104369 from the Australian Research Council (ARC). M.J.C. is supported by ARC Future Fellowship FT120100145.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611445113/-/DCSupplemental.
References
- 1.Beddoe T, et al. Antigen ligation triggers a conformational change within the constant domain of the αβ T cell receptor. Immunity. 2009;30(6):777–788. doi: 10.1016/j.immuni.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 2.Kim ST, et al. The αβ T cell receptor is an anisotropic mechanosensor. J Biol Chem. 2009;284(45):31028–31037. doi: 10.1074/jbc.M109.052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuhns MS, Davis MM. Disruption of extracellular interactions impairs T cell receptor-CD3 complex stability and signaling. Immunity. 2007;26(3):357–369. doi: 10.1016/j.immuni.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Liu B, Chen W, Evavold BD, Zhu C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell. 2014;157(2):357–368. doi: 10.1016/j.cell.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Z, Janmey PA, Finkel TH. The receptor deformation model of TCR triggering. FASEB J. 2008;22(4):1002–1008. doi: 10.1096/fj.07-9331hyp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martínez-Martín N, et al. Cooperativity between T cell receptor complexes revealed by conformational mutants of CD3ε. Sci Signal. 2009;2(83):ra43. doi: 10.1126/scisignal.2000402. [DOI] [PubMed] [Google Scholar]
- 7.Aivazian D, Stern LJ. Phosphorylation of T cell receptor ζ is regulated by a lipid dependent folding transition. Nat Struct Biol. 2000;7(11):1023–1026. doi: 10.1038/80930. [DOI] [PubMed] [Google Scholar]
- 8.DeFord-Watts LM, et al. The CD3 ζ subunit contains a phosphoinositide-binding motif that is required for the stable accumulation of TCR-CD3 complex at the immunological synapse. J Immunol. 2011;186(12):6839–6847. doi: 10.4049/jimmunol.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deford-Watts LM, et al. The cytoplasmic tail of the T cell receptor CD3 ε subunit contains a phospholipid-binding motif that regulates T cell functions. J Immunol. 2009;183(2):1055–1064. doi: 10.4049/jimmunol.0900404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gil D, Schamel WW, Montoya M, Sánchez-Madrid F, Alarcón B. Recruitment of Nck by CD3 ε reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell. 2002;109(7):901–912. doi: 10.1016/s0092-8674(02)00799-7. [DOI] [PubMed] [Google Scholar]
- 11.Xu C, et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3ε cytoplasmic tyrosine-based motif. Cell. 2008;135(4):702–713. doi: 10.1016/j.cell.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Cordoba SP, Dushek O, van der Merwe PA. Basic residues in the T-cell receptor ζ cytoplasmic domain mediate membrane association and modulate signaling. Proc Natl Acad Sci USA. 2011;108(48):19323–19328. doi: 10.1073/pnas.1108052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brazin KN, et al. Structural features of the αβTCR mechanotransduction apparatus that promote pMHC discrimination. Front Immunol. 2015;6:441. doi: 10.3389/fimmu.2015.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman DM. Electrostatic fasteners hold the T cell receptor-CD3 complex together. Mol Cell. 2003;11(1):5–6. doi: 10.1016/s1097-2765(03)00016-9. [DOI] [PubMed] [Google Scholar]
- 15.Kuhns MS, Davis MM. TCR signaling emerges from the sum of many parts. Front Immunol. 2012;3:159. doi: 10.3389/fimmu.2012.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhns MS, Davis MM, Garcia KC. Deconstructing the form and function of the TCR/CD3 complex. Immunity. 2006;24(2):133–139. doi: 10.1016/j.immuni.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Lee MS, et al. A mechanical switch couples T cell receptor triggering to the cytoplasmic juxtamembrane regions of CD3ζζ. Immunity. 2015;43(2):227–239. doi: 10.1016/j.immuni.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Call ME, Wucherpfennig KW. The T cell receptor: Critical role of the membrane environment in receptor assembly and function. Annu Rev Immunol. 2005;23:101–125. doi: 10.1146/annurev.immunol.23.021704.115625. [DOI] [PubMed] [Google Scholar]
