Significance
In contrast to animals, plants lack a segregated germline. Instead, all plant tissue is derived from small clusters of meristematic cells that throughout development give rise to vegetative tissues and only late in development to reproductive organs. Hence, mutations acquired during vegetative growth can be passed on to offspring; it is generally believed that older plants will acquire more such mutations over their lifetimes due to replication during cell division. Here we measure the number of DNA replications occurring in the cell lineage leading to gametes and demonstrate that the number of replications is independent of life span, suggesting that older plants may not be passing on more mutations to their offspring relative to younger plants.
Keywords: mutation rate, shoot apical meristem, germline, mismatch repair, telomeres
Abstract
In plants, gametogenesis occurs late in development, and somatic mutations can therefore be transmitted to the next generation. Longer periods of growth are believed to result in an increase in the number of cell divisions before gametogenesis, with a concomitant increase in mutations arising due to replication errors. However, there is little experimental evidence addressing how many cell divisions occur before gametogenesis. Here, we measured loss of telomeric DNA and accumulation of replication errors in Arabidopsis with short and long life spans to determine the number of replications in lineages leading to gametes. Surprisingly, the number of cell divisions within the gamete lineage is nearly independent of both life span and vegetative growth. One consequence of the relatively stable number of replications per generation is that older plants may not pass along more somatically acquired mutations to their offspring. We confirmed this hypothesis by genomic sequencing of progeny from young and old plants. This independence can be achieved by hierarchical arrangement of cell divisions in plant meristems where vegetative growth is primarily accomplished by expansion of cells in rapidly dividing meristematic zones, which are only rarely refreshed by occasional divisions of more quiescent cells. We support this model by 5-ethynyl-2′-deoxyuridine retention experiments in shoot and root apical meristems. These results suggest that stem-cell organization has independently evolved in plants and animals to minimize mutations by limiting DNA replication.
In contrast to most animals, plants lack a developmentally defined germline. Instead, gametes are derived late in plant development following variable periods of vegetative growth (1). An important consequence of this developmental strategy is that somatic mutations acquired during vegetative growth can be transmitted to the next generation (2). Numerous studies have been conducted in attempts to understand whether and how somatic mutations contribute to fitness and evolution in plants (3–8). DNA replication during cell division is hypothesized to be a leading cause of genetic mutation (9–11), and mutation rates are highly correlated with genome duplications in many taxa (12–16). Thus, a critical impediment to the studies examining the role of somatic mutation in plant genome evolution is the lack of knowledge on the number of cell divisions separating a zygote from its gametes, a characteristic termed “cell depth” (17), and how that number changes with vegetative growth. To our knowledge, estimates of gametic cell depth in plants are limited to calculations based on mitotic index and growth rates (5, 18) or total cell numbers and DNA content (19), which provide no information on correlations between cell depth and development.
In contrast to the paucity of knowledge on cell depth, cell lineage analyses have been conducted in multiple plant species. The primary origin of all above-ground tissues of a plant is a dome-like structure named the shoot apical meristem (SAM). These cell-fate analyses have demonstrated that stem cells within the SAM do not have predetermined fates but give rise to organs in a probabilistic way based on their location within this stem-cell niche (20–23). In Arabidopsis, it is estimated that two to four genetically effective cells (GEC) in the dry seed are the progenitors of late rosettes as well as flowers (20, 21, 24). In late-flowering mutants that undergo prolonged vegetative growth, the extra leaves produced are derived from these two to four cells, and not from expanded growth of cells normally responsible for earlier leaves (25). Similar results have been reported in maize mutants that undergo additional vegetative growth (26), suggesting that this is a conserved feature of plant growth. It is generally accepted that over longer periods of growth, divisions of the genetically effective cells will increase the cell depth in the SAM before flowering and gametogenesis, resulting in an increase in the number of somatically acquired mutations transmitted to offspring (6).
Here we report quantitative analysis of germline DNA replications in Arabidopsis and test whether the number of replications increases with prolonged vegetative growth. We used two independent methodologies, based on intrinsic properties of DNA replication. First, we measured loss of telomeric DNA due to the end-replication problem in telomerase mutants, which are incapable of maintaining telomeres. Second, we measured accumulation of mutations due to polymerase misincorporation in mutants deficient for mismatch repair. This analysis showed that the number of DNA replications increased only slightly under long-lived conditions, demonstrating that the cell depth of gametes is not linearly proportional to the vegetative growth period.
Results
Telomere Shortening During Life Span of Arabidopsis tert Mutants.
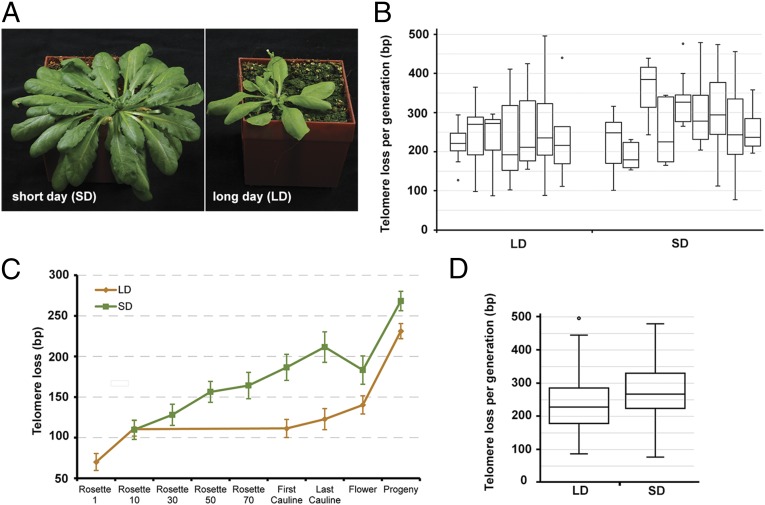
The vegetative growth period in Arabidopsis can be modulated by day length (Fig. 1A; SI Appendix, Table S1). Under long-day conditions, (LD, 16 h daylight) plants flowered in 35 d after producing 10 leaves; those grown under short-day conditions (SD, 8 h daylight) took 91 d to flower after producing 75 leaves. To measure the number of germline cell divisions in these plants, we took advantage of the end-replication problem. Telomeres, the ends of eukaryotic chromosomes, are unable to be fully replicated by conventional DNA polymerases (SI Appendix, Fig. S1). Instead, they are extended by the enzyme telomerase (27). Disruption of the Arabidopsis gene encoding the catalytic subunit of telomerase, TERT, leads to a gradual loss of telomeric DNA that is directly proportional to the number of rounds of DNA replication (SI Appendix, Fig. S1) (28, 29).