- 19.Call ME, Pyrdol J, Wiedmann M, Wucherpfennig KW. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell. 2002;111(7):967–979. doi: 10.1016/s0092-8674(02)01194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Call ME, et al. The structure of the ζζ transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell. 2006;127(2):355–368. doi: 10.1016/j.cell.2006.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnaud J, et al. The interchain disulfide bond between TCR α β heterodimers on human T cells is not required for TCR-CD3 membrane expression and signal transduction. Int Immunol. 1997;9(4):615–626. doi: 10.1093/intimm/9.4.615. [DOI] [PubMed] [Google Scholar]
- 22.Call ME, Pyrdol J, Wucherpfennig KW. Stoichiometry of the T-cell receptor-CD3 complex and key intermediates assembled in the endoplasmic reticulum. EMBO J. 2004;23(12):2348–2357. doi: 10.1038/sj.emboj.7600245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sussman JJ, et al. Failure to synthesize the T cell CD3-ζ chain: Structure and function of a partial T cell receptor complex. Cell. 1988;52(1):85–95. doi: 10.1016/0092-8674(88)90533-8. [DOI] [PubMed] [Google Scholar]
- 24.Dietrich J, Geisler C. T cell receptor ζ allows stable expression of receptors containing the CD3γ leucine-based receptor-sorting motif. J Biol Chem. 1998;273(41):26281–26284. doi: 10.1074/jbc.273.41.26281. [DOI] [PubMed] [Google Scholar]
- 25.Berry R, et al. Structure of the chicken CD3εδ/γ heterodimer and its assembly with the αβT cell receptor. J Biol Chem. 2014;289(12):8240–8251. doi: 10.1074/jbc.M113.544965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng J, Call ME, Wucherpfennig KW. The assembly of diverse immune receptors is focused on a polar membrane-embedded interaction site. PLoS Biol. 2006;4(5):e142. doi: 10.1371/journal.pbio.0040142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng J, Garrity D, Call ME, Moffett H, Wucherpfennig KW. Convergence on a distinctive assembly mechanism by unrelated families of activating immune receptors. Immunity. 2005;22(4):427–438. doi: 10.1016/j.immuni.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garrity D, Call ME, Feng J, Wucherpfennig KW. The activating NKG2D receptor assembles in the membrane with two signaling dimers into a hexameric structure. Proc Natl Acad Sci USA. 2005;102(21):7641–7646. doi: 10.1073/pnas.0502439102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu C, Call ME, Wucherpfennig KW. A membrane-proximal tetracysteine motif contributes to assembly of CD3δε and CD3γε dimers with the T cell receptor. J Biol Chem. 2006;281(48):36977–36984. doi: 10.1074/jbc.M607164200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett. 1999;314(1-2):141–151. [Google Scholar]
- 31.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Im W, Lee MS, Brooks CL., 3rd Generalized born model with a simple smoothing function. J Comput Chem. 2003;24(14):1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 33.Gratkowski H, Lear JD, DeGrado WF. Polar side chains drive the association of model transmembrane peptides. Proc Natl Acad Sci USA. 2001;98(3):880–885. doi: 10.1073/pnas.98.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou FX, Merianos HJ, Brunger AT, Engelman DM. Polar residues drive association of polyleucine transmembrane helices. Proc Natl Acad Sci USA. 2001;98(5):2250–2255. doi: 10.1073/pnas.041593698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Call ME, Wucherpfennig KW. Common themes in the assembly and architecture of activating immune receptors. Nat Rev Immunol. 2007;7(11):841–850. doi: 10.1038/nri2186. [DOI] [PubMed] [Google Scholar]
- 36.Campbell KS, Bäckström BT, Tiefenthaler G, Palmer E. CART: A conserved antigen receptor transmembrane motif. Semin Immunol. 1994;6(6):393–410. doi: 10.1006/smim.1994.1049. [DOI] [PubMed] [Google Scholar]
- 37.Fuller-Espie S, Hoffman Towler P, Wiest DL, Tietjen I, Spain LM. Transmembrane polar residues of TCR β chain are required for signal transduction. Int Immunol. 1998;10(7):923–933. doi: 10.1093/intimm/10.7.923. [DOI] [PubMed] [Google Scholar]
- 38.Kunjibettu S, Fuller-Espie S, Carey GB, Spain LM. Conserved transmembrane tyrosine residues of the TCR β chain are required for TCR expression and function in primary T cells and hybridomas. Int Immunol. 2001;13(2):211–222. doi: 10.1093/intimm/13.2.211. [DOI] [PubMed] [Google Scholar]