Fig. 1.
Telomere shortening in Arabidopsis tert mutants. (A) Plants grown in SD (Left) or LD (Right) show dramatic differences in vegetative growth. (B) Loss of telomeric DNA per generation within individual plant families grown at SD and LD. Box plots indicate median and 25th and 75th percentile from 6 to 29 telomeres. (C) Average loss of telomeric DNA at each developmental stage. Rosette leaf number for LD plants is precise, whereas for SD plants it is only approximate (Materials and Methods). Error bars represent SEM from three LD and five SD plants (SI Appendix, Fig. S2). (D) Loss of telomeric DNA per generation in all plants grown at LD (n = 125) and SD (n = 96). Box plots indicate median and 25th and 75th percentile. The LD and SD datasets are significantly different (P = 0.0014, two-tailed t test).
To quantify the number of DNA replications per generation in plants grown under short- and long-lived conditions, we compared telomere length of progeny with their parents in seven LD and nine SD families. We measured the length of telomeres from up to six individual chromosome arms in one cotyledon of each-TERT deficient plant. These values then served as a baseline for comparison with telomere length measured in cotyledons from its progeny (Fig. 1B). In eight families (three LD and five SD) we also sampled DNA throughout the course of development of parental plants to determine the amount of telomeric DNA lost from a particular chromosome arm during different periods of plant growth (Fig. 1C; SI Appendix, Fig. S2). As expected, telomeres gradually shorten during plant development, supporting the view that cell depth increases as organs are sequentially generated from the SAM (Fig. 1B). On the developmental timescale, telomere loss was most pronounced between flowers and the cotyledons of a plant’s progeny, which likely reflects an increased requirement for cell divisions due to the bottleneck represented by germ-cell differentiation and embryogenesis. The average decline of telomere length over the entire life cycle ranged from 187 to 366 bp between individual families, with a higher variation in SD families (Fig. 1B). This may reflect more stochasticity in the frequency of germline cell divisions when plants are grown longer under the SD conditions. Nevertheless, despite the threefold difference in vegetative growth period and an up to 8 times higher number of leaves produced, the average loss of telomeric DNA in SD versus LD over one generation is minimal (238 nt and 275 nt, respectively; Fig. 1D). Using previous estimates of Arabidopsis G-overhangs of 20–30 nt (30) and models of telomere shortening (SI Appendix, Fig. S1) (31), we estimate that Arabidopsis undergoes roughly 34 germline cell divisions in LD and 39 in SD. Thus, although the telomere shortening data imply a small increase in cell depth in the functional germline of SD plants, the difference was lower than we expected based on the dramatic difference in life span and vegetative tissue produced.
Formation of de Novo Mutations in Mismatch Repair-Deficient Arabidopsis.
We next wanted to confirm this result through an independent method. Another intrinsic feature of DNA replication is the introduction of mutations due to nucleotide misincorporation, and analysis of these mutations has been used to track cell depth in mammals (17). Hence, estimates of per-generation mutation rates in SD and LD plants should be proportional to the number of replications in the functional germline. The frequency of spontaneous mutations is normally low, as DNA polymerase errors are repaired by the mismatch repair (MMR) machinery. Genome-wide studies in yeast and bacteria revealed that MMR deficiency results in a 10- to 200-fold increase in the mutation rate (32, 33). Therefore, we decided to determine the rate of replication errors in MMR-null plants. MSH2 (MutS protein homolog 2) is an essential MMR component, and previous work with msh2 mutants demonstrated that these plants have a strong mutator phenotype (34).
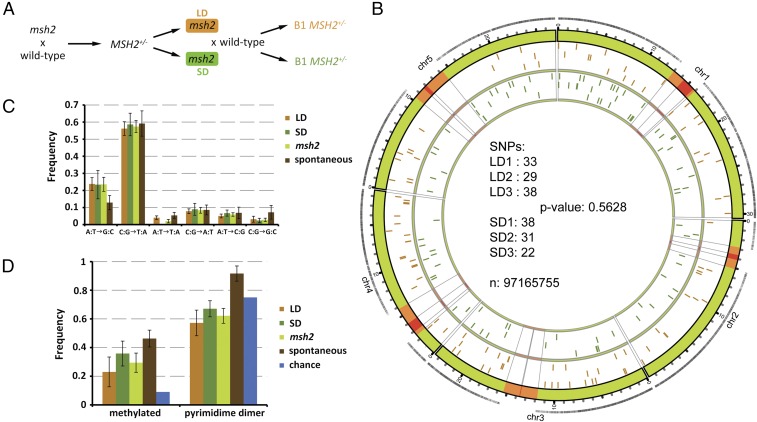
We crossed msh2 mutant plants to wild-type Col-0 to obtain an MMR-proficient F1 founder plant and sequenced it twice to 120× and 140× coverage. Mutant progeny of this plant were grown under either SD or LD conditions and then backcrossed to wild-type siblings to obtain MMR-proficient B1 plants (Fig. 2A). These plants were then sequenced with a minimum of 95× coverage. This strategy allows for the detection of de novo mutations that had occurred within one sexual generation in the functional germline in SD and LD plants and avoids chimeric somatic mutations caused by ongoing mutagenesis in the MMR-deficient background. In this approach, only half of the de novo mutations are transmitted to the B1 generation, where they occur in a heterozygous context.
Fig. 2.
Analysis of de novo mutations in LD- and SD-grown msh2 plants. (A) Genetic pedigree of msh2 plants analyzed in this study. Orange and green boxes indicate generation grown at LD and SD, respectively. (B) Number of SNPs in three LD and SD B1 plants and their chromosomal location; n equals the size of our masked genome. Orange regions on the chromosomes represent pericentromeric regions, and red regions the centromeres. Black ticks on the outer orbital correspond to sequence coverage after masking. Orange ticks in the inner orbital represent location of SNPs in LD plants and green ticks in SD plants. (C) Distribution of SNP classes identified and compared with spontaneous mutations (35). (D) Ratio of C:G transitions known to be in methylated DNA (57) or in dipyrimidine dimer contexts.
Our sequence processing focused on calling accuracy at the expense of reference coverage. One hundred base-pair paired-end reads were generated on an Illumina HiSeq Analyzer, and uniquely aligned reads were mapped to The Arabidopsis Information Resource 10 reference genome. Following stringent filtering, the reference genome was masked for repetitive and low coverage regions at the single-nucleotide level for each individual. The intersection of these masks was then used as the final mask for analysis so that the genomes would be directly comparable. Despite these rigorous filters, the final mask contains 81.6% of the reference genome, largely due to the high sequence coverage. Single-nucleotide polymorphisms (SNPs) were then categorized based on the ratio of the most frequent nucleotide call at each site; following verification of heterozygous SNPs in the founding line, SNPs were considered heterozygous when this ratio fell between 0.4 and 0.6. SNPs that were present in more than one sample were excluded as having likely arisen in the original founder line. We verified 65 random SNPs by Sanger sequencing. None of the 65 SNPs were present in the founder DNA sample, and 64 were confirmed, resulting in a false-positive rate of 1.5%. We detected no deficit of nonsynonymous relative to synonymous mutations in general, nor between the LD and SD populations (Fisher’s exact test, P = 0.875 and 0.520, respectively), suggesting that the mutation profiles were not significantly affected by selection under either growth condition (SI Appendix, Fig. S3).
In total, we identified 191 new SNPs in six B1 plants descending from msh2 mutants (Fig. 2B; SI Appendix, Table S2). B1 progeny of msh2 plants grown in LD had an average of 33.3 ± 4.5 SNPs (n = 3) whereas SD-derived B1 plants had 30.3 ± 8.0 SNPs (n = 3) (Fig. 2B). There was no significant difference between the two growth conditions (Fisher’s exact test, P = 0.5628). Recently, the spectrum of spontaneous mutations in Arabidopsis was determined following 30 sexual generations (35). This spectrum is highly biased toward C:G→T:A transitions, a phenomenon that has been explained by the combined effects of deamination of methylated cytosines and UV-induced mutagenesis at dipyrimidine dimers. Although C:G transitions were the largest class of mutations in our study (Fig. 2C), we observed a significant twofold increase in A:T transitions relative to the spontaneous mutation rate (Fisher’s exact test, P = 0.03953). A similar shift toward A:T transitions has also recently been observed in MMR-deficient Escherichia coli (32). There was no significant difference in the ratio of A:T transitions between LD and SD grown plants (Fisher’s exact test, P = 0.729), nor did we observe any significant differences between the other mutation classes, as might be expected if environment or metabolic rate had a large influence on the mutational spectrum that we observed. We further examined the C:G transitions for bias in methylation and dipyrimidine dimer contexts (Fig. 2D). There were no significant differences between LD and SD plants in the number of C:G transitions at methylated sites (Fisher’s exact test, P = 0.08495) or between the msh2 mutants and spontaneous events (Fisher’s exact test, P = 0.2198). UV-induced mutations consist primarily of C:G transitions at dipyrimidine dimer sites, and spontaneous mutations at these sites occur more often than expected by chance. If the mutational spectrum that we observed was due to UV light, we would expect an overrepresentation of C:G transitions at these sites. There was no difference in this class between LD and SD (Fisher’s exact test, P = 0.4814), and C:G transitions at these sites occurred less often than expected by chance (Fisher’s exact test, P = 0.02649) and significantly less often than observed spontaneously (Fisher’s exact test, P = 8.557e-05). We conclude that the mutations that we detected are unlikely to be caused by failure to repair UV damage. In summary, we can detect no difference between the mutation profile of SD- and LD-grown plants, suggesting that the mutations that we detected are not being dramatically influenced by environmental or metabolic factors.
From the SNPs that we identified, we calculate a mutation rate of 7 × 10−7/bp/generation, which is 100-fold higher than the rate of spontaneous mutations in wild-type Arabidopsis (35). When combined with cell division estimates from our telomeric data, this equals a mutation rate of 1.7 × 10−8/bp/replication, which is at the high end of estimates of DNA polymerase fidelity (36). Thus, we conclude that the de novo mutations that we detected are primarily due to replication errors. Detection of a similar mutation rate per generation in SD- and LD-grown plants supports the data on loss of telomeric DNA and provides further evidence that the number of replications in the functional germline is not coupled to either vegetative growth or life span.
Long-Term 5-Ethynyl-2′-Deoxyuridine Retention in Arabidopsis Meristems.