- 39.Brooks AJ, et al. Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science. 2014;344(6185):1249783. doi: 10.1126/science.1249783. [DOI] [PubMed] [Google Scholar]
- 40.Lu C, et al. Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor. Mol Cell Biol. 2010;30(22):5432–5443. doi: 10.1128/MCB.00742-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu J, et al. The structure of a receptor with two associating transmembrane domains on the cell surface: Integrin αIIbβ3. Mol Cell. 2009;34(2):234–249. doi: 10.1016/j.molcel.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng X, Im W. NMR observable-based structure refinement of DAP12-NKG2C activating immunoreceptor complex in explicit membranes. Biophys J. 2012;102(7):L27–L29. doi: 10.1016/j.bpj.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Birnbaum ME, et al. Molecular architecture of the αβ T cell receptor–CD3 complex. Proc Natl Acad Sci USA. 2014;111(49):17576–17581. doi: 10.1073/pnas.1420936111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar R, et al. Increased sensitivity of antigen-experienced T cells through the enrichment of oligomeric T cell receptor complexes. Immunity. 2011;35(3):375–387. doi: 10.1016/j.immuni.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 45.Schamel WW, et al. Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J Exp Med. 2005;202(4):493–503. doi: 10.1084/jem.20042155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molnár E, et al. Cholesterol and sphingomyelin drive ligand-independent T-cell antigen receptor nanoclustering. J Biol Chem. 2012;287(51):42664–42674. doi: 10.1074/jbc.M112.386045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swamy M, et al. A cholesterol-based allostery model of T cell receptor phosphorylation. Immunity. 2016;44(5):1091–1101. doi: 10.1016/j.immuni.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 48.Sun ZY, et al. Solution structure of the CD3εδ ectodomain and comparison with CD3εγ as a basis for modeling T cell receptor topology and signaling. Proc Natl Acad Sci USA. 2004;101(48):16867–16872. doi: 10.1073/pnas.0407576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Touma M, et al. The TCR C β FG loop regulates α β T cell development. J Immunol. 2006;176(11):6812–6823. doi: 10.4049/jimmunol.176.11.6812. [DOI] [PubMed] [Google Scholar]
- 50.Natarajan A, et al. Structural model of the extracellular assembly of the TCR-CD3 complex. Cell Rep. 2016;14(12):2833–2845. doi: 10.1016/j.celrep.2016.02.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He Y, et al. Identification of the docking site for CD3 on the T cell receptor β chain by solution NMR. J Biol Chem. 2015;290(32):19796–19805. doi: 10.1074/jbc.M115.663799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuhns MS, et al. Evidence for a functional sidedness to the αβTCR. Proc Natl Acad Sci USA. 2010;107(11):5094–5099. doi: 10.1073/pnas.1000925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livnah O, et al. Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science. 1999;283(5404):987–990. doi: 10.1126/science.283.5404.987. [DOI] [PubMed] [Google Scholar]
- 54.Das DK, et al. Force-dependent transition in the T-cell receptor β-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc Natl Acad Sci USA. 2015;112(5):1517–1522. doi: 10.1073/pnas.1424829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu KH, Butte MJ. T cell activation requires force generation. J Cell Biol. 2016;213(5):535–542. doi: 10.1083/jcb.201511053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y, et al. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc Natl Acad Sci USA. 2016;113(20):5610–5615. doi: 10.1073/pnas.1600163113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Criscitiello MF, Ohta Y, Saltis M, McKinney EC, Flajnik MF. Evolutionarily conserved TCR binding sites, identification of T cells in primary lymphoid tissues, and surprising trans-rearrangements in nurse shark. J Immunol. 2010;184(12):6950–6960. doi: 10.4049/jimmunol.0902774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodríguez-Tarduchy G, Sahuquillo AG, Alarcón B, Bragado R. Apoptosis but not other activation events is inhibited by a mutation in the transmembrane domain of T cell receptor β that impairs CD3ζ association. J Biol Chem. 1996;271(48):30417–30425. doi: 10.1074/jbc.271.48.30417. [DOI] [PubMed] [Google Scholar]