We next asked how plants maintain reduced cell depth in the functional germline irrespective of their generation time and the number of organs initiated by cell divisions in the SAM. Studies in adult stem-cell niches of mammals suggest the presence of reserve quiescent stem cells, which only rarely divide and are able to repopulate more mitotically active stem cells that support tissue differentiation and regeneration (37). Such a hierarchical organization of cell proliferation, where a population of mitotically active stem cells is occasionally replaced by divisions of a quiescent stem cell, can greatly reduce cell depth within a tissue. The SAM of Arabidopsis is a dome-shaped structure with two distinct zones, the central and peripheral zones (CZ and PZ, respectively), which are differentiated by their position and mitotic index (38). Products of division with the CZ are displaced to the PZ where mitotic rates are increased and organ primorida are formed (39, 40).
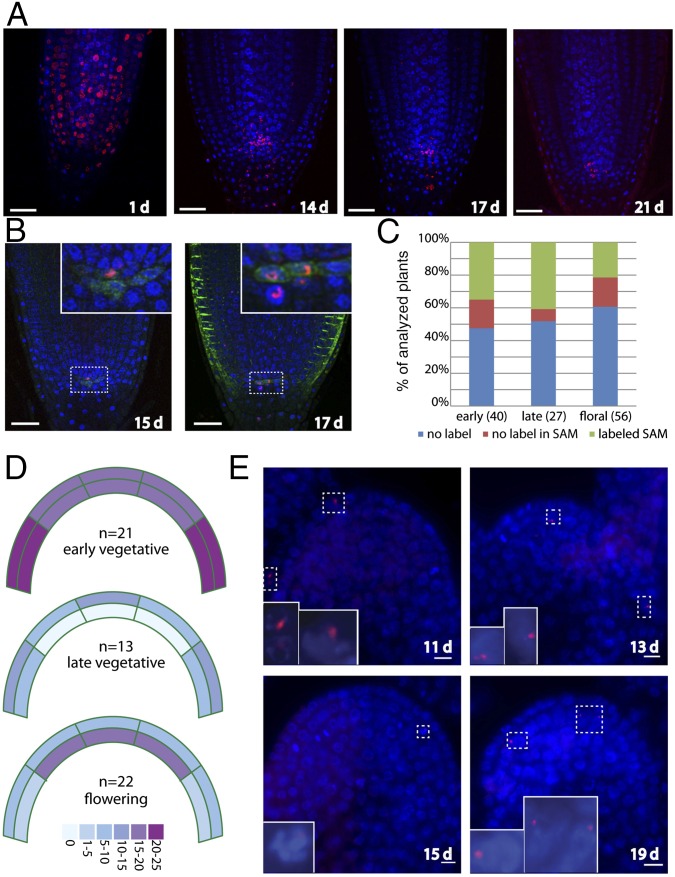
We reasoned that the reduced cell depth in the functional germline could be achieved by a similar hierarchical arrangement of cell divisions in plant meristem. According to this model, exceedingly rare divisions of cells from the center of the CZ would produce a daughter that is pushed to the border between the CZ and the PZ where it could undergo more rapid divisions to generate the transit-amplifying cells in the PZ. Real-time analysis of divisions within the SAM has identified leaf-progenitor cells abutting the CZ, but the short time frame in which real time imaging is practical has inhibited the identification of leaf progenitors in the CZ and their turnover (41). An alternative approach to testing this model is pulse labeling the nuclei of meristematic stem cells followed by an extended time-course analysis of the retention and redistribution of the DNA label. Although a number of pulse-labeling studies have been conducted in plants, the aim of these studies was generally to measure mitotic activity in different cell types over relatively short time frames of a few days. Our approach was to determine how long labeled cells could be retained within meristems from early development to flowering. We began by studying the more experimentally tractable root apical meristem (RAM). We shortly incubated developing roots with the DNA analog EdU and followed the DNA labeling for up to 3 wk (Fig. 3A). After a period of 1 wk, EdU labeling was visible at the base of the root, indicating that these cells had differentiated and ceased proliferation shortly after labeling. There was generally no labeling along the entire body of the root, but strong EdU signal was visible in several meristematic cells in most roots throughout the 3-wk duration of the experiment (Fig. 3A). Interestingly, cells within the RAM were not labeled immediately after the pulse, but became labeled several days thereafter. This suggests that cells can retain the label for periods of at least several days before it is incorporated into DNA (Fig. 3A, compare 1 d and 14 d). Although we did observe labeling of the quiescent center, marked by WOX5:GFP (42), at later time points, most of the labeled nuclei corresponded to root initials (Fig. 3B). We conclude that, in the root, some stem cells maintain an extremely low proliferation profile while still producing significant amounts of growth. These data are consistent with 3H-thymidine labeling and metaphase accumulation studies across several plant species, indicating that the duration of the cell cycle in the quiescent center can reach up to 520 h (43, 44).
Fig. 3.
Retention of label-retaining cells within plant meristems. (A) EdU labeling time course. Roots were pulse-labeled with EdU for 1 h, 2 d after imbibition and transferred to plates lacking EdU. Cells that rapidly proliferated lost labeled chromosomes, whereas those that divided more slowly retained the label. Blue: DAPI; Red: EdU. (B) Colocalization of EdU-labeled nuclei with a WOX5:GFP (green) reporter that labels quiescent center (QC). The majority of the colocalized signals were located above the WOX5-labeled QC (Left), although QC cells were also labeled (Right). (C) Frequency of labeling of plants in the SAM. Plants were pulse-labeled with EdU for 1 h and then transferred to soil for 6–10 d (early vegetative), 11–14 d (late vegetative), or 13–19 d (flowering) at which time they were fixed, embedded in paraffin, and sectioned. Plants were scored as stained if EdU staining was visible anywhere in the aerial portion of the plant. (D) Heat map indicating the percentage of labeled plants at each stage with EdU-positive cells within the indicated zones of the SAM. (E) Examples of EdU-labeled SAMs with the days after the initial pulse indicated. (Top Left) Labeling within the L1 layer. (Bottom Left) Labeled cell in the L2 layer entering mitosis. (Top Right) Central section of a floral meristem with labeled cells in the L2 layer. (Bottom Right) Inflorescence meristem with several cells labeled in the L2 layer.