- 59.Sahuquillo AG, Roumier A, Teixeiro E, Bragado R, Alarcón B. T cell receptor (TCR) engagement in apoptosis-defective, but interleukin 2 (IL-2)-producing, T cells results in impaired ZAP70/CD3-ζ association. J Exp Med. 1998;187(8):1179–1192. doi: 10.1084/jem.187.8.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teixeiro E, Fuentes P, Galocha B, Alarcon B, Bragado R. T cell receptor-mediated signal transduction controlled by the β chain transmembrane domain: Apoptosis-deficient cells display unbalanced mitogen-activated protein kinases activities upon T cell receptor engagement. J Biol Chem. 2002;277(6):3993–4002. doi: 10.1074/jbc.M107797200. [DOI] [PubMed] [Google Scholar]
- 61.Teixeiro E, et al. Different T cell receptor signals determine CD8+ memory versus effector development. Science. 2009;323(5913):502–505. doi: 10.1126/science.1163612. [DOI] [PubMed] [Google Scholar]
- 62.Utz U, Banks D, Jacobson S, Biddison WE. Analysis of the T-cell receptor repertoire of human T-cell leukemia virus type 1 (HTLV-1) Tax-specific CD8+ cytotoxic T lymphocytes from patients with HTLV-1-associated disease: Evidence for oligoclonal expansion. J Virol. 1996;70(2):843–851. doi: 10.1128/jvi.70.2.843-851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharma P, et al. Production of disulfide-stabilized transmembrane peptide complexes for structural studies. J Vis Exp. 2013;(73):e50141. doi: 10.3791/50141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salzmann M, Wider G, Pervushin K, Wüthrich K. Improved sensitivity and coherence selection for [15N,1H]-TROSY elements in triple resonance experiments. J Biomol NMR. 1999;15(2):181–184. doi: 10.1023/a:1008358030477. [DOI] [PubMed] [Google Scholar]
- 65.Kay LE, Ikura M, Tschudin R, Bax A. Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins. 1990. J Magn Reson. 2011;213(2):423–441. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 66.Keller R. The Computer Aided Resonance Assignment Tutorial. Cantina; Goldau, Switzerland: 2004. [Google Scholar]
- 67.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J Comput Chem. 2008;29(11):1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 69.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23(3):327–341. [Google Scholar]
- 70.Woodcock HL, 3rd, et al. Interfacing Q-Chem and CHARMM to perform QM/MM reaction path calculations. J Comput Chem. 2007;28(9):1485–1502. doi: 10.1002/jcc.20587. [DOI] [PubMed] [Google Scholar]
- 71.Jo S, Kim T, Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One. 2007;2(9):e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu EL, et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J Comput Chem. 2014;35(27):1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: The Langevin piston method. J Chem Phys. 1995;103(11):4613–4621. [Google Scholar]
- 75.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. J Chem Phys. 1994;101(5):4177–4189. [Google Scholar]
- 76.Steinbach PJ, Brooks BR. New spherical-cutoff methods for long-range forces in macromolecular simulation. J Comput Chem. 1994;15(7):667–683. [Google Scholar]
- 77.Essmann U, et al. A smooth particle mesh Ewald method. J Chem Phys. 1995;103(19):8577–8593. [Google Scholar]
- 78.Kjer-Nielsen L, et al. The 1.5 Å crystal structure of a highly selected antiviral T cell receptor provides evidence for a structural basis of immunodominance. Structure. 2002;10(11):1521–1532. doi: 10.1016/s0969-2126(02)00878-x. [DOI] [PubMed] [Google Scholar]
- 79.Arnett KL, Harrison SC, Wiley DC. Crystal structure of a human CD3-ε/δ dimer in complex with a UCHT1 single-chain antibody fragment. Proc Natl Acad Sci USA. 2004;101(46):16268–16273. doi: 10.1073/pnas.0407359101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kjer-Nielsen L, et al. Crystal structure of the human T cell receptor CD3 ε γ heterodimer complexed to the therapeutic mAb OKT3. Proc Natl Acad Sci USA. 2004;101(20):7675–7680. doi: 10.1073/pnas.0402295101. [DOI] [PMC free article] [PubMed] [Google Scholar]