We next asked whether rarely dividing cells are also retained in the SAM. In contrast to the root, labeling efficiency was generally much lower, likely due to poor uptake of the EdU, as only half of the plants were labeled (SI Appendix, Fig. S4). Nevertheless, of the labeled plants, 50–80% had cells in the SAM that retained EdU (Fig. 3C). We monitored labeling in the SAM at early (6–10 d) and late (11–14 d) vegetative time points and after the transition to flowering (13–21 d). Remarkably, we detected EdU-retaining cells in the SAM even 21 d after labeling (Fig. 3 C–E), demonstrating the existence of slowly dividing cells within this otherwise highly proliferative organ. Although clusters of labeled cells were visible in the roots, in the shoot experiments we generally observed only a few labeled cells that were scattered throughout the SAM (Fig. 3E). Interestingly, these cells did not exclusively colocalize with the CZ, but were also detected in the highly dividing PZ (Fig. 3E). As it is generally accepted that embryos are derived from the L2 layer of the SAM, we quantified the labeled cells in the L1 and L2 layers. At early time points, labeling throughout all sectors of the L1 and L2 was visible. At late vegetative time points, we observed no labeling in the central area of the L2 although, interestingly, labeled cells were visible in this area after the transition to flowering. It is tempting to speculate that the labeling that we observed at late time points in the L2 is due to these cells having undergone the first division since the initial pulse, thereby incorporating EdU into their DNA at the transition to flowering. Nonetheless, our observation of cells that retained EdU in the highly proliferative root and SAMs over the entire vegetative Arabidopsis life cycle is consistent with a hierarchical model of cell divisions as proposed in animal stem-cell niches.
Discussion
In this study we used telomere dynamics in telomerase-null plants and the frequency of de novo mutation occurrence in MMR-deficient mutants to assess the number of cell divisions in the plant cell lineage. The vast majority of research on cell division in plants is focused on following divisions over relatively short time frames. This analysis is an experimentally derived estimate of division rates in the cell-line lineage that gives rise to progeny encompassing an entire plant generation. Our data suggest that prolonged vegetative growth does not lead to a proportional increase in cell depth in the cell lineage that eventually leads to the gametes. Hence, at least in Arabidopsis, there is no proportional relationship between plant age and the number of somatic mutations transmitted to the next generation. We believe these data provide critical parameters for our understanding of plant development and molecular evolution.
As previously mentioned, multiple experiments on the SAM of different plant species have demonstrated common organizational features of angiosperm meristems (20–23, 45, 46). At the embryonic stage, SAM cells do not have a predetermined fate, but instead display probabilistic fates likely based on their location in the SAM. Cells low in the PZ give rise to small sectors of the earliest leaves, whereas cells in the CZ give rise to larger sectors of vegetative tissue and flowers. Along with knowledge of mitotic index rates across the meristem, a general model has been accepted where slow divisions of cells in the CZ replace cells in the PZ that are recruited for organ formation (1). Under conditions of prolonged growth, vegetative expansion is accomplished by descendants of the genetically effective cells in the CZ (25, 26). Under the conditions in our study, SD plants produced seven times more vegetative tissue than their LD counterparts. Thus, continual replacement of cells from the PZ by divisions of the GEC should result in an increase in the cell depth of the gametes. As this result was not observed in our experiments, we propose that the increased vegetative growth observed under these conditions is accomplished primarily through expansion of cells within the SAM not destined for the germline. This expansion would occur through amplification of the early progenitors of the two to four GEC, whereas the GEC may remain largely quiescent throughout the vegetative growth period before returning to division at the transition to flowering, when increased mitotic activity has been observed across the meristem in both Arabidopsis and Silene latifolia (47, 48). This hypothesis is supported by our observation of EdU-retaining cells within the L2 layer of the SAM at the transition to flowering. A recent study describing patterns of cell division in the SAM that precede the formation of axillary meristems showed that such a model also applies to plant branching (49), a key determinant of architecture of perennial plants. This study provides strong support to the idea that the organization of the SAM minimizes the number of cell divisions required throughout development, even in long-lived perennials.
As the structure of the SAM is conserved across the angiosperms, these results have important implications for our understanding of variation in the rates of molecular evolution across the plant kingdom. It is now generally accepted that perennials have lower rates of molecular evolution than annuals (15), and more recently it has been demonstrated that taller plants have lower rates of molecular evolution (50). In the MMR-deficient plants, where mutations are accumulated primarily due to replication errors, the rate of molecular evolution over time in SD plants is more than threefold lower than LD plants, a difference entirely accounted for by the length of the generation time. Although multiple factors are likely involved in determining the mutation rate (11, 51), replication errors are an intrinsic feature of DNA replication and must therefore play some role in determining the rate of molecular evolution. It has therefore been proposed that differences in rates of mitosis can account for the observed differences in rate variation across lineages, with larger plants having overall lower rates of mitosis (50). The data presented here suggest that it may be the near-quiescent nature of cells within the SAM that ultimately gives rise to the next generation that accounts for the observed differences in molecular evolution rates.
This organization of the SAM parallels emerging models of stem-cell dynamics in many human tissues, where populations of quiescent and mitotically active stem cells can reside within the same stem-cell niche (37). Whereas the mitotically active cells would be responsible for growth, the quiescent stem cells could repopulate the mitotically active stem cells in response to DNA damage or proliferation exhaustion. This suggests that, similar to many of the observed analogies between plant and animal stem-cell niches (1), evolutionary pressure to reduce replication-dependent errors has resulted in independent evolution of similar developmental strategies in both animal and plant stem-cell niches.
Materials and Methods
Plant Materials and Growth Conditions.
All plant lines used in this study—tert-1 (At5g16850) (52) and msh2-1 (At3g18524, SALK_002708 obtained from Nottingham Arabidopsis Stock Centre) (53, 54)—have been previously described. For LD conditions, plants were germinated on soil and grown in 16 h light at 21 °C and 60% relative humidity. For SD conditions, plants were germinated on soil and grown in 8 h light at 21 °C and 60% relative humidity.
Telomere-Length Analysis.
Tissue for telomere analysis was harvested when plants began to set seed (near the end of the life cycle). For LD grown plants, the first and last rosette leaves were easily identified; for SD grown plants, counting rosette leaves was much more difficult, and the leaf number reported is a rough estimate that may vary by ±5.
Telomere length of individual chromosomes was determined by Primer Extension Telomere Repeat Analysis (PETRA) (55). PETRA products were separated in a 1.2% (wt/vol) agarose gel and detected by Southern hybridization with a [32P]ATP-labeled (TTTAGGG)4 probe. To obtain accurate measures across the gel, a DNA ladder (1-kb+ ladder; Fermentas) was loaded in every fifth lane. Autoradiograms were produced with Molecular Imager FX (Bio-Rad) and analyzed with ImageQuant software (Bio-Rad). To specifically test the error associated with the three different steps of PETRA reaction, we analyzed one set of DNA samples seven times (SI Appendix, Fig. S5). Telomeres from homologous chromosomes in Arabidopsis are not always of the same length (SI Appendix, Fig. S6). In these cases, loss of telomeric DNA was always calculated from the nearest length telomere in the cotyledon. Furthermore, telomere changes that were identified to be caused by recombinational processes and not the end replication problem were excluded (SI Appendix, Fig. S6B).
Genome Sequencing.
A total of 300–500 ng of genomic DNA was fragmented by sonication with a Bioruptor (Diagenode); the peak of fragment sizes centered around 500 bp. End-repair of sheared DNA fragments, A-tailing, and adapter ligation were performed with the NEXTflexTM DNA Sequencing Kit (BioScientific). Adaptor-ligated DNA was size-selected on 1.5% (wt/vol) low melt agarose (Peqlab) gels and stained with SybrGold (Invitrogen). DNA fragments ranging from 300 to 600 bp were excised and purified with the Zymoclean Gel DNA Recovery Kit (Zymo Research). The paired-end DNA libraries were amplified by PCR for 10–12 cycles with NEXTflex-supplied PCR primers using a KAPATM Library Amp kit (Peqlab). Libraries were then sequenced on Illumina HiSeq Analyzers with a 100-base read length.
Sequence Analysis.
Analysis pipeline used for analysis of de novo mutations and mutation rates is described in SI Appendix.
EdU Labeling and Image Acquisition.
EdU labeling and detection was performed according to published protocol (56). For roots, Z-stacks were acquired with a confocal laser-scanning microscope (Carl Zeiss LSM 780 microscope) using a 40× oil immersion objective. Pictures were acquired and analyzed using ZEN2011 software (Carl Zeiss). Z-stacks were deconvoluted with Huygens deconvolution software (SVI). For the time course of the Col-0 images (Fig. 3B), the Z-stack of the Alexa488 channel, which corresponds to the EdU labeling, was merged using maximum intensity projection. This projection was then overlaid with a single-image layer of the DAPI channel, representative of a middle section of the root tip. Brightness in individual channels over the entire image was adjusted to improve visibility.
For shoot sections, images were acquired with an Axio-Imager Z2 using a 40× oil immersion objective.
Supplementary Material
Acknowledgments
We thank Nick Fulcher for helpful discussion. Sequencing was performed at the Next Generation Sequencing unit of the Vienna BioCenter Core Facilities (www.vbcf.ac.at/); microscopy and Sanger sequencing were conducted with the help of the IMP/IMBA Core Facilities. This work was funded by the Austrian Academy of Sciences; by the Austrian Science Fund (Grant Y418-B03); by the Ministry of Education, Youth and Sports of the Czech Republic under the project CEITEC 2020 (Project LQ1601); and by the European Research Council (Grant ERC 268962–MAXMAP).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited at BioSample (www.ncbi.nlm.nih.gov/sra/?term=SRP041456) under accessions SAMN02737223, SAMN02737224, SAMN02737225, SAMN02737226, SAMN02737227, SAMN02737228, SAMN02737229, SAMN02737230, and SAMN02737231.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609686113/-/DCSupplemental.
References
- 1.Sablowski R. Plant and animal stem cells: Conceptually similar, molecularly distinct? Trends Cell Biol. 2004;14(11):605–611. doi: 10.1016/j.tcb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Klekowski EJ. 1988. Mutation, Developmental Selection, and Plant Evolution (Columbia University Press, New York), pp xi, 373 pp.
- 3.Ally D, Ritland K, Otto SP. Aging in a long-lived clonal tree. PLoS Biol. 2010;8(8):e1000454. doi: 10.1371/journal.pbio.1000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobiwash K, Schultz ST, Schoen DJ. Somatic deleterious mutation rate in a woody plant: Estimation from phenotypic data. Heredity (Edinb) 2013;111(4):338–344. doi: 10.1038/hdy.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cloutier D, Rioux D, Beaulieu J, Schoen DJ. Somatic stability of microsatellite loci in Eastern white pine, Pinus strobus L. Heredity (Edinb) 2003;90(3):247–252. doi: 10.1038/sj.hdy.6800221. [DOI] [PubMed] [Google Scholar]
- 6.Klekowski EJ, Godfrey PJ. Aging and mutation in plants. Nature. 1989;340(6232):389–391. [Google Scholar]
- 7.O’Connell LM, Ritland K. Somatic mutations at microsatellite loci in western Redcedar (Thuja plicata: Cupressaceae) J Hered. 2004;95(2):172–176. doi: 10.1093/jhered/esh024. [DOI] [PubMed] [Google Scholar]
- 8.Petit RJ, Hampe A. Some evolutionary consequences of being a tree. Annu Rev Ecol Evol Syst. 2006;37:187–214. [Google Scholar]
- 9.Arnheim N, Calabrese P. Understanding what determines the frequency and pattern of human germline mutations. Nat Rev Genet. 2009;10(7):478–488. doi: 10.1038/nrg2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baer CF, Miyamoto MM, Denver DR. Mutation rate variation in multicellular eukaryotes: Causes and consequences. Nat Rev Genet. 2007;8(8):619–631. doi: 10.1038/nrg2158. [DOI] [PubMed] [Google Scholar]
- 11.Gaut B, Yang L, Takuno S, Eguiarte LE. The patterns and causes of variation in plant nucleotide substitution rates. Annu Rev Ecol Evol Syst. 2011;42:245–266. [Google Scholar]
- 12.Bromham L. The genome as a life-history character: Why rate of molecular evolution varies between mammal species. Philos Trans R Soc Lond B Biol Sci. 2011;366(1577):2503–2513. doi: 10.1098/rstb.2011.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mooers AO, Harvey PH. Metabolic rate, generation time, and the rate of molecular evolution in birds. Mol Phylogenet Evol. 1994;3(4):344–350. doi: 10.1006/mpev.1994.1040. [DOI] [PubMed] [Google Scholar]
- 14.Ohta T. An examination of the generation-time effect on molecular evolution. Proc Natl Acad Sci USA. 1993;90(22):10676–10680. doi: 10.1073/pnas.90.22.10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith SA, Donoghue MJ. Rates of molecular evolution are linked to life history in flowering plants. Science. 2008;322(5898):86–89. doi: 10.1126/science.1163197. [DOI] [PubMed] [Google Scholar]
- 16.Thomas JA, Welch JJ, Lanfear R, Bromham L. A generation time effect on the rate of molecular evolution in invertebrates. Mol Biol Evol. 2010;27(5):1173–1180. doi: 10.1093/molbev/msq009. [DOI] [PubMed] [Google Scholar]
- 17.Reizel Y, et al. Cell lineage analysis of the mammalian female germline. PLoS Genet. 2012;8(2):e1002477. doi: 10.1371/journal.pgen.1002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romberger JA, Hejnowicz Z, Hill JF. 1993. Plant Structure: Function and Development: A Treatise on Anatomy and Vegetative Development, with Special Reference to Woody Plants (Springer, Berlin), pp xix, 524 pp.
- 19.Otto SP, Walbot V. DNA methylation in eukaryotes: Kinetics of demethylation and de novo methylation during the life cycle. Genetics. 1990;124(2):429–437. doi: 10.1093/genetics/124.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furner IJ, Pumfrey JE. Cell fate in the shoot apical meristem of Arabidopsis thaliana. Development. 1992;115(3):755–764. [Google Scholar]
- 21.Irish VF, Sussex IM. A fate map of the Arabidopsis embryonic shoot apical meristem. Development. 1992;115(3):745–753. [Google Scholar]
- 22.Jegla DE, Sussex IM. Cell lineage patterns in the shoot meristem of the sunflower embryo in the dry seed. Dev Biol. 1989;131(1):215–225. doi: 10.1016/s0012-1606(89)80053-3. [DOI] [PubMed] [Google Scholar]
- 23.McDaniel CN, Poethig RS. Cell-lineage patterns in the shoot apical meristem of the germinating maize embryo. Planta. 1988;175(1):13–22. doi: 10.1007/BF00402877. [DOI] [PubMed] [Google Scholar]
- 24.Meinke DW, Sussex IM. Isolation and characterization of six embryo-lethal mutants of Arabidopsis thaliana. Dev Biol. 1979;72(1):62–72. doi: 10.1016/0012-1606(79)90098-8. [DOI] [PubMed] [Google Scholar]
- 25.Furner IJ, Ainscough JFX, Pumfrey JA, Petty LM. Clonal analysis of the late flowering fca mutant of Arabidopsis thaliana: Cell fate and cell autonomy. Development. 1996;122(3):1041–1050. doi: 10.1242/dev.122.3.1041. [DOI] [PubMed] [Google Scholar]
- 26.Dudley M, Poethig RS. The effect of a heterochronic mutation, Teopod2, on the cell lineage of the maize shoot. Development. 1991;111(3):733–739. doi: 10.1242/dev.111.3.733. [DOI] [PubMed] [Google Scholar]
- 27.Watson JM, Riha K. Telomeres, aging, and plants: From weeds to Methuselah—A mini-review. Gerontology. 2011;57(2):129–136. doi: 10.1159/000310174. [DOI] [PubMed] [Google Scholar]
- 28.Huffman KE, Levene SD, Tesmer VM, Shay JW, Wright WE. Telomere shortening is proportional to the size of the G-rich telomeric 3′-overhang. J Biol Chem. 2000;275(26):19719–19722. doi: 10.1074/jbc.M002843200. [DOI] [PubMed] [Google Scholar]
- 29.Riha K, McKnight TD, Griffing LR, Shippen DE. Living with genome instability: Plant responses to telomere dysfunction. Science. 2001;291(5509):1797–1800. doi: 10.1126/science.1057110. [DOI] [PubMed] [Google Scholar]
- 30.Riha K, McKnight TD, Fajkus J, Vyskot B, Shippen DE. Analysis of the G-overhang structures on plant telomeres: Evidence for two distinct telomere architectures. Plant J. 2000;23(5):633–641. doi: 10.1046/j.1365-313x.2000.00831.x. [DOI] [PubMed] [Google Scholar]
- 31.Soudet J, Jolivet P, Teixeira MT. Elucidation of the DNA end-replication problem in Saccharomyces cerevisiae. Mol Cell. 2014;53(6):954–964. doi: 10.1016/j.molcel.2014.02.030. [DOI] [PubMed] [Google Scholar]
- 32.Lee H, Popodi E, Tang H, Foster PL. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci USA. 2012;109(41):E2774–E2783. doi: 10.1073/pnas.1210309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zanders S, et al. Detection of heterozygous mutations in the genome of mismatch repair defective diploid yeast using a Bayesian approach. Genetics. 2010;186(2):493–503. doi: 10.1534/genetics.110.120105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffman PD, Leonard JM, Lindberg GE, Bollmann SR, Hays JB. Rapid accumulation of mutations during seed-to-seed propagation of mismatch-repair-defective Arabidopsis. Genes Dev. 2004;18(21):2676–2685. doi: 10.1101/gad.1217204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ossowski S, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327(5961):92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18(1):148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327(5965):542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fletcher JC. Shoot and floral meristem maintenance in Arabidopsis. Annu Rev Plant Biol. 2002;53:45–66. doi: 10.1146/annurev.arplant.53.092701.143332. [DOI] [PubMed] [Google Scholar]
- 39.Laufs P, Grandjean O, Jonak C, Kiêu K, Traas J. Cellular parameters of the shoot apical meristem in Arabidopsis. Plant Cell. 1998;10(8):1375–1390. doi: 10.1105/tpc.10.8.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwiatkowska D. Flowering and apical meristem growth dynamics. J Exp Bot. 2008;59(2):187–201. doi: 10.1093/jxb/erm290. [DOI] [PubMed] [Google Scholar]
- 41.Reddy GV, Heisler MG, Ehrhardt DW, Meyerowitz EM. Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development. 2004;131(17):4225–4237. doi: 10.1242/dev.01261. [DOI] [PubMed] [Google Scholar]
- 42.Xu J, et al. A molecular framework for plant regeneration. Science. 2006;311(5759):385–388. doi: 10.1126/science.1121790. [DOI] [PubMed] [Google Scholar]
- 43.Phillips HL, Torrey JG. The quiescent center in cultured roots of Convolvulus arvensis L. Am J Bot. 1971;(58):665–671. [Google Scholar]
- 44.Clowes FAL. Rates of mitosis in a partially synchronous meristem. New Phytol. 1962;(61):111–118. [Google Scholar]
- 45.Gifford EM., Jr Incorporation of tritiated thymidine into nuclei of shoot apical meristems. Science. 1960;131(3397):360. doi: 10.1126/science.131.3397.360. [DOI] [PubMed] [Google Scholar]
- 46.Johri MM, Coe EH., Jr Clonal analysis of corn plant development. I. The development of the tassel and the ear shoot. Dev Biol. 1983;97(1):154–172. doi: 10.1016/0012-1606(83)90073-8. [DOI] [PubMed] [Google Scholar]
- 47.Jacqmard A, Gadisseur I, Bernier G. Cell division and morphological changes in the shoot apex of Arabidopsis thaliana during floral transition. Ann Bot (Lond) 2003;91(5):571–576. doi: 10.1093/aob/mcg053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zluvova J, Janousek B, Vyskot B. Immunohistochemical study of DNA methylation dynamics during plant development. J Exp Bot. 2001;52(365):2265–2273. doi: 10.1093/jexbot/52.365.2265. [DOI] [PubMed] [Google Scholar]
- 49.Burian A, Barbier de Reuille P, Kuhlemeier C. Patterns of stem cell divisions contribute to plant longevity. Curr Biol. 2016;26(11):1385–1394. doi: 10.1016/j.cub.2016.03.067. [DOI] [PubMed] [Google Scholar]
- 50.Lanfear R, et al. Taller plants have lower rates of molecular evolution. Nat Commun. 2013;4:1879. doi: 10.1038/ncomms2836. [DOI] [PubMed] [Google Scholar]
- 51.Bromham L. Why do species vary in their rate of molecular evolution? Biol Lett. 2009;5(3):401–404. doi: 10.1098/rsbl.2009.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fitzgerald MS, et al. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc Natl Acad Sci USA. 1999;96(26):14813–14818. doi: 10.1073/pnas.96.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leonard JM, Bollmann SR, Hays JB. Reduction of stability of Arabidopsis genomic and transgenic DNA-repeat sequences (microsatellites) by inactivation of AtMSH2 mismatch-repair function. Plant Physiol. 2003;133(1):328–338. doi: 10.1104/pp.103.023952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 55.Heacock M, Spangler E, Riha K, Puizina J, Shippen DE. Molecular analysis of telomere fusions in Arabidopsis: Multiple pathways for chromosome end-joining. EMBO J. 2004;23(11):2304–2313. doi: 10.1038/sj.emboj.7600236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kazda A, Akimcheva S, Watson JM, Riha K. Cell proliferation analysis using EdU labeling in whole plant and histological samples of Arabidopsis. Methods Mol Biol. 2016;1370:169–182. doi: 10.1007/978-1-4939-3142-2_13. [DOI] [PubMed] [Google Scholar]
- 57.Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152(1-2):352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.